ABSTRACT
Soft-ripened cheeses (SRCs) are at a higher risk for the growth of the foodborne pathogen Listeria monocytogenes due to favorable moisture content and pH compared to other cheeses. L. monocytogenes growth is not consistent across SRCs, however, and may be affected by physicochemical and/or microbiome characteristics of the cheeses. Therefore, the purpose of this study was to investigate how the physicochemical and microbiome profiles of SRCs may affect L. monocytogenes growth. Forty-three SRCs produced from raw (n = 12) or pasteurized (n = 31) milk were inoculated with L. monocytogenes (103 CFU/g), and the pathogen growth was monitored over 12 days at 8°C. In parallel, the pH, water activity (aw), microbial plate counts, and organic acid content of cheeses were measured, and the taxonomic profiles of the cheese microbiomes were measured using 16S rRNA gene targeted amplicon sequencing and shotgun metagenomic sequencing. L. monocytogenes growth differed significantly between cheeses (analysis of variance [ANOVA]; P < 0.001), with increases ranging from 0 to 5.4 log CFU (mean of 2.5 ± 1.2 log CFU), and was negatively correlated with aw. Raw milk cheeses showed significantly lower L. monocytogenes growth than pasteurized-milk cheeses (t test; P = 0.008), possibly due to an increase in microbial competition. L. monocytogenes growth in cheeses was positively correlated with the relative abundance of Streptococcus thermophilus (Spearman correlation; P < 0.0001) and negatively correlated with the relative abundances of Brevibacterium aurantiacum (Spearman correlation; P = 0.0002) and two Lactococcus spp. (Spearman correlation; P < 0.01). These results suggest that the cheese microbiome may influence the food safety in SRCs.
IMPORTANCE Previous studies have identified differences in L. monocytogenes growth between SRCs, but no clear mechanism has yet been elucidated. To the best of our knowledge, this is the first study to collect a wide range of SRCs from retail sources and attempt to identify key factors associated with pathogen growth. A key finding in this research was the positive correlation between the relative abundance of S. thermophilus and the growth of L. monocytogenes. The inclusion of S. thermophilus as a starter culture is more common in industrialized SRC production, suggesting that industrial production of SRC may increase the risk of L. monocytogenes growth. Overall, the results of this study further our understanding of the impact of aw and the cheese microbiome on the growth of L. monocytogenes in SRCs, hopefully leading toward the development of SRC starter/ripening cultures that can prevent L. monocytogenes growth.
KEYWORDS: Listeria monocytogenes, soft-ripened cheese, cheese microbiome
INTRODUCTION
Listeria monocytogenes is a psychrophilic foodborne pathogen, and one of the leading causes of foodborne-related deaths in the developed world (1). Due to its ability to persist in the food processing environment (2), L. monocytogenes is a common contaminant of ready-to-eat foods, resulting in many foodborne outbreaks associated with soft cheeses (3).
Soft-ripened cheeses (SRCs) have a moisture on a fat-free basis (MFFB) of greater than 67% (4, 5) and can be either mold ripened (e.g., Brie and Camembert) or smear ripened (i.e., washed rind cheeses). Due to a favorable aw and pH in these cheeses, L. monocytogenes contamination can grow to dangerous levels, both during the ripening period (6, 7) and at the retail establishment (8). Various biocontrol strategies have been investigated for controlling the growth of this foodborne pathogen in these cheeses (e.g., bacteriocins and bacteriophages), but none have been able to completely prevent the growth of L. monocytogenes over the whole shelf-life of the cheese (3).
The growth rate of L. monocytogenes is not consistent across all SRCs, however, with certain washed-rind SRCs showing complete inhibition against L. monocytogenes growth despite favorable conditions of aw and pH (9). Previous research has suggested an effect of the milk treatment (10), ripening culture (11–14), and/or the production/concentration of organic acids (15–17), but no clear mechanism has yet been elucidated.
With the advent of high-throughput sequencing (HTS) technology, researchers have been able to investigate the microbial communities of cheese using metataxonomic (i.e., 16S rRNA gene targeted amplicon sequencing) and shotgun metagenomic methods (18). Of importance is that (i) finished cheeses possess a nontrivial number of microbial taxa not inoculated by the cheesemaker (19–21), (ii) the final cheese microbiota is affected by the microbiome of the cheese processing facility (22), and (iii) many of the taxa present in the cheese processing facility and finished cheeses are also present in the dairy farm environment (23). Recent research has also highlighted the interaction between the cheese microbiome and L. monocytogenes in Gouda cheese made with unpasteurized milk (24). A variety of studies have been conducted to determine the microbial taxa associated with L. monocytogenes inhibition in surface-ripened cheeses (13–15, 25), but no clear mechanism has yet been elucidated.
The growth of L. monocytogenes in cheeses may also be affected through functional mechanisms of the native microbiota that are independent of taxonomy, such as the competition for nutrients (26–28) or the production of inhibitory compounds. Inhibitory compounds that affected the growth of L. monocytogenes include bacteriocins, which are ribosomally synthesized, antimicrobial peptides produced by bacteria that often target closely related taxa (29), other antimicrobial proteins, such as the metalloprotease pseudoalterin (30), which was recently identified in members of the cheese microbiome (31), and organic acids, particularly lactic and acetic acids, which have been identified as important mechanisms for the inhibition of L. monocytogenes in ripened cheeses (15–17).
The objectives of the current study were to (i) compare the growth of L. monocytogenes in a variety of SRCs at refrigerated temperatures, (ii) determine how much variation in L. monocytogenes growth can be explained by physicochemical differences (e.g., pH and aw), and (iii) use a combination of viable plate counts, 16S rRNA gene targeted amplicon sequencing, and metagenomic sequencing to investigate microbial community differences between cheeses, with the goal of identifying microbial species or strains associated with L. monocytogenes inhibition.
RESULTS
Differential growth of L. monocytogenes across SRCs.
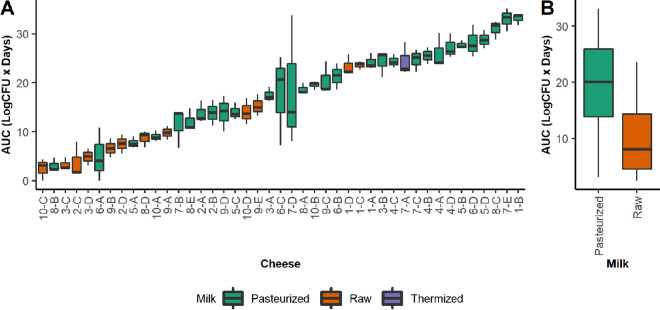
The growth of L. monocytogenes at 8°C in the cheeses was modeled over 12 days and measured by both the area under the growth curve (AUC) (Fig. 1A) and the total increase in pathogen concentration over the incubation period (see Table S1 in the supplemental material). The AUCs of L. monocytogenes growth differed significantly across cheeses (analysis of variance [ANOVA]; P < 0.0001), with the total increase in pathogen ranging from no growth to as high as a 5.4 log CFU increase over the refrigerated incubation period and a mean increase of 2.5 ± 1.2 log CFU. Of the 43 cheeses assessed, 6, 7, 20, and 10 cheeses were assigned to the no-, low-, medium-, and high-growth groups, respectively (Table S1).
FIG 1.
Box plots summarizing of the area under the curve (AUC) of Listeria monocytogenes growth across the individual cheeses (A) and treatment of the milk used for cheesemaking (B). Each box is colored based on milk treatment. Since only one cheese was produced using thermized milk, it was not included in the comparison between milk treatments.
On average, cheeses made with raw milk showed significantly lower growth of L. monocytogenes than cheeses made with pasteurized milk (t test; P = 0.001) (Fig. 1B). Furthermore, no raw milk cheeses allowed more than a 3.5 log CFU increase in L. monocytogenes concentration over the incubation period (Table S1). It should be noted, however, that L. monocytogenes growth in some individual pasteurized-milk cheeses was lower than that in some raw milk cheeses (Fig. 1A). No significant difference in levels of L. monocytogenes growth was observed between washed- and bloomy-rind cheeses (t test; P = 0.70).
High-throughput sequencing of the microbial communities of SRCs.
Metataxonomic analysis based on 16S rRNA gene targeted amplicon sequencing was used to survey the microbial composition of 39 of the cheeses used in this study (Table S1). A total of 3,896,539 reads from 150 samples were recovered after sequence assembly and quality filtering, with an average of 25,977 ± 20,552 reads per sample grouped into 223 operational taxonomic units (OTUs). After rarefaction to 3,000 reads per sample, 135 samples and 222 OTUs remained. An average of 30 ± 15 OTUs was observed for each cheese.
Metagenomic sequencing was used to conduct species- and strain-level taxonomic analysis on a subset of 15 cheeses used in this study (Table S1). A total of 137,937,980 paired-end reads from 58 samples remained after quality filtering. Not including two sample replicates that failed sequencing (<100 reads), the average quality-filtered read count per sample was 2,463,175 ± 941,408. During taxonomic analysis, 102,471,448 reads were assigned to 7,201 species, with 99,170,141 of those reads being assigned to 6,841 species within the kingdom Bacteria. After rarefaction to an even depth (729,719 reads per sample replicate), 7,121 species were still present across the 56 samples, with 6,783 of those being from the kingdom Bacteria. An average of 2,268 ± 1,247 species of bacteria were observed in each sample.
Physicochemical, microbial, and α-diversity analysis of SRC with respect to L. monocytogenes growth.
The pH and aw of cheeses are summarized in Table 1. The average pH across the tested cheeses was 6.9 ± 0.5 and ranged from 5.5 to 7.7. The aw of cheeses ranged from 0.95 to 0.99, with a mean value of 0.97 ± 0.01. Raw milk cheeses showed a significantly lower average water activity than cheeses made with pasteurized milk (Wilcoxon test; n = 42; P = 0.03), and aw was significantly correlated with the growth of L. monocytogenes in cheeses (Spearman’s rank correlation; ρ = 0.417; P = 0.005). No significant difference in pH was observed between milk treatments, and no significant correlation was observed between pH and the growth of L. monocytogenes in the cheeses.
TABLE 1.
