ABSTRACT
The study of fusobacterial virulence factors has dramatically benefited from the creation of various genetic tools for DNA manipulation, including galK-based counterselection for in-frame deletion mutagenesis in Fusobacterium nucleatum, which was recently developed. However, this method requires a host lacking the galK gene, which is an inherent limitation. To circumvent this limitation, we explored the possibility of using the hicA gene that encodes a toxin consisting of a HicAB toxin-antitoxin module in Fusobacterium periodonticum as a new counterselective marker. Interestingly, the full-length hicA gene is not toxic in F. nucleatum, but a truncated hicA gene version lacking the first six amino acids is functional as a toxin. The toxin expression is driven by an rpsJ promoter and is controlled at its translational level by using a theophylline-responsive riboswitch unit. As a proof of concept, we created markerless in-frame deletions in the fusobacterial adhesin radD gene within the F. nucleatum rad operon and the tnaA gene that encodes the tryptophanase for indole production. After vector integration, plasmid excision after counterselection appeared to have occurred in 100% of colonies grown on theophylline-added plates and resulted in in-frame deletions in 50% of the screened isolates. This hicA-based counterselection system provides a robust and reliable counterselection in wild-type background F. nucleatum and should also be adapted for use in other bacteria.
IMPORTANCE Fusobacterium nucleatum is an indole-producing human oral anaerobe associated with periodontal diseases, preterm birth, and several cancers. Little is known about the mechanisms of fusobacterial pathogenesis and associated factors, mainly due to the lack of robust genetic tools for this organism. Here, we showed that a mutated hicA gene from Fusobacterium periodonticum expresses an active toxin and was used as a counterselection marker. This hicA-based in-frame deletion system efficiently creates in-frame deletion mutations in the wild-type background of F. nucleatum. This is the first report to use the hicA gene as a counterselection marker in a bacterial genetic study.
KEYWORDS: HicA, toxin-antitoxin, counterselection marker, Fusobacterium nucleatum, in-frame deletion system
INTRODUCTION
Fusobacterium nucleatum is a Gram-negative, indole-producing microorganism naturally isolated from the human oral cavity and is especially abundant within the subgingival plaque biofilm composed of more than 400 bacterial species (1, 2). Using three major surface adhesin proteins, FadA (3, 4), RadD (5, 6), and Fap2 (7, 8), F. nucleatum physically interacts with the host and many other early or late colonizers in oral biofilm, making its presence tightly associated with the onset and development of periodontal disease (2, 9, 10). In addition, evidence has shown that F. nucleatum plays an essential role in extraoral diseases like preterm birth (11, 12) and colorectal cancer (13–16). To better understand the role of this bacterium in various disorders, it is necessary to have comprehensive knowledge of F. nucleatum’s biology and pathogenesis. Such knowledge has been dramatically expanded in the past 15 years due to many genetic tools being adapted for use in studies of this organism. There are two ways to perform genetic studies of F. nucleatum. One way is to use the Tn5-based mutagenesis to produce many random gene mutants in one single assay (7, 17). Another example is to use the defined genetic mutation that makes a single mutant each time. The defined mutations are usually engineered using three methods, (i) plasmid insertion duplication mutagenesis (5, 18), (ii) allelic exchange mutagenesis with an antibiotic cassette (3), and (iii) markerless in-frame deletion (17, 19). The first two methods are easy to carry out and were primarily employed in early studies of F. nucleatum. But both have an obvious drawback, frequently creating undesirable polar effects on downstream genes. In addition, the limited number of available selectable markers limits the number of mutations in a strain. However, these issues could be overcome by developing a markerless in-frame deletion system.
The in-frame deletion procedure is a two-step process. First, two similarly sized fragments that are homologous to sequences flanking the target locus are PCR amplified. The two fragments are ligated adjacent to each other in a suicide vector. This construct is subsequently integrated onto the chromosome via homologous recombination and selected using a positive selection pressure, often with antibiotic resistance. Then, the selected simple recombinants are grown without any selective pressure to allow second recombination events and plasmid loss. Next, plasmid-excision/loss recombinants are screened for a second recombination event that occurs between the duplicated homologous chromosomal region, which is created after the initial plasmid recombination event. Since the second recombination event is rare, screening these recombinants that have undergone the double crossover is time-consuming. Counterselection strategies are often introduced to facilitate this process to create markerless in-frame deletions (20).
For this purpose, we introduced galK as a counterselection marker in F. nucleatum (17). The galK gene encodes galactokinase, a key enzyme in galactose metabolism. This enzyme phosphorylates d-galactose to generate galactose-1-phosphate and can also convert 2-deoxy-d-galactose (2-DG) into 2-deoxygalctose-1-phosphate, which is not further metabolized and is toxic to the cell. F. nucleatum carries a copy of the galK gene. To use galK for counterselection, a galK-minus recipient strain was constructed. An in-frame deletion vector was generated in which the galK gene from F. nucleatum or Clostridium acetobutylicum was cloned, and a strong and constitutive promoter controlled its expression. If the recipient strain lacks galactose kinase (galK) and phosphate uridylyltransferase (galT), galactose can be used instead of the more expensive 2-deoxy-d-galactose molecule for selection (19). The galK-based counterselection system consists of a galK minus or galKT minus recipient strain and an in-frame deletion vector, which results in a strong negative selection in F. nucleatum. This shows that all viable strains, when selected with 2-deoxy-d-galactose, undergo the second recombination to excise the plasmid. This system successfully generated many fusobacterial in-frame deletions in our lab and other groups (6, 17, 19, 21). However, the apparent limitation of this system is the modified mutant recipient strain requirement. The intermediate product, UDP-galactose from galactose utilization, is vital in cell envelope maintenance in bacteria (22, 23). The use of galactose present in a culture medium or animal hosts by F. nucleatum could change its physiology, as it would potentially module the function of Fap2, an important adhesin that binds galactose and is responsible for cell-cell coaggregation and cell adhesion (7, 8). Consistently, we identified several cases in which the recipient background impacted the phenotypes of our markerless in-frame deletion strains in our studies (our unpublished data). Thus, a new counterselection marker is urgently needed to replace galK for genetic studies in F. nucleatum.
The HicAB is a type II toxin-antitoxin (TA) module that is ubiquitously present in bacteria (24) and is involved in bacterial persistence and chronic infections (25). HicA is a type of RNase that can cause cell growth to stop when expressed without cognate antitoxin HicB. There has only been one study on the TA system in the genus Fusobacterium, and it is related to a plasmid addition system (Txf-Axf) (26). We tried to explore the possibility of using one of the hicA genes from fusobacterial species as a counterselection marker.
In this study, we report the development of a HicA toxin gene-based in-frame deletion system in F. nucleatum. We tested this system to construct two markerless in-frame deletions in the fusobacterial adhesin gene radD and indole-producing gene tnaA. Following counterselection, plasmid loss occurred in every colony tested, while half of the screened isolates grown on plates with theophylline were in-frame deletion mutants. To our knowledge, this is the first report of using a hicA as a counterselection in bacteria.
RESULTS
A putative mutated HicA toxin from F. periodonticum is functional in F. nucleatum.
In previous studies, we used the galK gene as a counterselection marker to efficiently create in-frame deletion mutations in F. nucleatum (17). However, the galK-based allelic exchange system has an obvious limitation: the host cell must be galK minus. Without this gene, bacteria were unable to utilize galactose, which is crucial for survival and virulence in certain conditions. This can affect the characteristics of the resulting mutant derivatives. To address this issue, in this study, we sought to find a new counterselection marker to replace the galK gene for genetic manipulations in F. nucleatum.
