Foreman et al. find CD30 is preferentially expressed on granuloma T cells in Mtb-infected macaques. In mice, CD30 drives T cell differentiation and is required for host survival of Mtb infection, showing T cell immunity to Mtb requires CD30-dependent co-stimulation.
Abstract
Control of Mycobacterium tuberculosis (Mtb) infection requires generation of T cells that migrate to granulomas, complex immune structures surrounding sites of bacterial replication. Here we compared the gene expression profiles of T cells in pulmonary granulomas, bronchoalveolar lavage, and blood of Mtb-infected rhesus macaques to identify granuloma-enriched T cell genes. TNFRSF8/CD30 was among the top genes upregulated in both CD4 and CD8 T cells from granulomas. In mice, CD30 expression on CD4 T cells is required for survival of Mtb infection, and there is no major role for CD30 in protection by other cell types. Transcriptomic comparison of WT and CD30−/− CD4 T cells from the lungs of Mtb-infected mixed bone marrow chimeric mice showed that CD30 directly promotes CD4 T cell differentiation and the expression of multiple effector molecules. These results demonstrate that the CD30 co-stimulatory axis is highly upregulated on granuloma T cells and is critical for protective T cell responses against Mtb infection.
Introduction
Mycobacterium tuberculosis (Mtb) is a leading cause of global mortality due to infectious disease. The current vaccine for Mtb infection, Bacillus Calmette-Guérin (BCG), is administered neonatally and provides protection against severe forms of tuberculosis (TB) in children, but it does not provide sufficient protection in adolescents and adults to drastically reduce the global burden TB. Over the past few years, clinical vaccine trials using BCG re-vaccination or an adjuvanted fusion protein have demonstrated ∼40–50% protection (Nemes et al., 2018; Van Der Meeren et al., 2018), and several vaccine studies in non-human primates (NHPs) have afforded extraordinary levels of protection, showing that a highly effective vaccination for TB is achievable (Darrah et al., 2020; Dijkman et al., 2019; Hansen et al., 2018; Kaushal et al., 2015; Sharpe et al., 2016; White et al., 2015). A better understanding of the mechanisms of host protection against Mtb infection would improve our ability to develop novel vaccination as well as host-directed therapeutic strategies for TB.
Control of Mtb infection requires the generation of T cells that can traffic to pulmonary granulomas, but little is known about the basic biology of granuloma T cells (i.e., the mechanisms regulating the generation, differentiation, homing, and effector functions of protective granuloma T cells). CD4 T cell responses to mycobacterial infections in humans, NHPs, and mice are primarily comprised of IFNγ-producing effector cells. However, there are differences in the quality of CD4 T cell differentiation observed in response to mycobacterial infection in C57Bl/6 mice compared to macaques and humans. In mice, Mtb-specific CD4 T cells develop into highly polarized Th1 cells, with some cells even differentiating into terminally differentiated Th1 effector cells. In contrast, these terminally differentiated cells are not observed in NHPs or humans (Kauffman et al., 2018b). Instead, in primate hosts, Mtb-specific T cells mostly differentiate into atypical IFNγ-producing cells referred to as Th1* cells that are characterized by a mixture of Th1 and Th17-like features (Acosta-Rodriguez et al., 2007; Burel et al., 2018; Lindestam Arlehamn et al., 2013; Singhania et al., 2021). Another key difference between the most common mouse models and primates is in the pulmonary lesions that develop at sites of bacterial infection. Specifically, mice do not develop typical granulomatous lesions characterized by a well-organized macrophage core circumscribed by lymphocyte cuff. While efforts are underway to improve on the classic mouse model by changing the dose or strain of bacteria or strain of mouse (Plumlee et al., 2021), NHPs are currently the most powerful animal model for investigating granuloma T cell responses in a setting that closely mirrors that seen in humans.
Here we identify unique properties of Mtb granuloma T cells. We compare gene expression profiles of FACS-purified CD4 and CD8 T cells from the blood, bronchoalveolar lavage (BAL), and individual granulomas of Mtb-infected rhesus macaques and identify homing receptors, transcription factors, cytokines, and co-stimulatory molecules that distinguish granuloma T cells from those appearing in circulation and non-granulomatous tissue. To identify a core granuloma T cell signature, we focused on granuloma-enriched genes that were shared between CD4 and CD8 T cells and whose expression occurred independent of granuloma bacterial loads. This analysis identified the TNF superfamily CD30 as a key molecule of interest in granuloma T cells. Importantly, we show in mice that expression of CD30 on CD4 T cells drives their differentiation and expression of multiple effector molecules and is required for host survival of Mtb infection. Together with our previous findings demonstrating that CD30 ligand, CD153, is also required for CD4 T cell–mediated control of Mtb infection (Du Bruyn et al., 2021; Riou et al., 2020; Sallin et al., 2018), these data point to a central role for the CD30/CD153 co-stimulatory axis in CD4 T cell–mediated control of Mtb infection.
Results
RNA sequencing (RNAseq) of FACS-purified Mtb granuloma T cells in NHP
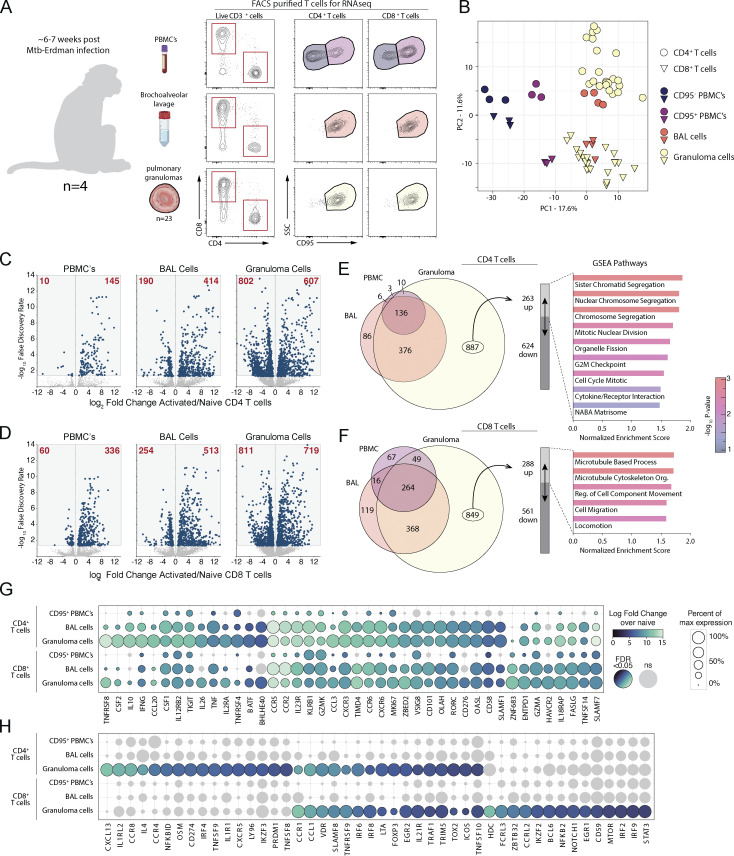
Rhesus macaques were bronchoscopically exposed to 40–80 CFU of Mtb-Erdman-mCherry and euthanized 6–7 wk after infection for tissue harvest. Eight different populations of T cells were FACS-purified under biosafety level 3 (BSL3) conditions. Non-naive CD95high CD4 and CD8 T cells were FACS-purified from peripheral blood mononuclear cells (PBMCs), BAL, and individual granulomas. Naive T cells are not found in the BAL and granulomas, so naive CD4 and CD8 T cells (CD95low) were isolated from PBMC to serve as a comparator for CD95high T cells isolated from each of the three tissues (Fig. 1 A and Table S1). These purified cell populations were each subjected to RNAseq. Principal component 1 (PC1) discriminated PBMC from lung samples and naive from non-naive T cells, whereas PC2 separated CD4 and CD8 T cells (Fig. 1 B). CD95high blood T cells clustered apart from granuloma and BAL T cells, which scored similarly in PC1 and PC2. In comparison with naive T cells, granuloma CD4 and CD8 T cells had the largest number of differentially expressed genes (DEGs) followed by BAL T cells and then CD95high PBMC T cells, which had the lowest number of DEGs (Fig. 1, C and D; and Tables S2 and S3). We next performed gene set enrichment analysis (GSEA) on the upregulated DEGs unique to granuloma T cells and gene sets associated with cell division for CD4 T cells and with cell migration for CD8 T cells were significantly overrepresented (Fig. 1, E and F).
Figure 1.
Gene expression pattern in rhesus macaque Mtb granuloma T cells. (A) Study schematic showing CD4 and CD8 T cells FACS-purified from blood, BAL, and 23 pulmonary granulomas isolated from four rhesus macaques infected with a low dose of Mtb for subsequent transcriptomic analysis. (B) Principal component analysis of the top 7,000 genes expressed. (C and D) Log2 fold change of gene expression of activated over naive T cells from (C) CD4 and (D) CD8 T cells with number of genes indicated in red. (E and F) Proportional Venn diagrams of significant genes differentially regulated from the three tissue compartments with GSEA of the upregulated genes specific only to granulomas from (E) CD4 and (F) CD8 T cells. (G and H) Curated list of genes demonstrated as relative percent expression with indicated significant fold change shown by color to demonstrate genes (G) upregulated in activated T cells compared to (H) genes significantly upregulated only in T cells isolated from granulomas.
We manually curated lists of genes that were preferentially upregulated in BAL and granuloma T cells (Fig. 1 G) or that were statistically significantly upregulated only in granuloma T cells (Fig. 1 H). We found several key homing receptors including CCR5, CCR2, CXCR3, CCR6, and CCR1 were preferentially expressed by granuloma homing T cells. CCR8 and CCR4 were upregulated only on granuloma CD4 T cells. We also found cytokines and chemokines that are preferentially expressed in BAL and granuloma T cells including GM-CSF, IL-10, IFNγ, M-CSF, IL-26, TNF, CCL20, CCL3, and CCL1. Upregulation of CXCL13, IL4, and oncostatin M (OSM) was unique to CD4 T cells in granulomas. We also noted several key transcription factors that were preferentially expressed in granuloma T cells: BATF, BHLHE40, ZBED2, RORC, vitamin D receptor (VDR), IRF6, IRF8, EGR2, and TOX2. Interestingly, PRDM1 was only found in granuloma CD4 T cells, and ZBTB32, IKZF2, BCL6, EGR1, IRF2, IRF9, and STAT3 were only found upregulated in granuloma CD8 T cells. Lastly, we observed several TNF superfamily molecules preferentially expressed in granulomas, including TNFRSF8 (CD30), TNFRSF4 (OX-40), TNFRSF9 (4-1BB), LTA, and TNFSF10 (TRAIL). TNFSF9 (4-1BB-L) and TNFSF8 (CD30L/CD153) were only found on granuloma CD4 T cells.