Summary of physicochemical and microbial characteristics of cheeses, and their correlation with the growth of L. monocytogenes in the respective cheeses
| Characteristic | n | Milk treatmenta |
All cheeses | Range | Corrb | P value | |
|---|---|---|---|---|---|---|---|
| Pasteurized | Raw | ||||||
| Physicochemical | |||||||
| pH | 39 | 6.8 ± 0.6 A | 7.1 ± 0.3 B | 6.9 ± 0.5 | 5.5–7.7 | −0.057 | 0.732 |
| aw | 43 | 0.97 ± 0.01 A | 0.96 ± 0.01 B | 0.97 ± 0.01 | 0.95–0.99 | 0.417 | 0.005 |
| Organic acid concn (μg/mg) | |||||||
| Lactic acid | 8 | —c | — | 2.42 ± 1.28 | 1.11–4.73 | 0.167 | 0.703 |
| Citric acid | 9 | — | — | 3.86 ± 1.80 | 0.734–5.93 | −0.267 | 0.493 |
| Viable plate counts (log CFU/g) | |||||||
| TSA | 43 | 8.2 ± 1.1 A | 9.0 ± 0.5 B | 8.4 ± 1.0 | 4.8–9.6 | −0.430 | 0.004 |
| M17 | 42 | 8.0 ± 1.2 | 8.5 ± 0.5 | 8.1 ± 1.1 | 4.1–9.5 | 0.103 | 0.514 |
| MRS-5.4 | 43 | 6.7 ± 1.6 | 7.2 ± 0.9 | 6.8 ± 1.4 | 3.0–8.9d | −0.128 | 0.413 |
| α diversity | |||||||
| Observed OTUs | 39 | 30 ± 15 A | 45 ± 8 B | 34 ± 15 | 5–65 | −0.394 | 0.013 |
| Shannon diversity | 39 | 1.08 ± 0.61 A | 1.81 ± 0.35 B | 1.26 ± 0.63 | 0.12–2.23 | −0.292 | 0.072 |
Values in the same row followed by different letters were significantly different between milk treatments (Wilcoxon test; P < 0.05).
Corr, Spearman’s rank correlation (ρ).
—, insufficient samples for each individual milk treatment group.
The starting point of the range, 3.0, is below the reliable limit of enumeration (~3.4 log CFU/g).
Of the organic acids assessed, only lactic and citric acids were consistently detected across the tested cheeses, with mean concentrations of 2.42 ± 1.28 μg/mg and 3.86 ± 1.80 μg/mg for lactic and citric acids, respectively (Table 1). As shown in Table 1, no significant correlation was observed between either of these two organic acids and the growth of L. monocytogenes (Spearman’s rank correlation; P > 0.5). Malic acid was detected in only two cheeses: one each from the low- and no-growth groups.
The total aerobic microbial count (TAMC) was enumerated for all cheeses by spread plating on tryptic soy agar (TSA) and incubating at 30°C (Table 1). The mean TAMC across all cheeses was 8.4 ± 1.0 log CFU/g and ranged from 4.8 log CFU/g to 9.6 log CFU/g. On average, TAMC was significantly higher in raw milk cheeses than in pasteurized milk cheeses, with mean TAMCs of 9.0 ± 0.5 log CFU/g and 8.2 ± 1.1 log CFU/g for each milk treatment, respectively (Wilcoxon test; n = 42; P = 0.003). Furthermore, a significant negative correlation was observed between the TAMC and the growth of L. monocytogenes in the cheeses (Spearman’s rank correlation; ρ = −0.43, P = 0.004).
The viable plate count was also used to enumerate total lactic acid bacteria (TLAB) on both M17 (aerobic) and MRS-5.4 (anaerobic) agars (Table 1). The average viable plate count on M17 agar was 8.1 ± 1.1 log CFU/g across cheeses, with a range of 4.1 log CFU/g to 9.5 log CFU/g. On MRS-5.4 agar, the average viable plate count across cheeses was 6.8 ± 1.4 log CFU/g, and it ranged from below the limit of enumeration (~3.4 log CFU/g) to 8.9 log CFU/g. No significant difference in TLAB on either medium was observed between cheeses made with raw milk and those made with pasteurized milk (Wilcoxon test; n = 42; P > 0.4), nor were there any significant correlations observed between TLAB and the growth of L. monocytogenes (Table 1).
Analysis of the α diversity of the cheeses using 16S rRNA gene targeted amplicon sequencing showed differences in both the richness and evenness of microbial communities across the cheeses (Table 1; see Fig. S1 in the supplemental material). The number of observed OTUs (richness) in each cheese ranged from 5 to 65, with an average of 34 ± 15, and was significantly negatively correlated with L. monocytogenes growth in the individual cheeses (Spearman’s rank correlation; ρ = −0.394; P = 0.013). On average, cheeses made with raw milk had a greater number of observed OTUs than cheeses made with pasteurized milk (Wilcoxon test; P < 0.004), but the cheese with the highest richness in the study was made with pasteurized milk (Fig. S1A). Of note, there was no significant correlation observed between microbial richness and TAMC (r = 0.24; P = 0.13). The Shannon diversity index of cheeses (evenness) was greater in raw milk cheeses than in pasteurized milk cheeses (Wilcoxon test; P < 0.001), but no significant correlation was observed between the Shannon diversity index of cheeses and the growth of L. monocytogenes.
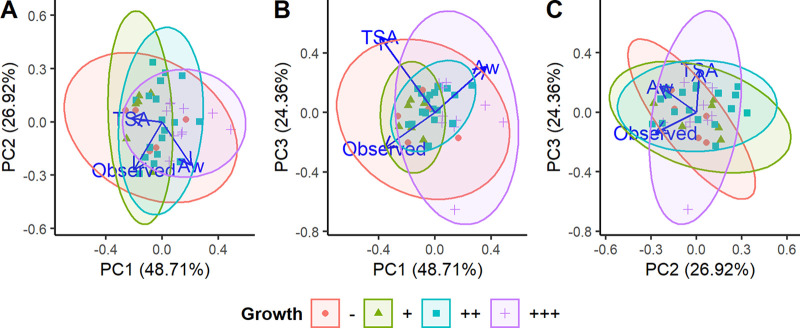
Principle-component analysis was conducted to assess the combined predictive power of the three metrics that were significantly correlated with L. monocytogenes growth in cheese (i.e., aw, TAMC, and observed richness) (Fig. 2). Despite the inclusion of all three correlated metrics, no significant separation was observed between the growth categories (permutational multivariate analysis of variance [PERMANOVA]; P = 0.061). Furthermore, multiple linear regression combining the three metrics explained only 26% of the observed variation (R2adj = 0.264; n = 39; P = 0.003).
FIG 2.
Principal-component analysis of total aerobic microbial count (TSA), water activity (aw), and total observed OTUs measured for each cheese. Graphs show the relationship between principal component 1 (PC1) and PC2 (A), PC1 and PC3 (B), and PC2 and PC3 (C). Cheeses are colored and surrounded by ellipses based on growth category.
Taxonomic analysis of SRCs relative to L. monocytogenes growth.
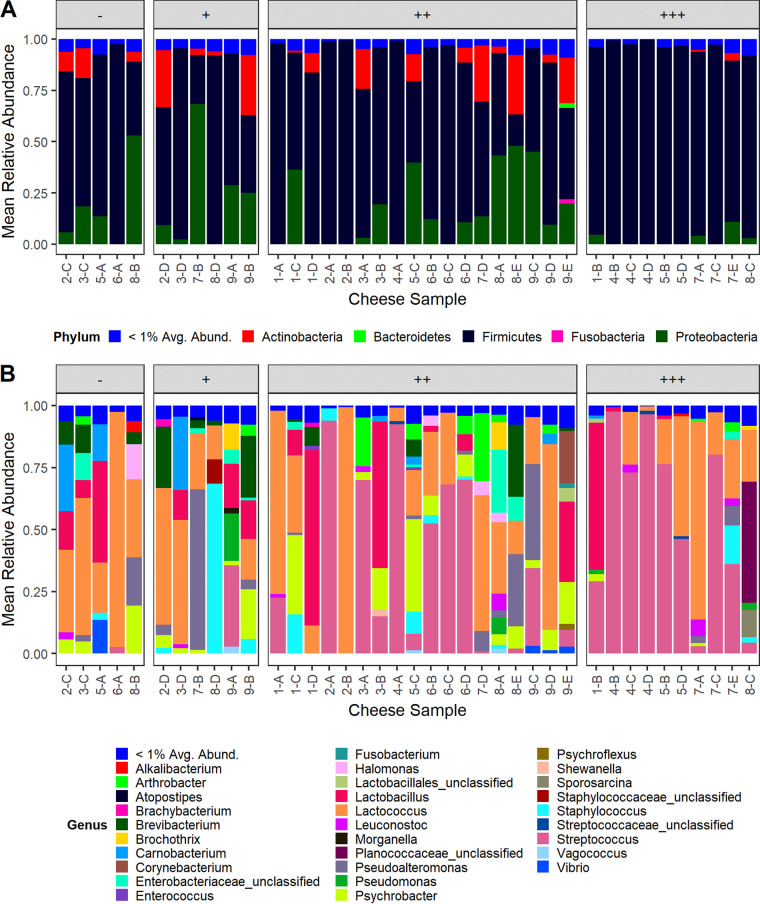
Taxonomic analysis at the phylum level is summarized in Fig. 3A. Firmicutes was the dominant phylum observed, representing 78.1% ± 23.4% of the observed reads across the tested cheeses. Proteobacteria and Actinobacteria made up the bulk of the remaining taxa, with 15.2% ± 18.4% and 6.6% ± 10.0% of the remaining reads in each cheese, respectively. Small amounts of the phyla Bacteroidetes, Fusobacteria, and Deinococcus-Thermus were also observed. Most individual cheeses were dominated by the Firmicutes phylum, but three of the tested cheeses had Proteobacteria as their most abundant phylum.
FIG 3.
Mean relative abundance of phyla (A) and genera (B) representing greater than 1% of total reads across all cheeses as measured by 16S rRNA targeted amplicon sequencing. Cheeses are grouped by growth category.
