Abstract
Background and Objectives:
SARS-CoV-2 variants of concern (VOC) and interest (VOI) pose a significant threat to public health because the rapid change in the SARS-CoV-2 genome can alter viral phenotypes such as virulence, transmissibility and the ability to evade the host response. Hence, SARS-CoV-2 quantification techniques are essential for timely diagnosis and follow-up. Besides, they are vital to understanding viral pathogenesis, antiviral evaluation, and vaccine development.
Materials and Methods:
Five isolates of SARS-CoV-2: D614G strain (B.1), three VOC (Alpha, Gamma and Delta), and one VOI (Mu) were used to compare three techniques for viral quantification, plaque assay, median tissue culture infectious dose (TCID50) and real-time RT-PCR.
Results:
Plaque assay showed viral titers between 0.15 ± 0.01×107 and 1.95 ± 0.09×107 PFU/mL while viral titer by TCID50 assay was between 0.71 ± 0.01×106 to 4.94 ± 0.80×106 TCID50/mL for the five SARS-CoV-2 isolates. The PFU/mL titer obtained by plaque and the calculated from TCID50 assays differed by 0.61 log10, 0.59 log10, 0.59 log10 and 0.96 log10 for Alfa, Gamma, Delta, and Mu variants (p≤0.0007), respectively. No differences were observed for the D614G strain. Real-time PCR assay exhibited titers ranging from 0.39 ± 0.001×108 to 3.38 ± 0.04×108 RNA copies/μL for all variants. The relation between PFU/mL and RNA copies/mL was 1:29800 for D614G strain, 1:11700 for Alpha, 1:8930 for Gamma, 1:12500 for Delta, and 1:2950 for Mu.
Conclusion:
TCID50 assay was comparable to plaque assay for D614G but not for others SARS-CoV-2 variants. Our data demonstrated a correlation among PFU/mL and E gene RNA copies/μL, units of measure commonly used to quantify the viral load in diagnostic and research fields. The results suggest that the proportion of infectious virions in vitro changes depending on the SARS-CoV-2 variant, being Mu, the variant reaching a higher viral titer with fewer viral copies.
Keywords: SARS-CoV-2 variants, Virus titer, Real-time reverse transcription-polymerase chain reaction, Plaque assay, Median tissue culture infectious dose assay
INTRODUCTION
Since it was defined as a pandemic, COVID-19 has become a global threat to public health, causing enormous physical, psychosocial and economic consequences (1). This disease is caused by SARS-CoV-2, an enveloped virus with an ssRNA (+) genome of approximately 30 kb (2, 3). Multiple variants of SARS-CoV-2 have been documented around the world. In September 2020, variants of interest (VOIs) and variants of concern (VOCs) began to be reported, with more distinctive substitutions than expected for this virus (2). VOIs are variants with genetic changes that have established, or suspected implications in epidemiology, antigenicity or virulence. In contrast, VOCs are variants with a demonstrated association with higher transmissibility, virulence, and lower diagnostic, vaccine or therapeutic efficacy (2, 3).
The World Health Organization (WHO) has identified five VOCs (Alpha, Beta, Gamma, Delta and Omicron) and two VOIs (Lambda and Mu) (3). Alpha (B.1.1.7), the first VOC reported, contains seventeen co-occurring non-synonymous mutations or deletions, including N501Y, 69-70del, Y144del and P681H, which are associated with increased viral transmission and immune evasion (4). Beta (B.1.351) has eight non-synonymous mutations in Spike protein (S) (such as K417N, E484K and N501Y) associated with increased binding to the human ACE2 (angiotensin-converting enzyme 2) receptor and three mutations in the N-terminal domain that may contribute to increased transmissibility (4). The Gamma (P.1) variant bears seventeen mutations (including K417T, E484K, and N501Y in S protein), accounting for greater transmissibility (between 1.7 and 2.4 times more transmissible than local non-P1 lineages) and a reduction from 21% to 46% in protective immunity caused by previous non-P1 lineages infections (5).
The SARS-CoV-2 Delta variant (B.1.617.2), has become one of the most problematic variants during the pandemic and is rapidly spreading world-wide (6). This variant contains mutations within S (L452R, T478K, P681R, and D614G) associated with increased binding affinity with cell receptor, viral infectivity, and virus replication, reduced antibody-mediated neutralization and cellular immune recognition, increased cell-to-cell fusion in the respiratory tract, potentially increasing transmissibility and pathogenicity in infected individuals (7). The most recent VOC, the omicron variant (B.1.1.529), includes 26–32 mutations in S, some of which may be associated with immune escape potential and higher transmissibility (8).
Concerning the VOIs, Mu (B.1.621) contains the insertion 146 N and several amino acid substitutions in the Spike protein (Y144T, Y145S, R346K, E484K, N501Y and P681H), and it has been associated with resistance to humoral response induced by natural infection and vaccination (2, 9). Due to the decline in the effectiveness of antibodies against these variants and the lack of vaccination coverage world-wide, SARS-CoV-2 continues to evolve and spread throughout the world (7, 9, 10).
