Abstract
Boron neutron capture therapy (BNCT), an experimental treatment for certain cancers, destroys only cells near the boron; however, there is a need to develop highly specific delivery agents. As nucleic acid aptamers recognize specific molecular targets, we investigated the influence of boronated nucleotide analogs on RNA function and on the systematic evolution of ligands by exponential enrichment (SELEX) process. Substitution of guanosine 5′-(α-P-borano) triphosphate (bG) for GTP or uridine 5′-(α-P-borano) triphosphate (bU) for UTP in several known aptamers diminished or eliminated target recognition by those RNAs. Specifically, ATP-binding aptamers containing the ζ-fold, which appears in several selections for adenosine aptamers, became inactive upon bG substitution but were only moderately affected by bU substitution. Selections were carried out using the bG or bU analogs with C8-linked ATP agarose as the binding target. The selections with bU and normal NTP yielded some ζ-fold aptamers, while the bG selection yielded none of this type. Non-ζ aptamers from bU and bG populations tolerated the borano substitution and many required it. The borano nucleotide requirement is specific; bU could not be used in bG-dependent aptamers nor vice versa. The borano group plays an essential role, as yet undefined, in target recognition or RNA structure. We conclude that the bG and bU nucleotides are fully compatible with SELEX, and that these analogs could be used to make boronated aptamers as therapeutics for BNCT.
INTRODUCTION
Boron neutron capture therapy (BNCT) is a therapeutic treatment currently under clinical and pre-clinical trials for certain skin and brain tumors (melanoma and glioblastoma multiforme) and for rheumatoid arthritis (1,2). Compounds containing the 10B isotope—usually sodium borocaptate or boronophenylalanine—are delivered to the affected tissue and irradiated with thermal or epithermal neutrons to form 11B. The excited nucleus ejects energetic 4He and 7Li nuclei, which destroy adjacent tissue (usually within 5–9 µm, about the diameter of one cell) (3). Ideally, BNCT avoids the systemic toxicity associated with most forms of chemotherapy and the low spatial resolution of gamma irradiation.
A significant challenge to developing BNCT is to design boron-containing molecules that specifically destroy tumor cells without damaging the surrounding tissue. Nucleic acid aptamers obtained using the systematic evolution of ligands by exponential enrichment (SELEX) protocol may be suitable for this purpose. Aptamers can serve as highly specific diagnostic or drug delivery agents by binding surface antigens that are specific for a given cell type (4,5). The SELEX method can be used to target purified proteins, cell membranes or whole cells. Individual aptamers from a complex pool selected to bind membranes from lyzed red blood cells (‘RBC ghosts’) specifically bound discrete targets on RBC ghosts, even though some of the binding targets were very rare on the membranes (6). Nuclease-stabilized aptamers to human neutrophil elastase (a serine protease that degrades the extracellular matrix in inflammatory responses) have been used to image sites of inflammation in whole animals (7). These same aptamers also protect against inflammatory tissue damage in a dose-dependent manner in a mouse model of acute lung inflammatory disease (8). Similarly, aptamers against human P-selectin (a cell-surface protein involved in recruiting neutrophils from circulation to sites of inflammation) block the binding of activated platelets to neutrophils in flow cytometry and hydrodynamic assays (9). Thus, it is reasonable to propose that aptamers could be used to deliver boron specifically to tumor cells for BNCT while avoiding non-tumor cells.
An essential element in the above therapeutic model is the availability of an appropriate means of incorporating boron into nucleic acid aptamers. Deoxynucleotide analogs with backbone boranophosphate or cyanoborano substitutions on the bases have been prepared for incorporation during PCR (10–12), for enzymatic sequencing with exonucleases (12) and for automated oligonucleotide synthesis using phosphoramidite chemistry (13). Incorporation of a single Sp boranophosphate linkage (which has a stereochemistry equivalent to the corresponding Rp thiophosphate analog) does not significantly alter the thermal stability of a short oligonucleotide duplex (14), and fully substituted oligodeoxynucleotides support RNase H-mediated cleavage of mRNA in antisense applications (15,16). Ribonucleoside analogs with 5′-O-(α-P-boranotriphosphate) substitutions have also been described (17,18) and are accepted as substrates by RNA polymerases (17).
To gain a better understanding of how boranophosphate-substituted ribonucleosides affect the SELEX process and RNA structure in general, we have measured target recognition by several known aptamers transcribed with α-boranophosphate analogs (bU or bG), and we have isolated two ATP-binding pools of boranophosphate-modified aptamers. Several selections of RNA aptamers to ATP yielded the same simple structural motif (19–21). This reproducibility has made ATP the target of choice in evaluating the use of modified nucleotides in SELEX experiments, such as fluorophore-labeled and biotinylated nucleotide analogs, to develop signaling aptamers as molecular sensors (22,23). In the presence of AMP, the purine-rich unpaired loop of the aptamer collapses into a highly organized ‘ζ-fold’ in which some nucleotides bind the adenine portion in a structure resembling a GNRA (where N represents any nucleotide and R represents either A or G) tetraloop and others contact the ribose hydroxyls (24,25). Additionally, the three-dimensional structure of the ζ-fold (24,25) and its susceptibility to specific mutations (26) are both well understood.