After adjusting for the false-discovery rate (FDR), significant correlations were observed between L. monocytogenes growth and two of the most abundant phyla: Firmicutes and Actinobacteria (Table 2). The relative abundance of the Firmicutes phylum was positively correlated with the growth of L. monocytogenes in the cheeses (Spearman rank correlation; ρ = 0.396; P = 0.013; FDR [q value] = 0.038), while the growth of L. monocytogenes was negatively correlated with the relative abundances of Actinobacteria (Spearman rank correlation; ρ = −0.475; P = 0.0023; q value = 0.014). The Proteobacteria phylum was also negatively correlated with the growth of L. monocytogenes (Spearman rank correlation; ρ = −0.351; P = 0.028; q value = 0.057), but this correlation was not significant after correcting for FDR.
TABLE 2.
Spearman rank correlations between the growth of L. monocytogenes and the 6 most relatively abundant phyla and 20 most abundant genera in the cheeses used in this study
| Taxonomy | Corra | P value | q valueb |
|---|---|---|---|
| Phlya | |||
| Actinobacteria | −0.474 | 0.002 | 0.014 |
| Firmicutes | 0.396 | 0.013 | 0.038 |
| Proteobacteria | −0.351 | 0.028 | 0.057 |
| Deinococcus-Thermus | 0.269 | 0.098 | 0.147 |
| Bacteroidetes | −0.213 | 0.193 | 0.231 |
| Fusobacteria | −0.165 | 0.316 | 0.316 |
| Genera | |||
| Streptococcus | 0.606 | <0.001 | 0.001 |
| Brevibacterium | −0.562 | <0.001 | 0.002 |
| Psychrobacter | −0.381 | 0.017 | 0.112 |
| Brachybacterium | −0.355 | 0.026 | 0.132 |
| Lactococcus | −0.310 | 0.055 | 0.220 |
| Leuconostoc | −0.293 | 0.070 | 0.234 |
| Pseudoalteromonas | −0.267 | 0.101 | 0.289 |
| Corynebacterium | −0.251 | 0.124 | 0.310 |
| Halomonas | −0.223 | 0.172 | 0.382 |
| Arthrobacter | −0.200 | 0.221 | 0.419 |
| Brochothrix | −0.195 | 0.234 | 0.419 |
| Pseudomonas | 0.188 | 0.251 | 0.419 |
| Sporosarcina | −0.154 | 0.350 | 0.539 |
| Carnobacterium | −0.136 | 0.408 | 0.566 |
| Staphylococcus | −0.132 | 0.424 | 0.566 |
| Vagococcus | −0.106 | 0.522 | 0.652 |
| Vibrio | −0.095 | 0.566 | 0.665 |
| Alkalibacterium | −0.044 | 0.790 | 0.878 |
| Enterococcus | 0.034 | 0.837 | 0.882 |
| Lactobacillus | 0.009 | 0.959 | 0.959 |
Corr, Spearman’s rank correlation.
FDR-adjusted P value.
Streptococcus and Lactococcus were the two most common genera observed, represented by 28.6% ± 35.1% and 29.1% ± 27.6% relative abundances in cheeses, respectively (Fig. 3B). At a finer taxonomic resolution, the overrepresentation of these two genera was shown to be the result of just one OTU each, with relative abundances in the cheeses of 28.5% ± 35.0% and 27.4% ± 26.7% for the dominant Streptococcus and Lactococcus OTUs, respectively (Fig. S2A and B). Other Firmicutes genera representing greater than 1% relative abundance included Lactobacillus, Psychrobacter, Pseudoalteromonas, Staphylococcus, Brevibacterium, Carnobacterium, and Arthrobacter. The overrepresentation of Streptococcus and Lactococcus notwithstanding, the most abundant genus in many of the cheeses was not either of those two. The genera with the highest relative abundance in some cheeses were Lactobacillus (4 cheeses), Psychrobacter (2 cheeses), Pseudoalteromonas (2 cheeses), Brevibacterium (2 cheeses), and an unidentified member of the Planococcaceae family (1 cheese).
After FDR adjustment, significant correlations were observed between L. monocytogenes growth and the relative abundance of the genera Streptococcus and Brevibacterium as summarized in Table 2. The relative abundance of Streptococcus was positively correlated with L. monocytogenes growth in cheeses (Spearman rank correlation; ρ = 0.606; q value = 0.001), while the growth of L. monocytogenes was negatively correlated with the relative abundance of Brevibacterium (Spearman rank correlation; ρ = −0.562; q value = 0.002).
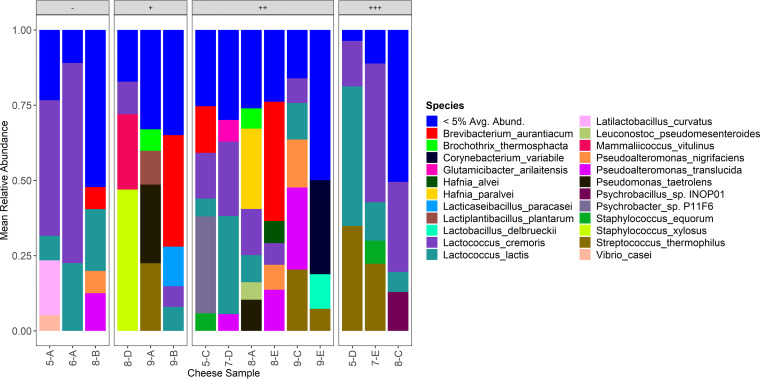
Shotgun metagenomic sequencing was used to conduct taxonomic analysis at the species level (Fig. 4). The two most abundant species were Lactococcus cremoris and Lactococcus lactis, which represented averages of 20.3% ± 19.0% and 13.7% ± 13.6% of reads in each cheese, respectively. Streptococcus thermophilus and Brevibacterium aurantiacum were also common, with averages of 7.6% ± 11.9% and 6.9% ± 13.3% of reads in each cheese, respectively. Other species representing >1% of total reads in cheeses included Pseudoalteromonas translucida, Staphylococcus xylosus, Pseudomonas taetrolens, Pseudoalteromonas nigrifaciens, Hafnia paralvei, Corynebacterium variabile, Mammaliicoccus vitulinus, Lacticaseibacillus paracasei, Lactilactobacillus curvatus, Lactiplantibacillus plantarum, Lactobacillus delbrueckii, Brochothrix thermosphacta, and an unidentified species of Psychrobacter. The most abundant species in most of the cheeses was either Lactococcus cremoris (4 cheeses) or Lactococcus lactis (3 cheeses). Of the remaining cheeses, two showed Brevibacterium aurantiacum as the species with the highest relative abundance, and Hafnia paralvei, Staphylococcus xylosus, Pseudomonas taetrolens, Pseudoalteromonas translucida, Corynebacterium variabile, and an unidentified species of Psycrhobacter were the most abundant species in one cheese each.
FIG 4.
Mean relative abundance of species representing greater than 5% of total reads across the cheeses using shotgun metagenomic sequencing. Cheeses are grouped by growth category.
β diversity and differential abundance.
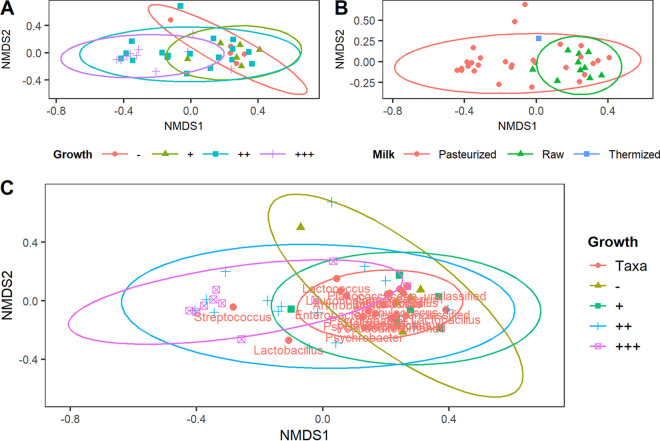
The β diversity of cheeses was measured using the weighted UniFrac distance method and is summarized in Fig. 5. A significant separation was observed between the L. monocytogenes growth categories (PERMANOVA; P = 0.002) (Fig. 5A). Specifically, the high-growth category was significantly different in microbial community structure from both the no- and low-growth categories (pairwise PERMANOVA; q value = 0.024). As shown in Fig. 5C, an OTU of the genus Streptococcus clustered exclusively with cheeses of the high- and medium-growth categories. One OTU from the Lactobacillus genus also clustered outside the no- and low-growth categories. A significant difference in community structure was also observed between cheeses made with raw versus pasteurized milk (PERMANOVA; P = 0.001) (Fig. 5B).
FIG 5.
Nonmetric multidimensional scaling ordination of cheeses based on weighted UniFrac distances calculated using all OTUs (A and B [stress = 0.15]) or the 20 most abundant OTUs (C [stress = 0.16]). Ellipses represent a multivariate t distribution for each growth category (A and C) or milk treatment (B). Ellipses are only presented for sample groups with ≥4 samples represented. Genera are labeled for the most common OTUs.
Differential abundance analysis of the 16S rRNA gene data identified five prevalent OTUs that were differentially abundant with respect to the growth of L. monocytogenes in the cheeses. The Spearman rank correlation between L. monocytogenes growth and these OTUs was then calculated (Table 3). An OTU of the genus Streptococcus was positively correlated with pathogen growth, while four other OTUs were negatively correlated with pathogen growth. Negatively correlated OTUs included two members of the genus Lactococcus, one Brevibacterium sp., and an unidentified member of the Brevibacteriaceae family. Scatterplot analysis (Fig. S3) confirmed that these correlations were not the result of outliers but represented true trends in the data. The positively correlated Streptococcus OTU (Otu00001) was one of the most abundant OTUs observed (Fig. S2A). The most abundant species of Streptococcus observed in the shotgun metagenomic sequencing data was S. thermophilus, implying this to be the species identity of Otu00001. Similarly, the negatively correlated OTU of Brevibacterium (Otu00003) is also a highly abundant OTU and was the most abundant OTU of the Actinobacteria phylum (Fig. S2D), suggesting it represents the species B. aurantiacum, which was the most abundant species of the Actinobacteria phylum in the shotgun metagenomic sequencing data. The OTUs from the Lactococcus genus that were negatively correlated with L. monocytogenes growth were not present in high relative abundance compared to other OTUs of Lactococcus (Fig. S2B). Therefore, it is unlikely that they represent L. lactis or L. cremoris, the most abundant species of Lactococcus observed in the shotgun metagenomic data. Other observed species of Lactococcus included L. raffinolactis, L. piscium, and L. garvieae, but there is not enough evidence to suggest which Lactococcus species may be represented by the OTUs in question.
