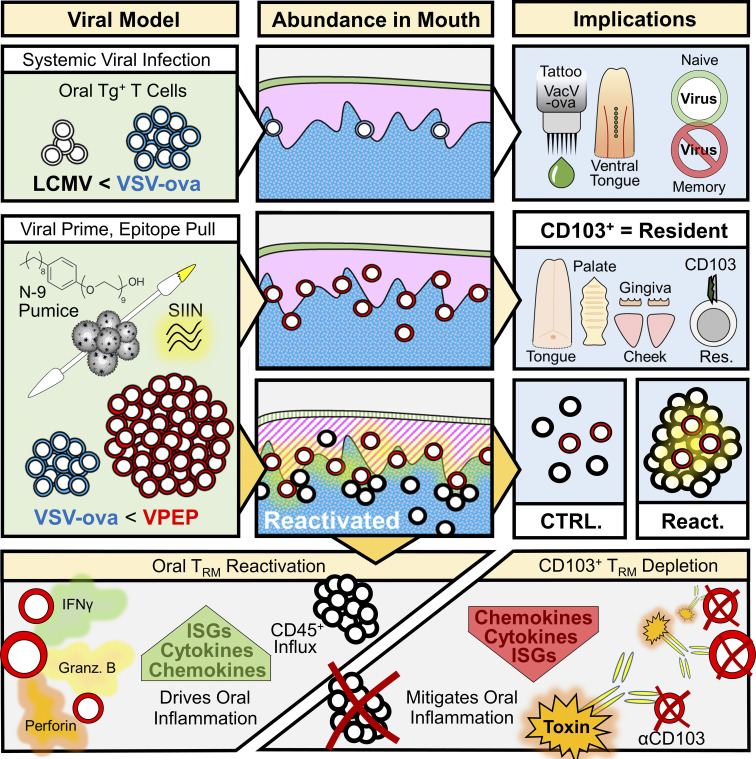
In the oral mucosa, Stolley et al. characterize resident memory T cell (TRM) distribution, ontogeny, role in protective immunity, and diverse functions, while describing strategies for locally augmenting or systemically depleting CD103+ TRM.
Abstract
The oral mucosa is a frontline for microbial exposure and juxtaposes several unique tissues and mechanical structures. Based on parabiotic surgery of mice receiving systemic viral infections or co-housing with microbially diverse pet shop mice, we report that the oral mucosa harbors CD8+ CD103+ resident memory T cells (TRM), which locally survey tissues without recirculating. Oral antigen re-encounter during the effector phase of immune responses potentiated TRM establishment within tongue, gums, palate, and cheek. Upon reactivation, oral TRM triggered changes in somatosensory and innate immune gene expression. We developed in vivo methods for depleting CD103+ TRM while sparing CD103neg TRM and recirculating cells. This revealed that CD103+ TRM were responsible for inducing local gene expression changes. Oral TRM putatively protected against local viral infection. This study provides methods for generating, assessing, and in vivo depleting oral TRM, documents their distribution throughout the oral mucosa, and provides evidence that TRM confer protection and trigger responses in oral physiology and innate immunity.
Graphical Abstract
Introduction
The oral cavity constitutes a first line of exposure for commensals and pathogens accessing the alimentary canal and respiratory tract, and is colonized by several diverse and dynamic microbial communities which directly interface with the oral epithelium and dental tissues (Dewhirst et al., 2010; Human Microbiome Project Consortium, 2012). Often in homeostatic balance with this microbiota, the oral mucosa is vulnerable to infection by viruses including members of the Herpesviridae family, human papilloma virus (HPV), Coxsackie viruses, and coronaviruses; many of which are transmissible through an oral route (Grinde, 2013; Hairston et al., 2003; Huang et al., 2021; La Rosa et al., 2021; Santosh and Muddana, 2020). Oral viruses may initiate, amplify, or sustain immune responses against the oral microbiota, and often re-emerge under conditions of immunosuppression or prolonged stress (Epstein and Chow, 1999; Ives and Bertke, 2017; Padgett et al., 1998). A site of physiologic extremes, the oral mucosa endures daily fluctuations in temperature, hydration, and significant epithelial stress through mastication (Moutsopoulos and Konkel, 2018). The mouth is also highly susceptible to inflammatory disease, with dysregulated immune responses implicated in several oral conditions including aphthous ulcers (canker sores; Dudding et al., 2019; Lewkowicz et al., 2011), oral manifestations of autoimmune diseases (including Sjogren’s syndrome and pemphigus vulgaris; Singh and Cohen, 2012; Zhou et al., 2021), oral premalignant and malignant lesions (Feller et al., 2013), and progressive forms of periodontitis (Balaji et al., 2021; Cardoso and Arosa, 2017; Hajishengallis et al., 2020). Indeed, the interface between the tooth and gingiva is the only location in the human body where a hard structure breaches a soft tissue covering (Hand and Frank, 2014), and this distinctive anatomy is a site of persistent microbial-driven immune stimulation. Critical for antiviral and antitumor immunity, unrestrained CD8+ T cell responses can perpetuate pathogenic inflammation in response to dietary antigens, environmental antigens, and self-antigens (Clark, 2015; Masopust and Soerens, 2019; Riding and Harris, 2019; Tang et al., 2012). CD8+ T cell biology has begun to be well characterized in many non-lymphoid tissues (NLTs), where phenotype and function vary depending on anatomic location (Crowl et al., 2022; Lin et al., 2023; Milner et al., 2020). The mouth encompasses a truly unique yet functionally organized constellation of absorptive, secretory, and chemosensory tissues, yet comparatively little is known about how CD8+ T cells integrate within this extreme environment to mediate local immunosurveillance and tissue homeostasis.
Memory CD8+ T cells expedite pathogen detection and clearance employing one of three immunosurveillance modalities. Recirculating central memory T cells constitutively surveil secondary lymphoid organs (including spleen and LNs). Effector memory T cells patrol blood with some extending their surveillance to NLTs via continuous recirculation (Mueller and Mackay, 2016). In contrast to their itinerant counterparts, resident memory T cells (TRM) remain parked within NLTs where their sentinel positioning affords rapid pathogen sensing at primary sites of infection (Ariotti et al., 2014; Schenkel et al., 2014; Schenkel et al., 2013). TRM populate most anatomic compartments; however, their differentiation state is influenced by instructive cues that vary from one location to another (Lin et al., 2023; Szabo et al., 2019). Environmental contributions to TRM differentiation may include the juxtaposition of a local microbiota, oxygen and nutrient availability, repetitive mechanical tissue damage, the rate of tissue self-renewal, and the architectural features of the tissue itself. Therefore, rather than a monolithic subset, TRM represent a collection of non-recirculating cells whose biology must be considered on a tissue-by-tissue basis. Paradigms in mucosal immunology and TRM biology were largely established in a few select tissues (Crowl et al., 2022; Gaffen and Moutsopoulos, 2020), and an understanding of the ontogeny, distribution, and functional implications of CD8+ TRM in the oral mucosa and periodontium remains practically unknown. A more thorough understanding of how CD8+ TRM form and function in the mouth will allow us to contextualize their role in oral health and disease and may uncover new therapeutic targets to harness or inhibit their immunostimulatory potential.
This study was inspired by a motivation to uncover basic principles of antiviral immunity in the oral mucosa. Here, we report a novel viral-prime, epitope-pull (VPEP) model, introduced on the readily accessible mucosal surfaces of the mouth, for locally augmenting oral antiviral CD8+ TRM populations. Leveraging this approach, we document CD8+ TRM distribution throughout diverse structures of the mouth, validate CD103 as a canonical marker of oral residency in both specific pathogen–free (SPF) and non-SPF mice, and profile the immunostimulatory functions of oral TRM upon cognate antigen re-exposure. Intentional deployment of oral CD8+ TRM inflammatory functions elicited a potent local anti-pathogen state including fortification of oral cellular immunity and transcriptional amplification of antiviral-, interferon-stimulated-, and chemotactic-genes. Periodontitis-relevant genes were similarly invoked upon oral TRM reactivation. Utilizing toxin-conjugated antibodies against CD103, we developed an in vivo depletion model to characterize the extent by which transcriptional and inflammatory changes induced upon oral antigen re-exposure were catalyzed by CD103+ TRM. Collectively, our study demonstrates antiviral TRM can be locally augmented in the oral mucosa, describes their biogeography throughout the mouth, and reveals that transcriptional and inflammatory changes driven by oral TRM reactivation can be effectively abrogated by their in vivo depletion.
Results
Systemic viral infections establish protective CD8+ T cell immunity in the oral mucosa
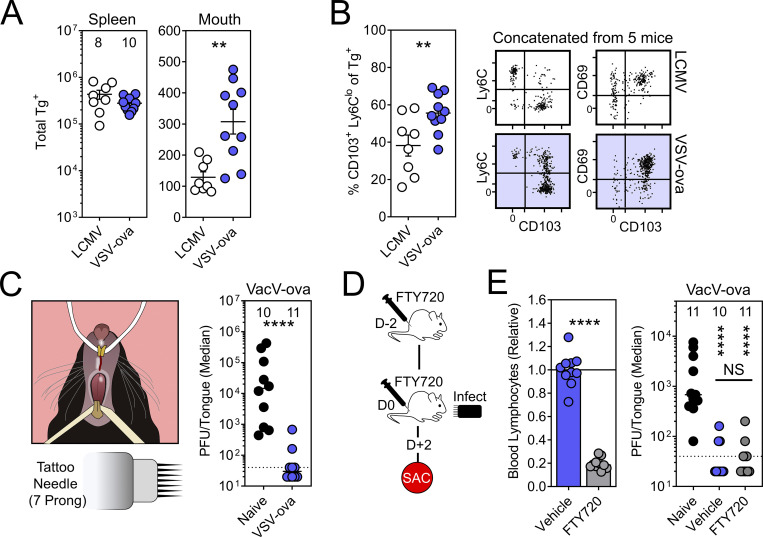
We wished to establish a model for studying oral CD8+ TRM, first testing whether systemically delivered viral infections can generate memory CD8+ T cells within the oral mucosa. Lymphocytic choriomeningitis virus (LCMV) and vesicular stomatitis virus expressing OVA (VSV-ova) were compared. For this, 5 × 104 naive congenially distinct TCR-Tg+ CD8+ T cells (P14 T cells recognizing the H-2Db/gp33-41 epitope from LCMV or OT-I T cells recognizing the H-2Db/SIINFEKL epitope from OVA) were transferred into recipient mice. 1 d later, mice were infected with 2 × 105 PFU LCMV Armstrong (LCMV-Arm) i.p. or 1 × 106 PFU VSV-ova i.v. 6–8 wk later, oral tissues (including tongue, buccal mucosa, gingiva, and hard/soft palate) were isolated, pooled, and processed for flow cytometry. VSV-ova/OT-I T cells outperformed LCMV-Arm/P14 T cells both in the magnitude of total Tg+ T cells isolated from the oral mucosa (Fig. 1 A), as well as the percentage of those Tg+ T cells expressing canonical markers of tissue residency (CD103+, CD69+, and Ly6Clo; Fig. 1 B). Tongues of naive mice, or those challenged >30 d earlier with VSV-ova, were then infected with VacV-ova (vaccinia virus expressing OVA) by puncturing the ventral lingual mucosa with a seven-prong tattoo needle dipped immediately prior in neat VacV-ova stock (3.99 × 107 PFU/ml; Fig. 1 C, left). We chose the ventral tongue as our challenge site as it can be sufficiently exteriorized for infection and contains a relatively thin non-keratinized epithelium. We chose vaccinia virus because it naturally infects via the oral mucosa (Shannon et al., 2021), recombinant virus expressing OVA was available, and CD8+ T cells have been shown to protect against this pathogen (Hickman et al., 2015; Hickman et al., 2008). While all naive mice showed detectable viral titers 48 h after infection, mice previously challenged with VSV-ova were protected from lingual VacV-ova infection (Fig. 1 C, right). FTY720 treatment, which sequesters many circulating T cells in secondary lymphoid organs (Jiang et al., 2012; Pan et al., 2017; Turner et al., 2014; Zens et al., 2016) and inhibits the entry of peripheral T cells into afferent lymphatics (Ledgerwood et al., 2008), did not compromise viral clearance (Fig. 1, D and E), which is compatible with the interpretation that local TRM conferred protection.
Figure 1.
TRM-phenotype cells populate the oral mucosa following systemic viral infections and contribute to local immunity. (A) Mice received 5 × 104 congenically labeled P14 or OT-I T cells 1 d prior to systemic infection with LCMV-Arm or VSV-ova. Enumeration of Tg+ T cells in spleen (left) and oral tissue (right). Numbers above bars indicate replicates per group from two independent experiments. (B) Phenotype of Tg+ T cells in the oral mucosa following systemic LCMV-Arm or VSV-ova infection. Flow cytometry plots are concatenated from five mice from one of two independent experiments (gated on intravascular-staining negative Tg+ CD8+ T cells). (C) Left: Cartoon of mouse restraint for lingual infections. Right: Tongue viral titers 48 h after infection of the ventral lingual mucosa with VacV-ova in naive (black) vs. VSV-ova memory mice (blue). Dotted line represents limit of detection (40 PFU/tongue). (D) Experimental strategy and time course. Mice received two doses of FTY720 or vehicle control and were sacrificed (SAC) 48 h after lingual challenge with VacV-ova. (E) Left: Relative lymphocyte abundance in blood 48 h after treatment with vehicle control or FTY720. Right: VacV-ova viral titers in tongues of naive (black) or VSV-ova memory mice treated with either vehicle control (blue) or FTY720 (gray). Numbers indicate replicates per group from two independent experiments (N ≥ 4 mice/group/experiment). Error bars in A and B represent mean ± SEM. Data in C and D show median. All dots represent individual mice. **, P < 0.01; ****, P < 0.0001 as determined by an unpaired Student’s t test (A, B, and E; left) with an additional Mann–Whitney test (C) or Kruskal–Wallis multi-comparison test (E; right).
