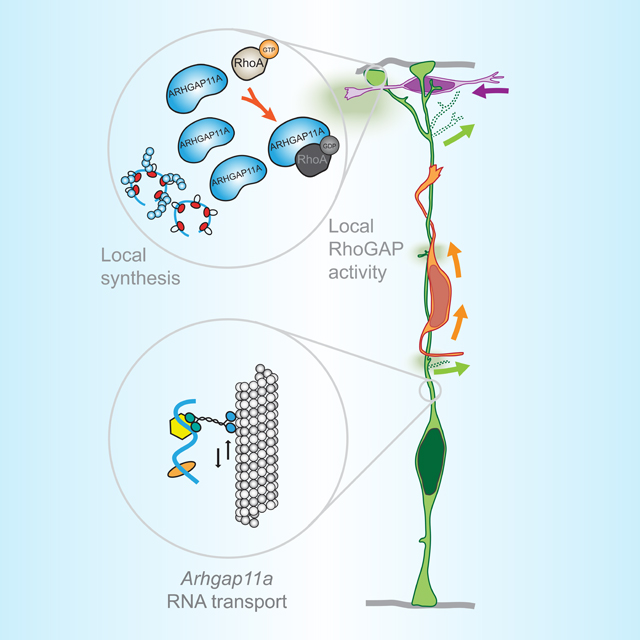
Summary
mRNA localization and local translation enable exquisite spatial and temporal control of gene expression, particularly in polarized, elongated cells. These features are especially prominent in radial glial cells (RGCs), which are neural and glial precursors of the developing cerebral cortex and scaffolds for migrating neurons. Yet the mechanisms by which subcellular RGC compartments accomplish their diverse functions are poorly understood. Here, we demonstrate that RNA localization and local translation of the RhoGAP ARHGAP11A in basal endfeet of RGCs controls their morphology and mediates neuronal positioning. Arhgap11a transcript and protein exhibit conserved localization to RGC basal structures in mice and humans, conferred by the 5′UTR. Proper RGC morphology relies upon active Arhgap11a mRNA transport and localization to basal endfeet, where ARHGAP11A is locally synthesized. This translation is essential for positioning interneurons at the basement membrane. Thus, local translation spatially and acutely activates Rho signaling in RGCs to compartmentalize neural progenitor functions.
Keywords: mRNA localization, local translation, Arhgap11a, radial glial endfeet, interneuron positioning neuronal migration
eTOC
Pilaz et al. demonstrate Arhgap11a mRNA undergoes active transport and local translation in radial glial basal endfeet, which is essential for proper endfeet morphology. Further, Arhgap11a non-cell autonomously impacts positioning of interneurons and excitatory neurons. This demonstrates for the first time, functions of local translation in radial glial progenitors.
Graphical Abstract
Introduction
In eukaryotes, subcellular RNA localization and local translation allow cells to temporally and spatially control functions that rely on dynamic and complex proteomes. In highly polarized cells, such as neurons and migrating fibroblasts, mRNA localization plays a pivotal role in local cytoskeletal regulation and hence local morphology 1,2. Most localization studies use cultured cells or mechanical axotomy, limiting an understanding of events in vivo. Notably, the developing and adult brain contains some of the most highly polarized and elongated cell types found in animals. These features are especially prominent in radial glial cells (RGCs), which control cortical development by acting as neural stem cells to generate neurons and then astrocytes, and by scaffolding radial neuron migration 3–8.
RGCs are bipolar with basal processes emanating from cell bodies in the ventricular zone (VZ) and radially traversing the cortex to form basal endfeet at the pia and apical endfeet at the ventricle 9,10. The basal process can be extremely long, reaching several hundred microns in the mouse and centimeters in humans. The apical and basal endfeet encounter unique niches, with the latter including interneurons, Cajal-Retzius neurons and excitatory neurons 11. Basal endfeet are tightly connected to the basal lamina, forming a barrier between the brain and the overlying meninges, composed of fibroblasts and blood vessels 12–14. Along the basal process and at endfeet, dynamic filopodia-like protrusions extend and retract, which is postulated to influence signaling and neuronal migration 15. Further, as development proceeds, basal endfeet become more complex in number 16; basal structures are also notably complex in humans 17–19. Disruptions to RGC morphology can have disastrous consequences on the architecture of the mature cortex, causing cobblestone malformation and lissencephaly 20,21. Thus, RGC morphology and subcellular compartmentalization are central to cortical development. Yet, we know surprisingly little about the cellular and molecular mechanisms mediating proper morphology, dynamics, and function of distal RGC basal structures.
RGC endfeet are major sites of mRNA localization and are enriched for transcripts encoding cytoskeletal and signaling regulators, including GTPase regulators 22,23. Notably, the ubiquitous Rho GTPase is essential for cortical development 24, but whether and how modulation of localized Rho activity controls RGCs is unknown. Arhgap11a encodes a RhoA-specific GAP, which promotes GTP hydrolysis and therefore inactivates the small GTPase RhoA 25. Hence, loss of Arhgap11a is predicted to increase RhoA signaling. Arhgap11a has essential roles in modulating the cytoskeleton, including mediating cytokinesis 26, cell invasion 27,28, and neurite outgrowth 29. In this light, local synthesis of a Rho regulator, such as ARHGAP11A, could help dictate subcellular morphology and function of RGCs. However, the requirement of local translation of any transcript in RGCs has never been examined.
In this study, we tested whether mRNA localization and local translation of the RhoGAP ARHGAP11A in endfeet mediates RGC morphology and cortical development. We show that both Arhgap11a mRNA and protein subcellularly localize to RGC basal endfeet and basal processes during cortical development in mice and humans. Consistent with a role in these structures, Arhgap11a depletion from RGCs disrupts basal process and endfeet morphology. This causes a non-cell autonomous impairment of excitatory neuron migration and inhibitory neuron positioning. We show the Arhgap11a 5′UTR is critical for its active transport in the basal process and local translation in basal endfeet. Importantly, impaired RGC morphology is rescued only when Arhgap11a localizes to endfeet and when RhoGAP activity is intact. We further find that rescuing morphology recovers proper position of interneurons along the basement membrane. Altogether, our study establishes critical, new in vivo requirements of subcellular mRNA localization in neural stem cells of the developing brain.
Results
Arhgap11a mRNA and protein subcellularly localize to RGC basal endfeet midway through cortical development
In this study we asked whether the morphology and functions of RGCs are mediated by subcellular targeting of specific mRNAs and their local translation (Figure 1A). From our prior study, we noted that RGC basal endfeet are significantly enriched for mRNAs encoding cytoskeletal and GTPase signaling regulators 22. We therefore sought to investigate functions of transcripts associated with these cellular processes. Arhgap11a stood out as an outstanding candidate, given its established role in cytoskeletal regulation, neurite outgrowth, and GTPase signaling 25,26,28,29.
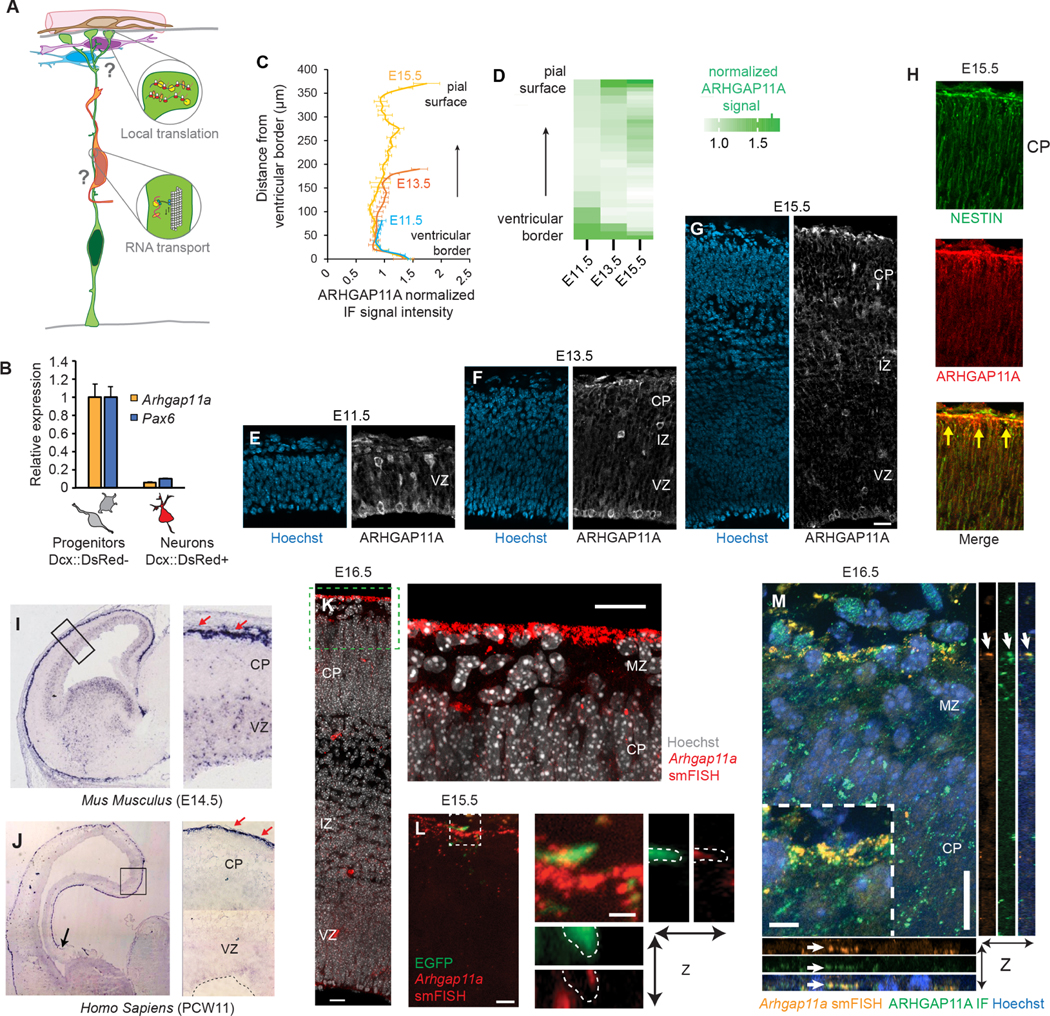
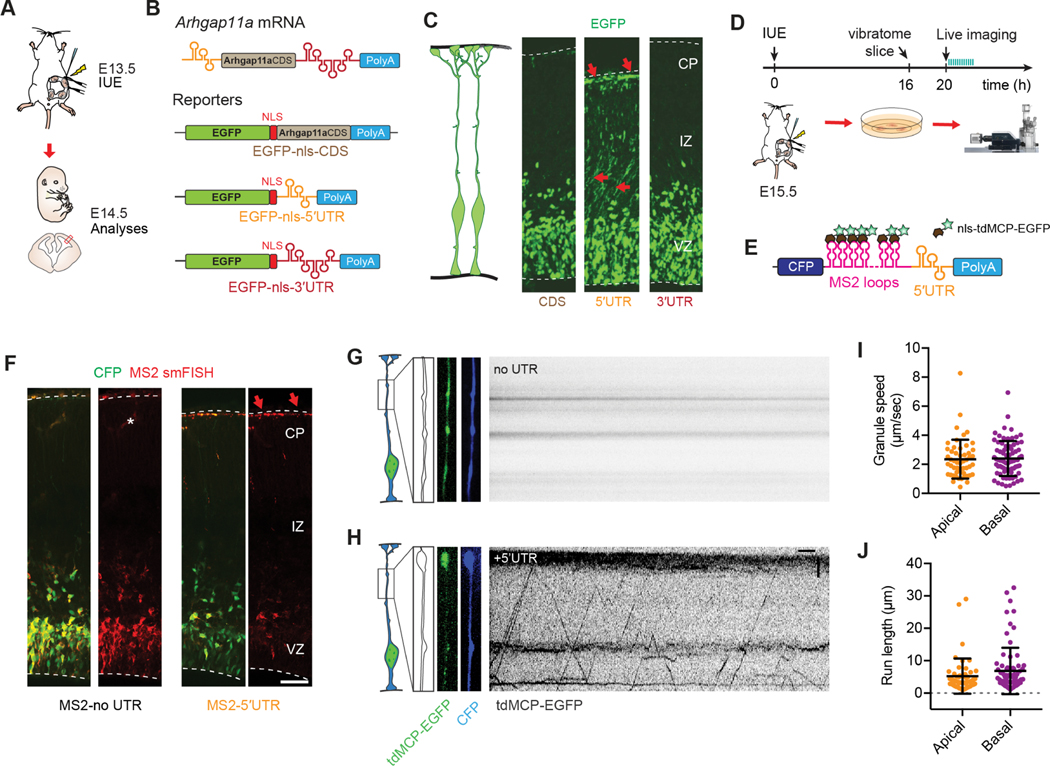
Figure 1. Subcellular localization of Arhgap11a mRNA and protein to RGC basal processes and endfeet during cortical development.
(A) Cartoon of a radial glial progenitor (RGC, green) with mRNA transport along the basal process and local translation in endfeet. Question marks reflect goal of the present study: what is the role of mRNA subcellular localization and translation in RGCs and for positioning of excitatory neurons (orange), migratory interneurons (purple), and cajal-retzius neurons (blue)?
(B) qPCR analyses of Arhgap11a mRNA levels in E14.5 sorted embryonic cortical cells. (n=4 brains, 3 technical replicates)
(C,D) Quantification of ARHGAP11A Immunofluorescence in E11.5, E13.5 and E15.5 brains. (E-G) Immunofluorescence of ARHGAP11A (grey) and Hoechst (blue) in E11.5 (E), E13.5 (F) and E15.5 (G) brains.
(H) Immunofluorescence of ARHGAP11A (red) at E15.5, showing expression in NESTIN positive RGCs (green) with overlap (yellow signal) in basal process and endfeet at the pial surface (yellow arrows).
(I,J) In situ hybridization of Arhgap11a mRNA (purple signal), showing strong enrichment at the pia where RGC basal endfeet reside (red arrows) at E14.5 (I) and in GW11 human fetal brains (J).
(K,L) smFISH in situ hybridization depicting Arhgap11a mRNA (red) at the pia at E15.5 (K) and in EGFP+ RGC basal endfeet (brains electroporated one day earlier) (L). Right panels, magnified areas highlighted in left panels (K, L) and maximum intensity projections of a z-stack (L).
(M) smFISH and immunofluorescence targeting Arhgap11a mRNA (red) and protein (green), respectively highlights colocalization (arrows) in RGC basal endfeet.
VZ: ventricular zone, CP: cortical plate, IZ: intermediate zone, smFISH: single molecule fluorescent in situ hybridization. Scale bars: C-E, 20 μm; I, 20 μm; J, left panel: 5 μm, right panel: 1μm; M, 20 μm.
