Abstract
Background
Despite circumstantial evidence for aerosol and fomite spread of SARS-CoV-2, empirical data linking either pathway with transmission are scarce. Here we aimed to assess whether the presence of SARS-CoV-2 on frequently-touched surfaces and residents' hands was a predictor of SARS-CoV-2 household transmission.
Methods
In this longitudinal cohort study, during the pre-alpha (September to December, 2020) and alpha (B.1.1.7; December, 2020, to April, 2021) SARS-CoV-2 variant waves, we prospectively recruited contacts from households exposed to newly diagnosed COVID-19 primary cases, in London, UK. To maximally capture transmission events, contacts were recruited regardless of symptom status and serially tested for SARS-CoV-2 infection by RT-PCR on upper respiratory tract (URT) samples and, in a subcohort, by serial serology. Contacts' hands, primary cases' hands, and frequently-touched surface-samples from communal areas were tested for SARS-CoV-2 RNA. SARS-CoV-2 URT isolates from 25 primary case-contact pairs underwent whole-genome sequencing (WGS).
Findings
From Aug 1, 2020, until March 31, 2021, 620 contacts of PCR-confirmed SARS-CoV-2-infected primary cases were recruited. 414 household contacts (from 279 households) with available serial URT PCR results were analysed in the full household contacts' cohort, and of those, 134 contacts with available longitudinal serology data and not vaccinated pre-enrolment were analysed in the serology subcohort. Household infection rate was 28·4% (95% CI 20·8–37·5) for pre-alpha-exposed contacts and 51·8% (42·5–61·0) for alpha-exposed contacts (p=0·0047). Primary cases' URT RNA viral load did not correlate with transmission, but was associated with detection of SARS-CoV-2 RNA on their hands (p=0·031). SARS-CoV-2 detected on primary cases' hands, in turn, predicted contacts' risk of infection (adjusted relative risk [aRR]=1·70 [95% CI 1·24–2·31]), as did SARS-CoV-2 RNA presence on household surfaces (aRR=1·66 [1·09–2·55]) and contacts' hands (aRR=2·06 [1·57–2·69]). In six contacts with an initial negative URT PCR result, hand-swab (n=3) and household surface-swab (n=3) PCR positivity preceded URT PCR positivity. WGS corroborated household transmission.
Interpretation
Presence of SARS-CoV-2 RNA on primary cases' and contacts' hands and on frequently-touched household surfaces associates with transmission, identifying these as potential vectors for spread in households.
Funding
National Institute for Health Research Health Protection Research Unit in Respiratory Infections, Medical Research Council.
Introduction
SARS-CoV-2 transmission mostly occurs in households,1 yet our understanding of the routes of transmission remains an area of ongoing scientific debate. Despite public guidance on self-isolation, facemask wearing, handwashing, and surface and air decontamination, there is very little empirical evidence for the pathways through which the virus spreads. Modelling and epidemiological observations implicate viral spread through large respiratory droplets that settle on surfaces from where virus is transferred to contacts' mucosal membranes, known as fomite spread, as well as airborne spread through smaller, aerosolised droplets directly inhaled by contacts.2
Although previous studies have documented the presence of SARS-CoV-2 RNA on household surfaces,3, 4 as well as in exhaled and ambient air from hospitalised patients with COVID-19,5, 6 its detection on potential vectors has not hitherto been shown to correlate with transmission. This gap in knowledge hampers accurate modelling of disease transmission and development of evidence-based public health guidance,7, 8 which remain important given the ability of SARS-CoV-2 to spread through highly vaccinated populations.9, 10 As isolation restrictions for household contacts have now been lifted, understanding the risk factors and pathways of transmission within households has become increasingly important to prevent the spread of SARS-CoV-2.
Research in context.
Evidence before this study
Extensive literature on SARS-CoV-2 transmission has been published, with different types of studies (observational prospective cohort studies, retrospective cohort studies, modelling, case-control, etc) evaluating transmission in different scenarios and populations. Although most transmission globally occurs in households, the vectors and pathways of household SARS-CoV-2 transmission remain poorly understood and there is ongoing scientific debate as to the predominant modes of transmission. We searched for the terms “SARS-CoV-2” AND “household” AND “transmission” with “pathways” OR “surface” OR “hand” OR “fomite” OR “droplet” in PubMed for articles published from database inception up until Nov 2, 2022, in English. After omitting studies with different settings or study populations (hospitalised populations, health-care workers, schools, care homes, etc), we identified few studies assessing SARS-CoV-2 in household settings. Several systematic and non-systematic reviews have been published on household infection rate, often denoted as secondary attack rate, and some studies have evaluated the contribution of demographic, clinical, and epidemiological factors to infection (sex, age, vaccinations status, co-infection, comorbidities, relation to the primary case, household characteristics, etc). At the time of writing, only six studies reported the presence of viral RNA on household surfaces, but none have correlated this presence to risk of infection among contacts. Additionally, we found no studies analysing the presence of viral RNA on primary cases' or contacts' hands, which might be important vectors of transmission.
Added value of this study
We prospectively enrolled symptomatic and asymptomatic household contacts recently exposed to their primary cases, and serially performed upper respiratory tract swabs RT-PCR testing, and when consented, collected blood samples for serial serology. Additionally, we also took swabs from frequently touched household surfaces, and primary cases' and contacts' hands, and tested them for presence of SARS-CoV-2 RNA. Our prospective study design and dense longitudinal testing of contacts enabled us to maximally capture secondary COVID-19 cases among household contacts. Epidemiological linkage and whole-genome sequencing of primary and secondary cases' SARS-CoV-2 isolates confirmed household transmission. Presence of SARS-CoV-2 RNA on primary cases' hands, household surfaces, and contacts' hands strongly correlated with contacts' risk of infection. To our knowledge, this study provides the first empirical data linking detection of SARS-CoV-2 on candidate vectors of transmission. This study also identified primary cases' hands, household surfaces, and contacts' hands as potential vectors of transmission. Aerosol transmission was not investigated in this study and therefore cannot be excluded as a potential route of household spread.
Implications of all the available evidence
This study, to our knowledge, is the first to identify a correlation between SARS-CoV-2 RNA on candidate vectors and infection of contacts. These longitudinal empirical data from a real-world community setting substantially advance our understanding of SARS-CoV-2 household spread, which is the setting of most transmission globally. Our findings support interventions such as frequent handwashing, surface cleansing, physical distancing, reducing direct contact, and use of masks to reduce transmission in households.
Risk factors, vectors, and pathways for transmission can only be rigorously assessed by maximally and prospectively capturing transmission to susceptible household contacts. However, large-scale data from national testing services rely on contacts requesting tests, usually because they are symptomatic.11 Moreover, almost all contacts' infection rate estimates use upper respiratory tract (URT) PCR testing at a single timepoint to detect or exclude infection,12 limiting the ability to detect new secondary infections.13
We therefore prospectively recruited susceptible household contacts recently exposed to PCR-positive COVID-19 primary cases. We serially sampled both symptomatic and asymptomatic contacts' URT for PCR testing and, when consented, blood for serial serology. We tested for the presence of SARS-CoV-2 RNA on primary cases' and contacts' hands and frequently-touched surfaces. We aimed to assess whether the presence of SARS-CoV-2 on one or more of these candidate vectors would prospectively predict SARS-CoV-2 transmission.
Methods
Study design and participants
Participants were enrolled in two longitudinal community-based observational studies in Greater London, UK: Integrated Network for Surveillance, Trials and Investigations into COVID-19 Transmission (INSTINCT), and Assessment of Transmission And Contagiousness of COVID-19 in Contacts (ATACCC). Ethics approval was obtained from the Health Research Authority (20/NW/0231).