CD103 expression identifies CD8+ TRM in the oral mucosa
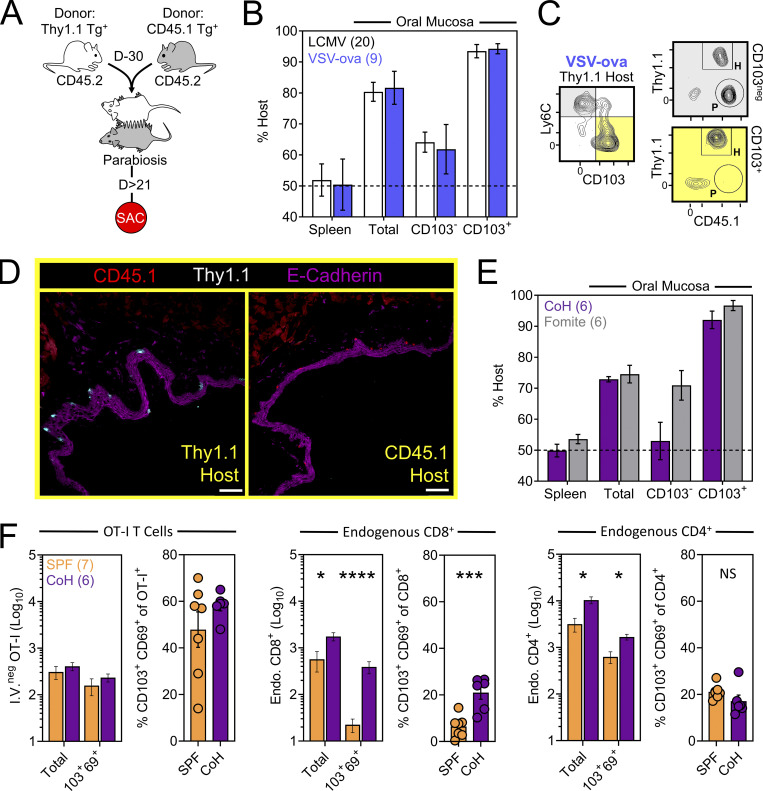
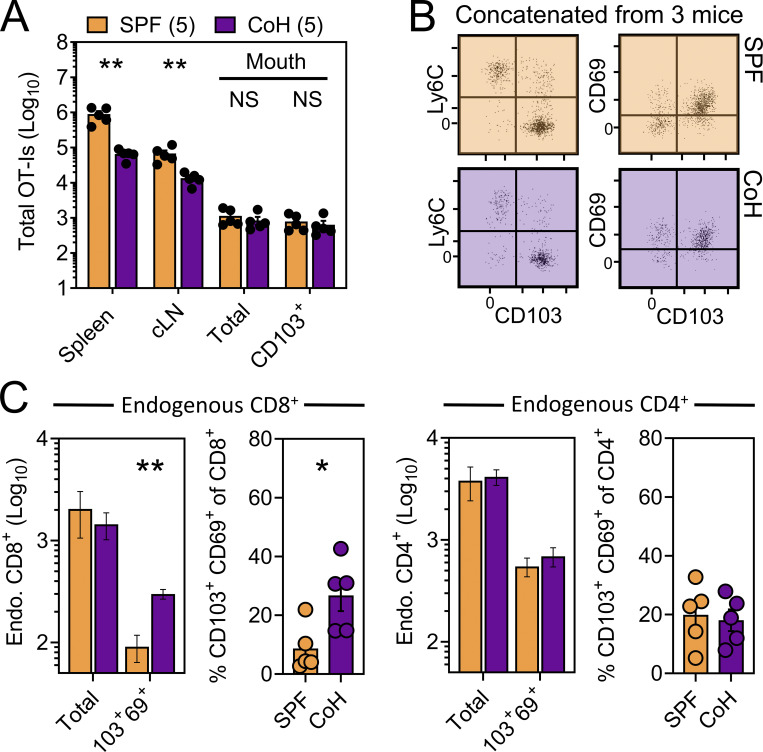
We next asked whether TRM-phenotype cells populating the oral mucosa following systemic viral infection were truly residents (non-recirculating cells as shown by a migration assay), and if so, how that property might correlate with surface phenotype. CD45.1+ and Thy1.1+ OT-I T cells were transferred into separate mice followed by systemic infection with VSV-ova. Parallel experiments employed a similar strategy using CD45.1+ and Thy1.1+ P14 T cells followed by LCMV-Arm infection. 30 d later, mice underwent parabiosis surgery, and remained conjoined for an additional >21 d upon which spleens and oral mucosa were isolated and assessed for the presence of OT-I or P14 T cells of host or partner origin (Fig. 2 A). Within spleens, Tg+ T cells were fully equilibrated between parabiotic partners, indicating a high degree of T cell recirculation. In contrast, total Tg+ T cells isolated from the oral mucosa were strongly host-biased, indicating residence. CD103 expression further refined identification of oral TRM, although some CD103neg OT-I T cells were also TRM (Fig. 2, B and C). Residency in the oral mucosa was confirmed histologically, focusing on the ventral lingual mucosa (underside of the tongue; oral infection site in Fig. 1) in VSV-ova immune parabionts (Fig. 2 D). We next utilized a previously described “dirty” mouse model to address the preponderance of CD8+ T cell residency in a murine oral mucosa with more diverse microbial experience than SPF mice (Beura et al., 2016). For this, congenically distinct SPF mice were cohoused with mice purchased from a pet shop, or exposed to their soiled bedding, for >60 d within the University of Minnesota’s ABSL3 facility. Mice were parabiosed and remained conjoined for >21 d. Consistent with observations in LCMV-Arm and VSV-ova immune parabionts (Fig. 2, B and C), CD103 remained a faithful marker of oral residency among dirty mouse CD8+ T cells of unknown specificity (Fig. 2 E). Thus, CD103 expression identifies CD8+ TRM in the oral mucosa irrespective of microbial experience. To better understand how cohousing impacted the abundance of pre-established oral antiviral T cells and endogenous CD8+ and CD4+ T cells of unknown specificity, VSV-ova immune mice were cohoused for >60 d with mice purchased from a pet shop. Here we observed little impact on the abundance and phenotype oral OT-I T cells following cohousing (Fig. 2 F, left), whereas total endogenous and TRM-phenotype (CD103+ CD69+) CD8+ T cells in the mouth were increased (Fig. 2 F, middle) relative to SPF mice. Endogenous CD4+ T cells were also more numerous in cohoused vs. SPF oral tissue; however, the percentage of CD103+ CD69+ CD4+ T cells in the mouth remained identical (Fig. 2 F, right). Collectively, these data indicate that the number of memory T cells in the oral mucosa is flexible and reflects microbial experience (Wijeyesinghe et al., 2021).
Figure 2.
CD103+ memory CD8+ T cells in the oral mucosa are TRM. (A) Parabiosis strategy to determine the degree by which Tg+ T cells in the oral mucosa following systemic viral infections are resident. (B) Percent host among P14 T cells and OT-I T cells in the spleen and oral mucosa (comparing CD103neg vs. CD103+ subsets) following systemic LCMV-Arm (white) or VSV-ova (blue) infection. Numbers indicate parabiotic mice per group from at least two independent experiments. (C) Representative flow cytometry highlighting host vs. partner OT-I T cells among CD103neg Ly6Chi and CD103+ Ly6Clo CD8+ T cells within the oral mucosa of a Thy1.1+ VSV-ova immune parabiont. H = Host-derived cells, P = Partner-derived cells. (D) Representative histology of the ventral lingual mucosa from VSV-ova immune parabionts. Scale bar represents 75 µm. (E) Percent host among endogenous CD8+ T cells within spleen and oral mucosa (comparing CD103neg vs. CD103+ subsets) of parabiotic mice cohoused (purple; CoH) or exposed to the soiled bedding (gray; fomite) of pet shop mice for >60 d prior to surgery. Numbers represent individual parabiotic mice per group from cohoused or fomite-exposed parabiont cohorts analyzed on sperate days. (F) Left: Total i.v.neg OT-I T cells (left) and percentage of CD103+ CD69+ OT-I T cells isolated from the oral mucosa of VSV-ova immune mice housed under SPF conditions (orange) or cohoused with pet shop mice (purple) for >60 d. Middle: Enumeration of endogenous and TRM-phenotype CD8+ T cells in SPF vs. cohoused mice. Right: Enumeration of endogenous and TRM-phenotype CD4+ T cells in SPF vs. cohoused mice. Numbers in F indicate replicates per group from two independent experiments. In all graphs, error bars represent mean ± SEM. All dots represent individual mice. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 as determined by an unpaired Student’s t test between the relevant comparisons.
VPEP durably augments oral TRM
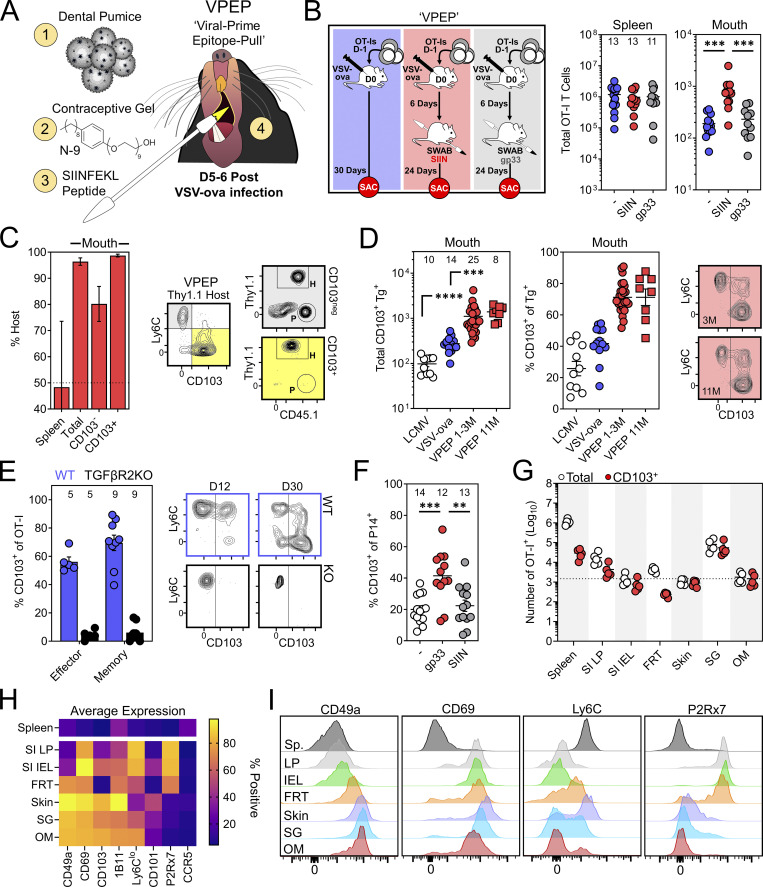
TRM are inefficiently isolated from many tissues, and low memory CD8+ T cell yields from the oral mucosa can be an impediment to their intensive characterization. Indeed, only ∼300 OT-I T cells could be isolated from the oral mucosa following systemic VSV-ova infection (Fig. 1 A). To facilitate the generation of a more tractable population of oral TRM to manipulate and study, we devised a prime-pull strategy termed VPEP. This approach also addresses the question of whether additional cognate antigen delivered within the oral mucosa during the effector phase of CD8+ T cell responses would increase the establishment of local memory. OT-I T cells were primed through i.v. VSV-ova infection. 5–6 d later, the oral surfaces of anesthetized mice were swabbed with SIINFEKL peptide (SIIN) dissolved in a slurry of fine dental pumice (a mechanical irritant) and contraceptive gel containing nonoxynol-9 (N-9; a chemical irritant; Fig. 3 A). This approach was chosen because (1) N-9 has been used to experimentally stimulate nonspecific inflammation at mucosal surfaces (Mackay et al., 2012), (2) the known abrasive effects of pumice, and (3) the mixture of pumice and N-9 forms a thick paste that durably adheres to the oral surfaces, increasing contact time between peptide and the epithelium. Compared to systemic VSV-ova infection alone, VPEP yielded ∼fourfold more memory OT-I T cells that could be isolated from the oral mucosa for flow-cytometric analysis (Fig. 3 B, red vs. blue). This augmentation was antigen dependent, as swabbing with an irrelevant gp33 peptide failed to enhance T cell numbers in the oral mucosa above VSV-ova infection alone (Fig. 3 B, gray vs. blue). Thus, local antigen recognition potentiates memory CD8+ T cell formation in the oral mucosa. Parabiosis surgery confirmed oral OT-I T cells generated through VPEP were TRM (Fig. 3 C). Once established, TRM after VPEP were durably maintained for at least 10 additional months without loss of CD103 expression (Fig. 3 D). VPEP-elicited oral TRM were also stably maintained following 2 mo of cohousing with pet shop mice (Fig. S1). We next tested whether TGFβ signaling, important for the induction of CD103 expression in the gut, skin, and corneal epithelium, was similarly required for TRM differentiation in the mouth (Casey et al., 2012; Hirai et al., 2019; Loi et al., 2022; Mackay et al., 2013; Zhang and Bevan, 2013). WT and TGFβRII−/− OT-I T cells were co-transferred and expanded through VPEP in congenically mismatched recipient mice. WT OT-I T cells robustly expressed CD103 within 12 d of infection while their TGFβRII−/− counterparts did not (Fig. 3 E). Next, to address whether VPEP could be adapted to other systemic viral infections, mice challenged with LCMV-Arm were orally swabbed 5–6 d later with gp33 peptide (here the relevant peptide) in pumice and N-9. 30 d later, more CD103+ P14 T cells could be isolated from the oral mucosa of gp33-swabbed mice compared to mice infected with LCMV-Arm, alone or in combination with oral SIIN peptide swabbing (here the irrelevant antigen; Fig. 3 F). Finally, we performed a broader phenotypic analysis to address whether VPEP-elicited oral TRM expressed unique combinations of cell-surface markers compared to putative TRM isolated from other barrier tissues. Additionally, OT-I T cells were enumerated in the mouth and other NLTs after VPEP to provide a reference point on their general abundance in the oral mucosa compared to other TRM-dominated locations. Consistent with our previous observations (Fig. 1 B), oral OT-I T cells were largely CD103+. Strikingly, more CD103+ OT-I T cells could be isolated from the mouth than the entirety of the small intestine intraepithelial compartment or female reproductive tract (Fig. 3 G). Thus, despite its comparatively small size, the oral mucosa contains a relatively high density of CD103+ TRM after VPEP. Oral OT-I T cells phenocopied those isolated from the salivary glands and skin (Fig. 4 H) yet were most dissimilar to OT-I T cells isolated from the gut, particularly with respect to CD49a and P2Rx7 (Fig. 4 I). We also found that VPEP-elicited OT-I TRM lacked expression of CCR5, recently identified as a marker of IL-17–producing CD4+ TRM in the human oral mucosa (Woodward Davis et al., 2019). Collectively, oral memory CD8+ T cells express unique phenotypic signatures likely reflecting their distinctive tissue microenvironment.
Figure 3.
A VPEP strategy generates preternaturally abundant oral TRM. (A) VPEP approach. The oral surfaces are swabbed with SIIN peptide dissolved in fine dental pumice and contraceptive gel containing N-9. (B) Quantification of OT-I T cells in spleens and oral mucosa of mice given systemic VSV-ova alone (blue), VPEP (red; SIIN), or VPEP with irrelevant gp33 peptide (gray; gp33). Numbers above bars indicate replicates per group from three independent experiments (N ≥ 3 mice per group). (C) Percent host OT-I T cells in spleens and oral mucosa of VPEP parabionts 21 d after surgery (left). Right: Representative flow cytometry of CD8+ T cells isolated from the oral mucosa of a Thy1.1+ VPEP parabiont. Data represent two pairs of VPEP parabionts. (D) Enumeration (left) and percent CD103+ of Tg+ T cells (middle) in the oral mucosa in mice 1 mo after infection with systemic LCMV-Arm (white), systemic VSV-ova alone (blue), given VPEP (red circles), or given VPEP and aged an additional 10 mo (red squares). Right: Representative flow cytometry of oral OT-I T cells from VPEP mice infected 3 or 11 mo prior. (E) Left: Percentage of WT and TGFβRII−/− OT-I T cells expressing CD103 in the oral mucosa 4 and 24 d after VPEP (10 or 30 d after VSV-ova infection). Numbers indicate replicates per group from the two timepoints analyzed. Right: Representative flow cytometry plots. (F) Mice were infected with LCMV-Arm ± oral swabbing with relevant (gp33) or irrelevant (SIIN) peptide. Percentage of P14 T cells expressing CD103 is plotted. Numbers indicate replicates per group from three independent experiments (N ≥ 3 mice per group). (G) Enumeration of total i.v.neg OT-I T cells and CD103+ OT-I T cells from the indicated locations. Hashed line denotes the average number of CD103+ OT-I T cells isolated from the oral mucosa. Data are representative of five VPEP memory mice. SI LP, small intestine lamina propria; SI IEL, small intestine intraepithelial compartment; FRT, female reproductive tract; SG, salivary glands; OM, oral mucosa. (H) Heatmap showing expression patterns of several canonical TRM markers among total i.v.neg OT-I T cells isolated from the indicated tissues. (I) Representative histograms showing expression patterns of CD49a, CD69, Ly6C, and P2Rx7 among OT-I T cells isolated from the indicated tissues. In all graphs, error bars represent mean ± SEM. All dots represent individual mice. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 as determined by an unpaired Student’s t test between the relevant comparisons.
