Abstract
Nononcogenic somatic mutations are thought to be uncommon and inconsequential. To test this, we analyzed 43,693 National Heart, Lung and Blood Institute Trans-Omics for Precision Medicine blood whole genomes from 37 cohorts and identified 7131 non-missense somatic mutations that are recurrently mutated in at least 50 individuals. These recurrent non-missense somatic mutations (RNMSMs) are not clearly explained by other clonal phenomena such as clonal hematopoiesis. RNMSM prevalence increased with age, with an average 50-year-old having 27 RNMSMs. Inherited germline variation associated with RNMSM acquisition. These variants were found in genes involved in adaptive immune function, proinflammatory cytokine production, and lymphoid lineage commitment. In addition, the presence of eight specific RNMSMs associated with blood cell traits at effect sizes comparable to Mendelian genetic mutations. Overall, we found that somatic mutations in blood are an unexpectedly common phenomenon with ancestry-specific determinants and human health consequences.
Somatic mutations are common in blood from healthy individuals and have germline genetic causes and health consequences.
INTRODUCTION
As humans age, their dividing cells acquire mutations. Except for the rare cases when a mutation occurs in a driver gene—possibly leading to cancer (1)—the vast majority of these somatic mutations do not alter protein coding sequence. Therefore, they are believed to have little effect on cellular function. However, this assumption has not been rigorously tested and may not be true, as noncoding regions can actively regulate gene expression through enhancers, splicing, genomic structure, and chromatin configuration.
If nononcogenic somatic mutations affect physiology, then the blood cells would be one of the cell types most affected. Erythrocytes and immune cells are constantly regenerated by hematopoietic stem cells (HSCs), which have accumulated a lifetime of mutations, and these blood cells are responsible for distributing oxygen, mediating immune responses, and regulating clotting—all of which could be affected by somatic mutations. Studies have found seemingly noncancerous clonal expansion of HSCs in healthy individuals, suggesting that somatic mutation is common (2–8). They also found that individuals with clonal hematopoiesis have increased risk of not only blood cancer but also an array of nonmalignant aging diseases, including heart disease, suggesting that somatic mutations can affect physiology. However, these studies only assessed coding variation. Recent efforts to identify noncoding somatic mutations have been performed only in a limited number of people, such as the recent Pan-Cancer Analysis of Whole Genomes (PCAWG) analysis, which included only 2658 genomes (9).
Here, using 43,693 deeply sequenced (38×) whole genomes from the National Heart, Lung and Blood Institute Trans-Omics for Precision Medicine (TOPMed) initiative (10), we developed a catalog of somatic mutations derived from peripheral blood samples of individuals from the general population and identify a class of recurring mutations across individuals, which we term recurrent non-missense somatic mutations (RNMSMs). We then conducted the first large-scale investigation of their germline determinants and health consequences.
RESULTS
Identifying recurrent somatic mutations
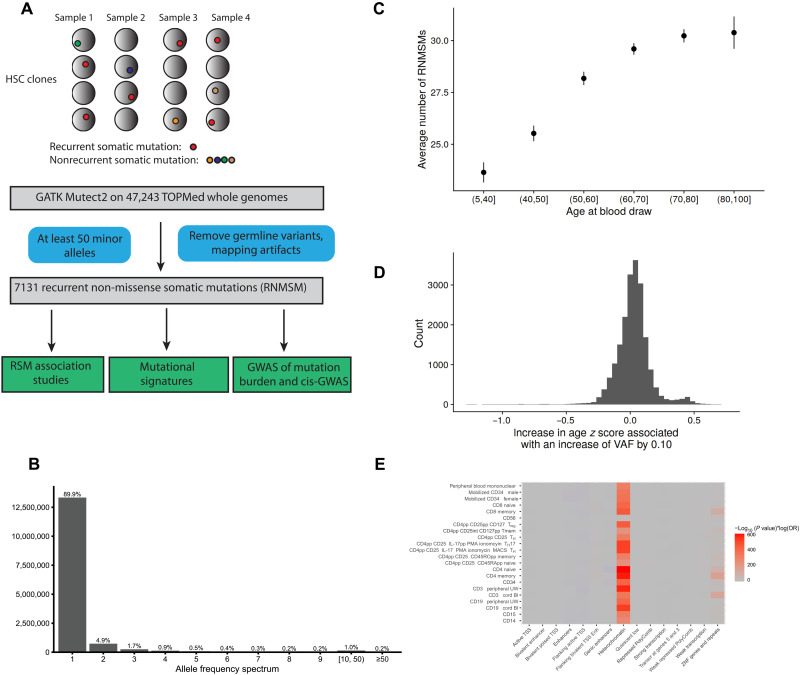
In total, we used the whole-genome data of 47,243 individuals from 37 cohorts included in TOPMed, which were sequenced at seven distinct genome sequencing centers (tables S1 to S3), and identified somatic point mutations in their peripheral blood by running GATK Mutect2 (11). We applied stringent variant filtering procedures to deplete our call set of sequencing artifacts and germline variants (Fig. 1A, Methods, and text S1). We also excluded all calls with a variant allele fraction (VAF) of >35%, defined as the percentage of reads that are alternative at a given site. Of the 14,841,388 somatic variants identified, most (89.9%) were only observed in one individual (Fig. 1B). These singleton variants are likely passenger mutations on a clonally expanded HSC because, at the read depth sequenced, it is unlikely that single variants would be detected unless they were present in a large enough proportion of peripheral blood cells (5, 12).
Fig. 1. Identification of recurrent somatic mutations from TOPMed whole genomes.
(A) Identification of RNMSMs. (B) The allele frequency spectrum of somatic mutations. These represent the frequency of sets of variants with the given allele count. (C) The average number of RNMSMs stratified across age bins with 95% CIs. (D) The histogram of the slopes of the individual RNMSMs with age. (E) Enrichments of chromHMM annotations across blood epigenomes from Roadmap Epigenomics. Enrichments are defined as the log odds ratio (OR) multiplied by the −log10 (P value) from the Fisher’s exact test. Treg, regulatory T cell; TH, T helper cell; IL-17pp, interleukin-17pp; PMA, phorbol 12-myristate 13-acetate; MACS, magnetic-activated cell sorting; Tmem, T memory.
Intriguingly, we identified a small subset of somatic genetic variants that are recurrently mutated and were neither missense nor nonsense mutations, which we call RNMSMs (synonymous mutations are retained). We then further refined our RNMSM calls with additional germline and sequencing artifact filters. We first excluded high-quality germline variants that were identified in the TOPMed germline single-nucleotide polymorphism (SNP) calls (13). Next, using the TOPMed germline structural variant calls (14), we also excluded all mutations that overlapped with a common [minor allele fraction (MAF) > 10%] germline duplication, as such events may result in artifactual somatic variant calls. As sequencing artifacts may have similar signatures to somatic mutations, we also excluded parts of the genome that were not uniquely mappable, and we excluded mutations with mapping artifact signatures (text S1). We then excluded samples where age at blood draw was not available, resulting in an analysis set of 43,693 samples. We focused our analysis on the RNMSMs that occurred at least 50 times and in at least five distinct cohorts to deplete for cohort-specific sequencing artifacts. This yielded a set of 7131 RNMSMs (table S4) or ~0.05% of all variants. The average number of RNMSMs per sample was 28.6, with a median of 25, a minimum of 1, a maximum of 163, and an SD of 16.1.
The RNMSM burden was positively associated with age at the time of blood draw [in units of decades, β = 1.4, 95% confidence interval (CI) (1.3, 1.5), P value = 5.3 × 10−135; Fig. 1, C and D] likely because older people have had more time to accumulate mutations. The association between RNMSM burden and age was attenuated by the inclusion of a cohort indicator covariate [in units of decades, β = 0.20, 95% CI (0.08, 0.32), P value = 8.4 × 10−4], but it is still unlikely to occur by chance.
We then experimentally validated a subset of the RNMSMs by calling somatic variants from 46 individuals not sequenced as part of TOPMed using 30× whole-genome sequencing (WGS) of paired blood and heart tissue (Methods). We observed that 219 of the RNMSMs detected in the TOPMed study were present in the 46 blood samples (table S5), with at least one alt-allele present in the data. We observed that 266 mutations were present, for an average of 5.8 mutations per sample. The low number of replicated RNMSMs likely reflects the younger age of the validation samples (mean of 50.6 years old in validation and mean of 61.3 in discovery cohorts), the decreased diversity compared to TOPMed, and the lower sequencing depth.
We then further validated the RNMSMs by estimating their Mendelian concordance. We reasoned that RNMSMs should have high rates of Mendelian discordance among trios because they are not germline variants. Using 4103 trios in TOPMed, we calculated the Mendelian genotype discordance among Mendelian-informative meiosis. We then categorized RNMSMs as Mendelian discordant if there were at least two Mendelian discordances present and the genotype discordance was greater than 2%. We observed that 71.4% (n = 5094) of the RNMSMs were Mendelian discordant (table S6).
To assess whether the RNMSMs were spurious signatures of rare germline duplications, we used the TOPMed structural variant calls to estimate the co-occurrence of the individual RNMSM genotypes with overlapping germline duplications and deletions. We observed that 9% (n = 616) of the RNMSMs co-occurred frequently (>80% of mutations) with overlapping germline duplications and deletions (table S7), suggesting that most RNMSMs are unlikely to tag germline structural variants.
To test whether the RNMSMs could be identified in another population-scale sequencing cohort, we cross-referenced the RNMSMs with the site list of the gnomAD 3.1.2 release (15). Because samples overlap between TOPMed and gnomAD, we used the non-TOPMed allele frequencies reported in the site list. We observed that 6386 of the RNMSMs were reported in the gnomAD site list (table S8). Collectively, these observations highlight that the RNMSMs have been depleted of germline variants and sequencing artifacts and can be detected in other cohorts.
To characterize the function of the RNMSMs, we annotated them using Variant Effect Predictor (VEP) (16). We observed that the RNMSMs were enriched in intergenic regions (twofold increase; fig. S1). We then calculated the enrichment of RNMSMs in cell type–specific chromatin signatures using the Roadmap Epigenomics catalog (17) (see Methods) and found strong enrichment in heterochromatin across blood cell types (Fig. 1E). This is consistent with a previous study that found that 40% of variance in cancer mutation density can be attributed to variation in H3K9me3, a histone mark associated with heterochromatin (18). This association may be mediated by diminished DNA repair activity in heterochromatin (19).
To identify potential causes of RNMSMs, we conducted a mutation signature analysis (see Methods) and found that the mutational signature SBS5 was the only contributing factor. SBS5 is the result of a clock-like mutational process, meaning that the burden of SBS5 mutations correlates with age and may be associated with tobacco smoking and nucleotide excision repair deficiency (20).
We then asked whether the RNMSMs could be explained by clonal hematopoiesis of indeterminate potential (CHIP) (21) or were an independent phenomenon. We stratified the distribution of RNMSMs based on CHIP carrier status (8). We observed a very modest increase in RNMSM number among the CHIP carriers [fig. S2; β = 0.84, 95% CI (0.43, 1.25), P value = 5.6 × 10−5], suggesting that RNMSMs are not only explained by clonal driver mutation phenomena.