SARS-CoV-2 PCR-positive, symptomatic primary cases (defined as the first detected PCR-positive case in the household) and their contacts were identified through the national contact tracing system (National Health Service Test & Trace) and invited to participate by UK Health Security Agency (UKHSA) staff between Aug 1, 2020, and March 31, 2021. The study period included the pre-alpha (September to December 2020) and alpha (B.1.1.7; December, 2020, to April, 2021) waves of SARS-CoV-2, when vaccination coverage was low and social distancing policies were in place. Contacts were deemed household contacts if resident at the same address as their primary case.
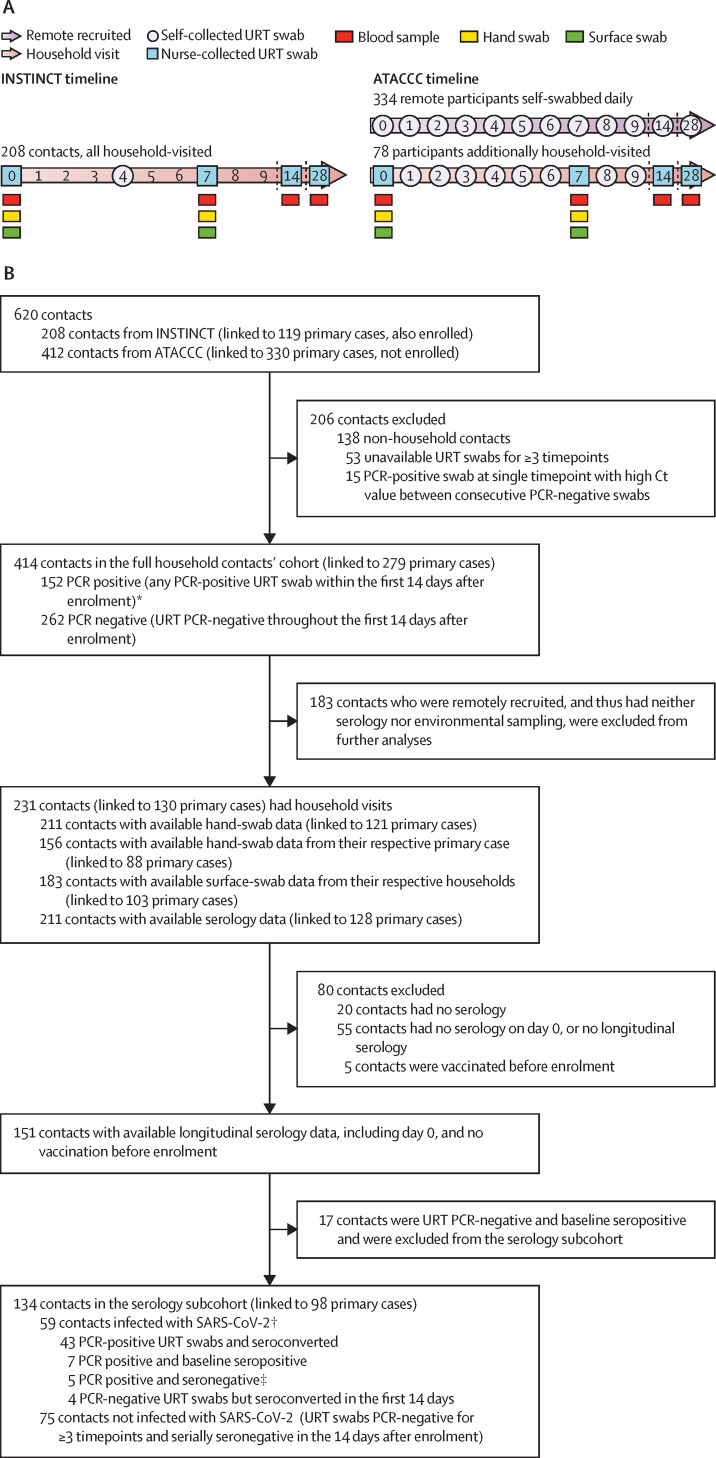
In the INSTINCT study, primary cases and their household contacts were enrolled within 5 days (median of 3 days [IQR 2–4]) of primary case symptom onset and visited by study staff on the day of enrolment (day 0), day 7, and day 28, and on day 14 if any contact had tested PCR positive. URT swabs were collected at all visits as shown in figure 1A . On study day 4, participants took an additional self-taken URT swab. In the ATACCC study, household and non-household contacts were recruited within 5 days (median of 3 days [IQR 2–4]) of their primary case symptom onset to perform self-taken daily URT swabs for 14 days and, if still PCR positive, up to day 28 after enrolment; self-taken swabs were delivered by courier to the UKHSA laboratory daily. Primary cases were not enrolled. Most ATACCC contacts were recruited remotely; and the remaining contacts who consented to household visits underwent additional sampling according to figure 1. In all INSTINCT and visited ATACCC households, surface swabs (sampled from the most frequently touched surfaces in communal areas as reported by the household residents) and hand swabs were taken by trained nurses at enrolment (day 0) and day 7. Serology samples were taken on day 0, day 7, day 14, and day 28. All participants provided sociodemographic, clinical, and epidemiological data via questionnaires on the day of enrolment.
Figure 1.
INSTINCT and ATACCC studies recruitment timelines (A), and inclusion criteria, exposure, and SARS-CoV-2 infection status of contacts included in the analyses (B)
ATACCC=Assessment of Transmission And Contagiousness of COVID-19 in Contacts. Ct=cycle threshold. INSTINCT=Integrated Network for Surveillance, Trials and Investigations into COVID-19 Transmission. URT=upper respiratory tract. WGS=whole-genome sequencing. *WGS was possible for 35 of the 152 PCR-positive household contacts, where URT swabs from both the contact and their respective epidemiologically-linked primary case were PCR positive, available, and had a viral load of more than 200 copies per mL. †URT swabs from 21 (36%) of 59 contacts in the serology subcohort deemed infected underwent WGS analysis alongside URT swabs from their primary case (appendix p 12). ‡Insufficient longitudinal serological data among contacts who were PCR positive (no serology after day 7).
Participants were free to leave the study prematurely if they wished or to omit some sociodemographic characteristic information. Among the visited participants, not all agreed to give all types of samples (serological, surface swabs, and hand swabs).
Two contacts cohorts were defined for analysis: the full household contacts' cohort, consisting of all household contacts with longitudinal (≥3) URT samples, and the serology subcohort, consisting of the contacts within the full cohort who also had longitudinal serology data (ie, serological data available at day 0 and at least one other timepoint), were known to be unvaccinated before enrolment, and were seronegative at enrolment. Households with co-primary cases (concurrent PCR positivity) were excluded.
Contacts within the full household contacts' cohort were defined as being PCR positive (positive URT swab within the first 14 days after enrolment) or PCR negative if all URT swabs (≥ three) were negative throughout the first 14 days after enrolment. Hand-swab and surface-swab PCR positivity was identically assigned (ie, 5 RNA copies per mL). Contacts within the serology subcohort were classified as infected if they were PCR positive or seroconverted within the first 14 days after enrolment, or both, and uninfected if they remained both PCR negative and seronegative. Further details on laboratory results definitions are available in the appendix (pp 2–4).
