Dear Editor,
Several subvariants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant have evolved very quickly and are also evolving continuously. Notably, some significant subvariants such as BF.7, BA.2, BA.2.75, BA.2.75.2, BQ, XBB, etc., are potentially expanding in the brief period of current times1,2. These significant subvariants were circulating throughout the globe, eventually became dominant, and are creating surges in coronavirus disease 2019 (COVID-19) cases from time to time in different countries across the world. They have acquired some exclusive characteristics like enhanced immune evasion, higher transmissibility, additional spike mutations, distinct viral signatures, epidemiological, and clinical features, and are leading to vaccine breakthrough infections and reinfections3,4. Recently, it has become very apparent that BQ, and XBB family members are noteworthy subvariants, which were described as the ‘variant soup’ by the scientists and are posing fears of unpredictable surges in some cases this winters5. Researchers have tried to analyze and unfold the receptor-binding domain (RBD) mutations over time6. These new subvariants, BQ and XBB, are currently overgrowing in different countries, owing to which COVID-19 cases are increasing significantly in Europe, North America, India, Singapore, and other countries5,7.
Among the ‘variants soups’, XBB and its offspring, such as XBB.1 and XBB.1.5, are playing a significant role in the recent surges observed in different nations. XBB is a recombinant sublineage, and the offspring XBB.1 resulted from recombination between two lineages of BA.2, which are BA.2.75 and BJ.1(BA.2.10.1) and sublineages (Fig. 1A). Researchers have reported that the subvariant is increasing very fast in the USA, Singapore, France, and India,7. These sublineages have several unique mutations that help them acquire antibody escape mechanisms and overpower the protection rendered by neutralizing antibodies and monoclonal antibodies (mAbs).
Figure 1.
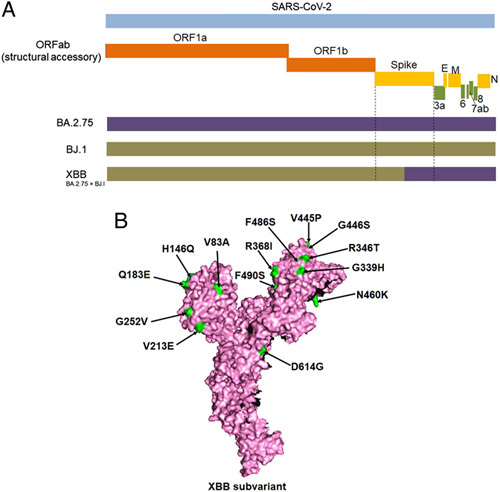
Origin and significant spike mutations of the XBB subvariant. (A) the schematic diagram depict the recombinant origin of the XBB subvariant, which was resulted from recombination between two lineages of BA.2, which are BA.2.75 and BJ.1(BA.2.10.1). (B) Significant spike mutations of the XBB subvariant including receptor-binding domain (RBD) mutations.
The newly emerged Omicron sublineages XBB and XBB.1 were first identified in India in August 2022. Subsequently, these spread rapidly in several Asian countries and are now predominant in Singapore and India, as well as other countries in Asia, and are rising worldwide. The spike (S) protein of XBB has 14 mutations additionally found in BA.2, precisely nine mutations in the RBD regions (Fig. 1B). XBB.1 contained an additional mutation (G252V) in comparison to XBB. The wide range of S-protein mutations in these two sublineages signifies major concerns about the compromising efficacy of existing vaccines and antibodies (mAbs, neutralizing antibodies) in use as therapeutics against COVID-198. The mutations may cause antigenic shifts in newly emerging sublineages that facilitate the vaccine escape phenomena5.
Previously, it was noted that the substitution mutation in Omicron S-protein R346T confers therapeutic antibody resistance3,9. Researchers have also evidenced that the XBB and XBB.1 share the R346T and N460K mutations, holding the evolutionary convergence targeted to the escape of antibodies engaged to the S-protein regions10. The mutations N460K and F486S show resistance to the RBD region’s mAbs (classes 1 and 2). Moreover, the mutations F490S, R346T, G446S, and V455P are considered for revealing resistance to the mAbs of RBD class 3. It has also been noted that the substitution of V445P in XBB and XBB.1 might employ a comparable effect as the mutation of K444T. This mutation is responsible for the steric hindrance as well as the breaking of the hydrogen bond with mAbs, resulting in the incapability of the antibody neutralization mechanism8.