By substituting UTP or GTP with bU or bG, we sought to answer four questions. First, do backbone boranophosphates perturb RNA structure? Understanding the general effects of these substitutions will help in the design and development of boronated aptamers as antitumor agents. Second, how does inclusion of these nucleotides affect the outcome of in vitro selections for aptamers? Perturbations that either facilitate or antagonize target binding will necessarily force the selections towards or away from a particular outcome. Third, for those RNAs selected in the presence of boronated analogs, how important are the borons in establishing RNA structure or function; i.e. are the BH3 groups required or merely tolerated? Finally, how does the binding specificity of boronated aptamers compare with that of normal aptamers? The use of bNTP-modified aptamers is predicated on the assumption that their specificity will be comparable with that of aptamers derived from normal NTPs. We find that substituting specific bases with their boranophosphate analogs has variable effects on aptamer activity ranging from moderate to strong, and that including boronated nucleotide analogs in transcripts from the beginning of a selection is likely to yield aptamers suitable for testing in BNCT.
MATERIALS AND METHODS
RNAs and pools
About 1 × 1014 different sequences (165 pmol) of a double-stranded DNA pool containing 70 random nucleotides [(70N)] and primer-binding sites were transcribed in vitro to yield RNA of the form 5′-gggaaaagcgaaucauacacaaga(70N)gggcauaagguauuuaauuccaua-3′. In vitro transcriptions were carried out under standard conditions (substituting 1 mM of either bU or bG in place of UTP or GTP in two of the three selections) using either the Ampliscribe kit from Epicentre or purified components (New England Biolabs) (27). Uniformly radiolabeled transcripts (118 nt for pools) were purified on denaturing polyacrylamide gels (urea–Tris–borate–EDTA) prior to use. MALDI-TOF mass spectrometry of a particular single-sequence transcript gave a signal that was shifted 2 mass units for each position that was supposed to contain boron, as expected. A sample of 3–5 nmol of RNA was used in the first cycle of each selection and 100–1000 pmol in subsequent cycles.
Affinity immobilization of RNAs
During screens of activity ± boron, RNA was heated to 72°C for 2 min, then cooled to room temperature in the presence of binding buffer (50 mM Bis–Tris pH 6.4, 200 mM NaCl, 10 mM MgCl2). After allowing the RNA to fold for 10 min it was applied to 100 µl bed vol of the appropriate affinity matrix (Sigma) that had been equilibrated with binding buffer. Matrices contained ATP immobilized through the adenine C-8 position (for S20, wt204, and all RNAs from this selection, unless specified otherwise), FAD immobilized through the ribose hydroxyls (for F8.35.82), or CoA immobilized through the sulfur (for 70SA#6 and 70PSA#5). Fractions (100 µl) were collected during the flow through, six washes with buffer and six elutions with 5 mM ATP in binding buffer. Uniformly radiolabeled RNA in each fraction was monitored by Cherenkov counting before combining the elution fractions.
Selection of ATP-binding aptamers
During the selection, C8-linked ATP–agarose (Sigma) was washed with a minimum of 1500 µl binding buffer before being gently mixed with the folded RNA for 1 h at room temperature. The mixture was filtered through glass wool that had been pre-wetted with binding buffer in a 1 cc syringe, then the RNA was washed and affinity eluted as above. After elution, excess ATP was removed by buffer exchange in a Microcon-10 (Amicon) before ethanol precipitation, reverse transcription and PCR amplification in preparation for the next round of selection. In the last two rounds of each selection the stringency was increased by washing 10 times with 100 µl binding buffer. After either seven (normal nucleotides), eight (bU) or 10 (bG) rounds of selection the pools were cloned into plasmids for sequencing.
Functional boundary determinations
Deletion/selection experiments were performed as described previously (28) to determine the 5′-most and 3′-most nucleotides required for target recognition. Transcripts were labeled at the 5′-end with [γ-32P]ATP (ICN) using T4 polynucleotide kinase, or at the 3′-end by annealing an oligonucleotide with three overhanging thymidines on its 5′-end and filling in the overhang with [α-32P]dATP (ICN) using the Klenow fragment (exo–) of Escherichia coli DNA polymerase I (NEB) (29). Partially hydrolyzed RNA ladders were generated by incubating RNA in 50 mM sodium carbonate buffer at pH 9.0, 1 mM EDTA and 0.48 mg/ml tRNA for 10 min at 90°C (pH 8.0, 80°C for boronated samples, which were more susceptible to spontaneous hydrolysis 5′ to the boronated nucleotides). Partially hydrolyzed, end-labeled RNA was heated to 72°C for 2 min and then layered onto a bed of preformed resin. RNA from the wash fractions or specifically eluted with free ATP was ethanol precipitated and analyzed by gel electrophoresis on 12% denaturing polyacrylamide gels alongside size ladders of alkaline hydrolyzed RNA and T1 nuclease-digested RNA. T1 nuclease ladders were generated with 0.1 U enzyme (Epicentre) for 10 min at 50°C.