When beginning our search for a likely candidate, we considered the toxin-antitoxin modules. The toxin-antitoxin systems are widespread in bacteria and typically consist of two components, a toxin that inhibits cell growth and an antitoxin that counteracts the cognate toxin (27). The toxin gene mazF in the toxin-antitoxin MazEF modules from Escherichia coli has been used for counterselection in various bacteria when mazF expression was in highly adjustable controls, often controlled by an inducible promoter (28–33). We first attempted to use mazF as a counterselection marker in F. nucleatum but were unsuccessful. Then, we switched our attention to the HicA toxin gene in the HicAB toxin-antitoxin module. The HicAB system has been well studied in E. coli (34), Burkholderia pseudomallei (35), and Streptococcus mutans (36). HicA toxins are typically smaller than MazF toxins. MazF in E. coli has 111 amino acids, and HicA has 58. HicA degrades both mRNA and transfer mRNA, halting protein translation and cell growth. MzaF only cleaves mRNA. Coexpression of HicB was able to counteract HicA-induced bacterial growth arrest (36, 37). Since our lab is interested in the biological roles of toxin-antitoxin systems in fusobacterial species, we plan to use an endogenous HicA homolog from the genus Fusobacterium and test if it could be employed as a counterselection marker. This way, the codon bias issue can be efficiently avoided during the ectopic expression of the candidate marker gene in F. nucleatum, a low G+C bacterium. Protein expression in a host bacterium is heavily influenced by GC content.
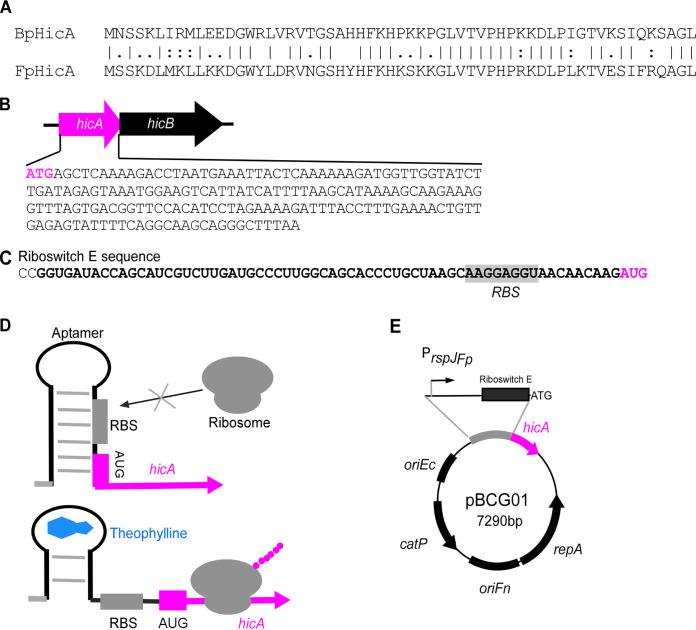
First, we identified hicA gene homologs by performing BLAST searches with the hicA gene in B. pseudomallei (BphicA) as a query against the genome sequence of Fusobacterium periodonticum, another oral bacterium. A hicA homolog was found in F. periodonticum and was named FpHicA (see Fig. 1A). It encodes a putative 59-amino-acid polypeptide by the gene of FperA3_010100004605 (https://img.jgi.doe.gov) with 61% identity and 80% similarity to BpHicA. The downstream gene of FphicA is hicB, which encodes a putative antitoxin protein for neutralization of HicA toxicity (Fig. 1B). Notably, the genome of F. nucleatum subsp. nucleatum ATCC 23726, our genetic working strain, contains a putative 63-amino-acid peptide HicA homolog (FnHicA) encoded by the gene HMPREF0397_0209 (https://img.jgi.doe.gov). However, FnHicA shares only 39% identity and 54% similarity with FpHicA. To use the FphicA gene as a counterselection marker in our in-frame deletion plasmids, the low sequence homology of the FphicA gene to the FnhicA gene locus would not affect the initial integration of our in-frame deletion constructs.
FIG 1.
Construction of E. coli-Fusobacterium shuttle plasmid pBCG01 expressing putative HicA toxin under the control of riboswitch E-based inducible system. (A) Protein alignment of toxin HicA from Burkholderia pseudomallei (BpHicA) with HicA encoded by FperA3_010100004605 (https://img.jgi.doe.gov) in Fusobacterium periodonticum ATCC 33693 (FpHicA) using the ClustalW program. (B) Genetic organization of the hicA-hicB toxin-antitoxin pair in F. periodonticum and the whole DNA sequence of the hicA gene. (C) The mRNA sequence of the riboswitch E is shown with an aptamer sequence emboldened, ribosome binding site (RBS) sequence highlighted, and start codon red colored. (D) Schematic diagrams of a functional model of the theophylline-responsive riboswitch. Without theophylline, the riboswitch forms a stem-loop structure that conceals the RBS in the mRNA transcript. The riboswitch alters its confirmation when theophylline binds to the aptamer, resulting in the release of the RBS and the initiation of translation of hicA. (E) E. coli-Fusobacterium shuttle plasmid pBCG01 containing the hicA gene under the control of the rpsJ promoter from F. periodonticum. The RBS in the rpsJ promoter region was replaced by the riboswitch E element. ColE1 RNA II origin (oriEc), the pFN1 origin (oriFn), repA gene, and catP gene are indicated. The oriEc allows replication of the shuttle plasmid in E. coli, and the plasmid replicates in F. nucleatum, requiring oriFn and repA.
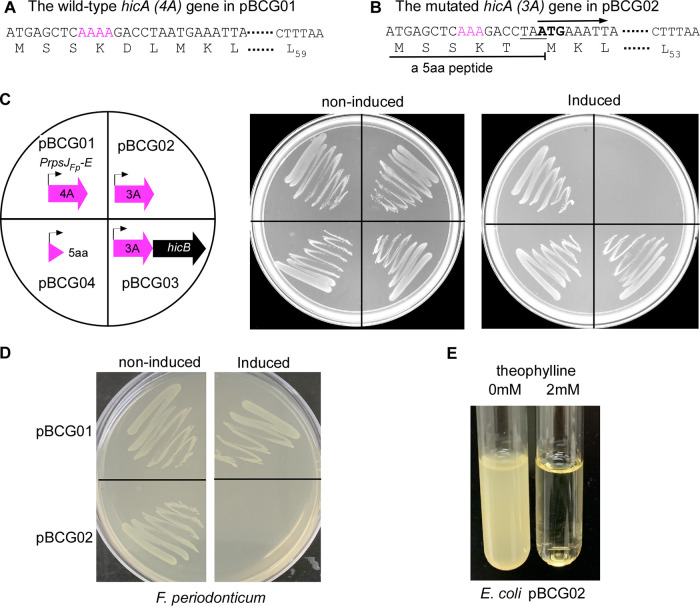
Next, we wanted to express the hicA gene in F. nucleatum ectopically and test the toxicity of the FpHicA protein. To do so, the Gibson assembly cloning was performed to construct a pCWU6-based (17) plasmid that expresses the hicA gene. The rpsJ gene promoter was proven as a strong, constitutive promoter in F. nucleatum (6). Based on this finding, we hypothesized that the promoter of the rpsJ gene in F. periodonticum (FperA3_010100002588; https://img.jgi.doe.gov) would be similar. As a result, we used the rpsJFp promoter to drive hicA expression. To control the putative HicA expression level, a well-characterized synthetic theophylline-responsive riboswitch E (38) (Fig. 1C) was inserted in between the rpsJfp promoter and the hicA start codon (Fig. 1E). This riboswitch E unit is functional in Gram-positive and Gram-negative bacteria (38, 39). It is comprised of an aptamer and a synthetic ribosome binding site (RBS). Theoretically, transcription of the riboswitch under the control of the rpsJFp promoter occurs constitutively during cell growth. However, the synthetic RBS located downstream of the aptamer is sequestered via pairing with the stem of this riboswitch, resulting in a translational stop. The binding of ligand theophylline exposes the RBS to the translation machine by altering the downstream base pairing. Consequently, the hicA gene translation begins when the ribosome binds to the RBS (Fig. 1D). We wanted to obtain a plasmid that ectopically expresses the hicA gene under the control of the riboswitch E element. Four positive E. coli clones generated by the Gibson assembly procedure were randomly chosen to culture for plasmid isolation. Before the four plasmids were verified by DNA sequencing, we transformed them individually into the F. nucleatum ATCC 23726 strain by electroporation. Cells were then plated on an agar plate with thiamphenicol. The colonies formed after 3 days of growth in an anaerobic chamber. One colony from each plasmid transformation plate was picked up to grow in tryptic soy broth (TSB) supplemented with 1% Bacto peptone plus 0.25% freshly made cysteine (TSPC) medium. The growth of these transformants was similar to the wild-type strain when hicA gene expression was not induced with theophylline (data not shown). Interestingly, only one transformant did not grow when 2 mM theophylline was provided; the other three grew the same with or without the theophylline (Fig. 2C and data not shown). Subsequently, DNA sequencing revealed that the three plasmids are identical and each contains a complete hicA gene. Surprisingly, the plasmid conferring cell growth arrest had a point mutation in a “4A” sequence (Fig. 2A). There were no errors in the promoter region, riboswitch E unit, or other parts of the hicA gene in the four sequenced plasmids. An adenine base missed in the 4A sequence (Fig. 2B), subsequently referred to as hicA(3A), caused a frameshift and introduced a premature stop codon at codon 6. It presumably produced a five-amino-acid peptide (MSSKT), starting its translation from the first ATG codon when the inducer was added. By closely examining the mutated hicA sequence, we found that the introduced stop codon TAA had one nucleotide overlapping with a second ATG codon, codon 7 of the wild-type hicA gene (Fig. 2A). This ATG at codon seven is postulated to allow hicA(3A) mRNA to restart its translation, creating a shorter HicA protein lacking the first six amino acids (Fig. 2B). In other words, it is possible