Abstract
Influenza A virus (IAV)’s frequent genetic changes challenge vaccine strategies and engender resistance to current drugs. We sought to identify conserved and essential RNA secondary structures within IAV’s genome predicted to have greater constraints on mutation in response to therapeutic targeting. We identified and genetically validated a RNA structure (PSL2) that mediates in vitro packaging and in vivo disease, and is conserved across all known IAV isolates. A PSL2-targeting locked nucleic acid (LNA) administered 3 days after, or 14 days before, a lethal IAV inoculum provided 100% survival in mice, lead to the development of strong immunity to rechallenge with a ten-fold lethal inoculum, evaded attempts to select for resistance, and retained full potency against neuraminidase inhibitor (NAI)-resistant virus. Using an analogous approach to target SARS-CoV-2, prophylactic administration of LNAs specific for highly conserved RNA structures in the viral genome protected hamsters from efficient transmission of the SARS-CoV-2 USA_WA1/2020 variant. These findings highlight the potential applicability of this approach to any virus of interest via a process we term “programmable antivirals”, with implications for antiviral prophylaxis and post-exposure therapy.
Introduction
Influenza A virus (IAV) is a segmented RNA virus that causes major morbidity and mortality worldwide. Current antiviral therapies target viral proteins that frequently mutate, rendering many such therapies inadequate1–3. Despite a breadth of knowledge about the viral lifecycle, knowledge of the RNA secondary structure of the genome is limited. Research on other RNA viruses has revealed genomic RNA to be capable of playing many important roles in viral lifecycles beyond merely encoding amino acid sequences, suggesting that viral RNA structural elements could be promising therapeutic targets4,5. To the extent these RNA structural elements are both essential and highly conserved, these features could reduce the degree of freedom for mutations that are compatible with virus function. This, in turn, could translate into a high barrier for resistance to therapeutics designed to disrupt these RNA structures. In IAV, genome packaging is one such critical juncture in which RNA structure might serve a central function.
The IAV genome consists of eight single-stranded negative-sense viral RNA (vRNA) segments that encode a minimum of 14 known viral proteins6. The vRNA, together with nucleoprotein (NP) and the heterotrimeric polymerase complex, comprised of PB2, PB1, and PA proteins, forms the complete viral ribonucleoprotein (vRNP)7. To be fully infectious, IAV virions must incorporate at least one of each segment’s vRNP8. Current paradigm supports a selective packaging method whereby the eight vRNPs are selected in a hierarchal manner mediated by unique, segment-specific packaging signals present in the terminal and central coding regions of each vRNA that allow for discrimination between the latter9–11. Each vRNP interacts with at least one other partner vRNP to form a supramolecular complex12 likely maintained by intersegment RNA-RNA and/or protein-RNA interactions hypothesized to guide the packaging process8,13. The mechanism mediating this selection and arrangement, however, is poorly understood. Curiously, packaging signals exist in regions of high nucleotide conservancy that strongly suppress synonymous codon usage14–16. Conservation of primary sequence beyond what is required for protein coding suggests the potential for maintenance of RNA structures possessing biological functionalities. Certain synonymous mutations within the polymerase gene, PB2, not only affect its own packaging, but also the incorporation of other segments11,14,17.
We hypothesized that PB2’s dominant role in the packaging process might be facilitated by non-protein elements encoded by the PB2 vRNA, including structured RNA elements. To test this hypothesis, we first mapped the RNA secondary structure within PB2 that mediates packaging, and then genetically validated this structure’s role in the viral life cycle in vitro and IAV pathogenesis in vivo. With this model, we demonstrate proof-of-concept for a new class of antiviral therapeutics that can efficiently disrupt packaging and prevent and treat otherwise lethal IAV disease in vivo, as well as enable the development of strong functional immunity in mice, with a high barrier to resistance. Moreover, we hypothesized that an analogous approach can be applied to any RNA virus of interest, and the recent COVID-19 pandemic provided an opportunity for us to successfully test this hypothesis by targeting SARS-CoV-2 in vitro and in vivo.
Results
SHAPE identifies conserved candidate PB2 packaging signal
To search for structured RNA domains in IAV segments, we first applied selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE)18 and computational modeling to IAV segment PB2 genomic vRNA. In vitro transcribed full-length (−)-sense PB2 vRNA from strain A/Puerto Rico/8/1934 (H1N1) PR8 was folded in solution19 and interrogated using an electrophilic SHAPE reagent that preferentially reacts with nucleotides existing in flexible, single-stranded states18(Fig. 1). This analysis revealed that much of the 2341-nt vRNA is largely unstructured (Supplementary Fig. 1a), as described in previous genome analyses15,20. However, these previous studies did not analyze the last 80 nucleotides of PB2’s terminal coding regions. SHAPE-guided modeling suggested several areas in this terminal region that contain stable RNA secondary structures, most notably a stem-loop motif, named herein as Packaging Stem-Loop 2 (PSL2) (Fig. 1a and Supplementary Fig. 1b, nucleotides 34–87). This region includes a set of nucleotides that were previously implicated in segment PB2 packaging through mutational analysis via an unidentified mechanism (Fig. 1a,b, see circled nucleotides, and Supplementary Table 1)11,14,16. Supporting the hypothesis that these prior mutations act through disruption of PSL2 structure, SHAPE analysis of the mutants yielded different conformations that all abrogated the wild-type PSL2 structure (Fig. 1c, and Supplementary Fig. 2). The 60-nucleotide region encompassing PSL2 displays near 100% sequence conservation at the single nucleotide level between representative seasonal as well as pandemic IAV strains of different subtypes and species origins (Fig. 1d, Supplementary Fig. 1c). Further analysis revealed that this high degree of conservation extends to all known IAV isolates available in public databases (Supplementary Fig. 1d), suggesting the existence of a strict biologic requirement to maintain an intact PSL2 structure. Indeed, the PSL2 stem-loop structure was recovered in SHAPE-guided modeling of full-length PB2 RNA across diverse species and subtypes including the highly pathogenic avian H5N1 and pandemic 1918 H1N1 strains (Fig. 1e).
Figure 1. SHAPE-determined RNA secondary structures of wild-type PB2 and packaging mutant vRNAs.
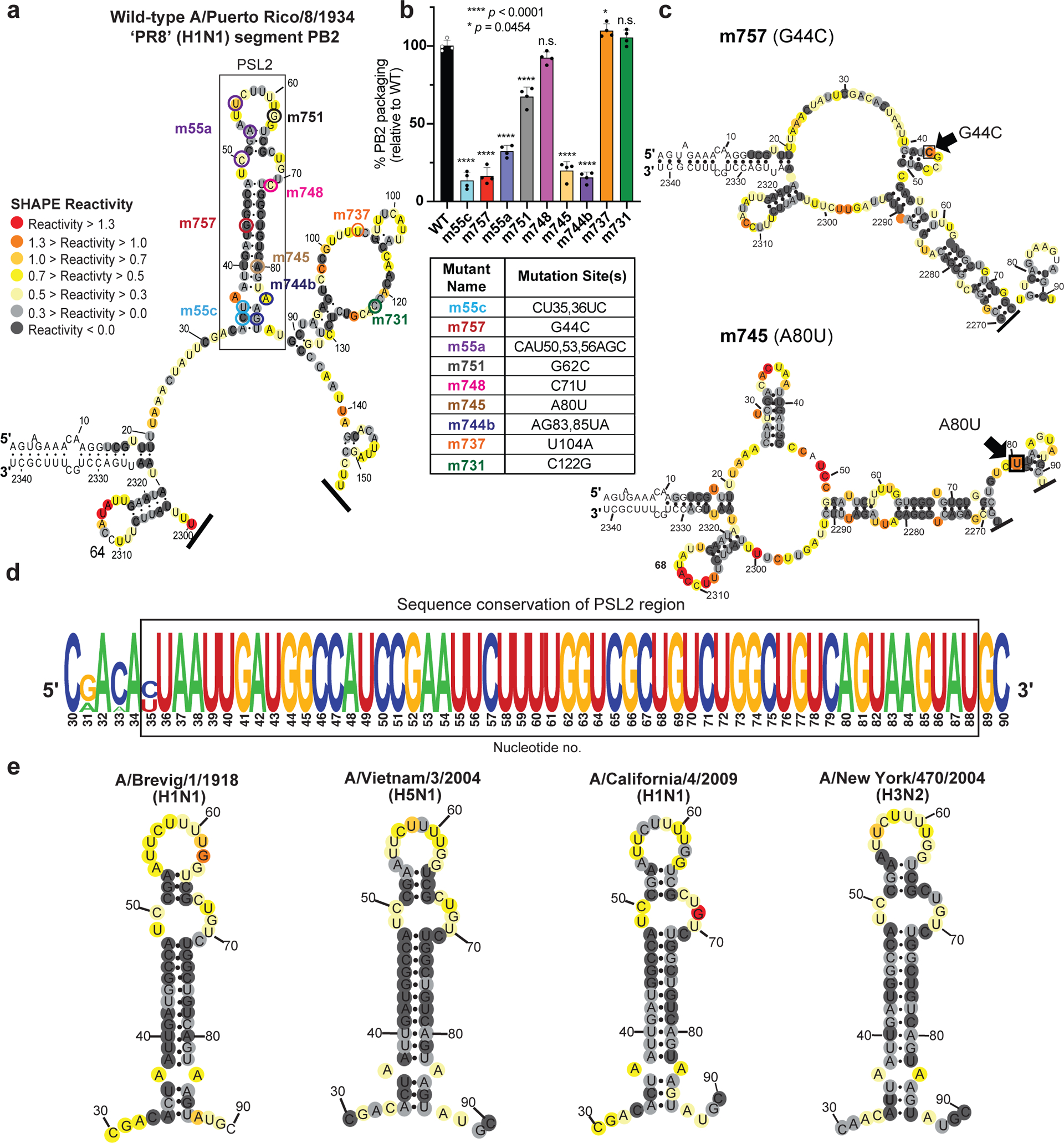
SHAPE-chemical mapping performed on full-length (−)-sense PB2 vRNAs. Colors denote SHAPE reactivity, which is proportional to the probability that a nucleotide is single-stranded. All structures are truncated to highlight the 5′ termini sequence structure. (a) SHAPE-predicted wild-type PB2 RNA secondary structure from strain A/Puerto Rico/8/1934 “PR8” (H1N1). Color-coded circles correspond to nucleotides sites where synonymous mutations were reported to affect PB2 packaging11,14. (b) Packaging efficiency of synonymous mutants in (a), determined by RT-qPCR. Results representative of two independent experiments with biological replicates, each performed in triplicate. Statistical analysis was performed using one-way ANOVA with Dunnett’s multiple comparisons test against the WT mean by GraphPad Prism 9 software (n=4). Error bars represents mean ± standard deviation (s.d.); **** p < 0.0001; * p = 0.0454, n.s. = not significant. Box below indicates mutant name and corresponding nucleotide change. Nucleotide numbering shown in the genomic (−)-sense orientation. (c) SHAPE-mapped structures of PB2 packaging-defective mutant vRNAs, m757 (G44C) and m745 (A80U) indicating loss of PSL2’s RNA secondary structure. Black arrowheads and boxed nucleotides denote site(s) of synonymous mutation. (d) Web-logo representation of the PSL2 region conservation across IAV strains and diverse influenza A viral subtypes (weblogo.berkeley.edu). The overall height represents sequence conservation at that nucleotide position, while the symbol height within each position indicates the relative frequency of each nucleotide at that site. Black box denotes PSL2 region. Sequences included in the alignment: pandemic A/Brevig Mission/1/1918 (H1N1), pandemic A/California/04/2009 (H1N1), seasonal human A/New York/470/2004 (H3N2), A/Puerto Rico/8/1934 (H1N1), highly pathogenic avian A/Vietnam/03/2004 (H5N1), avian A/mallard/Maryland/14OS1154/2014 (H6N1), pandemic A/Hong Kong/8/1968 (H3N2), and seasonal human A/New York/312/2001 (H1N1) (see Supplementary Fig. 1d). RNA nucleotides are numbered in (−) -sense orientation. (e) SHAPE-mapped structures of full-length wild-type PB2 vRNA from pandemic and highly pathogenic strains, including different subtypes to modern human strains: 1918 pandemic (A/Brevig Mission/1/1918 (H1N1)), highly-pathogenic avian (A/Vietnam/1203/2004 (H5N1)), 2009 pandemic ‘swine’ (A/California/04/2009 (H1N1)), and Fujian-like human seasonal virus, A/New York/470/2004 (H3N2)
M2 validates PSL2 and predicts novel packaging mutants
To further test the SHAPE analysis of the PSL2 RNA structure and to uncover additional informative mutations needed for in vivo tests, we applied multidimensional chemical mapping methods to the PSL2 segment. Mutate-and-Map (M2) analysis couples systematic mutagenesis with high-throughput chemical mapping to produce accurate base-pair inferences and interactions of RNA domains21. By sequentially mutating RNA one nucleotide at a time with its Watson-Crick complement and measuring the impact this mutagenesis has on chemical reactivity, pair-wise correlations between close and distant residues can be established. First, M2 measurements confirmed disruption of the chemical reactivity pattern upon systematic mutation of each PSL2 stem residue, including changes at nucleotides previously identified to be relevant for PB2 packaging (Fig. 2, see noted fields)11,14. Automated computational analysis based on these M2 data recovered the SHAPE-guided PSL2 structure with high confidence (Fig. 2, Supplementary 3), further validating our structural model. Second, as predictive tests, we designed compensatory mutations to restore base pairings—albeit not the native sequence—in the wild-type structure that were disrupted by the initial packaging-defective mutations (Supplementary Fig. 4). These mutation-rescue variants restored the PSL2 SHAPE pattern, providing in vitro validation of the modeled structure at base-pair resolution and suggesting sequence variants to test the role of PSL2 structure in vivo.
Figure 2. 2-Dimensional Mutate-and-Map (M2) analysis and empiric validation of PSL2 motif.
