Abstract
Both a leaky gut (a barrier defect of the intestinal surface) and gut dysbiosis (a change in the intestinal microbial population) are intrinsic to sepsis. While sepsis itself can cause dysbiosis, dysbiosis can worsen sepsis. The leaky gut syndrome refers to a status with which there is an increased intestinal permeability allowing the translocation of microbial molecules from the gut into the blood circulation. It is not just a symptom of gastrointestinal involvement, but also an underlying cause that develops independently, and its presence could be recognized by the detection, in blood, of lipopolysaccharides and (1→3)-β-D-glucan (major components of gut microbiota). Gut-dysbiosis is the consequence of a reduction in some bacterial species in the gut microbiome, as a consequence of intestinal mucosal immunity defect, caused by intestinal hypoperfusion, immune cell apoptosis, and a variety of enteric neuro-humoral-immunity responses. A reduction in bacteria that produce short-chain fatty acids could change the intestinal barriers, leading to the translocation of pathogen molecules, into the circulation where it causes systemic inflammation. Even gut fungi might be increased in human patients with sepsis, even though this has not been consistently observed in murine models of sepsis, probably because of the longer duration of sepsis and also antibiotic use in patients. The gut virobiome that partly consists of bacteriophages is also detectable in gut contents that might be different between sepsis and normal hosts. These alterations of gut dysbiosis altogether could be an interesting target for sepsis adjuvant therapies, e.g., by faecal transplantation or probiotic therapy. Here, current information on leaky gut and gut dysbiosis along with the potential biomarkers, new treatment strategies, and future research topics are mentioned.
Keywords: faecal transplantation, gut dysbiosis, leaky gut, lipopolysaccharides, microbiome, sepsis
Introduction
Sepsis is a common syndrome with high mortality and morbidity [1]. Despite recent reductions in sepsis mortality rates, sepsis continues to account for approximately 20% of global deaths, with a staggering 60% mortality rate in patients with septic shock [2,3]. Bacterial infection is the most common cause of sepsis, but most of the clinical manifestations of severe infection caused by bacteria, fungi, viruses and parasitic infection, such as leptospirosis, aspergillosis, dengue shock syndrome and severe malaria, are surprisingly similar, and include cardiovascular dysfunction, resulting in low blood pressure and poor tissue perfusion, renal injury, resulting in anuria, and pulmonary dysfunction, resulting in hypoxemia [4–7]. These similarities imply the possible operation of a predominant innate immune response, i.e., the rapid immune response, rather than adaptive immunity, i.e., the late specific response [8]. The role of microbial molecules not produced by the host (pathogen-associated molecular patterns [PAMPs]) and molecules from the host’s cells (damage-associated molecular patterns [DAMPs]) are distinguished from regular immune homeostasis by innate immunity during sepsis [9]. An adaptive immunity, orchestrated by T and B lymphocytes, along with antibodies, is equally important [10]. Some of PAMPs and DAMPs with the sources and main pattern recognition receptors are listed in Table 1 [11–13].
Table 1. Common PAMPs and DAMPs in sepsis.
| Pathogen-associated molecular patterns (PAMPs) | ||
|---|---|---|
| Name | Possible sources* | Main PRR**/*** |
| Lipopolysaccharide | Gram-negative bacteria | TLR4# |
| Lipoteichoic acid | Gram-positive bacteria in gut | TLR2 |
| Flagellin | Filamentous bacteria | TLR5 |
| Lipoarabinomannan | Non-tuberculous mycobacteria | TLR2 |
| Triacyl lipoproteins | Several bacteria | TLR1, TLR2 |
| CpG motifs | Bacteria and viruses | TLR9 |
| β-Glucan | Fungi | Dectin-1, TLR2, TLR4 |
| Zymosan (beta glucan with mannans) | Fungi | TLR2, Several Dectins |
| Double-stranded RNA (dsRNA) | Viruses | RIG-I, MDA5, PKR (cytosolic receptors) ## |
| Single-stranded RNA ssRNA | Viruses | TLR7, TLR8 |
| Host damage-associated molecular patterns (DAMPs) | ||
|---|---|---|
| Name | Possible sources | Main PRR* |
| RNA | Ruptured host cells | TLR3, TLR7, TLR8, RIG-I |
| MDA5 | ||
| DNA (host and mitochondria) | Ruptured host cells | TLR9, TLR4 |
| Histones | Ruptured host cells | TLR2, TLR4 |
| High mobility group box-1 (HMGB1) and S100 proteins | Secreted from immune cells or cell rupture | TLR2, TLR4, RAGE ### |
| Heat shock proteins (HSPs) | Host cells with stress | TLR2, TLR4, CD91 |
*, mostly in gut or respiratory tract; **, PRR, pattern recognition receptors; ***, mostly on myeloid cells; #, TLR, Toll-like receptor; ##, RIG-I, retinoic acid-inducible gene I, MDA5, RIG-I-like receptor dsRNA helicase enzyme, PKR, Protein Kinase R (an interferon-induced kinase); ###, RAGE, receptor for advanced glycation end products.
The importance of PAMPs in sepsis implicates the gastrointestinal tract as an endogenous reservoir of several groups of organisms, including prokaryotes, i.e., bacteria and archaea, eukaryotes, i.e., fungi, and viruses, mostly bacteriophages, which are jointly referred to as ‘gut microbiota’. These organisms are separated from the host by only a single layer of enterocytes containing tight junction molecules [14,15]. During sepsis, enterocytes experience hyperpermeability caused by several factors, including intestinal hypoperfusion, enterocyte apoptosis, a systemic cytokine storm, and gut dysbiosis, which could promote the translocation of microbial molecules from the gut into systemic circulation. This is often referred to as the ‘leaky gut’ [16,17], which is a factor that might be associated with enhanced systemic inflammation in several conditions, either with regular activities (vigorous exercise, high amount of chili, some drugs and stress) [18–20] or pathogenic conditions (autoimmune diseases, infections, obesity and uremia) [21–24]. There are differences in the pathophysiology of leaky gut in these diseases. For example, gut permeability damage in systemic lupus erythematosus (a common autoimmune disease) is possibly due to immune-complex deposition in the gut and the adverse effects of some medications, including nonsteroidal anti-inflammatory drugs (NSAIDs), corticosteroids, and disease-modifying antirheumatic drugs (DMARDs) [14]. Meanwhile, the stress-induced leaky gut is a result of stress hormone-induced immune alteration with autonomic nervous system (gut–brain axis) [25] and both impacts of lupus and stress finally cause gut dysbiosis and leaky gut. The enhanced gut permeability that is sufficiently severe to allow the translocation of viable bacteria, and especially some invasive bacteria, from the gut to the systemic circulation, could be a cause of sepsis, frequently referred to as ‘gut-derived sepsis’ [26,27]. The microbiota, local immunity and integrity in the gut are important factors for the maintenance of the gut microenvironment; therefore, manipulations of these factors might be beneficial in sepsis treatment. Despite increasing knowledge on leaky gut and gut dysbiosis in sepsis, the clinical translation of this information to patients is still very less. Although the alteration of gut bacteria during sepsis is well-known, the exploration of sepsis-induced alteration in fungi and viruses in the gut is recently increasing which might uncover new interesting aspects. Then, the collection of current data on this topic might facilitate interest in the use of some parameters and treatments in real clinical practice. Hence, this review summarizes the potential impact of the gut microbiome, in terms of bacteria, fungi and viruses, on the course of sepsis, and explores currently proposed adjuvant therapies, including faecal transplantation or probiotic therapy.
A leaky gut leads to the presence of microbial molecules in the systemic circulation
A single layer of epithelial cells with a surface area of about 32 m2 lines the gastrointestinal (GI) surface and is held together by epithelial tight junctions (TJs). This layer functions as the first stage of the intrinsic mucosal defence system and serves as a selective physical barrier between the host and microbial molecules [28,29]. The TJ complex does not allow the passage of molecules larger than 3.6 Å or 0.6 kDa through the normal paracellular passage (the space between the proximity of enterocytes). The larger molecules are transported through the gut epithelial cells by several transcytosis mechanisms, including clathrin-mediated endocytosis, micropinocytosis and caveolin-mediated endocytosis [30,31]. Some microbial-derived molecules, such as p-cresol (a gut-derived uremic toxin derived from protein fermentation by gut bacteria), are small enough to pass through the normal gut barrier [32], whereas other molecules, such as lipopolysaccharide (LPS) from Gram-negative bacteria and (1→3)-β-D-glucan (BG) from fungi (the most abundant and second most abundant organisms in the gut) or microbial DNA, are too large to cross the barrier [33]. However, although large intact bacterial DNAs (i.e., the genome) with molecular sizes of 100 to 15,000 kilobase pairs (kbp) (6.5 × 104–9.8 × 106 kDa) are too large to pass through the gut barrier, DNA molecules are rapidly broken down into bacteria-free DNA through several processes (depurination and deamination) into pieces approximately 100 bp (65 kDa) in size (i.e. similar in size to LPS and BG) [34]. Hence, the detection of these PAMPs (LPS, BG and bacteria-free DNA) could be useful indirect markers of leaky gut. Alternatively, the oral administration of a non-absorbable carbohydrate and its subsequent detection in blood or urine is a well-known direct test for leaky gut [35,36]. However, the necessity of oral administration and intact intestinal peristalsis limits the use of this procedure only to patients in non-moribund conditions.
Local intestinal injury with a large surface area does not surprisingly induce leaky gut, as demonstrated in mice treated with a low concentration of dextran sulphate solution (DSS), a substance that directly causes TJ injury. The intestinal symptoms of leaky gut progress from asymptomatic to overt diarrhoea [37] or acute pancreatitis with endotoxemia [38]. In parallel, high-abundance PAMPs detected in serum, including in DSS-administered mice, are indicators of leaky gut [39]. Interestingly, leaky gut in DSS-administered mice is demonstrable with a fluorescein isothiocyanate (FITC)-dextran assay. In humans, the detection of some non-absorbable carbohydrates in urine after an oral administration is demonstrated, even without abdominal symptoms (diarrhoea or abnormal stool consistency) [40], implies a possible asymptomatic leaky gut. As such, the current hypothesis is that ‘a physiologic leaky gut (leaky gut without a significant adverse effect)’ may exist, as is observed in blood microbiome analyses with the presence of DNA from anaerobic gut bacteria that are usually not present in the blood circulation [34]. Although the abundance of DNA in the blood of healthy control mice is very low or non-detectable, the DNA amplification processes used in bacteriome analysis can detect low amounts of DNA. Notably, the regular repairing process of the ‘physiologic leaky gut’ should not produce intestinal fibrosis due to the prominent self-renewal property of the enterocytes [41]. However, intestinal fibrosis can be developed in case of severe overt chronic inflammation as reported in inflammatory bowel disease (ulcerative colitis) [42].
Further possible evidence for a physiologic leaky gut is the detection of serum BG in some healthy people, especially with the Fungitell assay (Associates of Cape Cod, Inc.), as BG is a major component of fungi that is foreign molecule to the host, with a normal range (less than 60 pg/ml) that possibly reflects a leaky gut in healthy individuals (detectable serum BG without detrimental condition) [17,35,36]. As such, BG is the natural polysaccharide consisting of sequential D-glucose moieties linked by β-(1→3)-glycosidic bonds with other structural varieties depending on the sources, such as BG from fungi is composed of β-(1→6)-linked branches from the β-(1→3) backbone [43]. Proinflammatory impacts of BG, especially in synergy with LPS, are frequently mentioned [44–48].
By contrast, endotoxaemia should not be detectable in a healthy host, despite a possible low level of leaky gut, perhaps due to several LPS neutralization actions, such as deacylation and dephosphorylation by acyl-oxy-acyl hydrolase and alkaline phosphatase, respectively [49–51]. Notably, no enzymatic reaction exists for BG neutralization [52]. Hence, LPS and BG in serum, in the absence of other obvious sources, are interesting leaky gut biomarkers that are more practical for clinical use compared with the standard oral carbohydrate administration. However, the level of LPS and BG in serum not only depends on the severity of a leaky gut, but also correlates with increased numbers of Gram-negative bacteria and fungi in the gut. In animal models, several conditions lead to an increase in Gram-negative bacteria (Bacteroides and Proteobacteria) and possibly LPS in gut contents, including sepsis, DSS-induced mucositis, uraemia, obesity and fungal administration [39,48,53–55], while an enhanced content of faecal fungi (and BG) in the gut is possible after antibiotic use, intestinal inflammation (inflammatory bowel disease; IBD), and alcohol consumption [44,45,56–58]. Thus, using the actual quantity levels of LPS and BG to determine the severity of leaky gut is difficult; however, they might be useful for qualitatively indicating gut barrier damage.
