Abstract
Among urological cancers, renal cancer has the highest fatality rate. In a previous pan-cancer study of the METTL family, we observed a stronger association between the METTL family members and the risk of renal cancer compared to other cancers. Among these members, METTL7A, a potential methyltransferase, was identified as a protective factor, although its role and mechanism in renal cancer remain unclear. In this study, we utilized public databases to examine the expression of METTL7A in renal cancer tissues and normal tissues and found that METTL7A expression was much lower in renal cancer tissues. We also noticed a link between low METTL7A expression and poor prognosis for patients. According to the results of our functional enrichment analysis, METTL7A may have a role in immunological functions in renal cancer. METTL7A expression was strongly linked with the degrees of immune cell infiltration and expression of numerous immunological components. METTL7A had significantly different effects on the survival times of renal cancer patients with high or low immune infiltration. Our findings suggest that METTL7A may be used as both a prognostic biomarker and an immunological target for kidney cancer. In conclusion, our study sheds light on the importance of METTL7A in renal cancer and emphasizes the potential of targeting METTL7A as a novel therapeutic strategy for kidney cancer.
Keywords: METTL7A, Renal cancer, Prognostic biomarker, Immune infiltration, Methyltransferase
1. Introduction
Renal cancer has the highest fatality rate among all urological cancers [1], with over 40% of patients being unable to achieve tumor remission due to ineffective medical care [1,2]. Despite numerous immune-targeted investigations, clinical trial results have been inconclusive [3], highlighting the urgent need to identify novel immune-relevant genes to complement classical targets for kidney cancer treatment.
Many studies have demonstrated that RNA modification, specifically the N6-methyladenosine (m6A) [4], is crucial to the onset and progression of cancer [5]. The role of m6A in the clinical diagnosis and treatment prediction of kidney renal clear cell carcinoma (KIRC) and kidney renal papillary cell carcinoma (KIRP) has garnered considerable attention recently [6,7]. These studies suggest that targeting m6A methyltransferase could potentially contribute to clinical treatment and prognosis diagnosis for renal cell carcinoma. The METTL family, including METTL3 and METTL14, have been found to possess m6A RNA methyltransferase activity [[8], [9], [10], [11], [12]]. Our previous study revealed that the METTL family is closely linked to the risk of kidney cancer (Supplementary Figure S1), indicating a possible unique function in renal cancer. Interestingly, our research also found a strong negative link between METTL7A and the risk ratio of KIRC and KIRP, suggesting that it could be a protective factor in these cancers (Supplementary Figure S1). METTL7A also shows methyltransferase activity and is linked to the m6A methylation of lncRNAs [13]. According to several studies, METTL7A is down-regulated in tumors and is linked to patients' bad prognoses for hepatocellular carcinoma (HCC) and lung adenocarcinoma (LUAD), raising the possibility that METTL7A functions as a tumor suppressor gene or has other beneficial effects in human [14,15]. In methotrexate-treated cells, METTL7A knockout significantly reduced cell viability, destroyed cloning, and increased cell apoptosis, while its overexpression promotes cell viability and reduces cell apoptosis [16]. Despite these discoveries, it is yet unknown how METTL7A contributes to renal cancer.
The purpose of this research is to investigate the functions of METTL7A in renal cancer. Initially, the expression and prognostic significance of METTL7A were analyzed in various subtypes of kidney cancer using the Cancer Genome Atlas (TCGA) database. Subsequently, we looked into the molecular mechanisms and pathways associated with METTL7A's functions and assessed its impact on the immune microenvironment. Finally, we identified the potential METTL7A transcription factors in renal carcinoma.
2. Materials and method
2.1. UALCAN
UALCAN (http://ualcan.path.uab.edu/) is a database that allows users to mine and analyze data from TCGA, the Clinical Proteomic Tumor Analysis Consortium (CPTAC), and other public datasets [17]. The UALCAN database was used in this study to look into METTL7A expression as well as its relationship to various clinical and pathological characteristics of KIRC, including gender, cancer stage, age, race, cancer subtype, Body Mass Index (BMI), and lymph node metastasis status. Additionally, the protein expression and phosphorylation levels of chosen genes in tumorous and normal tissues were examined using UALCAN.
2.2. Kaplan-Meier Plotter
The Kaplan-Meier plotter database (http://kmplot.com/analysis/) is based on microarray and RNA-sequencing data sets from the Gene Expression Omnibus (GEO), European Genome-phenome Archive (EGA), and TCGA [18]. We assessed the impact of METTL7A mRNA expression on KIRC patients with high or low levels of immune cell infiltration using the Kaplan-Meier Plotter module “Pan-cancer RNA-seq”.
2.3. Linkedomics
Linkedomics database (http://www.linkedomics.org) provides a visual platform for evaluating multi-omics and clinical data from TCGA for 32 cancer types [19]. The co-expressed proteins of METTL7A were collected from the Linkedomics database, and the 50 genes that had the most positive and negative correlations with METTL7A were shown in a heat map. Simultaneously, using the “LinkInterpreter” module, the Gene Set Enrichment Analysis (GSEA) was performed on these METTL7A co-expressed proteins to discover the biological processes and pathways in which METTL7A was engaged.
2.4. Gene expression profiling interactive analysis (GEPIA)
GEPIA (http://gepia.cancer-pku.cn/index.html) is a website for gene expression analysis, which contains data on 9736 tumors and 8587 normal samples from the TCGA and Genotype-Tissue Expression (GTEx) databases [20]. In this work, the module “Expression DIY” of GEPIA was used to examine the gene expression correlation between METTL7A and other genes in KIRC and normal neighboring kidney tissue samples. The X-axis is based on METTL7A, and the Y-axis shows different genes. The Spearman method was used to get the correlation coefficient.
2.5. CIBERSORT
CIBERSORT (https://cibersort.stanford.edu/) is a well-known computational resource for assessing immune cell composition based on the validated leukocyte gene signature matrix containing 547 genes and 22 distinct categories of human immune cells. The CIBERSORT algorithm was utilized to quantify the fraction of infiltrating immune cells within malignancies in our study.