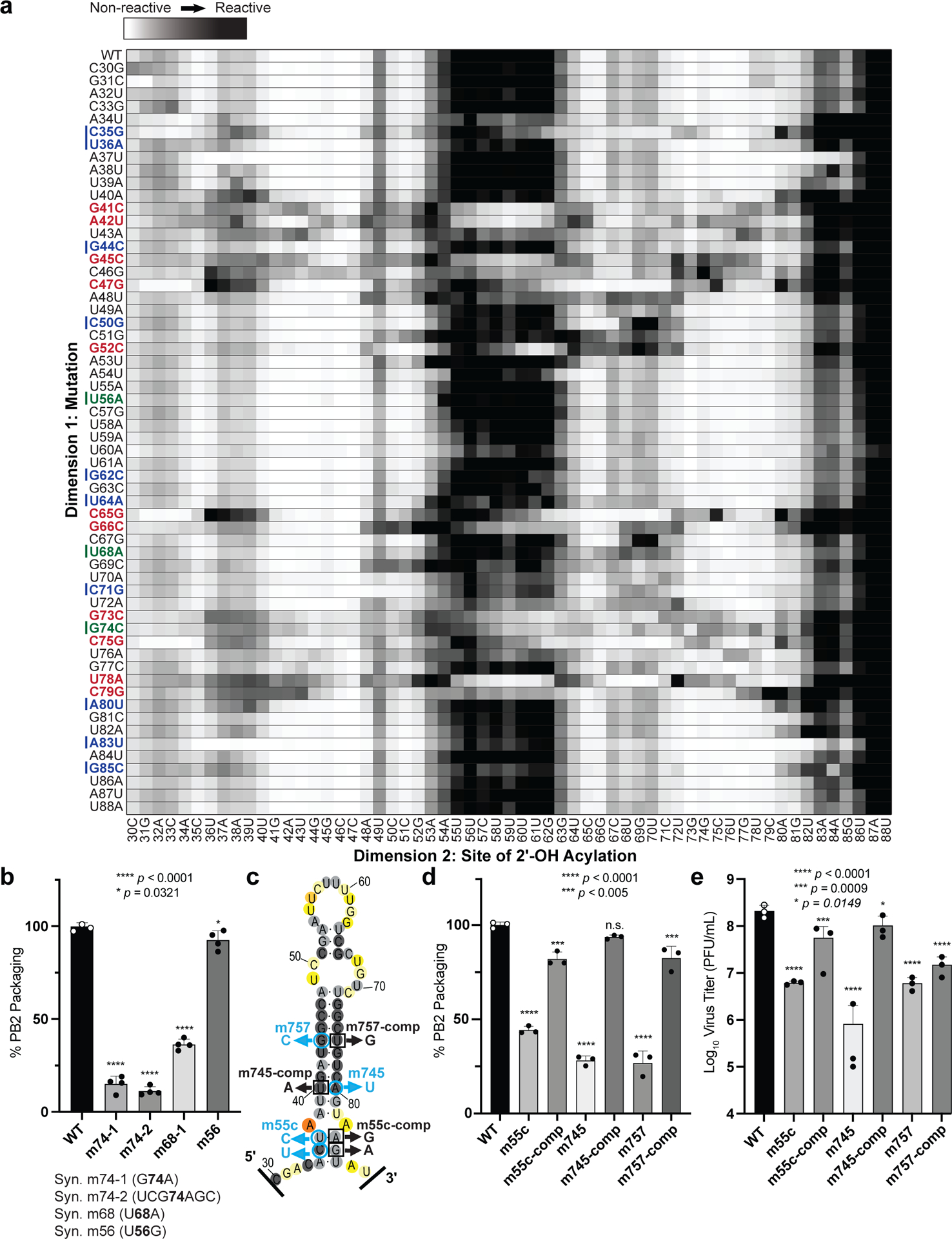
(a) Systematic single nucleotide mutation and mapping of resulting chemical accessibility reveals interactions in the three-dimensional structure of the RNA. Chemical accessibilities, plotted in grayscale (black, highest SHAPE reactivity), across 59 single mutations at single-nucleotide resolutions of PSL2 element from PR8 strain segment PB2. Reactivity peaks (left to right) correspond to nucleotides from the 5′ to 3′ end of the PB2 RNA. Nucleotides corresponding to known packaging mutation sites11 are indicated on left in blue. Red bolded mutations denote packaging-defective mutant sites predicted by M2 analysis. Green bolded mutations indicate synonymous mutant sites analyzed in (b). (b) Packaging efficiencies of M2-identified synonymous mutants read out by RT-qPCR. Packaging efficiency represents the percentage of mutant PB2 vRNA packaging relative to parental wild-type PB2. Results from two independent experiments in biological duplicate and technical triplicate (n=4). **** p < 0.0001, * p = 0.0321. (c) Previously described synonymous mutants (m757, m745, m55c) are mapped onto PSL2 structure. Compensatory, non-synonymous mutations m55c-comp, m745-comp, and m757-comp were designed at sites predicted to restore wild-type PSL2 structure based on SHAPE and mutate-and-map chemical analyses. Black boxed nucleotides denote compensatory mutation sites. (−)-sense vRNA orientation is shown. (d) Packaging efficiencies of packaging-defective and compensatory mutant viruses. For compensatory mutations where a non-synonymous change was required, a wild-type PB2 protein expression plasmid was co-transfected during virus rescue. Values given as percentage of PB2 vRNA packaging relative to wild-type PR8 virus. Results from three independent experiments (n=3), assays performed in triplicate. **** p < 0.0001, *** p < 0.0005, n.s. = not significant. (e) Virus titer determined by plaque assay. Results in PFU / mL, plaque assays in triplicate (n=3). **** p < 0.0001, *** p = 0.0009, ** p = 0.0149. All error bars represent mean ± s.d. All statistical analysis were performed by ordinary one-way ANOVA using Dunnett’s multiple comparisons test against WT computed in GraphPad Prism 9 software.
To test whether the PSL2 stem-loop structure observed in solution was relevant to virus packaging in the cellular milieu, the same nine synonymous mutations reported by Gog et al. (2007) and Marsh et al. (2008) (Fig. 1a,b and Supplementary Table 1) as well as four new synonymous mutations characterized by M2 analysis (Fig. 2a,b) were cloned into plasmids containing the PR8 PB2 gene11,14,16. The packaging efficiencies of the nine previously known mutants, now in the PR8 background, were comparable to those originally described in the WSN33 virus11 (Fig. 1b and Supplementary Table 1). Of these, mutants m55c, m757, m745, and m744b, were predicted to show the most significant impairment based on their location within PSL2’s stem regions (Fig. 1a–c and Supplementary Fig. 2). In contrast, published mutations that have no effect on PB2 packaging (e.g. m731) mapped to the unstructured apical loop or fell outside of PSL2 and did not alter its structural integrity (Supplementary Fig. 5a), while mutations with minor effects on virus packaging showed only minor alterations to the structure (Supplementary Fig. 5b)11. The three novel synonymous mutants (m74–1, m74–2, and m68) identified by M2-analysis as having a significant effect on in vitro PSL2 structure (Fig. 2a, see green-marked nucleotides) showed significant loss in PB2 packaging, whereas mutation sites that resulted in negligible change in SHAPE reactivity compared to wild-type PSL2 (e.g., m56), gave wild-type-like packaging efficiency levels (Fig. 2b). The strong correlation of structure disruption with in cellulo packaging efficiency observed across these mutants supports a role of PSL2 structure in virus packaging.
Compensatory mutations restore PSL2 structure and function
To investigate the functional role of PSL2 in IAV genome packaging, compensatory mutations designed to restore the wild-type stem-loop structure destroyed by the packaging-defective mutations (Fig. 2c, Supplementary Fig. 4) were cloned into PR8 expression plasmids to generate mutant rescue viruses. The compensatory mutations rescued not only the virus packaging for segment PB2 (Fig. 2d), but also other segments previously reported to be affected by the deleterious mutations, consistent with the proposed hierarchal role of PB2 in IAV packaging (Supplementary Fig. 6a)10,11,17. In addition to recovering PB2 packaging, the compensatory mutations gave complete or near-complete rescue of the virus titer loss caused by the defective mutations (Fig. 2e and Supplementary Fig. 6b). Some non-synonymous compensatory mutations were able to restore PB2 packaging better than others (e.g. m745-comp compared to m757-comp). This possibly reflects incomplete restoration of PB2 protein function through exogenous addition (Fig. 2d, e and Supplementary Fig. 6) since for non-synonymous mutations, we also expressed WT PB2 protein to mitigate the possibility of any impairment in PB2 protein function.
We performed computational enumeration and multidimensional mutation-rescue22 (M2R) experiments in order to identify additional successful PSL2-defective and compensatory mutant pairs (Fig. 3a, b and Supplementary Fig. 7). Successful mutation-rescue was defined when each single mutation alone disrupted the SHAPE-mapped wild-type PSL2 structure, while the double compensatory mutations recovered its structure (Fig. 3a, b see boxed electropherograms). Although most successful partners required non-synonymous changes, we discovered a single mutation-rescue pair of synonymous substitutions that obviated wild-type PB2 protein addition (Fig. 3b, c and Supplementary Fig. 7). Making each mutation alone (m52 and m65) resulted in severe packaging defects and, in the case of the m52 mutation, virus titer loss exceeding 4 log10—an extreme impairment beyond the 1–2 log10 that has been previously reported11,14 for packaging-defective viruses (Figs. 3d, e and Supplementary Figs. 6 and 8). When introduced together into a doubly mutated m52/65-compensatory virus that restored PSL2 structure, albeit with an altered sequence, the compensatory mutations restored both packaging efficiency and virus titer to wild-type levels.
Figure 3. Mutate-Map-Rescue analysis reveals novel PB2 packaging-defective and compensatory mutant partners.
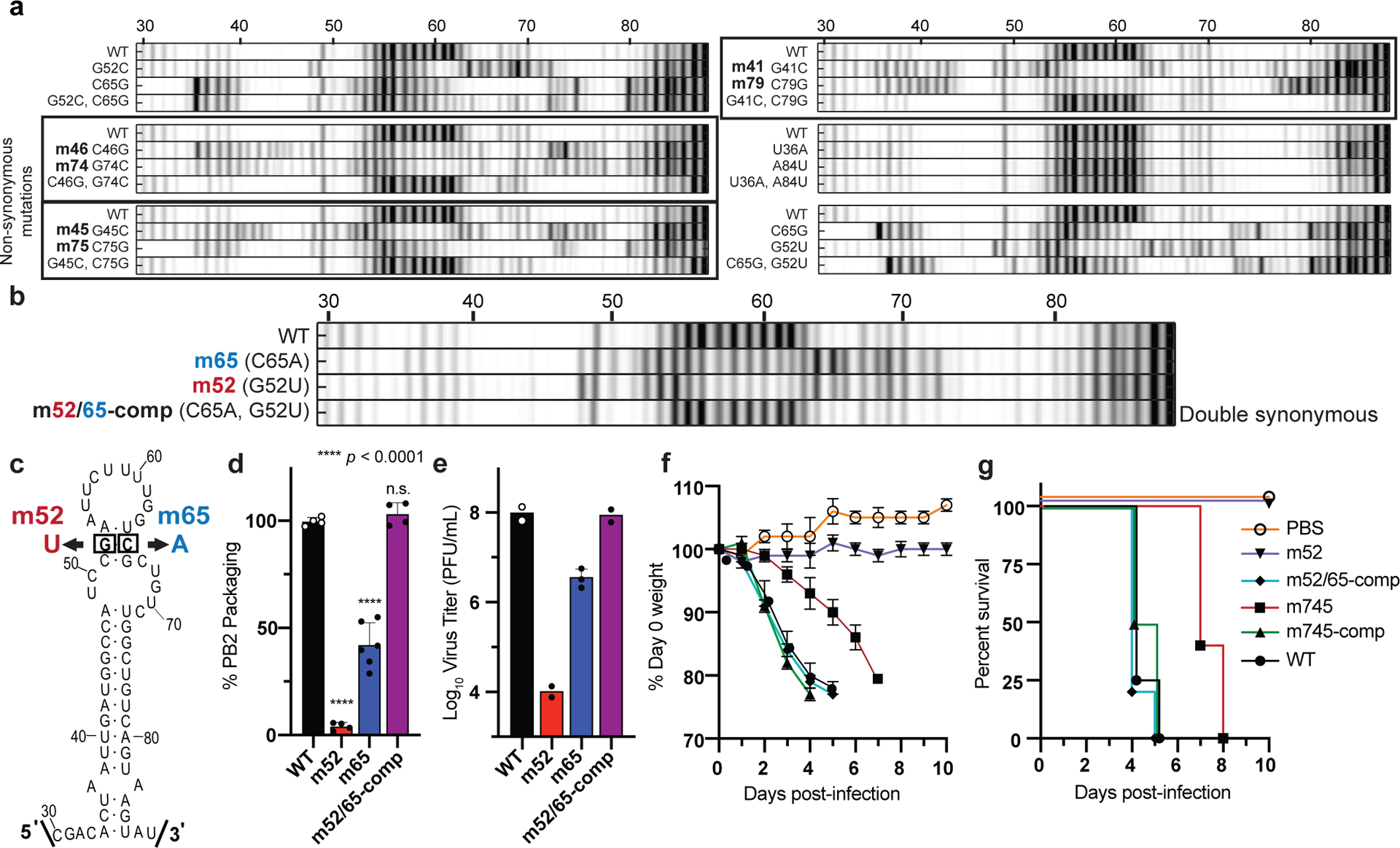
(a) Electropherograms from systematic single nucleotide mutation SHAPE chemical mapping with rescue (Mutate-Map-Rescue)21 analysis of individual and compensatory double mutations to test base-pairings from 1D-data-guided models and to identify predicted successful synonymous PSL2-defective and compensatory mutant pairs. Chemical accessibilities, plotted in grayscale (black = highest SHAPE reactivity), across 59 single mutations at single-nucleotide resolutions of PSL2 element from PR8 strain segment PB2. Reactivity peaks (left to right) correspond to nucleotides from the 5′ to 3′ end of the PB2 RNA. See Supplementary Fig. 7 for complete set of Mutate-Rescue pairs. (b) Electropherogram of successful double synonymous mutant pair determined by Mutate-Map-Rescue analysis. (c) Mutational design of single mutants m52 (G52U) and m65 (C65A), and the double m52/65-comp rescue pair on the PSL2 structure. (d) Packaging efficiency of the synonymous single and double mutant Mutate-Rescue pair. Values given as a percentage of PB2 vRNA packaging relative to WT PR8 virus. Results represent two independent experiments with biological duplicates performed in technical triplicate (n=4), except for m65 performed in biological triplicate (n=6). Statistical analysis by ordinary one-way ANOVA using Dunnett’s multiple comparisons test against WT; * p < 0.0001, n.s. = not significant. (e) Viral titer of the supernatants collected in (d) in PFU / mL, plaque assays in biological duplicate (n=2), except for m65 (n=3), and performed in technical triplicate. (f-g) Percent Day 0 weight and survival of mice infected with single PSL2-disrupting, and compensatory PSL2-restoring double-mutant viruses. Six- to eight-week-old female BALB/c mice (n=6) were intranasally infected with PR8 wild-type (WT) virus, packaging-defective single mutant viruses, m52 and m745, compensatory double mutant viruses, m52/65-comp and m745-comp, or PBS control. (f) Percent Day 0 weight. (g) Kaplan-Meier survival plot of the individual cohorts depicted in (f). All error bars represent mean ± s.d. Statistics and graphs for all figures were generated in GraphPad Prism 9 software.