Observing leaky gut is not surprising after acute or chronic diarrhoea from any causes (infection, immune-mediated diseases, and DSS) [53,59–61] due to direct damage to the TJ. However, the pathophysiology of systemic inflammation-induced leaky gut might involve inflammation-induced paracellular enterocyte permeability (as demonstrated by LPS injection models) [62] and/ or stress-induced gut dysbiosis [63,64]. Indeed, LPS injection triggers the production of serum cytokines, which can affect every cell in the body, including enterocytes, and cytokine activation worsens enterocyte integrity, as demonstrated by the reduced transepithelial electrical resistance in enterocytes after incubation with pro-inflammatory cytokines [65]. In addition, neuro-hormonal disturbances in response to stress (and depression), especially the enhancement of catecholamine, can alter the bacterial composition in the gut, in part, due to catecholamine iron chelation that facilitates the growth of iron-metabolizing bacteria [66]. The activation of enteric neurons by corticotropin-releasing factors in immune cells (macrophages and mast cells) can also alter the microbial control mechanism in the gut [67]. It is also interesting to note that there is a balance of the immune responses, referred to as the ‘counter anti-inflammatory response’, during the hyper-inflammatory activity in severe systemic inflammation, especially in sepsis, the imbalance homeostasis of immune regulation seems to induce either hyper-inflammatory septic shock or immune exhaustion (an increased susceptibility to secondary infection) [68–70] that might be able to cause enterocyte injury and leaky gut, perhaps, with different processes. More studies on this topic would be interesting.
Leaky gut and gut dysbiosis
The balance between the host immune activities and micro-organisms in the gut leads to the specific characteristic of gut microbiota in different hosts as genetic-based immune responses and gut microenvironmental aspects (diets and regular activities) might be different among individuals. As such, an alteration of immune activities in the host, due to aging, antibiotics, foods, or the new onset of some systemic diseases possibly results in a change in gut microbiota [71–73]. For example, the depletion of macrophages or splenectomy in the host reduces the microbicidal activity against some gut organisms leading to gut dysbiosis [64,74] and the selective microbicidal activities of different antibiotics induce some different dysbiosis in the host [75,76]. In contrast, gut dysbiosis might induce some changes in immune responses that possibly affect intestinal integrity. As such, gut dysbiosis induced by oral administration of pathogenic bacteria or fungi facilitates a direct invasion of enterocytes and activates the more prominent immune responses leading to a more severe leaky gut than the presence in the host with lesser harmful microbes [37,45,77]. Notably, the presence of gut fungi alters the composition of gut bacteria through several mechanisms, such as a selection of bacteria that can digest some molecules on fungal cell walls or bacteria with fungal toxin resistance [39,74]. Hence, immune activities, both local intestinal immunity and systemic immune responses, affect gut dysbiosis and vice versa that can cause defects in the intestinal barrier (leaky gut) through the damage by immune responses (enterocytes are the bystanders from microbicidal immunity) and/ or from the invasiveness of the pathogenic microbes.
During sepsis, there was an alteration in immune responses and gut dysbiosis with several sepsis factors that enhances intestinal barrier defect. For sepsis-induced immune responses, hyper-inflammatory cytokines, death of immune cells from the overwhelming immune activities and stress hormone-activated intestinal immunity [8,78,79] that might affect the normal balance between the host immunity and microbes. In sepsis-induced gut dysbiosis, an abundance of the high virulence organisms in the gut during sepsis might be increased because these bacteria usually have several factors against the harsh microenvironment, while normal microbiota mostly demonstrated a lack of these factors [80]. Moreover, several defects during sepsis, for example, gut hypoperfusion from systemic vasodilatation and/or sepsis-induced cardiomyopathy, intestinal hypomotility and gut mucosal disruption [81] also directly induce gut barrier defect and leaky gut. Hence, enterocyte hyperpermeability in sepsis is caused by several factors, including intestinal hypoperfusion, enterocyte apoptosis, systemic cytokine storm and gut dysbiosis that could promote the translocation of microbial molecules from the gut into the blood circulation (leaky gut or gut leakage) [16,17].
Although the endotoxemia and circulating cell-free DNAs (cf-DNAs) observed in bacterial sepsis might be derived from dead bacteria in the blood, some LPS molecules might be correlated with translocation from the gut into the blood circulation (gut translocation). Better evidence of leaky gut during sepsis comes from the presence of endotoxemia and glucanemia (serum BG) without bacteraemia during viral sepsis, such as is observed with dengue and coronavirus disease 2019 (COVID-19) with high disease severity [35,82–84]. Although a mixed bacterial–viral infection is possible, antibiotics (and anti-fungal) are not necessary for most of these patients with severe viral sepsis. Additionally, the administration of bacterial lysate also containing bacterial DNA during leaky gut induction by DSS in mice increases the level of cf-DNAs in the blood [34], implying a possible gut translocation during sepsis. One interesting finding is that leaky gut could be a cause and/or consequence of bacterial sepsis because (i) the severe gut barrier defect induces viable bacterial translocation and bacteraemia, as indicated by DSS-induced sepsis [64,77], and (ii) the damage to the enterocyte TJ during sepsis facilitates leaky gut [45]. In both situations, the leaky gut enhances systemic inflammation through innate immunity responses, especially through macrophages and neutrophils [40,46,85]. Similarly, gut dysbiosis (the imbalance of gut microbiota associated with an unhealthy outcome) can be a cause and/ or a consequence of bacterial sepsis due to the importance of gut microbiota in the maintenance of intestinal integrity [86].
Currently, several methods (multi-sugar probe, LPS, BG and other molecules) [87] are available for leaky gut measurements, but performing these measurements during sepsis is challenging because of the limitations in oral carbohydrate administration to critically ill patients, the possible differences in the abundance of LPS and BG in the gut contents, Gram-negative bacteraemia (which limits the use of LPS as a leaky gut marker) and the unclear clinical usefulness of several molecules (zonulin, fatty acid binding protein and others). Due to high susceptibility to the leaky gut during sepsis, a quantitative test of leaky gut might not be necessary, and qualitative tests for leaky gut (such as BG) with dysbiosis indicators (such as the abundance of Firmicutes, Bacteroides and Proteobacteria by microbiome analysis or polymerase chain reaction [PCR]) might be adequate for clinical use. Although species differences detected by microbiome analysis are more informative, the differences in phylum levels using PCR with selected primers might be less expensive and more suitable for real clinical use. More studies on this topic are warranted.
The intestinal bacterial microbiome
Because bacteria are the most predominant organisms in the gut, most of the ‘gut dysbiosis’ mentioned in the studies predominantly refers to bacterial dysbiosis. The normal gut microbiota includes a predominance of Firmicutes (Bacillota) (mostly Gram-positive bacteria with obligate aerobes or facultative anaerobes) and Bacteroides (mostly Gram-negative anaerobes that are pathogens in some situations) [88]. Firmicutes are the most prominent bacteria in the healthy gut, in part due to the conversion of complex carbohydrates into short-chain fatty acids (SCFAs, particularly butyrate), which are important growth factors for the gut epithelium. Bacteroides are the most dominant Gram-negative bacteria in the gut and possibly represent a major source of LPS in the intestine [89]. The ratio of Firmicutes/ Bacteroides could serve as a biomarker for the health of the gut barrier, as this is lower in several conditions, including infection, DSS colitis, post-splenectomy, macrophage depletion, obesity, uraemia, iron overload and sepsis [24,48,55,77,90,91], and an increased Firmicutes/Bacteroides ratio is reported in IBD [92,93]. Despite the benefits of SCFA production by most Firmicutes bacteria (such as the probiotic strains of lactobacilli and enterococci), some groups (such as a subset of clostridial species) are pathogens that might induce gut barrier damage [94,95]. Likewise, several species of Bacteroides bacteria supply nutrients to other microbial residents and reduce pathogens in the gut, despite the possible pathogenicity of other Bacteroides [88]. Proteobacteria (Pseudomonadota), a major phylum of Gram-negative bacteria (including a wide variety of pathogens), is another bacterial phylum that frequently shows increases during gut dysbiosis [96–98]. Thus, both increases and decreases in the Firmicutes/Bacteroides ratio with increased Proteobacteria indicate gut dysbiosis; however, more studies are warranted before adopting this ratio for clinical use.
The normal gut microbiota is vulnerable to the microenvironment, as the oral administration of bacteria or fungi causes leaky gut from an increase in pathobionts [37,45], while leaky gut due to DSS induces dysbiosis through gut mucosal inflammation [53]. Intestinal inflammation might therefore be another factor that induces gut dysbiosis, as the oral administration of Candida albicans in control mice does not alter faecal microbiota patterns, while C. albicans gavage in septic mice after cecal ligation and puncture (CLP) surgery or DSS-colitis increased the proportion of Gammaproteobacteria (a group of pathogenic bacteria, including Pseudomonas aeruginosa) [39,53]. Indeed, intestinal inflammation from several causes, including some diets (high-fat diets), drugs (non-steroidal anti-inflammatory drugs; NSAIDs) and stresses (heavy exercise), can reduce mucin production (mucin barrier) and increase the number of pro-inflammatory cells (and mediators), resulting in a selection of some groups of bacteria that are more resistant to host immunity (mostly the highly virulent pathogenic bacteria) [18,19,99–101]. Conversely, the reduction in immune responses, such as macrophage depletion, also possibly increases some bacteria that are naturally controlled by intestinal macrophages and causes gut dysbiosis [74].
Due to the vulnerability of gut microbiota, some host characteristics might be theoretically classified as sepsis-vulnerable features. This could occur in individuals with a lower abundance of SCFA-producing bacteria, a genetic deficiency in the normal gut barrier (production of mucin and anti-microbial peptides; AMPs), or in those with malnutrition or immunodeficiency, as the faecal microbiome is a sensitive biomarker for these conditions [102,103]. For example, Mucin 2 deficient (Muc2-/-) mice develop colitis at 6 months of age, with increases in Firmicutes/Bacteroidetes and some Proteobacteria (Desulfovibrio and Escherichia) [104]. A defect in AMPs is mentioned in IBD-induced dysbiosis [105], and children with severe acute malnourishment demonstrate increased Proteobacteria and decreased Bacteroides in faeces [106,107]. Therefore, a reduction in Firmicutes or a low Firmicutes/Bacteroides ratio might be an indicator of low numbers of SCFA-producing bacteria and might represent a characteristic of susceptibility to gut-derived sepsis because of the easier gut invasion of pathogenic bacteria [93,108]. However, detection of the possible adverse bacterial groups in healthy individuals might not be clinically significant because of the other intact protective factors (such as mucin and intestinal immunity). Moreover, the organismal molecules from a transient leaky gut, even a severe one, might be quickly neutralized by several processes similar to those occurring in the physiological leaky gut. Therefore, measurements of the leaky gut at several time points might be necessary to identify a representative and clinically significant leaky gut in real patients, as this might differ from animal models that have less fluctuation in conditions.
Our experiments have indicated that spontaneous bacteraemia in some acute uremic mice after 48 h bilateral nephrectomy is possibly caused by intestinal apoptosis, which leads to severe leaky gut [90], again implying the importance of the gut barrier. Although the prediction of sepsis susceptibility by gut dysbiosis alone, or perhaps by the reduction in Firmicutes (or increases in Bacteroides and Proteobacteria) without leaky gut measurement, might provide limited information, several reports support some predictive properties of dysbiosis. For example, the depletion of Rosburia (phylum Firmicutes) and increases in Prevotella (Phylum Bacteroides) in the gut are identified risk factors for stroke-associated pneumonia and chronic obstructive pulmonary disease (COPD), respectively [109,110], while increases in Klebsiella variicola and Enterobacteriaceae (phylum Proteobacteria) are associated with sepsis cardiomyopathy [111]. Notably, some bacterial metabolites, mostly derived from the digestion of nutrients (such as polyamines), are small enough to pass through the normal gut barrier; however, the impact of these molecules in sepsis is not as clear as that of the larger microbial molecules (LPS, BG and cf-DNA) [112,113].
