ABSTRACT
Despite a reduction of Salmonella contamination on final poultry products, the level of human salmonellosis cases attributed to poultry has remained unchanged over the last few years. There needs to be improved effort to target serovars which may survive antimicrobial interventions and cause illness, as well as to focus on lessening the amount of contamination entering the processing plant. Advances in molecular enumeration approaches allow for the rapid detection and quantification of Salmonella in pre- and postharvest samples, which can be combined with deep serotyping to properly assess the risk affiliated with a poultry flock. In this study, we collected a total of 160 boot sock samples from 20 broiler farms across four different integrators with different antibiotic management programs. Overall, Salmonella was found in 85% (68/80) of the houses, with each farm having at least one Salmonella-positive house. The average Salmonella quantity across all four complexes was 3.6 log10 CFU/sample. Eleven different serovars were identified through deep serotyping, including all three key performance indicators (KPIs; serovars Enteritidis, Infantis, and Typhimurium) defined by the U.S. Department of Agriculture—Food Safety and Inspection Service (USDA-FSIS). There were eight multidrug resistant isolates identified in this study, and seven which were serovar Infantis. We generated risk scores for each flock based on the presence or absence of KPIs, the relative abundance of each serovar as calculated with CRISPR-SeroSeq (serotyping by sequencing the clustered regularly interspaced palindromic repeats), and the quantity of Salmonella organisms detected. The work presented here provides a framework to develop directed processing approaches and highlights the limitations of conventional Salmonella sampling and culturing methods.
IMPORTANCE Nearly one in five foodborne Salmonella illnesses are derived from chicken, making it the largest single food category to cause salmonellosis and indicating a need for effective pathogen mitigation. Although industry has successfully reduced Salmonella incidence in poultry products, there has not been a concurrent reduction in human salmonellosis linked to chicken consumption. New efforts are focused on improved control at preharvest, which requires improved Salmonella surveillance. Here, we present a high-resolution surveillance approach that combines quantity and identity of Salmonella in broiler flocks prior to processing which will further support improved Salmonella controls in poultry. We developed a framework for this approach, indicating that it is possible and important to harness deep serotyping and molecular enumeration to inform on-farm management practices and to minimize risk of cross-contamination between flocks at processing. Additionally, this framework could be adapted to Salmonella surveillance in other food animal production systems.
KEYWORDS: Salmonella, CRISPR-SeroSeq, food safety, broiler, risk, quantification, serovars
INTRODUCTION
Salmonella is a leading bacterial foodborne pathogen in the United States, responsible for an estimated 1.35 million infections and 420 deaths annually (1). According to the Interagency Food Safety Analytical Collaboration (IFSAC) report on foodborne illness source attribution estimates for 2019, 17% of Salmonella illnesses were attributed to chicken (2). Salmonella effectively colonizes the gastrointestinal tract of chickens, and contamination of carcasses and parts can occur during slaughter and cutting up into parts (3–5).
There are over 2,600 Salmonella serovars, defined by their unique combination of O (somatic) and H (flagellar) antigens. Genetic differences among serovars give rise to very diverse phenotypes and allow for different serovars to thrive in different hosts and environments, express virulence factors, and respond to mitigations (6), as well as affect their ability to colonize poultry (7–9). For instance, until 2020, Salmonella enterica serovar Kentucky was the most common serovar isolated from broilers at processing by the U.S. Department of Agriculture—Food Safety and Inspection Service (USDA-FSIS), but it rarely causes illness in the United States (10–14). Conversely, serovar Enteritidis is a human pathogen and is often linked to poultry-related foodborne outbreaks (1). It should be noted that serovar Kentucky is still the most common serovar found in preharvest sampling, accounting for nearly 90% of all Salmonella isolates identified (11), and is able to grow better than other serovars (7). Because only half of serovars identified in carcasses at processing belong to Kentucky, it is likely that this serovar is more susceptible to antimicrobial interventions used in broiler processing. Most recently, USDA-FSIS has listed three serovars—Enteritidis, Typhimurium, and Infantis—as key performance indicators (KPIs) because they are the most frequent serovars found in poultry that also cause significant foodborne illness in humans (15).
Despite several mitigation strategies during broiler processing and an overall reduction in Salmonella in poultry over the last several years, there has not been a concurrent reduction in human Salmonella cases that are linked to poultry (11, 16). Therefore, there is an increasing focus on Salmonella control in preharvest broiler flocks to reduce the burden of Salmonella on broilers arriving for slaughter, and this creates a concurrent need for improved Salmonella sampling methodology preharvest.
Recent work has demonstrated that Salmonella organisms often occur in poultry as mixed populations of different serovars (11, 17–20). This serovar complexity within a single sample is often not realized when conventional Salmonella isolation protocols are used (e.g., as described in the USDA-FSIS Microbiological Laboratory Guide [MLG] [21]), as they typically culminate in selection and characterization of one to three colonies from selective indicator agar (17, 20, 22). In this case, the colonies serotyped are likely to represent the most abundant serovar(s) in the mixed population of serovars present (or those that grow best under the selective enrichment conditions used), leaving the less abundant serovars undetected. This can be detrimental, especially when the undetected serovars are of human clinical importance. Deep serotyping using CRISPR-SeroSeq (serotyping by sequencing the clustered regularly interspaced palindromic repeats) has overcome this challenge by identifying multiple serovars present in a mixed Salmonella population (20). CRISPR-SeroSeq is an amplicon-based next-generation sequencing methodology that interrogates the sequence of both CRISPR arrays, CRISPR1 and CRISPR2, in Salmonella by identifying the presence of CRISPR spacers. Since Salmonella CRISPR spacer sequences correlate with serovar identity (23–25), the resulting sequence information can be used to identify and quantify the relative frequency of multiple serovars in a single sample. Further, approximately 10% of Salmonella enterica subsp. enterica serovars are polyphyletic, and CRISPR content can also separate these serovars (26–30). Most recently, this approach has shown that 9 out of 10 times, when serovar Infantis is present in preharvest environmental poultry samples, it is outnumbered by other serovars (11). This is important because serovar Infantis is classified as a KPI by USDA-FSIS (15).
In addition to serotyping, Salmonella quantification is a critical, yet elusive, metric to assess Salmonella risk in broiler flocks. Flocks with higher Salmonella levels confer a greater risk, and high levels may exceed the antimicrobial intervention capabilities used during processing (31). It is likely that both serovar and quantity contribute to risk of Salmonella contamination in the final product, though precise impacts of each are unknown (e.g., a high load of a serovar that is more susceptible to in-plant interventions could be eliminated more easily than the same load of a tolerant serovar). Traditionally, quantification has been accomplished using a most-probable-number (MPN) method. A previous study using MPN analysis showed that higher Salmonella levels in broiler flocks before processing were associated with higher levels at the slaughter plant (32). MPN is a culture-based approach that takes 3 days to complete and is limited in its throughput. More recently, commercial quantitative PCR (qPCR) assays have become available that enable rapid Salmonella quantification. Sampling methods like environmental sampling of broiler litter, or via boot socks, can be an efficient indicator of Salmonella levels (32, 33); of importance, the latter has also been used to assess the complexity of serovar populations by CRISPR-SeroSeq (11, 20).
Broiler production in the United States is vertically integrated within complexes, and an integrator can own multiple complexes. A typical broiler complex encompasses parental (breeder) and broiler flocks, a hatchery, and a slaughter processing facility and also produces the feed for both breeders and broilers. Salmonella subtypes found in breeders can pass downstream through broiler production and later be found at processing (34). Salmonella management approaches, such as breeder vaccination, are performed on all flocks within a complex (35, 36). Whether or not a company uses antibiotics, or restricts use to certain antibiotic classes, is also managed at the complex level. For example, some companies choose to not use any antibiotics (no antibiotics ever [NAE]), while others might restrict use to those antibiotic classes not used in humans (no antibiotics important to human medicine [NAIHM]). Given the risk to public health regarding antibiotic resistance, it is important to understand how these management approaches may contribute to the prevalence of resistant Salmonella in broiler production. It is not known how diverse Salmonella populations are among broiler farms in the same complex or how much on-farm Salmonella levels vary within a complex. An average complex processes one million broilers a week, arising from 75 to 100 broiler farms (each with four to six houses). Given that broiler chicks come from the same breeder flocks, and all flocks within the same complex are subject to the same Salmonella vaccination schemes, Salmonella populations may be similar across broiler farms within a complex. Conversely, there are other sources from which Salmonella can be introduced to commercial broiler flocks, including rodents, insects, and service personnel or farmers moving between houses or farms (37, 38). Rodents and other pests can be limited geographically and may give rise to different Salmonella populations among farms within the same complex. Given these complex factors that may influence Salmonella found in broilers and the increased concern regarding Salmonella control preharvest, the objective of this study was to investigate Salmonella serovar populations and levels in commercial 80 broiler flocks across 20 total farms supplying four different broiler integrator complexes, under two different antibiotic management practices.
