Abstract
KMT2A-rearranged acute lymphoblastic infant leukemia (KMT2A-r iALL) is associated with outsize risk of relapse and relapse mortality. We previously reported strong upregulation of the immediate early gene EGR3 in KMT2A::AFF1 iALL at relapse; now we provide analyses of the EGR3 regulome, which we assessed through binding and expression target analysis of an EGR3-overexpressing t(4;11) cell culture model. Our data identify EGR3 as a regulator of early B-lineage commitment. Principal component analysis of 50 KMT2A-r iALL patients at diagnosis and 18 at relapse provided strictly dichotomous separation of patients based on the expression of four B-lineage genes. Absence of B-lineage gene expression translates to more than two-fold poorer long-term event-free survival. In conclusion, our study presents four B-lineage genes with prognostic significance, suitable for gene expression-based risk stratification of KMT2A-r iALL patients.
Subject terms: Acute lymphocytic leukaemia, Acute lymphocytic leukaemia
Introduction
Seventy to eighty percent of iALL are KMT2A-rearranged (KMT2A-r); an association with inferior survival was noted and independently confirmed [1, 2]. 49% of KMT2A-r iALL possess the translocation t(4;11)(q21;23) generating the fusion oncogenes KMT2A::AFF1 and AFF1::KMT2A [3]. On-treatment relapse is frequent and mortality is high [1, 2, 4]. Despite the successful implementation of targeted therapies in pediatric leukemia, a relapse-preventing iALL therapy remains to be established.
Our group recently reported the immediate early gene Early Growth Response 3 (EGR3) as relapse-associated, with 100-fold increased EGR3 gene expression levels at the time of relapse compared to primary diagnosis [5]. This prompted us to investigate the role of EGR3 in the context of KMT2A::AFF1 iALL in more detail.
EGR3 belongs to the family of C2H2 zinc finger transcription factors with high structural and functional relation to EGR1 and EGR2 [6, 7]. EGR proteins act as direct transcriptional activators and repressors, with the ability to recruit NAB proteins as co-regulators [8–11]. Functionally, the EGR genes were related to neuronal development [12–16], hematopoietic stem cell quiescence [17, 18], and development of T and B cells [19–21].
Here we explore the EGR3 regulome of KMT2A::AFF1 proB-ALL in detail through integration of data derived from massive analysis of cDNA ends-sequencing (MACE-Seq) and chromatin immunoprecipitation DNA-sequencing (ChIP-Seq) of an EGR3-overexpression SEM cell model. Our study identifies EGR3 as a regulator of early B-lineage specification and commitment. Additionally, gene expression and principal component analysis of 50 KMT2A-r iALL patients at diagnosis and 18 at relapse provides strictly bimodal clustering of patients based on the expression of the identified B-lineage genes. Absence of B-lineage gene expression translates to dismal outcome with more than two-fold poorer long-term event-free survival.
Methods
Cell culture
Establishment of the cell lines SEM::EGR3 and SEM::mock with Doxycycline-inducible transgene expression is described in our former study [5]. SEM cells were cultivated under sterile conditions and maintained in RPMI 1640 (RPMI-HA, Capricorn Scientific) supplemented with 10% FBS (FBS-11A, Capricorn Scientific), 2 mM L-glutamine (STA-B, Capricorn Scientific), 100 U/ml penicillin and 100 μg/ml streptomycin (PS-B, Capricorn Scientific), preheated to 37 °C prior to use. Cells were kept at 37 °C in 5% CO2 and a relative humidity of 95%. SEM cells were passaged twice a week keeping a density of ~1-3 × 106 cells/mL. Cell lines are tested on a regular basis.
Gene expression analysis using qRT-PCR of patient samples
Informed consent was obtained for all patients through the respective study center, which provided patient RNA. Analyzed patients displayed a proB phenotype and were diagnosed between 0 and 12 months of age (infants). RNA was extracted from peripheral blood at the day of diagnosis (dx cohort, n = 50) or relapse (rel cohort, n = 18) by the respective study center. cDNA was synthesized out of 1 µg RNA using random hexamer N6 primers and SuperScript™ II Reverse Transcriptase (18064071, invitrogen). The dx cohort is composed of KMT2A::AFF1 iALL cases (Table 1), the rel cohort is composed of different KMT2A-r iALLs (Table 2). IGHM, CD79A, BLK, and PTPN6 gene expression was measured as technical triplicates using qRT-PCR (StepOnePlus system) and ΔCT mean values were calculated using GAPDH expression as a reference. Relative expressions were calculated as ratio (reference/target) = 2CT(GAPDH) - CT(target) = 2−ΔCT. If available, clinical outcome was provided by the study centers. Used oligonucleotides are described in Table 3. The EGR3 expression data were already assessed during our former study with the same cohorts [5]. Pearson correlations were calculated using GraphPad Prism software.
Table 1.
Patient characteristics of the dx cohort (n = 50).
| Patient no. | Patient age [months] | Treatment protocol | sex | Time to event (last follow-up) [months] | % blasts | EGR3 ΔCT Mean | IGHM ΔCT Mean | CD79A ΔCT Mean | BLK ΔCT Mean | PTPN6 ΔCT Mean | HOXA9ΔCT Mean |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 5.0 | Interfant-06 | F | (40.1) | 49 | 8.1850 | 3.9578 | 2.6439 | 5.1517 | 3.8854 | 13.3592 |
| 2 | 6.0 | / | F | / | 82 | 11.2758 | 4.1454 | 2.2853 | 4.3700 | 2.1229 | 3.9204 |
| 3 | 2.0 | Interfant-06 | F | (23.6) | 46 | 9.3618 | 4.8963 | 2.7340 | 4.4007 | 4.4306 | 12.9546 |
| 4 | 6.0 | Interfant-06 | M | (17.1) | 90 | 11.1637 | 9.1284 | 7.8716 | 9.8465 | 10.1343 | 11.5783 |
| 5 | 10.0 | / | M | / | 90 | 8.8214 | 5.4178 | 6.2042 | 10.2556 | 9.6764 | 3.5974 |
| 6 | 2.0 | Interfant-06 | M | (83.6) | 95 | 11.5618 | 6.2522 | 6.9626 | 8.1896 | 8.8812 | 4.9698 |
| 7 | 3.0 | Interfant-06 | F | (5.6) | 96 | 8.5196 | 5.8631 | 2.8374 | −1.4396 | 2.4810 | 11.0093 |
| 8 | 3.0 | Interfant-06 | F | (1.2) | 71 | 8.2271 | 4.9521 | 5.1072 | −0.4361 | 4.2495 | 4.9163 |
| 9 | 3.0 | Interfant-06 | M | (2.5) | 85 | 8.0114 | 4.5490 | 3.3648 | −0.8208 | 2.8412 | 4.5976 |
| 10 | 6.0 | Interfant-06 | F | 9.5 (10.7) | 98 | 12.3136 | 8.2192 | 8.1777 | 8.7455 | 8.1889 | 5.0818 |
| 11 | 0.1 | / | F | / | 96 | 11.2989 | 11.8824 | 9.8611 | 10.2911 | 9.2719 | 8.6964 |
| 12 | 1.0 | / | M | / | 98 | 11.6236 | 11.8944 | 8.6850 | 9.7199 | 9.8527 | 12.3443 |
| 13 | 12.0 | / | F | / | 90 | 9.1826 | 4.9885 | 1.9742 | 3.1891 | −0.3517 | 2.2824 |
| 14 | 4.0 | / | F | / | n/a | 11.4695 | 6.3329 | 3.2521 | 4.4310 | 1.3435 | 11.4825 |
| 15 | 2.0 | Interfant-06 | M | 6.0 (6.0) | 90 | 13.3440 | 4.6279 | 2.6102 | 4.9218 | 1.1705 | 4.6973 |
| 16 | 5.5 | Interfant-06 | M | 9.5 (15.0) | 94 | 9.5330 | 9.9035 | 5.8136 | 6.3955 | 4.2272 | 6.2129 |
| 17 | 1.5 | Interfant-06 | F | 10.0 (13.0) | 92 | 6.2132 | 11.4431 | 8.3688 | 8.4548 | 6.1221 | 6.0703 |
| 18 | 3.0 | Interfant-06 | F | 6.0 (11.5) | 87 | 9.4723 | 9.2393 | 7.5200 | 8.7426 | 4.7647 | 2.0906 |
| 19 | 2.0 | Interfant-06 | F | 10.5 (19) | 96 | 7.2056 | 2.4819 | 2.0453 | −1.9534 | 2.4821 | 7.8537 |
| 20 | 1.5 | Interfant-06 | M | 26.5 (41.0) | 96 | 11.2026 | 5.8739 | 4.8320 | 0.2568 | 6.2901 | 4.4627 |
| 21 | 6.5 | Interfant-06 | M | 24.0 (32.8) | 43 | 8.7817 | 6.5124 | 3.1900 | 0.3659 | 3.1614 | 11.2673 |
| 22 | 6.0 | Interfant-06 | F | 19.5 (23.5) | 94 | 10.0553 | 6.4983 | 2.9073 | −1.3699 | 3.5784 | 11.5891 |
| 23 | 12.0 | Interfant-06 | F | 32.0 (60.6) | 92 | 8.0600 | 6.9642 | 4.0856 | 3.3604 | 1.6443 | 11.1226 |
| 24 | 9.0 | Interfant-06 | M | (84.9) | 93 | 7.0569 | 3.3029 | 1.1508 | 2.1274 | 1.1842 | 8.7665 |
| 25 | 2.0 | Interfant-06 | F | (118.8) | 93 | 10.2925 | 4.4468 | 1.7712 | 2.0883 | 0.4488 | 4.6017 |
| 26 | 3.0 | Interfant-06 | M | 8.0 (108.5) | 96 | 8.7375 | 7.0230 | 3.4687 | 4.0864 | 2.6431 | 13.0872 |
| 27 | 5.5 | Interfant-06 | M | (9.0) | 95 | 9.1105 | 5.8320 | 2.9365 | 4.0461 | 1.5317 | 5.9626 |
| 28 | 5.0 | Interfant-06 | M | 17.0 (33.0) | 97 | 5.0456 | 5.0784 | 2.0327 | 4.8259 | 1.2226 | 5.9922 |
| 29 | 2.0 | Interfant-06 | F | 56.0 (58.6) | 90 | 9.3271 | 3.7605 | 3.9045 | 2.4719 | 4.2582 | 5.3297 |
| 30 | 1.5 | Interfant-06 | F | (146.1) | 88 | 9.8910 | 3.3500 | 2.1610 | 0.6500 | 2.7175 | 5.8358 |
| 31 | 8.3 | Interfant-06 | F | (109.3) | 81 | 6.5075 | 1.0567 | 0.0773 | −0.7910 | 1.6433 | 2.2195 |
| 32 | 8.2 | Interfant-06 | M | (95.9) | 92 | 7.2074 | 1.7896 | 2.8559 | 0.3826 | 0.8519 | 1.7726 |
| 33 | 10.7 | Interfant-06 | M | 8.8 (8.8) | 85 | 2.4138 | -2.0719 | 0.3777 | −0.1263 | 1.5011 | 2.6953 |
| 34 | 1.7 | Interfant-06 | F | (91.5) | 93 | 6.2540 | 1.4646 | 1.9100 | 1.0432 | 2.1243 | 3.8579 |
| 35 | 11.7 | Interfant-06 | F | (73.2) | 95 | 10.9273 | 4.7296 | 1.2813 | 3.3225 | 2.8393 | 2.9311 |
| 36 | 10.4 | Interfant-06 | F | (75.9) | 92 | 10.4144 | 4.0039 | 2.3851 | 3.3520 | 2.9626 | 5.7541 |
| 37 | 2.4 | Interfant-06 | F | (73.8) | 95 | 9.8438 | 5.2626 | 2.1926 | 4.1514 | 3.4033 | 13.3331 |
| 38 | 4.5 | Interfant-06 | M | 16.1 (63.5) | 87 | 11.4891 | 4.6039 | 1.4411 | 3.5708 | 2.5554 | 4.6562 |
| 39 | 1.1 | Interfant-06 | F | 3.3 (15.7) | 93 | 11.9464 | 6.0799 | 2.8532 | 4.2890 | 4.2377 | 12.9071 |
| 40 | 5.3 | Interfant-06 | M | (58.7) | 94 | 10.3705 | 4.6305 | 2.2531 | 3.8496 | 3.0655 | 5.4284 |
| 41 | 6.9 | Interfant-06 | F | (63.2) | 16 | 9.7053 | 5.4245 | 1.3355 | 3.4142 | 5.0462 | 9.2472 |
| 42 | 5.3 | Interfant-06 | M | (60.7) | 88 | 9.5640 | 3.2046 | −0.1283 | 2.8862 | 1.5220 | 3.4737 |
| 43 | 0.6 | Interfant-06 | F | 5.0 (11.9) | 97 | 8.6364 | 5.2214 | 1.5813 | 3.6253 | 2.8227 | 7.9122 |
| 44 | 1.5 | Interfant-06 | M | 17.3 (18.3) | 98 | 9.5745 | 6.0122 | 0.6954 | 3.0358 | 3.6119 | 8.1333 |
| 45 | 4.4 | Interfant-06 | M | 6.6 (10.8) | 82 | 10.2528 | 4.5355 | 0.2110 | 3.4874 | 2.4696 | 4.2993 |
| 46 | 9.1 | Interfant-06 | F | 39.7 (45.2) | 71 | 9.3985 | 5.1434 | 1.5751 | 4.8219 | 2.9164 | 3.1663 |
| 47 | 0.5 | Interfant-06 | F | 8.7 (13.3) | 93 | 9.9358 | 10.9029 | 7.9047 | 7.4532 | 8.7790 | 6.2701 |
| 48 | 6.1 | AIEOP-BFM ALL 2017 | F | 6.6 | n/a | 9.4741 | 11.2982 | 8.5621 | 6.0722 | 10.3424 | 12.6734 |
| 49 | 6.1 | AIEOP-BFM ALL 2017 | F | 8.3 | n/a | 10.5529 | 11.9934 | 8.9069 | 9.1727 | 8.9088 | 4.5847 |
| 50 | 10.6 | / | M | / | n/a | 5.1199 | 10.1187 | 9.3696 | 7.7286 | 11.0893 | 5.0774 |
Samples were taken from PB at the day of diagnosis. Gene expression values are indicated as the ΔCT mean of technical triplicates.
