Abstract
The pandemic caused by COVID-19 and the emergence of new variants of SARS-CoV-2 have generated clinical and epidemiological impacts on a global scale. The use of strategies for monitoring viral circulation and identifying mutations in genomic regions involved in host interaction are important measures to mitigate viral dissemination and reduce its likely complications on population health. In this context, the objective of this work was to explore the potential of high-resolution melting (HRM) analysis combined with one-step real-time reverse transcription PCR in a closed-tube system, as a fast and convenient method of screening for SARS-CoV-2 mutations with possible implications on host-pathogen interactions. The HRM analyses allowed the distinction of the Gamma, Zeta, Alpha, Delta, and Omicron variants against the predecessors (B.1.1.28, B.1.1.33) of occurrence in Brazil. It is concluded that the molecular tool standardized here has the potential to optimize the genomic surveillance of SARS-CoV-2, and could be adapted for genomic surveillance of other pathogens, due to its ability to detect, prior to sequencing, samples suggestive of new variants, selecting them more assertively and earlier for whole genome sequencing when compared to random screening.
Keywords: COVID-19, SARS-CoV-2, Variants, HRM, Genomic sequencing
1. Introduction
More than three years have passed since the first case of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infection was identified in Wuhan, China (12/2019) (Shu and McCauley, 2017). A particularity that has been observed in the Coronavirus Disease 2019 (COVID-19) pandemic is the recurring fluctuations in different geographic regions, related to periods of higher incidence and transmission rates, alterations in infectivity in different age groups, as well as the emergence of new viral phenotypes and changes in the severity of disease symptoms (Diaz and Vergara, 2021, Iftimie et al., 2021; Shu and McCauley, 2017). Despite the significant variability that has been experienced during the COVID-19 pandemic, the SARS-CoV-2 genome has a low mutation rate compared to other RNA viruses since coronaviruses encode an enzyme that corrects some of the errors made during the replication process (Lauring and Hodcroft, 2021). It is estimated that SARS-CoV-2 lineages accumulate about 1–2 nucleotide mutations monthly (Duchene et al., 2020). Although most mutations do not cause significant changes in viral properties, depending on the genomic region affected, impactful changes in transmission rates, receptor affinity, or pathogenicity can be triggered (Martin et al., 2021). In addition, the global circulation of the virus for long periods of time, in a diversity of populations and physiological conditions, favors the emergence of variants, the process of viral multiplication in which one genome differs from another by one or more mutations (Grubaugh et al., 2020). It is also suggested that the capacity of viral recombination in coinfections as a mechanism of genetic variability has a great impact on the emergence of variants and on its infecting potential in humans.
SARS-CoV-2 is an enveloped, non-segmented, positive-sense RNA with approximately 30 Kb, consisting of coding and non-coding regions. Its genome is polycistronic and contains 14 open reading frames (ORFs). Located on the terminal region of the viral genome are the coding ORFs of accessory proteins and four structural proteins: spike, envelope, membrane, and nucleocapsid. In particular, the coding region of the spike gene has aroused great genomic and immunological interest, due to the interaction between this protein and host cells, and its involvement in the development of the neutralizing immune response (Romano et al., 2020).
Large-scale genomic sequencing has been widely used around the world to monitor the evolution of SARS-CoV-2 genetic diversity (WHO, 2021). However, whole genome sequence analysis is expensive, requires long data-acquisition times, and computational power. On the other hand, in the last two decades, a molecular method based on DNA intercalation of saturating dyes, known as high-resolution melting (HRM), has grown in popularity due to its versatility for genotyping, scanning mutation, and sequence matching (Reed et al., 2007). After PCR amplification, analysis of melting curves is monitored by a fluorescent dye that does not interfere with PCR, saving time by not requiring process separation. In addition to being fast, HRM analyses are inexpensive because they do not require covalently labeled probes, combined with the popularization of real-time thermal cyclers, which are also cost-effective and convenient as they do not require post-PCR processing (Reed et al., 2007, Slinger et al., 2007).
In this context, we present a strategy for tracking viral mutations in genomic regions that impact host-pathogen interactions using HRM analyses. This cost-effective and rapid assay has the potential to optimize genomic surveillance through sequencing by pre-selecting candidate samples for further investigation, rather than relying on random selection. By optimizing the efficiency of genomic surveillance, approaches of this nature can contribute to improving public health responses and should be better encouraged and explored.