To examine the role of Arhgap11a in RGCs, we assessed its expression during mouse cortical development. Cortical neurogenesis in mice occurs between E11 and E18.5 and in humans, at gestational week (GW) 7–24 30–32. We used E14.5 Dcx-DsRed transgenic mice 33 together with FACS and quantitative PCR (qPCR) to measure Arhgap11a mRNA expression in both progenitors (DsRed-negative) and neurons (DsRed-positive) (Figures 1B,S1A). Notably, Arhgap11a was expressed in progenitors but absent from newborn excitatory neurons. Likewise, in single-cell RNA-seq datasets of the developing mouse and human cortex, Arhgap11a was highly enriched in RGCs but absent from post-mitotic excitatory and inhibitory neurons (Figure S1B–D) 34–36. These data demonstrate that Arhgap11a expression in the developing cerebral cortex is largely specific to progenitors, including RGCs.
Next, we used immunohistochemistry to evaluate the expression pattern of ARHGAP11A protein in the developing mouse cortex. At E11.5, ARHGAP11A localized within the germinal zones (Figures 1C,D,E). However, strikingly, beginning at E13.5, ARHGAP11A protein became enriched near the pial surface, at presumptive RGC endfeet and along basal processes (Figure 1C,D,F). This pial localization of ARHGAP11A was especially visible at later stages (Figure 1C,D,G). We verified ARHGAP11A expression and localization in RGCs by co-staining with the RGC intermediate filament marker NESTIN at E15.5 (Figure 1H). In contrast, ARHGAP11A did not co-localize with Reelin-positive Cajal-Retzius neurons at the pia, nor was it expressed in interneurons (Figure S1C–F). This timing of ARHGAP11A localization to RGC basal processes and endfeet coincides with the onset of increased branching of endfeet 16, suggesting that this RhoGAP could locally influence RGC basal morphology.
We next examined Arhgap11a mRNA localization over the course of corticogenesis, to determine if its spatial and temporal pattern matched that of the protein. Towards this end, we used traditional as well as single-molecule fluorescent in situ hybridization (smFISH). At E14.5, Arhgap11a mRNA was significantly enriched at the pial surface compared to the VZ (Figure 1I, S1G,H). Reinforcing the specific expression of Arhgap11a in RGCs, Arhgap11a co-localized with EGFP-labeled RGCs (introduced one day earlier via in utero electroporation, IUE) but was notably absent from Tuj1-positive neurons (Figure S1G,H). At E15.5 and E16.5, Arhgap11a localization at the pia was especially prominent. smFISH confirmed Arhgap11a mRNA expression in EGFP-labeled RGC basal endfeet (Figure 1K,L). Arhgap11a RNA and protein co-localized at the pia, further evidencing this expression pattern (Figure 1M).
Given this striking localization pattern of Arhgap11a to RGC endfeet, and the conserved expression within RGCs of mice and humans (Figure S1B–D), we next tested whether Arhgap11a mRNA is also present in the basal endfeet region of the human developing cortex. Thus, we employed in situ hybridization on post-conceptional week 11 tissue sections. ARHGAP11A mRNA exhibited conserved robust enrichment at the pia in human fetal cortices (Figure 1J), in line with a previous report 37. Altogether, these data demonstrate that Arhgap11a RNA and protein exhibit concordant subcellular localization in a developmentally controlled fashion to RGC basal processes and endfeet.
Arhgap11a is critical for RGC basal process morphology and non-cell autonomous control of radial neuronal migration
ARHGAP11A localization to RGC basal processes and endfeet coincides developmentally with increasing morphological complexity of these structures 16. Further, excitatory neurons rely on the integrity of RGC basal processes to migrate to the cortical plate 38–40. Given that ARHGAP11A has established functions in modulating RhoA signaling and cytoskeletal morphology 25,26,29, we hypothesized that Arhgap11a could regulate basal process morphology and thus influence excitatory neuron migration. To evaluate these possibilities, siRNAs targeting the 3′UTR of Arhgap11a mRNA were introduced by IUE together with a membrane-localized EGFP reporter driven by an RGC-specific promoter (pGLAST-EGFP-CAAX, Figure 2A). We manipulated Arhgap11a expression at E15.5, reasoning that alterations to basal process complexity should be evident at this stage 16. Using siRNAs, we effectively depleted Arhgap11a in RGCs both at the mRNA and protein level, in RGC cell bodies and basal endfeet (Figure 2B,C, bins 1 and 10, respectively). Quantification of Arhgap11a RNA punctae per endfoot further validated this finding (Figure 2D). Furthermore, we observed little FISH signal in the IZ or CP within the control, thus reinforcing the specific expression of Arhgap11a in RGCs but not neurons. Finally, immunofluorescence showed a drastic reduction in ARHGAP11A protein levels in the electroporated area (Figure 2E).
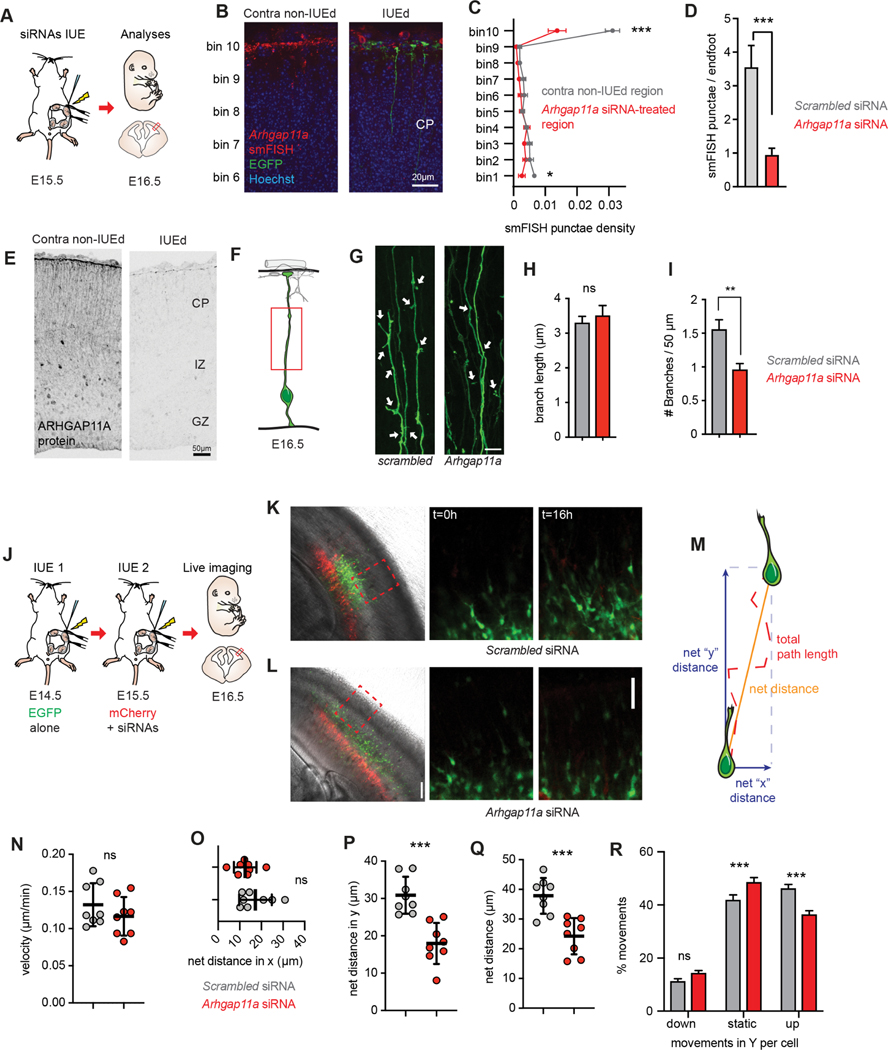
Figure 2. Arhgap11a controls RGC basal process morphology and non-cell autonomously controls radial migration of excitatory neurons.
(A) Schematic overview of the experiments in (B-I).
(B-E) Arhgap11a mRNA is depleted from endfeet in the Arhgap11a siRNA electroporated region (IUE, green), evidenced by smFISH (red) (B) and immunofluorescence (E). (C) Binned quantification of Arhgap11a smFISH punctae in electroporated and contralateral non-electroporated regions. Bin 1 is apical lining the ventricle, and Bin 10 is adjacent to the meninges.
(D) Quantification of Arhgap11a smFISH punctae in electroporated RGC endfeet.
(F) Cartoon of regions analyzed in RGC basal processes.
(G) EGFP electroporated RGCs depicting reduced branches (arrows) along the basal process following Arhgap11a knockdown.
(H) Quantification of the length of branches along the RGC basal process. (Scrambled: n=101 branches, 3 brains, 3 independent experiments; Arhgap11a: n=72 branches, 3 brains, 3 independent experiments, unpaired t-test with Welch’s correction)
(I) Quantification of the density of branches along the RGC basal process. (Scrambled: n=112 cells, 6 brains, 5 independent experiments, Arhgap11a: n=99 cells, 5 brains, 4 independent experiments, unpaired t-test with Welch’s correction)
(J) Schematic overview of the experiments in (J-R) aimed at testing the impact of Arhgap11a depletion in RGCs on neuronal migration. Sequential IUEs were performed to label neurons (EGFP, green) and RGCs (red) at E16.5 when analysis is performed.
(K,L) Representative images showing electroporated regions (left) and position of migrating of neurons (green) at the beginning (t=0 hrs, middle) and end of the live-imaging experiment (t=16 hrs, right).
(M) Neuronal migration parameters analyzed.
(N-R) Quantification of velocity of neuronal migration (N), net-distance in X trajectory (O), net distance travelled in Y trajectory (P) and compiled distance (Q). (R) Arhgap11a knockdown in RGCs non-cell autonomously causes neurons to undergo more static movements and fewer movements toward the cortical plate (up). (Scrambled and Arhgap11a: n=8 brains, 2 independent experiments, unpaired t-tests)
siRNAs: small interfering RNAs, IUE: in utero electroporation, CP: cortical plate, IZ: intermediate zone. *: p-value<0.05. **: p-value<0.01. ***: p-value<0.001. Scale bars: B: 20 μm, D,J-K right panels 50 μm, F: 10 μm, J-K left panels: 100 μm. Bar plots, means +/− SE.
We next examined morphology of the Arhgap11a-depleted RGC basal process. For this we generated 3D reconstructions of either scrambled (control) or siRNA-treated EGFP+ basal processes to quantify the density and length of extensions in the basal process (Figure 2F–I). There was no impact of Arhgap11a depletion upon the length of small extensions (average extensions were 3.3 μm and 3.5 μm for control and Arhgap11a conditions, respectively) (Figure 2H). However, we noted a significant decrease in the density of cellular extensions in brains transfected with Arhgap11a siRNAs compared to control (Figure 2I). This data demonstrates that Arhgap11a regulates branching along the RGC basal process.
The integrity of the RGC basal process is paramount for proper radial migration of excitatory neurons from the ventricular zone to the cortical plate 38,40. Indeed, ectopic basal process branching can alter patterns of migrating neurons, including speed, directionality, and pausing 39. Therefore, we assessed whether aberrant RGC morphology induced by Arhgap11a loss impacts neuron migration in a non-cell-autonomous manner. To do this we used a paradigm relying on sequential in utero electroporation on consecutive days (Figure 2J). E14.5 brains were IUE’d with an EGFP plasmid to label newborn cells that would become migratory neurons one day later (E15.5). At E15.5 we then performed an IUE in the identical location using scrambled or Arhgap11a siRNAs and membrane-bound mCherry to manipulate and label the RGC scaffold which EGFP+ neurons migrate upon. Importantly, since Arhgap11a is not expressed in migrating excitatory neurons (Figures 1,S1), this allowed us to quantify migration of WT neurons along an Arhgap11a-deficient RGC scaffold. At E16.5, we performed overnight (16h) live imaging of EGFP+ migrating neurons along mCherry+ RGCs in organotypic brain slices, focusing on migration within the SVZ and IZ (Figure 2K,L,Movies S1,S2).
From live imaging, several parameters of neuronal migration were quantified: average speed, maximum speed, total distance, net distance, net distance in the Y and X axes (in the radial or tangential dimensions, respectively), and fraction of time spent mobile including towards the CP or VZ (Figures 2M–R,S2A–G). Loss of Arhgap11a in RGCs had no impact upon average velocity or maximum velocity, total path length (distance), and net distance of neurons traveled in the X axis (Figures 2N,O,S2C,D). However, there was a significant 35% decrease in the net migrated distance and a particularly striking 42% decrease in the net distance in the Y axis (Figure 2P,Q). Additionally, radially migrating neurons were more immobile (static) and showed decreased propensity to move towards their final destination in the CP (Figure 2R). Thus, Arhgap11a loss does not impair the speed or ability of neurons to migrate per se, but significantly reduces the efficiency and trajectory of radial migration. This was further corroborated by analysis of the orientation of neurons at the beginning of the live imaging experiments, when EGFP+ neurons were mostly located in the SVZ and lower IZ. Arhgap11a loss increased tangential orientation of those neurons (Figure S2E–G). Altogether, these results show that Arhgap11a expression in RGCs non-cell autonomously controls radial migration of pyramidal neurons along basal processes; this broadly links Arhgap11a-mediated RGC morphology to neuronal migration.
Given these defects in radial migration, we next asked if acute Arhgap11a loss from RGCs affects cell composition and cortical lamination 41. We assessed brains which had been electroporated at E15.5 with either scrambled siRNAs or Arhgap11a siRNAs. A 2-day knockdown of Arhgap11a had no striking impact on progenitor proliferation; indeed, 40% of control and mutant IUE’d cells were Ki67-positive and RGC (Sox2+) and IP (Tbr2+) fractions were normal (Figure S3A–D). Consistent with this, at P0 we noted no significant difference in the distribution of electroporated cells, nor in the density of cells, between conditions (Figure S3E–H). Although we cannot rule out earlier roles in cell fate specification, these data suggest that acute Arhgap11a knockdown does not grossly influence neurogenesis.
We next assessed whether the radial migration deficiency was associated with altered laminar organization in the P0 cortex (Figure S4A). We first quantified the distribution of neurons born several days prior to Arhgap11a siRNA treatment (layers V and VI) (Figure S4A). Tbr1+ layer VI neuron distribution was similar between knockdown and control mice (Figure S4B,C). In comparison, CTIP2 layer V neurons showed a slight but significant altered distribution (Figure S4D,E). This is consistent with the expectation that, at the time of acute RGC knockdown, virtually all layer VI neurons completed their migration while some layer V neurons are still migrating. We then examined the distribution of neurons with peak genesis one day prior to, or simultaneous with electroporation (layer IV and II/III). Compared with scrambled control, fewer ROR-ß+ layer IV neurons localized to superficial layers in Arhgap11a knockdown brains (Figures S4F,G). LHX2+ layer II/III neurons were also altered in their distribution (Figures S4H,I). Notably, the distribution of Ctip2+ and ROR-ß+ neurons in P21 Arhgap11a-deficient cortices was also significantly altered, particularly layer IV neurons (Figure S4J–N). This phenotype is striking given that it resulted from an acute embryonic knockdown about 25 days earlier. Altogether, these results indicate that acute Arhgap11a knockdown in RGCs cause subtle but significant impairment of the laminar organization of cortical neurons at both P0 and P21. As Arhgap11a is not expressed in excitatory neurons (Figure S1B–F), this suggests that neuronal organization is influenced by non-cell autonomous requirements of Arhgap11a in the RGC scaffold.