% blasts Blast percentages measured at diagnosis in PB, F female, M male.
Table 2.
Patient characteristics of the rel cohort (n = 18).
| Patient no. | Age at primary diagnosis [months] | KMT2A-r partner | sex | EGR3 ΔCT Mean | IGHM ΔCTMean | CD79A ΔCTMean | BLK ΔCTMean | PTPN6 ΔCTMean |
|---|---|---|---|---|---|---|---|---|
| REZ1 | 1.3 | AFF1 | F | 2.8330 | 3.6274 | 2.5294 | 6.2860 | 4.2386 |
| REZ2 | 2.9 | MLLT1 | M | 4.1774 | 8.3719 | 6.2586 | 7.3628 | 4.5660 |
| REZ3 | 3.8 | AFF1 | F | 4.3014 | 11.1143 | 7.5835 | 9.1950 | 10.0947 |
| REZ4 | 2.5 | AFF1 | F | 6.6574 | 12.0503 | 10.1751 | 11.9828 | 9.3876 |
| REZ5 | 1.2 | AFF1 | F | 1.1889 | 10.5701 | 8.1192 | 9.9020 | 9.5213 |
| REZ6 | 10.7 | MLLT1 | M | 5.1781 | 5.0754 | 3.5868 | −0.4993 | 3.8987 |
| REZ7 | 4.3 | MLLT1 | M | 3.7298 | 6.0183 | 5.0341 | −0.4709 | 6.2223 |
| REZ8 | 3.6 | MLLT3 | F | 4.7424 | 3.6338 | 4.7103 | 0.1660 | 3.3973 |
| REZ9 | 9.1 | AFF1 | F | 2.2762 | 10.9410 | 7.9419 | 8.6553 | 9.9990 |
| REZ10 | 0.7 | AFF1 | F | 1.6308 | 13.4663 | 9.1956 | 10.7877 | 10.0112 |
| REZ11 | 4.4 | AFF1 | M | 6.5653 | 10.6022 | 9.1016 | 10.0768 | 11.0481 |
| REZ12 | 4.2 | AFF1 | M | 5.0302 | 5.1254 | 3.0619 | 4.9896 | 1.7865 |
| REZ13 | 2.0 | AFF1 | F | 2.5849 | 7.4593 | 4.5334 | 4.2896 | 1.5519 |
| REZ14 | 3.1 | AFF1 | F | 8.4397 | 10.9710 | 7.2141 | 9.6779 | 1.5646 |
| REZ15 | 7.6 | AFF1 | F | 7.8336 | 4.8100 | 5.3276 | 6.5240 | 4.4725 |
| REZ16 | 6.7 | AFF1 | M | 9.5416 | 9.4965 | 5.6740 | 8.0000 | 4.4870 |
| REZ17 | 5.3 | AFF1 | M | 9.8667 | 5.2598 | 3.3857 | −0.1229 | 5.3876 |
| REZ18 | 7.2 | AFF1 | M | 6.7247 | 5.9524 | 3.2850 | 0.2192 | 4.3320 |
Samples were taken from PB at relapse diagnosis. Gene expression values are indicated as the ΔCT mean of technical triplicates.
F female, M male.
Table 3.
Oligonucleotides used in this study.
| Name | Sequence (5′ -> 3′) | Application |
|---|---|---|
| GAPDH_fwd | TTGCCCTCAACGACCACTTT | qRT-PCR |
| GAPDH_rev | TGGTCCAGGGGTCTTACTCC | qRT-PCR |
| IGHM_fwd | CATCCTGACCGTGTCCGAAG | qRT-PCR |
| IGHM_rev | TGGCCCACAGGTTCTCAAAG | qRT-PCR |
| CD79A_fwd | ACCGAATCATCACAGCCGAG | qRT-PCR |
| CD79A_rev | CAACCCGAGCTTCTCGTTCT | qRT-PCR |
| BLK_fwd | GACAGTGAATACACGGCCCA | qRT-PCR |
| BLK_rev | CCCGCCCATAAGTGACAACT | qRT-PCR |
| PTPN6_fwd | GAGGCGCAGTACAAGTTCATC | qRT-PCR |
| PTPN6_rev | GTTCCCGTACTCCGACTCCT | qRT-PCR |
Flow cytometry of SEM::EGR3 and SEM::mock
Cells were blocked using Human BD Fc BlockTM (BD) and stained with FACS antibodies (BD) according to the manufacturer’s protocol. Cells were analyzed using a BD FACSVerseTM. Cells of interest were gated out of all cells using FSC-H and FSC-A. Subsequently, single cells were gated and assessed for CD19, IgM and CD79A protein surface expressions with gates set using fluorescence minus one (FMO) controls. Flow cytometric analysis was performed with four biological replicates of SEM::mock and SEM::EGR3 on a BD FACSAria™ III Cell Sorter. FACS plots were created using FlowJo™ Software. Antibodies used for flow cytometry are described in Table 4.
Table 4.
Antibodies used in this study.
| Antibody | Source | Identifier |
|---|---|---|
| PE Mouse Anti-Human CD79a Monoclonal Antibody | BD | BD Biosciences Cat# 563777, RRID: AB_2738423 |
| BV421 Mouse Anti-Human IgM Monoclonal Antibody | BD | BD Biosciences Cat# 562618, RRID: AB_2737681 |
| APC-H7 Mouse Anti-Human CD19 Monoclonal Antibody | BD | BD Biosciences Cat# 560177, RRID: AB_1645470 |
Massive analysis of cDNA ends-sequencing (MACE-Seq)
MACE-Seq is 3′ single end mRNA sequencing enabling high resolution transcription profiling of RNA extracted from three biological replicates of SEM::mock and SEM::EGR3 48 h after induction with Doxycycline 1 µg/mL. Screen tape analysis of RNA was performed using the bioanalyzer Agilent 2200 TapeStation assessing the RNA integrity number (RIN). RIN values above 8.5 were considered as tolerable. MACE-Seq of extracted RNAs was performed by GenXPro GmbH. MACE-Seq data are available at GEO with accession code GSE225710.
Chromatin immunoprecipitation DNA-sequencing (ChIP-Seq)
ChIP-Seq data of SEM::EGR3 immunoprecipitated with an α-FLAG antibody in comparison to input were already obtained during our former study (GSE205652). ChIP-Seq data are available at GEO with accession code GSE205652.
Statistics and data analysis
Appropriate statistical tests were performed within DeSeq2, GSEA, or BETA plus algorithms in context of differential expression and gene set enrichment analyses of MACE-Seq data or binding and expression target analysis of ChIP- and MACE-Seq data, respectively. The level of significance is indicated by p values (DeSeq2), false discovery rates (GSEA) or rank products (BETA plus). Used expression and binding data meet the demands of respective algorithms and associated statistical tests in terms of normality and equal-variance assumptions. The phenotypic populations assessed by flow cytometry were compared using two-tailed t tests performed using GraphPad Prism 9.5.0 software. Principal component analyses (PCA) were conducted as singular value decompositions using ClustVis [22] with applied unit variance scaling for rows.
Sample sizes were chosen depending on experimental context. qRT-PCR was performed as technical triplicates of 68 patient samples in total. MACE-Seq was performed using three biological triplicates of SEM::mock and SEM::EGR3 RNA. Flow cytometric analysis was performed using four biological replicates of SEM::mock and SEM::EGR3.
Gene set enrichment analysis (GSEA) was conducted using GSEA 4.2.3 according to the developer’s protocol [23].
Integration of MACE-Seq and ChIP-Seq data as well as transcription factor motif scanning was performed using the online tool BETA plus according to the developer’s protocol [24].
The 3890 up- and 3107 downregulated direct EGR3 target genes identified with BETA were uploaded to the PANTHER 17.0 classification system [25]. The functional classification considering protein class, biological function/gene ontology (GO) term, and chromosomal location was plotted using Microsoft Excel as net plots.
Survival analysis was performed using GraphPad Prism 9.4.1. Event-free survival (EFS) was defined as the time from diagnosis to first failure including induction failure, relapse, death, or second malignant neoplasm according to the Interfant-99 protocol [2]. Time was censored at last follow-up if no events were observed. Curves were computed with the Kaplan–Meier estimator, standard errors (SE) with the Greenwood formula, and curves were compared with the log-rank test.
Results
Overexpression of EGR3 in the KMT2A::AFF1 proB cell line SEM upregulates B-lineage specification and commitment genes
We recently identified EGR3 as a relapse-associated factor in KMT2A-r iALL, whose gene expression is ~100-fold increased in patients at relapse (rel) compared to primary diagnosis (dx) [5]. Surprisingly, re-analysis of the Leukemia MILE study data involving 2096 patient samples using the online database BloodSpot revealed B cell malignancies to be characterized by a decreased EGR3 gene expression in comparison to healthy bone marrow (BM) (Supplementary Fig. 1A) [26, 27]. Regarding healthy hematopoietic cells, the BloodSpot DMAP dataset demonstrates low EGR3 expression in proB cells which strongly elevates with differentiation to naïve B cells and their progeny (Supplementary Fig. 1B) [28].