We observed a small number of RNMSMs in genes that are mutated in clonal hematopoiesis and hematological malignancy, including intronic RNMSMs in TET2 (rs1341404863, rs1428954790), JAK2 (rs1223087352), and ASXL2 (rs1416952509). Among acute myeloid leukemia (AML) genes, we observed a variant downstream of BCL6 (rs9878379) with 194 mutations and an intronic variant of FLT3 (rs1242299801). rs9878379 was previously reported in the International Cancer Genome Consortium (ICGC) cancer cohort in six cancer types (22) but has previously been unreported in a noncancer population cohort. BCL6 is a proto-oncogene that is a frequently mutated in B cell lymphomas (23).
To estimate the noncancer population prevalence of RNMSMs that occur in cancer genomes, we cross-referenced the RNMSMs with the cancer genome somatic calls released by the PCAWG (24). We observed that 81 of the somatic mutations called in the PCAWG cancer genomes were present among the RNMSMs (table S9). These include (rs1379713562, COSV71733298), a synonymous mutation SBSN that was mutated in 191 genomes, and an intronic variant (rs1473587176) in PTER with a notable allele frequency (930 mutated alleles, 1% MAF). SBSN expression has recently been described as a potential biomarker in myelodysplastic syndromes (25). We observed that 23 of the PCAWG-RNMSMs had at least 100 mutations in our cohort, indicating that a subset of mutations in cancer genomes is prevalent in a population cohort that was not ascertained for cancer.
RNMSMs and genetic ancestry
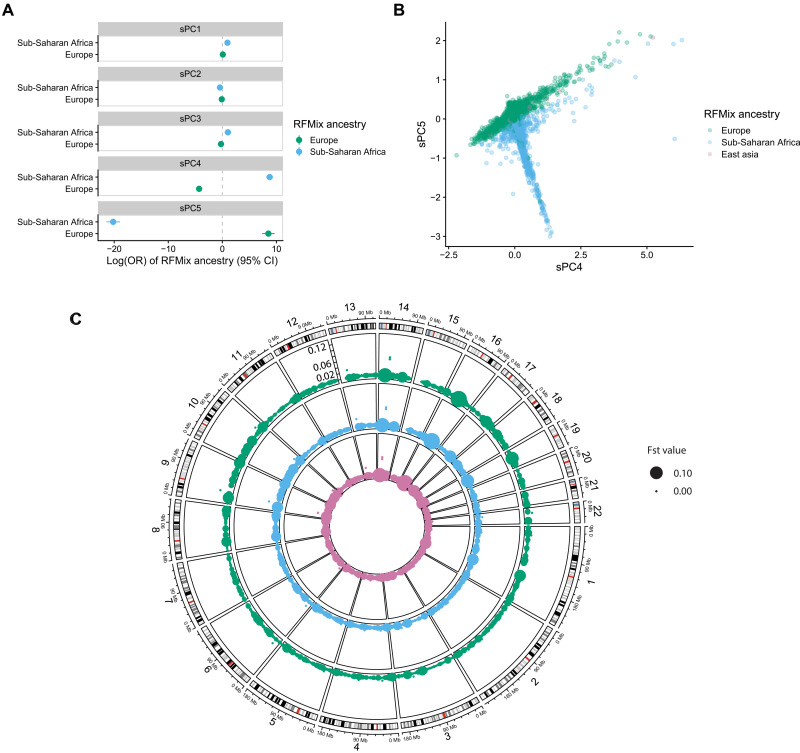
We then performed principal components analysis (PCA) on the somatic variant matrix of VAFs to characterize the underlying structure of RNMSMs (Methods). We estimated the first 50 components (26) and observed that the first three axes of variation explain >92% of the variance, indicating strong shared underlying structure among RNMSMs. We observed that the first somatic principal component (sPC1) was strongly associated with overall RNMSM burden (Rsq = 94%, P value <2 × 10−16). As PCs of germline variants associate with genetic ancestry (27–29), we then asked whether sPCs had similar properties. As previously described, the global genetic ancestry of TOPMed samples has been estimated with RFMix (13). Using this resource, we labeled samples according to their largest global ancestry contribution. We then conducted a multinomial logistic regression of global ancestry on the first five sPCs, including a study indicator as a covariate. We observed that sPCs were strongly associated with ancestry (Fig. 2, A and B, and table S10). We then calculated fixation index (Fst) values (Methods) among the somatic variants to estimate the degree of population differentiation at individual mutations. We observed that 55 of the somatic mutations had Fst estimates > 0.05 (Fig. 2C and table S11). These ancestry associations suggested that RNMSMs may associate with germline variants common in particular ancestries. This is consistent with previously known associations between genetic ancestry and DNA replication timing (30)—DNA recombination hotspots (31). The association between clonal phenomena and genetic ancestry has been previously observed in both CHIP (8) and mosaic chromosomal alterations (32). Therefore, we sought to establish whether there were common germline genetic variants associated with acquiring RNMSMs.
Fig. 2. Somatic genotype PCs are associated with genetic ancestry.
(A) Global ancestry labels, as estimated from RFMix, are regressed on the first five sPCs in a multinomial regression with a study indicator included as a covariate. The ORs are estimated with East Asian ancestry as the reference level. (B) A scatterplot of sPC4 and sPC5, with colors indicated by the RFMix global ancestry label. (C) A circular genome plot where the angle indicates genomic position, and the radius within a given track indicates the allele frequency, which ranges from 0 to 0.12. A separate track is plotted for the allele frequencies computed separately in European, Sub-Saharan African, and East Asian genomes, respectively, which are colored by the legend indicated in (B). The size of points indicates the fixation index (Fst) estimate, where larger points have larger Fst values.
Germline genetic basis of RNMSMs
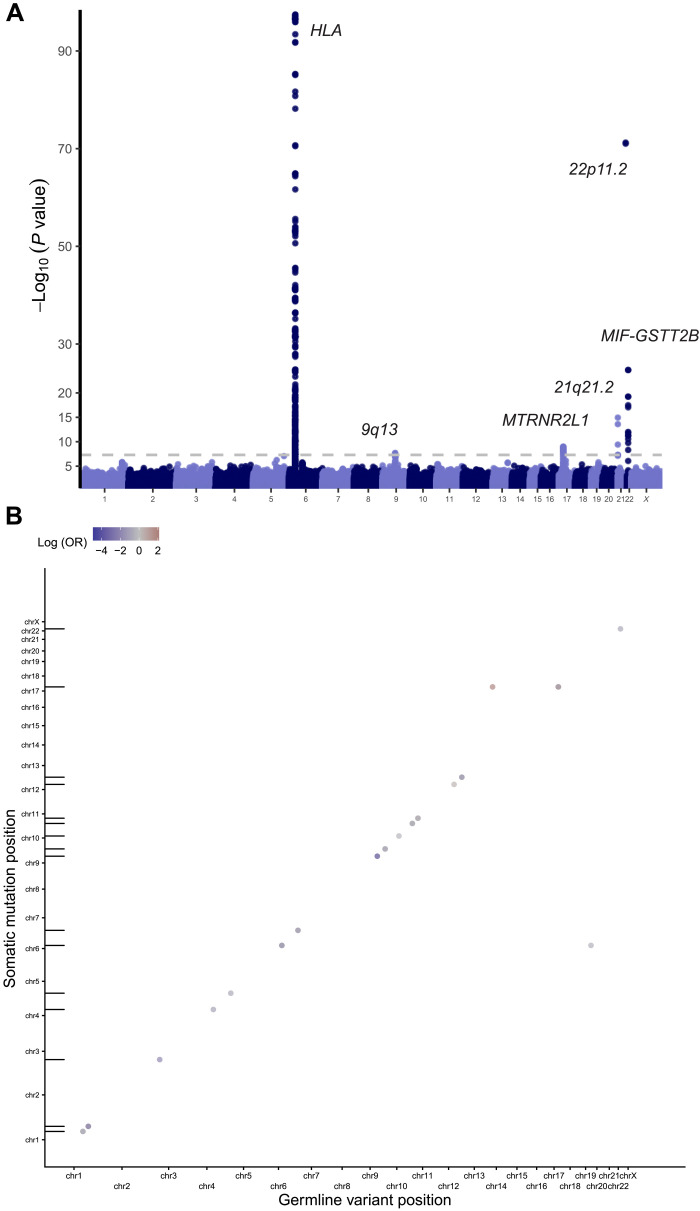
We performed a multi-ancestry genome-wide association study (GWAS) of the RNMSM burden using the scalable and accurate implementation of generalized mixed model (SAIGE) (33) and identified five genome-wide significant loci, all of which collectively highlight the influence of immune function on RNMSM burden (Fig. 3A). The strongest signal was observed in the HLA region at rs9271735, which is a common (MAF 28%) variant and is 2.5 kb upstream of the transcription start site (TSS) of HLA-DQA1, whose protein plays an essential role in antigen presentation. The A allele was associated with a 0.09 SD increase in RNMSM burden (P value = 3.8 × 10−98). Given that there is extensive linkage disequilibrium at the HLA locus, this variant likely tags a specific HLA haplotype. To ensure that this signal was not a consequence of population stratification, we then performed both European and African ancestry–specific GWAS (Methods; fig. S3). We observed that rs9271735 was genome-wide significant in both ancestry-specific GWAS (African ancestry, P value = 4.9 × 10−18; European ancestry, P value = 5.1 × 10−40), indicating that the association is unlikely to be a consequence of population stratification. Consistent with the possible role of the adaptive immune system in surveillance of HSCs for excessive mutation, a recent report showed that HSCs in humans are antigen-presenting cells (34).
Fig. 3. Germline determinants of RNMSM burden.
(A) Manhattan plot from GWAS of RNMSM burden, computed using SAIGE. Germline variants included had a minor allele count ≥ 600 and were distinct from the set of RNMSMs. (B) Genetic determinants of individual RNMSMs are primarily in “cis,” where cis is defined as within 2 Mb of the RNMSM. Genomic coordinates of the associated linkage disequilibrium (LD)-clumped germline variants are plotted on the x axis, and coordinates of the RNMSMs are plotted on the y axis. Points falling along the diagonal indicate cis associations. Tick marks along the y axis indicate positions of the RNMSMs from the LD-clumped associations.
The MIF-GSTT2B locus was another hit, with the leading variant being rs5760124, a common (MAF 43%) intergenic variant 31.3 and 35.5 kb from the TSSs of GSTT2B and MIF, respectively. The T allele was associated with a 0.04 SD increase in RNMSM burden (P value = 1.8 × 10−25). It is an expression quantitative trait locus (eQTL) in whole blood (35) for MIF, DDT, and DDTL, and the T allele increases expression for all three (normalized effect sizes of 0.80, 0.40, and 0.25; P values of 2.6 × 10−60, 3.5 × 10−55, and 3.6 × 10−14). It is also a protein quantitative trait locus (pQTL) for GSTT2B in blood plasma (36) and a splicing quantitative trait locus (sQTL) for DDT in whole blood (35). Consistent with the role of rs5760124 in altered hematopoiesis, the T allele is also associated with increased neutrophil counts (37). MIF is a proinflammatory cytokine that regulates macrophage function (38), with DDT and DDTL being its functional homologs. MIF has been shown to inhibit p53 in murine models (39), facilitating cell proliferation. Increased expression of MIF promotes inflammation, which may result in accelerated aging of HSCs (40), providing fertile substrate for the acquisition of aging-associated clonal phenomena.
