Abstract
Combatting the ever-increasing threat from invasive fungal pathogens faces numerous fundamental challenges, including constant human exposure to large reservoirs of species in the environment, the increasing population of immunocompromised or immunosuppressed individuals, the unsatisfactory efficacy of current antifungal drugs and their associated toxicity, and the scientific and economic barriers limiting a new antifungal pipeline. DectiSomes represent a new drug delivery platform that enhances antifungal efficacy for diverse fungal pathogens and reduces host toxicity for current and future antifungals. DectiSomes employ pathogen receptor proteins – C-type lectins – to target drug-loaded liposomes to conserved fungal cognate ligands and away from host cells. DectiSomes represent one leap forward for urgently needed effective pan-antifungal therapy. Herein, we discuss the problems of battling fungal diseases and the state of DectiSome development.
Keywords: Fungal pathogens, Dectins, Antifungals, Targeted drug delivery, Therapeutics
Graphical Abstract
1. Introduction
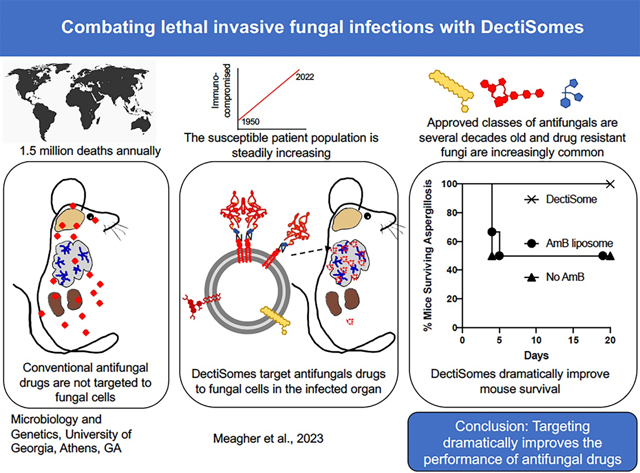
This review discusses the design and development of DectiSomes, a pan-antifungal drug delivery system with exceptional potential to improve clinical outcomes for patients with life-threatening fungal infections. DectiSomes are anti-infective drug-loaded liposomes targeted to pathogenic cells by host receptor proteins that recognize diverse pathogens. DectiSomes concentrate anti-infective drugs on fungal cells and reduce off-target drug exposure of healthy organs, tissues, and host cells (Fig. 1). The long-term goals behind this new technology are (1) to lower the effective systemic dose of antifungal drugs needed to control fungal pathogens, (2) to decrease the frequency and duration of drug treatments, (3) to overcome most forms of dose-dependent drug resistance and multidrug resistance by increasing the local concentration of antifungals at infection sites, and (4) to reduce drug toxicity to patients through combinations of the various properties of DectiSomes.
Fig. 1. DectiSomes are drug-loaded liposomes coated with carbohydrate recognition domains of mammalian pathogen receptors.
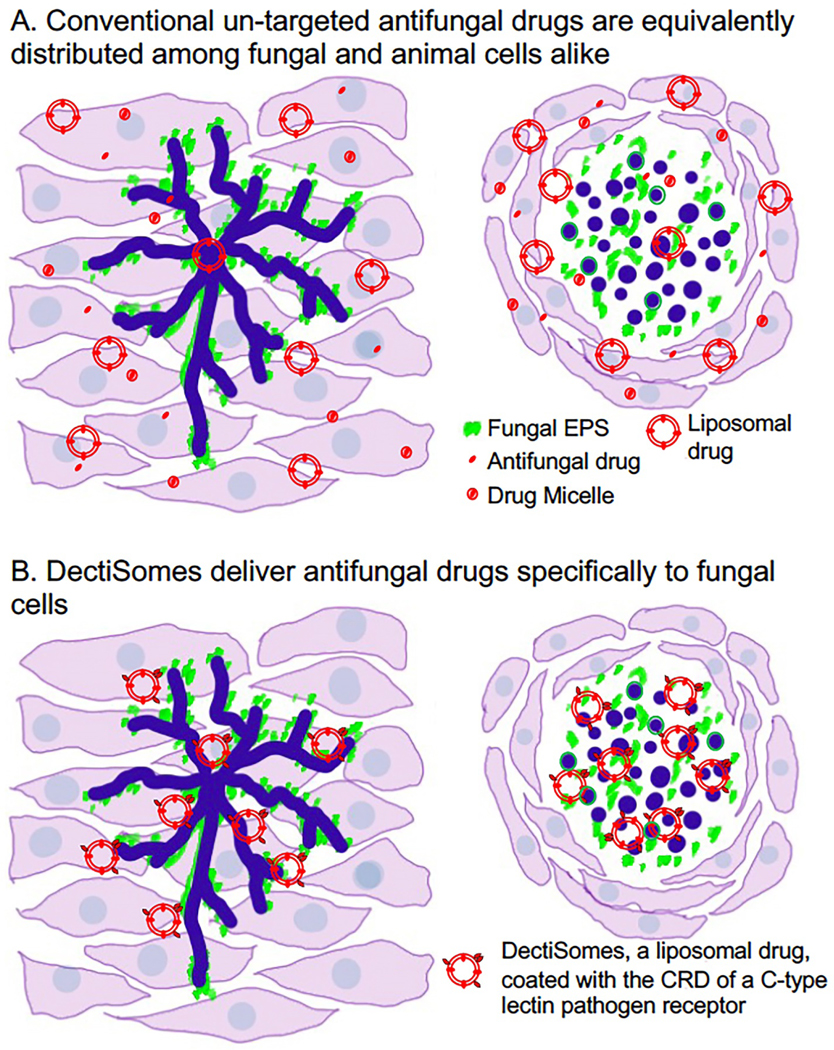
(A). Un-targeted antifungal drugs are equivalently distributed among fungal and animal cells alike. Soluble antifungal drugs and antifungals packaged in micelles or liposomes (e.g., AmBisome®) have no special affinity for fungal pathogens. (B). DectiSomes deliver antifungal drugs specifically to fungal cells. Because they are coated with the CRD of pathogen receptors such as the dectins, DectiSomes have a strong affinity for the carbohydrates in the cell walls and EPS matrices of fungal cells and no particular affinity for animal cells. Concentrating antifungals at sites of infection reduces the effective dose. Targeted delivery should also reduce the number of drug treatments needed to clear fungal infections and hence reduce host toxicity. The left side of each image of A & B models a filamentous fungal infection (e.g., Aspergillus in the lungs) and the right side an infection by fungal cells with yeast morphology (e.g., Cryptococcus in a mucoid cyst in the brain). Fungal cells are shown in dark blue, cognate ligand carbohydrates in cell wall and EPS in green, and antifungal drugs in red.
Before discussing DectiSomes (Section 3.0), we will summarize the complexity of problems posed by invasive pathogenic fungi, including fungal cell properties that constrain antifungal drug strategies and the limitations of current clinically-approved antifungal drugs. To place some bounds on this enormous topic, we will focus most of our discussion on four common pathogens causing aspergillosis, candidiasis, cryptococcosis, and mucormycosis: Aspergillus fumigatus, Candida albicans, Cryptococcus neoformans, and Rhizopus delemar (a.k.a., R. oryzae).
2. The public health threat posed by deadly invasive fungal infections
2.1. The human toll from invasive fungal infections
Globally, there are several million individuals with invasive fungal infections (IFIs) and approximately 1.5 million associated deaths annually [1,2,3,4,5,6]. Estimates of infected global populations with ongoing acute invasive infections made in 2017 suggest there are approximately 750,000 cases of candidiasis, 300,000 cases of aspergillosis, 220,000 cases of cryptococcosis, and 900,000 cases of mucormycosis [2]. The COVID-19 pandemic has driven a dramatic increase in invasive fungal infections caused by species of Candida, Aspergillus, Cryptococcus, and Mucormycosis. The increase of IFIs by Covid-19 infections is attributed in part by heavily compromised patients’ lungs and innate immune systems and in part by the use of steroids to suppress tissue damage caused by anti-viral inflammation [7,8,9,10,11]. Invasive mechanical ventilation used for severe cases is also well-correlated with higher rates of fungal co-infections [12,13], although the cause-and-effect relationship is hard to establish for patients with this advanced level of disease. Even before the impact of the COVID-19 pandemic, the annual medical costs in the U.S. alone from invasive candidiasis, aspergillosis, cryptococcosis, and mucormycosis, were estimated at 3 billion, 1.5 billion, 200 million, and 120 million dollars, respectively [14].
In addition to those with serious cases of COVID-19, patients at high risk of developing life-threatening fungal infections include those with various other lung diseases and individuals with weakened immune systems such as AIDS and diabetes patients. The at-risk population has been steadily increasing for several decades due to growing numbers of patients on immunosuppressants as part of their therapies for some cancers, for stem cell and organ transplants, for medical device implants, and for inflammatory diseases [4,15]. Healthy individuals can also acquire serious fungal infections such as coccidioidomycosis (valley fever), histoplasmosis, blastomycosis, or cryptococcosis caused by Cryptococcus gattii, which is responsible for the ongoing outbreak in the Pacific Northwest.
The mortality rates for these diseases, even after antifungal drug therapy, are high and vary widely depending on the vulnerability of the patient population and medical resource availability: (1) 46% to 75% for invasive candidiasis [3,16,17,18,19], (2) 30% to 95% for pulmonary aspergillosis [3,15,18,20,21,22], (3) 10% to 90% for cryptococcal meningitis [3,23,24,25], and (4) 20% to 99% for pulmonary mucormycosis even when combined with surgery [26,27,28].
2.2. Fungi vs animals, a civil war between relatives
The fungal kingdom is estimated to have diverged from their last common ancestor with animals about 1,100 to 1,200 million years ago (MYA)[29,30,31,32]. Although this divergence is ancient, animals and fungi are the two most closely related kingdoms in the tree of life. They have much in common, including highly conserved structures of their organelles (e.g., lipid membranes, nucleus, endoplasmic reticulum, golgi, mitochondria, and lysosomes) and a common basic gene set and its descending biochemistry (e.g. electron transport chain, ribosomes, spliceosomes). Thus, we are not afforded the advantages given to antibacterial drug development that can target more divergent cellular structures and biochemistry, which presents a major challenge for the development of antifungal drugs with high selectivity.
Mammals have evolved hundreds of new genes and proteins in the innate and adaptive immune systems many of which help restrict and/or clear fungal infections. Upon initial fungal infection, the adaptive immune system produces low affinity fungal pathogen specific antibodies, some of which recognize glycan and glycolipid components of cell walls and exopolysaccharide matrices [33,34,35,36,37]. The production of pathogen specific high affinity IgG antibodies may take months. Although humoral adaptive immunity plays a role in the battle against fungal pathogens, cell mediated innate and adaptive immunity play the dominant role in controlling fungal infections [38,39,40].
Pathogen recognition receptors (PRRs) recognize fungal pathogens and signal the innate immune response. Many PRRs are produced constitutively, while some are induced upon infection. They are primarily expressed on the surface of dendritic cells. Their cognate ligands are fungal oligoglycans, glycolipids, glycoproteins, or other fungal extracellular components. There are more than two dozen members in the C-type-lectin (CTL) receptor superfamily recognizing diverse oligoglycans with potential as targeting proteins [41]. Thirteen of the human CTL receptors recognize twenty genera of fungal pathogens, essentially all of the best characterized fungal pathogens [42,43]. At the onset of this project, the four C-type lectin receptors that we now study, Dectin-1, Dectin-2 (Mincle), DC-SIGN, and Dectin-3 (MCL), were known to recognize 19, 7, 6 and 2 of these 20 genera (Table 2) [42]. The number of fungal species that are recognized by C type lectins is currently underrepresented given the limited literature on less common fungal pathogens and their extracellular carbohydrates. Toll-like receptors such as TLR2 and TLR4 also recognize oligoglycans and lipopolysaccharides [44,45]. Upon binding to their cognate ligands, these and other PRRs detect and signal the presence of an ongoing infection, triggering innate resistance mechanisms [46,47]. It appears that PRRs are often concentrated in plasma membrane microdomains (a.k.a. lipid rafts) of immune cells to enhance their effectiveness [48]. Clustering of PRRs makes functional sense, because PRRs generally bind their cognate fungal ligands most efficiently as homo- or heterodimers or multimers and may act coordinately [49,50].
Table 2.
Cognate ligands of Dectin-1, Dectin-2, Dectin-3, and DC-SIGN isoform DCS12.
| C-Type Lectin / Gene/organism | Properties of CRD and stalk/neck regions a | Well defined glycan ligands | Pathogenic fungal genera recognized out of 20 [42] |
|---|---|---|---|
| Dectin-1 / CLEC7A / Mouse | 20,247 Da, 177 a.a., 32.8% HR, 14.1% BR, pI 6.77, OD280 2.2 |
branched beta-1,3-glucan polymers of at least 8 glucose units. Glucan phosphate > scleroglucan > laminarin > branched nonasaccharied, branched heptasaccharide > heptasaccharide [50,222] | 19/20 [42] |
| Dectin-2 / CLEC6A / Mouse | 19,486 Da, 166 a.a., 35.9% HR, 11.4 BR, pI 5.58, OD280 3.0 | high-manno- and mannopyranose oligo-saccharides. Mannan-alpha-1–6-mannan > mannan-alpha-1–2-mannan > Mannoglycan mannose and Man9GlcNAc2> Man8 GlcNAc2 > Man7GlcNAc2 [223,224,225] | 7/20 [42] |
| Dectin-3, MCL / CLEC4D / Mouse | 20,810 Da, 176 a.a., 37.5% HR, 14.2% BR, pI 5.89, OD280 2.9 | alpha-1,2-mannans, glucuronoxylomannan GXM, trehalose 6,6’-dimycolate TDM, fatty acid moieties of glycolipids [201,205,226,227] | 2/20 [42,198,227] |
| DC-SIGN DCS12 isoform/CD209 / human | 22,648 Da, 199 a.a., 37.7% HR, 10.1% BR, pI 4.93, OD280 2.45 | long chain mannose-rich oligosaccharides Man10>Man5>Man2 and for fucosylated blood type antigens (e.g., the Lewis x trisaccharide), lipomannans and glactomannans often found in protein conjugates. Man9GlcNAc2 > Mannan-alpha-1,3 (mannan-alpha-1,6)mannan> mannose [227,228,229,230,231] | 6/20 [42] |
Molecular weight in Daltons (Da), number of amino acid residues (a.a.), % hydrophobic residues (HR), % basic residues (BR), predicted Isoelectric point (pI), optical density of reduced protein at A280 at 1.0/mg/mL (OD280). These parameters were obtained from the National Center for Biotechnology Information or computed using https://web.expasy.org/protparam/andhttps://www.peptide2.com/N_peptide_hydrophobicity_hydrophilicity.php/
The highly evolved mammalian immune system is encoded by more than 1,600 genes and composed of even more proteins due to the expression of different protein isoforms [51,52]. Many of these genes are in gene families derived from gene duplication and subsequent evolution of paralogs for specialized functions. For example, the C-type lectin family members recognize different cognate glycan ligands, but are evolved from a single ancient origin. For most individuals with healthy immune systems, this gene set is sufficient to combat the daily exposure to invasive fungal pathogens. In addition, nutrient immunity (e.g. restriction of free iron) provides additional protection from fungal infections [53,54,55,56,57].
In comparison, fungal pathogens have smaller genome sizes and limited numbers of virulence related genes (VRGs) with which to resist the host immune system and/or to attack animal hosts. There are obligate pathogens with contracted haploid genomes as small as 2.9 Megabase pairs (Mbp) harboring only approximately 3,000 protein-coding genes, smaller than that of E. coli [58]. Most opportunistic fungal pathogens, including Candida, Aspergillus, Cryptococcus, and Mucor species such as Rhizopus, have haploid genomes on the order of 20 to 50 Mbp, carrying approximately 7000 to 13,000 protein-encoding genes. About 20% of their genomes likely encode essential genes and 10–20% encode virulence-related genes [59,60,61]. That said, successful fungal pathogens have evolved various mechanisms for evading and resisting host innate and adaptive immunity. This includes phase transitions in Candida albicans and Coccidioides immitis, capsule production in Cryptococcus neoformans, melanization in Aspergillus fumigatus, α-(1,3)-glucan biosynthesis in some Histoplasma capsulatum isolates, and mycotoxin production in Rhizopus delemar and Talaromyces marneffei. All these fungi produce degradative enzymes (e.g., proteases, lipases) to assist their colonization and invasion. Their cell wall and extensive extracellular matrices also present a formidable barrier for the host. These host-resistance mechanisms, particularly in opportunistic fungal pathogens, may have evolved under selective pressure in their natural ecosystems [62].
In addition, fungi have evolved various means to adapt to different environmental niches, some of which make them highly successful in colonizing animal or human hosts. Here are two examples. (1) Fungi use three fundamentally different methods to solubilize and take up iron from their environment – electrochemical reduction of ferric (III) iron to ferrous (II) iron, the chelation of ferric iron, or the extraction of heme bound iron– and there is a wide variation in how these three mechanisms are employed [63,64,65]. The utilization of the various mechanisms for managing iron appear to be distributed stochastically among both ancient phyla in the fungal tree of life and even among recently diverged family members such as C. albicans and S. cerevisiae [64,65,66]. (2) Sterols are incorporated into bilipid membranes to impart membrane flexibility and fluidity. The property replacing cholesterol with ergosterol in the bilipid cell membranes, which is often thought of as fundamental to fungi and a common target of antifungal drugs, is in fact, only found in half of the fungal phyla or subphyla [67]. Cholesterol, Stigmatast-7-enol, 24-ethyl cholesterol, are among sterols employed instead of ergosterol. Which sterol is employed in the fungal membrane is only partially correlated with fungal pathology [67]. Fortunately, most human fungal pathogens synthesize and utilize ergosterol in their membranes and other forms of sterols are close enough in structure to ergosterol, that we expect them to be similarly impacted by the antifungal Amphotericin B (AmB).