that the production of truncated HicA protein is functional in F. nucleatum and able to cease cell growth, but not its full-length version. As the produced MSSKT peptide is small, it may act as an antimicrobial peptide (AMP) that kills or prevents bacterial growth (40). To exclude the possibility of the MSSKT peptide acting as an AMP, the gene region only synthesized with the five-amino-acid peptide was amplified from the mutated plasmid pBCG02 and recloned in pCWU6. Its expression was under the control of the rpsJFP promoter and the riboswitch E unit (pBCG04), the same as those in pBCG01 and pBCG02. Expectedly, in the presence of inducers, the expression of the short peptide did not affect bacterial growth (Fig. 2C, pBCG04). The hicB gene could counteract the toxicity of mutated hicA(3A). As the hicB was coexpressed with mutated hicA, fusobacterial growth arrest was reversed (Fig. 2C, pBCG03) to normal levels when grown on agar plates with 2 mM theophylline (Fig. 2C). We were interested in knowing if the mutated hicA gene was functional in its native host, F. periodonticum, as well. For that purpose, the pBCG02 was transformed into F. periodonticum by electroporation, and one transformant was further cultured and streaked on the plates. In the presence of 2 mM inducer theophylline, cell growth was stopped entirely. In contrast, the cells of F. periodonticum with pBCG01 containing the wild-type hicA gene showed no signs of a growth defect when hicA gene expression was induced (Fig. 2D). In addition, we showed that induction of the mutated hicA gene prevented cell growth of E. coli in the presence of 2 mM theophylline (Fig. 2E). Together, these data suggested that the putative mutated HicA with 53 amino acids functions as a toxin in our tested bacteria.
FIG 2.
A putative shorter HicA protein that lacks the first six amino acids is toxic in Fusobacterium nucleatum and other bacteria. (A) The plasmid pBCG01 contains an intact hicA gene, indicating the presence of a sequence comprised of four consecutive adenine bases (4A) after the start codon (red colored). The wild-type hicA putatively encodes a 59-amino-acid protein. (B) An adenine base was missing from the 4A sequence, resulting in the hicA(3A) gene during construction of pBCG01. We later named that mutated construct pBCG02. This point mutation causes a frameshift resulting in a premature stop codon (TAA, underlined) at codon six and generates a five-amino-acid peptide (MSSKT). The hicA(3A) mRNA then restarts its translation from the new initiation site (ATG, emboldened), giving a putative shorter HicA protein lacking the first six amino acids. (C) Growth of F. nucleatum strain 23726 harboring the plasmid construct pBCG01, pBCG02, pBCG03, or pBCG04 on TSPC agar plates supplemented with thiamphenicol plus 0 (noninduced) or 2 mM (induced) theophylline and incubated at 37°C in an anaerobic chamber for 2 days. Schematic representations of the constructs are shown in the left diagram. (D) Growth of F. periodonticum containing the plasmid pBCG01 or pBCG02 on TSPC agar plates under the uninduced or induced condition with theophylline after 2 days of growth in an anaerobic chamber. (E) Overnight E. coli containing pBCG02 was diluted 1:1,000 and aliquoted into two tubes with 6 mL LB broth plus an indicated concentration of theophylline after 18 h of growth with shaking (200 rpm) at 37°C. These experiments were performed more than three times, and a representative result is presented.
Use of mutated hicA as a counterselection marker for in-frame deletion in F. nucleatum.
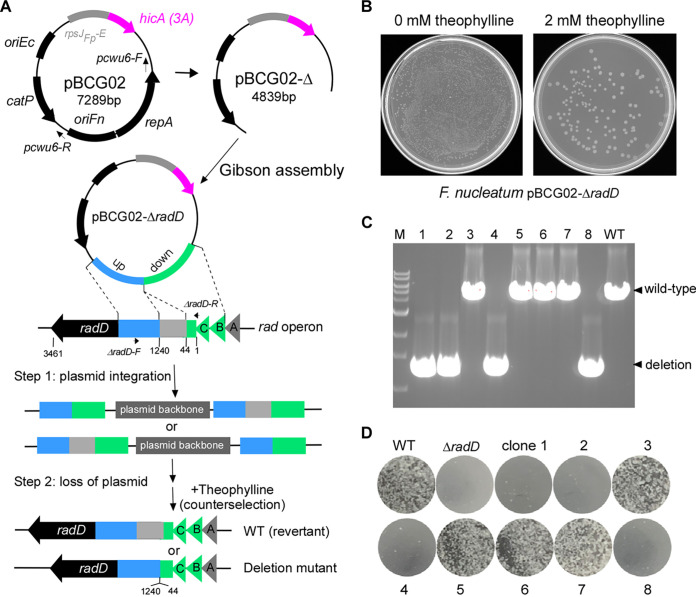
The ectopic expression of the putative HicA(3A) inhibited fusobacterial cell growth, indicating that the hicA(3A) gene might be suitable for counterselection in F. nucleatum. To test this possibility, an allelic exchange vector named pBCG02-ΔradD (Fig. 3A) was constructed to delete the gene region coding for the amino acids in positions between 44 and 1240 of the RadD protein. Our data indicated that the RadD (44 to 1240) region contains a motif critical for RadD activity (our unpublished data). The RadD protein is a large surface adhesin that mediates F. nucleatum’s physical interactions with some oral bacteria, mostly Gram-positive, early colonizers in the human mouth (5). These aggregated interactions are critical for dental plaque formation. An inverse PCR linearized the pBCG02 with primer pair pCWU6-F/pCWU6-R to remove repA and oriFN (Fig. 3A). The RepA and oriFN elements are required for pCWU6-based plasmid replication in F. nucleatum (18). Without them, the shuttle plasmid pBCG02 becomes a suicide vector with a positive selection marker (catP) and a negative selection marker [hicA(3A)]. The same length of upstream and downstream DNA sequences flanking the region—responsible for the synthesis of the RadD (44 to 1240)—was amplified. An overlapping PCR then fused the upstream and downstream fragments. Gibson assembly was used to clone the fused fragment into the linearized pBCG02 that lacks repA and oriFN. The resulting in-frame deletion construct was transformed into E. coli C2987. Positive colonies were confirmed by colony PCR using the primer pair ΔradD-F/ΔradD-R. Positive colonies showed a 1.34-kb insert and were further selected for plasmid preparation and sequencing. The transfer of the allelic exchange plasmid pBCG02-ΔradD into F. nucleatum ATCC 23726 was traditionally performed via the electroporation procedure that our lab used previously (41). The selection of recombinant vectors to integrate into the bacterial chromosome was performed by thiamphenicol, a chloramphenicol derivative. One randomly selected thiamphenicol-resistant integration colony was chosen for theophylline-negative selection based on putative HicA(3A) toxicity. This strain was grown in the absence of antibiotics overnight and then diluted 1:1,000 the following day before plating cells on the TSPC agar plates with 2 mM theophylline. To estimate the frequency of plasmid excision, a plate without an inducer was used to grow the uncounterselected cells as a control. As shown in Fig. 3B, the theophylline-negative selection only allowed a small fraction of the cells to survive. Based on the ratio of theophylline-resistant cells to total cells (cells grown on plates without selection; see Fig. 3B, the left plate without an inducer), we estimated that approximately 1 in 2,000 cells underwent a second recombination to lose the integrated plasmid. Eight randomly selected theophylline-resistant colonies were confirmed for antibiotic sensitivity to screen for possible in-frame deletion mutants. Next, colony PCR was used to detect their genotypes using the primer pair ΔradD-F/ΔradD-R, illustrated in Fig. 3A. As shown in Fig. 3C, four of the eight colonies generated amplicons of approximately 1.4 kb, indicating an in-frame deletion. The remaining four colonies produced the expected wild-type amplicons of approximately 5.0 kb. The coaggregation assay was also used to confirm the four generated radD mutant strains and four wild-type revertant strains. We expected coaggregation defects in these mutant strains because this region of RadD (44 to 1240) is located in the passenger domain, which is responsible for F. nucleatum coaggregation with oral bacterium Actinomyces oris (6). This was indeed the case (Fig. 3D). Clones 1, 2, 4, and 8 showed coaggregation defects with A. oris, whereas all wild-type revertants (clones 3, 5, 6, and 7) exhibited aggregation similar to the wild-type strain. Finally, two of the in-frame deletion mutant clones (clones 1 and 4) were also sequenced and found to exhibit the expected in-frame deletion that the radD gene only lacks in the desired deletion region by amplicon sequencing.