Figure S1.
VPEP-established oral TRM are durably maintained following cohousing with pet shop mice. (A) Enumeration of OT-I T cells isolated from the indicated tissues of VPEP mice following >2 mo of cohousing (CoH) with pet shop mice. Data represent two independent experiments with N = 2–3 mice per experiment. (B) Representative flow cytometry plots concatenated from three mice from one of two independent experiments, highlighting phenotype of oral OT-I T cells in SPF vs. cohoused VPEP memory mice. (C) Left: Enumeration of total endogenous CD8+ T cells and percentage of endogenous CD8+ T cells expressing CD103 and CD69 in VPEP mice housed under SPF conditions (orange) or cohoused for >2 mo with pet shop mice (purple). Right: Enumeration of total endogenous CD4+ T cells and percentage of endogenous CD4+ T cells expressing CD103 and CD69 in VPEP mice housed under SPF conditions or cohoused for >2 mo. All dots represent individual mice. *, P < 0.05; **, P < 0.01 as determined by an unpaired Student’s t test between SPF and CoH groups.
Figure 4.
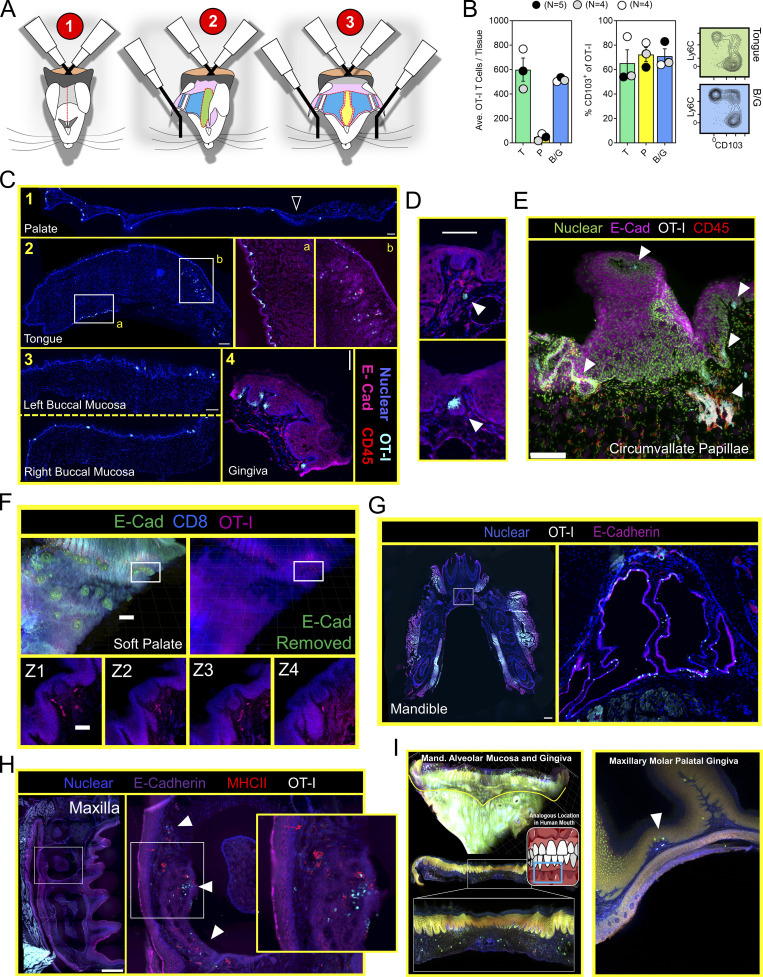
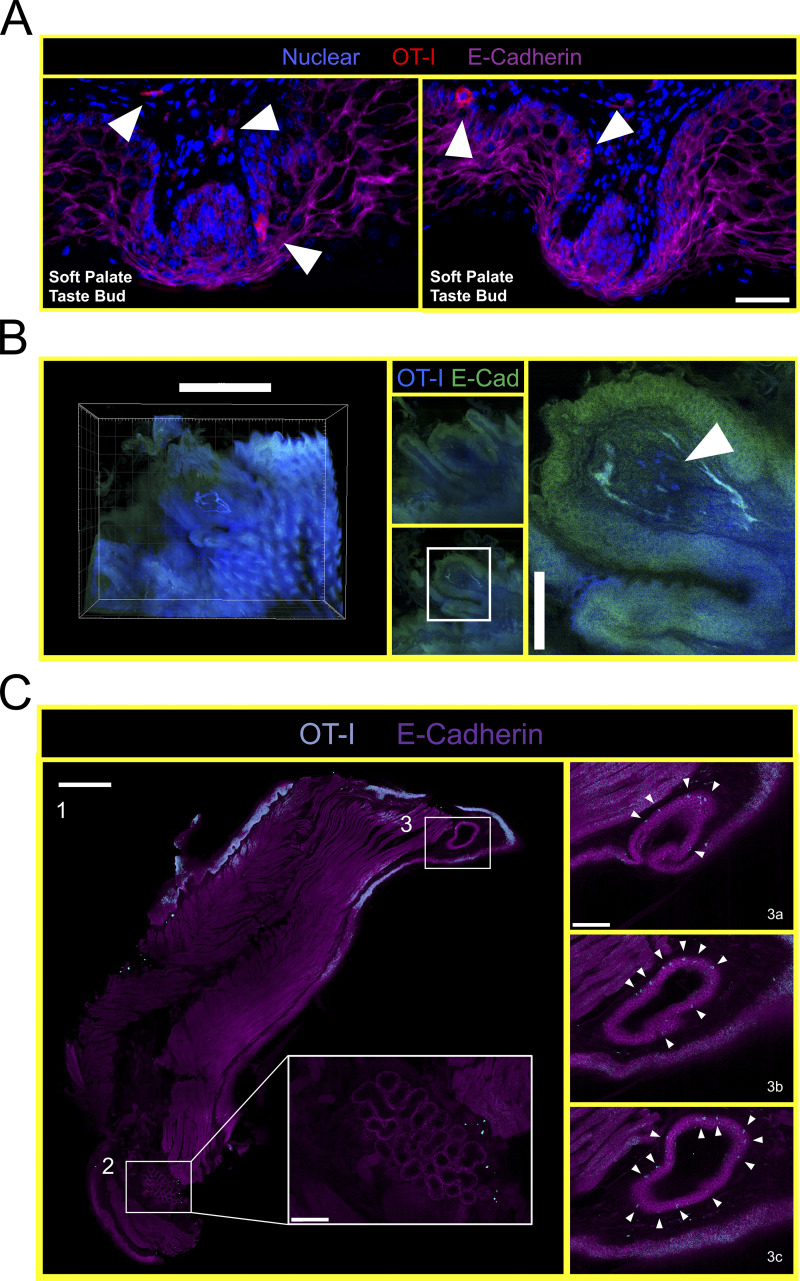
Mouth TRM are broadly distributed and associated with taste buds. (A) Stepwise process for isolating murine oral tissues including tongue (T; green), palate (P; yellow), and buccal mucosa/gingiva (BM/G; blue). Lateral ovoid shapes in 1 represent the masseter muscles. (B) Individual oral tissues from N ≥ 4 VPEP mice pooled by tissue type into a single sample for flow cytometric analysis. Left: Average number of OT-I T cells recovered from the indicated tissue. Right: Percentage of OT-I T cells expressing CD103, and representative flow cytometry highlighting tongue and buccal mucosa. Data from three independent experiments are shown. (C) IF microscopy of (1) hard and soft palate, (2) tongue, (3) left and right buccal mucosa, and (4) gingiva from VPEP memory mice >90 d after infection. All scale bars represent 200 µm, except in 2 (tongue) where the scale bar represents 500 µm. (D) OT-I T cells associated with taste buds in the soft palate. Two separate examples are shown with scale bar representing 100 µm. Colors reflect staining panel in C. (E) IF microscopy of the circumvallate papillae. Scale bar denotes 100 µm. Arrowheads reflect location of OT-I T cells in VPEP memory mice. (F) Ce3D imaging of the soft palate showing OT-I T cells in close proximity to taste buds directly posterior to the eighth palatine ridge. Scale bar in top left panel depicts 200 µm and scale bar in Z-stacks represent 50 µm. (G) IF image of a whole decalcified mandible from a mouse that received VPEP >30 d prior. Inset highlights OT-I T cells within Wharton’s (submandibular) duct. Lingual muscles intentionally obscured to better visualize periodontium and salivary ducts. Scale bar represents 500 µm. (H) OT-I T cells within maxillary gingiva of VPEP memory mouse, with corresponding magnified images highlighting gingiva surrounding the maxillary second molar. Scale bars in left panel represent 500 µm. Arrowheads denote clusters of OT-I T cells. (I) Ce3D imaging of mandibular (left) and maxillary (right) gingiva showing the presence of OT-I T cells (green) established through VPEP. Analogous location in human mouth shown for mandibular gingiva example. Yellow line denotes mucogingival junction. Arrowhead denotes location of OT-I T cells. All images are representative of at least three histological sections, per indicated tissue location, per mouse, of three or more individual VPEP mice. Taste buds in D and F were identified based on location and crescent morphology.
Mouth TRM occupy diverse oral structures
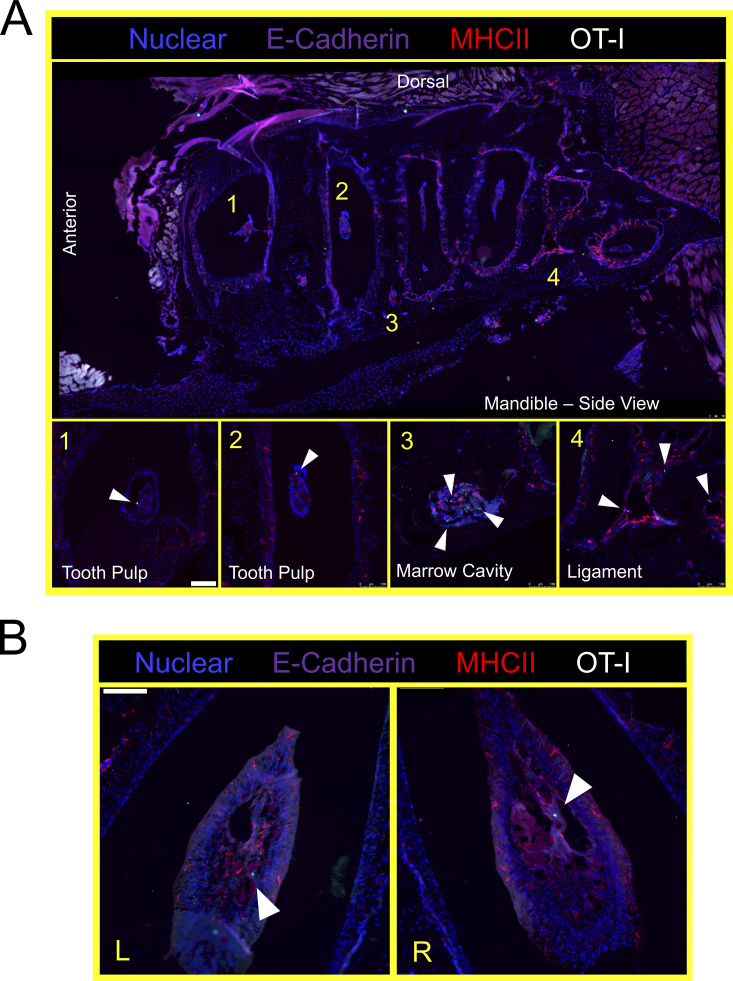
Thus far, experiments were performed by pooling and processing the tongue, buccal mucosa (cheek), gingiva, and palate into a single “oral mucosa” sample for flow cytometry. We next addressed the contribution of individual oral structures to the overall abundance of oral T cells. Oral tissues were isolated from groups of VPEP mice and pooled by tissue type (e.g., tongues from four mice were concatenated into a single “tongue” sample). Due to its small size, gingiva was processed with buccal mucosa. Our stepwise process for isolating individual oral structures is depicted in Fig. 4 A. Across three independent experiments (four or five pooled tissues per experiment), we found that the tongue and buccal mucosa/gingiva represented the principal reservoirs of memory OT-I T cells in the mouth. The palate (which was collected in a manner preventing nasal-associated lymphoid tissue contamination; Bittner-Eddy et al., 2017), contained a detectable, but comparatively small, oral TRM population that could be isolated for flow cytometry (Fig. 4 B, left graph). In each location, ∼60–80% of isolated OT-I T cells expressed CD103 (Fig. 4 B, right graph and representative flow cytometry). We next took a histological approach to visualize memory OT-I T cell distribution in diverse oral structures and tissue microenvironments. VPEP was performed. 3 mo later, OT-I T cells were broadly dispersed within the palate, buccal mucosa, and gingiva where they closely associated with the epithelial basement membrane, rarely extending into the stratified squamous epithelium (Fig. 4 C, panels 1, 3, 4). In contrast, their distribution was more regionalized in the tongue, where OT-I T cells primarily localized to the ventral epithelium (Fig. 4 C, panel 2 and 2a inset for a magnified image) and within mixed serous and mucus glands in the posterior dorsum (Fig. 4 C, panel 2 and 2b inset). Notably, we routinely observed OT-I T cells in proximity to isolated taste buds of the soft palate (Fig. 4 D and Fig. S2 A) and within and surrounding the circumvallate papillae of the tongue (Fig. 4 E). We further utilized Clearing-enhanced three-dimentional imaging (Ce3D) to document their association with taste buds directly posterior to the eighth palatine ridge (the junction of the hard and soft palate, denoted by arrow in Fig. 4 C, panel 1; Fig. 4 F and Video 1). Ce3D was also used to document their positioning within the circumvallate papillae (Fig. S2 B) and within minor salivary glands and ducts in the buccal mucosa (Fig. S2 C). We wished to image the floor of the mouth. However, our methods to expose the oral tissues for isolation required we cut the mandible lengthwise (Fig. 4 A). Therefore, intact mandibles from separate VPEP mice were decalcified, cryosectioned, and imaged by immunofluorescent (IF) microscopy. The sublingual mucosa contained abundant OT-I T cells within the epithelial lining of the submandibular duct (Fig. 4 G). OT-I T cells were also seen within dental pulp and ligaments surrounding the mandibular molar roots (Fig. S3). Decalcification/cryosectioning and Ce3D imaging was further used to visualize oral TRM distribution in maxillary and mandibular gingiva (Fig. 4, H and I), where its small size precluded thorough characterization in thin histological sections. Here we observed dense accumulations of OT-I T cells. In conclusion, TRM were broadly distributed throughout the oral mucosa and periodontium, and while we did not pursue quantitative immunofluorescence microscopy, the images made it quite evident that cell isolation for flow cytometry grossly underestimated the true number of OT-I T cells in the mouth following VPEP (as has also been observed in other TRM-dominated locations; Steinert et al., 2015; Watanabe et al., 2015; Wijeyesinghe et al., 2021).
