ABSTRACT
The bacterium Leptothrix cholodnii generates cell chains encased in sheaths that are composed of woven nanofibrils. The nanofibrils are mainly composed of glycoconjugate repeats, and several glycosyltransferases (GTs) are required for its biosynthesis. However, only one GT (LthA) has been identified to date. In this study, we screened spontaneous variants of L. cholodnii SP6 to find those that form smooth colonies, which is one of the characteristics of sheathless variants. Genomic DNA sequencing of an isolated variant revealed an insertion in the locus Lcho_0972, which encodes a putative GT family 8 protein. We thus designated this protein LthB and characterized it using deletion mutants and antibodies. LthB localized adjacent to the cell envelope. ΔlthB cell chains were nanofibril free and thus sheathless, indicating that LthB is involved in nanofibril biosynthesis. Unlike the ΔlthA mutant and the wild-type strain, which often generate planktonic cells, most ΔlthB organisms presented as long cell chains under static conditions, resulting in deficient pellicle formation, which requires motile planktonic cells. These results imply that sheaths are not required for elongation of cell chains. Finally, calcium depletion, which induces cell chain breakage due to sheath loss, abrogated the expression of LthA, but not LthB, suggesting that these GTs cooperatively participate in glycoconjugate biosynthesis under different signaling controls.
IMPORTANCE In recent years, the regulation of cell chain elongation of filamentous bacteria via extracellular signals has attracted attention as a potential strategy to prevent clogging of water distribution systems and filamentous bulking of activated sludge in industrial settings. However, a fundamental understanding of the ecology of filamentous bacteria remains elusive. Since sheath formation is associated with cell chain elongation in most of these bacteria, the molecular mechanisms underlying nanofibril sheath formation, including the intracellular signaling cascade in response to extracellular stimuli, must be elucidated. Here, we isolated a sheathless variant of L. cholodnii SP6 and thus identified a novel glycosyltransferase, LthB. Although mutants with deletions of lthA, encoding another GT, and lthB were both defective for nanofibril formation, they exhibited different phenotypes of cell chain elongation and pellicle formation. Moreover, LthA expression, but not LthB expression, was influenced by extracellular calcium, which is known to affect nanofibril formation, indicating the functional diversities of LthA and LthB. Such molecular insights are critical for a better understanding of ecology of filamentous bacteria, which, in turn, can be used to improve strategies to control filamentous bacteria in industrial facilities.
KEYWORDS: filamentous bacterium, Leptothrix, insertion sequence, next-generation sequencing, sheath formation, sheathless variant
INTRODUCTION
Microorganisms are widely utilized for the remediation of water resources in water purification and wastewater treatment plants, where their innate biological function and metabolism are harnessed to remove contaminants. For example, due to their ability to deposit iron oxides to extracellular polymers, iron-oxidizing bacteria such as Leptothrix and Gallionella are used to remove iron from groundwater in water purification plants (1–4). In contrast to their usefulness, unexpected overgrowth of filamentous bacteria such as Leptothrix, Sphaerotilus, and Thiothrix clogs water distribution systems and causes filamentous bulking of activated sludge (5–7). Therefore, a better understanding of bacterial filamentous growth is important for the proper control of their use in industrial settings (8, 9). However, little is known about the regulation of cell chain elongation in filamentous bacteria.
Leptothrix species, which are closely related to Sphaerotilus, are aerobic, sheath-forming, filamentous, and Gram-negative species that inhabit Fe-rich groundwater, seeps, and streams (10). They form biomats and pellicles (specialized types of biofilms) on submerged solid surfaces and at the air-liquid interface, respectively. Planktonic Leptothrix cells move around in aquatic environments using monopolar flagella (9–12) and attach to favorable surfaces using hair-like appendages called nanofibrils (13). Once attached, the founder cells then initiate cell division to generate chains of daughter cells that are encased in microtubular sheaths composed of entangled nanofibrils. The elongating cell chains interweave with each other using their sheaths and eventually generate biomats consisting of porous networks of cell chains (14, 15). Under static conditions, planktonic cells that move up to the air-liquid interface are trapped by small cell clusters, and the net-like pellicles are formed by randomly docking small cell clusters (9). Nanofibrils play important roles in this process by reducing cell adsorption to the air-liquid interface and enabling cells to move toward each other and stick together (9). Therefore, the regulation of nanofibril formation and thus sheath formation is important for the fundamental understandings of the ecology of Leptothrix.
In general, the cell envelope of Gram-negative bacteria consists of the inner and outer membranes, which are separated by the thin peptidoglycan cell wall called the periplasm (16). In these bacteria, polysaccharide synthesis begins with the transfer of a sugar phosphate onto a lipid carrier, undecaprenyl phosphate, on the cytoplasmic face of the inner membrane. The lipid-linked sugar repeats are then built by the sequential actions of various glycosyltransferases (GTs). The resulting sugar repeat is flipped across the inner membrane, polymerized in periplasmic spaces, and then transported to the outer membrane (17, 18). Leptothrix nanofibrils are mainly composed of a glycoconjugate repeat [→4)-α-GalA-(1→4)-β-GlcNAc-(1→3)-β-GalNAc-(1→4)-α-GalNAAc-(1→4)-α-GalNAc-(1→]n with modifications such as cysteine residue addition (19, 20), implying that several GTs and glycan-processing enzymes are required for their biosynthesis. Approximately 30 GT-encoding genes are annotated in the reference genome of Leptothrix cholodnii SP6. Therefore, it is challenging to systematically disrupt these genes to investigate their mutant phenotypes. To date, only LthA, a GT that is homologous to SthA in Sphaerotilus, has been shown to be required for nanofibril biosynthesis in the Leptothrix (9, 21). Thiol and amino groups in this glycoconjugate repeat are involved in nanofibril entanglement and interaction with iron oxide particles, respectively (3, 22), indicating that glycoconjugates anchoring these functional groups play ecologically important roles for strengthening sheaths.
Intercellular gaps within elongating cell chains and cell escape from the elongating pole(s) are observed during cell filament elongation (13). These characteristics of cell chains suggest that local environmental conditions, such as the availability of dissolved nutrients or minerals, could affect Leptothrix behavior. Indeed, the removal of extracellular calcium ions (Ca2+) was demonstrated to result in the breakage of elongating cell chains (12). Recently, filamentous growth of Sphaerotilus was shown to be regulated by bacterial signaling molecules released in activated sludge (23). These findings imply that filamentous growth associated with sheath formation in the Sphaerotilus-Leptothrix group is regulated by environmental stimuli. We thus hypothesized that the activity and/or expression of enzymes involved in nanofibril biosynthesis might be under the control of such extracellular signals.
Isolation of spontaneous mutants conferring selectable phenotypes is routinely employed in microbiological research (24). Bacterial genomes are widely recognized as being remarkably stable across generations. However, bacterial populations often generate genetic variations, including base substitutions, insertions, deletions, genome rearrangements, and the transfer of exogenous and endogenous DNA, to increase genetic diversity in the presence of exogenous stresses such as antibiotics (25). A number of studies, including metagenome analyses, have revealed that various mobile genetic elements (MGEs), including transposons, viruses, and integrative plasmids, are located in bacterial genomes (26). MGEs that typically encode diverse families of transposases and integrases facilitate the movement of genetic material among bacterial chromosomes (27). Generally, insertion sequences composed of MGEs exhibit short terminal inverted repeat (IR) sequences of 10 to 40 bp. The IRs are further divided into two elements: inverted repeats are involved in transposase binding, while target repeats locating at the terminal 2 to 3 bp of the IR sequence are involved in the cleavage and strand transfer reactions that lead to element transposition (28).
In L. cholodnii, sheathless variants can be isolated from morphologically abnormal colonies that form after repeated culturing (5 to 15 times) (29, 30). These findings indicate that these spontaneous mutations in the L. cholodnii genome might be caused by active MGEs. However, the mutation point(s) and MGEs in these mutants remain unclear. In this study, we isolated a novel sheathless mutant using the “step-by-step” culturing strategy. By comparing the genomic data for the mutant to those for its parental strain, we were able to easily determine the critical mutation for a given phenotype. We thus identified lthB, a GT-encoding gene that was responsible for sheath formation, and further characterized its deletion mutant.
RESULTS AND DISCUSSION
Isolation of a variant that forms smooth colonies due to a lack of sheath formation.
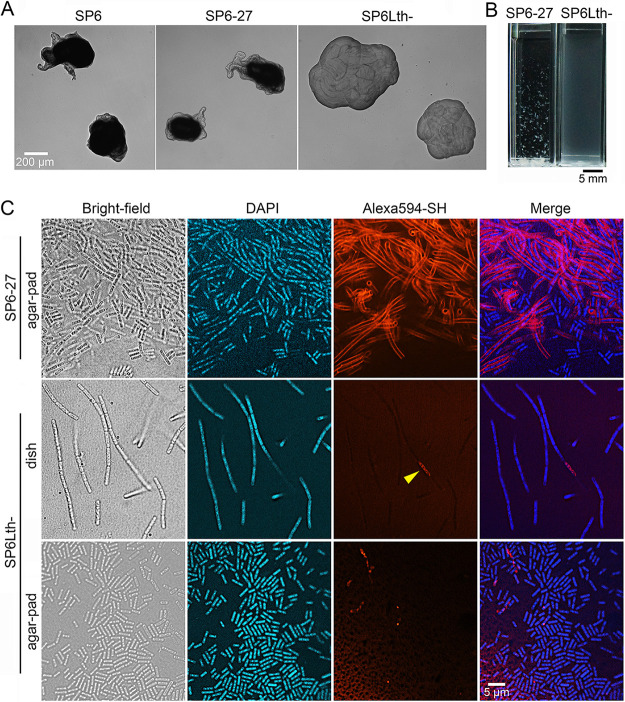
Sheathless variants of Leptothrix cholodnii have been obtained by repeated culturing 5 to 15 times (29, 30), suggesting that spontaneous mutations frequently occur in L. cholodnii during repeated culture. However, the mutation points in these spontaneous variants remain unclear. In this study, we used SP6 to implement a similar cyclic strategy consisting of cycles of liquid culturing, plating, and single-colony isolation, to elicit variants that formed smooth colonies. The formation of smooth colonies is a characteristic of sheathless mutants, referred to here as the Lth− phenotype. The step-by-step culturing facilitated the identification of critical mutations responsible for the phenotypes that we describe in this study. After 28 cycles, we successfully obtained a candidate (referred to here as SP6Lth−) that formed smooth colonies (Fig. 1A, right). In contrast, a single colony that was isolated after 27 cycles (referred to here as SP6-27) formed rough colonies (Fig. 1A, middle) that was similar to that of wild-type SP6 cells (Fig. 1A, left). However, SP6-27 cells may carry spontaneous mutations that are not responsible for the Lth− phenotype.
