Abstract
With the advent of long-read sequencing, previously unresolvable genomic elements are being revisited in an effort to generate fully complete reference genomes. One such element is ribosomal DNA (rDNA), the highly conserved genomic region that encodes rRNAs. Genomic structure and content of the rDNA are variable in both prokarya and eukarya, posing interesting questions about the biology of rDNA. Here, we consider the types of variation observed in rDNA – including locus structure and number, copy number, and sequence variation – and their known phenotypic consequences. With recent advances in long-read sequencing technology, incorporating the full rDNA sequence into reference genomes is within reach. This knowledge will have important implications for understanding rDNA biology within the context of cell physiology and whole-organism phenotypes.
Functions of the rDNA locus
Ribosome biogenesis is a universal and essential cellular process. Ribosomes consist of ribosomal proteins and rRNAs; the latter of which perform both structural and catalytic functions. The high cellular demand for ribosomes necessitates high abundance of rRNAs; 86% of total cellular RNA in bacteria and 60% of active transcription in eukaryotes are dedicated to rRNA [1,2]. To support such high levels of rRNA production, the genetic loci that encode the rRNAs (rDNA) are often found in many copies; typically arranged as tandem repeats in eukaryotes. The importance of rDNA is not limited to ribosome biogenesis, and rDNA (see Glossary) is also implicated in genome organization, genome stability, and regulation of replication and transcription [3–5]. Despite its large size and impact on cellular physiology and proliferation, rDNA is often absent from reference genomes.
In spite of the fundamental essentiality of the rRNAs, variation in the structure and sequence of the rDNA is ubiquitous (Figure 1, Key figure). The rRNA genes differ between species not only in genomic location and organization, but also in sequence of both coding and associated noncoding regions. Many rDNA properties are understudied – in no small part because some features are more easily assessed than others (e.g., rDNA locus position is more easily visualized than copy number within a locus) – and the interaction of different types of variation is even more rarely studied. A holistic knowledge of rDNA variation is nevertheless important; sequence variation, copy number variation, and locus number variation may well interact in affecting phenotype. For example, differing rDNA locus numbers drive speciation events in fish [6], while differing rDNA copy numbers affect lung cancer risk in smokers [7]. Questions of how copy number distribution or sequence variant distribution across multiple rDNA loci plays into either of these phenomena remain unexplored. A specific phenomenon that could feasibly interface between two or more categories of rDNA variation is nucleolar dominance, in which whole rDNA arrays are silenced in a heritable fashion in plants [8].
Variation of rDNA structure across kingdoms
Although rRNA genes are highly conserved across all kingdoms, eukaryotic and prokaryotic rDNA architecture differs. In bacteria, the ribosome consists of a small 30S subunit, containing a 16S rRNA, and a large 50S subunit, containing 23S and 5S rRNAs. These three rRNAs are encoded in rRNA or rrn operons in bacterial genomes, which are present in one to 15 copies per genome [9], with one being the most common. Copies encoding all three rRNAs are scattered throughout the genome and not necessarily identical in sequence. By contrast, the eukaryotic ribosome consists of a small 40S subunit, containing the 16–18S rRNA, and a 60S subunit, containing the 5S, 5.8S, and 25–28S rRNAs (Figure 2). The 18S, 5.8S, and 28S are typically encoded in a cotranscribed operon, known as the 45S in humans, while the 5S is encoded and transcribed separately. The 45S operon is commonly structured as a tandem array and a given genome may encode one or more 45S arrays on one or more chromosomes. In some species, such as the yeast Saccharomyces cerevisiae, the 5S is encoded next to the 45S in each unit of an array, an arrangement termed linked or L-type rDNA. This contrasts to S-type rDNA arrangements, in which the 5S rRNA gene is present in its own separate tandem array; S-type rDNA is found in many organisms, including humans and flies. L-type arrangements are less common, and observed in <5% of plant species and select arthropods and crustaceans [10,11].
Figure 2. Anatomy of a ribosomal DNA (rDNA) repeat unit.
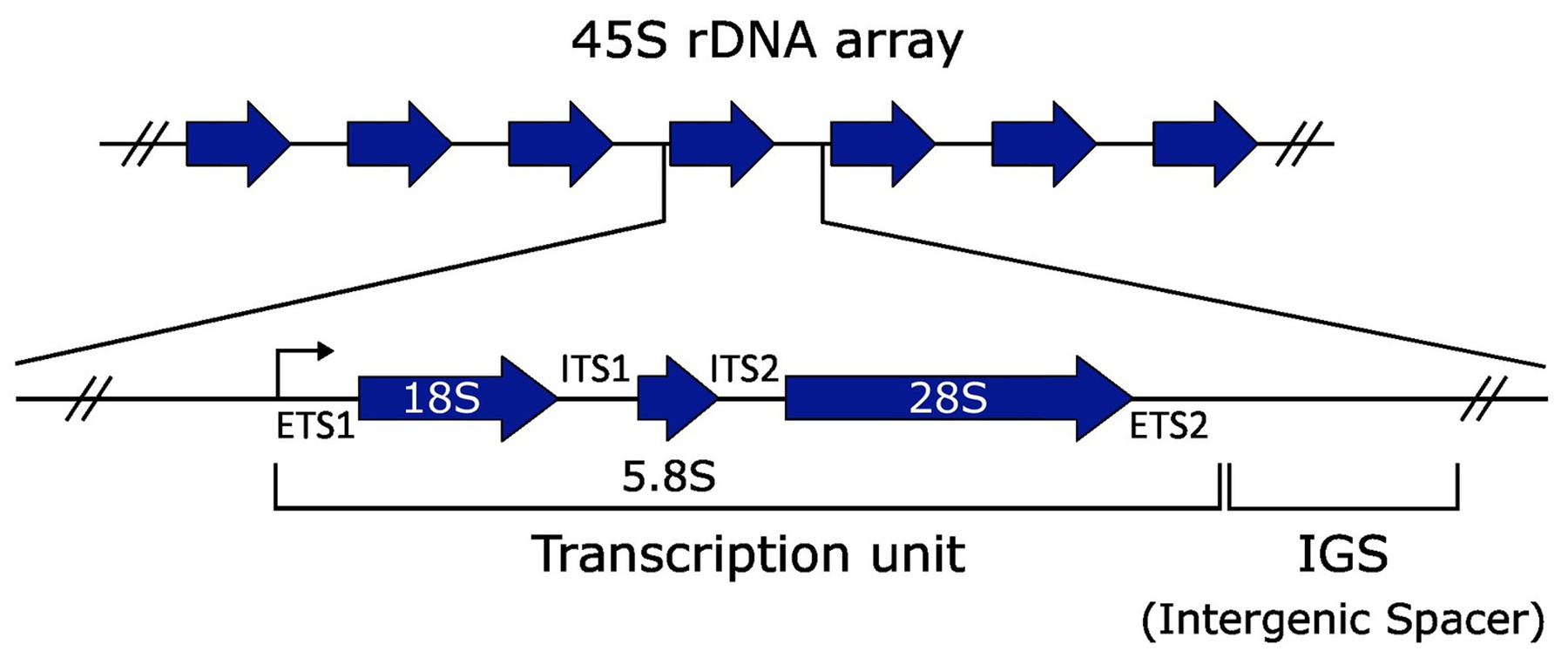
Example of a 45S rDNA repeat unit from an organism with S-type rDNA. The repeat units are represented in a tandem orientation. Each repeat unit contains a transcription unit, consisting of the 18S, 5.8S, and 28S rRNAs that are cotranscribed with two external transcribed spacers (ETS1 and ETS2) and two internal transcribed spacers (ITS1 and ITS2) that are removed post-transcriptionally. Each repeat unit also has an intergenic spacer (IGS) that is not part of the rRNA transcription unit, and may contain regulatory sequences, enhancers of the rRNA promoter, pseudogenes, origins of replication, and small repeat sequences such as Alu and short interspersed nuclear element (SINE) elements, depending on the organism.