Figure S2.
VPEP-elicited OT-I T cells surveil taste buds of the soft palate, circumvallate papillae, and minor salivary glands and ducts in the buccal mucosa. (A) Immunofluorescence images of two separate soft palate taste buds located immediately distal to the eighth palatine ridge. Blue = nuclear, red = OT-I, pink = E-Cadherin. Scale bar represents 50 µm. Taste buds identified based on location and crescent morphology. Arrowheads denote location of OT-I T cells. (B) Ce3D imaging of OT-I T cells within the circumvallate papillae of a VPEP memory mouse. Scale bars in left and right panels represent 500 and 100 µm, respectively. Arrowhead denotes location of OT-I T cells. (C) Ce3D on isolated buccal mucosa >30 d after VPEP. Individual 2-D slices from overall 3-D structure are shown in all panels. (1) Entire cleared buccal mucosa slice, highlighting a minor salivary gland (bottom of image; 2) and a salivary duct (top of image; 3). Scale bar represents 500 µm. (2) Enlarged image of buccal mucosa minor salivary gland. Scale bar represents 100 µm. (3a–c) Magnified images of buccal mucosa salivary duct, showing three different images through the Z plane. Scale bar represents 100 µm. Arrowheads denote OT-I T cells. Ce3D images are representative of at least two VPEP memory mice in which Ce3D was performed.
Video 1.
TRM in proximity to soft palate taste buds. Ce3D imaging of VPEP-elicited OT-I T cells in the vicinity of isolated taste buds of the soft palate. Palatal tissue from a VPEP memory mouse was made optically transparent as previously described (Li et al., 2017). Cleared tissue was imaged by tiling Z-stacks with confocal laser scanning microscopy (Stelleris), and 3D reconstructions were produced using Imaris (Bitplane) software v9.2.1. Movement through the Z-stack is viewed by 60 µm extended sections at 10 frames per second. For orientation, the eighth palatine ridge is visible at 0:00:00, and isolated taste buds are visible between 0:00:00–0:00:02. Taste buds are identified by crescent morphology. Red, OT-I CD8+ T cells; blue, nonspecific labeling of the epithelium. Corresponding still images from this video are displayed in Fig. 4 E. Images are representative of two Ce3D soft palate specimens from VPEP memory mice.
Figure S3.
VPEP-elicited OT-I T cells surveil the murine periodontium. (A) Lateral view of decalcified mandibula. Magnified images 1 and 3 are from the indicated locations, but from different tissue slices of the same tissue. (B) Aerial view of decalcified mandibular molar dental pulp in which OT-I T cells are present. L and R denote left and right second molars. Arrowheads denote location of OT-I T cells. Scale bars in A and B represent 100 µm.
Oral TRM reactivation fortifies cellular immunity in the mouth and draining cervical lymph nodes (cLNs)
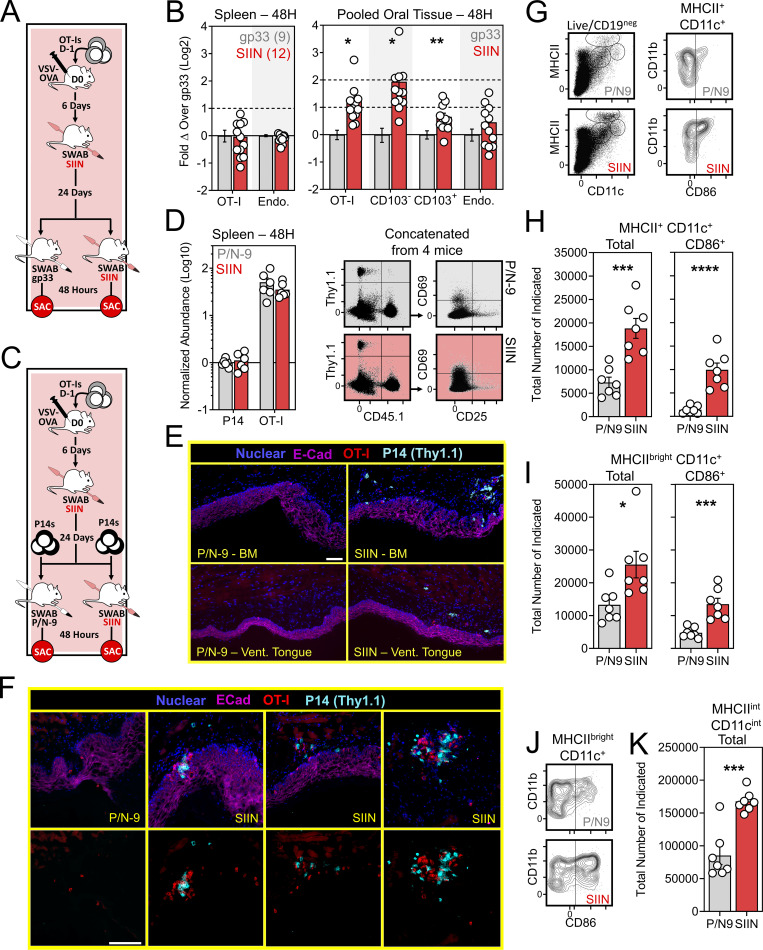
Our VPEP model demonstrates that local antigen recognition by mouth infiltrating effector CD8+ T cells promotes TRM establishment. We next asked whether oral memory T cells could be locally reactivated, and if so, how that might impact cellular immunity in the mouth. At least 30 d after VSV-ova infection, VPEP memory mice were re-exposed to cognate antigen by swabbing with SIIN peptide in pumice and N-9. Control VPEP memory mice were orally swabbed with gp33 peptide in pumice and N-9. 2 d after swabbing, CD8+ T cells were characterized and enumerated in the oral mucosa. (Fig. 5 A). Mice swabbed with SIIN peptide showed a pronounced increase in total oral OT-I T cells, largely attributed to an ∼fourfold increase in OT-I T cells expressing a circulating (CD103neg Ly6Chi) phenotype (Fig. 5 B). No changes in OT-I T cell abundance (or CD25 and CD69 expression; data not shown) were observed in spleen. To investigate whether oral T cell reactivation recruited circulating cells into the oral mucosa, 5 × 105 congenically distinct memory P14 T cells (generated from systemic LCMV-Arm infection) were adoptively transferred into VPEP memory mice 1 d prior to oral swabbing with SIIN in pumice and N-9, or pumice and N-9 alone (P/N-9; Fig. 5 C). When analyzing spleen 48 h after swabbing, transferred P14 T cells and OT-I T cells were found at comparable frequencies and both expressed a largely non-activated phenotype (CD25negCD69neg; Fig. 5 D). These data suggested that oral peptide exposure triggered local, rather than systemic, antigen-specific T cell reactivation. When analyzing the oral mucosa, only SIIN treatment elicited recruitment of bystander memory P14 T cells (Fig. 5 E), where they showed biased accumulation around OT-I T cell aggregates (Fig. 5 F). Control treated mice were completely devoid of transferred P14 T cells within the oral mucosa by histology, and OT-I T cell aggregates were not evident. Thus, oral T cell reactivation locally augments T cell immunity in the mouth, at least in part through mobilization of memory T cells from circulation into the oral mucosa. We next wished to assess how oral TRM reactivation impacted the abundance and phenotype of antigen presenting cells in the mouth-draining cLN. VPEP memory mice were orally swabbed with P/N-9 or P/N-9 containing SIIN peptide. 2 d later, we observed an increase in the number of conventional (MHCII+ CD11c+) dendritic cells (DCs) and migratory phenotype (MHCIIbright CD11c+) DCs within cLNs only when upstream TRM were previously reactivated (Fig. 5, G and I). MHCIIint CD11cint macrophages within cLNs were similarly increased following SIIN swabbing of the oral mucosa (Fig. 5, G and K). Moreover, DCs expressed elevated levels of the T cell costimulatory molecule CD86 in cLNs draining TRM-reactivated oral mucosa (Fig. 5, G and J). These observations align with previous reports that TRM reactivation induces DC maturation and migration from NLTs into draining LNs (Schenkel et al., 2014).
Figure 5.
Oral T cell antigen sensing bolsters cellular immunity in the mouth. (A) Experimental design. (B) Relative abundance of OT-I T cells and endogenous CD8+ T cells in spleens and oral mucosa of VPEP memory mice orally swabbed with gp33 (gray) or SIIN (red) 48 h earlier. Data represent fold change over the indicated phenotype in gp33 swabbed mice. Numbers indicate replicates per group from two independent experiments with ≥4 mice per group. (C) Experimental strategy to assess the ability of oral T cell reactivation to recruit circulating memory T cells into the oral mucosa. (D) Comparison of transferred P14 T cells and OT-I T cells abundance and phenotype in spleens of gp33- or SIIN-swabbed mice 48 h later. Concatenated flow cytometry plots are shown from one of two independent experiments (right). (E) IF imaging of buccal mucosa (top) and ventral lingual mucosa (bottom) from mice orally swabbed 48 h prior with P/N-9 or SIIN in irritants. (F) Higher magnification images of buccal mucosa 48 h after swabbing with P/N-9 or SIIN peptide. Nuclear and E-Cadherin staining removed from bottom panels to better visualize T cell aggregates. Images in E and F are representative of at least three sections, per tissue, per mouse, of six individual mice per group over two independent experiments. (G) Representative flow cytometry plots showing MHCII- and CD11c-expressing subsets in the cLNs of VPEP memory mice exposed 48 h prior to oral swabbing with P/N-9 or P/N-9 containing SIIN (left). CD86 expression among MHCII+ CD11c+ cLN DCs following the indicated treatment (right). (H) Quantification of total MHCII+ CD11c+ DCs (left) and those expressing CD86 (right). (I) Quantification of total MHCbright CD11c+ DCs (left) and those expressing CD86 (right). (J) Representative flow cytometry showing CD86 upregulation on cLN MHCIIbright CD11c+ DCs following the indicated treatment. (K) Enumeration of cLN macrophages (MHCIIint CD11cint) 48 h after oral swabbing with P/N-9 or P/N-9 containing SIIN. Data in G–K are representative of two independent experiments. Scale bars in E and F represents 100 µm. Dots in B, D, H, I, and K represent individual mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 as determined by an unpaired Student’s t test between the relevant comparisons.
Oral TRM peptide reactivation triggers a local antimicrobial state and changes in gene expression related to mucosal sensing, olfaction, and oral inflammation
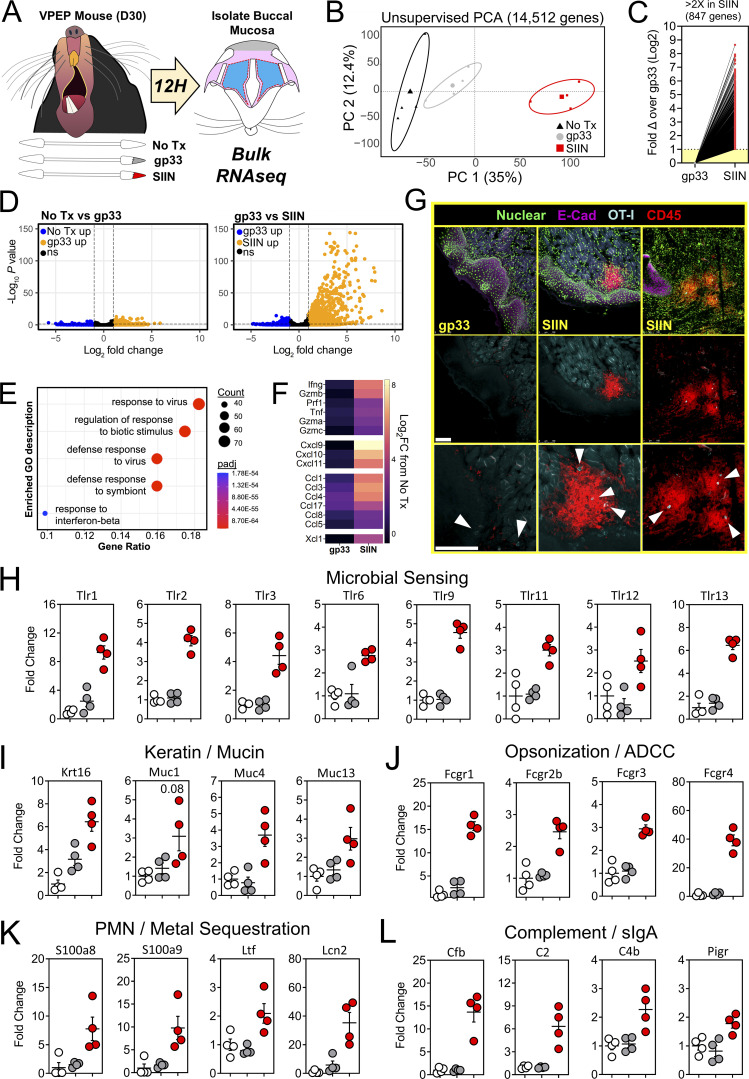
We took a bulk RNA sequencing (RNAseq) approach to better understand the tissue-wide transcriptional changes induced by oral T cell reactivation, including genes that may be involved in cellular recruitment into the oral mucosa. Groups of VPEP memory mice were matched for OT-I T cell frequency in blood (data not shown). Experimental groups included those left untreated (No Tx) or orally swabbed with gp33 or SIIN peptide dissolved in pumice and N-9 (gp33 and SIIN, respectively). 12 h later, total RNA was isolated from buccal mucosa (Fig. 6 A). Bulk RNAseq and principal component analysis (PCA) revealed that all three treatment groups clustered independently, with samples from SIIN-treated mice most transcriptionally dissimilar (Fig. 6 B). Oral swabbing with gp33 peptide (swabbing control) provoked few transcriptional changes, with only 74 statistically upregulated genes compared to untreated mice (Log2 fold change >1, false discovery rate [FDR] < 0.05; Fig. 6 D, left). These genes (and the 27 significantly downregulated genes) reflect the adjuvant effect of oral P/N-9 swabbing (Fig. 6 D, left, and Table S1). In contrast, oral swabbing with SIIN peptide induced robust transcriptional changes, with 847 statistically upregulated genes (Log2 fold change >1, FDR < 0.05) compared to the gp33 group (Fig. 6, C and D, right). Gene ontogeny enrichment analysis revealed an antimicrobial gene signature (Fig. 6 E), including upregulation of genes encoding T cell effector molecules interferon-γ (Ifng), perforin (Prf1), and granzymes A, B, and C (Gzma, Gzmb, Gzmc). We also observed induction of several chemokine genes in SIIN-treated mice (Fig. 6 F). Therefore, we used immunohistochemistry to test whether oral TRM reactivation could stimulate general leukocyte recruitment and/or reorganization within the oral mucosa. We found that CD45+ (pan leukocyte marker) cells were discernably increased in the buccal mucosa of SIIN-swabbed VPEP memory mice 8 h later, where they aggregated around OT-I T cells adjacent to the oral epithelium and within the lamina propria (Fig. 6 G). Minor salivary glands in the buccal mucosa were also visibly inflamed upon SIIN swabbing (Fig. S4 A). We further examined the transcriptome for differentially expressed genes (DEGs) with a known or potential role in host microbial sensing and mucosal barrier defense. In SIIN-treated buccal mucosa, we found the significant induction of genes for TLRs (Fig. 6 H), keratin and mucus production (Fig. 6 I), opsonization (Fig. 6 J), metal sequestration (Fig. 6 K), complement activation and transcytosis of mucosal IgA (Fig. 6 L).
