Abstract
Background:
During cardiomyocyte (CM) maturation, the centrosome, which functions as a microtubule organizing center (MTOC) in CMs, undergoes dramatic structural reorganization where its components reorganize from being localized at the centriole to the nuclear envelope. This developmentally programmed process, referred to as centrosome reduction, has been previously associated with cell cycle exit. However, understanding of how this process influences CM cell biology, and whether its disruption results in human cardiac disease, remains unknown. Here, we study this phenomenon in an infant with a rare case of infantile dilated cardiomyopathy (iDCM) who presented with left ventricular ejection fraction of 18% and disrupted sarcomere and mitochondria structure.
Methods:
We performed an analysis beginning with an infant patient who presented with a rare case of iDCM. We derived induced pluripotent stem cells (iPSCs) from the patient to model iDCM in vitro. We performed whole exome sequencing on the patient and his parents for causal gene analysis. CRISPR/Cas9 mediated gene knockout and correction in vitro were used to confirm whole exome sequencing results. Zebrafish and Drosophila models were utilized for in vivo validation of causal gene. Matrigel mattress technology and single cell RNA-sequencing were used to further characterize iDCM-CMs.
Results:
Whole-exome sequencing and CRISPR/Cas9 gene knockout/correction identified the centrosomal protein rotatin (RTTN) as the causal gene underlying the patient’s condition, representing the first time a centrosome defect has been implicated in a non-syndromic dilated cardiomyopathy (DCM). Genetic knockdowns in zebrafish and Drosophila confirmed an evolutionarily conserved requirement of RTTN for cardiac structure and function. Single cell RNA-sequencing of iDCM-CMs showed impaired maturation of iDCM-CMs, which underlie the observed CM structural and functional deficits. Interestingly, we also observed persistent localization of the centrosome at the centriole, contrasting with expected programmed perinuclear reorganization, which led to subsequent global microtubule network defects. Finally, we identified a small molecule that restored centrosome reorganization, and improved the structure and contractility of iDCM-CMs.
Conclusions:
This study is the first to demonstrate a case of human disease caused by a defect in centrosome reduction. Further, we uncovered a novel role for RTTN in perinatal cardiac development, and identified a potential therapeutic strategy for centrosome-related iDCM. Future study aimed at identifying variants in centrosome components may uncover additional contributors to human cardiac disease.
Keywords: Infantile Dilated Cardiomyopathy, Rotatin, Centrosome, Cardiomyocyte, iPSC
Introduction:
Infantile dilated cardiomyopathy (iDCM) is a rare condition of multiple etiologies which causes significant morbidity and mortality1–5. Of the studied infantile cardiomyopathies, dilated cardiomyopathy is the most prevalent, responsible for approximately 50% of infantile/pediatric cardiomyopathy cases2. Such cases are particularly difficult to manage, and many will necessitate heart transplant as the final management approach2. Given the lack of donor hearts available, particularly in the infant demographic, there is thus a significant need to better understand the etiology of iDCM for development of tools for identification and targeted approaches for effective therapeutics.
Previous studies have suggested that 27–54% of iDCM cases have a genetic component5–8. Common mutations in genes related to sarcomere structure, such as MYH7, MYBPC3 and TNNT2, nuclear envelope, such as LMNA, and cytoskeleton, such as DES and DMD, have described previously4–8. Increased widespread genetic testing abilities have begun to help identify novel classes of genes suspected to be responsible for iDCM, greatly expanding our understanding of the etiology of this condition.
Here, we describe a male infant who presented with infantile dilated cardiomyopathy (iDCM) with a left ventricular ejection fraction of 18% (Figure S1; Supplemental Clinical Information). He underwent a successful heart transplant and does not presently exhibit any neurocognitive or neuromuscular deficits. The infant was born to non-consanguineous parents without a family history of cardiomyopathy. An endomyocardial biopsy and transmission electron microscopy (TEM) of the infant’s heart revealed cardiomyocytes with shortage of myofilament structures, pleomorphic mitochondria, and indistinct Z-lines (Figure 1a). Given that multiple aspects of cardiac structure and function were significantly affected shortly after birth in this case of iDCM, we were interested in further investigating whether this patient’s condition had a genetic etiology.
Figure 1. CMs derived from the iDCM patient’s iPSCs recapitulate cardiac defects.
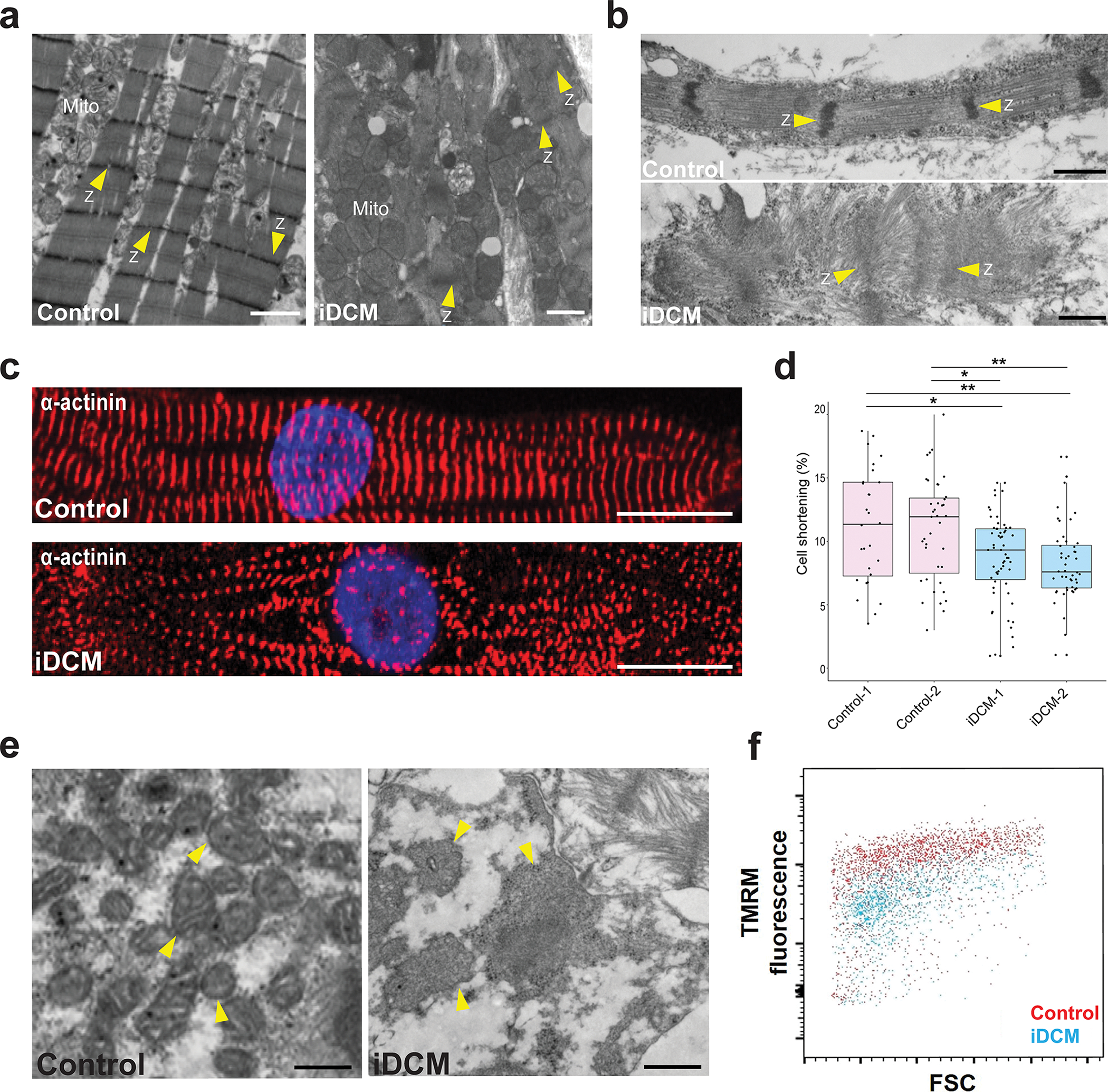
a, TEM images of hearts. Left, TEM of normal heart showing well-organized myofilaments, distinct Z-lines (Z; arrowheads) and mitochondria (Mito) with distinct cristae. Right, EM of the iDCM patient’s heart showing severely disorganized myofibrils with indistinct Z-lines and dysmorphic mitochondria (Mito) without appreciable cristae. Scale bar = 1 μm. b, iPSC-CM from a healthy control exhibiting organized myofilaments with distinct Z-lines (Z; arrowheads) (top), whereas the iPSC-CM derived from the iDCM patient (bottom) exhibit disorganized myofibrils without distinct Z-lines (Z). Scale bar = 0.5 μm. c, Representative immunofluorescence images of α-actinin staining of control (top) and iDCM (bottom) CMs. Scale bar = 10 μm. d, Compared with control-CMs derived from 2 independent healthy control iPSC lines (Control-1, −2), iDCM-CMs from 2 independent iPSC lines (iDCM-1, −2) exhibited significantly reduced cell shortening (~11% vs ~8%). p value by one-way ANOVA and post-hoc Tukey *p < 0.01, **p < 0.05. n = 28 Control 1-CMs, 39 Control 2-CMs, 59 iDCM 1-CMs, 48 iDCM 2-CMs. Center line = median; whiskers = 1.5IQR. e, TEM images of iPSC-CMs focused on mitochondria. Left, mitochondria of control iPSC-CMs are easily distinguishable and have distinct cristae, whereas mitochondria (arrowheads) from iDCM-CMs appear larger and swollen, without clear cristae. Scale bar = 0.5 μm. f, Fluorescence-activated cell sorting analysis demonstrating that iDCM-CMs have quantitatively lower TMRM uptake and hence lower mitochondrial membrane potentials than control iPSC-CMs. TEM images of CMs were acquired at 45 days of differentiation. Other studies were done at 35 days of differentiation.