2.3. Fungal pathogens are dispersed throughout the fungal tree of life.
The several million fungal species are separated into several phyla and perhaps 15 or more subphyla. Some of these groups of fungi date their divergence from a common fungal ancestor near to the origin of the fungal kingdom [29]. Furthermore, fungal pathogens of animals/humans appear to have emerged numerous times among nearly all the various fungal subphyla. Hence, within the fungal kingdom, fungal pathogens do not have a common phylogenetic origin that distinguishes them from other members of the kingdom [68]. Many opportunistic pathogens appear to have evolved first as degraders of plant material and animal waste. For example, the basal subphyla Chytridiomycota (e.g., the amphibian pathogen Batrachochytrium dendrobatidis) and Blastocladiomycota (e.g., the mosquito pathogens, Coelomomyces spp.) split off from the rest of the fungal tree as much as 800 MYA, way before the emergence of insects and amphibians 360 to 500 MYA. The Saccharomycotina appear to have split off of the main fungal tree of life only 350 million years ago [29], which could have been concordant with the explosion in the number of land animal hosts at that time. One of our investigated pathogens, C. albicans, is classified as a commensal fungus associated with endothermic animals. As to our other three pathogens of interest, A. fumigatus belongs to the subphylum Pezizomycotina (a sister subphylum to the Saccharomycotina) in the phylum Ascomycota, C. neoformans belongs to the subphyla Agaricomycotina in the phylum Basidiomycota, and R. delemar to the subphyla Mucormycotina in the phylum Zygomycota, which split off from the main fungal tree 350, 400, and 600 MYA, respectively [29]. In any case, it is clear that fungal pathogens and “groups” of pathogens have ancient and diverse origins. They have had the time and genetic potential to evolve distinct and sometimes convergent strategies of virulence to different animal hosts.
2.4. Environmental reservoirs of fungal pathogens
Knowledge about environmental reservoirs of opportunistic fungal pathogens can be used to limit the exposure of high-risk individuals to pathogens, assist diagnosis, or restrict the emergence of drug resistant strains. Opportunistic fungal infections primarily occur when the host’s health is compromised. Studies on environmental reservoirs of human fungal pathogens have accelerated recently because of the ability to perform rapid genotyping of SNPs or haplotypes, whole genome sequencing of a large numbers of isolates, or metagenomics on environment/host samples. Here are a few examples pointing the direction forward.
A. fumigatus is one of the better elucidated examples of the important role played by environmental reservoirs of an opportunistic fungal pathogen [69,70,71]. This fungus commonly lives off of decaying plant materials [72]. Most infections occur from inhalation of airborne spores and begins as pulmonary aspergillosis in an individual with compromised immunity. Hospital acquired infections are rare except under special circumstances such as disturbances to contaminated air handling systems [73]. Phylogenetic comparisons of A. fumigatus isolated from clinical and environmental settings around the globe using, for example, genetic polymorphisms in short tandem repeats [74,75] or whole genome sequencing [70,76], revealed that most genetic branches of the tree contain both clinical and field isolates. In other words, clinical strains are typically derived from natural local environmental reservoirs.
Infection of high-risk individuals by various pathogenic species of Mucormycota, with Rhizopus spp. being the most common [77], come primarily from inhalation of spores derived from native soils, dust, animal excreta and decaying plant materials [78]. The prevalence of particular Mucor species varies dramatically across different geographic locations [78]. Until the COVID-19 pandemic, pulmonary mucormycosis was considered a relatively rare fungal disease across the globe, but reports surged with progression of the pandemic [10,79,80]. The burden of spores of mucor species in soil, dust, animal excreta, and the air appears to be higher in tropical countries, increasing the risk of exposure and infection [81]. Consistent with this distribution, COVID associated mucormycosis remained relatively rare in most of northern Europe, but southern France with its tropical climate had a higher proportion of cases [82]. A vastly disproportionate number of cases of COVID-associated mucormycosis were reported in India [7,78,83], likely reflecting a large environmental reservoir of mucor species and the large agriculture population with higher exposure due to various agricultural practices [84]. A metagenomic study should begin to define and quantify the sources of Mucor pathogens.
Candida albicans has long been considered a commensal pathogen inhabiting the bodies of warm-blooded animals and humans. It was thought to be most frequently transmitted directly or indirectly among its hosts. Yet standing against this view, human clinical isolates are (1) distributed among highly genetically divergent clades. (2) It grows at a wide range of temperatures such as those in the natural environment, whereas well-adapted commensal pathogens are expected to have temperature growth optima near human body temperature of 37oC. (3) C. albicans has been isolated from soil, water, and plant material, and some environmental isolates are closely related to human pathogens [85,86,87,88,89]. A recent analysis of three C. albicans strains isolated from the bark of exceptionally old oak trees in an ancient English woodland pasture revealed that two tree isolates are more closely related to clinical isolates of C. albicans than most of the other clinical isolates are to each other. The data suggests that Candida albicans could be an opportunistic pathogen with more recent environmental origins [89]. These data prompt another question that we can begin to answer using metagenomics: what fraction of invasive fungal disease cases arise from environmental sources vs those from transmission within human populations.
As the classification of commensal and opportunistic pathogen may depend upon finding the suitable host, Casadevall [90] introduced the concept of the “pathogen potential” of a microbe. Pathogen potential may be expressed as a continuum of values that take into account many properties including the fraction of individuals that become infected at a defined inoculum, mortality, communicability, and the time from infection to disease.
In the last decade, Candida auris has emerged as a problematic life-threatening invasive pathogen. It is generally spread within healthcare facilities and causes harm to patients who are already seriously ill [91]. Initial hospital-associated outbreaks often occur from multidrug resistant strains, making it difficult to control [92,93,94]. Seemingly simultaneous outbreaks have occurred on multiple continents and most often from genetically distinct geographically-defined clades. As fewer than one in two thousand patients were positive for C. auris, when first admitted to the hospital, this fungus is unlikely to be a common human commensal. Strains sensitive and multidrug resistant were recently isolated from coastal marine environments on the remote Andaman and Nicobar Islands, hundreds of miles from the coast of Southwestern India [95,96]. Sequencing of the ribosomal DNA internal transcribed spacer regions demonstrated that they were distantly related to hospital isolates. More detailed analysis of global environmental isolates will yield even more useful information about the reservoirs of multidrug resistant C. auris isolates. This study also raises the interesting question: what selective forces might have led to multidrug resistance in environmental reservoirs.
In summary, data on Aspergillus and Mucor species as opportunistic pathogens with large environmental reservoirs fit conventional wisdom, while the latest genomic and environmental distribution data on C. albicans and C. auris challenge concepts of purely anthropological reservoirs for these species. Here are a few conclusions. First, we are in the early stages of understanding the environmental sources and origins of all fungal pathogens, and metagenomic studies on many pathogens will be informative. Second, some fungal pathogens are likely to be more pervasively distributed in the natural environment than previously suspected. Third, restricting human exposure to fungal pathogens will be difficult to achieve, but it has been possible with the appropriate construction of hospital facilities. Finally, the problem of nearly ubiquitous exposure of many human populations to environmental sources of drug resistant fungal pathogens, emphasizes the need for better pan-antifungal therapeutics.
2.5. Conserved components of fungal cell walls and exopolysaccharide matrices contribute both to their protection and vulnerability
Fungal cell walls and exopolysaccharide matrices vary widely among pathogens from different phyla and subphyla and they contribute to fungal survival in distinct hostile host environments [97,98,99]. The cell walls of nearly all fungal pathogens share a chitin layer adjacent to their membrane, with one exception being Pneumocystis spp. The most common remaining components in the cell wall include different glycans, glycolipids, and glycoproteins that are stacked upon each other in different orders [100]. Variations in layering, length of glycan oligomers, chemical crosslinking of sugars, and other chemical modifications of all these components contribute to the complex presentation of different fungal cell wall components to the host immune system [100,101,102,103]. As a particular example, an oligomannan matrix lies outside of the oligoglucan matrix in the C. albicans cell wall [100]. Although 50% of the Candida cell wall is composed of oligoglucans, the outer oligomannan layer effectively masks oligoglucans from recognition by the host innate and adaptive immune systems [104,105,106], including binding by the oligoglucan specific C-type lectin Dectin-1 and by oligoglucan-specific antibodies [105,107,108].
The matrices of exopolysaccharide (EPS) contribute to fungal pathogenicity by promoting adherence to host tissues or medical devices [98,99,109,110] and confer fungal resistance to host immune attack and antifungal drugs [110,111,112,113,114]. The EPS often extends long distances from the cell (e.g., tens to hundreds of microns). For example, the capsule of C. neoformans, which includes the proximal components of a highly extended EPS, may comprise 80% to 90% of the cell volume [115,116] and is copiously shed from cells. It is the basis for the rapid and sensitive diagnosis of cryptococcosis used clinically. A primary issue confronting the biomedically relevant analysis of EPS is that its composition appears to vary widely dependent upon the fungal growth environment, the strain genetic background, and the developmental stage of the fungus. The general composition of EPS matrices overlaps among divergent groups of pathogens in containing oligoglucans, oligomannans, glycolipids, and mannoproteins. But the EPS of subsets of fungal pathogens also contain other novel crosslink variants, distinct oligoglycans, novel glycoproteins, and DNA that appear to contribute to adherence and pathology [109,111,117,118,119,120,121].
These diverse polymers and particularly the oligoglycans are the cognate ligands recognized by families of PRRs (e.g., C-type lectins) as vertebrates launch an immune response. The large subset of the most conserved polymers recognized by Dectin-1, Dectin-2, Dectin-3, and DC-SIGN are central to the successful targeting of DectiSomes to diverse fungal pathogens in our pan-antifungal strategy.
2.6. Limited classes of antifungal drugs
There is an overwhelming need for new and effective antifungal drugs [122,123]. Currently, there are four major classes of small molecule antifungal drugs in common clinical use to treat invasive fungal infections: triazoles, echinocandins, polyenes, and nucleotide analog antimetabolites [123,124,125,126]. Most of the antifungal drugs listed on the CDC fungal disease web site [3] or in market reports on antifungal drugs [124] as commonly used in the clinic to treat aspergillosis, candidiasis, cryptococcosis, mucormycosis and Pneumocystis pneumonia were first reported to have antifungal activity in vitro and/or in animal models several decades ago (Table 1). Very few new antifungals have made it to the clinic in the last two decades [124]. Amphotericin B (AmB), perhaps the most commonly used therapeutic drug against a wide range of fungal pathogens, was already used clinically in the 1950s [127,128,129,130,131]. Among proposed mechanisms of AmB’s antifungal activity, is that it binds to ergosterol and other membrane sterols and thereby interferes with fungal plasma membrane integrity, which leads to cell death [132]. Liposomal AmB (e.g., AmBisome®) is still one of most widely used first line antifungal drugs, in spite of AmB’s toxicity. The improved efficacy of liposomal AmB over other AmB formulations was first reported in the 1980s [132] and it was approved by the FDA for clinical use in 1997 [133]. Liposomal AmB is commonly used as the standard of comparison when new drugs are being put through the gauntlet of testing. The antimetabolite 5-fluorocytosine (5-FC) was first shown effective against fungi in 1968 [134,135], although it is not used in monotherapy but in combination with other antifungals such as AmB. After being taken up by fungi, 5-FC is metabolized first to 5-fluorouracile (5FU) and then to 5-fluorouridine 5’-triphosphate, which interferes with fungal RNA synthesis and hence protein synthesis. The azoles, and particularly itraconazole [136,137,138] and fluconazole (FLZ) [139,140,141], were introduced as antifungals in the mid 1980s. Azoles interfere with the synthesis of ergosterol and other membrane sterols. Liposomal formulations of FLZ have reported advantages over solubilized FLZ [142,143,144]. However, to be effective at killing fungal cells, azole drugs must be delivered at much higher doses (e.g., hundreds of mg/kg) than most other antifungals. Although the candin family of antifungals were discovered in the 1970s [145,146], newer echinocandin analogs such as caspofungin, micafungin, and anidulafungin are only used in the clinic since early 2000s. They all interfere with fungal beta-glucan synthesis. The laboratory efficacy of anidulafungin was first reported in the mid 1990, and FDA approval for its clinical use was granted in 2006 [147]. Compared to AmB and azoles, echinocandins are effective against a more limited set of fungal pathogens and not at all against Cryptococcus.
Table 1.
First reported experimental evidence for the antifungal efficacy of drugs in clinical use against invasive fungal infections
| Drug class | Antifungal | Date | Reference |
|---|---|---|---|
| Polyenes | Nystatin | 1951 | [149,150] |
| Amphotericin B (AmB) | ~1956 | [129,130,131] | |
| Liposomal AmB | ~1983 | [132,151] | |
| Nucleotide antimetabolites | Flucytosine (5-flurocytosine) | ~1968 | [134,135] |
| Azoles | Sulfamethoxazole | 1974 | [152,153] |
| Itraconazole | ~1984 | [136,137,138], | |
| Fluconazole (FLZ) | ~1985 | [139,140,141] | |
| Voriconazole | ~1996 | [154,155], | |
| Posaconazole | ~1996 | [156,157] | |
| Isavuconazole | ~2006 | [158,159] | |
| Echinocandins | Papulacandin B | 1977 | [146] |
| Anidulafungin (AFG) | ~1996 | [160,161] | |
| Caspofungin (CAS) | ~1997 | [162,163] | |
| Micafungin | ~1999 | [164] |
Subsequent to the introduction of these classes of antifungals, newer chemical analogs were introduced, which often show modest improvement in efficacy against a limited number of species [148]. Fortunately, a number of novel classes of antifungals are now in clinical development [122], and hopefully some will be applied in the clinic in the near future. Nonetheless, there is an urgent need to dramatically improve the performance of existing and future classes of antifungal drugs. This is the major goal driving our efforts to develop DectiSomes.
2.7. Antifungal drug resistance
Increasing numbers of drug resistant fungal pathogens have become a serious problem facing public health and food security [165,166,167]. Resistance has been reported for all antifungal drugs and resistance to some classes of drugs in all major genera of human fungal pathogens [168]. This problem is exacerbated by the fact that the natural environment is a source of some, if not most, drug resistant fungi. Here are examples to illustrate our emerging understanding of the problem.
The environmental reservoirs of Aspergillus spp. appear to play a major role in the epidemiology of aspergillosis caused by drug resistant isolates [70,76,169]. Azoles such as fluconazole, posaconazole, and isavuconazole (Table 1) target fungal cytochrome P450s in A. fumigatus, Cyp51A and CYP51B (the equivalent of Erg11 in yeasts), and hence block sterol and particularly ergosterol synthesis [170]. More and more patients appear to have been infected by pre-existing Cyp51 mutant azole resistant strains (i.e., not from the clinic) [71]. Tandem repeat mutations in the Cyp51A promoter such as TR34 and TR46 cause protein overexpression [171]. Amino acid substitutions such as V46F, L98H, Y121F, V172M, and T289A are either known or molecularly modeled to reduce Cyp51 protein binding to azoles [172,173,174]. In some clinically resistant strains, these regulatory and structural gene mutations are often found together (e.g., TR34/L98H and TR46/Y121F/T289A). A. fumigatus strains with these same combinations of resistance alleles have now been found in agricultural soils from North and South America, Africa, Europe and Asia [70,71,76,175]. Whole genome sequence based phylogenetic analysis reveals distinct clades of A. fumigatus strains with subsets of particular azole resistance mutations. [70,76]. Hence, the emerging view is that agricultural practices, including the widespread and extended use of azole pesticides, have contributed to the selection and subsequent global dispersal of azole-resistant Aspergillus strains now found in the clinic.
Multidrug resistant (MDR) fungi arise from several mechanisms. Specific combinations of specific drug resistance mutant genes can lead to MDR. But more commonly, MDR fungi result from mutations leading to overexpression of individual members of the large ABC transporter gene family. When overexpressed, ABC transporters more efficiently pump out multiple classes of antifungals resulting in MDR phenotypes [176]. The genomes of most opportunistic fungal pathogens such as A. fumigatus [177], C. albicans [93,178], C. neoformans [179] and R. delemar [180] encode a few to several dozen ABC transporters [176]. Gene amplification of just one ABC transporter can lead to MDR.
2.8. Host toxicity of antifungal drugs
After extended or recurrent treatments at effective doses that control or clear fungal infections, essentially all antifungals become toxic to humans. For example, amphotericin B formulations, nucleotide analog antimetabolites (e.g., 5-FC), triazoles, and echinocandins are hepatoxic and/or nephrotoxic [181,182]. Amphotericin B is one of the most toxic, earning the name “Amphoterrible” [183]. The toxicity is believed mainly due to its affinity to cholesterol, the human equivalent of ergosterol. AmB is incorporated into animal cell membranes, resulting in loss of osmotic integrity and causing anemia, nephrotoxicity, and cardiotoxicity [132]. Yet, AmB remains one of the most effective and widely employed first line antifungals, because it is a fungicidal broad-spectrum drug capable of rapidly reducing or clearing fungal burden. Liposomal formulations of AmB such as AmBisome® improve performance and show somewhat reduced nephrotoxicity and infusion toxicity as compared to detergent solubilized micellar AmB [184,185,186,187,188,189].
The azoles must be delivered at high mg/kg doses to be effective, but even one of the best tolerated azoles, fluconazole, causes hepatotoxicity [181,190]. Because the azoles inhibit important host liver P450s, drug-drug interactions are a major impediment for azole drugs in patients that are already under multiple medications. For example, by inhibiting the P450-dependent turnover of other life-saving medications commonly taken by immunosuppressed patients, azoles increase the toxicity of other drugs [191,192,193]. Echinocandins may be the least toxic [183], but they may result in facial swelling, rash, fever and some alterations in hepatic enzyme activity [194]. Furthermore, they are not effective against Cryptococcus either in vitro or in vivo [195]. The antimetabolite 5-FC, once being converted to 5-FU, a common anticancer drug, interferes with bone marrow synthesis of lymphocytes [196] with the potential outcome of fatal anemia [197].
In conclusion, employing most currently available antifungal formulations and treatment regimens required to be effective anti-infectives, some patients may suffer from severe drug toxicity. This potential violation of the Hippocratic injunction is a major dilemma for prescribing physicians and likely complicates and delays their decision tree for treatment.
3.0. DectiSomes
The numerous problems posed by pathogenic fungi and the limitations of current therapeutic antifungal drug strategies to control them led directly to our team’s invention of DectiSomes. DectiSomes are novel class of drug-delivery vehicle that may address the major limitations of current antifungal therapies, including patient toxicity and poor fungal clearance. DectiSomes are comprised of three major components (Fig. 2): (1) a lipid nanoparticle (e.g., a liposome), (2) an anti-infective therapeutic (e.g., AmB), and (3) a C-type lectin polypeptide (e.g., Dectin-1) that serves to target the drug-loaded liposomes to fungal pathogens inside the host and away from host cells.
