Abstract
Objectives
To evaluate the genomic epidemiology of SARS-CoV-2 from Venezuelan migrants living in Colombia.
Methods
This study sequenced SARS-CoV-2 from 30 clinical specimens collected from Venezuelan migrants. Genomes were compared with the Wuhan reference genome to identify polymorphisms, reconstruct phylogenetic relationships and perform comparative genomic analyses. Geographic, sociodemographic and clinical data were also studied across genotypes.
Results
This study demonstrated the presence of six distinct SARS-CoV-2 lineages circulating among Venezuelan migrants, as well as a close relationship between SARS-CoV-2 genomic sequences obtained from individuals living in the Venezuelan-Colombian border regions of La Guajira (Colombia) and Zulia (Venezuela). Three clusters (C-1, C-2 and C-3) were well supported by phylogenomic inference, supporting the hypothesis of three potential transmission routes across the Colombian-Venezuelan border. These genomes included point mutations previously associated with increased infectivity. A mutation (L18F) in the N-terminal domain of the spike protein that has been associated with compromised binding of neutralizing antibodies was found in 2 of 30 (6.6%) genomes. A statistically significant association was identified with symptomatology for cluster C2.
Conclusion
The close phylogenetic relationships between SARS-CoV-2 genomes from Venezuelan migrants and from people living at the Venezuela-Colombian border support the importance of human movements for the spread of COVID-19 and for emerging virus variants.
Keywords: Venezuelan-Colombian border, SARS-CoV-2, L18F, Lineages, Mutations
INTRODUCTION
The socioeconomic, political and healthcare crisis of the last two decades has resulted in violence, poverty and the massive migration of Venezuelans to neighboring countries (Daniels 2020). This has driven further economic and social instability and catalyzed the overflow of infectious diseases such as malaria, dengue, tuberculosis, HIV, and most recently, Coronavirus Disease 2019 (COVID-19) caused by Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) (Grillet et al. 2019; Rodríguez-Morales et al. 2019b, 2019a; Paniz-Mondolfi et al. 2020a, 2020b). The inability of the precarious Venezuelan healthcare service to meet the acutely increased needs due to the COVID-19 pandemic impelled almost a doubling in the number of Venezuelans crossing to neighboring countries, particularly Brazil and Colombia (Daniels 2020; Standley et al. 2020). In fact, compared to the nearly 800,000 Venezuelans that migrated to Colombia in 2017, almost 1.5 million Venezuelans have crossed the border as a result of the current pandemic (Fernández-Niño et al. 2020; García Pinzón and Mantilla 2020).
Recently reported evidence from genomic sequencing of SARS-CoV-2 from clinical specimens indicated spread of the virus between Venezuela and Colombia, despite closure of the border with Colombia on 14 March 2020 in an effort to curb the influx of COVID-19 (Paniz-Mondolfi et al. 2020a). This SARS-CoV-2 spread suggests that the illegal transit between Venezuela and Colombia continued, particularly in the departments of Cundinamarca, Norte de Santander and La Guajira, which border the western Venezuelan states with the highest number of COVID-19 cases (Táchira and Zulia) (Estadisticas-Venezuela-COVID-19. 2021).
In order to address concerns regarding the high transmission rate (Paniz-Mondolfi et al. 2020a), spread of new variants (e.g., P.1) (Instituto Nacional de Salud (INS) 2021) and the possibility of reinfection (Lee et al. 2020; Mulder et al. 2020), a robust analysis was performed of the epidemiological landscape of SARS-CoV-2 across the Colombian-Venezuelan border. The findings for SARS-CoV-2 genomes obtained from Venezuelans inhabiting Colombia (predominantly from La Guajira) are reported, including: genomic monitoring of lineages circulating at the Colombian-Venezuelan border and analysis of related sociodemographic and clinical data.
METHODS
Study population and ethical considerations
This study sequenced and analyzed SARS-CoV-2 from 30 clinical nasopharyngeal specimens collected between 05 May and 05 July 2020 from Venezuelan migrants living in seven departments of Colombia (Ven_Col): Amazonas, Atlántico, Boyacá, Cundinamarca, La Guajira, Magdalena, and Norte de Santander. The authorization and use of biological material and associated epidemiological information followed previously described ethical criteria (Paniz-Mondolfi et al. 2020a). This study was performed following the Declaration of Helsinki and its later amendments, and all patient data were anonymized to minimize risk to participants. The epidemiological and clinical characteristics of Ven_Col genomes are presented in Supplementary Table 1.
