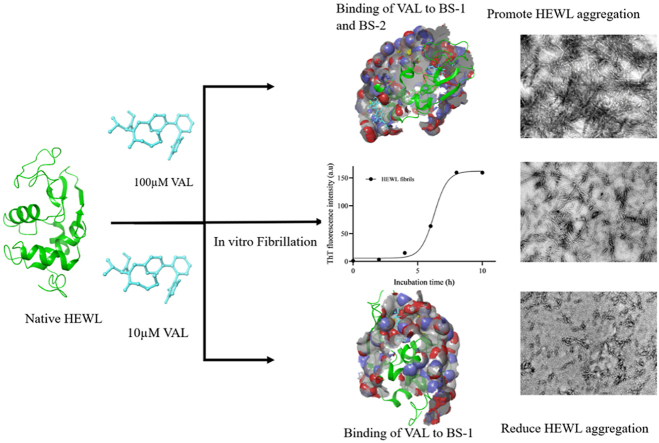
Abstract
Protein misfolding can result in amyloid fiber aggregation, which is associated with various types of diseases. Therefore, preventing or treating abnormally folded proteins may provide therapeutic intervention for these diseases. Valsartan (VAL) is an angiotensin II receptor blocker (ARB) that is used to treat hypertension. In this study, we examine the anti-aggregating effect of VAL against hen egg-white lysozyme (HEWL) amyloid fibrils through spectroscopy, docking, and microscopic analysis. In vitro formation of HEWL amyloid fibrils was indicated by increased turbidity, RLS (Rayleigh light scattering), and ThT fluorescence intensity. 10 μM VAL, amyloid/aggregation was inhibited up to 83% and 72% as measured by ThT and RLS respectively. In contrast, 100 μM VAL significantly increases the fibril aggregation of HEWL. CD spectroscopy results show a stabilization of HEWL α-helical structures in the presence of 10 μM VAL while the increase in β-sheet was detected at 100 μM concentration of VAL. The hydrophobicity of HEWL was increased at 100 μM VAL, suggesting the promotion of aggregation via its self-association. Steady-state quenching revealed that VAL and HEWL interact spontaneously via hydrogen bonds and van der Waals forces. Transmission electron microscopy (TEM) images illustrate that the needle-like fibers of HEWL amyloid were reduced at 10 μM VAL, while at 100 μM the fibrils of amyloid were increased. Additionally, our computational studies showed that VAL could bind to two binding sites within HEWL. In the BS-1 domain of HEWL, VAL binds to ASN59, ILE98, ILE58, TRP108, VAL109, SER50, ASP52, ASN59, ALA107, and TRP108 residues with a binding energy of −9.72 kcal mol−1. Also, it binds to GLU7, ALA10, ALA11, CYS6, ARG128, and ARG14 in the BS-2 domain with a binding energy of −5.89 kcal mol−1. VAL, therefore, appears to have dual effect against HEWL aggregation. We suggest that VAL stabilizes HEWL's aggregation-prone region (APR) at 10 μM, preventing aggregation. Also, we assume that at 100 μM, VAL occupies BS-2 beside BS-1 and destabilizes the folding structure of HEWL, resulting in aggregation. Further studies are needed to investigate the mechanism of action and determine its potential side effects.
Keywords: Valsartan, Lysozyme amyloidosis, Fluorescence, Circular dichroism, Molecular modeling
Graphical abstract
1. Introduction
The formation of amyloid fibrils occurs both in vitro and in vivo under abnormal conditions [[1], [2], [3]]. Amyloid fibril formation has been linked to neurodegenerative diseases and systematic diseases, such as Alzheimer's disease, Parkinson's disease, and type II diabetes [[4], [5], [6], [7], [8]]. Various types of amyloid-associated diseases are caused by the accumulation of a specific protein [9]. In its native state, each amyloid-forming protein differs from the others in terms of sequence and folding structure. All proteins, however, share the same structural morphology after fibrillation, so amyloid fibrillation can be considered a general property of all proteins [6,8]. Amyloid fibrils are unbranched proteins that are predominantly made up of β-sheet structures with solvent-exposed grooves [3,10]. Each strand is connected by loops and stabilized by intermolecular hydrogen bonds [11]. Amyloid fibrils are formed when the natively folded or intrinsically disordered protein undergoes significant restructuring [12].
It is known that human lysozyme (HL) displays different structural changes that lead to the formation of amyloid fibrils under certain conditions [13]. Of the 130 amino acid residues of HL, TRP64 and TRP109 are confirmed as the most dominant fluorescent markers in the binding site [14]. Lysozyme amyloidosis is a hereditary non-neuropathic disease, it may associate with renal dysfunction, sicca syndrome, hepatic rupture, gastrointestinal tract hemorrhage, lymphadenopathy, purpura, and petechiae [15]. Lysozyme amyloidosis occurs because of a single-point mutation in the lysosomal gene. It causes the extracellular plaque aggregation of human lysozyme amyloid to occur in large quantities in different organs such as the gastrointestinal tract, kidneys, lymph nodes, liver, spleen, and blood vessels [16]. The symptoms and severity of these diseases have been associated with the physical appearance of mature fibrils. However, the oligomers may have a considerable level of toxicity [17]. The analysis of lysosomal fibril data revealed that the core of this fibril consists of particular amino acid residues [15]. Therefore, targeting these amino acids may block fibril formation [18]. Until now there is no effective treatment for lysosomal amyloidosis. Therefore, great scientific efforts toward the development of treatments that prevent lysosomal amyloid fibrils formation are needed [19].
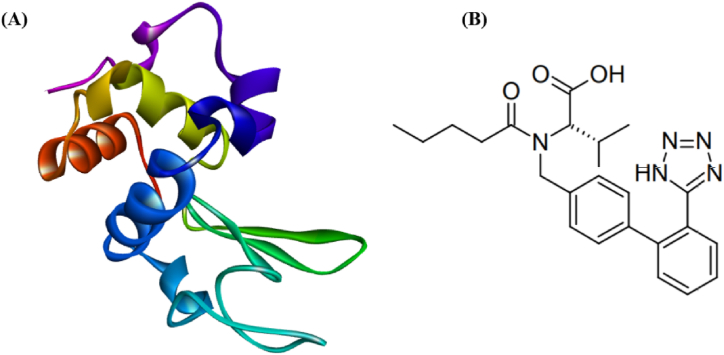
A HEWL extensively studied for its ability to form fibrils and amorphous aggregates [19]. This globular protein, made up of 129 amino acids and four disulfide bridges, is folded into a single polypeptide chain (Fig. 1A). In vitro, HEWL produces amyloid fibrils that are identical morphologically to those produced by HL. As HEWL is readily available in aqueous media and is easily soluble, it is an ideal model for lysosomal aggregation and amyloidosis studies [15].
Fig. 1.
(A) HEWL folded structure. (B) Chemical structure of VAL [20].
Angiotensin II receptor blockers (ARB) were first investigated as antihypertension medications in the 1990s. ARB such as VAL prevents angiotensin II from binding to the angiotensin II type 1 (AT1) receptor, which causes arteries to vasodilate. Thus, lowering blood pressure [21]. A previous study by Zhao et al. shows that VAL can prevent the oligomerization of the Aβ peptide in vitro [22]. In another study, VAL was found to improve cognitive function and reduced Aβ aggregation in mice [23]. According to Barthold et al. ARBs can prevent the onset of Alzheimer's disease [24]. Accordingly, we hypothesized that VAL has anti-amyloidogenic effects on HEWL aggregation (Fig. 1B).
2. Materials and methods
2.1. Materials
HEWL (Lot #: L6876, purity ≥90%), 8-Anilino-1-naphthalene sulfonic acid (ANS), and Thioflavin-t (ThT) was provided from Sigma-Aldrich (St. Louis, MO 68178, USA). VAL was purchased from MedChemExpress (MCE) (Monmouth junction, NJ 08852, USA). Other chemicals were obtained from Sigma-Aldrich (St. Louis, MO 68178, USA). All experimental solutions were prepared using double-distilled water purified by the Milli-Q system and were of analytical grade. SENTRON INTEGRATED SENSOR TECHNOLOGY 2001 pH meter (Texas City, TX, USA) was used to assess the pH. All fluorescence measurements were conducted using Agilent Technologies Cary Eclipse-Fluorescence Spectrophotometer (Santa Clara, USA).
2.2. In vitro fibrillation of HEWL
100 μM of HEWL in the absence and presence of 10 μM and 100 μM VAL were prepared immediately before the experiment by dissolving HEWL in glycine buffer, pH 2.0 (100 mM glycine/HCl, 100 mM NaCl) and filtered by 0.45 μm filter syringe. The concentration of HEWL was determined using a molar extinction coefficient of 37970 M−1cm−1 at 280 nm [25]. VAL was prepared in absolute DMSO and filtered by 0.45 μm filter syringe. The VAL stock solution was subjected to further dilution with Milli-Q to meet the required concentrations. To induce in vitro formation of amyloid fibrils, HEWL samples were incubated for 10 h at 55 °C under constant agitation (600 rpm). At different time points, HEWL solutions were aliquoted after being vortexed to well-distribute the amyloid fibrils [26]. The concentration of HEWL was fixed at 10 μM for all experimental assays.