Surveys of specificity and binding conditions
For the survey of elution by ATP analogs, column-bound RNA was washed with 10 bed vol binding buffer, then eluted with 6 bed vol of buffer containing 0.5 mM analog, followed by elution with 6 bed vol of buffer containing 5 mM ATP. Elution values reported indicate the relative amount of RNA removed from the resin by the analog relative to the sum of the total RNA removed during these 12 elution steps.
RESULTS
Retrofitting ‘normal’ aptamers with boranophosphates affects target recognition to variable degrees
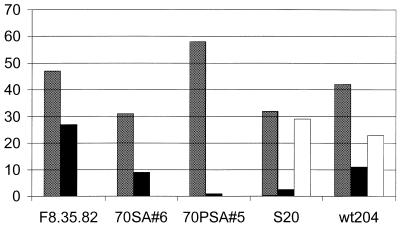
To determine the extent to which boranophosphate substitutions enhance or interfere with aptamer function, five RNA aptamers that were originally selected with normal NTPs were transcribed either with all normal NTPs or in the presence of three NTPs and either bG or bU (Fig. 1). The chosen aptamers recognize flavin adenine dinucleotide (FAD) (‘F8.35.82’) (30), coenzyme A (CoA) (‘70SA6’ and ‘70PSA5) (D.Saran, J.Frank and D.Burke, manuscript in preparation) or adenosine [‘S20’ (21) and ‘wt204’ (this work)]. Each pair of transcripts was applied to the cognate resins and eluted with its respective target. Target recognition by each of the boronated aptamers was impaired relative to that of the ‘normal’ transcripts, but the degree of the impairment varied with the nucleotide and with the aptamer (Fig. 2). Incorporating bG into the 82-nucleotide FAD-binding aptamer F8.35.82 was only moderately inhibitory, as was incorporation of bU into the adenosine-binding aptamer S20. Transcripts of the CoA-binding aptamer 70SA6 carrying bG retained just over a quarter of the CoA-binding activity of the non-boronated form. In contrast, bG incorporation into either the CoA-binding aptamer 70PSA5 or the adenosine-binding aptamer S20 eliminated target recognition for both RNAs. Boranophosphate substitutions can, therefore, exert considerable influence over RNA structure and function. While some aptamers can be readily retrofitted, others do not tolerate the substitution. In particular, the ζ-motif of the ATP-binding aptamer tolerates bU substitution but not bG substitution.
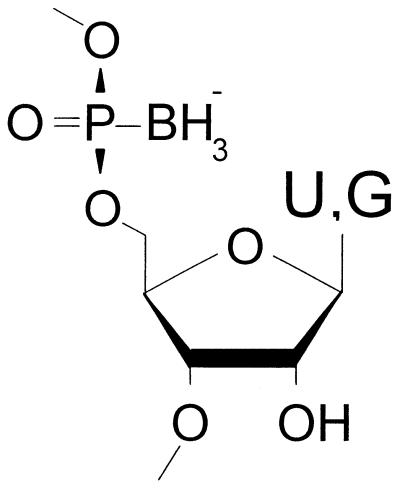
Figure 1.
Boranophosphate-substituted nucleotides used in this study, shown in Sp configuration.
Figure 2.
The effect of including boronated nucleotides on target binding for aptamers that had originally been selected with normal NTPs to bind various nucleotide cofactor targets. y-Axis is percent RNA eluted from corresponding affinity resins when RNAs were transcribed with normal NTPs (shaded), bG in place of GTP (black) or bU in place of UTP (white). Aptamer wt204 is the 65 nt element identified through boundary analysis of an ATP-binding aptamer from the present selection. Only the last two of this set were assayed with bU. Identities of other aptamer and resins are given in the text.
In vitro selection of boronated aptamers
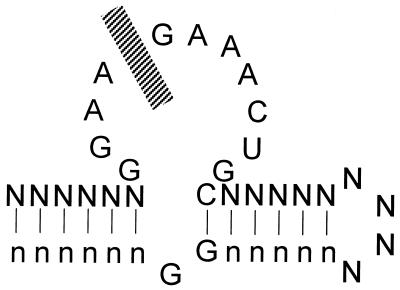
Given the variable but consistently negative impact of uniform boranophosphate incorporation into pre-selected aptamers, we used the SELEX method to generate aptamers that would at least tolerate the boranophosphate substitutions, if not prefer the substitution. Affinity resins that immobilize adenosine cofactors through the C8 position have elicited the same structural element, known as the ‘ζ-motif’ (24), in several independent experiments (19–21) (Fig. 3). A single DNA pool with 70 random nucleotides was transcribed with normal nucleotides or with bG or bU substitutions. All three selections returned pools that bound and eluted from the affinity resin to an equivalent extent after seven (normal nucleotides), eight (bU) or 10 cycles (bG). When these pools were cloned and sequenced, a recognizable ζ-motif (including potential secondary structure elements) was found in just over half of the sequences in the bU pool (39 out of 68 total bU isolates) and the control pool [37 out of 68 total wild-type (wt) isolates] (Fig. 4). Both pools contained a distribution of sequences, with more diversity in the ‘wt’ pool. In contrast, the ζ-motif was absent from the bG pool, consistent with the interference noted in Figure 2. The bG pool contained instead two dominant sequence families (14 of 34 total isolates for one family and seven for the other), four minor families (comprising four, two, two and two isolates) and three unique sequences (Table 1). All three pools show virtually identical base composition. Thus, the presence of the boronated nucleotide does not create additional selection pressures that either significantly prejudice or favor its inclusion (e.g. by perturbing RNA synthesis or reverse transcription).
