Abstract
Advancements in deep learning and computer vision provide promising solutions for medical image analysis, potentially improving healthcare and patient outcomes. However, the prevailing paradigm of training deep learning models requires large quantities of labeled training data, which is both time-consuming and cost-prohibitive to curate for medical images. Self-supervised learning has the potential to make significant contributions to the development of robust medical imaging models through its ability to learn useful insights from copious medical datasets without labels. In this review, we provide consistent descriptions of different self-supervised learning strategies and compose a systematic review of papers published between 2012 and 2022 on PubMed, Scopus, and ArXiv that applied self-supervised learning to medical imaging classification. We screened a total of 412 relevant studies and included 79 papers for data extraction and analysis. With this comprehensive effort, we synthesize the collective knowledge of prior work and provide implementation guidelines for future researchers interested in applying self-supervised learning to their development of medical imaging classification models.
Subject terms: Medical imaging, Computer science, Scientific data
Introduction
The utilization of medical imaging technologies has become an essential part of modern medicine, enabling diagnostic decisions and treatment planning. The importance of medical imaging is exemplified by the consistent rate of growth in medical imaging utilization in modern healthcare1,2. However, as the number of medical images relative to the available radiologists continues to become more disproportionate, the workload for radiologists continues to increase. Studies have shown that an average radiologist now needs to interpret one image every 3–4 s to keep up with clinical workloads3–5. With such an immense cognitive burden placed on radiologists, delays in diagnosis and diagnostic errors are unavoidable6,7. Thus, there is an urgent need to integrate automated systems into the medical imaging workflow, which will improve both efficiency and accuracy of diagnosis.
In recent years, deep learning models have demonstrated diagnostic accuracy comparable to that of human experts in narrow clinical tasks for several medical domains and imaging modalities, including chest and extremity X-rays8–10, computed tomography (CT)11, magnetic resonance imaging (MRI)12, whole slide images (WSI)13,14, and dermatology images15. While deep learning provides promising solutions for improving medical image interpretation, the current success has been largely dominated by supervised learning frameworks, which typically require large-scale labeled datasets to achieve high performance. However, annotating medical imaging datasets requires domain expertize, making large-scale annotations cost-prohibitive and time-consuming, which fundamentally limits building effective medical imaging models across varying clinical use cases.
Besides facing challenges with training data, most medical imaging models underperform in their ability to generalize to external institutions or when repurposed for other tasks16. The inability to generalize can be largely due to the process of supervised learning, which encourages the model to mainly learn features heavily correlated with specific labels rather than general features representative of the whole data distribution. This creates specialist models that can perform well only on the tasks they were trained to do17. In a healthcare system where a myriad of opportunities and possibilities for automation exist, it is practically impossible to curate labeled datasets for all tasks, modalities, and outcomes for training supervised models. Therefore, it is important to develop strategies for training medical artificial intelligence (AI) models that can be fine-tuned for many downstream tasks without curating large-scale labeled datasets.
Self-supervised learning (SSL), the process of training models to produce meaningful representations using unlabeled data, is a promising solution to challenges caused by difficulties in curating large-scale annotations. Unlike supervised learning, SSL can create generalist models that can be fine-tuned for many downstream tasks without large-scale labeled datasets. Self-supervised learning was first popularized in the field of natural language processing (NLP) when researchers leveraged copious amounts of unlabeled text scraped from the internet to improve the performance of their models. These pre-trained large language models18,19 are capable of achieving state-of-the-art results for a wide range of NLP tasks, and have shown the ability to perform well on new tasks with only a fraction of the labeled data that traditional supervised learning techniques require. Motivated by the initial success of SSL in NLP, there is great interest in translating similar techniques of SSL to computer vision tasks. Such work in computer vision has already demonstrated performance for natural images that is superior to that achieved by supervised models, especially in label-scarce scenarios20.
Reducing the number of manual annotations required to train medical imaging models will significantly reduce both the cost and time required for model development, making automated systems more accessible to different specialties and hospitals, thereby reducing workload for radiologists and potentially improving patient care. While there is already a growing trend in recent medical imaging AI literature to leverage SSL (Fig. 1), as well as a few narrative reviews21,22, the most suitable strategies and best practices for medical images have not been sufficiently investigated. The purpose of this work is to present a comprehensive review of deep learning models that leverage SSL for medical image classification, define and consolidate relevant terminology, and summarize the results from state-of-the-art models in relevant current literature. We focus on medical image classification tasks because many clinical tasks are based on classification, and thus our research may be directly applicable to deep learning models for clinical workflows. This review intends to help inform future modeling frameworks and serve as a reference for researchers interested in the application of SSL in medical imaging.
Fig. 1. Timeline showing the number of publications on deep learning for medical image classification per year, found by using the same search criteria on PubMed, Scopus and, ArXiv.
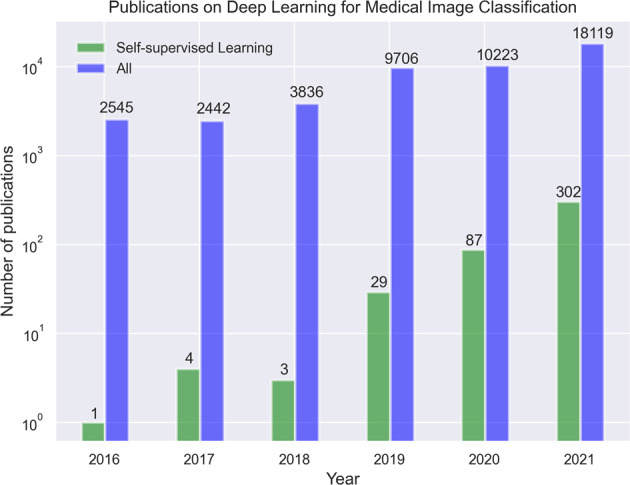
The figure shows that self-supervised learning is a rapidly growing subset of deep learning for medical imaging literature.
Terminology and strategies in self-supervised learning
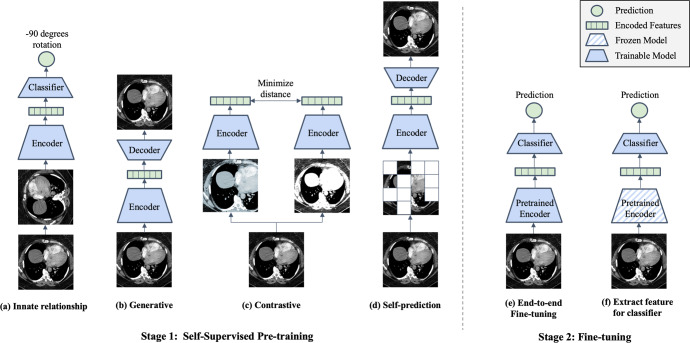
Here we provide definitions for different categorizations of self-supervision strategies, namely innate relationship, generative, contrastive, and self-prediction (Fig. 2)23.
Fig. 2. Illustration of different self-supervised learning and fine-tuning strategies.
During Stage 1 a model is pre-trained using one or more of the following self-supervised learning strategies: (a) Innate relationship SSL pre-trains a model on a hand-crafted task by leveraging the internal structure of the data, (b) Generative SSL learns the distribution of training data, enabling reconstruction of the original input (c) Contrastive SSL forms positive pairs between different augmentations of the same image and minimizes representational distances of positive samples (d) Self-prediction augments or masks out random portions of an image, and reconstructs the original image based on the unaltered parts of the original image. During Stage 2, the pre-trained model can be fine-tuned using one of the following strategies: (e) end-to-end fine-tuning of the pre-trained model and classifier, or (f) train a classifier that uses extracted features from the SSL pre-trained model.
Innate relationship SSL is the process of pre-training a model on a hand-crafted task, which can leverage the internal structure of the data, without acquiring additional labels. In the most general sense, innate relationship models perform classification or regression based on the hand-crafted task instead of optimizing based on the model’s ability to reconstruct (generative and self-prediction) or represent the original image (contrastive). Specifically, these methods are optimized using classification or regression loss derived from the given task. Pre-training the model on such a hand-crafted task makes the model learn visual features as a starting point. However, innate relationship SSL can lead to visual features that are effective only for the hand-crafted task but have limited benefits for the downstream task. Examples of innate relationship for visual inputs include predicting image rotation angle24, solving jigsaw puzzles of an image25, or determining the relative positions of image patches26.
Generative models, popularized through the advent of traditional autoencoders27, variational autoencoders28 and generative adversarial networks (GANs)29, are able to learn the distribution of training data, and thereby reconstruct the original input or create new synthetic data instances. By using readily available data as the target, generative models can be trained to automatically learn useful latent representations without the need for explicit labels, and they thus constitute a form of self-supervision. Early work that leverages generative models for self-supervised learning rely on autoencoders, where an encoder converts inputs into latent representations and a decoder reconstructs the representation back to the original image30. Subsequently, these models are optimized based on how closely the reconstructed images resemble the original image. More recent work has explored utilizing GANs for generative self-supervised learning, with improvement in performance over prior work31,32.
Contrastive self-supervised methods are based on the assumption that variations caused by transforming an image do not alter the image’s semantic meaning. Therefore, different augmentations of the same image constitute a so-called positive pair, while the other images and their augmentations are defined to be negative pairs in relation to the current instance. Subsequently a model is optimized to minimize distance in latent space between the positive pairs and push apart negative samples. Separating representations for positive and negative pairs can be based on arbitrary distance metrics incorporated into the contrastive loss function. One pioneering contrastive-based method is SimCLR20, which outperformed supervised models on ImageNet benchmark using 100 times fewer labels. However, SimCLR requires a very large batch size to perform well, which can be computationally prohibitive for most researchers. To reduce the large batch size required by SimCLR to ensure enough informative negative samples, Momentum Contrast (MoCo) introduced a momentum encoded queue to keep negative samples33. More recently, a subtype of contrastive self-supervised learning called instance discrimination, which includes methods such as DINO34, BYOL35 and SimSiam36, further eliminates the need for negative samples. Instead of contrastive augmented pairs from the same image, several studies have explored contrasting clustering assignments of augmented versions of the same image37–39.
Self-prediction SSL is the process of masking or augmenting portions of the input and using the unaltered portions to reconstruct the original input. The idea of self-prediction SSL originated from the field of NLP, where state-of-the-art models were pre-trained using the Masked Language Modeling approach by predicting missing words in a sentence18,19. Motivated by the success in NLP, early work in the field of computer vision made similar attempts by masking out or augmenting random patches of an image and training Convolutional Neural Networks (CNNs) to reconstruct the missing regions as a pre-training strategy40 but only with moderate success. Recently, the introduction of Vision Transformers (ViT) allowed computer vision models to also have the same transformer-based architecture. Studies such as BERT Pre-Training of Image Transformers (BEiT) and Masked Auto-encoders (MAE), which combine ViT with self-prediction pre-training objective, achieve state-of-the-art results when fine-tuned across several natural image benchmarks41,42. Similar to generative SSL, self-prediction models are optimized using the reconstruction loss. The key difference between self-prediction and generative SSL methods is that self-prediction applies masking or augmentations only to portions of the input image, and uses the remaining, unaltered portions to inform reconstruction. On the other hand, generative based SSL applies augmentations on the whole image and subsequently reconstruct the whole image.