We then asked whether genetic variation associates with individual RNMSMs rather than RNMSM burden. Cis-acting genetic variation has been previously reported to associate with somatic mutations including JAK2 V617F (41–43) and somatic chromosomal mosaicism (44, 45). We focused on the 82 most frequent RNMSMs and performed association analyses between each germline variant in a 2-Mb region surrounding each RNMSM. To identify trans RNMSM regulators, we also tested each of the RNMSM cis regions with all the other RNMSMs. In total, we examined 6724 possibly overlapping windows for association (82 2-Mb regions × 82 RNMSMs) and used 382,359 germline variants (31,353,438 total germline variant–RNMSM pairs).
We identified 3094 associations at P value < 5 × 10−9, comprising 2783 germline variants and 79 loci (tables S12 and S13). Consistent with prior observations regarding somatic structural variants, such as mosaic chromosomal abnormalities (mCAs) (44), the vast majority (99.8% or 3088 of 3094) of significant hits were in cis (Fig. 3B).
Nine of 2783 germline variants altered protein coding sequences, including eight missense variants and one splice donor variant. The C allele of rs3737737, a missense variant in MAST2, strongly reduced the odds of acquiring a mutation in cis [chr1-46207599-T-C, odds ratio (OR) = 0.26, P value = 8.7 × 10−12]. MAST2 is a serine/threonine kinase that regulates interleukin-12 synthesis in macrophages (46). The G allele of rs3737744, a missense variant in NSUN4, was associated with increased odds of acquiring the same mutation (chr1-46207599-T-C, OR = 1.27, P value = 8.4 × 10−11). Both rs3737744 and rs3737737 have been previously associated with altered hematocrit levels in GWAS of blood cell counts (37).
We then examined noncoding associations and identified four variants (rs116227346, rs1485836, rs7152548, and rs113359887) with a scaled combined annotation dependent depletion (CADD) (47) score ≥ 20, indicating that they are predicted to be among the 1% most deleterious mutations. According to chromHMM (48) annotations, rs116227346 is in an enhancer in 24 epigenomes, including CD14-positive monocytes and common myeloid progenitors. rs1485836 lies in a region of heterochromatin in a highly conserved sequence (PhyloP = 2.5) 173 kb from the TSS of GRIK2. Among other hits, the C allele of rs1707302 was also associated with reduced odds of mutated chr1-46207599-T-C (OR = 0.71, P value = 3.6 × 10−18). rs1707302 is 2.2 kb from the TSS of PIK3R3 and is an eQTL in monocytes of PIK3R3 (49) and also an sQTL of MUTYH in whole blood (35). MUTYH, which is involved in base excision repair, has recently been shown to alter somatic mutation rates in normal human cells (50) and alter the germline mutation rate in mice (51). The C allele of rs116750427, an intronic variant of PAX5, was associated with reduced odds of acquiring a mutated chr9-37358292-A-G (OR = 0.61, P value = 5.7 × 10−10). The C allele is common in African ancestry genomes [3.5% in 1000 Genomes (52)] but unobserved in other ancestries. PAX5 is a transcription factor that regulates lineage commitment in lymphoid progenitors and regulates VDJ recombination (53). The G allele of rs112886224, a variant upstream of MDM2, was associated with reduced odds of acquiring a mutated chr12-68547943-G-T. MDM2 is a proto-oncogene that negatively regulates p53 (54). Together, these genetic association analyses reveal that inherited genetic variation shapes somatic mutations through three distinct mechanisms. First, inherited genetic mutations can result in an increased likelihood of somatic mutations nearby through a hypermutable local milieu. Second, germline genetic variants can lead to alterations in adaptive immune cell function, as suggested by the HLA signal. Third, germline genetic variants can promote an inflammatory state, which may lead to accelerated aging of HSCs, as suggested by the MIF-GSTT2B signal.
Phenotypic consequences of RNMSMs
We then sought to establish whether RNMSMs have any human health consequences. We developed an RNMSM association study (RSMAS) procedure, where we examined the association between each somatic variant with a given phenotype. To mitigate confounding, we included an extensive covariate adjustment set, including separate PCs from both the germline genetic variation (gPCs) and the somatic variation (sPCs), as well as measures of sample sequencing depth and contamination estimates (Methods). As previous reports have implicated mosaicism of driver mutations with altered hematopoiesis (8), and our germline genetic studies highlighted genes important in blood cell function, we focused our RSMAS on 15 blood cell traits (Methods).
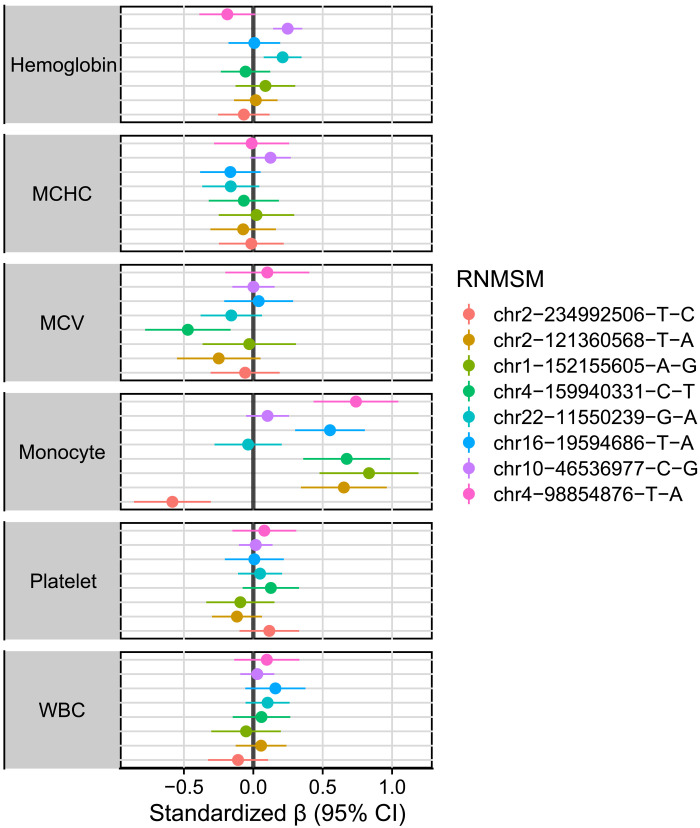
We performed 106,955 trait-variant associations between 15 blood cell traits and the 7131 RNMSMs. To adjust for multiple testing, we first identified the effective number independent RNMSMs using the eigenvalue ratio measure (55). The effective number of independent tests (4598) is lower than the number of tested RNMSMs (7131) because of the correlation structure present, resulting in a significance threshold of 1.1 × 10−5 for each individual blood trait. To estimate the false discovery rate of these associations, we also computed q values (56). Eight significant associations were detected at a q value < 5% threshold, and four were detected with a P value < 1.1 × 10−5 (Fig. 4). Of the eight q value significant variant-trait pairs, six were RNMSM-monocyte count associations, and one association was observed for hemoglobin and hematocrit, respectively. The eight RNMSMs were all rare, each with fewer than 100 mutations. The effect size of these associations is large compared to common germline variant disease associations: An increase in VAF of 0.1 in rs1319097708 is associated with an increase in monocyte count by half an SD [β = 0.55, 95% CI (0.301, 0.804), q value = 0.036] and explains 0.21% of the variance in monocyte counts.
Fig. 4. RNMSMs associate with blood cell traits.
Associations between eight RNMSMs (q values < 0.05) with harmonized blood cell traits in 2996 individuals. Units correspond to an increase of an SD of the phenotype associated with an increase in the VAF value of an RNMSM by 0.10. WBC, white blood cell; MCHC, mean corpuscular hemoglobin concentration; MCV, mean corpuscular volume.
DISCUSSION
In this study, we characterized the spectrum of somatic variation in blood in 43,693 WGS individuals from 37 studies that comprise the TOPMed WGS dataset. We observed that most somatic variants are individually very rare. However, there were 7131 non-missense mutations present in more than 50 genomes, corresponding to an MAF of >0.05%. RNMSM burden increased with age. These variants were highly enriched in heterochromatin. A notable number of these were differentially present in individuals of different genetic ancestries, due in part to ancestry-specific cis and trans QTL variants that associate with RNMSMs. The RNMSM-QTLs implicated diverse mechanisms including lymphoid lineage commitment, inflammatory cytokine production, and DNA repair. Eight RNMSMs were significantly associated with blood cell traits.
These findings permit several conclusions. First, nononcogenic somatic variation is a common phenomenon. Although the majority of somatic variants are individually very rare, a subset are quite common. Every sample had at least one RNMSM mutation. Analysis of other somatic mutations in blood cells, such as clonal hematopoiesis, suggests that they reoccur as a consequence of mutation rate and fitness (57). Given the limited functional consequence of RNMSMs, we identify three possible causes of this phenomenon. First, they reoccur at sites with high mutation rates in hematopoietic cells. This suggests an etiology where multiple HSCs acquire the same mutation rather than a single clone acquiring an RNMSM and then clonally expanding. Second, a subset of RNMSMs occur in B cell or T cell progenitors that subsequently expand clonally. Third, a subset of RNMSMs may cooperate with driver mutations to promote clonal expansion.
Second, the spectrum of somatic variation in humans is associated with genetic ancestry. Concordantly, two recent reports have indicated that germline variation is associated with the acquisition of somatic copy number variants in both the U.K. Biobank (45) and BioBank Japan (32), and they observed considerable differences in the prevalence of specific mCAs. Our observation extending this to somatic point mutations has implications for our understanding of cancer biology and its treatment. Recent reports have indicated that tumor mutational burden, a biomarker of response to cancer immunotherapy, is associated with germline variation (58). Differences in predisposition toward mutational burden is derived in part from differences in germline variation mediating somatic variation, highlighting a potential disparity in immunotherapy outcomes and an important area for future research.
Third, noncoding somatic variants associate with blood cell traits. We also introduce RSMAS, an analogous analysis to a GWAS on germline variants. GWAS has underscored the importance of regulatory biology in complex trait variation. In contrast to germline association studies, little is known about regulatory somatic variation. Despite the enrichment of germline loci in regulatory regions, most somatic analyses have focused on the acquisition and identification of oncogenic drivers rather than sets of variants with modest effects that may collectively contribute nontrivial variation to phenotypes. We anticipate that this method will yield novel insights in other phenotypes and tissues.