rDNA arrays vary in chromosomal location and the number of loci per genome. rDNA can be located in terminal (subtelomeric), pericentromeric, or interstitial positions within the chromosome [11,12]. In animals, the 45S is most frequently found terminally, whereas the 5S shows less bias in positioning [12]. Even closely related species differ in rDNA array number and location. For example, species of the Mus subgenus Mus have six to 40 rDNA loci located across 40 diploid chromosomes, with one species carrying rDNA on every chromosome [13]. Different numbers of rDNA loci may have phenotypic consequences, as in certain fish, in which increased numbers of rDNA loci are found in individuals living in polluted waters [14]. Even in humans, where rDNA arrays are present on five chromosomes (13, 14, 15, 21, and 22) and thus 10 diploid locations, up to four of these locations lack measurable rDNA sequence in some samples [15]. rDNA arrays also exhibit structural variation such as palindromic and inverted rDNA repeat units, the prevalence of which is still under debate. Some reports found substantial structural variation [16,17], while recent long-read sequencing studies have reported that most sequential rDNA units are in the same orientation [18,19]. The questions of how frequently rDNA locus loss or nontandem arrangements occur have not yet been explored in depth, due to technical limitations.
rDNA sequence variation
The sequence of the rDNA repeat unit is highly homogeneous between individuals of a species [20]. This high sequence conservation is maintained by concerted evolution, driven by intrachromosomal homologous recombination and gene conversion. Nevertheless, sequence variation exists. For example, in Arabidopsis thaliana, there are numerous subtypes of each rRNA gene; some of which are expressed and others are not [21]. Beyond germline variation in rDNA sequence, rounds of mitotic cell division can result in de novo variation in the rDNA, found in both healthy and cancerous tissues in humans [22,23]. In mice, humans, and plants, rRNA coding variants can have tissue-specific expression, suggesting that some variants affect ribosome function [24–26]. In bacteria, rRNA coding variants are differentially expressed under conditions of nutritional stress [27]. The existence of condition- and tissue- specific rRNA variants has clear implications for rRNA evolution and human disease [28]. However, the full extent to which rDNA sequence variation contributes to human diseases is unclear because rDNA geno-types are not considered in genotype–phenotype analyses; in fact, rDNA remains excluded from the human reference genome.
Most identified rDNA sequence variation occurs in the noncoding intergenic spacer (IGS). For example, primates have similarly sized intergenic spacers (24–30 kb) but the content of their IGS differs, such as the number of Alu or short interspersed nuclear element (SINE) elements or the presence of pseudogenes [94,95]. IGS length variants are common between strains or isolates of a species and may affect the rRNA gene promoter or enhancers in ways that change rRNA expression levels among the strains [96]. For example, among the individual plants of a largely isogenic A. thaliana population, 18 distinct IGS length variants are present [97]. Further, in humans, a 2-kb length variant in the IGS may stratify by population [19], the functional consequence of which is still unknown. Apart from their potential biological functions, polymorphisms in rDNA have played a crucial role in computationally assembling these arrays from long-read sequencing data [18,98].
Mobile genetic elements also contribute to rDNA sequence variation: retrotransposons disrupt rRNA genes in many organisms (Figure 1) [34]. The R1 and R2 retrotransposons are non-long terminal repeat (non-LTR) retrotransposons that insert into the 28S rRNA in many species of arthropods [35,36]. Anywhere from <5% to >80% of rDNA repeat units are disrupted among Drosophila species [37–39]. De novo insertion of retrotransposons changes the number of functional rDNA units, producing variation among individuals in a population [40]. These retrotransposon interruptions may not be completely without function: transcription of R2 retrotransposons in Drosophila melanogaster are required for maintaining wild-type rDNA copy number [29]. Further study into the mechanisms by which retrotransposons disrupt rDNA function but maintain rDNA copy number should help discern more generally how rDNA copy number is established and maintained at a certain, background-specific level.
Sequence variation is not limited to the 45S and is readily studied in organisms with separate 5S and 45S arrays because the 5S array is typically much smaller than the 45S. In humans, the 5S repeat unit is 2.3 kb, compared with the 43 kb of a 45S repeat unit [30], implying average array lengths of ~616 kb (~268 copies), while a given 45S array is expected to be 387 kb to 3.612 Mb [18,31]. Full-length capture of human 5S arrays with long-read sequencing is therefore feasible. Contiguous sequencing of the entire repetitive rDNA arrays is needed to fully characterize differences in individual repeat units, their locations and their orientations across the array. The 5S has been completely assembled in nematode species, where 5S repeat units with unique insertions or deletions were identified and mapped in Caenorhabditis elegans and Caenorhabditis briggsae [32]. The shorter length of the human 5S also makes it more amenable to copy number estimation by methods such as pulsed-field gel electrophoresis, an approach that is frequently not feasible for the 45S.
Prevalence of rDNA copy number variation
Compounding variation in rDNA locus number is variation in the total number of rDNA copies per genome. Total repeat copy number and distribution of these copies across multiple arrays vary among and within species. rDNA copy number estimation remains challenging. Short read sequencing is commonly used to estimate rDNA copy number, but results are highly sensitive to library preparation methods and prone to batch effects [31,33]. Other approaches include relative quantification with qPCR or quantitative hybridization, which have limited resolution. Droplet digital PCR, offering improved resolution and replicability compared with traditional PCR, has been used for rDNA copy number estimation in yeast and humans [22,41]. Regardless of measurement technique, rDNA copy number estimates require orthogonal validation.