Accordingly, the inducible EGR3 overexpression SEM cell model (SEM::EGR3) of our previous study represents the cellular identity of leukemic blasts in relapsed patients, with a proB phenotype, a strongly increased EGR3 gene expression, and the KMT2A::AFF1 genotype. To ascertain the role of EGR3 in disease progression and relapse, we performed massive analysis of cDNA ends-sequencing (MACE-Seq) of SEM::EGR3 and the corresponding empty vector control cell line SEM::mock. MACE-Seq and differential gene expression analysis using DeSeq2 identified 10,645 differentially expressed genes between SEM::EGR3 and SEM::mock, referred to as the EGR3 transcriptome. 6082 genes were up- and 4563 downregulated (log2> or <0, p < 0.05). A set of B-lineage genes was found to be strongly overexpressed (IGHM (log2fc = 11.42), CD79A (log2fc = 6.84), BLK (log2fc = 8.55), PTPN6 (log2fc = 7.16), CD22 (log2fc = 6.42), CD19 (log2fc = 5.21, IGLL1 (log2fc = 8.91).
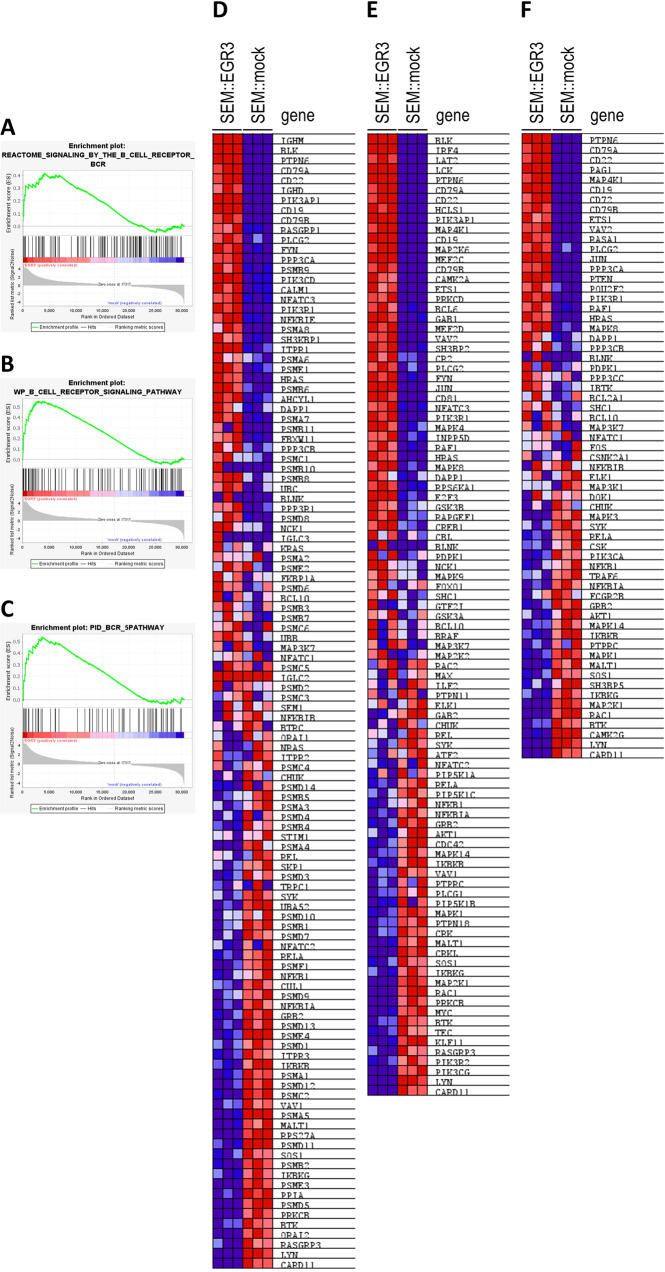
Gene set enrichment analysis (GSEA) was used to functionally characterize the EGR3 transcriptome considering 13,938 gene sets, of which 9910 were significantly enriched in the phenotype EGR3 at a nominal p value below 0.01. ‘Signaling by the B cell receptor’ (Reactome, R-HSA-983705) was identified as the highest ranked gene set (NES = 2.96, FDR = 0.050) (Table 5, Fig. 1A, D). This was corroborated by the B cell receptor (BCR) signaling gene sets of Wiki Pathways (NES = 2.73, FDR = 0.050) (Fig. 1B, E) and the Pathway Interaction Database (NES = 2.61, FDR = 0.050) (Fig. 1C, F) being at ranks four and 18, respectively [29–31]. The top scored upregulated genes in the three sets were IGHM, CD79A, BLK, PTPN6, CD22, and CD19 (Fig. 1D–F). Importantly, also the pre-BCR surrogate light chain gene IGLL1 was found to be upregulated upon EGR3 overexpression (log2fc = 8.91, p = 5.03 × 10−16). In summary, GSEA identified genes involved in BCR signaling as important targets of the EGR3 induced transcriptional profile.
Table 5.
Top fifty gene sets enriched in phenotype EGR3.
| rank | gene set | size | ES | NES | FDR q-val | rank at max |
|---|---|---|---|---|---|---|
| 1 | REACTOME_SIGNALING_BY_THE_B_CELL_RECEPTOR_BCR | 114 | 0.41 | 2.96 | 0.05 | 4040 |
| 2 | LI_WILMS_TUMOR_VS_FETAL_KIDNEY_1_DN | 164 | 0.41 | 2.79 | 0.05 | 1819 |
| 3 | GNF2_PTPRC | 67 | 0.62 | 2.74 | 0.05 | 2657 |
| 4 | WP_B_CELL_RECEPTOR_SIGNALING_PATHWAY | 97 | 0.55 | 2.73 | 0.05 | 3716 |
| 5 | GOBP_POSITIVE_REGULATION_OF_INTERFERON_BETA_PRODUCTION | 37 | 0.52 | 2.73 | 0.05 | 2366 |
| 6 | GOBP_ATP_METABOLIC_PROCESS | 201 | 0.38 | 2.71 | 0.05 | 4185 |
| 7 | HP_MYOCLONUS | 348 | 0.42 | 2.71 | 0.05 | 4350 |
| 8 | YAP1_UP | 43 | 0.51 | 2.7 | 0.05 | 4532 |
| 9 | GOBP_INTERFERON_BETA_PRODUCTION | 55 | 0.51 | 2.67 | 0.05 | 2366 |
| 10 | REACTOME_DDX58_IFIH1_MEDIATED_INDUCTION_OF_INTERFERON_ALPHA_BETA | 68 | 0.46 | 2.66 | 0.05 | 2201 |
| 11 | ALCALA_APOPTOSIS | 85 | 0.41 | 2.66 | 0.05 | 2052 |
| 12 | DER_IFN_BETA_RESPONSE_UP | 103 | 0.43 | 2.66 | 0.05 | 3809 |
| 13 | BROWNE_INTERFERON_RESPONSIVE_GENES | 64 | 0.57 | 2.66 | 0.05 | 3071 |
| 14 | GNF2_VAV1 | 35 | 0.58 | 2.66 | 0.05 | 3195 |
| 15 | HP_BRITTLE_HAIR | 40 | 0.52 | 2.65 | 0.05 | 1355 |
| 16 | GOBP_DEFENSE_RESPONSE_TO_SYMBIONT | 257 | 0.43 | 2.63 | 0.05 | 3422 |
| 17 | NGUYEN_NOTCH1_TARGETS_DN | 82 | 0.5 | 2.62 | 0.05 | 3342 |
| 18 | PID_BCR_5PATHWAY | 63 | 0.54 | 2.61 | 0.05 | 3716 |
| 19 | HP_CHOREOATHETOSIS | 99 | 0.41 | 2.61 | 0.05 | 3007 |
| 20 | REACTOME_SARS_COV_INFECTIONS | 355 | 0.34 | 2.61 | 0.05 | 6457 |
| 21 | HP_MEMORY_IMPAIRMENT | 139 | 0.43 | 2.61 | 0.05 | 3896 |
| 22 | GOBP_RESPONSE_TO_VIRUS | 347 | 0.44 | 2.59 | 0.05 | 3494 |
| 23 | HALLMARK_INTERFERON_ALPHA_RESPONSE | 96 | 0.46 | 2.56 | 0.05 | 4121 |
| 24 | GOBP_REGULATION_OF_INTRACELLULAR_TRANSPORT | 320 | 0.35 | 2.56 | 0.05 | 6271 |
| 25 | HP_NEOPLASM_OF_THE_LIVER | 124 | 0.49 | 2.55 | 0.05 | 5154 |
| 26 | WP_APOPTOSIS | 85 | 0.46 | 2.55 | 0.05 | 3921 |
| 27 | REACTOME_TAK1_DEPENDENT_IKK_AND_NF_KAPPA_B_ACTIVATION | 43 | 0.53 | 2.54 | 0.05 | 2041 |
| 28 | HP_ECTOPIC_CALCIFICATION | 209 | 0.44 | 2.54 | 0.05 | 3466 |
| 29 | HP_ABNORMAL_CNS_MYELINATION | 307 | 0.36 | 2.53 | 0.05 | 6718 |
| 30 | BOQUEST_STEM_CELL_DN | 206 | 0.51 | 2.52 | 0.05 | 2711 |
| 31 | HP_WEAKNESS_DUE_TO_UPPER_MOTOR_NEURON_DYSFUNCTION | 469 | 0.39 | 2.52 | 0.05 | 4528 |
| 32 | HP_ACUTE_LEUKEMIA | 105 | 0.41 | 2.52 | 0.05 | 5158 |
| 33 | REACTOME_RHO_GTPASE_CYCLE | 436 | 0.37 | 2.52 | 0.05 | 4897 |
| 34 | HOEBEKE_LYMPHOID_STEM_CELL_UP | 92 | 0.44 | 2.51 | 0.05 | 2556 |
| 35 | GOBP_REGULATION_OF_VIRAL_GENOME_REPLICATION | 82 | 0.45 | 2.51 | 0.05 | 3466 |
| 36 | REACTOME_INTERLEUKIN_1_FAMILY_SIGNALING | 144 | 0.39 | 2.51 | 0.05 | 2041 |
| 37 | REACTOME_CYTOSOLIC_SENSORS_OF_PATHOGEN_ASSOCIATED_DNA | 62 | 0.41 | 2.51 | 0.05 | 2201 |
| 38 | WANG_TARGETS_OF_MLL_CBP_FUSION_UP | 43 | 0.49 | 2.51 | 0.05 | 4346 |
| 39 | GYORFFY_DOXORUBICIN_RESISTANCE | 33 | 0.65 | 2.51 | 0.05 | 2036 |
| 40 | HALLMARK_ALLOGRAFT_REJECTION | 179 | 0.55 | 2.51 | 0.05 | 3064 |
| 41 | GOBP_ACTIVATION_OF_GTPASE_ACTIVITY | 109 | 0.49 | 2.51 | 0.05 | 2641 |
| 42 | GNF2_MYD88 | 59 | 0.5 | 2.49 | 0.05 | 2926 |
| 43 | MODULE_256 | 58 | 0.46 | 2.49 | 0.05 | 4870 |
| 44 | HP_CHRONIC_DIARRHEA | 75 | 0.48 | 2.48 | 0.05 | 3992 |
| 45 | HP_ABNORMALITY_OF_CARDIOVASCULAR_SYSTEM_ELECTROPHYSIOLOGY | 449 | 0.4 | 2.48 | 0.05 | 3898 |
| 46 | YAGI_AML_WITH_T_8_21_TRANSLOCATION | 351 | 0.39 | 2.48 | 0.05 | 5568 |
| 47 | HP_PREMATURE_GRAYING_OF_HAIR | 39 | 0.51 | 2.48 | 0.05 | 1796 |
| 48 | HP_CNS_DEMYELINATION | 39 | 0.61 | 2.48 | 0.05 | 4378 |
| 49 | HP_SHORTENING_OF_ALL_DISTAL_PHALANGES_OF_THE_FINGERS | 39 | 0.52 | 2.48 | 0.05 | 5023 |
| 50 | GOBP_REGULATION_OF_GENERATION_OF_PRECURSOR_METABOLITES_AND_ENERGY | 122 | 0.45 | 2.48 | 0.05 | 4933 |
ES enrichment score, NES nominal ES, FDR false discovery rate.