This study has several limitations. First, our variant-calling procedures are limited by the sensitivity of the sequencing used. Some sequencing artifacts have similar properties to somatic mutations, and a subset of somatic mutations are difficult to distinguish from sequencing and mapping artifacts. We observed that our variant calling required extensive filtering to exclude likely artifacts. Refined somatic variant calling processes for single-tissue cohorts remain an area of future research. At 38× coverage, we are also insensitive to low VAF (<5%) variants (8). However, previous reports suggest that these presently undetected variants have little bearing on disease (59). This suggests that variants present in our RNMSMs are likely enriched for disease associations relative to the undetected class of low VAF RNMSMs. Second, as our compendium is derived from a complex collection of cohorts, ethnicities, and disease sampling ascertainments, our results are limited by our relatively incomplete knowledge of potential environmental confounders. Third, somatic variants are more challenging to interpret than germline variants, as causal inference is difficult to achieve in cross-sectional analyses, where date of mutation acquisition is unknown. In the absence of longitudinal or other study designs that permit causal inference in this setting, we cannot fully characterize the direction of causality in associations between RNMSMs and phenotypes. Fourth, as few other population cohorts with whole-genome somatic variant calls exist, it is difficult to replicate our observations in other contexts. Although most RNMSMs were found in gnomAD, a subset of the most frequent RNMSMs were not observed in a small, paired heart-blood tissue cohort. Further research is needed to fully characterize the technical factors that contribute to differences between paired-tissue and single-tissue somatic variant calling. Fifth, because of the absence of somatic variants in catalogs of molecular QTLs, the functional consequence of the blood-associated RNMSMs is unclear. Sixth, although we observed only a modest association with CHIP, it is possible that other forms of clonal hematopoiesis that we were unable to detect at our sequencing depth may contribute to the genesis of RNMSMs.
In conclusion, our compendium of recurrently mutated somatic variants demonstrates that this class of genetic variation is widely present throughout the human genome and has germline genetic determinants and phenotypic consequences. Our observations expand at scale our understanding of the spectrum of acquired human genetic variation and its origin. Future efforts are required to extend these observations to other human tissues.
METHODS
WGS processing, variant calling, and CHIP annotation
BAM files were remapped and harmonized through the functionally equivalent pipeline (60). SNPs and indels were discovered across TOPMed and were jointly genotyped across samples using the GotCloud pipeline (13, 61). A support vector machine (SVM) filter was trained to discriminate between high- and low-quality variants. Variants were annotated with snpEff 4.3 (62). Sample quality was assessed through Mendelian discordance, contamination estimates, sequencing converge, and among other quality control metrics. Samples were aligned to GRCh38.
Putative somatic SNPs were called with GATK Mutect2 (11), which searches for sites with at least two alt-reads present. Mutect2 then performs a local haplotype reassembly and applies several variant quality filters. We used a “panel of normals” to filter sequencing artifacts and used an external reference of germline variants to exclude germline calls. We deployed this pipeline on Google Cloud using Cromwell (63).
Samples were annotated as CHIP carriers if the Mutect2 output contained at least one variant in a curated list of leukemogenic driver mutations with at least three alt-reads supporting the call (7, 8). We expanded the list of driver mutations to include those in recently identified CHIP genes (64).
We defined RNMSMs as those occurring in at least 50 distinct samples and occurring in at least five TOPMed cohorts, which would indicate a prevalence similar to those of the most frequently mutated CHIP genes. We used cyvcf2 (65) to parse the Mutect2 VCFs and encoded each variant in an int64 value using the variant key encoding (66). We developed a Python application to perform the recurrent somatic variant identification and filtering. We stored the VAFs values in a sparse matrix using the Python Sparse module. For a given individual and sample, we stored the VAF of the mutation if present and, otherwise, coded the value as 0. We performed extensive filtering to exclude germline variants and sequencing artifacts (text S1).
RNMSM validation in paired tissue samples
We gathered paired blood and heart tissue samples from 46 donors from Vanderbilt University Medical Center. We sequenced the whole genomes of the 92 samples at ~30× coverage and called somatic variants using the Illumina DRAGEN small variant caller in paired tumor/normal mode. We examined variants that were marked as either “PASS” or “weak evidence” as indicating replication for the RNMSMs of our discovery cohort. We limited our analysis to only those variants that were present in the blood tissues with at least one minor allele. We then examined the intersection between the RNMSM site list from our discovery cohort with the validation samples using bcftools (67).
VEP annotation and enrichment
We annotated the RNMSMs using VEP (16) using the “regulatory” and CADD plugins. We filtered the annotations using the “flag_pick” command line option. As a comparison set, we applied the same VEP command to all PASS variants from chromosome 22 of the TOPMed Freeze 8 binary variant call format (BCF) files. We then compared the proportions of variants with a given annotation using a proportion Z test.
Roadmap Epigenomics enrichment
We downloaded the hg38 mnemonic files from the Roadmap Epigenomics project derived from the chromHMM 15-state model applied to 127 epigenomes (48, 68). We then constructed a searchable index over the 1905 (51 * 127) annotations using GIGGLE (69). We used GIGGLE to calculate the genome-wide enrichments of the RNMSMss across the 1905 annotations. GIGGLE implements the enrichment hypothesis testing using a Fisher’s exact test.
Mutational signature analysis
We estimated the contributions of the v3 COSMIC mutational signatures to the RNMSMs using SignatureAnalyzer (70). We set “nruns” = 50 and the “a” hyperparameter to 15.
PCA on RNMSMs
We computed the partial singular value decomposition (SVD) of the sparse matrix of VAFs using the augmented implicitly restarted Lanczos bidiagonalization algorithm as implemented in the IRLBA R package (26). We normalized the VAF matrix by column centering and scaling by the VAF SD for the given RNMSM. We computed the first 50 nonzero components among the left and right singular vectors.
Fst calculation
We stratified samples into European, Sub-Saharan Africa, and East Asian ancestry groups based on their RFMix global ancestry estimate, which was defined as the largest global ancestry estimate for each sample. Within each strata, we computed the allele frequencies of all RNMSMs separately. We then computed the RNMSM variance within each strata k as vark = 2AFk(1 − AFk). We then computed the weighted average of the stratified variance estimates, , where ck indicates the prevalence of group k. We calculated , where vartot is the binomial variance of the RNMSM computed across all samples.
Single variant association analyses on RNMSM burden
Single variant association for each variant in Freeze 8 with a minor allele count (MAC) > 20 was performed with SAIGE (33) using the TOPMed Encore analysis server. We defined the quantitative RNMSM burden phenotype by counting the RNMSM mutations per sample and then applying an inverse normal transformation. We included age at blood draw, genotype-inferred sex, study, average sample depth, sample contamination from VerifyBamID (71), sPCs 4-5, and the first 10 genetic ancestry PCs as covariates. We declared variants from this analysis as significant if their P value was less than 5 × 10−8.
We also computed European ancestry– and Sub-Saharan ancestry–specific GWAS by first stratifying the samples based on their RFMix global ancestry label. We then repeated the same GWAS analysis within each strata separately.
As the TOPMed germline variant call set intersects with the somatic calls, we took additional filtering measures to exclude somatic variants from the germline variant calls. First, we excluded any variant that was also called as an RNMSM. Second, we then created a database of variants where either the allelic-balance z score exceeded 5.0, the SVM filter was less than −0.25, or the −log10 Hardy-Weinberg equilibrium (HWE) P value was greater than 5.0. We then excluded any variant in the summary statistics where at least one of these criteria was true. Last, we included only variants where the minor allele count was at least 600.
Single variant association analyses on individual RNMSMs
We selected the 80 most frequent RNMSMs and performed association analyses between each germline variant in a 2-Mb region surrounding each RNMSM using PLINK2 (72) with the “first-fallback” option, which defaults to standard logistic regression to compute statistics but switches to firth logistic regression when convergence fails. To identify trans RNMSM regulators, we also tested each of the RNMSM cis regions with all the other RNMSMs. In total, we examined 6724 possibly overlapping windows for association (82 2-Mb regions × 82 RNMSMs). We assessed significance at 5 × 10−9. As with the analysis of RNMSM burden, we then filtered the summary statistics to exclude somatic variants from the germline variant calls and to exclude any variant with a MAC less than 700.
Association analyses with RNMSMs (RSMAS)
For quantitative outcomes, we computed test statistics using linear regression on the inverse normal transformed outcome. We computed a marginal analysis where each RNMSM was analyzed in a separate regression. Each RNMSM was included as a vector of VAF values. We included the first 10 germline ancestry PCs, the first 10 sPCs, an indicator for study, genotype-inferred sex, average sample depth, and average sample contamination as covariates. We calculated q values (56) on the resultant test statistics within each outcome. We implemented this procedure in a bespoke R package, somaticWAS.
Blood cell traits
Blood cell counts and indices were selected for analysis including hemoglobin, hematocrit, red blood cell count, white blood cell count, basophil count, eosinophil count, neutrophil count, lymphocyte count, monocyte count, platelet count, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, mean corpuscular volume, mean platelet volume, and red cell distribution width. The phenotypes were collected by each cohort and were centrally harmonized by the TOPMed Data Coordinating Center. Before analysis, each blood cell trait was separately inverse normal transformed.
Code availability
Code availability can be found at Zenodo archives: 10.5281/zenodo.7484723 and 10.5281/zenodo.7484713
Acknowledgments
WGS for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung, and Blood Institute (NHLBI). See the Supplementary Materials for study omics support information. Centralized read mapping and genotype calling, along with variant quality metrics and filtering, were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1; contract HHSN268201800002I). Phenotype harmonization, data management, sample identity quality control, and general study coordination were provided by the TOPMed Data Coordinating Center (R01HL-120393; U01HL-120393; contract HHSN268201800001I). We thank the studies and participants who provided biological samples and data for TOPMed. The full study-specific acknowledgments are included in the Supplementary Materials. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services. We wish to acknowledge the contributions of the consortium working on the development of the NHLBI BioData Catalyst ecosystem.
Funding: This work was supported by National Institutes of Health grant 3R01HL-117626-02S1, National Institutes of Health contract HHSN268201800002I, National Institutes of Health grant R01HL-120393, National Institutes of Health grant U01HL-120393, National Institutes of Health contract HHSN268201800001I, National Institutes of Health grant DP5-OD029586 (A.G.B.), Burroughs Wellcome Foundation Career Award for Medical Scientists (A.G.B. and S.J.), and NHLBI BioData Catalyst Fellowship (J.S.W).
Author contributions: Conceptualization: J.S.W. and A.G.B. Methodology: J.S.W., T.W.B., G.R.A., A.V.S., H.M.K., and A.G.B. Investigation: J.S.W. Project administration: C.A.L., J.G.B., and J.A.B. Sample and funding acquisition: K.D.T., X.G., A.R.S., J.R.O., J.P.L., E.B., K.C.B., N.C., E.E.K., R.J.F.L., M.F., S.R., B.E.C., F.D.G., Z.C., W.J.G., R.K., L.G., R.P.S., B.M.P., J.C.B., E.K.S., J.H.Y., D.Q., S.T.W., J.L.-S., D.L.D., N.D.P., B.I.F., D.W.B., M.H.C., R.S.V., A.D.J., R.M., L.R.Y., L.C.B., S.K., J.H., R.K., S.R.H., N.L.S., K.L.W., D.K.A., M.R.I., H.T., A.C., L.M.R., Y.G., M.d.A., J.I.R., S.S.R., A.W.M., B.A.K., J.M.J., M.M.W., B.S.C., R.D., J.E.C., J.B., H.G., S.X., L.K.W., D.A.M., X.L., V.O., S.M., C.C.G., Y.-D.I.C., W.-J.L., M.B.S., D.D., D.R., C.A., C.K., P.D., and A.P.R. Supervision: T.W.B., G.R.A., A.V.S., H.M.K., A.J., R.M., P.N., S.J., A.P.R., and A.G.B. Writing—original draft: J.S.W. and A.G.B. Writing—review and editing:: K.D.T., X.G., A.R.S., J.R.O., J.P.L., E.B., K.C.B., N.C., E.E.K., R.J.F.L., M.F., S.R., B.E.C., F.D.G., Z.C., W.J.G., R.K., L.G., R.P.S., B.M.P., J.C.B., E.K.S., J.H.Y., D.Q., C.A.L., J.G.B., J.A.B., S.T.W., J.L.-S., D.L.D., N.D.P., B.I.F., D.W.B., M.H.C., R.S.V., A.D.J., R.A.M., L.R.Y., L.C.B., S.K., J.H., R.K., S.R.H., N.L.S., K.L.W., D.K.A., M.R.I., H.T., A.C., L.M.R., Y.G., M.d.A., J.I.R., S.S.R., A.W.M., B.A.K., J.M.J., M.M.W., B.S.C., R.D., J.E.C., J.B., H.G., S.X., L.K.W., D.A.M., X.L., V.O., S.M., C.C.G., Y.-D.I.C., W.-J.L., M.B.S., D.D., D.R., C.A., C.K., P.D., A.P.R., T.W.B., G.R.A., A.V.S., H.M.K., A.D.J., R.M., P.N., S.J., and A.P.R.