FIG 1.
Isolation of a spontaneous mutant, SP6Lth−. (A) Colony morphology of SP6 (left), SP6-27 (middle), and SP6Lth− (right) cells on MSVP plates incubated for 3 days. (B) Images of SP6-27 and SP6Lth− cells from 2-day-old shaking cultures. (C) Fluorescent staining of nanofibril sheaths and DNA with Alexa Fluor 594-SH and DAPI, respectively, in SP6-27 and SP6Lth− cells cultured under shaking conditions. Stained cells were imaged in liquid culture in glass-bottom dishes (middle) or after being spotted and dried on agar pads (top and bottom). The arrowhead indicates intracellular fluorescence.
In a shaking culture, SP6-27 cells generated aggregates composed of fibrous filaments. In contrast, SP6Lth− cells did not aggregate but spread throughout the culture medium (Fig. 1B), similar to the cloudy medium that was previously observed when the sheathless ΔlthA::kanR mutant was cultured (9). To verify the absence of sheath structure around the periphery of SP6Lth− cells, cultures that had undergone 2 days of shaking were stained with the fluorescent reagent Alexa Fluor 594 C5-maleimide (hereafter Alexa Fluor 594-SH) (12, 19) and DAPI (4′,6-diamidino-2-phenylindole) to visualize nanofibril sheaths and DNA, respectively. As expected, except for a few cells, no fluorescence from Alexa Fluor 594-SH was observed around the SP6Lth− cells (Fig. 1C, middle, arrowhead). Most SP6Lth− cell chains were broken into multiple single cells when spotted and dried on an agar pad (Fig. 1C, bottom), which strongly suggests that these cell chains failed to generate sheaths. In contrast, SP6-27 cells persisted as cell chain aggregates, even on agar pads. Strong fluorescence enclosing cell chains was also observed (Fig. 1C, top), suggesting that these SP6-27 cell chains were still encased in sheaths. The lack of sheath structure around the SP6Lth− cells was also confirmed by atmospheric scanning electron microscopy (ASEM) (see Fig. S1 [right] in the supplemental material). In contrast, SP6-27 cell chains were encased in sheath structures (Fig. S1, left, red dashed lines). Together, these results clearly indicate that a spontaneous mutation(s) that arose during culturing of SP6-27 cells produced the SP6Lth− variant, which lacked nanofibril sheaths and hence formed smooth colonies.
Determination of the mutation point(s) critical for the Lth− phenotype by comparing genomic DNA sequences.
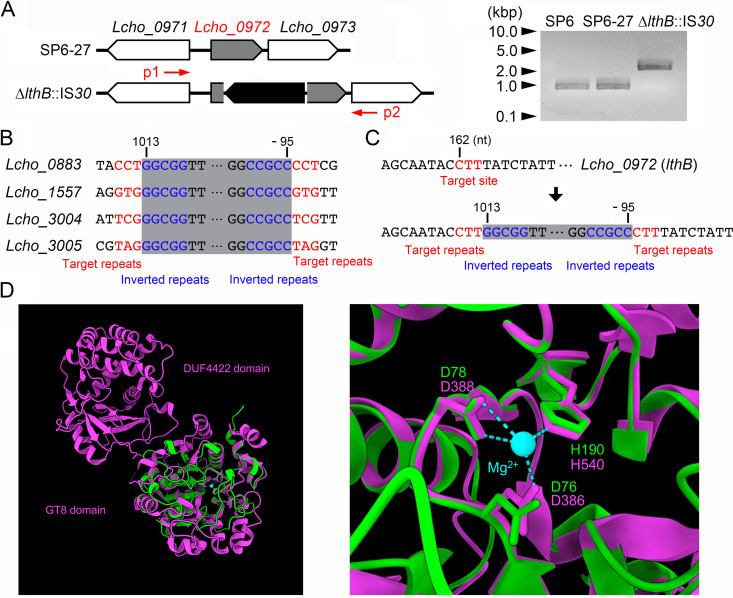
To identify the spontaneous mutation point(s) responsible for the characteristics that were observed in the SP6Lth− cells, we used next-generation sequencing (NGS) to analyze the genomic DNA (gDNA) isolated from SP-6Lth− cells and their parental SP6-27 cells. Compared to the reference SP6 genome, breseq software identified several point mutations even in the SP6-27 genome (Table S1). Since the phenotype of the parental SP6-27 cells was similar to the wild-type phenotype, we concluded that these point mutations were not critical for the Lth− phenotype, and we therefore ignored them. Compared to the SP6-27 genome, only an insertion sequence (IS) element encoding an IS30 family transposase into the Lcho_0972 locus was found in the SP6Lth− genome (Fig. 2A, left; Tables S1 and S2). In the reference SP6 genome, four copies of the same IS element are present at Lcho_0883, Lcho_1557, Lcho_3004, and Lcho_3005. These elements have conserved short terminal 5-bp inverted repeats and nonconserved 3-bp target repeats (Fig. 2B, in red and blue, respectively), which were also found in the flanking regions of the IS element inserted into Lcho_0972 in SP6Lth− cells (Fig. 2C). These results indicated that one copy of the IS element was duplicated and inserted into Lcho_0972 during culture of the SP6-27 cells. In agreement with the NGS data, the PCR product from the SP6Lth− genome (~2.0 kbp) that was amplified from a region between Lcho_0971 and Lcho_0973 was larger than the corresponding PCR products amplified from SP6 and SP6-27 genomes (~0.9 kbp) (Fig. 2A, right; red arrows [p1 and p2] in Fig. 2A, left).
FIG 2.
Determination of an insertion sequence within the Lcho_0972 gene, which encodes a putative glycosyltransferase. (A) (Left) Schematic of an insertion sequence within the Lcho_0972 (lthB) gene. A primer set is indicated by red arrows. (Right) PCR confirmation of an insertion into the Lcho_0972 gene using gDNA isolated from SP6, SP6-27, and SP6Lth− cells. Size markers are indicated on the left. (B) Loci containing identical insertion sequences in the reference SP6 genome. Numbers indicate the nucleotide position relative to the start codon. (C) Detailed sequence of the Lcho_0972 gene before (top) and after (bottom) the insertion. Numbers indicate nucleotide position relative to the start codon of Lcho_0972 and the transposase, respectively. (D) Superposition of the Lcho_0972 protein (green) and WbbM (magenta). Structural alignments show that the conserved aspartic acid and histidine residues in the Lcho_0972 protein are predicted to be located around the Mg2+ ion, as in WbbM. The right image is focused on the active center of GT8 with the cofactor Mg2+.
Lcho_0972 encodes a conserved hypothetical protein that is annotated in GenBank (31). Interestingly, the Pfam (32) and InterPro (33) databases predict that this hypothetical protein belongs to glycosyltransferase family 8 (GT8), which is known to transfer diverse sugars onto lipo-oligosaccharides, proteins, inositol, oligosaccharides, or polysaccharide acceptors (34, 35). The enzyme activity of GT8 enzymes require the interaction of the conserved DxD motif and C-terminus histidine residues with a divalent metal ion (Mn2+ or Mg2+), which acts as an essential cofactor (36–38). We compared amino acid sequences between the Lcho_0972 protein and other GT8 enzymes, whose structures were experimentally determined, and found that these residues are conserved in the Lcho_0972 protein (Table S3; Fig. S2). Recently, a tool for the prediction of three-dimensional structure from a primary amino acid sequence was provided by the AlphaFold protein structure database (39). We thus compared the structure between the Lcho_0972 protein and WbbM bound with Mg2+. A structural alignment showed that the conserved aspartic acid and histidine residues in the Lcho_0972 protein were predicted to be situated around the Mg2+ ion, similar to WbbM (Fig. 2D). These data support our claim that the Lcho_0972 protein is a GT8 enzyme. We therefore designated the Lcho_0972 protein LthB, another glycosyltransferase required for nanofibril formation in SP6, and refer to the SP6Lth− strain as the lthB::IS30 strain.
Generation of anti-LthB antibody and analyses of LthB expression and localization in SP6 cells.