RESULTS
In this study, 80 broiler houses (flocks) from 20 farms within four broiler complexes were sampled to assess differences in Salmonella prevalence, quantity, and serovar population diversity (Fig. 1). Two pairs of boot sock samples were collected from each of the 80 broiler houses for a total of 160 samples. Each boot sock pair was individually cultured to determine Salmonella presence by colony isolation, and a house was considered positive when at least one pair of boot socks tested positive. Salmonella prevalences by complex, farm, and pairs of boot socks are presented in Table 1. Overall, Salmonella was found in 85% (68/80) of the houses, with average prevalences of 95% each (19/20) for complexes 2 and 4 and 80% (16/20) and 70% (14/20) for complexes 1 and 3, respectively (Table 1). All farms had at least one Salmonella-positive house. Variance component analysis was initially performed using an intercept-only multilevel mixed-effects logistic regression model with Salmonella presence as the binary response variable. Nested random effects were specified as complex: farm: house. There was no significant contribution to the variance associated with complex; however, farm (nested within complex) and house (nested within farm) represented 21.9% and 74.4% of the total variance (intracluster correlations [ICC]), respectively. Therefore, farm- and house-level effects explained ~22 and 74% of the variance of Salmonella prevalence, respectively. In other words, this analysis suggests that Salmonella mitigations in broiler production would have less utility at the complex level and should be focused at the house and farm levels.
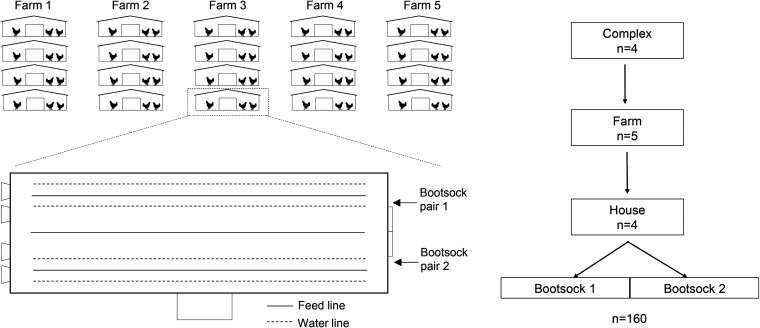
FIG 1.
Schematic of sampling procedure. At each of four complexes, five farms were visited and on each farm, the first four houses were sampled with premoistened boot socks (n = 80 houses; n = 160 samples). One pair was worn to walk along one side of the house between the feed and water lines. A second pair was worn and the process repeated on the opposite side of the house.
TABLE 1.
Prevalence of Salmonella in broiler flocks from four complexes
| Complex | Farm | Prevalence, % (no. of positive houses/totala) | No. of positive boot sock pairs/totalb,c |
|---|---|---|---|
| 1 | 1 | 50 (2/4) | 4/8 |
| 2 | 50 (2/4) | 4/8 | |
| 3 | 100 (4/4) | 8/8 | |
| 4 | 100 (4/4) | 8/8 | |
| 5 | 100 (4/4) | 8/8 | |
| Mean | 80 | 80.00 AB (P = 0.002) | |
| 2 | 1 | 100 (4/4) | 5/8 |
| 2 | 100 (4/4) | 7/8 | |
| 3 | 75 (3/4) | 6/8 | |
| 4 | 100 (4/4) | 8/8 | |
| 5 | 100 (4/4) | 8/8 | |
| Mean | 95 | 85.00 A (P = 0.162) | |
| 3 | 1 | 75 (3/4) | 6/8 |
| 2 | 100 (4/4) | 8/8 | |
| 3 | 75 (3/4) | 5/8 | |
| 4 | 25 (1/4) | 1/8 | |
| 5 | 75 (3/4) | 5/8 | |
| Mean | 70 | 62.50 B (P = 0.004) | |
| 4 | 1 | 100 (4/4) | 7/8 |
| 2 | 75 (3/4) | 5/8 | |
| 3 | 100 (4/4) | 7/8 | |
| 4 | 100 (4/4) | 5/8 | |
| 5 | 100 (4/4) | 7/8 | |
| Mean | 95 | 77.50 AB (P = 0.52) |
Number of Salmonella-positive houses out of total houses sampled per farm.
Number of Salmonella-positive boot sock pairs out of total boot sock pairs (complex-level comparisons [P = 0.06; SEM = 6.09]).
Means with various letters differ at a P value of <0.05.
We initially designed this study to examine potential Salmonella differences between complexes with different antibiotic management programs: complexes 1 and 2 were under NAE production, and complexes 3 and 4 were under NAIHM production. Adding the fixed effect of NAIHM versus NAE to the model above failed to reduce the ICC of farm and house by a substantive amount. Considering these two practices, the ICC for farm and house became 19.1% and 73.7%, respectively. The effect of this complex-level management practice was nonsignificant (P = 0.178), though the observed effect was toward lower odds of Salmonella on the NAIHM versus NAE complexes. The marginal mean estimates for Salmonella prevalence were 0.83 (95% confidence interval [CI], 0.72, 0.95) in NAE houses and 0.69 (95% CI, 0.55, 0.84) in NAIHM houses. Collectively, these findings show that different antibiotic management practices do not significantly impact Salmonella prevalence in broiler production and confirm that differences on the house and farm levels are stronger drivers for Salmonella presence.
The unadjusted likelihood ratio Chi-square statistics were found to be significant when comparing NAE versus NAIHM for single resistance to chloramphenicol (P = 0.006), gentamicin (P = 0.012), streptomycin (P = 0.022), and sulfamethoxazole-trimethoprim (P = 0.012). When incorporated in a multilevel mixed logistic regression model adjusting for clustering by farm and house, these differences were no longer significant (P > 0.05). Similarly, the unadjusted likelihood ratio Chi-square statistic indicated that the difference in prevalence of multidrug resistance (MDR; five different MDR phenotypes) was significant between NAE and NAIHM (P = 0.036). After adjusting for clustering by farm and house, this was no longer significant (P > 0.05). To summarize, due to the strong effect of house and farm, we were unable to attribute the prevalence of overall, single-drug-resistant, or multidrug-resistant Salmonella to either NAE or NAIHM poultry production.
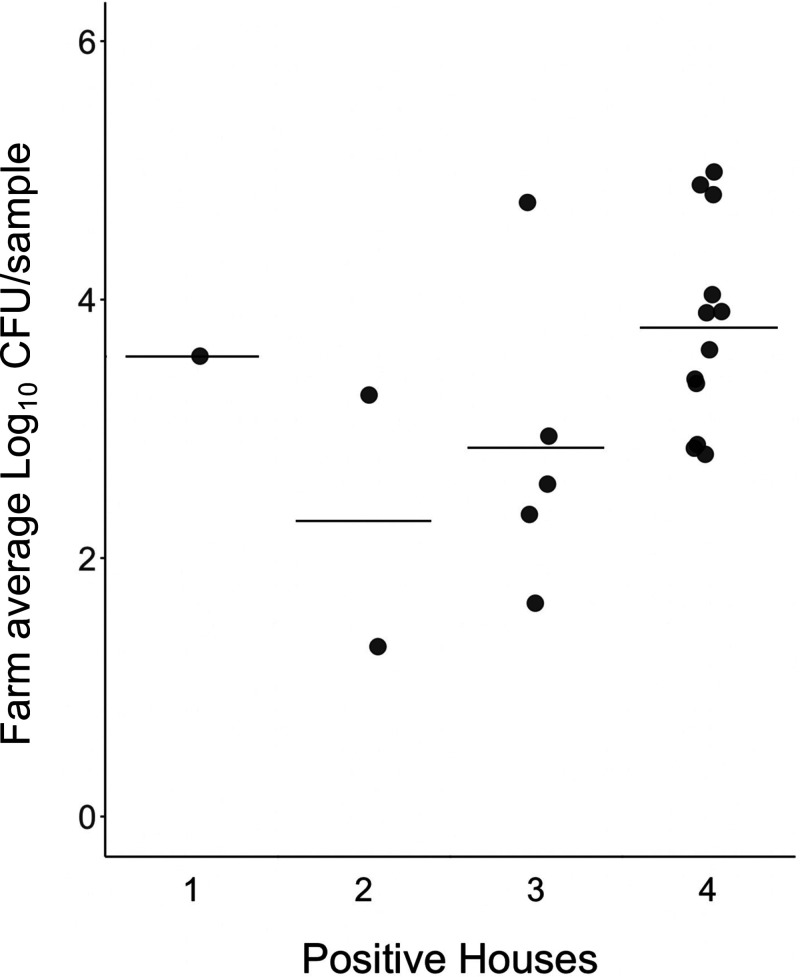
We performed molecular enumeration on each pair of Salmonella-positive boot socks to estimate Salmonella quantity. Using linear regression, there was no significant correlation between Salmonella level and the number of positive houses (P = 0.10; R2 = 0.15) (Fig. 2). This is likely due to the low number of instances where only one house (n = 1) or two houses (n = 2) on a farm were positive and the variability across those three instances. Ignoring the baseline level, the houses on farms with four positive houses had higher Salmonella levels (farm average, 3.78 log10 CFU/sample; n = 12 farms) than those farms where three houses were positive (farm average, 2.85 log10 CFU/sample; n = 5 farms).