Arhgap11a is essential for RGC basal endfeet morphology and positioning of cortical interneurons
RGC basal process morphology is dynamic and complex not only in the IZ but also near the pial surface in the marginal zone (MZ) 15,16. Indeed, morphological analyses of RGCs show that endfeet number is constant through E14.5 and increases by E16.5 16. Moreover, consistent with others’ findings 15 we observed by live imaging E16.5 embryonic brain slices, that basal processes can be highly dynamic in the MZ (Movie S3). ARHGAP11A localization to distal regions of RGC basal processes and endfeet coincides developmentally with this RGC dynamic behavior and endfeet complexity16 (Figure 1C–H). Therefore, we tested whether Arhgap11a regulates basal process complexity in the MZ.
Towards this, we employed 3D reconstructions of either control or siRNA-treated membrane-bound-EGFP+ basal processes at the pia, introduced by IUE at E15.5. We specifically quantified the RGC arbor and branching of basal processes (>5μm long42) in the MZ (Figure 3A–C). In RGCs transfected with scrambled siRNAs, we observed extensive branches emanating from the basal process in the MZ, as described previously (Figure 3C–E) 15,16. In contrast, Arhgap11a siRNA knockdown induced a significant decrease in both branching complexity and total number of branches. This effect persisted after 48h, albeit to a lesser extent (Figure S5A–C). To orthogonally validate this phenotype, we used Arhgap11a germline knockout mice (Figure S5F–H). Analysis of endfeet of E16.5 KO brains showed significant reductions in complexity, paralleling that seen in the siRNA-depleted brains (Figure S5I–K). Given these mice are germline deficient in Arhgap11a, we opted to use siRNA-treated brains for further analysis of acute phenotypes.
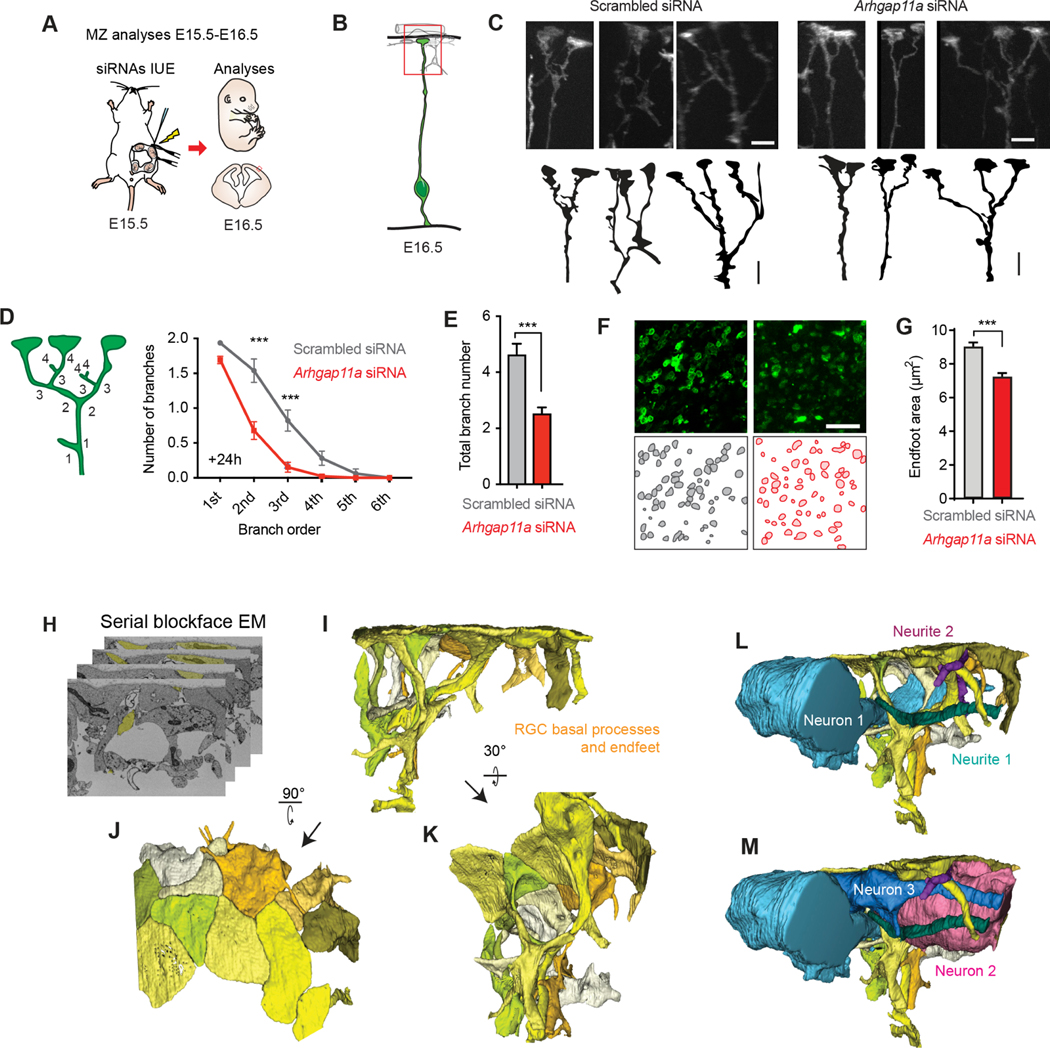
Figure 3. Arhgap11a promotes RGC basal process and endfeet complexity and interneuron numbers in the marginal zone.
(A) Schematic overview of the experiments in (B-F) which examine acute impact of RGC knockdown upon RGC basal process and endfeet in the marginal zone (MZ).
(B) Region analyzed in the experiments.
(C) Representative images showing basal process and endfoot complexity in the MZ in IUE’d RGCs. Tracing of images is below.
(D-E) Method to define branch orders in RGC basal processes in MZ (D, left) and quantification of branch complexity (D, right), and average total branch number per RGC (E). (Scrambled: n=78 cells, 6 brains, 4 independent experiments, Arhgap11a: n=78 cells, 5 brains, 4 independent experiments, two-way ANOVA, Sidak post-hoc analyses to compare branch order).
(F,G) Representative images (F) for quantification of endfoot-basal lamina contact area in the MZ (G). (Scrambled: n=351 endfeet, 6 brains, 4 independent experiments, Arhgap11a: n=351 endfeet, 6 brains, 4 independent experiments, Mann-Whitney test)
(H-M) 3D reconstructions of the MZ niche using serial blockface electronic microscopy shows tight interactions between presumed (blue, pink) interneurons, cajal retzius neurons, and RGC basal processes and endfeet (yellow).
siRNAs: small interfering RNAs, IUE: in utero electroporation, MZ: marginal zone, EM: electronic microscopy, *: p-value<0.05. ***: p-value<0.001. Individual data points represent different brains. Scale bars: C: 5 μm, M, O: 25 μm. Bar plots, means +/− SEM.
We next evaluated whether Arhgap11a influences basal endfoot area contacting the basal lamina. To do this we generated endfoot preparations from electroporated brains, which included meninges, basal lamina and RGC basal endfeet22. Imaging basal endfeet with an en-face view, we quantified an average basement membrane coverage of ~9μm2 by control RGC endfeet (Figure 3F,G). However, this coverage significantly diminished by 25% in RGC endfeet depleted of Arhgap11a. Of note, there was no observable impact of Arhgap11a knockdown on the number of endfeet per RGC (Figure S5D). Altogether, these results show that Arhgap11a is essential for RGC morphology in the MZ.
This raises the question of what is the impact of these morphology defects upon cortical architecture at the pia? To first assess this question, we used serial-blockface electron microscopy (SBFEM) to visualize RGC basal processes and endfeet structure as well as their interactions with the surrounding MZ niche (Figure 3H–M). 3D reconstructions of RGCs highlighted that basal endfeet tile the basement membrane, forming a tight interface with the basement membrane (Figures 3H,I). RGC basal processes and endfeet also directly contacted surrounding cells and neurites of the MZ. For example, we reconstructed tangentially directed cells (presumably interneurons or Cajal Retzius neurons) contacting a multitude of RGC basal processes and branches (Figure 3L,M).
Given that Arhgap11a controls RGC morphology in the MZ, we asked whether depleting Arhgap11a in RGCs could non-cell autonomously affect resident cells of the MZ, including Cajal Retzius neurons and interneurons 43–49. We introduced scrambled or Arhgap11a siRNAs into RGCs using IUE at E15.5, and analyzed cell density in the MZ 24h later, focusing on the region containing EGFP+ RGC basal processes and endfeet (Figures 4A,B). Strikingly, compared to control, Arhgap11a knockdown in RGCs significantly reduced the number of Hoechst+ cells lining the LAMININ+ basement membrane (Figure 4C,D).
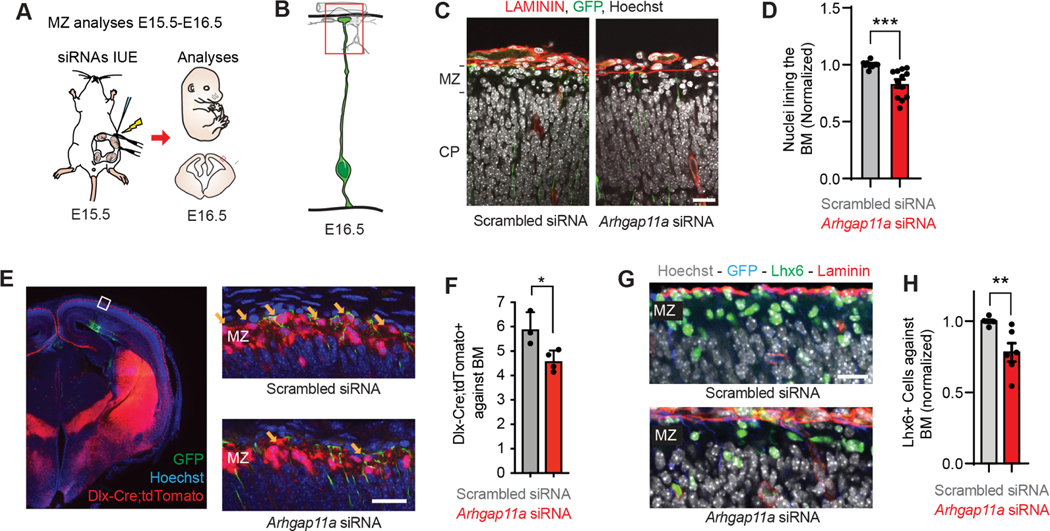
Figure 4. Arhgap11a influences interneuron positioning in the marginal zone.
(A) Schematic overview of the experiments in (B-H) which examine acute impact of RGC knockdown upon interneuron positioning in the marginal zone (MZ).
(B) Region analyzed in the experiments.
(C) Immunofluorescence depicting nuclei (white, Hoechst) and laminin (red) in GFP (green) electroporated regions.
(D) Quantification of the number of nuclei lining the basement membrane (BM), across a region of analysis. (Scrambled: n=9 brains, 5 independent experiments, Arhgap11a: n=4 brains, 4 independent experiments, unpaired t-test)
(E) Immunofluorescence depicting Hoechst+ nuclei (blue), tdTomato+ interneurons (red) in GFP (green) electroporated region, with higher magnification images on right. Yellow arrows, tdTomato+ interneurons located against the basement membrane.
(F) Quantification of Tomato+ interneurons lining the BM in indicated brains. (Scrambled: n=3 brains, 1 experiment, Arhgap11a: n=4 brains, 1 experiment, unpaired t-test)
(G) Immunofluorescence depicting Hoechst+nuclei (blue), Laminin (red), Lhx6+ interneurons (green) in GFP (blue) electroporated region.
(H) Quantification of Lhx6+ interneurons lining the BM in indicated brains. (Scrambled: n=6 brains, 4 experiments, Arhgap11a: n=6 brains, 3 experiments, Mann-Whitney test)
siRNAs: small interfering RNAs, IUE: in utero electroporation, MZ: marginal zone, EM: electronic microscopy, *: p-value<0.05. ***: p-value<0.001, Individual data points represent different brains. Scale bars: C: 5 μm, M, O: 25 μm. Bar plots, means +/− SEM.
We then quantified interneurons, which migrate tangentially from the ganglionic eminences into the cortex via 3 streams, including along the MZ. Interneurons which migrate through the MZ invade the CP, along the way forming axons that extend from layer 1 50. This interneuron population is thus critical for cortical circuitry. We employed IUE of siRNAs and EGFP in Dlx-Cre; Ai14 animals, in which interneurons express tdTomato (Figure 4E,F). Strikingly, Arhgap11a knockdown in RGCs significantly decreased the number of interneurons lining the basement membrane, where endfeet are located (Figure 4F). Consistent with this, we quantified fewer LHX6+ interneurons lining the basement membrane, following acute Arhgap11a knockdown Figure 4G,H). In contrast, there was no obvious alteration in CALRETININ+ cells, which labels small numbers of interneurons but at this stage mainly labels Cajal Retzius neurons 44 (Figure S5L,M). These results demonstrate that Arhgap11a acts non-cell autonomously within RGCs to promote the position of a population of interneurons in the MZ. Taken together with the discovery that basal process complexity is associated with excitatory neuron migration (Figures 2, S2,S3), these findings broadly link RGC morphology in basal structures to non-cell autonomous positioning of neurons.
Arhgap11a mRNA is actively transported to RGC basal endfeet via a 5′UTR cis element
Our data indicate that Arhgap11a is essential for RGC basal morphology and neuronal migration. This begs the question of how Arhgap11a controls these RGC subcellular compartments, and specifically basal endfeet architecture. Notably, both Arhgap11a mRNA and protein are subcellularly enriched and co-localize in RGC endfeet (Figure 1), as seen for other transcripts 22. This suggests that RNA transport and local translation of Arhgap11a may enable rapid and specific control of RGC basal endfoot morphology.
Active mRNA transport depends on cis localization elements within mRNAs 51. Therefore, we first determined the sequence(s) within Arhgap11a mRNA that confers endfoot localization. To this end, we generated three reporters containing EGFP with a nuclear localization sequence (NLS-EGFP), together with either the Arhgap11a (i) 5′UTR (527 bp), (ii) coding sequence (2,942 bp) or (iii) 3′UTR (1,057 bp). To test for position-independent localization capacity, these elements were included at the 3′ end of the reporter, followed by a poly(A) sequence (Figure 5A,B). The reporters were introduced by IUE at E13.5 and E14.5 brains were collected. As previously shown 23, EGFP reporters which are not transported are nuclear, whereas EGFP localization in the basal process and endfeet indicates transport from the RGC cell body and translation in basal endfeet. Using this strategy, we discovered that the endfoot RNA localization sequence of murine Arhgap11a mRNA resides in its 5′UTR and not in its coding or 3′UTR sequences (Figure 5A–C). This was further confirmed by in situ hybridization targeting the EGFP mRNAs in electroporated brains (Figure S6A,B).