Within species, wild strains of the laboratory model organisms such as S. cerevisiae and C. elegans vary substantially in their rDNA copy numbers (Figure 3). Human rDNA copy number variation is commonly estimated as 200–600 45S rDNA copies (per haploid genome) (Figure 3). Some studies have estimated minima as low as nine rDNA copies and maxima as high as 1500 rDNA copies [25,31,42], although later evidence suggests some of these extremes may be artifactual [31]. rDNA copy number variation reportedly stratifies by population [25].
Figure 3. Ribosomal DNA (rDNA) copy number variation is common and is associated with various phenotypes.
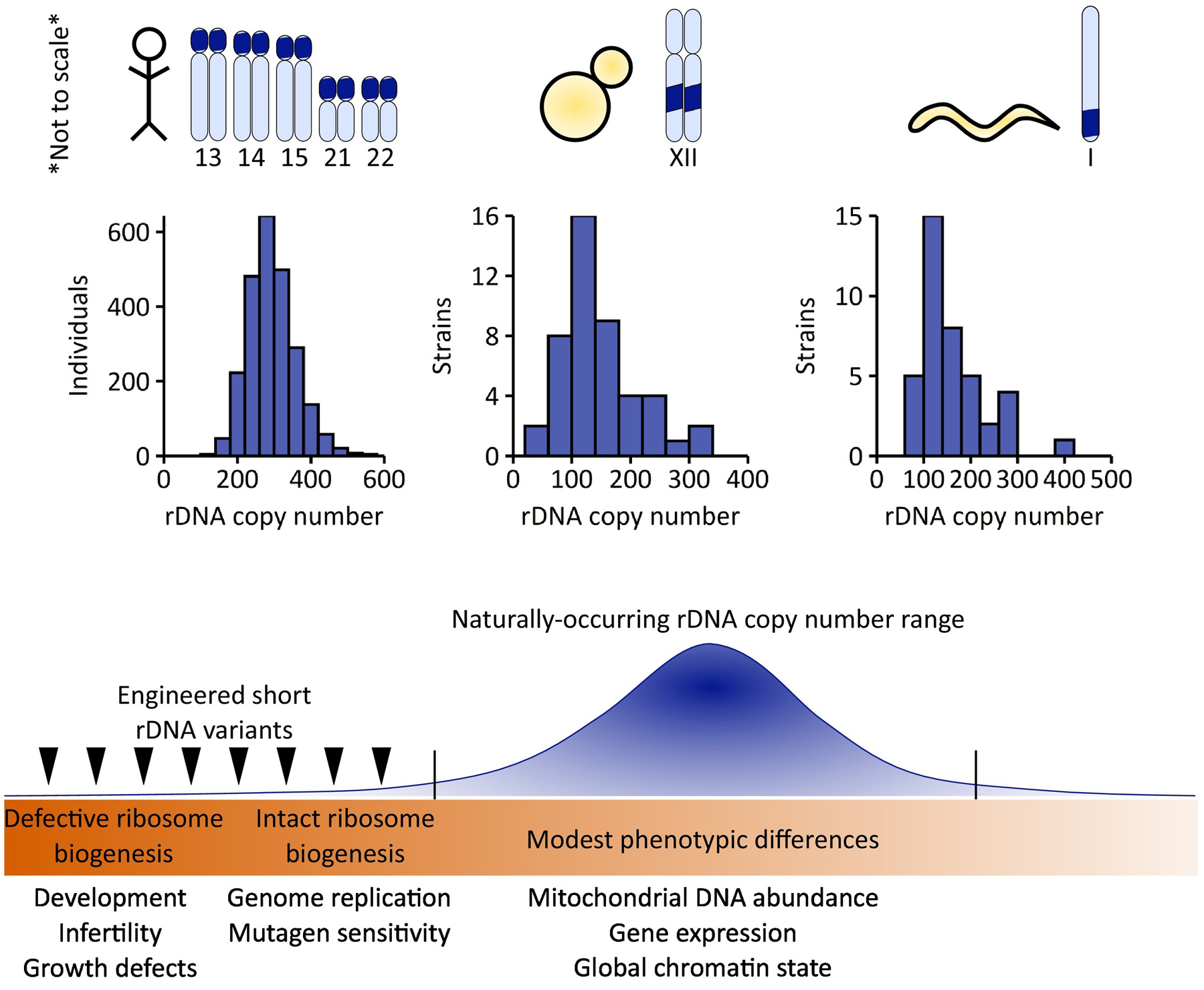
rDNA copy number varies between individual humans [31], strains of yeast [33], and wild isolates of Caenorhabditis elegans [93]. Organisms with insufficient rDNA copies to support ribosome biogenesis have developmental delays (flies, worms, and chickens) and in those organisms that do develop to adulthood, reduced or abolished fertility (flies and worms) [51,55]. Growth defects are observed in yeast with too few rDNA copies, and yeast with low rDNA copy number but no ribosome biogenesis defects have defects in genome replication [5].
In multicellular organisms, examples of tissue-specific copy number variation have been observed. The most widely known example is one of rDNA copy number increasing to meet ribosome biogenesis needs in oocytes of Xenopus laevis and other cold-blooded organisms. Across animal species, high levels of rDNA amplification are found in oocytes of ray-finned fish, caecilians, salamanders, frogs, crocodiles, and turtles [43,44]. Thus far, there is no evidence in animals for large rDNA copy number differences between somatic tissues within an individual, having been examined in both chickens and mice [22,45].
While rDNA copy number variation is common, there are cellular mechanisms that limit changes to rDNA copy number or restore rDNA copy number after copy loss. Unequal sister chromatid exchange is suppressed at the rDNA in yeast, reducing the likelihood of rDNA copy number changing during mitosis [46,47]. Also in yeast, rDNA copy number reduction can be rescued by amplification of extrachromosomal rDNA circles and reinsertion into the genome, restoring wild-type rDNA copy number over many generations [48–50]. In D. melanogaster, recovery of rDNA copies occurs quickly over fewer generations, through rDNA magnification [51], a process that uses unequal sister chromatid exchange [52–54]. It is currently unknown if similar mechanisms are in place to maintain rDNA copy number in human cells.