Phylogenomic analysis and genetic diversity
The molecular detection, sequencing, assembly and bioinformatic analysis of the 30 Ven_Col SARS-CoV-2 genomes were performed according to previously reported methodology (Paniz-Mondolfi et al. 2020a). Molecular typing was performed using the Phylogenetic Assignment of Named Global Outbreak LINeages ‘Pangolin’ tool (Rambaut et al. 2020). The Ven_Col genomes were compared with 10 SARS-CoV-2 genomes previously reported from Venezuela (Loureiro et al. 2021; Paniz-Mondolfi et al. 2020a), 277 reported from Colombia, and 2,641 representative genomes from all available data from countries in North America, Europe, Asia, Oceania, and South America hosted in the GISAID (Global Initiative on Sharing All Influenza Data) (Hadfield et al. 2018) and GenBank databases (Loureiro et al. 2021); these last genomes were included in a category called 'Other groups'. The complete dataset of 2,958 genomes was aligned, the untranslated regions were trimmed, phylogenetic relationships were constructed, and single nucleotide polymorphisms (SNP) were extracted following previously reported methods (Muñoz et al. 2021; Ramírez et al. 2021). Nucleotide variation was characterized by comparing the 30 Ven_Col genomes with the Wuhan reference sequence (NC_045512), using UGENE v.33.0 software (Okonechnikov et al. 2012).
Phylogeographic analysis
Latitude and longitude data from a publicly available meta-data of Colombian genomes deposited into GISAID and from the 30 Ven_Col genomes sequenced in this study were mapped across their location in Colombian departments using the QGIS Geographic Information System Open Source Geospatial Foundation software (v3.16, http://qgis.osgeo.org).
Statistical analysis
A descriptive analysis of the clinical and epidemiological data was performed. For continuous values (age), normality hypotheses were evaluated using the Shapiro-Wilk test. As data showed a normal distribution, this variable was summarized in terms of mean and standard deviation. Qualitative variables were summarized in frequencies and proportions according with the cluster groups. Since a normal distribution was obtained for the continuous variable of age, a parametric ANOVA test was performed to explore potential differences in ages between cluster groups. Chi-Square test was performed to identify potential relationships between categorical variables and the cluster groups. Statistical analyses were carried out using R software (R Core Team 2019). All tests of significance were two-tailed, and P-values < 0.05 were considered statistically significant.
RESULTS
Phylogenomic analysis and genomic diversity
Six different PANGO lineage groups were identified among the 30 Ven_Col genomes that were analyzed (Supplementary Table 1). The B.1 lineage was the most frequently found (14 genomes, 47%) followed by B.1.111 (seven genomes, 23%) and B.1.420 lineages (six genomes, 20%); single isolates of B.1.1.237, B.1.1.161 and B.1.1.255 lineages were recovered. Overall, five of the six lineages were represented among the reported genome lineages from Colombian individuals: B.1, B.1.111, B.1.420, B.1.1.161, and B.1.1.237. Of note, there have been no reported Colombian isolates that correspond with B.1.1.255 lineages as of 10 May 2021 (Instituto Nacional de Salud (INS) 2021).
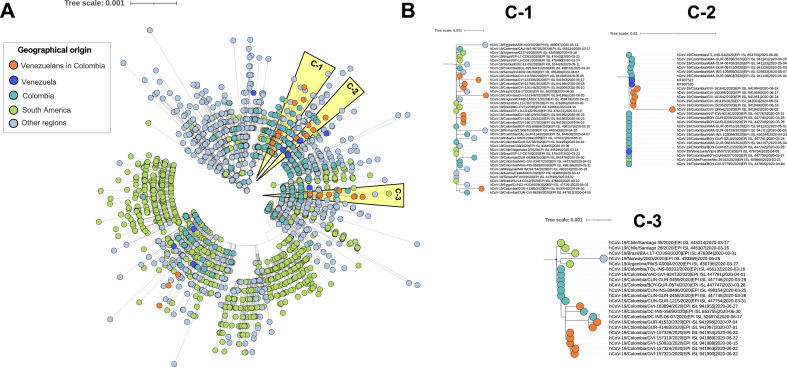
The phylogeny revealed heterogeneity within the SARS-CoV-2 sequences globally ( Figure 1 A). The analysis revealed that Ven_Col SARS-CoV-2 genomes (23 genomes, 76%) were positioned into three different clusters (C-1 to C-3), which lead to the hypothesis that there were three potential transmission routes across the Colombian-Venezuelan border. The first transmission route was supported by cluster C-1, which includes various subgroups; nine of the analyzed Ven_Col genomes were included in four of these subgroups. Four of these genomes were closely related with one genome from Serbia, three with genomes from Brazil, and the last two with genomes from Colombia, Egypt, England, and Iraq. Interestingly, this cluster did not include any genomes from Venezuela. The second transmission route (cluster C-2) could be further divided into two subgroups, one of them included six of the Ven_Col genomes, with a close phylogenetic relationship with two genomes (MT907521, MT907520) from the bordering Venezuelan department of Zulia and with Colombian genomes. Finally, the third transmission route (cluster C-3), which was subdivided in various subgroups like C-1, included eight Ven_Col genomes; five of these genomes shared sequence similarity between them and three genomes were related to Colombian genomes, mainly genomes from Bogotá and Cundinamarca (Figure 1B). Additionally, a relationship between the clustering and the SARS-CoV-2 lineages was observed.