Figure 5. Arhgap11a mRNA is actively transported to radial glial basal endfeet via a 5′UTR element.
(A,B) Schematic overview (A) of the strategy used in (B, C) to determine the endfoot localization element in Arhgap11a mRNA using a 1-day electroporation of indicated reporter constructs (B). (C) EGFP-nls localizes to RGC basal endfeet only when the Arhgap11a 5′UTR is present, but not in CDS alone or containing 3′UTR.
(D, E) Schematic overview (D) of the strategy used in (F-J) to visualize transport of Arhgap11a mRNA reporters (E) in RGC basal processes.
(F) smFISH (red) targeting MS2 stem-loop RNA sequences shows Arhgap11a 5′UTR induces RNA localization from cell bodies to RGC basal process and endfeet (CFP, green).
(G,H) Kymographs showing absence (G) and presence (H) of MS2-tagged mRNA transport in RGC basal process over a 1-min period, in no UTR and 5′UTR, respectively.
(I,J) Quantification of similar average speeds of MS2-tagged mRNA transport in RGC basal processes (I) and average run lengths (J) in both apical and basally directed movements. n=126 EGFP+ punctae, 11 cells, 2 brains, 2 independent experiments.
IUE: in utero electroporation, CDS: coding sequence, UTR: untranslated region, nls: nuclear localization signal, tdMCP: tandem MS2-coat protein, CFP: cyan fluorescent protein, CP: cortical plate, IZ: intermediate zone, VZ: ventricular zone. Scale bars: F: 50μm, G,H: horizontal axis: 5sec, vertical axis: 5μm.
Having determined the sequence element of Arhgap11a sufficient for its localization to endfeet, we next asked if Arhgap11a is actively transported to endfeet. To visualize Arhgap11a mRNA transport, we live imaged mRNAs in RGC basal processes within organotypic brain slices as previously 22. This technique uses a reporter plasmid with CFP and a 3′UTR containing 24 MS2 stem loops and a second plasmid containing the nuclear localized tdMCP-EGFP protein with high affinity to RNA MS2 stem loops (Figure 5D,E)52. Co-transfecting both plasmids allows the indirect visualization of mRNAs. Two different reporters encoding CFP were generated: one control sequence with MS2 loops only (MS2-no UTR) and the Arhgap11a 5′UTR sequence downstream of the MS2 loops (MS2–5′UTR). These reporters were transfected into RGCs by IUE at E15.5 and organotypic slices were generated at E16.5. In brains electroporated with either control MS2-noUTR or the MS2–5′UTR, we observed diffuse EGFP+ in the nucleus and cytoplasm of CFP+ RGCs, demonstrating the efficiency of the technique (Figure 5F). smFISH targeting MS2 loops in fixed electroporated brain sections corroborated localization of the 5′UTR reporter to basal endfeet.
Having established these tools, we then performed live imaging of electroporated EGFP+ mRNAs in the basal process of E16.5 RGCs within organotypic brain slices. This allowed us to visualize active directed transport of MS2–5′UTR EGFP+ mRNAs (Figure 5D,E,G,H,Movie S4). Kymograph analyses highlighted bidirectional movements, with frequent changes of orientation in the 5′UTR reporter but not the no UTR (Figure 5G,H). 60% of the observed movements were directed towards the basal endfeet (Figure S6C). We measured average speeds of 2.5 μm/s and tracked single uninterrupted movements of up to 32 μm, with an average of 6 μm (Figure 5I,J). These speeds and processivity are consistent with active microtubule-dependent transport, and similar to those observed previously for other mRNA reporters 22. Altogether, these results demonstrate that Arhgap11a mRNA is actively transported to basal endfeet, arguing that its subcellular localization may ultimately impact basal endfeet.
ARHGAP11A subcellular localization in RGC basal processes and endfeet relies upon local translation in the basal endfeet
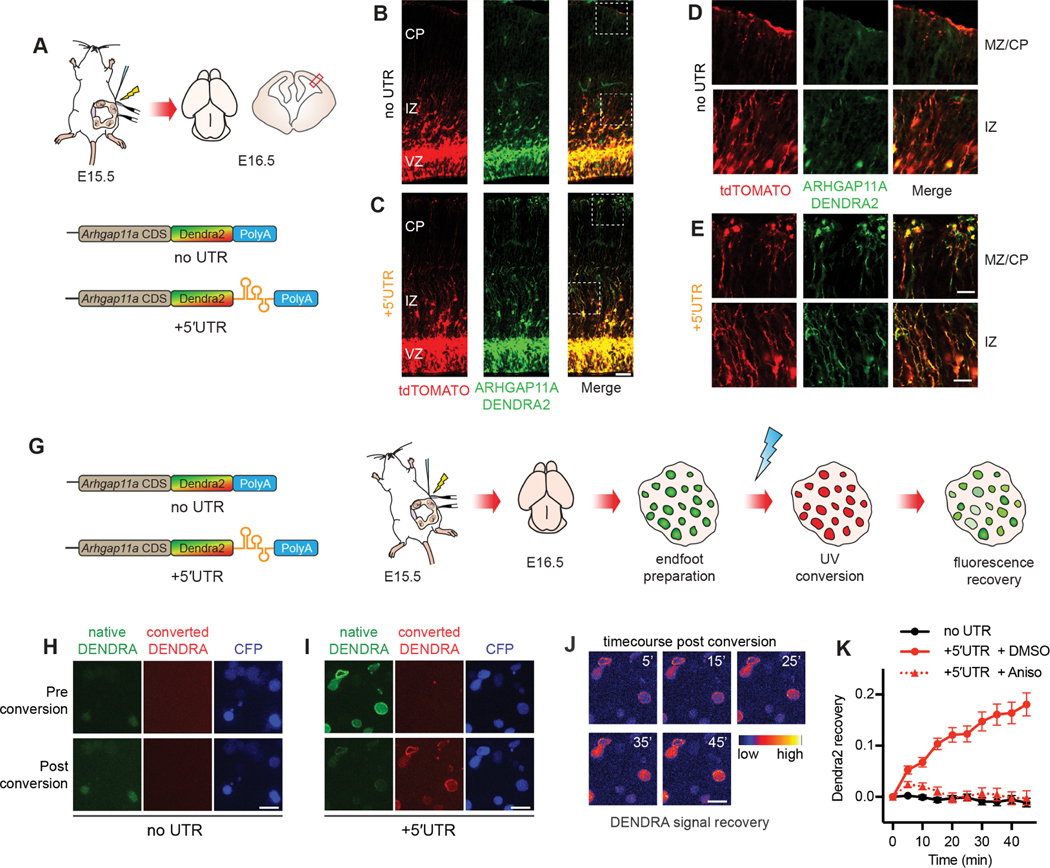
The presence of ARHGAP11A protein in the basal process and endfeet could be explained by at least two possibilities: synthesis of ARHGAP11A in the soma followed by protein transport into the basal process and endfeet, and/or local production of ARHGAP11A within endfeet. Given the active transport of Arhgap11a mRNA along the basal process and enrichment in endfeet, we predicted the latter. To test this directly, we generated reporters expressing ARHGAP11A full-length protein fused to the fluorescent photoconvertible protein DENDRA2. Two reporters were generated: a control producing an mRNA devoid of any localization sequence (Dendra2-no UTR) and another including the 5′UTR of Arhgap11a sufficient for RNA localization at the endfoot (Dendra2–5′UTR, Figure 6A). DENDRA2 fluorescent signal was analyzed in fixed E16.5 brains after IUE at E15.5. smRNA FISH targeting Dendra sequence confirmed the transport of the 5′UTR reporter to basal endfeet (Figure S7A–C). Expression of either the control or 5′UTR reporter resulted in DENDRA2 signal in RGC cell bodies (Figure 6B,C), recapitulating endogenous protein expression (Figures 1C–H). However, DENDRA2 signal in RGC basal endfeet was only evident in brains electroporated with the 5′UTR reporter (Figure 6B–E). In these brains, ARHGAP11A localization was also evident along the basal processes within the IZ and CP (Figure 6D–E). This localization was not context dependent as placement of the 5′UTR in either the 5′ or 3′ part of the reporter led to similar enriched localization to basal endfeet (Figure S7D). This strongly argues that ARHGAP11A protein localization within endfeet and along the basal process relies upon subcellular localization of the transcript.
Figure 6. ARHGAP11A protein localization to RGC basal processes and endfeet relies on local translation of Arhgap11a mRNA in basal endfeet.
(A) Schematic overview of the strategy used in (B-E) to test if Arhgap11a mRNA localization mediates ARHGAP11A expression in RGC basal endfeet and basal processes.
(B-E) Immunofluorescence of tdTomato electroporated RGCs (red) and ARHGAP11A fusion reporter (green) containing no UTR (top) or 5′UTR (bottom). High magnification images (D,E) reflect ARHGAP11A protein localizes to RGC basal endfeet (MZ/CP) and basal processes (IZ) only with Arhgap11a 5′UTR.
(G) Schematic overview of the strategy used in (H-K) to visualize local translation of Arhgap11a in RGC basal endfeet.
(H-I) Images showing ARHGAP11A-DENDRA fluorescence in RGC basal endfeet pre (top) and post- (bottom) photoconversion in no UTR (H) or 5′UTR (I) conditions.
Time course showing recovery of native DENDRA signal in the +5′UTR condition, as psuedocolored using indicated scale (time, min).
(K) Quantification of positive recovery of native DENDRA signal in RGC basal endfeet only with 5′UTR +DMSO (red, solid line) relative to both the no UTR condition (black) and anisomycin treatment (red, dotted line). no UTR: n=27 endfeet, 2 brains, 2 independent experiments, 5′UTR + DMSO: n= 62 endfeet, 3 brains, 3 independent experiments, n=69 endfeet, 3 brains, 3 independent experiments, two-way ANOVA interaction time x condition: p value < 0.0001. UTR: untranslated region, CDS: coding sequence, Aniso: anisomycin. Scale bars: B,C: 100 μm; D,E: 20 μm; H-J: 5 μm. Graph, average values +/− SEM.
Next, we used live imaging to directly test if ARHGAP11A can be locally synthesized within endfeet. Using the DENDRA2 constructs described above together with CFP to identify electroporated endfeet, we employed live imaging using ex vivo endfoot preparations consisting of isolated basal endfeet connected to the basement membrane, and overlying meninges, as previously described22,53 (Figure 6G). UV-exposure was used to photoconvert DENDRA2 signal in isolated endfeet from green to red (Figure 6H,I). Subsequently, the recovery of green fluorescence was monitored over 45 minutes. Since basal endfeet were completely disconnected from the cell body, any new green fluorescence is a robust readout of de novo local protein synthesis. No recovery was noted in the negative control lacking the 5′UTR (Figure 6K). In contrast, green fluorescence increased in endfeet expressing Dendra2-5′UTR over 45 minutes (Figure 6J,K). Importantly, these kinetics were completely abolished in the presence of translation inhibitor anisomycin (Figure 6K). Altogether, these data demonstrate that active Arhgap11a mRNA transport and local translation in endfeet enable subcellular expression of ARHGAP11A protein in RGC basal structures.
Locally synthesized ARHGAP11A controls basal process morphology through RhoGAP activity which influences interneuron positioning
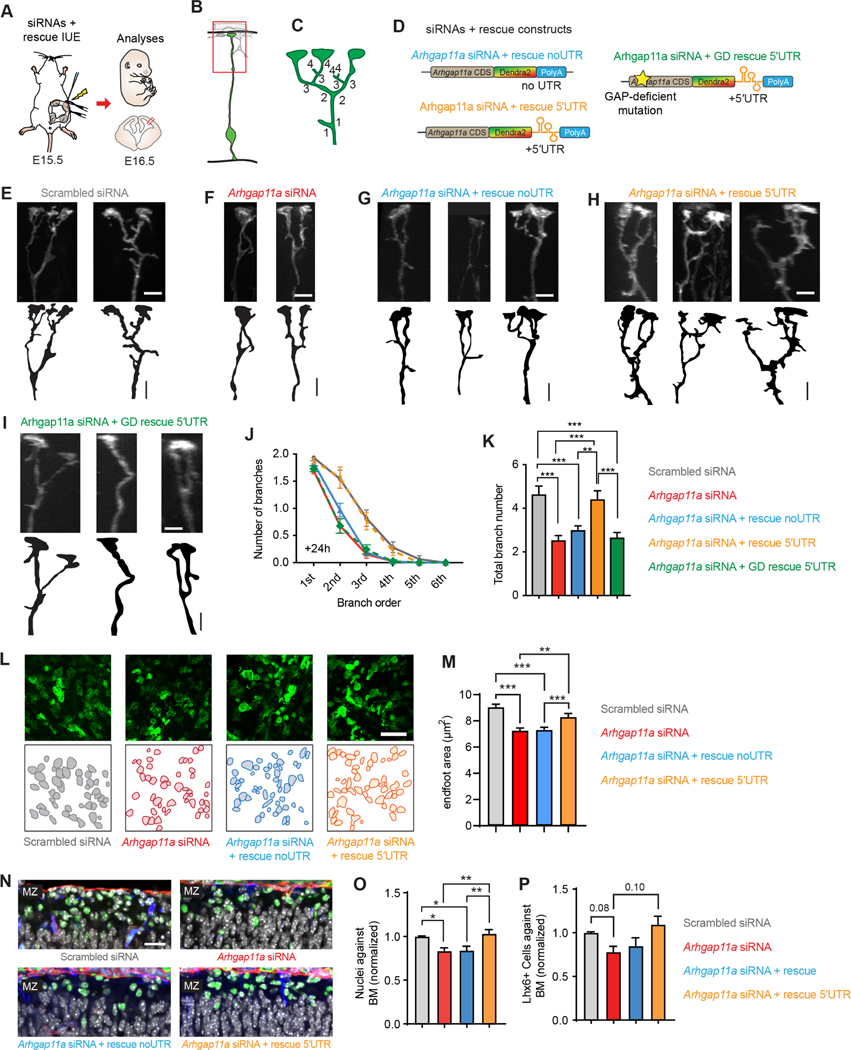
ARHGAP11A mRNA and protein localizes to RGC basal endfeet coincident with increasingly complex endfoot morphology. Given this, and the observation that mRNA is actively transported and locally translated during development, we postulated that local synthesis of this RhoGAP controls endfoot morphology and thus impacts interneuron positioning along the basal membrane. To test this, we used IUE at E15.5 to introduce Arhgap11a siRNAs together with Arhgap11a full length protein either with or without the 5′UTR (Figure 7A–D). Importantly, both Arhgap11a cDNAs were resistant to the transfected siRNAs. Compared to siRNA knockdown alone, co-expression of siRNAs with full length Arhgap11a did not restore RGC basal process complexity in the MZ (Figure 7E–G,J,K). In contrast, introduction of Arhgap11a-5′UTR completely restored the RGC morphology phenotype (Figure 7E,F,H,J,K). Further, while non-localized Arhgap11a failed to rescue reduced endfoot area, localized ARHGAP11A did (Figure 7L,M). Of note, expression of Arhgap11a with or without the 5′UTR alone, but without siRNA, did not affect RGC branching complexity in the MZ, indicating there is no overt overexpression phenotype (Figure S8A–C). These findings demonstrate that subcellular localization of Arhgap11a via its 5′UTR is critical for proper basal process morphology.