Unique features of Mtb granuloma T cells
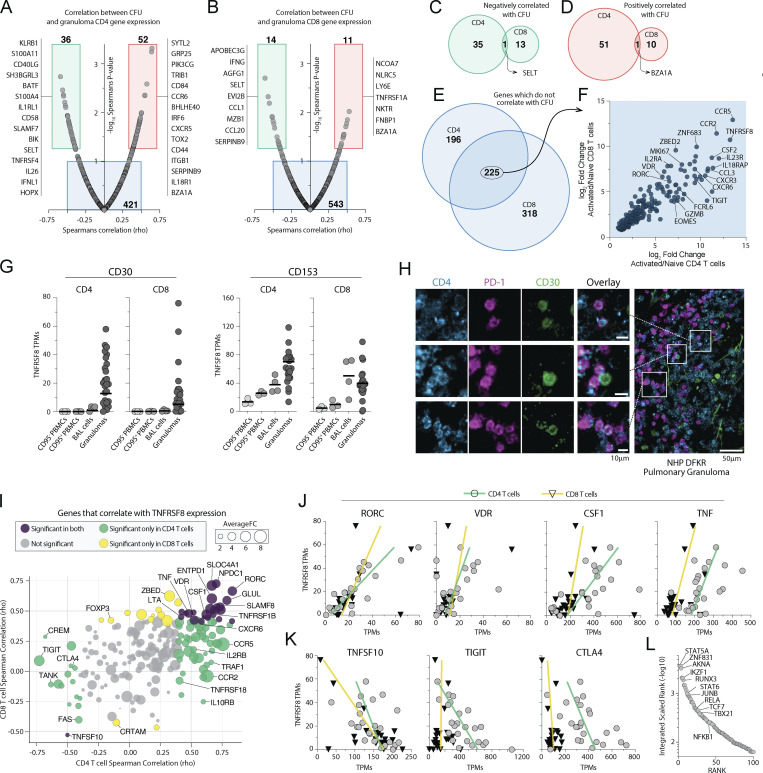
We next analyzed correlations between gene expression levels in T cells and bacterial loads in individual granulomas. In CD4 T cells, we found 52 genes significantly positively correlated and 36 genes significantly negatively correlated with bacterial burdens (Fig. 2 A and Table S4). KLRB1, S100A11, CD40LG, BATF, and S100A4 were among the top negative correlates with CFU, and SYTL2, GPR25, CCR6, and BHLHE40 were the strongest positive correlates (Fig. 2 A). In CD8 T cells, 11 genes positively and 14 genes negatively correlated with bacterial burden (Fig. 2 B and Table S5). IFNG, CCL1, and CCL20 were among the genes in CD8 T cells that negatively correlated with CFU, and NLRC5 and LY6E were among the positive correlates.
Figure 2.
TNFRSF8 (CD30) is highly upregulated in granuloma CD4 and CD8 T cells. (A and B) Gene expression correlation with individual granuloma CFU demonstrating genes positively and negatively correlated with bacterial burden in (A) CD4 and (B) CD8 T cells with list of curated genes displayed next to box indicating significance. (C–E) Venn diagrams of genes shared between CD4 and CD8 T cells which (C) negatively correlate, (D) positively correlate, or (E) do not correlate with granuloma CFU. (F) Log2 fold change of the genes which are independent of granuloma bacterial burden and shared between CD4 and CD8 T cells. (G) TPM expression levels of both CD30 and CD153 in blood, BAL, and granulomas. (H) Immunohistochemical staining for CD30 expression on distinct subset of CD4 T cells in a granuloma. Scale bars represent 10 μm in small panels and 50 μm in the zoomed image. (I) Gene correlations with Tnfrsf8 expression in both CD4 and CD8 T cells showing average fold change between both CD4 and CD8 T cells from granulomas. (J and K) Linear correlations of genes (J) positively correlated and (K) negatively correlated with CD30 expression. (L) Transcription factor enrichment analysis of genes significantly correlated with Tnfrsf8 expression.
We next set out to identify a set of granuloma T cell core genes. We focused on genes that do not correlate with bacterial loads, as it is possible that genes which correlated with CFU were only differentially expressed in granulomas because they are regulated by inflammatory processes or antigen stimulation ongoing at the site of infection. Moreover, we speculated that genes important in the identity of granuloma T cells would be shared between CD4 and CD8 T cells. Thus, we next examined the overlap in genes between CD4 and CD8 T cells that positively correlated, negatively correlated, or did not correlate with CFU (Fig. 2, C–E). The genes in CD4 and CD8 T cells that correlated with CFU were largely different. Among the genes that positively or negatively correlated with bacterial loads, there was very little overlap between CD4 and CD8 T cells. Only a single gene was shared between the genes in CD4 and CD8 T cells that positively correlated with CFU. Similarly, a single gene was shared between the genes in CD4 and CD8 T cells that negatively correlated with CFU. On the other hand, there was a substantial overlap between CD4 and CD8 T cell genes that did not correlate with CFU (Fig. 2 E). We next asked what genes in this subset of non-correlated, shared genes were the most differentially expressed on granuloma T cells over naive T cells. CCR5, TNFRSF8 (CD30), and CCR2 were the top three genes. CSF2 (GM-CSF) was the top cytokine in the list. CXCR3 and CXCR6 were also found to be among the top genes most differentially expressed. Notably, the transcription factors ZNF683 (Hobit), ZBED2, VDR, RORC, and EOMES were also found. The cytokine receptors IL18RAP, IL23R, and IL2RA were also highly differentially expressed in both in both CD4 and CD8 T cells in granulomas independent of bacterial loads.
TNFRSF8 (CD30) expression distinguishes granuloma T cells
We have previously shown that CD153/CD30L (TNFSF8) is required for CD4 T cell–mediated control of Mtb infection (Sallin et al., 2018), so we were surprised to find that expression of its receptor, CD30 (TNFRSF8), was one of the most distinguishing features of granuloma T cells second only to CCR5. Indeed, both the ligand and receptor are highly expressed by activated T cells in granulomas (Fig. 2 G). Although we observed CD153 mRNA in sorted CD8+ cells, we previously showed Mtb-specific CD8 T cells express very little CD153 protein as detected by flow cytometry (Sallin et al., 2018). In contrast, Mtb-specific CD4 T cells in macaque granulomas express low baseline levels of CD153 and further upregulate it after peptide restimulation. Imaging thick-section tissue explants of freshly isolated granulomas showed that CD30 staining was detected on a subset of granuloma T cells (Fig. 2 H). Therefore, we next looked for genes that correlated with CD30 in granuloma CD4 and CD8 T cells (Fig. 2 I; and Tables S6 and S7). RORC, VDR, CSF1, and TNF all positively correlated with CD30 expression in both CD4 and CD8 T cells (Fig. 2 J). In contrast, CTLA4, TIGIT, and TNFSF10 (TRAIL) negatively correlated with CD30 (Fig. 2 K). To ask if there is a common upstream transcriptional regulator of the genes that correlate with CD30, we performed integrated transcription factor rank analysis. This identified STAT5A, ZNF831 (Dybska et al., 2021), or AKNA (Siddiqa et al., 2001) as candidates that may contribute to the control of CD30-associated genes (Fig. 2 L). In summary, these data revealed that CD30 is highly upregulated on a subset of granuloma CD4 and CD8 T cells and correlates with two transcription factors previously associated with anti-mycobacterial immunity (RORC and VDR), macrophage acting cytokines (TNF and M-CSF) and correlates inversely with co-inhibitory receptors (CTLA4 and TIGIT).
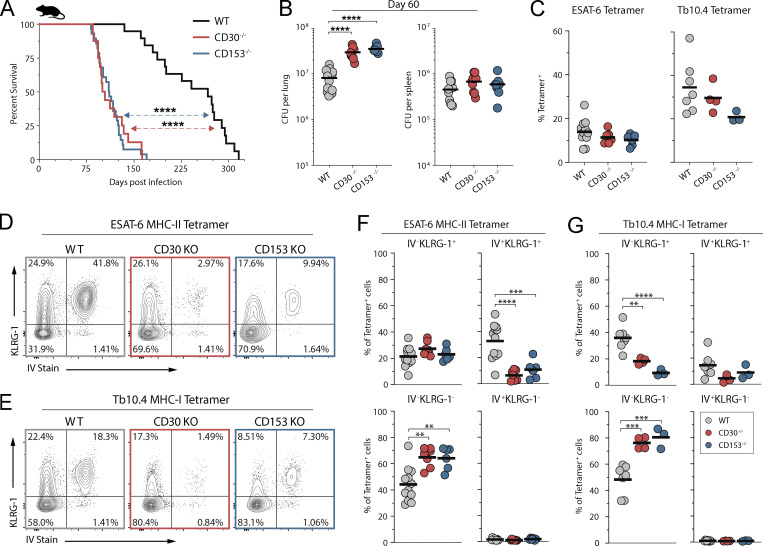
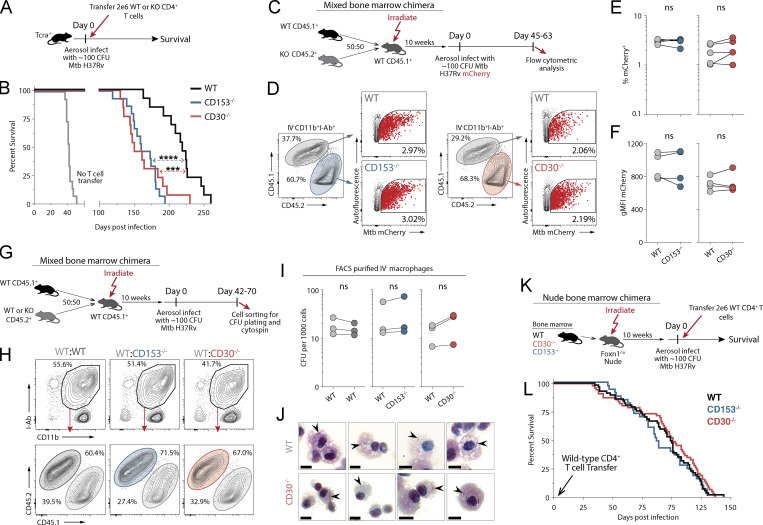
CD30-deficient mice succumb early to Mtb infection
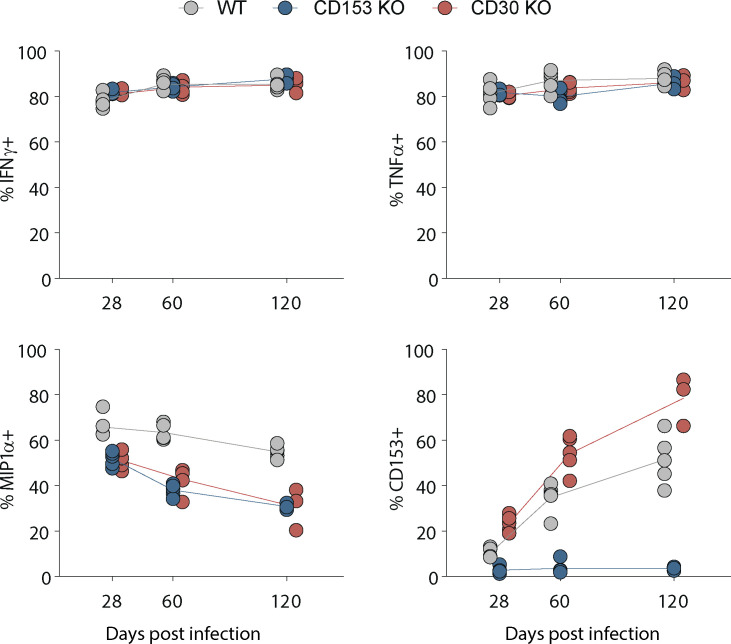
We next examined the role of CD30 in control of Mtb infection in the murine model. Mice deficient in either CD30 or CD153 succumbed to Mtb infection before WT mice and had higher lung bacterial loads at day 60 after infection (Fig. 3, A and B). I-Ab/ESAT-64-17 tetramer+ CD4 T cells and Kb/TB10.3/474-88 tetramer+ CD8 T cells were present in similar frequencies in the lungs of WT, CD30−/−, or CD153−/− mice, indicating that the CD30/CD153 axis is not required for the accumulation of T cells in the lungs during Mtb infection in mice (Fig. 3 C). We also did not observe any major defect in the ability of CD4 T cells in CD153 or CD30-deficient mice to produce IFNγ, TNF, or MIP1α after in vitro peptide restimulation (Fig. S1). Mtb-specific T cells in mice can be subdivided by their location in the lung parenchyma versus vasculature using the intravascular stain (iv) and differentiation state based on KLRG1 (Sakai et al., 2014). KLRG1+iv+ T cells are terminally differentiated cells and do not adoptively transfer protection against Mtb infection. The iv− cells in the parenchyma can be subdivided based on KLRG1 expression. KLRG1+iv− cells typically bear markers of an intermediate differentiation state and the KLRG1−iv− cells the least, but both iv− subsets can mediate control of Mtb infection in mice (Clemmensen et al., 2021; Moguche et al., 2015; Sallin et al., 2017; Woodworth et al., 2017). CD30- and CD153-deficient mice both displayed increased frequencies of KLRG1−iv− cells and decreased frequencies of KLRG1+iv+ Mtb-specific CD4 and CD8 T cells (Fig. 3, D–G). Collectively, these data show that mice deficient in the TNF(R)SF8 pathway succumb to Mtb infection early with increased pulmonary bacterial burdens despite normal frequencies of antigen-specific T cells and increased frequencies of protective T cells, indicating that CD30 may be required for the protective function, not generation, of iv− T cells.