Figure 1.
Phylogenomic relationships of evaluated SARS-CoV-2 genomes in the global context. A dataset with 2,958 genomes (30 genomes from Venezuelans in Colombia (Ven_Col), 10 from Venezuela, 277 from Colombia, and 2,641 from other countries) was analyzed from the alignment with a length of 5,209 positions, corresponding to the whole genome single-nucleotide polymorphisms (SNP) excluding untranslated regions. A) Maximum likelihood tree from whole genome SNPs for the complete dataset. Clusters consisting of 23 of the 30 Ven_Col genomes are highlighted in orange (C-1 to C-3), which are depicted in B) at a greater magnification to visualize all genomes included in each cluster. These clusters support the hypothesis of three potential transmission routes across the Colombian-Venezuelan border.
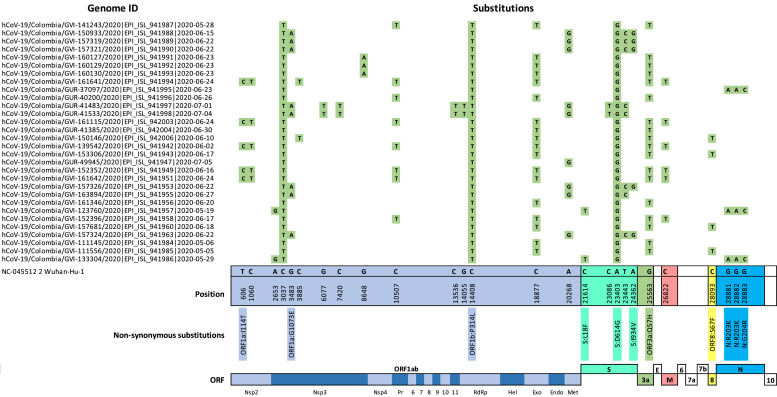
Moreover, 26 polymorphic sites that distinguished the 30 Ven_Col genomes from the Wuhan reference sequence were found ( Figure 2 ). There were three substitutions that were shared across all 30 of the Ven_Col genomes: 1) a synonymous substitution in ORF1a (C3037T); 2) a non-synonymous substitution in ORF1b (C14408T), which resulted in the P314L substitution in the RdRp protein; and 3) a non-synonymous substitution in the S gene, which resulted in the D614G mutation in the spike protein. In addition, the synonymous substitution in ORF1b (C18877T) and non-synonymous substitution in ORF3a (G22563T: Q57H) was found to be concurrent in > 60% of the Ven_Col sequences. Of note, two (6.6%) of the Ven_Col genomes encoded a non-synonymous substitution (C21614T) in the N-terminal domain of the S gene, which resulted in the L18F mutation.
Figure 2.
Nucleotide diversity between 30 Ven_Col genomes. Single nucleotide polymorphisms (SNP) found in the 30 Ven_Col SARS-CoV-2 isolates (Genome ID) compared with the Wuhan reference sequence (NC_045512). The positions of each SNP that is represented in ≥ 5% of all genomes are shown. If the SNP is a non-synonymous substitution, the corresponding amino acid change is indicated below the nucleotide position. All SNPs are annotated across an annotated SARS-CoV-2 genome.