2.6. Statistical analysis
The hazard ratio (HR) of METTL7A expression in renal cancer was calculated using a univariate cox regression model, and independent prognostic factors were found using multivariate cox regression models. The log-rank test was used for the survival analysis, and a P value < 0.05 was regarded as statistically significant.
3. Results
3.1. The reduced expression of METTL7A in renal cancer
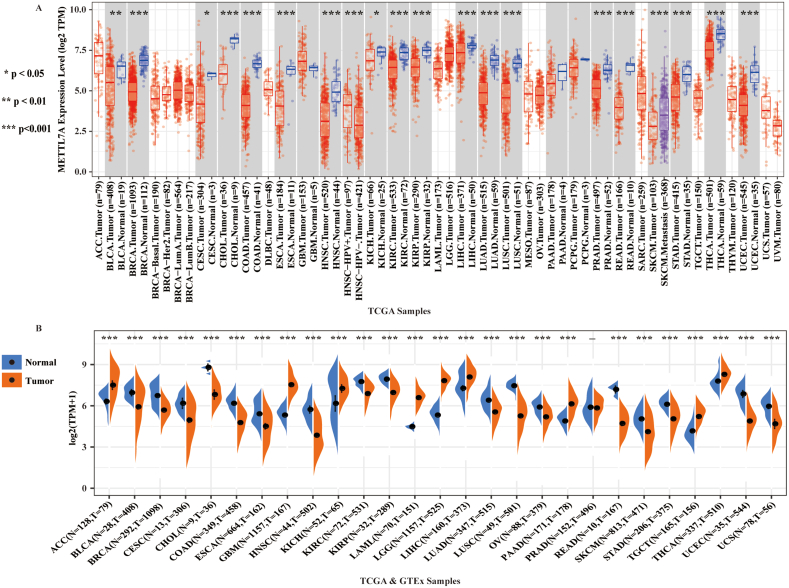
Using data from TCGA database, we examined the mRNA expression of METTL7A in 33 distinct kinds of human malignancies to look into its possible involvement in carcinogenesis. Compared with adjacent normal tissues, we discovered that the expression of METTL7A was significantly downregulated in a variety of tumor tissues (18 out of 33), including bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangio carcinoma (CHOL), colon carcinoma (COAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), gastric adenocarcinoma (STAD), thyroid carcinoma (THCA) and uterine corpus endometrial carcinoma (UCEC) (Fig. 1A). In the other 15 cancers, there was no significant change in the expression of METTL7A or lack of corresponding normal tissues for comparison. Using the GTEx and TCGA databases, we examined the mRNA expression of METTL7A in human cancer. Our findings demonstrated that, as compared to normal tissues from the GTEx database, the expression of METTL7A was considerably down-regulated in malignant tissues of 17 types of cancer from TCGA (Fig. 1B). We found that METTL7A expression was reduced in both KIRC and KIRP subtypes when we narrowed our focus to renal cancer (as consistent in both figures). Overall, our findings indicated that METTL7A is down-regulated in most malignancies, suggesting its potential involvement in carcinogenesis.
Fig. 1.
The mRNA expression of METTL7A in multiple cancers. (A) The mRNA expression of METTL7A in multiple cancers as analyzed using the data sets from TCGA. (B) The mRNA expression of METTL7A in multiple cancers as analyzed using the data sets from TCGA and GTEx. *: p-value <0.05; **: p-value <0.01; ***: p-value <0.001.
3.2. METTL7A expression in different subgroups of KIRC patients
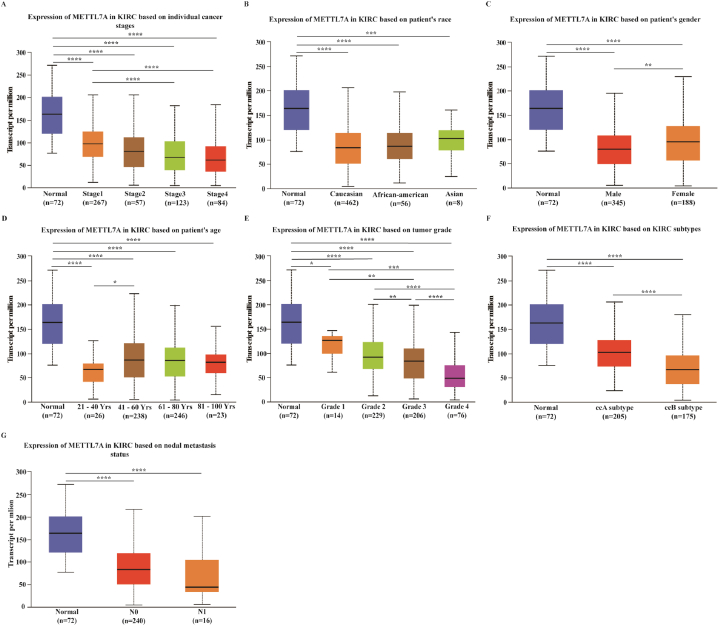
We selected KIRC as our model for the current investigation because previous results indicated a decrease in the expression of METTL7A in KIRC, which is the most prevalent form of kidney cancer. We examined the mRNA expression of METTL7A in different groups of KIRC patients with various clinical characteristics using the UALCAN database. According to our research, METTL7A expression drastically reduced with the progression of tumor stage, with stages 3 and 4 displaying lower expression than stage 1 (p < 0.05) (Fig. 2A). In terms of race, Caucasian, African-American and Asian KIRC patients had significantly lower levels of METTL7A expression in tumorous tissues compared to adjacent normal tissues (Fig. 2B). METTL7A expression was considerably down-regulated in male and female KIRC samples than in normal control samples, with lower expression observed in males than in females (Fig. 2C). In kidney cancer tissues from patients of various ages (21–40, 41–60, 61–80, and 81–100), METTL7A expression levels were substantially lower than in healthy neighboring tissues, and they were significantly lower in patients aged 21–40 than in those aged 41–60 (Fig. 2D). METTL7A expression was down-regulated in grades 1, 2, 3, and 4 of KIRC patients relative to normal controls. And as tumor grade increased, METTL7A expression became considerably down-regulated (Fig. 2E). As for molecular subtypes (ccA, ccB), METTL7A expression was downregulated in the tumor tissues of the two most prevalent KIRC molecular subtypes, with the ccB subtype (greater malignancy) displaying lower METTL7A expression than the ccA subtype, implying that METTL7A could be linked to KIRC malignancy (Fig. 2F). The expression of METTL7A in tumor tissues was found to be lower in patients with lymph node metastasis classified as N0 and N1 than in nearby normal tissues. Furthermore, METTL7A expression in N1 was lower than in N0 (Fig. 2G). Additionally, we investigated the protein expression of METTL7A in various patient groups (Supplementary Figure S2). The findings demonstrated that primary tumor tissue had considerably lower levels of METTL7A protein expression than normal tissues (p < 0.0001). As the stage progressed, there was a downward trend in METTL7A expression. Our findings demonstrate a correlation between METTL7A expression and tumor stage, grade, N stage and malignancy, implying that METTL7A may contribute to the growth of renal cancer.