Competing interests: P.N. has received grants from Amgen, Apple, Boston Scientific, and Novartis; has received consulting income from Apple, Blackstone Life Sciences, Novartis, Foresite Labs, and Genentech; and has spousal employment at Vertex, all unrelated to the present work. R.K. is a consultant for Regeneron. J.H.Y. is a consultant for Bridge Biotherapeutics. L.M.R. is a consultant for the TOPMed Administrative Coordinating Center (through WeStat). S.J., A.G.B., and P.N. are cofounders, equity holders, and on the scientific advisory board of TenSixteen Bio. H.M.K. and G.R.A. are employees of Regeneron Pharmaceuticals and owns stock and stock options for Regeneron Pharmaceuticals. M.H.C. has received grant support from Bayer. All other authors declare that they have no competing interests.
Data and materials availability: All TOPMed WGS data used in this analysis are available to researchers for purposes of reproducing or extending the analysis via the dbGaP accession numbers listed in table S1. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S3
Variant Calling Supplementary Text
TOPMed Cohort Acknowledgements
Legends for tables S1 to S13
Other Supplementary Material for this manuscript includes the following:
Table S1 to S13
REFERENCES AND NOTES
- 1.K. Yizhak, F. Aguet, J. Kim, J. M. Hess, K. Kübler, J. Grimsby, R. Frazer, H. Zhang, N. J. Haradhvala, D. Rosebrock, D. Livitz, X. Li, E. Arich-Landkof, N. Shoresh, C. Stewart, A. V. Segrè, P. A. Branton, P. Polak, K. G. Ardlie, G. Getz, RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 364, eaaw0726 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.P. Desai, N. Mencia-Trinchant, O. Savenkov, M. S. Simon, G. Cheang, S. Lee, M. Samuel, E. K. Ritchie, M. L. Guzman, K. V. Ballman, G. J. Roboz, D. C. Hassane, Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat. Med. 24, 1015–1023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.S. Abelson, G. Collord, S. W. K. Ng, O. Weissbrod, N. Mendelson Cohen, E. Niemeyer, N. Barda, P. C. Zuzarte, L. Heisler, Y. Sundaravadanam, R. Luben, S. Hayat, T. T. Wang, Z. Zhao, I. Cirlan, T. J. Pugh, D. Soave, K. Ng, C. Latimer, C. Hardy, K. Raine, D. Jones, D. Hoult, A. Britten, J. D. McPherson, M. Johansson, F. Mbabaali, J. Eagles, J. K. Miller, D. Pasternack, L. Timms, P. Krzyzanowski, P. Awadalla, R. Costa, E. Segal, S. V. Bratman, P. Beer, S. Behjati, I. Martincorena, J. C. Y. Wang, K. M. Bowles, J. R. Quirós, A. Karakatsani, C. La Vecchia, A. Trichopoulou, E. Salamanca-Fernández, J. M. Huerta, A. Barricarte, R. C. Travis, R. Tumino, G. Masala, H. Boeing, S. Panico, R. Kaaks, A. Krämer, S. Sieri, E. Riboli, P. Vineis, M. Foll, J. McKay, S. Polidoro, N. Sala, K.-T. Khaw, R. Vermeulen, P. J. Campbell, E. Papaemmanuil, M. D. Minden, A. Tanay, R. D. Balicer, N. J. Wareham, M. Gerstung, J. E. Dick, P. Brennan, G. S. Vassiliou, L. I. Shlush, Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 559, 400–404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.S. Jaiswal, P. Fontanillas, J. Flannick, A. Manning, P. V. Grauman, B. G. Mar, R. Coleman Lindsley, C. H. Mermel, N. Burtt, A. Chavez, J. M. Higgins, V. Moltchanov, F. C. Kuo, M. J. Kluk, B. Henderson, L. Kinnunen, H. A. Koistinen, C. Ladenvall, G. Getz, A. Correa, B. F. Banahan, S. Gabriel, S. Kathiresan, H. M. Stringham, M. I. McCarthy, M. Boehnke, J. Tuomilehto, C. Haiman, L. Groop, G. Atzmon, J. G. Wilson, D. Neuberg, D. Altshuler, B. L. Ebert, Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 26, 2488–2998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.G. Genovese, A. K. Kähler, R. E. Handsaker, J. Lindberg, S. A. Rose, S. F. Bakhoum, K. Chambert, E. Mick, B. M. Neale, M. Fromer, S. M. Purcell, O. Svantesson, M. Landén, M. Höglund, S. Lehmann, S. B. Gabriel, J. L. Moran, E. S. Lander, P. F. Sullivan, P. Sklar, H. Grönberg, C. M. Hultman, S. A. McCarroll, Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.M. Xie, C. Lu, J. Wang, M. D. McLellan, K. J. Johnson, M. C. Wendl, J. F. McMichael, H. K. Schmidt, V. Yellapantula, C. A. Miller, B. A. Ozenberger, J. S. Welch, D. C. Link, M. J. Walter, E. R. Mardis, J. F. Dipersio, F. Chen, R. K. Wilson, T. J. Ley, L. Ding, Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 20, 1472–1478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.S. Jaiswal, P. Natarajan, A. J. Silver, C. J. Gibson, A. G. Bick, E. Shvartz, M. McConkey, N. Gupta, S. Gabriel, D. Ardissino, U. Baber, R. Mehran, V. Fuster, J. Danesh, P. Frossard, D. Saleheen, O. Melander, G. K. Sukhova, D. Neuberg, P. Libby, S. Kathiresan, B. L. Ebert, Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N. Engl. J. Med. 377, 111–121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.A. G. Bick, J. S. Weinstock, S. K. Nandakumar, C. P. Fulco, E. L. Bao, S. M. Zekavat, M. D. Szeto, X. Liao, M. J. Leventhal, J. Nasser, K. Chang, C. Laurie, B. B. Burugula, C. J. Gibson, A. E. Lin, M. A. Taub, F. Aguet, K. Ardlie, B. D. Mitchell, K. C. Barnes, A. Moscati, M. Fornage, S. Redline, B. M. Psaty, E. K. Silverman, S. T. Weiss, N. D. Palmer, R. S. Vasan, E. G. Burchard, S. L. R. Kardia, J. He, R. C. Kaplan, N. L. Smith, D. K. Arnett, D. A. Schwartz, A. Correa, M. de Andrade, X. Guo, B. A. Konkle, B. Custer, J. M. Peralta, H. Gui, D. A. Meyers, S. T. McGarvey, I. Y.-D. Chen, M. B. Shoemaker, P. A. Peyser, J. G. Broome, S. M. Gogarten, F. F. Wang, Q. Wong, M. E. Montasser, M. Daya, E. E. Kenny, K. E. North, L. J. Launer, B. E. Cade, J. C. Bis, M. H. Cho, J. Lasky-Su, D. W. Bowden, L. A. Cupples, A. C. Y. Mak, L. C. Becker, J. A. Smith, T. N. Kelly, S. Aslibekyan, S. R. Heckbert, H. K. Tiwari, I. V. Yang, J. A. Heit, S. A. Lubitz, J. M. Johnsen, J. E. Curran, S. E. Wenzel, D. E. Weeks, D. C. Rao, D. Darbar, J.-Y. Moon, R. P. Tracy, E. J. Buth, N. Rafaels, R. J. F. Loos, P. Durda, Y. Liu, L. Hou, J. Lee, P. Kachroo, B. I. Freedman, D. Levy, L. F. Bielak, J. E. Hixson, J. S. Floyd, E. A. Whitsel, P. T. Ellinor, M. R. Irvin, T. E. Fingerlin, L. M. Raffield, S. M. Armasu, M. M. Wheeler, E. C. Sabino, J. Blangero, L. K. Williams, B. D. Levy, W. H.-H. Sheu, D. M. Roden, E. Boerwinkle, J. E. Manson, R. A. Mathias, P. Desai, K. D. Taylor, A. D. Johnson, P. L. Auer, C. Kooperberg, C. C. Laurie, T. W. Blackwell, A. V. Smith, H. Zhao, E. Lange, L. Lange, S. S. Rich, J. I. Rotter, J. G. Wilson, P. Scheet, J. O. Kitzman, E. S. Lander, J. M. Engreitz, B. L. Ebert, A. P. Reiner, S. Jaiswal, G. Abecasis, V. G. Sankaran, S. Kathiresan, P. Natarajan, Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature 586, 763–768 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.E. Rheinbay, M. M. Nielsen, F. Abascal, J. A. Wala, O. Shapira, G. Tiao, H. Hornshøj, J. M. Hess, R. I. Juul, Z. Lin, L. Feuerbach, R. Sabarinathan, T. Madsen, J. Kim, L. Mularoni, S. Shuai, A. Lanzós, C. Herrmann, Y. E. Maruvka, C. Shen, S. B. Amin, P. Bandopadhayay, J. Bertl, K. A. Boroevich, J. Busanovich, J. Carlevaro-Fita, D. Chakravarty, C. W. Y. Chan, D. Craft, P. Dhingra, K. Diamanti, N. A. Fonseca, A. Gonzalez-Perez, Q. Guo, M. P. Hamilton, N. J. Haradhvala, C. Hong, K. Isaev, T. A. Johnson, M. Juul, A. Kahles, A. Kahraman, Y. Kim, J. Komorowski, K. Kumar, S. Kumar, D. Lee, K.-V. Lehmann, Y. Li, E. M. Liu, L. Lochovsky, K. Park, O. Pich, N. D. Roberts, G. Saksena, S. E. Schumacher, N. Sidiropoulos, L. Sieverling, N. Sinnott-Armstrong, C. Stewart, D. Tamborero, J. M. C. Tubio, H. M. Umer, L. Uusküla-Reimand, C. Wadelius, L. Wadi, X. Yao, C.-Z. Zhang, J. Zhang, J. E. Haber, A. Hobolth, M. Imielinski, M. Kellis, M. S. Lawrence, C. von Mering, H. Nakagawa, B. J. Raphael, M. A. Rubin, C. Sander, L. D. Stein, J. M. Stuart, T. Tsunoda, D. A. Wheeler, R. Johnson, J. Reimand, M. Gerstein, E. Khurana, P. J. Campbell, N. López-Bigas; PCAWG Drivers and Functional Interpretation Working Group; PCAWG Structural Variation Working Group, J. Weischenfeldt, R. Beroukhim, I. Martincorena, J. S. Pedersen, G. Getz; PCAWG Consortium , Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 578, 102–111 (2020).32025015 [Google Scholar]
