Abstract
Vishva Dixit recounts his favorite discoveries after 30-plus years studying the proteins that allow infected, damaged, or obsolete cells to die.
As a child growing up in a village in Kenya in the 50s and 60s, I had vivid dreams of exploring and finding new lands. As an adult, the realm of biomedicine provided the exhilaration, joy, trepidation, and uncertainty of exploring the unknown. My career had a meandering start. I completed medical school in Kenya and then pathology training and a fellowship in biochemistry at Washington University in St. Louis, Missouri. Subsequently, I have had two jobs—the first one in academia at the University of Michigan, Ann Arbor and the second in industry, working at Genentech in South San Francisco. I have been blessed with many discoveries, but two stand out: Illuminating how death receptors signal and finding the noncanonical inflammasome pathway. This article is a personal recollection of the ups and downs surrounding these two discoveries.
DEATH RECEPTOR SIGNALING
As a pathologist studying blood coagulation in the late 1980s, I was ready to shift fields to something new and exciting. Apoptosis, an immunologically silent form of cell death, looked promising. Pathologists Andrew Wyllie, Alastair Currie, and John Kerr had recently characterized apoptosis morphologically in mammalian tissues, and evidence for an underlying genetic program was emerging. I wondered whether I could tease apart the signaling pathway eliciting the apoptotic demise.
I had a modicum of biochemistry training, but I lacked a facile and scalable cell culture system for triggering apoptosis in a synchronized manner. For a time, finding such a system felt like a fool’s errand. Shin Yonehara and Peter Krammer solved that problem in 1989 when they described a monoclonal antibody that induced apoptosis in cultured cells. The race was now on to identify the cell surface receptor that the antibody engaged.
The formidable Shigekazu Nagata cloned and characterized the receptor in 1991, thereafter named Fas/Apo-1/CD95. The receptor was highly similar in sequence to the major receptor for tumor necrosis factor (TNF), which could also be engaged in particular cell lines to induce apoptosis. For simplicity, I will dub these the “death receptors.”
Intriguingly, the cytosolic portion of the death receptors did not reveal an obvious mode for signaling apoptosis. Neither death receptor had an enzymatic domain, such as a kinase, nor a binding motif for posttranslational modifications. Renowned molecular biologist David Goeddel nonetheless noted that they both contained a conserved cytosolic segment of approximately 80 residues. Mutation of these residues disabled the death receptors and prevented them from signaling apoptosis, leading his group to name this segment the “death domain” (abbreviated DD) in 1993. I now had a system to tackle the “death pathway” downstream of death receptors.
I had some major competition in the form of Nagata, Krammer, and Goeddel, but I decided to throw caution to the wind. This was my opportunity to pursue my childhood dream of finding something completely new. I had come too far not to swing for the fences.
I reasoned that the only way to battle the titans in their arena was to find inhibitors of cell death. At the time, I was convinced that sheer grit and determination would yield inhibitors from among the thousands of commercially available chemicals—that is, the Sigma-Aldrich catalog. These inhibitors would be the tools that would allow me to illuminate the mysterious components downstream from death receptors.
The hard work proved the easy part. A bright and ambitious MD/PhD student, Muneesh Tewari, and technician, Karen O’Rourke, helped establish the system. We induced apoptosis easily using recombinant TNF from Genentech or Fas agonist antibody from Yonehara. The ensuing mayhem under the microscope was enthralling to watch. The cells underwent an unforgettable and violent dance of death. The nucleus condensed, membranes blistered, and then the cell fragmented into membrane-enclosed sacs, known as apoptotic bodies. However, our efforts to identify a chemical inhibitor of apoptosis floundered. Whatever we tested either did not work or had only a marginal effect. We wondered whether components of the pathway might have built-in redundancies. If so, the project was dead.
Meanwhile, seemingly disparate lines of investigation were providing important clues to the cell death field. In 1992, Roy Black and Nancy Thornberry described a most unusual cysteine protease, which they called interleukin-1 (IL-1)–converting enzyme or ICE. Uncommonly specific, ICE cleaved IL-1 after aspartic acid residues. It was also remarkably similar in sequence to the protein encoded by the Caenorhabditis elegans death gene, ced3, that Robert Horvitz had identified in 1986. Horvitz’s laboratory showed in 1993 that ced3 encoded a protease with substrate specificity and properties similar to ICE.