Phylogeographic analysis
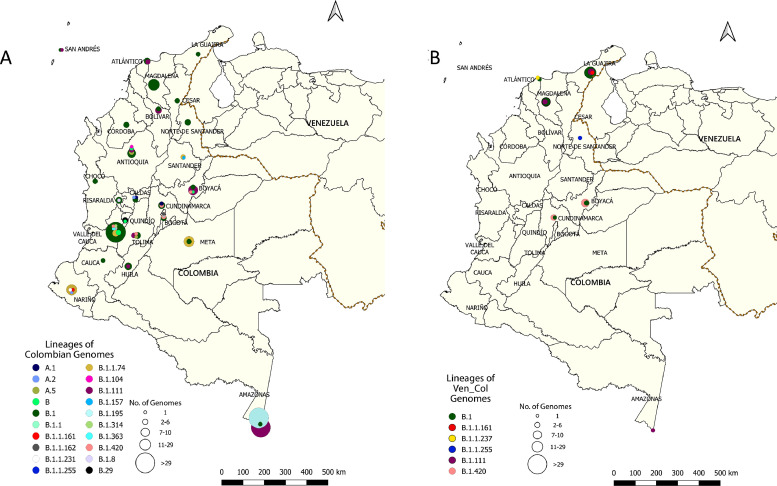
In order to gain greater insight into transmission dynamics at a geographic level, the lineages of both Colombian and Ven_Col SARS-CoV-2 isolates across Colombian departments were compared (Figure 3 ). Overall, the entire dataset included Colombian genomes from 24 departments (Figure 3A) and Ven_Col genomes (Figure 3B) from seven departments (Supplementary Table 2). Of the seven departments with Ven_Col isolates, three (Amazonas, Cundinamarca and Boyacá) had isolates whose lineages were also represented among Colombian genomes in the same department. However, within the other four departments with Ven_Col genomes there were Ven_Col isolates whose lineages were not represented among genomes from Colombian patients in the same department. Specifically, these Ven_Col isolates mapped to lineages B.1.1.255 (Norte de Santander); B.1.111 (Magdalena); B.1.1.237 (Atlántico); B.1.1.161 and B.1.111 (La Guajira).
Figure 3.
Geographic distribution of Colombian and Ven_Col SARS-CoV-2 isolates in Colombia. Using geographical coordinates for sequenced genomes, quantities of A) Colombian and B) Ven_Col isolates characterized by evolutionary lineage are depicted as circles across Colombian departments. Circles are proportional in size (see scale) to the number of genomes sequenced that correspond to a given color-coded PANGO lineage.
Epidemiological analysis
To determine if there were epidemiologic differences among the phylogenetic clusters of Ven_Col genomes, clinical data across 23 participants that comprised each of the three clusters and six that did not make up a cluster (‘OUT’ group) were evaluated. Specifically, seven clinical variables across the cohort were compared (Table 1 ). Of the 30 participants, 60% (n = 18) were female and 40% (n = 12) were male. The average age was 35.23 years (± 15.53); no statistically significant differences in age were found between the three phylogenetic clusters (ANOVA, P = 0.71).
Table 1.
Association of sociodemographic and clinical data with phylogenetic clusters of Venezuelan SARS-CoV-2 isolates.
| Cluster |
|||||||
|---|---|---|---|---|---|---|---|
| Variable | Category | C1 | C2 | C3 | OUT | TOTAL | P-value (Chi-Squared) |
| Gender | Female | 6 (33.3%) | 3 (16.7%) | 7 (38.9%) | 2 (11.1%) | 18 (100%) | 0.150 |
| Male | 3 (25%) | 4 (33.3%) | 1 (8.3%) | 4 (33.3%) | 12 (100%) | ||
| Hospitalization | No | 7 (28.0%) | 5 (20.0%) | 8 (32.0%) | 5 (20%) | 25 (100%) | 0.473 |
| Yes | 2 (40%) | 2 (40%) | 0 (0.0%) | 1 (20,0%) | 5 (100%) | ||
| Health Status | Asymptomatic | 4 (40.0%) | 0 (0%) | 5 (50.0%) | 1 (10.0%) | 10 (100%) | 0.043 |
| Symptomatic | 4 (21.1%) | 7 (36.8%) | 3 (15.8%) | 5 (26.3%) | 19 (100%) | ||
| Respiratory Symptoms | No | 5 (33.3%) | 1 (6.7%) | 6 (40.0%) | 3 (20.0%) | 15 (100%) | 0.128 |
| Yes | 4 (26.7%) | 6 (40.0%) | 2 (13.3%) | 3 (20.0%) | 15 (100%) | ||
| Comorbidities | No | 7 (33.3%) | 3 (14,3%) | 5 (23.8%) | 6 (28.6%) | 21 (100%) | 0.139 |
| Yes | 2 (22.2%) | 4 (44.4%) | 3 (33.3%) | 0 (0.0%) | 9 (100%) | ||
| Department | Amazonas | 0 (0.0%) | 1 (100%) | 0 (0.0%) | 0 (0.0%) | 1 (100%) | < 0.01 |
| Atlántico | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 2 (100%) | 2 (100%) | ||
| Boyacá | 0 (0.0%) | 0 (0.0%) | 6 (100%) | 0 (0.0%) | 6 (100%) | ||
| Cundinamarca | 1 (33.3%) | 0 (0.0%) | 2 (67.7%) | 0 (0.0%) | 3 (100%) | ||
| La Guajira | 3 (27.3%) | 5 (45.5%) | 0 (0.0%) | 3 (27.3%) | 11 (100%) | ||
| Magdalena | 5 (83.3%) | 1 (16.7%) | 0 (0.0%) | 0 (0.0%) | 6 (100%) | ||
| Norte de Santander | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 1 (100 %) | 1 (100%) | ||
Among the other clinical variables, it was found that the department varied among the three clusters (P < 0.01). In the north in La Guajira, eight (27%) specimens clustered within the C-1 (n = 3) and C-2 (n = 5) clusters; the remaining three did not cluster in any of the C-1 to C-3 groups (Table 1). There were six (20%) specimens in the neighboring Magdalena that also clustered within C-1 (n = 5) and C-2 (n = 1) groups, and two specimens (6.7%) from Atlántico that did not cluster to the C-1 to C-3 groups. Further south, only C-3-clustering genomes were found in Boyacá (six, 20%) and Cundinamarca (two, 6.7%). The lone specimen (3.3%) from Norte de Santander did not cluster with any of the C-1 to C-3 groups. Finally, a lone specimen (3.3%) was identified in C-2 in the southernmost department of Amazonas.