To ensure that any loss or subsequent rescue of virus production caused by these mutations was not due to defects in replication or translation, each mutation-rescue pair was tested in transfection-based replicon assays. All mutant PB2 proteins and vRNAs were produced at comparable wild-type levels (Supplementary Figs. 9 and 10). As an orthogonal means of testing packaging efficiency, we also assessed the effect of select mutations on vRNA packaging by denaturing RNA gel which recapitulated our earlier packaging findings (Supplementary Fig. 11c, Figs. 2d, 3d, and Supplementary Fig. 6).
To test the relevance of the PSL2 structure in an in vivo model, BALB/c mice were intranasally infected with either wild-type or mutant PR8 viruses harboring point mutations predicted to disrupt or restore PSL2 structure. Mice infected with the PSL2-disrupting mutations—m745 mutant strain (20% packaging efficiency) or the severely defective mutant virus, m52 (<4% packaging efficiency)—showed reduced or no clinical signs of illness, respectively, either in weight loss or survival as compared to the PBS control (Fig. 3f, g). In contrast, inclusion of compensatory mutations that restore PSL2 structure rescued virus pathogenicity: animals infected with m52/65-comp and m745-comp, displayed comparable mortality profiles as mice infected with wild-type PR8 (Fig. 3f, g). To the best of our knowledge, these are the first data indicating that packaging-defective viruses are attenuated in vivo and a genomic IAV RNA secondary structure mediates influenza disease progression.
Designing anti-IAV LNA therapeutics targeting PSL2 structure
Given the strong evolutionary conservation of PSL2 structure (Fig. 1d, e and Supplementary Fig. 1c, d), we postulated that PSL2-targeted therapeutics could possess broad-spectrum activity across IAV subtypes and strains. Nine antisense oligonucleotides (ASO) with modified locked nucleic acid (LNA) bases23 were designed against PSL2 to disrupt various regions of the overall RNA secondary structure (Fig. 4a). Two LNAs, LNA8 and LNA9, are identical in sequence to LNA6 and LNA7, respectively, but possess 6–7 unmodified (non-locked) DNA nucleotide “gapmers” optimized for RNase-H activation that can degrade RNA in RNA-DNA hybrids24. First, to assess the impact that LNA binding has on PSL2 RNA secondary structure, toeprinting and SHAPE chemical mapping were performed on PB2 vRNA in the presence of the LNAs. Sequences encoded in LNAs 6–9, corresponding to binding sites on the right 3′ side of the stem-loop structure (Fig. 4a), exhibited the greatest ability to bind and disrupt the wild-type PSL2 structure (Supplementary Fig. 12).
Figure 4. Locked Nucleic Acids (LNAs) targeting PSL2 RNA structure display potent antiviral activity in vitro.
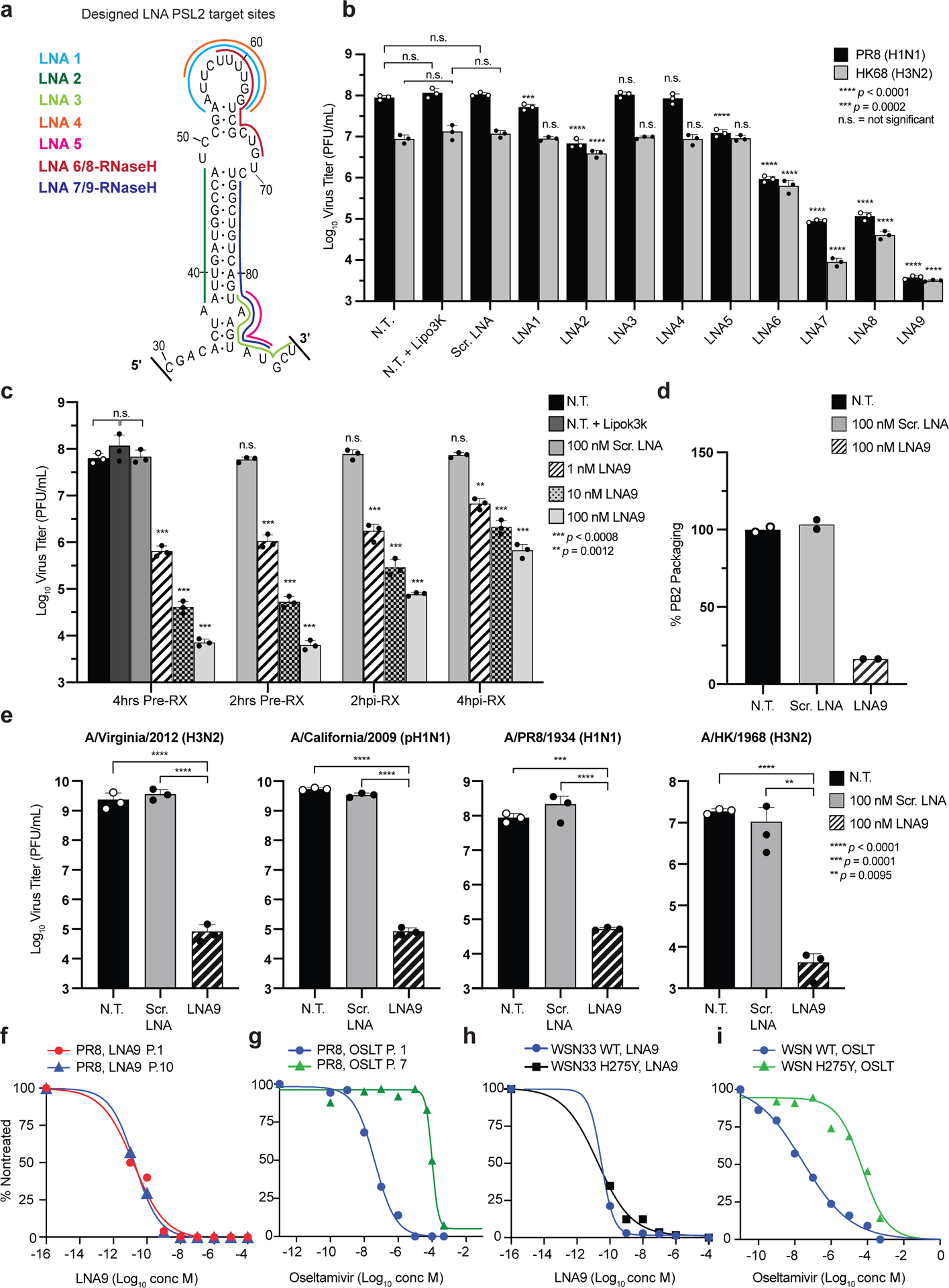
(a) Regions of PSL2 targeted by indicated LNAs. (b) Antiviral screen of LNAs transfected into MDCK cells and infected 4 hours later with PR8 (H1N1) or A/Hong Kong/8/68 (H3N2) virus (0.01 MOI) and viral titers determined 48 hours post infection (n=3). Statistics performed by unpaired ordinary one-way ANOVA (PR8 and HK68) with Dunnett’s multiple comparisons. (c) Antiviral efficacy as a function of time of LNA addition (n=3), analyzed as in (b). Statistics by 2-way ANOVA with Dunnett’s multiple comparisons test against non-treated +Lipo3k (N.T.). (d) PB2 vRNA (PR8) packaging efficiency of viruses treated with 100 nM LNA9 or Scr. LNA control. Values given as a percentage of PB2 vRNA packaging in comparison to non-treated wild-type PR8 virus; readout by qPCR. Results from two biological replicates (n=2), assays performed in technical triplicates. (n=6). (e) LNA9 efficacy against multiple IAV strains in MDCK cells pretreated with 100 nM of indicated LNAs. Analyzed as in (b). Statistics as described in (c) against the N.T. control. (f) In vitro selection for drug resistance to LNA9 with escalating concentrations of LNA and the sensitivity of passaged virus in response to drug treatment. EC50s determined at the indicated passage (P) numbers. Results expressed as a percentage of nontreated virus titer. (g) In vitro selection of PR8 virus selected with oseltamivir carboxylate (OSLT). OSLT-treated PR8 virus and drug sensitivity determined by plaque reduction assay. The number of viral plaques with each drug concentration was normalized against the nontreated control to determine the EC50. (h, i) In vitro sensitivity of wild-type WSN33 (H1N1) and neuraminidase (NA) inhibitor-resistant (WSN H275Y NA mutant) virus to LNA9 (h) or OSLT (i). EC50 values were computed using a nonlinear regression model with variable slope. Statistics for all graphs performed in GraphPad Prism 9 software. All error bars represent mean ± s.d. * Statistical P value as indicated in each panel.
To test LNA-mediated targeting of PSL2 across different viral subtypes, MDCK cells were treated with LNA prior to infection with either PR8 (H1N1) virus or the tissue culture-adapted A/Hong Kong/8/1968 (HK68) (H3N2) virus, and virus inhibition was assessed by plaque assay (Fig. 4b). As predicted by the mutational and LNA chemical mapping experiments (Fig. 2, Supplementary Fig. 12), LNAs directed against only the top loop of PSL2 (LNA1, LNA4), and LNAs solely targeting the 3¢ base of PSL2 (e.g., LNAs 3 and 5) had minimal effect on viral titer. In contrast, nucleotide coverage of both the top loop and middle bulge by LNA6 resulted in greater than 2 log10 titer deficits for PR8 (Fig. 4a, b). LNA8, the RNase-H activated copy of LNA6, produced even greater antiviral activity against both viruses of up to 3 logs10. Notably, LNA9, the RNase-H activated copy of LNA7, possessed the strongest antiviral capacity, inhibiting virus production by over 4 logs10 and 3 logs10 against PR8 and HK68, respectively. While a labeled version of LNA9 could be clearly visualized in cells harboring vRNPs (Supplementary Fig. 13), no off-target effect of LNA9 on steady state levels of viral protein, vRNA, cRNA, or cellular toxicity after 24 hours was observed (Supplementary Fig. 14).
Having identified a potent candidate LNA, we next investigated the treatment time-course and concentration parameters of LNA9’s antiviral activity when administered prophylactically at 2- and 4-hours pre-infection, and therapeutically at 2- and 4-hours post-infection. Cells pre-treated with the LNA had the most potent antiviral response (greater than 4 logs10) and displayed strong virus inhibition even at the lowest tested concentration (1 nM) (Fig. 4c). There was a trend towards decreasing antiviral activity as the time of administration post-infection increased, but even at the latest tested time point of addition, greater than 2 logs10 suppression of viral titer was achieved. Similar efficacy was seen in the presence of high MOI (Supplementary Fig. 15). Furthermore, LNA9 treatment resulted in a dramatic loss of PB2 packaging compared to controls (Fig. 4d), an effect that resembled the mutational studies and extended to the 2009 pandemic virus (pH1N1) and 2012 H3N2 viruses, indicating the potential for broad-spectrum antiviral activity.
Selection of IAV variants with escalating drug pressure
To test the hypothesis that the high conservation of PSL2 reflects a biologic constraint against mutation of this structure, and that this could translate into a high barrier to the development of resistance for PSL2-targeting therapeutics, we determined the susceptibility of wild-type PR8 virus to LNA9 under conditions designed to promote the development of resistance over serial virus passaging (Fig. 4f). In parallel, we performed analogous experiments using the neuraminidase inhibitor (NAI) oseltamivir carboxylate (OSLT, Tamiflu™). After seven virus passages in the presence of escalating drug concentrations, the EC50 of OSLT increased from 4.1 nM to 100 μM—a greater than 20,000x fold increase (Fig. 4g). In comparison, after 10 virus passages in the presence of LNA9, the EC50 showed no significant change and remained in the picomolar range of 16 to 22 pM (Fig. 4f). To date, we have yet to be able to select for viral mutations capable of generating resistance to LNA9. Moreover, LNA9 was equally effective (EC50 = 18 pM) against a virus strain (A/WSN/33 (H1N1)) containing the H275Y mutation (N1 numbering system) that confers resistance to OSLT, while high-level resistance to OSLT was confirmed, with an EC50 of 53 μM (Fig. 4h, i). These results extend the therapeutic capabilities of LNA9 and provide strong support for potential treatment of NAI-resistant viruses with PSL2-targeting LNAs.
PSL2-targeted LNAs protect mice from lethal IAV infections
As a proof-of-concept to assess the in vivo efficacy of prophylactic LNA treatment against PSL2, BALB/c mice were intranasally (I.N.) administered a single 20 μg dose of LNA9 or Scr. LNA one day, or three days, prior to infection with a lethal dose of PR8 virus. The untreated control mice experienced dramatic weight loss and were humanely sacrificed by days 5 and 6. In contrast, a single I.N. dose of LNA9 was completely protective when administered one day, and even three days, prior to viral infection (Fig. 5a), and showed significantly reduced virus titers in the lungs (> 2.5 log10 virus reduction) compared to the Scr. LNA control at 72 hours post-infection (Supplementary Fig. 16). In addition to the well-known benefits of LNA antisense gapmer technology that enables recruitment of RNaseH to degrade the targeted RNA23,24, LNA ASOs have also been reported to show dramatic, long-lasting effects (even >1 month) after the last administered dose in a variety of disease models25–28. We hypothesized that PSL2-targeted LNAs might similarly possess long-term prophylactic effects due to the nuclease-resistant property conferred by the phosphorthioate backbone present in LNAs.