Figure 3.
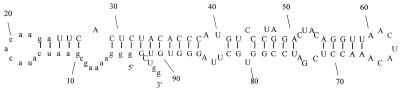
The ζ-motif ATP aptamer. N, any nucleotide; n, any nucleotide that pairs with N. Stippled bar represents an AMP moiety intercalated between A10 and G11.
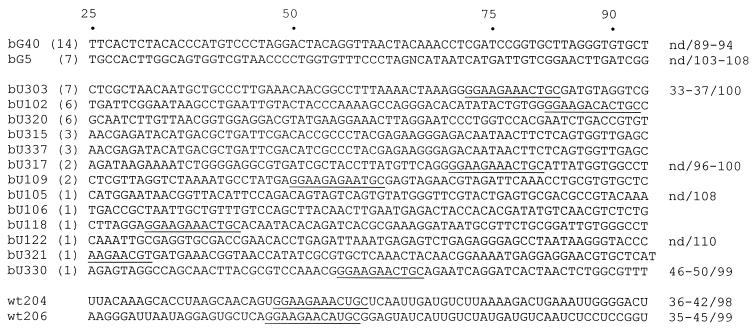
Figure 4.
Sequences from selections with bG or bU substitutions or with normal nucleotides. Nucleotide numbers are given above the sequences (primer-binding sequences not shown). Functional boundaries, where measured, are given to the right of the corresponding sequences, with the form (5′-boundary)/(3′-boundary), including uncertainty in the measured boundary; nd, not determined. The signature sequence of the ζ-motif is underlined (the motif from bU321 is formed in part by the 5′-primer-binding sequence). The number of occurrences of nearly identical sequences is given in parentheses next to the sequence name. The complete sequence data set is available at http://ernie.chem.indiana.edu/dhburke/all_seqs.html.
Table 1. Nucleotide compositions of the three selected pools.
| Nucleotide | bG selection | bU selection | wt selection | |||
|---|---|---|---|---|---|---|
| Number | Percent | Number | Percent | Number | Percent | |
| A | 486 | 22.1 | 1075 | 22.6 | 1061 | 22.7 |
| C | 546 | 24.9 | 1244 | 26.2 | 1254 | 26.8 |
| G | 482 | 21.9 | 1025 | 21.5 | 967 | 20.6 |
| T | 589 | 26.8 | 1410 | 29.6 | 1395 | 29.8 |
| N | 96 | 4.4 | 6 | 0.1 | 6 | 0.1 |
| Total | 2199 | 4760 | 4680 | |||
Nucleotides that were incorporated into RNA transcripts as analogs during the selection (bU and bG) are indicated in bold.
Requirement for boron
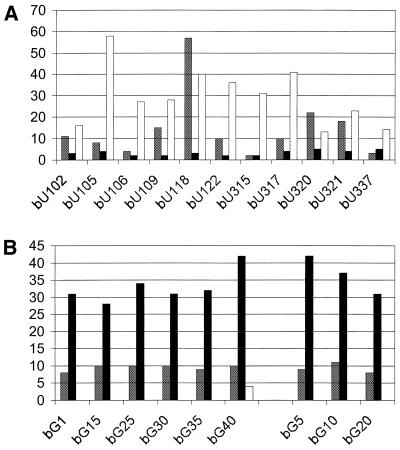
Eleven isolates from the bU selection were transcribed either with normal NTPs or with bU, and all 22 transcripts were assayed for their abilities to bind and elute from the ATP-affinity resin. This was done to differentiate sequences that require bU from those that simply tolerate its presence. Both sets of RNA displayed a wide range of ATP-binding ability (60 to <5%). Only three aptamers (bU118, bU320 and bU321) bound well both with and without bU, but for most (8 of 11), elution percentages were significantly higher when the RNA contained bU (Fig. 5A). Four aptamers (bU105, bU106, bU315 and bU337) bound the resin well when transcribed with bU but were indistinguishable from background when transcribed with normal nucleotides. Nine independent isolates from the two main sequence groups of the bG selection (bG1, bG15, bG25, bG30, bG35 and bG40 in one group, and bG5, bG10 and bG20 in the other) were similarly tested upon transcription with either GTP or bG. Each bound to and eluted from the ATP resin very well when the RNA contained bG, but all were inactive with normal G (Fig. 5B). Thus, all aptamers from both selections at least tolerate BH3 in the backbone, and several require its presence.