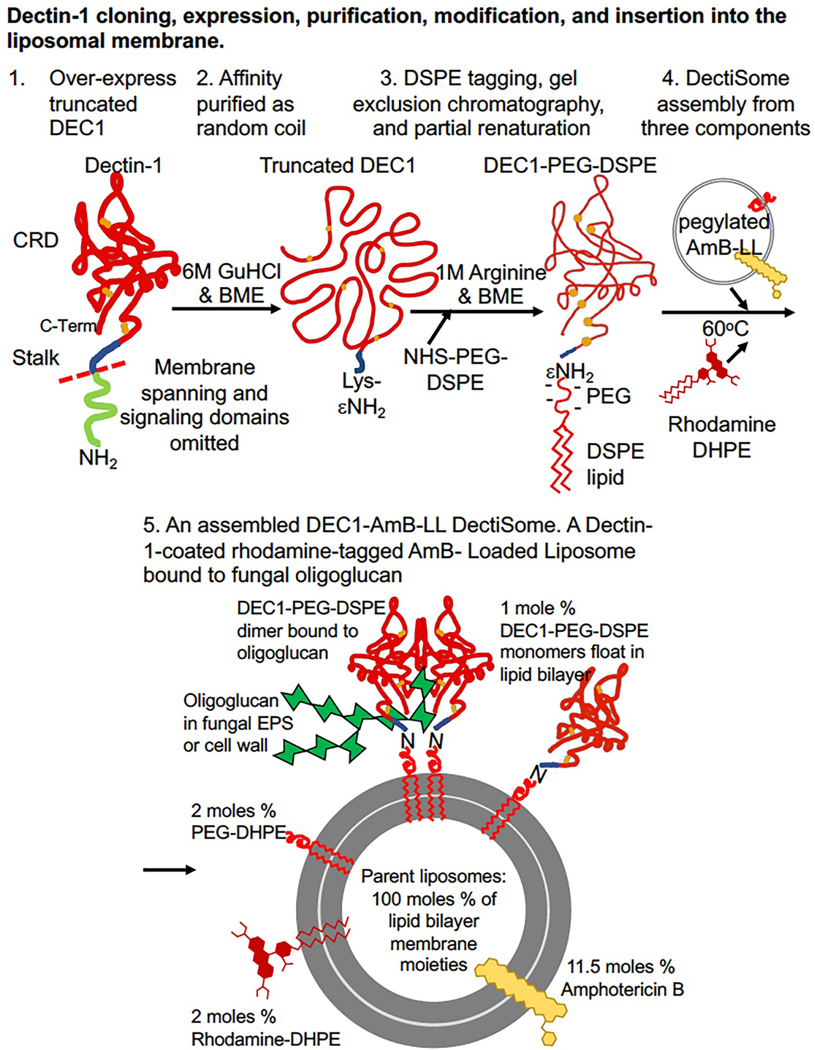
Fig. 2. Construction of a Dectin-1-CRD coated Amphotericin B loaded pegylated liposome, DEC1-AmB-LL.
DEC1-AmB-LL serves as an example DectiSome. (A). Steps involved in Dectin-1 cloning, expression, purification, modification, and insertion into the liposome membrane are shown. 1. Synthetic DNA encoding a truncated fragment of Dectin-1 (DEC1) including the CRD and stalk region and N-terminal his6 and lysine (Lys) tags, but lacking the membrane spanning and signaling domain, was cloned into an E. coli plasmid expression vector. 2. Dectin proteins from expressing E. coli cells were extracted into 6 M guanidine hydrochloride (GuHCl) with beta-mercaptoethanol (BME) generating random coiled protein with its six reduced cysteine residues. The random coiled protein was affinity purified on Ni(II) resin. 3. A pegylated lipid moiety, PEG-DSPE, was coupled to the Lys tag in the denatured proteins. Coupling reagents and GuHCl were removed by gel exclusion chromatography into a 1 M Arginine BME crowding buffer that is proposed to partially renature the protein and improve its final renaturation in biological buffers. 4. DEC1-AmB-LLs were assembled by incubating a pegylated analog of AmBisome®, AmB-LLs, the lipid modified DEC1 protein, and lipid modified rhodamine at 60°C, the transition temperature of these liposomes. When these DectiSomes are diluted into biological buffers, the liposomal DEC1 is proposed to reform three cystine bridges necessary for proper protein folding. (B). A DEC1-AmB-LL is shown binding its cognate ligand an oligoglucan (green). The typical structure and composition of a DectiSome are indicated. Based on the initial lipid moieties of an un-pegylated liposome representing 100 moles percent of lipid, the proportion of each additional reagent is shown and its mole percent indicated, diluting the unmodified liposomal lipids to 83.5 moles percent. Each 100 nm diameter liposome contains approximately 1,500 molecules of Dectin-1, 3,000 of Rhodamine-DHPE, 3,000 of PEG-DHPE, and 17,000 of AmB. The DEC1-PEG-DSPE monomers are free to float in the liposome membrane and form active dimers that bind Dectin-1’s cognate oligoglucans [107]. Alternate functional AmB-loaded DectiSomes coated with the CRDs of Dectin-2, Dectin-3 and DC-SIGN have been constructed similarly [215,217,220]. Dectin-2 coated liposomes containing Anidulafungin instead of AmB have also been assembled by related methods [218].
3.1. C-type lectins as targeting proteins
There are more than two dozen members of the CTL superfamily recognizing diverse oligoglycans [41]. Among these, the four C-type lectins we have employed as liposome targeting proteins, namely Dectin-1, Dectin-2, Dectin-3 and DC-SIGN, are encoded by murine CLEC7A, CLEC6A, and CLEC4D and by human CD209, respectively. Each has an extracellular Carbohydrate Recognition Domain (CRD) that recognizes their cognate ligands, a stalk or neck region, a membrane spanning domain, and intracellular signaling domain. The native forms of the four C-Type lectins are expressed and often co-expressed on the surface of several types of leukocytes and most often dendritic cells [198,199,200,201,202]. They act as receptors to signal an early innate immune response to Candida, Aspergillus, Cryptococcus, and Rhizopus species [200,201,203,204,205,206]. They respond rapidly upon binding to fungal cells, cell membrane fragments, or soluble polysaccharides [204]. DEC1−/− and DEC2−/− null mice are more susceptible to most fungal infections than wild type [199,201,204,207,208,209,210]. DEC3−/− null mice are deficient in stress signaling in response to C. neoformans infection, but are not more susceptible to pulmonary infection [211]. Human dendritic cell DC-SIGN participates in internalizing C. albicans and A. fumigatus cells and in signaling responses to infection [212,213,214].
The physical properties of the CRD and stalk or neck regions of these four C-type lectins are given in Table 2 along with examples of the known synthetic cognate ligands. They are of similar size, 19 to 23 kilo-Daltons (kDa). Their CRD regions have modestly homologous amino acid sequences with 32 to 53% identity of residues for any pairwise comparison. They have six similarly placed cystine residues that are necessary to form three 3-disulfide crosslinks that stabilize the characteristic C-type CRD folded structure [50].
DectiSomes must deliver anti-infective drugs in close proximity to fungal cells in order to increase the local concentration of the drug if they are to be more effective than untargeted drug delivery methods. Further, to be worthy of clinical development, they must target diverse fungal pathogens regardless of fungal cell morphologies in their hosts (Fig. 1). As originally conceived, DectiSomes were designed to target oligosaccharides in both fungal cell walls and EPS matrices. Our engineered DectiSomes employ the CRDs and all or part of the stalk regions as targeting polypeptides, leaving off their membrane spanning and signaling domains. The stalk regions (blue lines in Fig. 2) give flexibility to the presentation of the CRDs on the liposome surface so that they may orient appropriately to form functional dimers or multimers as they bind their cognate glycans. There are multiple transcript isoforms of human DC-SIGN that incorporate different combinations of 8 possible neck repeats. We engineered and tested two isoforms of human DC-SIGN. DCS12 includes neck repeats 1 and 2. DCS12 is an endogenous isoform that we found was superior in binding to several fungal species relative to another isoform with neck repeats 7 and 8 (DCS78) [215]. Other combinations of stalk regions have not yet been explored. We chose to work with the human DC-SIGN gene, because in mice there is no clear ortholog but a family of related genes and proteins (e.g., SIGNR1, CD209-like proteins). The known ligands of these four C-type lectins encompass most of the known glycan components of the cell walls and EPS matrices of essentially all fungal pathogens, including Aspergillus, Candida, Cryptococcus, and Mucor spp. [42]. In practice, however, we discovered that the vast majority of Dectin-1, Dectin-2, Dectin-3, and DC-SIGN targeted DectiSomes bind highly efficiently to EPS matrices and significantly less to cell walls. This binding pattern could be due to limited access to the cell wall ligands, extensive chemical crosslinking of otherwise cognate ligands, or both. Although DectiSomes bind primarily to fungal EPS matrices, they do efficiently improve antifungal drug efficacy both in vitro and in vivo relative to untargeted antifungals [107,215,216,217,218,219,220]. Hence, the physical distance of EPS matrices from fungal cell bodies does not appear to significantly limit DectiSome efficacy. In contrast to our primarily targeting the EPS, a recent study used a related strategy to link the chitin binding domain of a chitinase to a lipid carrier and link this construct to the surface of AmB-loaded liposomes [221]. Chitin is the glycan layer closest to the fungal plasma membrane and is briefly exposed during hyphal tip growth. These chitin targeted liposomes bind efficiently to the hyphal tips of the ascomycete Trichoderma viride, which results in modestly improved antifungal activity in vitro.
3.2. Construction of a DectiSome
The structure of a typical DectiSome is illustrated for a Dectin-1 targeted AmB-loaded liposome, DEC1-AmB-LL, in Fig. 2.5. Dectin-2, Dectin-3 and DC-SIGN targeted liposomal reagents have nearly identical compositions to this example. Each 100-nanometer diameter DEC1-AmB-LL has approximately 1,500 molecules of Dectin-1 and 3,000 molecules of Rhodamine B (Fig. 2.5). Each DEC1-AmB-LL has approximately 16,500 molecules of AmB incorporated into its membrane or 11 moles percent AmB relative to moles of liposomal lipid. Hence, our liposomes have the same size and AmB content as commercial AmBisome®. One other difference between DectiSomes and AmBisome® is the presence of pegylated lipid (5 moles %), which is not found in AmBisome®. Surface pegylation protects liposomes from opsonization and phagocytosis, extends drug half-life in vivo, and improves overall drug efficacy [232,233,234]. Currently, pegylation is widely used in commercial liposomal drugs such as Moderna’s COVID-19 mRNA vaccine [235]. Most of our work has been done starting with AmB loaded, rhodamine tagged, pegylated liposomes (AmB-LLs) that were coated with the various targeting polypeptides (DectiSomes). AmB-LLs are then used as controls, comparing their performance to, for example, DEC1-AmB-LLs. In DectiSome binding experiments in vitro we also included bovine serum albumin coated liposomes, BSA-AmB-LLs, and found that a protein coating alone did not skew our results.
3.3. DectiSomes are a new class of immunoliposome
DectiSomes represent a new class of cell-targeted therapy. The most commonly employed targeting proteins for drug loaded liposomes have been monoclonal antibodies, antibody fragments, or single chain antibodies that enable immunoliposomes to bind an antigen on a specific pathogenic cell type or types and most commonly to cancer cells [236]. The specific and focused delivery of immunoliposomes generally improves cell-type specificity and drug effectiveness by 3- to 10-fold over passive delivery [237,238] and hence, reduces host toxicity [236,239]. In a pioneering study published in 1998, AmB-loaded pegylated liposomes (PEG-L-AmBs) were coated with a monoclonal antibody (34A) specific for the vessel wall of pulmonary capillary cells [240]. 34A-PEG-L-AmBs reduced the burden of A. fumigatus cells in the mouse lungs 2.5-fold and significantly improved mouse survival, relative to the untargeted PEG-L-AmB control [240]. While they were not targeting fungal cells, this study suggested that bringing AmB in closer proximity to the infection site improves efficacy.
The CTLs are part of the mammalian immune system and similar to antibodies they recognize foreign ligand structures. Therefore, DectiSomes are functional analogs of immunoliposomes. However, they are structurally distinct. By employing the small pathogen receptor CRDs of C type lectins (~20 kDa) to target liposomes instead of antibodies (IgGs are typically 145 kDa), we appear to have circumvented many of the limitations and problems associated with immunoliposomes (see Section 3.8).
3.4. Engineering, production, and attachment of C-type lectin polypeptides to the surface of liposomes
The extracellular CRD and stalk regions of the C-type lectins lacking their membrane domains were originally called “soluble” isoforms, because they are more soluble than the full-length proteins containing the hydrophobic membrane spanning domains and the signaling domains. However, these truncated isoforms are still highly hydrophobic (Table 2), aggregation prone, and are functionally unstable in normal biological buffers over periods of hours or days. They are generally produced in animal cells at less than 1 mg/L of media. These problems of insolubility, aggregation, and low yields have kept the price of commercial sources of human or murine Dectin-1 (DEC1) and Dectin-2 (DEC2) (e.g., Abcam, SinoBiological) high (typically $300 to $5,000 U.S. dollars per 100 micrograms). The dectins prepared from E. coli or from animal cells are usually made more soluble by tethering the CRD and stalk region to moderately large peptide carriers such as the 56-amino-acid-long B1 domain of streptococcal protein G [241] or more commonly to the 232-amino-acid-long Fc constant region of IgG1 antibody [203,242,243]. Fortunately, glycosylation status of dectins does not appear to affect their ligand binding [244,245,246]. In the last few years, we and others [224,226] overcame most of the yield, solubility, and renaturation problems associated with producing large quantities of the C-type lectin CRD polypeptides in bacteria by optimizing their codon preferences for E. coli and manipulating them in their denatured random coil form until late stages in their production and biochemical manipulation. Once manipulated to the surface of a DectiSome, we found them to be much more stable.
Fig. 2 summarizes the key steps in making a DectiSome. In our constructs, we added a 6 a.a. residue histidine tag and a 11 a.a. residue glycine-serine flexible spacer to the gene construct (Fig. 2.1)[107,215,216,220]. The cost of making each polypeptide is almost negligible. Extracting the polypeptides into 6 M guanidine hydrochloride (GuHCl) was essential to high yields, whereas much less soluble protein was recovered from E. coli cell pellets using 8 M urea. After affinity purification on Ni+ resin (Fig. 2.2) [107,215,216,220], the lysine tag is reacted with NHS-PEG-DSPE to make for example DEC1-PEG-DSPE (Fig. 2.3). The excess of reagents and toxic GuHCl were removed by gel exclusion chromatography into a 1 M arginine crowding buffer with reducing agent, which partially renature the proteins [247,248]. Pegylated liposomes carrying an antifungal drug such as Amphotericin B loaded liposomes (AmB-LL) were produced separately [107] and were the template for constructing most DectiSomes. The lipophilic DSPE moiety of DEC1-PEG-DSPE and the lipophilic DHPE moiety of Rhodamine-DHPE are inserted into the membrane of the AmB-LLs (Fig. 2.4) by briefly incubating at 60oC or 37oC to form the final product DectiSome (Fig. 2.5). Once loaded onto liposomes, all four types of DectiSomes, DEC1-AmB-LLs, DEC2-AmB-LLs, DEC3-AmB-LLs, and DCS12-AmB-LLs, retain functionally stable protein for several months with the periodic addition of fresh reducing agent. Bovine serum albumin coated BSA-AmB-LLs were produced similarly except that GuHCl and arginine were omitted from the various buffers. The microgram quantity of BSA was equivalent to that of the C-type lectins, so that the total amount of protein coating on these control liposomes was similar. The final preparations of each type of liposome or DectiSome were highly concentrated for AmB (e.g., ~900 uM AmB and ~1.4 mg/mL C-type lectin) such that they typically had to be diluted 100− to 5,000-fold into PBS or liposome dilution buffers or fungal cell growth media prior to their use in most test experiments. Calcium aids in the folding and stability of the glycan binding pocket of the CRD, even for DEC1, which was once thought to be calcium independent in its binding to beta-glucans [50,224]. In practice, we have not observed any significant measurable loss of activity for DectiSomes diluted into PBS relative to that diluted into buffers with high calcium concentrations (e.g., 2 to 10 mM CaCl2). However, we normally dilute liposomes just before using them, which may explain this lack of dependence on added calcium.
3.5. Validation pipeline
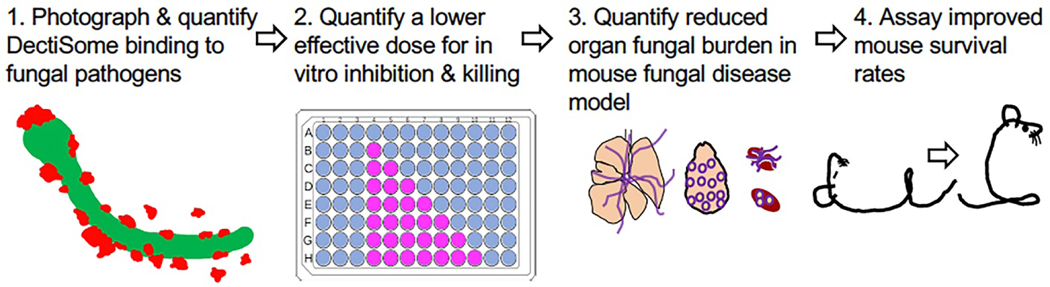
Once antifungal drug-loaded rhodamine-fluorescent liposomes (e.g., AmB-LLs) are coated with the various CTLs or BSA, we follow the four steps in the validation pipeline illustrated in Fig. 3. 1. We determine if a particular C-type lectin increases fluorescent liposome binding to the pathogen in question magnitude more strongly than AmB-LL or BSA-AmB-LL controls. 2. If there is efficient binding, we go on to confirm that this DectiSome dramatically lowers the effective dose of AmB relative to AmB-LLs or BSA-AmB-LLs for in vitro fungal cell growth inhibition and/or killing. 3. If the in vitro data are strong, we employ mouse models of invasive fungal infection and demonstrate a reduction in fungal burden in the effected organ(s), using DectiSomes delivering a very low dose of AmB, a concentration at which AmB-LLs are relatively ineffective. 4. If there is a significant reduction in fungal burden, we go on to demonstrate that DectiSomes improve animal survival relative to AmB-LLs. The same pipeline is used for DectiSomes delivering other antifungal drugs.
Fig. 3. Validation pipeline to demonstrate the improved efficacy of different classes of DectiSomes against diverse pathogens relative to untargeted antifungals.
Our validation pipeline for DectiSomes proceeds through four steps. (1) We characterize and quantify DectiSome binding to various and diverse fungal pathogens. (2) We quantify the effective dose of antifungal delivered by DectiSomes for inhibition and killing in vitro grown fungal cells. (3) Using mouse models of fungal disease, we quantify the effective dose of antifungal delivered by DectiSomes to reduce fungal burden in target organs. (4) Based on fungal burden experiments, we assay the dose(s) necessary to improve rates of mouse survival. Data are quantified relative to untargeted drug loaded liposomes.