TABLE 3.
Spearman rank correlations of Listeria monocytogenes growth with the abundances of differentially abundant OTUs identified using the ANCOM-BC algorithm
| OTU | Genus | Spearman correlation | q valuea |
|---|---|---|---|
| Otu00001 | Streptococcus | 0.606 | <0.0001 |
| Otu00003 | Brevibacterium | −0.561 | 0.0002 |
| Otu00046 | Unclassified member of Brevibacteriaceae | −0.614 | <0.0001 |
| Otu00058 | Lactococcus | −0.407 | 0.010 |
| Otu00061 | Lactococcus | −0.452 | 0.004 |
FDR-adjusted P value.
Of note, the OTUs of Streptococcus and Brevibacterium, which were differentially abundant based on L. monocytogenes growth, were also differentially abundant relative to milk treatment. The Streptococcus OTU (Otu00001) was present at greater relative abundance in pasteurized milk cheeses than in raw milk cheeses, whereas the Brevibacterium OTU (Otu00003) was more prevalent in raw milk cheeses than in pasteurized milk cheeses.
In contrast, no prevalent species were identified as being differentially abundant relative to L. monocytogenes growth in cheese in the metagenomic data. This lack of observed differential abundance may be the result of a loss of statistical power due to the reduced number of cheeses sampled. Therefore, species identity confirmation of the 16S rRNA gene data in relation to differential abundance could not be confirmed.
Identification of bacteriocin-encoding genes.
To investigate the possible presence of antimicrobial peptides among the cheeses, a BLAST search was used to identify bacteriocin-encoding genes in the respective metagenomic assemblies. A summary of the identified bacteriocin-encoding genes in each cheese is shown in Fig. 6.
FIG 6.
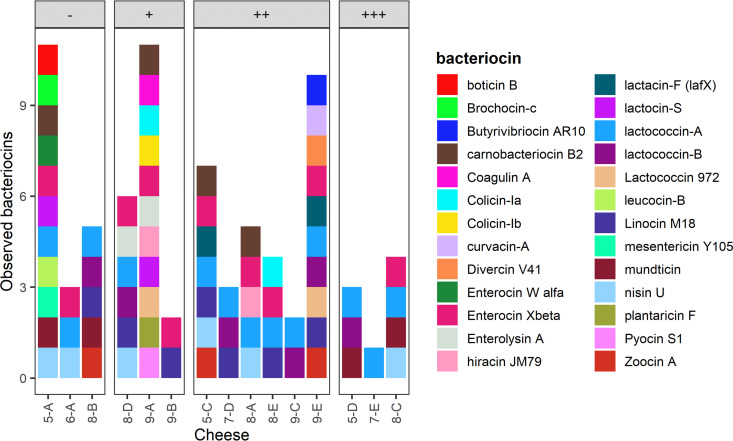
Number of unique bacteriocin-encoding genes observed in each cheese metagenome according to the BACTIBASE database. Cheeses are grouped by growth category.
The two most common bacteriocin-encoding genes observed were those for lactococcin-A and enterocin Xβ, which were identified in 13 and 10 cheeses, respectively. Furthermore, both lactococcin-A and enterocin Xβ were identified in cheeses from each growth category and milk treatment. Lactococcin-B and nisin U were also identified in each growth category and milk treatment, but were less common, with each being present in only six cheeses. Linocin M18 was also common, being observed in seven cheeses, but only from the no-, low-, and medium-growth categories.
A subset of the observed bacteriocin-encoding genes was observed strictly in the no- and/or low-growth categories of cheeses. Genes for production of boticin B, enterocin Wα, leucocin-B, and mesentericin Y105 were present only in cheeses from the no-growth category, while genes to produce coagulin A, colicin-Ib, enterolysin A, plantaricin F, and pyocin S1 were present only in cheeses from the low-growth category. Only lactocin-S was observed in both the no- and low-growth categories. It should be noted, however, that almost all the bacteriocin-encoding genes that were observed strictly in the no/low-growth categories were present in only a single cheese: one from each growth category. The only exceptions were enterolysin A and lactocin-S, which were each observed in two cheeses. Furthermore, all these single-hit bacteriocin genes were from the same cheese within their respective growth category.
DISCUSSION
Variations across L. monocytogenes growth in retail SRCs are correlated with aw and milk treatment.
This experiment investigated the growth of L. monocytogenes in SRCs at refrigerated temperatures and assessed whether differences in L. monocytogenes growth could be explained by physicochemical and/or microbial characteristics of the cheeses. As expected, L. monocytogenes growth was observed in most of the cheeses. Previous research has shown L. monocytogenes growth in SRCs at refrigerated temperatures during ripening (7), after ripening (9), and at retail establishments (8). Each of these previous studies showed levels of L. monocytogenes growth similar to or higher than the mean L. monocytogenes increase observed in the current study. Of interest to the current study was the variation in L. monocytogenes growth across the various cheeses tested, including multiple cheeses showing less than a 1 log CFU increase of L. monocytogenes, and two cheeses even showing less than a <0.5 log CFU increase of L. monocytogenes over the course of the experiment. This inhibition of L. monocytogenes growth is of note because SRCs, due to their favorable moisture content and near-neutral pH, are expected to promote the growth of L. monocytogenes (3). Previous studies have documented variations in L. monocytogenes growth in lab-prepared Camembert-style cheeses (10) and across a small sampling of washed-rind SRCs in Belgium (9), but to the best of our knowledge, this is the first study documenting considerable variations across a large sampling of SRCs from retail.
Two important physicochemical factors affecting L. monocytogenes growth in soft cheeses are pH and aw (3). In the current study, no correlation between L. monocytogenes and pH was observed. Since all cheese samples were purchased from retail, the ripening process had increased the pH back to near-neutral levels, removing the inhibitory barrier against L. monocytogenes associated with the early stages of cheesemaking (3, 32). On the other hand, a significant positive correlation was observed between pathogen growth and aw. Previous modeling data have suggested a stochastic effect on L. monocytogenes growth at aw values between 0.96 and 0.98 (33). At the population level, this stochastic effect would be extended to what proportion of viable cells will double at any point in time, thereby reducing the total rate of growth across the entirety of the L. monocytogenes population. These results confirm the importance of aw as the primary physicochemical factor associated with L. monocytogenes growth in SRC at retail establishments.
Despite the lack of correlation between L. monocytogenes growth and pH of SRC at retail establishments, the presence of weak organic acids (e.g., lactic acid) has been previously associated with L. monocytogenes inhibition in soft cheeses (16, 17). In the current study, however, no correlation was observed between the growth of L. monocytogenes and the concentration of lactic acid. During the ripening process, lactic acid is consumed by the ripening cultures (34), meaning that there may be insufficient concentrations of lactate present in ripened cheeses to have an inhibitory effect. Indeed, the concentration of lactic acid in the studies associating this acid with L. monocytogenes inhibition was 5 to 10 times higher than the levels observed in the cheeses from the current study (15, 17). Further studies investigating how lactate consumption during ripening affects L. monocytogenes growth in SRC are recommended.
Milk treatment was also associated with differences in the growth of L. monocytogenes, as pathogen growth, on average, was significantly lower in cheeses made with raw milk than in those produced with pasteurized or thermized milk, consistent with previous research (10, 33). The inhibition of L. monocytogenes in these previous studies was suggested to be an effect of increased microbial competition associated with the unpasteurized milk microbiota. In the current study, evidence for competition-based inhibition can be seen in the negative correlation between the growth of L. monocytogenes and both the TAMC and microbial richness in the cheeses. Interestingly, TAMC and the total number of observed OTUs in each cheese were not correlated, suggesting that each of these two factors may be inhibitory in its own way.
The relationship between L. monocytogenes growth and TAMC suggests that increasing concentrations of native microbes may be able to crowd out the contaminating pathogen, possibly through nonspecific competition for nutrients (27). The natural microbiotas of various food systems have previously been shown to inhibit the growth of L. monocytogenes (35), and microbial competition against L. monocytogenes has also been observed in other food systems, including apples (36), lettuce (37), and ready-to-eat meats (38).
Streptococcus thermophilus abundance positively correlates with L. monocytogenes growth in SRCs.
The relationship between microbial species richness and L. monocytogenes inhibition is less cut-and-dried, however, since the cheese with the highest microbial species richness was assigned to the medium-growth group, and 6 out of the 10 cheeses with the greatest species richness were assigned to either the medium- or high-growth groups. Similarly, while a recent study of apple packaging facilities found increasing occurrence of L. monocytogenes to be associated with a lower overall α diversity in the facility microbiome (39), several studies on cheese ripening consortia have shown that composition, and not species richness, was the most important factor related to antilisterial effects in the respective cheeses (40). A plausible explanation for the observed correlation between L. monocytogenes growth and reduced species richness in cheese could be that the relationship is confounded with the effect of milk treatment. Raw milk cheeses, which showed a lower average L. monocytogenes growth than pasteurized milk cheeses, had a higher average species richness. Furthermore, the three pasteurized milk cheeses with the highest richness (higher than many of the raw milk cheeses) were all members of the medium- and high-growth groups, suggesting that milk treatment and not species richness is a greater predictor of L. monocytogenes growth. While the species richness may not play a direct role, it is reasonable to suggest that increased richness reduces the opportunity for L. monocytogenes to gain a foothold due to a more varied use of nutrients by the native population.
16S rRNA gene targeted amplicon sequencing identified an average of 35 OTUs per cheese sample, which is similar to previous studies using this method to investigate the microbiota of SRCs (19, 25). Firmicutes were identified as the dominant phylum in the cheeses, which is consistent with previous studies investigating the microbial community of cheese (20, 23, 25). The overabundance of Firmicutes may be attributed to the fact that most microbial species in defined starter cultures used in cheesemaking (e.g., Lc. lactis, Lc. cremoris, and S. thermophilus) are members of the Firmicutes phylum (41), and the Streptococcus and Lactococcus genera have been suggested to be especially suited to the cheese environment (42). Indeed, Streptococcus and Lactococcus were the two most abundant genera observed in the current study.