Fig. 2.
METTL7A expression in different patient groups. Analysis was shown for (A) cancer stages, (B) race, (C) sex, (D) age, (E) grade, (F) subtype and (G) nodal metastasis. *: p-value < 0.05; **: p-value < 0.01; ***: p-value < 0.001.
3.3. METTL7A as one potential prognostic indicator for KIRC and KIRP
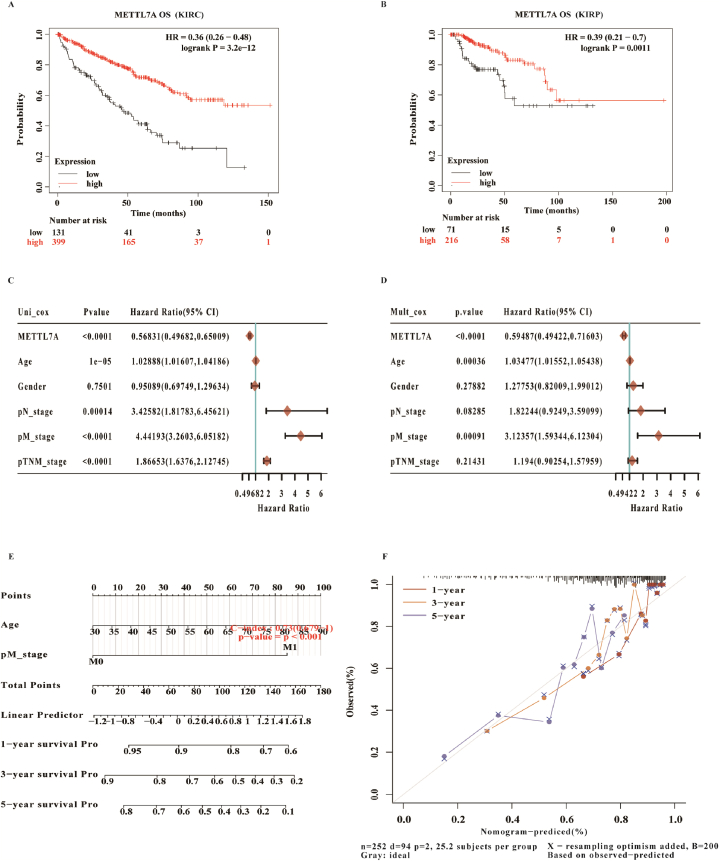
The relationship between METTL7A and renal cancer survival rates as well as its prognostic significance was then investigated. Based on Kaplan-Meier Plotter datasets, the overall survival (OS) of KIRC and KIRP patients with reduced METTL7A expression was poorer (Fig. 3A and B). Univariate and multivariate cox regression analysis of METTL7A expression in KIRC revealed that decreased METLL7A expression was a risk factor and a potential independent prognostic factor (Fig. 3C and D). The nomogram indicated that METTL7A could be combined with additional clinical markers to forecast KIRC development and prognosis (Fig. 3E and F). In addition, decreased METTL7A expression was also found to be a risk factor and possible independent prognostic factor for KIRP (Supplementary Figure S3A and B). METTL7A could also be used in conjunction with other clinical markers to forecast the course and prognosis of KIRP (Supplementary Figure S3C and D).
Fig. 3.
METTL7A as one potential independent prognostic factor in KIRC. (A, B) Survival curves showed that the lower expression of METTL7A was correlated with poorer OS in KIRC and KIRP. (C, D) Univariate and multivariate Cox regression analysis demonstrated that METTL7A was one putative independent prognostic factor for KIRC. (E, F) The nomogram demonstrates that METTL7A, when used in conjunction with other clinical markers, may forecast the 1-, 3-, and 5-year survival rates of KIRC patients.