3.6. DectiSomes target evolutionarily diverse fungal pathogens
We performed most of the proof-of-concept experiments on pan-fungal targeting of DectiSomes using A. fumigatus, C. albicans, C. neoformans, and R. delemar for the following reasons. (1) These species represent the genera that are responsible for the vast majority of mortality caused by mycoses and are themselves the most common clinical isolate from each genus [3]. (2) They have ancient and highly divergent evolutionary histories being members of the Ascomycota (Aspergillus, Candida), Basidiomycota (Cryptococcus), and Zygomycota (Rhizopus). (3) They represent distinct fungal morphotypes (yeast, dimorphic, and filamentous). (4) Most fungal species including these four are coated by a thick cell wall and exopolysaccharide matrix rich in glucans, mannans, and mannoproteins and/or glucuronoxylomannan (GXM), the targets of our fungal cell selective targeting technology [249,250,251,252,253,254]. (5) There are problems clearing patients of all four of these species making them most worthy of our focused efforts. For example, at least 90% of relapses of cryptococcal infection are caused by the original strains not cleared by the initial antifungal therapy [255]. Some classes of drugs are not effective against a broad spectrum of fungal species (e.g., echinocandins are ineffective against Cryptococcus and Rhizopus spp.). (6) These fungi are relatively tractable in the laboratory and there are well-established mouse disease models. In summary, in addition to their clinical importance, these four species reasonably represent the tremendous evolutionary distance and the distinct lifestyles and cell morphologies among human pathogens in the fungal kingdom and are good laboratory models.
3.7. Progress with DectiSomes
Following the four steps in the validation pipeline in Fig. 3, our progress is summarized in Table 3 and described in the following four corresponding sections.
Table 3.
Summary of the performance of DectiSomes, relative to untargeted control drug loaded liposomes
| A. In vitro assays | |||
|---|---|---|---|
| DectiSomes (References) | Pathogen | Increases liposome bindingb | Increases in vitro killing / inhibitionb |
| DEC1-AmB-LLs [107] [217] | Aspergillus fumigatus | >200-fold | >50-fold |
| Rhizopus delemar | >1,900-fold hyphae >120-fold germlings |
>70-fold | |
| Candida albicans | >130-fold | No data | |
| Cryptococcus neoformans | Yes, but not quatified | No data | |
| DEC1-AmBisomef [218] | C. albicans | >90-fold | No data |
| DEC2-AmB-LLs [220] | A. fumigatus | >50-fold | >30-fold |
| C. albicans | >100-fold | >90-fold | |
| C. neoformans | >150-fold | >10-fold | |
| DEC2-AmBisomef [218] | C. albicans | >25-fold | No data |
| DEC3-AmB-LLs [216] | C. albicans | yes | yes |
| R. delemar | yes | yes | |
| C. neoformans | yes | marginal | |
| DCS12-AmB-LLs (DC-SIGN isoform) [215] | A. fumigatus | >25-fold | >70-fold |
| C. albicans | >25-fold | >50-fold | |
| C. neoformans | >30-fold | >10-fold | |
| B. In vivo assays in murine models of invasive fungal diseases | |||
|---|---|---|---|
| Liposome, antifungal, mouse model (Reference) | Increase in liposome binding in effected organ | Reduction in fungal burden in effected organ | Increase in percent of surviving miced |
|
DEC1-AmB-LLs 0.2 mg/kg AmB |
|||
| Systemic candidiasis neutropenic model [218] | 24-fold in kidneysb | 6-fold in the kidneysb | 18% to 44% |
|
DEC2-AmB-LLs 0.2 mg/kg AmB |
|||
| Pulmonary aspergillosis neutropenic model [219] | 30-fold in lungsb | 40-fold in the lungsb | 0% to 58% |
| Pulmonary aspergillosis steroid model [219] | aN.D. | 20-fold in the lungsb | aN.D. |
| Systemic Candidiasis neutropenic model [218] | 50-fold kidneys | 12-fold kidneys | 8% to 58% |
|
DEC2-AFG-LLse 0.6 mg/kg AFG |
|||
| Systemic Candidiasis neutropenic model [218] | 9.8-fold kidneys | ||
|
DEC2-AmBisomef 0.2 mg/kg AmB |
|||
| Systemic Candidiasis neutropenic model [218] | 6.1-fold kidneys | ||
3.7.1. DectiSome binding to fungi.
We have shown that several types of DectiSomes, DEC1-AmB-LLs [107,217,218], DEC2-AmB-LLs [218,220], DEC3-AmB-LLs, and DCS12-AmB-LLs [215] bind specifically to three or four of our four fungal test species. We quantified the efficiency of red fluorescent DectiSome binding to our four test species by comparing number of patches of red fluorescence in large fields of fungal cells [107] or the total area of fluorescence associated with fungal cells [215,217,218,220] to that of fluorescent negative control liposomes, AmB-LLs and BSA-AmB-LLs. Some of our in vitro quantitative binding data are summarized in Table 3A. All four types of DectiSomes tested bound to the four target species of all developmental stages analyzed. DectiSomes showed from 25-fold to greater than 1,000-fold more efficient binding than AmB-LLs. Example images of various DectiSomes binding to our four test species at different developmental stages are shown in Fig. 4. DEC1-AmB-LLs bind to A. fumigatus swollen conidia and hyphae (Fig. 4A, 4B) and to R. delemar germlings and hyphae (Fig. 4C). DEC2-AmB-LLs bind to C. neoformans yeast cells (Fig. 4E) and C. albicans hyphae (Fig. 4F). DC-SIGN DCS12-AmB-LLs bind to C. neoformans (Fig. 4G) and C. albicans hyphae (Fig. 4H). These qualitative data illustrate the proximity of DectiSome binding to fungal cells. Although not shown, control liposomes do not show any significant binding [107,217,220,256,257]. DectiSomes mostly bind to fungal cell-associated EPS matrices and not to the fungal cell walls as might have been expected. In the case of Aspergillus and Rhizopus, we did detect the binding of some individual liposomes to their cell walls [107,217]. The limited binding to cell walls is not specific to our house made liposomes. When we coated commercial AmBisome® with a dectin and rhodamine B, these AmBisomes modified into DectiSomes also showed EPS binding (DEC1-Ambisome binding to C. albicans hyphae in Fig. 3D). The size of DectiSomes (~100 nm in diameter) may not be a main factor that prevent them from binding to their cognate ligands in cell walls, because our studies with rhodamine coupled Dectin-2 and DCS12 polypeptides [215,220], which are estimated to have rotational diameters of only a few nanometers, have shown similar specificity for EPS as their liposome labeled counterparts. Among possible explanations, it is likely that chemical crosslinking of cell wall glycans alters target ligands to limit binding.
Fig. 4. Example images of DectiSomes binding to multiple developmental stages of diverse fungal pathogens.
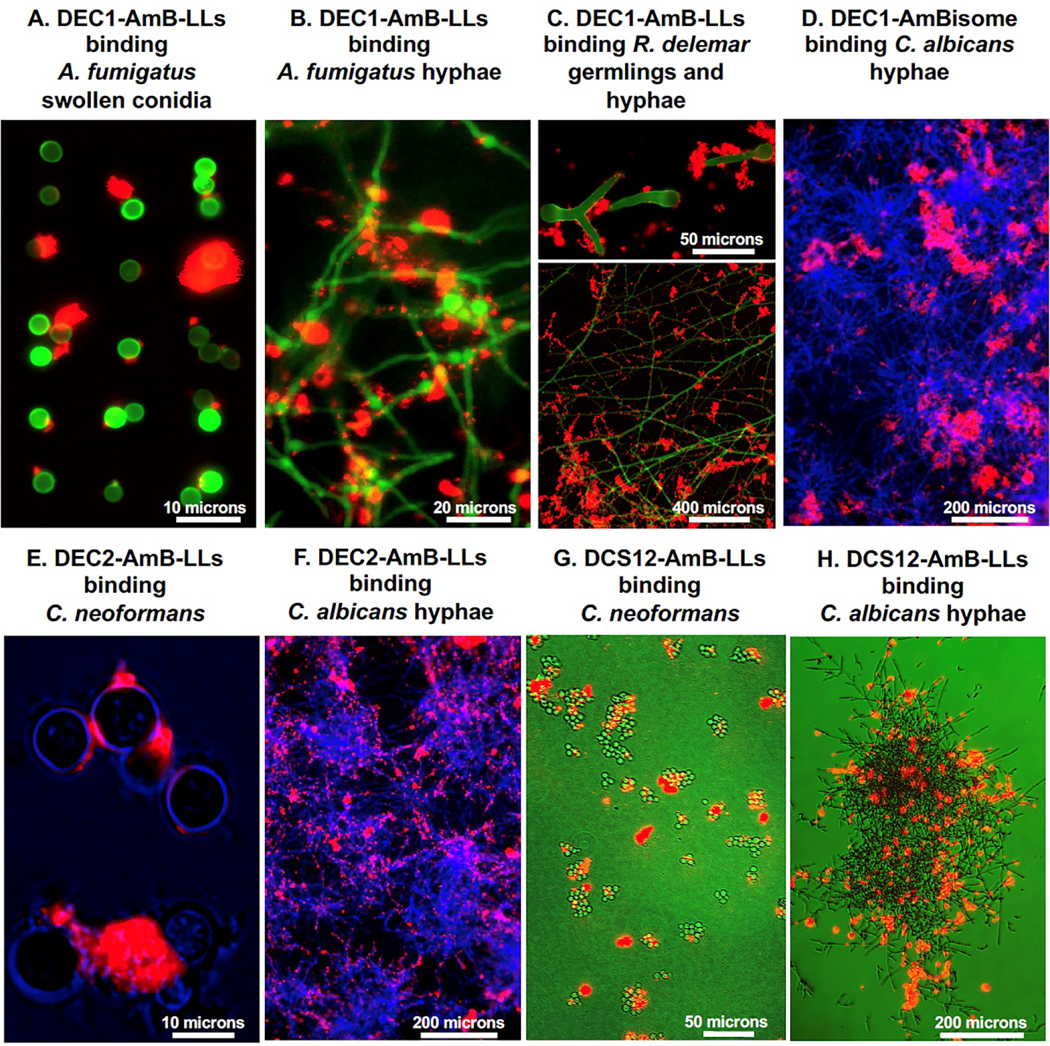
(A & B). DEC1-AmB-LLs binding to A. fumigatus swollen conidia and hyphae, respectively. (C). DEC1-AmB-LLs binding to R. delemar germlings and mature hyphae. (D). DEC1-AmBisome binding to C. albicans hyphae. (E). DEC2-AmB-LLs binding to C. neoformans yeast cells. (F). DEC2-AmB-LLs binding to C. albicans hyphae. (G. & H). DC-SIGN isoform coated DCS12-AmB-LLs binding to C. neoformans yeast cells and C. albicans hyphae. All liposomes were tagged with Rhodamine B and the red epifluorescence of their binding is shown in red. A, B. GFP fluorescently labeled cells. C, D, F, G. Cell chitin is stained with calcofluor white and their fluorescence is shown in green or blue. E, H. Cells illuminated with bright field, but colored blue and green, respectively. C and G. Cells were grown and imaged of the surface of agar. These are replicate images of cell staining not shown in the original publications [107,215,217,218,219,220]. Size bars indicate the degree of magnification.
DectiSomes have several advantages for demonstrating the specificity of C-type lectin binding to fungal pathogens over the protein tagged with one or two fluorophore molecules. First, each liposome contains 1,500 polypeptides, giving a greater chance of any one liposome binding to a glycan ligand through multiple binding interactions and likely stabilize binding via their avidity. Second, restricting the polypeptides to 2-dimensional liposome surface versus soluble monomeric proteins diffusing around in solution provides a greater chance for CTL oligomerization during ligand binding. Third, each DectiSome has ~3,000 rhodamine molecules, which enhance the fluorescent signal for each binding event.
3.7.2. In vitro inhibition and killing.
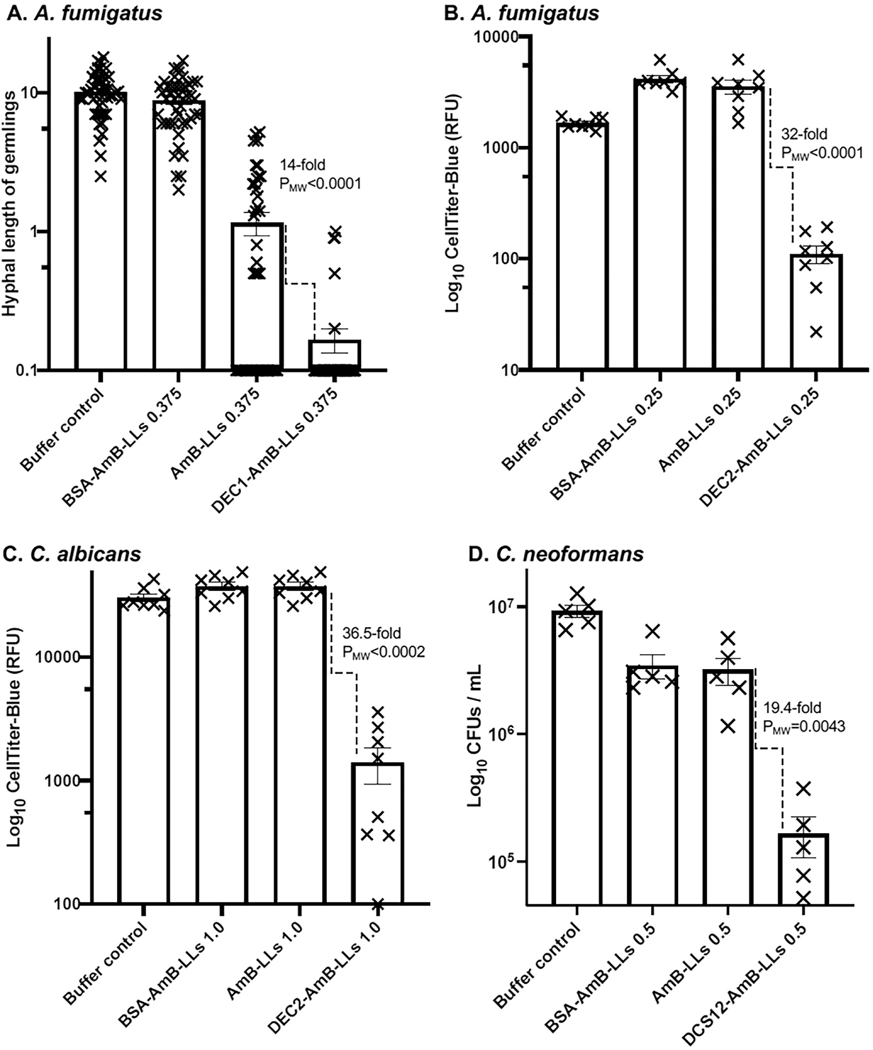
We have made robust measurements of in vitro inhibition and killing of all four fungal test species using multiple techniques. We have measured hyphal extension in germlings, electrochemical redox activity of live cells with Cell Titer Blue (CTB) reagent, colony forming units (CFUs), cell density with spectrophotometry, and live/dead viability staining with propidium iodide. Using any of these experimental approaches, DectiSomes out-performed both controls, AmB-LLs and BSA-AmB-LLs (Table 3). Fig. 5 shows a few examples: DEC1-AmB-LLs inhibiting hyphal extension from germlings of A. fumigatus (Fig. 5A), DEC2-AmB-LLs inhibiting redox activities of A. fumigatus (Fig. 5B) and C. albicans (Fig. 5C) using CTB reagent, and DCS12-AmB-LLs reducing CFUs of C. neoformans (Fig. 5D). Our published experiments used a variety of experimental designs with different lengths of drug exposure and growth period, and DectiSomes outperforming controls in all these experiments. The optimal drug concentration to resolve differences of DectiSomes from control BSA-AmB-LLs and AmB-LLs varied from 0.05 to 3.0 uM AmB. In each experiment, we observed that DectiSomes outperformed untargeted control liposomes, AmB-LLs and BSA-AmB-LLs, over about a 5- to 10-fold range in these AmB concentrations. When delivering AmB at higher concentrations, the control liposomes begin to significantly inhibit or kill the fungal cells, eliminating any ability to resolve the superior performance of DectiSomes.
Fig. 5. Examples of in vitro killing or growth inhibition experiments on fungal pathogens by DectiSomes.
(A). A hyphal length extension assay demonstrating DEC1-AmB-LLs selectively and quantitatively inhibited the in vitro growth A. fumigatus relative to untargeted control liposomes, AmB-LLs and BSA-AmB-LLs. (B). A Cell Titer Blue electrochemical reduction assay demonstrating DEC2-AmB-LLs inhibited the growth of or killed in vitro grown A. fumigatus. (C). A Cell Titer Blue assay demonstrating DEC2-AmB-LLs inhibited or killed in vitro grown C. albicans. (D). DC-SIGN isoform coated DCS12-AmB-LLs killed or inhibited the growth of C. neoformans in liquid media as quantified by counting colony forming units (CFUs). These data are from replicates of experiments not shown in the original publications [107,215,219,220].