The presence of Firmicutes was positively correlated with the growth of L. monocytogenes in the cheeses. But this correlation was driven primarily by the genus Streptococcus; particularly by a highly abundant OTU likely representing S. thermophilus. It should be noted that Lactococcus was negatively correlated with L. monocytogenes growth, implying that this observed trend is genus (or species) rather than phylum specific. The high relative abundance of Streptococcus in the cheese samples was unexpected since mesophilic starter cultures (i.e., Lactococcus spp.) are typically used in the production of Camembert and other soft, mold-ripened cheeses (34). Streptococcus has been previously observed as a dominant genus in cheeses where it was not included as a starter culture (25), which may indicate it as part of the nonstarter lactic acid bacteria (NSLAB). However, recent interest in the production of “stabilized” or “solubilized” SRCs has led to the addition of S. thermophilus as a starter culture (34). This stabilization process increases the shelf-life of the cheeses by controlling rate of acidification (through the inclusion of S. thermophilus), leading to a higher pH at draining, an increase in mineral content, and a firmer resulting paste (43). It is possible that many of the cheeses with the highest growth of L. monocytogenes were produced using the stabilized process (i.e., large-scale-production cheeses), meaning that the correlation between Streptococcus and L. monocytogenes growth is not necessarily causative, but that the high relative abundance of Streptococcus is merely indicative of a favorable environment for L. monocytogenes growth. If this is the case, it would mean that the stabilization process may produce cheeses that are at higher risk for L. monocytogenes growth than cheese produced by traditional methods. It should also be noted, however, that the S. thermophilus OTU had a significantly higher relative abundance in cheeses made with pasteurized milk than those made with raw milk, confounding this observed correlation. Furthermore, not all cheeses with high L. monocytogenes growth had high relative abundances of Streptococcus, but all were made with pasteurized milk. Additional targeted experiments should be conducted to assess the effect of the stabilization process on the growth of L. monocytogenes in the finished cheese.
Lactococcus abundance negatively correlates with L. monocytogenes growth in SRCs.
Despite the positive correlation between Firmicutes and L. monocytogenes growth, two OTUs of Lactococcus showed a significant negative correlation with growth of the pathogen (P < 0.026). A recent study of Belgian cheeses also found a negative correlation between the relative abundance of Lactococcus and the growth of L. monocytogenes in washed-rind SRCs (25). Bacteriocins produced by Lc. lactis have regularly been shown to inhibit the growth of L. monocytogenes in soft cheeses (3), but based on the relative abundance of the potentially inhibitory OTUs observed in this study, they are unlikely to represent Lc. lactis. Furthermore, the presence of genes encoding these bacteriocins (i.e., nisin of lactococcin) was also not associated with L. monocytogenes inhibition in the studied cheeses. Of the three other species of Lactococcus observed in this study, Lc. raffinolactis, Lc. piscium, and Lc garvieae, both Lc. piscium and Lc. garvieae have previously been shown to have antimicrobial action against L. monocytogenes. Strains of Lc. piscium isolated from modified-atmosphere-packaged salmon were inhibitory against L. monocytogenes in vitro (44) as well as in cooked shrimp (45). Similarly, Lc. garvieae from raw milk was previously credited with antilisterial activity observed in cheeses produced from the milk (14). A bacteriocin produced by select strains of Lc. garvieae, garvieacin Q, is known to inhibit L. monocytogenes (46) and has been identified in a strain collected from raw milk cheese (47). Further investigation into these two species for the inhibition of L. monocytogenes in soft cheeses is warranted.
Brevibacterium abundance negatively correlates with L. monocytogenes growth in SRCs.
The two other most abundant phyla were Proteobacteria, represented by species of Psychrobacter, Pseudoalteromonas, and Pseudomonas, and Actinobacteria, represented by species of Brevibacterium, Corynebacterium, and Arthrobacter. The large standard deviations in relative abundance associated with these two phyla, however, imply that these results are not consistent across all the cheeses and that these results should be analyzed with caution. Despite this intercheese variation in the relative abundances of these two phyla, these bacteria have all been previously identified as part of the cheese rind community (21). They are also known to populate the house microbiota in cheese production facilities (22, 48).
Actinobacteria were negatively correlated with L. monocytogenes growth, primarily due to the strong negative correlation between the growth of L. monocytogenes and an abundant OTU of Brevibacterium that was assumed to be B. aurantiacum. B. aurantiacum (previously B. brevis [49, 50]) has been shown to produce multiple antiliserial bacteriocins (51–53). One antilisterial bacteriocin associated with B. aurantiacum, linocin M18 (53), was identified in several cheeses from this study and was noticeably absent from any cheeses in the high-growth category. Additionally, linocin M18 was previous associated with L. monocyotogenes in SRCs (54).
Bacteriocins provide an attractive option for the control of L. monocytogenes since they are often found in bacteria already associated with the cheese microbiome (55). Therefore, it was relevant to assess the presence of bacteriocins in the cheese metagenomes relative to the growth of L. monocytogenes. A variety of bacteriocin-encoding genes were present solely in cheese metagenomes from the no- and low-growth categories, but most of these were only observed in a single cheese. The lack of multiple occurrences of these bacteriocin-encoding genes makes it impossible to assess whether they truly provide a protective effect against L. monocytogenes growth or their presence is merely a coincidence. That being said, three bacteriocins implied by the presence of their genes in metagenomics from the no-growth category, enterocin Wα (56), leucocin-B (57), and mesentericin Y105 (58), have all been demonstrated as inhibitory to L. monocytogenes. Since all three of these antilisterial bacteriocin-encoding genes were present in a single cheese, one or multiple genes could be responsible for the lack of L. monocytogenes growth. The only bacteriocin-encoding genes unique to the no/low-growth category that were present in multiple cheeses were those responsible for the production of enterolysin A and lactocin-S. Enterolysin A has shown only minimal inhibition against L. monocytogenes in vitro (59), which makes it an unlikely that it is driving the L. monocytogenes inhibition observed in these cheeses. To the best of our knowledge, no published work has been assessing the effect of lactocin-S against L. monocytogenes. Overall, there is little evidence to conclude that bacteriocins are having a meaningful effect on the growth of L. monocytogenes in the cheeses.
Care should be taken when analyzing the microbiome data presented, primarily due to the compositional nature of these data. Specifically, the negative-correlation bias associated with compositional data means that an observed (but not true) increase in one taxon may really be the result of a true decrease in the amount of another (60). In the context of this study, this means that negative correlations between L. monocytogenes growth and the relative abundance of Brevibacterium or Lactococcus could really be artifacts resulting from decreasing relative abundances of Streptococcus or vice versa. Another thing to consider with this piece of data is that the presence of L. monocytogenes in cheese can also affect the microbiome of the cheese itself (24), suggesting that the microbiomes observed in this study might not completely reflect the real-world scenario of L. monocytogenes contamination in the same cheeses. It is also important to acknowledge that the presence of a putative bacteriocin-encoding gene does not, in and of itself, demonstrate that this gene is expressed. Additionally, in vitro confirmation of the inhibitory effects of any expressed antimicrobial peptides is necessary, since mutations within bacteriocin peptides have been shown to affect their antimicrobial efficacy (61). Since the bacteriocin-encoding genes identified in this study could share as little as 50% sequence homology with those of the BACTIBASE database, it is possible that they could differ in their abilities to inhibit L. monocytogenes. Similarly, the identification of antimicrobial peptides is biased by the database used for alignment (62), meaning novel genes with antilisterial effects may go unnoticed. Therefore, further studies should be conducted to confirm the findings of the current study and to investigate the possible mechanisms behind these effects on L. monocytogenes growth.
Conclusion.
L. monocytogenes is capable of growing in SRCs at refrigerated temperatures, but the amount of growth varies across cheeses. Milk treatment, water activity, and microbial competition (i.e., TAMC and species richness) were predictive of L. monocytogenes growth but were not able to fully explain the total variation observed. Variation in the microbiome of SRCs may also affect, or at least predict, the growth of L. monocytogenes in the respective cheese. Of importance is a positive association between the relative abundance of S. thermophilus and L. monocytogenes, which may imply a higher food safety risk associated with industrialized cheese production processes. On the other hand, species of Brevibacterium and Lactococcus may provide a protective effect against the growth of L. monocytogenes. These results further our understanding of the effects of the cheese microbiome on L. monocytogenes growth and will lead to the development of safer SRCs by validating the efficacy of different microbiomes on specific L. monocytogenes strains in industrial applications.
MATERIALS AND METHODS
Inoculum preparation.
All assays were conducted using L. monocytogenes BCCDC-A3, which had been previously isolated from a cheese sample at a dairy plant in British Columbia. L. monocytogenes BCCDC-A3 was identified as a member of serogroup 4b/4d/4e using a multiplex PCR assay designed to differentiate between the major serovars of L. monocytogenes (63). For each biological replicate, an individual isolate of L. monocytogenes BCCDC-A3 was grown for 24 to 28 h at 37°C in tryptic soy broth (Becton Dickinson and Company, Franklin Lakes, NJ) with 0.6% yeast extract (Becton Dickinson and Company) (TSB-YE), while shaking, to achieve a concentration of ~109 CFU/mL. An aliquot of this culture was then diluted 100-fold into fresh TSB-YE and incubated at 8°C without shaking until early stationary phase (optical density at 600 nm [OD600] of ~0.900 [7 to 10 days]). These cold-adapted cultures were then washed three times with phosphate-buffered saline (PBS [pH 7.4]) (Alfa Aesar, Haverhill, MA) and resuspended in PBS to achieve a concentration of 105 CFU/mL.
Measuring the growth of L. monocytogenes in soft-ripened cheeses.