Figure 3.
CD30-deficient mice are susceptible to Mtb infection. (A) Survival curve of intact knockout mice after aerosol infection with ∼100 CFU of Mtb (n = 16–28 per group, three independent experiments shown together). (B) Bacterial burden in the lungs and spleens of mice on day 60 after infection (n = 10–12 per group). (C) Frequency of live CD4+CD44+ T cells that are ESAT-6 tetramer+ or live CD8+CD44+ T cells that are Tb10.4 tetramer+. (D and E) Example flow cytometry plots of T cells showing iv stain and expression of KLRG-1 in (D) ESAT-6 tetramer+ CD4 T cells and (E) Tb10.4 tetramer+ CD8 T cells. (F and G) Frequency of (F) ESAT-6 tetramer+ CD4 T cells and (G) Tb10.4 tetramer+ CD8 T cells expressing KLRG-1 within the vasculature versus parenchyma; (C–G) two independent experiments shown together with n = 3–12 per group. Statistical analysis was calculated by (A) Kaplan–Meier survival curve with log-rank test or (B, C, F, and G) one-way ANOVA with Tukey correction for multiple comparisons. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Figure S1.
WT, CD153−/−, and CD30−/− mice were euthanized at days 28, 60, and 120 after aerosol infection and T cells restimulated for 5 h with ESAT-61-20 peptide in the presence of brefeldin/monensin. Data shown are gated on CD4+CD154+ T cells to compare cytokine expression of antigen-specific CD4 T cells.
T cells require CD30 to mediate protection against Mtb infection in mice
To directly test whether there is a role for CD30 on T cells in control of Mtb infection, we next examined the ability of CD30-deficient CD4 T cells to transfer protection to Mtb-infected T cell–deficient mice (Fig. 4 A). T cell–deficient mice that received T cells from CD30−/− succumbed early to infection compared to mice receiving WT CD4 T cells (Fig. 4 B). As we previously showed (Sallin et al., 2018), transfer of CD153−/− CD4 T cells also failed to restore normal protection to T cell–deficient mice (Fig. 4 B). Thus, CD4 T cells surprisingly require both the receptor and ligand to mediate control of Mtb infection.
Figure 4.
T cells require CD30 to mediate protection against Mtb infection in mice. (A and B) Experiment schematic for CD4 T cell adoptive transfer (A) and the survival curve of Tcra−/− mice who received WT, CD30−/−, or CD153−/− CD4 T cells at the time of infection (B). Data represent three independent experiments with n = 4–5 per group per experiment. (C) Experiment schematic for MBMC mice to study the intrinsic roles of CD30 and CD153. (D) Example flow cytometry plots of live IV−CD11b+I-Ab+ macrophages from mice infected with mCherry-reporter Mtb. (E and F) Frequency of (E) and gMFI of (F) mCherry+ macrophages in MBMC mice in WT:CD30−/− and WT:CD153−/− chimeras. Data shown are representative of three independent experiments with n = 4–5 per group. (G) Experiment schematic for MBMC mice to study bacterial burdens on a per cell basis. (H) Example flow cytometry plots of live IV−CD11b+I-Ab+ macrophages FACS-sorted for CFU plating. (I) Quantification of the number of bacteria per 1,000 macrophages. Each point represents four to five mice pooled per group showing three independent experiments. (J) FACS-purified macrophages from MBMC mice were cytospin-mounted for subsequent acid-fast bacilli staining, demonstrating no increase in bacterial levels per cell. Arrowheads point to bacilli. Scale bars represent 5 μm. (K and L) Experiment schematic (K) for bone marrow reconstitution of athymic nude mice with subsequent CD4 T cell adoptive transfer at the time of infection with the (L) survival curve of mice where all hematopoietic cells except CD4 T cells are from WT, CD30−/−, or CD153−/− bone marrow donors. Data are representative of six independent experiments with n = 3–14 mice per group per experiment, for a total of 33–55 mice per group. Statistical analysis was calculated by (B and L) Kaplan–Meier survival curve with log-rank test or (E, F, and L) paired t tests. ***, P < 0.001; ****, P < 0.0001
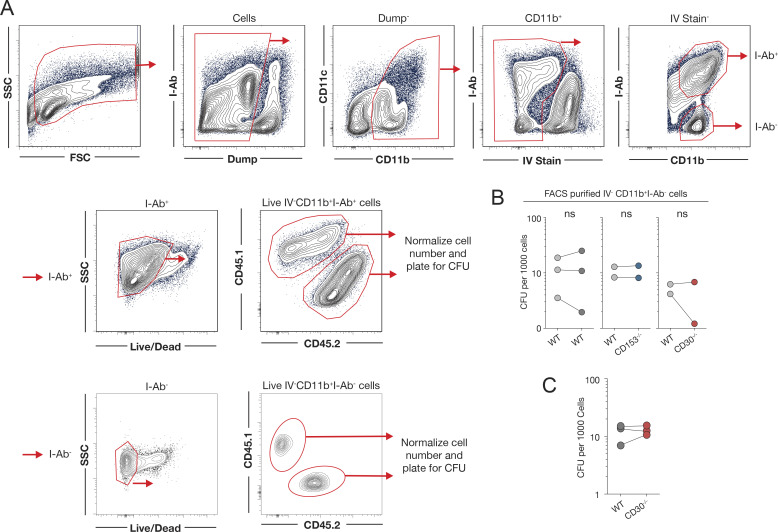
In our previous work that identified a key role for CD153 in CD4 T cell–mediated host resistance to Mtb infection, we speculated that CD153-expressing CD4 T cells may trigger CD30 on Mtb-infected macrophages (Sallin et al., 2018). Therefore, we next asked whether CD30 is also required on myeloid cells for control of Mtb infection. Groups of WT:CD30−/− and WT:CD153−/− mixed bone marrow chimeric (MBMC) mice were infected with an mCherry-reporter strain of Mtb, and myeloid cells were examined for bacterial loads 45–63 d after infection (Fig. 4 C). CD45.1+ WT and CD45.2+ CD30−/− parenchymal myeloid cells (iv− CD11b+ I-Ab+ cells) from the lungs of MBMC mice contained similar frequencies of mCherry+ Mtb-infected cells, and the infected cells had similar mean fluorescence intensities (Fig. 4, D–F). CD153−/− myeloid cells were also not different than WT myeloid cells in their bacterial loads. To further test whether there is difference in the ability of CD30- and CD153-deficient macrophages to control growth of Mtb infection in MBMC mice, I-Ab+ parenchymal myeloid cells were FACS-sorted and plated for enumeration of CFUs (Fig. S2 A and Fig. 4, G and H). There was no significant difference in the CFU per 1,000 plated myeloid cells in the WT:WT, WT:CD30−/−, or WT:CD153−/− MBMC mice (Fig. 4 I). Although they cannot present antigen to CD4 T cells, we also sorted and plated the I-Ab− parenchymal myeloid cells (comprising the rest of the CD11b+ cells) and found no role for CD153 or CD30 for suppressing Mtb growth in these cells (Fig. S2 B). FACS-purified WT and CD30−/− myeloid cells from MBMC mice were mounted on slides by cytospin, and bacteria were manually counted after acid-fast staining (Fig. S2 C and Fig. 4 J). There was no evident increase in bacteria per cell in WT or CD30−/− macrophages upon acid-fast staining (Fig. 4 J). These data show that CD30 on infected myeloid cells does not play a role in directly suppressing bacterial growth.
Figure S2.
Quantification of bacterial loads in FACS-purified cells from Mtb-infected MBMC mice. (A) Gating schema for FACS purification of parenchymal CD11b+I-Ab+ macrophages and CD11b+I-Ab− myeloid cells to plate for bacterial burden on a per cell basis. (B) Quantification of the number of bacteria per 1,000 parenchymal CD11b+I-Ab− myeloid cells. Each point represents four to five mice pooled per group, showing independent experiments. (C) Quantification of acid-fast bacilli from cytospin slides of sorted macrophages. (B and C) Statistical analysis was calculated using paired t tests.
