Abstract
The number of approved or investigational late phase viral vector gene therapies (GTx) has been rapidly growing. The adeno-associated virus vector (AAV) technology continues to be the most used GTx platform of choice. The presence of pre-existing anti-AAV immunity has been firmly established and is broadly viewed as a potential deterrent for successful AAV transduction with a possibility of negative impact on clinical efficacy and a connection to adverse events. Recommendations for the evaluation of humoral, including neutralizing and total antibody based, anti-AAV immune response have been presented elsewhere. This manuscript aims to cover considerations related to the assessment of anti-AAV cellular immune response, including review of correlations between humoral and cellular responses, potential value of cellular immunogenicity assessment, and commonly used analytical methodologies and parameters critical for monitoring assay performance. This manuscript was authored by a group of scientists involved in GTx development who represent several pharma and contract research organizations. It is our intent to provide recommendations and guidance to the industry sponsors, academic laboratories, and regulatory agencies working on AAV-based GTx viral vector modalities with the goal of achieving a more consistent approach to anti-AAV cellular immune response assessment.
Keywords: Adeno-associated virus, AAV, Cellular immune response, anti-AAV immunogenicity
Introduction
The past 25 years have witnessed numerous advances in the development of gene therapies as these products have moved from the investigational stage to commercial approval. With these advances has come the need to better understand the impact host immune responses may play in the safety, efficacy, and long-term durability of viral vectored gene therapies. Presently, gene therapy (GTx) is often developed to treat monogenic disorders by introducing DNA into cells to compensate for missing, poorly expressed, or mutated genes. The most common approach is to use a viral vector (e.g., adeno-associated virus, AAV) as a carrier for the transgene cassette which encodes a replacement for the deficient gene of interest which is delivered through a process referred to as vector transduction. Though some AAV serotypes have a notable “tropism” or propensity to deliver the transgene to a desired target tissue, transgenes are distributed to a variety of tissues following systemic administration of viral vectors. In these circumstances, tissue-specific or tissue-restricted promoter and enhancer elements incorporated into the transgene cassette are useful to ensure that the gene is expressed predominantly in the desired tissue. Alternatively, these promoter and enhancer elements may allow ubiquitous expression in a variety of cell or tissue types (if not in all cell types).
Gene therapies present unique challenges to immunogenicity assessment. In addition to the immune response to the viral carrier, treatment with GTx results in the expression of transgene proteins that have the potential to elicit immune responses. Immune responses to gene therapy products may compromise efficacy and patient safety. Thus, regulatory guidelines imply that immunogenicity should be monitored throughout drug development (1-4) and sufficient information should be provided to support label claims regarding durability, as needed.
The host immune system has many innate and adaptive mechanisms for contending with threats from intracellular pathogens. Most relevant to the long-term efficacy of AAV vector-mediated gene therapies may be the cytotoxic cell-mediated immune response. Cytotoxic lymphocytes most often consist of activated CD8 + T cells and natural killer (NK) cells; however, CD4 + cells, particularly of the Th1 phenotype, also have caused cytotoxicity under certain circumstances (5, 6). Both CD4 + and CD8 + T cells are Ag-specific and activated following recognition of Ag-derived peptides presented at the cell surface by the major histocompatibility complex (MHC; MHC class I presents to CD8 + T cells, and class II presents to CD4 + T cells) and represent the adaptive arm of the immune response. In clinical trials using intramuscular delivery, the rAAV-mediated transgene expression has been observed due to induction of regulatory T cells (Tregs). Both CD4 + and CD8 + T cells expressing forkhead box P3 (FOXP3; the transcription factor associated with Treg) have been associated with suppression of capsid-specific T cell responses. Additionally, the Treg-mediated cytokines can suppress the inflammatory effects of CD4 + and CD8 + T cells to enable AAV-mediated gene transduction (7, 8). Natural killer cells represent innate mechanism of response and target pathogen-infected or transformed cells through the expression of multiple families of activating and inhibitory surface receptors. Both T cells and NK cells can secrete interferon-γ (IFN-γ), tumor necrosis factor (TNF), and other pro-inflammatory cytokines upon stimulation. Importantly, IFN-γ secretion by stimulated CD4 + T cells may be more indicative of T cells helping the developing B cell (humoral or antibody) response and not necessarily associated with cellular cytotoxicity.
Due to the complex nature of gene therapy products, the evaluation of immunogenicity would include monitoring both humoral and cellular immune responses. Unlike protein therapeutics, it may be important to describe cellular immunity as part of the immunogenicity assessment for gene therapy products. Of many mechanisms of innate and adaptive immune response against AAV vector components, the cytotoxic cell-mediated immunogenicity could have the longest term pharmacodynamic impact on the efficacy of treatment. Data collected in early studies of systemic delivery of gene therapies for the treatment of liver diseases led to the hypothesis that AAV capsid protein-specific cellular immune responses, manifested in the form of cytotoxic CD8 + T cells, likely target transduced hepatocytes (9). Cytolysis of transduced cells then leads to the release of transaminases, including alanine aminotransferase (ALT) and ultimately loss of the transgene expression (9). The earliest clinical trials for AAV vector-mediated gene therapy for hemophilia B made the initial observation that an increase in liver transaminases could be correlated with a loss of transgene (factor IX) expression and that concomitant AAV capsid-specific cellular immune response could be detected temporally at related time points (10). This was the first study to suggest that immune suppression may be effective to ameliorate the immune response and prevent cell killing. This hypothesis was later put to the test in a second clinical trial, also in hemophilia B, where further observations were made related to increased ALT, loss of factor IX expression, and detection of cellular responses (11). Here, a tapering course of glucocorticoid (prednisolone) was associated with a decline in ALT and stabilization of protein expression. Similarly, AAV capsid-specific cellular responses were no longer detectable at later time points (11). Together, these data suggested that an AAV capsid-specific cellular immune response likely targeted transduced target tissue (e.g., liver) inducing cellular cytotoxicity, release of liver transaminases, and loss of transgene protein expression, symptoms that could be reduced by prednisolone treatment. It was proposed that cytotoxic T cell responses may eliminate cells containing material transferred by the gene therapy vector and cause cytotoxicity, although such responses may not be consequential for all gene therapies (12).
Measurement of cellular response to AAV capsid proteins could help to determine whether the cellular immune response is involved in certain adverse events (AEs) or loss of efficacy and evaluate potential strategies to reduce the cellular immune response and/or its impact. A risk-based immune monitoring strategy during clinical development can be proposed and shared as part of the pre-investigational new drug (IND) and IND meetings.
Previous work has shown pre-existing cellular immunity to the viral capsids in both human and non-human species (9). However, the predictive value of such a response on safety or transduction efficiency is not clear, and the value provided may be minimal. Implementation of a cellular immunity-based patient inclusion/exclusion criteria can be challenging and should include considerations related to indication type, patient medical history, and information about potential impact of cellular immunity against AAV on specific disease safety and efficacy. The manuscript is not intended to discuss the application of AAV-specific cellular immunity as part of patient treatment eligibility criteria, nor cellular immune responses against transgene proteins encoded by the AAV vector.
This manuscript aims to present considerations related to the value and methodologies for the monitoring of cellular immune responses against AAV capsid proteins during clinical studies. The review is focused on the application of the enzyme-linked immunosorbent spot (ELISpot) platform as it is currently the most broadly applied technology to investigate cellular immunity in patients while providing a comparison of ELISpot and other available technologies.
Analysis of Correlation Between Humoral and Cellular Immune Responses and Application to AAV GTx
Background Information on Humoral and Cellular Immune Response to AAV Viral Vectors
Humoral and cellular arms of the adaptive immune system are intricately connected and functionally complement each other. Antibody-based humoral immunity results in direct inactivation of infectious pathogens as well as blockage of cellular uptake of a pathogen, both of which contribute to the host immune defense. Cellular immune responses can eliminate and remove infected cells, thereby preventing further production and replication of infectious agents. An important mediator between cellular and humoral immune mechanisms is CD4 + helper T cells that provides essential signaling for effector CD8 + T cells and B cells differentiating into antibody-producing plasma cells. Because of this linkage, a pronounced humoral response may correlate with robust cellular immunity.
A possible use of animal models for the purpose of predicting clinical impact has been actively investigated although it should be noted that a robust cellular immune response to the AAV capsid has rarely been observed in monkeys. Studies in animal models of AAV-vectored gene transfer have often failed to recapitulate immune-related toxicities observed in clinical trials, indicating that the predictive power of preclinical studies to identify the immune-mediated limitations of in vivo gene transfer with AAV vector in the clinic is limited (13). Reported data implicating cytotoxic cellular immune responses in targeting AAV-transduced tissues (10, 11, 14, 15) have been difficult to replicate in both mice and non-human primates (NHP), the two small and large animal models commonly used to assess toxicity preclinically. In NHP, AAV capsid-specific cytotoxic T lymphocytes (CTL) derived from prior AAV8 infection were ineffective in targeting or eliminating AAV8-transduced hepatocytes (16), and in a separate study, AAV2-specific CTL were able to lyse transduced cell lines in vitro but were unable to target transduced hepatocytes in vivo (17). This suggests that cross presentation of AAV-derived peptides by transduced hepatocytes was not sufficient to cause their elimination by AAV2-specific CTL (18).
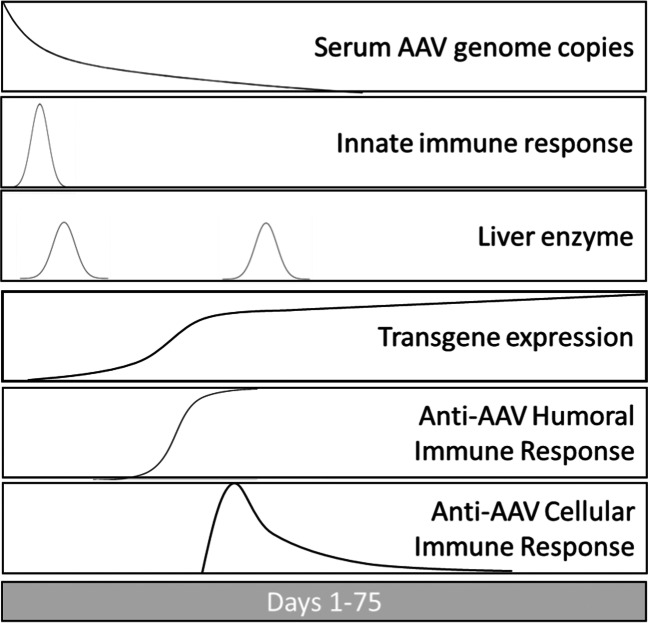
Much of what is known about humoral and cellular immune responses in humans comes from vaccine research. For many of the most effective licensed vaccines, the best correlates of protection from an infectious disease are sustained serum antibody titers to a pathogen and the development of persisting T cell and B cell memory (19, 20). Relatively high variability of protocols designed to detect cytotoxic T cells complicates direct comparison with humoral immune response. In addition, the biology of cellular immune response is highly complex. Effector T cells responsible for killing infected cells typically undergo three stages of development, including activation/expansion, contraction, and differentiation into memory cells. After the initial stage of activation and expansion, most effector T cells die, but a small percentage (under 10%) persists and matures into effector memory or central memory T cells (21). Unlike detection of antibody-producing plasma cells, there is a relatively narrow window for detection of activated T cells (see Fig. 1). Central memory T cells are less predominant and harder to detect. Effector memory T cells traffic in blood to non-immune tissues and are thought to be detected by most ELISpot protocols (22). Central memory T cells reside in lymphoid organs and are slow to respond to antigen.
Fig. 1.
Representative kinetics of systemic AAV vector administration, immune responses, and AAV gene expression. Each panel shows a representative curve after hypothetical AAV vector administration. The y-axis is expressed in arbitrary units; the x-axis begins at initial vector infusion and spans approximately 75 days. Serum genome copies are expected to decay after infusion. Innate immune responses are expected to peak soon after infusion. Liver enzyme elevation indicating liver damage, if expected, could occur after initial AAV exposure and/or after AAV gene expression and adaptive immune response
Clinical associations between humoral and cellular immune responses to AAV-based therapeutics have been assessed for several viral serotypes. Representative examples are briefly described below.
AAV Viral Vector Encoding a Micro-dystrophin Transgene
Duchenne muscular dystrophy patients were treated with AAV2.5 capsid vector containing micro-dystrophin encoding transgene (23). The vector was administered via intramuscular (IM) injection at doses ranging between 6 × 1011 and 3 × 1012 vector genomes (vg) accompanied with an immunosuppressive treatment. The laboratory-derived AAV2.5 vector version was generated by introducing mutations in the AAV2 capsid. During the study, samples collected from one of the trial subjects generated positive signals in the ELISpot assay, although the response appeared to be sporadic. There were no significant T cell responses to AAV2.5 capsid Ags detected in other patients. In addition to the ELISpot method, the presence of CD8 + cellular infiltrates was evaluated in muscle biopsies. Samples were stained with anti-CD8 + antibodies and showed no statistically significant difference in the counts of CD8 + cells between samples collected from AAV- and placebo-treated patients. The presence of pre-existing humoral immunity to AAV2.5 was not an exclusion criterion in this study with the rationale that the relevance of in vitro assays of vector neutralization before intramuscular administration is unknown due to a lack of previous clinical data. Samples collected from trial subjects were screened for neutralizing antibodies to wild-type AAV2, and the laboratory generated AAV2.5 serotype vectors using an in vitro transduction protocol. Pre-existing NAbs were detected in two subjects to both AAV2 and AAV2.5, with baseline titers of 1:800 and 1:100, respectively. Samples collected from other subjects were either negative (< 1:2) or produced low (1:4) titers. Significant increases in humoral responses were observed after AAV2.5 administration. NAb titers increased from weeks 2 to 6 and ranged from 50- to 1000-fold over the baseline titers. This included the subject that developed transient cellular immune response. Overall, no clear association between humoral and cellular anti-AAV2.5 responses was reported.