Figure 5. PSL2-targeted LNAs demonstrate potent antiviral activity in vivo.
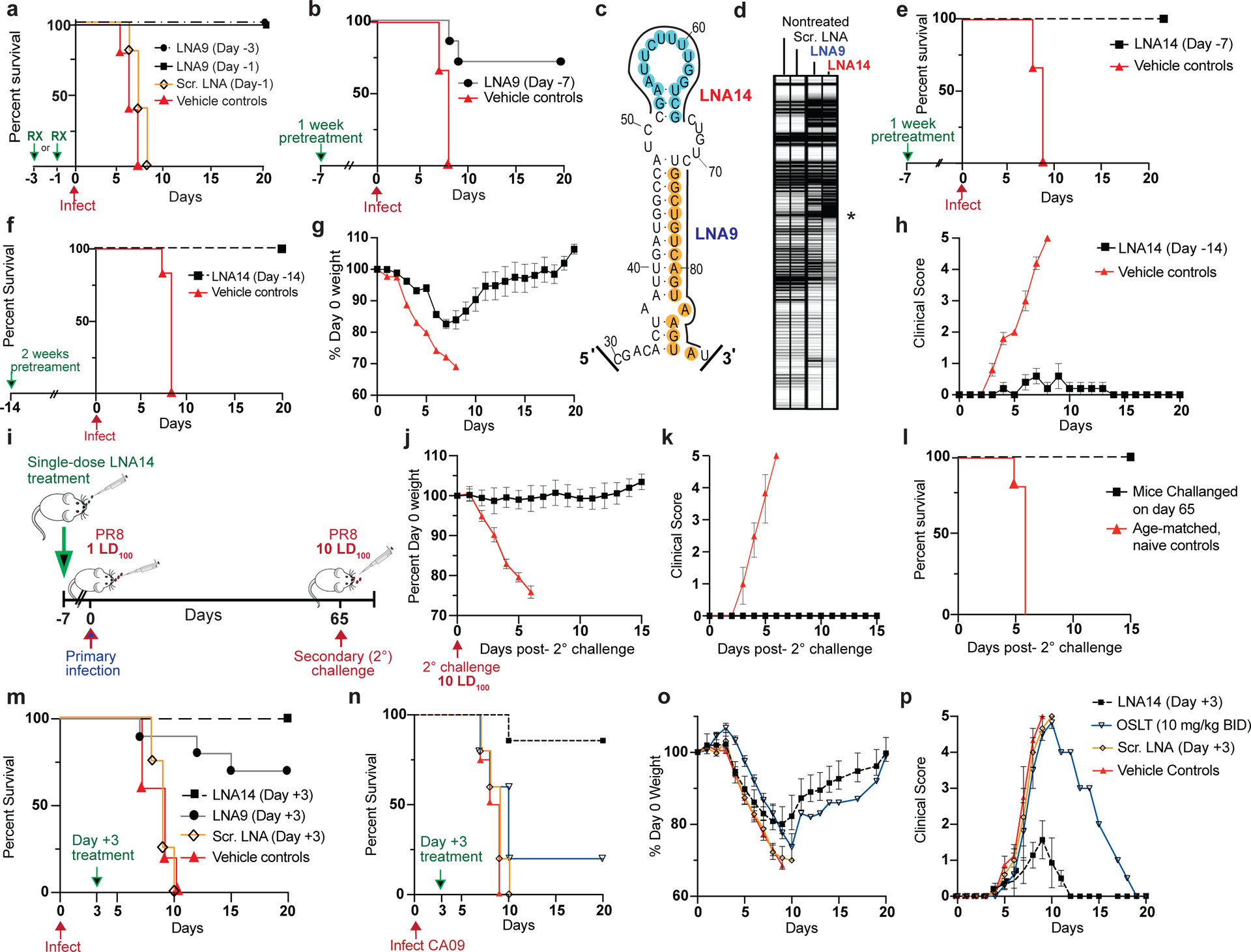
(a-b) Kaplan-Meier survival plots of mice intranasally administered a single dose of LNA9, Scr. LNA, or Vehicle (mock-treated) on the indicated days with respect to a subsequent lethal inoculum of wild-type PR8 virus (PR8); (a) 20 μg LNA9 (n=5); (b) 30 μg LNA9 (n=7) or vehicle control (n=5). (c) Target sites of LNA9 and newly designed, LNA14, mapped to the PSL2 structure. (d) Electrophoretic profile of SHAPE analysis performed on PR8 PB2 vRNA in the presence of non-treated, or 100nM Scr. LNA, LNA9, or LNA14 Asterisk marks site of reverse transcription (RT) stops indicating strength of LNA – RNA binding. (e-f) Kaplan-Meier survival plots of mice pretreated with a single IN dose of (e) 30 μg LNA14 (n=7) or vehicle (n=5), Day −7; (f) 40 μg of LNA14 or vehicle (n=6), Day −14, before receiving lethal PR8 inoculum. (g) Percent day 0 weights, and (h) clinical score, of mice from (f). Error bars show mean ± s.e.m. (i-l) Mice (n=7) from (e), who had received a single 30 μg intranasal dose of LNA14 one week prior to a first lethal inoculum (1 LD100) of PR8 and that all survived, were inoculated 65 days post-initial infection, along with age-matched naïve controls (n=10), with a 10-times lethal dose (10 LD100). (i) Challenge study timeline. (j) Weight loss, (k) clinical score; (l) Kaplan-Meier survival curve; error bars, mean ± s.d.. (m) Kaplan-Meier survival plot of mice (n=10/group) infected with a lethal dose of PR8, followed 3 days later with, a single intravenous dose of 30 μg of LNA14, LNA9, Scr. LNA, or vehicle control. (n-p) Mice were infected with a lethal dose of mouse-adapted A/California/04/2009 (pH1N1) virus (CA09), then treated intranasally with a single 30 μg dose of LNA14 (n=7/group), Scr. LNA (n=5), or Vehicle (n=4) on Day +3 post-infection. Mice given OSLT (n= 5) were treated by oral gavage BID for 5 days starting at Day +3. (n) Kaplan-Meier survival curve. (o) Percent day 0 weight. (p) Clinical score. All studies performed with 6–8 week-old female BALB/c mice Error bars in (o, p) indicate mean ± s.e.m.
To test this hypothesis, we administered a single, increased dose of LNA9 (30 μg) one week (Day −7) prior to infection with a lethal dose of IAV. While 100% of untreated mice succumbed to the infection, 70% of the LNA9 one-week pretreatment group were protected from lethality (Fig. 5b). To determine if our therapeutics could be further optimized for improved efficacy, we explored targeting other sequences in PSL2. While LNA9 targets nucleotides in the lower 3’ stem of PSL2, our mutational analyses suggested the importance of the 52–65 nucleotide pair in the upper stem (Fig. 3). We hypothesized that an LNA designed against these nucleotides (LNA14) might enhance efficacy (Fig. 4a). Indeed, SHAPE analysis indicated that LNA14 was an even more potent disruptor of PSL2 structure than LNA9 (Fig. 5c, d). In biological confirmation of this, mice treated with LNA14 were fully protected when given a single dose one week prior to infection with a lethal dose of IAV (Fig. 5e). When we increased the LNA dosage to 40 μg, and administered a single LNA14 treatment to mice two-weeks (Day −14) prior to IAV infection, the mock-treated mice succumbed to infection between days 7 and 8 due to severe disease, while the entire LNA14-treated cohort survived, with significantly lower disease scores, indicative of minor-to-undetectable disease symptoms, and reduced weight loss compared to controls (Fig. 5f–h, Supplementary Table 2).
We attribute the LNAs’ antiviral effect to their direct binding of PSL2, as opposed to non-specific activation of antiviral pathways. Indeed, neither LNA14, LNA9, or derivative molecules elicited activation of either interferon stimulated genes or the NF-kB pathway in lung or myeloid cells (Supplementary Figure 17).
Given the mild symptoms observed in LNA-treated and protected mice, we hypothesized that the resulting highly attenuated infection might be sufficient to enable mice receiving prophylactic LNA treatment to develop an effective immunization against a secondary infection through production of immunity. To test this hypothesis, mice from the one-week LNA14 pretreatment surviving cohort (Fig. 5e) were challenged alongside age-matched, naïve controls, sixty-five days post-primary infection with ten times the mouse lethal dose (10 LD100) of IAV (Fig. 5i). The secondary challenge had no effect on weight, clinical score, or survival of mice from the LNA14 pretreatment group (a total of 72 days since treatment, 65 days since primary infection, while the age-matched controls presented with rapid disease and were humanely sacrificed by Day 6 post-challenge infection (Fig. 5j–l). Blood and spleen cells were analyzed for the presence of virus-specific T-cells and the development of antibodies against viral proteins (Supplementary Fig. 18a,b) which demonstrated stimulation of both the cellular and humoral arms of the immune system, although their respective contributions to complete absence of symptoms following a 10-fold higher lethal inoculum await definition.
After demonstrating PSL2-targeted LNA efficacy in prophylactic models, we next tested its potential as a post-infection therapeutic. Due to the rapid onset of symptoms and illness in IAV infections, FDA-approved IAV therapeutics are most challenged when administered after 48 hours of disease onset29. In contrast, we hypothesized that anti-PSL2 LNAs, administered by either intravenous (I.V.) or intranasal (I.N.) routes, could treat IAV well after an infection has been established, To test this hypothesis, mice infected with a lethal dose of PR8 virus were treated with either LNA14, LNA9, Scr. LNA, or vehicle control by I.V. injection 3 days post-infection (Day +3) when mice typically become noticeably ill. While animals treated with controls rapidly succumbed to the infection, 65% of mice treated with LNA9 survived lethal infection, and all the LNA14-treated mice survived (Fig. 5m). Next, we tested the therapeutic efficacy of LNA14 via intranasal administration against the 2009 modern pandemic “swine” virus (A/California/04/2009 pH1N1; CA09), which has since become a dominant seasonal strain worldwide. Mice were infected with a lethal dose of CA09 then given a single intranasal dose of LNA14, Scr. LNA, or vehicle control 3 days post-infection. Oseltamivir phosphate was used as a therapeutic control and administered by oral gavage twice daily (BID) for 5 consecutive days starting at 3 days post-infection. The virus was 100% lethal against the Scr. LNA and vehicle control mice, and only 2/7 oseltamivir-treated mice were protected from lethal disease (Fig. 5n, o). In comparison, 6/7 mice in the LNA14-treated group were protected from lethality and collectively had much lower clinical scores than the other cohorts (Fig. 5p). Sampling of lungs taken 2 days after LNA14 treatment of CA09 infected mice (i.e., 5 days post-infection) showed significantly reduced virus titers (> 6 log10 virus reduction) compared to the controls (Supplementary Fig. 19). We also observed good dose proportionality in the lungs up to the highest dose tested to date (80 μg LNA14) Supplementary Fig. 20). Lowering the LNA14 dose down to 5 μg still resulted in full protection from death (Supplementary Fig. 21), while scrambled LNA offered no more protection than vehicle control.
Programmable antivirals against SARS-CoV-2 RNA structures
We hypothesized that we could leverage our approach to targeting IAV and similarly design potent LNAs against SARS-CoV-2. Briefly, we first performed sequence alignments of the entire 30 kilobase positive-sense RNA genome of SARS-CoV-2 and related betacoronaviruses to identify conserved sequence regions predicted to contain RNA secondary structures30. We then focused on highly conserved nucleotide stretches of sufficient length to enable antisense nucleotide attack, and high probability of structural stability to identify promising candidate antiviral targets. SHAPE yielded high resolution RNA secondary structure maps of these regions, and guided LNA design. Highly conserved targets included a stem-loop structure near the end of the SARS-CoV-2 genome (nts 28743–92; Fig. 6a) and the structured domain spanning the terminal 5′ UTR into the ORF1 coding sequence (nts 258–276; Fig. 6b). LNAs were vetted for their ability to disrupt these structures by chemical mapping of the LNA-RNA interaction (Fig. 6c, d).
Figure 6. Antisense targeting of highly conserved RNA secondary structures in SARS-CoV-2.