Lastly, we tested if there is a role for CD30 on any other hematopoietic cells in control of Mtb infection. To create a situation where T cells are normal, but all other hematopoietic cells lack CD30, we first lethally irradiated athymic nude mice and reconstituted them with WT, CD30−/−, or CD153−/− bone marrow cells. These reconstituted nude mice then were injected with WT CD4 T cells and infected with Mtb (Fig. 4 K). Mice that lacked the either CD30 or CD153 in all hemopoietic cells except CD4 T cells succumbed to infection at the same rate as nude mice reconstituted with WT bone marrow (Fig. 4 L). Thus, CD4 T cells require both CD30 and CD153 to control Mtb infection, and we could find no role for CD30 on any other cell types.
CD30 on T cells drives differentiation and effector molecule expression during Mtb infection
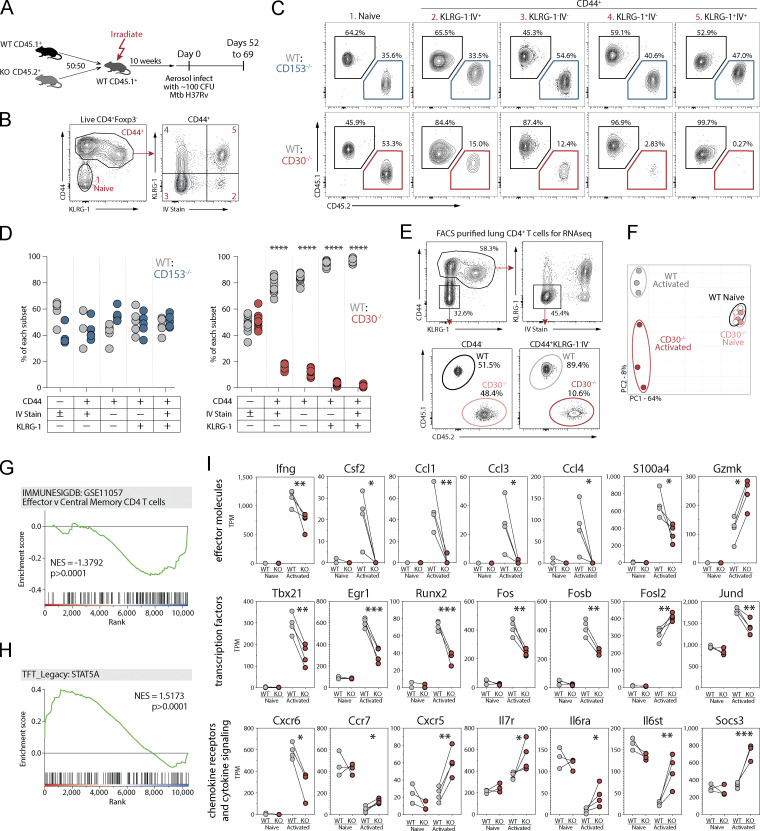
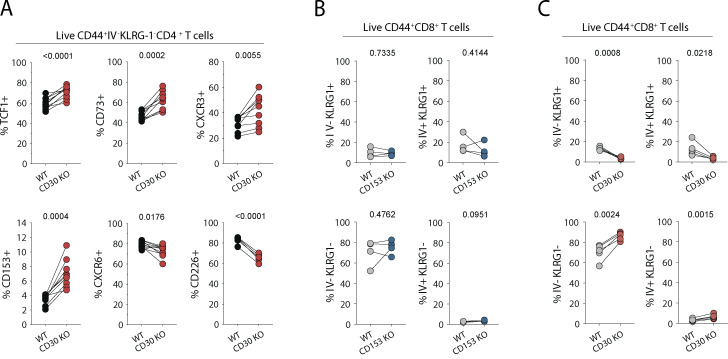
To test whether CD30 or CD153 signaling into CD4 T cells impacts their responses, we infected WT:CD30−/− or WT:CD153−/− MBMC mice with Mtb and analyzed the CD4 T cell response in the lungs (Fig. 5 A). First, we examined how these gene deficiencies impacted T cell differentiation based on the iv and KLRG1 expression as described above (Fig. 5, B and C). Naive T cells in both sets of chimeric mice were present in a 1:1 WT to KO ratio. However, the competitive setting of the MBMC mice exacerbated the phenotype of the CD30−/− T cells compared to what was observed in the intact WT versus CD30−/− mice. In WT:CD30−/− MBMC mice, the overall expansion of all CD30-deficient CD44high effector cells was defective relative to WT CD4 T cells in the same mouse, and the severity of the defect relative to WT cells worsened as the differentiation state of the cells progressed. That is, iv−KLRG1− cells displayed the least severe defect and iv+KLRG1+ cells the largest (Fig. 5, C and D). In contrast, while T cells in intact CD153−/− mice displayed an altered differentiation state, they mounted completely normal responses relative to their WT counterparts in the MBMC setting, indicating that when a source of CD153 is available to signal into CD30 on the CD153−/− T cells, their response returns to normal. Analysis of several other markers associated with differentiation further confirmed that CD30−/− T cells in the MBMC mice were less-differentiated (Fig. S3 A). Moreover, CD30−/− but not CD153−/− CD8 T cells also displayed defects in their differentiation (Fig. S3, B and C). Therefore, CD30, but not CD153, signals directly into T cells to drive their differentiation.
Figure 5.
CD30 signaling drives CD4 T cell differentiation and effector molecule expression during Mtb infection. (A) Experiment schematic showing MBMC mice to study the T cell–intrinsic roles of CD30 and CD153 in responses to Mtb infection. (B and C) Example flow cytometry plots and gating of CD4 T cell populations for analysis comparing WT:KO ratios within each of these populations. (D) Frequency of WT:CD153−/− and WT:CD30−/− cells within each of these T cell populations. Data shown are representative of four independent experiments with n = 4–10 mice per group per experiment. (E) Example flow cytometry plots for FACS purification of live IV−KLRG-1− CD44+CD4+ or CD44−CD4+ T cells for gene expression profiling. (F) Principal component analysis of the top 3,000 genes expressed in these populations from WT or CD30−/− CD4 T cells. Data shown are representative of three independent experiments with n = 5 mice per experiment pooled after FACS sorting. (G and H) GSEA of genes differentially expressed between WT and KO activated T cells in comparison to two gene sets showing WT cells highly enriched for STAT5a signaling, while KO cells are enriched for less effector-like gene expression. (I) Manually curated gene sets of CD30-regulated T cell effector molecules, transcription factors, chemokine receptors, and cytokine signaling molecules. Statistical analysis was calculated by (D) multiple t tests per row or (I) t tests. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Figure S3.
Phenotype of CD4 and CD8 T cells lacking CD30 and CD153. (A) Flow cytometric quantification of CD4 T cells from WT:CD30−/− MBMC mice euthanized 62–69 d after aerosol infection. Data are from two independent experiments with n = 5 each. (B and C) Comparison of CD8 T cell differentiation by IV stain and KLRG-1 expression in (B) WT:CD153−/− and (C) WT:CD30−/− MBMC mice. Statistical analysis was calculated using paired t tests with P value indicated above each graph.
Lastly, we investigated how CD30 signaling into CD4 T cells regulates their differentiation in Mtb-infected mice using RNAseq analysis. Since intact CD30−/− mice develop high Mtb bacterial loads which may introduce major caveats in a transcriptomic analysis, we again used WT:CD30−/− MBCM mice to ensure that the T cells we are comparing are exposed to the same infectious loads and inflammatory environment. Furthermore, since we have shown that there is a major difference in subsets of T cells that develop in the absence of CD30, we FACS-purified WT and CD30−/− iv−KLRG1− effector CD4 T cells from the lungs of MBMC mice (Fig. 5 E). Naive WT and CD30−/− CD4 T cells were also purified from the MBMC mice to serve as a comparator. Principal component analysis of the transcriptional profiles showed that WT or CD30−/− naive T cells were very similar, as expected, and PC1 largely discriminated naive from effector cells. However, the WT and CD30−/− effector cells separated from each other in PC2 (Fig. 5 F). GSEA of DEGs showed that CD30−/− CD4 T cells displayed a reduced effector memory T cell gene signature compared to their counterpart WT cells, again indicating that CD30 promotes CD4 T cell differentiation (Fig. 5 G; Tables S8 and S9). CD30−/− CD4 T cells also displayed a lack of a STAT5A gene signature (Fig. 5 H). This is consistent with the data from T cells purified from NHP granulomas showing that STAT5A was predicted to be an upstream regulator of CD30-correlated genes.
Several important effector molecules were defective in CD30−/− KLRG1−iv− effector CD4 T cells. IFNγ was significantly reduced, and GM-CSF, CCL1, CCL3, and CCL4 were almost completely lost in CD30-deficient T cells (Fig. 5 I). Interestingly, S100A4, the expression of which by CD4 T cells negatively correlated with bacterial loads in NHP granulomas was also significantly reduced in CD30−/− CD4 T cells. In contrast, CD30−/− CD4 T cells upregulated granzyme K to a higher level than WT CD4 T cells. CD30−/− CD4 T cells also displayed defective upregulation of several key transcription factors including Tbet, Egr1, and Runx2, and there was altered pattern of AP-1 family member expression (Fig. 5 I). CD30−/− CD4 T cells expressed lower levels of CXCR6, but increased levels of CCR7 and CXCR5 (Fig. 5 I). Lastly, there were several changes to the expression of molecules involved in cytokine signaling. CD30−/− effector CD4 T cells expressed higher levels of IL7R, IL6Ra, and Gp130. While SOCS3 was not upregulated in WT effector CD4 T cells compared to naive, CD30−/− effector CD4 T cells expressed elevated levels of this transcript. Collectively, these data show that in the mouse model of Mtb infection, CD30 promotes the differentiation of CD4 T cells to a more effector-like state and is required for the expression of several T cell effector cytokines and chemokines.
Discussion
Here we determined key features of T cells that home to Mtb granulomas in rhesus macaques. As expected, we found that genes associated with Th1* cells (i.e., CXCR3, CCR6, RORC, and IL23R) are highly enriched in granuloma T cells compared to T cells in the airways and blood. This is consistent with evidence that mycobacteria-specific T cells in humans polarize toward a Th1* phenotype, characterized by both Th1 and Th17-like markers, and the reports showing that humans with inborn errors in RORC and IL23R are susceptible to mycobacterial infection (Acosta-Rodriguez et al., 2007; Kushnareva et al., 2021; Lindestam Arlehamn et al., 2013; Nathan et al., 2021; Strickland et al., 2017). Additionally, we found that granuloma T cells were enriched for the expression CCR1, CCR2, CCR4, CCR5, CCR8, and CXCR5. This highlights several homing receptors that may be important for entry into granulomas and is consistent with our previous data indicating that multiple chemokine receptors collectively contribute to T cell homing into these lesions (Hoft et al., 2019).