Did death receptors activate an ICE-like protease? We realized that we had to test this notion quickly because the field was advancing swiftly. We knew that David Pickup and Guy Salvesen at Duke University had identified a potent ICE inhibitor in the poxvirus-encoded serpin called CrmA. Generously, they sent us an expression construct for CrmA.
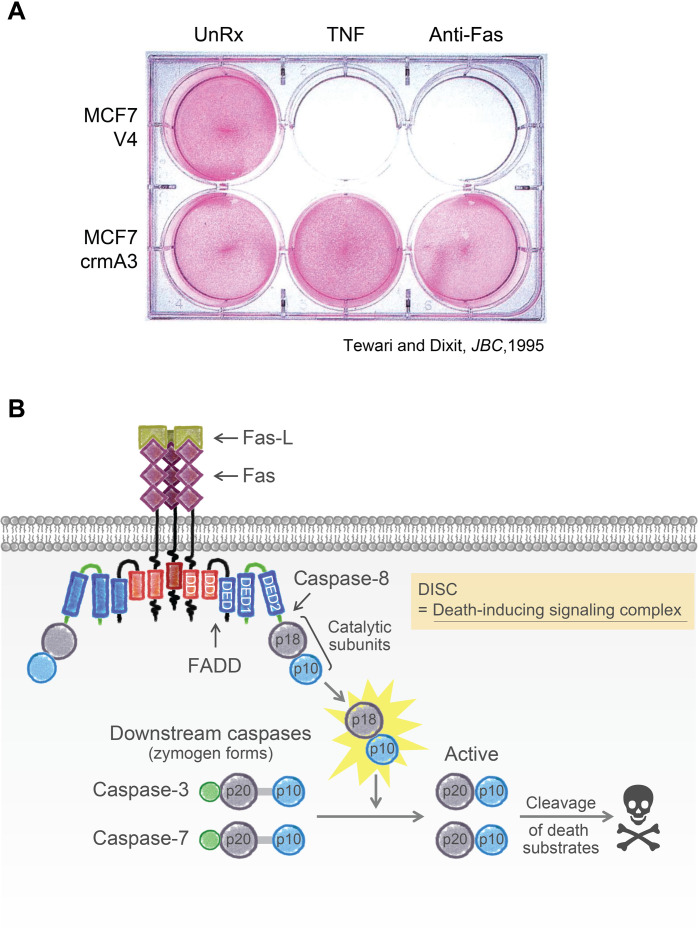
Late one evening, we struck gold. Peering into the microscope, like Howard Carter peering into Tutankhamun’s tomb at immense treasures, we watched CrmA completely inhibit apoptosis induced by death receptor agonists (Fig. 1A). The conclusion was unequivocal. Death receptors must be engaging ICE or an ICE-like protease to initiate apoptosis (1). In follow-up studies, Muneesh identified the ICE-like executioner protease and called it YAMA for the Hindu God of death (2). ICE and YAMA were founding members of a family of mammalian proteases called caspases and were eventually renamed caspase-1 and caspase-3, respectively. The name caspase denotes that they are cysteine proteases (c) that cleave after aspartic acid residues (aspase).
Fig. 1. Death receptor signaling.
(A) MCF7 breast cancer cells expressing poxvirus CrmA survive lethal challenge with TNF or agonist anti-Fas antibody. Reproduced from (1). (B) The DISC assembled by Fas ligand (Fas-L) is composed of the death receptor Fas, the adaptor molecule FADD, and pro–caspase-8. The DISC is cemented by homotypic interactions between DDs and DEDs. Activation of caspase-8 within the DISC elicits the p18 and p10 catalytic subunits (highlighted in yellow), which transmit the death signal by cleaving and activating downstream effector caspases, including YAMA/caspase-3. Cleavage of vital substrates leads to the demise of the cell.