2.3. Turbidity measurements
Measurements of turbidity between 350 and 450 nm can be used to study amyloid aggregate formation. Since protofibrils are too small to reflect light at 400 nm, turbidimetric measurements can only be used to detect mature fibrils [27,28]. It is easy to conduct turbidimetric tests with standard spectrophotometers, but they require a relatively high level of protein concentration [29,30]. Furthermore, this method cannot be used to detect oligomers or protofibrils, so it is not suitable for quantitative studies. Agilent Technologies Cary 60 UV–Vis Spectrophotometer (Santa Clara, USA) was used to measure the turbidity of HEWL with and without 10 μM and 100 μM VAL in a 1 cm path length cuvette at 350 nm with a path length of one cm. Additionally, HEWL fibrils were monitored for 10 h without VAL.
2.4. Rayleigh light scattering measurements
RLS of fibrillated HEWL with and without VAL was conducted at room temperature using Agilent Technologies Cary Eclipse - Fluorescence Spectrophotometer (Santa Clara, USA). The emission spectra were recorded between 300 nm and 600 nm after exciting the protein at 350 nm. The slit width of both excitation and emission was 2.5 nm [14].
2.5. ThT fluorescence assay
ThT stock solution was prepared from distilled water and filtered via 0.45 μm syringe. The concentration of ThT was determined using a molar extinction coefficient of 36000 M−1cm−1 at 412 nm [31]. For ThT fluorescence measurements, HEWL samples were incubated at pH 2.0 and 55 °C for 10 h with and without 10 m and 100 m VAL. HEWL fibrillation kinetics were analyzed by collecting aliquots of a mixture every 2 h for up to 10 h without VAL. Following this, HEWL samples were diluted in glycine/HCl buffer to a concentration of 10 μM. ThT was added to the 10 μM HEWL samples with and without VAL in a 1:1 M ratio and incubated in the dark at 25 °C for 30 min. The excitation wavelength was 440 nm, and the emission wavelength was from 450 to 600 nm, with a slit width of 5 nm.
2.6. ANS binding assay (hydrophobicity analysis)
ANS is a dye used to detffect hydrophobic patches on proteins' surfaces. ANS stock solution was prepared in ethanol and the concentration was determined using a molar extinction coefficient of 5000 M−1cm−1 at 350 nm [32]. 10 μM HEWL samples with and without 10 μM and 100 μM VAL were incubated in the dark at 25 °C for 30 min with a 50-fold molar excess of ANS prior to the measurements. The emission spectra were recorded from 400 nm to 600 nm after excitation at 380 nm with a slit width of 5 nm [33].
2.7. Far-UV circular dichroism (far-UV CD) measurement
Applied Photophysics Chirascan Plus spectrometer (United Kingdom) was used to investigate the change in the secondary structure of HEWL in the presence and absence of 10 μM and 100 μM of VAL. The CD spectra of HEWL were scanned in a cuvette of a path length equal to 0.1 mm between 196 nm and 250 nm at 25 °C. The percentage of secondary structure change was calculated by BeStSel [34].
2.8. Intrinsic fluorescence measurements
Intrinsic Fluorescence intensities of HEWL samples with and without 10 μM and 100 μM of VAL were measured using a spectrofluorometer. A fixed excitation wavelength of 295 nm was used, and the emission spectra ranged from 300 to 400 nm. Both excitation and emission were conducted with a 5 nm slit width.
2.9. Steady state fluorescence quenching measurements
Steady-state fluorescence quenching measurement was conducted at three different temperatures 298 K, 303 K, and 310 K to investigate the binding relationship between HEWL and VAL. The concentration of HEWL was fixed at 5 μM and the increase of the 1 mM VAL volume was chosen based on the quenching degree. The fluorescence intensity measurement was performed by exciting the protein at 295 nm and measuring its emission spectra at 300 nm–400 nm. For this binding experiment, 20 mM sodium phosphate buffer was used to prepare HEWL at pH 7.4, while VAL were prepared in 100% DMSO and then diluted with Milli-Q to the desired concentration. The data was analyzed according to the Stern-Volmer Equation (eq (1)) [35]:
| [1] |
where and represent fluorescence intensities of HEWL with and without VAL, represent VAL concentration, is the bimolecular rate constant of the quenching reaction, is the Stern-Volmer quenching constant, and represent the fluorescence lifetime of protein which is ∼10−8 s. Binding sites and binding constants will be obtained from the equation (eq (2)) [35]:
| [2] |
where represent the number of binding sites and is the binding constant. Change in entropy (ΔS°), enthalpy (ΔH°),and Gibbs free energy (ΔG°), were calculated using Van't Hoff equation (eq (3)), and Gibbs free energy equation, respectively (eq. (4)) [35]:
| [3] |
| [4] |
where T is the absolute temperature (K) and represents the gas constant which is equal to 1.987 cal mol−1 K−1.
2.10. Transmission electron microscopy (TEM) analysis
JEOL JEM1400 Transmission electron microscope operating at an accelerating voltage of 120 kV was used to obtain the micrographs of HEWL with/without VAL. Samples were dropped on a 300-mesh copper grid and were covered by carbon-stabilized formvar film and after 2 min the excess fluid was removed. Then, grids were negatively stained using a uranyl acetate solution.
2.11. Molecular modeling studies
2.11.1. Molecular docking
Further exploration of valsartan-HEWL interaction, molecular docking, and dynamic simulation has been conducted using AutoDock 4.2.6 software and NAMD [32,36]. First, all the binding sites of HEWL were predicted using the SiteMap program, followed by molecular docking [37]. The crystal structure of HEWL (6LYZ) was retrieved from Protein Data Bank (PDB) [32], whereas the three-dimensional (3D) structure of VAL (CID: 60846) was retrieved from PubChem. Water molecules were removed, and all hydrogen atoms were added. Following that, partial Kollman charges were added to the protein, and it was set to be rigid. The grid size was set for each site with 0.375 Å grid spacing. The size of the GA population was 150. Then all predicted sites were subjected to molecular dynamics simulation for 5 ns. Binding residues were visualized and identified using discovery studio 3.5.
2.11.2. Molecular dynamic simulation
Molecular Dynamic (MD) simulation of the free HEWL and its docked complex with VAL at BS-1 and BS-2 was performed with the Charmm-GUI and NAMD packages [38]. A cubic box was constructed using the NAMD force field. VAL was restrained at 300 K and equilibrated at NPT (constant number, pressure, and temperature), followed by NVT (constant number, volume, and temperature), at 1 bar and 300 K, respectively. Simulations of production were run at 300 K for 5 ns after equilibration. A computational analysis of the conformational changes of free HEWL and its docked complex with VAL at BS-1 and BS-2 was performed using Root-mean-square fluctuation (RMSF), Root-mean-square deviation (RMSD), and Radius of gyration (Rg).
3. Results and discussion
3.1. Turbidity measurements
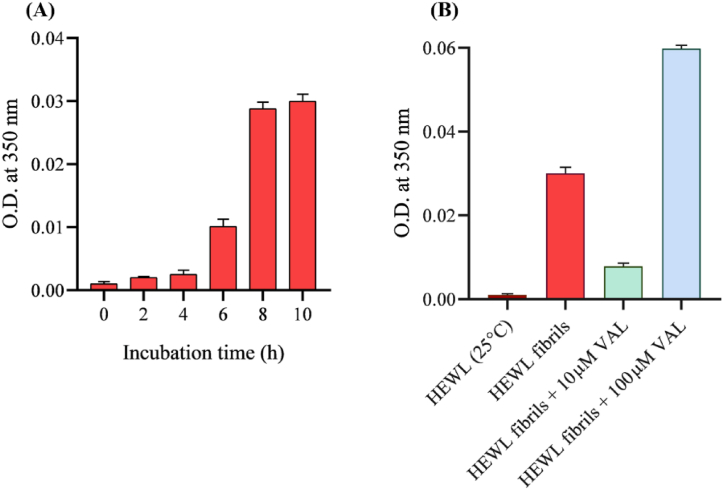
HEWL amyloid fibril formation was monitored via a turbidity assay at 350 nm [32]. As shown in Fig. 2A, increasing the absorbance with increasing time of incubation reflects the formation of aggregates and was more visible (precipitates) and significant after 10 h. The turbidity of HEWL aggregation was also measured with and without 10 μM and 100 μM VAL to test its behavior toward amyloid fibrils. 10 μM of VAL sharply reduces HEWL aggregation. Whereas unexpectedly higher concentration at 100 μM significantly induced fibril aggregation (Fig. 2B). Suggesting that VAL successively hinder the early formation of amyloid fibrils at 10 μM.
Fig. 2.
Turbidity measurements by recording O.D. at 350 nm. (A) HEWL incubated for 10 h at 55 °C with 600 rpm agitation. (B) 10 μM HEWL fibrils incubated for 10 h at 55 °C with 600 rpm agitation with and without 10 μM and 100 μM VAL.
3.2. Rayleigh scattering measurements
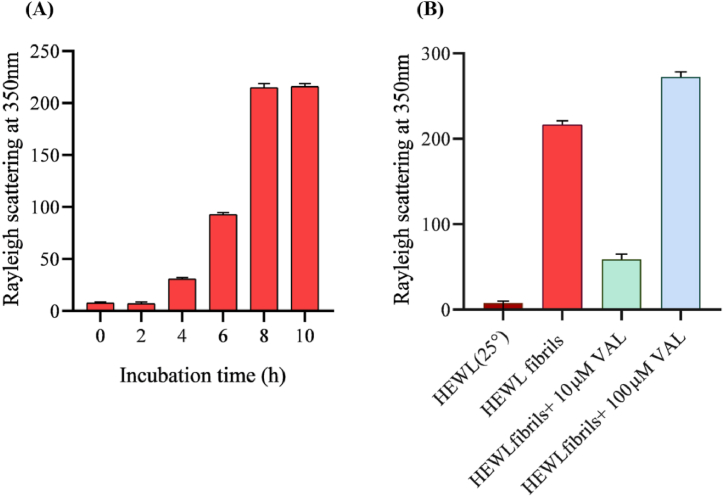
In this study, RLS was used to measure protein aggregation at 350 nm to investigate the anti-aggregation effect of VAL against HEWL fibrillation [39]. HEWL was incubated under the above-mentioned conditions to produce amyloid fibrils. Native HEWL at 25 °C shows little scattering, which indicates that amyloid fibrils are not present. When HEWL was co-incubated with 10 μM and 100 μM of VAL, different results were observed for aggregation. Light scattering increases with aggregation [40]. The results of the light scattering of HEWL fibrils with/without 10 μM and 100 μM VAL is illustrated in Fig. 3. VAL reduces the formation of amyloid fibrils by 72% at 10 μM. Comparatively, 100 μM VAL promotes fibril formation, resulting in a high spectra record. However, it appears that at a concentration of 10 μM, VAL inhibits the aggregation of HEWL.