Figure 6.
Oral TRM peptide reactivation triggers an oral inflammatory and antimicrobial state. (A) Buccal mucosa mRNA was isolated from untreated VPEP memory mice, or 12 h after swabbing with gp33 or SIIN peptide. (B) Unsupervised PCA plots showing clustering of No Tx (black), gp33 (gray), and SIIN (red) treatment groups. (C) Fold change of all significantly upregulated DEGs. (D) Volcano plots highlighting DEGs that are significantly (FDR P value <0.05) upregulated (>1log2 fold change; orange) or downregulated (<1log2 fold change; blue) between No Tx vs. gp33 (left) and gp33 vs. SIIN (right) treatment groups. N = 4 mice per group. (E) Gene Ontogeny (GO) enrichment analysis for genes upregulated (FDR P value <0.05) in SIIN- compared to gp33-treated buccal mucosa. (F) Log2 fold change (FC) of T cell effector molecule and chemokine genes comparing either gp33- or SIIN-swabbed mice to No Tx mice. (G) Immunofluorescence microscopy of buccal mucosa 8 h after swabbing with gp33 or SIIN peptide (top row). Two SIIN-swabbed mice are shown to portray heterogenous anatomic localization of CD45+ cell clusters upon oral TRM reactivation. Nuclear and E-Cadherin stains removed (middle, bottom). Bottom: Enlarged images with arrowheads highlighting OT-I T cells within inflammatory clusters. Scale bars represent 100 µm. Representative images are from at least three buccal mucosa sections, per mouse, of three or more individual mice per group. (H–L) Fold change in DEGs related to microbial sensing (H), keratin and mucus production (I), opsonization (J), neutrophil (PMN) degranulation and metal sequestration (K), and compliment activation and mucosal IgA secretion (L) in gp33-treated mice (gray) and SIIN-treated mice (red) relative to No Tx mice (white). Dots in H–L represent individual mice.
Figure S4.
Oral T cell reactivation induces minor salivary gland inflammation. (A) VPEP memory mice were orally swabbed with gp33 or SIIN peptide and buccal mucosa was isolated 8 h later. A pronounced increase in OT-I T cells (teal) and pan-leukocytes (red) was observed within minor salivary glands exclusively in SIIN-treated buccal mucosa. Bottom row: Nuclear and E-Cadherin staining removed to better visualize OT-I T cell (CD45.1) and pan-leukocyte (CD45.2) infiltration. (B) Left: Relative abundance of CD103+ and CD103neg OT-I T cells isolated from major salivary glands of VPEP mice receiving a single treatment with IgG-SAP or 103-SAP 1 wk prior. Right: Representative flow cytometry. All dots represent individual mice, with N = 10–11 mice per group from three independent experiments. Numbers above bar graphs represent mean fold change between the relevant comparisons. **, P < 0.01 as determined by an unpaired Student’s t test.
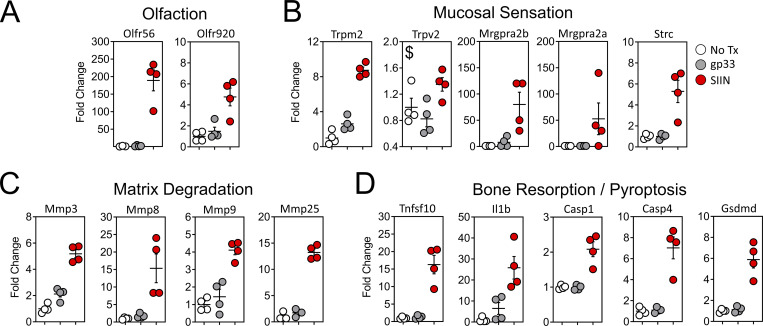
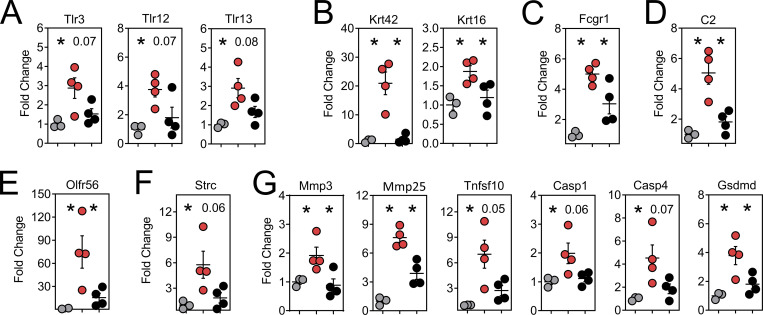
We searched for additional genes that may have unique roles in oral physiology and disease. Gustation and olfaction in the murine oral cavity primarily occur in the tongue and soft palate, with olfactory receptors recently shown to be functionally expressed in taste papillae (Malik et al., 2019). The buccal mucosa contains a smaller number of taste receptors; however, our transcriptomic data were limited to this tissue. Nevertheless, we observed an ∼200-fold induction of Olfr56, the predicted receptor for limonene (a major component of citrus fruit peel oil) within peptide reactivated buccal mucosa. Odorant receptor Olfr920 was also upregulated compared to gp33 treatment (Fig. 7 A). Genes for thermosensation (Trpm2, Trpv2) and neuropeptide binding (Mrgpra2a, Mrgpra2b) were also significantly upregulated, as was steriocillin (Strc), a structural component of sensory stereocilia found in the inner ear (Fig. 7 B). To our knowledge, the function of steriocillia in the oral mucosa has not yet been defined, although they are structurally related to gustatory hairs on the apical surface of taste receptor cells. Despite the buccal mucosa lacking any direct dental-supportive functions, we also identified genes etiologic to periodontal disease including Mmp8 and Mmp9, the main proteases in periodontitis and predictive biomarkers of disease onset and severity, when buccal TRM were reactivated (Al-Majid et al., 2018; Franco et al., 2017). Mmp3 and Mmp25, whose protein concentrations within gingival crevicular fluid of periodontitis patients increase proportionately with disease severity, were also upregulated after oral TRM reactivation in the cheek (Emingil et al., 2006; Reddy et al., 2012; Fig. 7 C). Genes promoting bone resorption via osteoblast cell death (Tnfsf10) and osteoclastogenesis (Il1b) were also significantly induced, as were genes regulating pyroptosis (Casp1, Casp4, Gsdmd); recently suggested as a pathway of exacerbated inflammation in periodontal disease (Sordi et al., 2021; Fig. 7 D).
Figure 7.
Oral peptide reactivation triggers local gene expression changes related to olfaction, mucosal sensing, and bone resorption. Fold change in DEGs relative to untreated VPEP memory mice (No Tx; white), 12 h after swabbing with gp33 (gray) or SIIN (red) in irritants. (A–D) Highlighted are genes involved in olfaction (A), mucosal sensation (B), extracellular matrix degradation (C), and bone resorption/pyroptosis (D). In all graphs, differences between gp33 vs. SIIN treatment groups are significant (FDR P value <0.05 and >1log2 fold change), except Trpv2 (a noxious temperature sensor; denoted with $ in B) where differences between gp33 vs. SIIN groups showed FDR P value <0.05 and a 1.63 fold change. All dots represent individual mice.
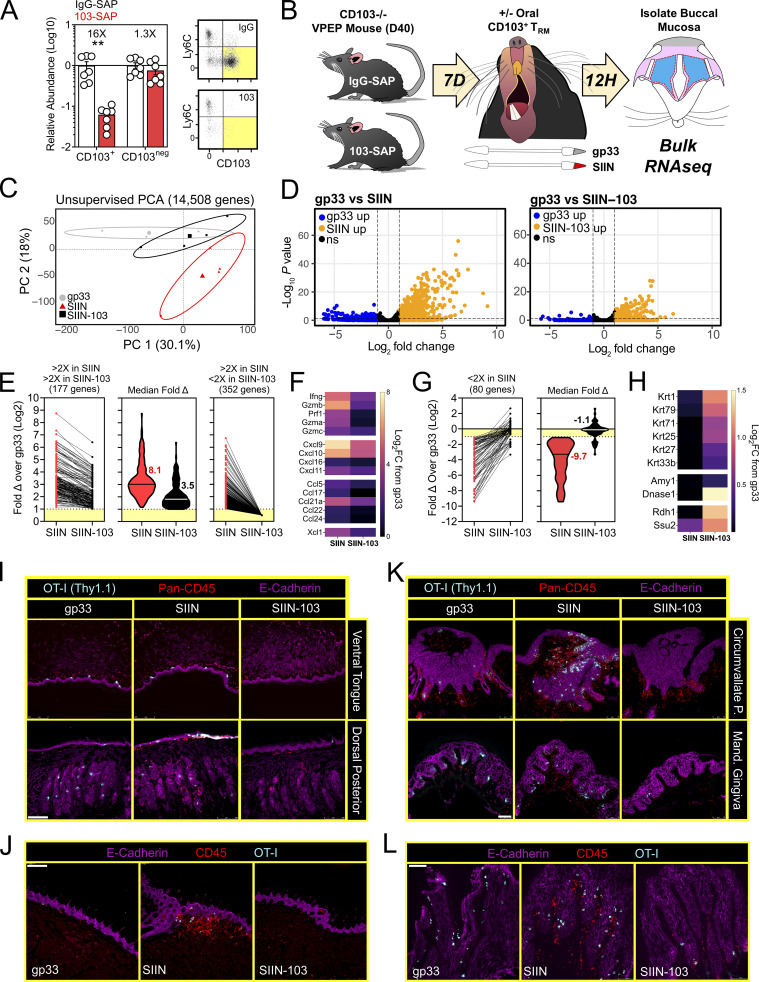
In vivo depletion of CD103+ TRM tempers local inflammatory responses in the oral mucosa
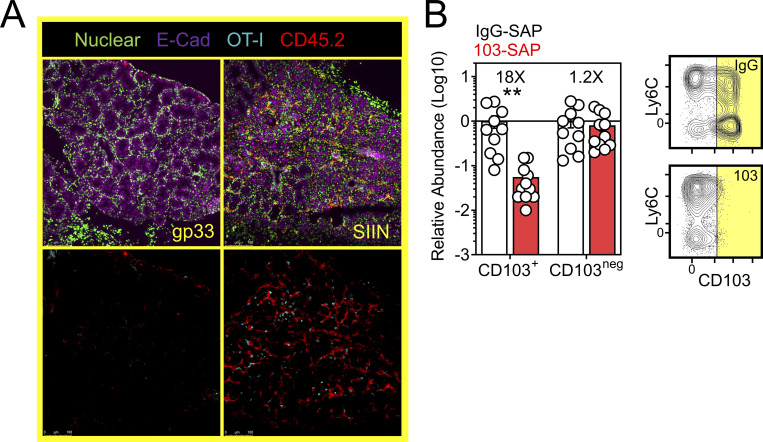
There is a paucity of approaches for eliminating specific populations of TRM in vivo (Stark et al., 2018). We sought to develop an experimental proof-of-principal approach for eliminating CD103+ TRM, comprising the majority of TRM in the oral mucosa after VPEP (Fig. 3 D and Fig. 4 B). However, DCs can also express CD103, as can regulatory T cells, resident natural killer cells, and γΔ T cells. Therefore, we developed an experimental approach to obviate the potential caveat of off-target effects on these and other CD103-expressing subsets, allowing us to isolate our analysis to antiviral TRM. For this, WT OT-I T cells were transferred and expanded through VPEP in CD103-deficient hosts, making OT-I T cells the only cell population capable of expressing CD103 in these mice. 30–40 d later, mice were injected i.p. with anti-CD103 antibodies conjugated to Saporin-toxin (103-SAP); a ribosome inhibiting protein. Control mice received Saporin-toxin-conjugated IgG2a (IgG-SAP). 7 d later, we observed a >90% reduction in CD103+ TRM within pooled oral mucosa (Fig. 8 A) and submandibular glands (Fig. S4 B). CD103neg OT-I T cells were unperturbed. There was a corresponding loss of Ly6Clo TRM following SAP-103 treatment. As CD103+ TRM are preferentially Ly6Clo, this indicated that SAP-103 treatment eliminated CD103+ TRM rather than simply preventing ex vivo flow cytometry staining for CD103. With this tool, we sought to address the extent to which transcriptional changes driven by oral T cell reactivation (Figs. 6 and 7) were triggered by CD103+ TRM rather than CD103neg TRM and/or recirculating cells. WT memory OT-I T cells were again established through VPEP in CD103-deficient hosts. Mice were split into three groups (memory OT-I T cell frequencies in blood were matched between groups to ensure that they were equivalent; data not shown). One group then received IgG-SAP, and oral TRM were exposed to irrelevant gp33 peptide 7 d later (negative control; gp33). A second group received IgG-SAP, and oral TRM were recalled 7 d later with SIIN peptide (positive control; SIIN). The third group received 103-SAP to therapeutically deplete CD103+ oral TRM 7 d prior to SIIN peptide recall (experimental group; SIIN-103). RNAseq was performed on buccal epithelium 12 h later (Fig. 8 B). PCA revealed that depletion of CD103+ cells altered TRM-driven changes in gene expression (Fig. 8 C). Comparing gp33 and SIIN-treated groups, 529 significantly upregulated genes (Log2 fold change >1, FDR < 0.05) were identified (Fig. 8 D, left). In contrast, only 177 significantly upregulated genes (Log2 fold change >1, FDR < 0.05) were identified between gp33 and SIIN-103–treated groups; a reduction of 352 genes (Fig. 8 D, right). Nearly every gene induced upon SIIN reactivation was significantly diminished (Fig. 8 E, left and middle) or expressed below a twofold cutoff (Fig. 8 E, right) in mice with prior CD103+ TRM depletion. Included among these were genes encoding T cell effector molecules and chemokines (Fig. 8 F), as well as genes for TLRs, keratin production, opsonization, complement activation, olfaction, and periodontitis (Fig. S5). Our RNAseq analysis also identified 80 genes significantly downregulated in SIIN-treated mice relative to gp33 controls that were re-expressed or fully restored upon CD103+ TRM depletion and SIIN swabbing (Fig. 8 G). Among these were genes for keratin production (Krt1, 2, 25, 27, 71, and 79), digestion (Amy1, Dnase1), and odontogenesis (Ssu2; Xiong et al., 2017; Fig. 8 H). Collectively, CD103+ TRM depletion effectively mitigated transcriptional changes induced upon cognate antigen swabbing of the oral mucosa. Tongues and gingiva not included in bulk RNAseq analysis were instead analyzed histologically. After SIIN swabbing in mice with an intact CD103+ TRM compartment, OT-I T cells and pan-leukocytes accumulated in the ventral lingual epithelium and posterior dorsum (Fig. 8, I and J), as well as the circumvallate papillae and gingiva (Fig. 8, K and L). In contrast, OT-I T cells were effectively absent, and pan-leukocyte cells were discernably reduced, when CD103+ TRM were depleted prior to oral peptide swabbing (Fig. 8, I and J, middle vs. right columns). Observations that most epithelial-localized OT-I T cells were eliminated following SAP-103 treatment is consistent with the notion that CD103+ TRM dominate the oral epithelial compartment (although CD103 is difficult to directly stain for in the oral mucosa by IF microscopy). Collectively, these data identify CD103+ TRM as critical catalysts of the transcriptional response to secondary antigen exposure in the oral mucosa.
Figure 8.