Figure 5.
Effect of including boronated nucleotides on binding to the ATP resin for aptamers selected with either (A) bU in place of UTP or (B) bG in place of GTP. Labels for y-axis and column shadings are as indicated in the legend to Figure 2.
Specificity of boron requirement
The above observations could result from either a non-specific effect of arbitrary BH3 incorporation or from a specific effect due to the precise location of the borano group. To distinguish between these models, the same 11 aptamers that had been tested with bU were transcribed with bG in place of GTP, and one aptamer from the bG selection was transcribed with bU in place of UTP. In every case, incorporation of the boron into sites not used in the selection destroyed all binding (Fig. 5). These aptamers fold into discrete structures that recognize ATP, with at least one of the BH3 groups participating directly in formation of that structure, in contacting the bound target or both.
Delimiting the binding sites
The limits of the RNA sequences required for binding were determined for nine aptamers: two from the ‘normal NTP’ selection (wt204 and wt206), five from the bU selection (bU105, bU122, bU303, bU317 and bU330) and two from the bG selection (bG5 and bG25). In brief, 5′-end-labeled RNA was partially hydrolyzed and passed over the ATP-affinity resin. Functional molecules were then eluted and separated by gel electrophoresis. The smallest end-labeled molecule recovered from the resin defines the 3′-most nucleotide required for activity. For four of these aptamers, 3′-labeled RNA was treated in a similar fashion to define the 5′-most required nucleotides. In each of these, sequences very near to the 5′-ends appeared to be necessary for activity. The locations of each of these functional boundaries are given in Figure 4.
Five of the aptamers for which boundaries were determined contained variations of the ζ-motif sequence, including potential helices consistent with those known to flank the ζ-turn structure (wt204, wt206, bU303, bU317 and bU330). In each case, the functional boundaries were located at the 5′- and 3′-ends of potential secondary structural elements flanking the canonical ζ-motif sequence. The 65 nt RNA fragment defined by the boundaries of aptamer wt204 showed strong binding to the ATP resin when transcribed with normal NTPs, slightly reduced binding with the bU substitution and little binding with the bG substitution (Fig. 2). This behavior is identical to that of the ζ-motif aptamer S20 described above. The 3′ functional boundary of bG40 lies near the end of the 70N random region. Deletion analysis from the 5′-end of bG40 gave variable results, with moderate binding observed with 10 nt removed (‘bG40Δ10’), but almost none with 1, 2 or 3 nt removed. The structure predicted for the RNA fragment extending from the original 5′-end through the 70N random region is shown in Figure 6. Small deletions from the 5′-end may release the loop nucleotides 4–9, allowing them to form alternative structures that interfere with binding, whereas the larger deletion in bG40Δ10 precludes these putative inactive structures.
Figure 6.
Predicted structure of bG40min, a truncated version of isolate bG40 identified through deletion and 3′ functional boundary analysis, as predicted by mfold (38). Lower case letters indicate the fixed primer-binding sequence.
Specificity of binding
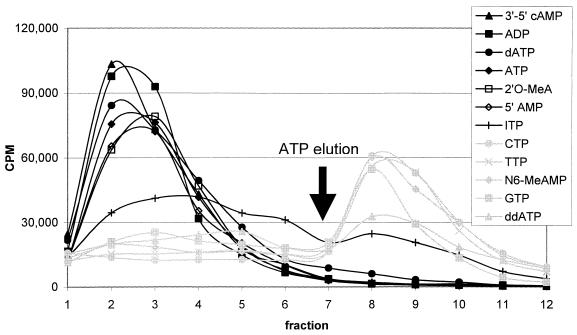
The eventual application of borano aptamers to BNCT requires that they bind preferentially to the target cell types. To determine the specificity of the aptamer–target interaction for aptamer bG40, RNA was transcribed with the bG substitution and allowed to bind to the ATP-affinity resin. After washing off non-specifically bound RNA, the bound RNA was challenged with low concentration (0.5 mM) of various ATP analogs and then the remaining RNA was removed by ATP in high concentration (5.0 mM). Recognition is defined by the ability of the analog to remove most of the RNA in the competitive elution prior to the strip with 5 mM ATP. With one exception, all analogs could be readily categorized as being ‘recognized’ or ‘not recognized’ by the aptamer (Fig. 7).
Figure 7.
Recognition of ATP analogs by aptamer bG40. Uniformly radiolabeled aptamer was bound to ATP resin and washed with binding buffer, then eluted with 0.5 mM analog (fractions 1–6), followed by elution with 5.0 mM ATP (fractions 7–12, after arrow). Analogs used are listed on the figure in the same order, from top to bottom, as the corresponding traces.