Fig. 3.
Rayleigh scattering measurements of 10 μM HEWL. (A) represents HEWL fibril formation incubated at pH2, 55 °C for 10 h. (B) Shows the effect of 10 μM and 100 μM VAL on HEWL aggregation after 10 hrs incubation at pH2, 55°C.
3.3. Effect of VAL on HEWL fibrils: ThT fluorescence assay
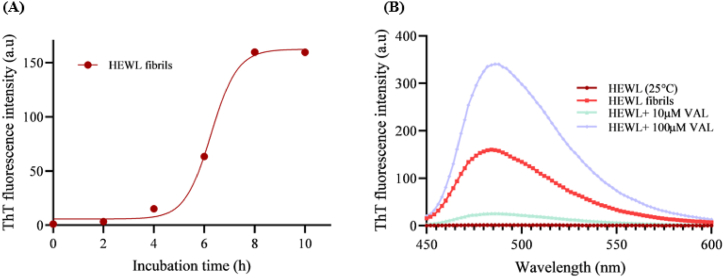
The binding of ThT dye into the β-sheet structure is a characterization of amyloid fibrillation [41]. To explore the effect of VAL toward HEWL amyloid fibrils, changes in the fluorescence intensities have been detected for fibrillated HEWL that is co-incubated with and without 10 μM and 100 μM VAL at 55 °C for 10 h. The ThT fluorescence intensity increases as the amyloid fibrils increase until it reaches the level of saturation [42]. Fig. 4A illustrates HEWL aggregation kinetics. As amyloid fibrils are formed in HEWL, ThT fluorescence intensity increases with time. In Fig. 4A, HEWL amyloid aggregation is shown to follow a sigmoidal curve with a lag phase, an growth phase, and a saturation phase. Native HEWL does not display significant increases in fluorescence intensity when compared to HEWL after 10 h incubation, due to its lack to β-sheets (Fig. 4B). A maximum peak of 485 nm is observed in HEWL fibrils [43]. The fluorescence intensities of HEWL fibrils were measured at 10 μM and 100 μM of VAL to investigate whether it influences HEWL fibrils growth. The dual effect of VAL can be seen in Fig. 4B. Co-incubation of HEWL with 10 μM VAL led to an 83% reduction of ThT intensity, thus in the number of amyloid aggregates. Whereas these aggregates were increased in the presence of 100 μM VAL. Proving the ani-aggregation activity of VAL at 10 μM.
Fig. 4.
The effect of VAL on HEWL fibrillation was measured by ThT fluorescence assay. (A) ThT fluorescence intensities of HEWL were monitored at different time points during the incubation period. (B) ThT fluorescence bar of HEWL in the presence and absence of 10 μM and 100 μM of VAL after incubation at 55 °C for 10 h.
3.4. Effect of VAL on the surface hydrophobicity of HEWL
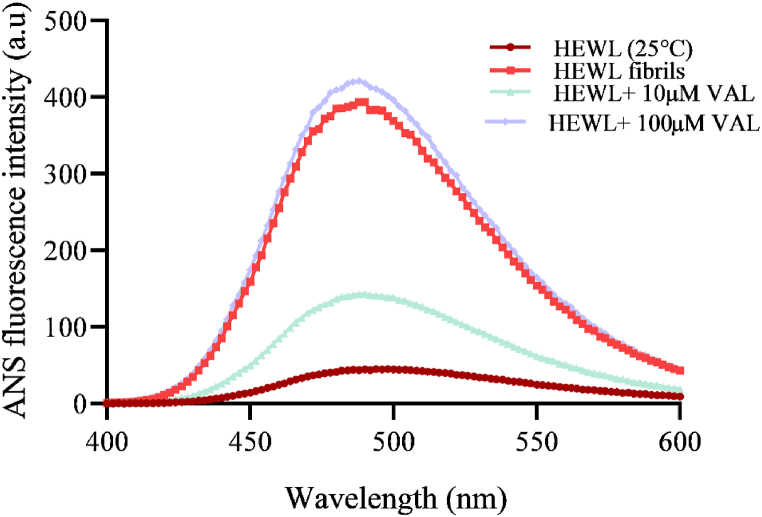
ANS fluorescence technique is conducted to elucidate protein folding and identify the presence of hydrophobic amino acids on the protein surface [44]. Upon binding to the hydrophobic group, ANS fluorescence intensity is increased. ANS fluorescence measurements were used to investigate the effect of VAL on surface hydrophobicity. In native HEWL the hydrophobic amino acids are present inside the folded conformation; thus, the ANS fluorescence intensity was insignificant. Whereas there was an increase in the fluorescence intensity in the case of fibrillated HEWL, due to the surface exposure of hydrophobic residues [45]. A concentration-dependent decrease and increase of ANS fluorescence have been observed on fibrillated HEWL that is co-incubated with different concentrations of VAL. In the presence of 10 μM VAL, ANS intensity is decreased, indicating the reduction of hydrophobic residues on the surface of HEWL. On the other hand, 100 μM of the same drug enhanced the ANS intensity which suggests the presence of high levels of amyloid fibrils (Fig. 5) [14]. Altogether these results suggest that in the presence of 10 μM VAL, HEWL forms fewer amyloid fibrils.
Fig. 5.
ANS fluorescence intensity of fibrillated 10 μM HEWL with and without 10 μM and 100 μM VAL.
3.5. Circular dichroism studies
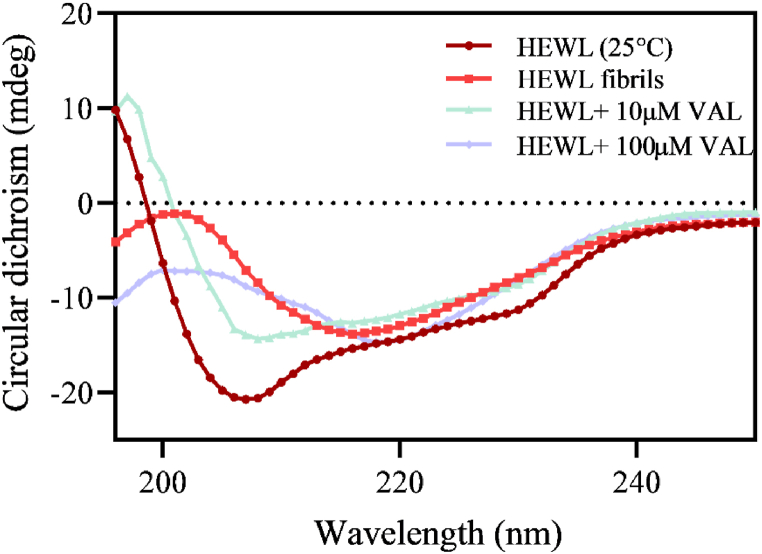
The CD spectroscopic measurement is extensively used in protein structural studies to investigate the changes in protein secondary structure [46,47]. The changes in the secondary structures after HEWL-VAL binding was measured using far-UV CD measurements. The typical α-helix structure of the native HEWL occurred at two negative bands: 208 nm and 222 nm. Moreover, HEWL amyloid fibrils shows a structural transition towered β-sheet secondary structure which is presented as a negative ellipticity at 218 nm CD spectrum and a positive band at 196 nm [48]. As shown in Fig. 6, compared to the native HEWL, fibrillated HEWL shows a negative peak at 218 nm suggesting the formation of amyloid aggregates. On the other hand, a significant decrease of β-sheet percent and increase in the content of α-helix occurred under the effect of 10 μM of VAL implying a significant correlation between the low concentration of VAL and the attenuation of β-sheet content. A remarkable increase of β-sheet content with red-shift was associated with 100 μM of VAL.
Fig. 6.
Far-UV CD spectra of 10 μM HEWL at 25 °C and HEWL incubated for 10 h at 55 °C in the presence and absence of 10 μM and 100 μM VAL.
The percentage of changes in the secondary structure with and without 10 μM and 100 μM VAL is summarized in Table 1. A slight red shift can be seen in the CD spectrum as a result of the broadening and decreasing intensity of the negative and positive bands. These events can be assigned to differential scattering phenomena and absorption flattening. Differential scattering phenomena refer to the relationship between the radiation and aggregation size, in which the scattering of incident radiation is proportional to β-sheet aggregation size. Therefore, the more extended the β-sheets, the more the loss in CD spectral intensity [49]. Moreover, absorption flattening occurred when the chromophore distributed non-randomly in the sample, i.e., different portions of the sample gave different absorptions [50]. According to CD spectroscopy measurement results, we found that VAL is effective in preventing the α-to-β transition of HEWL, only at 10 μM whereas the enhancement of this transition process occurs under the influence of high VAL concentration.