2. Materials and methods
2.1. RNA samples of SARS-CoV-2
SARS-CoV-2 RNA samples were obtained from the remaining material extracted for diagnosis of COVID-19 at FUNED (approved by the Research Ethics Committee with CAAE Number 32850420.4.0000.9507), the public central laboratory responsible for the diagnosis of COVID-19 in the state of Minas Gerais, Brazil. Samples were extracted by semi-automated and automated protocols, and real-time reverse transcription PCR (rRT-PCR) assays were performed using the Charité : SARS-CoV2 (E/RP) assay (Bio-Manguinhos/Fiocruz, Rio de Janeiro-Brazil) or the Allplex 2019-nCoV Assay (Seegene, Seoul-South Korea). In this study, samples from the states of Espirito Santo (ES), Paraná (PR), Rio de Janeiro (RJ), Rio Grande do Sul (RS), and Santa Catarina (SC) were included, in addition to samples from Minas Gerais (MG), since in 2021 FUNED joined the National Genetic Sequencing Network, created by the Brazilian Ministry of Health. In total, 115 positive samples (Dec/2020 to Dec/2022) were used in this study, in which all the samples had their sequence available for public access in the Global Initiative on Sharing Avian Influenza Data Bank (GISAID) (Supplementary Table S1), in addition to 16 clinical samples that tested negative for SARS-CoV-2.
2.2. PCR primers
Five primer pairs were selected from 98 primer pairs used for SARS-CoV-2 whole genome sequencing (Leung et al., 2021, ARTIC Network, 2020) and a primer pair (Var) was designed using Primer Express® Software Version 3.0 (Applied Biosystems, Foster City, CA, USA) ( Table 1). As a criterion for the selection of primers, amplicons were considered for regions of molecular signatures for variants and high impact of mutations, as well as genomic segments with high variability and previously described as regions of convergent mutations with positive implications for viral evolution. The primer coverage regions were visualized using the 6ZGG three-dimensional structure, available in the Protein Data Bank, as the model for observing the areas of interest in the protein. The Molecular Evolutionary Genetics Analysis Version 7.0 – MEGA7 (Kumar et al., 2016) and Multiple Sequence Alignment – CLUSTALW software allowed to identify the amplicons generated by the sequencing primers (S71 – S78) with coverage on the mutations of interest in the spike protein gene, and to generate the consensus sequence for the designed primers (Var).
Table 1.
Identification and sequences of forward and reverse primers used in HRM analyses.
| Primers |
Sequence (5 ´–3 ´) |
|
|---|---|---|
| Foward | Reverse | |
| S71 | ACAAATCCAATTCAGTTGTCTTCCTATTC | TGGAAAAGAAAGGTAAGAACAAGTCCT |
| S72 | ACACGTGGTGTTTATTACCCTGAC | ACTCTGAACTCACTTTCCATCCAAC |
| S75 | AGAGTCCAACCAACAGAATCTATTGT | ACCACCAACCTTAGAATCAAGATTGT |
| S76 | AGGGCAAACTGGAAAGATTGCT | ACACCTGTGCCTGTTAAACCAT |
| S78 | CAACTTACTCCTACTTGGCGTGT | TGTGTACAAAAACTGCCATATTGCA |
| Var | TTCTTATGGACCTTGAAGGAAAACA | AGAAGAATCACCAGGAGTCAAATAACT |
2.3. One-step real-time reverse transcription PCR
One-step rRT-PCR was performed using a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, USA) and the kit GoTaq® Probe 1-Step RT-qPCR System (Promega, Madison, USA). Amplifications were carried out in reaction mixtures containing 2 µl of viral RNA, 5 µl of GoTaq®Probe qPCR Master Mix (2X), 0.2 µl of GoScript™ RT Mix for 1-Step RT-qPCR (50X), 0.4 µl of 50 μM Syto™ 9 Green Fluorescent Nucleic Acid Stain (Invitrogen, Carlsbad, USA), 0.5 µl of 6 forward and reverse 10 µM primers (see Table 1) in singleplex reactions, and diethyl pyrocarbonate (DEPC) treated water (Sigma, St. Louis, USA) to 10 µl final volume. The cycling program consisted of a RT step at 50 °C for 30 min, initial denaturation at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. The amplifications were immediately followed by a melting program with initial denaturation at 95 °C for 15 s, cooling at 60 °C for 1 min and gradual heating (0.3 °C/s) to 95 °C. The fluorescence had its level collected at every 0.3 °C. Each 96-well reaction plate contained a negative RT-PCR control, where RNA was replaced by water. Melting curve analyses were performed using the High-Resolution Melt Software Version 3.0.1 (Applied Biosystems, Foster City, USA).
2.4. Library preparation and whole genome sequencing
The sequencing of SARS-CoV-2 positive samples was carried out using two different tecnologies, Ion Torrent (Thermofisher Scientific, Waltham, USA) and MiSeq (Illumina, San Diego, USA). For the Ion Torrent sequencing, viral RNA was converted to cDNA (complementary DNA) using the Superscript TM VILO cDNA Synthesis Kit (Invitrogen, Waltham, USA) and the libraries were prepared using the Ion AmpliSeq SARS-CoV-2 Panel (ThermoFisher Scientific, Waltham, USA). The normalized libraries were applied to a chip 318 and sequenced according to the manufacturer’s recommendations. For the MiSeq sequencing, cDNA was synthetized from RNA using the SuperScript IV TM Reverse Trascriptase Kit (Invitrogen, Waltham, USA), and then subjected to amplification of the complete genome of SARS-CoV-2 using a set of pathogen-specific primers designed by ARTIC Network (versions 3 and 4/4.1) (ARTIC Network, 2021, Quick, 2020). PCR products were cleaned up using AMPure XP beads (Beckman Coulter, Brea, USA). The libraries were prepared using the DNA Prep Kit (Illumina, San Diego, USA) and sequenced using the MiSeq Reagent Kit v3 (600 cycles) (Illumina, San Diego, USA), according to the manufacturer’s recommendations.