FIG 2.
Salmonella levels per house presented by the number of positive houses sampled on a farm. For each farm, the average log10 CFU of all positive boot socks was calculated and plotted according to the number of Salmonella-positive houses (out of four) on a farm. Horizontal bars show the mean Salmonella quantity per category. A linear regression showed no significant correlation between the number of houses positive on a farm and Salmonella levels (P = 0.10; R2 = 0.15).
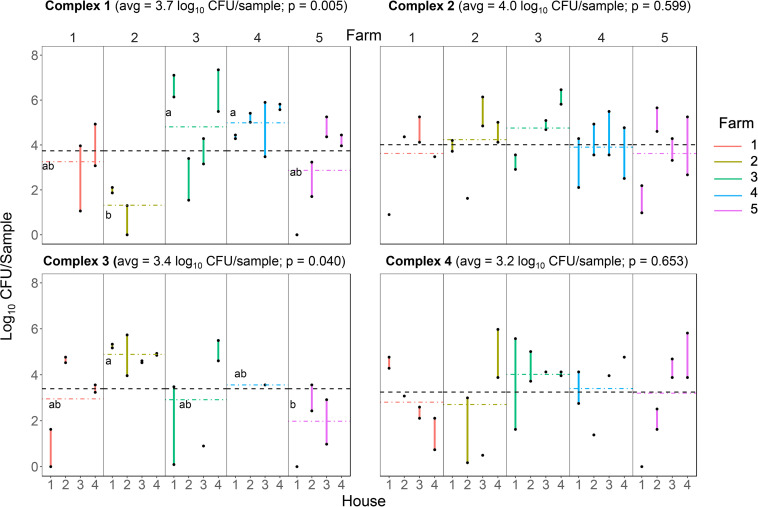
The average Salmonella quantity across all four complexes was 3.6 log10 CFU/sample (Fig. 3), and 58.8% of Salmonella-positive houses had levels above this value. Salmonella quantity between farms within a complex differed for complexes 1 and 3 (P = 0.005 and P = 0.040, respectively), while there was no difference between farms in complexes 2 and 4 (P = 0.599 and P = 0.653, respectively). In complex 1, farms 3 and 4 had higher Salmonella counts (averages, 4.81 and 4.99 log10 CFU/sample, respectively) and these were statistically different from that of farm 2 (average, 1.32 log10 CFU/sample) but not those of farms 1 and 5. There was a similar trend among the farms in complex 3, with farm 2 having the greatest average quantity (4.89 log10 CFU/sample); meanwhile, it differed only from farm 5 (1.98 log10 CFU/sample) and not from farm 1, 3, or 4. Importantly, of the 53 houses where both pairs of boot socks were Salmonella positive, 71.7% (38/53) had Salmonella levels that differed by 1 log10 CFU or greater between the two pairs.
FIG 3.
Salmonella quantity per sample in each house. Culture positive boot sock samples were quantified using the BAX SalQuant assay. The average Salmonella quantity across the four complexes was 3.6 log10 CFU/sample. Vertical colored bars connect the two Salmonella quantity values per house (one value per boot sock pair [black dots]). The horizontal colored lines reflect the average Salmonella quantity per farm, and the horizontal black line reflects the average per complex. Culture-positive samples that fell below the SalQuant threshold were scored as 0. One-way ANOVA was used to assess whether Salmonella quantity differed among farms in the same complex, and the letters indicate farms with statistically different Salmonella levels within each complex.
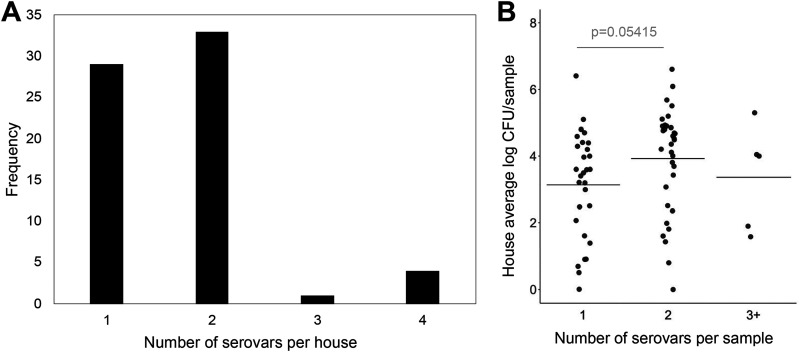
Deep serotyping by CRISPR-SeroSeq was used to profile Salmonella serovar populations in each house. In total, there were 68 positive houses; however, one sample failed to provide enough sequence reads for population analysis. Overall, 11 serovars were identified across the four complexes and 57% (38/67) of the houses that were Salmonella positive contained multiple serovars (Fig. 4A). Across the data set, an average of 2.7 serovars were identified per farm (range, 1 to 5), while an average of 1.7 serovars were identified per house (range, 1 to 4). Houses that contained two serovars tended to have higher Salmonella levels than houses with a single serovar (P = 0.054), despite similar numbers of houses for the two groups (33 and 29, respectively) (Fig. 4B).
FIG 4.
Higher Salmonella serovar complexity is associated with higher Salmonella levels. (A) A total of 43% (29/67) of Salmonella-positive houses contained a single serovar, while two serovars were found in the majority of houses (49% [33/67]). (B) The presence of a single serovar in a house is associated with a slightly lower Salmonella quantity than the presence of two serovars in a house (Welch’s two sample t test, P = 0.05415). Average house log10 CFU/sample was calculated as the average between two boot sock pairs from the same house.
In complex 1, we identified a total of five Salmonella serovars, including Enteritidis, Infantis, Kentucky, Schwarzengrund, and Typhimurium (Fig. 5, top gray-scale graphs). The average number of serovars per house was 1.56, with 56% (9/16 Salmonella-positive houses) having more than one serovar (range, 1 to 2). Each farm had a different Salmonella profile, with serovars Typhimurium and Schwarzengrund being the most abundant on farms 1 and 2, respectively, while farms 3, 4, and 5 had more heterogenous Salmonella populations. Farm 3, in particular, had a different serovar profile in each house and also had the highest total number of serovars (n = 4), while the other farms had a maximum of two total serovars. On farm 4, the relative abundance of serovar Typhimurium in houses 2 and 3 was much lower than that of serovar Kentucky, demonstrating the value of deep serotyping for identifying serovars of concern that are outnumbered by other serovars.
FIG 5.
Deep Salmonella serotyping and quantification to attribute risk scores to broiler flocks. The relative frequency of each serovar in Salmonella-positive samples was determined by CRISPR-SeroSeq. Data from the two pairs of boot socks per house were normalized and presented in a single data series as a grayscale heat map (top graph for each complex) showing the relative abundance of each serovar (see key). The USDA-FSIS KPI serovars (Enteritidis, Infantis, and Typhimurium) are in bold. Houses colored orange indicate where a KPI serovar was outnumbered by a non-KPI serovar and was not found by traditional colony isolation. The two houses in complex 2 shaded in light orange indicate where two serotypes (Kentucky and Typhimurium) were isolated. Salmonella was quantified following a 10-h selective preenrichment using the SalQuant BAX assay. The average log10 CFU per house is shown, and values for house averages of >3.6 log10 CFU/sample (the average Salmonella level per sample across all four complexes) are in bold. Total Salmonella quantity was multiplied by the relative abundance of each serovar to provide an estimated serovar quantity for each house (lower red [KPI] and blue [non-KPI] heat maps). Putative risk scores were attributed to each flock as described in Materials and Methods. Houses with the lowest risk (either 0 or 1) are highlighted in yellow as candidate flocks for processing first at slaughter. The suffixes (-I) for serovars Kentucky and Montevideo refer to polyphyletic lineages and are named as previously described (31, 32).