Table 1.
Secondary structural contents of HEWL incubated with and without 10 μM and 100 μM VAL as calculated from BeStSel.
| Type of sample | % of α-helix | % of β-sheet | % of Turn | %of Unordered |
|---|---|---|---|---|
| Native HEWL | 33 ± 4.87 | 12 ± 3.47 | 12.5 ± 1.80 | 42.4 ± 6.11 |
| HEWL Fibrils | 6.5 ± 4.59 | 26.3 ± 7.89 | 15 ± 2.16 | 52.2 ± 7.53 |
| HEWL Fibrils +10 μM VAL | 33.1 ± 4.59 | 13.4 ± 4.36 | 11.5 ± 1.65 | 42 ± 6.06 |
| HEWL Fibrils +100 μM VAL | 2.6 ± 1.83 | 26.5 ± 9.17 | 14.5 ± 2.09 | 56.4 ± 8.14 |
3.6. Intrinsic (tryptophan) fluorescence: conformational analysis
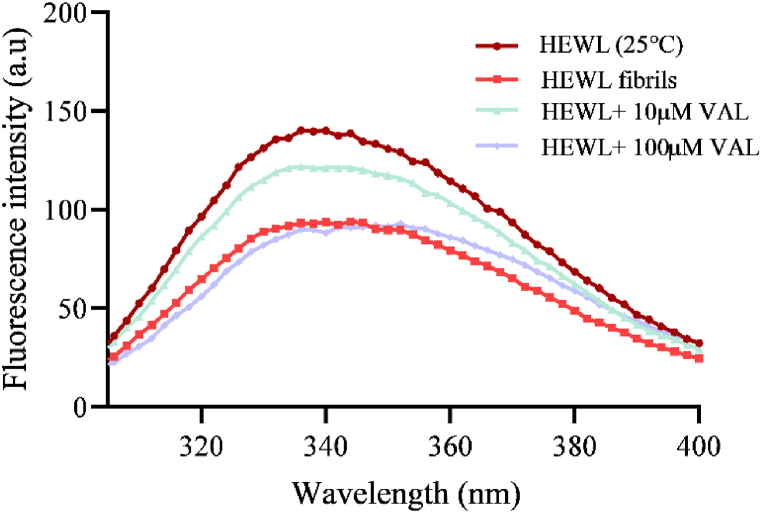
Protein intrinsic fluorescence can be used to evaluate the change in its functional, structural, and physicochemical properties [51]. Therefore, it can be used to monitor the formation of HEWL amyloid fibrils. According to Fig. 7, the fluorescence intensity of fibrillated HEWL compared to the native form was low. However, the fluorescence intensity has increased in the presence of 10 μM of VAL and decreased with a slight shift from 340 nm to 343 nm in the presence of 100 μM of VAL. The shifting occurred because of the exposure of TRP from the interior of the protein into solvent [52]. Also, the red-shift suggests the unfolding of HEWL which leads to more aggregation. The reduction of fluorescence intensity in presence of 10 μM of VAL indicates less exposure to TRP, and thus maintained the integrity of native HEWL in solution. Thus, the low mass of amyloid fibril formation. Furthermore, the quenching of intrinsic fluorescence indicates the interaction between HEWL and VAL.
Fig. 7.
Intrinsic fluorescence spectra of 10 μM HEWL incubated for 10 h at 55 °C in the presence and absence of 10 μM and 100 μM VAL.
3.7. Binding studies of Valsartan-HEWL interactions
3.7.1. Quenching measurement at different temperatures
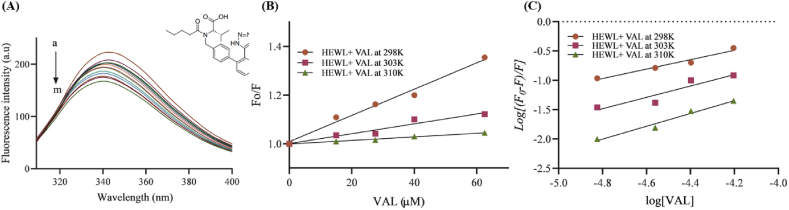
Fluorescence quenching spectroscopy is used to evaluate the interaction between small molecules and macromolecules such as proteins [53]. Aromatic amino acids tyrosine (TYR), phenylalanine (PHE), and tryptophan (TRP) are used to investigate conformational changes upon drug binding. In our study, this method has been utilized to analyze the interaction between HEWL and VAL at three different temperatures: 298 K, 303 K, and 310 K, to calculate the thermodynamic parameters. According to Fig. 8A, HEWL was excited to 295 nm, while emission spectra ranged from 300 to 400 nm. A strong emission peak occurred at 345 nm and shifted toward 340 nm after adding VAL. TRP fluorescence is sensitive to the change in environment polarity. Blue shift refers to the shift of the emission spectrum into lower wavelengths. The binding of VAL to HEWL increased the hydrophobicity in TRP surrounding environment, leading to 5 nm blue shift [54]. The concentration of VAL was increased gradually until 62.5 μM while the concentration of HEWL was fixed at 5 μM. A gradual quenching in the fluorescence intensity was observed, implying the interaction between VAL and HEWL.
Fig. 8.
(A) Quenching spectra of HEWL intrinsic fluorescence by elevating the concentration of VAL at 25 °C. The arrow indicates an increase in VAL concentration. VAL (a–m) = 0−65.2 μM and CHEWL = 5 μM. (B) Stern−Volmer plots of HEWL and VAL interaction at 298 K,303 K, and 310 K. (C) Plot of modified Stern−Volmer equation (eq (2)) regarding the fluorescence quenching of HEWL by VAL at 298 K,303 K, and 310 k.
3.7.2. Binding affinity and mechanism
The , and values of the interaction between HEWL and VAL were calculated by the Stern-Volmer Equation (eq (1)) (Fig. 8B). The value of was used to determine the quenching phenomenon [55]. As shown in Table 2, the and values were reduced with increasing temperature suggesting that the static quenching mechanism is the reason behind the decrease in the HEWL-VAL fluorescence emission. In static quenching VAL binds to HEWL and forms a ground-state complex [56]. reflects HEWL-VAL complex stability. The higher the value the stronger the complex stability. The value of VAL binding decrease with temperature elevation which implies that the HEWL-VAL complex has poor stability at high temperatures (Fig. 8C) [35].
Table 2.
Thermodynamic and binding parameters of HEWL-VAL at different temperatures obtained from fluorescence quenching experiments. aR2 for all values ranges from 0.98.
| T(K) | Ksv (L mol−1) | Kq(L mol−1 s−1) | Kb(M−1) | ΔH° (Kcal mol−1) | ΔS° (Kcal mol−1 K−1) | ΔG° (Kcal mol−1) |
|---|---|---|---|---|---|---|
| 298.15 | 5.40 × 109±0.13 | 5.40 × 1017±0.13 | 2.96 × 108±0.21 | −12.35 | −0.04 | −11.37 |
| 303.15 | 2.03 × 109±0.05 | 2.03 × 1017±0.05 | 1.82 × 108±0.27 | −11.35 | ||
| 310.15 | 0.67 × 109±0.01 | 0.67 × 1017±0.01 | 1.30 × 108±0.29 | −10.89 |
3.7.3. Thermodynamic parameters
Thermodynamic parameters such as ΔG°, ΔS°, and ΔH° have an important role in controlling protein-drug interaction and their calculation can help in evaluating the forces that dominate the HEWL-VAL complex formation [55]. Negative or low positive values of ΔH° with positive ΔS° are indicative of electrostatic interaction. Whereas negative ΔH° and ΔS° values are associated with van der Waals force, and hydrogen bonding [57]. Furthermore, positive ΔH° and ΔS° values are indicative of hydrophobic interaction [58]. Drug-protein interaction causes the displacement of surrounding solvent leading to a positive change of entropy for the system during the initial attachment of the protein to the drug. Then forces such as van der Waals, hydrophobic interaction, electrostatic interactions, and hydrogen bonding control the thermodynamic parameters. In the present study, ΔG° negative values at all temperatures imply that the HEWL-VAL interaction occurs spontaneously in nature. Moreover, the negative ΔH° and ΔS° values indicated that the HEWL- VAL interaction is derived by enthalpy rather than entropy and mainly depends on van der Waals interaction and hydrogen bonding (Table 2) [59].
3.8. Microscopy imaging of HEWL aggregates
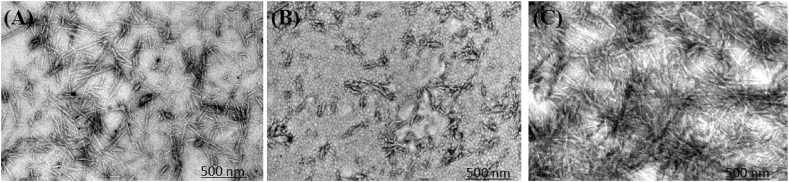
Taken all together, the above-mentioned results prove the dual effect of VAL on HEWL amyloid fibrillation. However, the TEM technique was used to visualize and verify these results. According to Fig. 9A, 10 hrs aged HEWL fibril clearly shows a large amount of amyloid fibril aggregate. However, compared to the fibrillated control 10 μM and 100 μM of VAL significantly inhibit and promote fibril formation, respectively (Fig. 9 B, and 9 C). Thus, our results significantly imply the dual behavior of VAL at low and high concentrations.
Fig. 9.
TEM images of HEWL fibrils with and without VAL. (A) 10 hrs. aged HEWL fibrils. (B) HEWL fibrils with 10 μM of VAL. (C) HEWL fibrils with 100 μM of VAL.