Phenotypic consequences of rDNA copy number variation
The many types of rDNA variation reviewed here would be of limited interest if there were no associated phenotypic differences. Recent years have seen increasing attention to the phenotypic consequences of rDNA copy number variation, particularly copy number reduction. Variation in rDNA copy number has reported association with global transcription, cancer, and aging (see ‘Human health relevance of rDNA copy number variation’). The most extreme reductions in rDNA copy number, unsurprisingly, have severe developmental phenotypes in model organisms. In C. elegans, complete deletion of the 45S rDNA produces worms that can complete embryo-genesis using maternally provided ribosomes but that arrest in the first larval stage [55]. The classic bobbed phenotype in D. melanogaster is the result of too few functional rDNA copies, presenting as tissue-specific defects, while more severe rDNA reduction in D. melanogaster is lethal. In yeast, low rDNA copy number increases DNA mutagen sensitivity and replication defects, while mutations in DNA replication machinery cause loss of rDNA copies [5,41]. Despite rDNA copy number reductions being the most well-studied copy number variants, if specific copy number thresholds exist for these developmental and tissue-specific phenotypes, these have not been defined consistently in any multicellular organism.
One potential impact of rDNA copy number variation is altered ribosome biogenesis. However, above a certain threshold of copies, differences in rDNA copy number largely do not correspond to differences in rRNA levels because only 50% or fewer rDNA copies are actively transcribed [56]. In some strains of D. melanogaster, as few as 10% of copies are transcriptionally active [57]. Variation in rDNA copy number may, however, affect tissue- or condition-specific regulation of ribosome biogenesis, including the example discussed earlier of amplification of rDNA circles during Xenopus oogenesis [58]. Ribosome biogenesis also increases in skeletal muscle in response to resistance exercise, and this increased ribosome biogenesis correlates with higher rDNA copy number [59,60]. Broad-scale coordinated assessments of rDNA copy number and tissue-specific rRNA levels, however, have not yet been performed.
Although rDNA copy number does not usually dictate rRNA abundance, the intriguing idea has been proposed that there are mechanisms coordinating stoichiometry of 5S and 45S gene copy number in humans and mice [61]. Such stoichiometry would suggest that complementary expansions and contractions of these arrays are somehow counted across many chromosomes to maintain a similar relative abundance of copies [42]. However, we recently demonstrated with thousands of newer, higher quality sequencing datasets that there is no meaningful or predictive covariation in 5S and 45S rDNA copy numbers in humans [31]. These results strongly support the notion that sequencing datasets must be carefully assessed for their appropriateness and utility in rDNA copy number estimation [31]. The question of what mechanisms coordinate stoichiometry of 5S and 45S rRNA transcripts therefore remains unresolved. 5S gene silencing is found in A. thaliana and X. laevis, but the extent of 5S silencing has not been studied in mammals [62]. Even in organisms with L-type rDNA – where copy numbers of the 45S and 5S genes are perforce the same – these genes are transcribed by separate RNA polymerases, leaving room for differential transcriptional regulation. Additionally, the 5S rRNA participates in cellular functions outside of cytoplasmic ribosomes. 5S rRNA associates with the tumor suppressor P53 in human cells, is imported into the mitochondria in some mammals, and accumulates in fish ovaries as a part of sex determination [63–65]. Having additional roles in the cell could mean that 5S rRNA levels do not have to be strictly equal to those of the 45S. The interplay between 5S-specific roles and ribosomal function remains to be investigated.
Human health relevance of rDNA copy number variation
Many human diseases have incompletely understood genetic underpinnings. Repetitive DNA regions such as the rDNA are underexplored sources of heritable variation with the potential to contribute to disease risk. A recent study determined that higher rDNA copy number, as measured in peripheral blood, associates with a higher risk of developing lung cancer in smokers [7]. This study was small (229 cancer cases) and does not report absolute rDNA copy numbers but opens the door to future studies of rDNA copy number and disease risk.
One of the most-reported associations of rDNA copy number variation with disease is with cancer. Multiple reports have observed reductions of 45S rDNA copy number in tumor samples [22,66–69]. It remains unresolved whether altered rDNA copy number is a cause or a consequence of cancer proliferation. In breast cancer, both gains and losses of rDNA copy number are found, so the changes are argued to be a consequence of increased genome instability [70]. Others have proposed that reductions in rDNA array size may facilitate rapid replication during cancer cell propagation [22]. These and other observations have positioned rDNA copy number assessment as relevant information in cancer therapy. The effectiveness of RNA polymerase I inhibitor CX-5461 as a cancer treatment depends on rDNA chromatin state [71]. A greater understanding of how rDNA copy number variation impacts the efficacy of RNA polymerase I inhibitors could therefore be valuable for personalized cancer treatments.
Other complex human diseases have been associated with rDNA copy number differences in a series of studies. For example, individuals with schizophrenia display both increased ribosome biogenesis and elevated rDNA copy number [72–74]. Aberrant regulation of rRNA biogenesis and rDNA amplification have been observed in patients with intellectual disability [75]. Individuals with Down syndrome, who have an extra rDNA array due to the additional copy of chromosome 21, have more active rDNA copies [76]. By contrast, rDNA copy number does not differ significantly in 1774 sibling pairs in which one sibling is affected by an autism spectrum disorder and the other is unaffected [31]. It is worth noting that the studies on schizophrenia, intellectual disability, and Down syndrome frequently had restricted sample sizes, and measurement of rDNA copy number was often performed with methods of limited resolution. Thus far, no causative relationship between rDNA copy number and a complex human disease has been identified.
As a final note, instability at the rDNA locus has implications for aging. rDNA instability is a hallmark of aging in yeast and can also be found in aging mammalian tissues. Instability can manifest in two different ways: with the formation of extrachromosomal rDNA circles (ERCs) [3,77], or through the loss of rDNA copies from breaks in the rDNA [78]. Accumulation of rDNA circles with age has been documented only in yeast. In D. melanogaster and humans, rDNA circles are present but do not accumulate with age [79,80]. In mammals, rDNA age associations present in contrasting studies as loss [81–84], no loss [85], and recently, even gains [86] in various tissues with age. Importantly, none of these mammalian studies were longitudinal, so rDNA copy numbers were not measured in the same individuals early and late in life. In Drosophila, old male flies exhibit heritable loss of rDNA copies in the germline [87], further evidence of the age-dependence of rDNA copy number, although whether or not similar loss of rDNA copies may be found in female flies or human germlines has not been determined.