3.7.3. Fungal burden and survival in mouse models of fungal diseases.
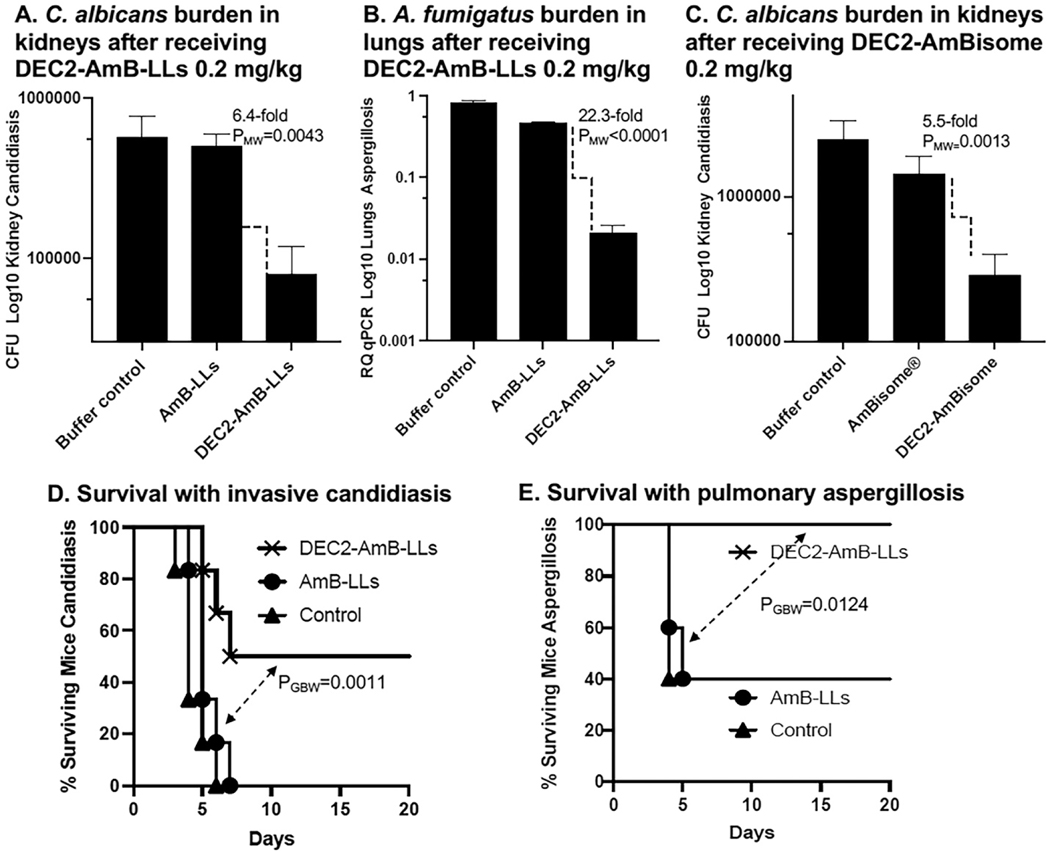
Our efforts to test DectiSomes in mouse models of invasive fungal diseases are in their early stages and far from complete. Nonetheless, the results so far suggest that the strong in vitro data (Fig. 4 & 5, Table 2) translate well to in vivo studies. Here are some examples of the results using mouse models of systemic candidiasis and pulmonary aspergillosis (Fig. 6). DEC2-AmB-LLs significantly reduced the fungal burden of C. albicans in the kidneys, the primary target organ of systemic candidiasis in this model (Fig. 6A) [218] and A. fumigatus in the lungs (Fig. 6B) [219]. It is worth noting that when we coated commercial un-pegylated AmBisome® with Dectin-2, DEC2-AmBisome preparations were significantly better at reducing the burden of C. albicans cells in the kidneys than AmBisome® (Fig. 6C) [218]. This result suggests that our method of CTL targeting may be effective for most bilipid membrane nanoparticles.
Fig. 6. Examples of fungal burden and survival studies employing DectiSomes.
(A). DEC2-AmB-LLs significantly reduced the fungal burden of C. albicans in the kidneys in an invasive candidiasis model relative to control AmB-LLs. (B). DEC2-AmB-LLs significantly reduced the fungal burden of A. fumigatus in the lungs in a pulmonary aspergillosis mouse model. (C). DEC2-AmBisome significantly reduced the fungal burden of C. albicans in the kidneys relative to AmBisome® in an invasive candidiasis mouse model. (D). DEC2-AmB-LLs significantly improved the survival of mice with invasive candidiasis relative to AmB-LLs. (E). DEC2-AmB-LLs significantly improved the survival of mice with pulmonary aspergillosis. (D & E). The % surviving mice is plotted vs days post-infection and mice were liposome treated on day 0 and day 1 respectively. One or two doses of DectiSomes and AmB-LLs delivering 0.2 mg AmB/kg mouse wight were used in each experiment. Buffer controls represent infected animals mock treated. A, C, D, E. Neutropenic immunosuppression mouse models. B. A steroid immunosuppression mouse model. A. B. C. PMW is the Mann-Whitney P test for non-parametric data. PGPW is the Gehan-Breslow-Wilcoxon P test of survival data. The data are from replicate experiments of those not shown in the original publications [215,218,219,274].
We performed a preliminary test of the advantage of employing pegylated liposomes in our system by comparing the performance of AmB-LLs to AmBisome® in a mouse model of invasive candidiasis. AmB-LLs reduced the fungal burden in the kidneys several fold more than AmBisome® delivering the same AmB concentration [218]. Although there are slight differences in other lipid membrane components between AmB-LLs and AmBisome® that might have contributed to the difference in efficacy, the result is consistent with the current concepts about the benefits of pegylation (more details at Section 3.2).
We have also used liposomes loaded with ten-moles percent anidulafungin (AFG-LLs), an echinocandin class antifungal [218]. Dectin-2 targeted DEC2-AFG-LLs were 10-fold more effective at reducing the kidney fungal burden in mouse candidiasis model than untargeted AFG-LLs. This result suggests that the performance of many different antifungals efficiently packaged in a liposome will be improved in a DectiSome.
3.7.4. Mouse survival rates.
DEC2-AmB-LL treatment resulted in 50% of mice surviving invasive candidiasis for 20 days relative to 100% mortality of the control mice treated with untargeted AmB-LLs (Fig. 6D) [218]. DEC2-AmB-LL treatment resulted in 100% of mice surviving pulmonary aspergillosis for 20 days relative to 40% of the control mice treated with AmB-LLs (Fig. 6E) [219]. In most of these experiments, only one to two doses with DectiSomes delivering AmB at 0.2 mg/kg mouse weight were used to produce such a significant effect. This is very low effective dose of AmB compared to what is commonly used to improve survival using detergent solubilized AmB-DOC or AmBisome® in mouse models (e.g., 5 to 20 mg/kg). When we treated human cells in culture with DectiSomes delivering high AmB concentrations of 15 to 30 uM, we found DectiSomes were of similar in toxicity to AmB-DOC, AmB-LLs, or BSA-AmB-LLs [107,220].
3.8. Summary of advantages and innovations associated with developing DectiSomes
DectiSomes offer many advantages as drug delivery system and several innovations have made this possible. 1. Because of the extreme breadth of cognate glycan structures recognized by the homodimers and heterodimers formed from a few CTLs, their potential as pan-antifungal targeting agents far exceeds that of any handful of monoclonal antibody reagents. For example, Dectin-1 binds diversely crosslinked beta-glucans with affinity constants (Kds) ranging from 2.6 mM to 2.2 pM, a billion-fold range [222]. By contrast, antibodies are generally high affinity reagents, specific for a restricted set of ligands or fungal developmental stages [258] and do not appear to provide the breadth of ligand specificities to be pan-antifungal targeting agents. Heterodimers, such as demonstrated by Dectin-2/Dectin-3 heterodimers, may expand the range of cognate ligands beyond that of their homodimers [201]. 2. The avidity created by having 1,500 CTLs on one liposome makes up for the weaker affinity (e.g., high Kds) of CTLs for some ligands. 3. Based on the surface area of a 100 nm diameter liposome (~31,400 nm2) and the Stokes radius of a 22 kDa CTL polypeptide (~2.6 nm), we estimate 1,500 to 2000 CTL molecules will fit on the surface of one liposome, whereas a 145 kDa antibody has a Stokes radius of 6.4 nm and thus only ~240 IgG molecules can fit. More targeting proteins should mean greater avidity for cognate ligands. 4. The CRD and stalk regions of CTLs are highly hydrophobic with a strong tendency to aggregate and precipitate, which is likely to have limited their use previously as drug targeting proteins. We developed simple rapid methods for producing and purifying these CTL polypeptides, working initially in random coil buffers and later in crowding buffers, which allowed long-term protein storage and transition of functional polypeptides into normal biological buffers and media. 5. We produce various CTL polypeptides at 20- to 50-fold lower cost than for equivalent mg or molar amounts of monoclonal antibodies. 6. Maintaining antibody integrity and function as they are conjugated to a lipid carrier for insertion into an immunoliposome is often a problem. We developed simple methods for modifying and attaching CTLs to liposomes, while maintaining full activity [107,216,220,256]. 7. Even after extensive efforts to “humanize” antibodies and antibody-related reagents, each has a unique new sequence, and hence, even after extensive humanization many are too immunogenic to be useful, whereas we expect the highly conserved human CTLs will be much less immunogenic. 8. The simplicity with which we manipulate the CRDs of these CTLs suggests they could be used to target not only liposomes of very different size and lipid composition, but almost any kind of nanoparticle drug delivery vehicle, each of which may offer their own therapeutic advantage [259].
3.9. Challenges to the clinical translation of DectiSomes
Our goal is to develop pan-antifungal DectiSomes worthy of commercial development to eventually benefit patients with invasive fungal diseases. While our in vitro and in vivo data are strong and encouraging, there are many scientific and technical questions that we wish to address before moving DectiSomes to clinical testing. (1) We need to test DectiSomes against more fungal species both in vitro and in vivo. Although we have in vitro data on four diverse fungal pathogens, DectiSomes have only been tested in animals against two pathogens (Table 3). (2) We have not yet examined the potential of combining multiple types of DectiSomes concurrently or heterodimeric DectiSomes against one pathogen. (3) We need to optimize DectiSomes for treatment of fungal diseases in various organs. For example, we have not yet examined mice with cryptococcal meningitis or pulmonary mucormycosis or fungal keratitis. AmBisome® is somewhat effective for treating patients with cryptococcal meningitis [260]. Liposomal AmB administering high concentrations of AmB (10 mg/kg) provides a transient reduction in the burden of C. neoformans in the mouse brain 24 hr after treatment [261]. But the blood brain barrier likely reduces the penetration of standard 100 nanometer liposomes into the brain. Because smaller nanoparticle particles are known pass the blood brain barrier much more readily [262,263], we plan to examine smaller DectiSomes. (4) We have examined DectiSomes loaded with AmB and AFG so far, but other antifungal drugs should also be tested (Table 1). Even given the relatively low cost of making DectiSomes, they will be more expensive to produce than detergent solubilized drug. They may not be cost effective for drugs delivered at high mg/kg/day doses such as the azoles. (5) In theory, increased local concentration of antifungals to the pathogen infection sites by DectiSomes should overcome the issues caused by dose-dependent drug resistant strains (or intermediate resistant strains). That said, we need to demonstrate experimentally that DectiSomes can indeed overcome intermediate dose-dependent resistance both in vitro and in vivo. (6) We have not yet fully tested DectiSome toxicity in animal cell models or potential immunogenicity of DectiSomes. This will be done when we have a better estimate of the range of effective doses and numbers of treatments in more mouse models of invasive fungal disease.
Besides the scientific and technical questions that we need to address, there are financial and physical constraints to bring DectiSomes to commercialization. On the positive side, the U.S. Federal Food and Drug administration has provided some incentives for investment in antifungal drug development [264,265]. One recent market study estimates the global markets for over the counter and prescription antifungal drugs are each in excess of 7 billion US dollars annually, and the market is growing steadily [124]. This same study estimates the investment opportunity in antifungals is in excess of 25 billion US dollars over 5 years, which should encourage investment into a dramatically improved antifungal therapeutic options such as DectiSomes [124]. In addition, academic laboratories continue to develop new antifungal agents [264,265,266,267,268,269,270].
On the other hand, the lack of commercial funding for development of novel anti-infectives is one major reason for the lack of new classes of FDA approved antifungal drugs [124]. There is strong aversion to investment on anti-infectives in general and in antifungals, in particular, by big pharma, because so few new antifungals survive the rigors of clinical testing and because of the high cost of clinical testing of relatively uncommon fungal infections. New antifungal candidates generally fail because of host toxicity in preclinical animal models or in clinical trials [271]. Lack of efficacy for clearing infections and/or human toxicity may not be evident until late stages of cinical development after hundreds of millions of dollars have already been spent [265,271,272]. Furthermore, while most FDA approved anticancer drugs are covered under Medicare and Medicaid [273], there is no such government policy to cover FDA approved antifungal drugs, dramatically reducing the market for antifungals. For all theses reasons, most of big pharmaceutical companies have divested themselves of antifugnal research and development programs [266]. Relatively few small companies are devoted to new antifungal drug development, in part, because of the cost to develop a single commerical product is so high [124,264,266]. However, with promising pre-clinical data, high selectivity and predicted host safety, and their broad spetrum (pan-antifungal therapy), DectiSomes may well become a poster child for commercial success of a new era of antifungal therapy.
4.0. Conclusions
DectiSomes are a pan-antifungal drug delivery system that has the potential to overcome many of the limitations of current antifungal therapies for the treatment of life-threatening invasive fungal infections. DectiSomes are functional analogs of immunoliposomes, but much simpler and much less expensive to construct. DectiSomes make use of the pan-fungal recognition properties of C-type lectins pathogen receptors to target antifungal drug loaded liposomes to diverse fungal pathogens. We have strong data showing DectiSomes efficiently bind and inhibit and/or kill four of the worst fungal pathogens in vitro and in vivo murine model data showing DectiSomes dramatically improved drug efficacy for reducing fungal burden in organs and increasing mouse survival over untargeted antifungal drugs. DectiSomes should provide dramatic improvements in drug efficacy in the clinic and repurpose most classes of existing or novel antifungal drugs.
Acknowledgments
We thank Valerie Maples (University of Georgia, Genetics) for suggesting the name DectiSome for our reagents.
Funding
The work is funded by the University of Georgia Research Foundation, Inc. (UGARF to SA and RBM), the National Institute of Allergy and Infectious Diseases (R21AI144498 and R21AI148890 to RBM and ZAL, R01AI162989 to RBM, SA, ZAL and XL), and the Georgia Research Alliance Ventures (to RBM). The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication and they are not responsible for the content of this article.
Footnotes
Declaration of Interest Statement
The authors have applied for patents on this technology.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Banerjee S, Denning DW & Chakrabarti A, One Health aspects & priority roadmap for fungal diseases : A mini-review, Indian J Med Res 153, 311–319 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bongomin F., Gago S., Oladele RO. & Denning DW., Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision, J Fungi (Basel) 3, 1–29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC, Fungal Diseases: Aspergillus, https://www.cdc.gov/fungal/diseases/aspergillosis/definition.html (2018).