Fig. 1. Gene set enrichment analysis (GSEA) of SEM::EGR3 and SEM::mock.
Enrichment plots and corresponding heatmaps of the gene sets ‘REACTOME-SIGNALING-BY-THE-B-CELL-RECEPTOR-BCR’ (A, D), ‘WP_B_CELL-RECEPTOR-SIGNALING-PATHWAY’ (B, E) and ‘PID_BCR_5PATHWAY’ (C, F).
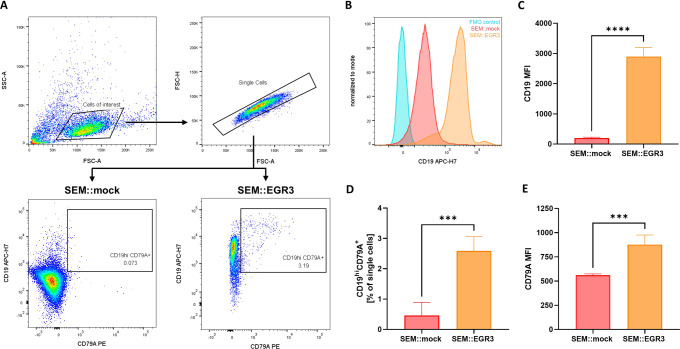
We assessed the surface expression of CD19 and CD79A in SEM::EGR3 and SEM::mock using flow cytometry, as the genes encoding these surface receptors were highly ranked and part of the core enrichment of the Reactome BCR signaling gene set (Fig. 2A). Remarkably, although the SEM cell line is per se CD19+, EGR3 overexpression resulted in an approximately ten-fold increase of the CD19 median fluorescence intensity (MFI) of the single cell population (p < 0.0001) (Fig. 2B, C). Furthermore, we observed a significant relative expansion of the CD19hiCD79A+ population of SEM::EGR3 (p = 0.0006) (Fig. 2D). This expansion of phenotypic B cells (CD19hiCD79A+) was characterized by a significant increase of the CD79A MFI compared to SEM::mock (p = 0.0008) (Fig. 2E).
Fig. 2. Flow cytometric analysis of SEM::EGR3 and SEM::mock.
A Gating strategy. Single cells were gated out of cells of interest for assessment of CD19 and CD79A surface expressions. Analyses were performed based on four biological replicates. B Exemplary histogram of CD19 fluorescence intensities of a full-fluorescence-minus-anti CD19 (FMO) control (blue), SEM::mock (red) and SEM::EGR3 (orange). Plots display the respective single cell populations and were normalized to mode. C CD19 median fluorescence intensities (MFI) of SEM::mock and SEM::EGR3 singlets. Significance was tested using a two-tailed t test (p < 0.0001). Error bars indicate standard deviation. D Mean percentages of the CD19hiCD79A+ population relative to singlets. Error bars indicate standard deviation and significance was tested using a two-tailed t test (p = 0.0006). E CD79A median fluorescence intensities (MFI) of the CD19hiCD79A+ populations of SEM::mock and SEM::EGR3. Significance was tested using a two-tailed t test (p = 0.0008). Error bars indicate standard deviation.
These results demonstrate that upregulation of CD19 and CD79A by EGR3 led to an increase of the receptors’ surface expressions. EGR3 overexpression induced B-lineage specification indicated by relative expansion of phenotypic B cells, thereby enabling functional BCR signaling as indicated by GSEA.
EGR3 and downstream intermediate factors transactivate B-lineage specification and commitment genes
To explore the EGR3 regulome in detail, we performed binding and expression target analysis using the open source application BETA plus [24]. The BETA software algorithm integrates transcription factor ChIP-Seq data with differential gene expression data to deduce direct target genes, and is a standard processing pipeline for transcription factor binding studies [24, 32].
BETA enabled integration of MACE-Seq transcriptome data with EGR3 chromatin immunoprecipitation DNA-sequencing (ChIP-Seq) data of SEM::EGR3 and SEM::mock. ChIP-Seq data were already obtained during our former study (GSE205652).
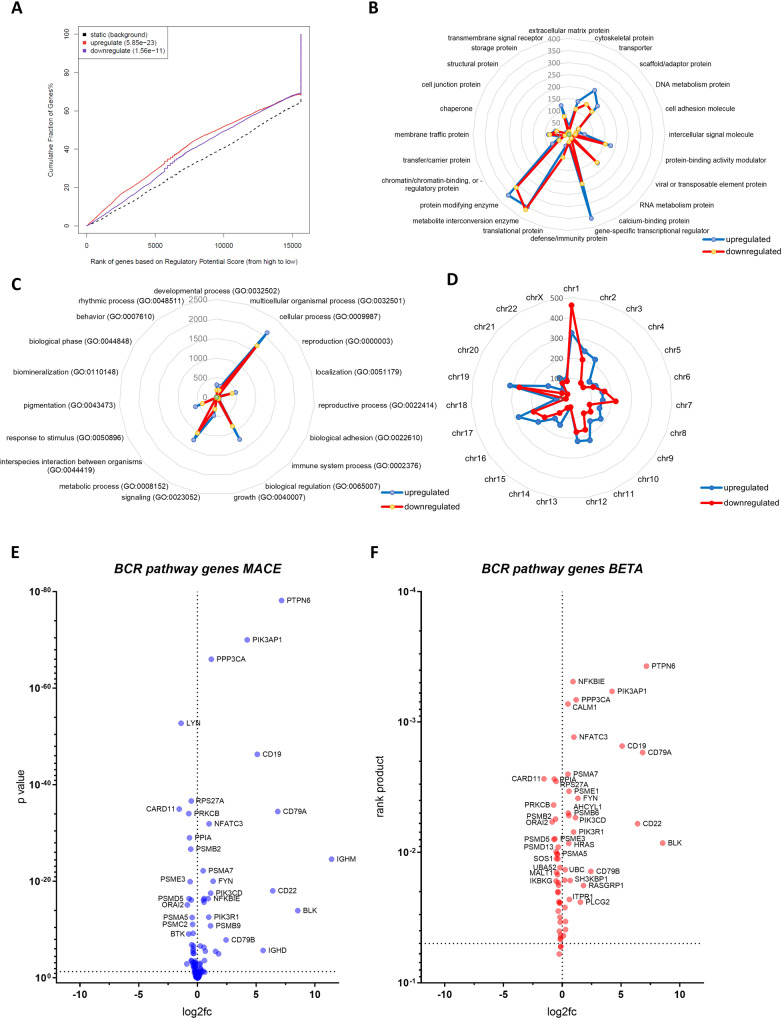
The BETA algorithm ranked genes according to the regulatory potential score and assigned to the cumulative percentage of genes. Plotting this assignment as a graph visualized that EGR3 owns a direct activating and repressive function, and thus, acts as a direct transactivator and -repressor in KMT2A::AFF1 proB-ALL (Fig. 3A).
Fig. 3. Integration of MACE-Seq and ChIP-Seq data revealed the EGR3 regulome of infant KMT2A::AFF1 ALL.
A Assignment of gene ranks to the cumulative fraction of genes using BETA identifies EGR3 as a direct activator and repressor of target genes. Analysis of the EGR3 regulome using the PANTHER database with up- and downregulated target genes subclassified considering their corresponding protein class (B), biological function/GO term (C) and chromosomal location (D). Numbers indicate the count of up- (blue) and downregulated (red) genes. E Volcano plot visualizing genes of the Reactome BCR geneset according to their differential expression indicated as the log2-fold change (log2fc) and p value (MACE-Seq data). F Volcano plot visualizing genes of the Reactome BCR geneset according to their differential expression (log2fc) and direct EGR3 regulation (rank product) (MACE-Seq and BETA data).
In total, 3890 directly upregulated and 3107 downregulated EGR3 target genes were identified, referred to as the EGR3 regulome. We used the PANTHER classification system [25] to functionally characterize the EGR3 regulome. This analysis revealed that EGR3 transactivates and -represses the same classes of genes (Fig. 3B), involved in the same biological functions (Fig. 3C) and located on the same chromosomes (Fig. 3D). Especially the class ‘gene-specific transcriptional regulator’ comprised more directly activated (n = 361) than repressed (n = 212) genes. Concordant with this, a higher number of transactivated (n = 1229) than -repressed (n = 840) genes were related to the Gene Ontology (GO) term ‘biological regulation’ (Fig. 3C).
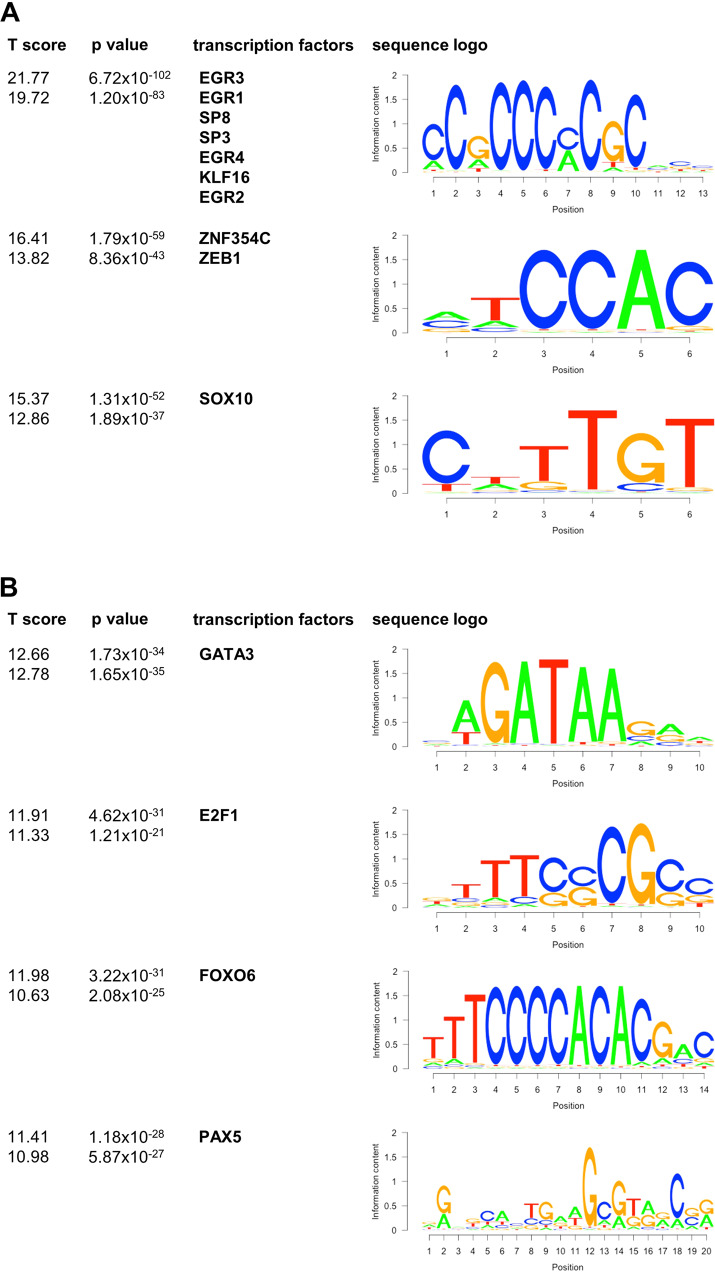
We compared the BCR pathway-related genes of the EGR3 transcriptome with those of the regulome (Fig. 3E, F) and identified 64 of 114 genes (56.1%) as direct EGR3 targets, including CD79A, BLK, PTPN6, CD19, and CD22 (Table 6). Thus, the remaining 50 genes including IGHM were indirect EGR3 targets and transactivated by unknown intermediate transcription factors. To uncover these transcription factors, a motif analysis of all differentially expressed genes (MACE-Seq data) and all direct EGR3 targets (ChIP-Seq data) was performed using BETA plus. As expected, the highest scoring and significant consensus sequence found in all up- and downregulated target genes matched that one of EGR3 and other transcription factors binding to the same or a highly similar motif including EGR1, EGR2, EGR4, KLF16, SP3, and SP8 (Fig. 4A). Lower scoring binding motifs of up- and downregulated genes were assigned to ZEB1, ZNF354C, and SOX10, indicating potential alternate regulation of EGR3 target genes by these transcription factors. Scanning for enriched motifs in differentially expressed but not direct EGR3-regulated genes identified a set of motif-assigned transcription factors. Of these were GATA3, FOXO6, and E2F1 strongly upregulated direct EGR3 targets, and PAX5 an indirect EGR3 target with strong differential expression (log2fc = 9.47, p = 4.95 × 10−41) (Fig. 4B). Especially PAX5 has been described as a mediator of B cell identity and B-lineage commitment [33–35]. Furthermore, analysis of the Leukemia MILE study data set using BloodSpot revealed the strongest gene correlations of PAX5 to be CD19 and CD79A. Concluding this, the binding and expression target analysis followed by motif scanning uncovered GATA3, FOXO6, E2F1, and PAX5 as intermediate factors in the EGR3 transcriptomic network, regulating B-lineage specification and commitment gene expression.