3.4. Possible regulating impact of METTL7A on renal carcinoma's immunological microenvironment
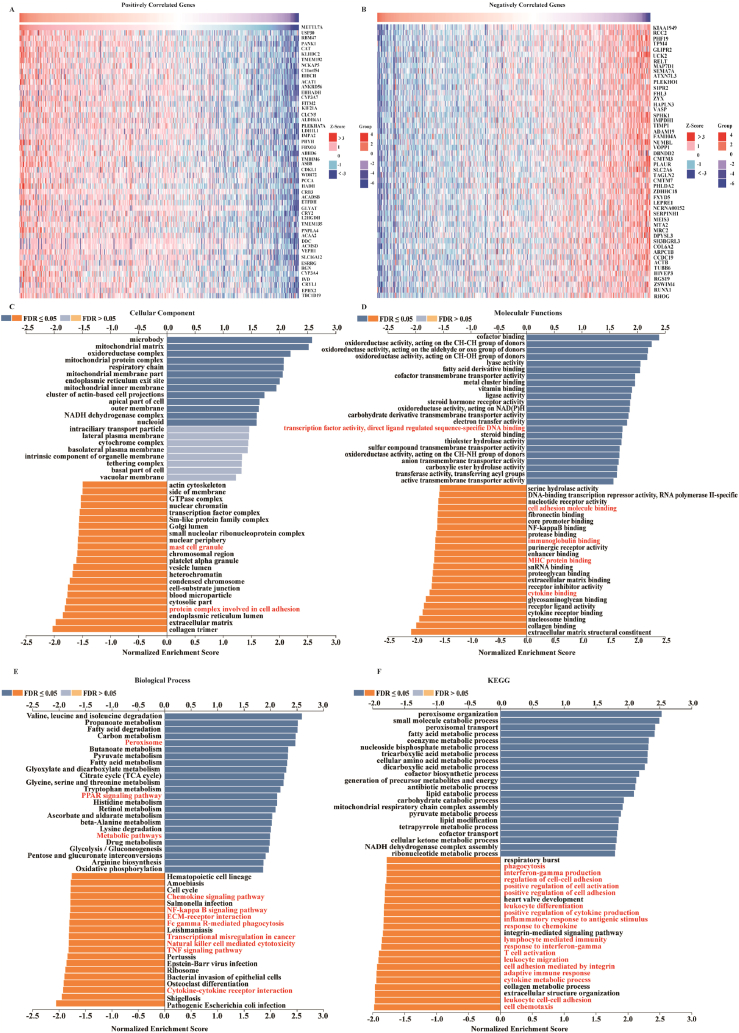
The function of METTL7A in kidney cancer has been largely unknown. To gain insight into its biological processes and pathways in KIRC, we screened the co-expressed mRNAs of METTL7A in KIRC using the Linkedomics database. Fig. 4A and B displayed the top 50 genes that have a positive or negative correlation with the expression of METTL7A in KIRC. Subsequently, we employed GSEA to investigate the co-expressed genes, and identified the 20 most significant terms for enrichment analysis of cellular component, molecular function, and biological process. And the top 20 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for METTL7A were shown (Fig. 4C, D, E, F). We found that the immune-relevant pathways like the NF-κB signaling pathway, chemokine signaling pathway, TNF signaling pathway and natural killer cell-mediated cytotoxicity were heavily enriched in these co-expressed genes, indicating that METTL7A may modulate the immune microenvironment of KIRC.
Fig. 4.
Enrichment analyses discovered a possible involvement of METTL7A in immune microenvironment regulation. (A) Heat maps showing the top 50 genes positively co-expressed with METTL7A in KIRC. (B) Heat maps showing the top 50 genes negatively co-expressed with METTL7A in KIRC. GSEA analysis of METTL7A in (C) CC categories, (D) MF categories, (E) BP categories, (F) and KEGG categories in KIRC.
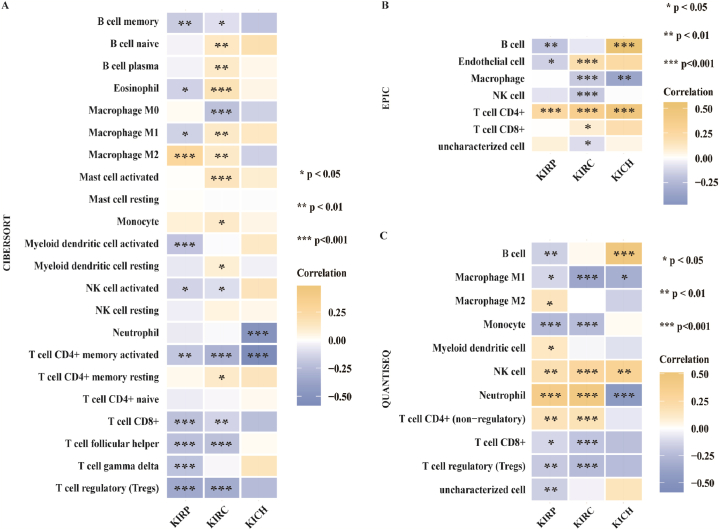
We further investigated the association between the expression of METTL7A and the degree of immune cell infiltration using CIBERSORT, EPIC and QUANTISEQ algorithms. The expression of METTL7A and regulatory T cells (Tregs) infiltration in KIRC and KIRP were shown to be significantly inversely correlated by CIBERSORT algorithm (Fig. 5A). In KIRC, KIRP, and KICH, METTL7A expression and CD4+T cell infiltration were found to be significantly positively correlated by EPIC analysis (Fig. 5B). Additionally, using the QUANTISEQ algorithm, the expression of METTL7A and the degree of NK cells and CD4+T cells infiltration in KIRC and KIRP were also found to be significantly positively correlated (Fig. 5C). These findings imply that the modulation function of METTL7A in the immune microenvironment of renal carcinoma may be mediated by immune cells.
Fig. 5.
The correlation between METTL7A expression and infiltration degrees of various immune cells in renal cancer. The correlation between METTL7A expression and immune cell infiltration as analyzed using (A) CIBERSORT, (B) EPIC and (C) QUANTISEQ algorithms.