Methods
Please refer to Supplemental Material for detailed methods and information on statistical tests.
Human iPSC lines were generated from healthy control subjects using an episomal approach and validated previously9–11. All protocols were approved by the Vanderbilt University Institutional Review Board and informed consent was given.
All data and methods used in analysis will be made available to any researcher upon request. scRNAsequencing data has been deposited in Gene Expression Omnibus (GSE184899) and full code can be found at: https://github.com/miyamoto5/cDCM.
Results
An iPSC model recapitulates iDCM phenotype
A male infant presented with infantile dilated cardiomyopathy (iDCM) with a left ventricular ejection fraction of 18% (Figure S1; Supplemental Clinical Information). He underwent a successful heart transplant and does not presently exhibit any neurocognitive or neuromuscular deficits. We first sought to test the hypothesis that this case of infantile dilated cardiomyopathy (iDCM) had a genetic etiology by deriving induced pluripotent stem cells (iPSCs) from the infant patient (Figure S2a,b) and generating iPSC-derived cardiomyocytes (CMs) (Figure S2c,d). Two independent iPSC lines were generated from the patient (Figure S2) and were compared to two independent iPSCs lines from healthy donors10–12. Compared with CMs from healthy donors, the iDCM-CMs exhibited profound sarcomere defects (Figure 1b), recapitulating the cardiac defects seen in the explanted heart (Figure 1a). Furthermore, immunofluorescence imaging demonstrated that iDCM-CMs had disorganized z-lines when compared with control-CMs (Figure 1c; Figure S3a–b). Cell area of iDCM-CMs was significantly decreased compared to Control-CMs (Figure S3c). The iDCM-CMs from two independent iPSC lines exhibited reduced contractility compared with controls (~8% shortening versus ~11%, respectively; p < 0.01; Figure 1d) on Matrigel mattress shortening analysis10.
To observe mitochondrial morphology of iDCM-CMs, we first visualized via TEM and we observed grossly dysmorphic mitochondria (Figure 1e). We then stained control- and iDCM-CMs with the vital mitochondrial dye MitoTracker. MitoTracker staining of healthy control-CMs revealed interconnected networks of elongated mitochondria. In contrast and consistent with our TEM findings, mitochondria in the iDCM-CMs were grossly abnormal, often appearing globular and punctate (Figure S3d). Staining with tetramethylrhodamine (TMRM), a vital dye that measures the mitochondrial membrane potential, indicated that iDCM-CMs had decreased mitochondrial membrane potential compared with control-CMs (Figure 1f). Additionally, proliferation of iDCM-CMs was increased when compared to control-CMs at differentiation day 21 and 36 (Figure S3e). This recapitulation of the patient’s phenotypic and functional deficits by patient-derived CMs indicates a genetic root cause for the patient’s iDCM.
Mutations in RTTN cause iDCM
To identify the causal mutation underlying this case of iDCM, we carried out whole-exome sequencing analysis of the patient and his parents. This analysis did not reveal pathogenic or likely pathogenic variants in any of the 123 genes known to be associated with cardiomyopathy (Supplemental Table 1). Given the lack of a family history of cardiomyopathy and the nonconsanguinity, we hypothesized that the causal mutation(s) were either a de novo autosomal dominant mutation or rare compound heterozygous recessive mutations. Based on this hypothesis, we filtered variants based on a read depth >4 for quality and excluded synonymous variants. This resulted in 258 indels and 13,632 SNPs as candidate mutations (Figure S4). Next, variants that were not rare (minor allele frequencies >0.1%) and variants where both parents were heterozygous carriers or either parent was homozygous were excluded, resulting in 37 indels and 215 SNPs as candidates. Finally, a three-fold filter of functional prediction (exclusion of benign prediction using polyphen2), expression in CMs or heart tissue and validation by Sanger sequencing yielded a single candidate gene, RTTN (encoding rotatin). The RTTN mutation consisted of an in-frame deletion (removing amino acid residues p.1921–1925) inherited from the mother and a G1321D missense mutation inherited from the father (Figure 2a, Figure S4, and Supplemental Table 2). We confirmed that the iPSCs derived from the patient were heterozygotes were 50/50 using Sanger Sequencing (Figure S4b). Further, we included a schematic and a model13 of Rotatin with annotations of predicted functional domains, armadillo domains, and polar domains (Figure S4c).
Figure 2. Compound heterozygous mutations in RTTN lead to iDCM phenotype.
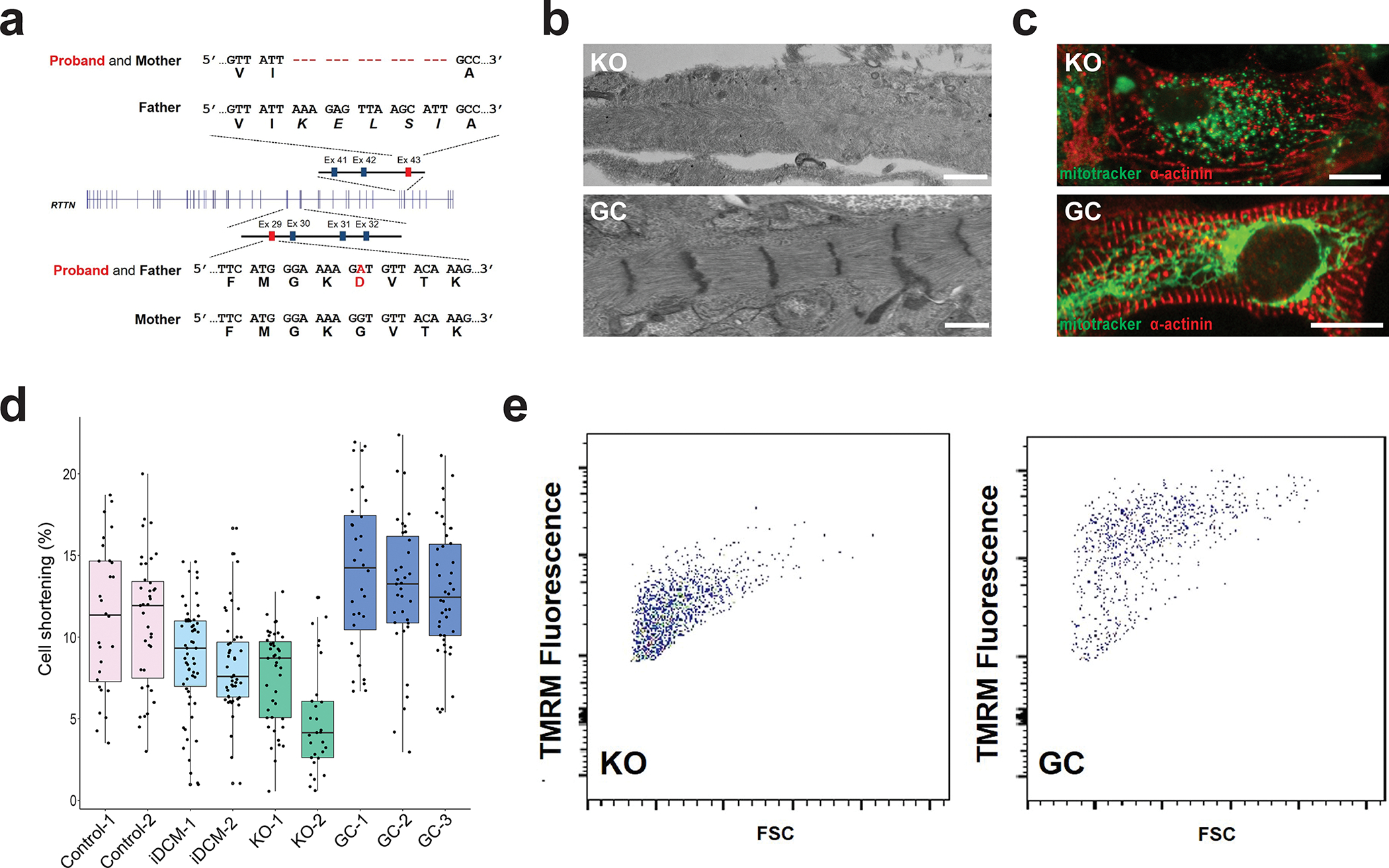
a, Whole-exome sequencing of the proband and his parents revealed that the patient was a compound heterozygote for a G1321D missense mutation from the father and an in-frame deletion (p.1921–1925) from the mother. b, RTTN knockout (KO) CMs displayed disorganized myofibrils with indistinct Z-lines (top), whereas the RTTN gene-corrected (GC) CMs (bottom) appear similar to the healthy control-CMs seen in Figure 1b. Scale bar = 0.5 μm. c, RTTN KO-CMs (top) displayed globular or punctate mitochondria, as visualized by MitoTracker (green), and disorganized myofilaments, as visualized by α-actinin staining (red). By contrast, RTTN GC-CMs (bottom) displayed networks of elongated mitochondria (green) and organized myofilaments, similar to healthy control-CMs. Scale bar = 20 μm. d, RTTN KO-CMs from two independent KO lines (KO-1, −2) displayed reduced cell shortening similar to the iDCM-CMs. By contrast, RTTN GC-CMs from three isogenic GC lines (GC-1, −2, −3) displayed restored shortening, comparable to healthy control-CMs. n = 28 Control 1-CMs, 39 Control 2-CMs, 59 iDCM 1-CMs, 48 iDCM 2-CMs, 43 KO 1-CMs, 29 KO 2-CMs, 32 GC 1-CMs, 35 GC 2-CMs. Center line = median; whiskers = 1.5IQR. e, FACS analysis of TMRM staining demonstrating reduced mitochondrial membrane potential in RTTN KO-CMs and restored mitochondrial membrane potential in RTTN GC-CMs.
