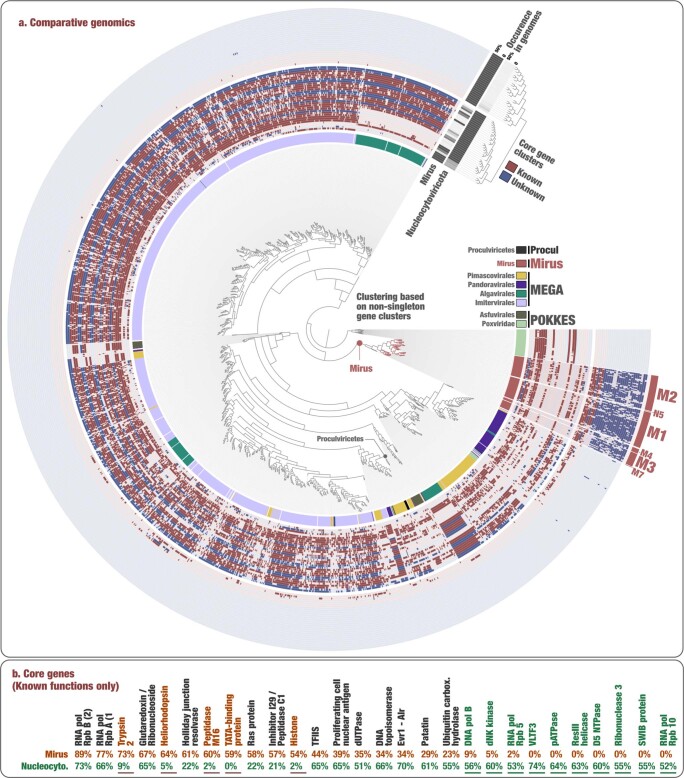
Extended Data Fig. 7. Functional clustering of abundant and widespread marine viruses within mirusviruses and Nucleocytoviricota.
In panel A, the inner tree is a clustering of ‘Mirusviricota’ and Nucleocytoviricota genomes >100 kbp in length based on the occurrence of all the non-singleton gene clusters (Euclidean distance), rooted with the Chordopoxvirinae subfamily of Poxviridae genomes. Rings of information display the main taxonomy of Nucleocytoviricota as well as the occurrence of 60 gene clusters detected in at least 50% of ‘Mirusviricota’ or Nucleocytoviricota. The 60 gene clusters are clustered based on their occurrence (absence/presence) across the genomes. Panel B displays the occurrence of gene clusters of known Pfam functions detected in at least 50% of ‘Mirusviricota’ or Nucleocytoviricota genomes.