AAV Viral Vector Encoding Lipoprotein Lipase Transgene
Humoral and cellular immune responses against alipogene tiparvovec (Glybera®) were evaluated in clinical trials (24). Alipogene tiparvovec (Glybera®) is an AAV1-based GTx therapeutic designed for the treatment of lipoprotein lipase (LPL) deficiency. The vector was administered via IM injection at doses ranging between 1 × 1011 and 1 × 1012 vector genome copies (gc) per kg (vc/kg) with or without immunosuppressive treatment, dependent on the study, started shortly before vector administration. Of the 26 patients admitted to the AT treatment, 15 had pre-existing anti-AAV1 antibodies. While all AT-treated patients developed anti-AAV1 antibody responses, variable and dose-dependent cellular immune response against alipogene tiparvovec (Glybera®) AAV1 therapeutic was reported (24-26). No apparent relationship was found between pre-existing AAV1 Abs and LPL expression after administration of the treatment: 7 of the 11 patients with pre-existing anti-AAV1 Abs had LPL expression in the biopsy vs. 4 of the 7 patients with no such Abs. Anti-AAV1 total Ab (TAb) response was detectable at 1 to 2 weeks after administration. Titers remained stable over the observation period (up to 52 weeks). There was no apparent difference in anti-AAV1 Ab response between studies and dose cohorts suggesting that dose or immunosuppressive regime did not influence anti-AAV1 TAb formation. Transient T cell activation was observed in several subjects, irrespective of whether immune suppression was applied. Moderate and non-persistent T cell responses to AAV1 capsid were observed in several subjects. Overall, no clear correlation was reported between pre-dose TAb-positive status and induction of CD8 + response in AT-treated patients.
AAV Viral Vector Encoding Factor VIII Transgene
Long et al. (27) assessed humoral and cellular immune response against the investigational AAV5-based therapeutic valoctocogene roxaparvovec (BMN 270, Roctavian®), intended for treatment of hemophilia A. The vector was administered via single infusion at doses ranging between 6 × 1012 and 6 × 1013 vgs per kg (vg/kg) accompanied by an immunosuppressive treatment started after vector administration. Patients were screened for pre-existing anti-AAV5 antibody using TAb and neutralizing (transduction inhibition TI) antibody detection methods. Only patients who screened double-negative were admitted to treatment. All subjects seroconverted to AAV5 TAb positive by week 8, the first time point assessed post-dosing. Titers remained stable during the observation period (up to 3 years). AAV5 TAb response was not associated with efficacy or safety signals. Peripheral blood mononuclear cell (PBMC) samples collected for up to 3 years following administration were tested for anti-AAV5 and transgene protein-specific cellular immune responses. All patients tested negative for the AAV5 cellular immune responses at baseline with several (4 of 15) becoming positive at a single time point post-administration while returning to negative status at all subsequent time points. Overall, no clear relationship between vector infusion-driven TAb response and T cell responses to AAV5 was reported.
In summary, these case studies suggest no definitive correlation between humoral and cellular (primarily CD8 + T cell-mediated with certain AAV serotypes) anti-AAV responses in clinical GTx studies.
Examples of Anti-AAV Cellular Immune Response Assessment in Clinical Studies
The most common adverse event reported following systemic administration of AAV gene therapies has been a transient increase in liver transaminases, ALT, and aspartate aminotransferase (AST), which is often managed using immune-suppressing corticosteroids (28). This has been the impetus for a widely held hypothesis, based on previously published gene therapy clinical trial data, that a capsid-specific cellular immune response may be stimulated by catabolized capsid antigens associated with transduced tissues (10). Consequently, these transduced tissues could become transient targets for cellular cytotoxicity, resulting in a release of transaminases and loss of transgene expression (10, 29-31). The time course for degradation and presentation of capsid antigen on MHC has not been well studied, but the inherent stability of the AAV capsid and the sequestration in perinuclear and nuclear compartments may indicate a wide window for targeting AAV-transduced cells (32). Corticosteroid treatment in some of these studies reduced detectable concentration of transaminases and was associated with a loss of detectable cellular immune response and stabilization of transgene expression. However, the cellular immune response may not be the sole cause of liver transaminase increases in all cases (27).
Openly available information about cellular responses in clinical trials is limited. Examples include development of cellular immune response in AAV vector-treated hemophilia A and B, LPL deficiency, alpha-1-antitrypsin (AAT) deficiency, early-onset severe retinal dystrophy, and Duchenne muscular dystrophy patients (9-11, 23, 24, 27, 33-36). Summary of case studies briefly described below is presented in Table I.
Table I.
Case Studies of Cellular Immunogenicity in Clinical AAV Modality-Based Gene Therapy Studies
| Indication | Name | AAV serotype | Route | Cellular immune response type | Impact in clinical investigations | Reference |
|---|---|---|---|---|---|---|
| Lipoprotein lipase (LPL) deficiency | Alipogene tiparvovec (Glybera®) | AAV1 | Intramuscular injections | Moderate and non-persistent | No direct correlation with transgene expression | (24, 25, 37, 38) |
| Hemophilia B | AAV-2-FIX | AAV2 | Hepatic artery | Detected in evaluated subjects | Associated with loss of transgene expression | (10) |
| Hemophilia B | scAAV2/8-LP1-hFIXco | AAV8 | Peripheral vein infusion | Transient and dose dependent | Associated with loss of transgene expression | (11) |
| Hemophilia A | Valoctocogene roxaparvovec (Roctavian®) | AAV5 | Intravenous infusion | Sporadic | None | (27) |
| Alpha-1 antitrypsin (AAT) deficiency | rAAV1-CB-hAAT | AAV1 | Intramuscular injections | Persistent | No direct correlation with transgene expression | (33) |
| Retinal dystrophy associated with retinal pigment epithelium-specific protein mutation (RPE65) | tgAAG76 (rAAV 2/2.hRPE65p.hRPE65) | AAV2 | Subretinal injection | None | None | (34, 35) |
| Duchenne muscular dystrophy patients | rAAV2.5-CMV-minidystrophin (d3990) | AAV2.5 | Intramuscular injections | None | None | (23) |
These clinical studies involving AAV vector carrying various transgenes reported development of cellular immune response to viral capsid proteins and evaluated association of such response with various clinical signals, including changes in liver transaminase concentrations and decline in transgene protein expression. These events were assessed for the ability of cell-mediated immune response to destroy AAV-transduced hepatocytes.
Immune responses against alipogene tiparvovec (Glybera®), an AAV1-based therapeutic evaluated for treatment of LPL deficiency, were assessed in several clinical trials (24). Cellular responses observed in patients treated with AT were variable and dose dependent (24, 25, 39). For some of the patients, a positive T cell response was reported at a single time point only, while for others, positive responses were reported at two or more time points. Response incidences seem to vary between studies and in relation to the patient pre-treatment anti-AAV humoral response status. Anti-AAV1 cellular immune response was evaluated pre-treatment and on days 14, 28, 42, 56, 84, 98, 182, and 273 and 1 year after treatment. Positive responses were observed as soon as day 14 after treatment. Importantly, transgene protein (LPL) expression in patients evaluated for the presence of AAV1-specific T cell response was similar between groups of patients who were positive and negative for the cellular response (24). In separate studies investigating alipogene tiparvovec (Glybera®), patients were treated with immunosuppressive agents including cyclosporine, mycophenolate mofetil, and methylprednisolone. Incidence of cellular response in studies with and without immunosuppression was considered as comparable, showing no correlation with the transgene protein expression levels. Adverse reactions reported during AT trials were mostly deemed as related to the administration procedure and were transient in nature. An increase in creatine phosphokinase and C-reactive protein was reported in one patient and was correlated with low level anti-AAV1 cellular response. This subject tested positive for an anti-AAV T cell response at some but not all assessments during the study. Robust transgene expression was found in biopsy samples collected from the subject.
Anti-AAV1 capsid-specific cellular immune responses were evaluated in patients treated with AAT transgene carrying AAV1 vector (33). The vector was administered via IM injection at doses ranging between 6.9 × 1012 and 6.0 × 1013 vgs. Sustained expression of AAT was detected 1 year after the initial dose in patients treated at the highest dose (3.0 × 1013 vector genome particles per patient) with all subjects testing positive for the development of anti-AAV1 capsid-specific cellular immune response. Testing was conducted on pre-treatment; days 14, 30, 45, 60, 75, and 90; and 1 year after treatment. Positive responses were reported as early as day 14. Patients remained positive for anti-AAV1 cellular immune activity on day 90 or later. Overall, persistent expression of transgene protein in patients positive for anti-capsid T cell immune response was reported.
A dose-dependent response level of cellular immune response against AAV8 capsid proteins was reported in a study where patients were treated with AAV8 carrying factor IX transgene (11). While no significant AAV8 capsid-specific T cell response was reported in patients treated at the low dose (2 × 1011 vg/kg), a significant incidence of response was observed at the intermediate dose level (6 × 1011 vg per kilogram). The presence of anti-AAV8-specific T cells was observed in patients treated at the highest dose tested in the study (2 × 1012 vg per kilogram) which, in some cases, coincided with the increase in liver enzyme levels. Importantly, for most of the patients, T cell responses significantly subsided or became negative by study week 20 or later. No hepatocellular injury was reported suggesting that presence and levels of circulating capsid-specific T cells were not sufficient for immune-mediated clearance of vector-transduced cells (11).
Lack of cellular immune response against AAV2-based vector was reported in studies aiming to develop treatment for early-onset, severe retinal dystrophy caused by mutations in the gene encoding retinal pigment epithelium-specific 65 kDa protein (RPE65) (34, 35) by subretinal administration of GTx. The vector was administered via uniocular subretinal injection at 5.96 × 1010 or 1 × 1011 vgs per injection accompanied by an immunosuppressive treatment. Due to local administration to an immune-privileged site, low systemic exposure was expected. ELISpot method demonstrated lack of specific T cell activation following trial subjects’ PBMC co-culture with the AAV2 vector (34) or with a library of AAV2 peptide pools (35).
Similarly, no substantial cellular immune response was reported against AAV2.5 capsid in a trial of Duchenne muscular dystrophy patients in which IM administration of the vector was accompanied by an immunosuppressive treatment (23). The laboratory-derived AAV2.5 vector version was generated by introducing a limited number of mutations in the AAV2 capsid. Some of the samples from several subjects generated positive signal in the ELISpot assay although overall response appeared to be intermittent.
Long et al. reported on assessment of cellular immune response against experimental AAV5-based therapeutic valoctocogene roxaparvovec (Roctavian®) (27). Samples collected during first in human phase 1/2 study were tested for the presence of anti-AAV5 and transgene protein-specific cellular immune responses using IFN-γ and TNF FluoroSpot assay. Samples were collected at baseline and generally every 2–4 weeks during the first 6 months, 4–8 weeks through the rest of year 1, and every 3 months thereafter. While all patients (n = 15) tested negative for the AAV5-specific cellular response at the baseline, 4 of the 15 patients tested positive at a single time point post-administration and negative at all subsequent time points. Positive responses were observed on weeks 8, 36, and 44 and past 18 months after treatment. Overall, sporadic positive cellular immune responses were reported in several of the treated patients with no definitive relationship with vector infusion, reported safety events, or impact on efficacy of treatment. Pairwise comparative analysis of IFN-γ and TNF-α positive and negative responses with liver enzyme (ALT, safety signal) and factor VIII (FVIII) (efficacy signal) activity showed no clear and apparent association. Observed cellular immune response may have been impacted by the administration of corticosteroids during the first several weeks after gene transfer in most of the subjects enrolled. Separately, Patton et al. (36) reported that in the phase 3 study for valoctocogene roxaparvovec (Roctavian®), a transition was made from a well-characterized, multiplexed FluoroSpot assay used in the phase ½ study to a validated, colorimetric, single-endpoint IFN-γ ELISpot assay to focus testing on the most established cytokine-based assessment used as a marker for cellular immune responses specific for AAV (36). It was proposed that ELISpot is generally appropriate for the assessment of CD8 + T cell anti-AAV responses in the clinic. Analysis of data generated for the first 16 subjects showed most tested positive for cellular immune response with a peak incidence at week 2 following dose administration with response declining over time.
At the time when this manuscript is written, there are several approved in vivo AAV gene therapies, Luxturna®, Zolgensma®, Roctavian®, Hemgenix®, and Upstaza® (40-43). Zolgensma® (onasemnogene abeparvovec-xioi) is indicated to be dosed at 1.1 × 1014 vg/kg by intravenous infusion (40). The recommended dose for Hemgenix® (etranacogene dezaparvovec) is 2 × 1013 gc/kg by intravenous infusion (43), while Luxturna® (voretigene neparvovec) is indicated to be dosed at 1.5 × 1011 vg by subretinal injection (44). The recommended dose for Roctavian® (valoctocogene roxaparvovec) is 6 × 1013 vg/kg by single intravenous infusion (42), and Upstaza® (eladocagene exuparvovec) is indicated to be administered at total dose of 1.8 × 1011 vg by bilateral intraputaminal infusion (45). The cellular immune response to the Upstaza® was not measured.
Zolgensma® and Luxturna® package inserts do not discuss cellular immune response to the vector (40, 41). In the US Food and Drug Administration (FDA) Clinical Review Document (46) for Zolgensma®, it was noted that four subjects in study CL-101 experienced elevated aminotransferases. For 3 subjects, the elevation of aminotransferase levels appeared to correlate with a greater T cell response to AAV9, as indicated by the increased number of spots forming cells per 1 × 106 PBMCs. In 1 subject, the elevation of aminotransferase levels was not associated with a greater T cell response to AAV9 (46). For subjects who did not experience elevation of aminotransferases, increased T cell responses to AAV9 were observed in some cases. The reviewer concluded that “Responses measured from T cells in the blood may not be well-correlated with T cell responses in the liver.” (46). The European Public Assessment Report (EPAR) notes that after treatment, T cell-mediated immunogenicity against AAV9 was detected in all patients as measured by ELISpot. In patients with the highest ELISpot values, similar responses in the CHOP-INTEND (Children’s Hospital of Philadelphia Infant Test of Neuromuscular Disorders) score and motor milestone achievement were observed as compared to patients with lower ELISpot values. This suggests that there was no identifiable relationship between baseline and post-baseline ELISpot signal and efficacy (47). It should be noted that all subjects except the first subject in study AVXS-101-CL-101 were pre-treated with prednisolone (1 mg/kg/day) to reduce the risk of immune-mediated liver toxicity (47).
During the development of Luxturna®, limited cytotoxic T cell responses to either AAV2 vector capsid or transgene product RPE65 were reported in a phase 3 study (44). The FDA’s Summary Basis of Approval (SBA) notes that in study 101, there were no T cell responses to AAV capsid or RPE65 (n = 12). In study 102, 6 out of the 11 subjects had a low response at a single time point. In study 301 (n = 21), there were 2 subjects with a low response at a single time point and one subject with a medium response at a single time point. In study 302 (n = 9), there were 3 subjects with a low response, 1 subject with a medium response, and 1 subject with a high response (44). However, the definition of what is a low, medium, or high response is not clear and will vary from sponsor to sponsor, particularly in the absence of a uniform approach to standardization of ELISpot assays. There is no mention of a cellular immune response in the Luxturna® EPAR (48).