In addition, it was also determined that the health status (e.g., symptomatic vs. asymptomatic) was statistically different among the clusters (Chi-squared test P < 0.05). Overall, from 29 individuals with health status data, 19 (65.5%) were collected from symptomatic versus 10 (34.5%) from asymptomatic people (Table 1). Among the symptomatic individuals, four (13.8%) were from C-1, seven (24.1%) were from C-2, three (10.3%) were from C-3, and five (17.2%) were from the OUT clusters. Indeed, when compared with sequences from non-C-2 clusters, the C-2 group included a greater number of symptomatic individuals (P = 0.0275). A logistic regression was performed that included significant variables identified during bivariate analysis (department and health status); however, no statistically significant differences were found.
DISCUSSION
Since the onset of the COVID-19 pandemic, SARS-CoV-2 has exponentially spread across the globe. In South America, Brazil and Colombia have been severely affected, with the highest recorded numbers of cases and deaths to date. In addition to the already volatile political and socioeconomic conditions in many South American countries (Paniz-Mondolfi et al. 2020a), this health emergency has driven inhabitants to migrate to bordering countries in search of stable and safer conditions. Such is the case with Venezuelan citizens who have increasingly migrated to Colombia, Brazil, Peru, and Ecuador over the last few years (Brito 2020). This human movement poses a hurdle to infection prevention efforts, as it potentiates the spread of infectious disease. Thus, in order to contain COVID-19 disease, it is vital to interrogate SARS-CoV-2 transmission events and viral evolution associated with its spread across countries.
The most recent epidemiological reports (as of 13 May 2021) indicate that some of Venezuela's highest infection rates occur in departments along the Colombian-Venezuelan border (Estadisticas-Venezuela-COVID-19 2021). However, given the socioeconomic, financial and clinical constraints in the countries, there is limited information on the origins and transmission of these viruses, as well as their relation to other infection events in Colombia and South America.
This study investigated the phylogenetic, phylodynamic and geographical characteristics of SARS-CoV-2 viral isolates distinctly recovered from 30 Venezuelan migrants in Colombia. The phylogenetic reconstruction based on whole-genome SNP variation enabled the SARS-CoV-2 viruses from the Ven_Col population to be classified into three distinct transmission clusters (Figure 1 ), which lead to the hypothesis that there are three potential transmission routes across the Colombian-Venezuelan border; cluster in which was evidenced a close ancestor-descendant relationship between Ven_Col genomes with genomes from close (Venezuela: C-1 and Colombia: C-3), and distant countries (Europa, Asia and Brazil) (Figure 1B). These findings describe the spread of SARS-CoV-2 from a Ven_Col population into Colombia via different routes, and also three potential divergence events of SARS-CoV-2. Likewise, the relationship observed between the SARS-CoV-2 lineages from the Ven_Col population and the geographical localization suggest that the COVID-19 cases in Colombia could be attributed to travel-related exposure (e.g. B.1 and B.1.111 lineage) or community transmission, as observed with the B.1.420 lineage.