2.5. Consensus sequences and lineage classification
The consensus sequences were generated using the Genome Detective tool (https://www.genomedetective.com/app) (Vilsker et al., 2019) and the ViralUnity pipeline (https://github.com/filiperomero2/ViralUnity). The SARS-CoV-2 reference genome EPI_ISL_402124 (GISAID) was used in the ViralUnity pipeline, and lineage assignments were performed using the Pangolin lineage classification software tool (https://github.com/cov-lineages/pangolin).
3. Results
The panel of primers selected in this study (S71, S72, S75, S76, S78 and Var) showed a surveillance coverage of 53% of the amino acids per Spike protein monomer. The S71/S72/VAR primers cover the N-terminal domain region, while the S75/S76 primers target the receptor-binding domain, and the S78 primer set covers the S1/S2 furin cleavage site.
The SARS-CoV-2 variants that the primer sets were effective in distinguishing and the mutations covered by them are described in Table 2.
Table 2.
Primers and mutations. Primers code used for HRM analysis, spike mutations covered by the generated amplicons and SARS-CoV-2 classification according to the Who and Pangolin designations.
| WHO designation | Pango lineage (s) | S71 | S72 | S75 | S76 | S78 | Var |
|---|---|---|---|---|---|---|---|
| – | B.1.1.28a | x | x | x | x | x | x |
| – | B.1.1.33a | x | x | x | x | x | x |
| Gamma | P.1b,d | L18F(21614 C>T) T20N(21621 C>A) P .26 S(21638 C>T) | D138Y (21974 G>T) | K417T (22812 A>C) | K417T, E484K (23012 G>A), N501Y (23063 A>T) |
H655Y (23525 C>T) |
R190S (22132 G>T) |
| Zeta | P.2c,d | x | x | x | E484K | x | x |
| Alpha | B.1.1.7b | x | H69_V70del (21766_21771delACATGT), S98F (21855 C>T), Y144del (21992_21994delTAT) |
x | N501Y | P681H (23604 C>A), T716I (23709 C>T) | x |
| Delta | B.1.617.2 +AY.b,e | T19R (21618 C>G) | G142D (21987 G>A) | x | L452R (22917 T > G), T478K (22995 C>A) | P681R | x |
| Omicron | BA.1b | x | A67V (21762 C>T) | G339D (22578 G>A), S371L (22673 T > C 22674 C>T), S373P (22679 T > C), S375F (22686 C>T), K417N (22813 G>T), N440K (22882 T > G), G446S (22898 G>A) | K417N, N440K, G446S, S477N (22992 G>A), T478K (22995 C>A), E484A (23013 A>C), T547K (23202 C>A) |
H655Y, N679K (23599 T > G), P681H |
x |
| Omicron | BA.2b | T19I (21618 C>T) | G142D (21987 G>A) | Same as BA.1, except G446S, and plus T376A (22688 A>G), D405N (22775 G>A) and R408S (22786 A>C), | Same as BA.1, except G446S, T547K, and plus Q493R (23040 A>G), Q498R (23055 A>G), N501Y and Y505H (23075 T > C) |
Same as BA.1 | V213G (22200 T > G) |
| Omicron | BA.4/BA.5b | Same as BA.2 | Same as BA.2 | Same as BA.2 | Same as BA.2, except Q493R, and plus L452R (22917 T > G), and F486V (23018 T > G) | Same as BA.1 | Same as BA.2 |
| Omicron | BQ.1b | Same as BA.2 | Same as BA.2 | Same as BA.2, except K417N, N440K, and plus R346T (22599 G>C |
Same as BA.2, except K417N, N440K, Q493R, and plus F486V (23018 T > G) |
Same as BA.2 | Same as BA.2 |
a Initially dominant lineages of SARS-CoV-2 in Brazil. Typical mutation profile represented by samples EPI_ISL_1182584 and EPI_ISL_1182608 for B.1.1.28 and B.1.1.33 respectively;
b Previously circulating variant of concern. Typical mutation profile represented by samples EPI_ISL_15773602, EPI_ISL_15819746, EPI_ISL_15773593, EPI_ISL_15773586, EPI_ISL_13821800, EPI_ISL_13821773, EPI_ISL_13821762 and EPI_ISL_16022456, for Alpha, Gamma, Delta (AY.99.2), Omicron BA.1, BA.2, BA.4.1, BA.5.2.1 and BQ.1 respectively;
c Previously circulating variant of interest. Typical mutation profile represented by sample EPI_ISL_1182582;
d The prefix P is an alias for B.1.1.28. Lineages;
e The prefix AY is an alias for B.1.617.2;
x No changes compared to the SARS-CoV-2 isolate Wuhan-Hu-1 (NC_045512.2) using Genome Detective Virus Tool (https://www.genomedetective.com/app/typingtool/virus/).