The Roctavian® EPAR states that treated patients were tested for cellular immune responses against AAV5 capsid and the FVIII transgene product using an IFN-γ ELISpot assay (42). AAV5 capsid cellular immune responses were detected beginning at week 2 following dose administration and often declined or reverted to negative over the first 52 weeks in most patients with available data. AAV5 capsid-specific cellular immune responses were associated with higher ALT values at matched time points. FVIII-specific responses were detected in fewer subjects, often sporadically at a single time point and reverting to negative in most patients. No association between FVIII cellular immune response and ALT or FVIII activity measures could be detected (42).
These examples demonstrate the need for a readily accessible and reliable biomarker strategy that will inform about the level of impact of cellular immunogenicity on safety and/or efficacy of AAV-based treatment. Serum transaminases and creatine kinase (CK) have been often viewed as indicators of liver and muscle damage, respectively, although the use of these safety biomarkers can be confounded by specific disease pathology. Efficacy biomarkers vary depending on the indication and may not be available for all conditions. It is difficult to draw a uniform inference regarding the impact of cellular immune responses on safety and efficacy of AAV-based treatment. Diversity of programs with available information, including diversity of the dose, use of immunosuppressive treatments, and route of administration, presents a barrier for making an overarching conclusion. An aligned strategy of sample collection is required to clearly demonstrate connection between cellular immune response and observed changes in biomarker levels. Without determining the cellular immune response-effect relationship, the selection of a threshold value for the cellular immune response with respect to a cause for concern, and potentially triggering an immunosuppressive treatment, may become arbitrary and not warranted. Access to data generated via aligned sampling can allow for a hypothesis-driven data analysis which may include understanding of the level of observed impact and associated mitigation strategy. A disconnect between clinical safety symptoms and the timing of relevant sample collection can impede the ability to perform a correlative analysis of the cellular immune response’s impact on clinical PD and efficacy.
Risk-Based Approach to Cellular Immune Response Assessment
The number of AAV-mediated gene therapies under investigation for the treatment of monogenic disorders has quickly increased over the past decade with many sponsored programs ranging across all stages of clinical development (49). Following several early studies in hemophilia B where cellular immune responses specific for the AAV capsid protein were associated with increased liver transaminases and a decline in transgene expression, the evaluation of cellular immune responses has been incorporated into many of clinical immunogenicity monitoring plans (10, 11, 14).
Cellular immunity assessment for gene therapy-based therapeutics can help identify if immune responses negatively impact safety and efficacy and, if so, what risk mitigation strategies may be employed. Often, patient populations are of young age, while high blood volumes are required for the testing. The evaluation of cellular immune responses in gene therapy trials should be based on a meaningful trigger where the analysis would foster further understanding of potential impact on safety and exposure/efficacy endpoints.
The following are hypothetical examples highlighting various risk levels associated with the development of cellular immune responses to AAV capsid proteins. Parameters to consider in the evaluation of cellular immune response risk in these case studies are as follows:
Route of administration, for example, intravenous (IV), intrathecal (IT), intravitreal (IVT), intramuscular (IM), and subretinal
Immune-privileged (versus immune robust) status of the site of administration
Dose (high or low)
Local site concentration vs. systemic concentration
Presence or absence of pre-existing TAb and NAb
Scenario 1
Subretinal administration of an AAV-based gene therapy to deliver a functional RPE65 gene will be considered a low risk because (a) the vector is administered to an artificial space that is created by the act of injection, (b) the dose is low (administered as a flat dose rather than a body weight adjusted dose and limited by dose volume and drug product concentration), and (c) the GTx is diluted by the time it reaches the circulation (i.e., the systemic exposure to the GTx is low). Consequently, the likelihood of developing a systemic humoral or cellular response is low. The eye is regarded as an immune-privileged space which does not mean that an immune response cannot be mounted in the eye or that inflammation does not occur, but that immune responses in the eye are different from those generated systemically. The anatomy of the eye is such that it is difficult for antibodies and immune cells to distribute from the systemic circulation to the eye (50, 51). These considerations, coupled with the extreme difficulty, if not infeasibility, to acquire ocular immune monitoring samples, would make the detection of a humoral or cellular immune response to an ocular gene therapy of low scientific value, and hence, such assessments may not be required (52, 53).
Scenario 2
AAV-based gene therapy intended primarily for treatment of pediatric patients, administered intramuscularly to replace a defective structural protein gene (e.g., dystrophin gene in muscle tissue), will be considered as moderate to high risk, as the vector is administered in an immune competent, highly vascularized space.
Scenario 3
AAV-based gene therapy intended primarily for treatment of adult patients, administered intravenously to replace a defective clotting factor gene in the liver, would be considered a high immunogenicity risk, as the viral vector is administered systemically to distribute via the circulation to the target tissue. In this case, the likelihood of cell-mediated immune responses is high. Previous clinical data have shown that cytolytic T cell activity, based on the mechanisms described earlier in the manuscript, tentatively eliminating cells transduced with AAV and elevated liver enzymes, can be expected (10, 28). High-risk potential for cellular response to capsid proteins may be expected for other AAV treatment administered systemically at high doses. Although liver-directed transfer is considered high risk, there are also studies indicating that targeting liver can induce tolerance to the transgene expressed protein (54, 55).
The decision to collect samples for the assessment of cellular immune responses may be made based upon several factors. Firstly, is there a scientific rationale (often based upon data from previous studies) for a role of the cellular immune response in safety and/or loss of efficacy of the gene therapy? Secondly, is there a scientific rationale for a cellular immune response in the blood to serve as a surrogate for a cellular immune response in a tissue (e.g., a cellular immune response in the eye for subretinally administered GTxs)? Thirdly, can a sufficient number of samples be collected to be able to assess the impact of the cellular immune response on safety and/or efficacy? Ideally, two or more samples should be collected prior to dosing in order to establish a baseline against which to evaluate the post-dose response. Several post-treatment samples should be collected with the sampling being relatively intensive in the first 4–6 weeks post-dose (i.e., when transaminase increases may be observed). The first post-treatment sample should ideally be collected before week 4; collection should continue at every 2 to 4 weeks during initial study period (e.g., 6 months). Sample collection frequency may be reduced to every 4 to 12 weeks during later phases of the study. Such a sample collection schedule is possible for adult patients. However, for pediatric patients, this may not be practical due to the large volume of blood that will be required. Therefore, for GTx administered by certain routes of administration (e.g., subretinal) and for certain patient populations (e.g., pediatric), default collection of samples for the assessment of a cellular immune response may not be necessary or warranted. Collected samples can be banked and tested based on the risk factors described below.
Factors that may trigger a decision to conduct cellular immunogenicity evaluations are listed below.
Lack of persistence of vector DNA in target tissues, unexpected clinical pharmacodynamics (PD), and unexpected lack of efficacy in the absence of detectable humoral immune response
Unexpected safety signals that cannot be explained by other assessments or require additional evaluation
Suspected presence of memory CTL prior to administration or re-administration of GTx vectors
Route of administration
Administration to non-immune-privileged site
Expected systemic exposure
High administered dose
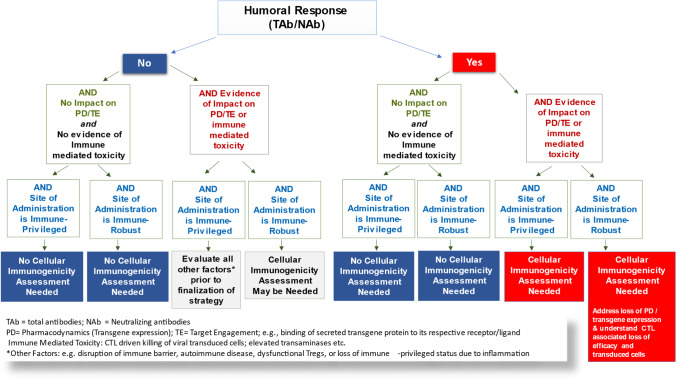
Together, these factors are summarized in a decision tree presented in Fig. 2.
Fig. 2.
Decision tree for implementation of cellular immunogenicity assays in AAV modality-based gene therapy clinical studies. Strategy is developed using a multi-factorial approach. Evidence of TAb and/or NAb response and immune tolerance status of site/tissue of administration are important factors to consider. Other parameters such as PD, lack of persistence of vector DNA in target tissue, and unexpected safety signals that may suggest evidence of immune-mediated toxicity should be evaluated prior to the decision to conduct assessment of cellular immune response in blood compartment
Unlike pre-existing serotype-specific humoral response assessed prior to treatment, cellular immunogenicity assessment is generally not considered relevant for the enrollment of study subjects or patients. Cellular immune response assessments to detect CTL responses against viral capsids have been performed for systemically administered viral vectors to explain adverse events such as elevated transaminases and loss of exposure or expression of transgene protein (56). The specifics of patient population and nature of the disease would be key drivers in developing a cellular immunogenicity strategy. As a critical factor, the state and severity of disease as well as the age of subjects, as studies are often conducted in pediatric populations, could limit ability for cellular immunogenicity assessment due to limitations in blood sample collection. Requirement for assessing of pre-existing cellular immunity was kept out of scope of this publication.
The assessment of cellular immune response to AAV capsid proteins has been performed to address cytotoxicity and loss of transduced cells as detailed above. Such adverse events were noted during systemic delivery of viral capsids to the muscle and liver. It is also evident that within tissues, transduced cells may either become inflamed or tolerogenic to the expressed protein (57, 58). A comprehensive risk assessment strategy to determine whether, when, and how to detect cellular immune responses to AAV vector would require a systematic look at the risk factors. This may begin with assessing cellular reactivity to AAV serotype in the target population to identify motifs in capsids that could potentially prime and activate innate response, tissue-specific risk and associated cell populations with Ag uptake, processing and presentation ability, and ability of T cells to be recruited to the tissue site and elicit an inflammatory or regulatory response. The disease state and patient demographics (age, population of origin, the type of gene defect leading to protein deficiency vs. splicing or point mutations) can all influence the strategy.
Analytical Methodologies
At the single cell level, cellular immunogenicity is typically assessed with ELISpot, a fluorescence-based ELISpot variant (FluoroSpot) or flow cytometry-based assays in which peripheral blood mononuclear cells (PBMC) are tested for ability to produce cytokines (most commonly IFN-γ) or other soluble factors (e.g., granzyme B) in response to specific Ags. While ELISpot detects one cytokine at a time, recent advancements with the FluoroSpot technology allow for the simultaneous detection of up to 4 analytes. Flow cytometry-based assays enable the simultaneous detection of even more cytokines, along with providing the ability to phenotype the cells. The complex features of flow cytometry methods come with increased demands for experimental and analytical expertise and validation. ELISpot, FluoroSpot, and flow cytometry assays are highly sensitive and can detect functional immune cells at very low frequencies. The “Analytical Methodologies” section describes these assays in more detail, focusing on the key elements for reliable assay performance and outcome. These key elements include (1) sample preparation, (2) Ags used to elicit and measure specific responses, and (3) critical assay performance parameters and controls. While the context for sample preparation and Ag formats are exchangeable between all assays, the assay performance section focusses on the ELISpot-based protocols as the presently most prevalent technology.
ELISpot and intracellular cytokine staining (ICS) assays have been employed for many years in the vaccine development field to identify antigen-specific immune responses at the single cell level (59-62). In the gene therapy space, the ELISpot and ICS methodologies have been adopted to study undesirable cellular immunogenicity against the gene delivery vector and the product expressed from the transgene (63). Both ELISpot and ICS protocols allow detection of cytokine-producing immune cells in very low abundance. The detection sensitivity of both assays is dependent on the background reactivity levels of tested immune cells that spontaneously produce cytokines and are majorly affected by procedures the cells are subjected to, as well as the reagents, including serum and antigens used for the functional assays as discussed in the following section (64). Table II summarizes advantages and limitations of ELISpot-, FluoroSpot-, and ICS-based assays.
Table II.
Comparative Overview of ELISpot, FluoroSpot, and Flow Cytometry-Based Staining Procedures for T Cell Analysis
| Assay | Features | Instrument and output | Advantages | Limitations | |
|---|---|---|---|---|---|
| ELISpot | Measures | Cytokine secretion on a single cell level | ELISpot reader: spot count, which equals the count of cytokine-secreting cells |
• High sensitivity (detects cells in very low abundance) • Signal stability allows for long-term read out • Easy to learn and to adapt to study requirements |
• Limited phenotypic information is accessible via additional steps, e.g., MHC blocking or isolation of cell subpopulations • Spot color saturation results in a low dynamic range, allows for limited correlation with the amount of cytokine secreted per cell • No reliable multiplexing feasible |
| Target markers | Wide range of relevant cytokines as well as other soluble analytes | ||||
| Methodology | Colorimetric detection of secreted cytokines on anti-cytokine Ab-coated membranes (spots) | ||||
| FluoroSpot | Measures | Cytokine secretion on a single cell level | FluoroSpot reader: spot count, which equals the count of cytokine-secreting cells |
• Same as for ELISpot • Multiplexing allows detection of up to 4 different analytes in one well, use of fewer number of cells when assessing multiple cytokines • RAWspot™ technology provides total spot number to correlate with total amount of cytokine secreted per cell (65) |
• Only limited phenotypic information accessible as for ELISpot |
| Target markers | Wide range of relevant cytokines as well as other soluble analytes | ||||
| Methodology | Fluorometric detection of secreted cytokines on anti-cytokine Ab-coated membranes (spots) | ||||
| Flow cytometry | Measures | Intracellular cytokine production, phenotype on a single cell level | Flow cytometer: percentage of cytokine-positive cells within phenotypic subsets |
• Multiplex staining with the possibility to assess a high number of cytokines and other markers within one sample • Identification of immune cell subsets secreting the target cytokine(s) |
• Complex assays demanding extensive training and expertise • High demands on panel development and assay validation • Demanding data analysis (gating process) |
| Target markers | Wide range of relevant cytokines and cellular markers | ||||
| Methodology | Detection of intracellularly enriched cytokines and cell surface markers using fluorescently conjugated reagents | ||||
IL-2, interleukin-2; IL-4, interleukin-4; IL-5, interleukin-5; IL-10, interleukin-10
ELISpot and FluoroSpot Analytical Platforms
ELISpot and FluoroSpot analytical platforms support the development of highly sensitive, microplate-based functional assays designed for the detection and enumeration of cytokine generation by immune cells. The initial method was developed based on standard ELISA assays to measure antibody secretion from B cells (66) and has since been broadly used to monitor adaptive immune responses in humans and animals. ELISpot assays have gained popularity as they can assess critical cellular immune-related activities such as IFN-γ secretion and granzyme B release and are adaptable to the evaluation of a variety of T cell, B cell, and innate immune cell functions.