Onward to the next mystery: How did death receptors activate caspases? In the early 1990s, researchers believed that receptors signaled either by acting as ion channels or by altering phosphorylation-dephosphorylation events. Two talented researchers in my laboratory, Arul Chinnaiyan and Marta Muzio, quickly found an entirely new mechanism by which receptors can signal. Arul found in 1995 that the DD, defined earlier by Goeddel, represented a homotypic protein-protein interaction motif. In other words, a DD in one protein can bind to the DD in another, thereby linking the proteins together. Arul showed that the DD in Fas recruits the Fas-associated DD (FADD) protein. His second discovery was that FADD has a second, related motif, which he called the death effector domain (DED). Both the DD and DED motifs in FADD are essential for transmitting the apoptotic signal that activates caspase-3 (3).
Marta’s mission was to identify the protein that interacts with the DED in FADD. Collaborating with the laboratories of Matthias Mann and Krammer, Marta showed in 1996 that the FADD DED binds to pro–caspase-8, which is the inactive form of the protease. Pro–caspase-8 has two DEDs in its pro-domain. The complex is composed of Fas, FADD, and pro–caspase-8, which they dubbed the death-inducing signaling complex (DISC) (4). David Wallach reached similar conclusions (5).
Later, in collaboration with Guy Salvesen, we showed that pro–caspase-8 is activated by induced proximity. Dimerization and autoprocessing of pro–caspase-8 within the DISC release active caspase-8, which then cleaves and activates caspase-3 and caspase-7. This caspase cascade unleashes precipitous proteolytic activity that dismantles the cell in an amazingly tidy and reproducible fashion (Fig. 1B).
I was heartened and invigorated by our unexpected success. We had identified a major gap in our understanding of how death receptors signal, tackled the problem head on, and used our excitement to allure other scientists to collaborate. I would rely on these insights as I made another major career shift, moving from academia to industry. In 1997, I took the job of Director of Molecular Oncology at Genentech, where my group found that cells could use another pathway to die that was distinct from apoptosis.
THE INFLAMMASOME
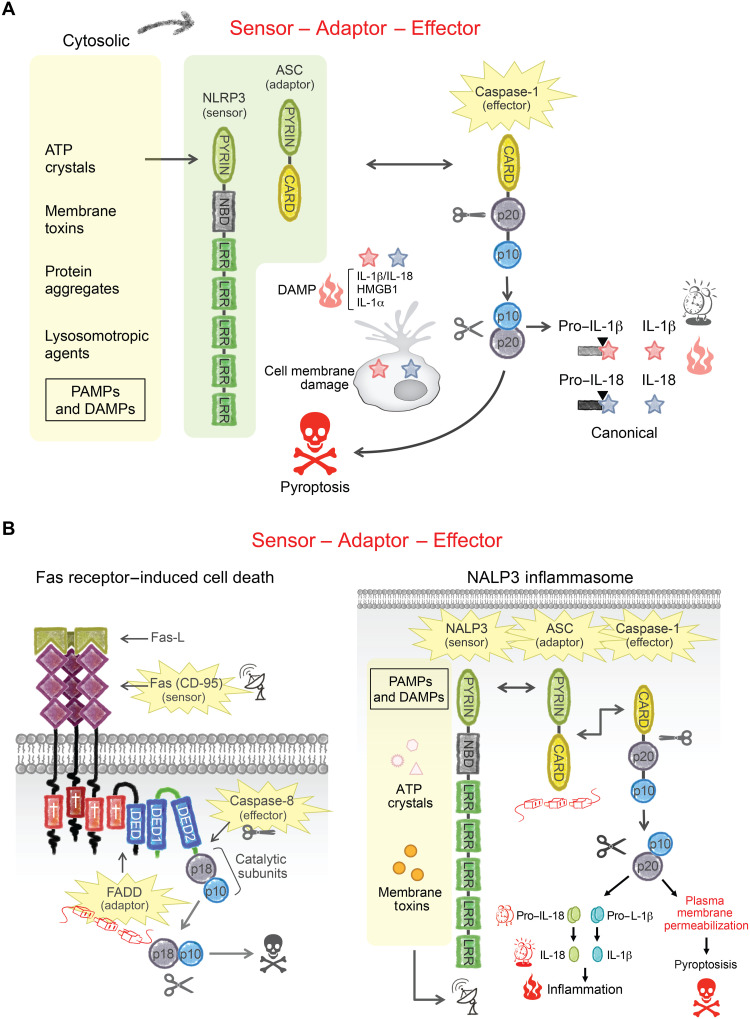
I have been a part of many exciting findings over the years, but I consider discovery of the noncanonical inflammasome with Nobuhiko Kayagaki in 2011 to be my other major contribution to molecular biology. In 2002, the late Jurg Tschopp coined the term inflammasome to describe the intracellular complex that activates caspase-1 when microorganisms or endogenous insults breach the cytoplasmic compartment. This signaling complex is composed of a sensor and an adaptor, which together form a platform that activates pro–caspase-1 (Fig. 2A). Inflammasome assembly is triggered by pathogen associate molecular patterns (PAMPs) derived from invading microorganisms or by endogenous insults, such as uric acid crystals, that act as danger-associated molecular patterns (DAMPs). Activation of pro–caspase-1 within the inflammasome, again