R346S/K mutations were detected in the sublineages of Omicron; specifically, the mutation in the position of R346 has been suggested to show a superior impact in the context of the other Omicron mutations according to the amino acid interaction networks model developed by Miller et al.8. However, the researchers have also pointed out that the mutation R346 has also been associated with the epitope network of S309 with other mutations8. In the position of R346, R→K is observed as a conservative mutation. However, different researchers, such as Liu et al.11 and Cameroni et al.12, have assessed the R346K+Omicron and noted no significant extra reduction in antibody neutralization.
The XBB and its offspring have acquired some exclusive characteristics, such as enhanced immune evasion, higher transmissibility, and additional spike mutations, which helped in the natural selection of the XBB, making them ‘great escapists and more transmissible’, a significant subvariant candidate of the Omicron family. Uraki and colleagues have tried to understand the current humoral immunity status of the XBB and BQ.1.1 by performing an experiment using three different groups using these isolates. The first group was made with the persons (n=20) who received third doses of the monovalent mRNA vaccines mRNA-1273 (Moderna) or BNT162b2 (Pfizer–BioNTech) both. The second group was made up of individuals who received four doses of the monovalent mRNA vaccine, mRNA-1273 BNT162b2. Another group was made up of individuals who received these three vaccine doses before the BA.2 infection (n=10). The researchers found a 50% reduction in the neutralization titer of the plasma samples against XBB and suggested that the subvariant efficiently evades the recently acquired humoral immunity induced by natural infection or the mRNA vaccine. They also concluded that clinical isolates of XBB have superior immune evasion capability compared to the subvariant of Omicron variants such as BA.2 and BA.57. Similarly, Kurhade et al.13 reported reduced neutralization activity of XBB.1 against the mRNA vaccine-induced immunity with different doses.
Recently, cases of the newly identified XBB.1.5 subvariant (an offshoot of XBB) have been seen significantly rising in the USA, and its prevalence is increasing in other countries; however, much is yet to be known about XBB.1.5. It is presently not clear whether it will lead to any high surge in COVID-19 cases or increased hospitalizations, and eventually might pose a global threat as a dominant subvariant outcompeting other previous subvariants in coming time14,15. It was reported that this recombinant subvariant has one additional mutation, F486P (substitution) a rarely seen amino acid change, in the spike protein, compared to XBB.1. The rarely seen mutation in XBB.1.5 might enhance its infectivity and facilitate opportunities for further evolutionary gains and acquiring new mutations14. XBB.1.5 has gained enhanced transmissibility and a strong immune evasion property. A recent report indicated that the robust ACE2 receptor-binding affinity and antibody evasion properties might significantly increase the transmissibility XBB.1.516. Antibody-evading mutations have allowed new lineages to overcome immune defenses gained from vaccination and previous infection waves with different variants and subvariants. One recent study has suggested that individuals who have been vaccinated with updated bivalent booster doses are better protected against the XBB, possibly including offspring such as XBB.1.5, than those who have not taken up such booster shots17.
In the current changing scenario, there is an urgent need to enhance surveillance and tracking of emerging novel Omicron subvariants, understand the protective levels of humoral and cellular immunity against these new subvariants, explore the beneficial effects of booster doses, and design newer and more efficacious vaccines and mAbs for the clinics for prophylaxis and treatment of COVID-9 patients. The global concerns of a surge in COVID-19 cases ahead need to be countered by promoting booster uptake and raising awareness against relaxed disease-mitigation measures and by adopting COVID-19 appropriate behavior and safety measures, including the wearing of face masks, hand washing, hygiene, and social distancing norms. Finally, we urge policymakers to implement adequate proactive control measures, formulate preparedness plans and appropriate infection prevention and control measures, and implement necessary strategies to prevent the transmission and spread of newly emerging Omicron subvariants so as to ameliorate the fears of a surge of a new COVID-19 pandemic.
Ethical approval
This article does not require any human or animal subjects to acquire such approval.
Sources of funding
This study received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author contribution
C.C. and K.D.: designed the study. C.C., M.B., H.C., A.I.: made the first draft. C.C., K.D., G.S.: updated the manuscript. C.C. and K.D.: reviewed the final draft. All authors have critically reviewed and approved the final draft and are responsible for the content and similarity index of the manuscript.
Conflicts of interest disclosure
All authors report no conflicts of interest relevant to this article.
Research registration unique identifying number (UIN)
Name of the registry: not applicable.
Unique identifying number or registration ID: not applicable.
Hyperlink to your specific registration (must be publicly accessible and will be checked): not applicable.