To confirm RTTN as the causal gene, we used CRISPR/Cas9 technology to generate knock-out (KO) and gene-corrected (GC) iPSCs (Figure S5a). For KO iPSCs, we designed a CRISPR/Cas9 single-guide RNA (sgRNA) targeting a protospacer-adjacent motif (PAM) site N-terminal to the region corresponding to p.G1321D in exon 29 of RTTN (Figure S5b). Using this sgRNA, we generated four RTTN KO lines from isogenic control iPSCs containing indels, which were then confirmed by Sanger sequencing (Figure S5c). To generate GC iPSCs, we designed another CRISPR/Cas9 sgRNA close to the p.G1321D missense mutation (Figure S5c) and the repair template to correct this mutation by homology-directed repair (Figure S5d). To ensure that the GC lines were indeed corrected, the repair template had a silent G→A mutation in the PAM site in order to prevent cleavage by Cas9 as a molecular signature for missense repair (Figure S5d). We generated several gene-corrected (GC) isogenic lines from the iDCM-1 iPSC line in which the variant adenine (A) at position n.3962 was corrected to the reference guanine (G) (Figure S5e). The KO and GC lines contained no alterations at any of the potential off-target sites (Supplemental Table 3 and 4), and each of the lines were confirmed for stemness (Figure S6a) and cardiomyogenic potential (Figure S6b).
Transmission electron microscopy ultrastructural studies of KO and GC-CMs at day 45 of cardiac differentiation revealed disorganized sarcomere structures with severely disrupted Z-lines in the KO-CMs when compared to control-CMs (Figure 1b; Figure 2b). By contrast, GC-CMs resembled the control-CMs, exhibiting well-organized sarcomere structures with intact Z-lines (Figure 1b; Figure 2b). Immunofluorescence analysis revealed that the KO-CMs exhibited sarcomere disarray and disorganized Z-lines similar to the iDCM-CMs (Figure 2c; Figure 1c), whereas sarcomeres and Z-lines were largely restored in the GC-CMs (Figure 2c; Figure 1c), similar to the control-CMs (Figure S7a–b). MitoTracker staining revealed dysmorphic, punctate mitochondria in the KO-CMs (Figure 2c), similar to those observed in the iDCM-CMs. In contrast, the GC-CMs exhibited interwoven networks of elongated mitochondria, as observed in the control-CMs (Figure 2c). Importantly, KO-CMs from two independent KO iPSC lines exhibited weakened cell shortening (~5–8%), similar to that observed in the iDCM-CMs (~8%) (Figure 2d). In contrast, cell shortening was restored in GC-CMs from three isogenic GC lines generated from the iDCM-1 iPSC line (Figure 2d) and was comparable to that of the control-CMs. Moreover, TMRM staining indicated a marked reduction in the mitochondrial membrane potential in the KO-CMs (Figure 2e; Figure S7b), comparable to that observed in the iDCM-CMs, whereas the mitochondrial membrane potential was restored in the GC-CMs (Figure 2e; Figure S7b). Additionally, the increased proliferation capacity observed in iDCM-CMs was seen in KO-CMs, while GC-CMs displayed normal levels of proliferation (Figure S7c). Taken together, these data demonstrate that RTTN is the causal gene for the sarcomere and mitochondrial defects observed in the iDCM patient.
Previous studies found that a distinct set of homozygous recessive mutations in RTTN cause congenital brain defects including microcephaly14–19. Although the iDCM patient had no neurocognitive deficits, we investigated whether there were defects in mitochondrial morphology and function in astrocytes and neurons derived from control and iDCM iPSCs20. We found no apparent morphological or mitochondrial membrane potential differences between iDCM and control iPSC- derived astrocytes and neurons (Figure S8a, b, c).
RTTN mutation leads to dilated cardiomyopathy in multiple in vivo systems
We next asked whether RTTN plays an evolutionarily conserved role in the heart through genetic knockdowns/knockouts in multiple in vivo systems. We first examined zebrafish embryos as they are particularly well suited for studying gene function during cardiovascular development21. Following knockdown of RTTN in developing zebrafish embryos, through morpholino (MO) and CRISPRi, which inactivates target gene transcription22, a significant percentage of embryos at 48 hours post-fertilization displayed a large pericardial edema (Vehicle = 8.4%; CRISPRi1 = 55.3%, CRISPRi2 = 57.9%, MO = 45.6%), a reliable marker of heart failure in zebrafish (Figure 3a). In addition, CRISPRi and MO-injected embryos exhibited abnormal developmental morphologies including abnormal heart looping (Figure S9a,b), in agreement with previous studies of global knockout of Rotatin in mice23. A small but significant number of fish exhibited microcephaly (Figure S9c,g). However, there were no significant increases in the occurrence of hydrocephaly, spinal curvature, or short body axis (Figure S9d–f). After verifying successful knockdown of RTTN in zebrafish embryos via RT-qPCR (Figure S10a,b), we were then interested in if sarcomere structure was disrupted in developing zebrafish embryos. Interestingly, upon staining with F-Actin, we saw regions of sarcomere malformation in knockdown embryos (both morpholino and CRISPRi), suggesting RTTN plays an important role in sarcomere development (Figure S10c). Use of a mitochondrial GFP tag in zebrafish suggested disruption of mitochondria in RTTN morphants, in line with our previous findings (Figure S10d)
Figure 3. RTTN is an evolutionarily conserved causal gene for iDCM.