Considering this background, a reliable, early, and accurate diagnosis is required to bring efficient medical support and control the spread of SARS-CoV-2 variants (11, 12). Detection of viral RNA by real-time RT-PCR (real-time reverse transcription-polymerase chain reaction) was the initial technique of choice for the COVID-19 diagnosis (13–14). Quantification of viral load in infected individuals has also been useful to assess disease progression and to monitor the efficacy of the therapy (13). Further, this technique allows detecting variants of concern and interest (14). However, despite its high sensitivity and specificity, this technique does not allow the quantification of infectious viruses, which is necessary for virological studies, particularly in experimental procedures such as the structural analysis of the viral particle, the understanding of viral pathogenesis, evaluation of antiviral agents and vaccine development or for improving the ability to determine and interpret infectious viral loads for current and future variants (12, 15–17). In this context, the plaque assay and the TCID50 assay (median tissue culture infectious dose) are cell-based techniques to determine the amount of infectious virus within a sample. Specifically, the plaque assay is the gold standard for quantifying replication-capable lytic virions (18, 19). Plaque assay consists of serial dilutions of the virus on adherent and susceptible cells. After the viral infection time, the liquid medium is replaced by a solid or semi-solid medium that restricts the virus spread to neighboring cells, forming a visible hole or “plaque” into the cell monolayer. Later, the cell monolayer can be stained with a dye and plaques are counted to achieve the titer; assuming that a single infectious virus particle causes each plaque is caused by a single infectious virus particle, the titer is reported as plaque-forming units (PFU) (18, 19).
On the other hand, the TCID50 assay determines the point at which 50% of cells in culture are infected and brings an approximate viral titer (18, 20). In contrast to plaque assay, the viral inoculum is replaced by a liquid medium after infection, and then the wells are observed and scored for the presence or absence of CPE (cytopathic effect) (20). The virus dilution at which 50% of the wells are infected is determined. The TCID50 titer is calculated based on Reed-Muench or Spearman-Kärber methods (17).
It has been reported that VOCs exhibit differences in plaque size, thermal stability, and infectivity in various cell lines (21–23). We set out to quantify the ancestral D614G strain (B.1) (24, 25), three VOCs (Alpha, Gamma and Delta) and one VOI (Mu) of SARS-CoV-2 using three techniques: Plaque assay, TCID50 assay, and real-time RT-PCR, to compare the results and describe the differences and similarities.
MATERIALS AND METHODS
Cells.
Vero E6 cells were donated by Instituto Nacional de Salud (Bogotá, Colombia). The cultures were incubated at 37°C, 5% CO2 atmosphere and a relative humidity. Vero E6 were propagated in Dulbecco's Modified Eagle Medium (DMEM, Sigma-Aldrich) supplemented with 2% heat-inactivated FBS, 2 mM L-glutamine (Gibco), 1.0% Penicillin-Streptomycin (Gibco) and 3.7 g/L sodium bicarbonate (Sigma-Aldrich).
Specimen collection and SARS-CoV-2 strain genotyping.
Nasopharyngeal swabs from Colombian patients were tested for SARS-CoV-2 by RT-PCR Berlin protocol at Grupo Inmunovirología, Universidad de Antioquia (Medellín, Colombia); then, positive samples were subjected to SARS-CoV-2 strain-specific mutations screening by using SARS-CoV-2 Variants RT-PCR kit (Vitro S.A): ORF1ab SGF3675-3677 deletion (ref sequence position: 11288-11296) was observed in Alpha and Gamma but not in D614G, Delta or Mu variants. Spike HV 69–70 deletion (ref sequence position: 21765-21770) was found in Alpha but not in D614G, Gamma, Delta nor Mu variants. Likewise, the Spike 157–158 deletion only observed so far in Delta variant was identified by using the Luna® Universal Probe One-Step RT-qPCR Kit (New England Biolabs, MA, USA) and the oligos S.21989-Fw 5’-GTTTATTACCACAAAAACAACAAAAG-3’, S.22083-Rv 5’-GGCTGAGAGACATATTCAAAAGTG-3’, and probe S.DEL157-P 5’-FAM-TGGATGGAAAGTG-GAGTTTATTCTAGT-MGB-3’, previously reported (14). First, retrotranscribing the RNA at 55°C for 10 minutes followed by 40 cycles of 94°C 10 seconds and alignment/extension step of 60°C for 30 seconds.
Later, SARS-CoV-2 strains were confirmed by next-generation sequencing using Nanopore technologies or Illumina MySeq platform following the ARTIC 12 protocol (https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye) at Corpogen Laboratory, Bogotá, Colombia. Gaps in Spike were filled out by Sanger sequencing using 3500 Series Genetic Analyzer (Thermo Fisher Scientific, Vilnius, Lithuania), and BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific) in Laboratorio Integrado de Medicina Especializada (LIME), Universidad de Antioquia, Medellin, Colombia.