Five serovars were detected across complex 2, with an average of 1.63 serovars per house (range, 1 to 2). Similar to the case with complex 1, 53% (10/19) of the Salmonella-positive houses contained multiple serovars. The serovar populations include Hadar, Kentucky, Senftenberg, Typhimurium, and Uganda. Serovar Kentucky was dominant across all houses except house 4 on farm 5, where serovar Hadar had a greater relative frequency. The largest number of serovars was found on farm 5 (four serovars) and the lowest was on farm 1 (where only serovar, Kentucky, was detected).
There was a higher number of serovars per house in complex 3 (1.77; range, 1 to 2) than in complexes 1 and 2, but complex 3 had the lowest prevalence of multiserovar populations (46% [6/13]) and the lowest Salmonella prevalence (70% [14/20 positive houses]) across the four complexes (Table 1). House 1 on farm 5 was culture positive; however, the CRISPR-SeroSeq run failed to provide enough reads to assess the Salmonella population. This was likely because the overall Salmonella content in this sample was low (i.e., no SalQuant value was detected). There were seven serovars, including Cerro, Enteritidis, Infantis, Kentucky, Montevideo, Mbandaka, and Typhimurium, identified across the farms, and some of the farms had similar Salmonella profiles. Farms 1, 2, and 3 were dominated by serovar Typhimurium, while serovar Kentucky was abundant on farms 4 and 5. Farms 2 and 4 had more complex Salmonella populations (i.e., five and four serovars across each farm, respectively), while only two serovars were found on farms 1 and 3.
Complex 4 also had seven serovars: Enteritidis, Kentucky, Montevideo, Mbandaka, Schwarzengrund, Senftenberg, and Typhimurium, with an average of 1.84 serovars across all houses (range, 1 to 4). Of the four complexes, complex 4 had the highest number of multiserovar populations (68% [13/19]); however, it also had the lowest prevalence of KPI serovars. Serovar Kentucky was dominant across all but three houses. Farms 1 and 3 each had four different serovars, while the rest of the farms had three serovars each.
Recently proposed regulatory frameworks for Salmonella in poultry include assessing Salmonella serovars and/or levels preharvest (39). This information could be used for directed processing (or logistical slaughter) by integrators whereby they would process lower-risk flocks before higher-risk flocks in order to minimize cross contamination during slaughter. In the absence of serovar-specific quantification assays (i.e., qPCR assays for individual serovars), we sought to estimate the representative quantity of each serovar using the deep serotyping data (because the relative frequencies of different serovars in a single sample differ) and the overall Salmonella quantification data. For each house, the house average log10 CFU/sample (averaged across both boot sock pairs) was multiplied by the relative frequency of each serovar identified (Fig. 5, lower red and blue heat maps). Overall, farms within complexes 1 and 3 had the highest levels of KPI serovars (darker red shading in Fig. 5). In terms of risk, farms in complex 4 were the lowest risk because the house average Salmonella quantity was 3.1 log10 CFU/sample, only two samples contained KPI serovars, and both of these were below house average, at 2 log10 CFU/sample. While complex 2 had the highest average Salmonella levels per house (3.9 log10 CFU/sample), these were largely attributed to serovar Kentucky.
To transform this information into a metric that integrators could use, a risk score was assigned to each house, based on the presence or absence of a KPI serovar and its quantity (Fig. 5, lower yellow and white boxes). Although all four houses on all five farms were Salmonella positive in complex 4, the serovars present and their quantity place 50% (10/20) of houses into the lowest risk categories. Houses that scored 3 or 4 all had at least one KPI present, with complex 1 having the greatest proportion of high-risk houses (65% [13/20]).
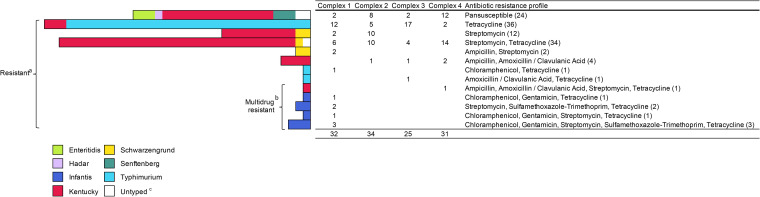
Kirby-Bauer disk diffusion assays were used to assess antimicrobial susceptibility in 122 Salmonella isolates (representing a single representative colony picked from each positive sample; see Table S1 in the supplemental material). There were eight MDR isolates identified in this study, and seven of these were serovar Infantis (Fig. 6). All MDR serovar Infantis isolates were recovered from complex 1 and included three isolates that were resistant to chloramphenicol, gentamicin, streptomycin, sulfamethoxazole-trimethoprim, and tetracycline. There were 36 isolates with resistance only to tetracycline; 33 of these (91.7%) were of serovar Typhimurium and were found in all four complexes. For serovar Kentucky, 75.4% (49/65) of the isolates were resistant to at least one antibiotic. Nearly two-thirds of antibiotic-resistant serovar Kentucky (32/49) were resistant to both streptomycin and tetracycline, and these too were found in all four complexes. Apart from one Schwarzengrund isolate, the streptomycin-tetracycline pattern was found only in serovar Kentucky isolates. All isolates belonging to serovars Enteritidis (3 isolates), Hadar (1), and Senftenberg (3) were pansusceptible.
FIG 6.
Distribution of antibiotic resistant Salmonella serovars. Kirby-Bauer disk diffusion assays were used to assess resistance to ampicillin, amoxicillin-clavulanic acid, ceftriaxone, gentamicin, streptomycin, tetracycline, ciprofloxacin, sulfamethoxazole-trimethoprim, and chloramphenicol. aUnadjusted likelihood ratio chi-square statistic assessing the association of Salmonella serovar by AMR phenotype: 212.4 (df = 84), P < 0.0001. bUnadjusted likelihood ratio chi-square statistic assessing the association of Salmonella serovar with AMR phenotype for MDR classification (≥3 classes of antibiotic): 48.8 (df = 7), P < 0.0001. c“Untyped” includes three isolates: (i) pansusceptible, Hadar and Kentucky were indistinguishable since both are O:8; (ii) streptomycin-tetracycline resistant, CRISPR-SeroSeq failed on both boot sock pairs, isolate typed as O:8; and (iii) pansusceptible, CRISPR-SeroSeq from this boot sock failed, isolate typed as O:7.
DISCUSSION
Salmonella contamination continues to significantly impact the poultry industry. Although the use of multihurdle Salmonella controls pre- and postharvest have resulted in successful reduction of Salmonella prevalence over the past few years (12), this has not translated to a reduction in number of human salmonellosis cases that are linked to poultry (2). In 2021, the USDA-FSIS launched new efforts to reduce Salmonella illnesses associated with poultry, and the proposed framework derived from this is to incentivize additional Salmonella control measures preharvest to reduce Salmonella load in birds arriving at processing facilities (39). Further, the agency aims to resolve the importance of Salmonella quantification upon surveillance, especially with regard to the most pathogenic serovars (15). Accordingly, a high-resolution surveillance approach that combines quantity and identity of Salmonella in broiler flocks prior to processing will further support improved Salmonella controls in poultry.
We investigated the prevalence, quantity, and serovar population dynamics of Salmonella in 80 broiler flocks at 20 broiler farms across four integrated broiler complexes. Across the four complexes, the Salmonella prevalence was 85% (range, 70 to 95%). This is in agreement with a comprehensive study of 55 broiler flocks by Berghaus and colleagues a decade ago showing that 93% of flocks were positive for Salmonella 48 h before processing (32). Deep serotyping of Salmonella-positive samples revealed the presence of low-abundance serovars, including those that were not isolated by selection of a single colony from a plate. Importantly, two or more serovars were found in nearly two-thirds of the houses. Conversely, deep serotyping of 134 parental breeder flocks within the same time frame showed that approximately one-third contained two or more serovars (11). That the prevalence of multiserovar Salmonella populations is greater in broiler houses may be a result of greater impetus for biosecurity management practices (e.g., changing boot covers between houses, on-farm rodent control) in the more valuable parental flocks. Parallel studies examining breeder and broiler flocks in the same complex would be needed to assess this difference.
Serovar Kentucky was the most prevalent serovar identified; it was found on 85% (17/20) farms, and four out of five times it was the most abundant serovar when present. This was expected, as Kentucky has been the prevalent serovar in broiler flocks in the United States for several years, although is of lesser human clinical importance (40, 41). This agrees with a recent study that compared Salmonella serovar prevalence in breeder flocks with that in processing plants in Georgia and found a higher prevalence of serovar Kentucky in breeders, with a significant reduction at processing (11). Missing pieces to that study were the data from its broiler flocks; however, the present study somewhat fills the gap, as we observed a similar abundance of serovar Kentucky prevalence in broiler flocks. Of the three KPI serovars, only serovar Typhimurium was found across all four complexes. Serovars Enteritidis and Infantis were found in complexes 1, 3, and 4 and complexes 1 and 3, respectively.