- 4.Low CY & Rotstein C, Emerging fungal infections in immunocompromised patients, F1000 Med Rep 3, 14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varshney A, Scott LJ, Welch RP, Erdos MR, Chines PS, Narisu N, et al. , Genetic regulatory signatures underlying islet gene expression and type 2 diabetes, Proc Natl Acad Sci U S A 114, 2301–2306 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG & White TC, Hidden killers: human fungal infections, Sci Transl Med 4, 165rv113 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Pal R, Singh B, Bhadada SK, Banerjee M, Bhogal RS, Hage N, et al. , COVID-19-associated mucormycosis: An updated systematic review of literature, Mycoses 64, 1452–1459 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salehi M, Ahmadikia K, Badali H. & Khodavaisy S, Opportunistic Fungal Infections in the Epidemic Area of COVID-19: A Clinical and Diagnostic Perspective from Iran, Mycopathologia 185, 607–611 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amin A, Vartanian A, Poladian N, Voloshko A, Yegiazaryan A, Al-Kassir AL, et al. , Root Causes of Fungal Coinfections in COVID-19 Infected Patients, Infect Dis Rep 13, 1018–1035 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song G, Liang G. & Liu W, Fungal Co-infections Associated with Global COVID-19 Pandemic: A Clinical and Diagnostic Perspective from China, Mycopathologia 185, 599–606 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Kholy NA, El-Fattah AMA & Khafagy YW, Invasive Fungal Sinusitis in Post COVID-19 Patients: A New Clinical Entity, Laryngoscope 131, 2652–2658 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paparoupa M, Aldemyati R, Roggenkamp H, Berinson B, Norz D, Olearo F, et al. , The prevalence of early- and late-onset bacterial, viral, and fungal respiratory superinfections in invasively ventilated COVID-19 patients, J Med Virol 94, 1920–1925 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gangneux JP, Dannaoui E, Fekkar A, Luyt CE, Botterel F, De Prost N, et al. , Fungal infections in mechanically ventilated patients with COVID-19 during the first wave: the French multicentre MYCOVID study, Lancet Respir Med 10, 180–190 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benedict K, Jackson BR, Chiller T. & Beer KD, Estimation of Direct Healthcare Costs of Fungal Diseases in the United States, Clin Infect Dis 68, 1791–1797 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaoutis TE., Heydon K., Chu JH., Walsh TJ. & Steinbach WJ., Epidemiology, outcomes, and costs of invasive aspergillosis in immunocompromised children in the United States, 2000, Pediatrics 117, e711–716 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Horn DL, Ostrosky-Zeichner L, Morris MI, Ullmann AJ, Wu C, Buell DN, et al. , Factors related to survival and treatment success in invasive candidiasis or candidemia: a pooled analysis of two large, prospective, micafungin trials, Eur J Clin Microbiol Infect Dis 29, 223–229 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Reboli AC, Rotstein C, Pappas PG, Chapman SW, Kett DH, Kumar D, et al. , Anidulafungin versus fluconazole for invasive candidiasis, N Engl J Med 356, 2472–2482 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Dragonetti G, Criscuolo M, Fianchi L. & Pagano L, Invasive aspergillosis in acute myeloid leukemia: Are we making progress in reducing mortality?, Med Mycol 55, 82–86 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Timsit JF, Azoulay E, Schwebel C, Charles PE, Cornet M, Souweine B, et al. , Empirical Micafungin Treatment and Survival Without Invasive Fungal Infection in Adults With ICU-Acquired Sepsis, Candida Colonization, and Multiple Organ Failure: The EMPIRICUS Randomized Clinical Trial, JAMA 316, 1555–1564 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Yang Q, Huang J. & Li L, Risk factors for invasive pulmonary aspergillosis and hospital mortality in acute-on-chronic liver failure patients: a retrospective-cohort study, Int J Med Sci 10, 1625–1631 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baddley JW, Andes DR, Marr KA, Kauffman CA, Kontoyiannis DP, Ito JI, et al. , Antifungal therapy and length of hospitalization in transplant patients with invasive aspergillosis, Med Mycol 51, 128–135 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Lestrade PP, Bentvelsen RG, Schauwvlieghe A, Schalekamp S, van der Velden W, Kuiper EJ, et al. , Voriconazole Resistance and Mortality in Invasive Aspergillosis: A Multicenter Retrospective Cohort Study, Clin Infect Dis 68, 1463–1471 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Chottanapund S, Singhasivanon P, Kaewkungwal J, Chamroonswasdi K. & Manosuthi W, Survival time of HIV-infected patients with cryptococcal meningitis, J Med Assoc Thai 90, 2104–2111 (2007). [PubMed] [Google Scholar]
- 24.Crabtree Ramirez B, Caro Vega Y, Shepherd BE, Le C, Turner M, Frola C, et al. , Outcomes of HIV-positive patients with cryptococcal meningitis in the Americas, Int J Infect Dis 63, 57–63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sloan DJ & Parris V, Cryptococcal meningitis: epidemiology and therapeutic options, Clin Epidemiol 6, 169–182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roden MM, Zaoutis TE, Buchanan WL, Knudsen TA, Sarkisova TA, Schaufele RL, et al. , Epidemiology and outcome of zygomycosis: a review of 929 reported cases, Clin Infect Dis 41, 634–653 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Zilberberg MD., Shorr AF., Huang H., Chaudhari P., Paly VF. & Menzin J., Hospital days, hospitalization costs, and inpatient mortality among patients with mucormycosis: a retrospective analysis of US hospital discharge data, BMC Infect Dis 14, 310 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouza E, Muñoz P. & Guinea J, Mucormycosis: an emerging disease?, Clinical Microbiology and Infection 12, 7–23 (2006). [Google Scholar]
- 29.Stajich JE, Berbee ML, Blackwell M, Hibbett DS, James TY, Spatafora JW, et al. , The fungi, Curr Biol 19, R840–845 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang DY, Kumar S. & Hedges SB, Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi, Proceedings. Biological sciences 266, 163–171 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor JW & Berbee ML, Dating divergences in the Fungal Tree of Life: review and new analyses, Mycologia 98, 838–849 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Kuramae EE, Robert V, Snel B, Weiss M. & Boekhout T, Phylogenomics reveal a robust fungal tree of life, FEMS Yeast Res 6, 1213–1220 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Wang F, Zhang C, Jiang Y, Kou C, Kong Q, Long N, et al. , Innate and adaptive immune response to chronic pulmonary infection of hyphae of Aspergillus fumigatus in a new murine model, J Med Microbiol 66, 1400–1408 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Misme-Aucouturier B, Touahri A, Albassier M, Jotereau F, Le Pape P. & Alvarez-Rueda N, Double positive CD4+CD8+ T cells are part of the adaptive immune response against Candida albicans, Hum Immunol 80, 999–1005 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Romani L, Mencacci A, Cenci E, Puccetti P. & Bistoni F, Neutrophils and the adaptive immune response to Candida albicans, Res Immunol 147, 512–518 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Gigliotti F, Garvy BA, Haidaris CG & Harmsen AG, Recognition of Pneumocystis carinii antigens by local antibody-secreting cells following resolution of P. carinii pneumonia in mice, J Infect Dis 178, 235–242 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Lopez-Ribot JL, Diez-Orejas R. & Gil C, Antibodies, Immunology of Fungal Infections, Brown G. &Netea MG ed. Springer Dordrecht, The Netherlands, 2007. pp. pages. [Google Scholar]
- 38.Casadevall A. & Pirofski L-A, A new synthesis for antibody-mediated immunity, Nature immunology 13, 21–28 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Annunziato F, Romagnani C. & Romagnani S, The 3 major types of innate and adaptive cell-mediated effector immunity, J Allergy Clin Immunol 135, 626–635 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Blanco JL. & Garcia ME., Immune response to fungal infections, Vet Immunol Immunopathol 125, 47–70 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Drickamer K. & Taylor ME, Recent insights into structures and functions of C-type lectins in the immune system, Curr Opin Struct Biol 34, 26–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goyal S, Castrillon-Betancur JC, Klaile E. & Slevogt H, The Interaction of Human Pathogenic Fungi With C-Type Lectin Receptors, Front Immunol 9, 1261–1285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plato A, Hardison SE & Brown GD, Pattern recognition receptors in antifungal immunity, Seminars in immunopathology 37, 97–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Netea MG, Van der Graaf C, Van der Meer JW & Kullberg BJ, Recognition of fungal pathogens by Toll-like receptors, Eur J Clin Microbiol Infect Dis 23, 672–676 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Netea MG, Van Der Meer J. & Kullberg BJ, Recognition of Fungal Pathogens by Toll-like Receptors, Immunology of Fungal Infections, Brown G. &Netea MG ed. Springer, Dordrecht, The Netherlands, 2007. pp. pages. [Google Scholar]
- 46.Barreto-Bergter E. & Figueiredo RT, Fungal glycans and the innate immune recognition, Frontiers in cellular and infection microbiology 4, 145–145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drummond RA, Gaffen SL, Hise AG & Brown GD, Innate Defense against Fungal Pathogens, Cold Spring Harb Perspect Med 5, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Souza TN, Valdez AF, Rizzo J, Zamith-Miranda D, Guimaraes AJ, Nosanchuk JD, et al. , Host cell membrane microdomains and fungal infection, Cell Microbiol 23, e13385 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Botos I, Segal, David M. & Davies, David R, The Structural Biology of Toll-like Receptors, Structure 19, 447–459 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown J, O’Callaghan CA, Marshall AS, Gilbert RJ, Siebold C, Gordon S, et al. , Structure of the fungal beta-glucan-binding immune receptor dectin-1: implications for function, Protein Sci 16, 1042–1052 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelley J, de Bono B. & Trowsdale J, IRIS: A database surveying known human immune system genes, Genomics 85, 503–511 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Abbas AR, Baldwin D, Ma Y, Ouyang W, Gurney A, Martin F, et al. , Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data, Genes Immun 6, 319–331 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Gerwien F, Skrahina V, Kasper L, Hube B. & Brunke S, Metals in fungal virulence, FEMS Microbiol Rev 42, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bairwa G., Hee Jung W. & Kronstad JW., Iron acquisition in fungal pathogens of humans, Metallomics 9, 215–227 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lax C, Perez-Arques C, Navarro-Mendoza MI, Canovas-Marquez JT, Tahiri G, Perez-Ruiz JA, et al. , Genes, Pathways, and Mechanisms Involved in the Virulence of Mucorales, Genes (Basel) 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hameed S, Hans S, Singh S. & Fatima Z, Harnessing Metal Homeostasis Offers Novel and Promising Targets Against Candida albicans, Curr Drug Discov Technol 17, 415–429 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Ibrahim AS, Gebermariam T, Fu Y, Lin L, Husseiny MI, French SW, et al. , The iron chelator deferasirox protects mice from mucormycosis through iron starvation, J Clin Invest 117, 2649–2657 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, et al. , Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi, Nature 414, 450–453 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Puértolas-Balint F, Rossen JWA, Oliveira dos Santos C, Chlebowicz MMA, Raangs EC, van Putten ML, et al. , Revealing the Virulence Potential of Clinical and Environmental Aspergillus fumigatus Isolates Using Whole-Genome Sequencing, Frontiers in Microbiology 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu OW, Chun CD, Chow ED, Chen C, Madhani HD & Noble SM, Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans, Cell 135, 174–188 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu T, Yao B. & Zhang C, DFVF: database of fungal virulence factors, Database : the journal of biological databases and curation 2012, bas032-bas032 (2012). [DOI] [PMC free article] [PubMed]
- 62.Siscar-Lewin S, Hube B. & Brunke S, Antivirulence and avirulence genes in human pathogenic fungi, Virulence 10, 935–947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Howard DH, Acquisition, transport, and storage of iron by pathogenic fungi, Clinical microbiology reviews 12, 394–404 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kornitzer D, Fungal mechanisms for host iron acquisition, Current Opinion in Microbiology 12, 377–383 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Kornitzer D. & Roy U, Pathways of heme utilization in fungi, Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1867, 118817 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Philpott CC, Iron uptake in fungi: A system for every source, Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1763, 636–645 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Weete JD., Abril M. & Blackwell M., Phylogenetic distribution of fungal sterols, PloS one 5, e10899-e10899 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heitman J, Microbial Pathogens in the Fungal Kingdom, Fungal biology reviews 25, 48–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Warris A. & Verweij PE, Clinical implications of environmental sources for Aspergillus, Medical Mycology 43, S59–S65 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Kang SE, Sumabat LG, Melie T, Mangum B, Momany M. & Brewer MT, Evidence for the agricultural origin of resistance to multiple antimicrobials in Aspergillus fumigatus, a fungal pathogen of humans, G3 (Bethesda) 12, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Burks C, Darby A, Gomez Londono L, Momany M. & Brewer MT, Azole-resistant Aspergillus fumigatus in the environment: Identifying key reservoirs and hotspots of antifungal resistance, PLoS Pathog 17, e1009711 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paulussen C, Hallsworth JE, Álvarez-Pérez S, Nierman WC, Hamill PG, Blain D, et al. , Ecology of aspergillosis: insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species, Microbial Biotechnology 10, 296–322 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haiduven D, Nosocomial aspergillosis and building construction, Medical Mycology 47, S210–S216 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Snelders E, Huis In ‘t Veld RA, Rijs AJ, Kema GH, Melchers WJ & Verweij PE, Possible environmental origin of resistance of Aspergillus fumigatus to medical triazoles, Appl Environ Microbiol 75, 4053–4057 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Y, Lu Z, Zhao J, Zou Z, Gong Y, Qu F, et al. , Epidemiology and Molecular Characterizations of Azole Resistance in Clinical and Environmental Aspergillus fumigatus Isolates from China, Antimicrob Agents Chemother 60, 5878–5884 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rhodes J, Abdolrasouli A, Dunne K, Sewell TR, Zhang Y, Ballard E, et al. , Population genomics confirms acquisition of drug-resistant Aspergillus fumigatus infection by humans from the environment, Nature Microbiology 7, 663–674 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jeong W, Keighley C, Wolfe R, Lee WL, Slavin MA, Kong DCM, et al. , The epidemiology and clinical manifestations of mucormycosis: a systematic review and meta-analysis of case reports, Clin Microbiol Infect 25, 26–34 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Prakash H. & Chakrabarti A, Global Epidemiology of Mucormycosis, J Fungi (Basel) 5, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Banerjee I, Robinson J, Asim M, Sathian B. & Banerjee I, Mucormycosis and COVID-19 an epidemic in a pandemic?, Nepal J Epidemiol 11, 1034–1039 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Salmanton-Garcia J., Koehler P., Kindo A., Falces-Romero I., Garcia-Rodriguez J., Racil Z., et al., Needles in a haystack: Extremely rare invasive fungal infections reported in FungiScope-Global Registry for Emerging Fungal Infections, J Infect 81, 802–815 (2020). [DOI] [PubMed] [Google Scholar]
- 81.Richardson MD & Rautemaa-Richardson R, Biotic Environments Supporting the Persistence of Clinically Relevant Mucormycetes, J Fungi (Basel) 6, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guemas E, Cassaing S, Malavaud S, Fillaux J, Chauvin P, Lelievre L, et al. , A Clustered Case Series of Mucorales Detection in Respiratory Samples from COVID-19 Patients in Intensive Care, France, August to September 2021, J Fungi (Basel) 8, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Prakash H, Skiada A, Paul RA, Chakrabarti A. & Rudramurthy SM, Connecting the Dots: Interplay of Pathogenic Mechanisms between COVID-19 Disease and Mucormycosis, J Fungi (Basel) 7, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Skaria J, John TM, Varkey S. & Kontoyiannis DP, Are Unique Regional Factors the Missing Link in India’s COVID-19-Associated Mucormycosis Crisis?, mBio 13, e0047322 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sautour M, Lemaître J-P, Ranjard L, Truntzer C, Basmaciyan L, Depret G, et al. , Detection and survival of Candida albicans in soils, Environmental DNA 3, 1093–1101 (2021). [Google Scholar]
- 86.Barnett JA, A history of research on yeasts 12: medical yeasts part 1, Candida albicans, Yeast 25, 385–417 (2008). [DOI] [PubMed] [Google Scholar]
- 87.Van Uden N, de Matos Faia M. & Assis-Lopes L, Isolation of Candida albicans from animal sources, J General Microbiology 15, 151–153 (1956). [DOI] [PubMed] [Google Scholar]
- 88.Bensasson D, mSphere of Influence: the Wild Genetic Diversity of Our Closest Yeast Companions, mSphere 4, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bensasson D, Dicks J, Ludwig JM, Bond CJ, Elliston A, Roberts IN, et al. , Diverse Lineages of Candida albicans Live on Old Oaks, Genetics 211, 277–288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Casadevall A, The Pathogenic Potential of a Microbe, mSphere 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.CDC, Candida auris, 2021.
- 92.Sabino R, Veríssimo C, Pereira ÁA & Antunes F, Candida auris, an agent of hospital-associated outbreaks: which challenging issues do we need to have in mind?, Microorganisms 8, 181 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wasi M., Khandelwal NK., Moorhouse AJ., Nair R., Vishwakarma P., Bravo Ruiz G., et al., ABC Transporter Genes Show Upregulated Expression in Drug-Resistant Clinical Isolates of Candida auris: A Genome-Wide Characterization of ATP-Binding Cassette (ABC) Transporter Genes, Frontiers in Microbiology 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rhodes J. & Fisher MC, Global epidemiology of emerging Candida auris, Curr Opin Microbiol 52, 84–89 (2019). [DOI] [PubMed] [Google Scholar]
- 95.Arora P, Singh P, Wang Y, Yadav A, Pawar K, Singh A, et al. , Environmental Isolation of Candida auris from the Coastal Wetlands of Andaman Islands, India, mBio 12, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Casadevall A, Kontoyiannis DP & Robert V, Environmental Candida auris and the Global Warming Emergence Hypothesis, mBio 12, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Briard B, Fontaine T, Kanneganti TD, Gow NAR & Papon N, Fungal cell wall components modulate our immune system, Cell Surf 7, 100067 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lagree K. & Mitchell AP, Fungal Biofilms: Inside Out, Microbiology spectrum 5, 10.1128/microbiolspec.FUNK-0024-2016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kernien JF, Snarr BD, Sheppard DC & Nett JE, The Interface between Fungal Biofilms and Innate Immunity, Front Immunol 8, 1968 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gow NAR, Latge JP & Munro CA, The Fungal Cell Wall: Structure, Biosynthesis, and Function, Microbiol Spectr 5, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xie X. & Lipke PN, On the evolution of fungal and yeast cell walls, Yeast 27, 479–488 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blatzer M, Beauvais A, Henrissat B. & Latge JP, Revisiting Old Questions and New Approaches to Investigate the Fungal Cell Wall Construction, Curr Top Microbiol Immunol 425, 331–369 (2020). [DOI] [PubMed] [Google Scholar]
- 103.Muszewska A, Pilsyk S, Perlinska-Lenart U. & Kruszewska JS, Diversity of Cell Wall Related Proteins in Human Pathogenic Fungi, J Fungi (Basel) 4, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bain JM, Louw J, Lewis LE, Okai B, Walls CA, Ballou ER, et al. , Candida albicans hypha formation and mannan masking of beta-glucan inhibit macrophage phagosome maturation, MBio 5, e01874 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Davis SE, Hopke A, Minkin SC Jr., Montedonico AE, Wheeler RT & Reynolds TB, Masking of beta(1–3)-glucan in the cell wall of Candida albicans from detection by innate immune cells depends on phosphatidylserine, Infect Immun 82, 4405–4413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang M, Solis NV, Marshall M, Garleb R, Zhou T, Wang D, et al. , Control of beta-glucan exposure by the endo-1,3-glucanase Eng1 in Candida albicans modulates virulence, PLoS Pathog 18, e1010192 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ambati S, Ferarro AR, Kang SE, Lin J, Lin X, Momany M, et al. , Dectin-1-Targeted Antifungal Liposomes Exhibit Enhanced Efficacy, mSphere 4, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen SM., Zou Z., Qiu XR., Hou WT., Zhang Y., Fang W., et al., The critical role of Dectin-1 in host controlling systemic Candida krusei infection, American journal of translational research 11, 721–732 (2019). [PMC free article] [PubMed] [Google Scholar]
- 109.Sheppard DC & Howell PL, Biofilm Exopolysaccharides of Pathogenic Fungi: Lessons from Bacteria, J Biol Chem 291, 12529–12537 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zarnowski R, Sanchez H, Covelli AS, Dominguez E, Jaromin A, Bernhardt J, et al. , Candida albicans biofilm-induced vesicles confer drug resistance through matrix biogenesis, PLoS Biol 16, e2006872 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mitchell KF, Zarnowski R. & Andes DR, The Extracellular Matrix of Fungal Biofilms, Adv Exp Med Biol 931, 21–35 (2016). [DOI] [PubMed] [Google Scholar]
- 112.de Chaves MA, Ferreira do Amaral T, Monteiro da Silva Rodrigues Coutinho N, Fernanda Andrzejewski Kaminski T, Teixeira ML, Flavio Souza de Oliveira L, et al. , Synergistic association of clioquinol with antifungal drugs against biofilm forms of clinical Fusarium isolates, Mycoses 63, 1069–1082 (2020). [DOI] [PubMed] [Google Scholar]
- 113.Sav H, Rafati H, Oz Y, Dalyan-Cilo B, Ener B, Mohammadi F, et al. , Biofilm Formation and Resistance to Fungicides in Clinically Relevant Members of the Fungal Genus Fusarium, J Fungi (Basel) 4, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mukherjee PK, Chandra J, Yu C, Sun Y, Pearlman E. & Ghannoum MA, Characterization of Fusarium keratitis outbreak isolates: contribution of biofilms to antimicrobial resistance and pathogenesis, Invest Ophthalmol Vis Sci 53, 4450–4457 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wear MP, Jacobs E, Wang S, McConnell SA, Bowen A, Strother C, et al. , Cryptococcus neoformans capsule regrowth experiments reveal dynamics of enlargement and architecture, J Biol Chem 298, 101769 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Maxson ME, Dadachova E, Casadevall A. & Zaragoza O, Radial mass density, charge, and epitope distribution in the Cryptococcus neoformans capsule, Eukaryot Cell 6, 95–109 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hamidi M, Okoro OV, Milan PB, Khalili MR, Samadian H, Nie L, et al. , Fungal exopolysaccharides: Properties, sources, modifications, and biomedical applications, Carbohydr Polym 284, 119152 (2022). [DOI] [PubMed] [Google Scholar]
- 118.Martins M, Uppuluri P, Thomas DP, Cleary IA, Henriques M, Lopez-Ribot JL, et al. , Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms, Mycopathologia 169, 323–331 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shopova I., Bruns S., Thywissen A., Kniemeyer O., Brakhage AA. & Hillmann F., Extrinsic extracellular DNA leads to biofilm formation and colocalizes with matrix polysaccharides in the human pathogenic fungus Aspergillus fumigatus, Front Microbiol 4, 141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zarnowski R, Westler WM, Lacmbouh GA, Marita JM, Bothe JR, Bernhardt J, et al. , Novel entries in a fungal biofilm matrix encyclopedia, MBio 5, e01333–01314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yu W, Chen G, Zhang P. & Chen K, Purification, partial characterization and antitumor effect of an exopolysaccharide from Rhizopus nigricans, Int J Biol Macromol 82, 299–307 (2016). [DOI] [PubMed] [Google Scholar]
- 122.Bouz G. & Doležal M, Advances in Antifungal Drug Development: An Up-To-Date Mini Review, Pharmaceuticals (Basel) 14, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Roemer T. & Krysan DJ, Antifungal drug development: challenges, unmet clinical needs, and new approaches, Cold Spring Harb Perspect Med 4, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Taylor P, Antifungal Drugs: Technologies and Global Markets, (Publishing, B., 2020). [Google Scholar]
- 125.Zheng YH, Ma YY, Ding Y, Chen XQ & Gao GX, An insight into new strategies to combat antifungal drug resistance, Drug Des Devel Ther 12, 3807–3816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nicola AM, Albuquerque P, Paes HC, Fernandes L, Costa FF, Kioshima ES, et al. , Antifungal drugs: New insights in research & development, Pharmacol Ther 195, 21–38 (2019). [DOI] [PubMed] [Google Scholar]
- 127.Donovick R, Gold W, Pagano JF & Stout HA, Amphotericins A and B, antifungal antibiotics produced by a streptomycete. I. In vitro studies, Antibiot Annu 3, 579–586 (1955). [PubMed] [Google Scholar]
- 128.Jambor WP, Steinberg BA & Suydam LO, Amphotericins A and B: two new antifungal antibiotics possessing high activity against deep-seated and superficial mycoses, Antibiot Annu 3, 574–578 (1955). [PubMed] [Google Scholar]
- 129.Louria DB, Feder N. & Emmons CW, Amphotericin B in experimental histoplasmosis and Cryptococcosis, Antibiot Annu 870–877 (1956). [PubMed]
- 130.Osswald H. & Seeliger HP, Animal experiments with antimycotic drugs. 1. Findings on the effects of mycostatin & amphotericin B on test models of mice infected with Candida albicans or Mucor pusillus, Arzneimittelforschung 8, 370–374 (1958). [PubMed] [Google Scholar]
- 131.Utz JP, Treger A, Mc CN & Emmons CW, Amphotericin B: intravenous use in 21 patients with systemic fungal diseases, Antibiot Annu 6, 628–634 (1958). [PubMed] [Google Scholar]
- 132.Cavassin FB, Baú-Carneiro JL, Vilas-Boas RR & Queiroz-Telles F, Sixty years of Amphotericin B: An Overview of the Main Antifungal Agent Used to Treat Invasive Fungal Infections, Infectious Diseases and Therapy 10, 115–147 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.FDA U., Ambisome (Amphotericin B), U.S. Food and Drug Admistration 050740, (1997).