3.9. Molecular modeling studies
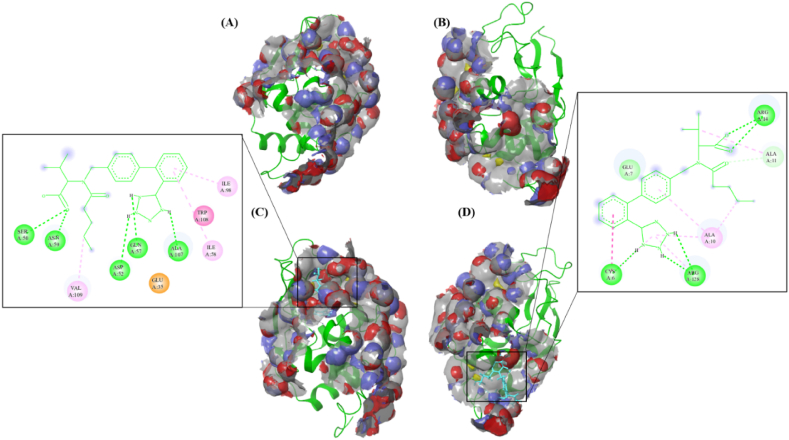
Five binding sites (BS) were predicted for HEWL using Schrodinger's SiteMap program [37]. Among these, only two sites were acceptable for VAL binding. These two sites have different sizes and have different positions. Molecular docking was conducted to explore the structural details and VAL-HEWL interaction mode at the atomic level within the predicted sites. Autodock software was used to generate a grid box for each site and then VAL was docked into these defined sites. Following that, the HEWL-VAL complex for each site was subjected to molecular dynamics simulation for 5 ns. The novel BS-2 is composed of LYS1, PHE3, GLY4, CYS6,GLU7,ALA10,ALA11,LYS13,ARG14,HIS15,SER86,ASP87,ILE88,THR89,GLY126,CYS127,ARG128,and LEU129. According to molecular docking results, VAL preferentially binds to BS-1 since the binding energy in BS-1 was lower (−9.72 kcal mol−1) than in BS-2 (−5.89 kcal mol−1). VAL binds to these sites in a concentration-dependent manner. Experimental evidence suggests that VAL binds to BS-1 at 10 μM, a 10-fold lower molar ratio than HEWL. In contrast, when VAL is present in 100 μM a 1:1 M ratio with HEWL, it binds first to BS-1, followed by BS-2, since BS-1 has low binding energy over BS-2. HEWL active site consists of the amino acid residues TRP62, GLU35, ASP52, and ALA107 [60]. The negative binding energy implies the spontaneous binding of VAL and HEWL [61]. Furthermore, the strength of HEWL-VAL binding stability increased via hydrogen bonding that forms directly between VAL and some amino acid residues in the pocket of BS-1 more than BS-2. Furthermore, TRP63 and TRP108 have reduced solvent accessibility due to VAL binding. This leads to the reduction of the intrinsic fluorescence intensity shown by the fluorescence quenching assay. According to previous studies, the HEWL residues that tend to form amyloid aggregation include the amino acids 25–33, 55–64,107-112, and 121–129, and the binding of 10 μM VAL to this sensitive region in BS-1 may prevent HEWL aggregation [62,63]. SiteMapping results are represented in Table 3. Predicted binding sites are illustrated in Fig. 10A, B. Whereas VAL-HEWL interaction profiles illustrated in Fig. 10C, D. Based on fluorescence studies, TRP residues interact with VAL in the HEWL binding pocket. The negative change in enthalpy indicates that hydrogen bonds and van der Waals interactions are the primary modes of VAL-HEWL interaction. Based on docking results, VAL interacts with HEWL by hydrophobic interactions and hydrogen bonds in both BS-1 and BS-2. Because VAL lies close to HEWL's TRP (TRP62 and TRP63) and TYR (TYR53) residues, it can quench HEWL's endogenous fluorescence [38].
Table 3.
Amino acid residues of BS-1 andBS-2 that interact with VAL as predicted by the SiteMap program. Binding energy was calculated using the Autodock program.
| Binding Site-1 | ||
|---|---|---|
| Amino acid residues | Interactions involved | Binding energy (kcal mol−1) |
| ILE58 | Hydrophobic Interactions | −9.72 |
| ILE 98 | ||
| TRP 108 | ||
| VAL 109 | ||
| ALA 107 | Hydrogen Bonds | |
| SER 50 | ||
| ASP 52 | ||
| GLN 57 | ||
| ASN 59 | ||
| Binding Site-2 | ||
| ALA 10 | Hydrophobic Interactions | −5.89 |
| CYS 6 | Hydrogen Bonds | |
| ARG 128 | ||
| ARG 14 | ||
| GLU 7 | ||
| ALA 11 | ||
Fig. 10.
HEWL binding sites (BS) as predicted by the SiteMap program. (A) BS-1, (B) BS-2. The binding sites are visualized at the top of the cartoon presentation of the HEWL surface. The surface of the binding sites is colored light grey with red and blue highlighting the hydrogen bond acceptor and donor spots respectively. Yellow coloring inside the binding sites indicates that the domains prefer hydrophobic interactions. Docking an interaction profile of HEWL and VAL in BS-1 (C), and BS-2 (D). Discovery Studio was used to visualize the binding residues at each site. In the 2D diagram, hydrogen bonds are represented by green color, and hydrophobic interactions are shown by pink color. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
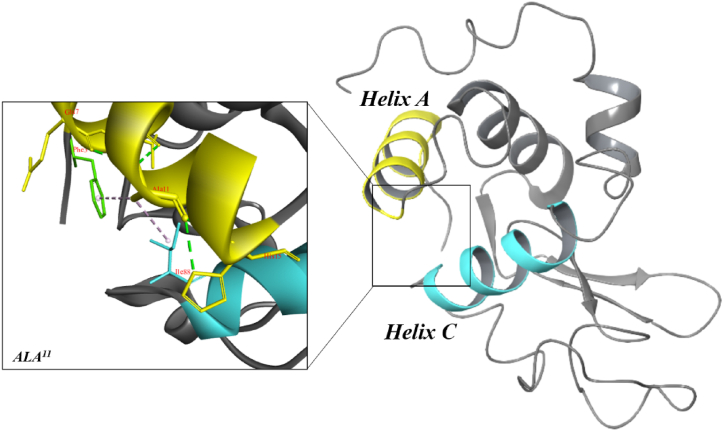
HEWL consists of two domains, the α-domain, and the β-domain. α-domain is made up of five helixes. Whereas the β-domain is composed of a central 310 helix, large loop, and triple-stranded β-sheet. The main catalytic groups at the active site are ASP52 and GLU35 [64,65]. A-helix ALA11 interacts with the C-helix ILE 88, as well as the A-helix HIS15, GLU7, and PHE3 (Fig. 11). Furthermore, HIS15 binds to THR89, and VAL92 via hydrogen bonds and hydrophobic interactions, respectively. The last two residues are in the C-helix. Although PHE3 is not part of the A-helix, it bonds to ILE88 via hydrogen and plays a crucial role in protein stabilization. C- and A-helix residues interact to help with protein packing and hold C- and A-helixes close together. As shown in Fig. 10D, VAL binds directly to ALA11 in BS-2 via hydrogen bonds in a 1:1 M ratio to HEWL. Thus, the C-helix becomes less packed and less stable, leading to unfolding [66]. Additionally, interactions between ALA11 and VAL may indirectly affect HIS15, THR89, and VAL92, altering the folding structure of HEWL. Also, this can affect the N-terminal stability and orientation of the A-helix. Small β-sheets connect the β-domain to the B-helix, and the stability of this sheet greatly affects the stability of the β-domain. The salt bridge forming between LYS1 and GLU7 is important for stabilizing this small β-sheet. It is important to note that the N-terminal domain, A-helixes, and B-helixes are highly organized, and any disruption in these regions will result in structural instability. In earlier studies, structural destabilization and protein aggregation have been reported to result from altering these regions. Furthermore, we found that VAL-GLU7 interactions in BS-2 could negatively affect the N-terminal β-sheet, A-helix, and B-helix interactions (Fig. 10 D). When the B-helix is destabilized, it can result in partial denaturation and then protein unfolding, which can increase the possibility of amyloid accumulation. In addition, experimental evidence indicates a significant role for the N-terminal β-sheet in protein unfolding and aggregation [[67], [68]].
Fig. 11.
The folding structure of HEWL. This is the folding structure of HEWL, with helix A represented by yellow color, and helix C represented by blue color. VAL binds to GLU7, and ALA11which are in helix A at a 1:1 M ratio with HEWL. VAL binding to these residues destabilizes the folding structure of HEWL. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.10. Structural stability of HEWL and HEWL–valsartan complex
To determine the stability of the HEWL-VAL complex, RMSD, RMSF, and Rg values were calculated. A 5ns MD simulation was conducted using CHARMM-GUI for free HEWL as well as HEWL-VAL complex to evaluate the stability of HEWL upon its binding to VAL at BS-1 and BS-2.