- 10.D. Taliun, D. N. Harris, M. D. Kessler, J. Carlson, Z. A. Szpiech, R. Torres, S. A. G. Taliun, A. Corvelo, S. M. Gogarten, H. M. Kang, A. N. Pitsillides, J. LeFaive, S. Lee, X. Tian, B. L. Browning, S. Das, A.-K. Emde, W. E. Clarke, D. P. Loesch, A. C. Shetty, T. W. Blackwell, Q. Wong, F. Aguet, C. Albert, A. Alonso, K. G. Ardlie, S. Aslibekyan, P. L. Auer, J. Barnard, R. G. Barr, L. C. Becker, R. L. Beer, E. J. Benjamin, L. F. Bielak, J. Blangero, M. Boehnke, D. W. Bowden, J. A. Brody, E. G. Burchard, B. E. Cade, J. F. Casella, B. Chalazan, Y.-D. I. Chen, M. H. Cho, S. H. Choi, M. K. Chung, C. B. Clish, A. Correa, J. E. Curran, B. Custer, D. Darbar, M. Daya, M. de Andrade, D. L. DeMeo, S. K. Dutcher, P. T. Ellinor, L. S. Emery, D. Fatkin, L. Forer, M. Fornage, N. Franceschini, C. Fuchsberger, S. M. Fullerton, S. Germer, M. T. Gladwin, D. J. Gottlieb, X. Guo, M. E. Hall, J. He, N. L. Heard-Costa, S. R. Heckbert, M. R. Irvin, J. M. Johnsen, A. D. Johnson, S. L. R. Kardia, T. Kelly, S. Kelly, E. E. Kenny, D. P. Kiel, R. Klemmer, B. A. Konkle, C. Kooperberg, A. Köttgen, L. A. Lange, J. Lasky-Su, D. Levy, X. Lin, K.-H. Lin, C. Liu, R. J. F. Loos, L. Garman, R. Gerszten, S. A. Lubitz, K. L. Lunetta, A. C. Y. Mak, A. Manichaikul, A. K. Manning, R. A. Mathias, D. D. McManus, S. T. McGarvey, J. B. Meigs, D. A. Meyers, J. L. Mikulla, M. A. Minear, B. Mitchell, S. Mohanty, M. E. Montasser, C. Montgomery, A. C. Morrison, J. M. Murabito, A. Natale, P. Natarajan, S. C. Nelson, K. E. North, J. R. O’Connell, N. D. Palmer, N. Pankratz, G. M. Peloso, P. A. Peyser, W. S. Post, B. M. Psaty, D. C. Rao, S. Redline, A. P. Reiner, D. Roden, J. I. Rotter, I. Ruczinski, C. Sarnowski, S. Schoenherr, J.-S. Seo, S. Seshadri, V. A. Sheehan, M. B. Shoemaker, A. V. Smith, N. L. Smith, J. A. Smith, N. Sotoodehnia, A. M. Stilp, W. Tang, K. D. Taylor, M. Telen, T. A. Thornton, R. P. Tracy, D. J. V. D. Berg, R. S. Vasan, K. A. Viaud-Martinez, S. Vrieze, D. E. Weeks, B. S. Weir, S. T. Weiss, L.-C. Weng, C. J. Willer, Y. Zhang, X. Zhao, D. K. Arnett, A. E. Ashley-Koch, K. C. Barnes, E. Boerwinkle, S. Gabriel, R. Gibbs, K. M. Rice, S. S. Rich, E. Silverman, P. Qasba, W. Gan, Trans-Omics for Precision Medicine (TOPMed) Program, TOPMed Population Genetics Working Group, G. J. Papanicolaou, D. A. Nickerson, S. R. Browning, M. C. Zody, S. Zöllner, J. G. Wilson, L. A. Cupples, C. C. Laurie, C. E. Jaquish, R. D. Hernandez, T. D. O’Connor, G. R. Abecasis, “Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program” (preprint, Genomics, 2019), doi: 10.1101/563866. [DOI]
- 11.K. Cibulskis, M. S. Lawrence, S. L. Carter, A. Sivachenko, D. Jaffe, C. Sougnez, S. Gabriel, M. Meyerson, E. S. Lander, G. Getz, Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.F. Zink, S. N. Stacey, G. L. Norddahl, M. L. Frigge, O. T. Magnusson, I. Jonsdottir, T. E. Thorgeirsson, A. Sigurdsson, S. A. Gudjonsson, J. Gudmundsson, J. G. Jonasson, L. Tryggvadottir, T. Jonsson, A. Helgason, A. Gylfason, P. Sulem, T. Rafnar, U. Thorsteinsdottir, D. F. Gudbjartsson, G. Masson, A. Kong, K. Stefansson, Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 130, 742–752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D. Taliun, D. N. Harris, M. D. Kessler, J. Carlson, Z. A. Szpiech, R. Torres, S. A. G. Taliun, A. Corvelo, S. M. Gogarten, H. M. Kang, A. N. Pitsillides, J. LeFaive, S. Lee, X. Tian, B. L. Browning, S. Das, A.-K. Emde, W. E. Clarke, D. P. Loesch, A. C. Shetty, T. W. Blackwell, A. V. Smith, Q. Wong, X. Liu, M. P. Conomos, D. M. Bobo, F. Aguet, C. Albert, A. Alonso, K. G. Ardlie, D. E. Arking, S. Aslibekyan, P. L. Auer, J. Barnard, R. G. Barr, L. Barwick, L. C. Becker, R. L. Beer, E. J. Benjamin, L. F. Bielak, J. Blangero, M. Boehnke, D. W. Bowden, J. A. Brody, E. G. Burchard, B. E. Cade, J. F. Casella, B. Chalazan, D. I. Chasman, Y.-D. I. Chen, M. H. Cho, S. H. Choi, M. K. Chung, C. B. Clish, A. Correa, J. E. Curran, B. Custer, D. Darbar, M. Daya, M. de Andrade, D. L. DeMeo, S. K. Dutcher, P. T. Ellinor, L. S. Emery, C. Eng, D. Fatkin, T. Fingerlin, L. Forer, M. Fornage, N. Franceschini, C. Fuchsberger, S. M. Fullerton, S. Germer, M. T. Gladwin, D. J. Gottlieb, X. Guo, M. E. Hall, J. He, N. L. Heard-Costa, S. R. Heckbert, M. R. Irvin, J. M. Johnsen, A. D. Johnson, R. Kaplan, S. L. R. Kardia, T. Kelly, S. Kelly, E. E. Kenny, D. P. Kiel, R. Klemmer, B. A. Konkle, C. Kooperberg, A. Köttgen, L. A. Lange, J. Lasky-Su, D. Levy, X. Lin, K.-H. Lin, C. Liu, R. J. F. Loos, L. Garman, R. Gerszten, S. A. Lubitz, K. L. Lunetta, A. C. Y. Mak, A. Manichaikul, A. K. Manning, R. A. Mathias, D. D. McManus, S. T. McGarvey, J. B. Meigs, D. A. Meyers, J. L. Mikulla, M. A. Minear, B. D. Mitchell, S. Mohanty, M. E. Montasser, C. Montgomery, A. C. Morrison, J. M. Murabito, A. Natale, P. Natarajan, S. C. Nelson, K. E. North, J. R. O’Connell, N. D. Palmer, N. Pankratz, G. M. Peloso, P. A. Peyser, J. Pleiness, W. S. Post, B. M. Psaty, D. C. Rao, S. Redline, A. P. Reiner, D. Roden, J. I. Rotter, I. Ruczinski, C. Sarnowski, S. Schoenherr, D. A. Schwartz, J.-S. Seo, S. Seshadri, V. A. Sheehan, W. H. Sheu, M. B. Shoemaker, N. L. Smith, J. A. Smith, N. Sotoodehnia, A. M. Stilp, W. Tang, K. D. Taylor, M. Telen, T. A. Thornton, R. P. Tracy, D. J. Van Den Berg, R. S. Vasan, K. A. Viaud-Martinez, S. Vrieze, D. E. Weeks, B. S. Weir, S. T. Weiss, L.-C. Weng, C. J. Willer, Y. Zhang, X. Zhao, D. K. Arnett, A. E. Ashley-Koch, K. C. Barnes, E. Boerwinkle, S. Gabriel, R. Gibbs, K. M. Rice, S. S. Rich, E. K. Silverman, P. Qasba, W. Gan; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium, G. J. Papanicolaou, D. A. Nickerson, S. R. Browning, M. C. Zody, S. Zöllner, J. G. Wilson, L. A. Cupples, C. C. Laurie, C. E. Jaquish, R. D. Hernandez, T. D. O’Connor, G. R. Abecasis, Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.M. M. Wheeler, A. M. Stilp, S. Rao, B. V. Halldórsson, D. Beyter, J. Wen, A. V. Mikhaylova, C. P. McHugh, J. Lane, M.-Z. Jiang, L. M. Raffield, G. Jun, F. J. Sedlazeck, G. Metcalf, Y. Yao, J. B. Bis, N. Chami, P. S. de Vries, P. Desai, J. S. Floyd, Y. Gao, K. Kammers, W. Kim, J.-Y. Moon, A. Ratan, L. R. Yanek, L. Almasy, L. C. Becker, J. Blangero, M. H. Cho, J. E. Curran, M. Fornage, R. C. Kaplan, J. P. Lewis, R. J. F. Loos, B. D. Mitchell, A. C. Morrison, M. Preuss, B. M. Psaty, S. S. Rich, J. I. Rotter, H. Tang, R. P. Tracy, E. Boerwinkle, G. Abecasis, T. W. Blackwell, A. V. Smith, A. D. Johnson, R. A. Mathias, D. A. Nickerson, M. P. Conomos, Y. Li, N. T.-O. for P. M. (TOPMed) Consortium, U. Þorsteinsdóttir, M. K. Magnússon, K. Stefansson, N. D. Pankratz, D. E. Bauer, P. L. Auer, A. P. Reiner, Whole genome sequencing identifies common and rare structural variants contributing to hematologic traits in the NHLBI TOPMed program (2021), p. 2021.12.16.21267871, doi: 10.1101/2021.12.16.21267871. [DOI]
- 15.K. J. Karczewski, L. C. Francioli, G. Tiao, B. B. Cummings, J. Alföldi, Q. Wang, R. L. Collins, K. M. Laricchia, A. Ganna, D. P. Birnbaum, L. D. Gauthier, H. Brand, M. Solomonson, N. A. Watts, D. Rhodes, M. Singer-Berk, E. M. England, E. G. Seaby, J. A. Kosmicki, R. K. Walters, K. Tashman, Y. Farjoun, E. Banks, T. Poterba, A. Wang, C. Seed, N. Whiffin, J. X. Chong, K. E. Samocha, E. Pierce-Hoffman, Z. Zappala, A. H. O’Donnell-Luria, E. V. Minikel, B. Weisburd, M. Lek, J. S. Ware, C. Vittal, I. M. Armean, L. Bergelson, K. Cibulskis, K. M. Connolly, M. Covarrubias, S. Donnelly, S. Ferriera, S. Gabriel, J. Gentry, N. Gupta, T. Jeandet, D. Kaplan, C. Llanwarne, R. Munshi, S. Novod, N. Petrillo, D. Roazen, V. Ruano-Rubio, A. Saltzman, M. Schleicher, J. Soto, K. Tibbetts, C. Tolonen, G. Wade, M. E. Talkowski, B. M. Neale, M. J. Daly, D. G. MacArthur, The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Ensembl Variant Effect Predictor | Genome Biology | Full Text; 10.1186/s13059-016-0974-4). [DOI] [PMC free article] [PubMed]
- 17.Roadmap Epigenomics Consortium, A. Kundaje, W. Meuleman, J. Ernst, M. Bilenky, A. Yen, A. Heravi-Moussavi, P. Kheradpour, Z. Zhang, J. Wang, M. J. Ziller, V. Amin, J. W. Whitaker, M. D. Schultz, L. D. Ward, A. Sarkar, G. Quon, R. S. Sandstrom, M. L. Eaton, Y.-C. Wu, A. R. Pfenning, X. Wang, M. Claussnitzer, Y. Liu, C. Coarfa, R. A. Harris, N. Shoresh, C. B. Epstein, E. Gjoneska, D. Leung, W. Xie, R. D. Hawkins, R. Lister, C. Hong, P. Gascard, A. J. Mungall, R. Moore, E. Chuah, A. Tam, T. K. Canfield, R. S. Hansen, R. Kaul, P. J. Sabo, M. S. Bansal, A. Carles, J. R. Dixon, K.-H. Farh, S. Feizi, R. Karlic, A.-R. Kim, A. Kulkarni, D. Li, R. Lowdon, G. Elliott, T. R. Mercer, S. J. Neph, V. Onuchic, P. Polak, N. Rajagopal, P. Ray, R. C. Sallari, K. T. Siebenthall, N. A. Sinnott-Armstrong, M. Stevens, R. E. Thurman, J. Wu, B. Zhang, X. Zhou, A. E. Beaudet, L. A. Boyer, P. L. De Jager, P. J. Farnham, S. J. Fisher, D. Haussler, S. J. M. Jones, W. Li, M. A. Marra, M. T. McManus, S. Sunyaev, J. A. Thomson, T. D. Tlsty, L.-H. Tsai, W. Wang, R. A. Waterland, M. Q. Zhang, L. H. Chadwick, B. E. Bernstein, J. F. Costello, J. R. Ecker, M. Hirst, A. Meissner, A. Milosavljevic, B. Ren, J. A. Stamatoyannopoulos, T. Wang, M. Kellis, Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.B. Schuster-Böckler, B. Lehner, Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature 488, 504–507 (2012). [DOI] [PubMed] [Google Scholar]