The adenosine base is required for the interaction, as changing the adenine base to G, T or C eliminates recognition. Methylation of the N6 amine also blocks recognition, while inosine triphosphate gave intermediate elution. The lone pair electrons of the N6 nitrogen may act as a hydrogen bond acceptor in the case of ATP, and be functionally replaced by the lone pairs of the keto oxygen in inosine. The methyl group of N6-methyladenosine may create a steric disruption that prevents formation of that hydrogen bond. The 3′-OH of the ribose also plays some role in the interaction, as ddATP is not recognized. As the RNA is eluted by 3′,5′-cAMP, the role of the 3′-OH is likely to be as a hydrogen bond acceptor. In contrast, the 2′-OH is not required, as 2′-O-methyl adenosine and 2′-dATP both efficiently elute the RNA. There is also no specificity for the triphosphate, since ATP, ADP and AMP all elute to equivalent degrees. Finally, bG40 was transcribed with either all normal nucleotides, or with the bU or bG substitution, and each RNA was applied to a biotin-affinity resin or to other adenosine cofactor-affinity resins (ATP, ADP, FAD or FMN) in which the targets were immobilized through the ribose hydroxyls. None of the forms of bG40 RNA bound to any of these resins, confirming both the importance of the ribose in the aptamer–ATP interaction and the specific requirement for bG. The pattern of target recognition exhibited by bG40 is comparable with that observed for other aptamers to nucleotide cofactors (19,21,28,31).
DISCUSSION
The data above show that aptamers containing significant numbers of boron can be made either by de novo selection or by ‘retrofitting’ pre-existing aptamers. Several aptamers selected without boron were severely weakened when transcribed in boronated form. Retrofitting existing aptamers with backbone BH3 modifications is a hit-or-miss proposition, sometimes facilitated by detailed knowledge of the structure of the unmodified complex. It appears to work best when the modified nucleotide is located in helical regions away from the binding core of the aptamer. This was the case for the ζ-fold aptamers, which tolerated bU incorporation (only one U residue in the binding core, and that one can be mutated without loss of activity; 21,26) but not bG incorporation (six G residues in the binding core, several of which make essential stacking or H-bonding interactions) (24,25). Helices that include boranophosphates are approximately isoenergetic with normal helices (14), so retrofitting existing aptamers in these positions may offer little structural disruption. Yet even if substitutions in helices do not interfere with RNA structure, the analog could cause mischief, particularly if a protein or small-molecule target normally contacts the non-bridging oxygen that is replaced with BH3.
De novo selection eliminates those ambiguities, virtually assuring that the surviving aptamers at least tolerate the presence of the modification. The ribonucleoside analogs with 5′-O-(α-P-boranotriphosphate) substitutions are fully compatible with the SELEX method. The resulting aptamers show no bias against the modified nucleotide, and the observed specificity is comparable with that of aptamers selected with normal nucleotides. Therefore, boronated aptamers could be developed for use in BNCT for anticancer treatments by targeting, for example, a protein or saccharide that is more abundant on tumor cells than on surrounding tissues.
While some of the aptamers selected in the presence of the boronated nucleotide were indifferent to its inclusion during transcription, others absolutely required the BH3 for activity. These borano groups appear to play some specific role in binding or structure, as they must be incorporated at specific positions: bG was not tolerated in aptamers selected with bU, and bU could not be used in aptamers that had been selected in the presence of bG. While most of these aptamers contain the modified nucleotide in 20–30 positions, we do not know at present which of these positions are required to carry the modifications. Aptamers and ribozymes selected with modified nucleotides often require those modifications for activity, and the technique of nucleotide analog interference mapping is based on the premise that at least some modifications are detrimental. Ribozymes for Diels–Alder condensation (32) and amide bond formation (33) that were selected to contain bulky aromatic substituents at position 5 of the uracils were later shown to be inactive when transcribed with UTP. Aptamers with sugar modifications (2′-fluoro-, 2′-amino and 2′-O-methyl) are appearing in the literature at an increasing rate, and most of these similarly required their modification. RNA aptamers do not usually function when generated as 2′-deoxy RNA (ssDNA) and vice versa. Counter examples have recently been reported in the form of a heme-binding G-quartet that is functional as either RNA or DNA (34) and some of the individuals from this selection for which inclusion of the boron was optional.
From a physicochemical point of view, it is curious that the BH3 group should be so significant. It does not greatly affect overall helical stability (14); like oxygen, it can carry a negative charge and serve as a hydrogen bond acceptor (35) and it is only slightly larger than the oxygen that it replaces (similar in size to a methyl group). On the other hand, the BH3 group is less electronegative than oxygen, and in ab initio calculations with model compounds it carries less negative charge (q = –1.28 for oxygen versus –0.87 for BH3) (36). As a result, oligodeoxynucleotides with boranophosphate substitutions display increased lipophilicity relative to normal oligonucleotides (37). The BH3 group of diisopropylboranophosphate makes hydrophobic contacts with aliphatic hydrogens in a crystal structure of that analog (36). In addition, the BH3 group is unlikely to chelate a divalent metal ion. Excluding metals from specific sites to avoid stabilizing alternative folds could be one mechanism for the BH3 dependence.