The growth of L. monocytogenes was monitored in 43 SRCs, comprised of 36 bloomy-rind and seven washed-rind cheeses (Table S1). Cheeses were purchased from local cheese retailers and selected to maximize the number of artisan-produced samples included. Information regarding style of production (i.e., milk treatment) was acquired from the label where possible or from the seller. Thirty cheeses were produced with pasteurized milk, and 12 were produced from raw milk. The remaining cheese was produced using thermized milk, which involves heating the milk at 57 to 68°C for 10 to 20 s (64). Cheeses were produced in France (n = 29), Canada (n = 12), and Denmark (n = 2). Each cheese was aseptically aliquoted into 3.0 ± 0.1-g subportions and divided into sterile 118-mL Whirl-Pak sample bags (Whirl-Pak, Madison, WI). For each biological replicate (n = 3), up to 10 subportions were inoculated with 30 μL of cold-adapted L. monocytogenes culture to achieve a concentration of ~103 CFU/g. Negative controls were inoculated with 30 μL of sterile PBS. Inoculated cheeses and negative controls were all incubated at 8°C until enumeration.
The growth of L. monocytogenes in each cheese was measured at least every second day as follows. One inoculated sample bag for each biological replicate was diluted 10-fold in sterile PBS and homogenized in a stomacher for 2 min at 230 rpm. The homogenized samples were then diluted in sterile PBS before spread plating on PALCAM agar (Neogen Corp., Lansing, MI). The plated samples were enumerated after incubation at 37°C for 24 h. Negative controls for each cheese were also enumerated at every second enumeration point to ensure the cheeses remained free of any countable L. monocytogenes.
For each biological replicate, the growth of L. monocytogenes was modeled over 12 days using a logistic regression model. For each sample, the growth was measured as both the area under the modeled growth curve (AUC) normalized to the starting concentration and the total increase in L. monocytogenes cells over the course of the experiment. For categorical analysis (e.g., principal component analysis), cheeses were grouped into four categories based on the total increase in L. monocytogenes modeled over the 12 days: no growth (<1 log CFU increase [−]), low growth (1 to 2 log CFU increase [+]), medium growth (2 to 3.5 log CFU increase [++]), and high growth (>3.5 log CFU increase [+++]).
Physicochemical and viable microbial analysis of cheeses.
The pH of each cheese was measured in triplicate from three different areas of the cheese (i.e., edge, rind, and core) using an Oakton pHTestr 50S Spear-Tip waterproof pocket tester (Cole-Parmer Canada Company, Montreal, QC, Canada). The water activity (aw) of each cheese was measured on triplicate 3.0 ± 0.1-g aliquots using an Aqualab series 3 water activity meter (Decagon Devices, Inc., Pullman, WA). Measurements of pH and aw were taken within 48 h of time zero.
For organic acid analysis, 20 g of each cheese was homogenized in 80 mL of high-performance liquid chromatography (HPLC)-grade water by bending for 30 s. The resulting homogenates were then centrifuged at 6,000 × g for 10 min and filtered through 0.2-μm-pore cellulose acetate membranes (VWR International, Mississauga, ON, Canada) before being subjected to HPLC analysis. Chromatography was conducted using an Agilent 1100 HPLC system with Nucleogel Ion 300 OA column (300 mm by 7.8 mm inside diameter [i.d.]) and refractive index detector as previously described (65). The concentrations of lactic, citric, and malic acids in the cheeses were determined from calibration curves prepared using authentic standards.
Total aerobic microbial count (TAMC) and total lactic acid bacteria (TLAB) were measured for each cheese (n = 3) by spread plating. At three different time points, uninoculated negative-control portions of cheese were prepared and homogenized as described above. For TAMC, appropriate dilutions were enumerated on tryptic soy agar (TSA) after incubation at 30°C for 48 h. TLAB dilutions were enumerated on both M17 agar and de Man, Rogosa, and Sharpe (MRS-5.4 [pH 5.4]) agar (HiMedia Laboratories, West Chester, PA). Samples on M17 agar were enumerated after aerobic incubation at 37°C for 48 h, while samples on MRS-5.4 agar were enumerated after anaerobic incubation at 37°C for 72 h.
DNA extraction from cheeses.
Total microbial DNA was extracted from up to four uninoculated 3.0 ± 0.1-g portions of each cheese as follows. Each cheese portion was diluted 10-fold in sterile PBS and homogenized as described above for L. monocytogenes enumeration. DNA extraction was then conducted on 1.0-mL aliquots of the homogenates using the DNeasy PowerFood microbial kit (Qiagen, Inc., Toronto, ON, Canada) following the manufacturer’s directions.
Library preparation and sequencing.
For 16S rRNA gene targeted amplicon sequencing, dual-indexed sequencing libraries were prepared as described previously (66). Briefly, a one-step 10-μL PCR was performed on a LabCyte Access workstation using Quanta 5PRIME HotMasterMix with 1 ng input DNA. Amplification was conducted using complete “fusion primers,” which included Illumina Nextera adaptors, indices, and sequences targeting the V4-V5 region of the 16S rRNA gene (515F, 5′-GTGYCAGCMGCCGCGGTAA-3′; 906R, 5′-CCGYCAATTYMTTTRAGTTT-3′) (67). The resulting amplicons were quantified using a Quant-iT PicoGreen double-stranded DNA (dsDNA) assay kit (Thermo Fisher Scientific, Ottawa, ON, Canada), before pooling 2 ng from each sample. The pooled library was then subjected to cleanup using the AmpureXP PCR cleanup protocol (Beckman Coulter Life Sciences; Mississauga, ON, Canada) and quantified using a PicoGreen assay. Sequencing of the pooled library was conducted using an Illumina MiSeq sequencer with reagent kit v.3 (600 cycles) following the manufacturer’s recommendations and with the addition of 10% PhiX.
For metagenomic sequencing, library preparation and sequencing were performed by the Sequencing and Bioinformatics Consortium at the University of British Columbia. Sequencing libraries were prepared using the Illumina DNA Prep library preparation kit following the manufacturer’s directions. The resulting libraries were pooled and loaded into a single NextSeq Mid output flow cell for sequencing. Paired-end 150-bp reads were generated using the Illumina sequencer, and raw base call data were converted to FastQ format using the bcl2fastq conversion software from Illumina.
Taxonomic and functional profiling of cheese microbiomes.
For the 16S rRNA gene targeted amplicon sequencing, the resulting sequences were processed using mothur software (v.1.43.0) (68) following the creators’ recommended guidelines. Briefly, paired-end sequences were assembled into contigs and screened to remove any contigs of improper length or containing ambiguous bases. The remaining sequences were aligned and classified using the SILVA database (v.132) (69) before removal of chimeric sequences or sequences from nonprokaryotic lineages. The sequences were then clustered into operational taxonomic units (OTUs) based on 97% similarity and quality filtered to remove any OTUs representing <0.005% of total reads as previously recommended (70). Finally, the samples were all rarefied to an even depth of 3,000 reads per sample.
For metagenomic sequence analysis, sequence data were processed using the READ_QC module of the MetaWRAP metagenomic wrapper suite (v.1.3.2) (71). Cleaned sequences were then classified using Kraken 2 (v.2.1.2) (72), and species-level abundance was estimated using Bracken (v.2.5) (73). Taxonomic analysis of the shotgun metagenomic sequence data was conducted using only species from the kingdom Bacteria.
The identification of putative bacteriocin-encoding genes in cheese metagenomes was assessed by aligning the metagenomic assemblies against the BACTIBASE database (74) using blastx (75). Metagenomic assemblies for each cheese were prepared from the quality-filtered reads using metaSPAdes (76) through the ASSEMBLY module of the MetaWRAP metagenomic wrapper suite (v.1.3.2) (71). Only hits to putative bacteriocin-encoding genes with an E value of <10−5 and similarity of >50% were retained. In cases where multiple hits to the BACTIBASE database were observed on a single contig, only the hit with the highest bit-score was retained.
Statistical analysis.
All data analysis was conducted using R software (v.4.1.0) R Foundation for Statistical Computing, Vienna, Austria [https://www.Rproject.org/]) with the assistance of the tidyverse collection of packages (v.1.3.1) (77). Growth modeling was achieved using the growthcurver package (v.0.3.1) (78). Taxonomic analysis was conducted using phyloseq (v.1.36.0) (79), except for permutational multivariate analysis of variance (PERMANOVA), which was conducted using the vegan software package (v.2.5–7) (80), and differential abundance analysis, which was conducted using the ANCOMBC package (v.1.2.2) (81). For β diversity analysis, dissimilarity between the microbial communities of cheeses was measured using the weighted UniFrac distance method (82). For differential abundance analysis, only OTUs with at least 0.1% relative abundance (~10 reads) in at least three different cheeses were included in order to prevent taxa from a single cheese from affecting the results. For all statistical analysis, including ANOVA, t test, Wilcoxon rank sum test, Spearman’s rank correlation, and PERMANOVA, the significance level (α) was set at 0.05. To correct for multiple comparisons where necessary, P values were adjusted using the false-discovery rate (FDR) method and are labeled as q values.
Data availability.
Sequence data are available at the NCBI Sequence Read Archive under BioProject ID PRJNA863305.
ACKNOWLEDGMENTS
We thank the BC Centre for Disease Control for providing the bacterial strain, Yvonne Ma for technical assistance, as well as Steven Hallam and Brandon Kieft at UBC for their guidance with metagenomic sequence analysis.
The research was funded by the UBC Faculty of Land and Food Systems PG no. 10R21629. Justin Falardeau was a recipient of a doctoral scholarship funded by the Canadian Dairy Commission and an NSERC CGS D Scholarship.
Footnotes
[This article was published on 28 March 2023 with a CC BY 4.0 copyright line (“Copyright © 2023 Falardeau et al. This is an open-access article distributed under the terms of the Creative Commons Attribution 4.0 International license.”). The authors elected to remove open access for the article after publication, necessitating replacement of the original copyright line, and this change was made on 4 April 2023.]
Supplemental material is available online only.
Contributor Information
Siyun Wang, Email: siyun.wang@ubc.ca.
Johanna Björkroth, University of Helsinki.