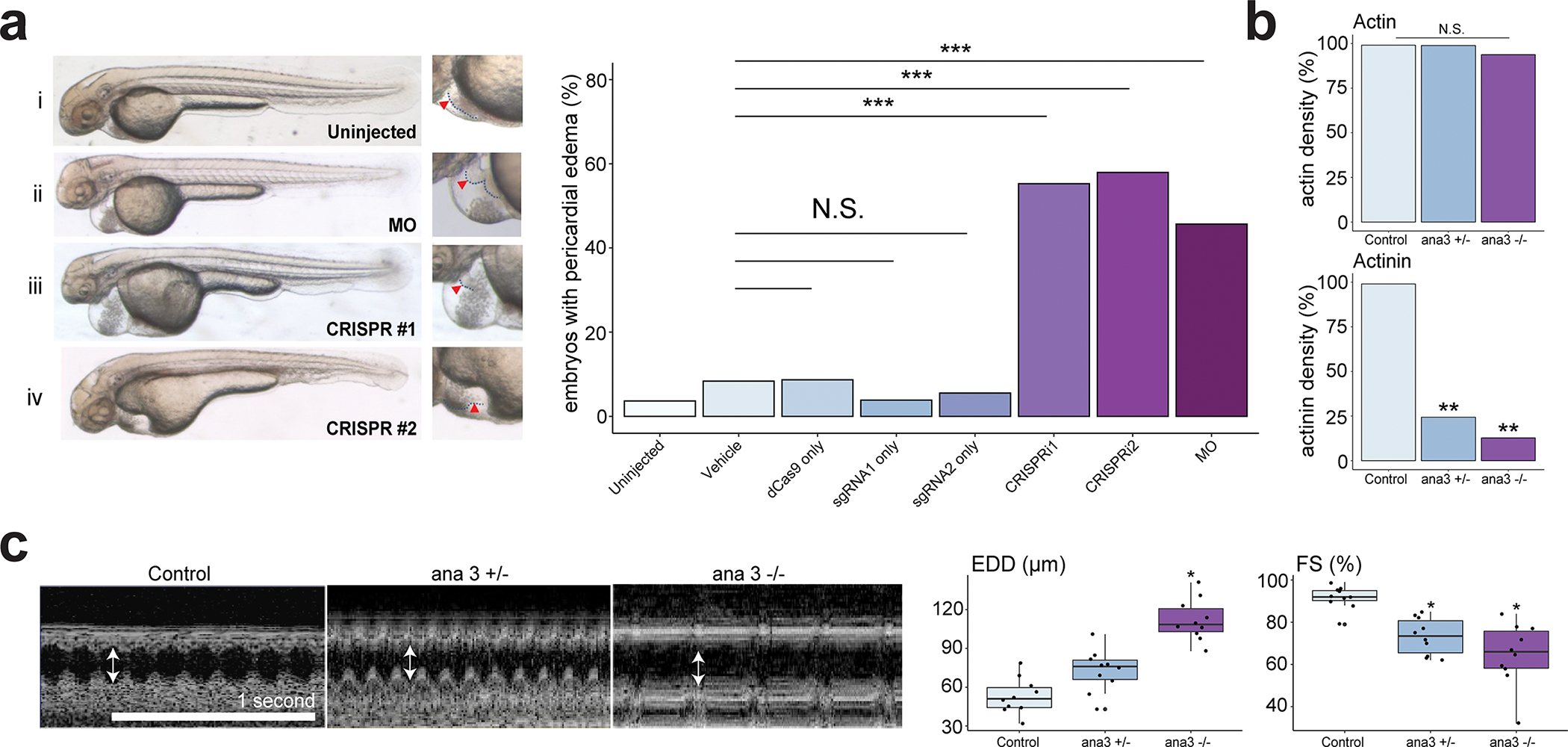
a, Left, representative images of embryos at 48 hpf. (i) uninjected embryo; (ii) embryo injected with translational blocking morpholino (MO); embryos injected with CRISPRi #1 (iii) and #2 (iv). Right, corresponding enlarged images of the heart. Arrowhead, outline of heart. Incidence of pericardial edema with impaired tail circulation in 48-hpf embryos. Results n ≥ 101 embryos from n ≥ 7 biological replicates using embryos from n ≥ 2 breeding pairs. Vehicle was 0.08% phenol red + PBS in ultrapure water. ***p <0.00017 by Fisher’s exact test with Bonferroni Correction; N.S. = not significant. b, Quantification of immunofluorescent imaging of actin and actinin in adult heart (segment A4) from control (w1118), ana3+/− and ana3−/− flies. Statistical results of normalized cardiac fiber density (N = 10). * P <0.05; ** P <0.01. c, OCT images for heart of control, ana3+/− and ana3−/− flies. Arrowheads indicate\ the end-diastolic diameter (EDD). Statistical analysis for EDD (μm) and percent fractional shortening (FS) obtained from the optimal computed tomography data. Each datapoint represents the average of measurements from three heartbeats randomly selected within a two-second time frame for each fly. Center line = median; whiskers = 1.5IQR for each genotype. *P <0.05.
Due to the co-occurrence of heart looping and pericardial edema in zebrafish, we used the Drosophila system, which has been established as an excellent model in which to study dilated cardiomyopathy24,25 and does not have a looped heart tube, to determine more specifically whether RTTN is associated with dilated cardiomyopathy. The human RTTN gene has a highly conserved and unique fly homolog called ana326. We obtained ana3 mutant flies with P-element insertions 3 BPs upstream of the initiating ATG, resulting in a null phenotype. The ana3 mutant flies were viable but died quickly after eclosion. Staining of structural actin with phalloidin determined that there was no significant difference between ana3 mutant and control embryos (Figure 3b; Figure S11a). Immunostaining for actinin, a protein localized normally in Z-lines, revealed that actinin structures were almost completely abolished (p <0.01) in both ana3 heterozygous and homozygous mutant Drosophila hearts (Figure 3b; Figure S11b). These findings demonstrate the importance of ana3 for heart structural development.
Given the severe structural deficits in cardiac filaments observed in the hearts of both ana3 mutants, we used optical coherence tomography to investigate whether these mutations affected cardiac contractile function. M-mode scans were selected at the maximum diastolic event for each genotype. Cross-sections of the heart tube obtained in this way showed a significantly increased end-diastolic dimension (EDD) in the ana3 homozygote fly hearts, but not in the ana3 heterozygote mutant hearts (Figure 3c). The M-mode orthogonal heart views provided precise and real-time measurements of the heart tube diameter and heart rate. Compared with control embryos, both ana3 heterozygous and homozygous mutant flies showed significantly reduced fractional shortening (FS) (Figure 3c).
In addition to the heart defects in Drosophila, we also observed that the brain lobes of homozygous ana3 deficient third instar larvae appeared smaller than those of control larvae (Figure S12), similar to the phenotype observed in patients with microcephaly due to RTTN variants14–19. Statistical analysis confirmed that ana3 homozygous larvae had significantly reduced brain volume compared with WT larvae (Figure S12). The ana3 heterozygous larvae showed only a slightly early ventral nerve cord dissociation phenotype (data not shown), suggesting that RTTN may have important functions in multiple organs. These data suggest evolutionarily conserved roles for RTTN in cardiac development, structure, and function.
Impaired maturation of iDCM-CMs revealed by scRNA-seq
Based on the developmental age (3 months, perinatal) of the patient when the iDCM first presented and the broad set of phenotypic defects modeled in our iPSC-CM system, we suspected that CM maturation might be involved in the etiology of the iDCM. ScRNA-seq has emerged as an extremely powerful tool to study and analyze CM maturation dynamics. Thus, we designed a scRNA-seq study aimed at testing the hypothesis that CM maturation is affected in iDCM. We performed scRNA-seq on d35 control and iDCM-CMs using the 10X chromium platform. After quality control and filtering of non-CMs (TNNT2− /MYH6− clusters) and cells undergoing the cell cycle, we began analysis of 4,671 control and 1,080 iDCM-CMs. We then performed differential gene testing of the control and iDCM cells, and found that 275 genes were significantly downregulated and 618 genes were significantly upregulated in the iDCM-CMs. We began our analysis of these genes by investigating the expression of a broad set of genes previously shown to be associated with and important to CM maturation that were consistently downregulated in iDCM-CMs (Figure 4a). To determine the affected pathways, we then performed gene ontology (GO) enrichment analysis on the iDCM-CMs and found that terms related to processes essential for heart maturation and function, such as sarcomere organization, cardiac muscle contraction, electron transport–coupled proton transport, and cardiac myofibril assembly, among others, were downregulated in iDCM-CMs (Figure 4b), while GO terms related to glycolysis, and apoptosis were enriched (Figure 4b). Finally, we used a recently published maturation scoring metric27, which provides an unbiased approach to determining the maturation status of CMs based on transcriptomic entropy of each CM. Our analysis showed an increase in transcriptomic entropy, corresponding to a decrease in maturity, of iDCM-CMs when compared with control-CMs (Figure 4c). Taken together, these data support that CM maturation is impaired in the iDCM-CMs.
Figure 4. RTTN mutation leads to impaired maturation via disrupted reorganization of the centrosome and microtubule network formation.