Table 6.
Genes of the set ‘signaling by the B cell receptor’ (Reactome, R-HSA-983705), their type of regulation by EGR3 and differential expression.
| gene | direct EGR3 target? | log2fc MACE-Seq | p value MACE-Seq | rp BETA |
|---|---|---|---|---|
| IGHM | no | 11.42362217 | 2.48921E-25 | n/a |
| BLK | yes | 8.55098816 | 1.10908E-14 | 8.46E-03 |
| PTPN6 | yes | 7.164737984 | 6.50833E-79 | 0.0003723 |
| CD79A | yes | 6.842767882 | 3.2689E-35 | 0.001712 |
| CD22 | yes | 6.416448966 | 8.85632E-19 | 0.006018 |
| IGHD | no | 5.59443973 | 1.92888E-06 | n/a |
| CD19 | yes | 5.097472391 | 4.62783E-47 | 0.001527 |
| PIK3AP1 | yes | 4.246158952 | 9.22432E-71 | 0.0005812 |
| CD79B | yes | 2.440657291 | 1.33431E-08 | 0.01395 |
| RASGRP1 | yes | 1.805660433 | 9.81104E-06 | 0.01795 |
| PLCG2 | yes | 1.549262561 | 3.06887E-06 | 0.02405 |
| FYN | yes | 1.335261599 | 8.88319E-21 | 0.00384 |
| PPP3CA | yes | 1.184342322 | 9.44927E-67 | 0.0006748 |
| PIK3CD | yes | 1.124084067 | 2.75855E-18 | 0.005402 |
| PSMB9 | no | 1.120899683 | 1.59578E-11 | n/a |
| NFATC3 | yes | 0.998713529 | 1.18837E-32 | 0.001304 |
| PIK3R1 | yes | 0.966311084 | 2.44716E-13 | 0.006969 |
| NFKBIE | yes | 0.914332634 | 3.75272E-17 | 0.0004891 |
| SH3KBP1 | yes | 0.677658035 | 2.64864E-06 | 0.01642 |
| PSMA6 | no | 0.628450298 | 0.062177736 | n/a |
| ITPR1 | yes | 0.608990433 | 0.000233393 | 0.02294 |
| PSME1 | yes | 0.579774438 | 3.5101E-17 | 0.003389 |
| PSMB6 | yes | 0.565638656 | 2.03686E-16 | 0.005221 |
| HRAS | yes | 0.553028624 | 4.35293E-07 | 0.008516 |
| AHCYL1 | yes | 0.516916343 | 6.89158E-17 | 0.004982 |
| CALM1 | yes | 0.491872592 | 0.039246507 | 0.000727 |
| PSMA7 | yes | 0.491383232 | 6.26317E-23 | 0.002513 |
| DAPP1 | no | 0.388597531 | 0.052583747 | n/a |
| FBXW11 | yes | 0.285022076 | 0.011951906 | 0.03383 |
| PPP3CB | yes | 0.27924157 | 0.031691017 | 0.03899 |
| UBC | yes | 0.253791938 | 2.26985E-07 | 0.01359 |
| PSMB8 | no | 0.250209285 | 0.011026312 | n/a |
| PPP3R1 | yes | 0.229316482 | 0.000922238 | 0.02644 |
| PSMC1 | no | 0.223758639 | 0.21214738 | n/a |
| PSMD8 | yes | 0.206878775 | 0.001486499 | 0.01632 |
| NCK1 | no | 0.177428109 | 0.185595712 | n/a |
| KRAS | no | 0.137742679 | 0.142867744 | n/a |
| PSMA8 | no | 0.12351416 | 0.636113579 | n/a |
| FKBP1A | yes | 0.113168534 | 0.057549916 | 0.04344 |
| PSME2 | no | 0.109161404 | 0.272267991 | n/a |
| PSMD6 | no | 0.099468329 | 0.227417909 | n/a |
| BCL10 | no | 0.097846109 | 0.427668399 | n/a |
| PSMB3 | no | 0.093466114 | 0.130863049 | n/a |
| PSMA2 | no | 0.092612935 | 0.73577585 | n/a |
| PSMB11 | no | 0.092497922 | 0.727664144 | n/a |
| PSMB10 | no | 0.091210982 | 0.717981181 | n/a |
| PSMB7 | no | 0.078594367 | 0.197210636 | n/a |
| UBB | no | 0.077804248 | 0.077068512 | n/a |
| PSMC6 | no | 0.07662523 | 0.396738012 | n/a |
| MAP3K7 | no | 0.053871719 | 0.520389098 | n/a |
| NFATC1 | no | 0.017988661 | 0.842017304 | n/a |
| BLNK | no | 0.012076394 | 0.962179165 | n/a |
| IGLC3 | no | 0.005188884 | 0.98330526 | n/a |
| PSMC5 | no | 0.002298519 | 0.966883202 | n/a |
| IGLC2 | no | 0 | 1 | n/a |
| PSMD2 | no | −0.000438565 | 0.99385484 | n/a |
| PSMC3 | no | −0.009374859 | 0.871548561 | n/a |
| SEM1 | no | −0.035848586 | 0.536641158 | n/a |
| BTRC | no | −0.043122119 | 0.743614989 | n/a |
| NFKBIB | no | −0.048602913 | 0.641683594 | n/a |
| ORAI1 | no | −0.050325263 | 0.689236497 | n/a |
| ITPR2 | no | −0.057315013 | 0.696661836 | n/a |
| NRAS | no | −0.067952655 | 0.403682448 | n/a |
| PSMC4 | no | −0.090473401 | 0.188421226 | n/a |
| CHUK | no | −0.09213649 | 0.463552091 | n/a |
| PSMD14 | no | −0.099389355 | 0.149921007 | n/a |
| REL | no | −0.105755893 | 0.567292607 | n/a |
| PSMB5 | yes | −0.117300018 | 0.071947041 | 0.0525 |
| PSMA3 | yes | −0.12765023 | 0.056413726 | 0.05304 |
| PSMA4 | no | −0.12765023 | 0.056413726 | n/a |
| STIM1 | no | −0.13504264 | 0.135062364 | n/a |
| PSMD4 | yes | −0.136908983 | 0.043599216 | 0.04312 |
| TRPC1 | no | −0.140211415 | 0.566506534 | n/a |
| PSMB4 | yes | −0.143316292 | 0.008444496 | 0.04541 |
| SKP1 | yes | −0.155405082 | 0.007980788 | 0.04654 |
| NFATC2 | no | −0.16313576 | 0.21867056 | n/a |
| SYK | no | −0.163962068 | 0.061590912 | n/a |
| PSMD3 | yes | −0.170338294 | 0.002588761 | 0.03421 |
| UBA52 | yes | −0.18111628 | 0.000640896 | 0.01305 |
| PSMD10 | no | −0.182449259 | 0.046626892 | n/a |
| PSMB1 | yes | −0.191173577 | 0.000313825 | 0.03138 |
| PSMD7 | yes | −0.193336641 | 0.001402521 | 0.02428 |
| RELA | yes | −0.219182607 | 0.005520858 | 0.02449 |
| PSMF1 | yes | −0.241001349 | 0.001520813 | 0.04021 |
| NFKB1 | no | −0.243914024 | 0.029935823 | n/a |
| PSMD9 | yes | −0.25415219 | 0.084691328 | 0.06018 |
| CUL1 | yes | −0.277874069 | 0.000645415 | 0.02387 |
| NFKBIA | no | −0.284994997 | 0.008752111 | n/a |
| GRB2 | yes | −0.310478144 | 8.1543E-06 | 0.01782 |
| PSMD13 | yes | −0.338660552 | 2.25353E-07 | 0.009095 |
| IKBKB | yes | −0.340568298 | 0.003982831 | 0.02814 |
| ITPR3 | yes | −0.345267237 | 0.002182622 | 0.01704 |
| PSMD1 | yes | −0.355786358 | 8.26092E-07 | 0.02033 |
| PSME4 | no | −0.356370366 | 7.30394E-06 | n/a |
| PSMD12 | yes | −0.369060374 | 2.31513E-06 | 0.01996 |
| PSMA1 | yes | −0.370940312 | 5.60831E-10 | 0.01119 |
| VAV1 | yes | −0.396708907 | 0.000758081 | 0.01034 |
| PSMC2 | yes | −0.406420999 | 8.04357E-12 | 0.01478 |
| PSMA5 | yes | −0.44324155 | 2.8338E-13 | 0.01024 |
| MALT1 | yes | −0.479264577 | 1.28722E-07 | 0.01421 |
| SOS1 | yes | −0.486278337 | 0.000293116 | 0.01117 |
| PSMD11 | yes | −0.517998908 | 7.00864E-17 | 0.009862 |
| RPS27A | yes | −0.518177519 | 2.15808E-37 | 0.002853 |
| IKBKG | yes | −0.518676703 | 0.000234099 | 0.01656 |
| PSMB2 | yes | −0.565388038 | 2.01317E-27 | 0.005551 |
| PSME3 | yes | −0.610618137 | 1.09492E-20 | 0.007852 |
| PPIA | yes | −0.673833501 | 9.15098E-30 | 0.002734 |
| PSMD5 | yes | −0.706954282 | 3.71916E-17 | 0.007929 |
| PRKCB | yes | −0.731810409 | 8.84403E-35 | 0.004322 |
| BTK | no | −0.742153419 | 7.76623E-10 | n/a |
| ORAI2 | yes | −0.847816906 | 7.16229E-16 | 0.005828 |
| RASGRP3 | no | −0.902579249 | 0.001205017 | n/a |
| LYN | no | −1.383822477 | 1.74706E-53 | n/a |
| CARD11 | yes | −1.549753074 | 9.97827E-36 | 0.002724 |
log2fc log2 fold change, rp rank product.