REFERENCES
- 1.Thakur M, Asrani RK, Patial V. 2018. Listeria monocytogenes: a food-borne pathogen, p 157–192. In Holban AM, Mihai A (ed), Foodborne diseases. Elsevier, New York, NY. [Google Scholar]
- 2.Ferreira V, Wiedmann M, Teixeira P, Stasiewicz MJ. 2014. Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J Food Prot 77:150–170. 10.4315/0362-028X.JFP-13-150. [DOI] [PubMed] [Google Scholar]
- 3.Falardeau J, Trmčić A, Wang S. 2021. The occurrence, growth, and biocontrol of Listeria monocytogenes in fresh and surface-ripened soft and semisoft cheeses. Compr Rev Food Sci Food Saf 20:4019–4048. 10.1111/1541-4337.12768. [DOI] [PubMed] [Google Scholar]
- 4.Canadian Dairy Information Centre. 2020. Glossary for the classification of cheeses. https://cheese-fromage.agr.gc.ca/glossary-lexique_eng.cfm?menupos=1.4. Accessed 7 July 2020.
- 5.Codex Alimentarius Commission. 1978. Codex general standard for cheese, CODEX STAN 283-1978. Codex Alimentarius Commission, Joint FAO/WHO Food Standards Programme, FAO, Rome, Italy. [Google Scholar]
- 6.Liu S, Puri VM, Demirci A. 2007. Spatial distribution of population of Listeria monocytogenes during manufacturing and ripening of Camembert cheese. J Food Saf 27:43–55. 10.1111/j.1745-4565.2007.00059.x. [DOI] [Google Scholar]
- 7.Lobacz A, Kowalik J, Tarczynska A. 2013. Modeling the growth of Listeria monocytogenes in mold-ripened cheeses. J Dairy Sci 96:3449–3460. 10.3168/jds.2012-5964. [DOI] [PubMed] [Google Scholar]
- 8.Lahou E, Uyttendaele M. 2017. Growth potential of Listeria monocytogenes in soft, semi-soft and semi-hard artisanal cheeses after post-processing contamination in deli retail establishments. Food Control 76:13–23. 10.1016/j.foodcont.2016.12.033. [DOI] [Google Scholar]
- 9.Gérard A, El-Hajjaji S, Van Coillie E, Bentaïb A, Daube G, Sindic M. 2020. Determination of the growth potential of Listeria monocytogenes in various types of Belgian artisanal cheeses by challenge tests. Food Microbiol 92:103582. 10.1016/j.fm.2020.103582. [DOI] [PubMed] [Google Scholar]
- 10.Gay M, Amgar A. 2005. Factors moderating Listeria monocytogenes growth in raw milk and in soft cheese made from raw milk. Lait 85:153–170. 10.1051/lait:2005010. [DOI] [Google Scholar]
- 11.Imran M, Desmasures N, Vernoux JP. 2010. From undefined red smear cheese consortia to minimal model communities both exhibiting similar anti-listerial activity on a cheese-like matrix. Food Microbiol 27:1095–1103. 10.1016/j.fm.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 12.Monnet C, Bleicher A, Neuhaus K, Sarthou AS, Leclercq-Perlat MN, Irlinger F. 2010. Assessment of the anti-listerial activity of microfloras from the surface of smear-ripened cheeses. Food Microbiol 27:302–310. 10.1016/j.fm.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 13.Retureau É, Callon C, Didienne R, Montel MC. 2010. Is microbial diversity an asset for inhibiting Listeria monocytogenes in raw milk cheeses? Dairy Sci Technol 90:375–398. 10.1051/dst/2010010. [DOI] [Google Scholar]
- 14.Saubusse M, Millet L, Delbès C, Callon C, Montel MC. 2007. Application of single strand conformation polymorphism—PCR method for distinguishing cheese bacterial communities that inhibit Listeria monocytogenes. Int J Food Microbiol 116:126–135. 10.1016/j.ijfoodmicro.2006.12.024. [DOI] [PubMed] [Google Scholar]
- 15.Callon C, Retureau E, Didienne R, Montel M-C. 2014. Microbial biodiversity in cheese consortia and comparative Listeria growth on surfaces of uncooked pressed cheeses. Int J Food Microbiol 174:98–109. 10.1016/j.ijfoodmicro.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Callon C, Saubusse M, Didienne R, Buchin S, Montel MC. 2011. Simplification of a complex microbial antilisterial consortium to evaluate the contribution of its flora in uncooked pressed cheese. Int J Food Microbiol 145:379–389. 10.1016/j.ijfoodmicro.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 17.Millet L, Saubusse M, Didienne R, Tessier L, Montel MC. 2006. Control of Listeria monocytogenes in raw milk cheeses. Int J Food Microbiol 108:105–114. 10.1016/j.ijfoodmicro.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Afshari R, Pillidge CJ, Dias DA, Osborn AM, Gill H. 2020. Cheesomics: the future pathway to understanding cheese flavour and quality. Crit Rev Food Sci Nutr 60:33–47. 10.1080/10408398.2018.1512471. [DOI] [PubMed] [Google Scholar]
- 19.Dugat-Bony E, Garnier L, Denonfoux J, Ferreira S, Sarthou AS, Bonnarme P, Irlinger F. 2016. Highlighting the microbial diversity of 12 French cheese varieties. Int J Food Microbiol 238:265–273. 10.1016/j.ijfoodmicro.2016.09.026. [DOI] [PubMed] [Google Scholar]
- 20.Quigley L, O'Sullivan O, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD. 2012. High-throughput sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Appl Environ Microbiol 78:5717–5723. 10.1128/AEM.00918-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolfe BE, Button JE, Santarelli M, Dutton RJ. 2014. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell 158:422–433. 10.1016/j.cell.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bokulich NA, Mills DA. 2013. Facility-specific “house” microbiome drives microbial landscapes of artisan cheesemaking plants. Appl Environ Microbiol 79:5214–5223. 10.1128/AEM.00934-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falardeau J, Keeney K, Trmčić A, Kitts D, Wang S. 2019. Farm-to-fork profiling of bacterial communities associated with an artisan cheese production facility. Food Microbiol 83:48–58. 10.1016/j.fm.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Salazar JK, Gonsalves LJ, Fay M, Ramachandran P, Schill KM, Tortorello ML. 2021. Metataxonomic profiling of native and starter microbiota during ripening of Gouda cheese made with Listeria monocytogenes-contaminated unpasteurized milk. Front Microbiol 12:642789. 10.3389/fmicb.2021.642789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gérard A, El-Hajjaji S, Burteau S, Fall PA, Pirard B, Taminiau B, Daube G, Sindic M. 2021. Study of the microbial diversity of a panel of Belgian artisanal cheeses associated with challenge studies for Listeria monocytogenes. Food Microbiol 100:103861. 10.1016/j.fm.2021.103861. [DOI] [PubMed] [Google Scholar]
- 26.Guillier L, Stahl V, Hezard B, Notz E, Briandet R. 2008. Modelling the competitive growth between Listeria monocytogenes and biofilm microflora of smear cheese wooden shelves. Int J Food Microbiol 128:51–57. 10.1016/j.ijfoodmicro.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 27.Mellefont LA, McMeekin TA, Ross T. 2008. Effect of relative inoculum concentration on Listeria monocytogenes growth in co-culture. Int J Food Microbiol 121:157–168. 10.1016/j.ijfoodmicro.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 28.van Gijtenbeek LA, Singer Q, Steffensen LE, Neuens S, Guldager HS, Bidstrup S, Høgholm T, Madsen MG, Glass K, Siedler S. 2021. Lacticaseibacillus rhamnosus impedes growth of Listeria spp. in cottage cheese through manganese limitation. Foods 10:1353. 10.3390/foods10061353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cleveland J, Montville TJ, Nes IF, Chikindas ML. 2001. Bacteriocins: safe, natural antimicrobials for food preservation. Int J Food Microbiol 71:1–20. 10.1016/s0168-1605(01)00560-8. [DOI] [PubMed] [Google Scholar]
- 30.Tang B-L, Yang J, Chen X-L, Wang P, Zhao H-L, Su H-N, Li C-Y, Yu Y, Zhong S, Wang L, Lidbury I, Ding H, Wang M, McMinn A, Zhang X-Y, Chen Y, Zhang Y-Z. 2020. A predator-prey interaction between a marine Pseudoalteromonas sp. and Gram-positive bacteria. Nat Commun 11:285. 10.1038/s41467-019-14133-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh AM, Macori G, Kilcawley KN, Cotter PD. 2020. Meta-analysis of cheese microbiomes highlights contributions to multiple aspects of quality. Nat Food 1:500–510. 10.1038/s43016-020-0129-3. [DOI] [PubMed] [Google Scholar]
- 32.Back JP, Langford SA, Kroll RG. 1993. Growth of Listeria monocytogenes in Camembert and other soft cheeses at refrigeration temperatures. J Dairy Res 60:421–429. 10.1017/s0022029900027758. [DOI] [PubMed] [Google Scholar]
- 33.Schvartzman MS, Belessi X, Butler F, Skandamis P, Jordan K. 2010. Comparison of growth limits of Listeria monocytogenes in milk, broth and cheese. J Appl Microbiol 109:1790–1799. 10.1111/j.1365-2672.2010.04807.x. [DOI] [PubMed] [Google Scholar]
- 34.Spinnler HE. 2017. Surface mold-ripened cheeses, p 911–928. In McSweeney PLH, Fox PF, Cotter PD, Everett D (ed), Cheese: chemistry, physics and microbiology, 4th ed. Elsevier, New York, NY. [Google Scholar]
- 35.Al-Zeyara SA, Jarvis B, Mackey BM. 2011. The inhibitory effect of natural microflora of food on growth of Listeria monocytogenes in enrichment broths. Int J Food Microbiol 145:98–105. 10.1016/j.ijfoodmicro.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 36.Leverentz B, Conway WS, Janisiewicz W, Abadias M, Kurtzman CP, Camp MJ. 2006. Biocontrol of the food-borne pathogens Listeria monocytogenes and Salmonella enterica serovar Poona on fresh-cut apples with naturally occurring bacterial and yeast antagonists. Appl Environ Microbiol 72:1135–1140. 10.1128/AEM.72.2.1135-1140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveira M, Viñas I, Anguera M, Abadias M. 2012. Fate of Listeria monocytogenes and Escherichia coli O157:H7 in the presence of natural background microbiota on conventional and organic lettuce. Food Control 25:678–683. 10.1016/j.foodcont.2011.12.002. [DOI] [Google Scholar]
- 38.Amézquita A, Brashears MM. 2002. Competitive inhibition of Listeria monocytogenes in ready-to-eat meat products by lactic acid bacteria. J Food Prot 65:316–325. 10.4315/0362-028x-65.2.316. [DOI] [PubMed] [Google Scholar]