In contrast to the intact gut barrier in dysbiosis before sepsis, sepsis leads directly to gut dysbiosis together with the leaky gut and allows the translocation of microbial molecules or viable microorganisms. The viable microbial translocation from the gut are mostly bacteria rather than fungi (Candida spp.), due to the larger size of fungi than bacteria. Reduced intestinal perfusion can be recognized in the early phase of sepsis with normal blood pressure (pre-shock stage), despite systemic vasodilatation (distributive shock) and myocardial depression (partly from hyper-cytokinaemia) [114,115], by a decrease in gut microcirculation as presented by sepsis-induced ileus [116]. Because ileus can be an early sign of systemic inflammation, either from infection (sepsis) or non-infection (multiple injury or multi-organ failure; MOF), but presents with normal blood pressure, the reduced gut perfusion in sepsis and MOF might occur very early in the natural course of diseases [117]. Among several factors associated with sepsis-induced intestinal disorders [81], gut hypoperfusion is an important factor that possibly results in (i) enterocyte damage (necrosis and apoptosis) with leaky gut and (ii) intestinal immunity defects (the death of immune cells) with decreased microbial control function and increased gut dysbiosis (the selection of only highly virulent bacteria). Sepsis is accompanied by apoptosis of all immune cells (neutrophils, macrophages, dendritic cells and lymphocytes), in part, due to the overwhelming immune activation by both PAMPs from the organisms and damage-associated molecular patterns (DAMPs) arising from the death of the host’s cells [118]. This immune cell apoptosis is one of the mechanisms that induce immune exhaustion (a reduced ability to prevent other infections, leading to secondary infections) [119]. Sepsis also causes dysfunctions in multiple organs (kidney, liver, lung, spleen and nervous system) and the damage to each organ can further affect gut dysbiosis. For example, kidney and liver damage during sepsis might lead to the excretion of accumulated metabolites (toxins) into the gut, and these could directly affect enterocytes and stimulate the growth of some bacteria (such as bacteria that can metabolize these toxins), resulting in dysbiosis with leaky gut [90,120]. Likewise, sepsis can possibly alter immune responses, such as elicitation of lung-produced type I interferons, that can directly alter the gut microbiome [121], thereby possibly reducing the numbers of obligate anaerobic bacteria and increasing the proportion of Proteobacteria [122]. Similarly, alteration of the neuro-immuno-endocrine axis during sepsis might also affect gut dysbiosis [113]. Hence, sepsis induces gut dysbiosis through effects on gut hypoperfusion, immune dysregulation and organ failure.
Interestingly, some similarities are evident between sepsis that arises from several different sources of infection. This is due, in part, to common factors among the critically ill and in systemic inflammatory response conditions, including the loss of possible beneficial bacteria and microbial diversity and an increase in pathogens [123,124]. For example, faecal microbiota in children with sepsis contain higher proportions of pathogens (Acinetobacter and Enterococcus) with fewer beneficial bacteria (Roseburia, Bacteroides, Clostridia, Faecalibacterium and Blautia), and these changes closely correlate with the clinical characteristics but show negative associations with the duration of antibiotics [125]. Similarly, depletion of Lachnospiraceae, Ruminococcaceae and Ruminococcus and an enhancement of Enterococcus are demonstrated in a systemic review of sepsis [126]. Severe viral infections (COVID-19, influenza and dengue) can also increase pathogen numbers, especially Gram-negative bacteria, during sepsis and facilitate gut translocation of LPS (endotoxaemia) or viable bacteria (bacteraemia), depending on the leaky gut severity, that worsen the severity of infection [127–129] (Figure 1).
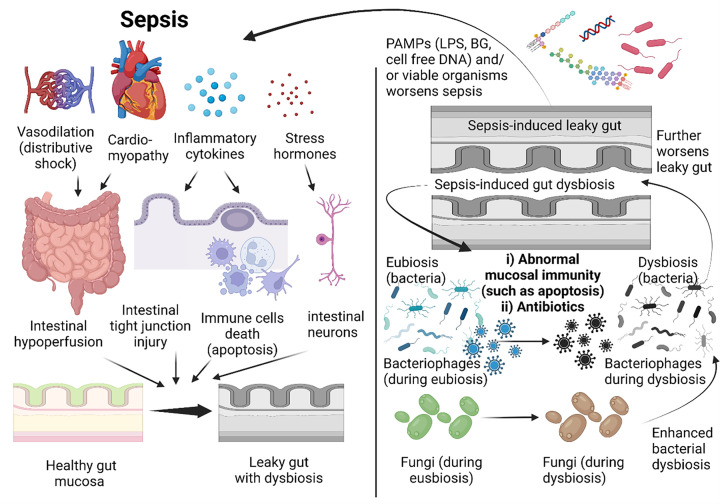
Figure 1. The alteration of all organisms (bacteria, fungi and phages) involved in sepsis and gut immunity.
Sepsis induces intestinal immunity defects, through intestinal hypoperfusion (vasodilatation and cardiomyopathy), immune cell apoptosis, the stress hormone (corticotropin)/enteric neuron-induced immune responses, and systemic inflammation, inducing gut dysbiosis (left side). In parallel, sepsis-induced gut dysbiosis, caused by intestinal immunity defect, antibiotics and alteration in fungi and phages, facilitates gut translocation of microbial molecules or viable organisms (leaky gut) causing systemic inflammation (right side) that worsen gut integrity and induce gut dysbiosis as a vicious cycle. Picture is created by BioRender.com.
The intestinal mycobiome
Despite the larger size of fungi (10–12 µm; Candida yeast) than bacteria (0.5–2 µm), fungi are the second most abundant organisms in the gut. As such, the abundance (by gene copies) is 1000-fold greater for bacteria (16S rRNA) than fungi (18S rRNA), with more than 3,500 bacterial species compared with 267 fungal species in the gut [29]. The bacterial community varies in quantity and composition from the stomach to the colon (102 vs 1011 cells/ gram content in the stomach and colon, respectively), whereas fungi seem to be localized mostly in the colon, with an average of 106 fungal cells per gram of colon content [130]. The predominant intestinal fungal mycobiota in healthy individuals are from the phyla Ascomycota (63%) (especially Candida albicans) and Basidiomycota (32%) [131], and the overgrowth of C. albicans commonly found in patients with bacterial sepsis arises in part due to antibiotic selective pressure [132]. Candida colonization in the gut is also an important risk factor for systemic candidiasis after bacterial sepsis [133]. Indeed, Candida colonization in the gut is very common in patients in intensive care units (ICUs+) [134,135], and Candida translocation from the gut into blood circulation is possible during bacterial sepsis [136,137]. Due to the lower abundance of fungi in mouse faeces than in human stool (positive culture is easier found from humans), the administration of C. albicans to mice is used to explore the importance of Candida in sepsis. Despite its lower abundance, the presence of Candida in the gut enhances some bacterial species (such as Pseudomonas spp.) [44,53], partly due to glucan digestion, as mixing glucan into the culture medium enhances the growth of isolated bacteria [39]. Interestingly, the fungal–bacterial interaction is complex and might depend on the time frame of the exposure, as the incubation of a clinical strain of Pseudomonas aeruginosa with C. albicans has no synergy on biofilm production, whereas the addition of the fungi onto Pseudomonas biofilms or onto cell lines facilitates more biofilm production [138,139]. Nevertheless, increases in Candida in the gut during sepsis possibly worsens the severity of bacterial sepsis through several pathways, including a higher translocation of BG (Candida increases the BG gut content), increases in invasive bacteria in the gut and direct injury to enterocytes (perhaps from the Candida germ tube or mucosal immune responses against fungi) [44,77]. Notably, the co-presentation of LPS and BG synergistically activates macrophage immune responses, in part through the simultaneous activation of TLR-4 and dectin-1 by LPS and BG, respectively [45,46,85].
Despite the lack of information regarding the gut mycobiota in patients with sepsis, septic mice demonstrate subtle changes in gut fungi (the abundance of fungal 18sRNA by PCR in sepsis is different from the control group), including a reduction of only Myrothecium spp. fungi that can produce some molecules against several harmful factors (some organisms and toxic substances) [15]. The differences in sepsis conditions between humans and mice [140,141] raise the possibility that gut fungi in patients with sepsis might be enhanced by several factors that differ from those in mice, such as the duration of sepsis (human patients survive longer than mice), antibiotic use (more potent in human conditions), intensive care unit (ICU) environment (nosocomial infections are likelier in patients in ICUs than in mice in controlled animal facilities) and naturally higher Candida levels in human faeces and underlying diseases (such as altered gut fungi in Type 2 diabetes) [142–144]. Based on the well-established increase in gut fungi in patients with IBD and alcohol ingestion [56–58], intestinal inflammation and reduced mucosal immunity might be important exacerbating factors for the enhancement of gut fungi associated with sepsis (systemic cytokine-induced intestinal barrier defects and apoptosis of immune cells) [65,118]. More exploration of gut fungi in patients with bacterial sepsis will be interesting. Of note, the identification of mycobiota at the phylum level might provide only limited information because Ascomycota predominate; therefore, faecal microbiome analysis might be necessary to explore fungal population in faeces.
The intestinal virobiome
Currently, viruses in the gut are not included as ‘gut microbiota’, as viruses are intracellular organisms and the presence of viruses in enterocytes will be categorized as a viral infection. However, bacteriophages, which are viruses (or genomes) of the gut bacteria, might be considered a group of viruses that can be found in the gut content and categorized as ‘gut microbiota’ because alteration in gut bacteria will automatically change the abundance of bacteriophages (or phages). Phages are specific to the species level of bacteria, partly because of different routes of entry, and phages of the same bacteria might have different responses to different bacterial isolates [145]. For example, the effective phages against P. aeruginosa from Person A might have no effect against P. aeruginosa from Person B. This will necessitate a tremendous accumulation of phage information (phage library) for any real clinical use [146].
The bacteriophage cycle is categorized into lysogenic and lytic patterns. The lysogenic cycle involves the insertion of viral genetic materials into the bacterial genome for replication together with the bacteria. These phages are referred to as ‘temperate phages or prophages’ and can be transferred to several bacterial generations without any viral gene expression. By contrast, the lytic cycle is a switch from the lysogenic phase to the release of new viral particles [147,148]. Because phages are one of the natural controls against bacteria [149] and because prophages can pass through several generations of bacteria before being induced (e.g. by stress) into lytic phages and killing the bacteria [150], any alteration in the bacterial microbiome during sepsis might automatically induce changes in virota (virome). Indeed, the faecal virota from septic mice demonstrates an alteration in the abundance of several groups of bacteriophages, including Myoviridae (in sham mice) and Podoviridae (in septic mice), which are components of several phage cocktails used in other studies [15]. The observation that viral particles isolated from faeces of a septic mouse can attenuate sepsis in another mouse [15] raises the possibility that bacterial stress during sepsis activates lytic phages that might be able to control some sepsis-induced pathogenic bacteria. Phages accumulating within the mucosal layer can be a barrier to bacterial invasion; however, bacteria that express phage-encoded proteins can show increased virulence (epithelial invasion, adhesion, antibiotic resistance, phagocytosis blockage and biofilm formation) and the transport of phages by transcytosis of phage particles and/or apical-basal transport may deliver phages into the circulation and enhance inflammatory responses [151,152]. Unfortunately, studies on gut virota (or phageomes), especially in sepsis, are scarce.
Adjunctive therapies
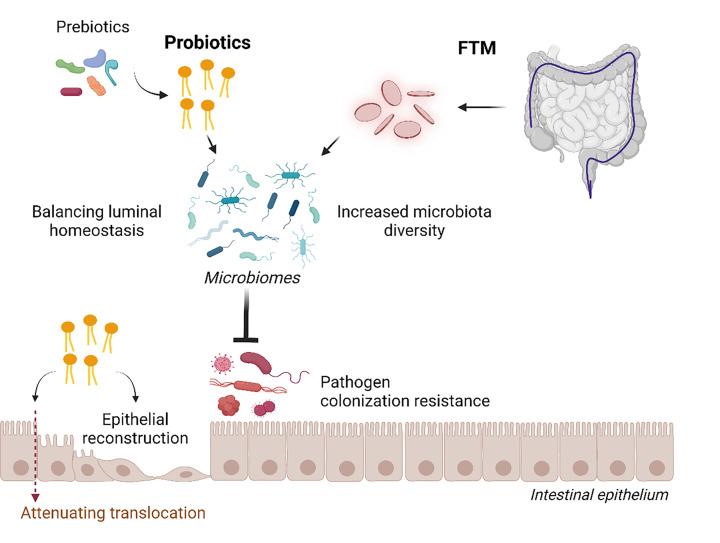
Due to the possible correlation between gut dysbiosis and sepsis severity, the manipulation of the gut microbiome (and gut barriers) might prevent gut-origin sepsis or attenuate sepsis severity by strengthening the gut barrier, reducing gut pathogens, reducing the PAMP content (LPS and BG) in the gut and eliciting direct anti-inflammatory responses. The normalization of gut microbiota by several methods, including faecal transplantation (administration of healthy microbiota), probiotics (beneficial bacteria) (Figure 2), prebiotics (probiotic-enhancing substances) and synbiotics (probiotics with prebiotics), has been tested in sepsis.