There are two main strategies for fine-tuning models that have been pre-trained using SSL (Fig. 2). If we consider any imaging model to be composed of an encoder part and a classifier part, then these two strategies are (1) end-to-end fine-tuning vs. (2) extract features from the encoder first and subsequently train an additional classifier. In end-to-end fine-tuning, all the weights of the encoder and classifier are unfrozen and can be adjusted through optimization using supervised learning in the fine-tuning phase. In the feature-extraction strategy, the weights of the encoder are kept frozen to extract features as inputs to the downstream classifier. While much previous work uses linear classifiers with trainable weights (also known as linear probing), any type of classifier or architecture can be used, including Support Vector Machines (SVMs) and k-nearest neighbor43. It is worth emphasizing that SSL is task agnostic, and the same SSL pre-trained model can be fine-tuned for different types of downstream tasks, including classification, segmentation, and object detection.
Results
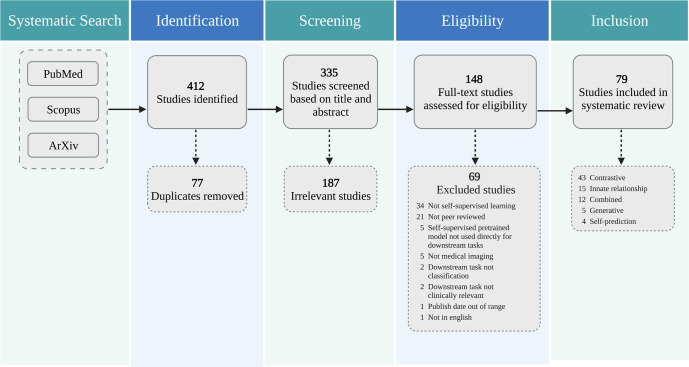
A total of 412 unique studies were identified through our systematic search. After removing duplicates and excluding studies based on title and abstract using our study selection criteria (see Methods), 148 studies remained for full-text screening. A total of 79 studies fulfilled our eligibility criteria and were included for systematic review and data extraction. Figure 3 presents a flowchart of the study screening and selection process. Table 1 displays the included studies and extracted data while Fig. 4 summarizes the statistics of extracted data.
Fig. 3. The PRISMA diagram for this review.
The authors independently screened all records for eligibility. Out of 412 studies identified from PubMed, Scopus, and ArXiv, 79 studies were included in the systematic review.
Table 1.
Overview of studies included in our systematic review.
| SSL strategy | Year | First author | Imaging modality | Clinical domain | Outcome/Task | Combined methods | SSL framework | Strategy for fine-tuning (freeze layers, end-to-end) | Metrics | SSL performance | Supervised performance | Relative difference in SSL and supervised performance |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Combined | 2020 | Behzad Bozorgtabar111 | Chest X-ray | Radiology | Chest abnormality | Generative, Contrastive | Autoencoder, MoCo (modified), other | Extract features from encoder -> Calculate “anomaly score” using KNN | AUROC | 0.917 | 0.861 | 0.065 |
| Combined | 2020 | Wan-Ting Hsieh112 | MRI | Radiology | Cognitive Impairment and Alzheimer’s disease | Generative, Contrastive | Autoencoder, Multimodal contrastive | Extract features from encoder -> SVM | Accuracy | 0.594 | – | – |
| Combined | 2020 | Jianbo Jiao53 | Ultrasound | Obstetrics & Gynecology | Standard plane detection | Contrastive, Innate Relationship | Constrastive learning, other | End-to-end | F1 | 0.726 | 0.725 | 0.001 |
| Combined | 2021 | Yu Tian113 | Colonoscopy | Gastroenterology | Gastrointenstinal abnormality | Contrastive, Innate Relationship | Contrastive Learning, Augmentation Prediction, Patch Position Prediction | End-to-end using unsupervised abnormality detection methods | AUROC | 0.972 | – | – |
| Combined | 2021 | Fatemeh Haghighi72 | CT | Radiology | Lung nodule | Generative, Innate Relationship, Self-prediction | Autoencoder, patch pseudo label prediction, perturbed image restoration | End-to-end | AUROC | 0.985 | 0.943 | 0.045 |
| Combined | 2021 | Stefan Cornelissen71 | Endoscopy | Gastroenterology | Barett’s esophagus | Self-prediction, Generative | GAN, Other | Extract features from encoder -> MLP | Accuracy | 0.838 | 0.792 | 0.058 |
| Combined | 2021 | Xiaomeng Li70 | Fundus Image | Ophthalmology | Pathologic Myopia | Contrastive, Innate Relationship | Multi-view contrastive learning, rotation prediction | Extract features from encoder -> KNN | AUROC | 0.991 | 0.98 | 0.011 |
| Combined | 2021 | Alex Fedorov114 | MRI | Radiology | Alzheimer’s disease | Generative, Contrastive | Autoencoder, SimCLR | Extract features from encoder -> Linear classifier | AUROC | – | – | – |
| Combined | 2021 | Jiahong Ouyang115 | MRI | Radiology | Cognitive Impairment and Alzheimer’s disease | Contrastive, Generative | Longitudinal Neighborhood Embedding, Autoencoder | End-to-end | Accuracy | 0.836 | 0.794 | 0.053 |
| Combined | 2021 | Jing Ke68 | Whole Slide Image | Pathology | Colorectal cancer, stomach cancer and breast cancer | Generative, Contrastive | CycleGAN, Contrastive Learning, Clustering | Unclear | Accuracy | 0.91 | – | – |
| Combined | 2021 | Pengshuai Yang84 | Whole Slide Image | Pathology | Colorectal cancer and healthy tissue types | Generative, Contrastive | Contrastive learning, other | Extract features from encoder -> Linear classifier | Accuracy | 0.914 | 0.844 | 0.083 |
| Contrastive | 2020 | Hari Sowrirajan116 | Chest X-ray | Radiology | Pleural effusion | – | MoCo | Extract features from encoder -> Linear classifier | AUROC | 0.953 | 0.949 | 0.004 |
| Contrastive | 2020 | Hong-Yu Zhou117 | Chest X-ray | Radiology | Chest abnormality | – | Other | Unclear | AUROC | 0.893 | 0.879 | 0.016 |
| Contrastive | 2020 | Li Sun77 | CT | Radiology | COVID-19 | – | Contrastive learning | Extract features from encoder -> Linear classifier | Accuracy | 0.963 | 0.775 | 0.243 |
| Contrastive | 2020 | Philippe Burlina118 | Fundus Image | Ophthalmology | Diabetic retinopathy referral | – | Deep InfoMax | Extract local features -> DeepInfoMax | AUROC | 0.835 | 0.833 | 0.002 |
| Contrastive | 2020 | Xiaomeng Li119 | Fundus Image | Ophthalmology | Retinal disease | – | Multi-modal contrastive learning | Extract features from encoder -> KNN | AUROC | 0.986 | 0.98 | 0.006 |
| Contrastive | 2020 | Alex Fedorov120 | MRI | Radiology | Alzheimer’s disease | – | Mutual Information Maximization | Extract features from encoder -> Linear classifier | AUROC | 0.841 | 0.88 | −0.044 |
| Contrastive | 2020 | Nooshin Mojab121 | Whole Slide Image | Ophthalmology | Glaucoma | – | SimCLR | Extract features from encoder -> Linear Classifier | Accuracy | 0.923 | 0.904 | 0.021 |
| Contrastive | 2020 | Bin Li59 | Whole Slide Image | Pathology | Lung cancer and healthy tissue types | – | SimCLR | Extract features from encoder -> Multiple instance learning aggregator | AUROC | 0.963 | 0.726 | 0.326 |
| Contrastive | 2020 | Olivier Dehaene88 | Whole Slide Image | Pathology | Breast cancer | – | MoCo v2 | Extract features from encoder -> Multiple instance learner | AUROC | 0.987 | 0.829 | 0.191 |
| Contrastive | 2020 | Ozan Ciga85 | Whole Slide Image | Pathology | Colorectal cancer and healthy tissue types | – | SimCLR | End-to-end | F1 | 0.914 | 0.801 | 0.141 |
| Contrastive | 2021 | Colorado J Reed122 | Chest X-ray | Radiology | Chest abnormality | – | MoCo v2 | Extract features from encoder -> Linear classifier | – | – | – | – |
| Contrastive | 2021 | Fengbei Liu123 | Chest X-ray | Radiology | Chest abnormality | – | Contrastive learning (modified) | End-to-end | AUROC | 0.825 | – | – |
| Contrastive | 2021 | Heng Hao78 | Chest X-ray | Radiology | Pneumonia and COVID-19 | – | SimCLR | Extract features from encoder -> Gaussian process classifer | Sensitivity | 0.936 | 0.907 | 0.032 |
| Contrastive | 2021 | JinpengLi79 | Chest X-ray | Radiology | COVID-19 | – | SimCLR | Unclear | AUROC | 0.9 | 0.915 | −0.016 |
| Contrastive | 2021 | Matej Gazda124 | Chest X-ray | Radiology | Pneumonia | – | Contrastive Learning | Extract features from encoder -> Linear classifier | AUROC | 0.977 | – | – |
| Contrastive | 2021 | Nanqing Dong80 | Chest X-ray | Radiology | COVID-19 | – | MoCo | Extract features from encoder -> Linear classifier | Accuracy | 0.916 | 0.796 | 0.151 |
| Contrastive | 2021 | Nhut-Quang Nguyen125 | Chest X-ray | Radiology | Pneumonia detection | – | BYOL | Unclear | AUROC | 0.988 | 0.95 | 0.04 |
| Contrastive | 2021 | Shekoofeh Azizi93 | Chest X-ray | Radiology | Chest abnormality | – | SimCLR (modified) | End-to-end | AUROC | 0.773 | 0.763 | 0.013 |
| Contrastive | 2021 | Tuan Truong96 | Chest X-ray | Radiology | Pneumonia | – | SimCLR, SwAV, DINO | DVME (custom) attention based model | AUROC | 0.984 | 0.94 | 0.047 |
| Contrastive | 2021 | Xi Zhao126 | Chest X-ray | Radiology | Pneumonia | – | SimCLR (modified) | Extract features from encoder -> Linear classifer | AUROC | 0.889 | 0.84 | 0.058 |
| Contrastive | 2021 | Yen Nhi Truong Vu103 | Chest X-ray | Radiology | Pleural effusion | – | MoCo (modified) | End-to-end | AUROC | 0.906 | 0.858 | 0.056 |