through an induced proximity mechanism, has two consequences of crucial importance to innate immunity. Caspase-1 cleaves the immature forms of IL-1β and IL-18 to generate the active cytokines, and, in parallel, caspase-1 cleaves gasdermin D (GSDMD) to release a pore-forming fragment that kills the cell by perforating the membrane. This inflammatory form of cell death is termed pyroptosis. As a result, mature IL-1β and IL-18, which lack a signal sequence for secretion, can exit through the perforated cell membrane and thereby sound the inflammatory alarm to neighboring cells.
Fig. 2. Canonical inflammasome signaling.
(A) The canonical inflammasome is composed of an intracellular sensor, the adaptor ASC, and caspase-1. The prototypical sensor NLRP3 is activated by agents that disrupt the cytosolic compartment, including adenosine triphosphate, lysosomotropic agents, crystals, and protein aggregates. NLRP3 binds to ASC, which, in turn, binds to the pro–caspase-1 zymogen (closed scissors). PYRIN and CARD domains are homotypic protein-protein interaction motifs that hold the complex together. Active caspase-1 (open scissors) cleaves pro–IL-1β and pro–IL-18 into active IL-1β and IL-18, respectively. Proinflammatory DAMPs IL-1β, IL-18, IL-1α, and HMGB1 each lack a signal peptide but are released from the cell by pyroptosis, a lytic form of cell death. (B) The Fas DISC and NLRP3 inflammasome share a similar architecture; they each use related protein-protein interaction motifs to bring together a sensor, an adaptor, and a caspase.
The sensors that assemble inflammasomes recognize specific insults. The NLRC4 sensor, for example, detects Gram-negative intracellular pathogens, such as Salmonella and Shigella, by sensing associated PAMPs, including flagellin. A different sensor, NLPR3, responds to inflammatory mediators such as uric acid or the ionophore nigericin. Each sensor has a protein-protein interaction motif in the form of a caspase activation and recruitment domain (CARD) or a Pyrin domain. Both the CARD and Pyrin domains are structurally related to the previously discussed DD and DED. Pyrin domain sensors such as NLRP3 engage the CARD-containing pro–caspase-1 through the adaptor, ASC, which has both a Pyrin domain and a CARD. Thus, the DISC and the inflammasome share a common architecture (sensor-adaptor-caspase) and are cemented by related interaction motifs (Fig. 2B).
DISCOVERY OF THE NONCANONICAL INFLAMMASOME
Discovery of the noncanonical inflammasome started with a whimper rather than a bang. In 2010, we were testing various PAMPs to determine whether they activated the inflammasome in mouse bone marrow–derived macrophages (BMDMs). We noticed that cholera toxin subunit B (CTB) activated the inflammasome in cells from C57BL/6 mice but not from 129/SvEv mice (6). The finding was odd, but perhaps 129/SvEv mice lacked a known inflammasome component. Curiously, however, 129/SvEv BMDMs activated the inflammasome just fine in response to other PAMPs or DAMPs. Indeed, 129/SvEv cells expressed all the known sensors, the adaptor ASC, and caspase-1. We concluded that 129/SvEv cells must lack the specific sensor for CTB.