Long-read sequencing: the future of rDNA characterization
Long-read sequencing holds unique promise for understanding location, sequence variation, structural variation, and copy number of the rDNA. Currently, rDNA arrays are often represented in reference genomes as either a single rDNA reference copy or an unassembled scaffold (detached from chromosomal context). Efforts to fully assemble rDNA arrays using long-read sequencing are underway but still face challenges. Recently, the C. elegans 5S rDNA array was successfully assembled with long-read sequencing, although this study failed to assemble across the 45S due to insufficiently long reads and insufficient sequence variation between 45S rDNA copies [32]. A separate effort to recomplete the C. elegans genome applied both PacBio and Nanopore sequencing but also failed to obtain any reads spanning the 45S array or to accurately estimate 45S repeat unit number using read depth [88]. In A. thaliana, a combined approach of BAC cloning, short-read, and long-read sequencing was successful in assembling one of the two rDNA arrays in the genome [26].
Human rDNA sequencing poses a greater problem than in these model organisms, due to multiple loci and larger repeat unit sizes. Nevertheless, if each rDNA in the human genome could be assembled with long reads, the dual tasks of determining the total number of rDNA repeat units per genome and the number of rDNA repeat units per individual array could be accomplished. Recent efforts to do this for a functionally haploid human cell line have had commendable success, with two of the five arrays successfully assembled [18]. However, despite long reads being used to generate high-quality reference genomes for many other animal and human genomes, many of these studies fail to even mention assembling or locating the rDNA as a possible application [89,90]. Complete characterization of rDNA arrays has many fascinating applications for rDNA biology, including distinguishing between rDNA copy number variation caused by specific arrays or extrachromosomal variation, or cataloguing R1 and R2 retrotransposon distribution in D. melanogaster rDNA. As the accuracy and length of long-read sequencing continue to improve, this technology holds promise for rDNA copy number estimation of individual loci within an individual’s genome. Once accomplished, long read sequencing paired with additional techniques could be the key to finally integrating rDNA variation into the scholarship of genotype–phenotype association.
Concluding remarks
Variation in the rDNA is common, and the various types of rDNA variation are often considered separately in individual studies. Exciting advances have recently been made in determining how many loci carry rDNA in human cells, and in determining the extent of noncanonical rDNA repeat units present [15,18,19]. Sequence variation in rDNA is becoming increasingly appreciated, and functional consequences of rRNA coding variants are being explored in the context of specialized ribosomes [91,92]. Finally, copy number variation has received a lot of attention in recent years, especially in light of rDNA copy number changes found in various cancer types. Among these recent discoveries, it is particularly exciting that increased rDNA copy number associates with increased lung cancer risk in smokers [7], as this observation suggests that rDNA copy number may be a disease risk factor for cancer and possibly other diseases that are impacted by environmental factors.
Moving forward, long-read sequencing will enable a more holistic understanding of the structure, sequence, and copy numbers of rDNA sequences. There are many applications of having the rDNA fully assembled and sequenced(see Outstanding questions). For example, some rDNA sequence variants in the human genome are chromosome specific [18], so determining how many copies carry such a variant and when and where the variant arrays are expressed will be important in predicting the role of rRNA-coding variants. At this point, even the process of assembling the rDNA from long reads has resulted in novel computational methods for aligning these highly homogenous, repetitive sequences [18]. Taken together, the more we know about the rDNA loci and other previously inaccessible highly repetitive genomic regions, the better we will be able to understand fundamental biology and human health and disease.
Outstanding questions.
How many rDNA copies are truly necessary for full organismal development and optimal fitness?
Are there different thresholds of minimal rDNA copy numbers for specific tissues, conditions, or organismal stages?
Does rDNA copy number variation in the naturally occurring range affect organismal traits, such as fitness or development?
Are rDNA copy number changes drivers or a consequence of altered phenotypes or disease states, such as cancer?
What are the consequences of rDNA structural variation or genomic arrangement for organismal fitness? How do the types of rDNA variation interact with one another? For example, what is the distribution of rDNA copy number or coding sequence variation among different arrays in humans? Could the manner in which sequence variants are distributed along and among chromosomes impact phenotype, or perhaps heritability of this variation?
Key figure
The many types of ribosomal DNA (rDNA) variation may be accessible by long-read sequencing
Figure 1.
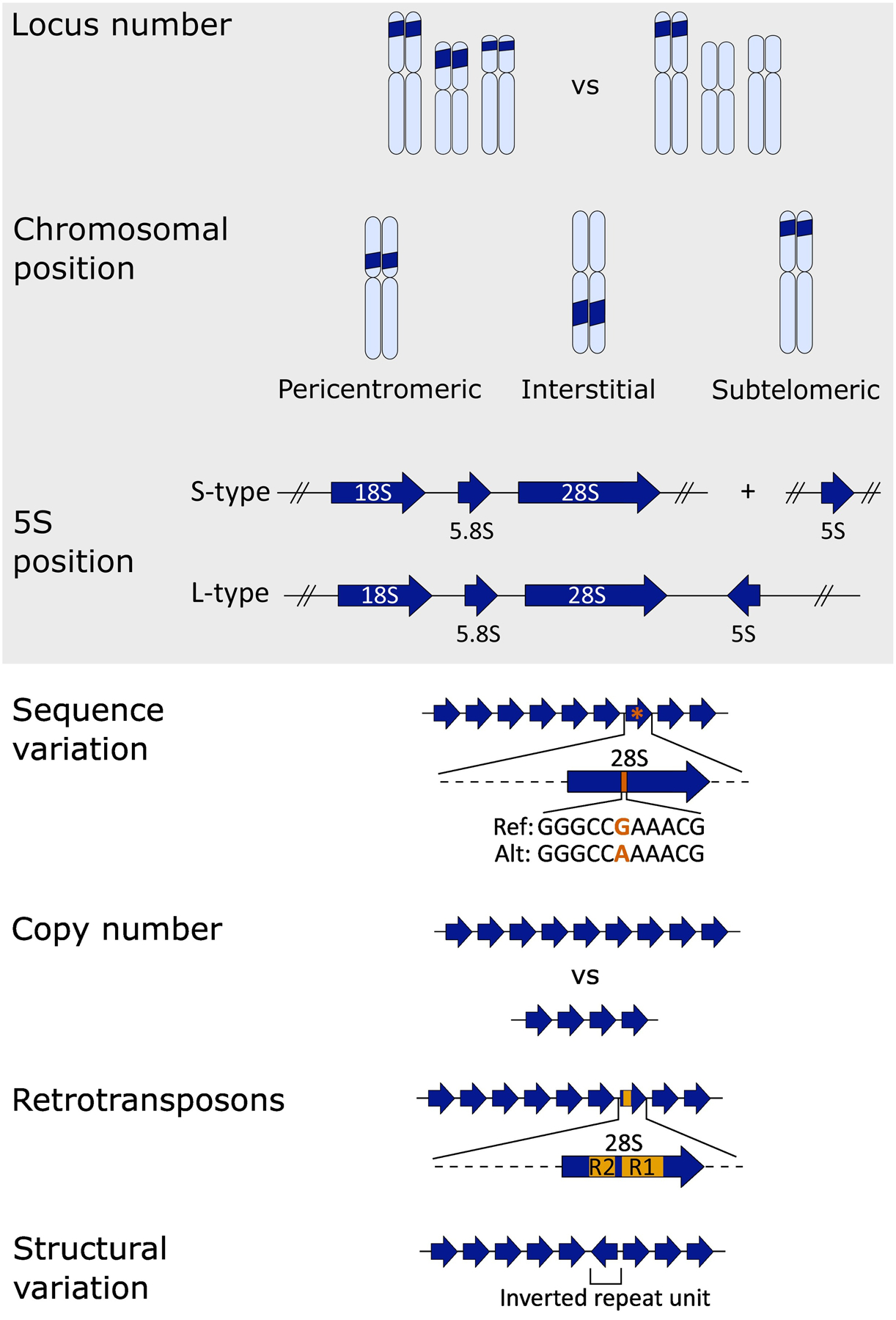
Some variation, such as the number of rDNA loci, chromosomal position of rDNA loci, and the position of the 5S with respect to the other rRNAs, have been readily ascertained for decades through use of in situ hybridization methods. Long-read sequencing has the potential to demystify rDNA variation types such as sequence variation, copy number variation, and structural variation – especially with regard to where different variants fall in the context of each rDNA array in the genome. In organisms with many rDNA arrays per genome (e.g., humans) long-read sequencing holds the potential to resolve the rDNA copy numbers of each array as well as the overall rDNA content per haploid genome, through ultralong reads that span whole arrays or by tiling long reads across the array using unique variants for anchoring.