ELISpot is typically performed using polyvinylidene difluoride (PVDF, preferred) or nitrocellulose (less favorable) membrane 96-well plates coated with a cytokine specific antibody. Pre-coated commercial kits are broadly available. Plates are commonly blocked with test media to reduce non-specific binding. Control and test stimulant solutions are added to the designated wells aiming to achieve the desired final concentration after subsequent PBMC sample addition. Assays are typically optimized on the cell number, concentration of stimulant, and other parameters (67). Typically, 200,000 to 400,000 of PBMC cells/well (for a 96-well plate) are used when analyzing antigen-specific responses in a T cell ELISpot assay. For the positive control stimulation, the number of cells per well can be reduced if a strong response is expected (67).
Cells are stimulated for approximately 24 h (e.g., to stimulate IFN-γ production) with the peptide pools from the test or control antigens. Due to the inherent variability of cell-based assays, samples are often assessed in triplicates. Cytokines (e.g., IFN-γ) secreted by responding T cell will bind to the anti-cytokine antibody pre-coated on the plate membrane. A detection antibody system is used to detect membrane captured cytokine via production of colored or fluorescence emitting spots that are detected by employing ELISpot and FluoroSpot instrumentation, respectively. Cytokine-secreting cells are represented by spots, which can be counted by automated reader instruments. Detailed harmonization guidelines for the performance of an IFN-γ ELISpot assay and the evaluation of ELISpot plates have been established in large, field-wide efforts (68, 69). The FluoroSpot technology offers the option of multiplexed analysis where multiple cytokines are detected in the assay. This approach allows for an assessment of cellular polyfunctionality; e.g., the analysis of 3 cytokines in one well can detect up to 7 subpopulations, while the analysis of 4 cytokines potentially detects 15 subpopulations of responding cells (70, 71). The rising number of different analytes in the assay does, however, increase the complexity of the assay development and qualification exercise, as well as the instrumentation platform used in the assay.
The major limitation of both ELISpot and FluoroSpot platforms is the high sample volume constraint that is based on the required number of cells needed for the analysis. When assessing multiple test conditions with samples plated in triplicate, there may be insufficient cells available to evaluate all test conditions, potentially necessitating establishing a sample testing priority plan.
Intracellular Cytokine Staining Flow Cytometry-Based Methodology
Intracellular cytokine staining is an alternative methodology for detecting antigen-specific cytokine production at a single cell level. In the ICS protocol, PBMCs are first incubated in the presence of a stimulating agent, e.g., a peptide pool, similar to what is done in an ELISpot-based protocol. Recognition of these peptide sequences by T lymphocytes results in the productions of IFN-γ and potentially other target cytokines in these cells. To prevent such cytokines from being secreted, brefeldin A (72) and/or monensin (73), which interfere with protein transport through the Golgi apparatus, are added to the stimulation cultures. Cytokines accumulated intracellularly are then identified by ICS. Stimulation cultures are usually performed overnight (~ 16 h), though the optimal incubation time depends on the nature of the stimulating antigen and analyte measured and needs to be determined during assay development.
Following the stimulation step, samples are incubated with a viability dye that will allow the exclusion of dead cells, which can non-specifically interact with Abs while also displaying increased autofluorescence (74, 75). This step is crucial in reducing background staining, thus increasing assay sensitivity. This is followed by blocking any Fc-receptors prior to incubating with fluorescently conjugated Abs to extracellular markers. Cells are washed, fixed, and permeabilized to allow for staining of intracellular molecules, such as the captured cytokines (e.g., IFN-γ). Other staining strategies exist and need to be evaluated during the assay development phase (76). As part of the assay development, all fluorescently conjugated Abs need to be carefully titrated (76) for the specific application (extracellular vs. intracellular staining, cell type, following the specific antigen stimulation culture conditions, etc.). The more markers are being interrogated in the assay, the more information regarding the identity of cytokine-producing cells can be obtained, and the higher the level of cytokine multiplexing that can be achieved. This allows for a more precise identification of the active players in an immune response. However, increasing the number of reagents used in an ICS assay will increase its complexity and thereby the effort and time required to appropriately troubleshoot the reagent panel during assay development to ensure that all analytes can be discerned unequivocally. Following a final thorough washing of cells, the samples are evaluated on a flow cytometer. Depending on the capabilities of the flow cytometer used, an ICS assay can include 40 + analytes, though it is advisable not to include more fluorescent reagents than necessary, as each additional reagent can result in reduced assay sensitivity for individual analytes. To deconvolute the fluorescent signatures obtained, single stained control samples are also acquired, which are necessary for the creation of a compensation matrix. This is then applied to all experimental samples, as well as biological controls, and allows for the separation of signals obtained from the different fluorescent molecules included in the assay. The resulting data are then analyzed by sequential gating, which first aims to exclude any unwanted events (such as cell aggregates, irrelevant cell types, and dead cells) before identifying cell types and subsets of interest. Within these, cytokine production is then investigated. The proper gating hierarchy is key to obtaining meaningful data.
Whereas both ELISpot and ICS yield a count for cytokine-secreting cells, ICS bears unique advantages, as it allows for the detection of multiple cytokines within a single cellular aliquot through multiplex staining. Additionally, ICS makes it possible to identify and enumerate T cell subpopulations that secrete the target cytokines. Furthermore, the presence and activation phenotype of Treg can be interrogated in parallel to assess their potential influence on effector T cells. This phenotyping feature is a key advantage of ICS over ELISpot. Larger cell numbers can be interrogated per well or tube than in an ELISpot or FluoroSpot assay, allowing for the analysis of cytokine production even within rare cellular subsets without the need of enriching them prior to ICS testing. The major limitation of the ICS methodology is the potential for non-specific binding, which results in reduced signal-to-noise ratio, as well as spillover spreading that can reduce detection sensitivity for specific fluorochromes and thereby the associated analyte (77). The goal of assay optimization strategies, as well as the use of appropriate controls, is to limit such effects, maximizing overall assay sensitivity (78).
PBMC Isolation, Freezing, and Thawing
Cellular immunogenicity assays that assess antigen-specific T cell activation are usually performed with either freshly isolated or cryopreserved PBMCs. In the clinical setting, where samples are collected from study subjects at multiple time points to monitor cell-mediated immune responses to the treatment, it is common to collect samples and analyze in batches to reduce potential run to run variability, assuming appropriate stability has been shown (79).
Blood samples are drawn into citrate or heparin-containing blood collection tubes for later isolation of PBMCs or into collection tubes specifically designed for immediate cell separation. Collection in heparin-containing tubes is viewed as favorable for functional studies (80), while blood collection into ethylenediaminetetraacetic acid (EDTA)-containing tubes may lead to reduced stability and is not favored (81). Blood samples are centrifuged to isolate PBMCs using a high-density medium such as polysucrose sodium metrizoate (e.g., Ficoll-Paque®, Lymphoprep™, and Histopaque®). Care needs to be taken to avoid inadvertent mixing of cell layers aiming to reduce processing inconsistencies. Detailed review of PBMC isolation and handling technique can be found elsewhere (63). Properly prepared and stored PBMC material can be retained for an extended period while remaining unresponsive to mock stimulation and consistently yielding a positive response to multiple stimuli (36, 82).
Current data suggest that PBMCs should ideally be isolated and frozen (or used in an assay) within approximately 8 h of blood sample collection to preserve cell functionality for optimal sensitivity (83) (84). Extended duration of blood sample storage prior to PBMC separation may result in granulocyte activation. This could lead to granulocyte co-separation with PBMCs in density gradient-based protocols and, to the extent that granulocytes survive cryopreservation, inaccurate PBMC cell counts later. Co-separated granulocytes may also bind to the ELISpot capture antibody reagent via their Fc receptor and disrupt the spot formation (85, 86). Furthermore, activated granulocytes release hydrogen peroxide and downregulate the CD3- ζ chain expression, both leading to decreased T cell functionality (87, 88). Overall, it is advised to consider timely PBMC isolation and preservation in liquid nitrogen at the site of blood collection or a nearby central laboratory, prior to their shipment and analysis at the bioanalytical laboratory. Preparation of PBMC samples within 8 h of blood sample collection may be challenging in a real clinical trial setting. It will be critical to understand the impact of specific preparation conditions on the quality of the resulting PBMCs. An attractive solution is offered by cell preparation tubes (e.g., Vacutainer® CPT™ (89)) that can be applied for both blood sample collection and cellular component isolation, enabling effective granulocyte separation by centrifugation directly at the clinical site. These tubes can then be shipped to the central laboratory for further processing.
To avoid cell loss, cryopreserved PBMC samples should be quickly thawed, preferably in a water bath at 37 °C and transferred to pre-heated wash media. Cell suspensions may be treated with nuclease to prevent cell clumping while considering that excessive exposure may result in cell death by apoptosis (90, 91). After pelleting cells by low-speed centrifugation and gentle washing by resuspension in wash media, PBMCs are examined for viability and total cell count (92). The viable cell count should be used for calculating cell dilutions for the assay to obtain the desired seeding density. It has been shown that the functionality of CD4 + and CD8 + T cells dramatically decreases with an increased number of apoptotic cells in a PBMC sample preparation as assessed by ELISpot (93, 94). It is recommended to only use PBMCs with a viability at or above 70% in functional assays (95, 96). It was reported that INF-γ ELISpot data obtained from samples containing > 18% apoptotic cells were erratic and did not reflect the true frequency of responder cells in the affected biological sample (93).
The challenges of limited sample volume, low cell viability, and the potential for high degree of apoptosis need to be recognized and proactively addressed. Mitigation strategies include the following:
Establishing and verifying laboratory procedures optimized for PBMC isolation and storage
Optimizing such protocols for the purposes of a specific trial
Introducing additional steps that may improve PBMC performance, for example, resting cells after thawing. Resting may reduce the number of apoptotic cells and most importantly resets T cell functionality to an improved, tissue-like state, although it has a potential to impact PBMC recovery (97-100).
Using viable cell count when calculating final cell dilution in the assay
Applying the lowest cell count of the linear range as established during assay validation, while ensuring that this cell number is sufficient to achieve the desired assay sensitivity
Establishing a priority plating strategy (step-by-step exclusion of wells from being tested, based on the priority of data)
Flagging samples with viability below 70% as reduced antigen-specific responses may be expected
Considering the use of an internal donor control reactivity (e.g., against CEF, a lyophilized mixture of peptides from cytomegalovirus (CMV), Epstein-Barr virus (EBV), and influenza virus) to compare sample functionality across time points
Antigen Quality and Preparation
Peptide pools are typically used as antigen presentation when evaluating cellular immunity by ELISpot or similar methods. Peptides are either purchased commercially or custom made based on the sequences of the viral vector. It is a common practice to prepare pools containing 15-amino acid (aa) peptides with an overlap of 10–12 aa (101, 102). Together, the appropriate peptide length and overlap decrease potential risks of overlooking stimulating amino acid sequences and enable for simultaneous detection of CD4 + and CD8 + T cell responses in the absence of information about patients’ human leukocyte antigen (HLA) type. Overlapping peptide pools spanning the entire protein sequence allow the presentation of all possible epitopes of that protein (103). Smaller sized peptide pools (< 30 peptides/pool) may exhibit a higher sensitivity in detecting responses than pools with higher number of peptides (104). In general, it is recommended to use peptide pools containing no more than 100 peptides.
As peptide pools are usually received in lyophilized form, material needs to be reconstituted using dimethyl sulfoxide (DMSO) and cell media with the understanding that the final concentration of DMSO in solutions applied to the cells should be below 0.5% and preferably at or below 0.1% to avoid toxicity. Peptides should be kept lyophilized, if possible, to avoid degradation resulting from e.g., oxidation of cysteine groups in the presence of DMSO. It is recommended to store peptide solutions in small aliquots at − 80℃ while avoiding repeated freeze/thaw cycles. The concentration of a peptide pool indicates the concentration of each peptide in that pool. Thus, adding a peptide pool for a final concentration of 5 µg/mL will add 5 µg/mL of each peptide contained in that pool. For functional assays, the final peptide concentration should typically be between 0.5 and 10 µg/mL.
The presence of contaminants in the peptide preparations may result in impacted assay performance and lead to false-positive responses. Contaminants may include residual solvents, hydrophobic protection groups and peptides, and neoepitopes arising due to missing capping during peptide synthesis with the latter presenting the biggest potential for false-positive signals in the assay. Experimental Controls.
Negative Control or Mock Stimulation
A negative control is used to define the assay limit of detection (LOD) and is essential for the determination of response acceptance. The negative control includes cells without stimuli (mock stimulation), aiming to assess the number of cells spontaneously producing cytokines detected in the assay. The mock stimulation used in the negative control test should contain an equivalent concentration of DMSO as contained in the samples stimulated with the antigen peptide pool preparations.
The use of an irrelevant antigen stimulation (e.g., irrelevant peptide pool expected to induce no stimulation) is not necessary although can be implemented during assay development and performance verification testing. Such pools should have a similar composition as the experimental (antigenic) peptide pools. Such peptide pools are commercially available.
A well-controlled experiment would include the following additional controls:
Positive Control
A positive control is used to demonstrate that PBMC preparation is functional, thus avoiding false-negative results. Positive control reagent can be applied to establish assay acceptance criteria (36). Positive control reagents used in ELISpot or similar methods are capable to induce polyclonal stimulation and include phytohaemagglutinin (PHA), concanavalin A (ConA), phorbol 12-myristate 13-acetate in combination with ionomycin (PMA/Iono), staphylococcal enterotoxin B (SEB), or anti-CD3 Abs (63). Generally, one positive control stimulation is sufficient to ensure PBMC functionality during study sample testing.
Background Control
The background (or medium only) monitors signals generated by assay reagents and media in the absence of cell material. A detected assay signal in the background control may be observed due to the presence of aggregated material in the antibody detector reagent. This can be avoided by filtering the antibody reagent prior to use in the assay. Commonly, three medium only wells per experimental run are sufficient where run includes one or more plates.
Internal Sample Control
An internal control that elicits a reliable and consistent peptide-specific response in longitudinal samples of individual patients is used to assess sample quality and ascertain consistency of antigenic responsiveness over time in a patient. Since its introduction, the commonly used control peptide pool CEF (105), comprising of well-defined class I-restricted peptides from CMV, EBV, and influenza, has been expanded to include more class I as well as class II peptides from a multitude of pathogens (CEFx) to elicit reliable T cell responses. It is of note that tested individuals can exhibit significantly different response levels against such a control peptide pool, including no response at all, though response levels should not differ widely over time for a given individual. If a patient demonstrates significant changes in reactivity against that internal control peptide pool over the time course of the study, it could be indicative of clinical or sample-related events.