Variant sequences were deposited in GISAID data base: B.1/D614G (EPI_ISL_536399), Alpha (EPI_ ISL_4549188), Gamma (EPI_ISL_4926393), Delta (EPI_ISL_5103929) and Mu (EPI_ISL_4005445).
Virus isolation and propagation.
A fraction of nasopharyngeal samples carrying SARS-CoV-2 D614G strain and Alpha, Gamma, Delta and Mu variants (100 μL diluted in 250 μL DMEM medium) were inoculated into a 75% confluent monolayer of Vero E6 cells. The cultures were daily monitored under the microscope to visualize cytopathic effect (CPE) appearance as previously described (26).
Later, the isolated viruses were propagated in Vero E6 cells. Briefly, 2.0×106 cells were seeded in T-75 cell culture flasks in DMEM supplemented with 2% FBS (Fetal bovine serum) for 24 h at 37°C, with 5% CO2. Then, cells were infected for 1 h with each sample in 2 mL of DMEM with 2% FBS. After incubation, the inoculum was removed, and 15 mL of fresh medium were added. Cells were incubated at 37°C, with 5% CO2, with daily observation until CPE appearance. CPE was observed 60 hours post-infection (h.p.i) as rounding and detachment. The supernatants were harvested, centrifuged at 2800 × g, 4°C, for 10 minutes, aliquoted, and stored at −80°C. The specificity of the variants was confirmed by real-time RT-PCR, using the oligo and probes described in item Specimen collection and SARS-CoV-2 strain genotyping. All experiments involving infectious SARS-CoV-2 were conducted within a biosafety level 3 laboratory, according to the conditions set out in Biosafety in Microbiological and Biomedical Laboratories (27).
Plaque assay.
SARS-CoV-2 was quantified by plaque assay on Vero E6 monolayers. Briefly, Vero E6 cells were seeded at a cell density of 1.0 × 105 cells/well in 24-well plates for 24 h, at 37°C, with 5% CO2. Tenfold serial dilutions of the viral stock were added to each well in duplicate (200 μL/well). Cells were incubated at 37°C, with 5% CO2 for 1 h. Viral inoculum was removed and 1 mL of semi-solid medium (1.5% carboxymethyl-cellulose) (Sigma-Aldrich, St. Louis, MO, USA), DMEM 1× with 2% FBS and 1% Penicillin-Streptomycin) was added to each well. Plates were incubated for 5 days at 37°C, with 5% CO2. After incubation, cells were washed twice with Phosphate Buffered Saline (PBS) (Lonza, Rockland, ME, USA). Then, cells were fixed/stained with 500 μL of 4% formaldehyde / 1% crystal violet solution for 30 min and washed twice with PBS. Plaques were counted, and the average number of plaques was multiplied by the reciprocal of the dilution and the inoculum volume. The result was expressed as Plaque Forming Units (PFU) per mL (PFU/mL) (15). For plaque assay quantification, five independent experiments were performed.
TCID50 assay.
SARS-CoV-2 was quantified by a TCID50 assay. This assay was performed on Vero E6 monolayers and revealed by crystal violet staining. Briefly, Vero E6 were seeded at a cell density of 1.2×104 cells/well in 96-well plates for 24 h, at 37°C with 5% CO2. Tenfold serial dilutions of the viral stock were added to each well in quadruplicate (50 μL/well). After 1 h of incubation, the inoculum was removed, and 150 μL of DMEM with 2% FBS were added to each well; cells were incubated for 5 days at 37°C, with 5% CO2. A control with non-infected cells was included in each plate. Later, cells were washed twice with PBS and fixed/stained with 200 μL of formaldehyde/crystal violet solution for 30 min. Then, cells were washed twice with PBS and allowed to air dry. After, the number of wells with CPE for each viral stock dilution was determined (CPE was considered positive when more than 30% of the cell monolayer was compromised) (18). TCID50/mL and the predicted value of PFU/mL (conversion factor 0.56 (17) were calculated with TCID50 calculator (Marco Binder, University of Heidelberg) based on Spearman - Kärber method (17). Five independent experiments were performed.
Real-time RT-PCR.
Viral RNA extraction was carried out from 100 μL of viral stock using the KingFisher™ Flex Purification System and the Mag-MAXTM Viral/Pathogen II (MVP II) Nucleic Acid Isolation Kit (Thermo Fisher Scientific, Vilnius, Lithuania), following the manufacturer instructions, with an elution volume of 50 μL. SARS-CoV-2 viral RNA was quantified using the Luna® Universal Probe One-Step RT-qPCR Kit (New England Biolabs, MA, USA). The reaction included 7 μL of viral RNA; the oligos and probe for the E gene and the conditions reported in the Berlin real-time RT-PCR protocol (28) with a modification, according to One-Step RT-qPCR Kit manufacturer's recommendations in a reverse transcription (55°C for 10 minutes) and in alignment/extension step (60°C for 30 seconds). The RT-PCR reactions were carried out in a CFX-96 Biorad thermal cycler (Biorad, CA, USA).