Highlights.
Ribosomal (r)DNA has historically been excluded from reference genomes because of its repetitive nature and large size.
Advances in long-read sequencing may be the key to fully integrated assessment of variation at the rDNA, including sequence, structural, and copy number variation.
rDNA variation may impact human health with possible roles in cancer and aging.
Glossary
- Ribosomal (r)DNA
the genetic region encoding rRNAs, found as a long tandem array in most eukaryotes.
- L-type rDNA
an arrangement of rDNA in eukaryotes, in which the 5S and 45S rRNA genes are found together within repeat units of an array.
- S-type rDNA
an arrangement of rDNA in eukaryotes, in which the 5S rRNA genes are encoded in a separate array from the 45S.
- Intergenic spacer (IGS)
the portion of the rDNA repeat unit that is not transcribed. It can contain the promoter and enhancers for the rRNA genes, in addition to origins of replication, non-rRNA genes, and noncoding RNAs.
Footnotes
Declaration of interests No interests are declared.
References
- 1.Warner JR (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci 24, 437–440 [DOI] [PubMed] [Google Scholar]
- 2.Bremer H and Dennis PP (2008) Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus Published online October 8, 2008. 10.1128/ecosal.5.2.3 [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi T (2008) A new role of the rDNA and nucleolus in the nucleus – rDNA instability maintains genome integrity. BioEssays 30, 267–272 [DOI] [PubMed] [Google Scholar]
- 4.Yu S and Lemos B (2016) A portrait of ribosomal DNA contacts with Hi-C reveals 5S and 45S rDNA anchoring points in the folded human genome. Genome Biol. Evol 8, 3545–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwan EX et al. (2021) Coordination of genome replication and anaphase entry by rDNA copy number in S. cerevisiae. bioRxiv Published online April 14, 2021. 10.1101/2021.02.25.432950 [DOI] [Google Scholar]
- 6.Symonová R et al. (2013) Genome differentiation in a species pair of coregonine fishes: an extremely rapid speciation driven by stress-activated retrotransposons mediating extensive ribosomal DNA multiplications. BMC Evol. Biol 13, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosgood HD III et al. (2019) Variation in ribosomal DNA copy number is associated with lung cancer risk in a prospective cohort study. Carcinogenesis 40, 975–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pikaard CS (2014) Nucleolar dominance. eLS Published online January 14, 2015. 10.1002/9780470015902.a0005976.pub2 [DOI] [Google Scholar]
- 9.Stoddard SF et al. (2015) rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 43, D593–D598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drouin G and de Sá MM (1995) The concerted evolution of 5S ribosomal genes linked to the repeat units of other multigene families. Mol. Biol. Evol 12, 481–493 [DOI] [PubMed] [Google Scholar]
- 11.Garcia S et al. (2017) Cytogenetic features of rRNA genes across land plants: analysis of the Plant rDNA database. Plant J. 89, 1020–1030 [DOI] [PubMed] [Google Scholar]
- 12.Sochorová J et al. (2018) Evolutionary trends in animal ribosomal DNA loci: introduction to a new online database. Chromosoma 127, 141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cazaux B et al. (2011) Are ribosomal DNA clusters rearrangement hotspots? A case study in the genus Mus (Rodentia, Muridae). BMC Evol. Biol 11, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Araújo da Silva F et al. (2019) Effects of environmental pollution on the rDNAomics of Amazonian fish. Environ. Pollut 252, 180–187 [DOI] [PubMed] [Google Scholar]
- 15.van Sluis M et al. (2020) NORs on human acrocentric chromo-some p-arms are active by default and can associate with nucleoli independently of rDNA. Proc. Natl. Acad. Sci. U. S. A 117, 10368–10377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caburet S et al. (2005) Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res. 15, 1079–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim J-H et al. (2018) Variation in human chromosome 21 ribosomal RNA genes characterized by TAR cloning and long-read sequencing. Nucleic Acids Res. 46, 6712–6725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nurk S et al. (2021) The complete sequence of a human genome. bioRxiv Published online May 27, 2021. 10.1101/2021.05.26.445798 [DOI] [Google Scholar]
- 19.Hori Y et al. (2021) The human ribosomal DNA array is composed of highly homogenized tandem clusters. Genome Res. 31, 1971–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eickbush TH and Eickbush DG (2007) Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics 175, 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandrasekhara C et al. (2016) Chromosome-specific NOR inactivation explains selective rRNA gene silencing and dosage control in Arabidopsis. Genes Dev 30, 177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu B et al. (2017) Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet. 13, e1006771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohashi R et al. (2020) Frequent germline and somatic single nucleotide variants in the promoter region of the ribosomal RNA gene in Japanese lung adenocarcinoma patients. Cells 9, 2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tseng H et al. (2008) Mouse ribosomal RNA genes contain multiple differentially regulated variants. PLoS ONE 3, e1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parks MM et al. (2018) Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci. Adv 4, eaao0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sims J et al. (2021) Sequencing of the Arabidopsis NOR2 reveals its distinct organization and tissue-specific rRNA ribosomal variants. Nat. Commun 12, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurylo CM et al. (2018) Endogenous rRNA sequence variation can regulate stress response gene expression and phenotype. Cell Rep. 25, 236–248.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parks MM et al. (2019) Implications of sequence variation on the evolution of rRNA. Chromosome Res. Int. J. Mol. Supramol. Evol. Asp. Chromosome Biol 27, 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson JO et al. (2021) The retrotransposon R2 maintains Drosophila ribosomal DNA repeats. bioRxiv Published online July 12, 2021. 10.1101/2021.07.12.451825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sørensen PD and Frederiksen S (1991) Characterization of human 5S rRNA genes. Nucleic Acids Res. 19, 4147–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall AN et al. (2021) Thousands of high-quality sequencing samples fail to show meaningful correlation between 5S and 45S ribosomal DNA arrays in humans. Sci. Rep 11, 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding Q et al. (2021) Genomic architecture of 5S rDNA cluster and its variations within and between species. bioRxiv Published online February 18, 2021. 10.1101/2021.02.17.431734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morton EA et al. (2019) Challenges and approaches to genotyping repetitive DNA. G3 Genes Genomes Genet. 10, 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujiwara H (2015) Site-specific non-LTR retrotransposons. In Mobile DNA III, pp. 1147–1163, John Wiley & Sons [Google Scholar]