Reference Sample
The assay trending control is used to attest to the assay performance over time (106). This control consists of PBMCs from a single healthy donor blood draw, frozen in aliquots, and tested with each experiment. The test is conducted using a mock stimulation (negative control), a mitogen stimulation (assay positive control), and an antigenic stimulation (e.g., peptide pool used for the internal control stimulation, such as CEF, CEFx, or similar). While reference samples are useful to monitor assay performance, it can be challenging to maintain sufficient amount of reference sample from the same donor for an entire study. Hence, bridging a new reference sample will become necessary and should ideally comprise 3 experimental runs.
ELISpot Assay Qualification Parameters
The following section reviews parameters that are critical for establishing a qualified ELISpot or FluoroSpot assay. There is some level of overlap with procedures followed during flow cytometry assay qualification with additional details found elsewhere (107-110).
Assay LOD and Screening Cut Point
ELISpot assay LOD, also referred to as screening cut point or method sensitivity, describes the noise level of the assay and hence represents the assay signal below which a response is reported as negative. Importantly, the LOD is a descriptive assay qualification parameter and is not applied as a treatment exclusion criterion for study subjects with high pre-treatment (background) assay reactivity level. High background reactivity signal can be mitigated by methods briefly described below.
When supporting research studies, it may be acceptable to define an arbitrary LOD value based on prior experience with the method. For example, LOD can be defined at 10 spot-forming units (SFU) per well containing 2 × 105 cells in a colorimetric ELISpot method. The suitability of such LOD should be confirmed by evaluating a small number of individual donor samples (e.g., 5–10) under non-stimulated conditions to ensure that background signal does not lead to false-positive sample score.
To determine sensitivity of an ELISpot method used in regulated studies, a statistically based LOD or screening cut point may be defined (36, 111). The statistically defined LOD value can be established based on an analysis of the frequency distribution of background signal produced by a collection of individual donor PBMC samples under non-stimulated conditions (111). Sufficient number of individual donor samples, ideally from treatment-naïve donors, should be evaluated. Typically, this assessment includes 30 or more individuals. It is understood that for rare conditions, obtaining a large number of individual disease state samples can be challenging. Use of commercially available PBMC samples from healthy donors is oftentimes viewed as suitable for LOD assessment.
As previously discussed, appropriate (e.g., > 70%) viability of PBMC samples should be confirmed in the assessment. Ideally, samples should be tested on several days by two operators to capture inter-run variability of the assay. During data analysis, the inter-quartile range method can be used to remove statistical outliers, and the Shapiro–Wilk test can serve to assess whether the sample responses are derived from a normal distribution. Logarithmic transformation may be used to achieve normality, if needed. For data sets that satisfy the assumption of a normal distribution, the 95% probability limit can be calculated by multiplying the mean SFU response under non-stimulated conditions across all samples tested during cut point assessment by a factor of 1.645 (the corresponding z-value from a normal distribution table). This will establish the LOD value with an estimated 5% false-positive rate. Alternatively, for a 1% false-positive rate, a factor of 2.326 could be used. If the signal distribution does not satisfy the assumption of a normal distribution, the empirical 95th or 99th percentile limits can be used as surrogates. Typical assay LOD value for a colorimetric ELISpot falls near 10 SFU/well, which corresponds to 50 SFU/million PBMCs assuming that 2 × 105 cells are plated per well.
Alternatively, an approach based on the median of background reactivity in non-stimulated PBMC samples can be applied. Using the distribution median avoids data distortion by outlier measurements. The twofold (or threefold, in case of very low background) over the median background value is then set as the assay LOD, the point below which even statistical differences between mock-stimulated and Ag-stimulated spot counts would not be considered a response since those differences are observed within the noise range of the assay (112). It is common that the assay LOD value established using the background median-based approach falls in a range similar to that established via the statistical approach, often between 5 and 12 SFU/well.
An important consideration for clinical or non-clinical studies accounts for potential differences in background reactivity levels between donor samples used during method qualification and study samples obtained from study relevant disease state population. While the assay LOD established during qualification provides a reference LOD value for assay performance, it may be useful to verify its suitability as screening cut point using available study sample data. This can be achieved by investigating study sample responses under non-stimulated conditions and applying the same methodologies described above. A study-specific screening cut point may be implemented if it differs substantially from the LOD value derived during pre-study method qualification. This could occur due to the circumstances related to participants’ health status (e.g., immune suppression or infection), varying reagents (e.g., freezing media), and differences in applied standard operating procedures (SOPs, e.g., sample processing for healthy donor samples used during qualification vs. during in-study testing), which may unexpectedly affect background reactivity levels.
It is acknowledged that a statistically defined LOD could be established based on background signal produced by non-Ag-responsive PBMCs under stimulated conditions, rather than based on background signal observed under non-stimulated conditions. However, due to pre-existing cellular immunity to AAV, identification of non-Ag responsive donors may be problematic. A potential approach to solve this challenge could be to utilize PBMC samples from treatment-naïve donors who were previously screened to be non-responsive to AAV peptides in non-qualified ELISpot or an orthogonal assay. Such donors may be identified in pre-study tests involving a large cohort of healthy treatment-naïve donors (30 to 50) whose PBMCs are tested against AAV peptide pools. The magnitude of background signals in non-Ag-responsive donors under stimulated conditions would be expected to mimic that of non-stimulated donor samples; thus, there is no clear advantage in pursuing this approach.
When pretesting healthy treatment-naïve donors for background reactivity levels, it is recommended to include testing against the AAV protein peptide pools. The observed response frequency against each peptide pool may serve as a barometer for pre-existing response frequency one has to expect within the study cohort at baseline.
Assay Confirmatory Parameter
Assay developers are oftentimes confronted with elevated non-specific background signal above LOD value that is observable in a fraction of PBMC samples under non-stimulated conditions. Such samples can be easily identified and removed as statistical outliers from the assay LOD (cut point) analysis described above. However, during clinical or non-clinical study sample testing, an additional confirmatory assay parameter may be needed to determine if the relative magnitude of a response under stimulated conditions should be considered as Ag-specific when compared to elevated background signal observed under non-stimulated conditions. For example, a study sample that generates ELISpot signal under both Ag-stimulated and non-stimulated conditions at 100 SFU/well, a value well above assay LOD of 10 SFU/well, may not be adequately reported as positive for Ag-specific reactivity. In contrast, a study sample that generates ELISpot signal of 100 SFU/well under non-stimulated conditions and 500 SFU/well under Ag-stimulated conditions should be reported as positive for the Ag-specific reactivity.
Hence, a pre-defined 2- to threefold increase in SFU above the background (mock treatment generated) signal can be helpful to account for potentially elevated non-specific response in some samples (111). Samples for which Ag stimulation results in signal above the assay LOD would be additionally assessed for the relative increase in SFU over the mock stimulation conditions.
Alternatively, two statistical approaches have been used to evaluate whether a relative increase in SFU between stimulated and non-stimulated conditions is considered significant for a test sample. The first approach uses statistical means during pre-study method qualification to establish a confirmatory cut point (CCP) based on the response ratio between Ag-stimulated conditions and mock response in treatment-naïve individual donors, as described (36). The subsequent use of the CCP should be restricted to clinical or non-clinical study samples that show elevated background signals (i.e., show a response > LOD under non-stimulated conditions) to preserve maximal sensitivity for samples with low background signal. Applying the CCP universally to all samples with responses above the LOD would likely eliminate otherwise acceptable low positive samples and thus under-report the immune reactivity in patients.
A second approach is based on response evaluation using the statistical distribution-free resampling (DFR) method. This method was specifically developed for response testing in ELISpot protocols (112). The DFR testing is a permutation test that does not rely on analysis of mean values and considers peculiarities of typical ELISpot data. Specifically, these are a small number of data points (which limits use of assumption-based analysis of data distribution), considerable level of variability, and varying level of background reactivity. Two slightly different tests are typically run simultaneously: (a) analysis that considers any statistically significant difference between signals generated under non-stimulated (background) and Ag-stimulated conditions (referred to as DFR (eq)) and (b) analysis that only considers differences that are greater than 2x (referred to as DFR (2x)). The DFR testing allows to detect positive response in the presence of high background reactivity. The determined LOD value (as assessed pre-study or adjusted based on available study data) continues to serve as a cutoff for the acceptance of tentatively positive responses whereas DFR data analysis confirms specificity of the response. While DFR data analysis accounts for the replicate variability, it is recommended to flag outlier replicates by determining their variance in relation to the median (112). An open webtool and the R code allowing to run DFR data analysis are available online (113).
Assay Specificity
Assay specificity refers to the ability of the method to distinguish positive responses to antigen-derived peptides of interest from negative responses to peptides derived from irrelevant antigens. Since donors of commercial PBMC samples are routinely screened to be human immunodeficiency virus (HIV)-negative, non-responsiveness of the PBMCs to HIV peptide stimulation could be used to confirm the absence of non-specific peptide responses during method characterization. Peptides derived from other viruses may be used if the serostatus of the donors is known to be negative.
A commonly used positive control stimulation in experiments that confirm specificity of an ELISpot method utilizes peptides derived from cytomegalovirus phosphoprotein of 65 kDa (CMVpp65), which generates responses in most human PBMC samples. Alternatively, CEF with similar broad reactivity (see the “Internal Sample Control” section) can be applied. These positive control stimulations could be run alongside the negative control peptides to fully demonstrate the discriminatory ability of the method to generate specific positive responses to peptides from antigens of interest.
Assay Low Limit of Quantification (LLOQ)
The LLOQ of the assay is the lowest amount of analyte that can be quantified with an acceptable precision. An LLOQ of an ELISpot method is commonly defined as spot count at which the coefficient of variability (CV) parameter becomes acceptable. At low spot counts even a minor change in the assay signal will lead to a significant increase in the %CV value. A typical LLOQ for an ELISpot protocol falls between 20 and 30 SFU/well (114). It is proposed that this LLOQ value can be broadly applied without a need to generate assay specific data set.
Assay Upper Limit of Quantification (ULOQ)
The ULOQ is related to the spot counting procedure and refers to the spot count at which single spots cannot be any more reliably distinguished due to the confluency issue. The ULOQ is affected by the size of spots (the larger the spots are, the lower is the ULOQ), the nature of the method (ELISpot has a limited dynamic range for spot saturation, while FluoroSpot offers an advantage of a larger dynamic range), and the spot counting method. Experiments where PBMC samples are tested in the presence of a strong stimulant (e.g., CEFx peptide pool, PHA) help to determine the assay ULOQ parameter. The ULOQ can range from about 200 SFU/well for an ELISpot method that produces large spots counted via image analysis to 5000 spots in a FluoroSpot assay.
Assay Precision
Precision is an important qualification parameter that describes the closeness of agreement between a series of measurements for the same sample. Assay precision is evaluated in an intra-assay (variability between replicate wells), inter-assay (variability between data obtained in different runs, days), and reproducibility tests (variability between different operators). The potential impact of sample handling on assay performance should be assessed where samples are prepared and tested by different operators. The suggested approach to evaluate assay precision includes 3 separate assessments of 3 individual samples with 3 different response levels (high, medium, low responders) analyzed in 3 replicates. Assessments are performed to determine intra-assay variability, inter-assay variability (same operator), and inter-operator variability (at least two operators that preferably will be involved in the study sample analysis). A precision below 50% %CV should be reached, and a %CV < 30% is considered as desirable. During data analysis, it is important to identify, flag, and report outlier values. The DFR-based method of ELISpot data analysis considers variability between sample replicates. Defining outliers in an IFN-γ ELISpot method has been described in detail elsewhere (112).
Assay Linearity
The main goal of the linearity assessment is to determine whether the assay response is linear over the desired range of the method. Linearity assessment may be conducted during assay development phase. In the linearity evaluation, a range of cell densities per well is tested to determine the resulting SFU signal. The SFU values are typically extrapolated to a million cells/well. The linearity of the assay is defined as the range of cell densities that produce similar SFU values when normalized per million cells in a well. When testing is done within the linear range of the assay, variable number of cells loaded per well will not negatively impact assay performance. To determine assay range of linearity, 3 sets of PBMC samples are serially diluted, typically over a range of 0.5 × 105–4 × 105 cells per well, and tested in the presence of control peptide pools, e.g., CEF or CMVpp65. Data analysis allows to determine the lowest cell number per well as one that provides reliable SFU value. Information about the lowest acceptable cell number per well can be critical when low cell recovery would not allow for plating of the desired number of cells. A common linear range for an IFN-γ ELISpot protocol is between 1.5 × 105 and 3.5 × 105 cells per well.
Assay Accuracy
Typically, no ELISpot assay standard is available to prepare quality control (QC) samples, and therefore, true accuracy of an assay cannot be defined. Assay accuracy may be evaluated via participation in proficiency panels. In such panels, participants test the same samples against the same antigens following laboratory-specific assay protocols and using internal laboratory reagents. Comparison of results provides a relative accuracy measure. An open proficiency panel program runs once a year and is accessible to everyone performing ELISpot analysis (115).
Assay Robustness
Robustness refers to the ability of the ELISpot method to withstand small but deliberate changes in experimental conditions, such as incubation times and reagent concentrations. Defining ranges of acceptable performance for each parameter during method characterization may provide for a certain degree of flexibility when executing the assay during sample analysis. For example, cell stimulation times for overnight incubation may vary between 20 and 24 h without impacting test results while results may be different, if cell stimulation proceeds for only 16 (36). Robustness studies assessing impact of plate drying time on colorimetric ELISpot method performance demonstrated a widely tolerated range varying between 20 h and 80 days. In contrast, substrate development time used in colorimetric ELISpot methods needs to be accurately timed and should be evaluated on a case-by-case basis.
Using PBMC samples with a known response profile, a series of robustness experiments can be performed under standard conditions or under varied robustness conditions. A robustness condition may be considered successfully qualified if the SFU/well result for responses above the assay LLOQ is similar as compared to the standard condition of the assay. For example, if the relative difference between signals generated under robustness condition and standard condition tests is less than 30% for an assay with typical precision characteristics, samples producing negative response under mock stimulation conditions or when using known non-stimulating peptides should also maintain their status.