Negative correlations between gene expression levels and bacterial burden may identify protective pathways and conversely positive correlations with CFU may identify molecules associated with susceptibility. The granuloma CD4 T cell gene whose expression most negatively correlated with CFU was KLRB1, which encodes CD161. Several recent studies have correlated CD161+ CD4 T cells with host protection against TB (Gela et al., 2022; Nathan et al., 2021; Yang et al., 2021). Our results indicate that this may reflect the enhanced ability of CD161 expressing CD4 T cells to traffic into Mtb lesions and highlight the importance of further studying this T cell subset. CD40L expression by CD4 T cells strongly inversely correlated with CFU, consistent with its known role in CD4 T cell:APC interactions. Unexpectedly, we found granuloma CD4 T cells express S100A11 and S100A4, and transcript levels of both of these genes inversely correlate with CFU. Although S100A11 and S100A4 may have multiple molecular interactions, both can be secreted and serve as RAGE ligands (Leclerc et al., 2009). S100A11 was recently shown to drive potent activation of human monocytes via binding to RAGE (Safronova et al., 2019), but its role in mycobacterial infection has not yet been evaluated. Expression of IL-26 by granuloma CD4 and CD8 T cells inversely correlated with bacterial loads. IL-26 is an inflammatory cytokine that can directly bind to bacteria, and it has been implicated in anti-mycobacterial immunity in humans (Dang et al., 2019; Hawerkamp et al., 2020). Our data further highlight the possibility that IL-26 may be a key host-protective cytokine. As expected, granuloma T cells were enriched for IFNG and TNF transcripts. However, IFNG expression in CD8 T cells, not CD4 T cells, inversely correlated with bacterial loads. Lastly, we also found that the transcription factor BATF was enriched in granuloma CD4 and CD8 T cells and inversely correlated with CFU. BATF plays a key role early in the priming of naive T cells (Kurachi et al., 2014) as well as promoting effector T cell responses during persistent antigenic stimulation (Chen et al., 2021; Seo et al., 2021). These data indicate that BATF-dependent transcription may also be important in the function or generation of protective T cells in Mtb infection.
Granuloma T cells were enriched for the expression of several other notable effector molecules that did not correlate with bacterial loads but may be important in the outcome of Mtb infection. T cells in granulomas expressed higher levels of both CSF2/GM-CSF and CSF1/M-CSF. GM-CSF production by T cells has been implicated in control of Mtb infection in murine models (Rothchild et al., 2014; Rothchild et al., 2017; Van Dis et al., 2022), but far less is known about the role of T cell–derived M-CSF during TB. Granuloma T cells expressed higher levels of IL-10 and OSM. IL-10 production by T cells regulates immune responses during TB (Harling et al., 2019; Moreira-Teixeira et al., 2017; Wong et al., 2020), while very little is known about the role of OSM in anti-mycobacterial immunity (O’Kane et al., 2008). Although multiple cytotoxic molecules were expressed in granuloma T cells, transcripts for granzyme K and granzyme A were higher in BAL and granuloma CD4 and CD8 T cells compared to memory cells in blood. This is consistent with the observation that granzyme K–expressing CD8 T cells are enriched in human pleural effusions from TB patients and indicate killer cells using these granzymes preferentially migrate into sites of infection (Cai et al., 2022). The transcription factors VDR, ZBED2, IRF6, IRF8, EGR2, and TOX were selectively expressed in granuloma T cells, but not correlated with CFU. Mechanistic experiments are needed to explore the role of these molecules in TB.
Expression of several immune genes by granuloma T cells positively correlated with bacterial loads. BHLHE40 is a transcription factor that has previously been observed in granuloma T cells in Mtb-infected macaques (Gideon et al., 2022), and variants of this gene have been associated with TB risk in humans (Shah et al., 2022). In mice, BHLHE40 has been shown to be a central regulator in the production of pro- versus anti-inflammatory T cells, as it promotes IFNγ while simultaneously inhibiting IL-10 production (Yu et al., 2018). In fact, deletion of BHLHE40 in CD4 T cells leads to early mortality in mice due to the over-production of IL-10 (Huynh et al., 2018). Thus, despite its positive correlation with bacterial loads, it is most likely a host-protective molecule in T cells. Similarly, CXCR5 expression in CD4 T cells positively correlated with bacterial loads, but CXCR5 has been shown to be important in host-protective CD4 T cell responses in mice, likely due to a role in the formation of tertiary lymphoid tissue aggregates (Slight et al., 2013). Moreover, as indicated above, CCR6 is a marker of Th1* and Th17 cells which likely play an important role in host protection, but we found that CCR6 expression by granuloma CD4 T cells positively correlated with CFU. This may indicate that T cells with these markers are better able to enter or survive in heavily infected granulomas.
We hypothesized that genes, which are selectively upregulated in granulomas, do not correlate with bacterial loads, and are also shared by both CD4 and CD8 T cells may contain a set of genes that define the core identity unique to granuloma T cells. Along with CCR2 and CCR5, the co-stimulatory molecule CD30 was at the top of the most DEGs in this list. Moreover, CD30 expression strongly correlated with the expression of several genes important in host resistance to Mtb infection: RORC, VDR, and TNF in both CD4 and CD8 T cells. We recently described a role for CD153, the sole ligand for CD30, in CD4 T cell–mediated host resistance to Mtb infection in mice monkeys and humans (Du Bruyn et al., 2021; Riou et al., 2020; Sallin et al., 2018). One hypothesis was that CD153-expressing T cells trigger CD30 on Mtb-infected myeloid cells. This major enrichment of CD30 on granuloma T cells prompted us to further explore the role of CD30 in T cell–mediated host resistance to Mtb infection. CD30-deficient mice succumbed early to Mtb infection, with similar kinetics to CD153-deficient mice. We were unable to find a role for CD30 in myeloid cells during Mtb infection. Instead, our data indicate that CD30 expression in CD4 T cells likely explains its role in host resistance. Moreover, CD30 signaling into CD4 T cells regulates their differentiation state and is required for the expression of multiple cytokine and chemokine effector molecules. In fact, CD30−/− effector CD4 T cells failed to upregulate GM-CSF, CCL1, CCL3, and CCL4. Due to the numerous effects of CD30 on CD4 T cells, it is not clear which specific defects in CD30−/− T cells lead to their inability to protect against Mtb infection.
The cells that provide CD153 co-stimulation into CD30 expressing CD4 T cells are not clear. However, as stated above control of Mtb infection requires both the receptor and the ligand on CD4 T cells. In MBMC mice, CD30-deficient CD4 T cells mount defective responses compared to their WT counterparts, whereas CD153-deficient CD4 T cells are normal, ruling out an autocrine interaction. It is possible that the CD153:CD30 axis mediates its effects through a T cell–to–T cell co-stimulatory interaction. In fact, there is evidence for this type of interaction in BCG-vaccinated mice (Tang et al., 2008). In such a model CD153-expressing CD4 T cells would drive the differentiation of CD30-expressing CD4 T cells. This hypothesis resembles the role of IL-2 in determining T cell differentiation fates, where during the early phase of T cell priming, IL-2–producing CD4 T cells remain less-differentiated and become Tfh cells, while IL-2–responding T cells are driven to differentiate further into non-Tfh lineages (DiToro et al., 2018). This hypothesis is further supported by the observation that STAT5A was predicted to be a regulator of the genes that correlated with CD30 in macaque granuloma CD4 T cells, and in mice, CD30 deficiency was associated with a loss of a STAT5A-driven gene signature. This suggests a potential connection between CD30 and STAT5 signaling. Future studies are needed to better understand the cellular mechanisms through which the CD30:CD153 axis generates host-protective CD4 T cell responses in Mtb infection.
This study identifies many features of macaque granuloma–homing T cells which may be useful in establishing correlates of protection to guide vaccine studies. It has also provided a set of candidate genes for further exploration of effector molecules, homing receptors, and transcriptional regulators that may be important in T cell-mediated immunity to Mtb infection. Moreover, our findings have revealed CD30 as a highly granuloma-specific T cell gene in Mtb-infected macaques, and subsequently showed that CD30 is required for protective CD4 T cell responses to Mtb infection in the mouse model. These results further establish the CD153:CD30 axis as critical for T cell responses at the site of mycobacterial infection.
Materials and methods
NHPs
3- to 4-yr-old animals were housed and maintained in biocontainment racks in accordance with the Animal Welfare Act in a BSL3 vivarium that is accredited by AAALAC International. Primates were bronchoscopically infected with 40–80 CFU of mCherry-reporter Mtb strain Erdman and cared for until termination of study 5 wk later. All animal procedures were approved by the National Institute of Allergy and Infectious Disease (NIAID) Division of Intramural Research (DIR) Animal Care and Use Committee under animal study LPD-25E. Other data from the rhesus macaques used in this study appeared in a previously study (Kauffman et al., 2018a).
FACS
For the NHP study, single-cell suspensions of PBMCs, BAL, or individual pulmonary granulomas were stained with antibodies (clone) CD11b (ICRF44), CD3 (SP34-2), CD4 (SK3), CD8 (RPA-T8), and CD95 (DX2) for 30 min at 4°C (Foreman et al., 2016; Kauffman et al., 2021). Samples were washed and stained with fixable viability dye eFluor780 from eBioscience/Thermo Fisher Scientific for 20 min at 4°C and thoroughly washed using 1× PBS with 1% FBS. Samples were sorted using a BD FACS Aria in a biosafety cabinet inside a BSL3 laboratory. Sorted cell populations were pelleted and stored in Trizol at −80°C until RNA isolation. RNA was isolated using Direct-zol RNA kit from Zymo Research according to the manufacturers protocol.
Sequencing library preparation and RNAseq
RNA was assessed for quality and degradation using the 2100 Bioanalyzer System from Agilent, with the product Bioanalyzer RNA 6000 pico assay. NHP library preparation was completed with the SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing from Takara Bio Inc. according to manufacturer’s protocol to create full-length cDNA using an input of 2 μl of total RNA for each sample. Quantification of cDNA was performed using Qubit 2.0 from Thermo Fisher Scientific with the Qubit dsDNA HS Assay Kit. The resultant full-length cDNA was used to create final sequencing libraries with the Nextera XT DNA Library Preparation Kit from illumina and the Nextera XT Index Kit v2 Set A from Illumina, using 150 pg of each cDNA product as input. Sequencing libraries were quality tested using the 2100 Bioanalyzer System with the Agilent High Sensitivity DNA Kit to verify library size and purity. Sequencing library quantification was performed using the Applied Biosystems 7900HT Fast Real-Time PCR System from Thermo Fisher Scientific and the KAPA Library Quantification Complete kit (ABI Prism) from Roche. Libraries were normalized to 2 nM concentration. Equimolar aliquots of 5 μl of each library were then pooled. The 2 nM library pool was then denatured and diluted to 1.8 pM, following the Illumina protocol “NextSeq System Denature and Dilute Libraries Guide” (Document #15048776 v02, January 2016). PhiX was added at 1% to serve as an internal control. The resultant final library pool was 1.8 pM final concentration with 1% PhiX spike-in. Mouse RNAseq library preparation was completed using SMARTer Stranded Total RNA-Seq Kit v2—Pico Input Mammalian, according to the manufacturer’s protocol, using a purified total RNA input estimated to be 20–30 ng. Paired-end sequencing was completed on an Illumina NextSeq 500 system, running Illumina NextSeq Control Software System Suite version 2.1.0 and RTA version 2.4.11. The final library pool was sequenced via 2 × 76 bp run configuration using the product NextSeq 500/550 High Output Kit v2.5 (150 cycles).