| Contrastive | 2021 | Zhanghexuan Ji60 | Chest X-ray | Radiology | Chest abnormality | – | Multimodal Contrastive, Text to Region Alignment | Unclear | AUROC | 0.932 | 0.91 | 0.024 |
| Contrastive | 2021 | Haohua Dong102 | CT | Radiology | Focal liver lesion | – | Contrastive learning (modified) | Extract features from encoder -> MLP | Accuracy | 0.854 | 0.836 | 0.022 |
| Contrastive | 2021 | Nahid Ul Islam127 | CT | Radiology | Pulmonary embolism | – | SeLa-v2 | End-to-end | AUROC | 0.957 | 0.947 | 0.011 |
| Contrastive | 2021 | Wenzhi Bao128 | CT | Radiology | Gastric cancer | – | SimSiam | End-to-end | AUROC | 0.975 | 0.95 | 0.026 |
| Contrastive | 2021 | Guo-Zhang Jian129 | Endoscopy | Gastroenterology | Helicobacter Pylori | – | SimCLR | Unclear | F1 | 0.9 | − | − |
| Contrastive | 2021 | Aakash Kaku130 | Fundus Image | Ophthalmology | Diabetic retinopathy | – | MoCo, MSE | End-to-end, and Extract features from encoder -> Lnear classifier | AUROC | 0.966 | 0.941 | 0.027 |
| Contrastive | 2021 | Baladitya Yellapragada131 | Fundus Image | Ophthalmology | Macular degeneration | – | Non-Parametric Instance Discrimination | Extract features from encoder -> Weighted KNN | Accuracy | 0.94 | 0.958 | −0.019 |
| Contrastive | 2021 | Shaked Perek132 | Mammogram | Radiology | Breast cancer | – | MoCo | End-to-end | AUROC | 0.754 | 0.68 | 0.109 |
| Contrastive | 2021 | Benoit Dufumier62 | MRI | Psychiatry | Schizophrenia and Bipolar | – | SimCLR (modified) | Extract features from encoder -> Linear classifier | AUROC | 0.968 | 0.942 | 0.028 |
| Contrastive | 2021 | Hongwei Li133 | MRI | Radiology | Brain tumor | – | Contrastive Learning (modified) | Extract features from encoder -> SVM | Sensitivity | 0.92 | 0.888 | 0.036 |
| Contrastive | 2021 | Siladittya Manna134 | MRI | Radiology | Knee injury | – | Constrastive learning (modified) | End-to-end | AUROC | 0.97 | 0.965 | 0.005 |
| Contrastive | 2021 | Sohini Roychowdhury135 | OCT | Ophthalmology | Eye disease | – | SimCLR | Unclear | F1 | 0.999 | – | – |
| Contrastive | 2021 | Zhihang Ren136 | Skin Image | Dermatology | Skin cancer | – | BYOL | Extract features from encoder -> Linear classifer | Accuracy | 0.744 | 0.679 | 0.096 |
| Contrastive | 2021 | Xiyue Wang137 | Ultrasound | Radiology | Kidney tumor | – | InfoNCE | Extract features from encoder -> Linear classifier | F1 | 0.829 | – | – |
| Contrastive | 2021 | Charlie Saillard138 | Whole Slide Image | Pathology | Microsatellite instability | – | MoCo | Extract features from encoder -> Multiple instance learning | AUROC | 0.88 | 0.82 | 0.073 |
| Contrastive | 2021 | Jiajun Li86 | Whole Slide Image | Pathology | Colorectal cancer and healthy tissue types | – | InfoNCE | Extract features from encoder -> Linear classifier | Accuracy | 0.952 | – | – |
| Contrastive | 2021 | Quan Liu139 | Whole Slide Image | Pathology | Skin cancer and healthy tissue types | – | Triplet loss (modified) | Extract features from encoder -> Linear classifier | Accuracy | 0.717 | 0.611 | 0.173 |
| Contrastive | 2021 | Xiyue Wang61 | Whole Slide Image | Pathology | Breast cancer | – | BYOL | End-to-end | AUROC | 0.978 | 0.963 | 0.016 |
| Contrastive | 2021 | Yash Sharmay140 | Whole Slide Image | Pathology | Celiac disease | – | SimCLR | Extract features from encoder -> Linear classifer | AUROC | 0.937 | 0.903 | 0.038 |
| Contrastive | 2021 | Antoine Spahr141 | X-ray | Radiology | Musculoskeletal abnormality | – | InfoNCE | End-to-end | AUROC | 0.78 | 0.701 | 0.113 |
| Contrastive | 2021 | Guang Li142 | X-ray | Radiology | Gastritis | – | Triplet Loss (modified) | Unclear | Sensitivity | 0.9 | 0.771 | 0.167 |
| Contrastive | 2022 | Belal Hossain81 | Chest X-ray | Radiology | COVID-19 | – | SwAV | End-to-end | Accuracy | 0.992 | 0.957 | 0.037 |
| Generative | 2020 | Johnathan Osin57 | MRI | Psychiatry | Psychiatric traits | – | Autoencoder (modified) | Extract features from encoder -> Linear classifer | Accuracy | − | − | − |
| Generative | 2020 | Qingyu Zhao58 | MRI | Radiology | Alzheimers disease | – | Autoencoder (modified) | End-to-end | Accuracy | 0.87 | 0.746 | 0.166 |
| Generative | 2021 | Jevgenij Gamper56 | Whole Slide Image | Pathology | Breast cancer | – | Multiple Instance Captioning | Unclear | Accuracy | 0.9 | 0.723 | 0.245 |
| Innate Relationship | 2019 | Antoine Rivail104 | OCT | Ophthalmology | AMD Progression | – | Siamese Network (modified) | Unclear | AUROC | 0.784 | 0.651 | 0.204 |
| Innate Relationship | 2019 | Richard Droste54 | Ultrasound | Obstetrics & Gynecology | Standard plane detection | – | Other | End-to-end | F1 | 0.766 | 0.67 | 0.143 |
| Innate Relationship | 2020 | Nicolas Ewen48 | CT | Radiology | COVID-19 | – | Predict horizontal flip | Train last layer, then End-to-End | AUROC | 0.861 | 0.785 | 0.097 |
| Innate Relationship | 2020 | Siladittya Manna50 | MRI | Radiology | ACL Tear | – | Jigsaw Puzzle | Replace part of the pretrained model with custom architecture | AUROC | 0.848 | – | – |
| Innate Relationship | 2020 | Jianbo Jiao49 | Ultrasound | Obstetrics & Gynecology | Standard plane detection | – | Reorder slices, Predict Translation | End-to-end | F1 | 0.757 | 0.67 | 0.13 |
| Innate Relationship | 2021 | Asmaa Abbas82 | Chest X-ray | Radiology | COVID-19 | – | Autoencoder (modified) | Freeze low-level layers, fine-tune high-level layers | Accuracy | 0.975 | 0.925 | 0.054 |
| Innate Relationship | 2021 | Anuja Vats44 | Colonoscopy | Gastroenterology | GI abnormality | – | Rotation Prediction | End-to-end | Accuracy | 0.6 | – | – |
| Innate Relationship | 2021 | Nicolas Ewen45 | CT | Radiology | COVID-19 | – | Rotation Prediction | Unclear | Accuracy | 0.867 | 0.9 | -0.037 |
| Innate Relationship | 2021 | Yujie Zhu46 | CT | Radiology | COVID-19 | – | Rotation Prediction, Patch Order Predicction | Unclear | – | – | – | – |
| Innate Relationship | 2021 | Yuting Long47 | CT | Radiology | Pneumonia | – | Rotation Prediction | Extract features from encoder -> Linear Classifier | Accuracy | 0.821 | 0.75 | 0.095 |
| Innate Relationship | 2021 | Fatemeh Taheri Dezaki55 | Echocardiogram | Radiology | Atrial fibrilation | – | Temporal Cycle-Consistency | Extract features from encoder -> Create similarity matrix -> CNN classifier | Accuracy | 0.787 | 0.609 | 0.292 |
| Innate Relationship | 2021 | Satrio Hariomurti Wicaksono51 | Light microscopy image | Obstetrics & Gynecology | Oocyte stage | – | Jigsaw Puzzle | Unclear | Accuracy | 0.919 | 0.911 | 0.009 |
| Innate Relationship | 2021 | Yen Nhi Truong Vu52 | Mammogram | Radiology | Malignancy | – | Jigsaw Puzzle | Unclear | AUROC | 0.962 | 0.954 | 0.008 |
| Innate Relationship | 2021 | Yuki Hashimoto90 | MRI | Psychiatry | Schizophrenia | – | Other | Extract features from encoder -> Linear classifier | Accuracy | 0.778 | – | – |
| Innate Relationship | 2021 | Chetan L. Srinidhi87 | Whole Slide Image | Pathology | Colorectal cancer and healthy tissue types | – | Other | End-to-end | F1 | 0.911 | 0.92 | −0.01 |
| Self-prediction | 2021 | Alex Tamkin143 | Chest X-ray | Radiology | Chest abnormality | – | SHED | Extract features from encoder -> Linear classifier | Accuracy | 0.745 | 0.681 | 0.094 |
| Self-prediction | 2021 | Junghoon Park67 | Chest X-ray | Radiology | COVID-19 | – | Inpainting, Local Pixel Shuffling | End-to-end | Accuracy | 0.986 | 0.975 | 0.011 |
| Self-prediction | 2021 | Ananya Jana65 | CT | Radiology | Liver fibrosis | – | Image Restoration | Extract features from encoder -> MLP | AUROC | 0.847 | – | – |
| Self-prediction | 2021 | Hai Zhong64 | MRI | Radiology | Heart failure | – | Pixel shuffling, Image Restoration | End-to-end | AUROC | 0.768 | – | – |
| Self-prediction | 2021 | Wonsik Jung66 | MRI | Radiology | Autism Spectrum Disorder | – | Masked Autoencoder | Unclear | AUROC | 0.76 | – | – |
| Self-prediction | 2021 | Chengcheng Liu63 | Ultrasound | Radiology | Gastrointenstinal tumor | – | Image Restoration | Extract features from second last layer -> Meta attention module | AUROC | 0.881 | 0.875 | 0.007 |
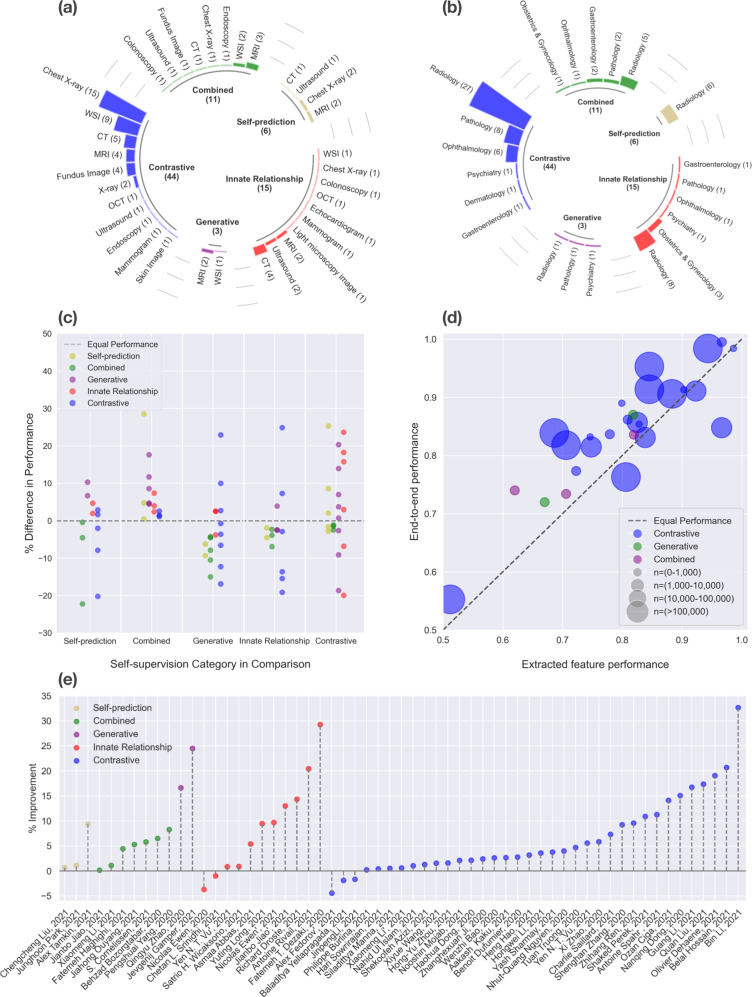
Fig. 4. Summary of extracted data from studies in our system review.
a Prevalence of different medical specialties split by self-supervised learning strategy. b Prevalence of different medical imaging modalities split by self-supervised learning strategy. c Relative performance difference between different types of self-supervised learning strategies on the same task. d Performance comparison between end-to-end fine-tuning vs. training a classifier using extracted features from pre-trained self-supervised models. e Relative difference in downstream task performance between self-supervised and non-self-supervised models.