Stability Assessment
Understanding PBMC sample stability is critical for successful implementation of an ELISpot method. Stability may be assessed by analysis of PBMC samples collected from the same donor after samples were stored under selected conditions (typically, in liquid nitrogen) for a specified period. Such an assessment could be incorporated as part of clinical testing. The donor should have known reactivity to particular Ag peptides of interest at the initial assessment, and this positive response profile should remain unaltered. In addition, one may require that the SFU/well values above the assay LLOQ remain within an acceptable relative difference as compared to signal generated by untreated (not subjected to storage conditions) sample (for example, within 30% for a typical assay) to successfully qualify a given stability period. Since cell lysis may appear during long-term sample storage, leading to an increased background signal, it is also important to confirm that responses under mock stimulation, or responses to stimulation with irrelevant peptides continue to be negative. The viability of the stored samples should also be evaluated and ideally remain above 80%. Optimization of conditions and media used during freezing and thawing of samples can help to maintain stability of PBMC material for a considerable time (months to years) (36, 116, 117).
Regulatory Position
Regulatory agencies have acknowledged the importance of monitoring the cellular immune response in clinical studies in recent guidance documents (1-4). FDA suggests that cellular immune responses may play an important role in both the safety and durability of a gene therapy treatment. Cellular immune responses, therefore, would be considered potential biomarkers for either safety-related adverse events or loss of transgene expression. Development, qualification, and in-study use of cellular immune response assays can take considerable time and regulatory planning. FDA recommends that these methods initially be “fit for purpose” and subsequently validated for clinical studies intended to provide primary evidence of effectiveness to support a marketing application (79). The frequency and length of testing for cellular immune responses may be determined on a case-by-case basis. In the 2020 “Human Gene Therapy for Hemophilia” Guidance, FDA recommends that sponsors consider assessing IFN-γ secretion from peripheral blood mononuclear cells by ELISpot assays for the detection of anti-vector and anti-coagulation factor T cell reactivity for short-term monitoring during the first 2 years of follow-up (118). More frequent monitoring may be appropriate if immune-mediated hepatic dysfunction is suspected, which could clarify potential mechanisms of hepatic dysfunction. In the 2021 “Human Gene Therapy for Neurodegenerative Diseases” Guidance, FDA suggests that immune responses to GTx products may pose important safety risks, such as by damaging the tissues transduced by viral vectors carrying a therapeutic transgene. FDA recommends sponsors monitor for systemic immune reactions and perform immunoassays measuring cellular and humoral immune responses to both the vector and the transgene-encoded protein (1).
Summary
From the immunological perspective, administration of AAV vectors mirrors a viral infection. Both gene therapy vectors and natural viruses are similar in how the viral antigens are presented, with the key difference being that viral vectors lack the requisite genes for viral replication (37, 38). Consequently, the only source for capsid antigen is the input vector at the time of administration. The intracellular events following transduction of target tissue led to translocation of the single-stranded DNA genome to the nuclease and degradation of the vector capsid protein (31). Proteolytic degradation of intracellular capsid proteins generates peptide fragments that can be loaded onto MHC class I or class II molecules for cell surface presentation to CD8 + and CD4 + T cells, respectively. The loading of exogenously derived peptides onto class I in this manner is considered atypical for tissue parenchymal cells but could occur through a process termed cross presentation particularly in specialized Ag presenting cells (65). Further, previous non-clinical studies have demonstrated that pre-existing AAV antibodies impact the outcome of gene transfer and an increased distribution of AAV to the spleen (119, 120). This off-target uptake into splenic macrophages, a subtype of professional APCs, suggests a greater likelihood for capsid peptide presentation to naïve CD8 + T cells by MHC Class I and de novo stimulation of cytotoxic effectors. Over time, the presentation of the GTx-derived AAV peptides on MHC proteins may decline. However, it is possible that peptides from endogenous AAV infections that occur months or even years post-dose may be presented on the MHCs. Consequently, at time points many months post-dose, it might become difficult to attribute an impact on safety or efficacy biomarkers directly to the gene therapy. Activated CD8 + T cells, NK cells, and CD4 + lymphocytes may exhibit cytotoxic activity against AAV-transduced cells of the host. Cytotoxic T cell activation by the antigen peptides presented on the surface of the cells or activation of the NK cells by other mechanisms may lead to expression of various cytokines and pro-inflammatory signals and destruction of the transduced cells. As an example, elevation of liver transaminases (ALT and AST), the most common adverse event reported after systemic administration of AAV therapies, is believed to be a consequence of capsid-specific cellular immune response against liver cells transduced with the AAV vector (10, 29-31). The observed spike in ALT and AST was interpreted as a consequence of destruction of transduced tissues by cytotoxic T cells. Administration of corticosteroids resulted in reduction of transaminase concentration in the blood, associated cellular immune response, and, importantly, steadying of the transgene expression. It should be noted that changes in serum transaminase concentrations may be caused by factors other than T cell response. There are few studies to investigate and establish direct correlation between anti-AAV cellular immunogenicity and ALT and AST response.
Similar to the methodologies applied when assessing humoral immune response against biotherapeutics, a risk-based approach should be considered when determining the value of assessment of cellular immune response to AAV capsid proteins. Based on considerations presented in this manuscript, several risk factors are proposed (see above). Additionally, evidence of memory CTL in patients previously treated with an AAV therapeutic and that are being considered for re-administration of an AAV vector should be evaluated.
Complexities related to the maturation of the immune system should also be considered. It should further be noted that the immune system in newborn infants is not as well developed as in older children or adults, and hence, a robust cellular immune response may not be mounted in newborns (121). Other challenges associated with generating robust data driven conclusions regarding the impact of cellular immunogenicity on safety and efficacy of AAV-based treatments include the complexity of collecting samples for cellular immune response assessment at appropriate time points to determine correlation with adverse observations, large sample volumes required for frequent cellular immune response evaluations, and complexity of associated sample handling and analysis. It should be appreciated that any sample collection, particularly collection of a large blood volume from certain types of patients, e.g., pediatric, may present a significant hurdle and inconvenience and should therefore be purpose driven and justified. Collection of fewer samples assigned for cellular immunogenicity assessment may result in an insufficient data package and make it challenging to show causal relationship between cellular immune response and evaluated clinical signal. In addition, should a sponsor decide that use of pre-existing cellular immunity is needed for patient treatment decisions, a companion diagnostic (CDx) or similar level protocol may be required once the treatment reaches the market. Given the nature of the cellular immune response assay, it will be challenging to develop and successfully implement such a CDx. In summary, sponsors should consider the benefits and risks of generating cellular immune response data for AAV gene therapies before initiating the development and implementation of cellular immune response detecting assays in support of clinical trials.
A robust and persistent humoral response against AAV vector is commonly expected and has been reported (27). The nature, likelihood, and type of cellular immune response are less predictable and potentially driven by many factors. Evaluation of correlation between humoral and cellular responses was performed using information available for the following examples: Duchenne muscular dystrophy treatment using AAV2.5 vector transgene (23); alipogene tiparvovec (Glybera®), an AAV1 vector treatment for lipoprotein lipase deficiency (24-26); and AAV5-based valoctocogene roxaparvovec (Roctavian®) intended for treatment of hemophilia A (27). An overall assessment suggests a lack of correlation between humoral and cellular responses, although it should be noted that examples of treatments reviewed in this manuscript describe products with potentially different amounts and types of impurities, different routes of administration, doses, immunosuppressive regiments, and methods used to assess the cellular immune response.
Analysis of publicly accessible information related to cellular response and its impact on treatment safety and efficacy leads us to make the following general conclusions regarding the value and triggers for such evaluation. We believe that the need for cellular immunogenicity testing should be driven by the expected mode of action of the potential impact. Such assessment should be viewed as one of the elements of treatment mitigation strategy and needs to be risk-based. Even if it is possible to show a temporal relationship between the cellular immune response and a change in a safety or efficacy biomarker, it is necessary to consider and test a mechanistic hypothesis to demonstrate cause and effect relationship. This mechanistic hypothesis will enable generation of hypotheses related to how the cellular immune response can be mitigated and hence the safety and efficacy profile of the gene therapy improved. For example, the ability of cellular immune responses measured in the blood compartment to serve as a surrogate for such responses in other tissues, e.g., the eye or liver, needs to be confirmed.
Several analytical platforms may be considered for evaluation of cellular immune response, including ELISpot, FluoroSpot, and ICS protocols. These methodologies have been successfully applied in the field of vaccine development with demonstrated ability to detect antigen-specific cytotoxic T cells in very low abundance. Currently, in the field of viral vector delivery gene therapy, ELISpot is the most applied technology. In this manuscript, we describe recommendations for the assay parameters that authors believe should be considered for assessment of ELISpot method performance. These include assessment of the assay LOD, precision, specificity, linearity, range of detection, accuracy, and robustness.
Industry-wide experiences with the assessment of cellular immune response against AAV vectors were applied, wherever appropriate, as the basis for the recommendations made in this manuscript. Our paper is a result of a consensus among multiple scientists involved with various aspects of AAV-based gene therapy development. It is hoped that recommendations outlined here will help clinicians, drug developers, and bioanalytical scientists to determine the need and value for cellular response evaluation. Proposed parameters for qualification of an ELISpot protocol are intended to promote harmonization across the industry. We would like to conclude that the relevance of evaluating the cellular immune response in blood or even evaluating the cellular response at all should be based, when possible, upon a cogent scientific hypothesis before embarking on developing cellular immune response assays or collecting samples in studies. The requirement to conduct cellular immune response measurements should be based on a risk assessment considering all possibly relevant parameters (patient population, ROA, etc.) and a structured data assessment strategy including sampling and biomarker selection. Assessment should be performed utilizing methods that are optimized for the parameters listed in the manuscript to enable further improvement of GTx product design and quality to reduce immune response triggers.
Acknowledgements
The authors would like to thank J.S. Havert for the assistance in creating Fig. 1.
Abbreviations
- AAT
Alpha-1-antitrypsin
- AAV
Adeno-associated virus
- Ab
Antibody
- AE
Adverse event
- Ag
Antigen
- ALT
Alanine aminotransferase
- AST
Aspartate aminotransferase
- CCP
Confirmatory cut point
- CDx
Companion diagnostic
- CEF
Lyophilized mixture of peptides from cytomegalovirus (CMV), Epstein-Barr (EBV), and influenza viruses
- CEFx
CEF plus peptides from a multitude of other pathogens
- CHOP-INTEND
Children’s Hospital of Philadelphia Infant Test of Neuromuscular Disorders
- CK
Creatine kinase
- CMV
Cytomegalovirus
- ConA
Concanavalin A
- CRO
Contract research organization
- CTL
Cytotoxic T lymphocytes
- CV
Coefficient of variability
- DFR
Distribution-free resampling
- DMSO
Dimethyl sulfoxide
- EBV
Epstein-Barr virus
- EDTA
Ethylenediaminetetraacetic acid
- ELISpot
Enzyme-linked immunosorbent spot
- EPAR
European Public Assessment Report
- FDA
US Food and Drug Administration
- FVIII
Factor VIII
- gc
Genome copies
- GTx
Gene therapy
- HIV
Human immunodeficiency virus
- HLA
Human leukocyte Ag
- ICS
Intracellular cytokine staining
- IFN-γ
Interferon-γ
- IL
Interleukin
- IM
Intramuscularly
- IND
Investigational new drug
- Iono
Ionomycin
- LLOQ
Lower limit of quantification
- LOD
Limit of detection
- LPL
Lipoprotein lipase
- MHC
Major histocompatibility complex
- NAbs
Neutralizing Abs
- NHP
Non-human primates
- NK cell
Natural killer cell
- PBMC
Peripheral blood mononuclear cell
- PD
Pharmacodynamics
- PHA
Phytohaemagglutinin
- PMA
Phorbol 12-myristate 13-acetate
- PVDF
Polyvinylidene difluoride
- QC
Quality control
- RPE65
Retinal pigment epithelium-specific 65 kDa protein
- SBA
Summary basis of approval
- SEB
Staphylococcal enterotoxin B
- SFU
Spot-forming units
- SOP
Standard operating procedure
- TAb
Total Ab
- TE
Target engagement
- TI
Transduction inhibition
- ULOQ
Upper limit of quantification
- vg
Vector genomes
Author Contribution
BG, MA, GB, MF, TH, MH, SJ, VJ, BL, YDM, AM, MM, RN, CV, and B.W. contributed equally in writing of the review.
Declarations
Conflict of Interest
The authors are employed by and receive compensation from companies that are involved in development of gene therapy modality therapeutics and are listed on the title page of the manuscript. The authors have no other relevant affiliations or financial involvements with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
Disclaimer
The views and conclusion presented in this paper are those of the authors and do not necessarily reflect the representative affiliation or company’s position on the subject.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.FDA: Human Gene Therapy for Neurodegenerative Diseases. Draft Guidance for Industry. https://www.fda.gov/media/144886/download (2021). Accessed 2022.
- 2.FDA: Human Gene Therapy for Hemophilia Guidance for Industry. https://www.fda.gov/media/113799/download (2020). Accessed.
- 3.FDA: Human Gene Therapy for Rare Diseases Guidance for Industry. https://www.fda.gov/media/113807/downlod (2020). Accessed.
- 4.FDA: Human Gene Therapy for Retinal Disorders Guidance for Industry. https://www.fda.gov/media/124641/download (2020). Accessed.