The linearity, repeatability, and PCR efficiency of this methodology were evaluated by extracting RNA and quantifying serial dilutions of the viral stocks. Two independent experiments in duplicate were performed for each strategy.
The number of viral RNA copies was calculated by extrapolating the cycle at which viral stock or its dilutions cross the fluorescence threshold (Ct) in a standard curve previously constructed with a serially diluted 3180 bp plasmid containing the E gene in a concentration of 2×109 copies/μL. The standard curve yield a coefficient of determination (r2) of 0.998.
Statistical analysis.
All data were reported as the media ± Standard Error (SE). The statistical tests were performed using GraphPad Prism Software version 7.02. Mann–Whitney U or Student t-test were performed to compare the viral titers obtained by plaque assay and predicted PFU/mL by TCID50 assay in each variant. The viral RNA copies of each viral stock were calculated by interpolating from the standard curve in a linear regression analysis. The correlation of the plaque assay and real-time RT-PCR was assessed using Pearson's correlation. A p-value ≤0.05 was considered statistically significant.
Ethics approval and consent to participate.
This study was carried out by keeping good records, practicing good data collection and management, transparency of data-sharing and realistic representation of study results.
RESULTS
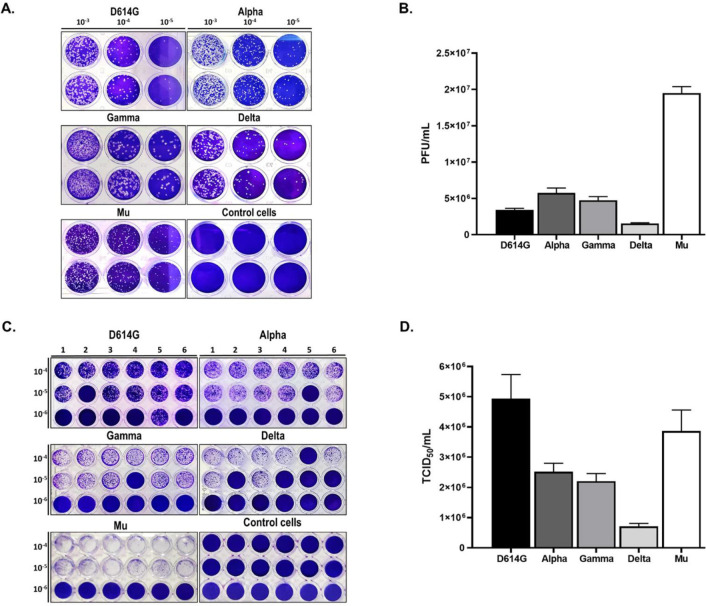
The mean values and standard error of PFU/mL titers obtained by plaque assay for five SARS-CoV-2 isolates were summarized in Table 1. The viral titers of the five SARS-CoV-2 isolates range from 0.15 ± 0.01×107 and 1.95 ± 0.09×107 PFU/mL (Fig. 1A–B). Concerning the TCID50 assay, used for quantifying the burden of viral infectious particles capable of producing a cytopathic effect in half of the infected cell cultures (29), the viral titers of the five isolates evaluated range between 0.71 ± 0.01×106 to 4.94 ± 0.80×106 TCID50/mL (Table 1 and Figs. 1C–D).
Table 1.
Viral titer of five SARS-CoV-2 isolates obtained by TCID50, plaque assay and real-time RT-PCR
| Virus | Viral titers | |||
|---|---|---|---|---|
|
| ||||
| PFU/mL (Plaque assay) | TCID50/mL | PFU/mL (predicted by TCID50 assay ) | E gene RNAcopies/μL | |
| D614G | 0.34 ± 0.02×107 | 4.94 ± 0.80×106 | 2.77 ± 0.45×106 | 3.38 ± 0.04×108 |
| Alpha | 0.58 ± 0.07×107 | 2.52 ± 0.28×106 | 1.40 ± 0.16×106 | 1.34 ± 0.03×108 |
| Gamma | 0.48 ± 0.05×107 | 2.21 ± 0.25×106 | 1.23 ± 0.14×106 | 0.85 ± 0.01×108 |
| Delta | 0.15 ± 0.01×107 | 0.71 ± 0.01×106 | 0.40 ± 0.05×106 | 0.39 ± 0.001×108 |
| Mu | 1.95 ± 0.09×107 | 3.87 ± 0.70×106 | 2.16 ± 0.39×106 | 1.15 ± 0.01×108 |
The Table summarizes the viral titer of D614G strain and four SARS-CoV-2 variants (Alpha, Gamma, Delta and Mu) obtained by plaque assay (PFU/mL), TCID50 assay (TCID50/mL) and real-time RT-PCR (E gene RNA copies/μL). Further, the PFU/mL predicted were calculated from the TCID50/mL, according to the Reed and Muench method. The values are expressed as mean ± SE (n=4–5).