Figure 7. Locally synthesized ARHGAP11A controls basal process morphology through GAP activity.
(A) Schematic overview of the strategy used in (B-K) to assess rescue of RGC endfeet morphology.
(B,C) Method to define branch orders in RGC basal processes in MZ.
(D) Rescue constructs used in experiments.
(E-I) Representative images showing basal process complexity at the level of the MZ in RGCs treated as indicated.
(J,K) Quantification of basal process and endfoot complexity at the level of the MZ in RGCs. (Scrambled: n=78 cells, 6 brains, 4 independent experiments, Arhgap11a: n=78 cells, 5 brains, 4 independent experiments, Arhgap11a + rescue: n= 97 cells, 6 brains, 4 independent experiments, Arhgap11a + rescue 5′UTR: n= 56 cells, 4 brains, 3 independent experiments, Arhgap11a + GD rescue 5′UTR: n=78 cells, 4 brains, 2 independent experiments, J, 2-way ANOVA: p-value<0.0001, K, One way ANOVA: p<0.0001, Tukey’s Post-Hoc comparisons)
(L,M) Quantification of endfoot-basal lamina contact area in RGCs. (Scrambled: n=351 endfeet, 6 brains, 4 independent experiments, Arhgap11a: n=351 endfeet, 6 brains, 4 independent experiments, Arhgap11a + rescue: n= 454 endfeet, 7 brains, 4 independent experiments, Arhgap11a + rescue 5′UTR: n= 200 endfeet, 4 brains, two independent experiments, Kruskal-Wallis test).
(N) Images of interneurons (LHX6, green) along the basement membrane (BM, red) in different genotypes.
(O, P) Quantification of the positioning of DAPI cells (white) and interneurons along the basement membrane. (Nuclei against BM: Scrambled: n=13 brains, 9 independent experiments, Arhgap11a: n=12 brains, 7 independent experiments, Arhgap11a + rescue: n=14 brains, 8 independent experiments, Arhgap11a + rescue 5′UTR: n= 13 brains, 8 independent experiments, One-way ANNOVA followed by Tukey Post-Hoc analyses; Lhx6+ cells against the BM: Scrambled: n=6 brains, 4 independent experiments, Arhgap11a: n=6 brains, 3 independent experiments, Arhgap11a + rescue: n=6 brains, 3 independent experiments, Arhgap11a + rescue 5′UTR: n= 8 brains, 4 independent experiments, Brown-Forsythe and Welch ANOVA followed by Dunnett T3 Post-Hoc analyses).
IUE: in utero electroporation, siRNA: small interfering RNA, CDS: coding sequence, UTR: untranslated region, GD: Rho-gap-deficient. **: p-value<0.01. ***: p-value<0.001. E-G: 5 μm. Graphs, average values +/− SEM.
Is RGC morphology dependent upon ARHGAP11A Rho GTPase function? To address this question, we generated a previously described GAP-deficient (GD) form of ARHGAP11A, 26. This GD-Arhgap11a-5′UTR reporter, contains a single missense mutation, R87A (Figure 7D). The transcript encoded by this mutant ARHGAP11A localized properly to endfeet (Figure S7C). However, like the no UTR construct, it also failed to rescue the siRNA-mediated basal process phenotype in the MZ (Figure 7I–K). This indicates local Rho GAP activity mediates RGC basal process complexity in the cortex.
Acute depletion of Arhgap11a from RGCs impaired their basal morphology and led to significantly fewer Hoechst+ cells and interneurons at the basement membrane (Figures 3 and 4). Given this, we asked whether rescuing RGC morphology at the pia was sufficient to also recover interneuron positioning. Re-introduction of full-length Arhgap11a, which lacked a localization element, failed to rescue both nuclei number and interneuron number (Figures 7N–P). This is consistent with the inability of non-localized Arhgap11a to rescue endfoot complexity and area (Figure 7K,M). In contrast, co-expression of Arhgap11a-5′UTR with the siRNAs recovered nuclei number as well as interneuron number. Altogether, these results demonstrate that RGCs employ mRNA transport and local protein synthesis of Arhgap11a at RGC endfeet to fine-tune basal process morphology and direct neurons to their proper position (Figure 8).
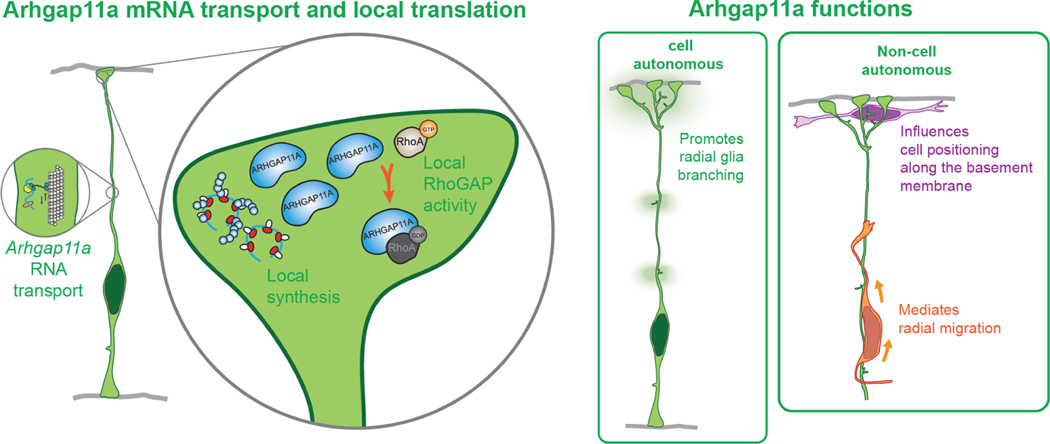
Figure 8. Model for major findings of this study.
Arhgap11a mRNA is actively transported in RGC basal process to basal endfeet. In basal endfeet local synthesis of ARHGAP11A protein enables expression in basal structures and local RhoGAP activity, thus promoting radial glia branching and interneuron position. Arhgap11a is non-cell autonomously required in RGCs for migration of excitatory neurons and positioning of inhibitory neurons.
Discussion
By controlling neurogenesis and neuronal position, RGCs are crucial for orchestrating development of the cerebral cortex. Yet how the polarized morphology of RGCs dictates these functions is poorly understood. Here, we demonstrate that RGCs and cortical architecture relies upon exquisite temporal and spatial control of gene expression via active mRNA transport and local translation. We show that RhoA GAP activity is spatially and acutely activated via local translation in RGC basal endfeet to control their morphology and non-cell autonomously influence interneuron positioning. Our study demonstrates, for the first time, that mRNA localization and local translation in RGCs is essential for brain development in vivo. This implicates a dynamic new gene regulatory mechanism by which progenitors shape brain development across species.
mRNA transport and local translation control RGC morphology
Using live and fixed ex vivo and in vivo imaging, we demonstrate that Arhgap11a mRNA undergoes active transport, subcellular localization, and local translation in RGC basal endfeet. This discovery adds Arhgap11a to a short but growing list of over 100 transcripts which we and others have shown are subcellularly localized to RGC endfeet 22,23,54,55. This further reinforces the finding that RGC basal processes are highways for active mRNA transport and exhibit subcellular translation at the pia 53,56.
To date the functions of any subcellularly localized transcript in endfeet was unknown. We address this significant gap by demonstrating that at least one function of subcellular RNA localization in RGCs is to mediate local morphology. This is based upon several lines of evidence. First, Arhgap11a mRNA and protein localization to basal endfeet and basal processes relies upon its 5’ UTR, and coincides with critical developmental stages when RGC complexity increases 16. Second, acute Arhgap11a depletion decreased RGC complexity in basal structures; importantly and strikingly, this phenotype was rescued only upon expression of endfoot-localized, functional Arhgap11a. Hence, RGC basal structures rely upon coordinated active mRNA transport, local translation, and local and acute Rho GTPase function.
Why do RGCs require protein production in endfeet rather than transport existing proteins from the cell body? Translation of an average protein takes about 1 minute, whereas transcription is 10 times longer 57. Thus, local expression is both highly efficient and energetically favorable, enabling rapid changes in RGC morphology as the brain grows radially and tangentially. Our previous data show that endfeet contain over 100 significantly enriched FMRP-bound mRNAs, including those encoding cytoskeletal and signaling regulators 22, suggesting this may be a widespread mechanism in RGCs. While not significantly enriched in this dataset, Arhgap11a could be a low-affinity FMRP target. Migrating fibroblasts and neuronal growth cones and spines also contain similar classes of localized transcripts 58–61, suggesting our findings may extend to other cell types. Beyond morphology, localized mRNAs in endfeet may also have additional functions, such as to promote cell fates or intra- and extracellular signaling 23.
How does ARHGAP11A protein synthesized in endfeet influence complexity along the basal process? Our data suggest that endfeet ARHGAP11A moves into the basal process either passively via diffusion or actively. Indeed, exogenous ARHGAP11A reporter localization to the basal process relied entirely on an endfoot localization element. We also observed sporadic enrichment of Arhgap11a mRNAs along the basal process, which could result from collective RNA trafficking to basal endfeet. Further, such enrichment could indicate local translation “hotspots” in the basal process, as proposed in axons 62.
Acute, spatial regulation of a Rho-GTPase modulator controls RGC morphology and neuronal positioning
We demonstrate that localized, acute expression of GTPase regulators mediates subcellular architecture. GTPases, including RhoA, can promote filopodial activity and morphology in RGC basal structures, ultimately influencing neuronal migration and neurogenesis 12,15,24,63–65. RhoA signaling via the microtubule regulator Memo, has also been linked to tiled distribution of RGC basal processes, hyperbranching, and migration defects 39,66. Consistent with these roles, Rho KO mice have heterotopia67. In contrast, consistent with the prediction that Arhgap11a loss induces excessive Rho signaling, Arhgap11a depletion reduces branching and impairs radial migration. Although acute knockdown did not drastically impact neurogenesis, we cannot rule out roles for Arhgap11a in cell fate at other stages. Thus, our findings are generally consistent with known Rho functions. Our data provide a new mechanism to understand how ubiquitously expressed Rho GTPases are locally and rapidly controlled in polarized cells of the nervous system.
Taken together, this suggest that both simplified and excessive RGC branching impair the orientation of radially migrating neurons, and ultimately influence lamination. We speculate that RGC basal process branches act as the “rungs of a ladder” for neurons. While the absence of rungs could impede neuronal movement along the basal process, overextended “rungs” could be misinterpreted by neurons as basal processes, thus deviating them away from their migratory path.
RGC morphology has been previously suggested to influence interneurons 43, however our study is the first to formally demonstrate a functional link. Indeed, our data argue that RGC morphology and local ARHGAP11A function in endfeet dictates interneuron positioning. This is reinforced by a recent study from our lab 68. Interneurons traverse the developing cortex mainly via the VZ and MZ and their ultimate destination is tied to their fate 69. The population of migrating interneurons in the MZ are specialized, developing into Martinotti cells which leave axons in layer I as they invade the CP 50. Of note, interneuron mis-positioning in the MZ was evident with just a one-day acute depletion of Arhgap11a from RGCs and perhaps more sustained manipulation of Arhgap11a in RGCs and thus, disruption of RGC morphology, could ultimately impact cortical circuitry.
A key question is how ARHGAP11A, as well as endfeet complexity, influence interneuron positioning at the basement membrane. As suggested by 3D-EM data, cells closer to the basal lamina maintain a strong interface with basal endfeet. Endfeet may represent a “lawn” on which interneurons are “crawling” between. There may be direct cell-cell interactions between RGC endfeet and migrating interneurons that regulate their migration or promote their adhesion and restriction in the MZ, as well as short or long-range signals. Our findings lay the groundwork for future studies using detailed live imaging and morphological studies to assess these possibilities. Of note, while we did not observe any positioning defects for Cajal Retzius neurons it is possible that some sub-types of CR neurons could be affected. However our data align with the timing of Cajal Retzius neuron migration which have already reached their final destination by E12.5, well before we acutely knockdown the expression of Arhgap11a 48.
Roles for subcellular RNA localization in human cortical development and disease
Given the conserved localization of ARHGAP11A in the human neocortex, we predict that this RhoGAP and more broadly, mRNA localization, is critical in human RGCs. In human neocortices, the RGC basal process is significantly longer than in mice (up to several millimeters) and a prominent feature of outer radial glia/basal radial glia (oRGs/bRGs) 70–72. Further, in primates, lamellar expansions decorate the basal process, and reside in close contact with migrating neurons 73. ARHGAP11A is highly expressed in both human RGCs and oRGs. Thus, localized pools of ARHGAP11A may promote human cortical development by influencing both RGC and oRG morphology.
During evolution, a hominid-specific partial duplication of ARHGAP11A led to the emergence of ARHGAP11B, which is linked to cerebral cortex expansion in humans. ARHGAP11B is also expressed in human neural progenitors and its forced expression promotes progenitor proliferation and neuron production 72,74,75. In constrast to ARHGAP11A, ARHGAP11B lacks both GTPase activity and the RNA motif for localizing to RGC endfeet, and indeed is not localized at the pia 37. It is interesting to consider whether ARHGAP11A might be modulated by ARHGAP11B activity in the cell body of human RGCs, a relationship seen for other human-specific duplications like SRGAP2 76 or NOTCH2NL 77,78.
In sum, our findings establish novel and essential roles for local translation in RGC integrity. It is important to note that defects in RGC scaffolds underlie diverse neurodevelopmental diseases including heterotopias and lissencephaly 13,79. This highlights the importance of investigating subcellular mRNA localization and local translation in RGCs towards understanding normal development and disease.
STAR Methods text
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Debra Silver (debra.silver@duke.edu).
Materials Availability
All unique/stable reagents generated in this study are available from the Lead Contact either without restriction or in some cases with a completed Materials Transfer Agreement.
Data and code availability
This study did not generate any unique code. Original/source data for figures in the paper are available from the corresponding authors on request.
Experimental Model and Subject Details
Mice
All experiments were performed in agreement with the guidelines from the Division of Laboratory Animal Resources from Duke University School of Medicine and approved by Duke IACUC. Plug dates were defined as embryonic day (E) 0.5 on the morning the plug was identified. All experiments were conducted in the C57BL/6J strain. The following mouse strains were used: Dlx-Cre (Tg(dlx5a-cre)1Mekk/J); Ai14-tdTomato (B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomatoHze/J) both from Jax labs). Arhgap11aem1(IMPC)J mice were generated by obtaining cryopreserved sperm from the KOMP consortium. These were used to produce germline knockout mice which were validated by qPCR and western analyses.
