Abstract
The use of the polymerase chain reaction (PCR) in molecular diagnostics has increased to the point where it is now accepted as the gold standard for detecting nucleic acids from a number of origins and it has become an essential tool in the research laboratory. Real-time PCR has engendered wider acceptance of the PCR due to its improved rapidity, sensitivity, reproducibility and the reduced risk of carry-over contamination. There are currently five main chemistries used for the detection of PCR product during real-time PCR. These are the DNA binding fluorophores, the 5′ endonuclease, adjacent linear and hairpin oligoprobes and the self-fluorescing amplicons, which are described in detail. We also discuss factors that have restricted the development of multiplex real-time PCR as well as the role of real-time PCR in quantitating nucleic acids. Both amplification hardware and the fluorogenic detection chemistries have evolved rapidly as the understanding of real-time PCR has developed and this review aims to update the scientist on the current state of the art. We describe the background, advantages and limitations of real-time PCR and we review the literature as it applies to virus detection in the routine and research laboratory in order to focus on one of the many areas in which the application of real-time PCR has provided significant methodological benefits and improved patient outcomes. However, the technology discussed has been applied to other areas of microbiology as well as studies of gene expression and genetic disease.
BACKGROUND
The polymerase chain reaction (PCR) (1,2) has been used as the new gold standard for detecting a wide variety of templates across a range of scientific specialties, including virology. The method utilises a pair of synthetic oligonucleotides or primers, each hybridising to one strand of a double-stranded DNA (dsDNA) target, with the pair spanning a region that will be exponentially reproduced. The hybridised primer acts as a substrate for a DNA polymerase (most commonly derived from the thermophilic bacterium Thermus aquaticus and called Taq), which creates a complementary strand via sequential addition of deoxynucleotides. The process can be summarised in three steps: (i) dsDNA separation at temperatures >90°C, (ii) primer annealing at 50–75°C, and (iii) optimal extension at 72–78°C (Fig. 1A). The rate of temperature change or ramp rate, the length of the incubation at each temperature and the number of times each set of temperatures (or cycle) is repeated are controlled by a programmable thermal cycler. Current technologies have significantly shortened the ramp times using electronically controlled heating blocks or fan-forced heated air flows to moderate the reaction temperature. Consequently, PCR is displacing some of the gold standard cell culture, antigenaemia and serological assays (3). Existing combinations of PCR and detection assays (called ‘conventional PCR’ here) have been used to obtain quantitative data with promising results. However, these approaches have suffered from the laborious post-PCR handling steps required to evaluate the amplicon (4).
Figure 1.
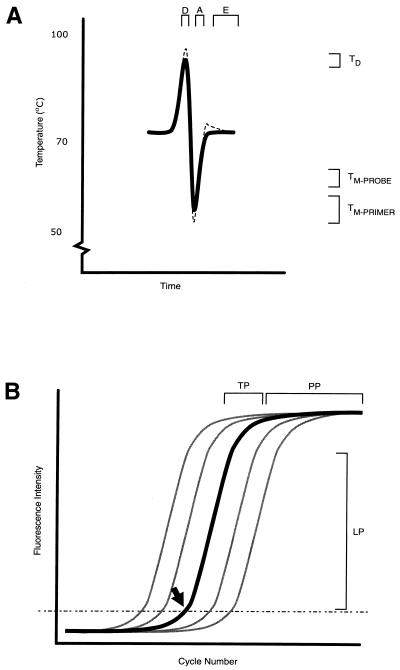
Kinetic amplification. (A) An idealised plot of temperature versus time during a single PCR cycle comprised of the denaturation (D), primer and probe annealing (A) and primer extension (E) steps. At the indicated optimal temperature ranges, dsDNA is denatured (TD), oligoprobes anneal (TM-PROBE) and finally the primers anneal as a precursor to their extension (TM-PRIMER). The actual temperature, shown as a dashed line, may overshoot the desired temperature to varying degrees, depending on the quality of the thermocycler employed. (B) The ideal amplification curve of a real-time PCR (bold), when plotted as fluorescence intensity against the cycle number, is a typical sigmoidal growth curve. Early amplification cannot be viewed because the detection signal is indistinguishable from the background. However, when enough amplicon is present, the assay’s exponential progress can be monitored as the rate of amplification enters a log-linear phase (LP). Under ideal conditions, the amount of amplicon increases at a rate of approximately one log10 every three cycles. As primers and enzyme become limiting and products inhibitory to the PCR accumulate, the reaction slows, entering a transition phase (TP), eventually reaching the plateau phase (PP) where there is little or no further increase in product yield. The point at which the fluorescence passes from insignificant levels to clearly detectable is called the threshold cycle (CT; indicated by an arrow), and this value is used in the calculation of template quantity during quantitative real-time PCR. Also shown are curves representing an optimal titration of template (grey), consisting of higher and lower starting template concentrations, which produce lower or higher CT values, respectively. Data for the construction of a standard curve are taken from the LP, which subsequently allows the concentration of unknown samples to be determined.