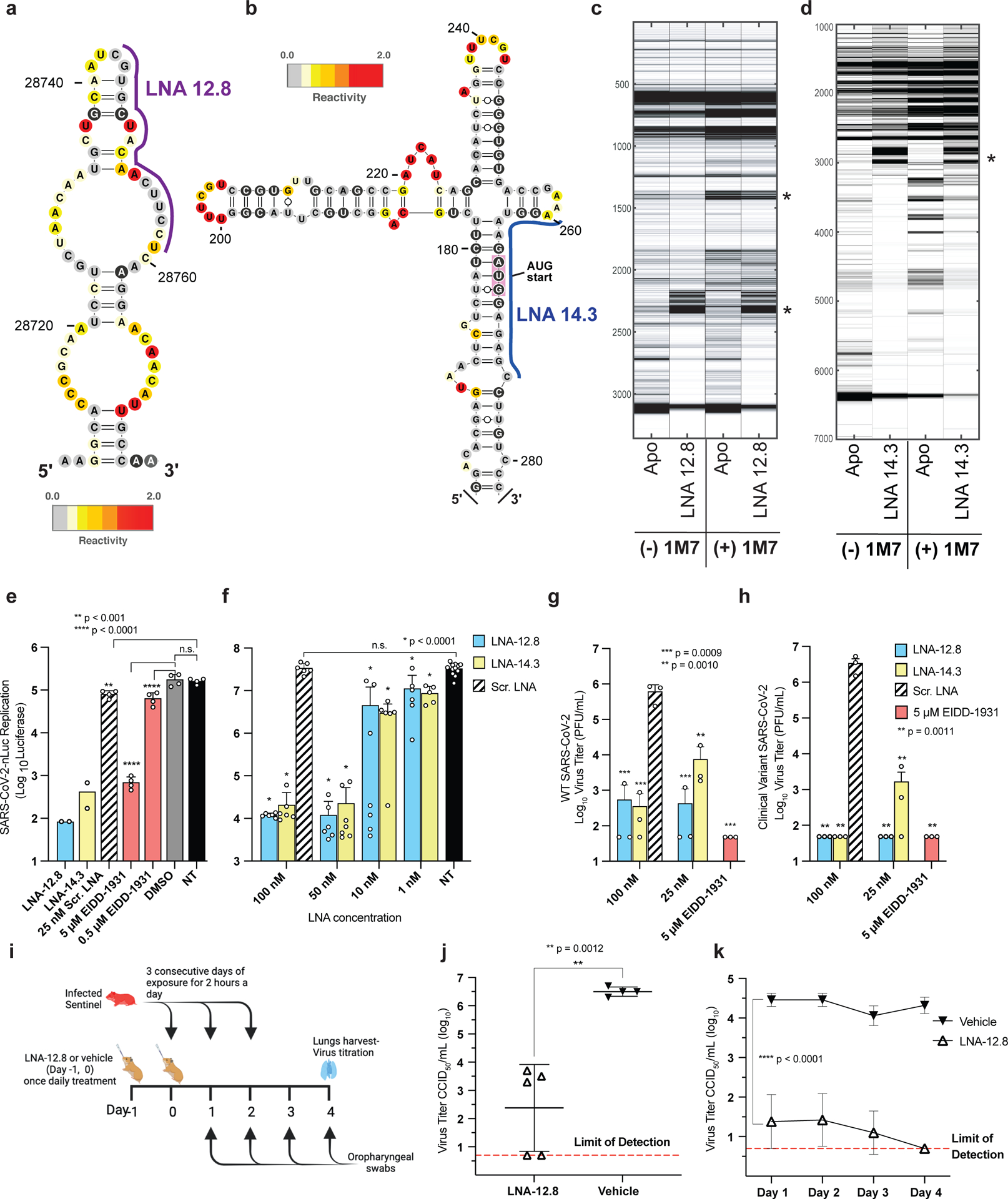
(a, b) Two SHAPE reactivity-derived prediction of RNA secondary structures within (+)-sense SARS-CoV-2 vRNA and LNAs designed against these targets. (a) nucleotide (nt) region 28743 – 28827; LNA-12.8 and (b) nt region 258 – 276; LNA-14.3 (c, d) Electrophoretic profile of SHAPE analysis performed in the presence or absence of the corresponding LNAs (e) In vitro antiviral activity of 25 nM LNAs against SARS-CoV-2-Nluc virus in Huh-7 cells (n=4). The nucleoside analog, EIDD-1931, was included as a positive control at 5 μM and 0.5 μM. (f) LNA dose response against SARS-CoV-2-Nluc virus in ACE2-TMPRSS2-Huh-7.5 cells, in biological replicates (n=6–7 for all treatment groups, except for NT controls where n=12). (g, h) Virus titers of supernatant collected from A549-hACE2 cells treated with LNA or EIDD-1931 followed by infection with either (g) wild-type SARS-CoV-2 or (h) a patient-isolated clinical variant of SARS-CoV-2 containing multiple mutations in the spike protein region including D614G, E484K, and N501Y present in variants of concern (VOC) (n=3). Statistical analysis in e, f, g and h was performed using ordinary one-way ANOVA with Dunnett’s multiple comparison tests between the controls as indicated. Error bars represent mean ± s.d., p values as indicated. (i-k) Prevention of SARS-CoV-2 transmission in Syrian hamsters. Ten-week-old female Syrian hamsters were pretreated with either 100 μg of LNA-12.8 (n=5) or vehicle (n=4) on Day −1 and Day 0 before exposure to SARS-CoV-2-infected sentinel hamsters for 2 hours per day for 3 consecutive days. Four days after the initial exposure, lungs were harvested, and virus titers determined by CCID50 in triplicate. (i) Experimental timeline. (j) Virus titer of SARS-CoV-2 in lungs (two-sided unpaired student’s t test) and (k) oropharyngeal swabs of LNA- or vehicle-treated hamsters (2-way ANOVA with multiple comparisons against the vehicle controls). The assay method detection limit is 0.7 log10 CCID50. Error bars represent mean ± s.d. Figure (i) was created with BioRender.com.
The LNAs were then screened against a SARS-CoV-2-Nluc reporter virus under BSL3 conditions (Fig. 6e). The nucleoside analog, EIDD-193131, was included as a positive control, and Scr. LNA, DMSO, and non-treated (N.T.) served as negative controls. Both LNAs against these target structures inhibited virus replication by as much as 3 log10, with the 25 nM LNA-12.8 outperforming the 5 μM EIDD control by nearly 1 log10 (Fig. 6e). Next, an LNA dose response experiment with expanded replicates was performed in Huh-7.5 cells that were modified to overexpress the human ACE2 (hACE2) and TMPRSS2 receptors to enable more efficient virus replication32 (Fig. 6f). Indeed, the viral replication signal was enhanced by nearly 2.5 log10 compared to Huh-7 cells in this system (Fig. 6e). Even at the higher virus burden, however, all tested LNA concentrations displayed statistically significant virus inhibition in a dose-dependent manner compared to the Scr. LNA controls and were nontoxic to the cells at concentrations 10x higher than the highest dose assayed for antiviral activity (Supplementary Fig. 22a). We next tested our LNAs against the fully intact wild-type SARS-CoV-2 virus, in a lung epithelial cell-line expressing human ACE2 (hACE2-A549 (Fig. 6g), which confirmed the previous Nluc virus results, whereby LNA treatment displayed > 3 log10 virus inhibition for both LNAs at 100 nM, and LNA-12.8 at 25 nM, and were non-toxic to cells tested up to 1 μM (Fig. 6h, Supplementary Fig. 22b).
We next pretreated hACE2-A549 cells with LNAs and then infected with a novel SARS-CoV-2 clinical isolate (SARS-CoV-2/human/USA/OH-UC-1/2020 ON642077) derived from a chronically infected cancer patient, whose sequence had evolved over a 7-month infection period (Fig. 6h). This clinical isolate harbors multiple mutations including L452R, D614G, E484A/Q, and N501Y in the spike protein receptor binding domain shown to be involved in antibody escape, enhancement of virus replication, and increased transmission found in variants of concern including Alpha, Beta, Delta, Mu, and Omicron33,34 (Supplementary Table 3). Despite numerous mutations throughout the viral spike protein coding region and elsewhere in the genome, the LNAs retained their high levels of potency against this variant: both 100 nM treatments, as well as the 25 nM LNA-12.8 treatment, demonstrated complete virus inhibition to the experimental limit of detection, equaling a 5 log10 drop in virus titer (Fig. 6h).
Given the high transmissibility of SARS-CoV-2, we sought to test the effectiveness of LNA treatment to prevent or mitigate SARS-CoV-2 transmission in a Syrian hamster model. In this system, hamsters were pretreated intranasally with a single 100 μg dose of our top SARS-CoV-2 LNA candidate, LNA-12.8, on two consecutive days, Day −1 and Day 0, before exposure to infected sentinel hamsters (Fig. 6i). Pretreated cohorts were repeatedly exposed to the infected sentinels for two hours a day for three consecutive days. Daily oropharyngeal swabs were collected. Four days post-initial exposure, lungs were collected and virus titers of LNA-12.8-treated versus vehicle-treated animals were compared (Fig. 6j). LNA-12.8 dramatically reduced viral titers by over 3 log10 compared to vehicle in exposed animals, with two hamsters containing no detectable virus in their lungs (Fig. 6j). As shown in Fig. 6k, viral titers in the oropharyngeal swabs showed parallel reductions in response to LNA treatment to the viral titer reductions observed in the lungs with all hamsters showing undetectable oropharyngeal viral titers by Day 4. Collectively, our identification, design, and targeting of highly conserved RNA secondary structures in IAV and SARS-CoV-2 are proof-of-principle evidence towards a new model of rapid and effective antiviral development.
Discussion
We describe here the discovery and characterization of an RNA stem-loop structure, PSL2, that serves as a packaging signal for genome segment PB2. PSL2 is conserved across all known influenza A isolates. Knowledge of PSL2’s RNA secondary structure helps explain previously discovered packaging-defective mutations and enabled rational design of more potently disrupting mutations. Compensatory mutations that restore PSL2’s structure (but not primary sequence) rescue virus packaging and titer loss in vitro and restore lethality in vivo, thus providing strong genetic validation of PSL2’s importance in influenza biology and disease. LNAs designed to disrupt PSL2 structure dramatically inhibit IAV in vitro against viruses of different strains and subtypes. In contrast to NAIs, like Tamiflu™, and the endonuclease inhibitor, Xofluza™, which are vulnerable to drug-resistant mutations35,36,37, our PSL2-targeting LNAs, exhibit a high barrier to the development of resistance, and are equally effective against wild-type and NAI-resistant viruses. In vivo, intranasal dosing of LNAs results in potent antiviral efficacy and prevents mortality in mice, even with a single dose administered two weeks prior to infection with a lethal IAV inoculum. Moreover, PSL2-targeting LNAs also enable the surviving mice to develop vigorous immunity. In therapeutic models, a single LNA dose given three days after infection provides complete protection from death. Together, these results have exciting implications for the development of a novel class of pan-genotypic anti-IAV therapies for prophylaxis of, treatment of established, and “just-in-time” universal vaccination against, an IAV infection. In addition, because traditional vaccines take weeks to provide full protection, a co-administered single dose of LNA14 could provide protection during this vulnerability window.
We envision several paths to the clinic for PSL2-targeted therapies like LNA14 against IAV, including aerosol delivery to outpatients or intravenous administration for severe hospitalized patients. Importantly, incorporating the virus’ RNA secondary structure into the LNA design allowed us to achieve far greater inhibition than ASOs designed against the same viral genomic sequence but which relied only on primary nucleotide sequence homology for their design38.
Although we initially focused on IAV, an analogous approach can be taken for virtually any virus of interest. Indeed, we were able to rapidly identify highly conserved RNA secondary structures in SARS-CoV-230 and then design appropriate LNAs leveraging the lessons learned in targeting IAV—a process we now term “programable antivirals.” Our lead LNAs demonstrated greater in vitro activity against SARS-CoV-2 than the nucleoside analog positive controls at 200x the LNA concentration. Moreover, the LNAs were highly effective against a clinical isolate harboring mutations that have been associated with decreased susceptibility to recent COVID-19 vaccines33, with titer reductions of up to 5 log10 at 25 nM concentrations of LNA. Finally, our top anti-SARS-CoV-2 LNA exhibited profound in vivo activity by successfully decreasing and in some cases completely preventing virus transmission in hamsters. Limitations of our study include the very preliminary nature of the SARS-CoV-2 data and the need for further experiments, including assessment of therapeutic application of the LNA (which is the subject of another manuscript).
Taken together, this programmable antiviral strategy now excitingly offers a highly adaptable potential solution to current and future viruses of concern, especially respiratory pandemic viruses. Their broad-spectrum potential can enable advanced stockpiling and prepositioning, with the ability to fill the void before a possible vaccine might be developed and deployed. Programmable antivirals can also offer protection against vaccine-resistant virus strains, as well as provide immediate protection during the months needed for vaccine-induced immunity to ramp up.
Online Methods
Cells and viruses.
HEK 293T (CRL-1573), VeroE6 (CRL-1586) and MDCK-NLB-2 (CCL-34) were obtained from American Type Culture Collection ‘ATCC’ (Manasass, VA) and were maintained according to ATCC instructions. A549-Dual (a549d-nfis) and THP1-Dual (thpd-nfis) were obtained from InvivoGen and maintained according to the the manufacturer instructions. All cell lines used in this report were routinely checked for mycoplama contamination (MycoAlert Mycoplasma Detection Kit, Lonza) and were authenticated by the respective vendors. All cell lines used for SARS-CoV-2 experimentation including Huh-7, ACE2-TMPRSS2-Huh-7.5, and ACE2-A549 cells were maintained at 37 °C in complete DMEM (Gibco) containing 10% FBS (Invitrogen), penicillin and streptomycin (Gibco), and HEPES buffer (Gibco). The ACE2-A549 cells were specially engineered to over-express the ACE2 receptor in human alveolar basal epithelial cells (A549), while the ACE2-TMPRSS2-Huh-7.5 cells were a kind gift by the Catherine Blish laboratory and engineered to overexpress both hACE2 and human TMPRSS2 receptors. A549-Dual and THP1-Dual cells were purchased from InvivoGen.
Wild-type influenza A/PR/8/34 (PR8) H1N1 virus (ATCC-VR-95) and the tissue-culture adapted PR8 virus (ATCC-VR-1469) were purchased from ATCC. PR8 mutant viruses were generated using an eight-plasmid reverse genetic system as previously described39. Tissue-cultured adapted influenza A/Hong Kong/8/68 (HK68) H3N2 virus (ATCC-VR-1679), A/Virginia/ATCC6/2012 (H3N2) virus (ATCC-VR-1811), A/Virginia/ATCC1/2009TC (H1N1) virus (ATCC-VR-1736), and A/Wisconsin/33 (H1N1) virus (VR-1520) were purchased from ATCC. A/California/4/2009 (pH1N1) virus was kindly gifted by Elena Govorkova from St. Jude Children’s Research Hospital (Memphis, USA). Viruses were grown and amplified in 10-day-old specific-pathogen-free research grade chicken embryos at 35°C (Charles River Laboratories; SPAEAS).
Wild-type recombinant SARS-CoV-2 was prepared and handled as described40. The recombinant SARS-CoV-2-Nluc virus is an authentic fully replicating virus in which ORF7a has been deleted and replaced with Nluc41. The SARS-CoV-2 clinical variant was isolated from a 54-year-old male with melanoma and lymphoma, who received a confirmed COVID-19 diagnosis by PCR in March 2020. Virus sequencing was performed on virus isolated from a nasopharyngeal swab sample in virus transport medium taken after the patient was infected for 7 months.
Plasmid constructs and cloning.