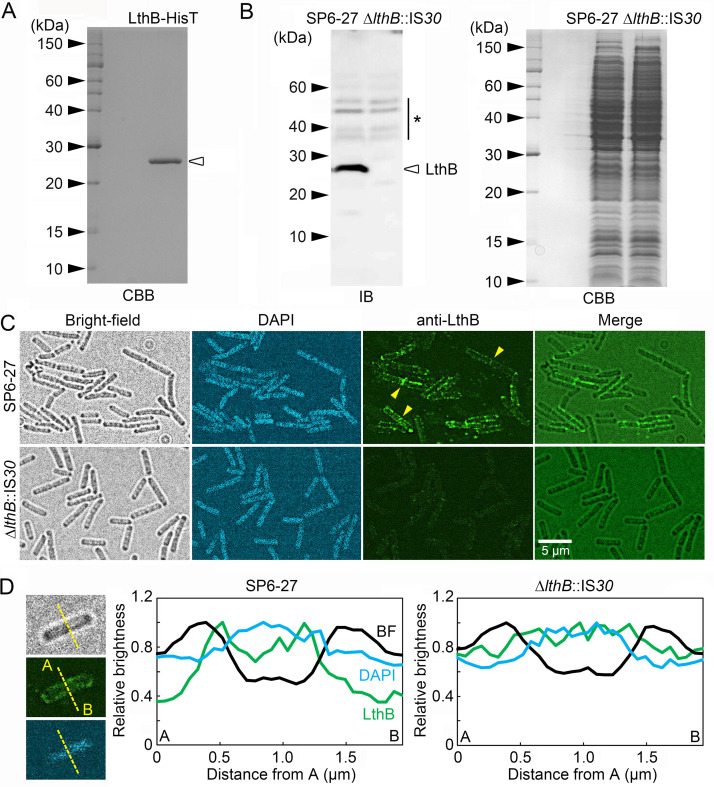
To detect LthB in SP6 cells using a polyclonal antibody, a His-tagged LthB protein (referred to here as LthB-HisT) with a predicted molecular weight of ~26 kDa was expressed in Escherichia coli cells and purified from their cell lysate using a resin that binds specifically to HisT (Fig. 3A). We then generated a polyclonal antibody against purified LthB-HisT in rabbits. The resulting serum was tested for specificity to LthB by immunoblotting using cell extracts from SP6-27 and ΔlthB::IS30 cells. A chemiluminescence image showed that a strong band at approximately 26 kDa was detected in SP6-27 cells, which had an intact lthB (Lcho_0972) locus (Fig. 3B, left). Despite the fact that nearly equal amounts of cell extract were loaded, no corresponding band was detected in ΔlthB::IS30 cells, which had an IS element at the lthB locus (Fig. 3B, right). Although several minor bands were also detected, these results indicate that a LthB-specific antibody was successfully generated.
FIG 3.
Immunoblotting and immunostaining of SP6-27 and ΔlthB::IS30 (SP6Lth−) cells using an anti-LthB antibody. (A) Purified LthB-HisT protein that was used to acquire rabbit anti-LthB antibodies. (B) Immunoblotting of LthB in cell extracts from SP6-27 and ΔlthB::IS30 cells using anti-LthB antibodies (left). CBB staining of an SDS-PAGE gel loaded with SP6-27 and ΔlthB::IS30 cell extracts, showing that almost the same amount was loaded (right). Size markers are indicated on the left of each gel. The asterisk indicates nonspecific bands. (C) Immunostaining of SP6-27 and ΔlthB::IS30 cells using anti-LthB antibodies. DNA was stained with DAPI. Fluorescence detected around septa between two connected cells is indicated by arrowheads. (D) Relative signal intensities of bright-field (black), DAPI (blue), and anti-LthB (green) at a cell cross section of SP6-27 (left) and ΔlthB::IS30 (right) cells. Average intensities from >15 cells were plotted.
The rabbit serum was then used for immunostaining to examine the localization of LthB in SP6-derivative cells. Strong pericellular fluorescent signals were observed in SP6-27 cells (Fig. 3C, top) but not in ΔlthB::IS30 cells (Fig. 3C, bottom). Relative fluorescent intensities at a cross-section of SP6-27 cells showed that the anti-LthB signal tended to be high between the peaks of the bright-field and DAPI signals, which represent the cell envelope and DNA-containing cytoplasm, respectively (Fig. 3D). Assuming that the glycoconjugate repeats of nanofibrils might be synthesized at the cytosolic side of the inner membrane, as was reported for other Gram-negative bacteria (17, 18), we surmised that the strong anti-LthB signal that was observed adjacent to the cell envelope was reasonable if LthB is required for nanofibril synthesis as a glycosyltransferase. However, further analyses are required to determine the detailed localization of LthB.
Confirmation of characteristics associated with lack of lthB.
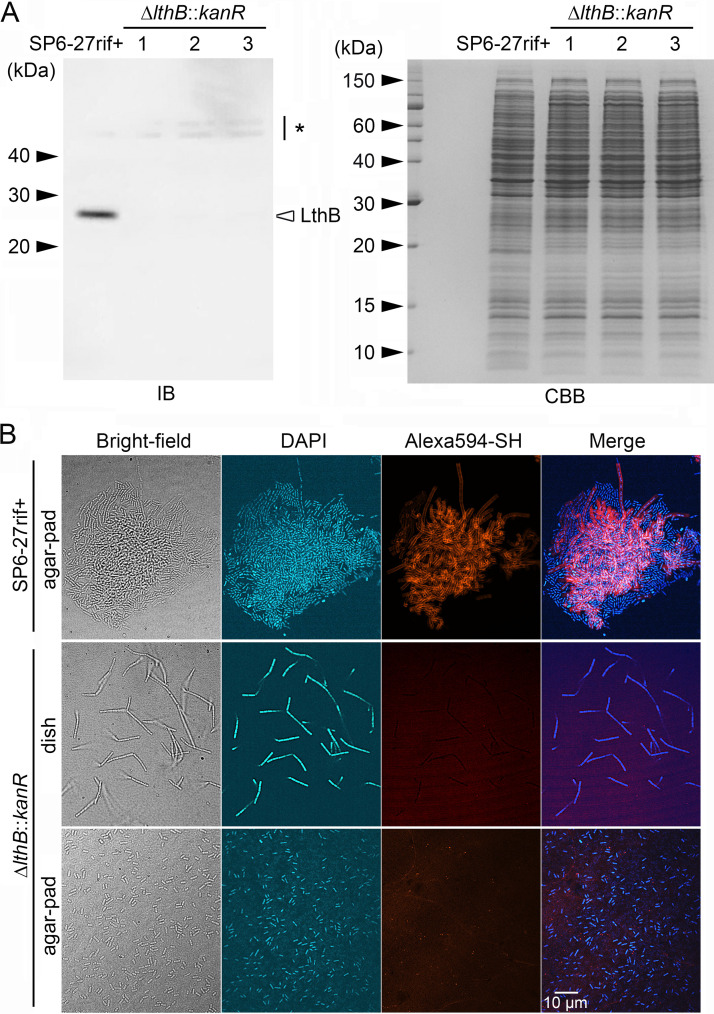
To confirm whether the Lth− phenotype in ΔlthB::IS30 cells is entirely dependent on the lack of the lthB gene, we next generated a deletion mutant from SP6-27 cells. Reverse transcription-PCR (RT-PCR) analysis using mRNA extracted from SP6, SP6-27, and ΔlthB::IS30 cells revealed that the expression of the Lcho_0973 gene, which is located downstream of the lthB (Lcho_0972) gene, was abrogated by the ΔlthB::IS30 mutation (Fig. S3A), suggesting that these genes are under the control of the same promoter. Therefore, both lthB (Lcho_0972) and Lcho_0973 were designed to be replaced by the kanamycin resistance gene (kanR) (Fig. S3B, top). For antibiotic selection after conjugation with E. coli, we first isolated a rifampicin-resistant variant of SP6-27 cells (SP6-27rif+). Gene replacement in this strain was then performed by plasmid transfer from E. coli. After acquiring candidate clones with the lthB (Lcho_0972)-Lcho_0973 deletion (referred to here as ΔlthB::kanR cells), the complete replacement of both Lcho_0972 and Lcho_0973 genes with kanR was confirmed in three independent candidates by colony PCR (Fig. S3B, bottom) and immunoblotting (Fig. 4A).
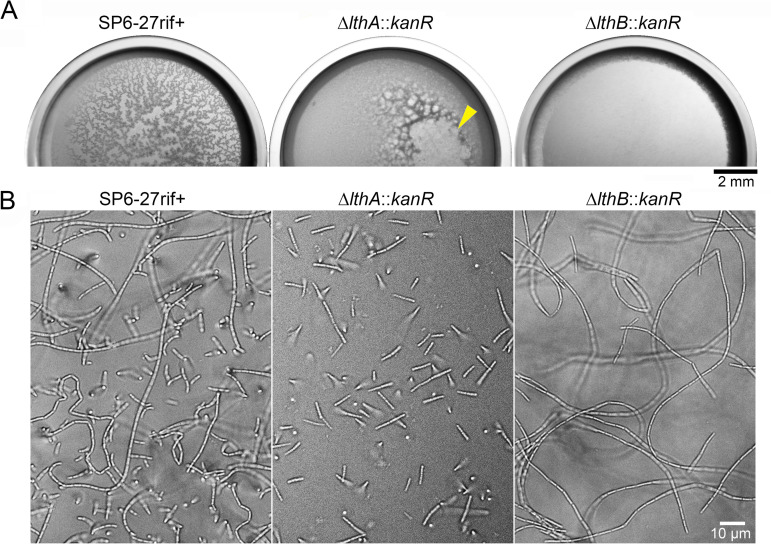
FIG 4.
Replacement of the Lcho_0972 (lthB) and Lcho_0973 genes with kanR in SP6-27 cells resulted in nanofibril-free cells that were similar to ΔlthB::IS30 cells. (A) Immunoblotting of LthB in cell extracts from SP6-27rif+ cells and their ΔlthB::kanR disruptants using an anti-LthB antibody (left). The asterisk on the right indicates nonspecific bands. An SDS-PAGE gel loaded with cell extracts from SP6-27rif+ cells and their ΔlthB::kanR disruptants was stained with CBB (right). Size markers are indicated on the left. (B) Fluorescent staining of nanofibril sheaths and DNA with Alexa Fluor 594-SH and DAPI, respectively, from SP6-27rif+ cells and their ΔlthB::kanR disruptant mutants under shaking conditions. Stained cells were imaged in liquid culture in glass-bottom dishes (middle) or after they were spotted and dried on agar pads (top and bottom).