Study covariates considered were sex assigned at birth, age (continuous and grouped in categories), ethnicity (grouped as White and non-White), BMI (grouped as <18·5, 18·5 to <25·0, 25·0 to <30·0, and ≥30·0 kg/m2), presence of one or more comorbidities, and COVID-19 vaccination pre-enrolment. COVID-19 vaccination pre-enrolment was defined as receiving one or more doses before enrolment. Household contacts were defined as contacts who were co-resident with the primary case. Relationship status to the primary case was assessed with predetermined categories: partner, parent to child aged 16 years or younger, child aged 16 years or younger, or sibling in same bedroom; parent to child older than 16 years, child older than 16 years, or sibling without shared bedroom; and housemate or residential employee. We used the 2019 Index of Multiple Deprivation (IMD), the UK Government's official measure of relative deprivation at a small area level (average population of 1500) across England,14 to assess household deprivation according to postcodes. Participants gave written informed consent.
Procedures
URT swabs were analysed by RT-PCR at Imperial College London, London, UK (INSTINCT)15 and UKHSA Colindale, London, UK (ATACCC) with comparable sensitivity and specificity (appendix pp 2–3). Serology was performed using a hybrid double antigen binding assay (DABA), as previously described (appendix pp 3–4).16 A DABA level of less than 1 indicated seronegativity and a DABA level of 1 or more indicated seropositivity. A DABA level of less than 1 at day 0 that increased by 3 in the binding ratio in the first 28 days after participant enrolment (if PCR positive) or in the first 14 days after enrolment (if PCR negative) indicated seroconversion. Variant status was assigned through S-gene target failure status (ATACCC; appendix p 5) and whole-genome sequencing (WGS; INSTINCT; appendix pp 5–6). Libraries for WGS were generated with the EasySeq RT-PCR SARS-CoV-2 (novel coronavirus) WGS kit v1 or v2 (Nimagen, Nijmegen, Netherlands) and an iSeq 100 device was used for sequencing (Illumina, San Diego, CA, USA). Hand swabs and surface swabs were tested for RNA presence at the Department of Infectious Disease, Imperial College London, London, UK (appendix p 4).3 PCR-positive hand and surface swabs with a cycle threshold (Ct) of less than 33 were subjected to viral culture in a mammalian cell line-based in vitro assay (appendix pp 4–5). Swabs were maintained at 4°C for 36–72 h until culture was performed, reflecting the time for transportation from residents' homes and laboratory storage while RT-PCR was performed. Further assay details are found in the appendix (pp 2–6).
Statistical analysis
The primary outcome was infection among contacts, defined as contacts becoming PCR positive in the full household contact cohort, or infected in the serology subcohort, as defined earlier. All analyses were performed with the data available for each covariate used. To identify confounding risk factors for contacts' infection, univariable associations between PCR positive or infected versus PCR negative or uninfected cases according to different covariates were evaluated using a modified χ2 test accounting for contacts' household clustering.17
Household infection rate was calculated as the proportion of contacts who became PCR positive or infected among all those tested.
To estimate the contribution of viral RNA presence on candidate vectors to contacts' risk of infection, adjusted relative risks (aRR) from multivariable, modified Poisson regression analyses were estimated, with an implemented generalised estimating equation model to account for household clustering and obtain robust standard errors.18 Binary indicators for infection or PCR positive versus PCR negative candidate vectors were used. The model was adjusted for significant demographic covariates and covariates associated with household characteristics identified with univariable analyses.
Analyses were performed on R version 4.0.5 (appendix p 8).
Role of the funding source
The funder of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
620 contacts of PCR-confirmed SARS-CoV-2-infected primary cases were recruited in London from Aug 1, 2020, to March, 31, 2021. After excluding non-household contacts (n=138), contacts without URT samples for three or more timepoints (n=53), and contacts with a PCR-positive swab at a single timepoint with high Ct value between consecutive PCR-negative swabs (n=15), 414 household contacts (from 279 households) with available serial URT PCR results were included in our analyses (full household contacts' cohort; figure 1; appendix p 9). 134 household contacts within the full cohort also had longitudinal serology data and were included in the serology subcohort.
No significant differences were observed in sex, age, ethnicity, or BMI between the 152 PCR-positive and 262 PCR-negative contacts from the full household contacts' cohort (table 1 ); although men had an increased risk of infection in the serology subcohort (p=0·047). In this predominantly vaccine-naive cohort, vaccinated contacts comprised a significantly higher proportion of PCR-negative cases (p=0·015) and participants with comorbidities were less likely to be PCR positive than those without (p=0·017; table 1).
Table 1.
Baseline demographic and clinical characteristics of household contacts exposed to PCR-confirmed SARS-CoV-2 primary cases, in the full household contacts' cohort and in the serial serology subcohort.
|
Full household contacts' cohort (n=414) |
Serial serology subcohort (n=134) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total (n=414) | PCR-positive (n=152) | PCR-negative (n=262) | χ2 test p value* | Total (n=134) | Infected with SARS-CoV-2† (n=59) | Not infected with SARS-CoV-2 (n=75) | χ2 test p value* | ||
| Sex assigned at birth | .. | .. | .. | 0·77 | .. | .. | .. | 0·047 | |
| Male | 200 (48%) | 75 (49%) | 125 (48%) | .. | 70 (52%) | 37 (63%) | 33 (44%) | .. | |
| Female | 214 (52%) | 77 (51%) | 137 (52%) | .. | 64 (48%) | 22 (37%) | 42 (56%) | .. | |
| Median age, years (IQR) | 34 (25–50) | 37 (25–49) | 33 (24–51) | 0·90 | 36 (24–51) | 36 (25–47) | 36 (24–52) | 0·54 | |
| Age group | .. | .. | .. | 0·36 | .. | .. | .. | 0·36 | |
| ≤16 years | 45 (11%) | 19 (12%) | 26 (10%) | .. | 12 (9%) | 5 (8%) | 7 (9%) | .. | |
| 17–29 years | 124 (30%) | 39 (26%) | 85 (32%) | .. | 43 (32%) | 19 (32%) | 24 (32%) | .. | |
| 30–49 years | 141 (34%) | 60 (39%) | 81 (31%) | .. | 43 (32%) | 23 (39%) | 20 (27%) | .. | |
| 50–69 years | 100 (24%) | 33 (22%) | 67 (26%) | .. | 35 (26%) | 11 (19%) | 24 (32%) | .. | |
| ≥70 years | 4 (1%) | 1 (1%) | 3 (1%) | .. | 1 (1%) | 1 (2%) | 0 (0%) | .. | |
| Ethnicity | .. | .. | .. | 0·53 | .. | .. | .. | 0·17 | |
| White | 325 (78%) | 122 (80%) | 203 (77%) | .. | 112 (84%) | 51 (86%) | 61 (81%) | .. | |
| Non-White | 73 (18%) | 24 (16%) | 49 (19%) | .. | 16 (12%) | 4 (7%) | 12 (16%) | .. | |
| Data missing | 16 (4%) | 6 (4%) | 10 (4%) | .. | 6 (4%) | 4 (7%) | 2 (3%) | .. | |
| BMI | .. | .. | .. | 0·12 | .. | .. | .. | 0·28 | |
| <18·5 kg/m2 | 8 (2%) | 2 (1%) | 6 (2%) | .. | 1 (1%) | 0 | 1 (1%) | .. | |
| 18·5 to <25·0 kg/m2 | 163 (39%) | 47 (31%) | 116 (44%) | .. | 48 (36%) | 16 (27%) | 32 (43%) | .. | |
| 25·0 to <30·0 kg/m2 | 111 (27%) | 45 (30%) | 66 (25%) | .. | 41 (31%) | 21 (36%) | 20 (27%) | .. | |
| ≥30·0 kg/m2 | 59 (14%) | 26 (17%) | 33 (13%) | .. | 13 (10%) | 4 (7%) | 9 (12%) | .. | |
| Data missing | 73 (18%) | 32 (21%) | 41 (16%) | .. | 31 (23%) | 18 (30%) | 13 (17%) | .. | |
| Presence of comorbidities‡ | .. | .. | .. | 0·017 | .. | .. | .. | 0·0038 | |
| Yes | 105 (25%) | 28 (18%) | 77 (29%) | .. | 22 (16%) | 3 (5%) | 19 (25%) | .. | |
| No | 298 (72%) | 121 (80%) | 177 (68%) | .. | 107 (80%) | 54 (92%) | 53 (71%) | .. | |
| Data missing | 11 (3%) | 3 (2%) | 8 (3%) | .. | 5 (4%) | 2 (3%) | 3 (4%) | .. | |
| COVID-19 vaccination pre-enrolment§ | .. | .. | .. | 0·015 | .. | .. | .. | NA | |
| Yes | 25 (6%) | 3 (2%) | 22 (8%) | .. | 0 | 0 | 0 | .. | |
| No | 382 (92%) | 144 (95%) | 238 (91%) | .. | 134 (100%) | 59 (100%) | 75 (100%) | .. | |
| Data missing | 7 (2%) | 5 (3%) | 2 (1%) | .. | .. | .. | .. | .. | |
Data are n (%) unless otherwise indicated. NA=not applicable.