Guarantor
Md. Aminul Islam, COVID-19 Diagnostic Lab, Department of Microbiology, Noakhali Science and Technology University, Noakhali 3814, Bangladesh
Data statement
The data in this correspondence article is not sensitive in nature and is accessible in the public domain. The data is therefore available and not of a confidential nature.
Provenance and peer review
Not commissioned, internally peer-reviewed.
Acknowledgments
All authors are thankful to their respective institutes and universities.
Footnotes
C.C. and M.B.: contributed equally for this study.
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Published online 14 March 2023
Contributor Information
Chiranjib Chakraborty, Email: drchiranjib@yahoo.com.
Manojit Bhattacharya, Email: mbhattacharya09@gmail.com.
Hitesh Chopra, Email: chopraontheride@gmail.com.
Md. Aminul Islam, Email: aminulmbg@gmail.com.
Gutulla Saikumar, Email: saikumarivri@gmail.com.
Kuldeep Dhama, Email: kdhama@rediffmail.com.
References
- 1. Chakraborty C, Bhattacharya M, Sharma AR, et al. The rapid emergence of multiple sublineages of Omicron (B.1.1.529) variant: dynamic profiling via molecular phylogenetics and mutational landscape studies. Journal of infection and public health 2022;15:1234–1258. [DOI] [PubMed] [Google Scholar]
- 2. Chakraborty C, Bhattacharya M, Dhama K. Cases of BA.2.75 and recent BA.2.75.2 subvariant of Omicron are increasing in India: is it alarming at the global level? Ann Med Surg (Lond) 2022;84:104963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Qu P, Evans JP, Faraone JN, et al. Enhanced neutralization resistance of SARS-CoV-2 Omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Cell host & microbe 2022;31:9–17. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mohapatra RK, Mahal A, Kutikuppala LS, et al. Renewed global threat by the novel SARS-CoV-2 variants ‘XBB, BF.7, BQ.1, BA.2.75, BA.4.6’: a discussion. Frontiers in Virology 2022;2:1–5. [Google Scholar]
- 5. Wang Q, Iketani S, Li Z, et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 2022;186:279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Callaway E. COVID ‘variant soup’ is making winter surges hard to predict. Nature 2022;611:213–214. [DOI] [PubMed] [Google Scholar]
- 7. Uraki R, Ito M, Furusawa Y, et al. Humoral immune evasion of the omicron subvariants BQ.1.1 and XBB. The Lancet Infectious diseases 2023;23:30–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miller NL, Clark T, Raman R, et al. Insights on the mutational landscape of the SARS-CoV-2 Omicron variant receptor-binding domain. Cell reports Medicine 2022;3:100527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sheward DJ, Kim C, Fischbach J, et al. Omicron sublineage BA.2.75.2 exhibits extensive escape from neutralising antibodies. The Lancet Infectious diseases 2022;22:1538–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bobe P, Kiger N, Kinsky RG. In vivo and in vitro immune responses of CBA/N (Xid) mice against allogeneic cells. Nephrologie 1987;8:147–153. [PubMed] [Google Scholar]
- 11. Liu L, Iketani S, Guo Y, et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 2022;602:676–681. [DOI] [PubMed] [Google Scholar]
- 12. Cameroni E, Bowen JE, Rosen LE, et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 2022;602:664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kurhade C, Zou J, Xia H, et al. Low neutralization of SARS-CoV-2 Omicron BA.2.75.2, BQ.1.1 and XBB.1 by parental mRNA vaccine or a BA.5 bivalent booster. Nature medicine 2022;29:344–347. [DOI] [PubMed] [Google Scholar]
- 14. Callaway E. Coronavirus variant XBB.1.5 rises in the United States – is it a global threat? Nature 2023;613:222–223. [DOI] [PubMed] [Google Scholar]
- 15. WHO. XBB.1.5 Rapid risk assessment. Accessed 11 January 2023. https://www.who.int/docs/default-source/coronaviruse/11jan2023_xbb15_rapid_risk_assessment.pdf
- 16. Yue C, Song W, Wang L, et al. Enhanced transmissibility of XBB.1.5 is contributed by both strong ACE2 binding and antibody evasion. bioRxiv 2023;23:278–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davis-Gardner ME, Lai L, Wali B, et al. Neutralization against BA.2.75.2, BQ.1.1, and XBB from mRNA Bivalent Booster. The New England journal of medicine 2023;388:183–185. [DOI] [PMC free article] [PubMed] [Google Scholar]