CD103+ TRM mediate transcriptional changes in the oral mucosa upon antigen sensing. (A) Relative abundance (left) and representative flow cytometry (right) of CD103+ and CD103neg OT-I T cells from the oral mucosa of VPEP memory mice treated with SAP-IgG or SAP-103 antibodies. Data represent three independent experiments (N = 2–3 mice per group per experiment). Flow cytometry plots are concatenated from two mice per group from one experiment. (B) OT-I T cells were expanded through VPEP in CD103−/− hosts. Treatment groups included gp33, SIIN, and SIIN-103, with groups established based on OT-I T cell frequencies in blood. 12 h after swabbing, buccal mucosa was isolated, and bulk RNAseq was performed. (C) Unsupervised PCA plots showing clustering of gp33 (gray), SIIN (red), and SIIN-103 (black) treatment groups. (D) Volcano plots highlighting DEGs significantly (FDR P value <0.05) upregulated (>1log2 fold change [FC]; orange) or downregulated (<1log2 FC; blue) between gp33- vs. SIIN-treated buccal mucosa (left) or gp33- vs. SIIN-103–treated buccal mucosa (right). N = 3 (gp33) or 4 (SIIN, SIIN-103) mice per treatment group. (E) Fold change over gp33 of all significantly upregulated DEGs in SIIN mice and their corresponding relative expression in SIIN-103 mice. (F) Heatmaps showing Log2 fold change of select T cell effector molecule and chemokine genes significantly upregulated in SIIN mice relative to gp33-treated mice, and their corresponding expression levels upon CD103+ TRM depletion and SIIN swabbing. (G) Fold change over gp33 of all significantly downregulated DEGs in SIIN mice and their corresponding relative expression in SIIN-103 mice. (H) Heatmaps showing Log2 fold change of select keratin, digestion, and odontogenesis genes significantly downregulated in SIIN mice relative to gp33 treated mice, and their corresponding expression levels upon CD103+ TRM depletion and SIIN swabbing. (I) Representative tongue histology from gp33-, SIIN-, or SIIN-103–treated mice. Images acquired at 10× magnification. Scale bar represents 250 µm. (J) Additional representative tongue histology as in I. Scale bar represents 200 µm. (K) Representative histology of circumvallate papillae (top) and gingiva (bottom) from gp33-, SIIN-, or SIIN-103–treated mice as in J. Images acquired at 20× magnification. Scale bar represents 75 µm. (L) Additional higher magnification image of gingiva as in K. Scale bar denotes 100 µm. Representative images are from at least three sections, per tissue, per mouse, of three to four individual mice per group. Dots in A represent individual mice. **, P < 0.01 in A determined by an unpaired Student’s t test between SAP-IgG- vs. SAP-103–treated mice.
Figure S5.
Genes related to microbial sensing, keratin production, opsonization, complement activation, and periodontitis induced upon oral TRM reactivation are blunted upon CD103+ TRM depletion. (A–G) Highlighted are genes for TLRs (A), keratin production (B), antibody-dependent cell-mediated cytotoxicity (C), complement (D), olfaction (E), steriocillin (F), and periodontitis (G). As in Fig. 8, gray circles represent gp33-treated animals, red circles represent SIIN-treated animals, black circles represent SIIN-103–treated animals. Differences between gp33 vs. SIIN and SIIN vs. SIIN-103 are all statistically significant (*, P < 0.05; as determined by a Student’s t test between the relevant comparisons) unless otherwise indicated, in which case P values are shown. Dots represent individual mice per group. N = 3 (gp33) or 4 (SIIN, SIIN-103) mice per treatment group.
Discussion
This study aimed to characterize basic mechanisms of CD8+ T cell immunosurveillance in the mouth. To facilitate these efforts, we developed an experimental approach to deplete CD103+ TRM, validating that a systemic depletion modality has the capacity to eliminate CD103+ TRM from NLTs including the oral mucosa and salivary glands. One potential outcome of such a strategy could be rapid repopulation of these compartments through homeostatic mechanisms; however, this was not observed. Moreover, depletion of CD103+ TRM from the oral mucosa had a functional consequence upon antigen re-exposure, namely, a marked reduction in proinflammatory gene signatures. Our study provides proof-of-principle data that TRM depleting agents, if developed, could be successful at mitigating T cell–driven NLT inflammation. While clinical applications of TRM-depleting therapies remain to be determined, our Saporin-conjugated antibody-mediated depletion approach provides a new tool for addressing basic science questions in experimental animal models which may be amenable to subset-specific immune cell depletion in other locations.
Mice offer several advantages for reductionist mechanistic approaches to interrogate the role of TRM in oral health and disease, but also several disadvantages, namely, a paucity of oral tissue and a lack of animal models facilitating the generation of abundant CD8+ T cells in the mouth to manipulate and study. VPEP, comprised of systemic viral infection and subsequent swabbing of the oral mucosa with virus-mimicking peptides, revealed local antigen recognition during the clonal expansion phase of the immune response potentiated oral TRM formation. Moreover, once established, VPEP-elicited oral TRM were durably maintained for at least 10 additional months. We observed enhanced TRM numbers and CD103 expression amongst Tg+ T cells in the oral mucosa following both systemic VSV-ova and LCMV-Arm infection when cognate antigen was provided. Thus, the enhancement of oral TRM through local antigen recognition appears to be a generalizable phenomenon. VPEP and its component parts (cotton swabs, dental pumice, N-9, peptides) are inexpensive, easily procured, and swabbing is quick and easy to perform. Vaccination strategies have recently targeted the establishment of TRM in specific barrier tissues, but not the mouth, which is prone to acute and chronic viral infections (including oncogenic viruses) and represents a staging ground for viral replication and transmission. A modified VPEP strategy in humans (for example, Vaccine-Prime, Epitope-Pull) may provide a means for broadly augmenting antiviral T cell populations in the mouth or within specific oral compartments. Here, oral swabbing with virus-mimicking peptides would follow primary immunization or boosting at distal sites. Further observations TGFβR signaling is critical for the induction of CD103 expression and oral TRM formation provides additional targetable pathways for their amplification or local depletion in the oral mucosa. Indeed, TRM-phenotype (CD45RO+ CD69+) T cells populate human oral tissues (Dutzan et al., 2016) and appear to be enriched in human periodontitis relative to healthy gingival tissue (Deli et al., 2020). Targeting CD103+ TRM depletion in the context of periodontitis may provide a means for alleviating excessive oral inflammation.
CD8+ T cells participate in anti-tumor immunity, and previous mouse studies have demonstrated the therapeutic potential of TRM reactivation in the treatment of solid cancers (Malik et al., 2017). Antiviral memory T cells abundantly infiltrated autochthonous and transplanted melanoma, where intratumoral deposition of T cell–reactivating peptides synergized with checkpoint blockade inhibitors (αPD-L1) to curtail tumor growth and promote tumor clearance (Rosato et al., 2019). The immunostimulatory potential of TRM within or surrounding oral cancers may similarly be exploited. Oral cancer currently represents the sixth most prevalent cancer globally, with HPV and alcohol/tobacco use representing major etiological factors (Rivera, 2015). Despite widespread HPV vaccination efforts and anti-smoking campaigns aimed at children and young adults, the incidence of oral cancer is increasing and is forecasted to claim ∼500,000 lives per year by 2030 (Ferlay et al., 2015). Removal of oral cancerous lesions can cause severe and permanent disfigurement and impact basic life functions. Alternative treatment options are needed. In the present study, we show that oral TRM reactivation induces regional upregulation of several antitumor genes. Moreover, peptide reactivation of oral TRM mobilized circulating memory T cells into the oral mucosa where they coalesced in inflammatory aggregates surrounding TRM. Deliberate reactivation of human oral TRM may provide a mechanism for directing neoantigen-specific or chimeric antigen receptor T cells into oral cancers, an approach which may be particularly tractable given the accessibility of the oral mucosa for swabbing.
We utilized conventional IF and Ce3D microscopy to better understand the distribution of antiviral TRM within disparate oral compartments. Dense accumulations of TRM were observed within the epithelial lining of the submandibular duct, as well as minor salivary glands in the tongue and buccal mucosa. Saliva released from these glands initiates alimentation, lubricates oral structures, buffers enamel-degrading acids, and harbors antimicrobial proteins and mucosal antibody. Leukocyte infiltration within minor salivary glands is often diagnostic of Sjogren’s syndrome in human labial mucosa biopsies, and we observed substantial minor salivary gland inflammation upon oral TRM reactivation in VPEP mice. Given their intimate association with salivary glands and ducts throughout the mouth, reactivation of oral TRM may indirectly modulate saliva production and/or the composition of antimicrobial proteins contained therein. Indeed, transcriptional analysis of TRM reactivated buccal mucosa revealed induction of several metal scavenging and compliment activation genes encoding proteins present in saliva. It is likely these observations reflect transcriptional changes within cells comprising minor salivary glands of the cheek. Nevertheless, that antiviral T cell reactivation can alter saliva-relevant gene expression may have significant clinical ramifications for diseases affecting saliva production. Also of note, TRM were commonly observed surrounding taste buds in the soft palate and tongue (including the circumvallate papillae where reactivation provoked discernable inflammation). Taste buds encompass an assemblage of 50–100 individual taste receptor cells (TRCs) that sense and transduce signals upon receptor ligation of proteins, carbohydrates, salt, acids, and bitter compounds in food. In TRCs, taste receptors localize to gustatory hairs; projections in the TRC plasma membrane, which traverse the taste pore into the mouth. This intentional breach in the oral epithelium may represent an access point for viruses infecting the mouth, and therefore TRM immunosurveillance of taste buds may reinforce protection at this particularly vulnerable mucosal location. Consequently, reactivation of taste bud adjacent TRM may impact gustation (taste) through mechanisms including TRC gene expression changes, synaptic signaling, or TRC death.
The mouth has long been considered a “canary in the coal mine” with respect to the overall health of the organism, with chronic oral inflammation predicting autoimmune and inflammatory conditions including diabetes mellitus, arthritis, atherosclerosis, and neurodegenerative disease (Cekici et al., 2014; Hajishengallis and Chavakis, 2021). A better understanding of oral mucosal immunology may provide reference and insight into its utility in representing other mucosal sites not so readily accessible in humans. This study demonstrates that mouth resident CD8+ T cells can potently contribute to oral inflammation, describes their diverse positioning throughout the oral mucosa and periodontium, pinpoints transcriptional changes catalyzed by oral CD103+ TRM upon cognate antigen sensing, and provides a framework and toolbox for future explorations into the fate and function of TRM within this complex and polyfunctional mucosal site. Subsequent studies will investigate the impact of oral TRM reactivation on experimental periodontitis, chemosensory biology, and oral cancer.
Materials and methods
Mice
C57BL/6J (B6) mice were purchased from the Jackson Laboratory and maintained under SPF conditions at the University of Minnesota. CD45.1+ OT-I (Hogquist et al., 1994) and Thy1.1+ P14 (Pircher et al., 1989) mice were fully backcrossed to C57BL/6J mice and maintained in our animal colony. CD103−/− mice were generated in-house from P14+ CD103−/− mice (Schön et al., 1999). TGFβRII−/− mice (Lucas et al., 2000) were kindly provided by Dr. Nu Zhang (University of Texas Health Science Center at San Antonio, San Antonio, TX, USA). Both female and male mice were used in experiments, except parabiosis studies where females were exclusively used due to aggression of male mice toward other males. All mice were used in accordance with the Institutional Animal Care and Use Committees guidelines at the University of Minnesota.
Adoptive transfers, infections, and plaque assays
Memory mice harboring P14 or OT-I T cells were generated by transferring 5 × 104 CD8+ T cells of either genotype into naive C57BL/6J mice via retro-orbital injection, followed by i.p. infection with 2 × 105 PFU LCMV-Arm or 1 × 106 PFU VSV-ova via tail vein injections 1 d later. For oral VacV-ova infections, mice were anesthetized with ketamine/xylazine and placed in a mouse-restrainer comprised of a bent paper clip (to hook maxillary incisors) and a stretched rubber band (to hook the mandibular incisors). Tongues were exteriorized with forceps and the ventral lingual mucosa was punctured with a seven-prong tattoo needle (Cheyenne; Softedge safety cartridge) dipped immediately prior in freshly thawed VacV-ova stock (3.99 × 107 PFU/ml). A needle depth of seven clicks (when placed in an adjustable Cheyene Hawk grip sleeve) was chosen based on previous experience. To assess viral titers, excised tongues were flash-frozen in 2 ml of PBS, thawed, mechanically dissociated using a tissuemizer homogenizer (Thermo Fisher Scientific), frozen and thawed a second time, and briefly centrifuged to 1,000 ×g to pellet debris. Homogenates were serial diluted 1:10–1:1,000 in PBS, and 500 μl of each dilution was plated on confluent 143 B cells in 6-well tissue culture plates, giving an effective virus detection range of 40–400,000 PFU/tongue. 90 min later, 2 ml of RP-10 media (RPMI, 10% FCS, 1× Penn-Strep) was added to each well, and plates were incubated for 48 h at 37°C prior to plaque enumeration using crystal violet (0.1% in 30% ethanol). Viral titers below the limit of detection (40 PFU/tongue) were assigned a value of 20 PFU/tongue.
VPEP model
N-9 has been used generate non-specific inflammation in the female reproductive tract of mice (Mackay et al., 2012). For experiments utilizing VPEP, peptide slurries were prepared in advance as follows: (1) a P/N-9 paste was prepared by dissolving 4 g of fine dental pumice (Dharma Research) in 10 ml of contraceptive gel containing N-9 (Gynol II, Emerson Healthcare or VCF, Apothecus Pharmaceutical Corp.); (2) gp33 or SIINFEKL peptide were dissolved in sterile H2O at a final concentration of 10 mg/ml; (3) 1 ml of peptide solution (10 mg/ml) was added to 4 ml of P/N-9 paste; and (4) peptide slurries were homogenized using a pointed-end micro lab spatula, aliquoted in 1.5 ml Eppendorf tubes, and frozen for subsequent use. For oral peptide swabbing, mice infected 5–6 d prior with VSV-ova or LCMV-Arm were anesthetized and orally swabbed with ∼35 μl of freshly thawed peptide slurry using a fine-tipped cotton applicator (Tamiya; triangular, extra small).
Intravascular labeling, cell isolation, and flow cytometry
Intravascular staining was used to discriminate cells present in the vasculature from tissue parenchyma, as previously described (Anderson et al., 2014). Briefly, 3 µg of biotinylated αCD8β antibody (YTS156.7.7) was injected retro-orbitally into mice 3 min prior to sacrifice. For oral tissue isolation, the oral mucosal surfaces were exposed through a longitudinal incision between the mandibular incisors and held open with bent 17G needles. The buccal mucosa and gingiva were carefully removed using fine forceps and the cutting edge of a bent insulin syringe. The tongue and hard/soft pallet were also collected avoiding nasal-associated lymphoid tissue contamination. Oral tissues were pooled into a single sample (unless otherwise stated), finely minced with scissors, and enzymatically digested in collagenase solution (RMPI, 5% FBS, 1 mM MgCl2, 1 mM CaCl2, 1× Hepes, 1× L-Glutamine) containing 0.5 mg/ml Collagenase IV (Sigma-Aldrich) and 1 µg/ml DNAse for 1H at 37°C before mechanical disruption via gentleMACS Dissociator (Miltenyi Biotec). Oral tissue homogenates were passed through a 70-µm filter and further enriched on a 44–67% Percoll gradient. Isolated mouse cells were stained with antibodies against TCRβ (H57-597), CD11b (M1/70), CD8α (53-6.7), CD8β (YTS156.7.7), CD90.1 (OX-7), CD90.1 (HIS51), CD45.1 (A20), CD103 (M290), CD69 (H1.2F3), CD25 (PC61), CD44 (IM7), CD62L (MEL-14), Ly6C (1A8), PD1 (RMP1-30), Integrin-β7 (FIB504), CD11c (N418), MHCII (M5-114), and CD86 (GL1). Ghost Dye Viability Dye (Tonbo Biosciences) was used to discriminate live vs. dead cells, and fluorescently conjugated streptavidin was used to detect intravascular staining antibodies. Cell counts were obtained using Polybead Polystyrene 10 µm microspheres (Polysciences) added directly to flow cytometry samples at a known microsphere concentration. Stained cell samples were acquired with LSRII or LSR Fortessa flow cytometers (BD) and analyzed with FlowJo software (TreeStar).