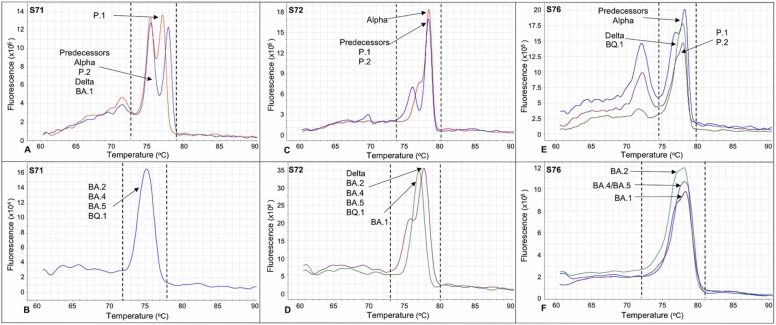
Based on the melting curves profiles of the amplicons generated by the sets of primers S71, S72 and S76, it was possible to distinguish important variants of concern and/or interest, such as the predecessors (B.1.1.28, B.1.1.33), Gamma, Zeta, Alpha, Delta and Omicron of occurrence in Brazil. As shown in Fig. 1A, the S71 primers generated amplicons with distinct profiles for the P.1 variant (red line), showing a double peak with greater proximity (Tm ∼75.0 °C ± 0.4 and ∼76.6 °C ± 0.4, ΔTm ∼1.6 °C ± 0.2) than the two peaks generated by the other analyzed variants (blue line), including the Omicron BA.1 (Tm ∼74.8 °C ± 0.5 and ∼77.2 °C ± 0.7, ΔTm ∼2.4 °C ± 0.2) or a single peak (Fig. 1B) for the other analyzed BA.2, BA.4, BA.5 and BQ.1 Omicron variants (∼Tm 75.2 °C ± 0.4). The S72 primers (Fig. 1C) made it possible to distinguish the Alpha variant (red line), which had a secondary peak (Tm ∼76.8 °C ± 0.2) separated by a discrete valley and with intensity close to the basal third of the main peak (Tm ∼78.3 °C ± 0.2), from the predecessors, and the P.1 and P.2 variants (blue line), which showed similar peaks with the main (Tm ∼77.8 °C ± 0.6) and secondary (Tm ∼75.8 °C ± 0.6) peaks being distanced by a more pronounced valley. The S72 primers also allowed BA.1 to be distinguished (Fig. 1D) (green line), demonstrating a single peak (Tm ∼76.7 °C ± 0.5), from the Delta and the BA.2, BA.4, BA.5 and BQ.1 Omicron variants analyzed (purple line), which showed peaks with proportionally similar intensities to the predecessor profile, but with a smooth valley between the secondary (Tm ∼75.5 °C ± 0.2) and main (∼77.7 °C ± 0.2) peaks. The S76 primers (Fig. 1E) allowed the P.1 and P.2 variants (purple line) to be distinguished, exhibiting a slight deformity in the distal region of the predominant peak (Tm ∼77.5 °C ± 0.6), from the predecessors, including the Alpha variant (blue line), which showed a double peak, with the secondary peak (Tm ∼76.9 °C ± 0.4) separated by a low amplitude valley near to the distal region of the main peak (Tm ∼78.2 °C ± 0.4), as well as Delta and Omicron BQ.1 variants (green line), which presented the predominant peak more symmetrical and funneled (Tm ∼77.7 °C ± 0.7). Additionally, the S76 primers (Fig. 1F) made it possible to distinguish the other analyzed BA.1, BA.2 and BA.4/BA.5 Omicron variants, displaying a single peak (Tm ∼77.7 °C ± 0.4). Despite the great similarity, small variations were observed in the apical region of the melting curve, with a slight deformity in BA.1 (purple line), as well as in BA.2, which showed a slightly less evident deformity (green line) or no deformity in BA.4/BA.5 Omicron variants (blue line).
Fig. 1.
Panel of typical HRM curves generated with primer sets S71, S72 and S76 for different variants of SARS-CoV-2.