- 39.Tan X, Chung T, Chen Y, Macarisin D, LaBorde L, Kovac J. 2019. The occurrence of Listeria monocytogenes is associated with built environment microbiota in three tree fruit processing facilities. Microbiome 7:115. 10.1186/s40168-019-0726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montel MC, Buchin S, Mallet A, Delbes-Paus C, Vuitton DA, Desmasures N, Berthier F. 2014. Traditional cheeses: rich and diverse microbiota with associated benefits. Int J Food Microbiol 177:136–154. 10.1016/j.ijfoodmicro.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 41.Parente E, Cogan TM, Powell IB. 2017. Starter cultures: general aspects, p 201–226. In McSweeney PLH, Fox PF, Cotter PD, Everett D (ed), Cheese: chemistry, physics and microbiology, 4th ed. Elsevier, New York, NY. [Google Scholar]
- 42.Kamimura BA, Cabral L, Noronha MF, Baptista RC, Nascimento HM, Sant'Ana AS. 2020. Amplicon sequencing reveals the bacterial diversity in milk, dairy premises and Serra da Canastra artisanal cheeses produced by three different farms. Food Microbiol 89:103453. 10.1016/j.fm.2020.103453. [DOI] [PubMed] [Google Scholar]
- 43.Batty D, Waite-Cusic JG, Meunier-Goddik L. 2019. Influence of cheese-making recipes on the composition and characteristics of Camembert-type cheese. J Dairy Sci 102:164–176. 10.3168/jds.2018-14964. [DOI] [PubMed] [Google Scholar]
- 44.Matamoros S, Pilet MF, Gigout F, Prévost H, Leroi F. 2009. Selection and evaluation of seafood-borne psychrotrophic lactic acid bacteria as inhibitors of pathogenic and spoilage bacteria. Food Microbiol 26:638–644. 10.1016/j.fm.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 45.Fall PA, Leroi F, Chevalier F, Guérin C, Pilet M-F. 2010. Protective effect of a non-bacteriocinogenic Lactococcus piscium CNCM I-4031 strain against Listeria monocytogenes in sterilized tropical cooked peeled shrimp. J Aquat Food Prod Technol 19:84–92. 10.1080/10498850.2010.486910. [DOI] [Google Scholar]
- 46.Tosukhowong A, Zendo T, Visessanguan W, Roytrakul S, Pumpuang L, Jaresitthikunchai J, Sonomoto K. 2012. Garvieacin Q, a novel class II bacteriocin from Lactococcus garvieae BCC 43578. Appl Environ Microbiol 78:1619–1623. 10.1128/AEM.06891-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flórez AB, Reimundo P, Delgado S, Fernández E, Alegría Á, Guijarro JA, Mayo B. 2012. Genome sequence of Lactococcus garvieae IPLA 31405, a bacteriocin-producing, tetracycline-resistant strain isolated from a raw milk cheese. J Bacteriol 194:5118–5119. 10.1128/JB.00975-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goerges S, Mounier J, Rea MC, Gelsomino R, Heise V, Beduhn R, Cogan TM, Vancanneyt M, Scherer S. 2008. Commercial ripening starter microorganisms inoculated into cheese milk do not successfully establish themselves in the resident microbial ripening consortia of a south German red smear cheese. Appl Environ Microbiol 74:2210–2217. 10.1128/AEM.01663-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gavrish EY, Krauzova VI, Potekhina NV, Karasev SG, Plotnikova EG, Altyntseva OV, Korosteleva LA, Evtushenko LI. 2004. Three new species of Brevibacteria, Brevibacterium antiquum sp. nov., Brevibacterium aurantiacum sp. nov., and Brevibacterium permense sp. nov. Microbiology 73:176–183. 10.1023/B:MICI.0000023986.52066.1e. [DOI] [PubMed] [Google Scholar]
- 50.Mounier J, Coton M, Irlinger F, Landaud S, Bonnarme P. 2017. Smear-ripened cheeses, p 955–996. In McSweeney PLH, Fox PF, Cotter PD, Everett DW (ed), Cheese: chemistry, physics and microbiology, 4th ed. Elsevier, New York, NY. [Google Scholar]
- 51.Maisnier-Patin S, Richard J. 1995. Activity and purification of linenscin OC2, an antibacterial substance produced by Brevibacterium linens OC2, an orange cheese coryneform bacterium. Appl Environ Microbiol 61:1847–1852. 10.1128/aem.61.5.1847-1852.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Motta AS, Brandelli A. 2002. Characterization of an antibacterial peptide produced by Brevibacterium linens. J Appl Microbiol 92:63–70. 10.1046/j.1365-2672.2002.01490.x. [DOI] [PubMed] [Google Scholar]
- 53.Valdés-Stauber N, Scherer S. 1994. Isolation and characterization of linocin M18, a bacteriocin produced by Brevibacterium linens. Appl Environ Microbiol 60:3809–3814. 10.1128/aem.60.10.3809-3814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eppert I, Valdés-Stauber N, Götz H, Busse M, Scherer S. 1997. Growth reduction of Listeria spp. caused by undefined industrial red smear cheese cultures and bacteriocin-producing Brevibacterium linens as evaluated in situ on soft cheese. Appl Environ Microbiol 63:4812–4817. 10.1128/aem.63.12.4812-4817.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trmčić A, Obermajer T, Rogelj I, Bogovič Matijašić B. 2008. Short communication: culture-independent detection of lactic acid bacteria bacteriocin genes in two traditional Slovenian raw milk cheeses and their microbial consortia. J Dairy Sci 91:4535–4541. 10.3168/jds.2008-1396. [DOI] [PubMed] [Google Scholar]
- 56.Sawa N, Wilaipun P, Kinoshita S, Zendo T, Leelawatcharamas V, Nakayama J, Sonomoto K. 2012. Isolation and characterization of enterocin W, a novel two-peptide lantibiotic produced by Enterococcus faecalis NKR-4-1. Appl Environ Microbiol 78:900–903. 10.1128/AEM.06497-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Makhloufi KM, Carré-Mlouka A, Peduzzi J, Lombard C, van Reenen CA, Dicks LMT, Rebuffat S. 2013. Characterization of leucocin B-KM432Bz from Leuconostoc pseudomesenteroides isolated from Boza, and comparison of its efficiency to pediocin PA-1. PLoS One 8:e70484. 10.1371/journal.pone.0070484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hechard Y, Derijard B, Letellier F, Cenatiempo Y. 1992. Characterization and purification of mesentericin Y105, an anti-Listeria bacteriocin from Leuconostoc mesenteroides. J Gen Microbiol 138:2725–2731. 10.1099/00221287-138-12-2725. [DOI] [PubMed] [Google Scholar]
- 59.Nilsen T, Nes IF, Holo H. 2003. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl Environ Microbiol 69:2975–2984. 10.1128/AEM.69.5.2975-2984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ. 2017. Microbiome datasets are compositional: and this is not optional. Front Microbiol 8:2224. 10.3389/fmicb.2017.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumariya R, Garsa AK, Rajput YS, Sood SK, Akhtar N, Patel S. 2019. Bacteriocins: classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb Pathog 128:171–177. 10.1016/j.micpath.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 62.Prakash T, Taylor TD. 2012. Functional assignment of metagenomic data: challenges and applications. Brief Bioinform 13:711–727. 10.1093/bib/bbs033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doumith M, Buchrieser C, Glaser P, Jacquet C, Martin P. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J Clin Microbiol 42:3819–3822. 10.1128/JCM.42.8.3819-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rukke EO, Sørhaug T, Stepaniak L. 2011. Heat treatment of milk: thermization of milk, p 693–698. In Fuquay JW, Fox PF, McSweeney PLH (ed), Encyclopedia of dairy sciences, 2nd ed. Academic Press, Cambridge, MA. [Google Scholar]
- 65.Wong DCJ, Lopez Gutierrez R, Dimopoulos N, Gambetta GA, Castellarin SD. 2016. Combined physiological, transcriptome, and cis-regulatory element analyses indicate that key aspects of ripening, metabolism, and transcriptional program in grapes (Vitis vinifera L.) are differentially modulated accordingly to fruit size. BMC Genomics 17:416. 10.1186/s12864-016-2660-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Comeau AM, Douglas GM, Langille MGI. 2017. Microbiome Helper: a custom and streamlined workflow for microbiome research. mSystems 2:e00127-16. 10.1128/mSystems.00127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parada AE, Needham DM, Fuhrman JA. 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 68.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO. 2014. The SILVA and “all-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 42:D643–D648. 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Uritskiy GV, DiRuggiero J, Taylor J. 2018. MetaWRAP—a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6:158. 10.1186/s40168-018-0541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu J, Breitwieser FP, Thielen P, Salzberg SL. 2017. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci 10.7717/peerj-cs.104. [DOI] [Google Scholar]
- 74.Hammami R, Zouhir A, Le Lay C, Ben Hamida J, Fliss I. 2010. BACTIBASE second release: a database and tool platform for bacteriocin characterization. BMC Microbiol 10:22. 10.1186/1471-2180-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 76.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. MetaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen T, Miller E, Bache S, Müller K, Ooms J, Robinson D, Seidel D, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H. 2019. Welcome to the Tidyverse. J Open Source Softw 4:1686. 10.21105/joss.01686. [DOI] [Google Scholar]
- 78.Sprouffske K, Wagner A. 2016. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 17:1016–1017. 10.1186/s12859-016-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McMurdie PJ, Holmes S. 2013. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin P, O’Hara RB, Simpson G, Solymos P, Stevens MHH, Szöcs E, Wagner H. 2020. vegan community ecology package version 2.5–7 November 2020. https://cran.r-project.org/web/packages/vegan/index.html.
- 81.Lin H, Peddada SD. 2020. Analysis of compositions of microbiomes with bias correction. Nat Commun 11:3514. 10.1038/s41467-020-17041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lozupone CA, Hamady M, Kelley ST, Knight R. 2007. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585. 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.02004-22-s0001.pdf, PDF file, 0.5 MB (527.4KB, pdf)
Data Availability Statement
Sequence data are available at the NCBI Sequence Read Archive under BioProject ID PRJNA863305.