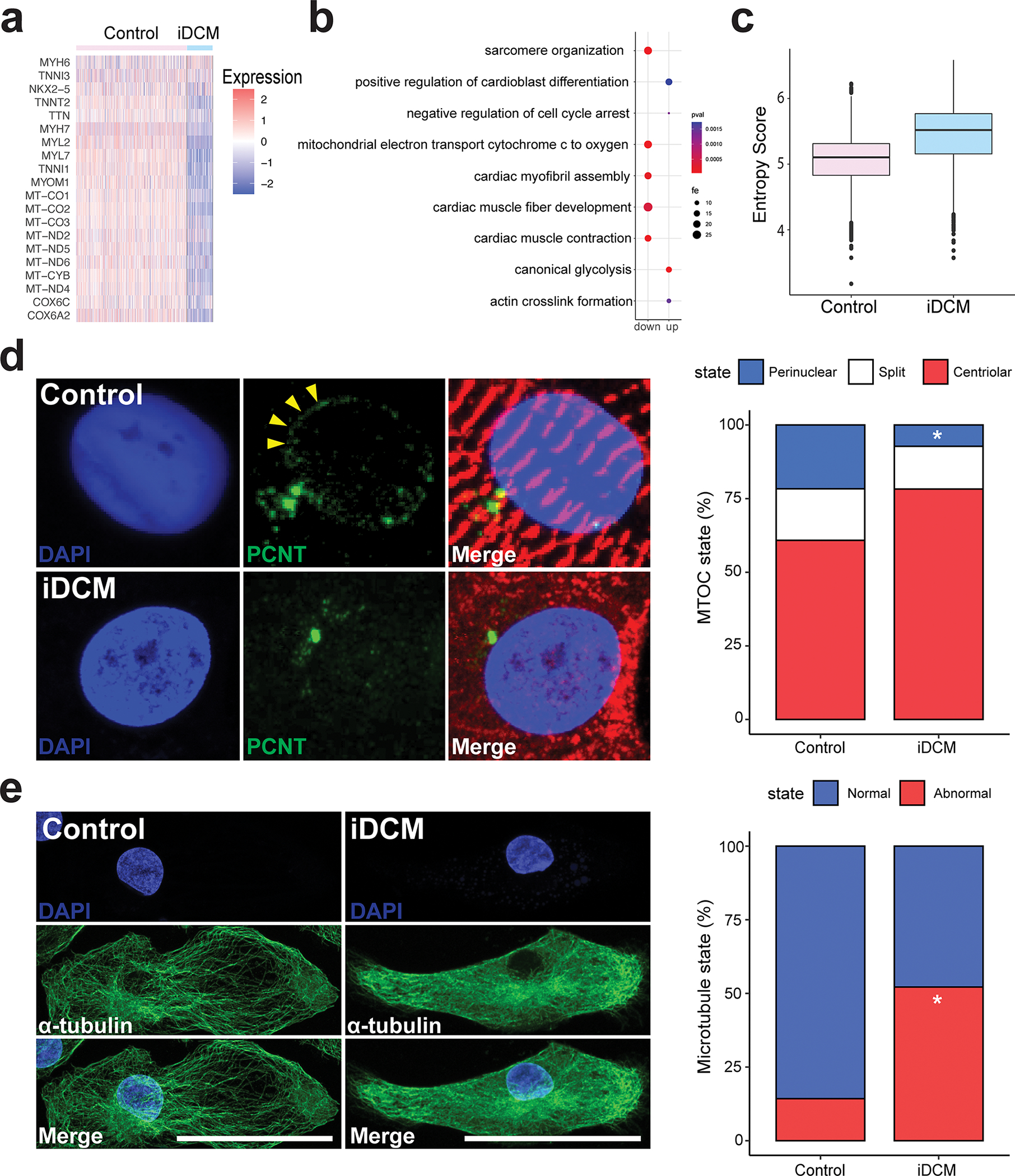
a, Heat map showing expression of genes related to CM maturation in control and iDCM-CMs. b, Dot plot of fold enrichment (fe) of gene ontology (GO) pathways. up = upregulated in iDCMs; down = downregulated in iDCMs. c, Shannon entropy score showing relative maturation score of control and iDCM-CMs. d, Top, In the control CM with a normally organized sarcomere (α-actinin, red), PCNT (green) is redistributed to the perinuclear region (arrowheads). Bottom, In the RTTN mutant (iDCM) CM with a disorganized sarcomere (α-actinin, red), PCNT (green) remains localized to the centrosome. Quantification of PCNT distribution in D36 control and iDCM-CMs by a blinded observer with categories of centriolar, split, or perinuclear. PCNT was perinuclear in 21.65% of control-CMs, whereas it was perinuclear in only 7.26% of iDCM-CMs. Perinuclear vs centriolar p = 0.0021 by two-tailed Fisher’s exact test. n = 97 control-CMs, 124 iDCM-CMs. e, Left, Control CM displaying a prominent network of thick microtubule (MT) fibers (α-tubulin, green) emanating from the perinuclear region, with an organized sarcomere (cardiac TnT, red). Right, by contrast, iDCM CM displayed thinner, shorter and fainter MT fibers without a clear organizing center, and a disorganized sarcomere. Quantification of CMs with grossly normal and abnormal MT network by a blinded observer. The MT network was grossly normal in 86% of control-CMs, whereas only 48% of iDCM-CMs had normal MT networks. n= 28 Control-CMs, 23 iDCM-CMs. p=0.006 by two-tailed Fisher’s exact test. Scale bar = 50 μm.
Defective centrosomal reduction in iDCM
During CM differentiation and maturation, the centrosome is known to undergo dramatic changes which include unpairing or “splitting” of the centriole, disappearance of certain pericentriolar proteins like CEP135, and the re-localization of other pericentriolar proteins, such as pericentrin (PCNT) and pericentriolar material 1 (PCM1), to form a MTOC at the nuclear envelope28–30. The transition of PCNT during CM differentiation involves a key isoform switch (from PCNT-B to PCNT-S), which contributes to postnatal cell cycle arrest through the preferential expression of PCNT-S in the perinuclear centrosome31.
Rotatin (encoded by RTTN) is known to be a centrosomal protein in both Drosophila (ana3) and humans, and has been previously shown to be involved in regulation of centrosome structure32,33. Based on this and the observed CM maturation defect in the iDCM-CMs, we then asked if developmentally programed centrosomal changes associated with CM maturation28–30, were disrupted in the iDCM-CMs. Although we could not localize rotatin to the centrosome using commercially available rotatin antibodies (see Supplementary Methods), we demonstrated its centriolar localization in HEK293 cells using SASY34, a polyclonal antibody directed against the N-terminus of rotatin (Figure S13). Additionally, using SASY, we detected rotatin in the centrosomes of iPSCs and CMs actively in mitosis (Figure S13). Based on SASY immunostaining, there were no differences in rotatin localization between iDCM and control cells.
As previously described29, in control iPSCs, PCNT colocalizes with the centriolar component γ-tubulin (Figure S14a), but as iPSC-CMs mature, both PCNT and PCM1 become largely perinuclear (Figure 4d Figure S14a,b), indicating that centrosome reduction. By contrast, in the iDCM-CMs, both PCNT and PCM1 localization remained mostly centriolar (Figure 4d; Figure S14b), suggesting that this process is impaired in RTTN mutant CMs. Importantly, we did not observe any significant changes in primary cilia length (Figure S14c). We then investigated whether reorganization of the centrosome is affected in KO-CMs and recovered in GC-CMs. Interestingly, we saw a decrease in perinuclear localization of the centrosome in KO-CMs when compared to control-CMs (21.6% in control-CMs compared to 6.6% in KO-CMs) and an increase in perinuclear centrosome in GC-CMs when compared to iDCM-CMs (7.3% in iDCM-CMs compared to 27.7% in GC-CMs) (Figure S15a). To determine if these changes in centrosome structure were linked to proper sarcomere development, we quantified the proportion of CMs that displayed concordant centrosome and sarcomere state (reduced centrosomes with mature sarcomeres and unreduced centrosomes with immature sarcomeres). Our quantification showed significant levels of concordance in control, iDCM, and GC-CMs (Figure S15b), showing that the process of centrosome reduction is linked to proper formation. Taken together, these results suggest that rotatin is essential for reorganization of the centrosome during cardiomyocyte development.
Following centrosomal changes in striated muscles, the nuclear envelope, which contains PCNT and PCM1, acts as the dominant microtubule organizing center (MTOC)30. Microtubules play a critical role in CMs, providing structural integrity, organizing sarcomere components, and serving as a mitochondrial highway system throughout the CM. As we observed defects in centrosomal reorganization in RTTN mutant CMs, we examined whether microtubule networks in iDCM-CMs were also disrupted. In iPSCs, we did not observe any gross differences in microtubule structures in control and iDCM-iPSCs immunostained for α-tubulin (Figure S16a). However, after cardiac differentiation, significant disruptions in the microtubule networks of the iDCM-CMs were frequently observed. Instead of the prominent meshwork of thick microtubule fibers emanating from the perinuclear region observed in control-CMs (Figure 4e), the iDCM-CMs displayed much thinner, shorter, and less distinct microtubule fibers without a clearly organized center (Figure 4e; Figure S16b–d). We also observed defects in the microtubule networks of ana3 Drosophila mutant hearts. Interestingly, the tubulin assembly pattern revealed by immunostaining for acetylated alpha-tubulin (Ac-tubulin) was partially lost in ana3+/− fly hearts and was severely disrupted in ana3−/− fly hearts compared with control fly hearts (Figure S11c). These results support the hypothesis that mutations in RTTN affect CM structure and function through impaired centrosome reduction and consequent disruption of the microtubule network (Figure S16e). Further, microtubule regrowth assays showed that iDCM-CMs retain the ability to regrow microtubule networks after cold mediated disassembly (Figure S17).