FIG 3.
Applying hicA(3A) as a counterselectable marker for gene deletion in F. nucleatum. (A) Graphic representation of producing the nonreplicative vector backbone pBCG02-Δ used to generate unmarked in-frame gene deletions in F. nucleatum via a two-step strategy. The primer pair (pCWU6-F/pCWU6-R, indicated by short arrows) was used to perform inverse PCR to remove the region, including the oriFn and repA. The radABCD operon is shown. Approximately 1.6-kb fragments homologous to the upstream (blue) and downstream (green) regions of the designed deletion region in radD were amplified by PCR. They have been ligated adjacently on the vector backbone pBCG02-Δ via the Gibson assembly procedure. The resulting plasmid, pBCG02-ΔradD, was used to generate an unmarked radD deletion (the gene deletion region coding for the amino acids in positions between 44 and 1240 of the RadD protein of 3,461 amino acids) mutant in the wild-type background by a two-step allelic exchange. (B) TSPC agar plate representative of the counterselection effect by putative HicA(3A) toxin expression in F. nucleatum. A 100-μL aliquot of 1,000-fold-diluted overnight culture of the pBCG02-pBCG02-ΔradD-integrated strain was plated on TSPC agar plates containing 0 or 2 mM inducer theophylline to select for cells in which the plasmid had been excised and lost. (C) Screen of radD deletion mutants by colony PCR with the primer pair ΔradD-F/ΔradD-R (indicated in panel A). Lanes 1 to 8, 10 μL of PCR products of eight colonies randomly selected from the TSPC agar plate. Four of the eight chosen random colonies were deletion mutants, while four reverted to wild type during the second recombination event. Bands of approximately 1.4 kb indicate radD deletion, whereas bands of about 5.0 kb indicate a wild-type (WT) genotype. (D) F. nucleatum wild-type, radD mutant strain, and 8 counterselection isolates from panel C were examined for coaggregation with A. oris MG-1. A representative result was presented after these experiments were carried out three times.
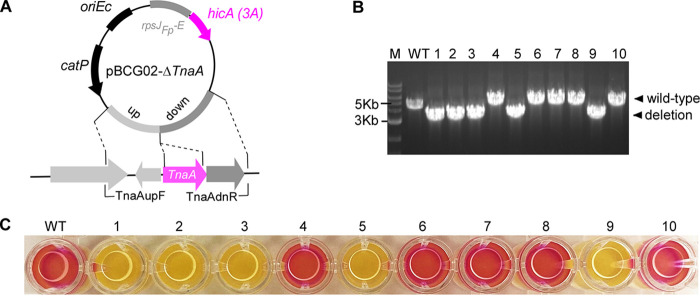
To further test this hicA(3A)-based counterselection system, we used it to make an in-frame deletion of the tnaA gene (HMPREF0397_1802; https://img.jgi.doe.gov/) in F. nucleatum. It has been reported that the tnaA gene encodes the enzyme tryptophanase that produces indole from tryptophan in E. coli (42). As a signaling molecule, indole plays an essential role in bacterial physiology, ecological balance, and human health (43). However, its biological function in F. nucleatum has yet to be explored. An allelic exchange cassette carrying the flanking regions of tnaA was generated by an overlapping PCR. The allelic exchange cassette was cloned into the linearized pBCG02 in which repA and oriFn were omitted. The generated suicide plasmid pBCG02-ΔtnaA (Fig. 4A) was then introduced into an F. nucleatum wild-type background strain by electroporation. The insertion obtained by homologous recombination of the plasmid into the chromosome was selected in the presence of thiamphenicol. A thiamphenicol-resistant colony was grown in TSPC without antibiotics overnight. The following day, aliquots of 1,000-fold diluted cultures were plated on TSPC agar plates containing 2 mM theophylline. After 3 days, 10 theophylline-resistant colonies were randomly chosen to check thiamphenicol sensitivity (100% efficiency). The colony PCR was then used to analyze the genotypes of the 10 colonies with primer pair TnaAupF and TnaAdnR (see Table 1) indicated in Fig. 4A. The results revealed that half of the colonies were the expected deletion mutant strains, and half were the wild-type revertant strain (Fig. 4B). Last, the tnaA deletion mutant was confirmed using the indole assay, which revealed that the wild-type cell and wild-type revertant cell (clones 4, 6,7, 8, and 10) developed red color, whereas the tnaA mutant (clones 1, 2, 3, 5, and 9) did not (Fig. 4C). This suggests that the tnaA homolog in F. nucleatum is responsible for indole production.
FIG 4.
Deletion of the indole-producing gene tnaA in F. nucleatum as hicA(3A) as a counterselectable marker. (A) Schematic drawing of pBCG02-ΔtnaA for the deletion of tryptophanase TnaA gene. Approximately 1.6-kb fragments homologous to the upstream (blue) and downstream (green) regions of the tnaA gene were ligated adjacently on the vector backbone pBCG02-Δ via the Gibson assembly procedure to create the vector pBCG02-ΔtnaA. The primer pair for colony PCR to screen ΔtnaA mutant strains is indicated by small arrows. (B) The genotype of tnaA mutant strains following hicA(3A) counterselection. Bands of approximately 3.2 kb indicate a tnaA deletion, whereas bands of about 5.0 kb indicate a wild-type genotype. WT represents the F. nucleatum wild type. (C) F. nucleatum wild type and 10 isolated colonies after counterselection from panel B were examined for indole production. Indole production was demonstrated by adding Kovacs’ reagent (100 μL) into 200 μL of individual culture in a 96-well plate, which reacts with the indole, giving a red color in wild-type genotype fusobacterial cultures. Three repeats of the studies were conducted, and one typical outcome is shown.
TABLE 1.