p values are given for each covariate; χ2 test accounting for household clustering was used for all categories for which data are available, except for median age, for which the Mann-Whitney test was used.
Contacts were considered infected if PCR-positive or, in the serology subcohort, if PCR-positive or seroconverted.
Presence of comorbidities was considered if the participant had one or more conditions (appendix p 7).
Vaccination pre-enrolment was defined as having received one or more doses before day 0.
Household-visited participants and remotely recruited participants had similar characteristics (appendix pp 13–15). The characteristics of primary cases who infected one or more of their recruited contacts were also similar to participants who did not infect their household contacts, except for age, with older primary cases resulting in more secondary infection events (appendix pp 16–17).
The overall household infection rate was 36·7% (95% CI 32·2–41·5) in the full cohort (table 2 ). 31 of the 152 PCR-positive contacts were defined contacts with incident infection as they became PCR positive after enrolment (as opposed to contacts with prevalent infection, who were PCR positive at enrolment), hence the household infection rate at the day of enrolment was lower than the overall HIR (29·2%, 25·0–33·8; data not shown). The number of days from primary case symptom onset to recruitment did not differ between contacts with prevalent and incident infection (median 3·00 days [IQR 2·00–4·00] vs 3·00 days [3·00–4·00]); Mann-Whitney U test p=0·98; data not shown). In the serial serology subcohort, after removal of 17 previously infected contacts (baseline seropositive) and reclassifying four PCR-negative contacts as incident infections (seroconverters between day 0 and day 14), the overall household infection rate was 44·0% (35·9–52·5; table 2).
Table 2.
SARS-CoV-2 household infection rate in the full household contacts' cohort and in the serial serology subcohort, stratified by type of exposure to the primary case and household characteristics
|
Full household contacts' cohort (n=414) |
Serial serology sub-cohort (n=134) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Number of PCR-positive/total number | Household infection rate (95% CI) | χ2 test p value* | Number infected/total number | Household infection rate (95% CI) | χ2 test p value* | |||
| Overall | 152/414 | 36·7% (32·2–41·5) | .. | 59/134 | 44·0% (35·9–52·5) | .. | ||
| Contacts' exposure to the primary case | ||||||||
| Relationship status to the primary case | <0·0001 | 0·0016 | ||||||
| Partner, parent to child aged ≤16 years, child aged ≤16 years, or sibling in same bedroom | 85/174 | 48·8% (41·5–56·2) | .. | 36/56 | 64·3% (51·2–75·5) | .. | ||
| Parent to child aged >16 years, child aged >16 years, or sibling without shared bedroom | 39/126 | 30·9% (23·5–39·5) | .. | 11/44 | 25·0% (14·6–39·4) | .. | ||
| Housemate or residential employee | 12/71 | 16·9% (9·9–27·2) | .. | 6/21 | 28·6% (13·8–50·0) | .. | ||
| Data missing | 16/43 | .. | .. | 6/13 | .. | .. | ||
| Primary case SARS-CoV-2 variant | 0·0047 | 0·0016 | ||||||
| Alpha (B.1.1.7) | 56/108 | 51·8% (42·5–61·0) | .. | 25/34 | 73·5% (56·9–85·4) | .. | ||
| Pre-alpha | 31/109 | 28·4% (20·8–37·5) | .. | 11/38 | 28·9% (17·0-44·8) | .. | ||
| Data missing | 65/197 | .. | .. | 23/62 | .. | .. | ||
| Primary case maximum-measured URT RNA viral load, viral copy number per mL | 0·83 | 0·64 | ||||||
| 5 to ≤4500 | 36/82 | 43·9% (33·7–54·7) | .. | 24/47 | 51·1% (37·2–64·7) | .. | ||
| 4500 to ≤12 000 000 | 35/76 | 46·0% (35·3–57·2) | .. | 25/44 | 56·8% (42·2–70·3) | .. | ||
| Data missing | 81/256 | .. | .. | 10/43 | .. | .. | ||
| Contacts' household exposure characteristics | ||||||||
| Number of people resident in the household, including the primary case | 0·22 | 0·44 | ||||||
| 2 people | 25/66 | 37·9% (27·1–49·9) | .. | 12/21 | 57·1% (36·5–75·5) | .. | ||
| 3–4 people | 90/224 | 40·2% (34·0–46·7) | .. | 29/71 | 40·8% (30·2–52·5) | .. | ||
| 5 or more people | 26/96 | 27·1% (19·2–36·7) | .. | 16/35 | 45·7% (30·5–61·8) | .. | ||
| Data missing | 11/28 | .. | .. | 2/7 | .. | .. | ||
| Ratio of the number of people resident in the household per number of bedrooms | 0·065 | 0·23 | ||||||
| ≤1 resident per bedroom | 70/217 | 32·2% (26·4–38·7) | .. | 27/69 | 39·1% (28·5–50·9) | .. | ||
| >1 resident per bedroom | 70/161 | 43·5% (36·1–51·2) | .. | 30/58 | 51·7% (39·2–64·1) | .. | ||
| Data missing | 12/36 | .. | .. | 2/7 | .. | .. | ||
| By household decile of Index of Multiple Deprivation | 0·68 | 0·51 | ||||||
| More deprived deciles, 1 to 5 | 71/200 | 35·5% (29·2–42·3) | .. | 29/71 | 40·8% (30·2–52·5) | .. | ||
| Least deprived deciles, 6 to 10 | 81/214 | 37·8% (31·6–44·5) | .. | 30/63 | 47·6% (35·8–59·7) | .. | ||
Household infection rate is given as the proportion of PCR-positive contacts in the full household contact cohort (n=414), and as the proportion of infected (PCR-positive or seroconverted) contacts in the serial serology subcohort (n=134). Primary case maximum-measured URT RNA viral load categories were divided to give similar number of contacts. URT=upper respiratory tract.
p values for each covariate were calculated with the χ2 test accounting for household clustering, applying for all categories where data was available.
Contacts' risk of infection increased with a closer relationship to the primary case, with the household infection rate ranging from 16·9% (95% CI 9·9–27·2) for non-related housemates to 48·8% (41·5–56·2) for primary-case partners or bedroom-sharing relatives (p<0·0001; table 2). The total number of residents per household was not associated with increased risk of infection; however, the point estimate proportion of infected contacts was higher in households with more than one resident per bedroom than in those with one resident per bedroom, although this difference was not significant (p=0·065; table 2). Household deprivation index (IMD deciles) did not significantly differ between the 152 PCR-positive and 262 PCR-negative contacts. Household infection rate was higher in contacts exposed to alpha (B.1.1.7) SARS-CoV-2 versus pre-alpha SARS-CoV-2 variants (table 2; 51·8% [42·5–61·0] vs 28·4% [20·8–37·5] in the full household contacts' cohort; p=0·0047 and 73·5% [56·9–85·4] vs 28·9% [17·0–44·8] in the serology subcohort; p=0·0016).