- 134.Pittillo RF & Ray BJ, Chemotherapeutic activity of 5-fluorocytosine against a lethal Candida albicans infection in mice, Appl Microbiol 17, 773–774 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shadomy S, Shadomy HJ, McCay JA & Utz JP, In vitro susceptibility of Cryptococcus neoformans to amphotericin B, hamycin, and 5-fluorocytosine, Antimicrob Agents Chemother (Bethesda) 8, 452–460 (1968). [PubMed] [Google Scholar]
- 136.Faergemann J, In vitro and in vivo activities of ketoconazole and itraconazole against Pityrosporum orbiculare, Antimicrob Agents Chemother 26, 773–774 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Van Cutsem J, Van Gerven F, Van de Ven MA, Borgers M. & Janssen PA, Itraconazole, a new triazole that is orally active in aspergillosis, Antimicrob Agents Chemother 26, 527–534 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Graybill JR & Ahrens J, R 51211 (itraconazole) therapy of murine cryptococcosis, Sabouraudia 22, 445–453 (1984). [DOI] [PubMed] [Google Scholar]
- 139.Humphrey MJ, Jevons S. & Tarbit MH, Pharmacokinetic evaluation of UK-49,858, a metabolically stable triazole antifungal drug, in animals and humans, Antimicrob Agents Chemother 28, 648–653 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Troke PF, Andrews RJ, Brammer KW, Marriott MS & Richardson K, Efficacy of UK-49,858 (fluconazole) against Candida albicans experimental infections in mice, Antimicrob Agents Chemother 28, 815–818 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Graybill JR, Sun SH & Ahrens J, Treatment of murine coccidioidal meningitis with fluconazole (UK 49,858), J Med Vet Mycol 24, 113–119 (1986). [DOI] [PubMed] [Google Scholar]
- 142.Singh M, Singh MP, Maiti SN, Gandhi A, Micetich RG & Atwal H, Preparations of liposomal fluconazole and their in vitro antifungal activity, J Microencapsul 10, 229–236 (1993). [DOI] [PubMed] [Google Scholar]
- 143.Arikan S, Ostrosky-Zeichner L, Lozano-Chiu M, Paetznick V, Gordon D, Wallace T, et al. , In vitro activity of nystatin compared with those of liposomal nystatin, amphotericin B, and fluconazole against clinical Candida isolates, J Clin Microbiol 40, 1406–1412 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schwarz JC, Kahlig H, Matsko NB, Kratzel M, Husa M. & Valenta C, Decrease of liposomal size and retarding effect on fluconazole skin permeation by lysine derivatives, J Pharm Sci 100, 2911–2919 (2011). [DOI] [PubMed] [Google Scholar]
- 145.van der Kaaden M, Breukink E. & Pieters RJ, Synthesis and antifungal properties of papulacandin derivatives, Beilstein J Org Chem 8, 732–737 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Traxler P., Fritz H. & Richter WJ., [On the structure of papulacandin B, a new antibiotic with antifungal activity (author’s transl) Helv Chim Acta 60, 578–584 (1977). [DOI] [PubMed] [Google Scholar]
- 147.FDA U, Eraxis (Anidulafungin) Injection, U.S. Food and Drug Admistration 021632 & 021948, (2006).
- 148.Davis SA, Vincent BM, Endo MM, Whitesell L, Marchillo K, Andes DR, et al. , Nontoxic antimicrobials that evade drug resistance, Nature Chemical Biology 11, 481–487 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Belicard P, [Ocular mycosis (hypopyon ulcer) cured with nystatin], Bull Soc Ophtalmol Fr 440–442 (1955). [PubMed]
- 150.Mangiaracine AB & Liebman SD, Fungus keratitis (Aspergillus fumigatus); treatment with nystain (mycostatin), AMA Arch Ophthalmol 58, 695–698 (1957). [DOI] [PubMed] [Google Scholar]
- 151.Lopez-Berestein G, Mehta R, Hopfer RL, Mills K, Kasi L, Mehta K, et al. , Treatment and prophylaxis of disseminated infection due to Candida albicans in mice with liposome-encapsulated amphotericin B, J Infect Dis 147, 939–945 (1983). [DOI] [PubMed] [Google Scholar]
- 152.Hughes WT, Feldman S. & Sanyal SK, Treatment of Pneumocystis carinii pneumonitis with trimethoprim-sulfamethoxazole, Can Med Assoc J 112, 47–50 (1975). [PMC free article] [PubMed] [Google Scholar]
- 153.Hughes WT, McNabb PC, Makres TD & Feldman S, Efficacy of trimethoprim and sulfamethoxazole in the prevention and treatment of Pneumocystis carinii pneumonitis, Antimicrob Agents Chemother 5, 289–293 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Barry AL & Brown SD, In vitro studies of two triazole antifungal agents (voriconazole [UK-109,496] and fluconazole) against Candida species, Antimicrob Agents Chemother 40, 1948–1949 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.George D, Miniter P. & Andriole VT, Efficacy of UK-109496, a new azole antifungal agent, in an experimental model of invasive aspergillosis, Antimicrob Agents Chemother 40, 86–91 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Perfect JR, Cox GM, Dodge RK & Schell WA, In vitro and in vivo efficacies of the azole SCH56592 against Cryptococcus neoformans, Antimicrob Agents Chemother 40, 1910–1913 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sugar AM & Liu XP, In vitro and in vivo activities of SCH 56592 against Blastomyces dermatitidis, Antimicrob Agents Chemother 40, 1314–1316 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Warn PA, Sharp A. & Denning DW, In vitro activity of a new triazole BAL4815, the active component of BAL8557 (the water-soluble prodrug), against Aspergillus spp, J Antimicrob Chemother 57, 135–138 (2006). [DOI] [PubMed] [Google Scholar]
- 159.Odds FC, Drug evaluation: BAL-8557--a novel broad-spectrum triazole antifungal, Curr Opin Investig Drugs 7, 766–772 (2006). [PubMed] [Google Scholar]
- 160.Ernst ME., Klepser ME., Wolfe EJ. & Pfaller MA., Antifungal dynamics of LY 303366, an investigational echinocandin B analog, against Candida ssp., Diagn Microbiol Infect Dis 26, 125–131 (1996). [DOI] [PubMed] [Google Scholar]
- 161.Bartlett MS, Current WL, Goheen MP, Boylan CJ, Lee CH, Shaw MM, et al. , Semisynthetic echinocandins affect cell wall deposition of Pneumocystis carinii in vitro and in vivo, Antimicrob Agents Chemother 40, 1811–1816 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Abruzzo GK, Flattery AM, Gill CJ, Kong L, Smith JG, Pikounis VB, et al. , Evaluation of the echinocandin antifungal MK-0991 (L-743,872): efficacies in mouse models of disseminated aspergillosis, candidiasis, and cryptococcosis, Antimicrob Agents Chemother 41, 2333–2338 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bartizal K, Gill CJ, Abruzzo GK, Flattery AM, Kong L, Scott PM, et al. , In vitro preclinical evaluation studies with the echinocandin antifungal MK-0991 (L-743,872), Antimicrob Agents Chemother 41, 2326–2332 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tomishima M, Ohki H, Yamada A, Takasugi H, Maki K, Tawara S, et al. , FK463, a novel water-soluble echinocandin lipopeptide: synthesis and antifungal activity, J Antibiot (Tokyo) 52, 674–676 (1999). [DOI] [PubMed] [Google Scholar]
- 165.Fisher MC, Hawkins NJ, Sanglard D. & Gurr SJ, Worldwide emergence of resistance to antifungal drugs challenges human health and food security, Science 360, 739–742 (2018). [DOI] [PubMed] [Google Scholar]
- 166.Wiederhold NP, Emerging Fungal Infections: New Species, New Names, and Antifungal Resistance, Clin Chem 68, 83–90 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yadav V. & Heitman J, On Fruits and Fungi: A Risk of Antifungal Usage in Food Storage and Distribution in Driving Drug Resistance in Candida auris, mBio 13, e0073922 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Berman J. & Krysan DJ, Drug resistance and tolerance in fungi, Nat Rev Microbiol 18, 319–331 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lofgren LA, Ross BS, Cramer RA & Stajich JE, Combined Pan-, Population-, and Phylo-Genomic Analysis of Aspergillus fumigatus Reveals Population Structure and Lineage-Specific Diversity, bioRxiv 2021.2012.2012.472145 (2022).
- 170.Warrilow AGS, Parker JE, Price CL, Rolley NJ, Nes WD, Kelly DE, et al. , Isavuconazole and voriconazole inhibition of sterol 14α-demethylases (CYP51) from Aspergillus fumigatus and Homo sapiens, Int J Antimicrob Agents 54, 449–455 (2019). [DOI] [PubMed] [Google Scholar]
- 171.Hagiwara D, Watanabe A, Kamei K. & Goldman GH, Epidemiological and Genomic Landscape of Azole Resistance Mechanisms in Aspergillus Fungi, Front Microbiol 7, 1382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Snelders E., Karawajczyk A., Verhoeven RJ., Venselaar H., Schaftenaar G., Verweij PE., et al., The structure-function relationship of the Aspergillus fumigatus cyp51A L98H conversion by site-directed mutagenesis: the mechanism of L98H azole resistance, Fungal Genet Biol 48, 1062–1070 (2011). [DOI] [PubMed] [Google Scholar]
- 173.Lescar J, Meyer I, Akshita K, Srinivasaraghavan K, Verma C, Palous M, et al. , Aspergillus fumigatus harbouring the sole Y121F mutation shows decreased susceptibility to voriconazole but maintained susceptibility to itraconazole and posaconazole, J Antimicrob Chemother 69, 3244–3247 (2014). [DOI] [PubMed] [Google Scholar]
- 174.Snelders E, Camps SM, Karawajczyk A, Rijs AJ, Zoll J, Verweij PE, et al. , Genotype-phenotype complexity of the TR46/Y121F/T289A cyp51A azole resistance mechanism in Aspergillus fumigatus, Fungal Genet Biol 82, 129–135 (2015). [DOI] [PubMed] [Google Scholar]
- 175.Chowdhary A, Sharma C, Kathuria S, Hagen F. & Meis JF, Azole-resistant Aspergillus fumigatus with the environmental TR46/Y121F/T289A mutation in India, J Antimicrob Chemother 69, 555–557 (2014). [DOI] [PubMed] [Google Scholar]
- 176.Khunweeraphong N. & Kuchler K, Multidrug Resistance in Mammals and Fungi-From MDR to PDR: A Rocky Road from Atomic Structures to Transport Mechanisms, Int J Mol Sci 22, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Paul S, Diekema D. & Moye-Rowley WS, Contributions of Aspergillus fumigatus ATP-binding cassette transporter proteins to drug resistance and virulence, Eukaryot Cell 12, 1619–1628 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Prasad R, Sharma M. & Rawal MK, Functionally Relevant Residues of Cdr1p: A Multidrug ABC Transporter of Human Pathogenic Candida albicans, Journal of Amino Acids 2011, 531412 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Winski CJ, Qian Y, Mobashery S. & Santiago-Tirado FH, An Atypical ABC Transporter Is Involved in Antifungal Resistance and Host Interactions in the Pathogenic Fungus Cryptococcus neoformans, mBio e0153922 (2022). [DOI] [PMC free article] [PubMed]
- 180.Kovalchuk A, Kohler A, Martin F. & Asiegbu FO, Diversity and evolution of ABC proteins in mycorrhiza-forming fungi, BMC Evol Biol 15, 249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Somchit N, Norshahida AR, Hasiah AH, Zuraini A, Sulaiman MR & Noordin MM, Hepatotoxicity induced by antifungal drugs itraconazole and fluconazole in rats: a comparative in vivo study, Hum Exp Toxicol 23, 519–525 (2004). [DOI] [PubMed] [Google Scholar]
- 182.Kyriakidis I, Tragiannidis A, Munchen S. & Groll AH, Clinical hepatotoxicity associated with antifungal agents, Expert Opin Drug Saf 16, 149–165 (2017). [DOI] [PubMed] [Google Scholar]
- 183.Yang YL, Xiang ZJ, Yang JH, Wang WJ, Xu ZC & Xiang RL, Adverse Effects Associated With Currently Commonly Used Antifungal Agents: A Network Meta-Analysis and Systematic Review, Front Pharmacol 12, 697330 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Allen U., Antifungal agents for the treatment of systemic fungal infections in children, Paediatrics & Child Health 15, 603–608 (2010). [PMC free article] [PubMed] [Google Scholar]
- 185.Dupont B, Overview of the lipid formulations of amphotericin B, J Antimicrob Chemother 49 Suppl 1, 31–36 (2002). [DOI] [PubMed] [Google Scholar]
- 186.Wong-Beringer A, Jacobs RA & Guglielmo BJ, Lipid formulations of amphotericin B: clinical efficacy and toxicities, Clin Infect Dis 27, 603–618 (1998). [DOI] [PubMed] [Google Scholar]
- 187.Tonin FS, Steimbach LM, Borba HH, Sanches AC, Wiens A, Pontarolo R, et al. , Efficacy and safety of amphotericin B formulations: a network meta-analysis and a multicriteria decision analysis, J Pharm Pharmacol 69, 1672–1683 (2017). [DOI] [PubMed] [Google Scholar]
- 188.Chastain DB, Giles RL, Bland CM, Franco-Paredes C, Henao-Martinez AF & Young HN, A clinical pharmacist survey of prophylactic strategies used to prevent adverse events of lipid-associated formulations of amphotericin B, Infect Dis (Lond) 51, 380–383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Faustino C. & Pinheiro L, Lipid Systems for the Delivery of Amphotericin B in Antifungal Therapy, Pharmaceutics 12, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Khoza S, Moyo I. & Ncube D, Comparative Hepatotoxicity of Fluconazole, Ketoconazole, Itraconazole, Terbinafine, and Griseofulvin in Rats, J Toxicol 2017, 6746989 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Billaud EM, Guillemain R, Berge M, Amrein C, Lefeuvre S, Louët AL-L, et al. , Pharmacological considerations for azole antifungal drug management in cystic fibrosis lung transplant patients, Medical Mycology 48, S52–S59 (2010). [DOI] [PubMed] [Google Scholar]
- 192.Godamudunage MP, Grech AM & Scott EE, Comparison of Antifungal Azole Interactions with Adult Cytochrome P450 3A4 versus Neonatal Cytochrome P450 3A7, Drug Metab Dispos 46, 1329–1337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Trösken ER, Fischer K, Völkel W. & Lutz WK, Inhibition of human CYP19 by azoles used as antifungal agents and aromatase inhibitors, using a new LC–MS/MS method for the analysis of estradiol product formation, Toxicology 219, 33–40 (2006). [DOI] [PubMed] [Google Scholar]
- 194.Grover ND, Echinocandins: A ray of hope in antifungal drug therapy, Indian J Pharmacol 42, 422 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Maligie MA & Selitrennikoff CP, Cryptococcus neoformans resistance to echinocandins: (1,3) beta-glucan synthase activity is sensitive to echinocandins, Antimicrobial agents and chemotherapy 49, 2851–2856 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Vermes A, Guchelaar HJ & Dankert J, Flucytosine: a review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions, J Antimicrob Chemother 46, 171–179 (2000). [DOI] [PubMed] [Google Scholar]
- 197.Meyer R. & Axelrod JL., Fatal aplastic anemia resulting from flucytosine, JAMA 228, 1573 (1974). [PubMed] [Google Scholar]
- 198.Preite NW, Feriotti C, Souza de Lima D, da Silva BB, Condino-Neto A, Pontillo A, et al. , The Syk-Coupled C-Type Lectin Receptors Dectin-2 and Dectin-3 Are Involved in Paracoccidioides brasiliensis Recognition by Human Plasmacytoid Dendritic Cells, Front Immunol 9, 464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wang T, Pan D, Zhou Z, You Y, Jiang C, Zhao X, et al. , Dectin-3 Deficiency Promotes Colitis Development due to Impaired Antifungal Innate Immune Responses in the Gut, PLoS Pathog 12, e1005662 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hole CR, Leopold Wager CM, Mendiola AS, Wozniak KL, Campuzano A, Lin X, et al. , Antifungal Activity of Plasmacytoid Dendritic Cells against Cryptococcus neoformans In Vitro Requires Expression of Dectin-3 (CLEC4D) and Reactive Oxygen Species, Infect Immun 84, 2493–2504 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhu LL, Zhao XQ, Jiang C, You Y, Chen XP, Jiang YY, et al. , C-type lectin receptors Dectin-3 and Dectin-2 form a heterodimeric pattern-recognition receptor for host defense against fungal infection, Immunity 39, 324–334 (2013). [DOI] [PubMed] [Google Scholar]
- 202.Norimoto A, Hirose K, Iwata A, Tamachi T, Yokota M, Takahashi K, et al. , Dectin-2 promotes house dust mite-induced T helper type 2 and type 17 cell differentiation and allergic airway inflammation in mice, Am J Respir Cell Mol Biol 51, 201–209 (2014). [DOI] [PubMed] [Google Scholar]
- 203.Sato K, Yang XL, Yudate T, Chung JS, Wu J, Luby-Phelps K, et al. , Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor gamma chain to induce innate immune responses, J Biol Chem 281, 38854–38866 (2006). [DOI] [PubMed] [Google Scholar]
- 204.Saijo S, Ikeda S, Yamabe K, Kakuta S, Ishigame H, Akitsu A, et al. , Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans, Immunity 32, 681–691 (2010). [DOI] [PubMed] [Google Scholar]