Innate relationship
Innate relationship was used in 15 out of 79 studies (Table 1). Nine of these studies designed their innate relationship pre-text task based on different image transformations, including rotation prediction44–47, horizontal flip prediction48, reordering shuffled slices49, and patch order prediction46,50–52. Notably, Jiao et al. pre-trained their models simultaneously with two innate relationship pre-text tasks (slice order prediction and geometric transformation prediction), and showed that a weight-sharing Siamese network out-performs a single disentanged model for combining the two pre-training objectives53. The remaining six studies designed clinically relevant pre-text tasks by exploiting the unique properties of medical images. For instance, Droste et al. utilized a gaze tracking dataset and pre-trained a model to predict sonographers’ gazes on ultrasound video frames with gaze-point regression54. Dezaki et al. employed temporal and spatial consistency to produce features for echocardiograms that are strongly correlated with the heart’s inherent cyclic pattern55. Out of all innate relationship based studies, ten compared performance to those of supervised pre-trained models and eight of them showed improvement in performance. On average, clinically relevant pre-text tasks achieved greater improvements in performance over transformation-based pre-text tasks, when compared to purely supervised methods (13.7% vs. 5.03%).
Generative
Generative SSL was used in 3 out of 79 studies (Table 1). Gamper et al. extracted histopathology images from textbooks and published papers along with the figure captions and devised an image captioning task for self-supervised pre-training, where a ResNet-18 was used for encoding images, and the representation was fed to transformers for image-captioning56. They were subsequently able to use the learned representations for a number of downstream histopathology tasks, including breast cancer classification. Osin et al.57 leveraged the chronology of sequential images in brain fMRI for self-supervised pre-training. Brain fMRI scans are typically acquired with subjects alternating between a passive and an active phase, where the subject is instructed to perform some task or receives some external stimulus. During the self-supervision phase, Osin et al. trained two networks: an autoencoder to generate the active fMRI image given the passive image, and an LSTM to predict the next active image. The representations learned during the self-supervision were then used to train a classifier to predict psychiatric traits such as post-traumatic stress disorder (PTSD). Finally, Zhao et al. use a generative approach with an autoencoder with an additional constraint that explicitly associates brain age to the latent representations for longitudinally acquired brain MRIs58. Of the three studies, two reported comparisons with purely supervised models and showed relative improvements of 16.6%58 and 24.5%56 with self-supervised learning.
Contrastive
The majority of the studies that remained after our full-text screening (44/79) used contrastive learning as their self-supervised pre-training strategy (Table 1). SimCLR, MoCo and BYOL were the three most used frameworks, applied in 13, 8, and 3 papers, respectively. Some papers leveraged medical domain priors to create specialized strategies for creating positive pairs. For pathology slices, Li et al. exploited that the neighborhood around a patch is likely to be similar, and used pre-clustering to find dissimilar patches59. In radiology, Ji et al. used multimodal contrastive learning by matching X-rays with corresponding radiology reports60. They extracted and fused the representations of the image and text modalities through both global image-sentence matching and local attention-based region-phrase matching. Wang et al. utilized both radiomic features and deep features from the same image to form positive pairs61. They also utilized the spatial information of the patches, by mining positive pairs from proximate tumor areas and negative pairs from distant tumor areas. Dufumier et al. (2021) used patient meta-data from MRI to form positive pairs62. Thirty-six studies compared contrastive SSL pre-training to supervised pre-training and reported an average improvement in performance of 6.35%.
Self-prediction
Self-prediction was used in six out of all included studies (Table 1). We consider studies that applied local-pixel shuffling as self-prediction since the augmentation operation, which shuffles the order of pixels, is applied only to a random patch of an image. Liu et al. used a U-net model to restore ultrasound images augmented with local-pixel shuffling, and they subsequently concatenated the outputs of the U-net encoder with featurized clinical variables (age, gender, tumor size) for the downstream prediction task63. Similarly, Zhong et al. designed three image restoration tasks on cine-MRI videos and used a U-net-like encoder-decoder architecture including skip connections to perform the image restoration64. Three different image restoration tasks were set up using local-pixel shuffling, within-frame pixel shuffling, and covering an entire video frame with random pixels. Jana et al. used an encoder-decoder architecture for image restoration of CT scans that were corrupted by swapping several small patches within a single CT slice65. Jung et al. created a functional connectivity matrix between pairs of region-of-interest in rs-fMRI for each subject, and created a masked auto-encoder task by randomly masking out different rows and columns of the matrix for restoration66. Two of the five studies compared their approach to models without self-supervised pre-training and reported slight relative improvements in performance of 1.12%67 and 0.690%63.
Combined approaches
Eleven studies found creative ways to combine different self-supervised learning strategies to pre-train their medical imaging models (Table 1). Over half of these studies (6/11) combined contrastive with generative approaches. With the exception of Ke et al.’s work68, which uses a CycleGAN for histopathology slide stain normalization, all studies utilized an autoencoder as their generative model when combined with contrastive strategies. A combination of contrastive and innate relationships was used in three studies. The innate relationship tasks range from augmentation prediction and patch positioning prediction69, rotation prediction70, and ultrasound video to speech correspondence prediction53. For the remaining two studies, Cornelissen et al. used a conditional GAN, and trained the generator network on endoscopic images of the esophagus to recolorize, inpaint and generate super-resolution images71. Because their tasks consisted of both inpainting (self-prediction) and super-resolution (generative), their approach was considered combined. Haghighi et al. combined three different SSL strategies (generative, innate relationship, self-prediction) by first training an auto-encoder and group instances with similar appearances based on the latent representations from the auto-encoder72. Then, the authors randomly cropped image patches at a fixed coordinate for all instances in the same group and assigned a pseudo label to the cropped patches at each coordinate. Finally, the cropped patches were randomly perturbed and a restoration autoencoder was trained simultaneously with a pseudo label classification objective. Eight of the studies that combined different strategies compared self-supervised pre-training with purely supervised approaches, all of which reported performance improvement (0.140–8.29%).
Discussion
This review aims to aggregate the collective knowledge of prior works that applied SSL to medical classification tasks. By synthesizing the relevant literature, we provide consistent definitions for SSL techniques, categorize prior work by their pre-training strategies, and provide implementation guidelines based on lessons learned from prior work. While five studies reported a slight decrease in performance (0.980–4.51%), the majority of self-supervised pre-trained models led to a relative increased performances of 0.216–32.6% AUROC, 0.440–29.2% accuracy, and 0.137–14.3% F1 score over the same model architecture without SSL pre-training, including both ImageNet and random initialization (Fig. 4e). In Fig. 4c we show a comparison of different SSL strategies on the same downstream task, which suggests that a combined strategy tends to outperform other self-supervised categories. However, it is important to note that combined strategies are typically the proposed method in the manuscripts, and thus publication bias might have resulted in this trend. In Fig. 4d we additionally plot the performance of the two main types of fine-tuning strategies on the same task, and the graph tends to indicate that end-to-end fine-tuning leads to better performance regardless of the dataset size. In the presence of relevant data, we recommend implementing self-supervised learning strategies for training medical image classification models since our literature review indicated that self-supervised pre-training generally results in better model performance, especially when annotations are limited (Table 1).
The types of medical images utilized for model development as well as the downstream classification task encompassed a wide range of medical domains and applications (Fig. 4a, b). The vast majority of the studies explored the clinical domain of radiology (47/79), of which 27 were focused on investigating abnormalities on chest imaging such as pneumonia, COVID-19, pleural effusion and pulmonary embolism (see Table 1). The choice of this domain is likely a combination of the availability of large-scale public chest datasets such as CheXpert73, RSPECT74, RadFusion75 and MIMIC-CXR76, as well as the motivation to solve acute or emerging healthcare threats, which was the case during the coronavirus pandemic45,46,48,67,77–83. The second most prevalent clinical domain was pathology (12/79). Similar to radiology, this field is centered around medical imaging in the form of whole slide images. The tasks were focused on histopathological classification, where the majority focused on colorectal cancer classification68,84–87. The remaining studies explored multiple other tasks and many focused on classification of malignant disorders such as breast cancer56,61,88, skin cancer89, and lung cancer59. A particularly interesting medical task that was explored was classification of psychiatric diseases or psychiatric traits using fMRI57,62,90. Current limited knowledge and understanding of possible imaging features arising in psychiatric diseases constitutes a major clinical challenge to making local annotations such as bounding boxes or segmentations on brain scans. In this case both Osin et al. and Hashimoto demonstrated that training a self-supervised framework could be beneficial to generate representative latent features of brain fMRIs before fine-tuning on image-level class labels57,90.
A majority of the included studies lacked strong baselines and ablation experiments. Even though 60 out of 79 studies compared their results with purely supervised baselines, only 33 studies reported comparisons with another self-supervised learning strategy. Of the 33 studies, 26 compared with a self-supervised category that differs from their best performing model. Among the SSL baselines, SimCLR was most frequently compared (16/26), followed by autoencoders (11/26) and MoCo (9/26). Furthermore, only 18 out of 79 studies indicated use of natural image pre-trained weights, either supervised or self-supervised, to initiate their model for subsequent in-domain self-supervised pre-training. Lastly, merely 13 studies compared performance between classification on extracted features to end-to-end fine-tuning, two of which did not report numerical results. Of the 11 studies that quantitatively reported performance, eight found end-to-end fine-tuning to outperform training a new classifier on extracted features (Fig. 4d). Since self-supervised learning for medical images is a promising yet nascent research area and the optimal strategies for training these models are still to be explored, researchers should systematically investigate different categories of self-supervised learning for their medical image datasets, in addition to fine-tuning strategy and pre-trained weights. Researchers should also test their newly developed strategies on multiple datasets, ideally on different modalities and medical imaging domains.