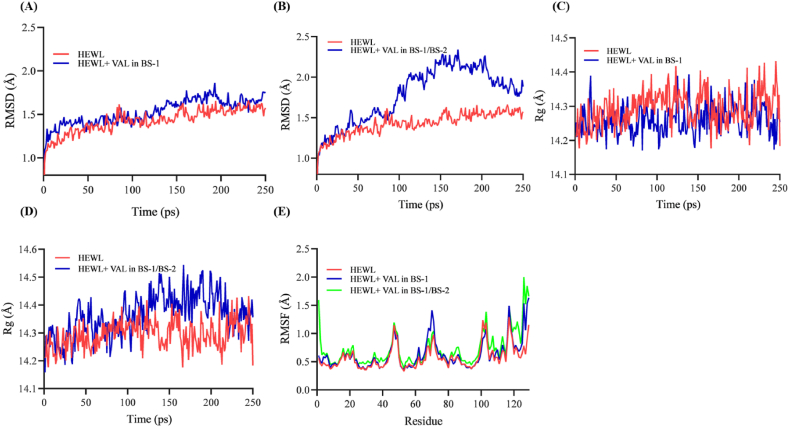
The RMSD is used to measure the equilibration process and dynamic stability of a protein structure after ligand binding. For the first 50 ps, the RMSD values of free and complex HEWLs increase due to protein structure optimization. Free HEWL backbone atoms reached equilibrium after 50 ps at 1.21 ± 0.16 Å and did not show significant change over the next 5 ns. A VAL-HEWL complex in BS-1 achieved equilibrium after 50 ps at 1.42 ± 0.17 with minimal change after 110 ps. In contrast, in the BS-2, the VAL-HEWL complex reached equilibrium after 50 ps 1.45 ± 0.30 with significant change after 90 ps. VAL-HEWL complexes in both BS achieve equilibrium at a higher RMSD value after binding to HEWL, as HEWL becomes less stable after binding to VAL. Fig. 12A and B shows that the values of RMSD of VAL-HEWL are higher than those of free HEWL, indicating that VAL affects HEWL's structural conformation. When VAL occupies BS-1 and BS-2 at a 1:1 M ratio with HEWL, however, more conformational changes and structural instability can be observed than when VAL binds to BS-1 at a 10-fold lower molar ratio than HEWL. For HEWL, HEWL-VAL complexes in BS-1, and HEWL-VAL complexes in BS-1/BS-2, the average RMSD was 1.42 ± 0.16 , 1.52 ± 0.17 , and 1,78 ± 0.30 , respectively, indicating structural destabilization of HEWL in the presence of VAL. However, at 100 μM VAL, significant destabilization occurs compared to 10 μM VAL. In BS-1, VAL binds to HEWL at a molar ratio less than HEWL after 110 ps and dissociates at 210 ps, as shown in Fig. 12A. At 250 ps, free HEWL resumes fluctuating. As a result, 10 μM of VAL does not significantly affect the structure of HEWL. In contrast, VAL occupies BS-1 and BS-2 at 100 μM, which is a 1:1 ratio with HEWL, and this binding occurs at 90ps with a large conformational change. Furthermore, it was unclear whether HEWL returned to its normal conformation (Fig. 12B). Longer simulation times are required to better understand this MD simulation. It is possible that BS-1 and BS-2 have different arrangements of amino acid residues, which could explain why high molar ratios of VAL bind faster than low concentrations. Furthermore, VAL has a different orientation and position in each BS, all of which could explain the early binding of VAL to BS-1/BS-2 in comparison to BS-1.
Fig. 12.
MD simulation analysis of free and complexed HEWL stability. Time evolutions of backbone RMSD of free and VAL-bounded HEW in BS-1(A) and BS-1/BS-2(B). The Rg of free and VAL-bounded HEW in BS-1(C) and BS-1/BS-2(D). (E) The RMSF of free and VAL-bound HEW in BS-1 and BS-1/BS-2.
During simulation, Rg diagrams were monitored to evaluate structural compression changes. According to the Rg analysis, HEWL and VAL-HEWL complexes are compressed throughout the simulation [69,70]. MD simulation uses Rg as an indicator of structural formation [43,71]. In Fig. 12D the VAL-HEWL complex occupying BS-1/BS-2 shows a sudden peak in its Rg profile, indicating structural transformation and greater exposure to the surface. On the other hand, the VAL-HEWL complex in BS-1 does not show any sudden drops or peaks (Fig. 12C) [43,72].
Cα RMSF analyses were conducted on each amino acid in HEWL to determine if VAL binding affected its flexibility. In Fig. 12E, the RMSF for each residue of the model at BS-1 and BS-1/BS-2 is shown for the 5 ns MD run. By comparing the RMSF profile of HEWL alon with the RMSF profile of VAL-bound HEWL. We found that both binding sites have the highest RMSF values at residues 46, 60–80, 102, 118, and 126 which indicates that these residues have a greater degree of flexibility. The structure of HEWL appears to undergo minor changes when VAL binds to it [43]. It is noteworthy that the Cα backbone of HEWL changes differently in BS-1 and BS-1/BS-2. In the novel BS-2, CYS6, GLU7, ARG14, and ARG128 fluctuate more than they do in BS-1. The results indicate that VAL only binds to BS-2 at high concentrations. The active pocket is formed by residues 3–12, 25–43, and 51–65, which are much less flexible than the other residues. These residues are more flexible when VAL occupies BS-1/BS-2 compared to free HEWL and VAL-HEWL complexes in BS-1. Furthermore, the APR in residues 25–33, 55–64,107-112, and 121–129 are more fluctuating at 100 μM VAL than at 10 μM, suggesting that these residues become more flexible at high VAL concentrations [51,63]. HEWL-VAL complexes have a higher fluctuation of their last four residues (THR118-LEU129) than HEWL alone. Despite this, SiteMapping did not detect direct interactions between these residues and VAL, except ARG128-VAL interaction at 100 μM. Perhaps VAL binding causes these C-terminal residues to become unstable. HEWL β-domain (61–78), is more flexible at 10 μM VAL and show higher fluctuation values than HEWL bounded 100 μM VAL. Destabilizing the β-domain may result in the formation of amyloid aggregate. The binding of VAL to this region helps stabilize it and therefore prevents the formation of amyloid aggregates [68]. Most residues in presence of 100 μM have greater RMSF values than free HEWL and HEWL bound to 10 μM VAL. As compared to free HEWL and HEWL bound to 10 μM, the C-terminal region of HEWL is more flexible in presence of 100 μM VAL. According to these observations, HEWL is more stable at 10 μM compared to 100 μM. 100 μM VAL increases the dynamics of the C–terminal strand and may initiate fibrillogenesis.
3.11. The dual effect of VAL against HEWL
In-situ results indicate that VAL affects HEWL aggregation in a dual manner. We assumed that HEWL might have two binding sites for VAL. SiteMapping demonstrated that VAL can bind to a novel site (BS-2) in HEWL with a lower affinity than its original binding site (BS-1) (Fig. 10A, B). VAL bind to BS-1 at a 10-fold lower concentration than HEWL. Because BS-1 has the APR, VAL can act to stabilize these regions and prevent aggregation (Fig. 10C). VAL, on the other hand, occupies the novel BS-2 beside BS-1 when it is present in a 1:1 M ratio with HEWL(Fig. 10D). Most amino acid residues in BS-2 are associated with the A-helix of HEWL, and it has been reported that HEWL's folding structure is stabilized by interactions between its N-terminal A-helix and C-helix (Fig. 11). Thus, VAL may disrupt the folding structure of HEWL by binding to these residues. HEWL is made up of two domains, the α-domain, and the β-domain. Both A-helix and B-helix lie within the α-domain [64,65]. An earlier study found that hydrophobic compounds inhibit insulin aggregation at 10 μM and 20 μM, but they promote it at 30 μM. According to the authors, this dual effect is the result of the acidic medium (pH 2.0) altering the native conformation of the protein, resulting in the dual effect [73].
A co-solute-dependent change in the solubility model has been reported recently. This model assumes that hydrophilic or hydrophobic compounds increase or decrease protein solubility, thereby inhibiting or promoting protein aggregation [74,75]. In this model, hydrophobic compounds shield proteins from the solution and promote protein precipitation, thus increasing aggregation. The interaction between HEWL and solution was not shielded by VAL when present at a concentration 10-fold lower than HEWL, however, this could be possible when VAL was present at a 1:1 M ratio. In all reaction mixtures containing 1% DMSO, hydrophobic VAL was prepared in DMSO, and this amount of DMSO had no significant effect on protein accumulation [76,77].
4. Conclusion
Our study investigates the anti-aggregating effects of VAL on HEWL amyloid aggregation. According to our findings, VAL inhibits amyloid fibrils at low concentrations and promotes them at high concentrations. This dual effect of VAL on HEWL fibrillation has been demonstrated by spectroscopic methods such as ThT, CD, and ANS. HEWL aggregation can be effectively reduced by 10 μM VAL, while 100 μM VAL can have the opposite effect. The TEM images show how HEWL fibrils change morphologically as VAL concentrations are changed. It may be possible to explain the dual effects by the existence of two HEWL binding sites for VAL. Based on the SiteMapping study, 10 μM VAL binds preferentially to BS-1 with the lowest energy, while 100 μM VAL binds BS-2 beside BS-1. According to our hypothesis, VAL produces dual action against HEWL by sequentially binding to BS-1 and BS-2. VAL may prevent aggregation by stabilizing the HEWL APR in BS-1 and preventing the formation of β-sheets. VAL, on the other hand, binds to two binding sites within HEWL and promotes amyloid formation. It was revealed from docking results that VAL binds to A-helix residues at high concentrations and destabilizes A-helix and C-helix interactions, thereby causing HEWL to aggregate. The docking and fluorescence quenching results indicate that hydrogen bonds and hydrophobic interactions are responsible for VAL-HEWL binding. According to our study, 10 μM of VAL significantly inhibited amyloid fibril formation. The mechanism of action and the side effects of the drug need to be further investigated. Moreover, the limitations of our study are that HEWL aggregates at acidic pH and extreme temperatures. Accordingly, folding and aggregation mechanisms may differ from physiologically aggregated proteins. Our findings would be enhanced by further revalidation in an in-vivo model system.