Small-molecule treatment rescues impaired maturation-related iDCM in the iPSC model
There are currently no therapeutic agents specifically for iDCM. We were therefore interested in testing whether ameliorating the CM maturation defect would be sufficient to rescue the structural and functional defects present in iDCM. As developmentally programmed centrosome changes are impaired in iDCM-CMs, we treated them with a small molecule, C1935 (Figure 5a) that was previously shown to induce centrosome reduction in CMs28. Interestingly, treatment with C19 facilitated improved initiation of the perinuclear MTOC in iDCM-CMs (Figure 5b). Moreover, the improved sarcomere formation (Figure 5c) in the C19-treated iDCM-CMs demonstrated that structural aspects of CM maturation were significantly improved. Excitingly, we also observed that contractility, a functional metric of CM maturation, was significantly improved in C19-treated iDCM-CMs (Figure 5d), with comparable levels of cell shortening in the treated iDCM-CMs and in healthy control-CMs. These results suggest that developmental centrosomal reorganizations are necessary for functional aspects of CM maturation to occur (Figure 5e). Though the extent to which centrosome reduction is affected in other cases of iDCM remains unknown, facilitation of this process seems to be sufficient to rescue CM defects in this particular case of centrosome-mediated iDCM, and insights gained from this opens the door for potential pharmacological treatments for iDCM in the future.
Figure 5. Pharmacological treatment to induce centrosomal reorganization restores CM structure and function in iDCM.
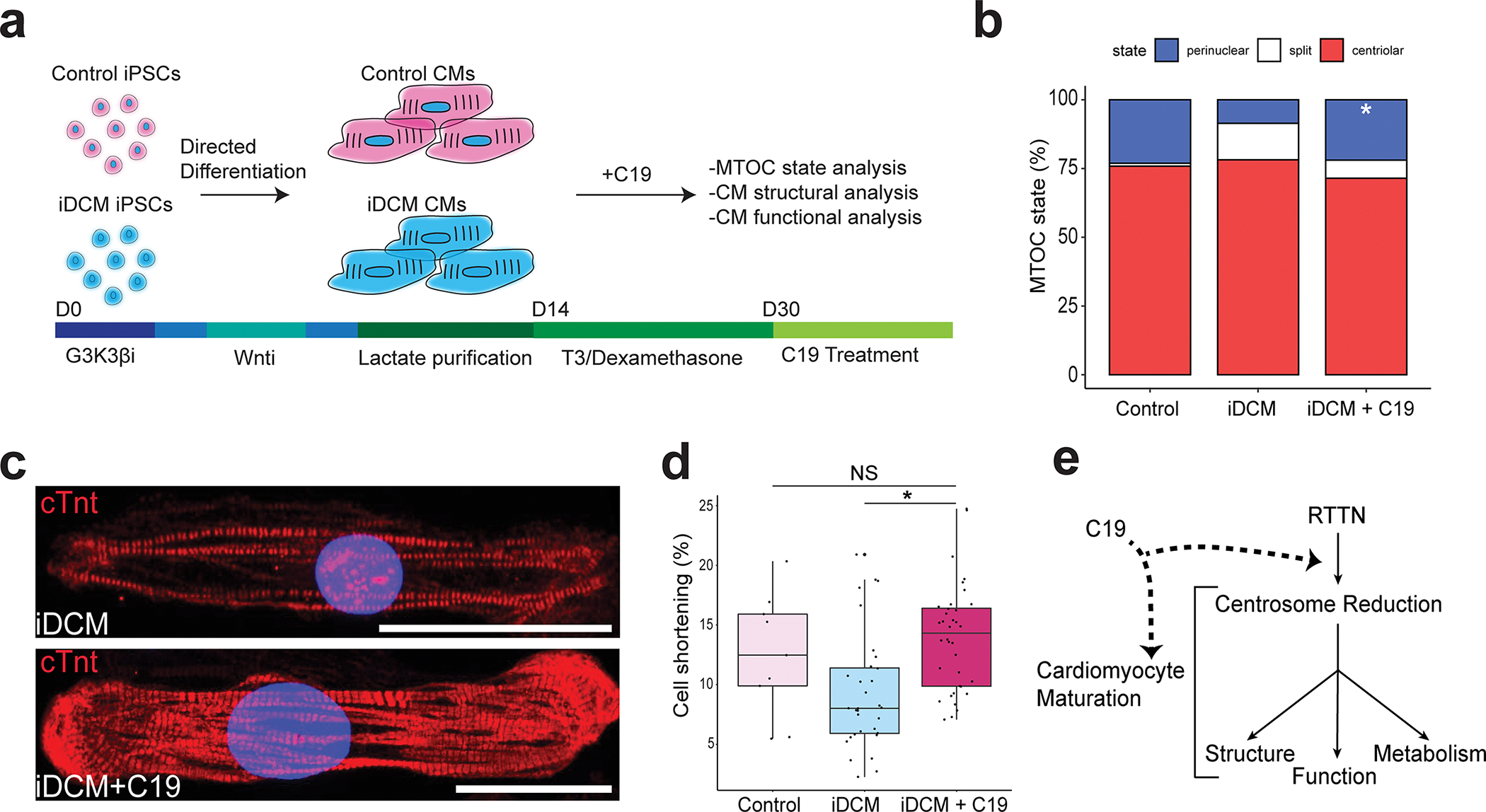
a, Schematic detailing C19 treatment at differentiation day 30 (d30) control-CMs and iDCM-CMs. b, Quantification of centriolar, split, and perinuclear MTOC localization in D32 CMs by a blinded observer. PCNT was perinuclear in 23.1% of control-CMs, 7.6% of iDCM-CMs, and 23.6% of iDCM+C19 CMs. p = 0.0082 by Fisher Exact test. c, Representative images of untreated iDCM-CMs (top) and iDCM-CMs treated with C19, which showed increased levels of sarcomere formation. Scale bar = 50 μm. d, Boxplots showing % cell shortening in control, D40 iDCM, and iDCM C19 treated CMs. n = 9 control, 32 iDCM, 36 iDCM + C19 from two differentiations. p value = 0.0006 by one-way ANOVA. Post hoc Tukey correction p value of iDCM vs iDCM + C19 = 0.001, control vs iDCM + C19 = 0.682 (NS). Center line = median; whiskers = 1.5IQR. e, Working model of CM maturation cascade showing RTTN-mediated centrosome reduction is upstream of other canonical maturation events.
Discussion:
Centrosomes are dynamic organelles that undergo a type of programmed reorganization, termed as centrosome reduction28, during cardiac differentiation. To our knowledge, this study is the first to report a defective centrosome component as the cause of nonsyndromic cardiomyopathy in humans (a syndromic cause of variable cardiomyopathy is observed in Alstrom syndrome36). Mutations in the centrosome component RTTN resulted in CM-specific defects in centrosomal reorganization during cardiac maturation. In the mutant CMs, the failure of the centrosome to reorganize and establish the nuclear envelope as the major microtubule organizing center results in a profoundly defective microtubule network (Figure 4e). As the microtubule cytoskeleton is critical for the maintenance of cellular structures 37–39, including vital roles in mitochondrial motility, fission, and fusion40–42, a defective microtubule network could be responsible for both the mitochondrial and sarcomeric defects observed in RTTN mutant CMs, resulting in defective mitochondrial respiration and contractility.
The precise role of RTTN in the cardiac centrosome is presently unknown, and it is unclear why the RTTN mutations described here only cause cardiac defects, in contrast to other published RTTN mutations, which cause CNS defects associated with centrosome amplification33,34,43, though there was at least one known patient carrying a RTTN mutation with a restrictive cardiomyopathy in addition to a co-occurring CNS defect34. Given the highly dynamic RTTN localization noted using SASY and the importance of alternative splicing in PCNT transition during cardiac differentiation31, we hypothesize that RTTN may likewise undergo substantial alternative splicing, resulting in tissue-specific isoforms with distinct roles in centrosome dynamics that may affect the development of specific organs. Although much attention has been paid to the association of centrosome reduction with cell cycle arrest, our study demonstrates that tissue-specific and developmentally programmed centrosome reorganization is essential for proper CM structure and function. As centrosome dynamics play fundamental roles in CM cell cycle exit, structure, and function, a better understanding of this tissue-specific, developmentally programmed process will be highly relevant to cardiac regenerative therapy efforts. Future studies in in vivo and cardiac organoid44 systems will be essential for expanding our understanding of centrosome reduction.
Supplementary Material
Clinical Perspective:
What is new?
In this study, we incorporate iPSCs, CRISPR/Cas9, whole exome sequencing, single cell RNA sequencing, and multiple in vivo models to uncover biological insights from a case of infantile dilated cardiomyopathy.
We identified compound heterozygous mutations in the centrosomal component rotatin (RTTN) as the causal gene for infantile dilated cardiomyopathy through dysregulating a process known as centrosome reduction in human cardiomyocytes.
Such defects in centrosome reduction have implications of broad cardiomyocyte biology – sarcomere structure, mitochondrial dynamics, cell cycle – and are caused by disorganization of microtubule networks, highlighting a non-canonical role of the centrosome in cardiomyocyte biology.
What are the clinical applications?