Immunohistochemistry
A pulmonary granuloma from a NHP infected with Mtb strain mCherry-reporter H37Rv strain was freshly isolated and embedded in 2% agarose for vibratome sectioning. Approximately 300-μm-thick sections were stained with antibodies CD4 (clone OKT4), PD-1 (clone EH12.2H7), and CD30 (clone Ber-H2) for 4–6 h at 4°C (Foreman et al., 2022). After thorough washes, the section was placed into a chamber slide and imaged using a Leica SP5 inverted confocal microscope from Leica Microsystems housed in a BSL3 laboratory. Static images were analyzed using Imaris Software from Bitplane.
Mice
B6.SJL-PtprcaPepcb/BoyJ (CD45.1; Congenic), C57BL/6J X B6.SJL-PtprcaPepcb/BoyJ (CD45.1/CD45.2; Congenic), B6.129S2-Tcratm1Mom/J (TCRa KO), and B6.Cg-Foxn1nu/J (nude) mice were obtained through a contract between NIAID and Taconic Farms. B6.129X1-Tnfsf8tm1Pod/J (CD153; KO) and B6.129P2-Tnfrsf8tm1Mak/J (CD30; KO) mice were purchased from Jackson Laboratory and were maintained and subsequently crossed at the NIAID animal facility. 6- to 12-wk-old mice were used in experiments with a conscious attempt to use both male and female sexes. Group sizes for each experiment are indicated in the legend of each respective figure. Mice were infected with ∼100 CFU of Mtb strain H37Rv using Glas-Col aerosol exposure system or 100 CFU of Mtb strain mCherry-reporter H37Rv (kindly provided by Dr. Clifton Barry III, chief of the Tuberculosis Research Section, Laboratory of Clinical Immunology and Microbiology, NIAID, Bethesda, MD, USA) by intratracheal instillation. Dose was confirmed by post-exposure plating of lung homogenates or/and serial dilutions of inoculum on 7H11 agar plates. For some studies, mice were sub-lethally irradiated, reconstituted with fresh bone marrow cells isolated from donor mice, and allowed to rest for ∼10 wk before infection with Mtb. MBMC mice were given 3–5 × 106 bone marrow cells from each congenic donor for a total of 6–10 × 106 cells. Nude mice were reconstituted with no <1 × 107 donor bone marrow cells. All animal procedures were approved by the NIAID DIR Animal Care and Use Committee under animal study LPD-24E.
Flow cytometry
Mice were intravenously injected with 2.5 mg of anti-CD45 antibody (clone 30-F11) and euthanized 3 min later. Single-cell suspensions of pulmonary cells were obtained after enzymatic dissociation with 1 mg/ml collagenase D from Roche Diagnostics, 1 mg/ml hyaluronidase, 1 mg/ml DNase I, and 1 mM aminoguanidine from Sigma-Aldrich for 45 min at 37°C. Tetramer staining was performed by incubation with 50 nM of dasatinib for 10 min then subsequent addition (bringing final dasatinib concentration to 25 nM) of I-Ab ESAT-64-17 tetramer for 1 h at 37°C. The H-2Kb TB10.44-11 tetramer was stained for 30 min at 4°C. Samples were stained with antibodies (clone) CD11b (M1/70), CD4 (RM4-5), CD44 (IM7), CD45 (30-F11), I-Ab (AF6-120.1), KLRG-1 (2F1/KLRG1), Ly6G (1A8), CD45.1 (A20), CD45.2 (104), Thy1.2 (53-2.1), TCRb (H57-597) and Fixable viability dye eFluor 780 from BD Biosciences, BioLegend, or eBioscience/Thermo Fisher Scientific for 30 min at 4°C. Intracellular antibody Foxp3 (FJK-16s) was stained after fixation and permeabilization with Fixation/Perm Solution Kit from BD Biosciences according to manufacturer’s protocol. Samples were analyzed using a BD Symphony and FlowJo software from BD Biosciences. Where specified, single-cell suspensions were stimulated with Mtb peptide ESAT-61-20 for 5 h in the presence of brefeldin and monensin.
FACS
Samples were processed as described above and subsequently sorted using a BD FACS Aria in a biosafety cabinet inside a BSL3 laboratory. Sorted macrophages were pelleted and resuspended in 1-ml of 1× PBS with 0.05% Tween 20. Serial dilutions were plated for determination of bacterial burden and CFU per cell back-calculated according to specific number of cells sorted. Alternatively, macrophages were resuspended in 1× PBS and cytospin mounted on slides before fixation and acid-fast staining for bacteria. Slides were imaged using Aperio VERSA from Leica Microsystems and image processed using QuPath software. Sorted CD4 T cell populations were pelleted, resuspended in RNAlater, and stored at −80°C until RNA isolation. RNA was isolated using a RNeasy Plus kit from Qiagen according to the manufacturer’s protocol.
CD4 T cell isolation and adoptive T cell transfer
Naive donor mice were 6- to 28-wk-old and sex-matched to recipient mice. Lymph nodes and spleen were collected from donor mice and processed to single-cell suspension using 100-μm filters. Bulk CD4 T cells were isolated using EasySep Mouse CD4 T cell Isolation kit from Stemcell Technologies according to the manufactures protocol. Approximately 2 × 106 of purified CD4 T cells were transferred by intravenous injection 1–6 d after aerosol infection.
Data and statistical analyses
Each figure legend describes the individual methods for statistical analysis of each graph. Briefly, all data were analyzed using FlowJo v10 (FlowJo LLC), Prism (GraphPad Software), or R (The R Foundation). Sequencing raw reads were processed and analyzed using DESeq2 and EdgeR packages and further analyzed using gene-set enrichment analysis. GSEA was run using Gene Set Enrichment Analysis version 4.2.3 with MSigDB human databases of hallmark, curated canonical pathways, and gene ontology biological process gene sets (Subramanian et al., 2005). Differential expression data were further analyzed using princomp R analysis package and custom data visualization scripts utilizing VennDiagram and ggplot2 packages. The integrated transcription factor analysis was performed with ChEA3 (Keenan et al., 2019). Bar graphs, dot plots, and survival curves were analyzed using a one-way ANOVA, unpaired Student’s t test, and paired Student’s t test, or Mantel–Cox rank test where appropriate. Significant values are denoted with asterisks, where * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001, or ns, non-significant.
Online supplemental material
Fig. S1 shows the production of IFNγ, TNF, MIP1α, and CD153 in WT, CD153−/−, and CD30−/− CD4 T cells after ESAT-61-20 peptide restimulation. Fig. S2 A shows the sorting strategy for the purification of WT and KO macrophages from the lungs of MBMC mice. Fig. S2 B shows the bacterial loads in the I-Ab− sorted cells. Fig. S2 C shows the number of acid-fast bacilli manually counted in FACS-sorted lung macrophages. Fig. S3 A shows the expression of differentiation markers in WT and CD30−/− CD4 T cells isolated from the lungs of MBMC mice (gated on parenchymal KLRG1-effector cells). Fig S3, B and C show the expression of KLRG1 and parenchymal localization of CD8 T cells in WT:CD153−/− and WT:CD30−/− MBMC mice. Table S1 shows the list of rhesus macaque granulomas used in this study. Tables S2 and S3 list the DEG in purified NHP CD4 and CD8 T cells respectively. Tables S4 and S5 list correlations between CFU and genes in CD4 and CD8 T cell, respectively. Tables S6 and S7 list correlations between TNFRSF8 and other genes in CD4 and CD8 T cells, respectively. Table S8 lists the genes with significant fold changes between CD44high versus CD44low WT and CD30−/− iv−KLRG1− CD4 T cells FACS-sorted from the lungs of MBMC mice. Table S9 lists the genes with significant fold changes between WT versus CD30−/− CD44high iv−KLRG1− CD4 T cells FACS sorted from the lungs of MBMC mice.
Supplementary Material
shows the list of rhesus macaque granulomas used in this study.
lists the DEGs in purified NHP CD4 T cells.
lists the DEGs in purified NHP CD8 T cells.
lists correlations between CFU and genes in CD4 T cells.
lists correlations between CFU and genes in CD8 T cells.
lists correlations between TNFRSF8 and other genes in CD4 T cells.
lists correlations between TNFRSF8 and other genes in CD8 T cells.
lists the genes with significant fold changes between CD44high versus CD44low WT and CD30−/− iv−KLRG1− CD4 T cells FACS-sorted from the lungs of MBMC mice.
lists the genes with significant fold changes between WT versus CD30−/− CD44high iv−KLRG1− CD4 T cells FACS-sorted from the lungs of MBMC mice.
Acknowledgments
We thank Dr. Rashida Moore and Temeri Wilder-Kofie for veterinary care of macaques.
This work was supported by the Intramural Research Program of the DIR/NIAID. T.W. Foreman, M.A. Sallin, K.D. Kauffman, and D.L. Barber are listed as inventors on a patent application related to this work.
Author contributions: T.W. Foreman and D.L. Barber designed the studies. T.W. Foreman, M.A. Sallin, K.D. Kauffman, and S. Sakai performed experiments. T.W. Foreman, C.E. Nelson, M.A. Sallin, and F. Otaizo-Carrasquero and T.G. Myers performed data analysis. T.W. Foreman, and D.L. Barber wrote the manuscript.
Data availability
RNAseq data of CD4 and CD8 T cells purified from blood, BAL, and granulomas of Mtb-infected rhesus macaques are publicly available under Gene Expression Omnibus accession number GSE227653. The RNAseq data of WT and TNFRSF8−/− CD4 T cells purified from Mtb-infected MBMC mice are publicly available under Gene Expression Omnibus accession number GSE228114. Other data are available in the published article and online supplemental material.