Fluorescent staining of nanofibrils by Alexa Fluor 594-SH revealed that ΔlthB::kanR cells in shaking cultures form short cell chains that lack sheath support (Fig. 4B, middle) and break into multiple single cells when spotted onto agar pads and air dried (Fig. 4B, bottom). In contrast, the cell chains of the parental SP6-27rif+ culture became entangled and formed small aggregates, which remained intact on the air-dried agar pad (Fig. 4B, top). These results showed phenotypic similarities between the SP6-27 and ΔlthB::IS30 cells (Fig. 1C). To confirm that the phenotypic change was caused by the loss of the lthB gene, we generated another Lcho_0973 deletion mutant (referred to here as the ΔLcho_0973::kanR mutant), which had the intact lthB (Lcho_0972) gene (Fig. S4A, top). Colony PCR was used to confirm the replacement of Lcho_0973 with kanR in the ΔLcho_0973::kanR candidates (Fig. S4A, bottom), and we found that all independently isolated ΔLcho_0973::kanR clones generated aggregates in shaking cultures (Fig. S4B). Furthermore, fluorescent staining of nanofibrils by Alexa Fluor 594-SH revealed that the cell chains of the ΔLcho_0973::kanR became entangled to form small aggregates, which persisted even after spotting onto the air-dried agar pad (Fig. S4D). Accordingly, we inferred that deletion of lthB, but not Lcho_0973, disrupted the glycoconjugate synthesis, which in turn resulted in sheathless cells exhibiting the Lth− phenotype.
Pellicle formation associated with planktonic-cell generation from cell chains.
We previously found that, under static conditions, planktonic cells move up to the air-water interface and generate porous pellicles (9, 12). In contrast, ΔflgAB::kanR cells, which lack flagellum-driven motility, failed to form pellicles. The sheathless mutant ΔlthA::kanR cells generated dense pellicles that were distinct from those generated by SP6 cells, indicating that the flagella that are used to swim toward the air-liquid surface and the nanofibril sheaths that are used to capture swimming cells are both critical for porous-pellicle formation (9). To examine whether ΔlthB mutant cells generated porous or dense pellicles, we inoculated ΔlthB mutant cells and their parental cells in dishes and cultured them under static conditions. Surprisingly, ΔlthB mutant cells did not generate pellicles in either static dishes or cuvette cultures (Fig. 5A; Fig. S5, right), while parental SP6-27 cells, SP6-27rif+ cells, and ΔLcho_0973::kanR cells formed porous pellicles at the same rate (Fig. 5A; Fig. S4C right; Fig. S5, left). For a while, ΔlthA::kanR cells formed dense pellicles that collapsed after thickening (Fig. 5A, middle, yellow arrowhead) (9). In a static glass-bottom dish culture, both parental SP6-27rif+ cells and ΔlthB::kanR cells elongated to form long cell chains (Fig. 5B, left and right, respectively). In contrast, ΔlthA::kanR cells divided into single cells and/or groups of several cells during elongation (Fig. 5B, middle), as reported previously (9). Notably, planktonic cells were observed in SP6-27rif+ and ΔlthA::kanR cultures but not in the ΔlthB::kanR culture (Fig. 5B). Moreover, ΔlthA::IS30 ΔlthB::kanR double mutant cells had a tendency to generate cell chains in static conditions and thus failed to form pellicles due to lower number of planktonic cells (Fig. S6). These results implied that deficient pellicle formation in ΔlthB::kanR and ΔlthA::IS30 ΔlthB::kanR cells could be due to the lack of planktonic cells, whose motility is required for movement toward the air-liquid interface.
FIG 5.
Different cell properties in ΔlthA and ΔlthB cells. (A) Pellicle formation in static dish cultures showing that ΔlthB::kanR cells failed to form pellicles. The arrowhead indicates the collapse of the ΔlthA::kanR pellicle. (B) Chain formation of SP6-27rif+, ΔlthA::kanR, and ΔlthB::kanR cells at the bottom surface of the glass.
Deficient cell division of ΔlthB cell chains.
To confirm that ΔlthB::kanR cells form long cell chains without the support of sheaths under static conditions, we stained nanofibril sheaths with Alexa Fluor 594-SH or fluorescein-NH2. Long cell chains of parental SP6-27rif+ cells in the static dish culture were clearly encased in sheaths (Fig. S7, top, and Fig. S8, top). In contrast, except for a few cells, there was no strong fluorescence around ΔlthB::kanR cell chains when they were stained with fluorescein-NH2 and Alexa Fluor 594-SH (Fig. S7, middle, and Fig. S8, middle). As was seen with ΔlthB::IS30 cells, these long ΔlthB::kanR cell chains broke into multiple single cells when spotted and air dried on agar pads (Fig. S7, bottom, and Fig. S8, bottom). Since there were propidium iodide (PI)-positive cells in ΔlthB::kanR cell chains under static culture conditions (Fig. S9), we inferred that Alexa Fluor 594- and fluorescein-positive cells in these cell chains could be an artifact of cell death rather than nanofibril formation. Indeed, Leptothrix cells that form cell chains often induce autolysis, presumably due to abrupt nutrient limitations (12, 40). Based on these results, we concluded that ΔlthB::kanR cells are able to generate long cell chains without the support of sheaths under static culture conditions. Thus, sheaths are not required for cell chain elongation.
Importantly, the mean lengths of single rod-shaped ΔlthB::kanR cells on agar pads were comparable to those of single SP6-27rif+ cells (Fig. 4B, top and bottom; Fig. S10A). This suggests that the peptidoglycan synthesis that is required for cell enlargement and cell division (41) was not impaired in ΔlthB::kanR cells. In addition, staining cell membranes with FM1-43 indicated the presence of septa produced at cell ends of the ΔlthB::kanR cell chains, comparable to the parental SP6-27rif+ cell chains (Fig. S10B). We therefore suggest that the deletion of lthB gene does not interfere with the cell division process. However, it remains possible that the loss of LthB, but not LthA, slowed the cleavage of the septum by the cell division complex (divisome) at the final stages of cell division (42, 43). In support of this possibility, we noticed that strong fluorescent signals from anti-LthB were often detected at cell-cell junctions (Fig. 3C, yellow arrowheads).
Expression of LthA and LthB under Ca2+ depletion.
We reported previously that breakage of elongating cell chains occurs via the removal of extracellular calcium ions (Ca2+) (12). However, low Ca2+ levels have been detected by elemental analysis of SP6 sheaths (44). We thus inferred that Ca2+ could be important for signal transduction, rather than the strengthening of the sheath structure (e.g., by nanofibril cross-linking). We therefore examined whether the expression of LthA and/or LthB was abrogated by Ca2+ depletion. To detect LthA expression, we generated a polyclonal antibody against a peptide mixture (the peptides consisted of 184 to 197 and 218 to 231 amino acids) in rabbits. The resulting immune serum was tested for specificity against LthA by immunoblotting using cell extracts from the ΔlthA::kanR strain and its parental rifampicin-resistant strain (SP6rif+). A chemiluminescence image shows that a band at approximately 55 kDa was detected in SP6rif+ cells, which have a wild-type Lcho_3510 (lthA) locus (Fig. S11, left, first lane). In contrast, no corresponding band was detected in ΔlthA::kanR cells, despite loading of nearly equal amounts of cell extracts (Fig. S11, left, second lane). We therefore concluded that the polyclonal antibody against LthA was successfully generated, although a strong nonspecific band was also observed (Fig. S11, right, asterisk). Next, SP6rif+ cells were cultured in medium that contained the Ca2+ chelator EGTA (MSVP+EGTA) or in MSVP without calcium chloride (MSVP−Ca) (Tables S4 and S5), and cell extracts were used for immunoblots. Although the expression of LthB under Ca2+-depleted conditions (+EGTA and −Ca) was comparable to that under control conditions (−EGTA and +Ca) (Fig. 6, right), the expression of LthA was abrogated by Ca2+ depletion (+EGTA and −Ca) (Fig. 6, left). The strong band (~30 kDa) that was expressed in cells cultured under the calcium-free conditions was considered nonspecific, because a similar band was detected in the ΔlthA::kanR cell extracts (Fig. S12). These results imply that the extracellular Ca2+ depletion affects the regulator(s) of LthA expression; thus, the loss of LthA expression abrogated nanofibril formation and resulted in cell chain breakage.
FIG 6.
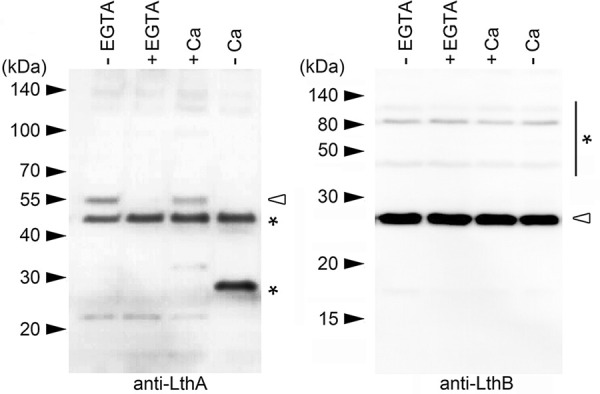
Abrogation of LthA expression by Ca2+ depletion. Immunoblots of cell extracts from SP6rif+ cells using anti-LthA (left) and anti-LthB (right) antibodies, respectively. SP6rif+ cells were cultured in EGTA-free (lane 1) and EGTA-containing (lane 2) MSVP for 6 h. SP6rif+ cells were also cultured in MSVP (lane 3) and Ca2+-free MSVP (−Ca) (lane 4) for 12 h. Size markers are indicated on the left. Asterisks indicate nonspecific bands.