When compared with a subset of global sequences, the Ven_Col genomes belonged to six distinct lineages across seven departments (Figure 3, Supplementary Table 1). Of the six lineages of the 30 Ven_Col genomes, it was observed that as of 10 May 2021, four of them had previously been reported in Colombia (Instituto Nacional de Salud (INS) 2021) and in other regions of South America. Specifically, the B.1.1.161 lineage had been reported in Ecuador, Brazil, Perú, and Chile. The B.1 and the B.1.111 lineages have been reported in at least seven different countries of South America, with B.1 being the most prevalent lineage in Colombia and Brazil (Resende et al. 2020; Ramírez et al. 2021). However, to date, this is the first report of the B.1.1.255 lineage in Colombia, which had previously only been described in Brazil and the United Kingdom (Hadfield et al. 2018) (https://www.gisaid.org/). Together, these results highlight the rapid global spread of SARS-CoV-2 to South America and across the bordering countries of Venezuela and Colombia.
To explore the differences between infections across Colombians and Venezuelans in Colombia, this study assessed overlap in lineages sampled across geographic areas (e.g., departments) (Figure 3). Lineages were represented among both Colombian and Ven_Col genomes in departments in central (Cundinamarca and Boyacá) and southern Colombia (Amazonas), which may reflect localized transmission events within each department affecting both Colombian citizens and migrant Venezuelans. Interestingly, it was found that distinct lineages existed among Colombians and Venezuelan migrants within each of four departments: Norte de Santander, Magdalena, Atlántico, and La Guajira. This may reflect cryptic transmission events within these departments that sampling has yet to recover; however, this may also be the result of transnational spread of SARS-CoV-2 by movement of infected individuals from Venezuela to Colombia or by movement of infected individuals from different countries to Venezuela and later to Colombia. The latter is further supported by the phylogenetically similar isolates at the northern Colombian (La Guajira)-Venezuelan (Zulia) departmental border (Figure 1B). This “viral transit” across borders has been reported in other South American border countries including Uruguay (Rivera) and Brazil (Elizondo et al. 2021) and Colombia and Brazil (Ramírez et al. 2021). Together, these highlight the role that human transit plays in the introduction and transmission of new viral variants, which further potentiates virus spread and evolution. Thus, additional, systematic studies are needed in this region to understand the current dissemination of SARS-CoV-2 lineages at a micro-geographical scale.
Among the Ven_Col isolates, the presence of the A23403G substitution across all 30 genomes was confirmed, which resulted in the D614G spike variant that is associated with increased infectivity (Plante et al. 2021; Zhou et al. 2021) (Figure 2). Moreover, this study also identified the non-synonymous substitution (C21614T) in two (6.6%) of the Ven_Col genomes analyzed. This resulted in the L18F mutation in the viral spike protein that has recently been characterized in the South African variant – 501Y.V2 – and which may confer increased viral replication and compromise neutralization by antibodies (Grabowski et al. 2021). While this mutation has been reported in isolates from other South American countries, including Colombia, Chile and Ecuador (Vilar and Isom 2021), recovery and further characterization of these isolates are vital to estimate the clinical and phylogenetic significance of this variant. Furthermore, identification of these variants is essential to inform understanding of viral fitness as spread throughout South America continues.
Given the precarious governmental and public health infrastructures in Colombia, and particularly Venezuela, it is important to note that constraints in available epidemiological information limited this study. Comparisons with isolates from Venezuela are desperately needed; however, after more than a year since the beginning of the pandemic, 12 genomes have been recovered from the country ((Loureiro et al. 2021; Paniz-Mondolfi et al. 2020). Moreover, given the sociopolitical and socioeconomic crises in Venezuela, the healthcare system has had limited diagnostic capacity, which likely severely underestimated the number of infected individuals. This presents a problem for any genomic surveillance study and further hinders any infection prevention efforts to curb further transmission within and across countries. The need for such studies is highlighted by the fact that although Colombia closed the border with Venezuela on 16 March 2020, Venezuelan residents have illegally crossed to Colombia. Moreover, in response to socioeconomic factors, these migrants have been forced to disperse throughout Colombia, migrate to surrounding countries, or even return to Venezuela amidst the pandemic (Fernández-Niño et al. 2020). This abrupt movement of people could potentiate transmission of events, and if continued, unchecked spread could further complicate management of the pandemic in already vulnerable communities and countries with limited vaccine availability across South America (Paniz-Mondolfi et al. 2020a, 2020b; Loyo et al., 2021).
Finally, it is important to note that the identification of circulating lineages and transmission routes in both countries could, in the future, be the basis for the development of studies aimed at analyzing the effectiveness of vaccines which, to date, have a global efficacy of > 90% (Pfizer–BioNTech (∼95%), Moderna (∼94%) and Sputnik V (∼92%)) except for Oxford-AstraZeneca (∼81%) (Baden et al. 2021; Doroftei et al. 2021).