Seven serovar Infantis isolates were identified by culture, and all were from complex 1. This was expected based on the CRISPR-SeroSeq profiles because where Infantis was present elsewhere (i.e., complex 3), it was at relatively low levels. All seven isolates were multidrug resistant, though the profiles differed among the isolates. The serovar Infantis genome is somewhat clonal, though the pESI plasmid, which confers much of the resistance observed in Infantis, is highly variable in its AMR gene content (42) (these isolates were not screened for the pESI plasmid). Complex 1 is an NAE complex, suggesting that in this instance, antibiotic use is not solely responsible for the abundance of serovar Infantis. Rather, this likely reflects the expansion of this serovar in poultry production over the last few years, as opposed to specific antibiotic management practices in a single complex.
There are few published studies that have enumerated Salmonella quantity in large numbers of broiler flocks, likely due to the cumbersome nature of performing MPN analysis. The availability of molecular assays for Salmonella quantification has resolved this limitation. Overall, we observed similar average Salmonella quantities across the four complexes, though there were significant differences between farms in the same complex and between houses on the same farm. Boot socks were used for poultry sample collection, as these are reported to be more sensitive than other sample types, such as fecal grabs or drag swabs (32, 33, 43). Furthermore, boot socks have become an industry standard (approved by the National Poultry Improvement Plan [44]) and are beneficial because they are an easy sample type to collect, are noninvasive, and can be used to sample a large area. There were 14 instances where a single boot sock pair was Salmonella positive while the other pair from the same house was negative (Table 1). Therefore, it is important to collect multiple samples in a single house to minimize false negatives. Similarly, where both pairs were positive, we observed considerable variability in Salmonella quantity between pairs.
The SalQuant assay depends on a 10-h selective preenrichment, and since there are medium biases for different serovars (45–47), it is possible that differences in serovar composition between sample pairs in one house may be responsible for the differences in Salmonella levels. To address this, we compared the quantification values between boot sock pairs in houses with a single serovar (data not shown). In 73.3% (11/15) of instances, there was a >1-log10 CFU/sample difference between boot socks from the same house, suggesting that any growth differences occurring between serovars during the preenrichment steps are not responsible for the Salmonella quantity variations we observed. Thus, sample-to-sample variability within a single house might be due to the intrinsic differences in Salmonella shedding and presence in different areas of the house, or it may suggest a need for an improved sampling method to quantify Salmonella in poultry house environments. Dust sampling is an attractive alternative Salmonella sampling method, though the presence of Salmonella in dust is influenced by litter moisture (48, 49).
Directed processing is used in the European Union, where on-farm Salmonella monitoring is mandatory and negative flocks are processed first (50, 51). The high prevalence of Salmonella in broiler farms in the United States, observed in this and other studies (for an example, see reference 32), precludes a directed processing strategy based on Salmonella presence alone. Nearly two-thirds (58.8%) of houses had above average Salmonella levels; therefore, the utility of Salmonella load as a single metric for directed processing is also limited. For example, across the 20 farms studied in this investigation, only two contained four houses that were either Salmonella negative or else had a house average quantity below 3.6 log10 CFU/sample (complex 1, farm 2, and complex 3, farm 5).
There is increasing evidence that some serovars, such as Infantis, can tolerate the antimicrobials that are used in processing (52). Further, because different serovars pose different risks to human health, there is a need to quantify individual serovars to attribute relevant risk to a flock. For example, in 23 houses with a house average of 3.6 log10 CFU/sample or higher, serovar Kentucky was the most abundant serovar. This serovar rarely causes human illness in the United States; thus, the risk of flocks containing this serovar is much lower than in a flock with the same Salmonella levels but with a more pathogenic serovar such as Enteritidis. Therefore, we proposed a risk profile based on estimated quantities of individual serovars that were derived from combining the Salmonella quantification and deep serotyping data. Across complexes 2 to 4, all but two farms had at least one house with a low risk (score, 0 or 1) and all had one farm that did not contain a high-risk house (score, 3 or 4), suggesting that with appropriate catching and transportation logistics, these integrators could implement a directed processing strategy. For complex 1, all houses on three farms scored 2 or higher (farms 3 to 5), suggesting the need for additional on-farm Salmonella controls. In such a real-world instance, depending on the slaughter schedule, it could be challenging under these parameters for the complex 1 integrator to make a choice between high- or low-risk flocks that were both ready to be slaughtered on the same day. Overall, using this approach, it was possible to identify a single farm in complexes 1, 3, and 4 where the majority of houses had low scores (score, 0 or 1; complex 1, farm 2, complex 3, farm 4, and complex 4, farm 1). There were at least three farms in each complex that included at least a single low-risk flock. The framework provided by this analysis may provide integrators the flexibility to implement directed processing. Future studies will be needed to determine the efficacy of such an approach to reduce Salmonella during poultry processing.
In addition to guiding processing decisions, the deep-surveillance approach described here can be used to assess Salmonella dynamics across multiple farms. Farms that routinely score as high risk are candidates for additional Salmonella controls (e.g., improved biosecurity). When birds from these farms are processed, it allows integrators the flexibility to adapt their in-plant interventions (e.g., adjusting peracetic acid concentrations as needed during carcass chilling when there is a particularly high load of a KPI serovar). This could be a cost-saving measure, where increased antimicrobials are used only when needed. Identifying farms where Salmonella levels are low and serovars of concern are absent will provide opportunities to identify practices that are associated with reduced Salmonella (or non-KPI) within a complex.
The presence of MDR Salmonella in preharvest samples is also a metric that could be added to a risk profile. Our analysis here focused on traditional phenotypic antimicrobial susceptibility testing on individual isolates and therefore only on the most prominent serovar(s) in a house. Serovars that are outnumbered can have markedly different antimicrobial resistance profiles and can contribute to the overall resistome within a sample (53). Newer sequencing-based methods, such as antimicrobial resistance target-enriched metagenomics (54), could provide additional information on the presence or enrichment of antibiotic resistance genes in a whole sample that could be incorporated into a risk profile. Data from this study show that the prevalence of resistant Salmonella is not driven at the complex level (i.e., NAE versus NAIHM management); rather, alternative factors that occur on the farm or house level contribute to the presence of antibiotic-resistant Salmonella in broilers. The presence of pansusceptible Salmonella did not differ across the two management practices, and MDR isolates were identified in only one complex (complex 1; NAE) and not the other NAE complex.
While the framework described here provides a deep surveillance of Salmonella preharvest, it is not logistically and economically feasible to implement this testing for every flock. It will likely have greater utility if used on a proportion of farms a few times a year to assess Salmonella across a complex. Understanding the longitudinal stability of the serovar populations within farms and houses would add to this understanding and aid in design of testing schemes. The present study has provided only a cross-sectional snapshot of the situation. Under the forthcoming regulatory landscape, a single assay that detects and quantifies KPI serovars rapidly (e.g., by qPCR) would have great utility. We earlier developed a serovar Infantis qPCR assay that can detect this serovar, even when outnumbered 1,000:1 by another serovar, showing that a serovar-specific qPCR assay will work on mixed serovar populations such as those resulting from enrichment cultures (55). The prevalence of Salmonella serovars over time in poultry is dynamic and often the result of serovar-specific mitigations such as vaccination (56–58). Thus, as is also recommended in the recent U.S. Department of Agriculture—National Advisory Committee on Microbiological Criteria for Foods (USDA-NACMCF) report, reevaluating serovars of concern every few years is important (59). Here, the utility of deep serotyping by methods such as CRISPR-SeroSeq is particularly apparent, as it can detect serovars before they become abundant and routine parallel surveillance can determine whether these serovars are increasing or decreasing.
This study presents a novel and timely approach to monitoring Salmonella in broiler flocks that can support long-term efforts to control this pathogen. While focused on poultry in this investigation, this framework could be adapted to Salmonella surveillance in other food production systems.
MATERIALS AND METHODS
Experimental design and sample collection.
We evaluated broiler flocks in four different complexes, each owned by a different integrator (Fig. 1). In each complex, five broiler farms were visited, for a total 20 farms, and four broiler houses were sampled at each farm. A total of 80 broiler flocks were sampled over a 5-week period to limit any variability due to seasonal effects. The flock ages ranged from 3 to 5 weeks (complex 1) and 4 to 5 weeks (complexes 2 to 4). Two pairs of boot socks moistened in buffered peptone water (BPW; Romer Labs, Newark, DE) were used for sampling each house, resulting in a total of 160 samples (Fig. 1). One pair was used to walk along the right side of the house and the other for the left side. Samples were collected by walking the entire length of the house between the feed and water lines, tending closest to the latter. The samples were kept in a cooler on ice and transported to the laboratory for analysis.