Fig. 1.
Titration of five SARS-CoV-2 isolates by Plaque and TCID50 assays experiments. Monolayers were infected with ten-fold serial dilutions of D614G strain and the SARS-CoV-2 variants (Alpha, Gamma, Delta and Mu). Plaque and TCID50 assays were revealed by crystal violet staining at 5 d.p.i. A control without infection (control cells) was included. A. Representative image of a plaque assay for D614G strain and Alpha, Gamma, Delta and Mu variants. B. The graph shows the viral titer (PFU/mL) of each isolated SARS-CoV-2 variant. C. Representative image of the TCID50 assay for five SARS-CoV-2 isolates. D. The viral titer (TCID50/mL) of each SARS-CoV-2 isolated were calculated. Bars with error bars represent the mean ± Standard Error (SE) of five independent experiments.
Based on TCID50 results, the predicted PFU/mL was calculated for each variant (Table 1). Statistically significant differences between the PFU/mL obtained by plaque assay and PFU/mL calculated from TCID50 were observed for Alpha (p=0.0007), Gamma (p<0.0001), Delta (p<0.0001), and Mu (p<0.0001) but no D614G (Fig. 2). Differences among the two quantification techniques were 0.61 log10, 0.59 log10, 0.59 log10, and 0.96 log10 for Alfa, Gamma, Delta, and Mu variants, respectively.
Fig. 2.
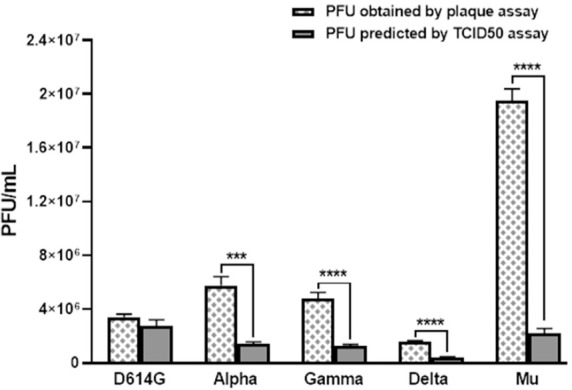
Comparison between SARS-CoV-2 titer (PFU/mL) by TCID50 and plaque assay. The viral titer (PFU/mL) predicted by TCID50 and obtained by plaque assay for D614G strain and the Alpha, Gamma, Delta and Mu variants. Bars with error bars represent the mean ± Standard Error (SE) of five independent experiments. t Student or Mann-Whitney U test were performed to compare the viral titers obtained by plaque assay and predicted PFU/mL by TCID50 assay in each variant. ***p ≤ 0.001, ****p ≤ 0.0001.
On the other hand, quantification of the gene E of each SARS-CoV-2 isolated was performed by real-time RT-PCR, obtaining titers from 0.39 ± 0.001×108 to 3.38 ± 0.04×108 E gene RNA/μL (Table 1). Further, these values were compared with the PFU/mL titers obtained by plaque assay. As shown in the Fig. 3, significant positive correlation between PFU/mL and RNA copies/μL was found for D614G strain (correlation coefficient, r = 0.999, p < 0.0001), Alpha (r = 0.999, p < 0.0001), Gamma (r = 0.999, p < 0.0001), Delta (r = 0.999, p < 0.0001) and Mu (r = 0.999, p < 0.0001) variants.
Fig. 3.
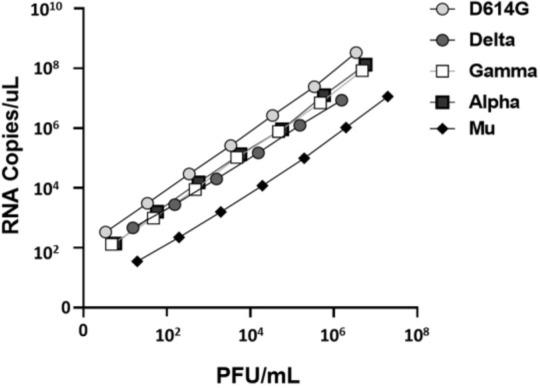
Correlation between RNA copies/μL and PFU/mL of five SARS-CoV-2 isolates. Serially diluted viral RNA extractions of five SARS-CoV-2 isolates (D614G strain and Alpha, Gamma, Delta and Mu variants) were quantified by real-time RT-PCR (n=4). The Ct values were extrapolated in the standard curve (r2=0.998) for calculating the E gene RNA copies/μL. Later, PFU/mL results from plaque assay and RNA copies of the E gene/μL were correlated.
Finally, the infectivity of each variant was measured by the number of infectious viruses (PFU/mL) per quantity of viral RNA (RNA copies/mL) (23, 30, 31). We found that the relation between PFU and viral RNA copies was 1:29800, 1:11700, 1:8930, 1:12500 and 1:2950 for D614G, Alpha, Gamma, Delta and Mu, respectively.