Human embryonic samples and in situ hybridization
The study using human fetal sample (9 wpc) was approved by three relevant Ethics Committees (Erasme Hospital, Université Libre de Bruxelles, and Belgian National Fund for Scientific Research FRS/FNRS) on research involving human subjects. Written informed consent was given by the parents in each case. In situ hybridization (ISH) on human fetal cortical sections using digoxigenin-labeled RNA probes was performed as described previously 77. The probe specifically recognizing exon11 and 12 of human ARHGAP11A, not ARHGAP11B, was prepared (2117–2657 of human ARHGAP11A (NM_014783.5)). Imaging was performed using a Zeiss Axioplan 2 and the intensity and contrast were modified using Fiji/ImageJ software. We confirmed the specificity of the signal produced by the anti-sense pan-ARHGAP11A probe by comparing with the virtual absence of the signal by the sense probe.
Histology
Mouse embryos were collected and dissected in cold phosphate buffer saline (PBS) and histology was performed as previously 22. Brain fixation was performed overnight by immersion in a 4% Paraformaldehyde 1X PBS solution. Following fixation, embryonic brains were washed twice in cold PBS for 20min. Cryoprotection was performed by overnight immersion in a 30% sucrose (w/v) PBS solution. Following cryoprotection, brain were transferred into OCT medium. Cryosections were generated using a cryostat and deposited on glass slides. The thickness of the sections varied depending on the purpose of the experiments (10–20μm for characterization of ARHGAP11A localization by immunofluorescence and in situ hybridization, 50μm for characterization of basal process morphology with 3D reconstructions, 30μm for analysis of cortical layering at post-natal stages).
Immunofluorescence was performed as described previously 22. Briefly, slides were left to thaw at room temperature (RT) for 10min. Then, they were washed by immersion in PBS for 10min, followed by permeablization with a 15–30min wash in 0.2–0.5% Triton-X (w/v) in PBS. Following this step, sections were washed once in PBS and blocking was performed with Mouse on Mouse (MOM, Vector Laboratories) blocking agent when using primary antibodies produced in the mouse, and 10% NGS for primary antibodies generated in any other species. Primary antibody incubation in PBS or MOM diluent was then performed overnight at 4C. Three 5–10-minute washes were then performed in PBS, followed by 30–60min RT incubation in a secondary antibody solution containing Hoechst. Before mounting was performed in Vectashield (Vector Laboratories) or Mowiol, three 5–10-minute washes were performed in PBS. The list of primary antibodies used in this study can be found in Star Methods. Cell counting was performed in FIJI (ImageJ), using the Cell counter plugin.
For binning analyses, X-Y coordinates were extracted from Cell counter data, and a script created in R 80 was used to assign punctae to specific bins, using the coordinates of the ventricular and pial borders as references. Bin numbers were reported in an Excel spreadsheet for analyses.
DNA constructs and siRNAs
pCAGGS-EX and pCAGGS-EGFP plasmids were kind gifts from Nicholas Gaiano 81, the pGLAST-EGFP-CAXX plasmid was kindly offered by Tarik Haydar82, and the pCAGGS-PB-mCherry-CAXX was generously given by Cagla’s Eroglu laboratory. EGFP-nls-CDS, EGFP-ns-5′UTR and EGFP-nls-3’UTR constructs were generated by cloning sequences of interest downstream of the EGFP sequence, using Arhgap11a cDNA cloned from mouse embryonic cortical cDNA as described previously23. The MS2-no UTR plasmid was described previously 22. We used a Gibson assembly strategy (NEB Hifi Builder) to clone Arhgap11a’s 5′UTR from the EGFP-nls-5′UTR construct into the MS2-no UTR plasmid downstream of the MS2 stem loops sequence. Similarly, we used Gibson assembly to generate the Dendra2-no UTR and the Dendra2–5’ UTR plasmids, cloning 2 fragments (Arhgap11a and Dendra2 coding sequences) or three fragments (Arhgap11a and Dendra2 coding sequences followed by Arhgap11a 5′UTR) into the EcoRI site of pCAGGS-EX, using EGFP-nls-CDS, Dendra2-no UTR 22, and EGFP-nls- 5′UTR as templates, respectively. siRNAs targeting the 3’UTR of Arhgap11a were purchased (siRNAflex, Qiagen), and a pool of 3 different siRNAs were used in these assays. These siRNAs were previously validated by the manufacturer.
In utero electroporation (IUE)
We performed IUE as described previously 22,83. Electroporation parameters were as follows: five consecutive 50ms electrical pulses spaced by 950ms, voltage varied from 40V to 60V depending on the embryonic stage at which the procedure was performed. Plasmids were produced using Qiagen or Sigma Endotoxin Free Maxi Prep kits and following the manufacturers’ instructions. Individual plasmid concentrations injected into lateral ventricles ranged from 0.5 to 1μg/ul. siRNAs were injected at a final concentration of 2.5μM.
Single molecule fluorescent in situ hybridization (smFISH)
In situ hybridization was performed following the protocol described by Takahashi and Osumi 84. Probes sequences are listed in Star Methods. The protocol used to reveal Arhgap11a, MS2 or Dendra2 mRNAs by smFISH was as previously 22. All buffers and solutions used for this protocol were previously treated with diethyl pyrocarbonate to quench RNA-ase activity. A pool of Stellaris probes targeting Arhgap11a and labeled with the Quasar570 fluorophore were purchased from Biosearch. MS2 and Dendra2 probes were described previously 22. For quantification of Arhgap11a mRNA density after IUE-mediated siRNA knockdown, 10μm mosaic Z-stacks covering the entire thickness of the electroporated and the corresponding region in the non-electroporated hemisphere were acquired using a 63X objective with a microscope equipped with Apotome technology (Zeiss). The electroporated region was evident based on the presence of EGFP+ cells. Coordinates of smFISH punctae were manually registered using the Cell Counter plugging in FIJI (ImageJ, over one thousand punctae were registered in non siRNA treated regions). A script created in R 80 was used to assign punctae to specific bins. Bin numbers were reported in an Excel spreadsheet.
qPCR analyses in Dcx-DsRed embryos
Cortices from E14.5 Dcx::DsRed embryos were isolated, incubated with 0.25% trypsin-EDTA solution for 10 min. at 37°C, dissociated into a single cell suspension, and sorted in a Sorter Astrios machine. Positive and negative cells were directly collected into RNA extraction buffer (RLT) supplemented with 1% β-Mercaptoethanol. Samples were vortexed and RNA was extracted using Qiagen RNeasy plus kit. cDNA was synthesized from RNA using Biogen iScript kit and qPCR was performed using either Sybr Green iTaq (BioRad) or TaqMan (Life Technologies) in an Applied Biosystems StepOne machine (Thermo Fisher Scientific). The following primers and TaqMan probes were used in the qPCR reaction: β-Actin (5’ Forward- AGATCAAGATCATTGCTCCT and 3’ Reverse-CCTGCTTGCTGATCCACATC), Pax6 (5’ Forward- TCTTTGCTTGGGAAATCCG and 3’ Reverse-CTGCCCGTTCAACATCCTTAG), Arhgap11a (5’ Forward-GCAGGTGTGCCAAGGCGAAGT and 3’ Reverse-TGCAAGTCGCCAACCAACACTTTCA)28, Gapdh (Mm99999915_g1), Tubb3 (Mm00727586_s1). Values were normalized to Gapdh (TaqMan) or β-Actin (Sybr Green) as loading control.
Live imaging
We performed and quantified live imaging of RNA trafficking, using methods identical to those described previously 22. For imaging of translation, following in utero electroporation of Dendra2 and pCAGGS-CFP plasmids at E15 in the afternoon, brains were dissected in cold 1x HBSS supplemented with 2.5 mM HEPES, 30 mM D-glucose, and 4mM NaHCO3 during the morning on the next day. We generated endfoot preparations as described previously 22, using tweezers to peel off the basement membrane together with connected endfeet from the surface of the brain. 2–4 endfoot preparations from brains electroporated with different plasmid conditions were mounted together in a 1mg/ml collagen solution supplemented with DMEM/F12 at the bottom of 35-mm glass bottom dishes (MatTek). A slab of 3% agarose gel was added on top of endfoot preparations to prevent their detachment from the bottom of the dish. Additional collagen was added to stabilize the preparation. Endfoot preparations were cultured in DMEM-F12 supplemented with B27 without vitamin A (Gibco), N2 (Gibco), 5% horse serum, 5% fetal bovine serum, 10ng/ml FGF and 20ng/ml EGF. Culture medium was added after a 15min incubation at 37C to ensure proper polymerization of collagen matrix. Endfoot preparations were left to equilibrate at 37C and 5% CO2 for 1–2h prior to live imaging. Live imaging was performed with a 100x/1.4 oil U PlanSApo objective mounted on an inverted spinning disk confocal microscope (Andor XD revolution spinning disk confocal microscope), equiped with a 37C and 5% CO2 incubation chamber. Following a one-hour equilibration of the incubation chamber, three 15μm Z-stacks per endfoot preparations were imaged in the blue, green and red channels with a 2μm resolution in the Z axis (pre-conversion acquisitions). This allowed the simultaneous recording of several conditions within one session and therefore minimized variability between imaging sessions. Positions were selected at locations where endfoot preparations presented minimal folding. Each position was then exposed to Arc lamp illumination at 10% intensity for 20 seconds with manual scanning in the Z-dimension. Each Z-stack was then acquired in the blue, green and red channels (post conversion acquisitions). Z stacks were then acquired every 5min for 45min in the green and blue channels (timecourse acquisitions). Following this, 40μM anisomycin treatment was performed for 20min and positions unaffected by the intial photoconversion were imaged using parameters described above (Aniso acquisitons). The FIJI software was used for the quantification of green fluorescence signal recovery over time. Z-projections were generated for all the Z-stacks. The ellipse-selection tool was used to generate regions of interests (ROIs) covering individual endfeet at each time point. Average green fluorescence intensity in endfeet was reported at each timepoint into an excel table where Dendra2 recovery was calculated using the following formula: (Dendra2 recovery)t = ((Dendra2 signal)t - (Dendra2 signal)t0) / (Dendra2 signal)t0.
Analyses of basal process branching in the MZ
Slides containing sections from brains electroporated with pGLAST-EGFP-CAXX, siRNAs and rescue plasmids were washed once in PBS for 10min, followed by a 15min wash in 0.25% Triton X in PBS (w/v) and one additional PBS wash for 15min. Vectashield was used to mount the slides with a coverslip and prevent bleaching of the EGFP signal. 20–40μm Z-stack images were acquired at a 0.2μm resolution in the Z dimension, using a 63X objective mounted on an epifluorescence microscope equipped with the Apotome technology (Zeiss). Analyses were performed in FIJI. Maximum intensity Z-projections of the Z stacks were generated to identify entire basal processes and endfeet. Rectangular regions of interests (ROIs) were traced around individual or small groups of basal processes. 3D projections of selected ROIs were generated with the following parameters deviating from the default settings: “axis of rotation”: Y axis, “rotation angle increment”: 5, “Interpolate”: checked. We used the Cell Counter plugin to count branches of each order, rotating the 3D projection in order to identify branches obscured by other basal processes. First order branches were those located further away from the pial surface (see Figure 4 for a depiction of branch orders). We used the Line tool to ensure that we counted branches >5μm only. The number of branches of each order was then reported into an Excel spreadsheet and the total number of branches was calculated as the sum of all the branches from different orders.
Analyses of endfoot area covering the basement membrane
These analyses were performed in fixed endfoot preparations from electroporated brains. These preparations were mounted in mounting medium on a glass slide, covered with a coverslip. We were careful to ensure that the EGFP+ endfeet were located immediately under the coverslip. Native EGFP fluorescence was acquired in Z-stacks with a confocal microscope. Using Maximum Intensity Z-projections, we employed the Polygon Selection tool in ImageJ to trace the outside limits of basal endfeet, and measured areas were reported into an Excel spreadsheet.
Analyses of basal process extensions in the IZ-low CP regions
These analyses were very similar to those performed in the upper CP/MZ regions, with the following modifications. Z-stack images of basal process in the IZ, low CP regions were acquired on a LSM 710 Zeiss confocal microscope. Here we did not assess the order of branches and a 5μm cutoff was not used. Instead we counted the total number of extensions emanating from the acquired region of the basal process and normalized this number to the length of basal process that we acquired and focusing on processes with a visualized length >50μm. Rotation of 3D projections were still used to visualize extensions masked by other structures.
Live imaging of neuron migration
A first in utero electroporation (IUE) of a pCAGGS-EGFP plasmid was performed at E14.5. 24h later, another IUE procedure was performed to transfect siRNAs together with a pCAGGS-PB-mCherryCAAX into the hemispheres transfected earlier with pCAGGS-EGFP. In the morning of the next day, 300μm-thick organotypic brain slices were generated as described previously85. Slices were cultured on cell culture inserts, and were surrounded with a 1mg/ml collagen solution supplemented with DMEM-F12. We used the same medium as the one described above for Dendra2 live imaging of basal endfeet. 4–6 slices transfected with different siRNA conditions were mounted on the same inserts. In the evening, live imaging was initiated for a total of 16h. Z-stack images of regions showing clear overlap between the first and second electroporations were acquired every 10min, using a 10x objective mounted on an inverted LSM 710 Zeiss confocal microscope, equipped with an incubation chamber (37C, 5% CO2). Analyses were performed in FIJI, using the Manual Tracking plugin. We focused on neurons located in the low IZ / SVZ region at t0h. Tracking results were reported in an Excel spreadsheet and the analyzed parameters are defined as follows. Net distances in Y and X: |Y0h-Y16h| and |X0h-X16h|, respectively. Net distance: Euclidean distance between neuron positions at t0h and t16h. Total path length: sum of distances traveled between each time points. Velocity: average travel speed between each time point. For analysis of movements in Y as “down”, “steady”, or “up”, we calculated the Y distance traveled from one time point to the next (Yt1-Yt2). “Down”, “steady” and “up” were defined as negative, null and positive Y distances, respectively.
Serial Block Face Electron Microscopy
For SBF-SEM, the samples then underwent a heavy metal staining protocol adapted from Deerinck (2010) after fixation. The tissues were washed in 0.1M sodium cacodylate pH7.4 and then a solution of 1.6 % potassium ferrocyanide with 2% osmium tetroxide buffered with 0.1M sodium cacodylate was added for 1h at RT. This is followed by filtered 10% thiocarbohydrazide (TCH) freshly prepared for 30min. Samples were then washed in distilled water and a secondary 2% osmium tetroxide incubation for 30min. The samples were then placed in 1% uranyl acetate at 4C overnight, washed in distilled water and then placed in freshly prepared lead aspartate solution for 30min at 60C. The samples were then dehydrated with cold ethanol, from 25% to 100% and then infiltrated with increasing concentrations of Durcupan resin in ethanol with several exchanges of 100% resin. The samples were finally embedded in 100% resin and left to polymerize at 60°C for 48 hours.