Traditional detection of amplified DNA relies upon electrophoresis of the nucleic acids in the presence of ethidium bromide and visual or densitometric analysis of the resulting bands after irradiation by ultraviolet light (5). Southern blot detection of amplicon using hybridisation with a labelled oligonucleotide probe is also time consuming and requires multiple PCR product handling steps, further risking a spread of amplicon throughout the laboratory (6). Alternatively, PCR–ELISA may be used to capture amplicon onto a solid phase using biotin or digoxigenin-labelled primers, oligonucleotide probes (oligoprobes) or directly after incorporation of the digoxigenin into the amplicon (7–12). Once captured, the amplicon can be detected using an enzyme-labelled avidin or anti-digoxigenin reporter molecule similar to a standard ELISA format.
The possibility that, in contrast to conventional assays, the detection of amplicon could be visualised as the amplification progressed was a welcome one (13). This approach has provided a great deal of insight into the kinetics of the reaction and it is the foundation of kinetic or ‘real-time’ PCR (Fig. 1B) (6,14–17). Real-time PCR has already proven itself valuable in laboratories around the globe, building on the enormous amount of data generated by conventional PCR assays.
The monitoring of accumulating amplicon in real time has been made possible by the labelling of primers, probes or amplicon with fluorogenic molecules, This chemistry has clear benefits over radiogenic oligoprobes that include an avoidance of radioactive emissions, ease of disposal and an extended shelf life (18).
The increased speed of real-time PCR is largely due to reduced cycle times, removal of post-PCR detection procedures and the use of fluorogenic labels and sensitive methods of detecting their emissions (19,20). The reduction in amplicon size generally recommended by the creators of commercial real-time assays may also play a role in this speed, however we have shown that decreased product size does not necessarily improve PCR efficiency (21).
The disadvantages of using real-time PCR in comparison with conventional PCR include the inability to monitor amplicon size without opening the system, the incompatibility of some platforms with some fluorogenic chemistries, and the relatively restricted multiplex capabilities of current applications. Also, the start-up expense of real-time PCR may be prohibitive when used in low-throughput laboratories. These shortcomings are mostly due to limitations in the system hardware or the available fluorogenic dyes or ‘fluorophores’, both of which will be discussed in more detail.
Because most of the popular real-time PCR chemistries depend upon the hybridisation of an oligoprobe to its complementary sequence on one of the strands of the amplicon, the use of more of the primer that creates this strand is beneficial to the generation of an increased fluorescent signal (22). Asymmetric PCR, as this is known, has been shown to produce improved fluorescence from a hairpin oligoprobe PCR (23) and we have found it directly applicable to other oligoprobe-hybridisation assays.
The most commonly used fluorogenic oligoprobes rely upon fluorescence resonance energy transfer (FRET; Fig. 2) between fluorogenic labels or between one fluorophore and a dark or ‘black-hole’ non-fluorescent quencher (NFQ), which disperses energy as heat rather than fluorescence. FRET is a spectroscopic process by which energy is passed between molecules separated by 10–100 Å that have overlapping emission and absorption spectra (24,25). Förster primarily developed the theory behind this process: the mechanism is a non-radiative induced-dipole interaction (26). The efficiency of energy transfer is proportional to the inverse sixth power of the distance (R) between the donor and acceptor (1/R6) fluorophores (27,28).
Figure 2.