- 19.C. L. Zheng, N. J. Wang, J. Chung, H. Moslehi, J. Z. Sanborn, J. S. Hur, E. A. Collisson, S. S. Vemula, A. Naujokas, K. E. Chiotti, J. B. Cheng, H. Fassihi, A. J. Blumberg, C. V. Bailey, G. M. Fudem, F. G. Mihm, B. B. Cunningham, I. M. Neuhaus, W. Liao, D. H. Oh, J. E. Cleaver, P. E. LeBoit, J. F. Costello, A. R. Lehmann, J. W. Gray, P. T. Spellman, S. T. Arron, N. Huh, E. Purdom, R. J. Cho, Transcription restores DNA repair to heterochromatin, determining regional mutation rates in cancer genomes. Cell Rep. 9, 1228–1234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.L. B. Alexandrov, J. Kim, N. J. Haradhvala, M. N. Huang, A. W. Tian Ng, Y. Wu, A. Boot, K. R. Covington, D. A. Gordenin, E. N. Bergstrom, S. M. A. Islam, N. Lopez-Bigas, L. J. Klimczak, J. R. McPherson, S. Morganella, R. Sabarinathan, D. A. Wheeler, V. Mustonen; PCAWG Mutational Signatures Working Group, G. Getz, S. G. Rozen, M. R. Stratton; PCAWG Consortium , The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D. P. Steensma, R. Bejar, S. Jaiswal, R. C. Lindsley, M. A. Sekeres, R. P. Hasserjian, B. L. Ebert, Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 126, 9–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.J. Zhang, R. Bajari, D. Andric, F. Gerthoffert, A. Lepsa, H. Nahal-Bose, L. D. Stein, V. Ferretti, The International Cancer Genome Consortium data portal. Nat. Biotechnol. 37, 367–369 (2019). [DOI] [PubMed] [Google Scholar]
- 23.A. Migliazza, S. Martinotti, W. Chen, C. Fusco, B. H. Ye, D. M. Knowles, K. Offit, R. S. Chaganti, R. Dalla-Favera, Frequent somatic hypermutation of the 5′ noncoding region of the BCL6 gene in B-cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 92, 12520–12524 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium , Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.M. Pribyl, S. Hubackova, A. Moudra, M. Vancurova, H. Polackova, T. Stopka, A. Jonasova, R. Bokorova, O. Fuchs, J. Stritesky, B. Salovska, J. Bartek, Z. Hodny, Aberrantly elevated suprabasin in the bone marrow as a candidate biomarker of advanced disease state in myelodysplastic syndromes. Mol. Oncol. 14, 2403–2419 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.J. Baglama, L. Reichel, Augmented implicitly restarted Lanczos Bidiagonalization methods. SIAM J. Sci. Comput. 27, 19–42 (2005). [Google Scholar]
- 27.J. Novembre, T. Johnson, K. Bryc, Z. Kutalik, A. R. Boyko, A. Auton, A. Indap, K. S. King, S. Bergmann, M. R. Nelson, M. Stephens, C. D. Bustamante, Genes mirror geography within Europe. Nature 456, 98–101 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.N. Patterson, A. L. Price, D. Reich, Population structure and eigenanalysis. PLOS Genetics 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.A. L. Price, N. J. Patterson, R. M. Plenge, M. E. Weinblatt, N. A. Shadick, D. Reich, Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 30.A. Koren, R. E. Handsaker, N. Kamitaki, R. Karlić, S. Ghosh, P. Polak, K. Eggan, S. A. McCarroll, Genetic variation in human dna replication timing. Cell 159, 1015–1026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.I. L. Berg, R. Neumann, K.-W. G. Lam, S. Sarbajna, L. Odenthal-Hesse, C. A. May, A. J. Jeffreys, PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat. Genet. 42, 859–863 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.C. Terao, A. Suzuki, Y. Momozawa, M. Akiyama, K. Ishigaki, K. Yamamoto, K. Matsuda, Y. Murakami, S. A. McCarroll, M. Kubo, P.-R. Loh, Y. Kamatani, Chromosomal alterations among age-related haematopoietic clones in Japan. Nature 584, 130–135 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.W. Zhou, J. B. Nielsen, L. G. Fritsche, R. Dey, M. E. Gabrielsen, B. N. Wolford, J. LeFaive, P. VandeHaar, S. A. Gagliano, A. Gifford, L. A. Bastarache, W.-Q. Wei, J. C. Denny, M. Lin, K. Hveem, H. M. Kang, G. R. Abecasis, C. J. Willer, S. Lee, Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 50, 1335–1341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.P. Hernández-Malmierca, D. Vonficht, A. Schnell, H. J. Uckelmann, A. Bollhagen, M. A. A. Mahmoud, S.-L. Landua, E. van der Salm, C. L. Trautmann, S. Raffel, F. Grünschläger, R. Lutz, M. Ghosh, S. Renders, N. Correia, E. Donato, K. O. Dixon, C. Hirche, C. Andresen, C. Robens, P. S. Werner, T. Boch, D. Eisel, W. Osen, F. Pilz, A. Przybylla, C. Klein, F. Buchholz, M. D. Milsom, M. A. G. Essers, S. B. Eichmüller, W.-K. Hofmann, D. Nowak, D. Hübschmann, M. Hundemer, C. Thiede, L. Bullinger, C. Müller-Tidow, S. A. Armstrong, A. Trumpp, V. K. Kuchroo, S. Haas, Antigen presentation safeguards the integrity of the hematopoietic stem cell pool. Cell Stem Cell 29, 760–775.e10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.GTEx Consortium , The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.B. B. Sun, J. C. Maranville, J. E. Peters, D. Stacey, J. R. Staley, J. Blackshaw, S. Burgess, T. Jiang, E. Paige, P. Surendran, C. Oliver-Williams, M. A. Kamat, B. P. Prins, S. K. Wilcox, E. S. Zimmerman, A. Chi, N. Bansal, S. L. Spain, A. M. Wood, N. W. Morrell, J. R. Bradley, N. Janjic, D. J. Roberts, W. H. Ouwehand, J. A. Todd, N. Soranzo, K. Suhre, D. S. Paul, C. S. Fox, R. M. Plenge, J. Danesh, H. Runz, A. S. Butterworth, Genomic atlas of the human plasma proteome. Nature 558, 73–79 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.M.-H. Chen, L. M. Raffield, A. Mousas, S. Sakaue, J. E. Huffman, A. Moscati, B. Trivedi, T. Jiang, P. Akbari, D. Vuckovic, E. L. Bao, X. Zhong, R. Manansala, V. Laplante, M. Chen, K. S. Lo, H. Qian, C. A. Lareau, M. Beaudoin, K. A. Hunt, M. Akiyama, T. M. Bartz, Y. Ben-Shlomo, A. Beswick, J. Bork-Jensen, E. P. Bottinger, J. A. Brody, F. J. A. van Rooij, K. Chitrala, K. Cho, H. Choquet, A. Correa, J. Danesh, E. Di Angelantonio, N. Dimou, J. Ding, P. Elliott, T. Esko, M. K. Evans, J. S. Floyd, L. Broer, N. Grarup, M. H. Guo, A. Greinacher, J. Haessler, T. Hansen, J. M. M. Howson, Q. Q. Huang, W. Huang, E. Jorgenson, T. Kacprowski, M. Kähönen, Y. Kamatani, M. Kanai, S. Karthikeyan, F. Koskeridis, L. A. Lange, T. Lehtimäki, M. M. Lerch, A. Linneberg, Y. Liu, L.-P. Lyytikäinen, A. Manichaikul, H. C. Martin, K. Matsuda, K. L. Mohlke, N. Mononen, Y. Murakami, G. N. Nadkarni, M. Nauck, K. Nikus, W. H. Ouwehand, N. Pankratz, O. Pedersen, M. Preuss, B. M. Psaty, O. T. Raitakari, D. J. Roberts, S. S. Rich, B. A. T. Rodriguez, J. D. Rosen, J. I. Rotter, P. Schubert, C. N. Spracklen, P. Surendran, H. Tang, J.-C. Tardif, R. C. Trembath, M. Ghanbari, U. Völker, H. Völzke, N. A. Watkins, A. B. Zonderman; VA Million Veteran Program, P. W. F. Wilson, Y. Li, A. S. Butterworth, J.-F. Gauchat, C. W. K. Chiang, B. Li, R. J. F. Loos, W. J. Astle, E. Evangelou, D. A. van Heel, V. G. Sankaran, Y. Okada, N. Soranzo, A. D. Johnson, A. P. Reiner, P. L. Auer, G. Lettre, Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell 182, 1198–1213.e14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.T. Calandra, T. Roger, Macrophage migration inhibitory factor: A regulator of innate immunity. Nat. Rev. Immunol. 3, 791–800 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R. A. Mitchell, H. Liao, J. Chesney, G. Fingerle-Rowson, J. Baugh, J. David, R. Bucala, Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53: Regulatory role in the innate immune response. Proc. Natl. Acad. Sci. 99, 345–350 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R. Bogeska, A.-M. Mikecin, P. Kaschutnig, M. Fawaz, M. Büchler-Schäff, D. Le, M. Ganuza, A. Vollmer, S. V. Paffenholz, N. Asada, E. Rodriguez-Correa, F. Frauhammer, F. Buettner, M. Ball, J. Knoch, S. Stäble, D. Walter, A. Petri, M. J. Carreño-Gonzalez, V. Wagner, B. Brors, S. Haas, D. B. Lipka, M. A. G. Essers, V. Weru, T. Holland-Letz, J.-P. Mallm, K. Rippe, S. Krämer, M. Schlesner, S. M. Freeman, M. C. Florian, K. Y. King, P. S. Frenette, M. A. Rieger, M. D. Milsom, Inflammatory exposure drives long-lived impairment of hematopoietic stem cell self-renewal activity and accelerated aging. Cell Stem Cell 29, 1273–1284.e8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A. V. Jones, A. Chase, R. T. Silver, D. Oscier, K. Zoi, Y. L. Wang, H. Cario, H. L. Pahl, A. Collins, A. Reiter, F. Grand, N. C. P. Cross, JAK2 haplotype is a major risk factor for the development of myeloproliferative neoplasms. Nat. Genet. 41, 446–449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O. Kilpivaara, S. Mukherjee, A. M. Schram, M. Wadleigh, A. Mullally, B. L. Ebert, A. Bass, S. Marubayashi, A. Heguy, G. Garcia-Manero, H. Kantarjian, K. Offit, R. M. Stone, D. G. Gilliland, R. J. Klein, R. L. Levine, A germline JAK2 SNP is associated with predisposition to the development of JAK2V617F-positive myeloproliferative neoplasms. Nat. Genet. 41, 455–459 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D. Olcaydu, A. Harutyunyan, R. Jäger, T. Berg, B. Gisslinger, I. Pabinger, H. Gisslinger, R. Kralovics, A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat. Genet. 41, 450–454 (2009). [DOI] [PubMed] [Google Scholar]
