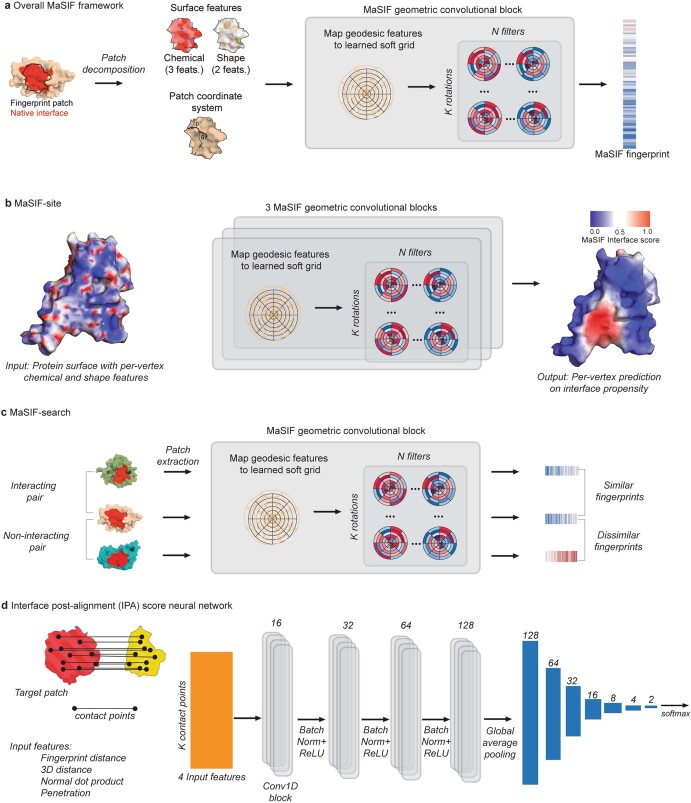
Extended Data Fig. 1. Overview of the neural network architectures used in the MaSIF protocols.
a, General MaSIF framework. Molecular surfaces are decomposed into patches which are annotated with chemical and shape features. The MaSIF network translates these input features into fingerprints that describe the original surface patch. b, MaSIF-site neural network. MaSIF-site predicts partner-independent protein interface propensities based on per-vertex chemical and shape features of the protein surface. c, MaSIF-search neural network. MaSIF-search embeds protein patches into a space where complementary patches are close to each other. The network was trained on discriminating interacting patches from non-interacting protein surface patches. The network uses MaSIF fingerprints to identify which are compatible and therefore to predict likely interacting proteins. d, Interface post-alignment (IPA) scoring neural network. The IPA scoring neural network enables the scoring of protein interfaces based on several input features: fingerprint distance between contacting points, 3D distance of corresponding points, normal dot product, and the distance between surface points in the seed and the closest atom in the target, which we call ‘penetration’.