Primers used in this study
| Primer | Sequencea | Use |
|---|---|---|
| J23119-F | GAAACAGCTATGACATGATTACGAATTCGAGCTCTGAGCTAGCTGTCAAACTGGCCGTCG | pBCG01 |
| J23119-R | AGTAATTTCATTAGGTCTTTTGAGCTCATCTTGTTGTTACCTCCTTAGCAGG | pBCG01 |
| hicA-F | GCTAAGGAGGTAACAACAAGATGAGCTCAAAAGACCTAATGAAATTACT | pBCG01 |
| hicA-R | GTATAAATGTTAGTGATGATTATAAAAATTAAAAGTATGAGGAGAACAAGCAAG | pBCG01 |
| PrpsJFP-F | GAAACAGCTATGACATGATTACGAATTCGAGCTAGCACCAGGTTTAGGGGCTG CTATAG | pBCG01 |
| PrpsJFP-R | GGCATCAAGACGATGCTGGTATCACCGGTTTAAATTAACTACATAAGATAATTCTTATG | pBCG01 |
| hicA(rpsJFP)-F | GAATTATCTTATGTAGTTAATTTAAACCGGTGATACCAGCATCGTCTTGATGCCCTTG | pBCG01 |
| hicA-R2 | AAGTATGAGGAGAACAAGCAAGGCATTAAAGCCCTGCTTGCCTG | pBCG01 |
| hicB-F | CAGGCAAGCAGGGCTTTAATGCCTTGCTTGTTCTCCTCATACTT | pBCG03 |
| hicB-R | GTATAAATGTTAGTGATGATTATAAAATTATTTTTCAATTCCTAATTCTTTT | pBCG03 |
| MSSKT-R | GTATAAATGTTAGTGATGATTATAAAATTAGGTCTTTGAGCTCATCTTGTTG | pBCG04 |
| pCWU6-F | GTCCTAGAACTAGTGAAATTTCCG | Removes repA and oriFn from pBCG02 |
| pCWU6-R | GTAGCTTCCATACATATCACACTC | Removes repA and oriFn from pBCG02 |
| TnaAupF | GAGTGTGATATGTATGGAAGCTACAGTAATATTGTTTGATGAAATTGAAAAGG | pBCG02-ΔtnaA |
| TnaAupR | TCTGCTTGTTGTTCAGGAGTTTCATCACCTAGGAACATAGCTGACCATTG | pBCG02-ΔtnaA |
| TnaAdnF | GCTATGTTCCTAGGTGATGAAACTCCTGAACAACAAGCAGAAGTTGCAAATG | pBCG02-ΔtnaA |
| TnaAdnR | CGGAAATTTCACTAGTTCTAGGACAGCAGCAAGCAATGCTCCAAGTGGACAAATATATATAG | pBCG02-ΔtnaA |
| RadDupF | GAGTGTGATATGTATGGAAGCTACATAAATTTAATTTAACTTATTAATTATAAGG | pBCG02-ΔradD |
| RadDupR | GCATCCTTACCTGACATTTTTACTGCTTCATCTGCATAAGAAAAACCACCATTAAT | pBCG02-ΔradD |
| RadDdnF | GATGAAGCAGTAAAAATGTCAGGTAAGGATGCTATTGGTATTT | pBCG02-ΔradD |
| RadDdnR | CGGAAATTTCACTAGTTCTAGGACTCCTGATTCATTTATTGCTTCACCATCTACT | pBCG02-ΔradD |
| ΔradD-F | GAGTTTCAGATGAAATGAATGCCCTACAAG | Confirms radD deletion |
| ΔradD-R | CTGCAGTAGTTGCTCCACCTTTTACAGCTG | Confirms radD deletion |
Underlined are primer sequences specific to the ends of the corresponding assembly.
DISCUSSION
The HicAB toxin-antitoxin system has been widely studied in bacteria. However, no HicA toxin has been used for counterselection to create markerless in-frame deletion mutations. HicA is an RNase, and its expression prevents cell growth in many bacteria (24, 34). In this study, we used a hicA gene from F. periodonticum and established a facile in-frame deletion system in F. nucleatum. The success of establishing this putative HicA toxin for counterselection in F. nucleatum has benefited from one unexpected finding. The expression of a full-length hicA gene, which encoded a putative 59-amino-acid HicA toxin, did not induce cell death; however, when inducing expression of a short form of putative HicA protein that lacks the first six amino acids, the cells died (Fig. 2C). In other words, the putative truncated version of HicA possesses a toxin activity. The finding of this active form of putative HicA was accidental. An unexpected point mutation in hicA occurred during plasmid constructions, allowing hicA mRNA translation to start from the second ATG, codon 7, in its original version of this gene (Fig. 2B). The presence of N-terminal six amino acids in the putative HicA inhibits its activity. Some proteins, like bacteriocins, must remove part of their N termini before being active (44, 45). For toxin proteins from all studied toxin-antitoxin systems, there is no such report that the removal of partial N-terminal sequences is needed for their activities. F. periodonticum may produce a nonfunctional premature HicA protein, which then requires an unknown mechanism to cleave its N-terminal partial sequence in order to be in an active state. Consistent with this possibility, the hicA expression in the hicB deletion mutants does not exhibit growth defects in E. coli (46), Haemophilus ducreyi (47), and Streptococcus mutans (36). Another possibility is that the translation of this gene in F. periodonticum may start from the second ATG codon—codon 7 of the gene annotated in the genome database (https://img.jgi.doe.gov)—but not the first ATG as its starting codon. All of these possibilities require further studies.
This work aimed to find a new gene to replace galK as a counterselection marker to generate markerless gene deletions in F. nucleatum. We showed that cell growth would be arrested entirely when the mutated hicA was induced in the wild-type background of F. nucleatum (Fig. 2C). Therefore, we attempted to use the truncated form of hicA gene as a counterselection maker in allelic exchange constructs for creating in-frame deletion mutations in F. nucleatum. The rpsJ promoter is a constitutive and robust promoter in many bacteria (6, 48). We chose to use the rpsJ promoter from F. periodonticum to drive the expression of the putatively active HicA toxin. Choosing the promoter from F. periodonticum, but not F. nucleatum, was to avoid the potential interference to the initial integration of our constructs due to the homology of this promoter fragment to the native rpsJ locus. A well-studied theophylline-responsive synthetic riboswitch was employed to control the active hicA gene expression. Six synthetic theophylline-responsible riboswitches were widely used in various bacteria (38, 39, 49). The riboswitch E was chosen to control hicA mRNA translation because only this riboswitch works and does not have a leaking issue when used in F. nucleatum (our unpublished data). This theophylline-sensing riboswitch tightly controls the expression of the active HicA protein. Since 2 mM theophylline could induce enough of the toxic protein to prevent cell growth, no step was carried out to optimize theophylline's concentration further. With this system, we successfully created the in-frame deletion for the fusobacterial adhesin protein RadD gene and the tnaA gene responsible for indole production in F. nucleatum.
The hicA(3A)-based system showed a reliable and robust counterselection exactly like galK-utilized counterselection. All colonies were thiamphenicol sensitive after counterselection, which indicated that 100% of colonies underwent loss of the integration plasmids once the putatively active HicA toxin protein was induced by 2 mM theophylline. Compared with the galK system, the hicA(3A)-dependent counterselection has an immediate advantage: it can work in the wild-type background of F. nucleatum. MazF has been used as a counterselection marker in other Gram-positive and Gram-negative bacteria (20). As we prepared the manuscript, a MazF-mediated counterselection strategy was reported in F. nucleatum (50). They use a tetracycline-inducible system to control the MazF expression in F. nucleatum. The expression of MazF is lethal. That allowed the counterselection in F. nucleatum due to MazF toxicity, and only cells lacking the integrated plasmid could grow. Undoubtedly, this system is excellent and easy to use in fusobacterial genetic study. But, it has one potential problem: many strains of F. nucleatum are tetracycline sensitive. We have found that even 20 ng/mL tetracycline or 80 ng/mL anhydrotetracycline discourages the growth of F. nucleatum ATCC 23726, and some are even more sensitive. That may limit the use of the tetracycline-induced MazF system for counterselection in some strains of F. nucleatum. In this aspect, the theophylline-sensed riboswitch is better than the tetracycline-inducible system since F. nucleatum’s growth is not affected by the presence of theophylline, even at high concentrations (data not shown).