Implementation guidelines
Definitive conclusions on the optimal strategy for medical images cannot be made since only a subset of studies made comparisons between different types of self-supervised learning strategies. Furthermore, the optimal strategy may be dependent on a number of factors including the specific medical imaging domain, the size and complexity of the dataset, and the type of classification task91,92. Due to this heterogeneity, we encourage researchers to compare multiple self-supervised learning strategies for developing medical image classification models, especially in limited data regimes. Although self-supervised pre-training can be computationally demanding, many models pre-trained in a self-supervised manner on large-scale natural image datasets are publicly available and should be utilized. Azizi et al. have shown that SSL pre-trained models using natural images tend to outperform purely supervised pre-trained models93 for medical image classification, and continuing self-supervised pre-training with in-domain medical images leads to the best results. More recently, Azizi et al. found that using generic and large-scale supervised pre-trained models, such as BigTransfer94, can also benefit subsequent domain-specific self-supervised pre-training, and ultimately improve model performance and robustness for different medical imaging modalities95. Truong et al. have demonstrated the effectiveness of combining representations from multiple self-supervised methods to improve performance for three different medical imaging modalities96.
It is worth noting that representations learned using certain SSL strategies can be relatively more linearly separable, while representations from other strategies can achieve better performance when more layers or the entire model are fine-tuned. For instance, for natural image datasets, MoCo outperforms MAE by training a linear model on extracted features (linear probing), while MAE achieves better performance than MoCo as the number of fine-tune layers increases41. Likewise, Cornelissen et al. demonstrated that using representations from earlier layers can improve downstream classification of neoplasia in Barrett’s Esophagus71. Factors such as the degree of domain shift between SSL pre-training data and downstream task data could also affect the linear separability of the representations. Based on our aggregated results, we found that end-to-end fine-tuning generally leads to better performance for medical images (Fig. 4c). However, due to the lack of ablation studies from current work, we cannot determine whether fine-tuning only later layers of the model could lead to better performance relative to end-to-end fine-tuning. Furthermore, even though self-supervised learning strategies generate label-efficient representations, the learning process typically requires a relatively large amount of unlabeled data. For instance, reducing the number of pre-training images from 250k to 50k typically leads to a more than 10.0% drop in accuracy for downstream tasks, while reducing from 1 M to 250k leads to a 2.00–4.00% decrease92. Curating large-scale medical image datasets from multiple institutions is often challenged by the difficulty of sharing patient data while preserving patient privacy. Nevertheless, using federated learning, Yan et al. have demonstrated the possibility of training self-supervised models with data from multiple healthcare centers without the need for explicitly sharing data, and have shown improvement in robustness and performance over models trained using data from only one institution97.
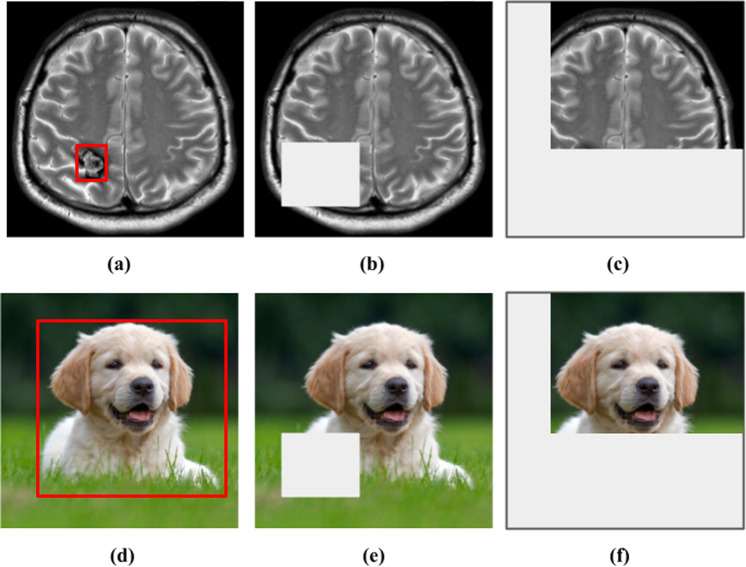
The field of self-supervised learning for computer vision is constantly and rapidly evolving. While many self-supervised methods have led to state-of-the-art results when fine-tuned on natural image datasets, how translatable these results are to medical datasets is unclear, mainly due to the unique properties of medical images. For instance, many contrastive-based strategies have been developed based on the assumption that the class-defining object is the main focus of an image, and thus variations caused by image transformations should not alter the image’s semantic meanings (Fig. 5). Therefore, these methods typically apply strong transformations to the original image and encourage the model to learn similar global representations for images with similar semantic meanings. However, the assumption made for natural images is not necessarily valid for medical images for two reasons. First, medical images have high inter-class visual similarities due to the standardized protocols for medical image acquisition and the homogeneous nature of human anatomy. Second, within the medical imaging domain, the semantic meaning of interest is rarely an object such as the anatomical organ, but is rather the presence or absence of pathological abnormalities within that organ or tissue. Many abnormalities are characterized by very subtle and localized visual cues, which can become ambiguous or obscured by augmentations (Fig. 5c). The random masking operation often utilized by self-prediction self-supervised learning methods may also alter a medical image’s semantic meaning by removing image regions with diseases or abnormalities (Fig. 5b). Recent work has demonstrated the benefit of using learned visual word masks98,99 or spatially constrained crops100,101 to encourage representational invariance with semantically more similar regions of an image. In a similar vein, we believe that augmentation strategies tailored for the nature of medical images during self-supervised learning is a research area that warrants further exploration.
Fig. 5. Examples of augmentations and transformations that alter the semantic meaning of medical images144 but not natural images145.
a The image shows a T2-weighted brain MRI with a cavernoma in the right parietal lobe (bounded in red). b and c Masking and cropping operations can obscure the cavernoma and alter the semantic meaning of the image, as the MRI-scan no longer exhibits any abnormality. d Image of a dog (bounded in red), (e) and (f) Masking and cropping operations do not obscure the dog nor alter the semantic meaning of the image.
The unique properties of medical images can be leveraged to design self-supervised learning methods more suitable for specific downstream tasks. For instance, instead of forming positive pairs with different augmented versions of the same image during contrastive learning, one can improve positive sampling according to the similarity between a patient’s clinical information. In fact, several studies have shown performance improvement when constructing positive pairs with slices from the same CT series102, images from the same imaging study103, images from the same patient93 and images from patients with similar age62. Future research should explore other strategies for defining positive pairs, such as leveraging patient demographics or medical history information. The unique attributes of medical images can also be utilized for creating relevant pre-text tasks. Rivail et al. proposed a self-supervised approach to model disease progression by estimating the time interval between pairs of optical coherence tomography (OCT) scans from the same patient104. Involving additional modalities during self-supervised learning has also been shown to improve a model’s performance when fine-tuned for downstream tasks. For example, Taleb et al. proposed a multimodal contrastive learning strategy between retinal fundus images and genetics data and showed improvement in performance over single modality pre-training105. Jiao et al. cleverly leveraged the correlation between fetal ultrasonography and the narrative speech of the sonographer to create a pre-text task for self-supervision, and subsequently used the learned representations for downstream standard plane classification on sonograms53. Furthermore, many medical imaging modalities have corresponding radiology reports that contain detailed descriptions of the medical conditions observed by radiologists. Several studies have utilized these medical reports to provide supervision signals during self-supervised learning and shown label efficiency for downstream tasks60,106. By leveraging radiology reports, Huang et al. demonstrated the model’s ability to localize chest abnormalities on chest x-rays without segmentation labels and revealed the possibility of zero-shot learning by converting the classification classes into textual captions and framing the image classification task as measuring the image-text similarity107. However, currently there are very few publicly available medical image datasets with corresponding radiology reports, largely due to the difficulties in preserving patient privacy. Therefore, these multi-modal self-supervised learning strategies are limited to implementation within a healthcare system until more datasets with medical image and report pairs are publicly released. Overall, the flexibility in creating self-supervised methods as well as the adaptability and transferability to multiple medical domains highlights the importance and utility of self-supervised techniques in clinical applications.
Limitations
For this review paper, publication bias can be a considerable limitation due to disproportionately reported positive results in the literature, which can lead to overestimation of the benefits of self-supervised learning. We also limited our search to only consider papers published after 2012, which excluded papers that applied self-supervised learning prior to the era of deep learning for computer vision108. Furthermore, we are unable to aggregate or statistically compare the effects of each self-supervised learning strategy on performance gain, since the included studies use different imaging modalities, report different performance metrics, and investigate different objectives. In addition, subjectivity may have been introduced when categorizing the self-supervised learning strategy in each paper, especially for studies that implemented novel, unconventional, or a mixture of methods. Lastly, our study selection criteria only included literature for the task of medical image classification, which limits the scope of this review paper, since we recognize that self-supervised pre-trained models can also be fine-tuned for other important medical tasks, including segmentation, regression, and registration.
Future research
Results from this systematic review have revealed that SSL for medical image classification is a growing and promising field of research across multiple medical disciplines and imaging modalities. We found that self-supervised pre-training generally improves performance for medical imaging classification tasks over purely supervised methods. We categorized the SSL approaches used in medical imaging tasks as a framework for methodologic communication and synthesized benefits and limitations from literature to provide recommendations for future research. Future studies should include direct comparisons of different self-supervised learning strategies using shared terminology and metrics whenever applicable to facilitate the discovery of additional insights and optimal approaches.
Methods
This systematic review was conducted based on the PRISMA guidelines109.
Search strategy
A systematic literature search was implemented in three literature databases: PubMed, Scopus and ArXiv. The key search terms were based on a combination of two major themes: “self-supervised learning” and “medical imaging”. Search terms for medical imaging were not limited to radiological imaging but were also broadly defined to include imaging from all medical fields, i.e., fundus photography, whole slide imaging, endoscopy, echocardiography, etc. Since we specifically wanted to review literature on the task of medical image classification, terms for other computer vision tasks such as segmentation, image reconstruction, denoising and object detection were used as exclusion criteria in the search. The search encompassed papers published between January 2012 and May 2021. The start date was considered appropriate due to the rising popularity of deep learning for computer vision since the 2012 ImageNet challenge. The complete search string for all three databases is provided in Supplementary Methods.
We included all research papers in English that used self-supervision techniques to develop deep learning models for medical image classification tasks. The research papers had to be original research in the form of journal articles, conference proceedings or extended abstracts. We excluded any publications that were not peer-reviewed. Applicable self-supervision techniques were defined according to the different types presented in the “terminology and strategies in self-supervised learning” section. We included only studies that applied the deep learning models to a downstream medical image classification task, i.e, it was not sufficient for the study to have developed a self-supervision model on medical images. In addition, the medical image classification task had to be a clinical task or clinically relevant task. For example, the downstream task of classifying the frame number in a temporal sequence of frames from echocardiography110 was not considered a clinically relevant task.