All PCR-positive surface swabs and primary cases' hand swabs, and most of the PCR-positive hand swabs from the contacts (16 [72%] of 22), were positive on the day of enrolment. This RNA detection on surface and hand swabs was simultaneous in time with the first detection of SARS-CoV-2 in prevalent PCR-positive contacts' respective URT swabs, at day 0. Of the 31 PCR-positive contacts with incident infection (ie, with initially PCR-negative URT swabs), 14 had household surface samples collected, of which three were positive—all were positive at day 0, before the respective contacts' URT swabs becoming PCR positive. 19 of 31 contacts with incident infection had hand swabs collected, of which seven were positive. In three contacts, hand swabs were already PCR positive before their respective URT swabs becoming positive. In the remaining four contacts, hand swabs were positive at the second time-point (day 7); hence, it is not possible to temporally resolve whether these contacts' hands became PCR positive before or after their URT swabs became PCR positive.
Although the maximum URT RNA viral load measured in primary cases did not correlate with the household infection rate (p=0·83; table 2; appendix p 10), primary cases with SARS-CoV-2 RNA detected on hand swabs (21 [24%] of 88 primary cases with hand swabs collected) had significantly higher URT RNA viral load than did those whose hand swabs were SARS-CoV-2 PCR-negative (p=0·031; appendix p 11). Presence of SARS-CoV-2 RNA on primary cases' hands was associated with SARS-CoV-2 infection in their contacts (aRR 1·70, 95% CI 1·24–2·31; table 3 ). Presence of SARS-CoV-2 RNA on primary cases' hands was also associated with viral RNA detection on their corresponding contacts' hands (3·11, 1·21–8·05; appendix pp 18–19), and presence of SARS-CoV-2 on contacts' hands was associated with their infection status—ie, having a PCR-positive UTR sample (2·06, 1·57–2·69; table 3). Among the three contacts with incident infection (with a PCR-negative initial URT sample) and positive hand swabs at day 0 before URT PCR positivity, SARS-CoV-2 RNA was first detected on their hands 1, 2, and 3 days before their first PCR-positive URT sample. One of these hand swabs was also positive on viral culture (attempted on ten hand swabs) and yielded sufficient RNA for WGS, which revealed indistinguishable sequence from that from the PCR-positive URT swab collected 1 day later. A primary case sample was unavailable for comparison.
Table 3.
Detection of SARS-CoV-2 on environmental surfaces, and on the hands of the contact and primary case, by PCR-status in the full household contacts' cohort, and by infection status in the serology analysis contacts' subcohort
|
Full household contacts' cohort (n=414) |
Serial serology subcohort (n=134) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Household contacts |
Multivariable analysis |
Household contacts |
Multivariable analysis |
||||||||
| Total (n=414) | PCR positive (n=152) | PCR negative (n=262) | χ2 test p value* | Adjusted relative risk (95% CI) | Total (n=134) | Infected with SARS-CoV-2† (n=59) | Not infected with SARS-CoV-2 (n=75) | χ2 test p value* | Adjusted relative risk (95% CI) | ||
| Primary cases' hand-swab SARS-CoV-2 PCR status | .. | .. | .. | 0·075 | .. | .. | .. | .. | 0·0037 | .. | |
| Positive | 33 (8%) | 20 (13%) | 13 (5%) | .. | 1·70 (1·24–2·31) | 19 (14%) | 17 (29%) | 2 (3%) | .. | 1·67 (1·23–2·28) | |
| Negative | 123 (30%) | 49 (32%) | 74 (28%) | .. | 1 (ref) | 65 (49%) | 31 (52%) | 34 (45%) | .. | 1 (ref) | |
| Data missing | 258 (62%) | 83 (55%) | 175 (67%) | .. | .. | 50 (37%) | 11 (19%) | 39 (52%) | .. | .. | |
| Contacts' hand-swab SARS-CoV-2 PCR status | .. | .. | .. | <0·0001 | .. | .. | .. | .. | <0·0001‡ | .. | |
| Positive | 22 (5%) | 20 (13%) | 2 (1%) | .. | 2·06 (1·57–2·69) | 16 (12%) | 16 (27%) | 0 (0%) | .. | 1·51 (1·07–2·14) | |
| Negative | 189 (46%) | 59 (39%) | 130 (50%) | .. | 1 (ref) | 103 (77%) | 40 (68%) | 63 (84%) | .. | 1 (ref) | |
| Data missing | 203 (49%) | 73 (48%) | 130 (50%) | .. | .. | 15 (11%) | 3 (5%) | 12 (16%) | .. | .. | |
| Environmental surface-swabs SARS-CoV-2 PCR status | .. | .. | .. | 0·044 | .. | .. | .. | .. | 0·0069 | .. | |
| Positive | 23 (5%) | 13 (9%) | 10 (4%) | .. | 1·66 (1·09–2·55) | 15 (11%) | 12 (20%) | 3 (4%) | .. | 1·78 (1·21–2·62) | |
| Negative | 160 (39%) | 50 (33%) | 110 (42%) | .. | 1 (ref) | 94 (70%) | 34 (58%) | 60 (80%) | .. | 1 (ref) | |
| Data missing | 231 (56%) | 89 (59%) | 142 (54%) | .. | .. | 25 (19%) | 13 (22%) | 12 (16%) | .. | .. | |
Data are n (%) unless otherwise indicated. Relative risk estimates from the multivariable analyses were calculated with a modified Poisson regression using robust standard errors, accounting for household clustering, and adjusting for contact's sex assigned at birth, comorbidity status, vaccination pre-enrolment status, relationship status to the primary case, and ratio number of residents in the household to number of bedrooms.
p values for each covariate were calculated with the χ2 test accounting for household clustering, applying for all categories where data are available.
Contacts in the serial serology subcohort were deemed infected if PCR-positive or if they seroconverted.
Fisher's exact test was used to calculate p values.
33 (10%) of 344 surface samples from 103 households were PCR positive on day 0 (appendix p 20). SARS-CoV-2 presence on one or more household surfaces was associated with increased risk of infection among contacts (aRR 1·66, 95% CI 1·09–2·55; table 3). Contacts were also substantially more likely to have detectable SARS-CoV-2 RNA on their hands if household surfaces were contaminated (3·77, 1·27–11·14; appendix pp 18–19). However, presence of viral RNA on surfaces did not correlate with its detection on primary cases' hands (1·30, 0·53–3·15; data not shown). No household surface swabs were positive for viral culture.
Each of the vectors of transmission identified in the full household contacts' cohort (primary cases' hands, household surfaces, and contacts' hands) was corroborated in the serology subcohort, with similar correlations with transmission (table 3; appendix pp 18–19).