Acknowledgments
AUTHORS CONTRIBUTIONS
Study design and conceptualization: LHP, MM, JDR, AEPM; Data collection: LHP, NB, MM, SC, CH, SG, CF, AR, LP, CEHP, LDN, MEG; Methodology: LHP, NB, MM, SC, CH, SG, CF, AR, LP, CEHP, LDN, ZK, AvdG, JG. Data analysis, curation and interpretation: LHP, NB, MM, SC, CH, SG, CF, AR, LP, CEHP, LDN, MEG, MMH, ASGR, VS, HvB, EMS, JDR, AEPM; Writing/Drafting: LHP, MM, MMH, JDR, AEPM; Critical revision of the article: LHP, MM, MMH, VS, HvB, EMS, JDR, AEPM. All authors have read and agreed to the published version of the manuscript.
ACKNOWLEDGEMENTS
We thank Dr. Martin Llewellyn and members from the Vector-borne disease control in Venezuela Network. Thanks to President and the University council for leading the strategic projects.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
FUNDING SOURCE
This study was partially funded by the UK Research and Innovation, Global Challenges Research Fund (EP/T003782/1), the University of Glasgow Small Grants Fund and Scottish Funding Council. This project was also funded by the Universidad del Rosario in the framework of its strategic plan RUTA2025.
ETHICAL APPROVAL
The study was approved by the Comité de Ética en Investigación de la Universidad del Rosario (CEI-UR) with the number DVO005 15 08-CV1400. By the health emergency under the law 9-1979, decrees 786-1990 and 2323-2006 the Colombian National Institute of Health (INS) is authorized to use biospecimens and associated epidemiological information without the written consent. The study followed the principles of the Declaration of Helsinki, and also all patient data were anonymized to minimize risk to participants.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2021.07.069.
Appendix. Supplementary materials
REFERENCES
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. The New England journal of medicine. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito M.O. COVID-19 in the Americas: Who's Looking After Refugees and Migrants? Ann Glob Health. 2020;86(1):69. doi: 10.5334/aogh.2915. Jun 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels J.P. Venezuelan migrants “struggling to survive” amid COVID-19. Lancet. 2020;395(10229):1023. doi: 10.1016/S0140-6736(20)30718-2. Mar 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroftei B., Ciobica A., Ilie O.D., Maftei R., Ilea C. Mini-Review Discussing the Reliability and Efficiency of COVID-19 Vaccines. Diagnostics. 2021;11(4) doi: 10.3390/diagnostics11040579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elizondo V., Harkins G.W., Mabvakure B., Smidt S., Zappile P., Marier C., et al. SARS-CoV-2 genomic characterization and clinical manifestation of the COVID-19 outbreak in Uruguay. Emerg Microbes Infect. 2021;10(1):51–65. doi: 10.1080/22221751.2020.1863747. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estadisticas-Venezuela|COVID-19. https://covid19.patria.org.ve/estadisticas-venezuela/ [WWW Document] (accessed 7820) 2021.
- Fernández-Niño J.A., Cubillos-Novella A., Bojórquez I., Rodríguez M. Recommendations for the response against COVID-19 in migratory contexts under a closed border: The case of Colombia. Biomedica. 2020;40(Supl. 2):68–72. doi: 10.7705/biomedica.5512. Oct 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Pinzón V., Mantilla J. Contested borders: organized crime, governance, and bordering practices in Colombia-Venezuela borderlands. Trends Organ Crime. 2020:1–17. doi: 10.1007/s12117-020-09399-3. Nov 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski F., Preibisch G., Giziński S., Kochańczyk M., Lipniacki T. SARS-CoV-2 Variant of Concern 202012/01 Has about Twofold Replicative Advantage and Acquires Concerning Mutations. Viruses [Internet] 2021;13(3) doi: 10.3390/v13030392. Mar 1Available fromhttp://dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillet M.E., Hernandez-Villena J.V., Llewellyn M.S., Paniz-Mondolfi A.E., Tami A., Vincenti-Gonzalez M.F., et al. Venezuela's humanitarian crisis, resurgence of vector-borne diseases, and implications for spillover in the region. The Lancet Infectious diseases. 2019;19(5):e149–ee61. doi: 10.1016/S1473-3099(18)30757-6. [DOI] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. Dec 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Instituto Nacional de Salud (INS). COVID-19 en Colombia [Internet]. Actualización No. 15 - Mayo 10 de 2021. 2021 [cited 2021 May 14]. Available from: https://www.ins.gov.co/Noticias/Paginas/coronavirus-genoma.aspx