We excluded studies that used semi-supervised learning or any amount of manually curated labels during the self-supervision step. We also excluded studies that investigated only regression or segmentation in their downstream tasks. Furthermore, we excluded any studies where the self-supervised pre-trained model was not directly fine-tuned for classification after pre-training. Studies that used non-human medical imaging data (i.e., veterinarian medical images) were also excluded.
Study selection
The Covidence software (www.covidence.org) was used for screening and study selection. After the removal of duplicates, studies were screened based on title and abstract, and then full texts were obtained and assessed for inclusion. Study selection was performed by three independent researchers (S.-C.H., A.P., and M.J.), and disagreements were resolved through discussion. In cases where consensus could not be achieved a fourth arbitrating researcher was consulted (A.S.C.).
Data extraction
For benchmarking the existing approaches we extracted the following data from each of the selected articles: (a) self-supervised learning strategy, (b) year of publication, (c) first author, (d) imaging modality, (e) clinical domain, (f) outcome/task, (g) combined method, (h) self-supervised framework, (i) strategy for fine-tuning, (j) performance metrics, (k) SSL performance, (l) supervised performance, and (m) difference in SSL and supervised performance (Table 1). We also computed the relative difference in performance between the supervised and self-supervised model on the p) full dataset and q) subset. We classified the specific self-supervised learning strategy based on the definitions in the section “Terminology and strategies in self-supervised learning”. We extracted AUROC whenever this metric was reported, otherwise we prioritize accuracy over F1 score and sensitivity. When the article contained results from multiple models (i.e., ResNet and DenseNet), metrics from the experiment with the best average performing self-supervised model were extracted. When the authors presented results from several clinical tasks, we chose tasks that best corresponded to the title and objective of the manuscript. If the tasks were deemed equal, we picked the task where the chosen SSL model had the highest performance. We picked supervised baseline with the same model architecture and pre-training dataset for performance comparison. If the author did not report performance from a supervised model that used the same pre-training dataset, preference was given to the ImageNet pre-trained model over a randomly initialized one. The pre-training dataset used by the self-supervised and supervised model are recorded in the Supplementary Table 1. When papers report results on many percentages of fine-tuning (i.e., 1%, 10%, 100%), we pick the lowest and highest to study the label-efficiency of self-supervised learning methods. We also provide in Supplementary Table 1 additional technical details including model architecture, dataset details, number of training samples, comparison to selected baselines and performance on subsets of data. These items were extracted to enable researchers to find and compare current self-supervised studies in their medical field or input modalities of interest.
Supplementary information
Acknowledgements
Research reported in this publication was supported by NIH grants R01 AR077604, R01 EB002524, R01 AR079431, R01 HL155410, R01 LM012966, and P41 EB027060; NIH contracts 75N92020C00008 and 75N92020C00021. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
S.-C.H. and A.P. are co-first authors who contributed equally to this study. Concept and design: S.-C.H. and A.P.. Study selection: S.-C.H. and A.P. Data extraction: S.-C.H., A.P., and M.J. Drafting of the paper: S.-C.H., A.P., and M.J. Critical revision of the paper for important intellectual content: S.-C.H., A.P., M.J., M.P.L., S.Y., and A.S.C. Supervision: S.Y. and A.S.C. share equal supervision.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary Information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shih-Cheng Huang, Anuj Pareek, Serena Yeung, Akshay S. Chaudhari.
Supplementary information
The online version contains supplementary material available at 10.1038/s41746-023-00811-0.
References
- 1.Hong AS, et al. Trends in Diagnostic Imaging Utilization among Medicare and Commercially Insured Adults from 2003 through 2016. Radiology. 2020;294:342–350. doi: 10.1148/radiol.2019191116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith-Bindman R, et al. Trends in Use of Medical Imaging in US Health Care Systems and in Ontario, Canada, 2000-2016. JAMA. 2019;322:843–856. doi: 10.1001/jama.2019.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonald RJ, et al. The effects of changes in utilization and technological advancements of cross-sectional imaging on radiologist workload. Acad. Radiol. 2015;22:1191–1198. doi: 10.1016/j.acra.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat. Rev. Cancer. 2018;18:500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dan Lantsman C, et al. Trend in radiologist workload compared to number of admissions in the emergency department. Eur. J. Radiol. 2022;149:110195. doi: 10.1016/j.ejrad.2022.110195. [DOI] [PubMed] [Google Scholar]
- 6.Alonso-Martínez JL, Sánchez FJA, Echezarreta MAU. Delay and misdiagnosis in sub-massive and non-massive acute pulmonary embolism. Eur. J. Intern. Med. 2010;21:278–282. doi: 10.1016/j.ejim.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Hendriksen JMT, et al. Clinical characteristics associated with diagnostic delay of pulmonary embolism in primary care: a retrospective observational study. BMJ Open. 2017;7:e012789. doi: 10.1136/bmjopen-2016-012789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunnmon JA, et al. Assessment of Convolutional Neural Networks for Automated Classification of Chest Radiographs. Radiology. 2019;290:537–544. doi: 10.1148/radiol.2018181422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajpurkar P, et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med. 2018;15:e1002686. doi: 10.1371/journal.pmed.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larson DB, et al. Performance of a Deep-Learning Neural Network Model in Assessing Skeletal Maturity on Pediatric Hand Radiographs. Radiology. 2018;287:313–322. doi: 10.1148/radiol.2017170236. [DOI] [PubMed] [Google Scholar]
- 11.Park A, et al. Deep Learning–Assisted Diagnosis of Cerebral Aneurysms Using the HeadXNet Model. JAMA Netw. Open. 2019;2:e195600–e195600. doi: 10.1001/jamanetworkopen.2019.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bien N, et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet. PLoS Med. 2018;15:e1002699. doi: 10.1371/journal.pmed.1002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteva A, et al. Prostate cancer therapy personalization via multi-modal deep learning on randomized phase III clinical trials. NPJ Digit. Med. 2022;5:71. doi: 10.1038/s41746-022-00613-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esteva A, et al. Development and validation of a prognostic AI biomarker using multi-modal deep learning with digital histopathology in localized prostate cancer on NRG Oncology phase III clinical trials. J. Clin. Orthod. 2022;40:222–222. [Google Scholar]
- 15.Esteva A, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zech JR, et al. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS Med. 2018;15:e1002683. doi: 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeCun, Y. & Misra, I. Self-supervised learning: The dark matter of intelligence. Meta AI https://ai.facebook.com/blog/self-supervised-learning-the-dark-matter-of-intelligence/ (2021). (Accessed: 17th February 2023).
- 18.Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. Preprint at https://arxiv.org/abs/1810.04805 (2018).
- 19.Brown, T., Mann, B. & Ryder, N. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. (2020).
- 20.Chen T, Kornblith S, Norouzi M, Hinton G. A Simple Framework for Contrastive Learning of Visual Representations. Proc. 37th Int. Conf. Mach. Learn., PMLR. 2020;119:1597–1607. [Google Scholar]
- 21.Krishnan, R., Rajpurkar, P. & Topol, E. J. Self-supervised learning in medicine and healthcare. Nat. Biomed. Eng. (2022). [DOI] [PubMed]
- 22.Shurrab S, Duwairi R. Self-supervised learning methods and applications in medical imaging analysis: a survey. PeerJ Comput. Sci. 2022;8:e1045. doi: 10.7717/peerj-cs.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lilian Weng, J. W. K. Self-Supervised Learning: Self-Prediction and Contrastive Learning. Adv. Neural Inf. Proc. Syst. (2021).
- 24.Gidaris, S., Singh, P. & Komodakis, N. Unsupervised Representation Learning by Predicting Image Rotations. Preprint at https://arxiv.org/abs/1803.07728 (2018).
- 25.Noroozi, M. & Favaro, P. Unsupervised Learning of Visual Representations by Solving Jigsaw Puzzles. Computer Vision–ECCV 2016, Part VI(2016).
- 26.Doersch, C., Gupta, A. & Efros, A. A. Unsupervised Visual Representation Learning by Context Prediction. 2015 IEEE International Conference on Computer Vision (ICCV) (2015).
- 27.Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Kingma, D. P. & Welling, M. Auto-Encoding Variational Bayes. Preprint at https://arxiv.org/abs/1312.6114 (2013).
- 29.Goodfello w, I. J. et al. Generative Adversarial Networks. Preprint at https://community.unix.com/uploads/short-url/oXsmq2VZ9hc2X6hwPRXZRMGbV20.pdf (2014).
- 30.Vincent, P., Larochelle, H., Bengio, Y. & Manzagol, P.-A. Extracting and composing robust features with denoising autoencoders. In Proceedings of the 25th international conference on Machine learning 1096–1103 (2008).
- 31.Donahue J. & Simonyan K. Large scale adversarial representation learning. Adv. Neural Inf. Process. Syst. (2019).
- 32.Donahue, J., Krähenbühl, P. & Darrell, T. Adversarial Feature Learning. Preprint at https://arxiv.org/abs/1605.09782 (2016).
- 33.He, K., Fan, H., Wu, Y., Xie, S. & Girshick, R. Momentum Contrast for Unsupervised Visual Representation Learning. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2020, 9729–9738 (2020).
- 34.Caron, M. et al. Emerging Properties in Self-Supervised Vision Transformers. Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), 2021, 9650–9660 (2021).
- 35.Grill J. B., Strub F., Altché F. & Tallec C. Bootstrap your own latent-a new approach to self-supervised learning. Adv. Neural Inf. Proc. Syst. 33, (2020).
- 36.Chen X, He K. Exploring Simple Siamese Representation Learning. Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit. (CVPR), 2021;2021:15750–15758. [Google Scholar]
- 37.Caron, M. et al. Unsupervised Learning of Visual Features by Contrasting Cluster Assignments. Adv. Neural Inf. Proc. Syst. 33, (2020).
- 38.Asano, Y. M., Rupprecht, C. & Vedaldi, A. Self-labelling via simultaneous clustering and representation learning. Preprint at https://arxiv.org/abs/1911.05371 (2019).
- 39.Gidaris, S., Bursuc, A., Komodakis, N., Perez, P. & Cord, M. Learning Representations by Predicting Bags of Visual Words. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2020).
- 40.Pathak D, Krahenbuhl P, Donahue J, Darrell T, Efros AA. Context Encoders: Feature Learning by Inpainting. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR) 2016;2016:2536–2544. [Google Scholar]
- 41.He K, et al. Masked Autoencoders Are Scalable Vision Learners. Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit. (CVPR) 2022;2022:16000–16009. [Google Scholar]
- 42.Bao, H., Dong, L. & Wei, F. BEiT: BERT Pre-Training of Image Transformers. Preprint at https://arxiv.org/abs/2106.08254 (2021).
- 43.Cunningham P, Delany SJ. k-Nearest Neighbour Classifiers - A Tutorial. ACM Comput. Surv. 2021;54:1–25. doi: 10.1145/3459665. [DOI] [Google Scholar]
- 44.Vats, A., Pedersen, M. & Mohammed, A. A Preliminary Analysis of Self-Supervision for Wireless Capsule Endoscopy. In 2021 9th European Workshop on Visual Information Processing (EUVIP) 1–6 (2021).