The results presented in this study support the use of HicA toxin activity for negative selection in F. nucleatum. The active HicA may work in many bacteria, as we already demonstrated that the induction of this putatively active HicA causes cell death in F. periodonticum and E. coli (Fig. 2D and E). As far as we know, this is the first time the hicA gene was used as a counterselection marker for bacterial genetic studies. This same strategy is also theoretically feasible in other bacteria that do not already possess an efficient negative selection system. Currently, we are working on this system in F. periodonticum to test if it can be widely applied in other fusobacterial species.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 2. F. nucleatum and F. periodonticum strains were grown in tryptic soy broth (TSB) supplemented with 1% Bacto peptone plus 0.25% freshly made cysteine (TSPC) or on TSPC agar plates in an anaerobic chamber filled with 80% N2, 10% H2, and 10% CO2. Hemin (5 μg/mL) and vitamin K3 (1 μg/mL) were added for F. periodonticum growth on TSPC agar plates. A. oris was grown in heart infusion broth (HIB). E. coli strains were grown in Luria-Bertani (LB) broth. Fusobacterial transformants with pBCG01, pBCG02, pBCG03, or pBCG04 were grown overnight in TSPC medium with 5 μg/mL thiamphenicol. Overnight cultures were directly streaked on TSPC agar with antibiotic selection, with or without 2 mM theophylline, after 72 h of incubation at 37°C in an anaerobic chamber. We prepared 40 mM of theophylline stock in a TSP medium. pREcas2 was purchased from the Synthetic Biology Research Center at the University of Nottingham, United Kingdom. Antibiotics used as needed were chloramphenicol (15 μg mL−1) and thiamphenicol (5 μg mL−1). Reagents were purchased from Sigma unless indicated otherwise.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| F. periodonticum 33693 | Parental strain | This study |
| F. nucleatum 23726 | Parental strain | 5 |
| F. nucleatum BG01 | ΔradD; an isogenic derivative of 23726 | This study |
| F. nucleatum ZP01 | ΔtnaA; an isogenic derivative of 23726 | This study |
| A. oris MG1 | Wild-type strain | 48 |
| E. coli C2987 | Cloning host | NEB |
| Plasmids | ||
| pCWU6 | E. coli-Fusobacterium shuttle vector, chloramphenicol /thiamphenicol resistance; cmr thiar | 18 |
| pRECas2 | The J23119 promoter incorporating the riboswitch element E controls Cas9 expression | 39 |
| pBCG01 | pCWU6 expressing wild-type hicA gene [hicA(4A)] from F. periodonticum 33693 under the control of the rpsJFP promoter and its ribosome binding site was replaced by riboswitch element E from pRECas2 | This study |
| pBCG02 | Derivative of pBCG01 expressing hicA with a point mutation [hicA(3A)]. The hicA(3A) mRNA is translated from the initiation site met-7, giving a shorter protein lacking the first six amino acids. | This study |
| pBCG03 | Derivative of pBCG02 coexpressing hicA(3A) and hicB, the cognate hicA antitoxin gene | This study |
| pBCG04 | Derivative of pBCG02 expressing the five amino acids peptide (MSSKT) alone | This study |
| pBCG02-ΔradD | Derivative of pBCG02 lacking repA and oriFn, deletion vector of radD Δ41-1240 | This study |
| pBCG02-ΔtnaA | Derivative of pBCG02 lacking repA and oriFn, deletion vector of tnaA | This study |
Plasmid construction.
All plasmids used in this study were constructed via Gibson assembly cloning, essentially performed according to the manufacturer’s instructions with NEBuilder HiFi DNA Assembly master mix. This method requires adjacent segments with at least 18-bp-long identical sequences on the ends. These identical sequences were generated via PCR with primers that contain a 5′ end identical to an adjacent segment and a 3′ end that anneals to the gene-of-interest sequence using a 2× Hot Start PCR master mix (TaKaRa; catalog no. R405A). The primers used for PCRs are listed in Table 1.
(i) pBCG01 and pBCG02. To create these plasmids, pREcas2 (39) was used as a DNA template to amplify the J23119 promoter region and the riboswitch E element with primers J23119-F/J23119-R. The primer pair hicA-F/hicA-R (Table 1) was used to PCR amplify the hicA gene (see gene identification no. FperA3_010100004605 in the F. periodonticum database at https://img.jgi.doe.gov/) with genome DNA of F. periodonticum ATCC 33693 as a template. The two amplicons were fused via an overlapping PCR, and the resulting PCR product was designated J23119-E-hicA. Then, the fragment encompassing the riboswitch E and hicA (E-hicA) was amplified with primers hicA(rpsJFP)-F and hicA-R. The primer pair PrpsJFP-F/PrpsJFP-R was used to amplify the promoter region of the rpsJ gene (FperA3_010100002588; https://img.jgi.doe.gov/) without its ribosome binding site (PrpsJFP) with the genome DNA of F. periodonticum ATCC 33693. The E-hicA and PrpsJFP amplicons were cloned into SacI- and HindIII-digested pCWU6 (17) by Gibson assembly and transformed into E. coli C2987 cells, as described previously (51). Four positive clones were chosen to culture for plasmid isolation, and the four plasmids were sent for sequencing. Three had the wild-type hicA DNA sequence, but one had a point mutation at the 4A sequence (see Fig. 2A and B). The plasmid that contains this point mutation was named pBCG02.
(ii) pBCG03. To test if the putative antitoxin HicB prevents the putative HicA(3A) toxicity in F. nucleatum, the hicB gene (FperA3_010100004610; https://img.jgi.doe.gov/) was placed immediately downstream of hicA, and the expression of the two genes was under the control of rpsJFP promoter and riboswitch E unit. The fragment, including PrpsJFP-E-hicA, was amplified with the primers PrpsJFP-F and hicA-R2 from pBCG02. Second, the primer pair hicB-F/hicB-R was used to PCR amplify the hicB gene. The two amplicons of PrpsJFP-E-hicA and hicB were mixed with SacI- and HindIII-cut pCWU6 in the Gibson assembly master mix solution to generate pBCG03.
(iii) pBCG04. With plasmid pBCG02 as a DNA template, the primer pairs PrpsJFP-F and MSSKT-R were used to amplify the fragment encompassing PrpsJFP-E and hicA(3A) region only, including the first ATG to the premature stop codon due to missing one “A” base in 4A sequence. The short hicA region codes a five-amino-acid peptide (MSSKT; see Fig. 2B). This amplicon was cloned into pCWU6 precut by SacI and HindIII via Gibson assembly to generate pBCG04.
All produced plasmids were confirmed by sequencing and then transformed into F. nucleatum ATCC 23726 or F. periodonticum ATCC 33693 with the electroporation procedure we previously published (41). In brief, cells of either F. nucleatum or F. periodonticum were harvested by centrifugation from a 100-mL stationary-phase culture. The cells were washed twice with sterile water and once with 10% glycerol and then resuspended in 3 mL of 10% glycerol and divided into 0.2-mL aliquots. Next, 1.0 µg purified plasmid DNA was added to a 0.2-mL aliquot of the above-described prepared fusobacterial cells in a precooled corvette (0.1-cm electrode gap; Bio-Rad) and left on ice for 10 min. The reactions were electroporated using a Bio-Rad Gene Pulser II set at 25 kV, 25µF, and 200 Ω. Immediately following electroporation, the cells were diluted in 1 mL of prereduced and prewarmed TSPC. The culture was plated on TSPC agar plates with 5 mg/ml thiamphenicol after being incubated anaerobically for 5 h without agitation.
Gene deletion based on hicA(3A) counterselection in F. nucleatum.