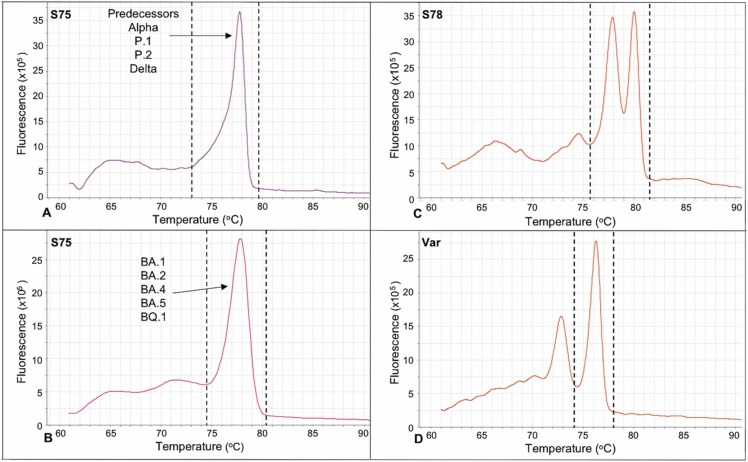
In relation to the other primers, the S75 presented a lower potential for variant discrimination, although it showed a slight difference between the Omicron variants (symmetric peak, Tm ∼77.3 °C ± 0.4) and its predecessors (peak with slight asymmetry in the left basal third, Tm ∼77.9 °C ± 0.4) ( Fig. 2 A and B). Finally, the S78 and Var primers also did not allow the differentiation of variants. The amplicons presented two predominant peaks (Tm ∼78.3 °C ± 0.4 and ∼80.7 °C ± 0.4) for the S78 (Fig. 2C) and a predominant peak (Tm ∼76.6 °C ± 0.5) for the Var primers (Fig. 2D), regardless of the variant.
Fig. 2.
Panel of typical HRM curves generated with primer sets S75, S78 and Var for different variants of SARS-CoV-2.
Regarding the universe of samples used, 13.0% (15/115) were unsuitable for HRM assays due to degradation, evidenced by atypical changes in the melting curves consistently repeated for different amplicons and/or late threshold (Ct) amplification cycles (>30). When considering the 87% (100/115) of eligible samples for HRM analysis, all of them presented conclusive results in agreement with the genomic sequencing. The comprehensive analysis of the concordance between the HRM primer set results and genomic sequencing (100/115), which included 16 SARS-CoV-2 negative clinical samples, revealed a sensitivity, specificity, accuracy, and Cohen's kappa value of 86.96% (79.59, 91.93*), 100% (80.64, 100*), 88.55% (81.97, 92.94*), and 0.619 (0.4612–0.7779) respectively. These values were calculated using OpenEpi, Version 3, an open-source code calculator. The values in parentheses represent the 95% lower-higher confidence interval, and an asterisk (*) indicates the Wilson score method.
4. Discussion
The pandemic spread of SARS-CoV-2 has resulted in the emergence of variants in which monitoring is crucial to support decision-making by health authorities around the world. Early identification of variants of interest/concern in a given population can be one of the most valuable mitigation measures for COVID-19, enabling the tracking and monitoring of infected individuals before massive spread.
Considering the immeasurable importance of complete sequencing for genomic surveillance but its high cost and time-consuming nature, fast and low-cost alternative methods for screening mutations are shown to be convenient strategies for optimizing the use of sequencing, by prior selection of potentially mutant-containing samples.
Compared to other SARS-CoV-2 genotyping methods such as whole genome sequencing, Sanger sequencing, single nucleotide polymorphism real-time PCR assays, and diagnostic screening nucleic acid amplification technique (NAAT)-based assays, among others (Koshikawa and Miyoshi, 2022, Miyoshi et al., 2022, WHO, 2022, Kiani et al., 2023), the HRM method presented in this study offers faster turnaround times and lower costs, making it a viable option for large-scale screening purposes. Here, we have explored the potential of HRM analysis combined with single-step rRT-PCR as a rapid method of screening for SARS-CoV-2 mutations, focusing on viral genomic regions of impact on interaction with the host. The one-step protocol used contributed to an assay with simple processing, saving time and protecting against risks of contamination and/or degradation due to reduced sample handling. Unlike other HRM methods that have used two-step reactions and short amplicons (Koshikawa and Miyoshi, 2022, Miyoshi et al., 2022, Kiani et al., 2023), for SARS-CoV-2 genotyping, our method demonstrates a higher coverage of mutations and an increased potential to detect new variants. In addition, this amplification strategy enabled the analysis of multiple targets in singleplex reactions, which would be challenging if an additional step for cDNA synthesis were required.
We emphasize our exploratory focus on the typing potential of HRM analyses by looking at the nuances of melting curve profiles generated by amplicons with higher coverage amplitudes, rather than focusing only on the melting temperatures of the predominant peaks generated for small amplicons, typically targeted in HRM analysis. While smaller amplicons can indeed amplify the effect of a mutation on melting temperature, our study has shown that larger amplicons covering a broader range of mutations effectively differentiate among variants. This approach would be less effective if focused solely on single-point mutations, given that SARS-CoV-2 is characterized by a set of mutations defining each variant. Furthermore, it is important to note that tracking single-point mutations would likely necessitate additional primer sets, emphasizing the advantage of our methodology. The selected primer panel presents a surveillance coverage of 53% of the amino acids per monomer of the Spike protein, the main protein subjected to selective pressure, host-pathogen interaction, and regions with convergent mutations and responsible for a higher rate of virus transmissibility (Martin et al., 2021, Shu and McCauley, 2017). The HRM analyses showed that the sets of primers S71, S72, and S76 were the most promising for distinguishing variants. The detection of SARS-CoV-2 lineages was based on the curve profile resulting from genomic changes accumulated in a given amplicon. These curve profiles showed some characteristics, such as single or double peaks with variations in the distance between them and deformations close to the base or to the peak of the melting curve.