Infantile dilated cardiomyopathy has multiple etiologies, and this study links a novel gene (RTTN) and a relatively unstudied developmental pathway (centrosome reduction) to human heart disease for the first time.
We highlight a multi-pronged approach to uncover the biological etiology of infantile dilated cardiomyopathy to identify therapeutic targets to overcome the defect – a paradigm we believe is applicable across disciplines and fields to discover cures to human disease.
Acknowledgments:
The authors thank members of the Hong lab for helpful discussions and Dr. Fern P. Finger for help in preparation of the manuscript. Author contributions – C.C.H. conceived and directed the project. Y.R.S, D.P.B., Y.W.C., M.M, C.C.S and C.C.H. conceptualized and designed the strategy. Y.W.C., A.R.C., U.S., M.K., T.K.F., and C.H.W. designed and performed iPSC maintaining and genome editing. Y.W.C., M.M., D.F. and K.W. performed mitochondrial staining and functional measurement. A.B.B., K.C.E., R.L., M.C., S.A., and M.D.N. designed constructs for reprogramming, generated iPSCs and differentiated them into neurons and astrocytes. C.H.W, M.S., and L.R.N. designed constructs for genome editing in zebrafish. Z.H. and H.L. carried out the Drosophila studies. S.K., K.C. and M.K. differentiated CMs and analyzed functional measurements and immunostaining. A.V.F. assisted with experiments for TEM. M.D.D. assisted with whole-exome sequencing. R.S. and G.M.S.M assisted with the SASY immunostaining. Y.W.C., C.H.W., M.M, Y.R.S., and C.C.H prepared the manuscript. C.C.H acquired funding for the project.
Funding sources:
NHLBI R01HL135129 (C.C.H)
MSCRF HP-00089001 (C.C.H)
AOA Carolyn L. Kuckein Student Research Fellowship (M.M)
Nonstandard Abbreviations and Acronyms:
- iDCM
Infantile Dilated Cardiomyopathy
- CM
Cardiomyocyte
- RTTN
Rotatin
- iPSC
Induced pluripotent stem cell
- MTOC
Microtubule organizing center
- KO
Knockout
- GC
Gene corrected
- scRNAseq
Single cell RNA sequencing
- PCNT
Pericentrin
Footnotes
Disclosures:
None
References
- 1.Lee TM, Hsu DT, Kantor P, Towbin JA, Ware SM, Colan SD, et al. Pediatric cardiomyopathies. Circ Res 2017;121:855–873. doi: 10.1161/CIRCRESAHA.116.309386/FORMAT/EPUB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lipshultz SE, Law YM, Asante-Korang A, Austin ED, Dipchand AI, Everitt MD, et al. Cardiomyopathy in Children: Classification and Diagnosis: A Scientific Statement From the American Heart Association. Circulation 2019;140:E9–E68. doi: 10.1161/CIR.0000000000000682/FORMAT/EPUB. [DOI] [PubMed] [Google Scholar]
- 3.Mehdiabadi NR, Choon B;, Sim B, Phipson B, Kalathur RKR, Sun Y, et al. Defining the Fetal Gene Program at Single-Cell Resolution in Pediatric Dilated Cardiomyopathy. Circulation 2022;146:1105–1108. doi: 10.1161/CIRCULATIONAHA.121.057763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan RS, Pahl E, Dellefave-Castillo L, Rychlik K, Ing A, Yap KL, et al. Genotype and Cardiac Outcomes in Pediatric Dilated Cardiomyopathy. J Am Heart Assoc 2022;11. doi: 10.1161/JAHA.121.022854/FORMAT/EPUB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Meulen MH, Herkert JC, den Boer SL, du Marchie Sarvaas GJ, Blom N, ten Harkel ADJ, et al. Genetic Evaluation of A Nation-Wide Dutch Pediatric DCM Cohort: The Use of Genetic Testing in Risk Stratification. Circ Genomic Precis Med 2022. doi: 10.1161/CIRCGEN.120.002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herkert JC, Abbott KM, Birnie E, Meems-Veldhuis MT, Boven LG, Benjamins M, et al. Toward an effective exome-based genetic testing strategy in pediatric dilated cardiomyopathy. Genet Med 2018;20:1374–1386. doi: 10.1038/GIM.2018.9. [DOI] [PubMed] [Google Scholar]
- 7.Long PA, Evans JM, Olson TM. Diagnostic Yield of Whole Exome Sequencing in Pediatric Dilated Cardiomyopathy. J Cardiovasc Dev Dis 2017;4. doi: 10.3390/JCDD4030011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasilescu C, Ojala TH, Brilhante V, Ojanen S, Hinterding HM, Palin E, et al. Genetic Basis of Severe Childhood-Onset Cardiomyopathies. J Am Coll Cardiol 2018;72:2324–2338. doi: 10.1016/J.JACC.2018.08.2171. [DOI] [PubMed] [Google Scholar]
- 9.Cadar AG, Feaster TK, Bersell KR, Wang L, Hong TT, Balsamo JA, et al. Real-time visualization of titin dynamics reveals extensive reversible photobleaching in human induced pluripotent stem cell-derived cardiomyocytes. Am J Physiol - Cell Physiol 2020;318:C163–C173. doi: 10.1152/ajpcell.00107.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feaster TK, Cadar AG, Wang L, Williams CH, Chun YW, Hempel JE, et al. Matrigel Mattress: A Method for the Generation of Single Contracting Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Circ Res 2015;117:995–1000. doi: 10.1161/CIRCRESAHA.115.307580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun YW, Balikov DA, Feaster TK, Williams CH, Sheng CC, Lee J-B, et al. Combinatorial polymer matrices enhance in vitro maturation of human induced pluripotent stem cell-derived cardiomyocytes. Biomaterials 2015;67:52–64. doi: 10.1016/j.biomaterials.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tidball AM, Neely MD, Chamberlin R, Aboud AA, Kumar KK, Han B, et al. Genomic instability associated with p53 knockdown in the generation of Huntington’s disease human induced pluripotent stem cells. PLoS One 2016;11. doi: 10.1371/journal.pone.0150372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021;596:583–589. doi: 10.1038/S41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavallin M, Bery A, Maillard C, Salomon LJ, Bole C, Reilly ML, et al. Recurrent RTTN mutation leading to severe microcephaly, polymicrogyria and growth restriction. Eur J Med Genet 2018;61:755–758. doi: 10.1016/j.ejmg.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Stouffs K, Moortgat S, Vanderhasselt T, Vandervore L, Dica A, Mathot M, et al. Biallelic mutations in RTTN are associated with microcephaly, short stature and a wide range of brain malformations. Eur J Med Genet 2018;61:733–737. doi: 10.1016/j.ejmg.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Alazami AM, Shamseldin H, Manning M, Hashem A, Caluseiu O, Tabarki B, et al. RTTN Mutations Cause Primary Microcephaly and Primordial Dwarfism in Humans. Am J Hum Genet 2015;97:862–868. doi: 10.1016/j.ajhg.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zakaria M, Fatima A, Klar J, Wikström J, Abdullah U, Ali Z, et al. Primary microcephaly, primordial dwarfism, and brachydactyly in adult cases with biallelic skipping of RTTN exon 42. Hum Mutat 2019;40:899–903. doi: 10.1002/humu.23755. [DOI] [PubMed] [Google Scholar]
- 18.Grandone A, Torella A, Santoro C, Giugliano T, del Vecchio Blanco F, Mutarelli M, et al. Expanding the phenotype of RTTN variations: a new family with primary microcephaly, severe growth failure, brain malformations and dermatitis. Clin Genet 2016;90:445–450. doi: 10.1111/cge.12771. [DOI] [PubMed] [Google Scholar]
- 19.Wambach JA, Wegner DJ, Yang P, Shinawi M, Baldridge D, Betleja E, et al. Functional characterization of biallelic RTTN variants identified in an infant with microcephaly, simplified gyral pattern, pontocerebellar hypoplasia, and seizures. Pediatr Res 2018;84:435–441. doi: 10.1038/s41390-018-0083-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joshi P, Bodnya C, Ilieva I, Neely MD, Aschner M, Bowman AB. Huntington’s disease associated resistance to Mn neurotoxicity is neurodevelopmental stage and neuronal lineage dependent. Neurotoxicology 2019;75:148–157. doi: 10.1016/j.neuro.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 2013;154:442. doi: 10.1016/J.CELL.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faisst AM, Alvarez-Bolado G, Treichel D, Gruss P. Rotatin is a novel gene required for axial rotation and left–right specification in mouse embryos. Mech Dev 2002;113:15–28. doi: 10.1016/S0925-4773(02)00003-5. [DOI] [PubMed] [Google Scholar]