- 35.Roiha H et al. (1981) Arrangements and rearrangements of sequences flanking the two types of rDNA insertion in D. melanogaster. Nature 290, 749–753 [DOI] [PubMed] [Google Scholar]
- 36.Burke WD et al. (1998) Are retrotransposons long-term hitchhikers? Nature 392, 141–142 [DOI] [PubMed] [Google Scholar]
- 37.Jakubczak JL et al. (1992) Turnover of R1 (type I) and R2 (type II) retrotransposable elements in the ribosomal DNA of Drosophila melanogaster. Genetics 131, 129–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malik HS and Eickbush TH (1999) Retrotransposable elements R1 and R2 in the rDNA units of Drosophila mercatorum: abnormal abdomen revisited. Genetics 151, 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raje HS et al. (2018) R1 retrotransposons in the nucleolar organizers of Drosophila melanogaster are transcribed by RNA polymerase I upon heat shock. Transcription 9, 273–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pérez-González CE and Eickbush TH (2002) Rates of R1 and R2 retrotransposition and elimination from the rDNA locus of Drosophila melanogaster. Genetics 162, 799–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salim D et al. (2017) DNA replication stress restricts ribosomal DNA copy number. PLoS Genet. 13, e1007006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibbons JG et al. (2015) Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc. Natl. Acad. Sci 112, 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian Q et al. (2001) Function of basonuclin in increasing transcription of the ribosomal RNA genes during mouse oogenesis. Development 128, 407–416 [DOI] [PubMed] [Google Scholar]
- 44.Davidian A et al. (2021) On some structural and evolutionary aspects of rDNA amplification in oogenesis of Trachemys scripta turtles. Cell Tissue Res. 383, 853–864 [DOI] [PubMed] [Google Scholar]
- 45.Ritossa FM et al. (1966) On the chromosomal distribution of DNA complementary to ribosomal and soluble RNA. Natl. Cancer Inst. Monogr 23, 449–472 [PubMed] [Google Scholar]
- 46.Kobayashi T et al. (2004) SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117, 441–453 [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi T and Ganley ARD (2005) Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309, 1581–1584 [DOI] [PubMed] [Google Scholar]
- 48.Mansisidor A et al. (2018) Genomic copy-number loss is rescued by self-limiting production of DNA circles. Mol. Cell 72, 583–593.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iida T and Kobayashi T (2019) RNA polymerase I activators count and adjust ribosomal RNA gene copy number. Mol. Cell 73, 645–654.e13 [DOI] [PubMed] [Google Scholar]
- 50.Iida T and Kobayashi T (2019) How do cells count multi-copy genes?: “Musical Chair” model for preserving the number of rDNA copies. Curr. Genet 65, 883–885 [DOI] [PubMed] [Google Scholar]
- 51.Ritossa FM (1968) Unstable redundancy of genes for ribosomal RNA. Proc. Natl. Acad. Sci. U. S. A 60, 509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tartof KD (1974) Unequal mitotic sister chromatid exchange as the mechanism of ribosomal RNA gene magnification. Proc. Natl. Acad. Sci 71, 1272–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohen S et al. (2005) Evidence for rolling circle replication of tandem genes in Drosophila. Nucleic Acids Res. 33, 4519–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aldrich JC and Maggert KA (2015) Transgenerational inheritance of diet-induced genome rearrangements in Drosophila. PLoS Genet 11, e1005148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cenik ES et al. (2019) Maternal ribosomes are sufficient for tissue diversification during embryonic development in C. elegans. Dev. Cell 48, 811–826.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gagnon-Kugler T et al. (2009) Loss of human ribosomal gene CpG methylation enhances cryptic RNA polymerase II transcription and disrupts ribosomal RNA processing. Mol. Cell 35, 414–425 [DOI] [PubMed] [Google Scholar]
- 57.Ye J and Eickbush TH (2006) Chromatin structure and transcription of the R1- and R2-Inserted rRNA genes of Drosophila melanogaster. Mol. Cell. Biol 26, 8781–8790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roger B et al. (2002) rDNA transcription during Xenopus laevis oogenesis. Biochem. Biophys. Res. Commun 290, 1151–1160 [DOI] [PubMed] [Google Scholar]
- 59.Figueiredo VC and McCarthy JJ (2018) Regulation of ribo-some biogenesis in skeletal muscle hypertrophy. Physiology 34, 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Figueiredo VC et al. (2021) Genetic and epigenetic regulation of skeletal muscle ribosome biogenesis with exercise. J. Physiol 599, 3363–3384 [DOI] [PubMed] [Google Scholar]
- 61.Gibbons JG et al. (2014) Ribosomal DNA copy number is coupled with gene expression variation and mitochondrial abundance in humans. Nat. Commun. Lond 5, 4850. [DOI] [PubMed] [Google Scholar]
- 62.Douet J and Tourmente S (2007) Transcription of the 5S rRNA heterochromatic genes is epigenetically controlled in Arabidopsis thaliana and Xenopus laevis. Heredity 99, 5–13 [DOI] [PubMed] [Google Scholar]
- 63.Nishimura K et al. (2015) Perturbation of ribosome biogenesis drives cells into senescence through 5S RNP-mediated p53 activation. Cell Rep. 10, 1310–1323 [DOI] [PubMed] [Google Scholar]
- 64.Diaz de Cerio O et al. (2012) 5S rRNA and accompanying proteins in gonads: powerful markers to identify sex and reproductive endocrine disruption in fish. Environ. Sci. Technol 46, 7763–7771 [DOI] [PubMed] [Google Scholar]