References
- Acosta-Rodriguez, E.V., Rivino L., Geginat J., Jarrossay D., Gattorno M., Lanzavecchia A., Sallusto F., and Napolitani G.. 2007. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 8:639–646. 10.1038/ni1467 [DOI] [PubMed] [Google Scholar]
- Burel, J.G., Lindestam Arlehamn C.S., Khan N., Seumois G., Greenbaum J.A., Taplitz R., Gilman R.H., Saito M., Vijayanand P., Sette A., and Peters B.. 2018. Transcriptomic Analysis of CD4(+) T Cells Reveals Novel Immune Signatures of Latent Tuberculosis. J. Immunol. 200:3283–3290. 10.4049/jimmunol.1800118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Y., Wang Y., Shi C., Dai Y., Li F., Xu Y., Zhang P., Kong F., Deng G., Wen Z., et al. 2022. Single-cell immune profiling reveals functional diversity of T cells in tuberculous pleural effusion. J. Exp. Med. 219:219. 10.1084/jem.20211777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Zander R.A., Wu X., Schauder D.M., Kasmani M.Y., Shen J., Zheng S., Burns R., Taparowsky E.J., and Cui W.. 2021. BATF regulates progenitor to cytolytic effector CD8+ T cell transition during chronic viral infection. Nat. Immunol. 22:996–1007. 10.1038/s41590-021-00965-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmensen, H.S., Dube J.Y., McIntosh F., Rosenkrands I., Jungersen G., Aagaard C., Andersen P., Behr M.A., and Mortensen R.. 2021. In Vivo Antigen Expression Regulates CD4 T Cell Differentiation and Vaccine Efficacy against Mycobacterium tuberculosis Infection. mBio. 12. e00226-21. 10.1128/mBio.00226-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang, A.T., Teles R.M., Weiss D.I., Parvatiyar K., Sarno E.N., Ochoa M.T., Cheng G., Gilliet M., Bloom B.R., and Modlin R.L.. 2019. IL-26 contributes to host defense against intracellular bacteria. J. Clin. Invest. 129:1926–1939. 10.1172/JCI99550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah, P.A., Zeppa J.J., Maiello P., Hackney J.A., Wadsworth M.H. II, Hughes T.K., Pokkali S., Swanson P.A. II, Grant N.L., Rodgers M.A., et al. 2020. Prevention of tuberculosis in macaques after intravenous BCG immunization. Nature. 577:95–102. 10.1038/s41586-019-1817-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkman, K., Sombroek C.C., Vervenne R.A.W., Hofman S.O., Boot C., Remarque E.J., Kocken C.H.M., Ottenhoff T.H.M., Kondova I., Khayum M.A., et al. 2019. Prevention of tuberculosis infection and disease by local BCG in repeatedly exposed rhesus macaques. Nat. Med. 25:255–262. 10.1038/s41591-018-0319-9 [DOI] [PubMed] [Google Scholar]
- DiToro, D., Winstead C.J., Pham D., Witte S., Andargachew R., Singer J.R., Wilson C.G., Zindl C.L., Luther R.J., Silberger D.J., et al. 2018. Differential IL-2 expression defines developmental fates of follicular versus nonfollicular helper T cells. Science. 361:361. 10.1126/science.aao2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Bruyn, E., Ruzive S., Lindestam Arlehamn C.S., Sette A., Sher A., Barber D.L., Wilkinson R.J., and Riou C.. 2021. Mycobacterium tuberculosis-specific CD4 T cells expressing CD153 inversely associate with bacterial load and disease severity in human tuberculosis. Mucosal Immunol. 14:491–499. 10.1038/s41385-020-0322-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybska, E., Adams A.T., Duclaux-Loras R., Walkowiak J., and Nowak J.K.. 2021. Waiting in the wings: RUNX3 reveals hidden depths of immune regulation with potential implications for inflammatory bowel disease. Scand. J. Immunol. 93:e13025. 10.1111/sji.13025 [DOI] [PubMed] [Google Scholar]
- Foreman, T.W., Mehra S., LoBato D.N., Malek A., Alvarez X., Golden N.A., Bucşan A.N., Didier P.J., Doyle-Meyers L.A., Russell-Lodrigue K.E., et al. 2016. CD4+ T-cell-independent mechanisms suppress reactivation of latent tuberculosis in a macaque model of HIV coinfection. Proc. Natl. Acad. Sci. USA. 113:E5636–E5644. 10.1073/pnas.1611987113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman, T.W., Nelson C.E., Kauffman K.D., Lora N.E., Vinhaes C.L., Dorosky D.E., Sakai S., Gomez F., Fleegle J.D., Parham M., et al. 2022. CD4 T cells are rapidly depleted from tuberculosis granulomas following acute SIV co-infection. Cell Rep. 39:110896. 10.1016/j.celrep.2022.110896 [DOI] [PubMed] [Google Scholar]
- Gela, A., Murphy M., Rodo M., Hadley K., Hanekom W.A., Boom W.H., Johnson J.L., Hoft D.F., Joosten S.A., Ottenhoff T.H.M., et al. 2022. Effects of BCG vaccination on donor unrestricted T cells in two prospective cohort studies. EBioMedicine. 76:103839. 10.1016/j.ebiom.2022.103839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gideon, H.P., Hughes T.K., Tzouanas C.N., Wadsworth M.H. II, Tu A.A., Gierahn T.M., Peters J.M., Hopkins F.F., Wei J.R., Kummerlowe C., et al. 2022. Multimodal profiling of lung granulomas in macaques reveals cellular correlates of tuberculosis control. Immunity. 55:827–846.e10. 10.1016/j.immuni.2022.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, S.G., Zak D.E., Xu G., Ford J.C., Marshall E.E., Malouli D., Gilbride R.M., Hughes C.M., Ventura A.B., Ainslie E., et al. 2018. Prevention of tuberculosis in rhesus macaques by a cytomegalovirus-based vaccine. Nat. Med. 24:130–143. 10.1038/nm.4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harling, K., Adankwah E., Güler A., Afum-Adjei Awuah A., Adu-Amoah L., Mayatepek E., Owusu-Dabo E., Nausch N., and Jacobsen M.. 2019. Constitutive STAT3 phosphorylation and IL-6/IL-10 co-expression are associated with impaired T-cell function in tuberculosis patients. Cell. Mol. Immunol. 16:275–287. 10.1038/cmi.2018.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawerkamp, H.C., van Geelen L., Korte J., Di Domizio J., Swidergall M., Momin A.A., Guzmán-Vega F.J., Arold S.T., Ernst J., Gilliet M., et al. 2020. Interleukin-26 activates macrophages and facilitates killing of Mycobacterium tuberculosis. Sci. Rep. 10:17178. 10.1038/s41598-020-73989-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoft, S.G., Sallin M.A., Kauffman K.D., Sakai S., Ganusov V.V., and Barber D.L.. 2019. The rate of CD4 T cell entry into the lungs during Mycobacterium tuberculosis infection is determined by partial and opposing effects of multiple chemokine receptors. Infect. Immun. 87:87. 10.1128/IAI.00841-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh, J.P., Lin C.C., Kimmey J.M., Jarjour N.N., Schwarzkopf E.A., Bradstreet T.R., Shchukina I., Shpynov O., Weaver C.T., Taneja R., et al. 2018. Bhlhe40 is an essential repressor of IL-10 during Mycobacterium tuberculosis infection. J. Exp. Med. 215:1823–1838. 10.1084/jem.20171704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman, K.D., Sakai S., Lora N.E., Namasivayam S., Baker P.J., Kamenyeva O., Foreman T.W., Nelson C.E., Oliveira-de-Souza D., Vinhaes C.L., et al. 2021. PD-1 blockade exacerbates Mycobacterium tuberculosis infection in rhesus macaques. Sci. Immunol. 6:6. 10.1126/sciimmunol.abf3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman, K.D., Sallin M.A., Hoft S.G., Sakai S., Moore R., Wilder-Kofie T., Moore I.N., Sette A., Arlehamn C.S.L., and Barber D.L.. 2018a. Limited pulmonary mucosal-associated invariant T cell accumulation and activation during Mycobacterium tuberculosis infection in rhesus macaques. Infect. Immun. 86:86. 10.1128/IAI.00431-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman, K.D., Sallin M.A., Sakai S., Kamenyeva O., Kabat J., Weiner D., Sutphin M., Schimel D., Via L., Barry C.E. III, et al. 2018b. Defective positioning in granulomas but not lung-homing limits CD4 T-cell interactions with Mycobacterium tuberculosis-infected macrophages in rhesus macaques. Mucosal Immunol. 11:462–473. 10.1038/mi.2017.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal, D., Foreman T.W., Gautam U.S., Alvarez X., Adekambi T., Rangel-Moreno J., Golden N.A., Johnson A.M., Phillips B.L., Ahsan M.H., et al. 2015. Mucosal vaccination with attenuated Mycobacterium tuberculosis induces strong central memory responses and protects against tuberculosis. Nat. Commun. 6:8533. 10.1038/ncomms9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan, A.B., Torre D., Lachmann A., Leong A.K., Wojciechowicz M.L., Utti V., Jagodnik K.M., Kropiwnicki E., Wang Z., and Ma’ayan A.. 2019. ChEA3: Transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 47:W212–W224. 10.1093/nar/gkz446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurachi, M., Barnitz R.A., Yosef N., Odorizzi P.M., DiIorio M.A., Lemieux M.E., Yates K., Godec J., Klatt M.G., Regev A., et al. 2014. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat. Immunol. 15:373–383. 10.1038/ni.2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnareva, Y., Mathews I.T., Andreyev A.Y., Altay G., Lindestam Arlehamn C.S., Pandurangan V., Nilsson R., Jain M., Sette A., Peters B., and Sharma S.. 2021. Functional analysis of immune signature genes in Th1* memory cells links ISOC1 and pyrimidine metabolism to IFN-γ and IL-17 production. J. Immunol. 206:1181–1193. 10.4049/jimmunol.2000672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc, E., Fritz G., Vetter S.W., and Heizmann C.W.. 2009. Binding of S100 proteins to RAGE: An update. Biochim. Biophys. Acta. 1793:993–1007. 10.1016/j.bbamcr.2008.11.016 [DOI] [PubMed] [Google Scholar]