- 44.P.-R. Loh, G. Genovese, R. E. Handsaker, H. K. Finucane, Y. A. Reshef, P. F. Palamara, B. M. Birmann, M. E. Talkowski, S. F. Bakhoum, S. A. McCarroll, A. L. Price, Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature 559, 350–355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.P.-R. Loh, G. Genovese, S. A. McCarroll, Monogenic and polygenic inheritance become instruments for clonal selection. Nature 584, 136–141 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.H. Zhou, H. Xiong, H. Li, S. E. Plevy, P. D. Walden, M. Sassaroli, G. D. Prestwich, J. C. Unkeless, Microtubule-associated serine/threonine kinase-205 kDa and Fc gamma receptor control IL-12 p40 synthesis and NF-kappa B activation. J. Immunol. 172, 2559–2568 (2004). [DOI] [PubMed] [Google Scholar]
- 47.P. Rentzsch, D. Witten, G. M. Cooper, J. Shendure, M. Kircher, CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47, D886–D894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.J. Ernst, M. Kellis, ChromHMM: Automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.L. Chen, B. Ge, F. P. Casale, L. Vasquez, T. Kwan, D. Garrido-Martín, S. Watt, Y. Yan, K. Kundu, S. Ecker, A. Datta, D. Richardson, F. Burden, D. Mead, A. L. Mann, J. M. Fernandez, S. Rowlston, S. P. Wilder, S. Farrow, X. Shao, J. J. Lambourne, A. Redensek, C. A. Albers, V. Amstislavskiy, S. Ashford, K. Berentsen, L. Bomba, G. Bourque, D. Bujold, S. Busche, M. Caron, S.-H. Chen, W. Cheung, O. Delaneau, E. T. Dermitzakis, H. Elding, I. Colgiu, F. O. Bagger, P. Flicek, E. Habibi, V. Iotchkova, E. Janssen-Megens, B. Kim, H. Lehrach, E. Lowy, A. Mandoli, F. Matarese, M. T. Maurano, J. A. Morris, V. Pancaldi, F. Pourfarzad, K. Rehnstrom, A. Rendon, T. Risch, N. Sharifi, M.-M. Simon, M. Sultan, A. Valencia, K. Walter, S.-Y. Wang, M. Frontini, S. E. Antonarakis, L. Clarke, M.-L. Yaspo, S. Beck, R. Guigo, D. Rico, J. H. A. Martens, W. H. Ouwehand, T. W. Kuijpers, D. S. Paul, H. G. Stunnenberg, O. Stegle, K. Downes, T. Pastinen, N. Soranzo, Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell 167, 1398–1414.e24 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.P. S. Robinson, L. E. Thomas, F. Abascal, H. Jung, L. M. R. Harvey, H. D. West, S. Olafsson, B. C. H. Lee, T. H. H. Coorens, H. Lee-Six, L. Butlin, N. Lander, R. Truscott, M. A. Sanders, S. V. Lensing, S. J. A. Buczacki, R. ten Hoopen, N. Coleman, R. Brunton-Sim, S. Rushbrook, K. Saeb-Parsy, F. Lalloo, P. J. Campbell, I. Martincorena, J. R. Sampson, M. R. Stratton, Inherited MUTYH mutations cause elevated somatic mutation rates and distinctive mutational signatures in normal human cells. Nat. Commun. 13, 3949 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.T. A. Sasani, D. G. Ashbrook, A. C. Beichman, L. Lu, A. A. Palmer, R. W. Williams, J. K. Pritchard, K. Harris, A natural mutator allele shapes mutation spectrum variation in mice. Nature 605, 497–502 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.A map of human genome variation from population scale sequencing. Nature 467, 1061–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.M. Fuxa, J. Skok, A. Souabni, G. Salvagiotto, E. Roldan, M. Busslinger, Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 18, 411–422 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.T. Iwakuma, G. Lozano, MDM2, An Introduction. Mol. Cancer Res. 1, 993–1000 (2003). [PubMed] [Google Scholar]
- 55.N. W. Galwey, A new measure of the effective number of tests, a practical tool for comparing families of non-independent significance tests. Genet. Epidemiol. 33, 559–568 (2009). [DOI] [PubMed] [Google Scholar]
- 56.J. D. Storey, The positive false discovery rate: A Bayesian interpretation and the q-value. Ann. Stat. 31, 2013–2035 (2003). [Google Scholar]
- 57.C. J. Watson, A. L. Papula, G. Y. P. Poon, W. H. Wong, A. L. Young, T. E. Druley, D. S. Fisher, J. R. Blundell, The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science 367, 1449–1454 (2020). [DOI] [PubMed] [Google Scholar]
- 58.X. Sun, A. Xue, T. Qi, D. Chen, D. Shi, Y. Wu, Z. Zheng, J. Zeng, J. Yang, Tumor mutational burden is polygenic and genetically associated with complex traits and diseases. Cancer Res. 81, 1230–1239 (2021). [DOI] [PubMed] [Google Scholar]
- 59.A. G. Bick, J. P. Pirruccello, G. K. Griffin, N. Gupta, S. Gabriel, D. Saleheen, P. Libby, S. Kathiresan, P. Natarajan, Genetic interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation 141, 124–131 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.A. A. Regier, Y. Farjoun, D. E. Larson, O. Krasheninina, H. M. Kang, D. P. Howrigan, B. J. Chen, M. Kher, E. Banks, D. C. Ames, A. C. English, H. Li, J. Xing, Y. Zhang, T. Matise, G. R. Abecasis, W. Salerno, M. C. Zody, B. M. Neale, I. M. Hall, Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat. Commun. 9, 4038 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.G. Jun, M. K. Wing, G. R. Abecasis, H. M. Kang, An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data. Genome Res. 25, 918–925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.P. Cingolani, A. Platts, L. L. Wang, M. Coon, T. Nguyen, L. Wang, S. J. Land, X. Lu, D. M. Ruden, A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.K. Voss, J. Gentry, G. Van der Auwera, Full-stack genomics pipelining with GATK4 + WDL + Cromwell. F1000Research 6, (2017). [Google Scholar]
- 64.E. M. Beauchamp, M. Leventhal, E. Bernard, E. R. Hoppe, G. Todisco, M. Creignou, A. Gallì, C. A. Castellano, M. McConkey, A. Tarun, W. Wong, M. Schenone, C. Stanclift, B. Tanenbaum, E. Malolepsza, B. Nilsson, A. G. Bick, J. S. Weinstock, M. Miller, A. Niroula, A. Dunford, A. Taylor-Weiner, T. Wood, A. Barbera, S. Anand, B. M. Psaty, P. Desai, M. H. Cho, A. D. Johnson, R. Loos; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium, D. G. MacArthur, M. Lek, D. S. Neuberg, K. Lage, S. A. Carr, E. Hellstrom-Lindberg, L. Malcovati, E. Papaemmanuil, C. Stewart, G. Getz, R. K. Bradley, S. Jaiswal, B. L. Ebert, ZBTB33 is mutated in clonal hematopoiesis and myelodysplastic syndromes and impacts RNA splicing. Blood Cancer Discov. 2, 500–517 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.B. S. Pedersen, A. R. Quinlan, cyvcf2: Fast, flexible variant analysis with Python. Bioinformatics 33, 1867–1869 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.VariantKey: A Reversible Numerical Representation of Human Genetic Variants | bioRxiv; https://biorxiv.org/content/10.1101/473744v3.
- 67.H. Li, B. Handsaker, A. Wysoker, T. Fennell, J. Ruan, N. Homer, G. Marth, G. Abecasis, R. Durbin; 1000 Genome Project Data Processing Subgroup , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Integrative analysis of 111 reference human epigenomes | Nature; https://nature.com/articles/nature14248. [DOI] [PMC free article] [PubMed]
- 69.R. M. Layer, B. S. Pedersen, T. DiSera, G. T. Marth, J. Gertz, A. R. Quinlan, GIGGLE: A search engine for large-scale integrated genome analysis. Nat. Methods 15, 123–126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.J. Kim, K. W. Mouw, P. Polak, L. Z. Braunstein, A. Kamburov, G. Tiao, D. J. Kwiatkowski, J. E. Rosenberg, E. M. Van Allen, A. D. D’Andrea, G. Getz, Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nat. Genet. 48, 600–606 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.G. Jun, M. Flickinger, K. N. Hetrick, J. M. Romm, K. F. Doheny, G. R. Abecasis, M. Boehnke, H. M. Kang, Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am. J. Hum. Genet. 91, 839–848 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.C. C. Chang, C. C. Chow, L. C. Tellier, S. Vattikuti, S. M. Purcell, J. J. Lee, Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S3
Variant Calling Supplementary Text
TOPMed Cohort Acknowledgements
Legends for tables S1 to S13
Table S1 to S13