Conclusion and outlook.
In this study, we used the step-by-step culturing strategy to screen spontaneous variants showing the Lth− phenotype, and used genomic sequencing analyses to determine their critical mutation(s). The NGS data suggested that the SP6 genome was relatively stable across generations; even after 28 cycles of repeated culture, only 7 point mutations and a single IS30 translocation were detected in the genome of the ΔlthB::IS30 strain. The stability of the SP6 genome suggests that the mutation(s) critical for the Lth− phenotype in previously isolated mutants such as SP6SL (29) might be elucidated by simple comparison of their genomic sequence with the reference.
Using genomic screening, we identified a spontaneous variant in which an IS30 element had been inserted into the lthB gene, which encodes a glycosyltransferase (GT) type 8 protein. Our findings show that the loss of LthA and LthB impairs nanofibril biosynthesis in L. cholodnii SP6, suggesting that the expression of these GTs is essential for the production of glycoconjugate repeats that make up nanofibrils. However, ΔlthA and ΔlthB cells exhibited different characteristics, including pellicle formation, which is closely associated with planktonic-cell generation. We observed that short ΔlthA cell chains broke into single cells, whereas long ΔlthB cell chains failed to generate single cells. Although further analyses are needed, we infer that LthB plays functional roles not only in nanofibril biosynthesis but also in cell division.
Consistent with these findings, extracellular Ca2+ depletion, which causes cell chain breakage, abrogated LthA expression but not LthB expression. These findings suggest that Ca2+ signaling may regulate nanofibril biosynthesis through the regulation of LthA expression. Although the role of Ca2+ in prokaryotes is largely elusive, LthA expression might be regulated by Ca2+-binding proteins (CaBPs), which have been shown to affect physiological processes in bacteria, including the maintenance of cell structure, motility, membrane transport, and cell differentiation (45).
The present findings provide a deeper understanding of the molecular mechanisms related to cell chain elongation that underpin the ecology of filamentous bacteria, which may contribute to the development of regulatory technologies for water treatment facilities in the future.
MATERIALS AND METHODS
Strains and culture conditions.
In this study, we used Leptothrix cholodnii SP-6 (ATCC 51168) (29) (referred to here as SP6) and its derivatives (Table 1). We cultured these strains by transferring frozen stock to an Minerals Salts Vitamins Pyruvate (MSVP) agar plate (Tables S4 and S5) (29), which was incubated at room temperature. After 4 to 7 days of culture, we transferred 1 to 3 single colonies into 25 mL of liquid MSVP medium, which was then incubated in a reciprocating shaker at 70 rpm for an additional 2 days at room temperature. For gene disruption in SP6 and its derivatives, we obtained spontaneous rifampin-resistant strains (SP6rif+, SP6-27rif+, and ΔlthB::IS30rif+) by successive transfer of SP6 cells to MSVP plates supplemented with rifampin (Rif [10, 15, and 20 mg/L]) (9). For the static condition, a sufficient amount of the 2-day-old shaking culture was inoculated into cuvettes (AS ONE, Osaka, Japan) or glass-bottom dishes (Iwaki, Tokyo, Japan) and incubated for 1 to 2 days. To culture SP6rif+ and ΔlthA::kanR cells under calcium-free conditions, 3 mM EGTA (Nacalai Tesque, Kyoto, Japan) was added to MSVP to chelate calcium ions. We also cultured these cells in a MSVP solution without calcium chloride (MSVP−Ca) (Tables S4 and S5) (12). The Escherichia coli strains SN1187 (NBRP E. coli at National Institute of Genetics) (46), S17I (our laboratory stock), and Rosetta 2(DE3) (Merck, Darmstadt, Germany) were used for plasmid manipulation, plasmid transfer via conjugation, and recombinant protein expression, respectively. E. coli carrying a plasmid was cultured in L broth (LB) medium (47) containing antibiotics such as ampicillin sodium and kanamycin sulfate (Km; Nacalai Tesque) for approximately 12 h in a reciprocating shaker at 30 to 37°C and 190 rpm.
TABLE 1.
Leptothrix cholodnii strains used in this study
| Strain | Description | Background | Reference |
|---|---|---|---|
| SP6 | Wild type | 29 | |
| SP6rif+ | Rifampicin resistant (undefined) | SP6 | 9 |
| SP6-27 | Several mutations not responsible for the Lth− phenotype (Tables S1 and S2) | SP6 | This study |
| SP6Lth− (ΔlthB::IS30) | ΔlthB::IS30 and several mutations not responsible for the Lth− phenotype (Tables S1 and S2) | SP6-27 | This study |
| SP6-27rif+ | Rifampicin resistant (undefined) | SP6-27 | This study |
| ΔlthB::IS30rif+ mutant | Rifampicin resistant (undefined) | ΔlthB::IS30 | This study |
| ΔlthA::kanR mutant | ΔlthA::kanR | SP6rif+ | 9 |
| ΔlthB::kanR mutant | ΔlthB::kanR | SP6-27rif+ | This study |
| ΔlthA::kanR ΔlthB::IS30 mutant | ΔlthA::kanR ΔlthB::IS30 | ΔlthB::IS30rif+ | This study |
| ΔLcho_0973::kanR mutant | ΔLcho_0973::kanR | SP6rif+ | This study |
Genomic DNA isolation from SP6, SP6-27, and SP6Lth− cells.
Genomic DNAs were isolated according to previously described methods for use in PCR and NGS (40). Briefly, SP6, SP6-27, and SP6Lth− cells that had been cultured for 2 days in 25 mL MSVP medium were collected by centrifugation, washed twice in TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 8.0) and suspended in 5 mL of TE buffer containing 20 μg/mL proteinase K (FujiFilm Wako, Osaka, Japan) and 0.6% sodium dodecyl sulfate (SDS; FujiFilm Wako). After incubation at 65°C for 1 h, 1 mL of 5 M NaCl and 800 μL of CTAB-NaCl solution (10% cetyltrimethylammonium bromide and 0.7 M NaCl) were added to the cell suspension, which was further incubated at 65°C for 1 h. To eliminate proteins, the cell suspension was sequentially extracted using an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) solution (FujiFilm Wako) and chloroform (Nacalai Tesque, Kyoto, Japan). To precipitate nucleic acids, the resulting supernatant was added to an equal volume of isopropanol (Nacalai Tesque), incubated at −20°C for 1 h, and then centrifuged at 7,000 × g for 20 min. The pellet was rinsed in 70% ethanol and air dried for 20 min. To eliminate RNA, the pellet was suspended in 500 μL of TE buffer containing 20 μg/mL RNase A (TaKaRa Bio, Inc., Shiga, Japan) and incubated at 37°C for 30 min, and DNA was extracted using phenol-chloroform-isoamyl alcohol (25:24:1) and chloroform. The ethanol-precipitated gDNA was dissolved in 50 μL of TE buffer.
Genome data.
The SP6 genome was downloaded from GenBank (accession no. GCA_000019785.1) and used as the reference sequence in NGS analyses. The accession numbers of the genes used for multiple sequence alignment and structural alignment are listed in Table S3.
Next-generation sequencing and analysis.
Next-generation sequencing was performed to determine the mutations responsible for the Lth− phenotype in SP6Lth− (ΔlthB::IS30) cells. Genomic DNA was isolated from SP6-27 and SP6Lth− (ΔlthB::IS30) cells as described above. Library preparation and DNA sequencing were performed by Bioengineering Lab. Co., Ltd. (Kanagawa, Japan). Whole-genome shotgun libraries were then constructed using an MGIEasy FS DNA library preparation set (MGI Tech, Shenzhen, China). After these libraries were circularized using an MGIEasy circularization kit, DNA nanoballs were prepared using a DNBSEQ-G400RS high-throughput sequencing set and were sequenced on a DNBSEQ-G400 instrument (paired end, 200-bp reads). The sequencing data were analyzed as follows. The software fastp (version 0.23.2) (48) was used to remove sequences with low-quality scores and trim adapters from raw fastq files, using the command “-q 20–adapter_sequence=AAGTCGGAGGCCAAGCGGTCTTAGGAAGACAA–adapter_sequence_r2 = AAGTCGGATCGTAGCCATGTCGTTCTGTGAGCCAAGGAGTTG.” Mutations from the reference L. cholodnii SP6 genome were predicted using breseq software (version 0.36.1) (49) with the default options selected. fastp and breseq software programs were downloaded from the BioContainers registry (50).
Sequence and structure alignment.
To confirm that LthB is a glycosyltransferase family 8 protein, a multiple-amino-acid-sequence alignment of LthB and other GT family 8 proteins was performed with T-Coffee software (version 13.41.0.28bdc39) (51) on the EMBL-EBI website (52) with the default options selected. Although WbbM has two domains (N terminus, DUF4422, and C terminus, GT8), only the C terminus GT8 domain (260 to 635) was used for multiple sequence alignment. A structural alignment of LthB and WbbM was performed using the Matchmaker tool in ChimeraX software (version 1.4) (53) with default options selected.
Plasmid construction.