Figure 2. The adjunctive therapy of prebiotics, probiotics and FMT in terms of intestinal permeability effects.
All of these strategies improve the balance of gut microbiota with increased organismal diversity that is beneficial to the host through reduced pathogenic microbes, strengthens the gut barrier and induces gut epithelial reconstruction.Picture is created by BioRender.com
Faecal microbiota transplantation
Several animal studies and case series have reported the ability of faecal microbiota transplantation (FMT) to attenuate sepsis severity, in part through the restoration of butyrate-producing bacteria, gut barrier strengthening, innate immunity enhancement, immune repertoire alteration and pathogen clearance; however, some studies have reported lethal bacteraemia [153]. Notably, the immune repertoire is a variety of receptors on T cells and B cells that has a large sequence diversity to recognize different organismal molecules as a part of the adaptive immune system [154] and innate immunity, for example, macrophages, is an important host response against pathogenic gut organisms [74]. Meanwhile, butyrate is an important short-chain fatty acid that is categorized as an enterocyte energy source and a factor of anti-inflammation and anti-malignancy [155]. Then, FMT administration seems to enhance the effectiveness of organismal control in the host through improved innate and adaptive immunity together with strengthened enterocyte integrity that will be beneficial in sepsis [156]. Clostridium difficile seems to be the first pathogen with FMT clinical implications. C. difficile is classified as a Gram-positive bacterial pathogenic cause of infectious colitis that frequently arises following excessive antibiotic use [157]. C. difficile contributes to complications of antibiotic therapy owing to its recurrent infections. Interestingly, the use of FMT by either oral pills or FMT colonoscopy in patients with recurrent C. difficile showed promising outcomes (96.2% and 96.1% of patients were cured after 12-week treatments by oral FMT and colonoscopy FMT, respectively) [158]. The more updated implications of FMT now involve its applications as cancer therapeutic. This potential as a therapy was first observed in mice with cancer but no microbiome, as these animals demonstrated a different response when treated with anticancer drugs, including cisplatin, cyclophosphamide, and anti-programmed cell death 1 protein (PD-1) immunotherapy [159,160]. These findings are also supported by evidence that Enterococcus faecalis is able to directly metabolize levodopa [161]. As such, using gut microbiota in conjunction with drugs could benefit the balance of gut microbes, thereby simultaneously suppressing gut pathogens during certain disease treatments. Nevertheless, in mid-2019, the U.S. Food and Drug Administration (FDA) announced that FMT therapy should be used with serious caution, based on a mortality case report of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli infection [162]. As a result, the FDA has posted a warning statement that in-depth screening for all resistant pathogens must be performed before FMT.
Probiotics
In contrast to the possibly severe side effects of FMT in sepsis treatment, the adverse effects of probiotics are usually minimal, as most probiotics are anaerobes and anaerobic bacteraemia is not normally severe and is easier to treat compared with aerobic bacteriaemia [163]. Probiotics consist of PAMPs; thus, gut translocation of probiotics or their components can activate innate immune responses. Therefore, the administration of probiotics to immunocompromised individuals or those of extreme age, critically ill or with severe leaky gut could cause bacteraemia [164,165]. In some conditions, with appropriate probiotics, the leaky gut might be advantageous because some relatively large beneficial molecules from probiotics might possibly be transported through the damaged gut barrier [54,55]. Probiotics potentiate colonization resistance through thier functions of reduced luminal pH, antimicrobial properties, and competing for nutrients and adhesion surface [36,37]. Indeed, some strains of Lactobacillus and Bifidobacterium produce some exopolysaccharides with immunomodulatory effects [166,167], while also reducing pathogens by nutrient competition, quorum sensing antagonists and production of substances that directly inhibit bacteria [168]. Several bacterial strains are choices for probiotics, but some bacteria might be more harmful than others. For example, enterococci can cause endocarditis in some conditions, while lactobacilli and Bifidobacterium are easily treatable [169]. Probiotics also enhance gut barrier function through mucin production and tight junction proteins. Now, probiotics are extended to other uses, including skin protection from various host pathogens, such as Staphylococcus, Corynebacterium and Propionibacterium, but this use can lead to the development of skin immune disorientation conditions, such as rosacea [170]. Interestingly, local application of probiotics improved skin colonization by Cutibacterium acnes [171]. In addition, oral forms of probiotics, such as Lactobacillus reuteri, demonstrated an ability to attenuate perifollicular inflammation by promoting a gut–brain–skin (GBS) axis [172].
Prebiotics
The rationale for the use of prebiotics in leaky gut syndrome is certain dietary components might promote the growth of certain gut bacteria strains that are closely associated with health benefits for the host [173]. Prebiotics are not only the food components non-digestible by the host that promote the fermenting bacteria in the colon [174] but also are nutrients degraded by the gastrointestinal microbiota that alter the microbiome's activity and composition [175]. Many kinds of dietary nutrients are termed prebiotics under these categorizations, especially the commercially available carbohydrate-based dietary fibers (polymers of monosaccharides), which are fermented by intestinal microorganisms. These nutrients are digested to produce several molecules, such as SCFAs and peptidoglycan, which affect the innate immune system [176]. Prebiotics may enhance insulin resistance and glucose tolerance [177] and reduce intestinal inflammation, endotoxemia, and cytokines which might be beneficial in sepsis. As such, desaminotyrosine (DAT) maintains mucosal immunological homeostasis and barrier integrity, and reduces mucosal inflammation in DSS-induced endotoxemia and septic shock in rodents [178]. Some prebiotics from the Chinese herbs, Xuanbai Chengqi decoction (XBCQ), also attenuates pulmonary infection in rodents through the improved gut barrier function and promoted survival [179,180]. In addition, Finger Millet arabinoxylan (FM-AX), a non-starch polysaccharide produced from cereals, demonstrates attenuates endotoxemia in mice via reduction of high-fat diet-induced leaky gut [181]. In human studies, prebiotics reduces the incidence of sepsis, mortality, and length of hospital stay in premature infants [182]. While the preparation cost for FMT and probiotics is usually high with sophisticated technology due to the management of the viable organisms, prebiotics preparation seems to be less expensive with, perhaps longer shelf life. However, prebiotics cannot promote the growth of bacteria that do not present in the gut, and most commercially available products are a combination of prebiotics with probiotics. Due to the less expensive process of preparation, the prebiotics, a single or in combination, selectively promotes the growth of beneficial bacteria that commonly found in the host in sepsis is interesting. More studies are warranted.
Conclusions
Gut leakage and changes in the intestinal microbiome in sepsis are the consequence of intestinal immunity defects caused by intestinal hypoperfusion, immune cell apoptosis, and enteric neuro-humoral-immunity responses. The increased abundance of pathogens in the bacterial microbiome associated with a leaky gut may result in the translocation of microbial molecules and even viable microorganisms, eventually worsening the course of sepsis. Despite several previous reviews on gut microbiota in sepsis [183–186], the collection of data on gut mycobiome (fungiome) and virobiome is usually limited to the non-sepsis condition [187–190] and the review of gut microbiome together with leaky gut in sepsis is still less. Here, the close correlation between gut microbiota (bacteria, fungi and viruses) and sepsis severity also suggests that attenuation of leaky gut and gut dysbiosis might be a target of future adjunctive therapies. Moreover, the role of virome, mycobiome, as well as novel metagenomics of microbial identification must be in the pipeline of the future research areas and are urgently needed fields.
Abbreviations
- DAMP
damage-associated molecular pattern
- DAT
desaminotyrosine
- ESBL
extended-spectrum β-lactamase
- FM-AX
Finger Millet arabinoxylan
- FMT
faecal microbiota transplantation
- GBS
gut–brain–skin
- NSAIDs
non-steroidal anti-inflammatory drugs
- XBCQ
Xuanbai Chengqi decoction
Contributor Information
Wiwat Chancharoenthana, Email: wiwat.cha@mahidol.ac.th.
Marcus J. Schultz, Email: marcus.j.schultz@gmail.com.
Asada Leelahavanichkul, Email: aleelahavanit@gmail.com.
Data Availability
Data sharing is not applicable to the review.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This research project is supported by Mahidol University (MU’s Strategic Research Fund): fiscal year 2023.
CRediT Author Contribution
Wiwat Chancharoenthana: Conceptualization, Resources, Data curation, Formal analysis, Funding acquisition, Validation, Visualization, Writing—original draft, Project administration, Writing—review & editing. Supitcha Kamolratanakul: Conceptualization, Resources, Data curation, Formal analysis, Writing—original draft, Project administration, Writing—review & editing. Marcus J. Schultz: Conceptualization, Supervision, Validation, Visualization, Writing—original draft, Writing—review & editing. Asada Leelahavanichkul: Conceptualization, Resources, Data curation, Formal analysis, Supervision, Validation, Visualization, Writing—original draft, Writing—review & editing.