- Lindestam Arlehamn, C.S., Gerasimova A., Mele F., Henderson R., Swann J., Greenbaum J.A., Kim Y., Sidney J., James E.A., Taplitz R., et al. 2013. Memory T cells in latent Mycobacterium tuberculosis infection are directed against three antigenic islands and largely contained in a CXCR3+CCR6+ Th1 subset. PLoS Pathog. 9. e1003130. 10.1371/journal.ppat.1003130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moguche, A.O., Shafiani S., Clemons C., Larson R.P., Dinh C., Higdon L.E., Cambier C.J., Sissons J.R., Gallegos A.M., Fink P.J., and Urdahl K.B.. 2015. ICOS and Bcl6-dependent pathways maintain a CD4 T cell population with memory-like properties during tuberculosis. J. Exp. Med. 212:715–728. 10.1084/jem.20141518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira-Teixeira, L., Redford P.S., Stavropoulos E., Ghilardi N., Maynard C.L., Weaver C.T., Freitas do Rosário A.P., Wu X., Langhorne J., and O’Garra A.. 2017. T cell-derived IL-10 impairs host resistance to Mycobacterium tuberculosis infection. J. Immunol. 199:613–623. 10.4049/jimmunol.1601340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan, A., Beynor J.I., Baglaenko Y., Suliman S., Ishigaki K., Asgari S., Huang C.C., Luo Y., Zhang Z., Lopez K., et al. 2021. Multimodally profiling memory T cells from a tuberculosis cohort identifies cell state associations with demographics, environment and disease. Nat. Immunol. 22:781–793. 10.1038/s41590-021-00933-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemes, E., Geldenhuys H., Rozot V., Rutkowski K.T., Ratangee F., Bilek N., Mabwe S., Makhethe L., Erasmus M., Toefy A., et al. 2018. Prevention of M. tuberculosis infection with H4:IC31 vaccine or BCG revaccination. N. Engl. J. Med. 379:138–149. 10.1056/NEJMoa1714021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Kane, C.M., Elkington P.T., and Friedland J.S.. 2008. Monocyte-dependent oncostatin M and TNF-alpha synergize to stimulate unopposed matrix metalloproteinase-1/3 secretion from human lung fibroblasts in tuberculosis. Eur. J. Immunol. 38:1321–1330. 10.1002/eji.200737855 [DOI] [PubMed] [Google Scholar]
- Plumlee, C.R., Duffy F.J., Gern B.H., Delahaye J.L., Cohen S.B., Stoltzfus C.R., Rustad T.R., Hansen S.G., Axthelm M.K., Picker L.J., et al. 2021. Ultra-low dose aerosol infection of mice with Mycobacterium tuberculosis more closely models human tuberculosis. Cell Host Microbe. 29:68–82.e5. 10.1016/j.chom.2020.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou, C., Du Bruyn E., Ruzive S., Goliath R.T., Lindestam Arlehamn C.S., Sette A., Sher A., Barber D.L., and Wilkinson R.J.. 2020. Disease extent and anti-tubercular treatment response correlates with Mycobacterium tuberculosis-specific CD4 T-cell phenotype regardless of HIV-1 status. Clin. Transl. Immunol. 9. e1176. 10.1002/cti2.1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothchild, A.C., Jayaraman P., Nunes-Alves C., and Behar S.M.. 2014. iNKT cell production of GM-CSF controls Mycobacterium tuberculosis. PLoS Pathog. 10. e1003805. 10.1371/journal.ppat.1003805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothchild, A.C., Stowell B., Goyal G., Nunes-Alves C., Yang Q., Papavinasasundaram K., Sassetti C.M., Dranoff G., Chen X., Lee J., et al. 2017. Role of granulocyte-macrophage colony-stimulating factor production by T cells during Mycobacterium tuberculosis infection. MBio. 8:8. 10.1128/mBio.01514-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safronova, A., Araujo A., Camanzo E.T., Moon T.J., Elliott M.R., Beiting D.P., and Yarovinsky F.. 2019. Alarmin S100A11 initiates a chemokine response to the human pathogen Toxoplasma gondii. Nat. Immunol. 20:64–72. 10.1038/s41590-018-0250-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, S., Kauffman K.D., Schenkel J.M., McBerry C.C., Mayer-Barber K.D., Masopust D., and Barber D.L.. 2014. Cutting edge: control of Mycobacterium tuberculosis infection by a subset of lung parenchyma-homing CD4 T cells. J. Immunol. 192:2965–2969. 10.4049/jimmunol.1400019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallin, M.A., Kauffman K.D., Riou C., Du Bruyn E., Foreman T.W., Sakai S., Hoft S.G., Myers T.G., Gardina P.J., Sher A., et al. 2018. Host resistance to pulmonary Mycobacterium tuberculosis infection requires CD153 expression. Nat. Microbiol. 3:1198–1205. 10.1038/s41564-018-0231-6 [DOI] [PubMed] [Google Scholar]
- Sallin, M.A., Sakai S., Kauffman K.D., Young H.A., Zhu J., and Barber D.L.. 2017. Th1 differentiation drives the accumulation of intravascular, non-protective CD4 T cells during tuberculosis. Cell Rep. 18:3091–3104. 10.1016/j.celrep.2017.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H., González-Avalos E., Zhang W., Ramchandani P., Yang C., Lio C.J., Rao A., and Hogan P.G.. 2021. BATF and IRF4 cooperate to counter exhaustion in tumor-infiltrating CAR T cells. Nat. Immunol. 22:983–995. 10.1038/s41590-021-00964-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J.A., Warr A.J., Graustein A.D., Saha A., Dunstan S.J., Thuong N.T.T., Thwaites G.E., Caws M., Thai P.V.K., Bang N.D., et al. 2022. REL and BHLHE40 variants are associated with IL-12 and IL-10 responses and tuberculosis risk. J. Immunol. 208:1352–1361. 10.4049/jimmunol.2100671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe, S., White A., Sarfas C., Sibley L., Gleeson F., McIntyre A., Basaraba R., Clark S., Hall G., Rayner E., et al. 2016. Alternative BCG delivery strategies improve protection against Mycobacterium tuberculosis in non-human primates: Protection associated with mycobacterial antigen-specific CD4 effector memory T-cell populations. Tuberculosis. 101:174–190. 10.1016/j.tube.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqa, A., Sims-Mourtada J.C., Guzman-Rojas L., Rangel R., Guret C., Madrid-Marina V., Sun Y., and Martinez-Valdez H.. 2001. Regulation of CD40 and CD40 ligand by the AT-hook transcription factor AKNA. Nature. 410:383–387. 10.1038/35066602 [DOI] [PubMed] [Google Scholar]
- Singhania, A., Dubelko P., Kuan R., Chronister W.D., Muskat K., Das J., Phillips E.J., Mallal S.A., Seumois G., Vijayanand P., et al. 2021. CD4+CCR6+ T cells dominate the BCG-induced transcriptional signature. EBioMedicine. 74:103746. 10.1016/j.ebiom.2021.103746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slight, S.R., Rangel-Moreno J., Gopal R., Lin Y., Fallert Junecko B.A., Mehra S., Selman M., Becerril-Villanueva E., Baquera-Heredia J., Pavon L., et al. 2013. CXCR5⁺ T helper cells mediate protective immunity against tuberculosis. J. Clin. Invest. 123:712–726. 10.1172/JCI65728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland, N., Muller T.L., Berkowitz N., Goliath R., Carrington M.N., Wilkinson R.J., Burgers W.A., and Riou C.. 2017. Characterization of Mycobacterium tuberculosis-Specific Cells Using MHC Class II Tetramers Reveals Phenotypic Differences Related to HIV Infection and Tuberculosis Disease. J. Immunol. 9:ji1700849. 10.4049/jimmunol.1700849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, C., Yamada H., Shibata K., Muta H., Wajjwalku W., Podack E.R., and Yoshikai Y.. 2008. A novel role of CD30L/CD30 signaling by T-T cell interaction in Th1 response against mycobacterial infection. J. Immunol. 181:6316–6327. 10.4049/jimmunol.181.9.6316 [DOI] [PubMed] [Google Scholar]
- Van Der Meeren, O., Hatherill M., Nduba V., Wilkinson R.J., Muyoyeta M., Van Brakel E., Ayles H.M., Henostroza G., Thienemann F., Scriba T.J., et al. 2018. Phase 2b controlled trial of M72/AS01E vaccine to prevent tuberculosis. N. Engl. J. Med. 379:1621–1634. 10.1056/NEJMoa1803484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dis, E., Fox D.M., Morrison H.M., Fines D.M., Babirye J.P., McCann L.H., Rawal S., Cox J.S., and Stanley S.A.. 2022. IFN-γ-independent control of M. tuberculosis requires CD4 T cell-derived GM-CSF and activation of HIF-1α. PLoS Pathog. 18. e1010721. 10.1371/journal.ppat.1010721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, A.D., Sarfas C., West K., Sibley L.S., Wareham A.S., Clark S., Dennis M.J., Williams A., Marsh P.D., and Sharpe S.A.. 2015. Evaluation of the immunogenicity of Mycobacterium bovis BCG delivered by aerosol to the lungs of macaques. Clin. Vaccin. Immunol. 22:992–1003. 10.1128/CVI.00289-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, E.A., Evans S., Kraus C.R., Engelman K.D., Maiello P., Flores W.J., Cadena A.M., Klein E., Thomas K., White A.G., et al. 2020. IL-10 Impairs Local Immune Response in Lung Granulomas and Lymph Nodes during Early Mycobacterium tuberculosis Infection. J. Immunol. 204:644–659. 10.4049/jimmunol.1901211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodworth, J.S., Cohen S.B., Moguche A.O., Plumlee C.R., Agger E.M., Urdahl K.B., and Andersen P.. 2017. Subunit vaccine H56/CAF01 induces a population of circulating CD4 T cells that traffic into the Mycobacterium tuberculosis-infected lung. Mucosal Immunol. 10:555–564. 10.1038/mi.2016.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, R., Peng Y., Pi J., Liu Y., Yang E., Shen X., Yao L., Shen L., Modlin R.L., Shen H., et al. 2021. A CD4+CD161+ T-cell subset present in unexposed humans, not Tb patients, are fast acting cells that inhibit the growth of intracellular mycobacteria involving CD161 pathway, perforin, and IFN-γ/Autophagy. Front. Immunol. 12:599641. 10.3389/fimmu.2021.599641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F., Sharma S., Jankovic D., Gurram R.K., Su P., Hu G., Li R., Rieder S., Zhao K., Sun B., and Zhu J.. 2018. The transcription factor Bhlhe40 is a switch of inflammatory versus antiinflammatory Th1 cell fate determination. J. Exp. Med. 215:1813–1821. 10.1084/jem.20170155 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the list of rhesus macaque granulomas used in this study.
lists the DEGs in purified NHP CD4 T cells.
lists the DEGs in purified NHP CD8 T cells.
lists correlations between CFU and genes in CD4 T cells.
lists correlations between CFU and genes in CD8 T cells.
lists correlations between TNFRSF8 and other genes in CD4 T cells.
lists correlations between TNFRSF8 and other genes in CD8 T cells.
lists the genes with significant fold changes between CD44high versus CD44low WT and CD30−/− iv−KLRG1− CD4 T cells FACS-sorted from the lungs of MBMC mice.
lists the genes with significant fold changes between WT versus CD30−/− CD44high iv−KLRG1− CD4 T cells FACS-sorted from the lungs of MBMC mice.
Data Availability Statement
RNAseq data of CD4 and CD8 T cells purified from blood, BAL, and granulomas of Mtb-infected rhesus macaques are publicly available under Gene Expression Omnibus accession number GSE227653. The RNAseq data of WT and TNFRSF8−/− CD4 T cells purified from Mtb-infected MBMC mice are publicly available under Gene Expression Omnibus accession number GSE228114. Other data are available in the published article and online supplemental material.