- Lee J.-S., Kim S.Y., Kim T.S., Hong K.H., Ryoo N.-H., Lee J., et al. Evidence of Severe Acute Respiratory Syndrome Coronavirus 2 Reinfection After Recovery from Mild Coronavirus Disease 2019. Clin Infect Dis [Internet] 2020 doi: 10.1093/cid/ciaa1421. Nov 21Available fromhttp://dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loureiro C.L., Jaspe R.C., D Angelo P., Zambrano J.L., Rodriguez L., Alarcon V., et al. SARS-CoV-2 genetic diversity in Venezuela: Predominance of D614G variants and analysis of one outbreak. PLoS One. 2021;16(2) doi: 10.1371/journal.pone.0247196. Feb 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyo E.S.L., Gonzalez M.J., Esparza J. Venezuela is collapsing without COVID-19 vaccines. Lancet. 2021;397(10287):1806. doi: 10.1016/S0140-6736(21)00924-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder M., van der Vegt D.S.J.M., Oude Munnink B.B., GeurtsvanKessel C.H., van de Bovenkamp J., Sikkema R.S., et al. Reinfection of SARS-CoV-2 in an immunocompromised patient: a case report. Clin Infect Dis [Internet]. 2020 doi: 10.1093/cid/ciaa1538. Oct 9; Available fromhttp://dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz M., Patiño L.H., Ballesteros N., Paniz-Mondolfi A., Ramírez J.D. Characterizing SARS-CoV-2 genome diversity circulating in South American countries: Signatures of potentially emergent lineages? Int J Infect Dis. 2021;105:329–332. doi: 10.1016/j.ijid.2021.02.073. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K., Golosova O., Fursov M. UGENE team. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28(8):1166–1167. doi: 10.1093/bioinformatics/bts091. Apr 15. [DOI] [PubMed] [Google Scholar]
- Paniz-Mondolfi A., Muñoz M., Florez C., Gomez S., Rico A., Pardo L., et al. SARS-CoV-2 spread across the Colombian-Venezuelan border [Internet] Infection, Genetics and Evolution. 2020;86 doi: 10.1016/j.meegid.2020.104616. Available fromhttp://dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniz-Mondolfi A.E., Sordillo E.M., Márquez-Colmenarez M.C., Delgado-Noguera L.A., Rodriguez-Morales A.J. The arrival of SARS-CoV-2 in Venezuela. Lancet. 2020;395(10236):e85–e86. doi: 10.1016/S0140-6736(20)31053-9. May 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592(7852):116–121. doi: 10.1038/s41586-020-2895-3. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. https://www.R-project.org/ (2018).
- Rambaut A., Holmes E.C., O'Toole Á., Hill V., McCrone J.T., Ruis C., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez J.D., Florez C., Muñoz M., Hernández C., Castillo A., Gomez S., et al. The arrival and spread of SARS-CoV-2 in Colombia. J Med Virol. 2021;93(2):1158–1163. doi: 10.1002/jmv.26393. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende P.C., Delatorre E., Gräf T., Mir D., do Couto Motta F., Appolinario L.R., et al. Genomic surveillance of SARS-CoV-2 reveals community transmission of a major lineage during the early pandemic phase in Brazil [Internet]. bioRxiv. 2020 [cited 2021 May 14]. p. 2020.06.17.158006. Available from: https://www.biorxiv.org/content/10.1101/2020.06.17.158006v1.abstract
- Rodríguez-Morales A.J., Bonilla-Aldana D.K., Morales M., Suárez J.A. Martínez-Buitrago E. Migration crisis in Venezuela and its impact on HIV in other countries: the case of Colombia. Ann Clin Microbiol Antimicrob. 2019;18(1):9. doi: 10.1186/s12941-019-0310-4. Mar 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Morales A.J., Suárez J.A., Risquez A., Delgado-Noguera L., Paniz-Mondolfi A. The current syndemic in Venezuela: Measles, malaria and more co-infections coupled with a breakdown of social and healthcare infrastructure. Quo vadis? Travel Med Infect Dis. 2019;27:5–8. doi: 10.1016/j.tmaid.2018.10.010. Jan. [DOI] [PubMed] [Google Scholar]
- Standley C.J., Chu E., Kathawala E., Ventura D., Sorrell E.M. Data and cooperation required for Venezuela's refugee crisis during COVID-19. Global Health. 2020;16(1):103. doi: 10.1186/s12992-020-00635-7. Oct 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar S., Isom D.G. One Year of SARS-CoV-2: How Much Has the Virus Changed? [Internet] Biology. 2021;10:91. doi: 10.3390/biology10020091. Available fromhttp://dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Thao T.T.N., Hoffmann D., Taddeo A., Ebert N., Labroussaa F., et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature. 2021;592(7852):122–127. doi: 10.1038/s41586-021-03361-1. Apr. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.