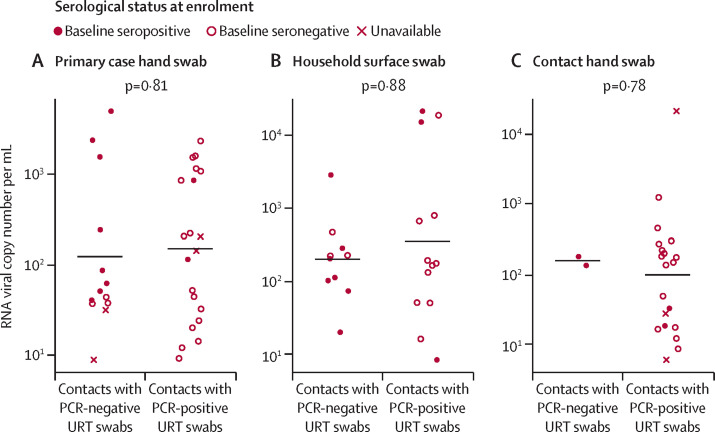
Of the 262 contacts whose URT samples remained PCR negative, 20 were linked to one or more PCR-positive swabs from household surfaces or residents' hands. Of the 152 contacts with PCR-positive URT samples, 42 were linked to one or more positive swabs from household surfaces or residents' hands (table 3, figure 2 ). We found no significant differences in the quantified RNA viral load in samples from any of the candidate vectors between the contacts with PCR-positive and PCR-negative URT samples (Figure 2).
Figure 2.
Measured SARS-CoV-2 RNA viral load in PCR-positive environmental samples, stratified by contacts' URT PCR status and baseline serological status
Only contacts linked to one or more PCR-positive environmental samples are depicted. (A) Primary cases' hand-swab RNA viral load (median 82·09 copies per mL, IQR 46·09–1082·29). (B) Household surface-swab RNA viral load (226·58, 101·38–602·51). (C) Contacts' hand-swab RNA viral load (117·66, 21·75–244·39). The measured environmental viral loads do not differ by contacts' URT PCR status, as shown by the p values in each panel (Mann-Whitney U test). All positive primary case hand swabs and surface swabs were collected on day 0, whereas contacts' hand swabs were positive on day 0 (n=16) and day 7 (n=6). Environmental samples with more than 5 RNA copies per mL were considered PCR positive. URT=upper respiratory tract.
18 of the 20 contacts with PCR-negative URT swabs that were linked to a PCR-positive household surface or residents' hand sample had serological data available, of whom 12 (67%) were seropositive at baseline (half of whom were recently vaccinated, including the two contacts whose URT samples remained PCR negative despite having PCR-positive hand swabs). By contrast, of the 37 (88%) of 42 contacts with PCR-positive URT samples who had serological results available, only five (13%) were seropositive at baseline (figure 2), significantly fewer than the contacts with PCR-negative URT samples linked to PCR-positive household surface or residents' hand samples (χ2 test p=0·0002).
Sensitivity analysis assessed the effect of incomplete recruitment of contacts within households on household infection rate (appendix p 9). 200 contacts were from 115 fully recruited households (ie, households where all residents were recruited), of which 72 were PCR positive and 128 PCR negative, giving a household infection rate of 36·0% (95% CI 29·7–42·9), very similar to that for the full household contacts' cohort (36·7%; table 2).
URT samples were collected from 72 of 152 PCR-positive household contacts alongside their epidemiologically linked primary case. Of these, 35 contacts and their 25 respective primary cases had adequate RNA viral load in their URT samples (ie, >200 genome copies per mL of virus transport media) to undergo WGS (figure 3 ; appendix p 12). Phylogenetic analysis indicated that all isolates clustered within their respective households (figure 3). In 20 of the 25 households, isolates from primary cases and contacts isolates were indistinguishable on WGS (figure 3). In three households, isolates from contacts had one or two additional non-synonymous mutations compared with isolates from the primary cases, and in two households, the isolates from the primary cases isolate had one non-synonymous mutation not detected in their contacts. These data indicate that the primary case–contact pairs were most likely infected with the same SARS-CoV-2 strain,19 strongly supporting household transmission as the source of infection in all infected contacts sequenced.
Figure 3.
Genomic evidence of SARS-CoV-2 transmission among households
Genomic analysis of 25 households (comprising 35 contacts and 25 of their respective epidemiologically linked primary cases) of non-synonymous mutations within the SARS-CoV-2 genome, to assess whether the same or different strains are present within the household. Genomic analyses were performed where upper respiratory tract swabs with viral loads of more than 200 copies per mL were available for both primary cases and contact(s).
Discussion
Rigorous assessment of risk factors and vectors of transmission necessitates maximising the capture of transmission events prospectively in epidemiologically well defined households. Our study design maximally captured transmission events by enrolling asymptomatic and symptomatic, recently exposed household contacts of newly diagnosed primary cases with COVID-19 and proactively detecting infection by repeated URT PCR testing. In a subcohort, longitudinal serology was conducted to detect contacts whose URT samples were PCR negative and seroconverted and exclude baseline seropositive, previously infected contacts. We also simultaneously tested for the presence of SARS-CoV-2 RNA on frequently touched household surfaces and residents' hands. Although other studies, with smaller sample sizes, have reported the presence of SARS-CoV-2 RNA on surfaces in households or other settings,3, 4, 20 this study is, to our knowledge, the first to identify correlations between SARS-CoV-2 RNA on candidate vectors with contacts' infection risk. These longitudinal empirical data from a real-world community setting with rigorously defined infection outcomes substantially advance our understanding of SARS-CoV-2 household spread, the setting of most transmission globally.
We report a household infection rate of 51·8% in the full household contacts' cohort and 73·5% in the serology subcohort for contacts exposed to alpha variant-infected primary cases, which is one of the highest overall household infection rates for SARS-CoV-2 reported in predominately unvaccinated and previously unexposed contacts.1 This finding probably reflects our rigorous, intensive sampling approach, which captured a higher proportion of transmission to susceptible contacts than previous studies.21, 22, 23
Unlike some previous studies,22, 24 but consistent with others,25 primary case SARS-CoV-2 RNA viral load in URT samples did not correlate with contacts' risk of infection in our cohort. Interestingly, however, primary cases' RNA viral load in URT samples did predict their risk of having a SARS-CoV-2 PCR-positive hand swab, which in turn was associated with transmission to their respective contacts. Presence of viral RNA on frequently touched household surfaces strongly associated with contacts' risk of having a PCR-positive hand swab and predicted contacts' risk of infection. Moreover, presence of SARS-CoV-2 RNA on contacts' hands correlated with presence of viral RNA on their respective primary cases' hands and with contacts' own risk of SARS-CoV-2 infection.
Interestingly, in six contacts with incident infection, three had SARS-CoV-2 PCR-positive hand swabs, and another three had PCR-positive surfaces in their households, before their URT swabs became PCR positive. Because the collection of URT swabs involved sampling both the nasal turbinates and pharynx, it is unlikely that SARS-CoV-2 infection was missed on earlier samples due to anatomical compartmentalisation.26 SARS-CoV-2 was cultured from one of these contacts' hand swabs, showing the presence of live, infectious virus on the hands of this individual. WGS confirmed that the viral isolates from the contact's hand and URT were indistinguishable. Conversely, in non-incident contacts who already had PCR-positive URT samples at enrolment, the corresponding PCR-positive hand and household surface swabs were also consistently positive at enrolment.
Among all contacts with one or more positive candidate vector samples, no differences were observed in the RNA viral load quantified on these vectors between contacts with PCR-positive and PCR-negative URT samples, suggesting that the source of the SARS-CoV-2 RNA on these vectors was unlikely to be retrograde contamination from the contacts' URT. Notably, contacts with PCR-negative URT samples and with viral RNA detected on their hands, their primary cases' hands, or household surfaces, predominantly had pre-existing immunity, as evidenced by being seropositive at enrolment. By contrast, very few contacts with PCR-positive URT samples linked to positive hand swabs or surface swabs in their households were seropositive at enrolment. This finding suggests that the presence of SARS-CoV-2 RNA on candidate vectors linked to uninfected contacts with PCR-negative URT samples might not have resulted in infection because of their pre-existing immunity.