The tissue samples embedded in resin were manually trimmed with a razor blade to expose the tissue on their surfaces, and then glued onto an aluminum SBF-SEM rivet with conductive epoxy (SPI Conductive Silver Epoxy) with the exposed tissue down. Specimens on the rivet were further trimmed with a razor blade to as small a size as possible (about 0.5mm), and block face was trimmed with a glass knife. Once tissue was exposed, semi thin sections 0.5μm were taken and stained with toluidine blue and viewed under a light microscope to check tissue orientation and condition. Then, the rivet with the sample was sputter coated with gold-palladium.
The image stacks were acquired in an automated fashion by using a high resolution scanning electron microscope (Merlin - Carl Zeiss, Germany) equipped with a 3View system (Gatan Inc., Pleasanton, CA, USA), and a back-scattered electron detector. The Digital Micrograph software (Gatan Inc.) was used to adjust the SEM imaging conditions and slicing parameters. The electron microscope was operated at the high-resolution mode with an acceleration voltage of 2 kV, current mode and in the high-vacuum mode. All images were taken with the following scanning settings at 80pA, dwell time = 2s; pixel sizes 5–7nm. On average 300 sections were obtained at 60nm thickness
Statistical analyses
All analyses were performed in a blinded fashion. Number of data points and statistical tests used for all the comparisons are indicated in the figure legends.
Supplementary Material
Movies S1. Time lapse imaging of untreated EGFP+ neurons migrating along a radial glia scaffold targeted with Scrambled siRNAs, Related to Figure 2.
Time in hh:mm. Dots and lines highlight the migration path of migrating neurons.
Movies S2. Time lapse imaging of untreated EGFP+ neurons migrating along a radial glia scaffold targeted with Arhgap11a siRNAs, Related to Figure 2.
Time in hh:mm. Dots and lines highlight the migration path of migrating neurons.
Movie S3. Time lapse imaging of the upper CP/MZ region of a RGC labeled with membrane-bound EGFP, Related to Figure 3.
Movie S4. Time lapse imaging showing the movement of MS2-labelled mRNAs (green) in radial glial cells within E16.5 mouse organotypic brain slices, Related to Figure 5.
These mRNAs include the Arhgap11a cis-regulatory sequence responsible for endfoot RNA localization in radial glia.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti Arhgap11a | Bethyl | Cat#A303–097A, RRID# AB_10890540 |
| Rabbit anti Arhgap11a | Abcam | Cat#ab113261, RRID# AB_10866587 |
| Mouse anti Nestin | BD biosciences | Cat#556309, RRID# AB_396354 |
| Rabbit anti Tbr1 | Abcam | Cat#ab31940, RRID# AB_2200219 |
| Rat anti Ctip2 | Abcam | Cat#ab18465, RRID# AB_2064130 |
| Mouse anti ROR-beta | R&D Systems | Cat#N7927, RRID# AB_1964364 |
| Rabbit anti Laminin | Millipore | Cat#AB2034, RRID# AB_91209 |
| Rabbit anti Calretinin | Swant | Cat#7697, RRID# AB_2721226 |
| Rat anti Sox2 | Thermo Fisher | Cat#14–9811-80, RRID# AB_11219070 |
| Rabbit anti Tbr2 | Abcam | Cat#ab183991, RRID# AB_2721040 |
| Rabbit anti Ki67 | Cell Signaling Technology | Cat#12202, RRID# AB_2620142 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | JAX | Cat#000664, RRID# IMSR_JAX:000664 |
| Mouse: Dlx-Cre (Tg(dlx5a-cre)1Mekk/J) | JAX | Cat#008199, RRID# IMSR_JAX:008199 |
| Mouse: Arhgap11aem1(IMPC)J | KOMP | Cat#MGI:5907256, RRID# MMRRC_042323-JAX |
| Oligonucleotides | ||
| siRNA: siRNA flex targetting Arhgap11a | Qiagen | Cat#SI00902111 |
| siRNA: siRNA flex targetting Arhgap11a | Qiagen | Cat#SI00902132 |
| siRNA: siRNA flex targetting Arhgap11a | Qiagen | Cat#SI00902118 |
| siRNA: All Stars Negative Control | Qiagen | Cat#1027281 |
| In situ hybridization probe target human ARHGAP11A |
This paper | 2117–2657 of human ARHGAP11A (NM_014783.5) |
| smFISH Probe Set: Arhgap11a | LGC Biosearch Technologies | Custom made |
| smFISH Probe Set: MS2 | LGC Biosearch Technologies | Pilaz et al., 2016 |
| smFISH Probe Set: Dendra | LGC Biosearch Technologies | Pilaz et al., 2016 |
| qPCR primer: Actb FW | Thermo Fisher | AGATCAAGATCATTGCTCCT |
| qPCR primer: Actb Rev | Thermo Fisher | CCTGCTTGCTGATCCACATC |
| qPCR primer: Pax6 FW | Thermo Fisher | TCTTTGCTTGGGAAATCCG |
| qPCR primer: Pax6 Rev | Thermo Fisher | CTGCCCGTTCAACATCCTTAG |
| qPCR primer: Arhgap11a FW | Thermo Fisher | GCAGGTGTGCCAAGGCGAAGT |
| qPCR primer: Arhgap11a Rev | Thermo Fisher | TGCAAGTCGCCAACCAACACTTTCA |
| Taqman probe: Tubb3 | Thermo Fisher | Cat#Mm00727586_s1, RRID# |
| Taqman probe: Gapdh | Thermo Fisher | Gapdh |
| Recombinant DNA | ||
| Plasmid: pCAGGS-GFP | Gift from Dr. Nicholas Gaiano |
Mizutani et al., 2007 |
| Plasmid: pGLAST-EGFP-CAXX | Gift from Dr. Tarik Haydar |
Gal et al., 2006 |
| Plasmid: pCAGGS-PB-mCherryCAXX | Gift from Dr. Cagla Eroglu | N/A |
| Plasmid: pCAGGS-EX | Gift from Dr. Nicholas Gaiano |
Mizutani et al., 2007 |
| Plasmid: pCAGGS-EGFP-nls | This paper | N/A |
| Plasmid: pCAGGS-EGFP-nls-Arhgap11a 3’UTR | This paper | N/A |
| Plasmid: pCAGGS-EGFP-nls-Arhgap11a 5’UTR | This paper | N/A |
| Plasmid: pCAGGS-EGFP-nls-Arhgap11a CDS | This paper | N/A |
| Plasmid: pCAGGS-Dendra2 | Dr. Debra Silver Lab | Pilaz et al., 2016 |
| Plasmid: pCAGGS-Dendra2-Arhgap11a-noUTR | This paper | N/A |
| Plasmid: pCAGGS-Dendra2-Arhgap11a-5’UTR | This paper | N/A |
| Plasmid: pCAGGS-5’UTRArhgap11a-Dendra2 | This paper | N/A |
| Plasmid: pCAGGS-Arhgap11a(GD)-Dendra-5’UTR | This paper | N/A |
| Plasmid: pCAGGS-CFP-MS2-noUTR | Dr. Debra Silver Lab | Pilaz et al., 2016 |
| Plasmid: pCAGGS-CFP-MS2-Arhgap11a 5’UTR | Dr. Debra Silver Lab | Pilaz et al., 2016 |
| Software and Algorithms | ||
| FIJI | NIH | https://ImageJ.NIH.gov/IJ |
| GraphPad Prism (9) | GraphPad | https://www.graphpad.com/scientificsoftware/prism/ |
Highlights.
Arhgap11a subcellularly localizes in radial glia of mouse and human cerebral cortex
Arhgap11a 5′UTR directs active transport and local translation in radial glia
Localized mRNA and RhoA-GAP activity controls radial glial endfoot morphology
Arhgap11a non-cell autonomously acts in radial glia to mediate interneuron position
Acknowledgements
The authors thank members of the Silver lab, Cagla Eroglu, and Denis Jabaudon for discussions and careful reading of the manuscript. This work was supported by NS110388, NS120667, NS083897, DST Spark grant (to DLS). We also thank the Duke EM, microscopy, and mouse core facilities.
Footnotes
Declaration of Interests
The authors confirm there are no competing interests.
Inclusion and Diversity
We support inclusive, diverse, and equitable conduct of research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Buxbaum AR, Haimovich G, and Singer RH (2015). In the right place at the right time: visualizing and understanding mRNA localization. Nat Rev Mol Cell Biol 16, 95–109. 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holt CE, Martin KC, and Schuman EM (2019). Local translation in neurons: visualization and function. Nat Struct Mol Biol 26, 557–566. 10.1038/s41594-019-0263-5. [DOI] [PubMed] [Google Scholar]
- 3.Noctor SC, Flint AC, Weissman TA, Dammerman RS, and Kriegstein AR (2001). Neurons derived from radial glial cells establish radial units in neocortex. Nature 409, 714–720. 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 4.Malatesta P, Hartfuss E, and Gotz M. (2000). Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development 127, 5253–5263. [DOI] [PubMed] [Google Scholar]
- 5.Rakic P. (1972). Mode of cell migration to the superficial layers of fetal monkey neocortex. The Journal of comparative neurology 145, 61–83. 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- 6.Belvindrah R, Graus-Porta D, Goebbels S, Nave K-A, and Müller U. (2007). Beta1 integrins in radial glia but not in migrating neurons are essential for the formation of cell layers in the cerebral cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience 27, 13854–13865. 10.1523/JNEUROSCI.4494-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva CG, Peyre E, and Nguyen L. (2019). Cell migration promotes dynamic cellular interactions to control cerebral cortex morphogenesis. Nat Rev Neurosci 20, 318–329. 10.1038/s41583-019-0148-y. [DOI] [PubMed] [Google Scholar]
- 8.Gadisseux JF, Kadhim HJ, van den Bosch de Aguilar P, Caviness VS, and Evrard P. (1990). Neuron migration within the radial glial fiber system of the developing murine cerebrum: an electron microscopic autoradiographic analysis. Brain Res Dev Brain Res 52, 39–56. 10.1016/0165-3806(90)90220-s. [DOI] [PubMed] [Google Scholar]
- 9.Rakic P. (2003). Elusive radial glial cells: Historical and evolutionary perspective. Glia 43, 19–32. 10.1002/glia.10244. [DOI] [PubMed] [Google Scholar]
- 10.Casingal CR, Descant KD, and Anton ES (2022). Coordinating cerebral cortical construction and connectivity: Unifying influence of radial progenitors. Neuron 110, 1100–1115. 10.1016/j.neuron.2022.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjornsson CS, Apostolopoulou M, Tian Y, and Temple S. (2015). It takes a village: constructing the neurogenic niche. Dev Cell 32, 435–446. 10.1016/j.devcel.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haubst N, Georges-Labouesse E, De Arcangelis A, Mayer U, and Gotz M. (2006). Basement membrane attachment is dispensable for radial glial cell fate and for proliferation, but affects positioning of neuronal subtypes. Development 133, 3245–3254. [DOI] [PubMed] [Google Scholar]
- 13.Myshrall TD, Moore SA, Ostendorf AP, Satz JS, Kowalczyk T, Nguyen H, Daza RA, Lau C, Campbell KP, and Hevner RF (2012). Dystroglycan on radial glia end feet is required for pial basement membrane integrity and columnar organization of the developing cerebral cortex. J Neuropathol Exp Neurol 71, 1047–1063. 10.1097/NEN.0b013e318274a128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeSisto J, O’Rourke R, Jones HE, Pawlikowski B, Malek AD, Bonney S, Guimiot F, Jones KL, and Siegenthaler JA (2020). Single-Cell Transcriptomic Analyses of the Developing Meninges Reveal Meningeal Fibroblast Diversity and Function. Dev Cell 54, 43–59 e44. 10.1016/j.devcel.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yokota Y, Eom TY, Stanco A, Kim WY, Rao S, Snider WD, and Anton ES (2010). Cdc42 and Gsk3 modulate the dynamics of radial glial growth, inter-radial glial interactions and polarity in the developing cerebral cortex. Development 137, 4101–4110. 10.1242/dev.048637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu X, Duan M, Song L, Zhang W, Hu X, Zhao S, and Chen S. (2015). Morphological changes of radial glial cells during mouse embryonic development. Brain Res 1599, 57–66. 10.1016/j.brainres.2014.12.039. [DOI] [PubMed] [Google Scholar]
- 17.Kalebic N, Gilardi C, Stepien B, Wilsch-Brauninger M, Long KR, Namba T, Florio M, Langen B, Lombardot B, Shevchenko A, et al. (2019). Neocortical Expansion Due to Increased Proliferation of Basal Progenitors Is Linked to Changes in Their Morphology. Cell Stem Cell 24, 535–550 e539. 10.1016/j.stem.2019.02.017. [DOI] [PubMed] [Google Scholar]
- 18.Fietz SA, Kelava I, Vogt J, Wilsch-Bräuninger M, Stenzel D, Fish JL, Corbeil D, Riehn A, Distler W, Nitsch R, and Huttner WB (2010). OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nature Neuroscience 13, 690–699. 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- 19.Hansen DV, Lui JH, Parker PRL, and Kriegstein AR (2010). Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561. 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 20.Myshrall TD, Moore SA, Ostendorf AP, Satz JS, Kowalczyk T, Nguyen H, Daza RAM, Lau C, Campbell KP, and Hevner RF (2012). Dystroglycan on radial glia end feet is required for pial basement membrane integrity and columnar organization of the developing cerebral cortex. Journal of neuropathology and experimental neurology 71, 1047–1063. 10.1097/NEN.0b013e318274a128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klingler E, Francis F, Jabaudon D, and Cappello S. (2021). Mapping the molecular and cellular complexity of cortical malformations. Science 371. 10.1126/science.aba4517. [DOI] [PubMed] [Google Scholar]
- 22.Pilaz LJ, Lennox AL, Rouanet JP, and Silver DL (2016). Dynamic mRNA Transport and Local Translation in Radial Glial Progenitors of the Developing Brain. Curr Biol 26, 3383–3392. 10.1016/j.cub.2016.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsunekawa Y, Britto JM, Takahashi M, Polleux F, Tan SS, and Osumi N. (2012). Cyclin D2 in the basal process of neural progenitors is linked to non-equivalent cell fates. EMBO J 31, 1879–1892. 10.1038/emboj.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cappello S, Bohringer CR, Bergami M, Conzelmann KK, Ghanem A, Tomassy GS, Arlotta P, Mainardi M, Allegra M, Caleo M, et al. (2012). A radial glia-specific role of RhoA in double cortex formation. Neuron 73, 911–924. 10.1016/j.neuron.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 25.Müller PM, Rademacher J, Bagshaw RD, Alp KM, Giudice G, Heinrich LE, Barth C, Eccles RL, Sanchez-Castro M, Brandenburg L, et al. (2018). Spatial Organization of Rho GTPase signaling by RhoGEF/RhoGAP proteins. bioRxiv. [Google Scholar]
- 26.Zanin E, Desai A, Poser I, Toyoda Y, Andree C, Moebius C, Bickle M, Conradt B, Piekny A, and Oegema K. (2013). A conserved RhoGAP limits M phase contractility and coordinates with microtubule asters to confine RhoA during cytokinesis. Dev Cell 26, 496–510. 10.1016/j.devcel.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai B, Zhang X, Shang R, Wang J, Yang X, Zhang H, Liu Q, Wang D, Wang L, and Dou K. (2018). Blockade of ARHGAP11A reverses malignant progress via inactivating Rac1B in hepatocellular carcinoma. Cell Commun Signal 16, 99. 10.1186/s12964-018-0312-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kagawa Y, Matsumoto S, Kamioka Y, Mimori K, Naito Y, Ishii T, Okuzaki D, Nishida N, Maeda S, Naito A, et al. (2013). Cell cycle-dependent Rho GTPase activity dynamically regulates cancer cell motility and invasion in vivo. PLoS One 8, e83629. 10.1371/journal.pone.0083629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, Zhou X, Wang J, Li Z, Kong X, Qian J, Hu Y, and Fang JY (2013). RhoGAPs attenuate cell proliferation by direct interaction with p53 tetramerization domain. Cell Rep 3, 1526–1538. 10.1016/j.celrep.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 30.Lodato S, and Arlotta P. (2015). Generating neuronal diversity in the mammalian cerebral cortex. Annu Rev Cell Dev Biol 31, 699–720. 10.1146/annurev-cellbio-100814-125353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller DJ, Bhaduri A, Sestan N, and Kriegstein A. (2019). Shared and derived features of cellular diversity in the human cerebral cortex. Curr Opin Neurobiol 56, 117–124. 10.1016/j.conb.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silbereis JC, Pochareddy S, Zhu Y, Li M, and Sestan N. (2016). The Cellular and Molecular Landscapes of the Developing Human Central Nervous System. Neuron 89, 248–268. 10.1016/j.neuron.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Qiu R, Tsark W, and Lu Q. (2007). Rapid promoter analysis in developing mouse brain and genetic labeling of young neurons by doublecortin-DsRed-express. J Neurosci Res 85, 3567–3573. 10.1002/jnr.21440. [DOI] [PubMed] [Google Scholar]