- 65.Smirnov A et al. (2011) Biological significance of 5S rRNA import into human mitochondria: role of ribosomal protein MRP-L18. Genes Dev. 25, 1289–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang M and Lemos B (2017) Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS Genet. 13, e1006994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Udugama M et al. (2018) Ribosomal DNA copy loss and repeat instability in ATRX-mutated cancers. Proc. Natl. Acad. Sci. U. S. A 115, 4737–4742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feng L et al. (2020) Ribosomal DNA copy number is associated with P53 status and levels of heavy metals in gastrectomy specimens from gastric cancer patients. Environ. Int 138, 105593. [DOI] [PubMed] [Google Scholar]
- 69.Lou J et al. (2021) Environmentally induced ribosomal DNA (rDNA) instability in human cells and populations exposed to hexavalent chromium [Cr (VI)]. Environ. Int 153, 106525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Valori V et al. (2019) Human rDNA copy number is unstable in metastatic breast cancers. Epigenetics 15, 85–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Son J et al. (2020) rDNA chromatin activity status as a biomarker of sensitivity to the RNA polymerase I transcription inhibitor CX-5461. Front. Cell Dev. Biol 8, 568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Veiko NN et al. (2003) Quantitation of repetitive sequences in human genomic DNA and detection of an elevated ribosomal repeat copy number in schizophrenia: the results of molecular and cytogenetic analyses. Mol. Biol 37, 349–357 [PubMed] [Google Scholar]
- 73.Krzyżanowska M et al. (2015) Ribosomal DNA transcription in the dorsal raphe nucleus is increased in residual but not in paranoid schizophrenia. Eur. Arch. Psychiatry Clin. Neurosci 265, 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chestkov IV et al. Abundance of ribosomal RNA gene copies in the genomes of schizophrenia patients. Schizophr. Res 197, 305–314 [DOI] [PubMed] [Google Scholar]
- 75.Kolesnikova IS et al. (2018) Alteration of rRNA gene copy number and expression in patients with intellectual disability and heteromorphic acrocentric chromosomes. Egypt. J. Med. Hum. Genet 19, 129–134 [Google Scholar]
- 76.Lyapunova NA et al. (2017) Viability of carriers of chromosomal abnormalities depends on genomic dosage of active ribosomal genes (rRNA genes). Russ. J. Genet 53, 703–711 [Google Scholar]
- 77.Sinclair DA and Guarente L (1997) Extrachromosomal rDNA circles – a cause of aging in yeast. Cell 91, 1033–1042 [DOI] [PubMed] [Google Scholar]
- 78.Warmerdam DO et al. (2016) Breaks in the 45S rDNA lead to recombination-mediated loss of repeats. Cell Rep. 14, 2519–2527 [DOI] [PubMed] [Google Scholar]
- 79.Cohen S et al. (2003) Extrachromosomal circular DNA of tandemly repeated genomic sequences in Drosophila. Genome Res. 13, 1133–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shoura MJ et al. (2017) Intricate and cell type-specific populations of endogenous circular DNA (eccDNA) in Caenorhabditis elegans and Homo sapiens. G3 Genes Genomes Genet 7, 3295–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johnson R and Strehler BL (1972) Loss of genes coding for ribosomal RNA in ageing brain cells. Nature 240, 412–414 [DOI] [PubMed] [Google Scholar]
- 82.Gaubatz JW and Cutler RG (1978) Age-related differences in the number of ribosomal RNA genes of mouse tissues. Gerontology 24, 179–207 [DOI] [PubMed] [Google Scholar]
- 83.Strehler BL et al. (1979) Loss of hybridizable ribosomal DNA from human post-mitotic tissues during aging: I. Age-dependent loss in human myocardium. Mech. Ageing Dev 11, 371–378 [DOI] [PubMed] [Google Scholar]
- 84.Zafiropoulos A et al. (2005) Preferential loss of 5S and 28S rDNA genes in human adipose tissue during ageing. Int. J. Biochem. Cell Biol 37, 409–415 [DOI] [PubMed] [Google Scholar]
- 85.Peterson CRD et al. (1984) Constancy of ribosomal RNA genes during aging of mouse heart cells and during serial passage of WI-38 cells. Arch. Gerontol. Geriatr 3, 115–125 [DOI] [PubMed] [Google Scholar]
- 86.Watada E et al. (2020) Age-dependent ribosomal DNA variations in mice. Mol. Cell. Biol 40, e00368–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu KL et al. (2018) Transgenerational dynamics of rDNA copy number in Drosophila male germline stem cells. eLife 7, e32421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yoshimura J et al. (2019) Recompleting the Caenorhabditis elegans genome. Genome Res. 29, 1009–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rhie A et al. (2021) Towards complete and error-free genome assemblies of all vertebrate species. Nature 592, 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beyter D et al. (2021) Long-read sequencing of 3,622 Icelanders provides insight into the role of structural variants in human diseases and other traits. Nat. Genet 53, 779–786 [DOI] [PubMed] [Google Scholar]
- 91.Xue S and Barna M (2012) Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol 13, 355–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Haag ES and Dinman JD (2019) Still searching for specialized ribosomes. Dev. Cell 48, 744–746 [DOI] [PubMed] [Google Scholar]
- 93.Thompson O et al. (2013) The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res. 23, 1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gonzalez IL et al. (1993) Fixation times of retroposons in the ribosomal DNA spacer of human and other primates. Genomics 18, 29–36 [DOI] [PubMed] [Google Scholar]
- 95.Agrawal S and Ganley ARD (2018) The conservation landscape of the human ribosomal RNA gene repeats. PLOS ONE 13, e0207531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weider LJ et al. (2005) The functional significance of ribosomal (r)DNA variation: impacts on the evolutionary ecology of organisms. Annu. Rev. Ecol. Evol. Syst 36, 219–242 [Google Scholar]
- 97.Havlová K et al. (2016) Variation of 45S rDNA intergenic spacers in Arabidopsis thaliana. Plant Mol. Biol 92, 457–471 [DOI] [PubMed] [Google Scholar]
- 98.Luttermann T et al. (2021) Establishment of a near-contiguous genome sequence of the citric acid producing yeast Yarrowia lipolytica DSM 3286 with resolution of rDNA clusters and telomeres. NAR Genomics Bioinforma. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]