- 24.Viswanathan MC, Kaushik G, Engler AJ, Lehman W, Cammarato A. A Drosophila melanogaster model of diastolic dysfunction and cardiomyopathy based on impaired troponin-T function. Circ Res 2014;114. doi: 10.1161/CIRCRESAHA.114.302028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolf MJ, Amrein H, Izatt JA, Choma MA, Reedy MC, Rockman HA. Drosophila as a model for the identification of genes causing adult human heart disease. Proc Natl Acad Sci U S A 2006;103:1394–1399. doi: 10.1073/pnas.0507359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stevens NR, Dobbelaere J, Wainman A, Gergely F, Raff JW. Ana3 is a conserved protein required for the structural integrity of centrioles and basal bodies. J Cell Biol 2009;187:355–363. doi: 10.1083/jcb.200905031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kannan S, Farid M, Lin BL, Miyamoto M, Kwon C. Transcriptomic entropy benchmarks stem cell-derived cardiomyocyte maturation against endogenous tissue at single cell level. PLOS Comput Biol 2021;17:e1009305. doi: 10.1371/JOURNAL.PCBI.1009305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ng DCH, Richards DK, Mills RJ, Ho UY, Perks HL, Tucker CR, et al. Centrosome Reduction Promotes Terminal Differentiation of Human Cardiomyocytes. Stem Cell Reports 2020;15:817–826. doi: 10.1016/j.stemcr.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zebrowski DC, Vergarajauregui S, Wu CC, Piatkowski T, Becker R, Leone M, et al. Developmental alterations in centrosome integrity contribute to the post-mitotic state of mammalian cardiomyocytes. Elife 2015;4. doi: 10.7554/eLife.05563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becker R, Leone M, Engel FB. Microtubule Organization in Striated Muscle Cells. Cells 2020;9:1395. doi: 10.3390/cells9061395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinfeldt J, Becker R, Vergarajauregui S, Engel FB. Alternative Splicing of Pericentrin Contributes to Cell Cycle Control in Cardiomyocytes. J Cardiovasc Dev Dis 2021, Vol 8, Page 87 2021;8:87. doi: 10.3390/JCDD8080087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sydor AM, Coyaud E, Rovelli C, Laurent E, Liu H, Raught B, et al. PPP1R35 is a novel centrosomal protein that regulates centriole length in concert with the microcephaly protein RTTN. Elife 2018;7. doi: 10.7554/eLife.37846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen HY, Wu CT, Tang CJC, Lin YN, Wang WJ, Tang TK. Human microcephaly protein RTTN interacts with STIL and is required to build full-length centrioles. Nat Commun 2017;8:1–14. doi: 10.1038/s41467-017-00305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vandervore LV, Schot R, Kasteleijn E, Oegema R, Stouffs K, Gheldof A, et al. Heterogeneous clinical phenotypes and cerebral malformations reflected by rotatin cellular dynamics. Brain 2019;142:867–884. doi: 10.1093/brain/awz045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basu D, Lettan R, Damodaran K, Strellec S, Reyes-Mugica M, Rebbaa A. Identification, Mechanism of Action, and Antitumor Activity of a Small Molecule Inhibitor of Hippo, TGF-b, and Wnt Signaling Pathways. Mol Cancer Ther 2014;13. doi: 10.1158/1535-7163.MCT-13-0918. [DOI] [PubMed] [Google Scholar]
- 36.Shenje LT, Andersen P, Halushka MK, Lui C, Fernandez L, Collin GB, et al. Mutations in Alström protein impair terminal differentiation of cardiomyocytes. Nat Commun 2014 51 2014;5:1–11. doi: 10.1038/ncomms4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muroyama A, Lechler T. Microtubule organization, dynamics and functions in differentiated cells. Dev 2017;144:3012–3021. doi: 10.1242/dev.153171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen CY, Caporizzo MA, Bedi K, Vite A, Bogush AI, Robison P, et al. Suppression of detyrosinated microtubules improves cardiomyocyte function in human heart failure. Nat Med 2018;24:1225–1233. doi: 10.1038/s41591-018-0046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ali H, Braga L, Giacca M. Cardiac regeneration and remodelling of the cardiomyocyte cytoarchitecture. FEBS J 2020;287:417–438. doi: 10.1111/febs.15146. [DOI] [PubMed] [Google Scholar]
- 40.Shen J, Zhang JH, Xiao H, Wu JM, He KM, Lv ZZ, et al. Mitochondria are transported along microtubules in membrane nanotubes to rescue distressed cardiomyocytes from apoptosis article. Cell Death Dis 2018;9. doi: 10.1038/s41419-017-0145-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu M, Kalyanasundaram A, Zhu J. Structural and biomechanical basis of mitochondrial movement in eukaryotic cells. Int J Nanomedicine 2013;8:4033–4042. doi: 10.2147/IJN.S52132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woods LC, Berbusse GW, Naylor K. Microtubules are essential for mitochondrial dynamics-fission, fusion, and motility-in Dictyostelium discoideum. Front Cell Dev Biol 2016;4:19. doi: 10.3389/fcell.2016.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kia SK, Verbeek E, Engelen E, Schot R, Poot RA, De coo IFM, et al. RTTN mutations link primary cilia function to organization of the human cerebral cortex. Am J Hum Genet 2012;91:533–540. doi: 10.1016/j.ajhg.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyamoto M, Nam L, Kannan S, Kwon C. Heart organoids and tissue models for modeling development and disease. Semin Cell Dev Biol 2021. doi: 10.1016/j.semcdb.2021.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc 2012;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hattori F, Chen H, Yamashita H, Tohyama S, Satoh YS, Yuasa S, et al. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nat Methods 2010;7:61–66. doi: 10.1038/nmeth.1403. [DOI] [PubMed] [Google Scholar]
- 47.Monici M. Cell and tissue autofluorescence research and diagnostic applications. Biotechnol Annu Rev 2005;11:227–256. doi: 10.1016/S1387-2656(05)11007-2. [DOI] [PubMed] [Google Scholar]
- 48.Chanthra N, Abe T, Miyamoto M, Sekiguchi K, Kwon C, Hanazono Y, et al. A Novel Fluorescent Reporter System Identifies Laminin-511/521 as Potent Regulators of Cardiomyocyte Maturation. Sci Rep 2020;10. doi: 10.1038/s41598-020-61163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cong L, Zhang F. Genome engineering using CRISPR-Cas9 system. Methods Mol Biol 2015;1239:197–217. doi: 10.1007/978-1-4939-1862-1_10. [DOI] [PubMed] [Google Scholar]
- 50.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo Y, Ye F, Sheng Q, Clark T, Samuels DC. Three-stage quality control strategies for DNA re-sequencing data. Brief Bioinform 2013;15:879–889. doi: 10.1093/bib/bbt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo Y, Zhao S, Sheng Q, Ye F, Li J, Lehmann B, et al. Multi-perspective quality control of Illumina exome sequencing data using QC3. Genomics 2014;103:323–328. doi: 10.1016/j.ygeno.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daniels J, Holmans P, Williams N, Turic D, McGuffin P, Plomin R, et al. A simple method for analyzing microsatellite allele image patterns generated from DNA pools and its application to allelic association studies. Am J Hum Genet 1998;62:1189–1197. doi: 10.1086/301816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Depristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–501. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang W, Zhang R, Xu X. Myofibrillogenesis in the developing zebrafish heart: A functional study of tnnt2. Dev Biol 2009;331:237–249. doi: 10.1016/J.YDBIO.2009.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Glenn NO, McKane M, Kohli V, Wen K-K, Rubenstein PA, Bartman T, et al. The W-Loop of Alpha-Cardiac Actin Is Critical for Heart Function and Endocardial Cushion Morphogenesis in Zebrafish. Mol Cell Biol 2012;32:3527–3540. doi: 10.1128/MCB.00486-12/ASSET/CB6EF5BF-433A-4932-A574-255E8AE0B6AF/ASSETS/GRAPHIC/ZMB9991096300010.JPEG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang J, Xu X. Immunostaining of Dissected Zebrafish Embryonic Heart. JoVE (Journal Vis Exp 2012:e3510. doi: 10.3791/3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han Z, Yi P, Li X, Olson EN. Hand, an evolutionarily conserved bHLH transcription factor required for Drosophila cardiogenesis and hematopoiesis. Development 2006;133:1175–1182. doi: 10.1242/dev.02285. [DOI] [PubMed] [Google Scholar]
- 62.Han Z, Olson EN. Hand is a direct target of Tinman and GATA factors during Drosophila cardiogenesis and hematopoiesis. Development 2005;132:3525–3536. doi: 10.1242/dev.01899. [DOI] [PubMed] [Google Scholar]
- 63.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive Integration of Single-Cell Data. Cell 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li G, Tian L, Goodyer W, Kort EJ, Buikema JW, Xu A, et al. Single cell expression analysis reveals anatomical and cell cycle-dependent transcriptional shifts during heart development. Dev 2019;146. doi: 10.1242/dev.173476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.TheGeneOntologyConsortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 2019;47. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: Tool for the unification of biology. Nat Genet 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