Fig. 4. Motif scanning of all differentially expressed genes.
A Motif scanning result of all direct EGR3 target genes. T score, p value, transcription factor and sequence logo were identified using BETA plus. Upper T score and p value apply to upregulated genes, lower T score and p value apply to downregulated genes. B Motif scanning result of differentially expressed but not directly EGR3-regulated genes. T score, p value, transcription factor and sequence logo were identified by BETA plus. Upper T score and p value apply to upregulated genes, lower T score and p value apply to downregulated genes.
IGHM, CD79A, BLK and PTPN6 expressions correlate strongly with each other and partly with EGR3 among 50 infant KMT2A::AFF1 proB-ALL patients
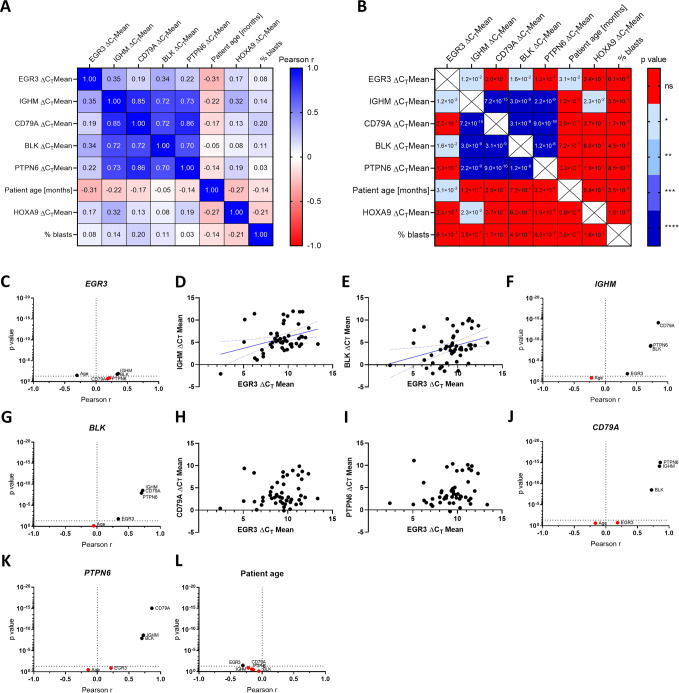
We aimed to investigate the regulation of B-lineage specification and commitment gene expression by EGR3 in patient material and therefore assessed the transcription levels of IGHM, CD79A, BLK, and PTPN6 in 50 infant KMT2A::AFF1 proB-ALL patients at diagnosis (Table 1). This patient cohort was already investigated in our recent study [5], from which we obtained the EGR3 expressions. cDNA of peripheral blood was used for quantitative real-time PCR (qRT-PCR) based gene expression measurement and subsequent Pearson correlation testing of ΔCT values was performed. The resulting Pearson correlation matrix demonstrated a very strong and highly significant correlation between the IGHM, CD79A, BLK, and PTPN6 gene expressions with Pearson r values above 0.70, suggesting their collective belonging to a distinct gene expression program (Fig. 5A, B).
Fig. 5. Pearson correlation testing of the diagnosis cohort.
Pearson correlation matrix indicating the Pearson r value (A) and p value (B) of all tested correlations. Volcano plots showing Pearson correlations of EGR3 (C), IGHM (F), BLK (G), CD79A (J), PTPN6 (K) and patient age (L). Correlations with p ≥ 0.05 were considered non-significant and marked in red. Patients plotted according to their IGHM/EGR3 (D), BLK/EGR3 (E), CD79A/EGR3 (H) and PTPN6/EGR3 (I) ΔCT mean expression values.
The transcription levels of EGR3 correlated less strongly with IGHM and BLK (Pearson r = 0.35 and 0.34, respectively) (Fig. 5C–G), and not significantly with CD79A and PTPN6 (Fig. 5C, H–K). Apart from that, higher patient age correlated with higher EGR3 expression (Pearson r = −0.31), whereas this was not the case for IGHM, CD79A, BLK, and PTPN6 (Fig. 5L).
These results confirm the B-lineage phenotype associated with engineered EGR3-overexpression suggesting direct causality between EGR3 expression and B-lineage specification. Furthermore, other factors are likely to influence the EGR3 target gene expression as well. The latter conclusion is concordant with the motif scan revealing that EGR1, EGR2, EGR4, KLF16, SP3, SP8, ZEB1, ZNF354C, and SOX10 bind the same genes as EGR3, suggesting these factors as possible co-regulators.
Low IGHM, CD79A, BLK, and PTPN6 expressions indicate a patient subgroup with inferior EFS
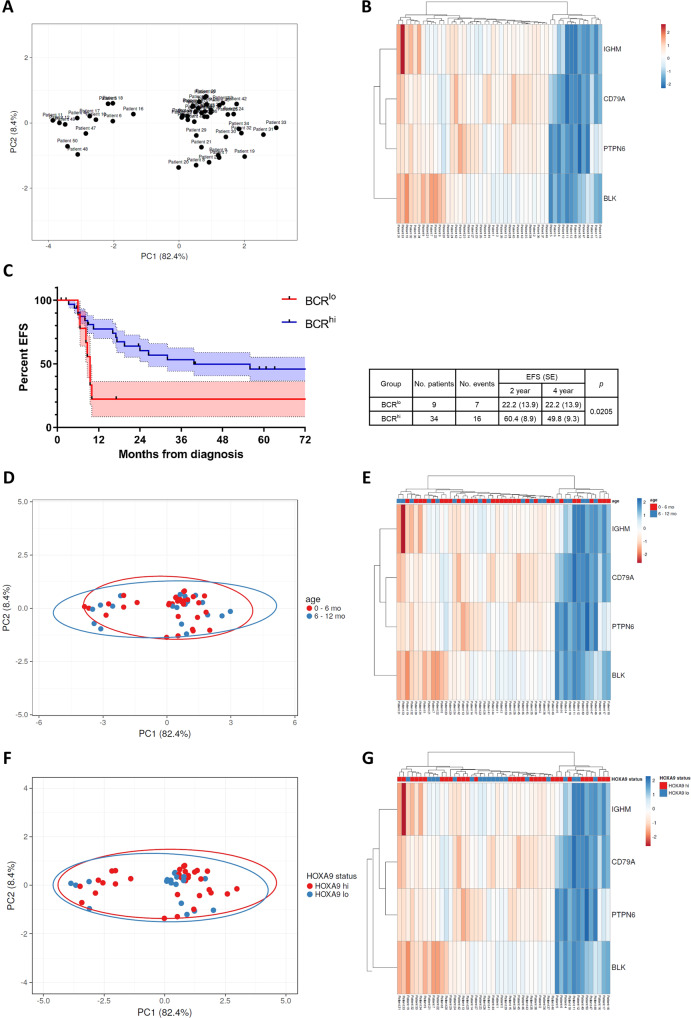
The strong correlation between the IGHM, CD79A, BLK, and PTPN6 gene expressions prompted us to analyze their distribution within the patient cohort. For that purpose, a principal component analysis (PCA) of the expression levels was conducted using the open-source tool ClustVis [22], uncovering a distinct bimodal distribution of patient clusters (Fig. 6A). The corresponding heatmap visualized that patients showed either a high (BCRhi, n = 37) or low (BCRlo, n = 13) expression of the analyzed B-lineage-related genes (Fig. 6B). This bimodal distribution points to differences in the B-lineage specification process, with BCRlo patients having a less committed B cell identity, indicated by the low expression of B-lineage-representing genes. Accordingly, the high expression of IGHM, CD79A, BLK, and PTPN6 in the BCRhi group indicates a more mature proB-cell phenotype. The strict bimodal clustering suggests that the development from BCRlo to BCRhi is rather a stepwise maturation process than a fluent transition, and presumably reflects the developmental stages of early and late proB cells.
Fig. 6. PCA and survival analysis of the diagnosis cohort.
A PCA of the diagnosis cohort among the IGHM, CD79A, BLK, and PTPN6 expressions. PC1 represents 82.4% and PC2 8.4% of total variance, respectively. B Heatmap of the PCA visualizing bimodal clustering of patients. C Kaplan–Meier curves and log-rank test of the BCRlo (red) and BCRhi (blue) patients. S.E.M. of each curve is indicated in bright color. D PCA of the diagnosis cohort among the IGHM, CD79A, BLK, and PTPN6 expressions with patients assigned to the age groups 0–6 months and 6–12 months. E Heatmap of the PCA visualizing bimodal clustering of patients is not affected by patient age at diagnosis. F PCA of the diagnosis cohort among the IGHM, CD79A, BLK, and PTPN6 expressions with patients assigned to HOXA status groups HOXA9 high (hi) and low (lo). G Heatmap of the PCA visualizing bimodal clustering of patients is not affected by HOXA status.
We compared the event-free survival (EFS) for all patients with available outcome data (n = 43) considering their assignment to the BCRlo (n = 9) or BCRhi (n = 34) group. Four-year-EFS of BCRlo patients was significantly poorer, reaching only 22.2 ± 13.9% compared to 49.8 ± 9.3% of the BCRhi group (p = 0.0205, Fig. 6C). Notably, all events within the BCRlo group occurred within the first 12 months from diagnosis. To test for age and HOXA status as possible confounders, we performed the same PCA analysis with patients assigned to age groups (0–6 months vs. 6–12 months) (Fig. 6D, E) and to HOXA9 status groups (HOXA9 high vs. low) (Fig. 6F, G). We observed that both factors did not affect dichotomous separation of patients as young and old as well as HOXA9 high and low groups overlap almost completely.
BCRlo vs. BCRhi clustering of patients is sustained at relapse
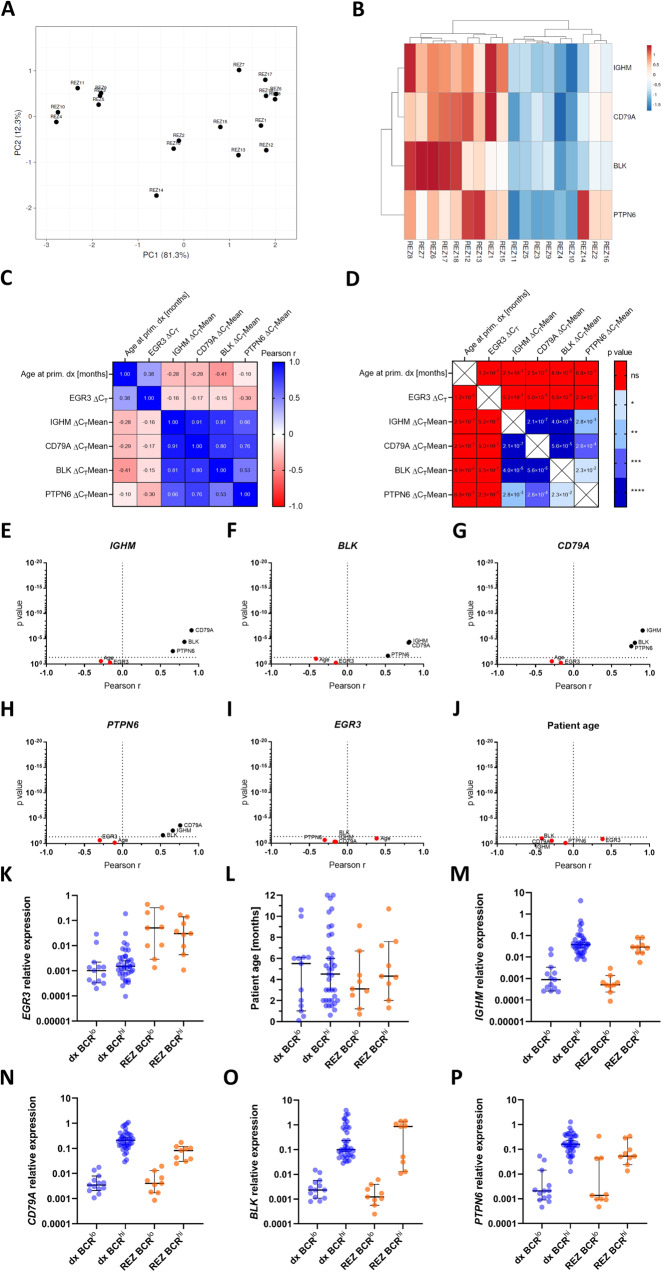
Considering the elevated level of EGR3 expression at relapse in infant KMT2A-r proB-ALL, we aimed to examine the IGHM, CD79A, BLK, and PTPN6 gene expressions at the time of relapse. To do so, we performed the same gene expression analysis with the relapse (rel) cohort of our former study. This cohort comprised 18 infant KMT2A-r proB-ALL patients (14 KMT2A::AFF1 cases) at the time of relapse, composing an independent cohort, not matched to the diagnosis (dx) cohort (Table 2).