Plasmids were used containing the wild-type PB2 segments from influenza viruses A/PuertoRico/8/34 (H1N1) [PR8], A/New York/470/2004 (H3N2) [NY470], A/New York/312/2001 (H1N1) [NY312], A/Brevig Mission/1/1918 (H1N1) [1918], A/California/04/2009 (H1N1) [CA09], and A/Vietnam/03/2004 (H5N1) [VN1203]. For the generation of PR8 packaging mutant vRNA, we utilized a Stratagene QuickChange XL site-directed mutagenesis kit (Stratagene) for mutagenesis of a pDZ plasmid containing the PB2 gene of PR839. Sequences of each mutated construct were confirmed by automated sequencing. The 8-plasmid pBD rescue system for A/WSN/33 (H1N1) was kindly donated by Andrew Mehle. The H275Y NA mutant was generated by QuickChange mutagenesis from the bidirectional pBD plasmids, as described above.
Reverse genetics and virus titrations.
Recombinant A/Puerto Rico/8/34 (PR8) virus and recombinant A/WSN/33 (WSN) virus were generated using eight-plasmid reverse genetic systems39. Briefly, 106 cells of a 293T/MDCK co-culture were Lipofectamine™ 3000 (Invitrogen™) transfected with 1 γ of one of each of the eight segments contained within plasmids that utilize a bidirectional dual Pol I/II promoter system for the simultaneous synthesis of genomic vRNA and mRNA. For rescue of compensatory PB2 mutant viruses where a non-synonymous change was required, a wild-type PB2 protein expression plasmid (Pol II) was co-transfected during virus rescue. Supernatants were collected 24 hours post-transfection. PR8 rescue viruses were then inoculated into the allantoic cavities of 10-day-old chicken embryos. WSN rescue viruses were passaged subsequent times on MDCK cells. Rescue of recombinant viruses was assessed by hemagglutination activity. Each newly rescued virus was further plaque titered and mutations were confirmed by sequencing of mutated genes. Plaque assays were carried out on confluent MDCK cells as described previously42. Hemagglutination (HA) assays were carried out in 96-well round-bottomed plates at room-temperature, using 50 μl of virus dilution and 50 μl of a 0.5% suspensions of turkey red blood cells (LAMPIRE® Biological Laboratories) in phosphate-buffered saline (PBS).
Isolation of packaged vRNAs.
To analyze packaged vRNA for PR8 mutated viruses, 10-day-old eggs were inoculated with approximately 1000 PFU of recombinant virus and incubated for 72 hours. Allantoic fluid was harvested, and supernatant was dual-clarified by low-speed centrifugation. Clarified supernatant was then layered on a 30% sucrose cushion and ultra-centrifuged at 30,000 RPM for 2.5 hours (Beckman Rotor SW41). Pelleted virus was resuspended in PBS and TRIzol (Invitrogen) extracted. Precipitated vRNA was resuspended in a final volume of 20 μl of 10 mM Tris-HCl (pH 8.0) and stored at −80 °C. Virus supernatant from LNA-treated cells was harvested 48 hours post-infection and subjected to low-speed centrifugation at 1000 RPM then 10,000 RPM. Isolation continued as indicated above.
qPCR analysis of packaged vRNAs.
Approximately 200 ng of extracted vRNA was reverse transcribed using a universal 3′ primer (5′-AGGGCTCTTCGGCCAGCRAAAGCAGG) and Superscript III reverse transcriptase (RT) (Invitrogen). The RT product was diluted approximately 10,000-fold and used as a template for quantitative PCR (qPCR). Separate PCRs were then carried out as previously described43 with segment-specific primers. The 10 μl reaction mixture contained 1 μl of diluted RT product, a 0.5 μM primer concentration, and SYBR Select Master Mix (Applied Biosystems) that included SYBR GreenER dye, 200 μM deoxynucleoside triphosphates, heat labile UDG, optimized SYBR Green Select Buffer, and AmpliTaq DNA polymerase UP enzyme. Relative vRNA concentrations were determined by analysis of cycle threshold values, total vRNA amount within a sample was normalized to the level of HA vRNA, and then percentages of incorporation were calculated relative to the levels of wild-type vRNA packaging. Viral packaging results represent the averaged levels of vRNA incorporation ± standard deviations derived from two independent virus purifications, with vRNA levels quantified in triplicate.
Denaturing RNA gel.
Extracted viral RNA (100–300 ng) was diluted with equal volume of NOVEX TBE Urea sample buffer and incubated at 70°C for 10 min prior to separation on a 6% TBE-Urea gel for 18 hours at a constant voltage of 80 volts. RNA from each sample was run in several dilutions to enable clear visualization of the genomic RNA without over saturation of the bands signal. The RNA was visualized by staining for 30 minutes in 0.5x TBE buffer supplemented with 0.5 mg/ml ethidium bromide followed by visualization in a GelDoc EZ Imager system (Bio-Rad). Silver staining was performed using the SilverXpress silver stain kit (Invitrogen) according to the manufacturer instruction. Silver staining and ethidium bromide staining were compared and shown to have the same linear range of detection (data not shown); ethidium bromide was selected for lane visualization due to a higher background signal in silver staining. The band intensity in each lane was determined using Image Lab Software (Bio-Rad) and analysis of the intensity of each genomic band relative to the total intensity of all genome segments was determined and normalized to the intensity of the HA band relative intensity.
Strand-specific RT-qPCR.
MDCK cells transfected with 1 mM LNA9 or Scrambled LNA were infected with PR8 virus at an MOI of 0.1 24 hours post-transfection. Eight hours post infection total cellular RNA was extracted in Trizol reagent (Invitrogen) and the RNA was purified using the Direct-Zol RNA mini-prep (Zymo Research) according to the manufacturer protocol. Reverse transcription and qPCR were performed according to44. cDNAs of the influenza viral RNA (vRNA) and complementary viral RNA (cRNA) were synthesized with tagged primers to add an 18–20 nucleotide tag that was unrelated to the influenza virus at the 5′ end (cRNAtag; 5′-GCT AGC TTC AGC TAG GCA TC-3′, vRNAtag; 5′-GGC CGT CAT GGT GGC GAA T-3′). Hot-start reverse transcription with the tagged primer was performed as described in Kawakami et al., 2011 using saturated trehalose. A 5.5 μl mixture containing 200 ng of total RNA sample and 10 pmol of tagged primer were heated for 10 min at 65°C, chilled immediately on ice for 5 min, and then heated again at 60°C. After 5 min, 14.5 μl of preheated reaction mixture [4 μl First Strand buffer (5 ×, Invitrogen), 1 μl 0.1 M dithiothreitol, 1 μl dNTP mix (10 mM each), 1 μl Superscript III reverse transcriptase (200 U/μl, Invitrogen), 1 μl RNasin Plus RNase inhibitor (40 U/μl, Promega) and 6.5 μl saturated trehalose] was added and incubated for 1 h. Real-time PCR (qPCR) was performed with PowerUp SYBR Green SuperMix (Applied Biosystems) on a BIORAD CFX96 Real-Time System. Seven microliters of a ten-fold dilution of the cDNA was added to the qPCR reaction mixture [10 μl SYBR Green SuperMix (2 ×), 1.5 μl forward primer (10 μM), 1.5 μl reverse primer (10 μM)]. The cycle conditions of qPCR were 95°C 10 min, followed by 40 cycles of 95°C 15 sec and 60°C for 1 min. qPCR primers were: PR8 segment 1 (PB2) cRNA, Forward: 5′-TCC ACC AAA GCA AAG TAG AAT GC-3′; Reverse: 5′-GCT AGC TTC AGC TAG GCA TCA GTA GAA ACA AGG TCG TTT TTA AAC-3′. PR8 segment 1(PB2) vRNA, Forward: 5′-GGC CGT CAT GGT GGC GAA TAG ACG AAC AGT CGA TTG CCG AAG C-3′, Reverse: 5′-AGT ACT CAT CTA CAC CCA TTT TGC-3′. PR8 segment 4 (HA) cRNA, Forward: 5′-CTG TAT GAG AAA GTA AAA AGC C-3′, Reverse: 5′-GCT AGC TTC AGC TAG GCA TCA GTA GAA ACA AGG GTG TTT TTC-3′. PR8 segment 4 (HA) vRNA, Forward: 5′-GGC CGT CAT GGT GGC GAA TAG GAT GAA CTA TTA CTG GAC CTT GC-3′, Reverse: 5′-TCC TGT AAC CAT CCT CAA TTT GGC-3′.
Animals.
All animal studies were performed in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and approved by the Stanford University Administrative Panel on Laboratory Animal Care and by the Utah State University Institutional Animal Care and Use Committee. Animals were housed in disposable cages connected to an Innorack® IVC ventilated rodent housing system under 12 hours light/dark cycle at 24°C and 25–30% humidity. Six- to eight-week-old healthy age-matched female BALB/c mice (Jackson Laboratories, Bar Harbor ME) were randomly separated into groups for infection/treatment or used as uninfected/non-treated controls. Treatment groups were not blinded to the investigators. Mice were identified with tag numbers throughout the experiment. Ten-week-old female golden Syrian hamsters (LVG strain, Charles River Laboratories) were separated into groups for exposure studies with an additional sentinel group that was inoculated directly with virus.
In vivo mouse infections.
Mice were lightly anesthetized with isoflurane and intranasally infected with 50 μl of virus preparation at 1 LD100—a concentration of approximately 1000 PFU for virus packaging mutant experiments and 900 PFU for LNA treatment experiments. Weights and clinical scores were assessed daily, and animals were humanely sacrificed when a clinical score of 5 was recorded (see Supplementary Table 2 for clinical score determination). Kaplan-Meier survival curves were generated using GraphPad Prism.
In vivo mouse IAV antiviral assays.
‘In vivo-ready’ LNAs were custom designed and ordered from Qiagen (formally Exiqon), and later by IDT. For intranasal delivery, in vivo-ready LNA was mixed in complexes with In vivo-JetPEI® transfection reagent (Polyplus) according to manufacturer’s protocol to the indicated final concentration in 50–75 μl of 5 % glucose solution. Mice were then lightly anesthetized with isoflurane and 50–75 μl of the solution was delivered intranasally. For retro-orbital delivery: ‘In vivo-ready’ LNA was mixed in complexes with In vivo-JetPEI® transfection reagent (Polyplus) according to manufacturer′s protocol to the indicated final concentration in 200 μl of 5 % glucose solution. Mice were then anesthetized, and the solution was delivered by retro-orbital injection. Oseltamivir phosphate (OSLT; Sigma Aldrich Cat. SML1606) was prepared in sterile water and administered to mice at a dose of 10 mg/kg bidaily by oral gavage (totaling 20 mg/kg/day), with 8 hours between dosing intervals.
Prevention of SARS-CoV-2 transmission in Syrian hamsters.
Infection of donor sentinel hamsters (non-treated): 6-week-old wild-type golden Syrian hamsters were intranasally infected with 1 × 104.3 CCID50 of SARS-CoV-2 (USA_WA1/2020 strain) in a 100 μl volume. LNA pretreatment group: 6-week-old golden Syrian hamsters (N=5) were pretreated by intranasal instillation with 200 μL volume containing 100 μg of LNA-12.8 on Day −1 and Day 0 before exposure to the SARS-CoV-2-infected sentinel hamsters. Vehicle-treated hamsters (N=4) received PBS by intranasal nebulization. LNA and vehicle-treated hamsters were co-housed and exposed to the infected sentinels for 2 hours per day for 3 consecutive days. Four days after the initial exposure, lungs were harvested, and virus titers were determined by CCID50 in triplicate. Statistical analysis was performed using an unpaired student’s t test by GraphPad Prism 9 software.
Locked Nucleic Acid (LNA) design and preparation.
Oligonucleotides containing locked nucleic acids (LNA) were custom synthesized from Exiqon (Vedbaek, Denmark), and later by IDT. Capitalized letters denote LNA. Lowercase letters denote typical (non-locked) DNA nucleotides. All oligonucleotides contain phosphorothioate internucleoside linkages. LNA 8 and 9 were designed as LNA gapmers to contain a stretch of 6–7 DNA nucleotides optimized for RNase-H recruitment. Sequences of all LNAs are shown below.
LNA 1: 5′ AccAaaAGaaT 3′
LNA 2: 5′ TggCcATcaaT 3′
LNA 3: 5′ TagCAtActtA 3′
LNA 4: 5′ CCAAAAGA 3′
LNA 5: 5′ CATACTTA 3′
LNA 6: 5′ CagaCaCGaCCaaAA 3′
LNA 7: 5′ TAcTtaCTgaCagCC 3′
LNA 8: 5′ AGAcacgaccaaAAG 3′
LNA 9: 5′ TACTtactgacaGCC 3′
LNA14: 5′ CGACcaaaagaATTC 3′
Scramble LNA (negative control): 5′ AACACGTCTATACGC 3′
SARS-CoV-2 directed LNAs:
LNA-12.8: 5′ AGGAagttgtagCACG 3′
LNA-14.3: 5′ GCTctccatcttaCCT 3′
In vitro LNA antiviral assays.
For all experiments, LNAs were reconstituted in RNAse-free water at 100 μM stock solutions, aliquoted and stored at −20 °C prior to single-use. Lipofectamine 3000® (Life Technologies) was used to transfect LNA into cells at indicated concentrations per manufacturer’s protocol. For IAV prophylactic antiviral assays, 106 MDCK cells were plated in 6-well plates 24 hours prior to being transfected with the indicated LNA. Cells were then infected at the indicated time points with 0.01 MOI of PR8 (H1N1) or HK68 (H3N2) virus. For post-infection therapeutic assessment, MDCK cells were infected with PR8 or HK68 prior to LNA transfection as described above. Forty-eight hours post-infection, supernatant was collected, and viral titer was determined by plaque assay in triplicate.