Author contribution statement
Nojood Altwaijry: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Ghaliah S. Almutairi: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Mohd Shahhnawaz Khan: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Majed S. Alokail: Contributed reagents, materials, analysis tools or data.
Nouf Alafaleq: Contributed reagents, materials, analysis tools or data. Rizwan Ali: Contributed reagents, materials, analysis tools or data.
Funding statement
The authors extend their appreciation to the Deputyship of Research and Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number IFKSURG-2-52.
Data availability statement
Data will be made available on request.
Declaration of interest's statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Nojood Altwaijry, Email: nojood@ksu.edu.sa.
Mohd Shahhnawaz Khan, Email: moskhan@ksu.edu.sa.
Nouf Alafaleq, Email: nalafaleg@KSU.EDU.SA.
Rizwan Ali, Email: Aliri@ngha.med.sa.
References
- 1.Nelson R., Eisenberg D. Structural models of amyloid-like fibrils. Adv. Protein Chem. 2006;73 doi: 10.1016/S0065-3233(06)73008-X. [DOI] [PubMed] [Google Scholar]
- 2.Eisenberg D., Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148(6) doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abedini A., et al. Atomic structures of fibrillar segments of hIAPP suggest tightly mated beta-sheets are important for cytotoxicity. Elife. 2016;5 doi: 10.7554/eLife.19273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sweeney P., et al. Protein misfolding in neurodegenerative diseases: Implications and strategies. Transl. Neurodegener. 2017;6(1) doi: 10.1186/s40035-017-0077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen F.E., Kelly J.W. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426(6968) doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 6.Stefani M. Protein folding and misfolding on surfaces. Int. J. Mol. Sci. 2008;9(12) doi: 10.3390/ijms9122515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobson C.M. Protein misfolding, evolution and disease. Trends Biochem. Sci. 1999;24(9) doi: 10.1016/S0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 8.Chiti F., et al. Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc. Natl. Acad. Sci. U. S. A. 1999;96(7) doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75 doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 10.Sawaya M.R., et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447(7143) doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 11.Jahn T.R., et al. The Common Architecture of cross-β amyloid. J. Mol. Biol. 2010;395(4) doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 12.Uversky V.N., Fink A.L. Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochimica et Biophysica Acta - Proteins and Proteomics. 2004;1698(2) doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Zhang R., et al. Lysozyme's lectin-like characteristics facilitates its immune defense function. Q. Rev. Biophys. 2017;50 doi: 10.1017/S0033583517000075. [DOI] [PubMed] [Google Scholar]
- 14.Nusrat S., et al. Anti-Parkinsonian L-Dopa can also act as anti-systemic amyloidosis—a mechanistic exploration. Int. J. Biol. Macromol. 2017;99 doi: 10.1016/j.ijbiomac.2017.03.028. [DOI] [PubMed] [Google Scholar]
- 15.Sattianayagam P.T., et al. Hereditary lysozyme amyloidosis - phenotypic heterogeneity and the role of solid organ transplantation. J. Intern. Med. 2012;272(1) doi: 10.1111/j.1365-2796.2011.02470.x. [DOI] [PubMed] [Google Scholar]
- 16.Swaminathan R., Ravi V.K., Kumar S., Kumar M.V.S., Chandra N. Lysozyme: a model protein for amyloid research. Advances in Protein Chemistry and Structural Biology. 2011;84 doi: 10.1016/B978-0-12-386483-3.00003-3. [DOI] [PubMed] [Google Scholar]
- 17.Kendi Celebi Z., et al. Kidney biopsy in AA amyloidosis: impact of histopathology on prognosis. Amyloid. 2017;24(3) doi: 10.1080/13506129.2017.1350158. [DOI] [PubMed] [Google Scholar]
- 18.Frare E., Mossuto M.F., Polverino de Laureto P., Dumoulin M., Dobson C.M., Fontana A. Identification of the core structure of lysozyme amyloid fibrils by Proteolysis. J. Mol. Biol. 2006;361(3) doi: 10.1016/j.jmb.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 19.Trexler A., Nilsson M. The Formation of amyloid fibrils from proteins in the lysozyme Family. Trexler. 2007;8(6) doi: 10.2174/138920307783018659. [DOI] [PubMed] [Google Scholar]
- 20.Parr M.K., Joseph J.F. NDMA impurity in valsartan and other pharmaceutical products: analytical methods for the determination of N-nitrosamines. J. Pharmaceut. Biomed. Anal. 2019;164 doi: 10.1016/j.jpba.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Odigboegwu O., Pan L.J., Chatterjee P. Use of antihypertensive drugs during Preeclampsia. Frontiers in Cardiovascular Medicine. 2018;5 doi: 10.3389/fcvm.2018.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao W., Wang J., Ho L., Ono K., Teplow D.B., Pasinetti G.M. Identification of antihypertensive drugs which inhibit amyloid-β protein oligomerization. J. Alzheim. Dis. 2009;16(1):49–57. doi: 10.3233/JAD-2009-0925. [DOI] [PubMed] [Google Scholar]
- 23.Chou C.L., Yeh H.I. The role of the renin-angiotensin system in amyloid metabolism of Alzheimer's disease. Acta Cardiol. Sin. 2014;30(2) [PMC free article] [PubMed] [Google Scholar]
- 24.Barthold D., Joyce G., Wharton W., Kehoe P., Zissimopoulos J. The association of multiple anti-hypertensive medication classes with Alzheimer's disease incidence across sex, race, and ethnicity. PLoS One. 2018;13(11) doi: 10.1371/journal.pone.0206705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh S., Pandey N.K., Dasgupta S. Crowded milieu prevents fibrillation of hen egg white lysozyme with retention of enzymatic activity. J. Photochem. Photobiol., B. 2014;138 doi: 10.1016/j.jphotobiol.2014.04.021. [DOI] [PubMed] [Google Scholar]
- 26.Wu J.W., et al. Carnosine's effect on amyloid fibril formation and induced cytotoxicity of lysozyme. PLoS One. 2013;8(12) doi: 10.1371/journal.pone.0081982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shirahama T., Cohen A.S. High-resolution electron microscopic analysis of the amyloid fibril. JCB (J. Cell Biol.) 1967;33(3) doi: 10.1083/jcb.33.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarrett J.T., Lansbury P.T. Seeding ‘one-dimensional crystallization’ of amyloid: a pathogenic mechanism in Alzheimer's disease and scrapie? Cell. 1993;73(6) doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 29.Dickens M.G., Franz K.J. A prochelator activated by hydrogen peroxide prevents metal-induced amyloid β aggregation. Chembiochem. 2010;11(1) doi: 10.1002/cbic.200900597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Necula M., Kayed R., Milton S., Glabe C.G. Small molecule inhibitors of aggregation indicate that amyloid β oligomerization and fibrillization pathways are independent and distinct. J. Biol. Chem. 2007;282(14) doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
- 31.Nilsson M.R. Techniques to study amyloid fibril formation in vitro. Methods. 2004;34(1) doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 32.Chaturvedi S.K., et al. Unraveling comparative anti-amyloidogenic behavior of pyrazinamide and D-Cycloserine: a mechanistic biophysical insight. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0136528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klunk W.E., Pettegrew J.W., Abraham D.J. Quantitative evaluation of Congo red binding to amyloid-like proteins with a beta-pleated sheet conformation. J. Histochem. Cytochem. 1989;37(8) doi: 10.1177/37.8.2666510. [DOI] [PubMed] [Google Scholar]
- 34.Micsonai A., et al. BeStSel: a web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gky497. W315–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin L., et al. Attenuation of human lysozyme amyloid fibrillation by ace inhibitor captopril: a combined spectroscopy, microscopy, cytotoxicity, and docking study. Biomacromolecules. May 2021;22(5):1910–1920. doi: 10.1021/acs.biomac.0c01802. [DOI] [PubMed] [Google Scholar]
- 36.Mollazadeh S., Sahebkar A., Shahlaei M., Moradi S. Nano drug delivery systems: molecular dynamic simulation. J. Mol. Liq. 2021;332 doi: 10.1016/j.molliq.2021.115823. [DOI] [Google Scholar]
- 37.Siposova K., Sedlak E., Kozar T., Nemergut M., Musatov A. Dual effect of non-ionic detergent Triton X-100 on insulin amyloid formation. Colloids Surf. B Biointerfaces. 2019;173 doi: 10.1016/j.colsurfb.2018.10.039. [DOI] [PubMed] [Google Scholar]
- 38.Zhan M., Guo M., Jiang Y., Wang X. Characterization of the interaction between gallic acid and lysozyme by molecular dynamics simulation and optical spectroscopy. Int. J. Mol. Sci. 2015;16(7) doi: 10.3390/ijms160714786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan A.N., Hassan M.N., Khan R.H. Gallic acid: a naturally occurring bifunctional inhibitor of amyloid and metal induced aggregation with possible implication in metal-based therapy. J. Mol. Liq. 2019;285 doi: 10.1016/j.molliq.2019.04.059. [DOI] [Google Scholar]