Salmonella isolation and quantification.
A total of 200 mL of BPW was added to each pair of boot socks, and these were stomached at 230 rpm for 2 min. A 60-mL aliquot of the homogenized culture was transferred to 60 mL of MP preenrichment medium (Hygiena, Camarillo, CA) and incubated at 42°C for 10 h for initial Salmonella recovery. At this time, Salmonella lysates were prepared following the manufacturer’s protocol (Hygiena) and stored at 4°C. Concurrently, 1 mL of the preenriched culture was added to 10 mL of tetrathionate broth (TT; VWR, Radnor, PA), followed by incubation at 37°C for 24 h before being plated onto XLT-4 (xylose–lysine–Tergitol-4) agar plates that were then incubated at 37°C for 24 h. At the same time as plating, 1 mL of the overnight TT culture was removed and placed into a microcentrifuge tube. These were centrifuged at 14,000 rpm for 3 min; after the supernatant was discarded, the pellets were stored at −20°C until deep serotyping. Up to three isolated H2S-positive colonies were selected, including those with different colony morphologies, where present. A single one of these (representing the most common colony morphology on the XLT-4 plate) was confirmed as Salmonella using O antigen antisera (see “Salmonella serogrouping” below). Salmonella was enumerated in culture-positive samples from the stored lysates using the BAX System SalQuant assay (Hygiena). Salmonella quantity was calculated using the regression curve provided with the assay. The limit of quantification (LOQ) for this assay is reported to be 1 log10 CFU/sample, and culture-positive samples that did not yield a value were scored and analyzed as 0 log10 CFU/sample.
DNA isolation and CRISPR-SeroSeq.
Total genomic DNA was isolated from frozen TT culture pellets using the Promega Genome Wizard kit (Promega, Madison, WI) following the manufacturer’s instructions. The DNA pellet was resuspended in 200 μL of molecular-grade water and stored at −20°C. DNA samples were diluted (1:20) in molecular-grade water, and 2 μL was used as a template in the first-step PCR for CRISPR SeroSeq analysis. This PCR amplifies Salmonella using primers that target the Salmonella CRISPR direct repeats (20). The PCR products were visualized by gel electrophoresis and purified using the AMPure system following the manufacturer’s instructions (Beckman Coulter, Indianapolis, IN). A total of 5 μL of the purified product was used as a template for a second PCR to add dual index sequences according to the Illumina Nextera protocol (Illumina, San Diego, CA). The products of the second PCR were visualized by gel electrophoresis and purified as described above, followed by pooling in approximately equimolar proportions. The library was sequenced using an Illumina NextSeq (Wright Labs, Huntingdon, PA) using a mid-throughput kit with 150 cycles. Two negative controls (nontemplate controls) for the first and second PCRs and one positive control of Salmonella Enteritidis with a known CRISPR profile were included in the library. After sequencing, CRISPR-SeroSeq analysis was performed as previously described using an R (version 4.04) script that parses sequence reads and uses BLAST to match sequence reads to a database of more than 145 Salmonella serovars, followed by writing the output to Microsoft Excel (11, 20). CRISPR spacers with fewer than 10 reads were not included in the analysis.
Risk scoring based on serovar profiles and Salmonella quantity.
Total Salmonella quantity (determined by BAX analysis) was multiplied by the relative abundance of each serovar (from CRISPR-SeroSeq data) to provide an estimated serovar quantity for each house. Putative risk scores were attributed to each flock as follows: 0, Salmonella negative; 1, KPI absent and average house Salmonella quantity of <3.6 log10 CFU/sample; 2, KPI absent and average house Salmonella quantity of >3.6 log10 CFU/sample, 3, KPI present and average house Salmonella quantity of <3.6 log10 CFU/sample; and 4, KPI present and average house Salmonella quantity of >3.6 log10 CFU/sample. House average of 3.6 log10 CFU/sample was used as the threshold because it was the average quantity across all four complexes.
Houses scored 0 or 1 are the lowest risk, as they are either Salmonella negative or have a non-KPI serovar average house Salmonella quantity below 3.6 log10 CFU/house.
Antibiotic susceptibility testing.
The Kirby-Bauer disk diffusion assay was used to determine the susceptibility of Salmonella isolates to nine different antibiotics: ampicillin (10 μg), amoxicillin-clavulanic acid (20/10 μg), ceftriaxone (30 μg), gentamicin (10 μg), streptomycin (10 μg), tetracycline (30 μg), ciprofloxacin (5 μg), sulfamethoxazole-trimethoprim (25 μg), and chloramphenicol (30 μg). Bacterial cultures were standardized to 0.5 McFarland and a bacterial lawn was plated onto Mueller-Hinton agar. After antibiotic disks were placed on the lawns, the samples were incubated for 18 to 22 h at 37°C. After incubation, zones of inhibition were measured and the CSLI interpretive standards were used to characterize isolates as susceptible, intermediate, or resistant (60). For subsequent analyses, intermediate resistance was recorded as susceptible. Isolates were considered multidrug resistant (MDR) if they were resistant to three or more classes of antibiotics.
Salmonella serogrouping.
Serogroups were determined by agglutination with O antigen serum purchased from BD Difco (Franklin Lakes, NJ): O:3 (includes serovar Senftenberg), O:4 (Typhimurium and Schwarzengrund), O:7 (Infantis), O:8 (Kentucky and Hadar), and O:9 (Enteritidis) on a single representative colony from each positive sample (n = 122 positive samples). Salmonella isolates were grown on tryptic soy agar (TSA). Serotypes were inferred using CRISPR-SeroSeq data and serogroup information. Serovars Typhimurium and Schwarzengrund never occurred in the same samples, averting inferential problems with the O:4 antigen. In the one house where serovars Kentucky and Hadar co-occurred, we were unable to determine the serotype from the serogroup analysis since both are group O:8 (indicated in Table S1).
Statistical analysis.
The molecular and serovar data were analyzed using R Studio (version 4.04) and SAS Studio v3.8 (SAS Institute Cary, NC). Salmonella prevalence and quantity by complex and boot socks were analyzed by analysis of variance (ANOVA), and the means were separated with Tukey’s HSD (honestly significant difference) test at a 5% level of significance (P < 0.05). For variance component analysis, Stata MP version 17.0 (College Station, TX) was used to construct a multilevel mixed-effect logistic regression analysis with random effects for complex, farm, and house. Contingency table analyses (unadjusted for clustering by complex: farm: house) were used to assess associations with single-drug and multidrug resistances, including of the use of nonmedically important antimicrobials at the house level. Multidrug resistance phenotypes were similarly explored with unadjusted and adjusted models, including using multinomial regression.
ACKNOWLEDGMENTS
This work was supported by a sponsored research agreement between the University of Georgia and Ancera, Inc., and also by a USDA-NIFA award to N.W.S. (2020-67017-30792).
We are grateful to the integrators who participated in this study.
Footnotes
Supplemental material is available online only.
Contributor Information
Nikki W. Shariat, Email: nikki.shariat@uga.edu.
Christopher A. Elkins, Centers for Disease Control and Prevention
REFERENCES
- 1.Tack DM, Ray L, Griffin PM, Cieslak PR, Dunn J, Rissman T, Jervis R, Lathrop S, Muse A, Duwell M, Smith K, Tobin-D’Angelo M, Vugia DJ, Zablotsky Kufel J, Wolpert BJ, Tauxe R, Payne DC. 2020. Preliminary incidence and trends of infections with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2016–2019. MMWR Morb Mortal Wkly Rep 69:509–514. 10.15585/mmwr.mm6917a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Interagency Food Safety Analytics Collaboration. 2021. Foodborne illness source attribution estimates for 2019 for Salmonella, Escherichia coli O157, Listeria monocytogenes, and Campylobacter using multi-year outbreak surveillance data, United States. https://www.cdc.gov/foodsafety/ifsac/pdf/P19-2019-report-TriAgency-508.pdf. Accessed 2 October 2022.