For SARS-CoV-2-Nluc assays: one day before transfection, Huh-7 or ACE2-TMPRSS2-Huh-7.5 were plated in 96-well clear-bottom plates to 60–70% confluency at the time of treatment with the LNA ASOs LNA-12.8, LNA-14.3 or Scrambled LNA (Scr. LNA). Lipofectamine 3000 (Life Technologies) was used to transfect LNA ASOs into cells at 25 nM or 100 nM final concentration, according to the manufacturer’s protocol. Cells were then infected with SARS-CoV-2 reporter virus expressing nanoluciferase (SARS-CoV-2-Nluc) at a multiplicity of infection of 0.3 for 1 h, after which the virus was removed and fresh medium was added. Recombinant SARS-CoV-2-Nluc is a fully replicating virus in which ORF7 has been deleted and replaced with Nluc. Thus, the measurement of Nluc expression is a surrogate marker of virus replication enabling the screening of antiviral compounds. A nucleoside analog NHC, EIDD-1931, with potent activity against SARS-CoV-2 was included as a positive control. A DMSO control was included as a mock-treated, negative control. Data were graphed and analyzed in Prism 8 and 9 by GraphPad. Statistical analysis of the data from each cell type was computed as an ordinary one-way analysis of variance using Dunnett’s multiple comparisons test against the DMSO control or scrambled LNA control from each cell type, where indicated.
SARS-CoV-2 plaque assay experiments were performed in 24-well plates: 24-well plates were seeded with 100K VeroE6 cells/well in DMEM, 10% FBS, 1X antibiotic/antimycotic 24 hours prior to Lipofectamine 3000 transfection of LNA. Scr. LNA was included as a negative control. Twelve hours post-transfection, LNA-transfection medium was removed, and cells were infected at a multiplicity of infection (MOI) of 0.01 with either recombinant wild-type SARS-CoV-2 (PMID: 32526206) or a SARS-CoV-2 clinical isolate derived from a nasopharyngeal swab taken from a chronically infected cancer patient 7 months after the initial confirmatory COVID-19 PCR test. After adding virus inoculum, plates were incubated at 37°C for 1h, after which input virus was removed, wells were washed with 1 mL medium, and 1mL “infection medium” (DMEM, 5% FBS, 1X antibiotic/antimycotic) was added. After 48 hours at 37°C, 200 μl of culture supernatant was collected and stored at −80°C until quantitation of infectious virus by plaque assay. Briefly, Vero E6 were seeded at 500K cells/well/2 mL in 6 well plates. Twenty-four hours later, samples were thawed at room temperature and serially diluted in PBS. Medium was removed from 6-well plates and serial dilutions were added to the plate, incubated for 1 hours at 37°C and then overlayed with DMEM, 5% FBS, 1X antibiotic/antimycotic, 0.9% agarose. 72 hours later, neutral red stain was added to each plate, incubated for 3 hours and plaques were counted. The number of plaque forming units per mL was generated using the following formula: PFU/mL = number of plaques × serial dilution factor × 5.
LNA-treatment and IAV packaging efficiency determination.
Briefly, T75 flasks of 80% confluent MDCK cells were transfected with 100 nM of Scrambled LNA (Scr. LNA), LNA9, or mock untreated by Lipofectamine 3000 transfection, according to manufacturer’s protocol. Twelve hours post-transfection, cells were infected with 0.01 MOI of wild-type TC-adapted PR8 virus. After 1 hour, virus was removed, and cells were washed with PBS. Forty-eight hours post-infection supernatants were collected, and RNA was isolated as described in isolation of packaged vRNAs and assay methods.
In vitro drug selection.
LNA9 selection: 80–90% confluent MDCK cells in 12-well plates were transfected in duplicate with a starting concentration of 0.01 nM (~1/2 EC50) LNA9 for Passage 1 by Lipofectamine transfection (see above). Twelve hours post-transfection, cells were washed with PBS and infected with 0.01 MOI of wild-type PR8 virus. After 1 hour incubation at 37 °C, cells were washed and virus growth media was added. Cells were incubated until 50 % CPE was evident (48–72 hours). Virus supernatant was harvested, low-speed centrifuge clarified, aliquoted, plaque titered and stored at −80°C. The virus supernatant was then continuously serially passaged in the presence of escalating concentrations of LNA9 (0.01 nM to 100 nM). If no CPE was evident, drug concentration was lowered and added virus concentration was increased until 50 % CPE occurred. OSLT selection: confluent MDCK cells in 12-well plates were infected with 0.01 MOI of PR8 virus. After adsorption for an hour, cells were washed with PBS, and OSLT (Sigma Aldrich Cat. No. Y0001340) was added to virus growth media at a starting concentration of 1 nM (~1/2 EC50). Drug selection proceeded as described above, with escalating concentrations of OSLT (0.01 nM to 250 μM) at each subsequent passage.
EC50 determination.
For LNA9, the 50% effective concentration (EC50) was defined as the concentration of drug effective in reducing the percent of virus titer to 50% of that for the no-drug control. In brief, the EC50 was determined by seeding 5 × 105 MDCK cells in each well of a 12-well plate and incubating overnight at 37°C under 5% CO2. Cells were then transfected with LNA9 as described above at concentrations from 0.01 nM to 10 μM. Plates were incubated at 37°C for 12 hours prior to infection with 0.01 MOI of wild-type PR8, serially passaged LNA-treated virus, WSN33 wild-type or WSN33 H275Y NAI-resistant virus. Forty-eight hours post-infection, supernatants were collected, centrifuge clarified, aliquoted and stored at −80°C. The viral titer for each drug dilution was performed by plaque assay in duplicate. The EC50 was the concentration of LNA9 yielding a percent titer of 50% of that without drug.
For OSLT, the EC50 was defined as the concentration of drug reducing the total percentage of plaques to 50% of that for the no-drug control, determined by plaque reduction assay1. Briefly, confluent MDCK cells in 12-well plates were infected with approximately 100 PFU of wild-type PR8, serially passaged OSLT-treated virus, WSN33 wild-type or WSN33 H275Y NAI-resistant virus and incubated for 1 hour at 37 °C. Cells were then washed with PBS, and a 50:50 mix of 1 % agarose to 2x virus growth DMEM containing varying concentrations of drug (0.1 nM to 1 mM) was added to the cells. Plates were harvested 72 hours later, stained with crystal violet, and plaques were counted. The EC50 was the concentration of OSLT reducing the total percentage of plaques to 50 % of that without drug. All results were plotted in GraphPad Prism to generate EC50 curves.
In vitro transcription of full-length IAV vRNA.
For each wild-type isolate (PR8, 1918, VN1203, NY470, NY312, and CA09) and PR8 packaging mutant clones, PB2 cDNA was amplified from plasmid using segment-specific primers under a T7 promoter. Amplified cDNA was gel-purified using an Invitrogen DNA gel kit. vRNAs were then produced by in vitro transcription, using T7-MEGAscript kit. vRNAs for SHAPE were purified by MEGAclear (Thermofisher, cat. no. AM1908) with purity and length verified by capillary electrophoresis.
sf-SHAPE 1D analysis of full-length IAV vRNA.
In vitro transcribed PB2 vRNA was folded (100 mM NaCl; 2.5 mM MgCl; 65 °C × 1’, 5′ cooling at room temperature, 37 °C for 20–30’) in 100 mM HEPES, pH=8. 2’ acylation with NMIA18 and reverse transcription (RT) primer extension were performed at 45 °Cx 1’, 52 °C × 25’, 65 °C × 5’, as previously described45. 6FAM was used for all labeled primers (primer sequences available upon request). Exceptions to these protocols were as follows: (i) RNA purification after acylation was performed using RNA C&C columns (Zymo Research), rather than ethanol precipitation; (ii) before and after SHAPE primer buffer was added, the mixture was placed at room temperature for 2–5 min, which enhanced RT transcription yields significantly; (iii) DNA purification was performed using Sephadex G-50 size exclusion resin in 96-well format then concentrated by vacuum centrifugation, resulting in a more significant removal of primer; and (iv) 2 pmol RNA was used in ddGTP RNA sequencing reactions.
The ABI 3100 Genetic Analyzer (50 cm capillaries filled with POP6 matrix) was set to the following parameters: voltage 15 kV, T = 60°C, injection time=15 s. The GeneScan program was used to acquire the data for each sample, which consisted of purified DNA resuspended in 9.75 μl of Hi-Di formamide, to which 0.25 μl of ROX 500 internal size standard (ABI Cat. 602912) was added. PeakScanner parameters were set to the following parameters: smoothing=none; window size=25; size calling=local southern; baseline window=51; peak threshold=15. Fragments 250 and 340 were computationally excluded from the ROX500 standard46. The data from PeakScanner were then processed into SHAPE data by using FAST (fast analysis of SHAPE traces), a custom algorithm developed in our lab19. FAST automatically corrects for signal differences due to handling errors, adjusts for signal decay, and converts fragment length to nucleotide position, using a ddGTP ladder as an external sizing standard and the local Southern method5,19. This algorithm embedded in the RNAstructure program is freely available at http://med.stanford.edu/glennlab/download.html.
RNAstructure parameters: slope and intercept parameters of 2.6 and −0.8 kcal/mol, were initially tried, as suggested47; however, we found that smaller intercepts closer to 0.0 kcal/mol (e.g. ~ −0.3) produced fewer less optimal structures (within a maximum energy difference of 10%). We speculate that this minor parameter difference may be due to the precise fitting achieved between experimental and control data sets by the automated FAST algorithm. FAST was written in ANSI C/C++ and is integrated into RNAstructure with FAST, which requires MFC (Microsoft Foundation Classes). RNA structures were drawn and colored using RNAViz 248 and finalized in Adobe Illustrator.
IAV PSL2 Construct design, RNA synthesis and chemical modification for Mutate-and-Map Experiments.
Double-stranded DNA templates were prepared by PCR assembly of DNA oligomers designed by an automated MATLAB script as previously described (available at https://primerize.stanford.edu/)49. Constructs for mutate-and-map (M2) includes all single mutants to Watson-Crick counterpart. Compensatory mutants for mutation/rescue were designed based on base-pairing in the proposed secondary structure22. In vitro transcription reactions, RNA purification and quantification steps were as described previously49. One-dimensional chemical mapping, mutate-and-map (M2), and mutation/rescue were carried out in 96-well format as described previously49–51. Briefly, RNA was heated up and cooled to remove secondary structure heterogeneity; then folded properly and incubated with SHAPE reagent (5 mg/mL 1-methyl-7-nitroisatoic anhydride (1M7))52; modification reaction was quenched and RNA were recovered by poly(dT) magnetic beads (Ambion) and FAM-labeled Tail2-A20 primer; RNA was washed by 70% ethanol (EtOH) twice and resuspended in ddH2O; followed by reverse transcription to cDNA and heated NaOH treatment to remove RNA. Final cDNA library was recovered by magnetic bead separation, rinsed, eluted in Hi-Di formamide (Applied Biosystems) with ROX-350 ladder, loaded to capillary electrophoresis sequencer (ABI3100). Data processing, structural modeling, and data deposition: The HiTRACE software package version 2.0 was used to analyze CE data (both MATLAB toolbox and web server available53,54). Trace alignment, baseline subtraction, sequence assignment, profile fitting, attenuation correction and normalization were accomplished as previously described55,56. Sequence assignment was accomplished manually with verification from sequencing ladders. Data-driven secondary structure models were obtained using the Fold program of the RNAstructure package version 5.457 with pseudo-energy slope and intercept parameters of 2.6 kcal/mol and −0.8 kcal/mol. 2-dimensional Z score matrices for M2 datasets, and helix-wise bootstrapping confidence values were calculated as described previously22,49. Z score matrices were used as base-pair-wise pseudofree energies with a slope and intercept of 1.0 kcal/mol and 0 kcal/mol. Secondary structure images were generated by VARNA58. These chemical mapping datasets, including one-dimensional mapping, mutate-and-map, and mutation/rescue have been deposited at the RNA Mapping Database (http://rmdb.stanford.edu)59, accession codes: PSL2IAV_1M7_0001, PSL2IAV_RSQ_0001.
SHAPE analysis of LNA-targeted IAV vRNA.
A truncated DNA template of PR8 virus segment PB2 containing nucleotides 1–88 was prepared by PCR assembly of DNA oligomers, and in vitro transcription reactions, RNA purification and quantification steps were as described previously49. One-dimensional SHAPE chemical mapping was performed in 96-well plate format as described above with the following exception: once RNA was denatured and refolded as described 100 nM of each prepared LNA was added to the folded RNA and incubated with 5 mg/mL of SHAPE reagent 1M7 (1-methyl-7-nitroisatoic anhydride). Modification quenching, RNA recovery, re-suspension, reverse transcription, cDNA sequencing and data processing were performed as described51.
SHAPE 1D analysis of nontreated and LNA-treated SARS-COV-2 RNA.
RNA was folded (0.5 M Na-HEPES, pH = 8; 90° C ×3′, 12′ cooling at room temperature, 50 °C × 20′, 12′ cooling at room temperate) in 100 mM MgCL2 with or without LNA, and with or without SHAPE reagent 1M7 (1-methyl-7-nitroisatoic anhydride) modification. RNA was purified and quenched with magnetic beads (0.5 M Na-MES pH = 6, FAM-A20 Tail 2 primer, 5 M NaCl, Ampure beads) and reverse transcribed at 48 °C for 40 min, followed by a 0.4 M NaOH acid quench to improve signal intensity. The resulting cDNA was resuspended in ROX Hi-Di-Formamide and diluted for capillary electrophoresis analysis. As an internal control, the RNA was created with GAGUA hairpins in 5′ and 3′ termini designed to be reactive in presence of 1M7. The results were analyzed using the HiTRACE method53 and standardized by the Kladwang et al. method56.