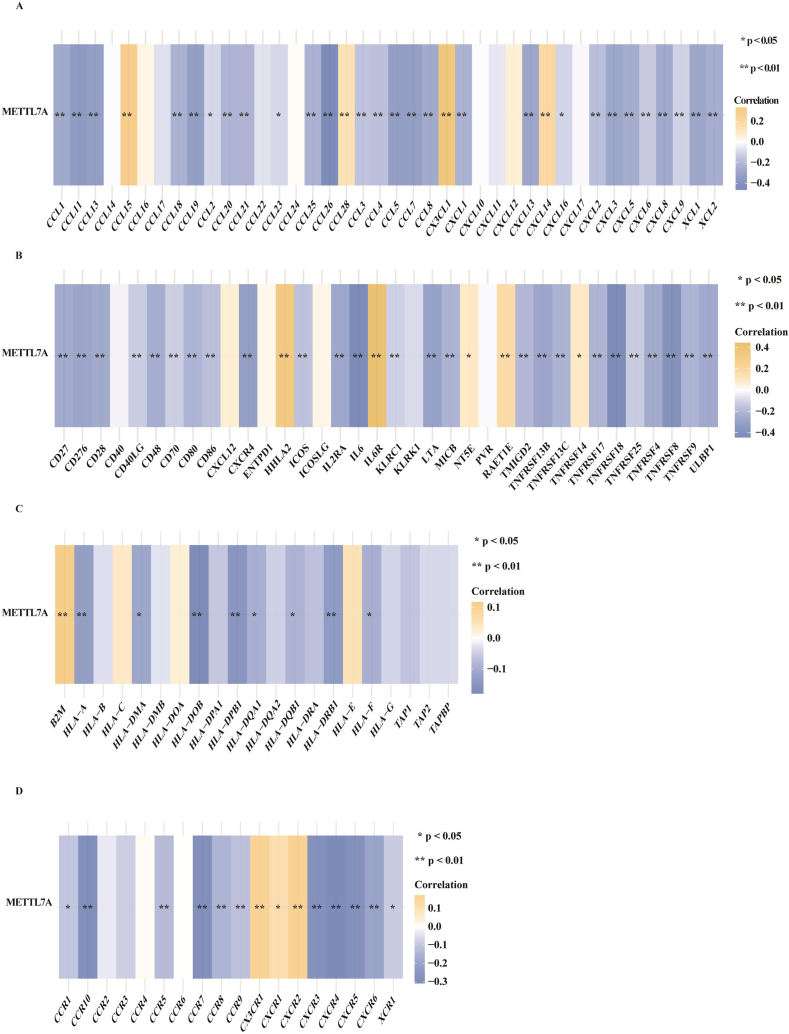
We also examined the expression relationships between METTL7A and various representative immune families, including chemokine (Fig. 6A), immunostimulators (Fig. 6B), major histocompatibility complex (MHC) (Fig. 6C) and immune receptors (Fig. 6D) in 530 KIRC patients to gain insight into the function of METTL7A in immune regulation. Their expression is frequently used to identify various kinds of immune cells, including B cells, T cells, CD8+ T cells, mast cells, monocytes, macrophages, neutrophils, eosinophils and dendritic cells. The findings revealed that METTL7A was inversely associated with most immune-related genes expression in KIRC, suggesting that METTL7A may control the immune response in KIRC.
Fig. 6.
The expression correlation between METTL7A and immune families. The expression correlation between METTL7A and (A) chemokine, (B) immunostimulator, (C) MHC molecule (D) and receptor using the data sets from TCGA.
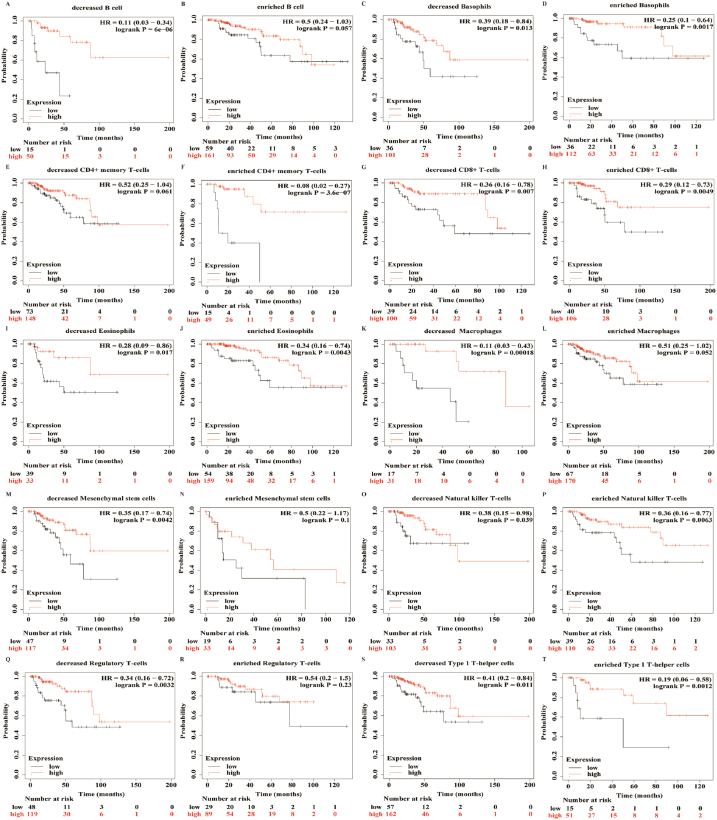
To investigate whether the role of METTL7A in renal carcinoma is associated with its involvement in the immune microenvironment, we used the Kaplan-Meier plotter database to examine the impact of METTL7A expression on the OS of KIRC and KIRP patients with high or low immune infiltration. We discovered that patients with high or low immunological infiltration experienced varied effects of METTL7A expression on overall survival (Fig. 7, Supplementary Figure S4). In KIRC, patients with low levels of type 1 T-helper cells, eosinophils, natural killer T-cells and type 2 T-helper cells had lower overall survival when METTL7A expression declined, and the effect of high levels of infiltration of these cells on OS was less pronounced than that of low levels (as indicated by the p-value). In contrast, high levels of basophil and CD8+ T cell infiltration were linked to poorer overall survival when METTL7A expression decreased, whereas the impact of low-level infiltration on overall survival was less pronounced than that of high-level infiltration (Supplementary Figure S4). Meanwhile, in KIRP, patients with low levels of infiltration of B cells, macrophages, mesenchymal stem cells, and regulatory T cells had poorer overall survival when METTL7A expression decreased, while high-level infiltration of these cells had no significant effect on overall survival. High levels of CD4+ memory T cell infiltration, on the other hand, were associated with poorer overall survival when METTL7A expression decreased, but low levels of infiltration had no significant impact on overall survival (Fig. 7). Our findings suggest that METTL7A can regulate immune cell infiltration and alter the prognosis of KIRC and KIRP patients. The down-regulation of METTL7A expression has different consequences on patients’ overall survival depending on the levels of immune cell infiltration (Fig. 7, Supplementary Figure S4), and this effect is more pronounced in KIRP than KIRC.