- 34.Telley L, Agirman G, Prados J, Amberg N, Fievre S, Oberst P, Bartolini G, Vitali I, Cadilhac C, Hippenmeyer S, et al. (2019). Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science 364. 10.1126/science.aav2522. [DOI] [PubMed] [Google Scholar]
- 35.Loo L, Simon JM, Xing L, McCoy ES, Niehaus JK, Guo J, Anton ES, and Zylka MJ (2019). Single-cell transcriptomic analysis of mouse neocortical development. Nat Commun 10, 134. 10.1038/s41467-018-08079-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323. 10.1126/science.aap8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Florio M, Heide M, Pinson A, Brandl H, Albert M, Winkler S, Wimberger P, Huttner WB, and Hiller M. (2018). Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. Elife 7. 10.7554/eLife.32332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elias LA, Wang DD, and Kriegstein AR (2007). Gap junction adhesion is necessary for radial migration in the neocortex. Nature 448, 901–907. 10.1038/nature06063. [DOI] [PubMed] [Google Scholar]
- 39.Nakagawa N, Plestant C, Yabuno-Nakagawa K, Li J, Lee J, Huang CW, Lee A, Krupa O, Adhikari A, Thompson S, et al. (2019). Memo1-Mediated Tiling of Radial Glial Cells Facilitates Cerebral Cortical Development. Neuron 103, 836–852 e835. 10.1016/j.neuron.2019.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belvindrah R, Graus-Porta D, Goebbels S, Nave KA, and Muller U. (2007). Beta1 integrins in radial glia but not in migrating neurons are essential for the formation of cell layers in the cerebral cortex. J Neurosci 27, 13854–13865. 10.1523/JNEUROSCI.4494-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, and Macklis JD (2013). Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci 14, 755–769. 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong HH, Lin JQ, Strohl F, Roque CG, Cioni JM, Cagnetta R, Turner-Bridger B, Laine RF, Harris WA, Kaminski CF, and Holt CE (2017). RNA Docking and Local Translation Regulate Site-Specific Axon Remodeling In Vivo. Neuron 95, 852–868 e858. 10.1016/j.neuron.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yokota Y, Gashghaei HT, Han C, Watson H, Campbell KJ, and Anton ES (2007). Radial glial dependent and independent dynamics of interneuronal migration in the developing cerebral cortex. PLoS One 2, e794. 10.1371/journal.pone.0000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka DH, Yanagida M, Zhu Y, Mikami S, Nagasawa T, Miyazaki J, Yanagawa Y, Obata K, and Murakami F. (2009). Random walk behavior of migrating cortical interneurons in the marginal zone: time-lapse analysis in flat-mount cortex. J Neurosci 29, 1300–1311. 10.1523/jneurosci.5446-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyoshi G, and Fishell G. (2011). GABAergic interneuron lineages selectively sort into specific cortical layers during early postnatal development. Cereb Cortex 21, 845–852. 10.1093/cercor/bhq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marín O, and Rubenstein JL (2001). A long, remarkable journey: tangential migration in the telencephalon. Nat Rev Neurosci 2, 780–790. 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- 47.Lavdas AA, Grigoriou M, Pachnis V, and Parnavelas JG (1999). The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. J Neurosci 19, 7881–7888. 10.1523/jneurosci.19-18-07881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teissier A, and Pierani A. (2021). Wiring of higher-order cortical areas: Spatiotemporal development of cortical hierarchy. Semin Cell Dev Biol 118, 35–49. 10.1016/j.semcdb.2021.05.010. [DOI] [PubMed] [Google Scholar]
- 49.Genescu I, and Garel S. (2021). Being superficial: a developmental viewpoint on cortical layer 1 wiring. Curr Opin Neurobiol 66, 125–134. 10.1016/j.conb.2020.10.003. [DOI] [PubMed] [Google Scholar]
- 50.Lim L, Pakan JMP, Selten MM, Marques-Smith A, Llorca A, Bae SE, Rochefort NL, and Marín O. (2018). Optimization of interneuron function by direct coupling of cell migration and axonal targeting. Nat Neurosci 21, 920–931. 10.1038/s41593-018-0162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, and Singer RH (2005). Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438, 512–515. 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 52.Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, and Singer RH (2011). A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods 8, 165–170. 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pilaz LJ, and Silver DL (2017). Moving messages in the developing brain-emerging roles for mRNA transport and local translation in neural stem cells. FEBS Lett 591, 1526–1539. 10.1002/1873-3468.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsunekawa Y, Kikkawa T, and Osumi N. (2014). Asymmetric inheritance of Cyclin D2 maintains proliferative neural stem/progenitor cells: a critical event in brain development and evolution. Dev Growth Differ 56, 349–357. 10.1111/dgd.12135. [DOI] [PubMed] [Google Scholar]
- 55.Dahlstrand J, Lardelli M, and Lendahl U. (1995). Nestin mRNA expression correlates with the central nervous system progenitor cell state in many, but not all, regions of developing central nervous system. Brain Res Dev Brain Res 84, 109–129. [DOI] [PubMed] [Google Scholar]
- 56.Pilaz LJ, McMahon JJ, Miller EE, Lennox AL, Suzuki A, Salmon E, and Silver DL (2016). Prolonged Mitosis of Neural Progenitors Alters Cell Fate in the Developing Brain. Neuron 89, 83–99. 10.1016/j.neuron.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shamir M, Bar-On Y, Phillips R, and Milo R. (2016). SnapShot: Timescales in Cell Biology. Cell 164, 1302–1302.e1301. 10.1016/j.cell.2016.02.058. [DOI] [PubMed] [Google Scholar]
- 58.Goering R, Hudish LI, Guzman BB, Raj N, Bassell GJ, Russ HA, Dominguez D, and Taliaferro JM (2019). FMRP promotes RNA localization to neuronal projections through interactions between its RGG domain and G-quadruplex RNA sequences. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, and Schuman EM (2012). The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron 74, 453–466. 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poulopoulos A, Murphy AJ, Ozkan A, Davis P, Hatch J, Kirchner R, and Macklis JD (2019). Subcellular transcriptomes and proteomes of developing axon projections in the cerebral cortex. Nature 565, 356–360. 10.1038/s41586-018-0847-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.D’Arcy BR, and Silver DL (2020). Local gene regulation in radial glia: Lessons from across the nervous system. Traffic 21, 737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim E, and Jung H. (2020). Local mRNA translation in long-term maintenance of axon health and function. Curr Opin Neurobiol 63, 15–22. 10.1016/j.conb.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 63.Li S, Jin Z, Koirala S, Bu L, Xu L, Hynes RO, Walsh CA, Corfas G, and Piao X. (2008). GPR56 regulates pial basement membrane integrity and cortical lamination. J Neurosci 28, 5817–5826. 10.1523/jneurosci.0853-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Radakovits R, Barros CS, Belvindrah R, Patton B, and Müller U. (2009). Regulation of radial glial survival by signals from the meninges. J Neurosci 29, 7694–7705. 10.1523/jneurosci.5537-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Neill AC, Kyrousi C, Klaus J, Leventer RJ, Kirk EP, Fry A, Pilz DT, Morgan T, Jenkins ZA, Drukker M, et al. (2018). A Primate-Specific Isoform of PLEKHG6 Regulates Neurogenesis and Neuronal Migration. Cell Rep 25, 2729–2741.e2726. 10.1016/j.celrep.2018.11.029. [DOI] [PubMed] [Google Scholar]
- 66.Zaoui K, Honoré S, Isnardon D, Braguer D, and Badache A. (2008). Memo-RhoA-mDia1 signaling controls microtubules, the actin network, and adhesion site formation in migrating cells. J Cell Biol 183, 401–408. 10.1083/jcb.200805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cappello S, Böhringer CRJ, Bergami M, Conzelmann K-K, Ghanem A, Tomassy GS, Arlotta P, Mainardi M, Allegra M, Caleo M, et al. (2012). A Radial Glia-Specific Role of RhoA in Double Cortex Formation. Neuron 73, 911–924. 10.1016/j.neuron.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 68.D’Arcy BR, Lennox AL, Musso CM, Bracher A, Escobar-Tomlienovich C, Perez-Sanchez S, and Silver DL (2022). Subcellular proteome of radial glia reveals non-muscle myosins control basal endfeet to mediate interneuron organization. bioRxiv, 2022.2011.2018.517018. 10.1101/2022.11.18.517018. [DOI] [Google Scholar]
- 69.Bartolini G, Sánchez-Alcañiz JA, Osório C, Valiente M, García-Frigola C, and Marín O. (2017). Neuregulin 3 Mediates Cortical Plate Invasion and Laminar Allocation of GABAergic Interneurons. Cell Rep 18, 1157–1170. 10.1016/j.celrep.2016.12.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hansen DV, Lui JH, Parker PR, and Kriegstein AR (2010). Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561. 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 71.Betizeau M, Cortay V, Patti D, Pfister S, Gautier E, Bellemin-Menard A, Afanassieff M, Huissoud C, Douglas RJ, Kennedy H, and Dehay C. (2013). Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron 80, 442–457. 10.1016/j.neuron.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 72.Florio M, Albert M, Taverna E, Namba T, Brandl H, Lewitus E, Haffner C, Sykes A, Wong FK, Peters J, et al. (2015). Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347, 1465–1470. 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- 73.Rakic P. (1972). Mode of cell migration to the superficial layers of fetal monkey neocortex. J Comp Neurol 145, 61–83. 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- 74.Heide M, Haffner C, Murayama A, Kurotaki Y, Shinohara H, Okano H, Sasaki E, and Huttner WB (2020). Human-specific ARHGAP11B increases size and folding of primate neocortex in the fetal marmoset. Science. 10.1126/science.abb2401. [DOI] [PubMed] [Google Scholar]
- 75.Kalebic N, Gilardi C, Albert M, Namba T, Long KR, Kostic M, Langen B, and Huttner WB (2018). Human-specific ARHGAP11B induces hallmarks of neocortical expansion in developing ferret neocortex. Elife 7. 10.7554/eLife.41241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Charrier C, Joshi K, Coutinho-Budd J, Kim JE, Lambert N, de Marchena J, Jin WL, Vanderhaeghen P, Ghosh A, Sassa T, and Polleux F. (2012). Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell 149, 923–935. 10.1016/j.cell.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Suzuki IK, Gacquer D, Van Heurck R, Kumar D, Wojno M, Bilheu A, Herpoel A, Lambert N, Cheron J, Polleux F, et al. (2018). Human-Specific NOTCH2NL Genes Expand Cortical Neurogenesis through Delta/Notch Regulation. Cell 173, 1370–1384 e1316. 10.1016/j.cell.2018.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fiddes IT, Lodewijk GA, Mooring M, Bosworth CM, Ewing AD, Mantalas GL, Novak AM, van den Bout A, Bishara A, Rosenkrantz JL, et al. (2018). Human-Specific NOTCH2NL Genes Affect Notch Signaling and Cortical Neurogenesis. Cell 173, 1356–1369 e1322. 10.1016/j.cell.2018.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lambert de Rouvroit C, and Goffinet AM (2001). Neuronal migration. Mech Dev 105, 47–56. [DOI] [PubMed] [Google Scholar]
- 80.R Development Core Team (2019). R: a language and environment for statistical computing (R Foundation for Statistical Computing; ). [Google Scholar]
- 81.Mizutani K, Yoon K, Dang L, Tokunaga A, and Gaiano N. (2007). Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 449, 351–355. [DOI] [PubMed] [Google Scholar]
- 82.Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, and Haydar TF (2006). Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci 26, 1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saito T, and Nakatsuji N. (2001). Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol 240, 237–246. [DOI] [PubMed] [Google Scholar]
- 84.Takahashi M, and Osumi N. (2002). Pax6 regulates specification of ventral neurone subtypes in the hindbrain by establishing progenitor domains. Development 129, 1327–1338. [DOI] [PubMed] [Google Scholar]
- 85.Pilaz LJ, and Silver DL (2014). Live imaging of mitosis in the developing mouse embryonic cortex. J Vis Exp. 10.3791/51298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movies S1. Time lapse imaging of untreated EGFP+ neurons migrating along a radial glia scaffold targeted with Scrambled siRNAs, Related to Figure 2.
Time in hh:mm. Dots and lines highlight the migration path of migrating neurons.
Movies S2. Time lapse imaging of untreated EGFP+ neurons migrating along a radial glia scaffold targeted with Arhgap11a siRNAs, Related to Figure 2.
Time in hh:mm. Dots and lines highlight the migration path of migrating neurons.
Movie S3. Time lapse imaging of the upper CP/MZ region of a RGC labeled with membrane-bound EGFP, Related to Figure 3.
Movie S4. Time lapse imaging showing the movement of MS2-labelled mRNAs (green) in radial glial cells within E16.5 mouse organotypic brain slices, Related to Figure 5.
These mRNAs include the Arhgap11a cis-regulatory sequence responsible for endfoot RNA localization in radial glia.
Data Availability Statement
This study did not generate any unique code. Original/source data for figures in the paper are available from the corresponding authors on request.