The S71 primer pair initially distinguished the P.1 variant from other circulating variants, including later differentiation of the Omicron BA.1 variant from other Omicron variants analyzed in this study. The S72 primers initially differentiated the Alpha variant from its predecessor, as well as later showed the differentiation of Omicron BA.1 in relation to the other Omicron and Delta variants analyzed. The S76 primer pair generated distinguishable melting curves for different SARS-CoV-2 variants, including predecessors/Alpha, P.1/P.2, Delta/Omicron BQ.1, and Omicron BA.1, BA.2, BA.4/BA.5. Certain mutations in these variants, such as E484K in P.1 and P.2, L452R and T478K in Delta, and G446S/T547K in Omicron BA.1, described in Table 2, contributed to the observed melting curve profiles. The Omicron BA.2, BA.4, BA.5, and BQ.1 variants showed even greater similarity, with additional mutations (Q498R, N501Y, and Y505H) in relation to the others commom with BA.1. The BQ.1 variant's melting curve profile was similar to the Delta variant, which may be due to the absence of K417N and N440K mutations, common to other Omicron variants.
It is noteworthy that S75 did not generate amplicons with great power to distinguish variants. These primers presented a profile with a single peak for most variants, without mutations, including P.1, despite the K417T mutation. The Omicron variants presented numerous mutations for this region, with a common characteristic of having a slightly wider and symmetrical base than their predecessors. Similarly, the mutations covered by S78 and Var did not induce marked variations in the profiles of the melting curves to distinguish the analyzed variants, despite the mutations observed for some variants and other peaks at lower temperature not considered for the characterization of these profiles. The S78 and Var primers cover the S1/S2 cleavage site and part of the amino-terminal domain, respectively. The similarities observed for the amplicons of these targets suggest that they could be important conserved regions potentially applicable to the development of immunotherapies and vaccines.
An interesting aspect of our approach to screening mutations by HRM analysis was the good agreement with typing by genomic sequencing (Cohen's kappa value = 0.619), which clearly evidenced the potential of this technique for the rapid distinction of variants at low cost. As the focus of this study was to demonstrate the applicability of HRM for screening mutants, rather than standardizing a diagnostic method, the limit of detection, cross-reactivity with other pathogens, and reproducibility were not determined. The purpose here was to work with known SARS-CoV-2 positive samples from COVID-19 diagnosis in order to screen potential mutants for whole genome sequencing. However, reproducibility could be indirectly estimated by the concordance of HRM analysis with observed sequencing for most samples analyzed. An important observation was the variation in Tm, likely due to some point mutations in different samples, in addition to the typical mutation pattern of the various variants (Table 2), without compromising, however, the characteristic shape of the melting curves.
We would like to emphasize that, in addition to the limited number of samples, one of the weaknesses, not only of our method but also of sequencing, is the need to work with samples with low Ct values, generally below 30, in order to obtain robust results. Other limitations include (i) reliance on trained technicians to analyze melting curve profiles to distinguish variants, as we currently do not have an algorithm to automate this process; and (ii) the challenge of differentiating between closely related variants exhibiting high mutation rates, such as Omicron variants. In these cases, the extensive accumulation of mutations may result in a less pronounced distinction between variants when using HRM analysis. Future studies could explore the development of algorithms to automate the analysis of melting curve profiles, which would improve efficiency and reduce the potential for human error, further increasing the applicability of this technique in a wide range of settings. Interestingly, some theories about the evolution of Omicron have been proposed in an attempt to explain the explosion of genomic alterations, different from the expected mutation rate for SARS-CoV-2. These theories include (i) infection in immunosuppressed individuals with possible establishment of a host environment favorable to the long-term adaptation of the virus (Mallapaty, 2022, Wei et al., 2021), (ii) accumulation of mutations in a non-human host, with a subsequent jump to humans (Mallapaty, 2022, Smyth et al., 2022, Wei et al., 2021), (iii) and undiagnosed cryptic spread in a population with insufficient viral surveillance (Mallapaty, 2022, Wei et al., 2021), in addition to recombination events between co-circulating variants and coinfection (González-Candelas et al., 2021, Ou et al., 2022).
Despite those limitations, it is worth mentioning that different authors, already cited here, have used HRM strategies for SARS-CoV-2 typing, reinforcing the applicability of this technique for this purpose.
Here, our approach consists of using HRM analysis not in isolation, but combined with sequencing to optimize its genomic surveillance and with a focus that goes beyond the usual analysis of melting temperatures from small genomic targets. We propose such a strategy of using HRM combined with genomic sequencing, instead of using it alone, understanding that the melting curve profile can be valuable for screening samples with alterations in the nucleotide sequence, and for early identification of mutants. However, all atypical profiles suggestive of gene variation must be submitted to sequencing to confirm the mutation, since it is not unlikely that different nucleotide changes could result in similar melting curve profiles, as we observed for the Omicron variants.