- 3.Harvey PC, Watson M, Hulme S, Jones MA, Lovell M, Berchieri AJ, Young J, Bumstead N, Barrow P. 2011. Salmonella enterica serovar Typhimurium colonizing the lumen of the chicken intestine grows slowly and upregulates a unique set of virulence and metabolism genes. Infect Immun 79:4105–4121. 10.1128/IAI.01390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mon KKZ, Zhu Y, Chanthavixay G, Kern C, Zhou H. 2020. Integrative analysis of gut microbiome and metabolites revealed novel mechanisms of intestinal Salmonella carriage in chicken. Sci Rep 10:4809. 10.1038/s41598-020-60892-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris GK, Wells JG. 1970. Salmonella contamination in a poultry-processing plant. Appl Microbiol 19:795–799. 10.1128/am.19.5.795-799.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng RA, Eade CR, Wiedmann M. 2019. Embracing diversity: differences in virulence mechanisms, disease severity, and host adaptations contribute to the success of nontyphoidal Salmonella as a foodborne pathogen. Front Microbiol 10:1368. 10.3389/fmicb.2019.01368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng Y, Pedroso AA, Porwollik S, McClelland M, Lee MD, Kwan T, Zamperini K, Soni V, Sellers HS, Russell SM, Maurer JJ. 2015. rpoS-regulated core genes involved in the competitive fitness of Salmonella enterica serovar Kentucky in the intestines of chickens. Appl Environ Microbiol 81:502–514. 10.1128/AEM.03219-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks LA, Bailey MA, Krehling JT, Chasteen KS, Macklin KS. 2021. A comparison of colonizing ability between Salmonella Enteritidis and Salmonella Heidelberg in broiler chickens challenged through feed administration. Foodborne Pathog Dis 18:784–789. 10.1089/fpd.2021.0016. [DOI] [PubMed] [Google Scholar]
- 9.Chadwick EV, MacKay L, Krehling JT, Macklin KS. 2020. Inoculation route and serotype alter Salmonella recovery in the broiler’s digestive tract. Avian Dis 64:467–470. 10.1637/aviandiseases-D-20-00012. [DOI] [PubMed] [Google Scholar]
- 10.Shah DH, Paul NC, Sischo WC, Crespo R, Guard J. 2017. Population dynamics and antimicrobial resistance of the most prevalent poultry-associated Salmonella serotypes. Poult Sci 96:687–702. 10.3382/ps/pew342. [DOI] [PubMed] [Google Scholar]
- 11.Siceloff AT, Doug W, Shariat NW. 2022. Regional Salmonella differences in United States broiler production from 2016 to 2020 and the contribution of multiserovar populations to Salmonella surveillance. Appl Environ Microbiol 88:e00204-22. 10.1128/aem.00204-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.United States Department of Agriculture—Food Safety and Inspection Service. Quarterly sampling reports on Salmonella and Campylobacter. https://www.fsis.usda.gov/science-data/data-sets-visualizations/microbiology/microbiological-testing-program-rte-meat-and-7. Accessed 12 January 2023.
- 13.Centers for Disease Control and Prevention. National Outbreak Reporting System (NORS). https://wwwn.cdc.gov/norsdashboard/. Accessed 12 January 2023.
- 14.United States Department of Agriculture—Food Safety and Inspection Service. 2010. A comparison of Salmonella serotype incidence in FSIS-regulated products and salmonellosis cases. https://www.fsis.usda.gov/sites/default/files/media_file/2021-02/Serotype_Incidence_and_Salmonellosis.pdf. Accessed 2 October 2022.
- 15.United States Department of Agriculture—Food Safety and Inspection Service. 2022. USDA FY 2022–2026 food safety key performance indicator. https://www.fsis.usda.gov/inspection/inspection-programs/inspection-poultry-products/reducing-salmonella-poultry/salmonella-0. Accessed 2 October 2022.
- 16.Thames HT, Sukumaran AT. 2020. A review of Salmonella and Campylobacter in broiler meat: emerging challenges and food safety measures. Foods 9:776. 10.3390/foods9060776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox NA, Berrang ME, House SL, Medina D, Cook KL, Shariat NW. 2019. Population analyses reveal preenrichment method and selective enrichment media affect Salmonella serovars detected on broiler carcasses. J Food Prot 82:1688–1696. 10.4315/0362-028X.JFP-19-166. [DOI] [PubMed] [Google Scholar]
- 18.Rasamsetti S, Berrang M, Cox NA, Shariat NW. 2021. Selective pre-enrichment method to lessen time needed to recover Salmonella from commercial poultry processing samples. Food Microbiol 99:103818. 10.1016/j.fm.2021.103818. [DOI] [PubMed] [Google Scholar]
- 19.Rasamsetti S, Berrang ME, Cox NA, Shariat NW. 2022. Assessing Salmonella prevalence and complexity through processing using different culture methods. Poult Sci 101:101949. 10.1016/j.psj.2022.101949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson CP, Doak AN, Amirani N, Schroeder EA, Wright J, Kariyawasam S, Lamendella R, Shariat NW. 2018. High-resolution identification of multiple Salmonella serovars in a single sample by using CRISPR-SeroSeq. Appl Environ Microbiol 84:e01859-18. 10.1128/AEM.01859-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.US Food Safety and Inspection Service. 2022. USDA-FSIS microbiological laboratory guide, Chapter 4 Isolation and Identification of Salmonella, version 12. US Food Safety and Inspection Service, Washington, DC. https://www.fsis.usda.gov/news-events/publications/microbiology-laboratory-guidebook. [Google Scholar]
- 22.Cason J, Cox N, Buhr R, Bourassa D, Richardson L. 2011. Probability of identifying different Salmonella serotypes in poultry samples. In International Poultry Scientific Forum. Southern Poultry Science Society, Mississippi State, MS. [Google Scholar]
- 23.Liu F, Barrangou R, Gerner-Smidt P, Ribot EM, Knabel SJ, Dudley EG. 2011. Novel virulence gene and clustered regularly interspaced short palindromic repeat (CRISPR) multilocus sequence typing scheme for subtyping of the major serovars of Salmonella enterica subsp. enterica. Appl Environ Microbiol 77:1946–1956. 10.1128/AEM.02625-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabre L, Zhang J, Guigon G, Le Hello S, Guibert V, Accou-Demartin M, De Romans S, Lim C, Roux C, Passet V, Diancourt L, Guibourdenche M, Issenhuth-Jeanjean S, Achtman M, Brisse S, Sola C, Weill F-X. 2012. CRISPR typing and subtyping for improved laboratory surveillance of Salmonella infections. PLoS One 7:e36995. 10.1371/journal.pone.0036995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shariat N, Timme RE, Pettengill JB, Barrangou R, Dudley EG. 2015. Characterization and evolution of Salmonella CRISPR-Cas systems. Microbiology (Reading) 161:374–386. 10.1099/mic.0.000005. [DOI] [PubMed] [Google Scholar]
- 26.Shariat N, Sandt CH, Dimarzio MJ, Barrangou R, Dudley EG. 2013. CRISPR-MVLST subtyping of Salmonella enterica subsp. enterica serovars Typhimurium and Heidelberg and application in identifying outbreak isolates. BMC Microbiol 13:254. 10.1186/1471-2180-13-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shariat N, Dudley EG. 2014. CRISPRs: molecular signatures used for pathogen subtyping. Appl Environ Microbiol 80:430–439. 10.1128/AEM.02790-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vosik D, Tewari D, Dettinger L, M’ikanatha NM, Shariat NW. 2018. CRISPR typing and antibiotic resistance correlates with polyphyletic distribution in human isolates of Salmonella Kentucky. Foodborne Pathog Dis 15:101–108. 10.1089/fpd.2017.2298. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen SV, Harhay DM, Bono JL, Smith TPL, Fields PI, Dinsmore BA, Santovenia M, Wang R, Bosilevac JM, Harhay GP. 2018. Comparative genomics of Salmonella enterica serovar Montevideo reveals lineage-specific gene differences that may influence ecological niche association. Microb Genom 4:e000202. 10.1099/mgen.0.000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Worley J, Meng J, Allard MW, Brown EW, Timme RE. 2018. Salmonella enterica phylogeny based on whole-genome sequencing reveals two new clades and novel patterns of horizontally acquired genetic elements. mBio 9:e02303-18. 10.1128/mBio.02303-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conner DE, Wakefield CB. 2006. Incidence of Salmonella in processed broilers following transportation in contaminated coops. In Memorias del XII European Poultry Conference, Verona, Italy. World’s Poultry Science Association, Beekbergen, The Netherlands. [Google Scholar]
- 32.Berghaus RD, Thayer SG, Law BF, Mild RM, Hofacre CL, Singer RS. 2013. Enumeration of Salmonella and Campylobacter spp. in environmental farm samples and processing plant carcass rinses from commercial broiler chicken flocks. Appl Environ Microbiol 79:4106–4114. 10.1128/AEM.00836-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crabb HK, Lee Allen J, Maree Devlin J, Matthew Firestone S, Reginald Wilks C, Rudkin Gilkerson J. 2018. Salmonella spp. transmission in a vertically integrated poultry operation: clustering and diversity analysis using phenotyping (serotyping, phage typing) and genotyping (MLVA). PLoS One 13:e0201031. 10.1371/journal.pone.0201031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liljebjelke KA, Hofacre CL, Liu T, White DG, Ayers S, Young S, Maurer JJ. 2005. Vertical and horizontal transmission of Salmonella within integrated broiler production system. Foodborne Pathog Dis 2:90–102. 10.1089/fpd.2005.2.90. [DOI] [PubMed] [Google Scholar]
- 35.Desin TS, Köster W, Potter AA. 2013. Salmonella vaccines in poultry: past, present and future. Expert Rev Vaccines 12:87–96. 10.1586/erv.12.138. [DOI] [PubMed] [Google Scholar]
- 36.Hofacre CL, Rosales AG, Da Costa M, Cookson K, Schaeffer J, Jones MK. 2021. Immunity and protection provided by live modified vaccines against paratyphoid Salmonella in poultry - an applied perspective. Avian Dis 65:295–302. [DOI] [PubMed] [Google Scholar]
- 37.Dale EL, Nolan SP, Berghaus RD, Hofacre CL. 2015. On farm prevention of Campylobacter and Salmonella: lessons learned from basic biosecurity interventions. J Appl Poult Res 24:222–232. 10.3382/japr/pfv016. [DOI] [Google Scholar]
- 38.Sanchez S, Hofacre CL, Lee MD, Maurer JJ, Doyle MP. 2002. Animal sources of salmonellosis in humans. J Am Vet Med Assoc 221:492–497. 10.2460/javma.2002.221.492. [DOI] [PubMed] [Google Scholar]
- 39.United States Department of Agriculture—Food Safety and Inspection Service. 2022. Proposed regulatory framework to reduce Salmonella illnesses attributable to poultry. https://www.fsis.usda.gov/inspection/inspection-programs/inspection-poultry-products/reducing-salmonella-poultry/proposed. Accessed 2 October 2022.