PCRs for plasmid construction were performed using PrimeSTAR Max DNA polymerase (TakaraBio, Kusatsu, Shiga, Japan). To obtain deletion mutants, two plasmids were constructed as described previously (Table 2) (9). To generate a plasmid for the replacement of lthB (Lcho_0972) and Lcho_0973 with the Km resistance gene kanR, ~1.5-kbp fragments located upstream of the lthB (Lcho_0972) gene and downstream of the Lcho_0973 gene were amplified by PCR from isolated SP6 gDNA (Fig. S3A) using the primer sets given in Table S6 (Lcho_0972_outF to Lcho0972_pUC18_R). The resulting fragments and the pUC-kanR plasmid were used to construct the lthB (Lcho0972)-Lcho0973::kanR cassette via overlap extension PCR, and the cassette was inserted into the pUC-mob plasmid using the iVEC system (9, 46). The sequence of the resulting plasmid (pUC-mob-Lcho_0972-0973::kanR) was confirmed by colony PCR and subsequent sequencing. Another plasmid (pUC-mob-Lcho_0973::kanR) (Fig. S4A) that was used for the disruption of only Lcho_0973 was constructed in the same way. The plasmid pUC-mob-lthA::kanR, which was previously constructed using the same method, was used to disrupt the lthA gene (Fig. S6A) (9). To express a recombinant LthB protein for use as an antigen in E. coli, an expression vector of the LthB-HisT fusion protein [pET24a(+)-LthB] (Table 2) was constructed as follows: an ~0.7-kbp fragment was PCR amplified from SP6 gDNA, and the primers Lcho_0972_F_NdeI_pET and Lcho_0972_R_XhoI_pET were cloned into the NdeI-XhoI gap of pET24a(+) (Table S6). The construction of the plasmid was confirmed by sequencing.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference |
|---|---|---|
| pUC-mob-lthB (Lcho0972)-Lcho_0973::kanR | Replacement of lthB (Lcho_0972) and Lcho_0973 with kanR | This study |
| pUC-mob-Lcho_0973::kanR | Replacement of Lcho_0973 with kanR | This study |
| pUC-mob-Lcho_lthA::kanR | Replacement of lthA (Lcho_3510) with kanR | 9 |
| pET24a(+)-LthB | Expression of LthB-HisT fusion protein in E. coli | This study |
Gene disruption by conjugation between E. coli S17I and rifampicin-resistant Leptothrix cells.
To obtain deletion mutants, plasmids were transferred through conjugation between E. coli S17I and SP6-derived cells as described previously (9, 54). We used spontaneous rifampicin-resistant mutants (referred to as SP6rif+, SP6-27rif+, and the lthB::IS30rif+ mutant) to select SP6-derived colonies on MSVP plates containing the antibiotics Km and Rif. After conjugation, the bacterial mixture suspension was spread and incubated on MSVP plates containing Km and Rif for approximately 1 week. Gene replacement with kanR in candidate colonies was confirmed by colony PCR using the primers Lcho_0972_outF, Km_Lcho0972_R, Lcho0972_Km_F, and Lcho_0972_outR for lthB (Lcho_0972)-Lcho_0973, primers Lcho_0973_outF, Km_Lcho0973_R, Lcho0973_Km_R, and Lcho_0973_outR for Lcho_0973, and primers Lcho_3510_F_out, Km_Lcho3510_2_R, Lcho3510_Km_1, and Lcho_3510_R_out for lthA (Lcho_3510) (Table S6; see also Fig. S3B, S4A, and S6A).
Imaging cell filament aggregates in culture dishes and cuvettes.
An Axio Observer Z1 microscope with an EC Plan Neofluar 5× lens objective (420331-9911-000; Carl Zeiss, Oberkochen, Germany) was used to image colonies on MSVP agar plates that showed different morphologies. To image elongating cell filaments, we inoculated the wild-type and mutant cells into 600 μL of MSVP in a polymer coverslip bottom dish (μ-Dish, 35 mm, quad; Ibidi GmbH, Gräfelfing, Germany) and then imaged them using an Axio Observer Z1 microscope with a C-Apochromat 40× water immersion lens objective (421767-9973-000; Carl Zeiss) at 10- to 15-min intervals. To image pellicle formation at the air-liquid interface, we incubated 2 mL of the culture in a 24-well dish and imaged it at 30-s intervals using an Axio Zoom.V16 microscope equipped with a heater unit (Tokai Hit, Fujinomiya, Japan). To image the floating pellicles horizontally, we incubated 3 mL of the culture in clear cuvettes and imaged them at 1-min intervals in a horizontal direction using equipment that has been described previously (9).
Fluorescent staining of nanofibrils encompassing cell surfaces.
To visualize nanofibrils that become entangled to form the sheath structure, 1 mL of SP6-derived cells were transferred to 1.5-mL microtubes and stained for approximately 10 min. Cells were incubated with Alexa Fluor 594 C5-maleimide (30 μM; Thermo Fisher Scientific, Waltham, MA, USA) (referred to here as Alexa Fluor 594-SH) (12) or NH2-reactive fluorescein (fluorescein labeling kit with NH2; Dojindo, Kumamoto, Japan) (13) for sheath staining and with DAPI (0.5 μg/mL; Dojindo) for DNA staining. The stained cells were washed twice with phosphate-buffered saline (PBS) buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and KH2PO4 [pH 7.4]), transferred onto agar pads, and air dried prior to observation. To image nanofibrils in liquid medium, cells in 1 mL of MSVP culture in 3.5 mm glass-bottom dishes were stained with Alexa Fluor 594-SH or NH2-reactive fluorescein combined with DAPI for approximately 10 min and then imaged using a SpinSR10 spinning disk confocal microscope (Olympus, Tokyo, Japan) with a 100× oil immersion lens objective (UAPON100XOTIRF; Olympus). PI solution (5 μg/mL; Thermo Fisher) and FM 1-43 dye (5 μg/mL; Thermo Fisher) were also used to detect dead cells and cell membranes, respectively. DAPI, fluorescein, and Alexa Fluor 594 were excited using lasers at 405 nm, 488 nm, and 561 nm, respectively, and detected with 417- to 477-nm, 500- to 550-nm, and 580.5- to 653.5-nm band pass filters, respectively.
Expression and purification of recombinant LthB proteins.
To express a recombinant LthB protein in E. coli, the pET24a(+)-LthB plasmid was introduced into the E. coli strain Rosetta 2(DE3) (Merck, Darmstadt, Germany). Expression of LthB-HisT protein was induced by incubation with 1 mM isopropyl β-d-thiogalactopyranoside (IPTG; Nacalai Tesque) for 12 h at 30°C. Cells were then collected by centrifugation at 7,000 × g for 5 min, washed once with PBS, and immediately frozen at −20°C. To purify the LthB-HisT protein, cells were suspended in 20 mM Tris-Cl (pH 8.0) containing a complete protease inhibitor cocktail (Merck), lysed using bead homogenization (FastPrep-24 5G; MP Biomedicals, Santa Ana, CA, USA), and centrifuged at 15,000 × g for 20 min. The resulting supernatant was incubated with Talon metal resin (TaKaRa Bio) for 2 h. After washing of the resin with wash buffer (20 mM Tris-Cl [pH 8.0], 10 mM imidazole) several times, the resin-bound fusion protein was eluted using elution buffer (20 mM Tris-Cl [pH 8.0], 100 mM imidazole) and dialyzed using Slide-A-Lyzer G2 dialysis cassettes (Thermo Fisher Scientific) in 20 mM Tris-Cl (pH 8.0).
Polyclonal antibodies.
To detect expression of LthA and LthB in SP6-derived cells, rabbit polyclonal antibodies for LthA and LthB were raised against peptides consisted of 184 to 197 and 218 to 231 amino acids in LthA, and purified LthB-HisT protein, respectively.
Immunostaining.
To analyze the cellular localization of LthB, we immunostained SP6-27 cells with the intact lthB gene, using ΔlthB::IS30 as the negative control. The staining procedure used here was very similar to that reported by Park et al. (55). Briefly, cells cultured in MSVP for 2 days were fixed in 7 mL of 3.5% glutaraldehyde (Nisshin-EM, Tokyo, Japan) in PBS buffer at room temperature for 30 min, then washed twice with PBS buffer. The resulting cells were resuspended in 70% ethanol and incubated at room temperature for 1 h for permeabilization. The cells were washed twice with TEG buffer (25 mM Tris-HCl [pH 8.0], 10 mM EDTA, 50 mM glucose), further incubated in TEG buffer containing 10 mg/mL lysozyme (FujiFilm Wako, Osaka, Japan), and then blocked in Blocking One solution (Nacalai Tesque) containing 0.05% Tween20 at room temperature for 1 h. For primary-antibody staining, cells were incubated with LthB antibody diluted 1:100 in Blocking One solution at room temperature for 1.5 h, followed by three washes with PBS buffer. For secondary-antibody staining, cells were incubated with Alexa Fluor Plus 488-conjugated anti-rabbit IgG (H+L) (Thermo Fisher Scientific) diluted 1:5,000 in Blocking One solution at room temperature for 1 h. Then, the cell suspensions were added to the DAPI solution, incubated for 5 min, and washed three times with PBS buffer. The resulting cell suspensions were transferred onto agar pads and air dried prior to imaging using a SpinSR10 microscope with a 100× oil immersion lens. DAPI and Alexa Fluor Plus 488 were excited at 405 and 488 nm using lasers and detected with 417- to 477-nm and 500- to 550-nm band pass filters, respectively.
Immunoblotting.
Immunoblotting was carried out on whole-cell extracts to detect expression of LthA and LthB in SP6-derived cells. Whole-cell extracts were prepared as follows. Cells were cultured in 25 mL MSVP, harvested by centrifugation, with sterile PBS, and suspended in 500 μL of the bacterial protein extraction reagent B-PER (Thermo Fisher Scientific) containing a complete protease inhibitor cocktail (Merck, Darmstadt, Germany). The cell suspensions were transferred to 2.0-mL microtubes containing approximately 0.8 g glass beads (Merck). After cells were lysed using FastPrep-24 5G, the cell suspensions were centrifuged at 7,000 × g for 15 min, and the resulting supernatants were used for immunoblotting. After separation by SDS-PAGE, separated proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon-P; Merck Millipore, Burlington, MA, USA).