On the other hand, it is possible to establish, as we have done here, panels with known melting curve profiles for different variants as they are identified, considering, when necessary, the combination of curve profiles presented by more than one set of primers. Thus, we show that HRM is a promising tool for screening samples with mutations, prior to sequencing, optimizing its use due to the greater probability of early detection of mutation profiles in a key genomic region for the host-pathogen interaction, before its spread in the population. This molecular approach can contribute to the prior stratification of samples to be sequenced, providing a quick estimate of circulating lineages and early identification of new variants which can even be extrapolated to other pathogens.
CRediT authorship contribution statement
Adriana de Souza Andrade: Formal analysis, Investigation, Methodology, Writing – original draft, Visualization. Eduarda Fernandes Freitas: Formal analysis, Investigation, Methodology, Visualization. Emerson de Castro Barbosa: Investigation, Writing – review & editing. Natália Rocha Guimarães: Investigation, Formal analysis. Felipe Campos de Melo Iani: Resources, Data curation. Alana Vitor Barbosa da Costa: Investigation, Formal analysis. André Felipe Leal Bernardes: Investigation, Formal analysis. Talita Emile Ribeiro Adelino: Investigation, Formal analysis. Ana Caroline Zampiroli Ataide: Investigation, Writing – review & editing. Tatiana Schäffer Gregianini: Resources, Investigation. Jônathas Dias Nunes: Resources, Investigation. Lorenzzo Lyrio Stringari: Resources, Investigation. Irina Nastassja Riediger: Resources, Investigation. Sandra Bianchini Fernandes: Resources, Investigation. Ronaldo de Jesus: Resources, Investigation. Vagner Fonseca: Resources, Investigation. Sérgio Caldas: Conceptualization, Data curation, Writing – review & editing, Supervision, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank all professionals at the Public Health Laboratories in Brazil and the General Coordination of Public Health Laboratories in Brazil (CGLab) for their contribution during the COVID-19 pandemic. This study was carried out with the financial support of Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) (APQ-00399–20/APQ-00251–21), Brazilian Ministry of Health, Fundação Ezequiel Dias (FUNED) and Pan American Health Organization / World Health Organization (PAHO/WHO).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jviromet.2023.114742.
Appendix A. Supplementary material
Supplementary material
.
References
- ARTIC Network, 2020. artic-network/primer-schemes: v1.1.1. https://doi.org/10.5281/ZENODO.4020380.
- ARTIC Network, 2021. SARS-CoV-2 V4.1 update for Omicron variant - Laboratory - ARTIC Real-time Genomic Surveillance [WWW Document]. URL 〈https://community.artic.network/t/sars-cov-2-v4–1-update-for-omicron-variant/342〉 (accessed 1.15.23).
- Diaz R.S., Vergara T.R.C. The COVID-19 second wave: A perspective to be explored. Brazilian Journal of Infectious Diseases. 2021;25:101537. doi: 10.1016/J.BJID.2020.101537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchene S., Featherstone L., Haritopoulou-Sinanidou M., Rambaut A., Lemey P., Baele G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020;6 doi: 10.1093/VE/VEAA061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Candelas F., Shaw M.A., Phan T., Kulkarni-Kale U., Paraskevis D., Luciani F., Kimura H., Sironi M. One year into the pandemic: short-term evolution of SARS-CoV-2 and emergence of new lineages. Infect. Genet. Evol. 2021;92 doi: 10.1016/J.MEEGID.2021.104869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N.D., Petrone M.E., Holmes E.C. We shouldn’t worry when a virus mutates during disease outbreaks. Nat. Microbiol. 2020;5:529–530. doi: 10.1038/S41564-020-0690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iftimie S., Lopez-Azcona A.F., Vallverdu I., Hernandez-Flix S., de Febrer G., Parra S., Hernandez-Aguilera A., Riu F., Joven J., Andreychuk N., Baiges-Gaya G., Ballester F., Benavent M., Burdeos J., Català A., Castañe E., Castañe H., Colom J., Feliu M., Gabaldo X., Garrido D., Garrido P., Gil J., Guelbenzu P., Lozano C., Marimon F., Pardo P., Pujol I., Rabassa A., Revuelta L., Rios M., Rius-Gordillo N., Rodriguez-Tomàs E., Rojewski W., Roquer-Fanlo E., Sabate N., Teixido A., Vasco C., Camps J., Castro A. First and second waves of coronavirus disease-19: A comparative study in hospitalized patients in Reus, Spain. PLoS One. 2021;16 doi: 10.1371/JOURNAL.PONE.0248029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani S.J., Ramshini M., Bokharaei-Salim F., Donyavi T., Eshrati B., Khoshmirsafa M., Ghorbani S., Tavakoli A., Monavari S.H., Ghalejoogh Z.Y., Abbasi-Kolli M. High resolution melting curve analysis for rapid detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants. Acta Virol. 2023:67. doi: 10.4149/av_2023_109. [DOI] [PubMed] [Google Scholar]