References
- 1.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M.et al. (2016) The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 315, 801–810 10.1001/jama.2016.0287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudd K.E., Johnson S.C., Agesa K.M., Shackelford K.A., Tsoi D., Kievlan D.R.et al. (2020) Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the Global Burden of Disease Study. Lancet 395, 200–211 10.1016/S0140-6736(19)32989-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincent J.L., Jones G., David S., Olariu E. and Cadwell K.K. (2019) Frequency and mortality of septic shock in Europe and North America: a systematic review and meta-analysis. Crit. Care 23, 196 10.1186/s13054-019-2478-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krishnan A. and Karnad D.R. (2003) Severe falciparum malaria: an important cause of multiple organ failure in Indian intensive care unit patients. Crit. Care Med. 31, 2278–2284 10.1097/01.CCM.0000079603.82822.69 [DOI] [PubMed] [Google Scholar]
- 5.Teparrukkul P., Hantrakun V., Day N.P.J., West T.E. and Limmathurotsakul D. (2017) Management and outcomes of severe dengue patients presenting with sepsis in a tropical country. PloS ONE 12, e0176233 10.1371/journal.pone.0176233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lepak A. and Andes D. (2011) Fungal sepsis: optimizing antifungal therapy in the critical care setting. Crit. Care Clin. 27, 123–147 10.1016/j.ccc.2010.11.001 [DOI] [PubMed] [Google Scholar]
- 7.Reddy P. (2022) Clinical approach to nosocomial bacterial sepsis. Cureus 14, e28601 10.7759/cureus.28601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makjaroen J., Thim-Uam A., Dang C.P., Pisitkun T., Somparn P. and Leelahavanichkul A. (2021) A comparison between 1 day versus 7 days of sepsis in mice with the experiments on LPS-activated macrophages support the use of intravenous immunoglobulin for sepsis attenuation. J. Inflamm. Res. 14, 7243–7263 10.2147/JIR.S338383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gentile L.F. and Moldawer L.L. (2013) DAMPs, PAMPs, and the origins of SIRS in bacterial sepsis. Shock 39, 113–114 10.1097/SHK.0b013e318277109c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Németh K., Leelahavanichkul A., Yuen P.S., Mayer B., Parmelee A., Doi K.et al. (2009) Bone marrow stromal cells attenuate sepsis via prostaglandin E(2)-dependent reprogramming of host macrophages to increase their interleukin-10 production. Nat. Med. 15, 42–49 10.1038/nm.1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yokoyama Y., Kino J., Okazaki K. and Yamamoto Y. (1994) Mycobacteria in the human intestine. Gut 35, 715–716 10.1136/gut.35.5.715-b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zivkovic S., Ayazi M., Hammel G. and Ren Y. (2021) For better or for worse: a look into neutrophils in traumatic spinal cord injury. Front Cell Neurosci. 15, 648076 10.3389/fncel.2021.648076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakayama H., Kurokawa K. and Lee B.L. (2012) Lipoproteins in bacteria: structures and biosynthetic pathways. FEBS J. 279, 4247–4268 10.1111/febs.12041 [DOI] [PubMed] [Google Scholar]
- 14.Charoensappakit A., Sae-Khow K. and Leelahavanichkul A. (2022) Gut barrier damage and gut translocation of pathogen molecules in lupus, an impact of innate immunity (macrophages and neutrophils) in autoimmune disease. Int. J. Mol. Sci. 23, 8223 10.3390/ijms23158223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chancharoenthana W., Sutnu N., Visitchanakun P., Sawaswong V., Chitcharoen S., Payungporn S.et al. (2022) Critical roles of sepsis-reshaped fecal virota in attenuating sepsis severity. Front Immunol. 13, 940935 10.3389/fimmu.2022.940935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fay K.T., Ford M.L. and Coopersmith C.M. (2017) The intestinal microenvironment in sepsis. Biochim. Biophys. Acta Mol. Basis Dis. 1863, 2574–2583 10.1016/j.bbadis.2017.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leelahavanichkul A., Worasilchai N., Wannalerdsakun S., Jutivorakool K., Somparn P., Issara-Amphorn J.et al. (2016) Gastrointestinal leakage detected by serum (1→3)-β-D-Glucan in mouse models and a pilot study in patients with sepsis. Shock 46, 506–518 10.1097/SHK.0000000000000645 [DOI] [PubMed] [Google Scholar]
- 18.Panpetch W., Visitchanakun P., Saisorn W., Sawatpanich A., Chatthanathon P., Somboonna N.et al. (2021) Lactobacillus rhamnosus attenuates Thai chili extracts induced gut inflammation and dysbiosis despite capsaicin bactericidal effect against the probiotics, a possible toxicity of high dose capsaicin. PloS ONE 16, e0261189 10.1371/journal.pone.0261189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhunyakarnjanarat T., Udompornpitak K., Saisorn W., Chantraprapawat B., Visitchanakun P., Dang C.P.et al. (2021) Prominent Indomethacin-Induced Enteropathy in Fcgriib Defi-cient lupus Mice: an impact of macrophage responses and immune deposition in gut. Int. J. Mol. Sci. 22, 1377 10.3390/ijms22031377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ribeiro F.M., Petriz B., Marques G., Kamilla L.H. and Franco O.L. (2021) Is there an exercise-intensity threshold capable of avoiding the leaky gut? Front. Nutr. 8, 627289 10.3389/fnut.2021.627289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Issara-Amphorn J., Somboonna N., Pisitkun P., Hirankarn N. and Leelahavanichkul A. (2020) Syk inhibitor attenuates inflammation in lupus mice from FcgRIIb deficiency but not in pristane induction: the influence of lupus pathogenesis on the therapeutic effect. Lupus 29, 1248–1262 10.1177/0961203320941106 [DOI] [PubMed] [Google Scholar]
- 22.Tungsanga S., Udompornpitak K., Worasilchai J., Ratana-Aneckchai T., Wannigama D.L., Katavetin P.et al. (2022) Candida Administration in 5/6 Nephrectomized Mice Enhanced Fibrosis in Internal Organs: An Impact of Lipopolysaccharide and (1→3)-β-D-Glucan from Leaky Gut. Int. J. Mol. Sci. 23, 15987 10.3390/ijms232415987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Udompornpitak K., Charoensappakit A., Sae-Khow K., Bhunyakarnjanarat T., Dang C.P., Saisorn W.et al. (2022) Obesity Exacerbates Lupus Activity in Fc Gamma Receptor IIb Deficient Lupus Mice Partly through Saturated Fatty Acid-Induced Gut Barrier Defect and Systemic Inflammation. J. Innate Immun. 1–22 10.1159/000526206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panpetch W., Somboonna N., Palasuk M., Hiengrach P., Finkelman M., Tumwasorn S.et al. (2019) Oral Candida administration in a Clostridium difficile mouse model worsens disease severity but is attenuated by Bifidobacterium. PloS ONE 14, e0210798 10.1371/journal.pone.0210798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madison A. and Kiecolt-Glaser J.K. (2019) Stress, depression, diet, and the gut microbiota: human-bacteria interactions at the core of psychoneuroimmunology and nutrition. Curr. Opin. Behav. Sci. 28, 105–110 10.1016/j.cobeha.2019.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deitch E.A. (2012) Gut-origin sepsis: evolution of a concept. Surgeon 10, 350–356 10.1016/j.surge.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alverdy J., Holbrook C., Rocha F., Seiden L., Wu R.L., Musch M.et al. (2000) Gut-derived sepsis occurs when the right pathogen with the right virulence genes meets the right host: evidence for in vivo virulence expression in Pseudomonas aeruginosa. Ann. Surg. 232, 480–489 10.1097/00000658-200010000-00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helander H.F. and Fändriks L. (2014) Surface area of the digestive tract - revisited. Scand. J. Gastroenterol. 49, 681–689 10.3109/00365521.2014.898326 [DOI] [PubMed] [Google Scholar]
- 29.Amornphimoltham P., Yuen P.S.T., Star R.A. and Leelahavanichkul A. (2019) Gut leakage of fungal-derived inflammatory mediators: part of a gut-liver-kidney axis in bacterial sepsis. Dig. Dis. Sci. 64, 2416–2428 10.1007/s10620-019-05581-y [DOI] [PubMed] [Google Scholar]
- 30.Vojdani A. (2013) For the assessment of intestinal permeability, size matters. Altern. Ther. Health. Med. 19, 12–24 [PubMed] [Google Scholar]
- 31.Dlugosz A., Winckler B., Lundin E., Zakikhany K., Sandström G., Ye W.et al. (2015) No difference in small bowel microbiota between patients with irritable bowel syndrome and healthy controls. Sci. Rep. 5, 8508 10.1038/srep08508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanholder R., De Smet R. and Lesaffer G. (1999) p-cresol: a toxin revealing many neglected but relevant aspects of uraemic toxicity. Nephrol. Dial. Transplant. 14, 2813–2815 10.1093/ndt/14.12.2813 [DOI] [PubMed] [Google Scholar]
- 33.Williams D., Trimble W.L., Shilts M., Meyer F. and Ochman H. (2013) Rapid quantification of sequence repeats to resolve the size, structure and contents of bacterial genomes. BMC Genomics 14, 537 10.1186/1471-2164-14-537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaewduangduen W., Visitchanakun P., Saisorn W., Phawadee A., Manonitnantawat C., Chutimaskul C.et al. (2022) Blood bacteria-free DNA in septic mice enhances LPS-induced inflammation in mice through macrophage response. Int. J. Mol. Sci. 23, 1907 10.3390/ijms23031907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chancharoenthana W., Kamolratanakul S., Ariyanon W., Thanachartwet V., Phumratanaprapin W., Wilairatana P.et al. (2022) Abnormal blood bacteriome, gut dysbiosis, and progression to severe dengue disease. Front Cell Infect. Microbiol. 12, 890817 10.3389/fcimb.2022.890817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chancharoenthana W., Leelahavanichkul A., Ariyanon W., Vadcharavivad S., Phatcharophaswattanakul S., Kamolratanakul S.et al. (2021) Leaky gut syndrome is associated with endotoxemia and serum (1→3)-β-D-glucan in severe dengue infection. Microorganisms 9, 2390 10.3390/microorganisms9112390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panpetch W., Chancharoenthana W., Bootdee K., Nilgate S., Finkelman M., Tumwasorn S.et al. (2018) Lactobacillus rhamnosus L34 Attenuates Gut Translocation-Induced Bacterial Sepsis in Murine Models of Leaky Gut. Infect. Immun. 86, e00700–e00717 10.1128/IAI.00700-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koh Y.Y., Jeon W.K., Cho Y.K., Kim H.J., Chung W.G., Chon C.U.et al. (2012) The effect of intestinal permeability and endotoxemia on the prognosis of acute pancreatitis. Gut Liver 6, 505–511 10.5009/gnl.2012.6.4.505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiengrach P., Panpetch W., Worasilchai N., Chindamporn A., Tumwasorn S., Jaroonwitchawan T.et al. (2020) Administration of Candida Albicans to Dextran Sulfate Solution Treated Mice Causes Intestinal Dysbiosis, Emergence and Dissemination of Intestinal Pseudomonas Aeruginosa and Lethal Sepsis. Shock 53, 189–198 10.1097/SHK.0000000000001339 [DOI] [PubMed] [Google Scholar]
- 40.Thim-Uam A., Surawut S., Issara-Amphorn J., Jaroonwitchawan T., Hiengrach P., Chatthanathon P.et al. (2020) Leaky-gut enhanced lupus progression in the Fc gamma receptor-IIb deficient and pristane-induced mouse models of lupus. Sci. Rep. 10, 777 10.1038/s41598-019-57275-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y. and Chen Y.G. (2020) Intestinal epithelial plasticity and regeneration via cell dedifferentiation. Cell Regen 9, 14 10.1186/s13619-020-00053-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Huang B., Jin T., Ocansey D.K.W., Jiang J. and Mao F. (2022) Intestinal fibrosis in inflammatory bowel disease and the prospects of mesenchymal stem cell therapy. Front. Immunol. 13, 835005 10.3389/fimmu.2022.835005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hiengrach P., Visitchanakun P., Finkelman M.A., Chancharoenthana W. and Leelahavanichkul A. (2022) More prominent inflammatory response to pachyman than to whole-glucan particle and oat-β-glucans in dextran sulfate-induced mucositis mice and mouse injection through proinflammatory macrophages. Int. J. Mol. Sci. 23, 4026 10.3390/ijms23074026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panpetch W., Somboonna N., Bulan D.E., Issara-Amphorn J., Worasilchai N., Finkelman M.et al. (2018) Gastrointestinal Colonization of Candida Albicans Increases Serum (1→3)-β-D-Glucan, without Candidemia, and Worsens Cecal Ligation and Puncture Sepsis in Murine Model. Shock 49, 62–70 10.1097/SHK.0000000000000896 [DOI] [PubMed] [Google Scholar]