- 5.Takeuchi A, Saito T. CD4 CTL, a cytotoxic subset of CD4(+) T cells, their differentiation and function. Front Immunol. 2017;8:194. doi: 10.3389/fimmu.2017.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aslan N, Yurdaydin C, Wiegand J, Greten T, Ciner A, Meyer MF, et al. Cytotoxic CD4 T cells in viral hepatitis. J Viral Hepat. 2006;13(8):505–514. doi: 10.1111/j.1365-2893.2006.00723.x. [DOI] [PubMed] [Google Scholar]
- 7.Arjomandnejad M, Sylvia K, Blackwood M, Nixon T, Tang Q, Muhuri M, et al. Modulating immune responses to AAV by expanded polyclonal T-regs and capsid specific chimeric antigen receptor T-regulatory cells. Mol Ther Methods Clin Dev. 2021;23:490–506. doi: 10.1016/j.omtm.2021.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mueller C, Chulay JD, Trapnell BC, Humphries M, Carey B, Sandhaus RA, et al. Human Treg responses allow sustained recombinant adeno-associated virus-mediated transgene expression. J Clin Invest. 2013;123(12):5310–5318. doi: 10.1172/jci70314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mingozzi F, Maus MV, Hui DJ, Sabatino DE, Murphy SL, Rasko JE, et al. CD8(+) T-cell responses to adeno-associated virus capsid in humans. Nat Med. 2007;13(4):419–422. doi: 10.1038/nm1549. [DOI] [PubMed] [Google Scholar]
- 10.Manno CS, Pierce GF, Arruda VR, Glader B, Ragni M, Rasko JJ, et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat Med. 2006;12(3):342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 11.Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, Linch DC, et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med. 2011;365(25):2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palaschak B, Marsic D, Herzog RW, Zolotukhin S, Markusic DM. An Immune-competent murine model to study elimination of AAV-transduced hepatocytes by capsid-specific CD8(+) T cells. Mol Ther Methods Clin Dev. 2017;5:142–152. doi: 10.1016/j.omtm.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martino AT, Markusic DM. Immune response mechanisms against AAV vectors in animal models. Mol Ther Methods Clin Dev. 2020;17:198–208. doi: 10.1016/j.omtm.2019.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nathwani AC, Reiss UM, Tuddenham EG, Rosales C, Chowdary P, McIntosh J, et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med. 2014;371(21):1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ertl HCJ, High KA. Impact of AAV capsid-specific T-cell responses on design and outcome of clinical gene transfer trials with recombinant adeno-associated viral vectors: an evolving controversy. Hum Gene Ther. 2017;28(4):328–337. doi: 10.1089/hum.2016.172. [DOI] [PubMed] [Google Scholar]
- 16.Jiang H, Couto LB, Patarroyo-White S, Liu T, Nagy D, Vargas JA, et al. Effects of transient immunosuppression on adenoassociated, virus-mediated, liver-directed gene transfer in rhesus macaques and implications for human gene therapy. Blood. 2006;108(10):3321–3328. doi: 10.1182/blood-2006-04-017913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, Hirsch M, Asokan A, Zeithaml B, Ma H, Kafri T, et al. Adeno-associated virus type 2 (AAV2) capsid-specific cytotoxic T lymphocytes eliminate only vector-transduced cells coexpressing the AAV2 capsid in vivo. J Virol. 2007;81(14):7540–7547. doi: 10.1128/jvi.00529-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Figueredo J, Calcedo R, Lin J, Wilson JM. Cross-presentation of adeno-associated virus serotype 2 capsids activates cytotoxic T cells but does not render hepatocytes effective cytolytic targets. Hum Gene Ther. 2007;18(3):185–194. doi: 10.1089/hum.2007.001. [DOI] [PubMed] [Google Scholar]
- 19.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17(7):1055–1065. doi: 10.1128/cvi.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plotkin SA, Plotkin SA. Correlates of vaccine-induced immunity. Clin Infect Dis. 2008;47(3):401–409. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 21.Kaech SM, Wherry EJ, Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol. 2002;2(4):251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 22.Calarota SA, Baldanti F. Enumeration and characterization of human memory T cells by enzyme-linked immunospot assays. Clin Dev Immunol. 2013;2013:637649. doi: 10.1155/2013/637649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bowles DE, McPhee SW, Li C, Gray SJ, Samulski JJ, Camp AS, et al. Phase 1 gene therapy for Duchenne muscular dystrophy using a translational optimized AAV vector. Mol Ther. 2012;20(2):443–455. doi: 10.1038/mt.2011.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferreira V, Petry H, Salmon F. Immune responses to AAV-vectors, the Glybera example from bench to bedside. Front Immunol. 2014;5:82. doi: 10.3389/fimmu.2014.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferreira V, Twisk J, Kwikkers K, Aronica E, Brisson D, Methot J, et al. Immune responses to intramuscular administration of alipogene tiparvovec (AAV1-LPL(S447X)) in a phase II clinical trial of lipoprotein lipase deficiency gene therapy. Hum Gene Ther. 2014;25(3):180–188. doi: 10.1089/hum.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mingozzi F, Meulenberg JJ, Hui DJ, Basner-Tschakarjan E, Hasbrouck NC, Edmonson SA, et al. AAV-1-mediated gene transfer to skeletal muscle in humans results in dose-dependent activation of capsid-specific T cells. Blood. 2009;114(10):2077–2086. doi: 10.1182/blood-2008-07-167510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long BR, Veron P, Kuranda K, Hardet R, Mitchell N, Hayes GM, et al. Early phase clinical immunogenicity of valoctocogene roxaparvovec, an AAV5-mediated gene therapy for hemophilia A. Mol Ther. 2021;29(2):597–610. doi: 10.1016/j.ymthe.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Au HKE, Isalan M, Mielcarek M. Gene therapy advances: a meta-analysis of AAV usage in clinical settings. Front Med (Lausanne) 2021;8:809118. doi: 10.3389/fmed.2021.809118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nathwani AC, Rosales C, McIntosh J, Rastegarlari G, Nathwani D, Raj D, et al. Long-term safety and efficacy following systemic administration of a self-complementary AAV vector encoding human FIX pseudotyped with serotype 5 and 8 capsid proteins. Mol Ther. 2011;19(5):876–885. doi: 10.1038/mt.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mingozzi F, High KA. Overcoming the host immune response to adeno-associated virus gene delivery vectors: the race between clearance, tolerance, neutralization, and escape. Ann Rev Virol. 2017;4(1):511–534. doi: 10.1146/annurev-virology-101416-041936. [DOI] [PubMed] [Google Scholar]
- 31.Colella P, Ronzitti G, Mingozzi F. Emerging issues in AAV-mediated in vivo gene therapy. Mol Ther Methods Clin Dev. 2018;8:87–104. doi: 10.1016/j.omtm.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson JS, Samulski RJ. Enhancement of adeno-associated virus infection by mobilizing capsids into and out of the nucleolus. J Virol. 2009;83(6):2632–2644. doi: 10.1128/jvi.02309-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brantly ML, Chulay JD, Wang L, Mueller C, Humphries M, Spencer LT, et al. Sustained transgene expression despite T lymphocyte responses in a clinical trial of rAAV1-AAT gene therapy. Proc Natl Acad Sci U S A. 2009;106(38):16363–16368. doi: 10.1073/pnas.0904514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bainbridge JW, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, et al. Effect of gene therapy on visual function in Leber's congenital amaurosis. N Engl J Med. 2008;358(21):2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 35.Hauswirth WW, Aleman TS, Kaushal S, Cideciyan AV, Schwartz SB, Wang L, et al. Treatment of Leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. Hum Gene Ther. 2008;19(10):979–990. doi: 10.1089/hum.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patton KS, Harrison MT, Long BR, Lau K, Holcomb J, Owen R, et al. Monitoring cell-mediated immune responses in AAV gene therapy clinical trials using a validated IFN-γ ELISpot method. Mol Ther Methods Clin Dev. 2021;22:183–195. doi: 10.1016/j.omtm.2021.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaudet D, Méthot J, Déry S, Brisson D, Essiembre C, Tremblay G, et al. Efficacy and long-term safety of alipogene tiparvovec (AAV1-LPLS447X) gene therapy for lipoprotein lipase deficiency: an open-label trial. Gene Ther. 2013;20(4):361–369. doi: 10.1038/gt.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaudet D, de Wal J, Tremblay K, Déry S, van Deventer S, Freidig A, et al. Review of the clinical development of alipogene tiparvovec gene therapy for lipoprotein lipase deficiency. Atheroscler Suppl. 2010;11(1):55–60. doi: 10.1016/j.atherosclerosissup.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Mingozzi F, Hasbrouck NC, Basner-Tschakarjan E, Edmonson SA, Hui DJ, Sabatino DE, et al. Modulation of tolerance to the transgene product in a nonhuman primate model of AAV-mediated gene transfer to liver. Blood. 2007;110(7):2334–2341. doi: 10.1182/blood-2007-03-080093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.FDA: Zolgensma. Prescribing Information https://www.fda.gov/media/126109/download (2022). Accessed 2022.
- 41.FDA: Luxturna. Prescribing Information. https://www.fda.gov/media/109906/download (2022). Accessed.
- 42.EMA: Valoctocogene roxaparvovec. https://www.ema.europa.eu/en/medicines/human/EPAR/roctavian-0 (2022). Accessed 30 October 2022.
- 43.FDA: HEMGENIX (etranacogene dezaparvovec-drlb) suspension, for intravenous infusion. Initial U.S. Approval: 2022. https://www.fda.gov/media/163467/download (2022). Accessed January 12 2023.
- 44.FDA: Luxturna. Approval History. https://www.fda.gov/media/110105/download (2018). Accessed.
- 45.EMA: Upstaza. Summary of product characteristics https://www.ema.europa.eu/en/documents/product-information/upstaza-epar-product-information_en.pdf (2022). Accessed April 2 2023.
- 46.FDA: Zolgensma. Clinical Review Documents. https://www.fda.gov/media/128116/download (2020). Accessed.
- 47.EMA: Zolgensma. Assessment report. https://www.ema.europa.eu/documents/assessment-report/zolgensma-epar-public-assessment-report_en.pdf (2020). Accessed.
- 48.EMA: Luxturna. Assessment Report. https://www.ema.europa.eu/en/documents/assessment-report/luxturna-epar-public-assessment-report_en.pdf (2019). Accessed April 2 2023.
- 49.Wang D, Tai PWL, Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov. 2019;18(5):358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dosmar E, Walsh J, Doyel M, Bussett K, Oladipupo A, Amer S, Goebel K. Targeting ocular drug delivery: an examination of local anatomy and current approaches. Bioengineering. 2022;9(1):41. 10.3390/bioengineering9010041. [DOI] [PMC free article] [PubMed]
- 51.Bucher K, Rodríguez-Bocanegra E, Dauletbekov D, Fischer MD. Immune responses to retinal gene therapy using adeno-associated viral vectors - implications for treatment success and safety. Prog Retin Eye Res. 2021;83:100915. doi: 10.1016/j.preteyeres.2020.100915. [DOI] [PubMed] [Google Scholar]
- 52.Gehrke M, Diedrichs-Möhring M, Bogedein J, Büning H, Michalakis S, Wildner G. Immunogenicity of novel AAV capsids for retinalgene therapy. Cells 2022;11(12):1881–97. 10.3390/cells11121881. [DOI] [PMC free article] [PubMed]
- 53.Ghoraba HH, Akhavanrezayat A, Karaca I, Yavari N, Lajevardi S, Hwang J, et al. Ocular gene therapy: a literature review with special focus on immune and inflammatory responses. Clin Ophthalmol. 2022;16:1753–1771. doi: 10.2147/opth.S364200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao O, Dobrzynski E, Wang L, Nayak S, Mingle B, Terhorst C, et al. Induction and role of regulatory CD4+CD25+ T cells in tolerance to the transgene product following hepatic in vivo gene transfer. Blood. 2007;110(4):1132–1140. doi: 10.1182/blood-2007-02-073304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sack BK, Herzog RW, Terhorst C, Markusic DM. Development of gene transfer for induction of antigen-specific tolerance. Mol Ther Methods Clin Dev. 2014;1:14013. doi: 10.1038/mtm.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ertl HCJ. Immunogenicity and toxicity of AAV gene therapy. Front Immunol. 2022;13:975803. doi: 10.3389/fimmu.2022.975803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herzog RW, Fields PA, Arruda VR, Brubaker JO, Armstrong E, McClintock D, et al. Influence of vector dose on factor IX-specific T and B cell responses in muscle-directed gene therapy. Hum Gene Ther. 2002;13(11):1281–1291. doi: 10.1089/104303402760128513. [DOI] [PubMed] [Google Scholar]
- 58.Kumar SRP, Hoffman BE, Terhorst C, de Jong YP, Herzog RW. The balance between CD8(+) T cell-mediated clearance of AAV-encoded antigen in the liver and tolerance is dependent on the vector dose. Mol Ther. 2017;25(4):880–891. doi: 10.1016/j.ymthe.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Streeck H, Frahm N, Walker BD. The role of IFN-gamma Elispot assay in HIV vaccine research. Nat Protoc. 2009;4(4):461–469. doi: 10.1038/nprot.2009.7. [DOI] [PubMed] [Google Scholar]
- 60.Slota M, Lim JB, Dang Y, Disis ML. ELISpot for measuring human immune responses to vaccines. Expert Rev Vaccines. 2011;10(3):299–306. doi: 10.1586/erv.10.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van der Burg SH, Kalos M, Gouttefangeas C, Janetzki S, Ottensmeier C, Welters MJ, et al. Harmonization of immune biomarker assays for clinical studies. Sci Transl Med. 2011;3(108):108ps44. doi: 10.1126/scitranslmed.3002785. [DOI] [PubMed] [Google Scholar]
- 62.De Rosa SC. Vaccine applications of flow cytometry. Methods. 2012;57(3):383–391. doi: 10.1016/j.ymeth.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Britten CM, Walter S, Janetzki S. Immunological monitoring to rationally guide AAV gene therapy. Front Immunol. 2013;4:273. doi: 10.3389/fimmu.2013.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Janetzki S. Elispot for Rookies (And Experts Too). Techniques in life science and biomedicine for the non-expert. Cham: Springer; 2016.