As for the dx cohort, a PCA revealed co-segregation of patients into a BCRlo and a BCRhi group (Fig. 7A). Contrasting the dx cohort, clustering of relapsed patients was less cohesive, with patients REZ2, REZ14, and REZ16 exhibiting low IGHM, CD79A, and BLK expressions, but increased PTPN6 gene expression levels reaching those of the BCRhi group (Fig. 7B).
Fig. 7. Patient clustering and Pearson correlation testing of the relapse cohort.
A PCA of the relapse cohort among the IGHM, CD79A, BLK, and PTPN6 expressions. PC1 represents 81.3% and PC2 12.3% of total variance, respectively. B Heatmap of the PCA visualizing clustering of patients. Pearson correlation matrix indicating the Pearson r value (C) and p value (D) of all tested correlations. Volcano plots showing Pearson correlations of IGHM (E), BLK (F), CD79A (G), PTPN6 (H) EGR3 (I) and patient age (J). Correlations with p ≥ 0.05 were considered non-significant and marked in red. Comparison of the BCRlo and BCRhi groups of the diagnosis and relapse cohorts regarding EGR3 relative expression (K), patient age (L), IGHM (M), CD79A (N), BLK (O) and PTPN6 (P) relative expressions. Bars indicate the median and 95% confidence interval.
Pearson correlation testing of the ΔCT values showed strong correlations between the IGHM, CD79A, BLK, and PTPN6 gene expressions, almost resembling the Pearson r and corresponding p values of the dx cohort (Fig. 7C–H). Contrasting the dx cohort, the rel cohort did not indicate a significant correlation for any of the aforementioned gene expressions with that of EGR3 (Fig. 7I). The transcription level of EGR3 was approximately 100-fold elevated at relapse compared to diagnosis, independent of BCRlo or BCRhi classification (Fig. 7K). Unlike EGR3, the expressions of IGHM, CD79A, BLK, and PTPN6 were not generally elevated at relapse, rather were the expression level differences between the BCRlo and BCRhi groups almost the same comparing the diagnosis and relapse cohorts (Fig. 7M–P). These results indicate that BCRlo vs. BCRhi clustering of patients was sustained at relapse and elevated EGR3 levels were not concomitant with IGHM, CD79A, BLK and PTPN6 gene expressions. This suggests a minor role for EGR3 in B-lineage-specific gene regulation at the time of relapse and demonstrates preservation of both developmental proB cell stages during relapse formation, although to a lesser extent.
Discussion
This study identified EGR3 as a regulator of B-lineage specification and commitment processes in the context of infant KMT2A::AFF1 acute lymphoblastic leukemia. This characterization is in line with the fact that EGR3 is generally downregulated in B cell malignancies according to the Leukemia MILE study [27] and upregulated in naïve and mature B cells referring to the Bloodspot DMAP dataset [26] (Supplementary Fig. 1B). Furthermore, the murine homologues Egr3 and Egr2 are required for B cell proliferation upon antigen receptor stimulation [20, 36]. Accordingly, our study and these data imply the limitation of EGR3 expression to be concomitant with a differentiation block of the B-lineage in hematologic malignancies.
That EGR3 is involved in B-lineage specification and commitment is further strengthened by the identification of PAX5 as an intermediate factor of the EGR3 regulome. PAX5 is a known activator of B cell identity regulating the gene expression of CD19, CD79B, and EBF1 [33, 37]. In addition, PAX5 is a regulator of B cell development [38] and B-lineage commitment [35, 39, 40]. In this regard, an important difference between EGR3 and PAX5 is that the latter not only binds DNA response elements but also regulates chromatin accessibility at target promoters [41]. This points to a collaborative effect of EGR3 and PAX5, regulating in part the same genes by different means.
Finally, CD19 is commonly accepted as a hallmark B cell commitment marker [42, 43], and we demonstrate direct transcriptional upregulation of CD19 by EGR3 resulting in approximately ten-fold increased surface expression of the corresponding protein (Fig. 2). Importantly, CD19-directed therapies including CAR T cells and Blinatumomab were reported to lead to lineage switch of infant KMT2A::AFF1 ALL [44–46]. This process has been shown to be accompanied by a loss of the B-lineage-specific transcription factors EBF1 and PAX5 [47]. As our data suggest a collaborative effect between PAX5 and EGR3, the role of EGR3 during lineage switch of KMT2A-r B ALL needs further investigation.
In contrast to primary diagnosis, elevated EGR3 expression at relapse was not accompanied by increased expression of B lineage-associated direct targets. Consequently, EGR3 expression at relapse does not indicate a more committed or mature proB cell identity, but could reflect external stress stimuli including response to chemotherapy or inflammatory mediators in the leukemic microenvironment as EGR3 is involved in rapid and transient stress responses and inflammatory signaling [48, 49].
Fetal pre-pro B cells are the earliest B-lineage-committed progenitors giving rise to proB cells [50]. In this context, both progenitor subtypes can be distinguished regarding their expression of B cell-associated genes including CD79A, CD19, PAX5, MME, EBF1, DNTT among many others [50]. As we worked with RNA extracted from peripheral blood, our gene expression analysis considered mainly the blast population. Therefore, it is very likely that clustering of patients into the BCRlo and BCRhi group at diagnosis and relapse represents an early vs. late proB state of the blast population, with BCRlo representing pre-proB cells and BCRhi proB cells. This interpretation is in line with the understanding of B-lineage commitment as a stepwise process, orchestrated by transcription factor-mediated gene regulatory networks [51, 52]. Furthermore, the inferior survival of the BCRlo group could be explained by decreased maturity of blasts within the proB cell state, likely to go along with elevated lineage plasticity, a hallmark of KMT2A-r leukemia and implicated in therapy resistance [53, 54]. On the other hand, the undifferentiated phenotype of the BCRlo group could indicate a more aberrant oncogenic signaling program interfering with B-lineage gene expression networks. Nevertheless, the identification of four B-lineage genes whose expression reflects outcome enables early gene expression-based risk-stratification of patients.
That we identified both developmental proB stages at diagnosis and relapse with similar gene expression differences indicates preservation of the blast cell identity from diagnosis through the minimal residual disease phase until relapse formation. Accordingly, it could well be that this cell identity is determined by the cell of origin, which is suggested to belong to the fetal liver-derived hematopoietic progenitor cell compartment [55–57]. However, further studies are needed to uncover mechanisms mediating the developmental arrest of blasts either in the pre-proB or proB stage.
In summary, analysis of the EGR3 regulome of infant KMT2A-r iALL identified EGR3 as a regulator of B-lineage commitment. Besides, our study presents four B-lineage genes with prognostic significance, suitable for gene expression-based risk stratification of KMT2A-r iALL patients.
Supplementary information
Acknowledgements
We thank Birthe Fedders for sample composition and sending, the Center for Biological Resources (CRB-cancer; BB-0033-00076), Robert Debré Hospital, France, and our lab colleagues for impactful discussions.
Author contributions
MK wrote the manuscript; MK, PL, HB, CM, MAR, and RM planned and interpreted experiments; MK performed GSEA and qRT-PCR; MK, TS, and MAR performed flow cytometry; PL conducted binding and expression target analysis; HB, CM, and PL helped to write the manuscript; JA, GC, CE, ACE, HC, MB, GiC, PDL, and MGV provided patient samples and outcome data, and helped to analyze clinical data; RM provided funding; all authors reviewed and approved the manuscript.
Funding
This work has been funded in part by DFG grant MA 1876/12-1 and Wilhelm Sander foundation grant 2020.070 to RM, and in part by ERA-NET TRANSCAN EJC, Foundation ARC, France to HC. Open Access funding enabled and organized by Projekt DEAL.
Data availability
All generated datasets are available from the corresponding authors on reasonable request. MACE-Seq data are available at GEO with accession code GSE225710. ChIP-Seq data are available at GEO with accession code GSE205652.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Marius Külp, Email: marius.kuelp@em.uni-frankfurt.de.
Rolf Marschalek, Email: rolf.marschalek@em.uni-frankfurt.de.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-023-01895-z.