- 40.United States Department of Agriculture—Food Safety and Inspection Service. Serotypes profile of Salmonella isolates from meat and poultry products January 1998 through December 2014. https://www.fsis.usda.gov/sites/default/files/media_file/2020-10/Salmonella-Serotype-Annual-2014.pdf. Accessed 2 October 2022.
- 41.Centers for Disease Control and Prevention. 2011. National Enteric Disease Surveillance: Salmonella annual report, 2011. https://www.cdc.gov/ncezid/dfwed/pdfs/salmonella-annual-report-2011-508c.pdf. Accessed 2 October 2022.
- 42.McMillan EA, Wasilenko JL, Tagg KA, Chen JC, Simmons M, Gupta SK, Tillman GE, Folster J, Jackson CR, Frye JG. 2020. Carriage and gene content variability of the pESI-like plasmid associated with Salmonella Infantis recently established in United States poultry production. Genes 11:1516. 10.3390/genes11121516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arnold ME, Mueller-Doblies D, Carrique-Mas JJ, Davies RH. 2009. The estimation of pooled-sample sensitivity for detection of Salmonella in turkey flocks. J Appl Microbiol 107:936–943. 10.1111/j.1365-2672.2009.04273.x. [DOI] [PubMed] [Google Scholar]
- 44.National Poultry Improvement Plan. 2014. National Poultry Improvement Plan and auxiliary provisions as of August 8, 2014 as found in the Code of Federal Regulations. Title 9, animals and animal products, parts 145–147 and part 56. https://poultryimprovement.org/documents/AuxiliaryProvisions.pdf. Accessed 2 October 2022.
- 45.Obe T, Berrang ME, Cox NA, House SL, Shariat NW. 2021. Comparison of selective enrichment and plating media for Salmonella isolation from broiler carcasses. J Food Saf 41:e12928. 10.1111/jfs.12928. [DOI] [Google Scholar]
- 46.Gorski L. 2012. Selective enrichment media bias the types of Salmonella enterica strains isolated from mixed strain cultures and complex enrichment broths. PLoS One 7:e34722. 10.1371/journal.pone.0034722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singer RS, Mayer AE, Hanson TE, Isaacson RE. 2009. Do microbial interactions and cultivation media decrease the accuracy of Salmonella surveillance systems and outbreak investigations? J Food Prot 72:707–713. 10.4315/0362-028x-72.4.707. [DOI] [PubMed] [Google Scholar]
- 48.Pal A, Bailey MA, Talorico AA, Krehling JT, Macklin KS, Price SB, Buhr RJ, Bourassa DV. 2021. Impact of poultry litter Salmonella levels and moisture on transfer of Salmonella through associated in vitro generated dust. Poult Sci 100:101236. 10.1016/j.psj.2021.101236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pal A, Jackson AP, Urrutia A, Macklin KS, Price SB, Buhr RJ, Bourassa DV. 2022. Research note: Bacterial composition of settled dust during growout of broiler chickens. Poult Sci 101:101602. 10.1016/j.psj.2021.101602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.European Commission. 2012. Commission regulation (EU) no 200/2012 of 8 March 2012 concerning a union target for the reduction of Salmonella Enteritidis and Salmonella Typhimurium in flocks of broilers. Off J Eur Communities 71:31–36. https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32012R0200. [Google Scholar]
- 51.a.v.e.c., COPA-COGECA. 2012. European poultrymeat industry guide. Guide of good hygiene practice for the prevention and control of microbiological infections focused on Salmonella control of chickens reared for meat—at slaughterhouses. https://www.adiveter.com/ftp_public/A1310709.pdf. Accessed 2 October 2022.
- 52.Drauch V, Ibesich C, Vogl C, Hess M, Hess C. 2020. In-vitro testing of bacteriostatic and bactericidal efficacy of commercial disinfectants against Salmonella Infantis reveals substantial differences between products and bacterial strains. Int J Food Microbiol 328:108660. 10.1016/j.ijfoodmicro.2020.108660. [DOI] [PubMed] [Google Scholar]
- 53.Siceloff AT, Ohta N, Norman KN, Loneragan GH, Norby B, Morgan SH, Shariat NW. 2021. Antimicrobial resistance hidden within multiserovar Salmonella populations. Antimicrob Agents Chemother 65:e00048-21. 10.1128/AAC.00048-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noyes NR, Weinroth ME, Parker JK, Dean CJ, Lakin SM, Raymond RA, Rovira P, Doster E, Abdo Z, Martin JN, Jones KL, Ruiz J, Boucher CA, Belk KE, Morley PS. 2017. Enrichment allows identification of diverse, rare elements in metagenomic resistome-virulome sequencing. Microbiome 5:142. 10.1186/s40168-017-0361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richards A, Hopkins B, Shariat N. 2020. Conserved CRISPR arrays in Salmonella enterica serovar Infantis can serve as qPCR targets to detect Infantis in mixed serovar populations. Lett Appl Microbiol 71:138–145. 10.1111/lam.13296. [DOI] [PubMed] [Google Scholar]
- 56.Foley S, Nayak R, Hanning I, Johnson T, Han J, Ricke S. 2011. Population dynamics of Salmonella enterica serotypes in commercial egg and poultry production. Appl Environ Microbiol 77:4273–4279. 10.1128/AEM.00598-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dórea FC, Cole DJ, Hofacre C, Zamperini K, Mathis D, Doyle MP, Lee MD, Maurer JJ. 2010. Effect of Salmonella vaccination of breeder chickens on contamination of broiler chicken carcasses in integrated poultry operations. Appl Environ Microbiol 76:7820–7825. 10.1128/AEM.01320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berghaus RD, Thayer SG, Maurer JJ, Hofacre CL. 2011. Effect of vaccinating breeder chickens with a killed Salmonella vaccine on Salmonella prevalences and loads in breeder and broiler chicken flocks. J Food Prot 74:727–734. 10.4315/0362-028X.JFP-10-542. [DOI] [PubMed] [Google Scholar]
- 59.United States Department of Agriculture—National Advisory Committee on Microbiological Criteria for Foods (NACMCF). 2022. Response to questions posed by the Food Safety and Inspection Service: enhancing Salmonella control in poultry products. https://www.fsis.usda.gov/sites/default/files/media_file/documents/NACMCF_Salmonella-Poultry_Response_for_Committee_Review.pdf. Accessed 2 October 2022.
- 60.Clinical and Laboratory Standards Institute. 2019. Performance standards for antimicrobial susceptibility testing, 29th ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.02035-22-s0001.xlsx, XLSX file, 0.02 MB (18.4KB, xlsx)