Fig. 7.
METTL7A might influence patients' overall survival by controlling immune cell infiltration. Kaplan-Meier survival curve of METTL7A in KIRP patients with immune cell infiltration at high or low levels. Decreased (A) and enriched (B) B cell; decreased (C) and enriched (D) basophils; decreased (E) and enriched (F) CD4+ memory T-cells; decreased (G) and enriched (H) CD8+ T-cells; decreased (I) and enriched (J) eosinophils; decreased (I) and enriched (J) eosinophils; decreased (K) and enriched (L) macrophages; decreased (M) and enriched (N) mesenchymal stem cells; decreased (O) and enriched (P) natural killer T-cells; decreased (Q) and enriched (R) regulatory T-cells; decreased (S) and enriched (T) type 1 T-helper cells.
3.5. Identification of SPI1 as a potential transcription factor for METTL7A in KIRC
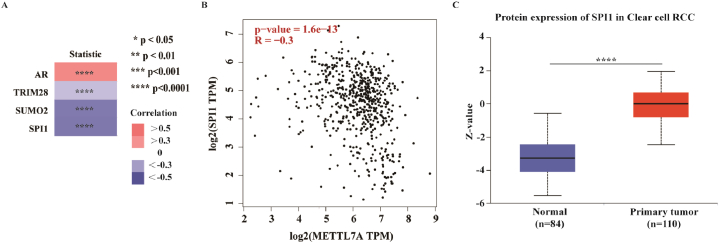
It has been demonstrated that transcription factors can influence gene transcription by attaching to the promoters of their target genes. To investigate the possible reasons for the downregulation of METTL7A in KIRC, we conducted a multi-omic analysis to identify the upstream transcription factors of METTL7A. Initially, four experimentally verified transcription factors for METTL7A in renal tissue were collected, including AR, SUMO2, SPI1 and TRIM28, from the hTFtarget database (Fig. 8A, Supplementary Table S1). It has been established that the expression of transcription factors is often associated with downstream targeting genes. We then analyzed all expression-related genes of METTL7A in KIRC (correlation coefficient >0.3 or - 0.3) to identify potential transcription factors using the GEPIA database (Supplementary Table S2). Among them, SPI1 expression had a negative correlation with METTL7A (correlation value = −0.3) (Fig. 8B). Additionally, KIRC tissue had a greater degree of SPI1 translation than normal kidney tissue (Fig. 8C). Based on these findings, SPI1 could be one of the potential transcription factors of METTL7A in KIRC, which might be responsible for the decreased METTL7A expression.
Fig. 8.
SPI1 as one potential transcription factor for METTL7A in KIRC. (A) Heat maps showing 4 experimentally-confirmed transcription factors of METTL7A in renal tissue basing on data from the hTFtarget database. (B) SPI1, a possible co-expressed upstream transcription factor, is significantly associated with METTL7A expression using GEPIA database. (C) Box plots showing the up-regulated protein expression of SPI1 in KIRC tissues versus normal adjacent tissues.
4. Discussion
Several studies have reported that KIRC is heavily infiltrated by immune cells [21,22]. The immune response has been linked to the spontaneous regression seen in up to 1% of KIRC patients [23]. Furthermore, KIRC was one of the first cancers to be evaluated for immunotherapy and has remained one of the most sensitive tumors to date [[24], [25], [26], [27]]. However, current immunotherapy modalities have not been entirely successful [3], highlighting the need to identify new therapeutic targets. Our study suggests that METTL7A may regulate the immune microenvironment in renal cancer patients, making it a potential immune target for therapy.
While surgery is often effective in treating early-stage renal cancer, up to one-third of cases may progress to metastasis [28,29]. Early-stage patients do not require high doses of chemotherapy or radiation, which may cause cellular damage. Conversely, advanced-stage patients have a poor prognosis, and aggressive treatment may weaken their immune systems. Therefore, identifying prognostic indicators to distinguish high-risk from low-risk patients is important for individualized care. Our research revealed that, in contrast to normal adjacent tissues, the expression of METTL7A was markedly lower in renal cancer tissues, and low expression was associated with poor overall survival. As a result, METTL7A could be a predictive factor for renal carcinoma, which could be useful for developing individualized treatment plans in the future.
METTL7A (methyltransferase like 7A) is a gene that encodes a protein thought to act as a methyltransferase in cells [30]. Previous studies have found that the downregulation of METTL7A in tumor tissues predicted a poor prognosis in HCC patients [15], and that inhibiting METTL7A might help choriocarcinoma cells respond to methotrexate (MTX)-based chemotherapy [16]. In the context of neutrophil degranulation, changes in METTL7A expression may compromise the integrity of the Golgi apparatus, resulting in abnormal neutrophil secretory granule production and altering the first defense barrier of the innate immune response in chronic schizophrenia [31]. Another study investigated the role of METTL7A in preserving the integrity of the Golgi, which may be crucial for neutrophil degranulation [32]. However, there is currently no research on the involvement of METTL7A in renal cancer or its modulation of the immune microenvironment.