- 205.Huang HR, Li F, Han H, Xu X, Li N, Wang S, et al. , Dectin-3 Recognizes Glucuronoxylomannan of Cryptococcus neoformans Serotype AD and Cryptococcus gattii Serotype B to Initiate Host Defense Against Cryptococcosis, Front Immunol 9, 1781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chamilos G, Ganguly D, Lande R, Gregorio J, Meller S, Goldman WE, et al. , Generation of IL-23 producing dendritic cells (DCs) by airborne fungi regulates fungal pathogenicity via the induction of T(H)-17 responses, PLoS One 5, e12955 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Saijo S, Fujikado N, Furuta T, Chung SH, Kotaki H, Seki K, et al. , Dectin-1 is required for host defense against Pneumocystis carinii but not against Candida albicans, Nat Immunol 8, 39–46 (2007). [DOI] [PubMed] [Google Scholar]
- 208.Taylor PR., Tsoni SV., Willment JA., Dennehy KM., Rosas M., Findon H., et al., Dectin-1 is required for beta-glucan recognition and control of fungal infection, Nat Immunol 8, 31–38 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ifrim DC, Quintin J, Courjol F, Verschueren I, van Krieken JH, Koentgen F, et al. , The Role of Dectin-2 for Host Defense Against Disseminated Candidiasis, J Interferon Cytokine Res 36, 267–276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Brown BR, Lee EJ, Snow PE, Vance EE, Iwakura Y, Ohno N, et al. , Fungal-derived cues promote ocular autoimmunity through a Dectin-2/Card9-mediated mechanism, Clin Exp Immunol 190, 293–303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Campuzano A, Castro-Lopez N, Wozniak KL, Leopold Wager CM & Wormley FL Jr., Dectin-3 Is Not Required for Protection against Cryptococcus neoformans Infection, PLoS One 12, e0169347 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Cambi A, Gijzen K, de Vries l J, Torensma R, Joosten B, Adema GJ, et al. , The C-type lectin DC-SIGN (CD209) is an antigen-uptake receptor for Candida albicans on dendritic cells, Eur J Immunol 33, 532–538 (2003). [DOI] [PubMed] [Google Scholar]
- 213.Cambi A, Netea MG, Mora-Montes HM, Gow NAR, Hato SV, Lowman DW, et al. , Dendritic cell interaction with Candida albicans critically depends on N-linked mannan, J Biol Chem 283, 20590–20599 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Serrano-Gómez D, Domínguez-Soto A, Ancochea J, Jimenez-Heffernan JA, Leal JA & Corbí AL, Dendritic Cell-Specific Intercellular Adhesion Molecule 3-Grabbing Nonintegrin Mediates Binding and Internalization of Aspergillus fumigatus Conidia by Dendritic Cells and Macrophages, The Journal of Immunology 173, 5635 (2004). [DOI] [PubMed] [Google Scholar]
- 215.Ambati S, Pham T, Lewis ZA, Lin X. & Meagher RB, DC-SIGN Targets Amphotericin B-Loaded Liposomes to Diverse Pathogenic Fungi, Fungal Biol and Biotech 8, 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Choudhury Q, Ambati S, Lewis Z, Lin X. & Meagher R, Dectin-3-targeted antifungal liposomes efficiently bind and kill diverse fungal pathogens, (In review). [DOI] [PMC free article] [PubMed]
- 217.Choudhury QJ, Ambati S, Lewis ZA & Meagher RB, Targeted Delivery of Antifungal Liposomes to Rhizopus delemar, J Fungi (Basel) 8, 1–14 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ambati S, Pham T, Lewis ZA, Lin X. & Meagher RB, DectiSomes-Glycan Targeting of Liposomal Amphotericin B Improves the Treatment of Disseminated Candidiasis, Antimicrob Agents Chemother 66, 1–13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Ambati S, Ellis EC, Pham T, Lewis ZA, Lin X. & Meagher RB, Antifungal Liposomes Directed by Dectin-2 Offer a Promising Therapeutic Option for Pulmonary Aspergillosis, mBio 12, 1–8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ambati S., Ellis EC., Lin J., Lin X., Lewis ZA. & Meagher RB., Dectin-2-Targeted Antifungal Liposomes Exhibit Enhanced Efficacy, mSphere 4, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Santoso P, Komada T, Ishimine Y, Taniguchi H, Minamihata K, Goto M, et al. , Preparation of amphotericin B-loaded hybrid liposomes and the inte chitin-binding proteins for enhanced antifungal activity, J Biosci Bioeng Pre, (2022). [DOI] [PubMed]
- 222.Adams EL, Rice PJ, Graves B, Ensley HE, Yu H, Brown GD, et al. , Differential high-affinity interaction of dectin-1 with natural or synthetic glucans is dependent upon primary structure and is influenced by polymer chain length and side-chain branching, J Pharmacol Exp Ther 325, 115–123 (2008). [DOI] [PubMed] [Google Scholar]
- 223.McGreal EP, Rosas M, Brown GD, Zamze S, Wong SY, Gordon S, et al. , The carbohydrate-recognition domain of Dectin-2 is a C-type lectin with specificity for high mannose, Glycobiology 16, 422–430 (2006). [DOI] [PubMed] [Google Scholar]
- 224.Feinberg H, Jegouzo SAF, Rex MJ, Drickamer K, Weis WI & Taylor ME, Mechanism of pathogen recognition by human Dectin-2, J Biol Chem 292, 13402–13414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Jones GH & Ballou CE, Studies on the structure of yeast mannan. II. Mode of action of the Arthrobacter alpha-mannosidase on yeast mannan, J Biol Chem 244, 1052–1059 (1969). [PubMed] [Google Scholar]
- 226.Furukawa A, Kamishikiryo J, Mori D, Toyonaga K, Okabe Y, Toji A, et al. , Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL, Proc Natl Acad Sci U S A 110, 17438–17443 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kottom TJ, Hebrink DM, Monteiro JT, Lepenies B, Carmona EM, Wuethrich M, et al. , Myeloid C-type lectin receptors that recognize fungal mannans interact with Pneumocystis organisms and major surface glycoprotein, Journal of medical microbiology 68, 1649–1654 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Holla A. & Skerra A, Comparative analysis reveals selective recognition of glycans by the dendritic cell receptors DC-SIGN and Langerin, Protein Eng Des Sel 24, 659–669 (2011). [DOI] [PubMed] [Google Scholar]
- 229.Sawettanai N, Leelayuwapan H, Karoonuthaisiri N, Ruchirawat S. & Boonyarattanakalin S, Synthetic Lipomannan Glycan Microarray Reveals the Importance of alpha(1,2) Mannose Branching in DC-SIGN Binding, J Org Chem 84, 7606–7617 (2019). [DOI] [PubMed] [Google Scholar]
- 230.Reina JJ, Diaz I, Nieto PM, Campillo NE, Paez JA, Tabarani G, et al. , Docking, synthesis, and NMR studies of mannosyl trisaccharide ligands for DC-SIGN lectin, Org Biomol Chem 6, 2743–2754 (2008). [DOI] [PubMed] [Google Scholar]
- 231.Feinberg H., Castelli R., Drickamer K., Seeberger PH. & Weis WI., Multiple modes of binding enhance the affinity of DC-SIGN for high mannose N-linked glycans found on viral glycoproteins, J Biol Chem 282, 4202–4209 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Allen TM & Hansen C, Pharmacokinetics of stealth versus conventional liposomes: effect of dose, Biochim Biophys Acta 1068, 133–141 (1991). [DOI] [PubMed] [Google Scholar]
- 233.Torchilin VP, Omelyanenko VG, Papisov MI, Bogdanov AA Jr., Trubetskoy VS, Herron JN, et al. , Poly(ethylene glycol) on the liposome surface: on the mechanism of polymer-coated liposome longevity, Biochim Biophys Acta 1195, 11–20 (1994). [DOI] [PubMed] [Google Scholar]
- 234.Immordino ML, Dosio F. & Cattel L, Stealth liposomes: review of the basic science, rationale, and clinical applications, existing and potential, Int J Nanomedicine 1, 297–315 (2006). [PMC free article] [PubMed] [Google Scholar]
- 235.Tenchov R, Understanding the nanotechnology in COVID-19 vaccines, CAS 1–15 (2021).
- 236.Gupta R, Gupta J. & Pathak A, Immunoliposomes: A Targeted Drug Delivery System for Cancer Therapeutics and Vaccination, Curr Pharm Biotechnol (2022). [DOI] [PubMed]
- 237.Mamot C, Drummond DC, Noble CO, Kallab V, Guo Z, Hong K, et al. , Epidermal growth factor receptor-targeted immunoliposomes significantly enhance the efficacy of multiple anticancer drugs in vivo, Cancer Res 65, 11631–11638 (2005). [DOI] [PubMed] [Google Scholar]
- 238.Koren E, Apte A, Jani A. & Torchilin VP, Multifunctional PEGylated 2C5-immunoliposomes containing pH-sensitive bonds and TAT peptide for enhanced tumor cell internalization and cytotoxicity, J Control Release 160, 264–273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wang D, Sun Y, Liu Y, Meng F. & Lee RJ, Clinical translation of immunoliposomes for cancer therapy: recent perspectives, Expert Opin Drug Deliv 15, 893–903 (2018). [DOI] [PubMed] [Google Scholar]
- 240.Otsubo T, Maruyama K, Maesaki S, Miyazaki Y, Tanaka E, Takizawa T, et al. , Long-circulating immunoliposomal amphotericin B against invasive pulmonary aspergillosis in mice, Antimicrob Agents Chemother 42, 40–44 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Dulal HP, Nagae M, Ikeda A, Morita-Matsumoto K, Adachi Y, Ohno N, et al. , Enhancement of solubility and yield of a beta-glucan receptor Dectin-1 C-type lectin-like domain in Escherichia coli with a solubility-enhancement tag, Protein Expr Purif 123, 97–104 (2016). [DOI] [PubMed] [Google Scholar]
- 242.Steele C, Rapaka RR, Metz A, Pop SM, Williams DL, Gordon S, et al. , The beta-glucan receptor dectin-1 recognizes specific morphologies of Aspergillus fumigatus, PLoS Pathog 1, e42 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mori D., Shibata K. & Yamasaki S., C-Type Lectin Receptor Dectin-2 Binds to an Endogenous Protein beta-Glucuronidase on Dendritic Cells, PLoS One 12, e0169562 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Ariizumi K, Shen GL, Shikano S, Xu S, Ritter R 3rd, Kumamoto T, et al. , Identification of a novel, dendritic cell-associated molecule, dectin-1, by subtractive cDNA cloning, J Biol Chem 275, 20157–20167 (2000). [DOI] [PubMed] [Google Scholar]
- 245.Hollmig ST, Ariizumi K. & Cruz PD Jr., Recognition of non-self-polysaccharides by C-type lectin receptors dectin-1 and dectin-2, Glycobiology 19, 568–575 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Hohl TM, Van Epps HL, Rivera A, Morgan LA, Chen PL, Feldmesser M, et al. , Aspergillus fumigatus triggers inflammatory responses by stage-specific beta-glucan display, PLoS Pathog 1, e30 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Chen J, Liu Y, Wang Y, Ding H. & Su Z, Different effects of L-arginine on protein refolding: suppressing aggregates of hydrophobic interaction, not covalent binding, Biotechnol Prog 24, 1365–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 248.Ejima D, Kohei Tsumoto K. & Arakawa T, Biotech Applications of Arginine: Novel Uses in Protein Production, Processing, Analysis, and Formulation, BioProcess Technical Dec., 20–28 (2005).
- 249.Bose I, Reese AJ, Ory JJ, Janbon G. & Doering TL, A yeast under cover: the capsule of Cryptococcus neoformans, Eukaryot Cell 2, 655–663 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Ikeda R, Saito F, Matsuo M, Kurokawa K, Sekimizu K, Yamaguchi M, et al. , Contribution of the mannan backbone of cryptococcal glucuronoxylomannan and a glycolytic enzyme of Staphylococcus aureus to contact-mediated killing of Cryptococcus neoformans, J Bacteriol 189, 4815–4826 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Panepinto JC, Komperda KW, Hacham M, Shin S, Liu X. & Williamson PR, Binding of serum mannan binding lectin to a cell integrity-defective Cryptococcus neoformans ccr4Delta mutant, Infect Immun 75, 4769–4779 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Gow NA, van de Veerdonk FL, Brown AJ & Netea MG, Candida albicans morphogenesis and host defence: discriminating invasion from colonization, Nat Rev Microbiol 10, 112–122 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Pradhan A, Avelar GM, Bain JM, Childers DS, Larcombe DE, Netea MG, et al. , Hypoxia Promotes Immune Evasion by Triggering beta-Glucan Masking on the Candida albicans Cell Surface via Mitochondrial and cAMP-Protein Kinase A Signaling, MBio 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Pietrella D, Corbucci C, Perito S, Bistoni G. & Vecchiarelli A, Mannoproteins from Cryptococcus neoformans promote dendritic cell maturation and activation, Infect Immun 73, 820–827 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Van Wyk M., Govender NP., Mitchell TG., Litvintseva AP. & Germs SA., Multilocus sequence typing of serially collected isolates of Cryptococcus from HIV-infected patients in South Africa, J Clin Microbiol 52, 1921–1931 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Ambati S, Pham T, Lewis ZA, Lin X. & Meagher RB, DC-SIGN targets amphotericin B-loaded liposomes to diverse pathogenic fungi, Fungal Biol Biotechnol 8, 22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Ambati S, Duan J, Duff E, Choi YH, Hartzell DL, Della-Fera MA, et al. , Gene expression in arcuate nucleus-median eminence of rats treated with leptin or ciliary neurotrophic factor, Biofactors 31, 133–144 (2007). [DOI] [PubMed] [Google Scholar]
- 258.Momany M, Lindsey R, Hill TW, Richardson EA, Momany C, Pedreira M, et al. , The Aspergillus fumigatus cell wall is organized in domains that are remodelled during polarity establishment, Microbiology 150, 3261–3268 (2004). [DOI] [PubMed] [Google Scholar]
- 259.Mitchell MJ, Billingsley MM, Haley RM, Wechsler ME, Peppas NA & Langer R, Engineering precision nanoparticles for drug delivery, Nature Reviews Drug Discovery 20, 101–124 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Jarvis JN, Lawrence DS, Meya DB, Kagimu E, Kasibante J, Mpoza E, et al. , Single-Dose Liposomal Amphotericin B Treatment for Cryptococcal Meningitis, N Engl J Med 386, 1109–1120 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Xu N, Gu J, Zhu Y, Wen H, Ren Q. & Chen J, Efficacy of intravenous amphotericin B-polybutylcyanoacrylate nanoparticles against cryptococcal meningitis in mice, Int J Nanomedicine 6, 905–913 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Betzer O, Shilo M, Opochinsky R, Barnoy E, Motiei M, Okun E, et al. , The effect of nanoparticle size on the ability to cross the blood–brain barrier: an in vivo study, Nanomedicine 12, 1533–1546 (2017). [DOI] [PubMed] [Google Scholar]
- 263.Ohta S, Kikuchi E, Ishijima A, Azuma T, Sakuma I. & Ito T, Investigating the optimum size of nanoparticles for their delivery into the brain assisted by focused ultrasound-induced blood–brain barrier opening, Scientific Reports 10, 18220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Tillotson J. & Tillotson GS, The Regulatory Pathway for Antifungal Drugs: A US Perspective, Clinical Infectious Diseases 61, S678–S683 (2015). [DOI] [PubMed] [Google Scholar]
- 265.Perfect JR, The antifungal pipeline: a reality check, Nat Rev Drug Discov 16, 603–616 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Wall G. & Lopez-Ribot JL, Current Antimycotics, New Prospects, and Future Approaches to Antifungal Therapy, Antibiotics (Basel) 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Hoenigl M, Sprute R, Egger M, Arastehfar A, Cornely OA, Krause R, et al. , The Antifungal Pipeline: Fosmanogepix, Ibrexafungerp, Olorofim, Opelconazole, and Rezafungin, Drugs 81, 1703–1729 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.McCart TP. & Pappa PG., Antifungal Pipeline, Front Cell Infect Microbiol 11, 732223 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Li D, She X. & Calderone R, The antifungal pipeline: the need is established. Are there new compounds?, FEMS Yeast Res 20, (2020). [DOI] [PubMed] [Google Scholar]
- 270.Gintjee TJ, Donnelley MA & Thompson GR 3rd, Aspiring Antifungals: Review of Current Antifungal Pipeline Developments, J Fungi (Basel) 6, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Guengerich FP, Mechanisms of drug toxicity and relevance to pharmaceutical development, Drug metabolism and pharmacokinetics 26, 3–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kola I. & Landis J, Can the pharmaceutical industry reduce attrition rates?, Nat Rev Drug Discov 3, 711–715 (2004). [DOI] [PubMed] [Google Scholar]
- 273.CMS.gov, Oral Anticancer Drugs - Policy Article A52479, 2022.
- 274.Qu Y, Locock K, Verma-Gaur J, Hay ID, Meagher L. & Traven A, Searching for new strategies against polymicrobial biofilm infections: guanylated polymethacrylates kill mixed fungal/bacterial biofilms, J Antimicrob Chemother 71, 413–421 (2016). [DOI] [PubMed] [Google Scholar]