- 45.Ewen, N. & Khan, N. Online Unsupervised Learning For Domain Shift In Covid-19 CT Scan Datasets. In 2021 IEEE International Conference on Autonomous Systems (ICAS) 1–5 (2021).
- 46.Zhu, Y. Self-supervised Learning for Small Shot COVID-19 Classification. In 2021 3rd International Conference on Information Technology and Computer Communications 36–40 (2021).
- 47.Long, Y. Pneumonia Identification with Self-supervised Learning and Transfer Learning. In Application of Intelligent Systems in Multi-modal Information Analytics 627–635 (2021).
- 48.Ewen, N. & Khan, N. Targeted Self Supervision For Classification On A Small Covid-19 Ct Scan Dataset. In 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) 1481–1485 (2021).
- 49.Jiao, J., Droste, R., Drukker, L., Papageorghiou, A. T. & Alison Noble, J. Self-supervised Representation Learning for Ultrasound Video. In 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI) (2020). [DOI] [PMC free article] [PubMed]
- 50.Manna S, Bhattacharya S, Pal U. Self-Supervised Representation Learning for Detection of ACL Tear Injury in Knee MR Videos. Pattern Recognit. Lett. 2022;154:37–43. doi: 10.1016/j.patrec.2022.01.008. [DOI] [Google Scholar]
- 51.Wicaksono, R. S. H., Septiandri, A. A. & Jamal, A. Human Embryo Classification Using Self-Supervised Learning. In 2021 2nd International Conference on Artificial Intelligence and Data Sciences (AiDAS) 1–5 (2021).
- 52.Vu YNT, Tsue T, Su J, Singh S. An improved mammography malignancy model with self-supervised learning. Med. Imaging 2021: Comput-Aided Diagn. 2021;11597:210–216. [Google Scholar]
- 53.Jiao, J. et al. Self-Supervised Contrastive Video-Speech Representation Learning for Ultrasound. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2020 534–543 (2020). [DOI] [PMC free article] [PubMed]
- 54.Droste, R. et al. Ultrasound Image Representation Learning by Modeling Sonographer Visual Attention. In International conference on information processing in medical imaging 2019 (2019). [DOI] [PMC free article] [PubMed]
- 55.Dezaki, F. T. et al. Echo-Rhythm Net: Semi-Supervised Learning For Automatic Detection of Atrial Fibrillation in Echocardiography. In 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) 110–113 (2021).
- 56.Gamper, J. & Rajpoot, N. Multiple Instance Captioning: Learning Representations from Histopathology Textbooks and Articles. In 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 16544–16554 (2021).
- 57.Osin, J. et al. Learning Personal Representations from fMRI by Predicting Neurofeedback Performance. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2020 469–478 (2020).
- 58.Zhao, Q., Liu, Z., Adeli, E. & Pohl, K. M. Longitudinal Self-Supervised Learning. Med. Image Anal. 71, (2021). [DOI] [PMC free article] [PubMed]
- 59.Li, B., Li, Y. & Eliceiri, K. W. Dual-stream Multiple Instance Learning Network for Whole Slide Image Classification with Self-supervised Contrastive Learning. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. (2021). [DOI] [PMC free article] [PubMed]
- 60.Ji, Z. et al. Improving Joint Learning of Chest X-Ray and Radiology Report by Word Region Alignment. In Machine Learning in Medical Imaging 110–119 (2021). [DOI] [PMC free article] [PubMed]
- 61.Wang, X. et al. TransPath: Transformer-Based Self-supervised Learning for Histopathological Image Classification. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2021 186–195 (2021).
- 62.Dufumier, B. et al. Contrastive Learning with Continuous Proxy Meta-data for 3D MRI Classification. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2021 58–68 (2021).
- 63.Liu C, et al. TN-USMA Net: Triple normalization-based gastrointestinal stromal tumors classification on multicenter EUS images with ultrasound-specific pre-training and meta attention. Med. Phys. 2021;48:7199–7214. doi: 10.1002/mp.15172. [DOI] [PubMed] [Google Scholar]
- 64.Zhong, H. et al. A Self-supervised Learning Based Framework for Automatic Heart Failure Classification on Cine Cardiac Magnetic Resonance Image. In 2021 43rd Annual International Conference of the IEEE Engineering in Medicine Biology Society (EMBC) 2887–2890 (2021). [DOI] [PubMed]
- 65.Jana, A. et al. Liver Fibrosis And NAS Scoring From CT Images Using Self-Supervised Learning And Texture Encoding. In 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) 1553–1557 (2021).
- 66.Jung, W., Heo, D.-W., Jeon, E., Lee, J. & Suk, H.-I. Inter-regional High-Level Relation Learning from Functional Connectivity via Self-supervision. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2021 284–293 (2021).
- 67.Park J, Kwak I-Y, Lim C. A Deep Learning Model with Self-Supervised Learning and Attention Mechanism for COVID-19 Diagnosis Using Chest X-ray Images. Electronics. 2021;10:1996. doi: 10.3390/electronics10161996. [DOI] [Google Scholar]
- 68.Ke, J., Shen, Y., Liang, X. & Shen, D. Contrastive Learning Based Stain Normalization Across Multiple Tumor in Histopathology. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2021 571–580 (Springer International Publishing, 2021).
- 69.Tian, Y. et al. Constrained Contrastive Distribution Learning for Unsupervised Anomaly Detection and Localisation in Medical Images. Preprint at https://arxiv.org/abs/2103.03423 (2021).
- 70.Li X, et al. Rotation-Oriented Collaborative Self-Supervised Learning for Retinal Disease Diagnosis. IEEE Trans. Med. Imaging. 2021;40:2284–2294. doi: 10.1109/TMI.2021.3075244. [DOI] [PubMed] [Google Scholar]
- 71.Cornelissen S. & van der Putten J. A.. Evaluating self-supervised learning methods for downstream classification of neoplasia In barrett’s esophagus. 2021 IEEE International Conference on Image Processing (ICIP), 66–70 (2021).
- 72.Haghighi, F., Taher, M. R. H., Zhou, Z., Gotway, M. B. & Liang, J. Transferable Visual Words: Exploiting the Semantics of Anatomical Patterns for Self-supervised Learning. IEEE Trans. Med. Imaging, (2021). [DOI] [PMC free article] [PubMed]
- 73.Irvin J, et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. Proc. AAAI Conf. Artif. Intell. 2019;33:590–597. [Google Scholar]
- 74.Colak E, et al. The RSNA Pulmonary Embolism CT Dataset. Radio. Artif. Intell. 2021;3:e200254. doi: 10.1148/ryai.2021200254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou, Y. et al. RadFusion: Benchmarking Performance and Fairness for Multimodal Pulmonary Embolism Detection from CT and EHR. Preprint at https://arxiv.org/abs/2111.11665 (2021).
- 76.Johnson, A. E. W. et al. MIMIC-CXR-JPG, a large publicly available database of labeled chest radiographs. Preprint at https://arxiv.org/abs/1901.07042 (2019). [DOI] [PMC free article] [PubMed]
- 77.Sun, L., Yu, K. & Batmanghelich, K. Context Matters: Graph-based Self-supervised Representation Learning for Medical Images. in Proc Conf AAAI Artif Intell. (2021). [PMC free article] [PubMed]
- 78.Hao, H., Didari, S., Woo, J. O., Moon, H. & Bangert, P. Highly Efficient Representation and Active Learning Framework for Imbalanced Data and its Application to COVID-19 X-Ray Classification. Conference on Neural Information Processing Systems (NeurIPS 2021) (2021).
- 79.Li J, et al. Multi-task contrastive learning for automatic CT and X-ray diagnosis of COVID-19. Pattern Recognit. 2021;114:107848. doi: 10.1016/j.patcog.2021.107848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dong, N. & Voiculescu, I. Federated Contrastive Learning for Decentralized Unlabeled Medical Images. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2021 378–387 (2021).
- 81.Hossain MB, Iqbal SMHS, Islam MM, Akhtar MN, Sarker IH. Transfer learning with fine-tuned deep CNN ResNet50 model for classifying COVID-19 from chest X-ray images. Inf. Med Unlocked. 2022;30:100916. doi: 10.1016/j.imu.2022.100916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abbas A, Abdelsamea MM, Gaber MM. 4S-DT: Self-Supervised Super Sample Decomposition for Transfer Learning With Application to COVID-19 Detection. IEEE Trans. Neural Netw. Learn Syst. 2021;32:2798–2808. doi: 10.1109/TNNLS.2021.3082015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang, S., Zou, B., Xu, B., Su, J. & Hu, H. An Efficient Deep Learning Framework of COVID-19 CT Scans Using Contrastive Learning and Ensemble Strategy. In 2021 IEEE International Conference on Progress in Informatics and Computing (PIC) 388–396 (2021).
- 84.Yang, P., Hong, Z., Yin, X., Zhu, C. & Jiang, R. Self-supervised Visual Representation Learning for Histopathological Images. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2021 47–57 (Springer International Publishing, 2021).
- 85.Ciga, O., Xu, T. & Martel, A. L. Self supervised contrastive learning for digital histopathology. Machine Learning with Applications7, (2022).
- 86.Li, J., Lin, T. & Xu, Y. SSLP: Spatial Guided Self-supervised Learning on Pathological Images. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2021 3–12 (2021).
- 87.Srinidhi, C. L., Kim, S. W., Chen, F.-D. & Martel, A. L. Self-supervised driven consistency training for annotation efficient histopathology image analysis. Med. Image Anal. 75, (2021). [DOI] [PubMed]
- 88.Dehaene, O., Camara, A., Moindrot, O., de Lavergne, A. & Courtiol, P. Self-Supervision Closes the Gap Between Weak and Strong Supervision in Histology. in ML4H 2020 NeurIPS workshop (2020).
- 89.Liu, Q. et al. SimTriplet: Simple Triplet Representation Learning with a Single GPU. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2021. Part II 24, 102–112 (2021).
- 90.Hashimoto Y, Ogata Y, Honda M, Yamashita Y. Deep Feature Extraction for Resting-State Functional MRI by Self-Supervised Learning and Application to Schizophrenia Diagnosis. Front. Neurosci. 2021;15:696853. doi: 10.3389/fnins.2021.696853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kotar, K., Ilharco, G. & Schmidt, L. Contrasting contrastive self-supervised representation learning pipelines. Proc. Estonian Acad. Sci. Biol. Ecol.
- 92.Cole E, Yang X, Wilber K, Aodha OM, Belongie S. When Does Contrastive Visual Representation Learning Work? Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit. (CVPR) 2022;2022:14755–14764. [Google Scholar]
- 93.Azizi, S., Mustafa, B., Ryan, F. & Beaver, Z. Big self-supervised models advance medical image classification. Proc. Estonian Acad. Sci. Biol. Ecol.
- 94.Kolesnikov, A. et al. Big Transfer (BiT): General Visual Representation Learning. Computer Vision–ECCV 2020: 16th European Conference, Part V 16. (2020).