Intrigued, we broadened our search, only to find that 129/Sv mice lack caspase-11, a caspase with substantial homology to caspase-1. A splicing mutation in the 129/Sv Caspase-11 gene alters the reading frame, leading to nonsense-mediated decay of the corrupted Caspase-11 transcript. This finding suggested that the inflammasome needed caspase-11 to respond to CTB. We sought confirmation of this unexpected dependence using mice deficient in either caspase-1 or caspase-11. However, we soon came to the unsettling realization that the original Caspase-1 knockout mouse, used in hundreds of publications, actually lacked both caspase-1 and caspase-11. Generated in Caspase-11–deficient 129/Sv embryonic stem cells, the Caspase-1 knockout allele was right next to the mutant Caspase-11, and the two alleles could not be easily segregated. Consequently, all conclusions derived using these mice would need to be reassessed. All of the reported phenotype(s) could be due to a deficiency in caspase-1, caspase-11, or both. I described these findings to a shocked audience at the Toll 2011 meeting in Italy. Many wondered just how far the field had been led astray by this oversight.
We hurried to generate a Caspase-11 knockout in the C57BL/6 mouse strain and also reconstituted expression of caspase-11 in the Caspase-1/−11 double knockout with a Caspase-11 transgene. The results using these mice were unequivocal. Caspase-11 is essential for inflammasome engagement by Gram-negative enteropathogens, including Escherichia coli and Citrobacter rodentium. We designated this new caspase-11–dependent signaling conduit the noncanonical inflammasome pathway (6).
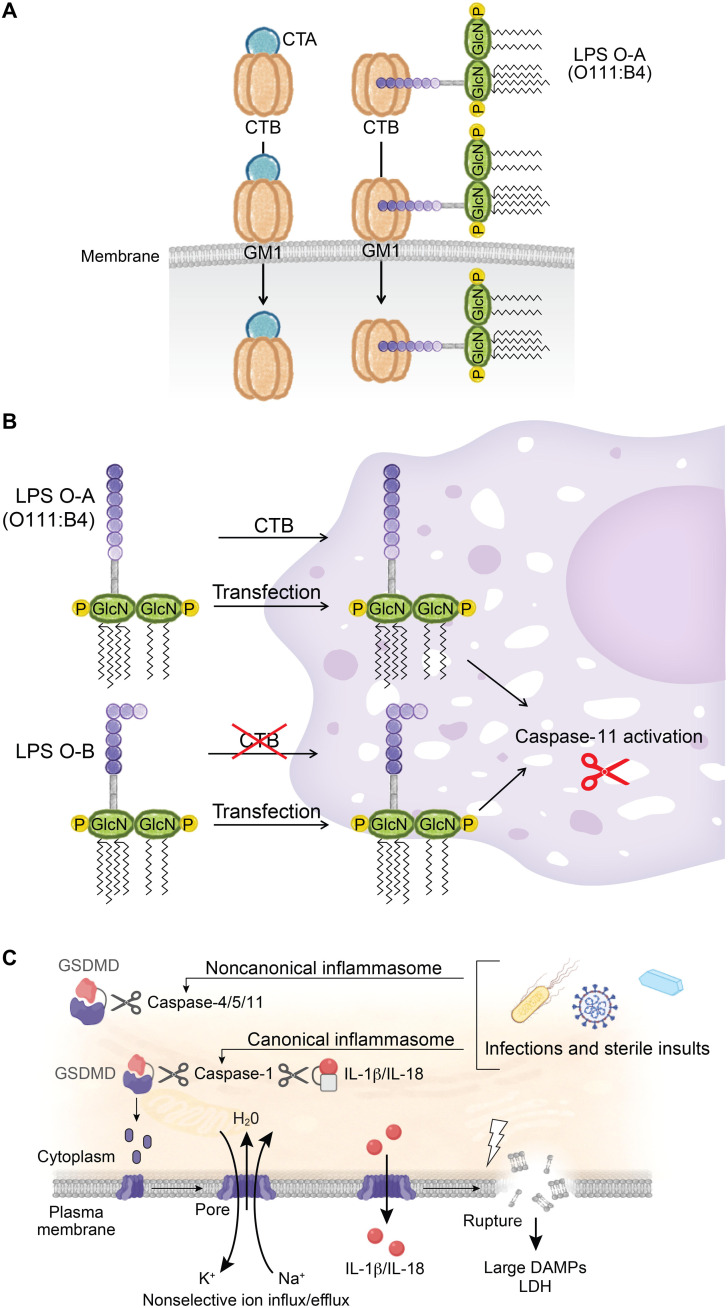
One question now loomed large: What component of Gram-negative organisms triggers the noncanonical pathway? We were excited to tackle this clinically relevant question. Runaway systemic inflammation associated with Gram-negative sepsis is responsible for millions of deaths every year. The molecular tool that we had in hand to engage the noncanonical pathway was CTB. We hoped to use it to reveal the precise trigger common to all Gram-negative bacteria, but it turned out to be an arduous, protracted, and most frustrating undertaking. The intricacies of the inflammasome assay foiled us. To obtain robust inflammasome activation, BMDMs must first be primed with an inflammatory