We used TBS-T (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.5% Tween 20) containing 5% skim milk (FujiFilm Wako) or Blocking One solution (Nacalai Tesque) for blocking and antibody dilution. Rabbit polyclonal anti-LthA or anti-LthB antibodies were used as primary antibodies (1:2,000 dilution), while an enhanced chemiluminescence (ECL) peroxidase-conjugated anti-rabbit antibody (Merck) was used as the secondary antibody (1:5,000 dilution). The ImmunoStar LD reagent (FujiFilm Wako) was used for chemiluminescence detection using a Fusion SL gel chemiluminescence documentation system (Vilber Lourmat Peqlab, Collégien, France).
To examine whether equal amounts of cell extracts were loaded, SDS-PAGE gels were stained with Coomassie brilliant blue (CBB) (FujiFilm Wako). After electrophoresis, gels were soaked in fixing solution (50% methanol and 10% acetic acid) and incubated for approximately 20 min with shaking, followed by staining in CBB solution (0.25% CBB, 5% methanol, and 7.5% acetic acid) for approximately 20 min. The CBB was destained from the gels using distilled water until the protein bands were clearly visible.
ASEM.
To directly visualize nanofibril distribution on SP6 cell surfaces, we used atmospheric scanning electron microscopy (ASEM; JASM-6200 instrument; JEOL, Tokyo, Japan) (13). We inoculated an ASEM membrane dish (35 mm in diameter; JEOL, Tokyo, Japan) with SP6-27 and ΔlthB::IS30 cells by transferring 3 mL of 2-day-old culture. After 4 h of incubation, the cells were washed in sterile PBS, and the remaining surface-attached cells were fixed with 1% (vol/vol) glutaraldehyde. After quenching in 50 mM ammonium chloride, the nanofibrils secreted from cell surfaces were stained with positively charged gold nanoparticles for visualization (Nanoprobes, Yaphank, NY, USA). To observe nanofibrils, the cells were further stained with 2% (vol/vol) phosphotungstic acid (TAAB Laboratories Equipment, Aldermaston, Berkshire, England). Finally, samples were soaked in 1% (vol/vol) ascorbic acid and observed using ASEM at an acceleration voltage of 30 kV (56).
mRNA extraction and RT-PCR.
To examine whether the expression of the Lcho_0973 gene was abrogated by the ΔlthB::IS30 mutation, RT-PCR was performed using mRNA extracted from SP6, SP6-27, and ΔlthB::IS30 cells. The ReliaPrep RNA cell miniprep system (Promega, Madison, WI, USA) was used to extract mRNA, according to the manufacturer’s instructions. The isolated mRNA was reverse transcribed using the ReverTra Ace qPCR master mix with the gDNA remover kit (Toyobo, Osaka, Japan), and the Lcho_0973 and Lcho_3274 genes were PCR amplified using the Lcho_973_ORF_F/Lcho_973_ORF_R and Lcho_3274_F/Lcho_3274_R primer sets, respectively (Table S6). The Lcho_3274 gene, which encodes a putative glutamate 5-kinase that is expected to be constitutively expressed, was used as an internal control.
Statistics.
To determine cell length in SP6-37rif+ and ΔlthB::kanR cells (Fig. S10A), the long axis of approximately 100 cells (n > 100) was measured in Adobe Photoshop 2022 (Adobe, San Jose, CA, USA). To plot bright-field, DAPI, and anti-LthB signal intensities in the SP6-27 and ΔlthB::IS30 cells (Fig. 3D), the intensities at the cross-section of approximately 15 cells (n > 15) were measured using ImageJ (National Institutes of Health, Bethesda, MD, USA). Statistical analyses were carried out using unpaired Welch’s t tests in R software version 4.2.2 (57).
Data availability.
The raw DNA sequencing data are accessible in the DDBJ Sequence Read Archive (accession no. DRA014967).
ACKNOWLEDGMENTS
We acknowledge that NBRP E. coli stocks at the National Institute of Genetics (NIG) were used to acquire the E. coli strain SN1187. We also thank Yoshiteru Hashimoto (University of Tsukuba) for providing the protocol for His-tagged protein expression from an expression vector based on pET24a(+) in Rosetta 2(DE3).
N.N., A.S.U., and T.K. are financially supported by the Japan Science and Technology Agency (JPMJER1502 and JPMJMI21G8), JSPS KAKENHI grant 21H01720, and a scholarship donation from Bridgestone Corporation, respectively.
T.K., T.Y., S.S., M.T., N.N., and A.S.U. designed the study. T.K., T.Y., E.O., and S.S. acquired the data. T.K., T.Y., and K.T. analyzed and interpreted the data. T.K., T.Y., S.S., and M.T. wrote and revised the manuscript. All authors agreed to submit the manuscript.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Tatsuki Kunoh, Email: kuno.tatsuki.gb@u.tsukuba.ac.jp.
Nobuhiko Nomura, Email: nomura.nobuhiko.ge@u.tsukuba.ac.jp.
Jeremy D. Semrau, University of Michigan—Ann Arbor
REFERENCES
- 1.Sawayama M, Suzuki T, Hashimoto H, Kasai T, Furutani M, Miyata N, Kunoh H, Takada J. 2011. Isolation of a Leptothrix strain, OUMS1, from ocherous deposits in groundwater. Curr Microbiol 63:173–180. 10.1007/s00284-011-9957-6. [DOI] [PubMed] [Google Scholar]
- 2.Kvartenko O, Sabliy L, Kovalchuk N, Lysytsya A. 2018. The use of the biological method for treating iron containing underground waters. J Water Land Dev 39:77–82. 10.2478/jwld-2018-0061. [DOI] [Google Scholar]
- 3.Kunoh T, Matsumoto S, Nagaoka N, Kanashima S, Hino K, Uchida T, Tamura K, Kunoh H, Takada J. 2017. Amino group in Leptothrix sheath skeleton is responsible for direct deposition of Fe(III) minerals onto the sheaths. Sci Rep 7:6498. 10.1038/s41598-017-06644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kunoh T, Kunoh H, Takada J. 2015. Perspectives on the biogenesis of iron oxide complexes produced by Leptothrix, an iron-oxidizing bacterium and promising industrial applications for their functions. J Microb Biochem Technol 7::419–426. 10.4172/1948-5948.1000249. [DOI] [Google Scholar]
- 5.Spring S. 2006. The genera Leptothrix and Sphaerotilus, p 758–777. In Prokaryotes: a handbook on the biology of bacteria, vol 5, 3rd ed. Springer, Berlin, Germany. [Google Scholar]
- 6.van der Waarde J, Krooneman J, Geurkink B, van der Werf A, Eikelboom D, Beimfohr C, Snaidr J, Levantesi C, Tandoi V. 2002. Molecular monitoring of bulking sludge in industrial wastewater treatment plants. Water Sci Technol 46:551–558. 10.2166/wst.2002.0533. [DOI] [PubMed] [Google Scholar]
- 7.Henriet O, Meunier C, Henry P, Mahillon J. 2017. Filamentous bulking caused by Thiothrix species is efficiently controlled in full-scale wastewater treatment plants by implementing a sludge densification strategy. Sci Rep 7:1430. 10.1038/s41598-017-01481-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams TM, Unz RF. 1985. Filamentous sulfur bacteria of activated sludge: characterization of Thiothrix, Beggiatoa, and Eikelboom type 021N strains. Appl Environ Microbiol 49:887–898. 10.1128/aem.49.4.887-898.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunoh T, Yamamoto T, Prasad M, Ono E, Li X, Sugimoto S, Iida E, Obana N, Takeda M, Nomura N, Utada AS. 2022. Porous pellicle formation of a filamentous bacterium, Leptothrix. Appl Environ Microbiol 88:e0134122. 10.1128/aem.01341-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Veen WL, Mulder EG, Deinema MH. 1978. The Sphaerotilus-Leptothrix group of bacteria. Microbiol Rev 42:329–356. 10.1128/mr.42.2.329-356.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furutani M, Suzuki T, Ishihara H, Hashimoto H, Kunoh H, Takada J. 2013. Assemblage of bacterial saccharic microfibrils in sheath skeleton formed by cultured Leptothrix sp. strain OUMS1. J Marine Sci Res Dev 3:001. 10.4172/2155-9910.S5-001. [DOI] [Google Scholar]
- 12.Kunoh T, Yamamoto T, Sugimoto S, Ono E, Nomura N, Utada AS. 2021. Leptothrix cholodnii response to nutrient limitation. Front Microbiol 12:691563. 10.3389/fmicb.2021.691563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunoh T, Morinaga K, Sugimoto S, Miyazaki S, Toyofuku M, Iwasaki K, Nomura N, Utada AS. 2020. Polyfunctional nanofibril appendages mediate attachment, filamentation, and filament adaptability in Leptothrix cholodnii. ACS Nano 14:5288–5297. 10.1021/acsnano.9b04663. [DOI] [PubMed] [Google Scholar]
- 14.Chan CS, McAllister SM, Leavitt AH, Glazer BT, Krepski ST, Emerson D. 2016. The architecture of iron microbial mats reflects the adaptation of chemolithotrophic iron oxidation in freshwater and marine environments. Front Microbiol 7:796. 10.3389/fmicb.2016.00796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunoh T, Nagaoka N, McFarlane IR, Tamura K, El-Naggar MY, Kunoh H, Takada J. 2016. Dissociation and re-aggregation of multicell-ensheathed fragments responsible for rapid production of massive clumps of Leptothrix sheaths. Biology (Basel) 5:32. 10.3390/biology5030032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodward L, Naismith JH. 2016. Bacterial polysaccharide synthesis and export. Curr Opin Struct Biol 40:81–88. 10.1016/j.sbi.2016.07.016. [DOI] [PubMed] [Google Scholar]