- 45.Panpetch W., Somboonna N., Bulan D.E., Issara-Amphorn J., Finkelman M., Worasilchai N.et al. (2017) Oral administration of live- or heat-killed Candida albicans worsened cecal ligation and puncture sepsis in a murine model possibly due to an increased serum (1→3)-β-D-glucan. PloS ONE 12, e0181439 10.1371/journal.pone.0181439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Issara-Amphorn J., Surawut S., Worasilchai N., Thim-Uam A., Finkelman M., Chindamporn A.et al. (2018) The Synergy of Endotoxin and (1→3)-β-D-Glucan, from Gut Translocation, Worsens Sepsis Severity in a Lupus Model of Fc Gamma Receptor IIb-Deficient Mice. J. Innate Immun. 10, 189–201 10.1159/000486321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sae-Khow K., Charoensappakit A., Visitchanakun P., Saisorn W., Svasti S., Fucharoen S.et al. (2020) Pathogen-associated molecules from gut translocation enhance severity of cecal ligation and puncture sepsis in iron-overload β-thalassemia mice. J. Inflamm. Res. 13, 719–735 10.2147/JIR.S273329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Panpetch W., Sawaswong V., Chanchaem P., Ondee T., Dang C.P., Payungporn S.et al. (2020) Candida administration worsens cecal ligation and puncture-induced sepsis in obese mice through gut dysbiosis enhanced systemic inflammation, impact of pathogen-associated molecules from gut translocation and saturated fatty acid. Front. Immunol. 11, 561652 10.3389/fimmu.2020.561652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bates J.M., Akerlund J., Mittge E. and Guillemin K. (2007) Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe 2, 371–382 10.1016/j.chom.2007.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zou B., Jiang W., Han H., Li J., Mao W., Tang Z.et al. (2017) Acyloxyacyl hydrolase promotes the resolution of lipopolysaccharide-induced acute lung injury. PLoS Pathog. 13, e1006436 10.1371/journal.ppat.1006436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feulner J.A., Lu M., Shelton J.M., Zhang M., Richardson J.A. and Munford R.S. (2004) Identification of acyloxyacyl hydrolase, a lipopolysaccharide-detoxifying enzyme, in the murine urinary tract. Infect. Immun. 72, 3171–3178 10.1128/IAI.72.6.3171-3178.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramendra R., Isnard S., Mehraj V., Chen J., Zhang Y., Finkelman M.et al. (2019) Circulating LPS and (1→3)-β-D-Glucan: A Folie à Deux Contributing to HIV-Associated Immune Activation. Front. Immunol. 10, 465 10.3389/fimmu.2019.00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panpetch W., Hiengrach P., Nilgate S., Tumwasorn S., Somboonna N., Wilantho A.et al. (2020) Additional Candida albicans administration enhances the severity of dextran sulfate solution induced colitis mouse model through leaky gut-enhanced systemic inflammation and gut-dysbiosis but attenuated by Lactobacillus rhamnosus L34. Gut Microbes 11, 465–480 10.1080/19490976.2019.1662712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tungsanga S., Katavetin P., Panpetch W., Udompornpitak K., Saisorn W., Praditpornsilpa K.et al. (2022) Lactobacillus rhamnosus L34 attenuates chronic kidney disease progression in a 5/6 nephrectomy mouse model through the excretion of anti-inflammatory molecules. Nephrol. Dial. Transplant. 37, 1429–1442 10.1093/ndt/gfac032 [DOI] [PubMed] [Google Scholar]
- 55.Ondee T., Pongpirul K., Visitchanakun P., Saisorn W., Kanacharoen S., Wongsaroj L.et al. (2021) Lactobacillus acidophilus LA5 improves saturated fat-induced obesity mouse model through the enhanced intestinal Akkermansia muciniphila. Sci. Rep. 11, 6367 10.1038/s41598-021-85449-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Underhill D.M. and Braun J. (2022) Fungal microbiome in inflammatory bowel disease: a critical assessment. J. Clin. Invest. 132, e155786 10.1172/JCI155786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sokol H., Leducq V., Aschard H., Pham H.P., Jegou S., Landman C.et al. (2017) Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048 10.1136/gutjnl-2015-310746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang A.M., Inamine T., Hochrath K., Chen P., Wang L., Llorente C.et al. (2017) Intestinal fungi contribute to development of alcoholic liver disease. J. Clin. Invest. 127, 2829–2841 10.1172/JCI90562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leelahavanichkul A., Panpetch W., Worasilchai N., Somparn P., Chancharoenthana W., Nilgate S.et al. (2016) Evaluation of gastrointestinal leakage using serum (1→3)-β-D-glucan in a Clostridium difficile murine model. FEMS Microbiol. Lett. 363, fnw204 10.1093/femsle/fnw204 [DOI] [PubMed] [Google Scholar]
- 60.Panpetch W., Phuengmaung P., Cheibchalard T., Somboonna N., Leelahavanichkul A. and Tumwasorn S. (2021) Lacticaseibacillus casei Strain T21 Attenuates Clostridioides difficile Infection in a Murine Model Through Reduction of Inflammation and Gut Dysbiosis With Decreased Toxin Lethality and Enhanced Mucin Production. Front Microbiol. 12, 745299 10.3389/fmicb.2021.745299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Michielan A. and D'Incà R. (2015) Intestinal permeability in inflammatory bowel disease: pathogenesis, clinical evaluation, and therapy of leaky gut. Mediators Inflamm. 2015, 628157 10.1155/2015/628157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hietbrink F., Besselink M.G., Renooij W., de Smet M.B., Draisma A., van der Hoeven H.et al. (2009) Systemic inflammation increases intestinal permeability during experimental human endotoxemia. Shock 32, 374–378 10.1097/SHK.0b013e3181a2bcd6 [DOI] [PubMed] [Google Scholar]
- 63.Zeng M.Y., Inohara N. and Nuñez G. (2017) Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 10, 18–26 10.1038/mi.2016.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thim-Uam A., Makjaroen J., Issara-Amphorn J., Saisorn W., Wannigama D.L., Chancharoenthana W.et al. (2022) Enhanced Bacteremia in Dextran Sulfate-Induced Colitis in Splenectomy Mice Correlates with Gut Dysbiosis and LPS Tolerance. Int. J. Mol. Sci. 23, 1676 10.3390/ijms23031676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tazuke Y., Drongowski R.A., Teitelbaum D.H. and Coran A.G. (2003) Interleukin-6 changes tight junction permeability and intracellular phospholipid content in a human enterocyte cell culture model. Pediatr. Surg. Int. 19, 321–325 10.1007/s00383-003-1003-8 [DOI] [PubMed] [Google Scholar]
- 66.Freestone P.P., Williams P.H., Haigh R.D., Maggs A.F., Neal C.P. and Lyte M. (2002) Growth stimulation of intestinal commensal Escherichia coli by catecholamines: a possible contributory factor in trauma-induced sepsis. Shock 18, 465–470 10.1097/00024382-200211000-00014 [DOI] [PubMed] [Google Scholar]
- 67.Million M. and Larauche M. (2016) Stress, sex, and the enteric nervous system. Neurogastroenterol. Motil. 28, 1283–1289 10.1111/nmo.12937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vu C.T.B., Thammahong A., Yagita H., Azuma M., Hirankarn N., Ritprajak P.et al. (2020) Blockade Of PD-1 Attenuated Postsepsis Aspergillosis Via The Activation of IFN-γ and The Dampening of IL-10. Shock 53, 514–524 10.1097/SHK.0000000000001392 [DOI] [PubMed] [Google Scholar]
- 69.Vu C.T.B., Thammahong A., Leelahavanichkul A. and Ritprajak P. (2022) Alteration of macrophage immune phenotype in a murine sepsis model is associated with susceptibility to secondary fungal infection. Asian Pac. J. Allergy Immunol. 40, 162–171 [DOI] [PubMed] [Google Scholar]
- 70.Sae-Khow K., Charoensappakit A., Chiewchengchol D. and Leelahavanichkul A. (2022) High-Dose Intravenous Ascorbate in Sepsis, a Pro-Oxidant Enhanced Microbicidal Activity and the Effect on Neutrophil Functions. Biomedicines 11, 51 10.3390/biomedicines11010051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vangay P., Johnson A.J., Ward T.L., Al-Ghalith G.A., Shields-Cutler R.R., Hillmann B.M.et al. (2018) US immigration westernizes the human gut microbiome. Cell 175, 962.e10–972.e10 10.1016/j.cell.2018.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ghosh T.S., Shanahan F. and O'Toole P.W. (2022) The gut microbiome as a modulator of healthy ageing. Nat. Rev. Gastroenterol. Hepatol. 19, 565–584 10.1038/s41575-022-00605-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ragonnaud E. and Biragyn A. (2021) Gut microbiota as the key controllers of “healthy” aging of elderly people. Immun. Ageing 18, 2 10.1186/s12979-020-00213-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hiengrach P., Panpetch W., Chindamporn A. and Leelahavanichkul A. (2022) Macrophage depletion alters bacterial gut microbiota partly through fungal overgrowth in feces that worsens cecal ligation and puncture sepsis mice. Sci. Rep. 12, 9345 10.1038/s41598-022-13098-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haak B.W., Lankelma J.M., Hugenholtz F., Belzer C., de Vos W.M. and Wiersinga W.J. (2019) Long-term impact of oral vancomycin, ciprofloxacin and metronidazole on the gut microbiota in healthy humans. J. Antimicrob. Chemother. 74, 782–786 10.1093/jac/dky471 [DOI] [PubMed] [Google Scholar]
- 76.Diamond E., Hewlett K., Penumutchu S., Belenky A. and Belenky P. (2021) Coffee consumption modulates amoxicillin-induced dysbiosis in the murine gut microbiome. Front. Microbiol. 12, 637282 10.3389/fmicb.2021.637282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Panpetch W., Phuengmaung P., Hiengrach P., Issara-Amphorn J., Cheibchalard T., Somboonna N.et al. (2022) Candida Worsens Klebsiella pneumoniae Induced-Sepsis in a Mouse Model with Low Dose Dextran Sulfate Solution through Gut Dysbiosis and Enhanced Inflammation. Int. J. Mol. Sci. 23, 7050 10.3390/ijms23137050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bantel H. and Schulze-Osthoff K. (2009) Cell death in sepsis: a matter of how, when, and where. Crit. Care 13, 173 10.1186/cc7966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Campos-Rodríguez R., Godínez-Victoria M., Abarca-Rojano E., Pacheco-Yépez J., Reyna-Garfias H., Barbosa-Cabrera R.E.et al. (2013) Stress modulates intestinal secretory immunoglobulin A. Front. Integr. Neurosci. 7, 86 10.3389/fnint.2013.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller W.D., Keskey R. and Alverdy J.C. (2021) Sepsis and the Microbiome: A Vicious Cycle. J. Infect. Dis. 223, S264–S269 10.1093/infdis/jiaa682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haussner F., Chakraborty S., Halbgebauer R. and Huber-Lang M. (2019) Challenge to the Intestinal Mucosa During Sepsis. Front. Immunol. 10, 891 10.3389/fimmu.2019.00891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saithong S., Worasilchai N., Saisorn W., Udompornpitak K., Bhunyakarnjanarat T., Chindamporn A.et al. (2022) Neutrophil Extracellular Traps in Severe SARS-CoV-2 Infection: A Possible Impact of LPS and (1→3)-β-D-glucan in Blood from Gut Translocation. Cells 11, 1103 10.3390/cells11071103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sirivongrangson P., Kulvichit W., Payungporn S., Pisitkun T., Chindamporn A., Peerapornratana S.et al. (2020) Endotoxemia and circulating bacteriome in severe COVID-19 patients. Intensive Care Med. Exp. 8, 72 10.1186/s40635-020-00362-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Koch R.M., Diavatopoulos D.A., Ferwerda G., Pickkers P., de Jonge M.I. and Kox M. (2018) The endotoxin-induced pulmonary inflammatory response is enhanced during the acute phase of influenza infection. Intensive Care Med. Exp. 6, 15 10.1186/s40635-018-0182-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saithong S., Saisorn W., Visitchanakun P., Sae-Khow K., Chiewchengchol D. and Leelahavanichkul A. (2021) A Synergy Between Endotoxin and (1→3)-Beta-D-Glucan Enhanced Neutrophil Extracellular Traps in Candida Administered Dextran Sulfate Solution Induced Colitis in FcGRIIB-/- Lupus Mice, an Impact of Intestinal Fungi in Lupus. J. Inflamm. Res. 14, 2333–2352 10.2147/JIR.S305225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gieryńska M., Szulc-Dąbrowska L., Struzik J., Mielcarska M.B. and Gregorczyk-Zboroch K.P. (2022) Integrity of the intestinal barrier: the involvement of epithelial cells and microbiota-a mutual relationship. Animals (Basel) 12, 145 10.3390/ani12020145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schoultz I. and Keita Å V. (2020) The intestinal barrier and current techniques for the assessment of gut permeability. Cells 9, 1909 10.3390/cells9081909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zafar H. and Saier M.H. Jr (2021) Gut Bacteroides species in health and disease. Gut Microbes 13, 1–20 10.1080/19490976.2020.1848158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Louis P., Young P., Holtrop G. and Flint H.J. (2010) Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ. Microbiol. 12, 304–314 10.1111/j.1462-2920.2009.02066.x [DOI] [PubMed] [Google Scholar]