DISCUSSION
SARS-CoV-2 VOCs and VOIs have been associated with increased transmissibility, virulence, and evasion of natural and vaccine-induced immunity, resulting in a significant challenge for COVID-19 pandemic control (24, 32). Previously, it has been reported that naturally occurring mutations in SARS-CoV-2 can substantially change its replication dynamics and infectivity in various cell lines (21). According to the above, we compared three different quantification techniques of SARS-CoV-2: plaque assay, TCID50 assay and real-time RT-PCR using five SARS-CoV-2 isolates: Alpha, Gamma, Delta and Mu variants and the ancestral D614G strain (B.1 lineage) to describe differences and similarities among them.
Plaque assay and TCID50 are cell culture-based methods used to estimate the titers for viruses causing cytopathogenic damage to infected cell cultures (33). Concerning SARS-CoV-2, our results showed differences among both quantification techniques between 0.59 log10 and 0.96 log10 for Alpha, Gamma, Delta and Mu variants. This result agrees with previous studies with three major Filovirus species in which lower titers by TCID50 assay than plaque assay in Vero E6 cells were obtained (18). The difference observed between the variants could be due to the amount of infectious virus present in the original sample from which the isolation was done or amino acid substi- tutions that impact in vitro measures of virulence, such as plaque size, cytopathic effect, and replication kinetics, as reported in other viral models (34).
Although plaque assay is the gold standard for determining concentrations of infectious lytic virions (35), this technique can be affected by several factors, including the choice of host cells, dilution ranges, amount of the semi-solid medium used for overlays, and the manual plaque counting (11, 15, 29). Similar to plaque assay, TCID50 assay can also be affected by several factors, including the number of cells, viral dilution, and the subjective visualization of the CPE (20, 36). The visualization of CPE is particularly sensitive in staining revealed techniques (20, 36). Further, both techniques are time-consuming and required to perform in a BSL-3 laboratory, limiting their use (37).
On the other hand, real-time RT-PCR is a reliable, rapid and robust method to estimate viral titers equivalents expressed as recorded Ct values (33). The results presented in this study showed a significant positive correlation between PFU/mL and RNA copies of the E gene/μL (correlation coefficient, r = 0.999, p < 0.0001) using different SARS-CoV-2 variants. This demonstrates the general applicability of using real-time RT-PCR to measure the concentration of viruses with higher sensitivity, speed and less subjectivity compared to the traditional culture-based methods for detecting viruses in clinical specimens (33, 38). Real-time PCR-based methods also show a reduced risk of amplicon carry-over contamination (33, 38).
Further, the results from our study showed that the relation between PFU and viral RNA could vary among viral strains or variants (relations between 1:2950 and 1:29800), which is consistent with previous reports (31, 39). Likewise, previous studies have reported that the PFU to genome ratio for SARS-CoV-2 was in the range 1:103–106 (30, 31). These findings reflect the complex and dynamic relationship between a replicating virus and the target cells. The difference among titer of the variants may result from producing defective, immature, or inactivated virus particles or free viral RNA within cells harboring an infection or by other factors such as fluctuating pH among original samples (38, 40).
Titration by plaque assay differs from molecular analysis for distinguishing infectious viral particles from non-infectious RNA, virtually present in every supernatant (29). According to the above, the real-time PCR assay is a useful addition to diagnostic procedures and monitoring of antiviral treatment; but can not replace the plaque assay for identifying infectious virus particles, when this quantification is required (33).
Our result showed that Mu had higher infectivity than the other variants evaluated, which could be related to its higher prevalence in Colombia, despite the circulation of other variants such as Alpha, Gamma, Delta and Lambda (41). This difference in the infectivity could be also correlated with a distinctive profile of synonymous and non-synonymous substitutions found in the Spike protein of this variant, including substitutions such as T95I, Y144T, Y145S in the N-terminal domain, and the insertion 146N in the N-terminal domain for which the implications in terms of infection, transmission and pathogenesis are still unknown (2).
It is important to mention that we use the first passage of each isolation to determine the behavior of each variant without generating changes associated with the adaptation of the virus in cell culture, considering that through passages, virus isolates acquire amino acid substitutions or deletions mainly in Spike, Envelope or non-structural proteins and, the relationship between viral genomic RNA and infectious viruses can also be altered (33, 42, 43).
CONCLUSION
TCID50 assay was comparable to plaque assay in D614G strain, while TCID50 for the Alpha, Gamma, Delta and Mu variants of SARS-CoV-2 showed significant differences between the two methods. Our data demonstrated a correlation among PFU/mL and E gene RNA/μL, units of measure commonly used to quantify the viral load in diagnostic and research fields. The results suggested that the proportion of infectious virions changes depending on the SARS-CoV-2 variant, with Mu reaching a higher viral titer with fewer viral copies. The infectivity should be investigated for future SARS-CoV-2 variants.