The potential modes of SARS-CoV-2 transmission are the subject of ongoing scientific controversy.2, 27 To our knowledge, our data are the first empirical evidence to correlate presence of SARS-CoV-2 RNA on candidate vectors with contacts' risk of infection. However, our findings do not prove causality. SARS-CoV-2 spread is believed to be predominantly through airborne transmission,28, 29, 30 which must be the sole route in several settings, including those involving long-range transmission.2 The transmission we observed could therefore have been exclusively airborne and the presence of SARS-CoV-2 RNA on residents' hands and household surfaces might reflect bystander contamination by large respiratory droplets that might be generated alongside infectious aerosols that mediate transmission. We did not systematically sample air in the households so we cannot exclude this possibility.
To demonstrate causality of either fomite or airborne transmission, a highly controlled artificial experiment would be required, in which SARS-CoV-2-naive, healthy, sentinel participants would be confined in an environment previously contaminated by an infectious-case. Experimental interventions testing the effects of complete air exchange or decontamination versus surface decontamination before the participants' confinement would then be correlated with their subsequent infection status. Although such an experimental design might confirm the route of transmission, it would not reflect the real-world setting of community transmission with its natural spectrum of environmental conditions and host factors.
Only one of our candidate vector samples, a contact's hand swab, was positive for viral culture. The very high proportion of SARS-CoV-2 RNA-positive but viral culture-negative swabs probably reflects viral degradation during the 36–72 h interval to transport the swabs from participants' homes and laboratory storage, while RT-PCR was performed, before they were subjected to in vitro cell culture. This explanation is supported by the recent observation that similar swabs processed in the same laboratory with the same cell culture assay by the same scientist, but within 12 h, yielded a much larger proportion of culture-positive samples.31
Our study has several limitations. We cannot exclude that in a proportion of primary case–contact pairs where the prevalent infected contact might have been asymptomatic or pre-symptomatic, the contact might have been the initial source of transmission, or that the contact might have been infected from outside of the household. These scenarios are unlikely for most of the households, because our assignment of case–contact pairs is underpinned by epidemiological linkage between each primary case and their respective contact(s), and further corroborated by WGS of SARS-CoV-2 isolates from primary cases and their contacts in all 25 households where this was possible. Similarly, although some of the contacts' infections could possibly have been tertiary rather than secondary to the primary case, we consider this unlikely because contacts were enrolled on average 3 days after symptom onset of the primary case. The probability of infection arising from outside the household was low because London was in a state of lockdown during our study, resulting in very limited social or occupational contact outside of households. A corollary of this point, however, is that our household infection rate estimate might not be generalisable to households in settings where societal non-pharmaceutical interventions are not in force with correspondingly less contact time between cohabitants.
We did not systematically collect behavioural information on hygiene practices and physical distancing, as preventative measures typically took place after primary case symptom onset, when exposure had already occurred, limiting the effect of such practices on reducing contacts' risk of infection. Additionally, the collection of such behavioural data would have been biased by the participants' conditioning of the underlying circumstances (eg, lockdown and participation in an observational study) and would not represent their usual practices.
The households that consented to participate might not be representative of the general population because non-White ethnicities and older age groups were under-represented. In some households, only a proportion of cohabitants were recruited; however, infection rate in households where all contacts were recruited approximated the infection rate in the full household contact cohort. Although the study could be biased by the difference between household-visited contacts and remotely recruited contacts, these groups were similar.
Our study began when the pre-alpha lineages were dominant in London and continued until the alpha variant became dominant. Our results cannot therefore be directly extrapolated to the more recent and more infectious variants. Interestingly, some recent evidence suggests that the omicron variant BA.1 is significantly more stable than ancestral SARS-CoV-2 on different surfaces, potentially increasing its propensity for transmission by the fomite route,27, 32 whereas other evidence suggests increased likelihood for aerosol transmission relative to earlier variants.33, 34
Our study provides the first empirical evidence to correlate presence of SARS-CoV-2 on candidate vectors with risk of infection in household contacts. These findings from a real-world community setting substantially advance our understanding of SARS-CoV-2 household transmission, the setting of most transmission globally. Our results also have practical implications and support interventions such as frequent handwashing, surface cleansing, physical distancing, reducing direct contact, and use of masks to curb transmission in households. As governments worldwide develop policies to manage successive new COVID-19 variants that transmit widely through vaccinated populations,10 alternative interventions to impede transmission are urgently needed. The straightforward public health interventions and messaging underpinned by our evidence provide a timely, pragmatic component of the future toolkit for living safely with COVID-19.
Data sharing
An anonymised, deidentified version of the dataset can be made available upon request to the corresponding author to allow all results to be reproduced.
Declaration of interests
NMF reports grants from the UK Medical Research Council, UK National Institute of Health and Care Research, UK Research and Innovation, Community Jameel, Janssen Pharmaceuticals, Bill & Melinda Gates Foundation, and Gavi, the Vaccine Alliance; consulting fees from the World Bank; payment or honoraria from the Wellcome Trust; travel expenses from WHO; advisory board participation for Takeda; and is a senior editor of the eLife journal. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
This work is supported by the NIHR Health Protection Research Units (HPRU) in Respiratory Infections (NIHR200927), in partnership with the UK Health Security Agency. JZ and WSB are supported by funding provided through PROTECT COVID-19 National Core Study and Genotype-to-Phenotype UK National Virology Consortium (G2P-UK), funded by the Medical Research Council (MRC; MR/W005611/1), which also partially funded this work. AS is supported by an NIHR academic clinical lectureship. PSF and MAC are supported by the UK Dementia Research Institute. JD is supported by the NIHR HPRU in Emerging and Zoonotic Infections. GPT is supported by the Imperial NIHR Biomedical Research Centre. NMF is supported by UK Research and Innovation (UKRI) MRC (Centre for Global Infectious Disease Analysis [MR/R015600/1] and MRC UKRI/DHSC NIHR COVID-19 rapid response call [MR/V038109/1]), NIHR HPRU in Modelling and Health Economics [NIHR200908], and philanthropic funding from Community Jameel. We thank all the participants who were involved in the study, UK Health Security Agency staff for facilitating recruitment into the study, and the staff of the Virus Reference Department for performing PCR and sequencing assays. We also thank Tamara Hopewell, Holly Grey, Jessica Grey, and Niamh Nichols for providing further logistical support. Finally, we thank the Molecular Diagnostics Unit at Imperial College London.