References
- 1.Pieters R, De Lorenzo P, Ancliffe P, Aversa LA, Brethon B, Biondi A, et al. Outcome of infants younger than 1 year with acute lymphoblastic leukemia treated with the interfant-06 protocol: results from an international phase III randomized study. J Clin Oncol. 2019;37:2246–56. doi: 10.1200/JCO.19.00261. [DOI] [PubMed] [Google Scholar]
- 2.Pieters R, Schrappe M, De Lorenzo P, Hann I, De Rossi G, Felice M, et al. A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet. 2007;370:240–50. doi: 10.1016/S0140-6736(07)61126-X. [DOI] [PubMed] [Google Scholar]
- 3.Meyer C, Burmeister T, Gröger D, Tsaur G, Fechina L, Renneville A, et al. The MLL recombinome of acute leukemias in 2017. Leukemia. 2018;32:273–84. doi: 10.1038/leu.2017.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driessen EMC, de Lorenzo P, Campbell M, Felice M, Ferster A, Hann I, et al. Outcome of relapsed infant acute lymphoblastic leukemia treated on the interfant-99 protocol. Leukemia. 2016;30:1184–7. doi: 10.1038/leu.2015.246. [DOI] [PubMed] [Google Scholar]
- 5.Külp M, Siemund AL, Larghero P, Dietz A, Alten J, Cario G, et al. The immune checkpoint ICOSLG is a relapse-predicting biomarker and therapeutic target in infant t(4;11) acute lymphoblastic leukemia. iScience. 2022;25:104613. doi: 10.1016/j.isci.2022.104613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beckmann AM, Wilce PA. Egr transcription factors in the nervous system. Neurochem Int. 1997;31:477–510. doi: 10.1016/S0197-0186(96)00136-2. [DOI] [PubMed] [Google Scholar]
- 7.O’Donovan KJ, Levkovitz Y, Ahn D, Baraban JM. Functional comparison of Egr3 transcription factor isoforms: identification of an activation domain in the N-terminal segment absent from Egr3beta, a major isoform expressed in brain. J Neurochem. 2000;75:1352–7. doi: 10.1046/j.1471-4159.2000.0751352.x. [DOI] [PubMed] [Google Scholar]
- 8.Desmazières A, Decker L, Vallat JM, Charnay P, Gilardi-Hebenstreit P. Disruption of Krox20–Nab interaction in the mouse leads to peripheral neuropathy with biphasic evolution. J Neurosci. 2008;28:5891–900. doi: 10.1523/JNEUROSCI.5187-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le N, Nagarajan R, Wang JYT, Svaren J, LaPash C, Araki T, et al. Nab proteins are essential for peripheral nervous system myelination. Nat Neurosci. 2005;8:932–40. doi: 10.1038/nn1490. [DOI] [PubMed] [Google Scholar]
- 10.Svaren J, Sevetson BR, Apel ED, Zimonjic DB, Popescu NC, Milbrandt J. NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol Cell Biol. 1996;16:3545–53. doi: 10.1128/MCB.16.7.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swirnoff AH, Apel ED, Svaren J, Sevetson BR, Zimonjic DB, Popescu NC, et al. Nab1, a corepressor of NGFI-A (Egr-1), contains an active transcriptional repression domain. Mol Cell Biol. 1998;18:512–24. doi: 10.1128/MCB.18.1.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis S, Bozon B, Laroche S. How necessary is the activation of the immediate early gene zif268 in synaptic plasticity and learning? Behav Brain Res. 2003;142:17–30. doi: 10.1016/S0166-4328(02)00421-7. [DOI] [PubMed] [Google Scholar]
- 13.Knapska E, Kaczmarek L. A gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK? Prog Neurobiol. 2004;74:183–211. doi: 10.1016/j.pneurobio.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Schneider-Maunoury S, Topilko P, Seitandou T, Levi G, Cohen-Tannoudji M, Pournin S, et al. Disruption of Krox-20 results in alteration of rhombomeres 3 and 5 in the developing hindbrain. Cell. 1993;75:1199–214. doi: 10.1016/0092-8674(93)90329-O. [DOI] [PubMed] [Google Scholar]
- 15.Williams J, Dragunow M, Lawlor P, Mason S, Abraham WC, Leah J, et al. Krox20 may play a key role in the stabilization of long-term potentiation. Mol Brain Res. 1995;28:87–93. doi: 10.1016/0169-328X(94)00187-J. [DOI] [PubMed] [Google Scholar]
- 16.Yamagata K, Kaufmann WE, Lanahan A, Papapavlou M, Barnes CA, Andreasson KI, et al. Egr3/Pilot, a zinc finger transcription factor, is rapidly regulated by activity in brain neurons and colocalizes with Egr1/zif268. Learn Mem. 1994;1:140–52. doi: 10.1101/lm.1.2.140. [DOI] [PubMed] [Google Scholar]
- 17.Cheng H, Hao S, Liu Y, Pang Y, Ma S, Dong F, et al. Leukemic marrow infiltration reveals a novel role for Egr3 as a potent inhibitor of normal hematopoietic stem cell proliferation. Blood. 2015;126:1302–13. doi: 10.1182/blood-2015-01-623645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min IM, Pietramaggiori G, Kim FS, Passegué E, Stevenson KE, Wagers AJ. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell. 2008;2:380–91. doi: 10.1016/j.stem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Carter JH, Lefebvre JM, Wiest DL, Tourtellotte WG. Redundant role for early growth response transcriptional regulators in thymocyte differentiation and survival. J Immunol. 2007;178:6796–805. doi: 10.4049/jimmunol.178.11.6796. [DOI] [PubMed] [Google Scholar]
- 20.Li S, Symonds ALJ, Zhu B, Liu M, Raymond MV, Miao T, et al. Early growth response Gene-2 (Egr-2) regulates the development of B and T cells. PLoS One. 2011;6:e18498. doi: 10.1371/journal.pone.0018498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogbe A, Miao T, Symonds ALJ, Omodho B, Singh R, Bhullar P, et al. Early growth response Genes 2 and 3 regulate the expression of Bcl6 and differentiation of T follicular helper cells. J Biol Chem. 2015;290:20455–65. doi: 10.1074/jbc.M114.634816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015;43:W566–570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S, Sun H, Ma J, Zang C, Wang C, Wang J, et al. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc. 2013;8:2502–15. doi: 10.1038/nprot.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, et al. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0) Nat Protoc. 2019;14:703–21. doi: 10.1038/s41596-019-0128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bagger FO, Kinalis S, Rapin N. BloodSpot: a database of healthy and malignant haematopoiesis updated with purified and single cell mRNA sequencing profiles. Nucleic Acids Res. 2019;47:D881–5. doi: 10.1093/nar/gky1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohlmann A, Kipps TJ, Rassenti LZ, Downing JR, Shurtleff SA, Mills KI, et al. An international standardization programme towards the application of gene expression profiling in routine leukaemia diagnostics: the Microarray Innovations in LEukemia study prephase. Br J Haematol. 2008;142:802–7. doi: 10.1111/j.1365-2141.2008.07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gillespie M, Jassal B, Stephan R, Milacic M, Rothfels K, Senff-Ribeiro A, et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022;50:D687–92. doi: 10.1093/nar/gkab1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martens M, Ammar A, Riutta A, Waagmeester A, Slenter DN, Hanspers K, et al. WikiPathways: connecting communities. Nucleic Acids Res. 2021;49:D613–21. doi: 10.1093/nar/gkaa1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, et al. PID: the pathway interaction database. Nucleic Acids Res. 2009;37:D674–9. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Höllbacher B, Balázs K, Heinig M, Uhlenhaut NH. Seq-ing answers: current data integration approaches to uncover mechanisms of transcriptional regulation. Comput Struct Biotechnol J. 2020;18:1330–41. doi: 10.1016/j.csbj.2020.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bullerwell CE, Robichaud PP, Deprez PML, Joy AP, Wajnberg G, D’Souza D, et al. EBF1 drives hallmark B cell gene expression by enabling the interaction of PAX5 with the MLL H3K4 methyltransferase complex. Sci Rep. 2021;11:1537. doi: 10.1038/s41598-021-81000-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calderón L, Schindler K, Malin SG, Schebesta A, Sun Q, Schwickert T, et al. Pax5 regulates B cell immunity by promoting PI3K signaling via PTEN down-regulation. Sci Immunol. 2021;6:eabg5003. doi: 10.1126/sciimmunol.abg5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–70. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 36.Li S, Miao T, Sebastian M, Bhullar P, Ghaffari E, Liu M, et al. The transcription factors Egr2 and Egr3 are essential for the control of inflammation and antigen-induced proliferation of B and T cells. Immunity. 2012;37:685–96. doi: 10.1016/j.immuni.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuxa M, Busslinger M. Reporter gene insertions reveal a strictly B lymphoid-specific expression pattern of Pax5 in support of its B cell identity function. J Immunol. 2007;178:3031–7. doi: 10.4049/jimmunol.178.5.3031. [DOI] [PubMed] [Google Scholar]
- 38.Horcher M, Souabni A, Busslinger M. Pax5/BSAP maintains the identity of B cells in late B lymphopoiesis. Immunity. 2001;14:779–90. doi: 10.1016/S1074-7613(01)00153-4. [DOI] [PubMed] [Google Scholar]
- 39.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–62. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 40.Revilla-I-Domingo R, Bilic I, Vilagos B, Tagoh H, Ebert A, Tamir IM, et al. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J. 2012;31:3130–46. doi: 10.1038/emboj.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McManus S, Ebert A, Salvagiotto G, Medvedovic J, Sun Q, Tamir I, et al. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J. 2011;30:2388–404. doi: 10.1038/emboj.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackson TR, Ling RE, Roy A. The origin of B-cells: human fetal B cell development and implications for the pathogenesis of childhood acute lymphoblastic leukemia. Front Immunol. 2021 [cited 2022 Dec 5];12. Available from: https://www.frontiersin.org/articles/10.3389/fimmu.2021.637975. [DOI] [PMC free article] [PubMed]
- 43.LeBien TW. Fates of human B-cell precursors. Blood. 2000;96:9–23. doi: 10.1182/blood.V96.1.9. [DOI] [PubMed] [Google Scholar]
- 44.Gardner R, Wu D, Cherian S, Fang M, Hanafi LA, Finney O, et al. Acquisition of a CD19-negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR-T-cell therapy. Blood. 2016;127:2406–10. doi: 10.1182/blood-2015-08-665547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mejstríková E, Hrusak O, Borowitz MJ, Whitlock JA, Brethon B, Trippett TM, et al. CD19-negative relapse of pediatric B-cell precursor acute lymphoblastic leukemia following blinatumomab treatment. Blood Cancer J. 2017;7:1–5. doi: 10.1038/s41408-017-0023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rayes A, McMasters RL, O’Brien MM. Lineage switch in MLL-rearranged infant leukemia following CD19-directed therapy. Pediatr Blood Cancer. 2016;63:1113–5. doi: 10.1002/pbc.25953. [DOI] [PubMed] [Google Scholar]
- 47.Tirtakusuma R, Szoltysek K, Milne P, Grinev VV, Ptasinska A, Chin PS, et al. Epigenetic regulator genes direct lineage switching in MLL/AF4 leukemia. Blood. 2022;140:1875–90. doi: 10.1182/blood.2021015036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baron VT, Pio R, Jia Z, Mercola D. Early Growth Response 3 regulates genes of inflammation and directly activates IL6 and IL8 expression in prostate cancer. Br J Cancer. 2015;112:755–64. doi: 10.1038/bjc.2014.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Healy S, Khan P, Davie JR. Immediate early response genes and cell transformation. Pharmacol Ther. 2013;137:64–77. doi: 10.1016/j.pharmthera.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 50.O’Byrne S, Elliott N, Rice S, Buck G, Fordham N, Garnett C, et al. Discovery of a CD10-negative B-progenitor in human fetal life identifies unique ontogeny-related developmental programs. Blood. 2019;134:1059–71. doi: 10.1182/blood.2019001289. [DOI] [PubMed] [Google Scholar]
- 51.Boller S, Grosschedl R. The regulatory network of B-cell differentiation: a focused view of early B-cell factor 1 function. Immunol Rev. 2014;261:102–15. doi: 10.1111/imr.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanz E, Muñoz-A N, Monserrat J, Van-Den-Rym A, Escoll P, Ranz I, et al. Ordering human CD34+CD10-CD19+ pre/pro-B-cell and CD19- common lymphoid progenitor stages in two pro-B-cell development pathways. Proc Natl Acad Sci USA. 2010;107:5925–30. doi: 10.1073/pnas.0907942107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen C, Yu W, Alikarami F, Qiu Q, Chen CH, Flournoy J, et al. Single-cell multiomics reveals increased plasticity, resistant populations, and stem-cell-like blasts in KMT2A-rearranged leukemia. Blood. 2022;139:2198–211. doi: 10.1182/blood.2021013442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao W, Kohler ME, Fry T, Ernst P. Does lineage plasticity enable escape from CAR-T cell therapy? Lessons from MLL-r leukemia. Exp Hematol. 2021;S0301-472X:00248–4. doi: 10.1016/j.exphem.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agraz-Doblas A, Bueno C, Bashford-Rogers R, Roy A, Schneider P, Bardini M, et al. Unraveling the cellular origin and clinical prognostic markers of infant B-cell acute lymphoblastic leukemia using genome-wide analysis. Haematologica. 2019;104:1176–88. doi: 10.3324/haematol.2018.206375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khabirova E, Jardine L, Coorens THH, Webb S, Treger TD, Engelbert J, et al. Single-cell transcriptomics reveals a distinct developmental state of KMT2A-rearranged infant B-cell acute lymphoblastic leukemia. Nat Med 2022;28:743–751. [DOI] [PMC free article] [PubMed]
- 57.Rice S, Jackson T, Crump NT, Fordham N, Elliott N, O’Byrne S, et al. A human fetal liver-derived infant MLL-AF4 acute lymphoblastic leukemia model reveals a distinct fetal gene expression program. Nat Commun. 2021;12:6905. doi: 10.1038/s41467-021-27270-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All generated datasets are available from the corresponding authors on reasonable request. MACE-Seq data are available at GEO with accession code GSE225710. ChIP-Seq data are available at GEO with accession code GSE205652.