It is unclear at this point which, if any, of these properties account for the BH3 requirement in the selected aptamers, or what causes the BH3-mediated interference noted above in the survey of aptamers selected with normal NTPs. However, it is clear that if boronated nucleotide analogs are present in transcripts from the beginning of a selection, their physicochemical properties become integrated into the architecture of the selected molecules to yield functional, boronated aptamers. A similar approach targeted to cancer cells could yield reagents suitable for testing in BNCT.
Acknowledgments
ACKNOWLEDGEMENTS
This material is based upon work supported by the National Science Foundation under grant no. 9896363 to D.H.B. and by the National Institutes of Health under grants no. AI45344 to D.H.B. and no. GM57693 to B.R.S.
REFERENCES
- 1.Barth R., Soloway,A., Goodman,J., Gahbauer,R., Gupta,N., Blue,T., Yang,W. and Tjarks,W. (1999) Boron neutron capture therapy of brain tumors: an emerging therapeutic modality. Neurosurger y, 44, 433–451. [DOI] [PubMed] [Google Scholar]
- 2.Hawthorne M.F. (1998) New horizons for therapy based on the boron neutron capture reaction. Mol. Med. Today, 4, 174–181. [DOI] [PubMed] [Google Scholar]
- 3.Coderre J. and Morris,G. (1999) The radiation biology of boron neutron capture therapy. Radiat. Res., 151, 1–18. [PubMed] [Google Scholar]
- 4.Romig T.S., Bell,C. and Drolet,D.W. (1999) Aptamer affinity chromatography: combinatorial chemistry applied to protein purification. J. Chromatogr. B Biomed. Sci. Appl., 731, 275–284. [PubMed] [Google Scholar]
- 5.Famulok M. and Mayer,G. (1999) Aptamers as tools in molecular biology and immunology. Curr. Top. Microbiol. Immunol., 243, 123–136. [DOI] [PubMed] [Google Scholar]
- 6.Morris K., Jensen,K., Julin,C., Weil,M. and Gold,L. (1998) High affinity ligands from in vitro selection: complex targets. Proc. Natl Acad. Sci. USA, 95, 2902–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlton J., Sennello,J. and Smith,D. (1997) In vivo imaging of inflammation using an aptamer inhibitor of human neutrophil elastase. Chem. Biol., 4, 809–816. [DOI] [PubMed] [Google Scholar]
- 8.Bless N., Smith,D., Charlton,J., Czermak,B., Schmal,H., Friedl,H. and Ward,P. (1997) Protective effects of an aptamer inhibitor of neutrophil elastase in lung inflammatory injury. Curr. Biol., 7, 887–880. [DOI] [PubMed] [Google Scholar]
- 9.Jenison R., Jennings,S., Walker,D., Bargatze,R. and Parma,D. (1998) Oligonucleotide inhibitors of P-selectin-dependent neutrophil-platelet adhesion. Antisense Nucleic Acid Drug Dev., 8, 265–279. [DOI] [PubMed] [Google Scholar]
- 10.Sergueev D., Hasan,A., Ramaswamy,M. and Shaw,B.R. (1997) Boranophosphate oligonucleotides: new synthetic approaches. Nucleosides Nucleotides, 16, 1533–1538. [Google Scholar]
- 11.Porter K., Tomasz,J., Huang,F., Sood,A. and Shaw,B.R. (1995) N7-cyanoborane-2′-deoxyguanosine 5′-triphosphate is a good substrate for DNA polymerase. Biochemistry, 34, 11963–11969. [DOI] [PubMed] [Google Scholar]
- 12.Porter K., Briley,J. and Shaw,B.R. (1997) Direct PCR sequencing with boronated nucleotides. Nucleic Acids Res., 25, 1611–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasan A., Li,H., Tomasz,J. and Shaw,B.R. (1996) Base-boronated dinucleotides: synthesis and effect of N7-cyanoborane substitution on the base protons. Nucleic Acids Res., 24, 2150–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Porter,K., Huang,F. and Shaw,B.R. (1995) Boron-containing oligodeoxyribonucleotide 14mer duplexes: enzymatic synthesis and melting studies. Nucleic Acids Res., 23, 4495–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rait V. and Shaw,B.R. (1999) Boranophosphates support the RNase H cleavage of polyribonucleotides. Antisense Nucleic Acids Drug Dev., 9, 53–60. [DOI] [PubMed] [Google Scholar]
- 16.Shaw B.R., Sergueev,D., He,K., Porter,K., Summers,J., Sergueeva,Z. and Rait,V. (2000) Boranophosphate backbone: a mimic of phosphodiesters, phosphorothioates and methyl phosphonates. Methods Enzymol., 313, 226–257. [DOI] [PubMed] [Google Scholar]