ACKNOWLEDGEMENTS
We want to thank Dr. Jaime Castellanos from Universidad El Bosque, Bogotá-Colombia, for kindly donate the plasmid containing E gene from SARS-CoV-2 and Instituto Nacional de Salud, Bogotá-Colombia for donate the Vero E6 cell line. This study was supported by Universidad de Antioquia and Universidad Cooperativa de Colombia. BPIN 2020000100131-SGR.
REFERENCES
- 1.Poudel K, Subedi P. Impact of COVID-19 pandemic on socioeconomic and mental health aspects in Nepal. Int J Soc Psychiatry 2020;66:748–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laiton-Donato K, Franco-Muñoz C, Álvarez-Díaz DA, Ruiz-Moreno HA, Usme-Ciro JA, Prada DA, et al. Characterization of the emerging B.1.621 variant of interest of SARS-CoV-2. Infect Genet Evol 2021;95:105038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. Tracking SARS-CoV-2 variants. 2021. Available from: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants [Google Scholar]
- 4.Singh J, Pandit P, McArthur AG, Banerjee A, Mossman K. Evolutionary trajectory of SARS-CoV-2 and emerging variants. Virol J 2021;18:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faria NR, Mellan TA, Whittaker C, Claro IM, Candido DDS, Mishra S, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021;372:815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baral P, Bhattarai N, Hossen ML, Stebliankin V, Gerstman BS, Narasimhan G, et al. Mutation-induced changes in the receptor-binding interface of the SARS-CoV-2 Delta variant B.1.617.2 and implications for immune evasion. Biochem Biophys Res Commun 2021;574:14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021;596:276–280. [DOI] [PubMed] [Google Scholar]
- 8.WHO. Enhancing Readiness for Omicron (B.1.1.529): Technical Brief and Priority Actions for Member States 2021. Available from: https://www.who.int/publications/m/item/enhancing-readiness-for-omicron-(b.1.1.529)-technical-brief-and-priority-actions-for-member-states)
- 9.Uriu K, Kimura I, Shirakawa K, Takaori-Kondo A, Nakada TA, Kaneda A, et al. Neutralization of the SARS-CoV-2 Mu Variant by Convalescent and Vaccine Serum. N Engl J Med 2021;385:2397–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaku Y, Kuwata T, Zahid HM, Hashiguchi T, Noda T, Kuramoto N, et al. Resistance of SARS-CoV-2 variants to neutralization by antibodies induced in convalescent patients with COVID-19. Cell Rep 2021;36:109385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendoza EJ, Manguiat K, Wood H, Drebot M. Two detailed plaque assay protocols for the quantification of infectious SARS-CoV-2. Curr Protoc Microbiol 2020;57(1):ecpmc105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schutten M, Niesters HG. Clinical utility of viral quantification as a tool for disease monitoring. Expert Rev Mol Diagn 2001;1:153–162. [DOI] [PubMed] [Google Scholar]
- 13.Cobo F. Application of molecular diagnostic techniques for viral testing. Open Virol J 2012;6:104–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yaniv K, Ozer E, Kushmaro A. SARS-CoV-2 variants of concern, Gamma (P.1) and Delta (B.1.617), sensitive detection and quantification in wastewater employing direct RT-qPCR. medRxiv 2021;07.14.21260495. [Google Scholar]
- 15.Jureka AS, Silvas JA, Basler CF. Propagation, inactivation, and safety testing of SARS-CoV-2. Viruses 2020;12:622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodinka RL. The clinical utility of viral quantitation using molecular methods. Clin Diagn Virol 1998;10:25–47. [DOI] [PubMed] [Google Scholar]
- 17.Wulff NH, Tzatzaris M, Young PJ. Monte Carlo simulation of the Spearman-Kaerber TCID50. J Clin Bioinforma 2012;2:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keiser PT, Anantpadma M, Staples H, Carrion R, Davey RA. Automation of infectious focus assay for determination of filovirus titers and direct comparison to plaque and TCID(50) assays. Microorganisms 2021;9:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shurtleff AC, Biggins JE, Keeney AE, Zumbrun EE, Bloomfield HA, Kuehne A, et al. Standardization of the filovirus plaque assay for use in preclinical studies. Viruses 2012;4:3511–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smither SJ, Lear-Rooney C, Biggins J, Pettitt J, Lever MS, Olinger GG., Jr. Comparison of the plaque assay and 50% tissue culture infectious dose assay as methods for measuring filovirus infectivity. J Virol Methods 2013;193:565–571. [DOI] [PubMed] [Google Scholar]