- Koshikawa T., Miyoshi H. High-resolution melting analysis to discriminate between the SARS-CoV-2 Omicron variants BA.1 and BA.2. Biochem. Biophys. Rep. 2022:31. doi: 10.1016/J.BBREP.2022.101306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/MOLBEV/MSW054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2-what do they mean. JAMA. 2021;325:529–531. doi: 10.1001/JAMA.2020.27124. [DOI] [PubMed] [Google Scholar]
- Leung K.S.S., Ng T.T.L., Wu A.K.L., Yau M.C.Y., Lao H.Y., Choi M.P., Tam K.K.G., Lee L.K., Wong B.K.C., Ho A.Y.M., Yip K.T., Lung K.C., Liu R.W.T., Tso E.Y.K., Leung W.S., Chan M.C., Ng Y.Y., Sin K.M., Fung K.S.C., Chau S.K.Y., To W.K., Que T.L., Shum D.H.K., Yip S.P., Yam W.C., Siu G.K.H. Territorywide study of early coronavirus disease outbreak Hong Kong China. Emerg. Infect. Dis. 2021;27:196–204. doi: 10.3201/EID2701.201543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. Where did Omicron come from? three key theories. Nature. 2022;602:26–28. doi: 10.1038/D41586-022-00215-2. [DOI] [PubMed] [Google Scholar]
- Martin D.P., Weaver S., Tegally H., San J.E., Shank S.D., Wilkinson E., Lucaci A.G., Giandhari J., Naidoo S., Pillay Y., Singh L., Lessells R.J., Gupta R.K., Wertheim J.O., Nekturenko A., Murrell B., Harkins G.W., Lemey P., MacLean O.A., Robertson D.L., de Oliveira T., Kosakovsky Pond S.L. The emergence and ongoing convergent evolution of the SARS-CoV-2 N501Y lineages. Cell. 2021;184:5189–5200. doi: 10.1016/J.CELL.2021.09.003. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H., Ichinohe R., Koshikawa T. High-resolution melting analysis after nested PCR for the detection of SARS-CoV-2 spike protein G339D and D796Y variations. Biochem. Biophys. Res. Commun. 2022;606 doi: 10.1016/j.bbrc.2022.03.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J., Lan W., Wu X., Zhao T., Duan B., Yang P., Ren Y., Quan L., Zhao W., Seto D., Chodosh J., Wu J., Zhang Q. Tracking SARS-CoV-2 Omicron diverse spike gene mutations identifies multiple inter-variant recombination events. BioRxiv. 2022;2022 doi: 10.1101/2022.03.13.484129. 03.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick, J., 2020. nCoV-2019 sequencing protocol v3 (LoCost). protocols.io [WWW Document]. protocols.io. URL 〈https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bp2l6n26rgqe/v3〉.
- Reed G.H., Kent J.O., Wittwer C.T. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics. 2007;8:597–608. doi: 10.2217/14622416.8.6.597. [DOI] [PubMed] [Google Scholar]
- Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis proofreading and final capping. Cells. 2020;9 doi: 10.3390/CELLS9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Eur. Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slinger R., Bellfoy D., Desjardins M., Chan F. High-resolution melting assay for the detection of gyrA mutations causing quinolone resistance in Salmonella enterica serovars Typhi and Paratyphi. Diagn. Microbiol. Infect. Dis. 2007;57:455–458. doi: 10.1016/J.DIAGMICROBIO.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Smyth D.S., Trujillo M., Gregory D.A., Cheung K., Gao A., Graham M., Guan Y., Guldenpfennig C., Hoxie I., Kannoly S., Kubota N., Lyddon T.D., Markman M., Rushford C., San K.M., Sompanya G., Spagnolo F., Suarez R., Teixeiro E., Daniels M., Johnson M.C., Dennehy J.J. Tracking cryptic SARS-CoV-2 lineages detected in NYC wastewater. Nat. Commun. 2022;2022:1–9. doi: 10.1038/s41467-022-28246-3. 13:1 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilsker M., Moosa Y., Nooij S., Fonseca V., Ghysens Y., Dumon K., Pauwels R., Alcantara L.C., vanden Eynden E., Vandamme A.M., Deforche K., de Oliveira T. Genome detective: an automated system for virus identification from high-throughput sequencing data. Bioinformatics. 2019;35:871–873. doi: 10.1093/BIOINFORMATICS/BTY695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Shan K.J., Wang W., Zhang S., Huan Q., Qian W. Evidence for a mouse origin of the SARS-CoV-2 Omicron variant. J. Genet. Genom. 2021;48:1111. doi: 10.1016/J.JGG.2021.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, E.C.D.C., 2022. Methods for The Detection and Characterisation of SARS-CoV-2 Variants-Second Update. European Centre for Disease Prevention and Control/World Health Organization Regional Office for Europe.
- WHO, 2021. Genomic sequencing of SARS-CoV-2: a guide to implementation for maximum impact on public health. URL 〈https://www.who.int/publications/i/item/9789240018440〉 (accessed 11.7.22).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material