- 18.Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem 75:39–68. 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 19.Takeda M, Makita H, Ohno K, Nakahara Y, Koizumi J. 2005. Structural analysis of the sheath of a sheathed bacterium, Leptothrix cholodnii. Int J Biol Macromol 37:92–98. 10.1016/j.ijbiomac.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Takeda M, Kondo K, Yamada M, Koizumi JI, Mashima T, Matsugami A, Katahira M. 2010. Solubilization and structural determination of a glycoconjugate which is assembled into the sheath of Leptothrix cholodnii. Int J Biol Macromol 46:206–211. 10.1016/j.ijbiomac.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki T, Kanagawa T, Kamagata Y. 2002. Identification of a gene essential for sheathed structure formation in Sphaerotilus natans, a filamentous sheathed bacterium. Appl Environ Microbiol 68:365–371. 10.1128/AEM.68.1.365-371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emerson D, Ghiorse WC. 1993. Role of disulfide bonds in maintaining the structural integrity of the sheath of Leptothrix-Discophora SP-6. J Bacteriol 175:7819–7827. 10.1128/jb.175.24.7819-7827.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu X, Chen C, Fu L, Cui B, Zhou D. 2023. Social network of filamentous Sphaerotilus during activated sludge bulking: identifying the roles of signaling molecules and verifying a novel control strategy. J Chem Eng 454:140109. 10.1016/j.cej.2022.140109. [DOI] [Google Scholar]
- 24.Robinson LJ, Cameron ADS, Stavrinides J. 2015. Spontaneous and on point: do spontaneous mutations used for laboratory experiments cause pleiotropic effects that might confound bacterial infection and evolution assays? FEMS Microbiol Lett 362:fnv177. 10.1093/femsle/fnv177. [DOI] [PubMed] [Google Scholar]
- 25.Schroeder JW, Yeesin P, Simmons LA, Wang JD. 2018. Sources of spontaneous mutagenesis in bacteria. Crit Rev Biochem Mol Biol 53:29–48. 10.1080/10409238.2017.1394262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schurch AC, Arredondo-Alonso S, Willems RJL, Goering RV. 2018. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin Microbiol Infect 24:350–354. 10.1016/j.cmi.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 27.Malachowa N, DeLeo FR. 2010. Mobile genetic elements of Staphylococcus aureus. Cell Mol Life Sci 67:3057–3071. 10.1007/s00018-010-0389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol Mol Biol Rev 62:725–774. 10.1128/MMBR.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emerson D, Ghiorse WC. 1992. Isolation, cultural maintenance, and taxonomy of a sheath-forming strain of Leptothrix discophora and characterization of manganese-oxidizing activity associated with the sheath. Appl Environ Microbiol 58:4001–4010. 10.1128/aem.58.12.4001-4010.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishihara H, Suzuki T, Hashimoto H, Kunoh H, Takada J. 2013. Initial parallel arrangement of extracellular fibrils holds a key for sheath frame construction by Leptothrix sp. strain OUMS1. Minerals 3:73–81. 10.3390/min3010073. [DOI] [Google Scholar]
- 31.Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2016. GenBank. Nucleic Acids Res 44:D67–D72. 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, Finn RD, Bateman A. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res 49:D412–D419. 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blum M, Chang HY, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, Richardson L, Salazar GA, Williams L, Bork P, Bridge A, Gough J, Haft DH, Letunic I, Marchler-Bauer A, Mi HY, Natale DA, Necci M, Orengo CA, Pandurangan AP, Rivoire C, Sigrist CJA, Sillitoe I, Thanki N, Thomas PD, Tosatto SCE, Wu CH, Bateman A, Finn RD. 2021. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 49:D344–D354. 10.1093/nar/gkaa977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breton C, Snajdrova L, Jeanneau C, Koca J, Imberty A. 2006. Structures and mechanisms of glycosyltransferases. Glycobiology 16:29R–37R. 10.1093/glycob/cwj016. [DOI] [PubMed] [Google Scholar]
- 35.Yin YB, Mohnen D, Gelineo-Albersheim I, Xu Y, Hahn MG. 2011. Glycosyltransferases of the Gt8 family. Ann Plant Rev 41:167–212. 10.1002/9781444391015.ch6. [DOI] [Google Scholar]
- 36.Clarke BR, Ovchinnikova OG, Sweeney RP, Kamski-Hennekam ER, Gitalis R, Mallette E, Kelly SD, Lowary TL, Kimber MS, Whitfield C. 2020. A bifunctional O-antigen polymerase structure reveals a new glycosyltransferase family. Nat Chem Biol 16:450–457. 10.1038/s41589-020-0494-0. [DOI] [PubMed] [Google Scholar]
- 37.Jiang YL, Jin H, Yang HB, Zhao RL, Wang SL, Chen YX, Zhou CZ. 2017. Defining the enzymatic pathway for polymorphic O-glycosylation of the pneumococcal serine-rich repeat protein PsrP. J Biol Chem 292:6213–6224. 10.1074/jbc.M116.770446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Persson K, Ly HD, Dieckelmann M, Wakarchuk WW, Withers SG, Strynadka NC. 2001. Crystal structure of the retaining galactosyltransferase LgtC from Neisseria meningitidis in complex with donor and acceptor sugar analogs. Nat Struct Biol 8:166–175. 10.1038/84168. [DOI] [PubMed] [Google Scholar]
- 39.Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, Zidek A, Green T, Tunyasuvunakool K, Petersen S, Jumper J, Clancy E, Green R, Vora A, Lutfi M, Figurnov M, Cowie A, Hobbs N, Kohli P, Kleywegt G, Birney E, Hassabis D, Velankar S. 2022. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 50:D439–D444. 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kunoh T, Suzuki T, Shiraishi T, Kunoh H, Takada J. 2015. Treatment of Leptothrix cells with ultrapure water poses a threat to their viability. Biology (Basel) 4:50–66. 10.3390/biology4010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Egan AJF, Cleverley RM, Peters K, Lewis RJ, Vollmer W. 2017. Regulation of bacterial cell wall growth. FEBS J 284:851–867. 10.1111/febs.13959. [DOI] [PubMed] [Google Scholar]
- 42.Egan AJF, Vollmer W. 2013. The physiology of bacterial cell division. Ann N Y Acad Sci 1277:8–28. 10.1111/j.1749-6632.2012.06818.x. [DOI] [PubMed] [Google Scholar]
- 43.Dai K, Xu YF, Lutkenhaus J. 1993. Cloning and characterization of Ftsn, an essential cell-division gene in Escherichia coli isolated as a multicopy suppressor of Ftsa12(Ts). J Bacteriol 175:3790–3797. 10.1128/jb.175.12.3790-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kunoh T, Nakanishi M, Kusano Y, Itadani A, Ando K, Matsumoto S, Tamura K, Kunoh H, Takada J. 2017. Biosorption of metal elements by exopolymer nanofibrils excreted from Leptothrix cells. Water Res 122:139–147. 10.1016/j.watres.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 45.Dominguez DC. 2004. Calcium signalling in bacteria. Mol Microbiol 54:291–297. 10.1111/j.1365-2958.2004.04276.x. [DOI] [PubMed] [Google Scholar]
- 46.Nozaki S, Niki H. 2019. Exonuclease III (XthA) enforces in vivo DNA cloning of Escherichia coli to create cohesive ends. J Bacteriol 201:e00660-18. 10.1128/JB.00660-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch ER, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 48.Akhter S, Aziz RK, Edwards RA. 2012. PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res 40:e126. 10.1093/nar/gks406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bai J, Bandla C, Guo J, Vera Alvarez R, Bai M, Vizcaino JA, Moreno P, Gruning B, Sallou O, Perez-Riverol Y. 2021. Biocontainers registry: searching bioinformatics and proteomics tools, packages, and containers. J Proteome Res 20:2056–2061. 10.1021/acs.jproteome.0c00904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Notredame C, Higgins DG, Heringa J. 2000. T-coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217. 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 52.Madeira F, Pearce M, Tivey ARN, Basutkar P, Lee J, Edbali O, Madhusoodanan N, Kolesnikov A, Lopez R. 2022. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res 50:W276–W279. 10.1093/nar/gkac240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettersen EF, Goddard TD, Huang CRC, Meng EEC, Couch GS, Croll TI, Morris JH, Ferrin TE. 2021. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci 30:70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bocioaga D, El Gheriany IA, Lion LW, Ghiorse WC, Shuler ML, Hay AG. 2014. Development of a genetic system for a model manganese-oxidizing proteobacterium, Leptothrix discophora SS1. Microbiology (Reading) 160:2396–2405. 10.1099/mic.0.079459-0. [DOI] [PubMed] [Google Scholar]
- 55.Park S, Reyer MA, McLean EL, Liu W, Fei J. 2019. An Improved method for bacterial immunofluorescence staining to eliminate antibody exclusion from the fixed nucleoid. Biochemistry 58:4457–4465. 10.1021/acs.biochem.9b00724. [DOI] [PubMed] [Google Scholar]
- 56.Sugimoto S, Okuda K, Miyakawa R, Sato M, Arita-Morioka K, Chiba A, Yamanaka K, Ogura T, Mizunoe Y, Sato C. 2016. Imaging of bacterial multicellular behaviour in biofilms in liquid by atmospheric scanning electron microscopy. Sci Rep 6:25889. 10.1038/srep25889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.R Core Team. 2022. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.01919-22-s0001.xlsx, XLSX file, 0.01 MB (10.2KB, xlsx)
Supplemental material. Download aem.01919-22-s0002.xlsx, XLSX file, 0.01 MB (13.6KB, xlsx)
Supplemental material. Download aem.01919-22-s0003.docx, DOCX file, 6.7 MB (28.8MB, docx)
Data Availability Statement
The raw DNA sequencing data are accessible in the DDBJ Sequence Read Archive (accession no. DRA014967).