To create deletion constructs for radD that eliminate the amino acids in positions between 44 and 1240, 1.6-kb fragments upstream and downstream of the radD interested region were amplified by PCR using primer pairs RadDupF/RadDupR and RadDdnF/RadDdnR (Table 1). For the tnaA deletion plasmid, 1.6-kb fragments upstream and downstream of tnaA were PCR amplified using primer pairs tnaAupF/tnaAupR and tnaAdnF/tnaAdnR. An overlapping PCR fused the upstream and downstream fragments. Each fused segment was then mixed in 10 μL Gibson assembly master mix solution with an equal amount of the pBCG02 vector backbone in which the repA and oriFn element were removed by inverse PCR with primer pair pCWU6-F/pCWU6-FR (see Fig. 3A). A 2× Hot Start PCR master mix from TaKaRa (catalog no. R405A) was used for high-fidelity PCR amplification of the pBCG02 backbone. The resulting plasmids pBCG02-ΔradD and pBCG02-ΔtnaA were further confirmed by DNA Sanger sequencing and transformed into F. nucleatum ATCC 23726 by electroporation (41), respectively.
Plasmid insertion into the bacterial chromosome by homologous recombination was selected by growth with TSPC liquid medium in the presence of 5 μg/mL thiamphenicol at 37°C. A 5-μg/mL thiamphenicol-resistant colony was inoculated overnight in TSPC without antibiotics. The following day, a 100-μL aliquot of 1,000-fold diluted cultures was plated on TSPC agar plates containing 2 mM inducer theophylline to select for cells in which the plasmid had been excised and lost. Approximately 8 to 10 growing colonies were restreaked on TSPC plates, checked for thiamphenicol sensitivity (100% efficiency), and screened by PCR amplification for the presence of the expected deletion in radD or tnaA.
Bacterial coaggregation.
Coaggregation assays were performed with wild-type, ΔradD, and 8 colonies from hicA(3A)-based counterselection of F. nucleatum and A. oris MG-1 as previously described (17). Briefly, stationary-phase cultures of bacterial strains were grown in TSPC or heart infusion broth (for MG-1) harvested by centrifugation, washed in coaggregation buffer (52), and suspended to an equal cell density of approximately 2 × 109 mL−1 based upon optical density at 600 nm (OD600) values. For coaggregation, 0.20-mL aliquots of Actinomyces and fusobacterial cell suspensions were mixed in a 24-well plate for a few minutes on a rotator shaker, and images were taken.
Indole assay.
Fusobacterial wild-type and 10 clones, obtained through hicA(3A) counterselection targeted toward in-frame deletion of the tnaA gene, were individually inoculated into a test tube containing 8 mL of TSPC medium. The tubes were incubated at 37°C for 24 h, and a 200-μL aliquot was taken from each culture and added to a 96-well plate. Following this, 100 μL of Kovacs’ reagent was added to each tube without shaking, and a resulting color change was observed. A positive indole test is indicated by the formation of a pink to red color in the reagent layer on top of the medium within a few seconds of adding the reagent, while a negative result appears yellow. The photograph was taken 10 min after the reagent was added.
Data availability.
Materials are available upon reasonable request with a material transfer agreement with UTHealth for bacterial strains or plasmids. The pBCG01 plasmid sequence and map can be accessed on Benchling at https://benchling.com/s/seq-VuTCpImziZSBrxT6otvO?m=slm-jUtGU7tIsA1ZnPAgUH1f.
ACKNOWLEDGMENTS
This work was supported by a National Institute of Dental and Craniofacial Research (NIDCR) grant (DE030895) to C.W.
C.W. conceived and designed all experiments. B.G.C. and P.Z. performed all experiments. B.G.C., P.Z., and C.W. analyzed data. C.W. wrote the manuscript with contribution and approval from all authors.
Contributor Information
Chenggang Wu, Email: chenggang.wu@uth.tmc.edu.
Nicole R. Buan, University of Nebraska-Lincoln
REFERENCES
- 1.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. 2001. Bacterial diversity in human subgingival plaque. J Bacteriol 183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolstad A, Jensen HB, Bakken V. 1996. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin Microbiol Rev 9:55–71. doi: 10.1128/CMR.9.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, Li M, Sojar HT, Genco RJ, Kuramitsu HK, Deng CX. 2005. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol 187:5330–5340. doi: 10.1128/JB.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fardini Y, Wang X, Témoin S, Nithianantham S, Lee D, Shoham M, Han YW. 2011. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity. Mol Microbiol 82:1468–1480. doi: 10.1111/j.1365-2958.2011.07905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaplan CW, Lux R, Haake SK, Shi W. 2009. The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol Microbiol 71:35–47. doi: 10.1111/j.1365-2958.2008.06503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu C, Chen YW, Scheible M, Chang C, Wittchen M, Lee JH, Luong TT, Tiner BL, Tauch A, Das A, Ton-That H. 2021. Genetic and molecular determinants of polymicrobial interactions in Fusobacterium nucleatum. Proc Natl Acad Sci USA 118:e2006482118. doi: 10.1073/pnas.2006482118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abed J, Emgård JEM, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, Mellul A, Chaushu S, Manson AL, Earl AM, Ou N, Brennan CA, Garrett WS, Bachrach G. 2016. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe 20:215–225. doi: 10.1016/j.chom.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parhi L, Abed J, Shhadeh A, Alon-Maimon T, Udi S, Ben-Arye SL, Tam J, Parnas O, Padler-Karavani V, Goldman-Wohl D, Yagel S, Mandelboim O, Bachrach G. 2022. Placental colonization by Fusobacterium nucleatum is mediated by binding of the Fap2 lectin to placentally displayed Gal-GalNAc. Cell Rep 38:110537. doi: 10.1016/j.celrep.2022.110537. [DOI] [PubMed] [Google Scholar]
- 9.Polak D, Shapira L, Weiss EI, Houri-Haddad Y. 2012. The role of coaggregation between Porphyromonas gingivalis and Fusobacterium nucleatum on the host response to mixed infection. J Clin Periodontol 39:617–625. doi: 10.1111/j.1600-051X.2012.01889.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Shi W, Song Y, Wang J. 2019. Metatranscriptomic analysis of an in vitro biofilm model reveals strain-specific interactions among multiple bacterial species. J Oral Microbiol 11:1599670. doi: 10.1080/20002297.2019.1599670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vander Haar EL, So J, Gyamfi-Bannerman C, Han YW. 2018. Fusobacterium nucleatum and adverse pregnancy outcomes: epidemiological and mechanistic evidence. Anaerobe 50:55–59. doi: 10.1016/j.anaerobe.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gauthier S, Tétu A, Himaya E, Morand M, Chandad F, Rallu F, Bujold E. 2011. The origin of Fusobacterium nucleatum involved in intra-amniotic infection and preterm birth. The J Maternal-Fetal & Neonatal Medicine 24:1329–1332. doi: 10.3109/14767058.2010.550977. [DOI] [PubMed] [Google Scholar]
- 13.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang J-Y. 2017. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170:548–563.e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, Jenab M, Prehn JHM, Hughes DJ. 2014. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis 33:1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 15.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. 2013. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennan CA, Garrett WS. 2019. Fusobacterium nucleatum—symbiont, opportunist and oncobacterium. Nat Rev Microbiol 17:156–166. doi: 10.1038/s41579-018-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu C, Al Mamun AAM, Luong TT, Hu B, Gu J, Lee JH, D’Amore M, Das A, Ton-That H. 2018. Forward genetic dissection of biofilm development by Fusobacterium nucleatum: novel functions of cell division proteins FtsX and EnvC. mBio 9:e00360-18. doi: 10.1128/mBio.00360-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haake SK, Yoder S, Gerardo SH. 2006. Efficient gene transfer and targeted mutagenesis in Fusobacterium nucleatum. Plasmid 55:27–38. doi: 10.1016/j.plasmid.2005.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casasanta MA, Yoo CC, Udayasuryan B, Sanders BE, Umaña A, Zhang Y, Peng H, Duncan AJ, Wang Y, Li L, Verbridge SS, Slade DJ. 2020. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci Signal 13:eaba9157. doi: 10.1126/scisignal.aba9157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reyrat J-M, Pelicic V, Gicquel B, Rappuoli R. 1998. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect Immun 66:4011–4017. doi: 10.1128/IAI.66.9.4011-4017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umaña A, Nguyen TT, Sanders BE, Williams KJ, Wozniak B, Slade DJ. 2022. Enhanced Fusobacterium nucleatum genetics using host DNA methyltransferases to bypass restriction-modification systems. J Bacteriol 204:e00279-22. doi: 10.1128/jb.00279-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaillancourt K, Bédard N, Bart C, Tessier M, Robitaille G, Turgeon N, Frenette M, Moineau S, Vadeboncoeur C. 2008. Role of galK and galM in galactose metabolism by Streptococcus thermophilus. Appl Environ Microbiol 74:1264–1267. doi: 10.1128/AEM.01585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neves AR, Pool WA, Solopova A, Kok J, Santos H, Kuipers OP. 2010. Towards enhanced galactose utilization by Lactococcus lactis. Appl Environ Microbiol 76:7048–7060. doi: 10.1128/AEM.01195-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makarova KS, Grishin NV, Koonin EV. 2006. The HicAB cassette, a putative novel, RNA-targeting toxin-antitoxin system in archaea and bacteria. Bioinformatics 22:2581–2584. doi: 10.1093/bioinformatics/btl418. [DOI] [PubMed] [Google Scholar]
- 25.Singh G, Yadav M, Ghosh C, Rathore JS. 2021. Bacterial toxin-antitoxin modules: classification, functions, and association with persistence. Curr Res Microb Sci 2:100047. doi: 10.1016/j.crmicr.2021.100047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachrach G, Haake SK, Glick A, Hazan R, Naor R, Andersen RN, Kolenbrander PE. 2004. Characterization of the novel Fusobacterium nucleatum plasmid pKH9 and evidence of an addiction system. Appl Environ Microbiol 70:6957–6962. doi: 10.1128/AEM.70.12.6957-6962.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jurėnas D, Fraikin N, Goormaghtigh F, Van Melderen L. 2022. Biology and evolution of bacterial toxin–antitoxin systems. Nat Rev Microbiol 20:335–350. doi: 10.1038/s41579-021-00661-1. [DOI] [PubMed] [Google Scholar]
- 28.Shi Y, Dong H, Zhang D. 2014. Application progress of mazF gene in genetic modification system. Biotechnology Bull 48–54. [Google Scholar]
- 29.Morimoto T, Ara K, Ozaki K, Ogasawara N. 2009. A new simple method to introduce marker-free deletions in the Bacillus subtilis genome. Genes Genet Syst 84:315–318. doi: 10.1266/ggs.84.315. [DOI] [PubMed] [Google Scholar]
- 30.Cheah YE, Albers SC, Peebles CA. 2013. A novel counter-selection method for markerless genetic modification in Synechocystis sp. PCC 6803. Biotechnol Progress 29:23–30. doi: 10.1002/btpr.1661. [DOI] [PubMed] [Google Scholar]
- 31.Van Zyl WF, Dicks LM, Deane SM. 2019. Development of a novel selection/counter-selection system for chromosomal gene integrations and deletions in lactic acid bacteria. BMC Molecular Biol 20:10. doi: 10.1186/s12867-019-0127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L, Liu H, Wang L, Tan X, Yang S. 2021. Synthetic counter-selection markers and their application in genetic modification of Synechococcus elongatus UTEX2973. Appl Microbiol Biotechnol 105:5077–5086. doi: 10.1007/s00253-021-11391-y. [DOI] [PubMed] [Google Scholar]
- 33.Lin Z, Deng B, Jiao Z, Wu B, Xu X, Yu D, Li W. 2013. A versatile mini-mazF-cassette for marker-free targeted genetic modification in Bacillus subtilis. J Microbiol Methods 95:207–214. doi: 10.1016/j.mimet.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 34.Jørgensen MG, Pandey DP, Jaskolska M, Gerdes K. 2009. HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J Bacteriol 191:1191–1199. doi: 10.1128/JB.01013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butt A, Higman VA, Williams C, Crump MP, Hemsley CM, Harmer N, Titball RW. 2014. The HicA toxin from Burkholderia pseudomallei has a role in persister cell formation. Biochemical J 459:333–344. doi: 10.1042/BJ20140073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie Z, Qi F, Merritt J. 2013. Development of a tunable wide-range gene induction system useful for the study of streptococcal toxin-antitoxin systems. Appl Environ Microbiol 79:6375–6384. doi: 10.1128/AEM.02320-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butt AT. 2013. Identification and characterisation of toxin-antitoxin systems (TA) in Burkholderia pseudomallei. PhD thesis. University of Exeter, Exeter, UK. [Google Scholar]
- 38.Topp S, Reynoso CMK, Seeliger JC, Goldlust IS, Desai SK, Murat D, Shen A, Puri AW, Komeili A, Bertozzi CR, Scott JR, Gallivan JP. 2010. Synthetic riboswitches that induce gene expression in diverse bacterial species. Appl Environ Microbiol 76:7881–7884. doi: 10.1128/AEM.01537-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canadas IC, Groothuis D, Zygouropoulou M, Rodrigues R, Minton NP. 2019. RiboCas: a universal CRISPR-based editing tool for Clostridium. ACS Synth Biol 8:1379–1390. doi: 10.1021/acssynbio.9b00075. [DOI] [PubMed] [Google Scholar]
- 40.Huan Y, Kong Q, Mou H, Yi H. 2020. Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front Microbiol 11:582779. doi: 10.3389/fmicb.2020.582779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peluso EA, Scheible M, Ton-That H, Wu C. 2020. Genetic manipulation and virulence assessment of Fusobacterium nucleatum. Curr Protoc Microbiol 57:e104. doi: 10.1002/cpmc.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li G, Young KD. 2013. Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology 159:402–410. doi: 10.1099/mic.0.064139-0. [DOI] [PubMed] [Google Scholar]
- 43.Lee J-H, Lee J. 2010. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev 34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 44.Zimina M, Babich O, Prosekov A, Sukhikh S, Ivanova S, Shevchenko M, Noskova S. 2020. Overview of global trends in classification, methods of preparation and application of bacteriocins. Antibiotics 9:553. doi: 10.3390/antibiotics9090553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darbandi A, Asadi A, Mahdizade Ari M, Ohadi E, Talebi M, Halaj Zadeh M, Darb Emamie A, Ghanavati R, Kakanj M. 2022. Bacteriocins: properties and potential use as antimicrobials. J Clin Lab Anal 36:e24093. doi: 10.1002/jcla.24093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Button JE, Silhavy TJ, Ruiz N. 2007. A suppressor of cell death caused by the loss of σE downregulates extracytoplasmic stress responses and outer membrane vesicle production in Escherichia coli. J Bacteriol 189:1523–1530. doi: 10.1128/JB.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeung A, Cameron DW, Desjardins M, Lee BC. 2011. Use of signature-tagged mutagenesis to identify virulence determinants in Haemophilus ducreyi responsible for ulcer formation. J Microbiol Methods 84:290–298. doi: 10.1016/j.mimet.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 48.Wu C, Ton-That H. 2010. Allelic exchange in Actinomyces oris with mCherry fluorescence counterselection. Appl Environ Microbiol 76:5987–5989. doi: 10.1128/AEM.00811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu C, Huang IH, Chang C, Reardon-Robinson ME, Das A, Ton-That H. 2014. Lethality of sortase depletion in Actinomyces oris caused by excessive membrane accumulation of a surface glycoprotein. Mol Microbiol 94:1227–1241. doi: 10.1111/mmi.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ponath F, Zhu Y, Cosi V, Vogel J. 2022. Expanding the genetic toolkit helps dissect a global stress response in the early-branching species Fusobacterium nucleatum. Proc Natl Acad Sci USA 119:e2201460119. doi: 10.1073/pnas.2201460119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 52.Cisar JO, Kolenbrander P, McIntire F. 1979. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect Immun 24:742–752. doi: 10.1128/iai.24.3.742-752.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Materials are available upon reasonable request with a material transfer agreement with UTHealth for bacterial strains or plasmids. The pBCG01 plasmid sequence and map can be accessed on Benchling at https://benchling.com/s/seq-VuTCpImziZSBrxT6otvO?m=slm-jUtGU7tIsA1ZnPAgUH1f.