- 40.Khan J.M., et al. Protonation favors aggregation of lysozyme with SDS. Soft Matter. 2014;10(15) doi: 10.1039/c3sm52435c. [DOI] [PubMed] [Google Scholar]
- 41.Bruggink K.A., Müller M., Kuiperij H.B., Verbeek M.M. Methods for analysis of amyloid-β aggregates. J. Alzheim. Dis. 2012;28(4) doi: 10.3233/JAD-2011-111421. [DOI] [PubMed] [Google Scholar]
- 42.Liu F., et al. Brazilin inhibits α-synuclein fibrillogenesis, disrupts mature fibrils, and Protects against amyloid-induced cytotoxicity. J. Agric. Food Chem. 2019;67(42) doi: 10.1021/acs.jafc.9b04558. [DOI] [PubMed] [Google Scholar]
- 43.Patel P., Parmar K., Patel D., Kumar S., Trivedi M., Das M. Inhibition of amyloid fibril formation of lysozyme by ascorbic acid and a probable mechanism of action. Int. J. Biol. Macromol. 2018;114 doi: 10.1016/j.ijbiomac.2018.03.152. [DOI] [PubMed] [Google Scholar]
- 44.Zaman M., et al. DNA induced aggregation of stem bromelain; A mechanistic insight. RSC Adv. 2016;6(44) doi: 10.1039/c6ra01079b. [DOI] [Google Scholar]
- 45.Haq S.K., Rasheedi S., Khan R.H. Characterization of a partially folded intermediate of stem bromelain at low pH. Eur. J. Biochem. 2002;269(1) doi: 10.1046/j.0014-2956.2002.02620.x. [DOI] [PubMed] [Google Scholar]
- 46.Nusrat S., et al. A comprehensive spectroscopic and computational investigation to probe the interaction of antineoplastic drug nordihydroguaiaretic acid with serum albumins. PLoS One. 2016;11(7) doi: 10.1371/journal.pone.0158833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zaman M., Ehtram A., Chaturvedi S.K., Nusrat S., Khan R.H. Amyloidogenic behavior of different intermediate state of stem bromelain: a biophysical insight. Int. J. Biol. Macromol. 2016;91 doi: 10.1016/j.ijbiomac.2016.05.107. [DOI] [PubMed] [Google Scholar]
- 48.Nusrat S., Khan R.H. Exploration of ligand-induced protein conformational alteration, aggregate formation, and its inhibition: a biophysical insight. Prep. Biochem. Biotechnol. 2018;48(1) doi: 10.1080/10826068.2017.1387561. [DOI] [PubMed] [Google Scholar]
- 49.Bustamante C., Tinoco I., Maestre M.F. Circular differential scattering can be an important part of the circular dichroism of macromolecules. Proc. Natl. Acad. Sci. U. S. A. 1983;80(12) doi: 10.1073/pnas.80.12.3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manzo G., et al. Enhanced amphiphilic profile of a short β-stranded peptide improves its antimicrobial activity. PLoS One. 2015;10(1) doi: 10.1371/journal.pone.0116379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He J., et al. Myricetin prevents fibrillogenesis of hen egg white lysozyme. J. Agric. Food Chem. 2014;62(39) doi: 10.1021/jf5025449. [DOI] [PubMed] [Google Scholar]
- 52.Duy C., Fitter J. How aggregation and conformational scrambling of unfolded states govern fluorescence emission spectra. Biophys. J. 2006;90(10) doi: 10.1529/biophysj.105.078980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nusrat S., et al. Interaction of catecholamine precursor L-Dopa with lysozyme: a biophysical insight. Int. J. Biol. Macromol. 2018;109 doi: 10.1016/j.ijbiomac.2017.11.107. [DOI] [PubMed] [Google Scholar]
- 54.Möller M., Denicola A. Protein tryptophan accessibility studied by fluorescence quenching. Biochem. Mol. Biol. Educ. 2002;30(3) doi: 10.1002/bmb.2002.494030030035. [DOI] [Google Scholar]
- 55.Anand U., Jash C., Boddepalli R.K., Shrivastava A., Mukherjee S. Exploring the mechanism of fluorescence quenching in proteins induced by tetracycline. J. Phys. Chem. B. 2011;115(19) doi: 10.1021/jp2008978. [DOI] [PubMed] [Google Scholar]
- 56.Lakowicz J.R., Weber G. Quenching of fluorescence by Oxygen. A probe for structural fluctuations in macromolecules. Biochemistry. 1973;12(21) doi: 10.1021/bi00745a020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ross P.D., Subramanian S. Thermodynamics of protein association reactions: forces Contributing to stability. Biochemistry. 1981;20(11) doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- 58.Al-Mehizia A.A., Bakheit A.H., Zargar S., Bhat M.A., Asmari M.M., Wani T.A. Evaluation of biophysical interaction between Newly Synthesized Pyrazoline Pyridazine Derivative and bovine serum Albumin by spectroscopic and molecular docking studies. Journal of Spectroscopy. 2019;2019 doi: 10.1155/2019/3848670. [DOI] [Google Scholar]
- 59.Li D., Ji B., Jin J. Spectrophotometric studies on the binding of Vitamin C to lysozyme and bovine liver catalase. J. Lumin. 2008;128(9) doi: 10.1016/j.jlumin.2008.01.010. [DOI] [Google Scholar]
- 60.Gálvez-Iriqui A.C., Plascencia-Jatomea M., Bautista-Baños S. Lysozymes: characteristics, mechanism of action and technological applications on the control of pathogenic microorganisms. Revista Mexicana de Fitopatología, Mexican Journal of Phytopathology. 2020;38(3) doi: 10.18781/r.mex.fit.2005-6. [DOI] [Google Scholar]
- 61.Malik S., Siddiqi M.K., Majid N., Masroor A., Moasfar Ali S., Khan R.H. Unravelling the inhibitory and cytoprotective potential of diuretics towards amyloid fibrillation. Int. J. Biol. Macromol. May 2020;150:1258–1271. doi: 10.1016/J.IJBIOMAC.2019.10.137. [DOI] [PubMed] [Google Scholar]
- 62.Chaudhary A.P., Vispute N.H., Shukla V.K., Ahmad B. A comparative study of fibrillation kinetics of two homologous proteins under identical solution condition. Biochimie. 2017;132 doi: 10.1016/j.biochi.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 63.Das T., Kolli V., Karmakar S., Sarkar N. Functionalisation of polyvinylpyrrolidone on gold nanoparticles enhances its anti-amyloidogenic propensity towards hen egg white lysozyme. Biomedicines. 2017;5(2) doi: 10.3390/biomedicines5020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jolles P. Lysozymes: model Enzymes in Biochemistry and Biology. Experientia Suppl. 1996;75 [Google Scholar]
- 65.Mine Y., Ma F., Lauriau S. Antimicrobial peptides Released by enzymatic Hydrolysis of hen egg white lysozyme. J. Agric. Food Chem. 2004;52(5) doi: 10.1021/jf0345752. [DOI] [PubMed] [Google Scholar]
- 66.Ramos J., et al. The impact of folding modes and deuteration on the atomic resolution structure of hen egg-white lysozyme. Acta Crystallogr D Struct Biol. 2021;77 doi: 10.1107/S2059798321010950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takano K., Isuchimori K., Yamagata Y., Yutani K. Effect of foreign N-terminal residues on the conformational stability of human lysozyme. Eur. J. Biochem. 1999;266(2) doi: 10.1046/j.1432-1327.1999.00918.x. [DOI] [PubMed] [Google Scholar]
- 68.Monhemi H., Tabaee S.S. The effects of mutation and modification on the structure and stability of human lysozyme: a molecular link between carbamylation and atherosclerosis. J. Mol. Graph. Model. 2020;100 doi: 10.1016/j.jmgm.2020.107703. [DOI] [PubMed] [Google Scholar]
- 69.Chun B.J., il Choi J., Jang S.S. Molecular dynamics simulation study of sodium dodecyl sulfate micelle: water penetration and sodium dodecyl sulfate dissociation. Colloids Surf. A Physicochem. Eng. Asp. 2015;474 doi: 10.1016/j.colsurfa.2015.03.002. [DOI] [Google Scholar]
- 70.Fixman M. Radius of gyration of polymer chains. J. Chem. Phys. 1962;36(2) doi: 10.1063/1.1732501. [DOI] [Google Scholar]
- 71.Stevens M.J., Hoh J.H., Woolf T.B. Insights into the molecular mechanism of membrane fusion from simulation: evidence for the association of splayed tails. Phys. Rev. Lett. 2003;91(18) doi: 10.1103/PhysRevLett.91.188102. [DOI] [PubMed] [Google Scholar]
- 72.Hashemzadeh H., Javadi H., Darvishi M.H. Study of Structural stability and formation mechanisms in DSPC and DPSM liposomes: a coarse-grained molecular dynamics simulation. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-58730-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Akbarian M., et al. Inhibitory effect of coumarin and its analogs on insulin fibrillation/cytotoxicity is depend on oligomerization states of the protein. RSC Adv. 2020;10(63) doi: 10.1039/d0ra07710k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.So M., et al. Polyphenol-solubility alters amyloid fibril formation of α-synuclein. Protein Sci. 2021;30(8) doi: 10.1002/pro.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sawada M., et al. Amyloid Formation of α-synuclein based on the solubility- and Supersaturation-dependent mechanism. Langmuir. 2020;36(17) doi: 10.1021/acs.langmuir.0c00426. [DOI] [PubMed] [Google Scholar]
- 76.Gao W., et al. Inhibition behavior of Sennoside A and Sennoside C on amyloid fibrillation of human lysozyme and its possible mechanism. Int. J. Biol. Macromol. 2021;178 doi: 10.1016/j.ijbiomac.2021.02.213. [DOI] [PubMed] [Google Scholar]
- 77.Zaidi F.K., Bhat R. Two polyphenols with diverse mechanisms towards amyloidosis: differential modulation of the fibrillation pathway of human lysozyme by curcumin and EGCG. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1860824. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.