stimulus that up-regulates the expression of a number of relevant components, including caspase-11 and IL-1β. Priming is typically accomplished with a Toll-like receptor (TLR) agonist. Perplexingly, we found that CTB robustly activated the inflammasome when we primed BMDMs with the TLR4 agonist lipopolysaccharide (LPS) but not when the TLR2 agonist Pam3CSK4 was used. By contrast, both methods of priming supported inflammasome activation by other stimuli. Further complicating matters, only priming with a specific LPS serotype, O111:B4, would then allow CTB to activate caspase-11.
We struggled mightily to make sense of the data. An important clue was that CTB binds to the hypervariable O-antigen polysaccharide of LPS serotype O111:B4 but does not bind to LPS serotypes with distinct O-antigens. We wondered whether CTB, which normally delivers the CTA subunit into cells, was merely the courier that delivered LPS O111:B4, carried over from the priming step, into the cytoplasm (Fig. 3A). It dawned on us that LPS, irrespective of serotype, might be able to trigger the noncanonical pathway provided that it could access the cytoplasm. Indeed, LPS introduced by transfection overcame serotype restriction (Fig. 3B). We concluded that LPS, a common component of Gram-negative organisms, is a trigger of the noncanonical pathway. Additional transfection studies revealed that the conserved lipid A portion of LPS is essential for activating caspase-11, whereas the hypervariable O-antigen portion is dispensable (7). Contemporaneously, Edward Miao’s laboratory also showed that cytosolic LPS activates caspase-11 (8).
Fig. 3. The non-canonical inflammasome.
(A) CTB binds to GM1 plasma membrane gangliosaccharides to deliver either CTA (left) or LPS serotype O111:B4 (right) into the cytoplasm. CTB binds to the hyperdivergent O-specific polysaccharide chain of LPS serotype O111:B4. (B) LPS serotype O-A (includes O111:B4) binds to CTB to access the cytosol. In contrast, LPS serotype O-B does not bind to CTB and therefore cannot gain access. Both serotypes, however, can be introduced by transfection. (C) Caspase-1 or caspase-11 cleaves GSDMD, releasing an N-terminal p30 fragment that oligomerizes into an approximately 20-nm pore. GSDMD pores collapse the electrochemical gradient and release DAMPs, including IL-1β and IL-18, into the extracellular milieu.