- 17.He K., Hasan,A. and Shaw,B.R. (1997) The synthesis of ribonucleoside 5′-O-(α-P-boranotriphosphates), separation of diastereomers and incorporation by RNA polymerases. Nucleic Acids Symp. Ser., 36, 159. [Google Scholar]
- 18.He K., Porter,K., Hasan,A., Briley,J. and Shaw,B.R. (1999) Synthesis of 5-substituted 2′-deoxycytidine 5′-(alpha-P-borano)triphosphates, their incorporation into DNA and effects on exonuclease. Nucleic Acids Res., 27, 1788–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sassanfar M. and Szostak,J.W. (1993) An RNA motif that binds ATP. Nature, 364, 550–553. [DOI] [PubMed] [Google Scholar]
- 20.Burgstaller P. and Famulok,M. (1994) Isolation of RNA aptamers for biological cofactors by in vitro selection. Angewandte Chemie-Int. Ed. English, 33, 1084–1087. [Google Scholar]
- 21.Burke D.H. and Gold,L. (1997) RNA aptamers to the adenosine moiety of S-adenosyl methionine: structural inferences from variations on a theme and the reproducibility of SELEX. Nucleic Acids Res., 25, 2020–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jhaveri S., Rajendran,M. and Ellington,A. (2000) In vitro selection of signaling aptamers. Nature Biotechnol., 18, 1293–1297. [DOI] [PubMed] [Google Scholar]
- 23.Kawazoe N. and Ito,Y. (1999) Extended in vitro selection for synthesis of novel molecular recognition oligonucleotide derivatives. Nucleic Acids Symp. Ser., 177–178. [DOI] [PubMed] [Google Scholar]
- 24.Dieckmann T., Suzuki,E., Nakamura,G.K. and Feigon,J. (1996) Solution structure of an ATP-binding RNA aptamer reveals a novel fold. RNA, 2, 628–640. [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang F., Kumar,R.A., Jones,R.A. and Patel,D.J. (1996) Structural basis of RNA folding and recognition in an AMP-RNA aptamer complex. Nature, 382, 183–186. [DOI] [PubMed] [Google Scholar]
- 26.Dieckmann T., Butcher,S.E., Sassanfar,M., Szostak,J.W. and Feigon,J. (1997) Mutant ATP-binding RNA aptamers reveal the structural basis for ligand binding. J. Mol. Biol., 273, 467–478. [DOI] [PubMed] [Google Scholar]
- 27.Mulligan J.F. and Uhlenbeck,O.C. (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol., 180, 51–62. [DOI] [PubMed] [Google Scholar]
- 28.Burke D.H. and Hoffman,D.C. (1998) A novel acidophilic RNA motif that recognizes coenzyme A. Biochemistry, 37, 4653–4663. [DOI] [PubMed] [Google Scholar]
- 29.Huang Z. and Szostak,J.W. (1996) A simple method for 3′-labeling of RNA. Nucleic Acids Res., 24, 4360–4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roychowdhury M., Lato,S.M., Shank,E.D. and Burke,D.H. (2002) Flavin recognition by an RNA aptamer targeted to FAD. Biochemistry, in press. [DOI] [PubMed] [Google Scholar]
- 31.Huizenga D.E. and Szostak,J.W. (1995) A DNA aptamer that binds adenosine and ATP. Biochemistry, 34, 656–665. [DOI] [PubMed] [Google Scholar]
- 32.Tasarow T.M., Tasarow,S.L. and Eaton,B.E. (1997) RNA-catalyzed carbon-carbon bond formation. Nature, 389, 54–57. [DOI] [PubMed] [Google Scholar]
- 33.Wiegand T.W., Janssen,R.C. and Eaton,B.E. (1997) Selection of RNA amide synthases. Chem. Biol., 4, 675–683. [DOI] [PubMed] [Google Scholar]
- 34.Travascio P., Bennet,A.J., Wang,D.Y. and Sen,D. (1999) A ribozyme and a catalytic DNA with peroxidase activity: active sites versus cofactor-binding sites. Chem. Biol., 6, 779–787. [DOI] [PubMed] [Google Scholar]
- 35.Zottola M., Pedersen,L., Singh,P. and Shaw,B.R. (1994) Borohydrides as novel hydrogen-bond acceptors. In Smith,D.A. (ed.), Modeling the Hydrogen Bond. American Chemical Society, Washington, DC, pp. 277–284
- 36.Summers J.S., Roe,D., Boyle,P.D., Colvin,M. and Shaw,B.R. (1998) Structural studies of a borane-modified phosphate diester linkage: ab initio calculations on the dimethylboranophosphate anion and the single-crystal structure of its diisopropylammonium salt. Inorgan. Chem., 37, 4158–4159. [DOI] [PubMed] [Google Scholar]
- 37.Li H., Hardin,C. and Shaw,B.R. (1996) Hydrolysis of thymidine boranomonophosphate and stepwise deuterium substitution of the borane hydrogens. 31P and 11B NMR studies. J. Am. Chem. Soc., 28, 6606–6614. [Google Scholar]
- 38.Diamond J., Turner,D. and Mathews,D. (2001) Thermodynamics of three-way multibranch loops in RNA. Biochemistry, 40, 6971–6981. [DOI] [PubMed] [Google Scholar]