- 95.Azizi, S. et al. Robust and Efficient Medical Imaging with Self-Supervision. Preprint at https://arxiv.org/abs/2205.09723 (2022).
- 96.Truong, T. et al. 158 54–74 (2021).
- 97.Yan, R. et al. Label-Efficient Self-Supervised Federated Learning for Tackling Data Heterogeneity in Medical Imaging. IEEE Transactions on Medical Imaging (2023). [DOI] [PMC free article] [PubMed]
- 98.Shi, Y., Siddharth, N., Torr, P. H. S. & Kosiorek, A. R. Adversarial Masking for Self-Supervised Learning. In International Conference on Machine Learning, 20026–20040. (2022).
- 99.Li, G. et al. SemMAE: Semantic-Guided Masking for Learning Masked Autoencoders. Preprint at https://arxiv.org/abs/2206.10207 (2022).
- 100.Van Gansbeke X, Vandenhende S. Revisiting contrastive methods for unsupervised learning of visual representations. Adv. Neural Inf. Process. Syst. 2021;34:16238–16250. [Google Scholar]
- 101.Peng, X., Wang, K., Zhu, Z. & Wang, M. Crafting better contrastive views for siamese representation learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 16031–16040 (2022).
- 102.Dong, H. et al. Case Discrimination: Self-supervised Feature Learning for the Classification of Focal Liver Lesions. In Innovation in Medicine and Healthcare 241–249 (2021).
- 103.Vu, Y. N. T. et al. MedAug: Contrastive learning leveraging patient metadata improves representations for chest X-ray interpretation. In Proceedings of Machine Learning Research 126, 1–14 (2021).
- 104.Rivail, A. et al. Modeling Disease Progression in Retinal OCTs with Longitudinal Self-supervised Learning. in Predictive Intelligence in Medicine 44–52 (2019).
- 105.Taleb A., Kirchler M. & Monti R. ContIG: Self-supervised Multimodal Contrastive Learning for Medical Imaging with Genetics. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 20908–20921 (2022).
- 106.Zhang, Y., Jiang, H., Miura, Y., Manning, C. D. & Langlotz, C. P. Contrastive Learning of Medical Visual Representations from Paired Images and Text. Preprint at https://arxiv.org/abs/2010.00747 (2020).
- 107.Huang, S.-C., Shen, L., Lungren, M. P. & Yeung, S. GLoRIA: A multimodal global-local representation learning framework for label-efficient medical image recognition. In 2021 IEEE/CVF International Conference on Computer Vision (ICCV) (2021).
- 108.Russakovsky O, et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015;115:211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 109.Page, M. J. et al. “The PRISMA 2020 statement: an updated guideline for reporting systematic reviews.” International journal of surgery. 88 (2021). [DOI] [PubMed]
- 110.Danu, M., Ciuşdel, C. F. & Itu, L. M. Deep learning models based on automatic labeling with application in echocardiography. In 2020 24th International Conference on System Theory, Control and Computing (ICSTCC) 373–378 (2020).
- 111.Bozorgtabar, B., Mahapatra, D., Vray, G. & Thiran, J.-P. SALAD: Self-supervised Aggregation Learning for Anomaly Detection on X-Rays. in Medical Image Computing and Computer Assisted Intervention – MICCAI2020 468–478 (Springer International Publishing, 2020).
- 112.Hsieh W-T, Lefort-Besnard J, Yang H-C, Kuo L-W, Lee C-C. Behavior Score-Embedded Brain Encoder Network for Improved Classification of Alzheimer Disease Using Resting State fMRI. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2020;2020:5486–5489. doi: 10.1109/EMBC44109.2020.9175312. [DOI] [PubMed] [Google Scholar]
- 113.Tian, Y. et al. Constrained Contrastive Distribution Learning for Unsupervised Anomaly Detection and Localisation in Medical Images. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2021 128–140 (2021).
- 114.Fedorov, A. et al. Tasting the cake: evaluating self-supervised generalization on out-of-distribution multimodal MRI data. In RobustML Workshop ICLR 2021 (2021).
- 115.Ouyang, J. et al. Self-supervised Longitudinal Neighbourhood Embedding. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2021 80–89 (Springer International Publishing, 2021). [DOI] [PMC free article] [PubMed]
- 116.Sowrirajan H, Yang J, Ng AY, Rajpurkar P. MoCo-CXR: MoCo Pre-training Improves Representation and Transferability of Chest X-ray Models. Proc. Mach. Learn. Res. 2021;143:727–743. [Google Scholar]
- 117.Zhou, H.-Y. et al. Comparing to Learn: Surpassing ImageNet Pre-training on Radiographs by Comparing Image Representations. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2020 398–407 (2020).
- 118.Burlina P, et al. Low-Shot Deep Learning of Diabetic Retinopathy With Potential Applications to Address Artificial Intelligence Bias in Retinal Diagnostics and Rare Ophthalmic Diseases. JAMA Ophthalmol. 2020;138:1070–1077. doi: 10.1001/jamaophthalmol.2020.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li X, Jia M, Islam MT, Yu L, Xing L. Self-Supervised Feature Learning via Exploiting Multi-Modal Data for Retinal Disease Diagnosis. IEEE Trans. Med. Imaging. 2020;39:4023–4033. doi: 10.1109/TMI.2020.3008871. [DOI] [PubMed] [Google Scholar]
- 120.Fedorov, A. et al. On self-supervised multi-modal representation learning: An application to Alzheimer’s disease. In IEEE 18th International Symposium on Biomedical Imaging (2021).
- 121.Mojab, N. et al. Real-World Multi-Domain Data Applications for Generalizations to Clinical Settings. In 2020 19th IEEE International Conference on Machine Learning and Applications (ICMLA) 677–684 (2020).
- 122.Reed, C. J. et al. Self-Supervised Pre-training Improves Self-Supervised Pre-training. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (2022).
- 123.Liu, F. et al. Self-supervised Mean Teacher for Semi-supervised Chest X-Ray Classification. In Machine Learning in Medical Imaging 426–436 (2021).
- 124.Gazda M, Gazda J, Plavka J, Drotar P. Self-supervised deep convolutional neural network for chest X-ray classification. IEEE Access. 2021;9:151972–151982. doi: 10.1109/ACCESS.2021.3125324. [DOI] [Google Scholar]
- 125.Nguyen, N.-Q. & Le, T.-S. A Semi-Supervised Learning Method to Remedy the Lack of Labeled Data. In 2021 15th International Conference on Advanced Computing and Applications (ACOMP) 78–84 (2021).
- 126.Zhao, X. & Zhou, S. Fast Mixing of Hard Negative Samples for Contrastive Learning and Use for COVID-19. In 2021 4th International Conference on Big Data Technologies 6–12 (2021).
- 127.Islam NU, Gehlot S, Zhou Z, Gotway MB, Liang J. Seeking an Optimal Approach for Computer-Aided Pulmonary Embolism Detection. Mach. Learn Med. Imaging. 2021;12966:692–702. doi: 10.1007/978-3-030-87589-3_71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bao, W., Jin, Y., Huang, C. & Peng, W. CT Image Classification of Invasive Depth of Gastric Cancer based on 3D-DPN Structure. In The 11th International Workshop on Computer Science and Engineering (WCSE 2021) 115–121 (2021).
- 129.Jian, G.-Z., Lin, G.-S., Wang, C.-M. & Yan, S.-L. Helicobacter Pylori Infection Classification Based on Convolutional Neural Network and Self-Supervised Learning. In 2021 the 5th International Conference on Graphics and Signal Processing 60–64 (2021).
- 130.Kaku, A., Upadhya, S. & Razavian, N. Intermediate layers matter in momentum contrastive self supervised learning. In 35th Conference on Neural Information Processing Systems (NeurIPS 2021). (2021).
- 131.Yellapragada B, Hornauer S, Snyder K, Yu S, Yiu G. Self-Supervised Feature Learning and Phenotyping for Assessing Age-Related Macular Degeneration Using Retinal Fundus Images. Ophthalmol. Retin. 2022;6:116–129. doi: 10.1016/j.oret.2021.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Perek, S., Amit, M. & Hexter, E. Self Supervised Contrastive Learning on Multiple Breast Modalities Boosts Classification Performance. In Predictive Intelligence in Medicine 117–127 (2021).
- 133.Li, H. et al. Imbalance-Aware Self-supervised Learning for 3D Radiomic Representations. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2021 36–46 (2021).
- 134.Manna, S., Bhattacharya, S. & Pal, U. Interpretive self-supervised pre-training: boosting performance on visual medical data. In Proceedings of the Twelfth Indian Conference on Computer Vision, Graphics and Image Processing 1–9 (2021).
- 135.Roychowdhury, S., Tang, K. S., Ashok, M. & Sanka, A. SISE-PC: Semi-supervised Image Subsampling for Explainable Pathology Classification. In 2021 43rd Annual International Conference of the IEEE Engineering in Medicine Biology Society (EMBC) 2806–2809 (2021). [DOI] [PubMed]
- 136.Ren, Z., Guo, Y., Yu, S. X. & Whitney, D. Improve Image-based Skin Cancer Diagnosis with Generative Self-Supervised Learning. In 2021 IEEE/ACM Conference on Connected Health: Applications, Systems and Engineering Technologies (CHASE) 23–34 (2021).
- 137.Zhao, Z. & Yang, G. Unsupervised Contrastive Learning of Radiomics and Deep Features for Label-Efficient Tumor Classification. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2021 252–261 (2021).
- 138.Saillard, C. et al. Self supervised learning improves dMMR/MSI detection from histology slides across multiple cancers. Preprint at https://arxiv.org/abs/2109.05819 (2021).
- 139.Liu, Q. et al. SimTriplet: Simple Triplet Representation Learning with a Single GPU. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2021 102–112 (2021).
- 140.Sharmay, Y., Ehsany, L., Syed, S. & Brown, D. E. HistoTransfer: Understanding Transfer Learning for Histopathology. In 2021 IEEE EMBS International Conference on Biomedical and Health Informatics (BHI) 1–4 (2021).
- 141.Spahr, A., Bozorgtabar, B. & Thiran, J.-P. Self-Taught Semi-Supervised Anomaly Detection On Upper Limb X-Rays. In 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) 1632–1636 (2021).
- 142.Li, G., Togo, R., Ogawa, T. & Haseyama, M. Triplet Self-Supervised Learning for Gastritis Detection with Scarce Annotations. In 2021 IEEE 10th Global Conference on Consumer Electronics (GCCE) 787–788 (2021).
- 143.Tamkin, A. et al. DABS: A Domain-Agnostic Benchmark for Self-Supervised Learning. In NeurIPS 2021 Datasets and Benchmarks Track (2021).
- 144.Hellerhoff. File:Kavernom rechts parietal 59M - MR - 001.jpg. Wikimediahttps://commons.wikimedia.org/wiki/File:Kavernom_rechts_parietal_59M_-_MR_-_001.jpg (2022).
- 145.Matio, H. File:Dog Breeds.jpg. Wikimedia Commonshttps://commons.wikimedia.org/wiki/File:Dog_Breeds.jpg (2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary Information files.