Gram-negative pathogens contain a plethora of PAMPs. Therefore, we needed to be sure that it was only LPS and not additional PAMPs that activate caspase-11. To definitively address this question, we used a strain of E. coli deficient in the penultimate step of LPS synthesis. Gratifyingly, this strain fails to activate caspase-11. We had proven that the noncanonical pathway was dedicated to the detection of intracellular LPS (7). This noncanonical pathway does not require the known LPS receptor TLR4 because the noncanonical inflammasome is fully functional in Tlr4 knockout mice. The 2011 Nobel Prize was awarded for the discovery of TLR4 as the LPS receptor (9), but our findings with caspase-11 have shown that there is another LPS sensor with a distinct function.
EPILOGUE
Our discovery of the noncanonical pathway as a cytosolic LPS sensor set off a gold rush that led to swift progress by our laboratory and others. Notably, this included the discovery of GSDMD as the downstream substrate for caspase-1 and caspase-11 (10, 11).
The exciting discoveries continue to this day. In 2021, we found that cell lysis following membrane rupture is not a passive event mediated by osmotic forces, as introductory textbooks of biology would have one believe, but rather actively accelerated by a membrane protein, NINJ1 (12).
What have I learned over my career other than persistence pays? Choose an important problem, let the data do the talking, and do experiments that challenge your preconceived notions.
REFERENCES
- 1.Tewari M., Dixit V. M., Fas- and tumor necrosis factor-induced apoptosis is inhibited by the poxvirus crmA gene product. J. Biol. Chem. 270, 3255–3260 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Tewari M., Quan L. T., O'Rourke K., Desnoyers S., Zeng Z., Beidler D. R., Poirier G. G., Salvesen G. S., Dixit V. M., Yama/CPP32 beta, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell 81, 801–809 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Chinnaiyan A. M., O'Rourke K., Tewari M., Dixit V. M., FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell 81, 505–512 (1995). [DOI] [PubMed] [Google Scholar]
- 4.Muzio M., Chinnaiyan A. M., Kischkel F. C., O'Rourke K., Shevchenko A., Ni J., Scaffidi C., Bretz J. D., Zhang M., Gentz R., Mann M., Krammer P. H., Peter M. E., Dixit V. M., FLICE, A novel FADD-homologous ICE/CED-3–Like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell 85, 817–827 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Boldin M. P., Goncharov T. M., Goltsev Y. V., Wallach D., Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell 85, 803–815 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Kayagaki N., Warming S., Lamkanfi M., Walle L. V., Louie S., Dong J., Newton K., Qu Y., Liu J., Heldens S., Zhang J., Lee W. P., Roose-Girma M., Dixit V. M., Non-canonical inflammasome activation targets caspase-11. Nature 479, 117–121 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Kayagaki N., Wong M. T., Stowe I. B., Ramani S. R., Gonzalez L. C., Akashi-Takamura S., Miyake K., Zhang J., Lee W. P., Muszyński A., Forsberg L. S., Carlson R. W., Dixit V. M., Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 341, 1246–1249 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Hagar J. A., Powell D. A., Aachoui Y., Ernst R. K., Miao E. A., Cytoplasmic LPS activates caspase-11: Implications in TLR4-independent endotoxic shock. Science 341, 1250–1253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravindran S., Profile of Bruce A. Beutler. Proc. Natl. Acad. Sci. U.S.A. 110, 12857–12858 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kayagaki N., Stowe I. B., Lee B. L., O’Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q. T., Liu P. S., Lill J. R., Li H., Wu J., Kummerfeld S., Zhang J., Lee W. P., Snipas S. J., Salvesen G. S., Morris L. X., Fitzgerald L., Zhang Y., Bertram E. M., Goodnow C. C., Dixit V. M., Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., Shao F., Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Kayagaki N., Kornfeld O. S., Lee B. L., Stowe I. B., O’Rourke K., Li Q., Sandoval W., Yan D., Kang J., Xu M., Zhang J., Lee W. P., McKenzie B. S., Ulas G., Payandeh J., Roose-Girma M., Modrusan Z., Reja R., Sagolla M., Webster J. D., Cho V., Andrews T. D., Morris L. X., Miosge L. A., Goodnow C. C., Bertram E. M., Dixit V. M., NINJ1 mediates plasma membrane rupture during lytic cell death. Nature 591, 131–136 (2021). [DOI] [PubMed] [Google Scholar]