Numerous publications have documented the roles and clinical importance of METTL family members in tumorigenesis. For example, METTL1 is linked to several human malignancies, including hepatocellular carcinoma and lung cancer [[33], [34], [35]]. METTL3 and METTL14 have been shown to promote oncogene expression and cancer development by methylating RNAs with m6A in leukemia and solid tumors [36,37]. METTL13 primarily mediates Ras-driven carcinogenesis of pulmonary and pancreatic epithelial malignancies [38]. METTL18 is also involved in the transformation of cancer cells, as discovered in a recent study [39]. These findings highlight the importance of METTLs in cellular processes of tumorigenesis. According to our prior research, the METTL family members have a closer association with the risk of renal cancer than other cancers, suggesting that METTL family may have unique roles in renal cancer (Supplementary Figure S1). Among these members, we discovered that METTL7A might function as a protector in kidney cancer, but its mechanism was uncertain. The impact and putative mechanism of METTL7A in renal cancer, providing a potential target for diagnostics and treatment.
T cells, including CD4+T cells and CD8+T cells, are the foundation of “immune surveillance” and are crucial in specifically inhibiting cancer cells in an antigen-specific manner [40,41]. CD8+ T cells and macrophages are known as anti-tumor effector cells, and CD4+ T cells can enhance their anticancer activity [42,43]. Therefore, CD4 + T cells are also directly related to preventing tumor growth. Tregs are the primary immunosuppressive cells in tumor immunity and can enter the tumor microenvironment (TME) to block the tumor suppressor immune response [44,45]. The accumulation of Tregs in TME results in an immune microenvironment that stimulates tumor growth out of control [46,47], promoting immune escape and cancer progression. Additionally, NK cells are specialized killer cells that support cancer immune surveillance [48,49]. In our study (Fig. 5), we discovered a negative correlation between Tregs and the expression of METTL7A in renal cancer while a positive correlation between the levels of CD4+, CD8+, and NK cell infiltration and METTL7A expression was found. This suggests that CD4+T cells, CD8+T cells, NK cells and Tregs may be involved in the mechanism of METTL7A's functions in tumor progression.
Although this study has advanced knowledge of METTL7A and renal cancer, limitations remain. Firstly, while we have examined the relationship between METTL7A and immune infiltration in renal carcinoma patients, further analysis of different subgroups would provide more clarity. Secondly, most of the studies on METTL7A in this paper were conducted at the gene level, and more detailed analysis at the protein level could strengthen the evidence. Thirdly, while this study has identified a link between METTL7A and immune cell infiltration levels in renal cancer patients, additional research is required to elucidate its molecular mechanism and its function in tumor growth and immune escape.
In conclusion, our study illuminates the function and probable mechanisms of METTL7A in carcinogenesis and its application as a predictive biomarker and immunological target for renal cancer.
Author contribution statement
Conceived and designed the experiments: Wei Zhang, Donge Tang. Performed the experiments: Wei Zhang, Lintai Li, Yumei Chen, Nan Hu. Analyzed and interpreted the data: Zhiqian Yang, Xiangnan Dong. Contributed reagents, materials, analysis tools or data: Zhiqian Yang, Wanxia Cai. Wrote the paper: Zhiqian Yang, Wei Zhang. Obtained funding Yong Dai, Wei Zhang. Study supervision: Fanna Liu, Lianghong Yin. All authors contributed to the article and approved the submitted version.
Funding statement
This work was supported by Guangdong Provincial High-level Clinical Key Specialties (No. SZGSP001), the science and technology plan of Shenzhen (No. JCYJ20210324113214039), Guangxi Key Laboratory of Metabolic Diseases Research (No. 20-065-76), and Key Renal Laboratory of Shenzhen (No. ZDSYS2015430161623417).
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interest's statement
We declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e15371.
Contributor Information
Fanna Liu, Email: tliufana@jnu.edu.cn.
Donge Tang, Email: tang.donge@szhospital.com.
Yong Dai, Email: daiyong@mail.sustech.edu.cn.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Bhatt J.R., Finelli A. Landmarks in the diagnosis and treatment of renal cell carcinoma. Nat. Rev. Urol. 2014;11(9):517–525. doi: 10.1038/nrurol.2014.194. [DOI] [PubMed] [Google Scholar]
- 2.Bray F., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Díaz-Montero C.M., Rini B.I., Finke J.H. The immunology of renal cell carcinoma. Nat. Rev. Nephrol. 2020;16(12):721–735. doi: 10.1038/s41581-020-0316-3. [DOI] [PubMed] [Google Scholar]
- 4.He R.Z., Jiang J., Luo D.X. The functions of N6-methyladenosine modification in lncRNAs. Genes Dis. 2020;7(4):598–605. doi: 10.1016/j.gendis.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma S., et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J. Hematol. Oncol. 2019;12(1):121. doi: 10.1186/s13045-019-0805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun Z., et al. Prognostic risk signature based on the expression of three m6A RNA methylation regulatory genes in kidney renal papillary cell carcinoma. Aging. 2020;12(21):22078–22094. doi: 10.18632/aging.104053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou J., et al. Gene signatures and prognostic values of m6A regulators in clear cell renal cell carcinoma - a retrospective study using TCGA database. Aging. 2019;11(6):1633–1647. doi: 10.18632/aging.101856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X., et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534(7608):575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 10.Liu J., et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10(2):93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pendleton K.E., et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169(5):824–835.e14. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Tran N., et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47(15):7719–7733. doi: 10.1093/nar/gkz619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z., et al. Induction of m(6)A methylation in adipocyte exosomal LncRNAs mediates myeloma drug resistance. J. Exp. Clin. Cancer Res. 2022;41(1):4. doi: 10.1186/s13046-021-02209-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan Y.Q., et al. Comprehensive analysis of the significance of METTL7A gene in the prognosis of lung adenocarcinoma. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.1071100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi L., et al. An RNA editing/dsRNA binding-independent gene regulatory mechanism of ADARs and its clinical implication in cancer. Nucleic Acids Res. 2017;45(18):10436–10451. doi: 10.1093/nar/gkx667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jun F., et al. Quantitative proteomic analysis identifies novel regulators of methotrexate resistance in choriocarcinoma. Gynecol. Oncol. 2020;157(1):268–279. doi: 10.1016/j.ygyno.2020.01.013. [DOI] [PubMed] [Google Scholar]