- 21.Yao H, Lu X, Chen Q, Xu K, Chen Y, Cheng M, et al. Patient-derived SARS-CoV-2 mutations impact viral replication dynamics and infectivity in vitro and with clinical implications in vivo. Cell Discov 2020;6:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong GU, Yoon GY, Moon HW, Lee W, Hwang I, Kim H, et al. Comparison of plaque size, thermal stability, and replication rate among SARS-CoV-2 variants of concern. Viruses 2021;14:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Despres HW, Mills MG, Shirley DJ, Schmidt MM, Huang ML, Roychoudhury P, et al. Measuring infectious SARS-CoV-2 in clinical samples reveals a higher viral titer: RNA ratio for Delta and Epsilon vs. Alpha variants. Proc Natl Acad Sci U S A 2022;119(5):e2116518119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khateeb J, Li Y, Zhang H. Emerging SARS-CoV-2 variants of concern and potential intervention approaches. Crit Care 2021;25:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 Spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020;182:812–827.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Díaz FJ, Aguilar-Jiménez W, Flórez-Álvarez L, Valencia G, Laiton-Donato K, Franco-Muñoz C, et al. Isolation and characterization of an early SARS-CoV-2 isolate from the 2020 epidemic in Medellín, Colombia. Biomedica 2020;40(Supl. 2):148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richmond JY, McKinney RW. (2009). Biosafety in microbiological and biomedical laboratories. 5th ed. U.S. Department of Health and Human Services Public. Health Service Centers for Disease Control and Prevention National Institutes of Health (CDC). U.S. [Google Scholar]
- 28.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020;25:2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brandolini M, Taddei F, Marino MM, Grumiro L, Scalcione A, Turba ME, et al. Correlating qRT-PCR, dPCR and viral titration for the identification and quantification of SARS-CoV-2: A new approach for infection management. Viruses 2021;13:1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uhteg K, Jarrett J, Richards M, Howard C, Morehead E, Geahr M, et al. Comparing the analytical performance of three SARS-CoV-2 molecular diagnostic assays. J Clin Virol 2020;127:104384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klimstra WB, Tilston-Lunel NL, Nambulli S, Boslett J, McMillen CM, Gilliland T, et al. SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients. J Gen Virol 2020;101:1156–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otto SP, Day T, Arino J, Colijn C, Dushoff J, Li M, et al. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr Biol 2021;31:R918–R929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jonsson N, Gullberg M, Lindberg AM. Real-time polymerase chain reaction as a rapid and efficient alternative to estimation of picornavirus titers by tissue culture infectious dose 50% or plaque forming units. Microbiol Immunol 2009;53:149–154. [DOI] [PubMed] [Google Scholar]
- 34.Abraham R, Manakkadan A, Mudaliar P, Joseph I, Sivakumar KC, Nair RR, et al. Correlation of phylogenetic clade diversification and in vitro infectivity differences among cosmopolitan genotype strains of Chikungunya virus. Infect Genet Evol 2016;37:174–184. [DOI] [PubMed] [Google Scholar]
- 35.Baer A, Kehn-Hall K. Viral concentration determination through plaque assays: using traditional and novel overlay systems. J Vis Exp 2014;(93):e52065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grigorov B, Rabilloud J, Lawrence P, Gerlier D. Rapid titration of measles and other viruses: optimization with determination of replication cycle length. PLoS One 2011;6(9):e24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith DR, Singh C, Green J, Lueder MR, Arnold CE, Voegtly LJ, et al. Genomic and virological characterization of SARS-CoV-2 variants in a subset of unvaccinated and vaccinated U.S. military personnel. Front Med (Lausanne) 2022;8:836658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bae HG, Nitsche A, Teichmann A, Biel SS, Niedrig M. Detection of yellow fever virus: a comparison of quantitative real-time PCR and plaque assay. J Virol Methods 2003;110:185–191. [DOI] [PubMed] [Google Scholar]
- 39.Mallm JP, Bundschuh C, Kim H, Weidner N, Steiger S, Lander I, et al. Local emergence and decline of a SARS-CoV-2 variant with mutations L452R and N501Y in the spike protein. medRxiv 2021.04.27.21254849. [Google Scholar]
- 40.Richardson J, Molina-Cruz A, Salazar MI, Black W., 4th Quantitative analysis of dengue-2 virus RNA during the extrinsic incubation period in individual Aedes aegypti. Am J Trop Med Hyg 2006;74:132–141. [PubMed] [Google Scholar]
- 41.Programa Nacional de Caracterización Genómica de SARS-CoV-2. Caracterización Genómica de SARS-CoV-2 por Muestreo Probabilístico en Colombia Segundo Muestreo. Instituto Nacional de Salud 2021; 1–10. [Google Scholar]
- 42.Pohl MO, Busnadiego I, Kufner V, Glas I, Karakus U, Schmutz S, et al. SARS-CoV-2 variants reveal features critical for replication in primary human cells. PLoS Biol 2021;19(3):e3001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Funnell SGP, Afrough B, Baczenas JJ, Berry N, Bewley KR, Bradford R, et al. A cautionary perspective regarding the isolation and serial propagation of SARS-CoV-2 in Vero cells. NPJ Vaccines 2021;6:83. [DOI] [PMC free article] [PubMed] [Google Scholar]