The INSTINCT and ATACCC study group investigators
Contributors
Contributor Information
INSTINCT and ATACCC study group:
Jessica Cutajar, Valerie Quinn, Sarah Hammett, Eimèar McDermott, Constanta Luca, Kristel Timcang, Jada Samuel, Samuel Bremang, Samuel Evetts, Lulu Wang, Sean Nevin, Megan Davies, Chitra Tejpal, Mohammed Essoussi, Anjeli V Ketkar, Giulia Miserocchi, Harriet Catchpole, Anjna Badhan, Simon Dustan, Isaac J Day Weber, Federica Marchesin, Michael G Whitfield, John Poh, and Alexandra Kondratiuk
Supplementary Material
References
- 1.Madewell ZJ, Yang Y, Longini IM, Jr, et al. Factors associated with household transmission of SARS-CoV-2 an updated systematic review and meta-analysis. JAMA Netw Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.22240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang CC, Prather KA, Sznitman J, et al. Airborne transmission of respiratory viruses. Science. 2021;373 doi: 10.1126/science.abd9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cordery R, Reeves L, Zhou J, et al. Transmission of SARS-CoV-2 by children to contacts in schools and households: a prospective cohort and environmental sampling study in London. Lancet Microbe. 2022;3:e814–e823. doi: 10.1016/S2666-5247(22)00124-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcenac P, Park GW, Duca LM, et al. Detection of SARS-CoV-2 on surfaces in households of persons with COVID-19. Int J Environ Rese Public Health. 2021;18 doi: 10.3390/ijerph18158184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams CM, Pan D, Decker J, et al. Exhaled SARS-CoV-2 quantified by face-mask sampling in hospitalised patients with COVID-19. J Infect. 2021;82:253–259. doi: 10.1016/j.jinf.2021.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou J, Otter JA, Price JR, et al. Investigating severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) surface and air contamination in an acute healthcare setting during the peak of the coronavirus disease 2019 (COVID-19) pandemic in London. Clin Infect Dis. 2020;73:e1870–e1877. doi: 10.1093/cid/ciaa905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cevik M, Marcus JL, Buckee C, Smith TC. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission dynamics should inform policy. Clin Infect Dis. 2021;73(suppl 2):S170–S176. doi: 10.1093/cid/ciaa1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Real LA, Biek R. Infectious disease modeling and the dynamics of transmission. Curr Top Microbiol Immunol. 2007;315:33–49. doi: 10.1007/978-3-540-70962-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of COVID-19 vaccines against the B.1.617.2 (delta) variant. N Engl J Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singanayagam A, et al. Community transmission and viral load kinetics of the SARS-CoV-2 delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: a prospective, longitudinal, cohort study. Lancet Infect Dis. 2022;22:183–195. doi: 10.1016/S1473-3099(21)00648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Accorsi EK, Qiu X, Rumpler E, et al. How to detect and reduce potential sources of biases in studies of SARS-CoV-2 and COVID-19. Eur J Epidemiol. 2021;36:179–196. doi: 10.1007/s10654-021-00727-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madewell ZJ, Yang Y, Longini IM, Jr, Halloran ME, Dean NE. Household transmission of SARS-CoV-2: a systematic review and meta-analysis. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.31756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kretzschmar ME, Rozhnova G, Bootsma MCJ, van Boven M, van de Wijgert JHHM, Bonten MJM. Impact of delays on effectiveness of contact tracing strategies for COVID-19: a modelling study. Lancet Public Health. 2020;5:e452–e459. doi: 10.1016/S2468-2667(20)30157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.GOV.UK; English indices of deprivation 2019: mapping resources. 2019. https://www.gov.uk/guidance/english-indices-of-deprivation-2019-mapping-resources
- 15.Rowan AG, May P, Badhan A, et al. Optimized protocol for a quantitative SARS-CoV-2 duplex RT-qPCR assay with internal human sample sufficiency control. J Virol Methods. 2021;294 doi: 10.1016/j.jviromet.2021.114174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosadas C, Khan M, Parker E, et al. Detection and quantification of antibody to SARS-CoV-2 receptor binding domain provides enhanced sensitivity, specificity and utility. J Virol Methods. 2022;302:114475. doi: 10.1016/j.jviromet.2022.114475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donner A, Banting D. Adjustment of frequently used chi-square procedures for the effect of site-to-site dependencies in the analysis of dental data. J Dent Res. 1989;68:1350–1354. doi: 10.1177/00220345890680091201. [DOI] [PubMed] [Google Scholar]
- 18.Zou G. A modified poisson regression approach to prospective studies with binary data. Am J Epidemiol. 2004;159:702–706. doi: 10.1093/aje/kwh090. [DOI] [PubMed] [Google Scholar]
- 19.Meredith LW, Hamilton WL, Warne B, et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis. 2020;20:1263–1272. doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shragai T, Pratt C, Castro Georgi J, et al. Household characteristics associated with surface contamination of SARS-CoV-2 and frequency of RT-PCR and viral culture positivity-California and Colorado, 2021. PLoS One. 2022;17 doi: 10.1371/journal.pone.0274946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis NM, Chu VT, Ye D, et al. Household transmission of SARS-CoV-2 in the United States. Clin Infect Dis. 2020;73:1805–1813. doi: 10.1093/cid/ciaa1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerami C, Popkin-Hall ZR, Rapp T, et al. Household transmission of severe acute respiratory syndrome coronavirus 2 in the United States: living density, viral load, and disproportionate impact on communities of color. Clin Infect Dis. 2022;74:1776–1785. doi: 10.1093/cid/ciab701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holt H, Talaei M, Greenig M, et al. Risk factors for developing COVID-19: a population-based longitudinal study (COVIDENCE UK) Thorax. 2022;77:900–912. doi: 10.1136/thoraxjnl-2021-217487. [DOI] [PubMed] [Google Scholar]
- 24.Marks M, Millat-Martinez P, Ouchi D, et al. Transmission of COVID-19 in 282 clusters in Catalonia, Spain: a cohort study. Lancet Infect Dis. 2021;21:629–636. doi: 10.1016/S1473-3099(20)30985-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian D, Lin Z, Kriner EM, et al. Ct values do not predict severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmissibility in college students. J Mol Diagn. 2021;23:1078–1084. doi: 10.1016/j.jmoldx.2021.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Killingley B, Mann AJ, Kalinova M, et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat Med. 2022;28:1031–1041. doi: 10.1038/s41591-022-01780-9. [DOI] [PubMed] [Google Scholar]
- 27.Hirose R, Itoh Y, Ikegaya H, et al. Differences in environmental stability among SARS-CoV-2 variants of concern: both omicron BA.1 and BA.2 have higher stability. Clin Microbiol Infect. 2022;28:1486–1491. doi: 10.1016/j.cmi.2022.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Azimi P, ZKeshavarz Z, Cedeno Laurent JG, Stephens B, Allen JG. Mechanistic transmission modeling of COVID-19 on the Diamond Princess cruise ship demonstrates the importance of aerosol transmission. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2015482118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenhalgh T, Jimenez JL, Prather KA, Tufekci Z, Fisman D, Schooley R. Ten scientific reasons in support of airborne transmission of SARS-CoV-2. Lancet. 2021;397:1603–1605. doi: 10.1016/S0140-6736(21)00869-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alsved M, Nygren D, Thuresson S, Medstrand P, Fraenkel CJ, Löndahl J. SARS-CoV-2 in exhaled aerosol particles from covid-19 cases and its association to household transmission. Clin Infect Dis. 2022;75:e50–e56. doi: 10.1093/cid/ciac202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J, Singanayagam A, Goonawardane N, et al. Viral emissions into the air and environment after SARS-CoV-2 human challenge: a phase 1, open label, first-in-human study. Lancet Microbe (in press). [DOI] [PMC free article] [PubMed]
- 32.Chin AWH, Lai AMY, Peiris M, Man Poon LL. Increased stability of SARS-CoV-2 omicron variant over ancestral strain. Emerg Infect Dis. 2022;28:1515–1517. doi: 10.3201/eid2807.220428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riediker M, Briceno-Ayala L, Ichihara G, et al. Higher viral load and infectivity increase risk of aerosol transmission for delta and omicron variants of SARS-CoV-2. Swiss Medical Weekly. 2022;152 doi: 10.4414/smw.2022.w30133. [DOI] [PubMed] [Google Scholar]
- 34.Lai J, Coleman KK, Tai SHS, et al. Exhaled breath aerosol shedding of highly transmissible versus prior severe acute respiratory syndrome coronavirus 2 variants. Clin Infect Dis. 2022;76:786–794. doi: 10.1093/cid/ciac846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
An anonymised, deidentified version of the dataset can be made available upon request to the corresponding author to allow all results to be reproduced.