Parabiosis and dirty mice
Parabiosis surgery was performed as previously described (Steinert et al., 2015). For dirty mice parabiosis experiments, mice were cohoused with mice purchased from a pet shop or exposed to their soiled bedding, for >60 d at University of Minnesota’s ABSL3 facility prior to parabiosis surgery. Tissues were evaluated ≥21 d after surgery.
FTY720 administration and in vivo SAP-conjugated antibody treatment
Mice were treated with FTY720 (Millipore) dissolved in 100% ethanol at a dose of 20 µg/mouse via i.p. injection. Vehicle control mice received i.p. injections with 200 μl of 100% ethanol. FTY720 or vehicle was administered 48 h prior to, and immediately preceding lingual VacV-ova infections. Saporin-conjugated IgG2A and αCD103 (M290) antibodies were purchased from Advanced Targeting Systems and administered at a dose of 5 µg/male mouse or 2 µg/female mouse in PBS via i.p. injection. Mice were rested for 7 d after SAP-conjugated antibody treatment before oral peptide swabbing.
IF and Ce3D microscopy
Harvested murine tissues were embedded in the optimum cutting temperature tissue freezing medium and frozen in a 2-methyl butane liquid bath. Frozen blocks were cut on a Leica cryostat to prepare 7-µm-thick sections. Slides were stained with antibodies against CD8β (YTS156.7.7), CD8α (53-6.7), CD45.1 (A20), Thy1.1 (Ox-7), CD45 (30-F11), CD45.2 (104), E-Cadherin (DECMA-1), and MHCII (M5/114). IF microscopy was performed using a Leica DM6000 B microscope. Counterstaining with Sytox Green (Thermo Fisher Scientific) or DAPI was carried out to detect nuclei where indicated. For Ce3D imaging, oral tissues were harvested and fixed with BD Cytofix/Cytoperm diluted 1:4 in PBS overnight at 4°C. Fixed tissues were incubated for ≥8 h in blocking buffer containing 1% BSA, 1:100 mouse FC block, and 0.3% Triton X-100 in PBS at 37°C. Tissues were then incubated with directly conjugated antibodies diluted 1:10–1:40 in blocking buffer for >2 d at 37°C on a shaker. Stained samples were washed with PBS containing 0.2% Triton X-100 and 1-thioglycerol (0.5%) for 12–24 h at 24°C or 37°C. Tissue was cleared using Ce3D as previously described (Li et al., 2017). In brief, N-methylacetamide prepared to 40% (vol/vol) in PBS was used to dissolve Histodenz to 86% (wt/vol) concentration (∼1.455 g Histodenz per 1 ml of 40% N-methyl-acetamide) inside a chemical fume hood, with the mixture incubated at 37°C, and dissolution was expedited using a stir bar and plate. Triton X-100 (0.1% vol/vol) and 1-thioglycerol (0.5% vol/vol) were added to the clearing solution. Stained tissues were placed in the Ce3D medium and incubated for ∼24 h at room temperature on a rotor. Stained and cleared tissue was mounted in fresh Ce3D medium with an adhesive silicone rubber spacer and tightly covered with a glass coverslip. Cleared tissue was imaged by tiling Z-stacks with confocal laser scanning microscopy on a Leica Stellaris 8. A 16× glycerol-immersion coverslip-corrected objective was used to capture images with an XY voxel size of 971 nm, 4 µm step size, and 1-A.U. pinhole. 3D reconstructions were done using Imaris (Bitplane) software v9.2.1.
RNAseq and analysis
Tissue isolation and RNA purification
For bulk RNAseq experiments, buccal mucosa was isolated directly into RNAlater (Thermo Fisher Scientific) 12 h following the indicated treatment. Tissue was homogenized in Trizol (Sigma-Aldrich) using a Tissuemiser homogenizer (Thermo Fisher Scientific), and RNA was extracted using a RNeasy mini kit (QIAGEN) per the manufacturer’s instructions. RNA integrity was assessed using capillary electrophoresis with the Agilent 2100 BioAnalyzer system (Agilent Technologies).
Library preparation, RNAseq, and data processing
Sequencing libraries were prepared using the Clontech SMARTer Stranded Total RNA-Seq Kit v2—Pico Input Mammalian kit. RNAseq (50-bp single-end) with the HiSeq 2500 Illumina was done at the University of Minnesota Genomics Center. The sequences were processed with the Collection of Hierarchical UMII-RIS Pipelines (CHURP, v.0.2.2) developed by the Research Informatics Solutions RIS group at the Minnesota Supercomputing Institute. The reference implementation of the CHURP package is available at https://github.com/msi-ris/CHURP.git (Baller et al., 2019). The Mouse mm10 reference was used for mapping the sequences.
Differential gene expression analyses
Within the CHURP pipeline, sequence reads were counted with featureCounts v.1.6.2 and the empirical analysis of DGE tool was used to identify differentially expressed genes between the experimental groups (Robinson et al., 2010). For volcano plot visualization of DEGs, genes were considered differentially expressed if they had an FDR P value <0.05 and an absolute fold change >2. For the Gene Ontology term enrichment analysis of genes upregulated in SIIN- vs. gp33-treated buccal mucosa genes were considered differentially expressed if they had an FDR P value <0.05.
PCA
To remove unexpressed genes in the dataset, genes with a row (gene) mean SD <0 and a row mean <0 across all of the samples were removed before analysis. The prcomp and factoextra in RStudio were used to perform the PCA and plot the first two principal components, respectively. PCA plot confidence ellipses represent the 95% confidence interval of the centroid along the two principal components for samples within each experimental group.
Pathway analyses and data visualization
The “clusterProfiler R” package was used to analyze and visualize enriched pathways for DEGs (Yu et al., 2012). The Bioconductor “EnhancedVolcano” package was used in R (version 4.1.3) to visualize DEGs between groups.
Statistical analysis
An unpaired Student’s t test was used when comparing two groups. In experiments comparing lingual infection viral titers, a Mann–Whitney tests or Kruskal–Wallis test (multi-comparison) was applied to account for non-Gaussian distribution in the data. All graphs represent mean ± SEM, except those plotting viral titers where median values are depicted. P < 0.05 was considered significant in all experimental analysis. Statistical analysis was done in Prism (GraphPad Software). Sample size was chosen based on previous experience. No sample exclusion criteria were applied, and investigators were not blinded.
Online supplemental material
Fig. S1 enumerates VPEP-elicited oral memory CD8+ T cells, as well as endogenous CD8+ and CD4+ T cells, following >60 d of cohousing with pet shop mice. Fig. S2 depicts the localization of VPEP-elicited oral TRM within and surrounding isolated taste buds of the soft palate, the circumvallate papillae, and minor salivary glands and ducts of the cheek. Fig. S3 shows VPEP-elicited oral TRM within dental supporting structures, bone marrow cavities, and tooth pulp. Fig. S4 highlights minor salivary gland inflammation upon oral TRM reactivation, as well as quantifies CD103+ CD8+ T cell depletion from major salivary glands upon SAP-103 treatment. Fig. S5 identifies genes related to microbial sensing, keratin production, opsonization, complement activation, and periodontitis induced upon oral TRM reactivation that are subsequently blunted upon CD103+ TRM depletion. Video 1 shows Ce3D imaging of VPEP-elicited OT-I T cells in the vicinity of isolated taste buds of the soft palate. Table S1 lists genes induced upon oral gp33 swabbing relative to untreated mice, representing the adjuvant effect of P/N-9 swabbing.
Supplementary Material
shows DEGs comparing No Tx to gp33 groups (adjuvant effect of P/N-9 swabbing).
Acknowledgments
We thank the University of Minnesota flow cytometry, microscopy, and genomics cores. The Minnesota Supercomputing Institute provided computing resources, informatics advice, and data storage. Dr. Rajaram Gopalakrishnan and members of the Oral Mucosal Immunology Consortium critically read and provided feedback on the manuscript. Dr. Jason Mitchell assisted with Ce3D imaging analysis. Hayley Scheubrein assisted with data analytics. Graphical abstract and figure illustrations are original artwork created by J.M. Stolley.
This work was supported by National Institutes of Health grants 5R01AI146032-03, 5R01AI084913-13, and 5R01AI150600-02 (D. Masopust), K99DE031014 and T90 DE 022732 (J.M. Stolley), and the Oral Mucosal Immunity Consortium Windsweep Farm pilot grant (J.M. Stolley and D. Masopust).
Author contributions: J.M. Stolley and D. Masopust acquired funding, conceived of the study, designed experiments, and wrote the manuscript; J.M. Stolley performed experiments and analyzed data; V. Joag performed Ce3D imaging; M.C. Scott and N.V. Gavil performed parabiosis surgeries; M.C. Scott and J.M. Stolley performed RNAseq analysis; A.J. Dale, T.S. Johnston, F. Saavedra, S. Lotfi-Emran, and A.G. Soerens assisted with experiments; E. Weyu and M.J. Pierson maintained animal colonies. F. Saavedra and M.C. Herzberg provided technical support and consulting. N. Zhang provided mice. V. Vezys provided input, mice, and reagents. All authors read, revised, and approved of the manuscript.
Data availability
The data underlying Figs. 6, 7, and 8 and Fig. S5 are openly available in the Gene Expression Omnibus under the accession number GSE229634.
References
- Al-Majid, A., Alassiri S., Rathnayake N., Tervahartiala T., Gieselmann D.-R., and Sorsa T.. 2018. Matrix metalloproteinase-8 as an inflammatory and prevention biomarker in periodontal and peri-implant diseases. Int. J. Dent. 2018:7891323. 10.1155/2018/7891323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, K.G., Mayer-Barber K., Sung H., Beura L., James B.R., Taylor J.J., Qunaj L., Griffith T.S., Vezys V., Barber D.L., and Masopust D.. 2014. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 9:209–222. 10.1038/nprot.2014.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariotti, S., Hogenbirk M.A., Dijkgraaf F.E., Visser L.L., Hoekstra M.E., Song J.-Y., Jacobs H., Haanen J.B., and Schumacher T.N.. 2014. T cell memory. Skin-resident memory CD8⁺ T cells trigger a state of tissue-wide pathogen alert. Science. 346:101–105. 10.1126/science.1254803 [DOI] [PubMed] [Google Scholar]
- Balaji, S., Cholan P.K., and Victor D.J.. 2021. An emphasis of T-cell subsets as regulators of periodontal health and disease. J. Clin. Transl. Res. 7:648–656. [PMC free article] [PubMed] [Google Scholar]
- Baller, J., Kono T., Herman A., and Zhang Y.. 2019. CHURP: A lightweight cli framework to enable novice users to analyze sequencing datasets in parallel. In. Proceedings of the Practice and Experience in Advanced Research Computing on Rise of theMachines (Learning), PEARC ’19. Association for Computing Machinery, New York, NY, USA. 1–5. 10.1145/3332186.3333156 [DOI] [Google Scholar]
- Beura, L.K., Hamilton S.E., Bi K., Schenkel J.M., Odumade O.A., Casey K.A., Thompson E.A., Fraser K.A., Rosato P.C., Filali-Mouhim A., et al. 2016. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 532:512–516. 10.1038/nature17655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner-Eddy, P.D., Fischer L.A., Tu A.A., Allman D.A., and Costalonga M.. 2017. Discriminating between interstitial and circulating leukocytes in tissues of the murine oral mucosa avoiding nasal-associated lymphoid tissue contamination. Front. Immunol. 8:1398. 10.3389/fimmu.2017.01398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso, E.M., and Arosa F.A.. 2017. CD8+ T cells in chronic periodontitis: Roles and rules. Front. Immunol. 8:145. 10.3389/fimmu.2017.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey, K.A., Fraser K.A., Schenkel J.M., Moran A., Abt M.C., Beura L.K., Lucas P.J., Artis D., Wherry E.J., Hogquist K., et al. 2012. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J. Immunol. 188:4866–4875. 10.4049/jimmunol.1200402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cekici, A., Kantarci A., Hasturk H., and Van Dyke T.E.. 2014. Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontol. 2000. 64:57–80. 10.1111/prd.12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, R.A. 2015. Resident memory T cells in human health and disease. Sci. Transl. Med. 7:269rv1. 10.1126/scitranslmed.3010641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowl, J.T., Heeg M., Ferry A., Milner J.J., Omilusik K.D., Toma C., He Z., Chang J.T., and Goldrath A.W.. 2022. Tissue-resident memory CD8+ T cells possess unique transcriptional, epigenetic and functional adaptations to different tissue environments. Nat. Immunol. 23:1121–1131. 10.1038/s41590-022-01229-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deli, F., Romano F., Gualini G., Mariani G.M., Sala I., Veneziano F., Bertero L., Cassoni P., and Aimetti M.. 2020. Resident memory T cells: Possible players in periodontal disease recurrence. J. Periodontal Res. 55:324–330. 10.1111/jre.12709 [DOI] [PubMed] [Google Scholar]
- Dewhirst, F.E., Chen T., Izard J., Paster B.J., Tanner A.C.R., Yu W.-H., Lakshmanan A., and Wade W.G.. 2010. The human oral microbiome. J. Bacteriol. 192:5002–5017. 10.1128/JB.00542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudding, T., Haworth S., Lind P.A., Sathirapongsasuti J.F., Tung J.Y., Mitchell R., Colodro-Conde L., Medland S.E., Gordon S., Elsworth B.. et al. 2019. Genome wide analysis for mouth ulcers identifies associations at immune regulatory loci. Nat. Commun. 10:1052. 10.1038/s41467-019-08923-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutzan, N., Konkel J.E., Greenwell-Wild T., and Moutsopoulos N.M.. 2016. Characterization of the human immune cell network at the gingival barrier. Mucosal Immunol. 9:1163–1172. 10.1038/mi.2015.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emingil, G., Kuula H., Sorsa T., and Atilla G.. 2006. Gingival crevicular fluid matrix metalloproteinase-25 and -26 levels in periodontal disease. J. Periodontol. 77:664–671. 10.1902/jop.2006.050288 [DOI] [PubMed] [Google Scholar]
- Epstein, J.B., and Chow A.W.. 1999. Oral complications associated with immunosuppression and cancer therapies. Infect. Dis. Clin. North Am. 13:901–923. 10.1016/s0891-5520(05)70115-x [DOI] [PubMed] [Google Scholar]
- Feller, L., Altini M., and Lemmer J.. 2013. Inflammation in the context of oral cancer. Oral Oncol. 49:887–892. 10.1016/j.oraloncology.2013.07.003 [DOI] [PubMed] [Google Scholar]