- 90.Panpetch W., Kullapanich C., Dang C.P., Visitchanakun P., Saisorn W., Wongphoom J.et al. (2021) Candida Administration Worsens Uremia-Induced Gut Leakage in Bilateral Nephrectomy Mice, an Impact of Gut Fungi and Organismal Molecules in Uremia. mSystems 6, e01187–e01220 10.1128/mSystems.01187-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Visitchanakun P., Panpetch W., Saisorn W., Chatthanathon P., Wannigama D.L., Thim-Uam A.et al. (2021) Increased susceptibility to dextran sulfate-induced mucositis of iron-overload β-thalassemia mice, another endogenous cause of septicemia in thalassemia. Clin. Sci. (Lond.) 135, 1467–1486 10.1042/CS20210328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Magne F., Gotteland M., Gauthier L., Zazueta A., Pesoa S., Navarrete P.et al. (2020) The firmicutes/bacteroidetes ratio: a relevant marker of gut dysbiosis in obese patients? Nutrients 12, 1474 10.3390/nu12051474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stojanov S., Berlec A. and Štrukelj B. (2020) The influence of probiotics on the firmicutes/bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms 8, 1715 10.3390/microorganisms8111715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fischer N. and Relman D.A. (2018) Clostridium difficile, aging, and the gut: can microbiome rejuvenation keep us young and healthy? J. Infect. Dis. 217, 174–176 10.1093/infdis/jix417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shin J.H., Gao Y., Moore J.H. 2nd, Bolick D.T., Kolling G.L., Wu M.et al. (2018) Innate immune response and outcome of clostridium difficile infection are dependent on fecal bacterial composition in the aged host. J. Infect. Dis. 217, 188–197 10.1093/infdis/jix414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shin N.R., Whon T.W. and Bae J.W. (2015) Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 33, 496–503 10.1016/j.tibtech.2015.06.011 [DOI] [PubMed] [Google Scholar]
- 97.Litvak Y., Byndloss M.X., Tsolis R.M. and Bäumler A.J. (2017) Dysbiotic Proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr. Opin. Microbiol. 39, 1–6 10.1016/j.mib.2017.07.003 [DOI] [PubMed] [Google Scholar]
- 98.Iebba V., Totino V., Gagliardi A., Santangelo F., Cacciotti F., Trancassini M.et al. (2016) Eubiosis and dysbiosis: the two sides of the microbiota. New Microbiol. 39, 1–12 [PubMed] [Google Scholar]
- 99.Lobionda S., Sittipo P., Kwon H.Y. and Lee Y.K. (2019) The Role of Gut Microbiota in Intestinal Inflammation with Respect to Diet and Extrinsic Stressors. Microorganisms 7, 271 10.3390/microorganisms7080271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Morishima S., Aoi W., Kawamura A., Kawase T., Takagi T., Naito Y.et al. (2021) Intensive, prolonged exercise seemingly causes gut dysbiosis in female endurance runners. J. Clin. Biochem. Nutr. 68, 253–258 10.3164/jcbn.20-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brown K., DeCoffe D., Molcan E. and Gibson D.L. (2012) Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients 4, 1095–1119 10.3390/nu4081095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mohandas S., Soma V.L., Tran T.D.B., Sodergren E., Ambooken T., Goldman D.L.et al. (2020) Differences in gut microbiome in hospitalized immunocompetent vs. immunocompromised children, including those with sickle cell disease. Front. Pediatr. 8, 583446 10.3389/fped.2020.583446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Barbara G., Feinle-Bisset C., Ghoshal U.C., Quigley E.M., Santos J., Vanner S.et al. (2016) The Intestinal microenvironment and functional gastrointestinal disorders. Gastroenterology, 1305–1318S0016-5085(16)00219-5 10.1053/j.gastro.2016.02.028 [DOI] [PubMed] [Google Scholar]
- 104.Wu M., Wu Y., Li J., Bao Y., Guo Y. and Yang W. (2018) The dynamic changes of gut microbiota in Muc2 deficient mice. Int. J. Mol. Sci. 19, 2809 10.3390/ijms19092809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ostaff M.J., Stange E.F. and Wehkamp J. (2013) Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol. Med. 5, 1465–1483 10.1002/emmm.201201773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Iddrisu I., Monteagudo-Mera A., Poveda C., Pyle S., Shahzad M., Andrews S.et al. (2021) Malnutrition and Gut Microbiota in Children. Nutrients 13, 2727 10.3390/nu13082727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kumar M., Ji B., Babaei P., Das P., Lappa D., Ramakrishnan G.et al. (2018) Gut microbiota dysbiosis is associated with malnutrition and reduced plasma amino acid levels: Lessons from genome-scale metabolic modeling. Metab. Eng. 49, 128–142 10.1016/j.ymben.2018.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.den Besten G., van Eunen K., Groen A.K., Venema K., Reijngoud D.J. and Bakker B.M. (2013) The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54, 2325–2340 10.1194/jlr.R036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xia G.H., Zhang M.S., Wu Q.H., Wang H.D., Zhou H.W., He Y.et al. (2021) Dysbiosis of Gut Microbiota Is an Independent Risk Factor of Stroke-Associated Pneumonia: A Chinese Pilot Study. Front Cell Infect. Microbiol. 11, 715475 10.3389/fcimb.2021.715475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li N., Dai Z., Wang Z., Deng Z., Zhang J., Pu J.et al. (2021) Gut microbiota dysbiosis contributes to the development of chronic obstructive pulmonary disease. Respir. Res. 22, 274 10.1186/s12931-021-01872-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen Y., Zhang F., Ye X., Hu J.J., Yang X., Yao L.et al. (2022) Association Between Gut Dysbiosis and Sepsis-Induced Myocardial Dysfunction in Patients With Sepsis or Septic Shock. Front Cell Infect Microbiol. 12, 857035 10.3389/fcimb.2022.857035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen J., Rao J.N., Zou T., Liu L., Marasa B.S., Xiao L.et al. (2007) Polyamines are required for expression of Toll-like receptor 2 modulating intestinal epithelial barrier integrity. Am. J. Physiol. Gastrointest. Liver Physiol. 293, G568–G576 10.1152/ajpgi.00201.2007 [DOI] [PubMed] [Google Scholar]
- 113.Geng Z.H., Zhu Y., Li Q.L., Zhao C. and Zhou P.H. (2022) Enteric nervous system: the bridge between the gut microbiota and neurological disorders. Front Aging Neurosci. 14, 810483 10.3389/fnagi.2022.810483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ackland G., Grocott M.P. and Mythen M.G. (2000) Understanding gastrointestinal perfusion in critical care: so near, and yet so far. Crit. Care 4, 269–281 10.1186/cc709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.van Haren F.M., Sleigh J.W., Pickkers P. and Van der Hoeven J.G. (2007) Gastrointestinal perfusion in septic shock. Anaesth. Intensive Care 35, 679–694 10.1177/0310057X0703500505 [DOI] [PubMed] [Google Scholar]
- 116.Overhaus M., Tögel S., Pezzone M.A. and Bauer A.J. (2004) Mechanisms of polymicrobial sepsis-induced ileus. Am. J. Physiol. Gastrointest. Liver Physiol. 287, G685–G694 10.1152/ajpgi.00359.2003 [DOI] [PubMed] [Google Scholar]
- 117.Overhaus M., Toegel S. and Bauer A.J. (2009) Interaction of hemorrhagic shock and subsequent polymicrobial sepsis on gastrointestinal motility. Shock 31, 382–389 10.1097/SHK.0b013e3181862ea4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Luan Y.Y., Yao Y.M., Xiao X.Z. and Sheng Z.Y. (2015) Insights into the apoptotic death of immune cells in sepsis. J. Interferon Cytokine Res. 35, 17–22 10.1089/jir.2014.0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Boomer J.S., Green J.M. and Hotchkiss R.S. (2014) The changing immune system in sepsis: is individualized immuno-modulatory therapy the answer? Virulence 5, 45–56 10.4161/viru.26516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zheng Z. and Wang B. (2021) The gut-liver axis in health and disease: the role of gut microbiota-derived signals in liver injury and regeneration. Front. Immunol. 12, 775526 10.3389/fimmu.2021.775526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou X. and Liao Y. (2021) Gut-lung crosstalk in sepsis-induced acute lung injury. Front. Microbiol. 12, 779620 10.3389/fmicb.2021.779620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Deriu E., Boxx G.M., He X., Pan C., Benavidez S.D., Cen L.et al. (2016) Influenza virus affects intestinal microbiota and secondary salmonella infection in the gut through type I interferons. PLoS Pathog. 12, e1005572 10.1371/journal.ppat.1005572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ravi A., Halstead F.D., Bamford A., Casey A., Thomson N.M., van Schaik W.et al. (2019) Loss of microbial diversity and pathogen domination of the gut microbiota in critically ill patients. Microb. Genom. 5, e000293 10.1099/mgen.0.000293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shimizu K., Ogura H., Goto M., Asahara T., Nomoto K., Morotomi M.et al. (2006) Altered gut flora and environment in patients with severe SIRS. J. Trauma. 60, 126–133 10.1097/01.ta.0000197374.99755.fe [DOI] [PubMed] [Google Scholar]
- 125.Du B., Shen N., Tao Y., Sun S., Zhang F., Ren H.et al. (2021) Analysis of gut microbiota alteration and application as an auxiliary prognostic marker for sepsis in children: a pilot study. Transl. Pediatr. 10, 1647–1657 10.21037/tp-21-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Woodall C.A., McGeoch L.J., Hay A.D. and Hammond A. (2022) Respiratory tract infections and gut microbiome modifications: A systematic review. PloS ONE 17, e0262057 10.1371/journal.pone.0262057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Farsi Y., Tahvildari A., Arbabi M., Vazife F., Sechi L.A., Shahidi Bonjar A.H.et al. (2022) Diagnostic, prognostic, and therapeutic roles of gut microbiota in COVID-19: a comprehensive systematic review. Front. Cell Infect. Microbiol. 12, 804644 10.3389/fcimb.2022.804644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sencio V., Barthelemy A., Tavares L.P., Machado M.G., Soulard D., Cuinat C.et al. (2020) Gut dysbiosis during influenza contributes to pulmonary pneumococcal superinfection through altered short-chain fatty acid production. Cell Rep. 30, 2934.e6–2947.e6 10.1016/j.celrep.2020.02.013 [DOI] [PubMed] [Google Scholar]
- 129.Lv L.-X., Jiang H.-Y., Yan R. and Li L. (2019) Interactions between gut microbiota and hosts and their role in infectious diseases. Infectious Microbes Dis. 1, 3–9 10.1097/IM9.0000000000000001 [DOI] [Google Scholar]
- 130.Richard M.L. and Sokol H. (2019) The gut mycobiota: insights into analysis, environmental interactions and role in gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 16, 331–345 10.1038/s41575-019-0121-2 [DOI] [PubMed] [Google Scholar]
- 131.Gouba N. and Drancourt M. (2015) Digestive tract mycobiota: a source of infection. Med. Mal. Infect. 45, 9–16 10.1016/j.medmal.2015.01.007 [DOI] [PubMed] [Google Scholar]
- 132.Samonis G., Kofteridis D.P., Maraki S., Alegakis D., Mantadakis E., Papadakis J.A.et al. (2005) Levofloxacin and moxifloxacin increase human gut colonization by Candida species. Antimicrob. Agents Chemother. 49, 5189 10.1128/AAC.49.12.5189.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vardakas K.Z., Michalopoulos A., Kiriakidou K.G., Siampli E.P., Samonis G. and Falagas M.E. (2009) Candidaemia: incidence, risk factors, characteristics and outcomes in immunocompetent critically ill patients. Clin. Microbiol. Infect. 15, 289–292 10.1111/j.1469-0691.2008.02653.x [DOI] [PubMed] [Google Scholar]
- 134.Hedderwick S.A., Lyons M.J., Liu M., Vazquez J.A. and Kauffman C.A. (2000) Epidemiology of yeast colonization in the intensive care unit. Eur. J. Clin. Microbiol. Infect. Dis. 19, 663–670 10.1007/s100960000348 [DOI] [PubMed] [Google Scholar]
- 135.Kumamoto C.A. (2011) Inflammation and gastrointestinal Candida colonization. Curr. Opin. Microbiol. 14, 386–391 10.1016/j.mib.2011.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Miranda L.N., van der Heijden I.M., Costa S.F., Sousa A.P., Sienra R.A., Gobara S.et al. (2009) Candida colonisation as a source for candidaemia. J. Hosp. Infect. 72, 9–16 10.1016/j.jhin.2009.02.009 [DOI] [PubMed] [Google Scholar]
- 137.Kollef M., Micek S., Hampton N., Doherty J.A. and Kumar A. (2012) Septic shock attributed to Candida infection: importance of empiric therapy and source control. Clin. Infect. Dis. 54, 1739–1746 10.1093/cid/cis305 [DOI] [PubMed] [Google Scholar]