- 17.Chandrashekar D.S., et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagy A., et al. Author Correction: validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-29514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasaikar S.V., et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–d963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Z., et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–w102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson R.H., et al. PD-1 is expressed by tumor-infiltrating immune cells and is associated with poor outcome for patients with renal cell carcinoma. Clin. Cancer Res. 2007;13(6):1757–1761. doi: 10.1158/1078-0432.CCR-06-2599. [DOI] [PubMed] [Google Scholar]
- 22.Yoshihara K., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janiszewska A.D., Poletajew S., Wasiutyński A. Spontaneous regression of renal cell carcinoma. Contemp. Oncol. 2013;17(2):123–127. doi: 10.5114/wo.2013.34613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbst R.S., et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Topalian S.L., et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012;366(26):2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Escudier B. Emerging immunotherapies for renal cell carcinoma. Ann. Oncol. 2012;23(Suppl 8):viii35–viii40. doi: 10.1093/annonc/mds261. [DOI] [PubMed] [Google Scholar]
- 27.Atkins M.B., Regan M., McDermott D. Update on the role of interleukin 2 and other cytokines in the treatment of patients with stage IV renal carcinoma. Clin. Cancer Res. 2004;10(18 Pt 2):6342s–6346s. doi: 10.1158/1078-0432.CCR-040029. [DOI] [PubMed] [Google Scholar]
- 28.Sánchez-Gastaldo A., et al. Systemic Treatment of Renal Cell Cancer: A Comprehensive Review. Cancer Treat. Rev. 2017;60:77–89. doi: 10.1016/j.ctrv.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Jonasch E., Gao J., Rathmell W.K. Renal cell carcinoma. BMJ. 2014;349:g4797. doi: 10.1136/bmj.g4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lichtenstein A.V., Kisseljova N.P. DNA methylation and carcinogenesis. Biochemistry. 2001;66(3):235–255. doi: 10.1023/a:1010249510906. [DOI] [PubMed] [Google Scholar]
- 31.Vera-Montecinos A., et al. Analysis of molecular networks in the cerebellum in chronic schizophrenia: modulation by early postnatal life stressors in murine models. Int. J. Mol. Sci. 2021;22(18) doi: 10.3390/ijms221810076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKinnon C.M., Mellor H. The tumor suppressor RhoBTB1 controls Golgi integrity and breast cancer cell invasion through METTL7B. BMC Cancer. 2017;17(1):145. doi: 10.1186/s12885-017-3138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma J., et al. METTL1/WDR4-mediated m(7)G tRNA modifications and m(7)G codon usage promote mRNA translation and lung cancer progression. Mol. Ther. 2021;29(12):3422–3435. doi: 10.1016/j.ymthe.2021.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian Q.H., et al. METTL1 overexpression is correlated with poor prognosis and promotes hepatocellular carcinoma via PTEN. J. Mol. Med. 2019;97(11):1535–1545. doi: 10.1007/s00109-019-01830-9. [DOI] [PubMed] [Google Scholar]
- 35.Okamoto M., et al. tRNA modifying enzymes, NSUN2 and METTL1, determine sensitivity to 5-fluorouracil in HeLa cells. PLoS Genet. 2014;10(9):e1004639. doi: 10.1371/journal.pgen.1004639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng X., et al. Role of N(6)-methyladenosine modification in cancer. Curr. Opin. Genet. Dev. 2018;48:1–7. doi: 10.1016/j.gde.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang P., Doxtader K.A., Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 2016;63(2):306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu S., et al. METTL13 methylation of eEF1A increases translational output to promote tumorigenesis. Cell. 2019;176(3):491–504.e21. doi: 10.1016/j.cell.2018.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Małecki J.M., et al. Human METTL18 is a histidine-specific methyltransferase that targets RPL3 and affects ribosome biogenesis and function. Nucleic Acids Res. 2021;49(6):3185–3203. doi: 10.1093/nar/gkab088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schreiber R.D., Old L.J., Smyth M.J. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331(6024):1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 41.Burnet F.M. Immunological surveillance in neoplasia. Transplant. Rev. 1971;7:3–25. doi: 10.1111/j.1600-065x.1971.tb00461.x. [DOI] [PubMed] [Google Scholar]
- 42.Busselaar J., et al. Helpless priming sends CD8(+) T cells on the road to exhaustion. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.592569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahrends T., et al. CD4(+) T cell help confers a cytotoxic T cell effector program including coinhibitory receptor downregulation and increased tissue invasiveness. Immunity. 2017;47(5):848–861.e5. doi: 10.1016/j.immuni.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 44.Lim W.C., et al. Human endothelial cells modulate CD4(+) T cell populations and enhance regulatory T cell suppressive capacity. Front. Immunol. 2018;9:565. doi: 10.3389/fimmu.2018.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Comito G., et al. Lactate modulates CD4(+) T-cell polarization and induces an immunosuppressive environment, which sustains prostate carcinoma progression via TLR8/miR21 axis. Oncogene. 2019;38(19):3681–3695. doi: 10.1038/s41388-019-0688-7. [DOI] [PubMed] [Google Scholar]
- 46.Nishikawa H., Sakaguchi S. Regulatory T cells in tumor immunity. Int. J. Cancer. 2010;127(4):759–767. doi: 10.1002/ijc.25429. [DOI] [PubMed] [Google Scholar]
- 47.Chaudhary B., Elkord E. Regulatory T cells in the tumor microenvironment and cancer progression: role and therapeutic targeting. Vaccines. 2016;4(3) doi: 10.3390/vaccines4030028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.López-Soto A., et al. Control of metastasis by NK cells. Cancer Cell. 2017;32(2):135–154. doi: 10.1016/j.ccell.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 49.Chiossone L., et al. Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol. 2018;18(11):671–688. doi: 10.1038/s41577-018-0061-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.