- Ferlay, J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., and Bray F.. 2015. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 136:E359–E386. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- Franco, C., Patricia H.-R., Timo S., Claudia B., and Marcela H.. 2017. Matrix metalloproteinases as regulators of periodontal inflammation. Int. J. Mol. Sci. 18:440. 10.3390/ijms18020440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffen, S.L., and Moutsopoulos N.M.. 2020. Regulation of host-microbe interactions at oral mucosal barriers by type 17 immunity. Sci. Immunol. 5:eaau4594. 10.1126/sciimmunol.aau4594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinde, B. 2013. Herpesviruses: Latency and reactivation - viral strategies and host response. J. Oral Microbiol. 5:22766. 10.3402/jom.v5i0.22766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand, A.R., and Frank M.E.. 2014. Fundamentals of Oral Histology and Physiology. https://www.wiley.com/en-us/Fundamentals+of+Oral+Histology+and+Physiology-p-9781118342916. [Google Scholar]
- Hairston, B.R., Bruce A.J., and Rogers R.S. III. 2003. Viral diseases of the oral mucosa. Dermatol. Clin. 21:17–32. 10.1016/s0733-8635(02)00056-6 [DOI] [PubMed] [Google Scholar]
- Hajishengallis, G., and Chavakis T.. 2021. Local and systemic mechanisms linking periodontal disease and inflammatory comorbidities. Nat. Rev. Immunol. 21:426–440. 10.1038/s41577-020-00488-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis, G., Chavakis T., and Lambris J.D.. 2020. Current understanding of periodontal disease pathogenesis and targets for host-modulation therapy. Periodontol. 2000. 84:14–34. 10.1111/prd.12331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman, H.D., Reynoso G.V., Ngudiankama B.F., Cush S.S., Gibbs J., Bennink J.R., and Yewdell J.W.. 2015. CXCR3 chemokine receptor enables local CD8(+) T cell migration for the destruction of virus-infected cells. Immunity. 42:524–537. 10.1016/j.immuni.2015.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman, H.D., Takeda K., Skon C.N., Murray F.R., Hensley S.E., Loomis J., Barber G.N., Bennink J.R., and Yewdell J.W.. 2008. Direct priming of antiviral CD8+ T cells in the peripheral interfollicular region of lymph nodes. Nat. Immunol. 9:155–165. 10.1038/ni1557 [DOI] [PubMed] [Google Scholar]
- Hirai, T., Zenke Y., Yang Y., Bartholin L., Beura L.K., Masopust D., and Kaplan D.H.. 2019. Keratinocyte-mediated activation of the cytokine TGF-β maintains skin recirculating memory CD8+ T cells. Immunity. 50:1249–1261.e5. 10.1016/j.immuni.2019.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogquist, K.A., Jameson S.C., Heath W.R., Howard J.L., Bevan M.J., and Carbone F.R.. 1994. T cell receptor antagonist peptides induce positive selection. Cell. 76:17–27. 10.1016/0092-8674(94)90169-4 [DOI] [PubMed] [Google Scholar]
- Huang, N., Pérez P., Kato T., Mikami Y., Okuda K., Gilmore R.C., Conde C.D., Gasmi B., Stein S., Beach M., et al. 2021. SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 27:892–903. 10.1038/s41591-021-01296-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium . 2012. Structure, function and diversity of the healthy human microbiome. Nature. 486:207–214. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives, A.M., and Bertke A.S.. 2017. Stress hormones epinephrine and corticosterone selectively modulate herpes simplex virus 1 (HSV-1) and HSV-2 productive infections in adult sympathetic, but not sensory, neurons. J. Virol. 91:e00582-17. 10.1128/JVI.00582-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X., Clark R.A., Liu L., Wagers A.J., Fuhlbrigge R.C., and Kupper T.S.. 2012. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature. 483:227–231. 10.1038/nature10851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa, G.R.M., Libra M., De Pasquale R., Ferlito S., and Pedullà E.. 2021. Association of viral infections with oral cavity lesions: Role of SARS-CoV-2 infection. Front. Med. 7:571214. 10.3389/fmed.2020.571214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledgerwood, L.G., Lal G., Zhang N., Garin A., Esses S.J., Ginhoux F., Merad M., Peche H., Lira S.A., Ding Y., et al. 2008. The sphingosine 1-phosphate receptor 1 causes tissue retention by inhibiting the entry of peripheral tissue T lymphocytes into afferent lymphatics. Nat. Immunol. 9:42–53. 10.1038/ni1534 [DOI] [PubMed] [Google Scholar]
- Lewkowicz, N., Kur B., Kurnatowska A., Tchorzewski H., and Lewkowicz P.. 2011. Expression of Th1/Th2/Th3/Th17-related genes in recurrent aphthous ulcers. Arch. Immunol. Ther. Exp. 59:399–406. 10.1007/s00005-011-0134-1 [DOI] [PubMed] [Google Scholar]
- Li, W., Germain R.N., and Gerner M.Y.. 2017. Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proc. Natl. Acad. Sci. USA. 114:E7321–E7330. 10.1073/pnas.1708981114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.H., Duong H.G., Limary A.E., Kim E.S., Hsu P., Patel S.A., Wong W.H., Indralingam C.S., Liu Y.C., Yao P., et al. 2023. Small intestine and colon tissue-resident memory CD8+ T cells exhibit molecular heterogeneity and differential dependence on Eomes. Immunity. 56:207–223.e8. 10.1016/j.immuni.2022.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi, J.K., Alexandre Y.O., Senthil K., Schienstock D., Sandford S., Devi S., Christo S.N., Mackay L.K., Chinnery H.R., Osborne P.B., et al. 2022. Corneal tissue-resident memory T cells form a unique immune compartment at the ocular surface. Cell Rep. 39:110852. 10.1016/j.celrep.2022.110852 [DOI] [PubMed] [Google Scholar]
- Lucas, P.J., Kim S.J., Melby S.J., and Gress R.E.. 2000. Disruption of T cell homeostasis in mice expressing a T cell-specific dominant negative transforming growth factor beta II receptor. J. Exp. Med. 191:1187–1196. 10.1084/jem.191.7.1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, L.K., Rahimpour A., Ma J.Z., Collins N., Stock A.T., Hafon M.-L., Vega-Ramos J., Lauzurica P., Mueller S.N., Stefanovic T., et al. 2013. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 14:1294–1301. 10.1038/ni.2744 [DOI] [PubMed] [Google Scholar]
- Mackay, L.K., Stock A.T., Ma J.Z., Jones C.M., Kent S.J., Mueller S.N., Heath W.R., Carbone F.R., and Gebhardt T.. 2012. Long-lived epithelial immunity by tissue-resident memory T (TRM) cells in the absence of persisting local antigen presentation. Proc. Natl. Acad. Sci. USA. 109:7037–7042. 10.1073/pnas.1202288109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, B., Elkaddi N., Turkistani J., Spielman A.I., and Ozdener M.H.. 2019. Mammalian taste cells express functional olfactory receptors. Chem. Senses. 44:289–301. 10.1093/chemse/bjz019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, B.T., Byrne K.T., Vella J.L., Zhang P., Shabaneh T.B., Steinberg S.M., Molodtsov A.K., Bowers J.S., Angeles C.V., Paulos C.M., et al. 2017. Resident memory T cells in the skin mediate durable immunity to melanoma. Sci. Immunol. 2:eaam6346. 10.1126/sciimmunol.aam6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust, D., and Soerens A.G.. 2019. Tissue-resident T cells and other resident leukocytes. Annu. Rev. Immunol. 37:521–546. 10.1146/annurev-immunol-042617-053214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner, J.J., Toma C., He Z., Kurd N.S., Nguyen Q.P., McDonald B., Quezada L., Widjaja C.E., Witherden D.A., Crowl J.T., et al. 2020. Heterogenous populations of tissue-resident CD8+ T cells are generated in response to infection and malignancy. Immunity. 52:808–824.e7. 10.1016/j.immuni.2020.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutsopoulos, N.M., and Konkel J.E.. 2018. Tissue-specific immunity at the oral mucosal barrier. Trends Immunol. 39:276–287. 10.1016/j.it.2017.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, S.N., and Mackay L.K.. 2016. Tissue-resident memory T cells: Local specialists in immune defence. Nat. Rev. Immunol. 16:79–89. 10.1038/nri.2015.3 [DOI] [PubMed] [Google Scholar]
- Padgett, D.A., Sheridan J.F., Dorne J., Berntson G.G., Candelora J., and Glaser R.. 1998. Social stress and the reactivation of latent herpes simplex virus type 1. Proc. Natl. Acad. Sci. USA. 95:7231–7235. 10.1073/pnas.95.12.7231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Y., Tian T., Park C.O., Lofftus S.Y., Mei S., Liu X., Luo C., O’Malley J.T., Gehad A., Teague J.E., et al. 2017. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature. 543:252–256. 10.1038/nature21379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pircher, H., Bürki K., Lang R., Hengartner H., and Zinkernagel R.M.. 1989. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 342:559–561. 10.1038/342559a0 [DOI] [PubMed] [Google Scholar]
- Reddy, N.R., Roopa D., Babu D.S.M., Kumar P.M., Raju C.M., and Kumar N.S.. 2012. Estimation of matrix metalloproteinase-3 levels in gingival crevicular fluid in periodontal disease, health and after scaling and root planing. J. Indian Soc. Periodontol. 16:549–552. 10.4103/0972-124X.106907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riding, R.L., and Harris J.E.. 2019. The role of memory CD8+ T cells in vitiligo. J. Immunol. 203:11–19. 10.4049/jimmunol.1900027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera, C. 2015. Essentials of oral cancer. Int. J. Clin. Exp. Pathol. 8:11884–11894. [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosato, P.C., Wijeyesinghe S., Stolley J.M., Nelson C.E., Davis R.L., Manlove L.S., Pennell C.A., Blazar B.R., Chen C.C., Geller M.A., et al. 2019. Virus-specific memory T cells populate tumors and can be repurposed for tumor immunotherapy. Nat. Commun. 10:567. 10.1038/s41467-019-08534-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santosh, A.B.R., and Muddana K.. 2020. Viral infections of oral cavity. J. Fam. Med. Prim. Care. 9:36–42. 10.4103/jfmpc.jfmpc_807_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel, J.M., Fraser K.A., Beura L.K., Pauken K.E., Vezys V., and Masopust D.. 2014. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 346:98–101. 10.1126/science.1254536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel, J.M., Fraser K.A., Vezys V., and Masopust D.. 2013. Sensing and alarm function of resident memory CD8⁺ T cells. Nat. Immunol. 14:509–513. 10.1038/ni.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schön, M.P., Arya A., Murphy E.A., Adams C.M., Strauch U.G., Agace W.W., Marsal J., Donohue J.P., Her H., Beier D.R., et al. 1999. Mucosal T lymphocyte numbers are selectively reduced in integrin alpha E (CD103)-deficient mice. J. Immunol. 162:6641–6649. 10.4049/jimmunol.162.11.6641 [DOI] [PubMed] [Google Scholar]
- Shannon, J.P., Vrba S.M., Reynoso G.V., Wynne-Jones E., Kamenyeva O., Malo C.S., Cherry C.R., McManus D.T., and Hickman H.D.. 2021. Group 1 innate lymphoid-cell-derived interferon-γ maintains anti-viral vigilance in the mucosal epithelium. Immunity. 54:276–290.e5. 10.1016/j.immuni.2020.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, N., and Cohen P.L.. 2012. The T cell in Sjogren’s syndrome: Force majeure, not spectateur. J. Autoimmun. 39:229–233. 10.1016/j.jaut.2012.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sordi, M.B., Magini R.S., Panahipour L., and Gruber R.. 2021. Pyroptosis-mediated periodontal disease. Int. J. Mol. Sci. 23:372. 10.3390/ijms23010372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, R., Wesselink T.H., Behr F.M., Kragten N.A.M., Arens R., Koch-Nolte F., van Gisbergen K.P.J.M., and van Lier R.A.W.. 2018. T RM maintenance is regulated by tissue damage via P2RX7. Sci. Immunol. 3:eaau1022. 10.1126/sciimmunol.aau1022 [DOI] [PubMed] [Google Scholar]
- Steinert, E.M., Schenkel J.M., Fraser K.A., Beura L.K., Manlove L.S., Igyártó B.Z., Southern P.J., and Masopust D.. 2015. Quantifying memory CD8 T cells reveals regionalization of immunosurveillance. Cell. 161:737–749. 10.1016/j.cell.2015.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo, P.A., Miron M., and Farber D.L.. 2019. Location, location, location: Tissue resident memory T cells in mice and humans. Sci. Immunol. 4:eaas9673. 10.1126/sciimmunol.aas9673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Y., Guan S.P., Chua B.Y.L., Zhou Q., Ho A.W.S., Wong K.H.S., Wong K.L., Wong W.S.F., and Kemeny D.M.. 2012. Antigen-specific effector CD8 T cells regulate allergic responses via IFN-γ and dendritic cell function. J. Allergy Clin. Immunol. 129:1611–1620.e4. 10.1016/j.jaci.2011.12.976 [DOI] [PubMed] [Google Scholar]
- Turner, D.L., Bickham K.L., Thome J.J., Kim C.Y., D’Ovidio F., Wherry E.J., and Farber D.L.. 2014. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 7:501–510. 10.1038/mi.2013.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, R., Gehad A., Yang C., Scott L.L., Teague J.E., Schlapbach C., Elco C.P., Huang V., Matos T.R., Kupper T.S., and Clark R.A.. 2015. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci. Transl. Med. 7:279ra39. 10.1126/scitranslmed.3010302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeyesinghe, S., Beura L.K., Pierson M.J., Stolley J.M., Adam O.A., Ruscher R., Steinert E.M., Rosato P.C., Vezys V., and Masopust D.. 2021. Expansible residence decentralizes immune homeostasis. Nature. 592:457–462. 10.1038/s41586-021-03351-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward Davis, A.S., Roozen H.N., Dufort M.J., DeBerg H.A., Delaney M.A., Mair F., Erickson J.R., Slichter C.K., Berkson J.D., Klock A.M., et al. 2019. The human tissue-resident CCR5+ T cell compartment maintains protective and functional properties during inflammation. Sci. Transl. Med. 11:eaaw8718. 10.1126/scitranslmed.aaw8718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, F., Ji Z., Liu Y., Zhang Y., Hu L., Yang Q., Qiu Q., Zhao L., Chen D., Tian Z., et al. 2017. Mutation in SSUH2 causes autosomal-dominant dentin dysplasia type I. Hum. Mutat. 38:95–104. 10.1002/humu.23130 [DOI] [PubMed] [Google Scholar]
- Yu, G., Wang L.-G., Han Y., and He Q.-Y.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zens, K.D., Chen J.K., and Farber D.L.. 2016. Vaccine-generated lung tissue-resident memory T cells provide heterosubtypic protection to influenza infection. JCI Insight. 1:85832. 10.1172/jci.insight.85832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, N., and Bevan M.J.. 2013. Transforming growth factor-β signaling controls the formation and maintenance of gut-resident memory T cells by regulating migration and retention. Immunity. 39:687–696. 10.1016/j.immuni.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H., Yang J., Tian J., and Wang S.. 2021. CD8+ T lymphocytes: Crucial players in sjögren’s syndrome. Front. Immunol. 11:602823. 10.3389/fimmu.2020.602823 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows DEGs comparing No Tx to gp33 groups (adjuvant effect of P/N-9 swabbing).
Data Availability Statement
The data underlying Figs. 6, 7, and 8 and Fig. S5 are openly available in the Gene Expression Omnibus under the accession number GSE229634.