- 138.Phuengmaung P., Somparn P., Panpetch W., Singkham-In U., Wannigama D.L., Chatsuwan T.et al. (2020) Coexistence of Pseudomonas aeruginosa With Candida albicans Enhances Biofilm Thickness Through Alginate-Related Extracellular Matrix but Is Attenuated by N-acetyl-l-cysteine. Front Cell Infect. Microbiol. 10, 594336 10.3389/fcimb.2020.594336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Phuengmaung P., Mekjaroen J., Saisorn W., Chatsuwan T., Somparn P. and Leelahavanichkul A. (2022) Rapid synergistic biofilm production of pseudomonas and candida on the pulmonary cell surface and in mice, a possible cause of chronic mixed organismal lung lesions. Int. J. Mol. Sci. 23, 9202 10.3390/ijms23169202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Laudanski K. (2021) Humanized Mice as a Tool to Study Sepsis-More Than Meets the Eye. Int. J. Mol. Sci. 22, 2403 10.3390/ijms22052403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen P., Stanojcic M. and Jeschke M.G. (2014) Differences between murine and human sepsis. Surg. Clin. North Am. 94, 1135–1149 10.1016/j.suc.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Al Bataineh M.T., Dash N.R., Bel Lassen P., Banimfreg B.H., Nada A.M., Belda E.et al. (2020) Revealing links between gut microbiome and its fungal community in Type 2 Diabetes Mellitus among Emirati subjects: A pilot study. Sci. Rep. 10, 9624 10.1038/s41598-020-66598-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Salamon D., Sroka-Oleksiak A., Gurgul A., Arent Z., Szopa M., Bulanda M.et al. (2021) Analysis of the Gut Mycobiome in Adult Patients with Type 1 and Type 2 Diabetes Using Next-Generation Sequencing (NGS) with Increased Sensitivity-Pilot Study. Nutrients 13, 1066 10.3390/nu13041066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jayasudha R., Das T., Kalyana Chakravarthy S., Sai Prashanthi G., Bhargava A., Tyagi M.et al. (2020) Gut mycobiomes are altered in people with type 2 Diabetes Mellitus and Diabetic Retinopathy. PloS ONE 15, e0243077 10.1371/journal.pone.0243077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Koskella B. and Meaden S. (2013) Understanding bacteriophage specificity in natural microbial communities. Viruses 5, 806–823 10.3390/v5030806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Salmond G.P. and Fineran P.C. (2015) A century of the phage: past, present and future. Nat. Rev. Microbiol. 13, 777–786 10.1038/nrmicro3564 [DOI] [PubMed] [Google Scholar]
- 147.Howard-Varona C., Hargreaves K.R., Abedon S.T. and Sullivan M.B. (2017) Lysogeny in nature: mechanisms, impact and ecology of temperate phages. Isme J 11, 1511–1520 10.1038/ismej.2017.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Erez Z., Steinberger-Levy I., Shamir M., Doron S., Stokar-Avihail A., Peleg Y.et al. (2017) Communication between viruses guides lysis-lysogeny decisions. Nature 541, 488–493 10.1038/nature21049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Czajkowski R., Jackson R.W. and Lindow S.E. (2019) Editorial: Environmental Bacteriophages: From Biological Control Applications to Directed Bacterial Evolution. Front. Microbiol. 10, 1830 10.3389/fmicb.2019.01830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Du Toit A. (2019) Phage induction in different contexts. Nat. Rev. Microbiol. 17, 126–127 10.1038/s41579-019-0150-4 [DOI] [PubMed] [Google Scholar]
- 151.Popescu M., Van Belleghem J.D., Khosravi A. and Bollyky P.L. (2021) Bacteriophages and the Immune System. Annu. Rev. Virol. 8, 415–435 10.1146/annurev-virology-091919-074551 [DOI] [PubMed] [Google Scholar]
- 152.Sinha A. and Maurice C.F. (2019) Bacteriophages: Uncharacterized and Dynamic Regulators of the Immune System. Mediators Inflamm. 2019, 3730519 10.1155/2019/3730519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Keskey R., Cone J.T., DeFazio J.R. and Alverdy J.C. (2020) The use of fecal microbiota transplant in sepsis. Transl. Res. 226, 12–25 10.1016/j.trsl.2020.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Monaghan T.M., Duggal N.A., Rosati E., Griffin R., Hughes J., Roach B.et al. (2021) A Multi-Factorial Observational Study on Sequential Fecal Microbiota Transplant in Patients with Medically Refractory Clostridioides difficile Infection. Cells 10, 3234 10.3390/cells10113234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Salvi P.S. and Cowles R.A. (2021) Butyrate and the Intestinal Epithelium: Modulation of Proliferation and Inflammation in Homeostasis and Disease. Cells 10, 1775 10.3390/cells10071775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cheng S., Ma X., Geng S., Jiang X., Li Y., Hu L.et al. (2018) Fecal Microbiota Transplantation Beneficially Regulates Intestinal Mucosal Autophagy and Alleviates Gut Barrier Injury. mSystems 3, e00137–e00218 10.1128/mSystems.00137-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kelly C.P. and LaMont J.T. (2008) Clostridium difficile–more difficult than ever. N. Engl. J. Med. 359, 1932–1940 10.1056/NEJMra0707500 [DOI] [PubMed] [Google Scholar]
- 158.Kao D., Roach B., Silva M., Beck P., Rioux K., Kaplan G.G.et al. (2017) Effect of Oral Capsule- vs Colonoscopy-Delivered Fecal Microbiota Transplantation on Recurrent Clostridium difficile Infection: A Randomized Clinical Trial. JAMA 318, 1985–1993 10.1001/jama.2017.17077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Iida N., Dzutsev A., Stewart C.A., Smith L., Bouladoux N., Weingarten R.A.et al. (2013) Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342, 967–970 10.1126/science.1240527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Panebianco C., Andriulli A. and Pazienza V. (2018) Pharmacomicrobiomics: exploiting the drug-microbiota interactions in anticancer therapies. Microbiome 6, 92 10.1186/s40168-018-0483-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Maini Rekdal V., Bess E.N., Bisanz J.E., Turnbaugh P.J. and Balskus E.P. (2019) Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science 364, eaau6323 10.1126/science.aau6323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Food U, Administration D . (2019) Fecal microbiota for transplantation: safety communication-risk of serious adverse reactions due to transmission of multi-drug resistant organisms, Food and Drug Administration (FDA), Silver Spring [Google Scholar]
- 163.Kunz A.N., Noel J.M. and Fairchok M.P. (2004) Two cases of Lactobacillus bacteremia during probiotic treatment of short gut syndrome. J. Pediatr. Gastroenterol. Nutr. 38, 457–458 10.1097/00005176-200404000-00017 [DOI] [PubMed] [Google Scholar]
- 164.Kumar R., Tripathi A.S., Sharma N., Singh G. and Mohapatra L. (2022) Is Regular Probiotic Practice Safe for Management of Sepsis? Chin. J. Integr. Med. 28, 185–192 10.1007/s11655-021-3334-5 [DOI] [PubMed] [Google Scholar]
- 165.Yelin I., Flett K.B., Merakou C., Mehrotra P., Stam J., Snesrud E.et al. (2019) Genomic and epidemiological evidence of bacterial transmission from probiotic capsule to blood in ICU patients. Nat. Med. 25, 1728–1732 10.1038/s41591-019-0626-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wu M.H., Pan T.M., Wu Y.J., Chang S.J., Chang M.S. and Hu C.Y. (2010) Exopolysaccharide activities from probiotic bifidobacterium: Immunomodulatory effects (on J774A.1 macrophages) and antimicrobial properties. Int. J. Food Microbiol. 144, 104–110 10.1016/j.ijfoodmicro.2010.09.003 [DOI] [PubMed] [Google Scholar]
- 167.Surayot U., Wang J., Seesuriyachan P., Kuntiya A., Tabarsa M., Lee Y.et al. (2014) Exopolysaccharides from lactic acid bacteria: structural analysis, molecular weight effect on immunomodulation. Int. J. Biol. Macromol. 68, 233–240 10.1016/j.ijbiomac.2014.05.005 [DOI] [PubMed] [Google Scholar]
- 168.Zorriehzahra M.J., Delshad S.T., Adel M., Tiwari R., Karthik K., Dhama K.et al. (2016) Probiotics as beneficial microbes in aquaculture: an update on their multiple modes of action: a review. Vet. Q. 36, 228–241 10.1080/01652176.2016.1172132 [DOI] [PubMed] [Google Scholar]
- 169.Ondee T., Pongpirul K., Janchot K., Kanacharoen S., Lertmongkolaksorn T., Wongsaroj L.et al. (2021) Lactiplantibacillus plantarum dfa1 Outperforms Enterococcus faecium dfa1 on Anti-Obesity in High Fat-Induced Obesity Mice Possibly through the Differences in Gut Dysbiosis Attenuation, despite the Similar Anti-Inflammatory Properties. Nutrients 14, 80 10.3390/nu14010080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Deng Z., Chen M., Xie H., Jian D., Xu S., Peng Q.et al. (2019) Claudin reduction may relate to an impaired skin barrier in rosacea. J. Dermatol. 46, 314–321 10.1111/1346-8138.14792 [DOI] [PubMed] [Google Scholar]
- 171.Muizzuddin N., Maher W., Sullivan M., Schnittger S. and Mammone T. (2012) Physiological effect of a probiotic on skin. J. Cosmet. Sci. 63, 385–395 [PubMed] [Google Scholar]
- 172.Arck P., Handjiski B., Hagen E., Pincus M., Bruenahl C., Bienenstock J.et al. (2010) Is there a ‘gut–brain–skin axis’? Exp. Dermatol. 19, 401–405 10.1111/j.1600-0625.2009.01060.x [DOI] [PubMed] [Google Scholar]
- 173.Herrema H., RG I.J. and Nieuwdorp M. (2017) Emerging role of intestinal microbiota and microbial metabolites in metabolic control. Diabetologia 60, 613–617 10.1007/s00125-016-4192-0 [DOI] [PubMed] [Google Scholar]
- 174.Zhang F., Xiao Y., Pan L., Yu L., Liu Y., Li D.et al. (2023) Effects of polysaccharides from Lyophyllum decastes (Fr.) Singer on gut microbiota via in vitro-simulated digestion and fermentation. Front Microbiol. 14, 1083917 10.3389/fmicb.2023.1083917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Davani-Davari D., Negahdaripour M., Karimzadeh I., Seifan M., Mohkam M., Masoumi S.J.et al. (2019) Prebiotics: definition, types, sources, mechanisms, and clinical applications. Foods 8, 92 10.3390/foods8030092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Vinolo M.A., Rodrigues H.G., Hatanaka E., Sato F.T., Sampaio S.C. and Curi R. (2011) Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J. Nutr. Biochem. 22, 849–855 10.1016/j.jnutbio.2010.07.009 [DOI] [PubMed] [Google Scholar]
- 177.Everard A., Lazarevic V., Derrien M., Girard M., Muccioli G.G., Neyrinck A.M.et al. (2011) Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes 60, 2775–2786 10.2337/db11-0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wei Y., Gao J., Kou Y., Liu M., Meng L., Zheng X.et al. (2020) The intestinal microbial metabolite desaminotyrosine is an anti-inflammatory molecule that modulates local and systemic immune homeostasis. FASEB J. 34, 16117–16128 10.1096/fj.201902900RR [DOI] [PubMed] [Google Scholar]
- 179.Huo J., Wang T., Wei B., Shi X., Yang A., Chen D.et al. (2022) Integrated network pharmacology and intestinal flora analysis to determine the protective effect of Xuanbai–Chengqi decoction on lung and gut injuries in influenza virus-infected mice. J. Ethnopharmacol. 298, 115649 10.1016/j.jep.2022.115649 [DOI] [PubMed] [Google Scholar]
- 180.Mu S., Zhang J., Du S., Zhu M., Wei W., Xiang J.et al. (2021) Gut microbiota modulation and anti-inflammatory properties of Xuanbai Chengqi decoction in septic rats. J. Ethnopharmacol. 267, 113534 10.1016/j.jep.2020.113534 [DOI] [PubMed] [Google Scholar]
- 181.Sarma S.M., Singh D.P., Singh P., Khare P., Mangal P., Singh S.et al. (2018) Finger millet arabinoxylan protects mice from high-fat diet induced lipid derangements, inflammation, endotoxemia and gut bacterial dysbiosis. Int. J. Biol. Macromol. 106, 994–1003 10.1016/j.ijbiomac.2017.08.100 [DOI] [PubMed] [Google Scholar]
- 182.Chi C., Buys N., Li C., Sun J. and Yin C. (2019) Effects of prebiotics on sepsis, necrotizing enterocolitis, mortality, feeding intolerance, time to full enteral feeding, length of hospital stay, and stool frequency in preterm infants: a meta-analysis. Eur. J. Clin. Nutr. 73, 657–670 10.1038/s41430-018-0377-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Adelman M.W., Woodworth M.H., Langelier C., Busch L.M., Kempker J.A., Kraft C.S.et al. (2020) The gut microbiome's role in the development, maintenance, and outcomes of sepsis. Crit. Care 24, 278 10.1186/s13054-020-02989-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kullberg R.F.J., Wiersinga W.J. and Haak B.W. (2021) Gut microbiota and sepsis: from pathogenesis to novel treatments. Curr. Opin. Gastroenterol. 37, 578–585 10.1097/MOG.0000000000000781 [DOI] [PubMed] [Google Scholar]
- 185.Sun T., Wang L. and Zhang H. (2022) Intestinal Microbiota in Sepsis. Intensive Care Res. 2, 1–7 10.1007/s44231-022-00001-8 [DOI] [Google Scholar]
- 186.Niu M. and Chen P. (2021) Crosstalk between gut microbiota and sepsis. Burns Trauma. 9, tkab036 10.1093/burnst/tkab036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Musumeci S., Coen M., Leidi A. and Schrenzel J. (2022) The human gut mycobiome and the specific role of Candida albicans: where do we stand, as clinicians? Clin. Microbiol. Infect. 28, 58–63 10.1016/j.cmi.2021.07.034 [DOI] [PubMed] [Google Scholar]
- 188.Zhang L., Zhan H., Xu W., Yan S. and Ng S.C. (2021) The role of gut mycobiome in health and diseases. Therap. Adv. Gastroenterol. 14, 17562848211047130 10.1177/17562848211047130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Spencer L., Olawuni B. and Singh P. (2022) Gut Virome: Role and Distribution in Health and Gastrointestinal Diseases. Front Cell Infect Microbiol. 12, 836706 10.3389/fcimb.2022.836706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Cao Z., Sugimura N., Burgermeister E., Ebert M.P., Zuo T. and Lan P. (2022) The gut virome: A new microbiome component in health and disease. EBioMedicine 81, 104113 10.1016/j.ebiom.2022.104113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to the review.