Peripheral blood mononuclear cells and splenocytes isolation:
Whole blood was collected from mice into heparinized tubes. The whole blood was overlaid on top Ficoll-Paque media and centrifuged at 400xg for 40 minutes at room temperature. The top layer containing plasma and platelets was removed and the peripheral blood mono-nuclear cells (PBMCs) at the interphase of the Ficoll layer were collected. The mononuclear cells were diluted in PBS and pelleted by centrifugation at 500 × g for 15 minutes following an additional wash with PBS. The pellet was suspended in PBS. Splenocytes were isolated by manual grinding the spleen over a 40 μm cell strainer. The cells were transferred several times through the strainer and processed further as described above. TruStain fcX (anti-mouse CD16/32) antibody specific for FcyR III/II (1 μg per 1e 6 cells) was used to block nonspecific staining, followed by staining with Zombie Aqua™ viability kit (Biolegend). Antibody staining was performed by mixing the cells with an antibody mix containing BD Horizon™ PE-CF594 Rat Anti-Mouse CD45 (BD Biosciences), PE/Cy7 anti-mouse CD3 (Biolegend), APC/Cy7 anti-mouse CD8a (Biolegend) and H-2Kd Influenza HA Tetramer-IYSTVASSL-PE (MBL International). All antibodies were diluted at a ratio of 2ml of each antibody per 1× 106 cells and HA tetramer was diluter at a ratio of 5 ml per 1× 106. Splenocytes were isolated from mouse spleen by immersing the spleen in HBSS with 10% FBS and washing with HBSS to remove blood. The spleen was placed in a 40 mm cell strainer and the tissue was mashed with a syringe plunger to break down the tissue and dislodge the cells. Following complete disruption of the organ the cells were run through the strainer into a conical tube. The cell suspension was pelleted by centrifugation at 350 × g for 10 minutes at 4°C. The pellet was resuspended in water for 20 seconds to disrupt red blood cells followed by addition of 2X PBS solution. The cells were pelleted and subjected to staining as described for PBMCs above.
Flow cytometry:
Following staining, the cells were fixed and subjected to flow cytometry using a BD LSR II flow cytometer (BD biosciences) equipped with 488 nm, 405 nm, 640 nm and 532 nm lasers. Data was collected using BD FACSDiva software (BD biosciences) and analyzed using FlowJo software (TreeStar). Gating strategy for positive tetramer cells was as follows: FSC-H/FSC-A gate used to collect cells and a gate for live cells was then generated. The live cells were gated for CD3+/CD45+ cells and the positive cells were gated to determine the CD8+/HA-Tetramer+ cells. Acquired data was analyzed using a FlowJo software (TreeStar).
LNA effect on interferon and NF-kB pathways.
THP1-Dual or A549-Dual cells (InvivoGen) were transfected in triplicates with LNA9, LNA14 or derivatives theroff, or poly(I:C) (R&D Systems) using Lipofectamine 2000 (ThermoFisher) according to the manufacturer protocol. Treatment with rcombinant hTNF-α (R&D Systems), recombinant hIFN-γ (R&D Systems), recombinant hIFN-α (R&D Systems) or LPS from O111:B4 E. Coli (Sigma Alderich) were used as positive control for activation of the two pathways. Supernatant from the cultures were collected 24–32 hours post transfection/treatment to measure luciferase signal or SEAP activity as indicators of interferon pathways stimulation or NF-kB pathway stimulation respectively. Results were normalized to reflect fold induction relative to NT cells.
IL-6 and TNF-α ELISA.
Supernatants from THP-1-Dual cells (InvivoGen) were transfected in triplicates with LNA9, LNA14 or poly (I:C) (R&D Systems). Treatment with recombinant hIFNγ (R&D Systems), recombinant hIFNα (R&D Systems) or LPS from O111:B4 E. Coli (Sigma Alderich) were used as positive control to indicteinduction of IL-6 or TNF-α secretion. Supernatants were collected 24 hours post transfection/treatment and assayed for TNF-α and IL-6 concentration by ELISA (ThermoFisher) accordign to the ELISA kit protocol.
Statistical analyses.
We expressed the data as the mean ± s.d. or ±s.e.m. where indicated. We used Student’s t-test (to compare two samples) or ANOVA (to compare multiple samples) as analyzed by GraphPad Prism (versions 8 and 9) for statistical analysis. We performed Kaplan-Meier log-rank tests for survival analyses. We considered all P values >0.05 not to be significant.
Supplementary Material
Acknowledgment
We thank Hong Jin and her team at MedImmune for the kind donation of chicken eggs and technical guidance at the start of the project. We acknowledge the Stanford in vitro BSL3 Service Center and its director Jaishree Garhyan for assistance with the SARS-CoV-2 research. We thank additional Das lab members, Ivan N. Zheludev, Ramya Rangan, and Hannah Wayment-Steele for meaningful discussions of the SARS-CoV-2 structures and design of original SARS-CoV-2 RNA constructs for SHAPE experiments. The work was supported in part by a Mona M. Burgess Stanford BIO-X Interdisciplinary Graduate Fellowship (R.J.H.), the National Institutes of Health (NIH) Graduate Training Grant 5T32AI007328-24 (R.J.H.), NIH Training Grant 5T32DK007056 (E.A.P.), and to J.S.G.: NIH research grants R56A1111460, U19A1109662, RO1AI132191, and U19AI171421, an influenza Harrington Scholar Innovator Grant, and W81XWH1810647 from USAMRAA, Department of Defense, Open Philanthropy and Good Ventures Fund, Fastgrants, a COVID-19 Harrington Scholar Innovator Grant, and the Dr. Tri Cao Nguyen Fund for Pandemic Preparedness. The work was also supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. Chemical mapping datasets for mutate-and-map and mutation/rescue experiments have been deposited at the RNA Mapping Database (http://rmdb.stanford.edu).
Footnotes
Competing Interests Statement
J.S.G., R.J.H., E.P. and M.E. are inventors on a patent pertaining to the materials presented in this article that has been filed with the U.S. Patent and Trademark Office by Stanford University.
Code availability statement
DNA oligomers automated design script is available as MATLAB script at https://primerize.stanford.edu/.
A code to analyze mutate and map data sets is available as MATLAB script for download at https://ribokit.github.io/HiTRACE/. A tutorial is available at http://hitrace.org/.
One-dimensional SHAPE with FAST algorithm embedded in the RNAstructure program is freely available at http://med.stanford.edu/glennlab/download.html.
Data availability statement
Data for 1-D SHAPE experiment is available in source data of this manuscript. Chemical mapping datasets for mutate-and-map and mutation/rescue experiments are available at http://rmdb.stanford.edu)80, accession codes: PSL2IAV_1M7_0001, PSL2IAV_RSQ_0001. All the data that support the findings of this study are available from the corresponding author upon request.
References
- 1.Memoli MJ, Hrabal RJ, Hassantoufighi A, Eichelberger MC & Taubenberger JK Rapid selection of oseltamivir- and peramivir-resistant pandemic H1N1 virus during therapy in 2 immunocompromised hosts. Clin Infect Dis 50, 1252–1255 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hai R, et al. Influenza A(H7N9) virus gains neuraminidase inhibitor resistance without loss of in vivo virulence or transmissibility. Nature communications 4, 2854 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayden FG & de Jong MD Emerging influenza antiviral resistance threats. The Journal of infectious diseases 203, 6–10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romero-Lopez C & Berzal-Herranz A Unmasking the information encoded as structural motifs of viral RNA genomes: a potential antiviral target. Reviews in medical virology 23, 340–354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pang PS, et al. Structural map of a microRNA-122: hepatitis C virus complex. J Virol 86, 1250–1254 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palese PS, M. L Fields Virology, (Lippincott Williams & Wilkins, 2007). [Google Scholar]
- 7.Compans RW, Content J & Duesberg PH Structure of the ribonucleoprotein of influenza virus. J Virol 10, 795–800 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noda T & Kawaoka Y Structure of influenza virus ribonucleoprotein complexes and their packaging into virions. Reviews in medical virology 20, 380–391 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutchinson EC, von Kirchbach JC, Gog JR & Digard P Genome packaging in influenza A virus. J Gen Virol 91, 313–328 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Gao Q, et al. The influenza A virus PB2, PA, NP, and M segments play a pivotal role during genome packaging. J Virol 86, 7043–7051 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marsh GA, Rabadan R, Levine AJ & Palese P Highly conserved regions of influenza a virus polymerase gene segments are critical for efficient viral RNA packaging. J Virol 82, 2295–2304 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fournier E, et al. A supramolecular assembly formed by influenza A virus genomic RNA segments. Nucleic Acids Res 40, 2197–2209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gavazzi C, et al. An in vitro network of intermolecular interactions between viral RNA segments of an avian H5N2 influenza A virus: comparison with a human H3N2 virus. Nucleic Acids Res 41, 1241–1254 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gog JR, et al. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res 35, 1897–1907 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss WN, Priore SF & Turner DH Identification of potential conserved RNA secondary structure throughout influenza A coding regions. Rna 17, 991–1011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang Y, Huang T, Ly H & Parslow TG Mutational analyses of packaging signals in influenza virus PA, PB1, and PB2 genomic RNA segments. J Virol 82, 229–236 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muramoto Y, et al. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J Virol 80, 2318–2325 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkinson KA, Merino EJ & Weeks KM Selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1, 1610–1616 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Pang PS, Elazar M, Pham EA & Glenn JS Simplified RNA secondary structure mapping by automation of SHAPE data analysis. Nucleic Acids Res 39, e151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Priore SF, Moss WN & Turner DH Influenza A virus coding regions exhibit host-specific global ordered RNA structure. PLoS One 7, e35989 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kladwang W & Das R A mutate-and-map strategy for inferring base pairs in structured nucleic acids: proof of concept on a DNA/RNA helix. Biochemistry 49, 7414–7416 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Tian S, Cordero P, Kladwang W & Das R High-throughput mutate-map-rescue evaluates SHAPE-directed RNA structure and uncovers excited states. Rna 20, 1815–1826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vester B & Wengel J LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry 43, 13233–13241 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Kurreck J, Wyszko E, Gillen C & Erdmann VA Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res 30, 1911–1918 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Straarup EM, et al. Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res 38, 7100–7111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staedel C, et al. Inhibition of Gastric Tumor Cell Growth Using Seed-targeting LNA as Specific, Long-lasting MicroRNA Inhibitors. Molecular therapy. Nucleic acids 4, e246 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Javanbakht H, et al. Liver-Targeted Anti-HBV Single-Stranded Oligonucleotides with Locked Nucleic Acid Potently Reduce HBV Gene Expression In Vivo. Molecular therapy. Nucleic acids 11, 441–454 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hillebrand F, et al. Gymnotic Delivery of LNA Mixmers Targeting Viral SREs Induces HIV-1 mRNA Degradation. International journal of molecular sciences 20(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.CDC.gov. Influenza Treatment. (Centers for Disease Control and Prevention, National Center for Immunization and Respiratory Diseases (NCIRD), cdc.gov, 2019). [Google Scholar]
- 30.Rangan R, et al. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: a first look. Rna 26, 937–959 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheahan TP, et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med 12(2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R, et al. Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses. Cell 184, 106–119 e114 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Variants of Concern. in SARS-CoV-2 Variant Classifications and Definitions., Vol. 2021 (National Center for Immunization and Respiratory Diseases (NCIRD), Division of Viral Diseases, Centers for Disease Control & Prevention., 2021). [Google Scholar]
- 34. https://covid.cdc.gov/covid-data-tracker/#published-sars-cov-2-sequences.
- 35.Bloom JD, Gong LI & Baltimore D Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328, 1272–1275 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takashita E, et al. Detection of influenza A(H3N2) viruses exhibiting reduced susceptibility to the novel cap-dependent endonuclease inhibitor baloxavir in Japan, December 2018. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin 24(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hurt AC, et al. Characteristics of a widespread community cluster of H275Y oseltamivir-resistant A(H1N1)pdm09 influenza in Australia. The Journal of infectious diseases 206, 148–157 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giannecchini S, Clausi V, Nosi D & Azzi A Oligonucleotides derived from the packaging signal at the 5’ end of the viral PB2 segment specifically inhibit influenza virus in vitro. Arch Virol 154, 821–832 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Hoffmann E, Neumann G, Kawaoka Y, Hobom G & Webster RG A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97, 6108–6113 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou YJ, et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell 182, 429–446 e414 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dinnon KH 3rd, et al. Publisher Correction: A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 590, E22 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Szretter KJ, Balish AL & Katz JM Influenza: propagation, quantification, and storage. Current protocols in microbiology Chapter 15, Unit 15G 11 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Marsh GA, Hatami R & Palese P Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J Virol 81, 9727–9736 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawakami E, et al. Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J Virol Methods 173, 1–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mortimer SA & Weeks KM Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution. Nat Protoc 4, 1413–1421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akbari A, et al. Improved DNA fragment length estimation in capillary electrophoresis. Electrophoresis 29, 1273–1285 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Deigan KE, Li TW, Mathews DH & Weeks KM Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci U S A 106, 97–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Rijk P, Wuyts J & De Wachter R RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics 19, 299–300 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Kladwang W, VanLang CC, Cordero P & Das R A two-dimensional mutate-and-map strategy for non-coding RNA structure. Nat Chem 3, 954–962 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kladwang W, Cordero P & Das R A mutate-and-map strategy accurately infers the base pairs of a 35-nucleotide model RNA. Rna 17, 522–534 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cordero P, Kladwang W, VanLang CC & Das R The mutate-and-map protocol for inferring base pairs in structured RNA. Methods Mol Biol 1086, 53–77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mortimer SA & Weeks KM A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. Journal of the American Chemical Society 129, 4144–4145 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Yoon S, et al. HiTRACE: high-throughput robust analysis for capillary electrophoresis. Bioinformatics 27, 1798–1805 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Kim H, Cordero P, Das R & Yoon S HiTRACE-Web: an online tool for robust analysis of high-throughput capillary electrophoresis. Nucleic Acids Res 41, W492–W498 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim J, et al. A robust peak detection method for RNA structure inference by high-throughput contact mapping. Bioinformatics 25, 1137–1144 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Kladwang W, et al. Standardization of RNA chemical mapping experiments. Biochemistry 53, 3063–3065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mathews DH, et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci U S A 101, 7287–7292 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Darty K, Denise A & Ponty Y VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics 25, 1974–1975 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cordero P, Lucks JB & Das R An RNA Mapping DataBase for curating RNA structure mapping experiments. Bioinformatics 28, 3006–3008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for 1-D SHAPE experiment is available in source data of this manuscript. Chemical mapping datasets for mutate-and-map and mutation/rescue experiments are available at http://rmdb.stanford.edu)80, accession codes: PSL2IAV_1M7_0001, PSL2IAV_RSQ_0001. All the data that support the findings of this study are available from the corresponding author upon request.