- 65.Jahnmatz P, Nyabundi D, Sundling C, Widman L, Mwacharo J, Musyoki J, et al. Plasmodium falciparum-specific memory B-cell and antibody responses are associated with immunity in children living in an endemic area of Kenya. Front Immunol. 2022;13:799306. doi: 10.3389/fimmu.2022.799306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Czerkinsky CC, Nilsson LA, Nygren H, Ouchterlony O, Tarkowski A. A solid-phase enzyme-linked immunospot (ELISPOT) assay for enumeration of specific antibody-secreting cells. J Immunol Methods. 1983;65(1–2):109–121. doi: 10.1016/0022-1759(83)90308-3. [DOI] [PubMed] [Google Scholar]
- 67.Janetzki S, Price L, Britten CM, van der Burg SH, Caterini J, Currier JR, et al. Performance of serum-supplemented and serum-free media in IFNgamma Elispot Assays for human T cells. Cancer Immunol Immunother. 2010;59(4):609–618. doi: 10.1007/s00262-009-0788-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Janetzki S, Panageas KS, Ben-Porat L, Boyer J, Britten CM, Clay TM, et al. Results and harmonization guidelines from two large-scale international Elispot proficiency panels conducted by the Cancer Vaccine Consortium (CVC/SVI) Cancer Immunol Immunother. 2008;57(3):303–315. doi: 10.1007/s00262-007-0380-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Janetzki S, Price L, Schroeder H, Britten CM, Welters MJ, Hoos A. Guidelines for the automated evaluation of Elispot assays. Nat Protoc. 2015;10(7):1098–1115. doi: 10.1038/nprot.2015.068. [DOI] [PubMed] [Google Scholar]
- 70.Janetzki S, Rueger M, Dillenbeck T. Stepping up ELISpot: multi-level analysis in FluoroSpot assays. Cells. 2014;3(4):1102–1115. doi: 10.3390/cells3041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Axelsson B. Detection and enumeration of cytokine-secreting cells by FluoroSpot. Methods Mol Biol. 2022;2386:81–99. doi: 10.1007/978-1-0716-1771-7_6. [DOI] [PubMed] [Google Scholar]
- 72.Klausner RD, Donaldson JG, Lippincott-Schwartz J. Brefeldin A: insights into the control of membrane traffic and organelle structure. J Cell Biol. 1992;116(5):1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jung T, Schauer U, Heusser C, Neumann C, Rieger C. Detection of intracellular cytokines by flow cytometry. J Immunol Methods. 1993;159(1–2):197–207. doi: 10.1016/0022-1759(93)90158-4. [DOI] [PubMed] [Google Scholar]
- 74.Metzger P, Kirchleitner SV, Koenig LM, Hörth C, Kobold S, Endres S, et al. Dying cells expose a nuclear antigen cross-reacting with anti-PD-1 monoclonal antibodies. Sci Rep. 2018;8(1):8810. doi: 10.1038/s41598-018-27125-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kozlova AA, Verkhovskii RA, Ermakov AV, Bratashov DN. Changes in autofluorescence level of live and dead cells for mouse cell lines. J Fluoresc. 2020;30(6):1483–1489. doi: 10.1007/s10895-020-02611-1. [DOI] [PubMed] [Google Scholar]
- 76.Maecker HT, Frey T, Nomura LE, Trotter J. Selecting fluorochrome conjugates for maximum sensitivity. Cytometry A. 2004;62(2):169–173. doi: 10.1002/cyto.a.20092. [DOI] [PubMed] [Google Scholar]
- 77.Nguyen R, Perfetto S, Mahnke YD, Chattopadhyay P, Roederer M. Quantifying spillover spreading for comparing instrument performance and aiding in multicolor panel design. Cytometry A. 2013;83(3):306–315. doi: 10.1002/cyto.a.22251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mahnke YD, Roederer M. Optimizing a multicolor immunophenotyping assay. Clin Lab Med. 2007;27(3):469–85.v. 10.1016/j.cll.2007.05.002. [DOI] [PMC free article] [PubMed]
- 79.FDA: Bioanalytical Method Validation Guidance for Industry. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/bioanalytical-method-validation-guidance-industry (2018). Accessed.
- 80.Hoffmeister B, Bunde T, Rudawsky IM, Volk HD, Kern F. Detection of antigen-specific T cells by cytokine flow cytometry: the use of whole blood may underestimate frequencies. Eur J Immunol. 2003;33(12):3484–3492. doi: 10.1002/eji.200324223. [DOI] [PubMed] [Google Scholar]
- 81.Thornthwaite JT, Rosenthal PK, Vazquez DA, Seckinger D. The effects of anticoagulant and temperature on the measurements of helper and suppressor cells. Diagn Immunol. 1984;2(3):167–174. [PubMed] [Google Scholar]
- 82.Bissoyi A, Nayak B, Pramanik K, Sarangi SK. Targeting cryopreservation-induced cell death: a review. Biopreserv Biobank. 2014;12(1):23–34. doi: 10.1089/bio.2013.0032. [DOI] [PubMed] [Google Scholar]
- 83.Kierstead LS, Dubey S, Meyer B, Tobery TW, Mogg R, Fernandez VR, et al. Enhanced rates and magnitude of immune responses detected against an HIV vaccine: effect of using an optimized process for isolating PBMC. AIDS Res Hum Retroviruses. 2007;23(1):86–92. doi: 10.1089/aid.2006.0129. [DOI] [PubMed] [Google Scholar]
- 84.Bull M, Lee D, Stucky J, Chiu YL, Rubin A, Horton H, et al. Defining blood processing parameters for optimal detection of cryopreserved antigen-specific responses for HIV vaccine trials. J Immunol Methods. 2007;322(1–2):57–69. doi: 10.1016/j.jim.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McKenna KC, Beatty KM, Vicetti Miguel R, Bilonick RA. Delayed processing of blood increases the frequency of activated CD11b+ CD15+ granulocytes which inhibit T cell function. J Immunol Methods. 2009;341(1–2):68–75. doi: 10.1016/j.jim.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 86.De Rose R, Taylor EL, Law MG, van der Meide PH, Kent SJ. Granulocyte contamination dramatically inhibits spot formation in AIDS virus-specific ELISpot assays: analysis and strategies to ameliorate. J Immunol Methods. 2005;297(1–2):177–186. doi: 10.1016/j.jim.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 87.Schmielau J, Finn OJ. Activated granulocytes and granulocyte-derived hydrogen peroxide are the underlying mechanism of suppression of t-cell function in advanced cancer patients. Cancer Res. 2001;61(12):4756–4760. [PubMed] [Google Scholar]
- 88.Zea AH, Rodriguez PC, Culotta KS, Hernandez CP, DeSalvo J, Ochoa JB, et al. L-Arginine modulates CD3zeta expression and T cell function in activated human T lymphocytes. Cell Immunol. 2004;232(1–2):21–31. doi: 10.1016/j.cellimm.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 89.Becton.Dickinson.and.Company: Vacutainer® CPT™ Cell Preparation Tube with Sodium Heparin. https://www.bdbiosciences.com/content/dam/bdb/products/global/blood-collection/blood-collection-tubes/362753_base/pdf/VDP40393.pdf (2020). Accessed.
- 90.Smith JG, Liu X, Kaufhold RM, Clair J, Caulfield MJ. Development and validation of a gamma interferon ELISPOT assay for quantitation of cellular immune responses to varicella-zoster virus. Clin Diagn Lab Immunol. 2001;8(5):871–879. doi: 10.1128/cdli.8.5.871-879.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weinberg A, Song LY, Wilkening C, Sevin A, Blais B, Louzao R, et al. Optimization and limitations of use of cryopreserved peripheral blood mononuclear cells for functional and phenotypic T-cell characterization. Clin Vaccine Immunol. 2009;16(8):1176–1186. doi: 10.1128/cvi.00342-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Altman SA, Randers L, Rao G. Comparison of trypan blue dye exclusion and fluorometric assays for mammalian cell viability determinations. Biotechnol Prog. 1993;9(6):671–674. doi: 10.1021/bp00024a017. [DOI] [PubMed] [Google Scholar]
- 93.Smith JG, Joseph HR, Green T, Field JA, Wooters M, Kaufhold RM, et al. Establishing acceptance criteria for cell-mediated-immunity assays using frozen peripheral blood mononuclear cells stored under optimal and suboptimal conditions. Clin Vaccine Immun. 2007;14(5):527–537. doi: 10.1128/cvi.00435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lenders K, Ogunjimi B, Beutels P, Hens N, Van Damme P, Berneman ZN, et al. The effect of apoptotic cells on virus-specific immune responses detected using IFN-gamma ELISPOT. J Immunol Methods. 2010;357(1–2):51–54. doi: 10.1016/j.jim.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 95.Disis ML, dela Rosa C, Goodell V, Kuan LY, Chang JC, Kuus-Reichel K, et al. Maximizing the retention of antigen specific lymphocyte function after cryopreservation. J Immunol Methods. 2006;308(1–2):13–8. doi: 10.1016/j.jim.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 96.Russell ND, Hudgens MG, Ha R, Havenar-Daughton C, McElrath MJ. Moving to human immunodeficiency virus type 1 vaccine efficacy trials: defining T cell responses as potential correlates of immunity. J Infect Dis. 2003;187(2):226–242. doi: 10.1086/367702. [DOI] [PubMed] [Google Scholar]
- 97.Römer PS, Berr S, Avota E, Na SY, Battaglia M, ten Berge I, et al. Preculture of PBMCs at high cell density increases sensitivity of T-cell responses, revealing cytokine release by CD28 superagonist TGN1412. Blood. 2011;118(26):6772–6782. doi: 10.1182/blood-2010-12-319780. [DOI] [PubMed] [Google Scholar]
- 98.Wegner J, Hackenberg S, Scholz CJ, Chuvpilo S, Tyrsin D, Matskevich AA, et al. High-density preculture of PBMCs restores defective sensitivity of circulating CD8 T cells to virus- and tumor-derived antigens. Blood. 2015;126(2):185–194. doi: 10.1182/blood-2015-01-622704. [DOI] [PubMed] [Google Scholar]
- 99.Kutscher S, Dembek CJ, Deckert S, Russo C, Körber N, Bogner JR, et al. Overnight resting of PBMC changes functional signatures of antigen specific T- cell responses: impact for immune monitoring within clinical trials. PloS One. 2013;8(10):e76215. doi: 10.1371/journal.pone.0076215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Santos R, Buying A, Sabri N, Yu J, Gringeri A, Bender J, et al. Improvement of IFNg ELISPOT performance following overnight resting of frozen PBMC samples confirmed through rigorous statistical analysis. Cells. 2014;4(1):1–18. doi: 10.3390/cells4010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kern F, Faulhaber N, Frömmel C, Khatamzas E, Prösch S, Schönemann C, et al. Analysis of CD8 T cell reactivity to cytomegalovirus using protein-spanning pools of overlapping pentadecapeptides. Eur J Immunol. 2000;30(6):1676–1682. doi: 10.1002/1521-4141(200006)30:6<1676::Aid-immu1676>3.0.Co;2-v. [DOI] [PubMed] [Google Scholar]
- 102.Kiecker F, Streitz M, Ay B, Cherepnev G, Volk HD, Volkmer-Engert R, et al. Analysis of antigen-specific T-cell responses with synthetic peptides–what kind of peptide for which purpose? Hum Immunol. 2004;65(5):523–536. doi: 10.1016/j.humimm.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 103.Maecker HT, Dunn HS, Suni MA, Khatamzas E, Pitcher CJ, Bunde T, et al. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J Immunol Methods. 2001;255(1–2):27–40. doi: 10.1016/s0022-1759(01)00416-1. [DOI] [PubMed] [Google Scholar]
- 104.Fiore-Gartland A, Manso BA, Friedrich DP, Gabriel EE, Finak G, Moodie Z, et al. Pooled-peptide epitope mapping strategies are efficient and highly sensitive: an evaluation of methods for identifying human T cell epitope specificities in large-scale HIV vaccine efficacy trials. PloS One. 2016;11(2):e0147812. doi: 10.1371/journal.pone.0147812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Currier JR, Kuta EG, Turk E, Earhart LB, Loomis-Price L, Janetzki S, et al. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. J Immunol Methods. 2002;260(1–2):157–172. doi: 10.1016/s0022-1759(01)00535-x. [DOI] [PubMed] [Google Scholar]
- 106.Cox JH, Ferrari G, Janetzki S. Measurement of cytokine release at the single cell level using the ELISPOT assay. Methods. 2006;38(4):274–282. doi: 10.1016/j.ymeth.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 107.Maecker HT, Rinfret A, D'Souza P, Darden J, Roig E, Landry C, et al. Standardization of cytokine flow cytometry assays. BMC Immunol. 2005;6:13. doi: 10.1186/1471-2172-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wood B, Jevremovic D, Béné MC, Yan M, Jacobs P, Litwin V. Validation of cell-based fluorescence assays: practice guidelines from the ICSH and ICCS - part V - assay performance criteria. Cytometry B Clin Cytom. 2013;84(5):315–323. doi: 10.1002/cyto.b.21108. [DOI] [PubMed] [Google Scholar]
- 109.Kagina BM, Mansoor N, Kpamegan EP, Penn-Nicholson A, Nemes E, Smit E, et al. Qualification of a whole blood intracellular cytokine staining assay to measure mycobacteria-specific CD4 and CD8 T cell immunity by flow cytometry. J Immunol Methods. 2015;417:22–33. doi: 10.1016/j.jim.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cossarizza A, Chang HD, Radbruch A, Acs A, Adam D, Adam-Klages S, et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition). Eur J Immunol. 2019;49(10):1457–973. 10.1002/eji.201970107. [DOI] [PMC free article] [PubMed]
- 111.Yang F, Patton K, Kasprzyk T, Long B, Gupta S, Zoog SJ, et al. Validation of an IFN-gamma ELISpot assay to measure cellular immune responses against viral antigens in non-human primates. Gene Ther. 2022;29(1–2):41–54. doi: 10.1038/s41434-020-00214-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moodie Z, Price L, Janetzki S, Britten CM. Response determination criteria for ELISPOT: toward a standard that can be applied across laboratories. Methods Mol Biol. 2012;792:185–196. doi: 10.1007/978-1-61779-325-7_15. [DOI] [PubMed] [Google Scholar]
- 113.RUnDFR: The RunDFR Web Tool. https://rundfr.fredhutch.org/ Accessed October 2022.
- 114.Maecker HT, Hassler J, Payne JK, Summers A, Comatas K, Ghanayem M, et al. Precision and linearity targets for validation of an IFNgamma ELISPOT, cytokine flow cytometry, and tetramer assay using CMV peptides. BMC Immunol. 2008;9:9. doi: 10.1186/1471-2172-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Immudex: T-cell ELISpot Proficiency Panel. https://www.immudex.com/services/proficiency-panels/t-cell-elispot-proficiency-panel/ (2022). Accessed.
- 116.Germann A, Schulz JC, Kemp-Kamke B, Zimmermann H, von Briesen H. Standardized serum-free cryomedia maintain peripheral blood mononuclear cell viability, recovery, and antigen-specific T-cell response compared to fetal calf serum-based medium. Biopreserv Biobank. 2011;9(3):229–236. doi: 10.1089/bio.2010.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li B, Yang C, Jia G, Liu Y, Wang N, Yang F, et al. Comprehensive evaluation of the effects of long-term cryopreservation on peripheral blood mononuclear cells using flow cytometry. BMC Immunol. 2022;23(1):30. doi: 10.1186/s12865-022-00505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.FDA: Human Gene Therapy for Hemophilia. Draft Guidance for Industry. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/human-gene-therapy-hemophilia (2018). Accessed 2019.
- 119.Wang L, Calcedo R, Bell P, Lin J, Grant RL, Siegel DL, et al. Impact of pre-existing immunity on gene transfer to nonhuman primate liver with adeno-associated virus 8 vectors. Hum Gene Ther. 2011;22(11):1389–1401. doi: 10.1089/hum.2011.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang L, Calcedo R, Wang H, Bell P, Grant R, Vandenberghe LH, et al. The pleiotropic effects of natural AAV infections on liver-directed gene transfer in macaques. Mol Ther. 2010;18(1):126–134. doi: 10.1038/mt.2009.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Semmes EC, Chen JL, Goswami R, Burt TD, Permar SR, Fouda GG. Understanding early-life adaptive immunity to guide interventions for pediatric health. Front Immunol. 2020;11:595297. doi: 10.3389/fimmu.2020.595297. [DOI] [PMC free article] [PubMed] [Google Scholar]