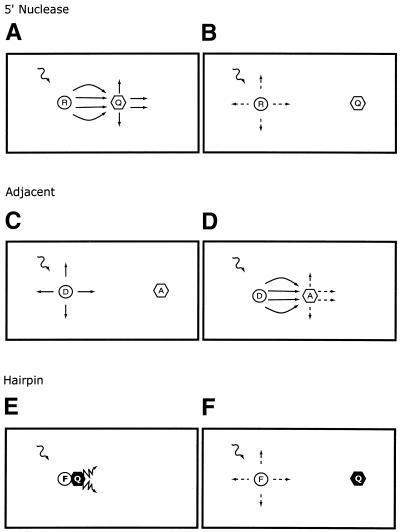
Fluorogenic mechanisms. When a 5′ nuclease probe’s reporter (R) and quencher (Q, open) are in close proximity as in (A), the quencher ‘hijacks’ the emissions that have resulted from excitation of the reporter by the instrument’s light source. The quencher then emits this energy (solid arrows). When the fluorophores are separated beyond a certain distance, as occurs upon hydrolysis as depicted in (B), the quencher no longer exerts any influence and the reporter emits at a distinctive wavelength (dashed arrows) which is recorded by the instrument. In the reverse process as depicted in (C) using adjacent oligoprobes, the fluorophores begin as separated entities. A signal from the acceptor (A) can only be generated when the donor (D) comes into close proximity as shown in (D). This occurs as the result of adjacent hybridisation of the oligoprobes to the target amplicon. In (E) another form of quenching is shown, caused by the intimate contact of labels attached to hairpin oligoprobes (molecular beacon, sunrise or scorpion). The fluorophore (F) and a NFQ (Q, closed) interact more by collision than FRET, disrupting each other’s electronic structure and directly passing on the excitation energy which is dissipated as heat (jagged, dashed arrows). When the labels are separated by disruption of the hairpin, as is the case in (F), the fluorophore is free to fluoresce (dashed arrows).
Post-amplification manipulation of the amplicon is not required for real-time PCR, therefore these assays are described as ‘closed’ or homogeneous systems. The advantages of homogeneous systems include a reduced result turnaround, minimisation of the potential for carry-over contamination and the ability to closely scrutinise the assay’s performance (29). In addition, real-time PCR has proven cost effective when implemented in a high throughput laboratory (30), particularly when replacing conventional, culture-based approaches to virus detection.
In the remainder of this article, the theory behind real-time PCR will be reviewed and its rapidly increasing use in the fields of human virology will be used as an illustration.
AMPLICON DETECTION
In the following section we will focus on the detection processes that discriminate real-time PCR from conventional PCR assays. There are five major chemistries currently in use, and they can be classified into amplicon sequence specific or non-specific methods of real-time PCR detection (31). Each of the chemistries has an associated nomenclature to describe the fluorescent labels; however, for general discussion, fluorophore will continue to be used to describe these moieties. Although this review focuses on the use of these chemistries in real-time applications, they can also be used as a label for end-point amplicon detection.
DNA-binding fluorophores
The basis of the sequence non-specific detection methods is the DNA-binding fluorogenic molecule. Included in this group are the earliest and simplest approaches to real-time PCR. Ethidium bromide (32), YO-PRO-1 (33,34) and SYBR® green 1 (35) all fluoresce when associated with dsDNA which is exposed to a suitable wavelength of light. This approach requires less specialist knowledge than the design of fluorogenic oligoprobes, is less expensive and does not suffer when the template sequence varies, which may abrogate hybridisation of an oligoprobe (36). Formation of primer-dimer (37) is common and, together with the formation of specific products, is strongly associated with entry of the PCR into the plateau phase (Fig. 1B) (38,39). Association of a DNA-binding fluorophore with primer-dimer or other non-specific amplification products can confuse interpretation of the results. Adding a short, higher temperature incubation after the extension step in which fluorescence data are acquired minimises the contribution of these products to the fluorescence signal (40). The problem of primer-dimer can also be addressed using software capable of fluorescent melting curve analysis. This method makes use of the temperature at which the dsDNA amplicon is denatured (TD; Fig. 1A). The shorter primer-dimer can be discriminated by its reduced TD compared with the full-length amplicon. Analysis of the melting curves of amplicon in the presence of SYBR green 1 has demonstrated that the practical sensitivity of DNA-binding fluorophores is limited by non-specific amplification at low initial template concentrations.
DNA binding fluorophores also increase the TD and broaden the melting transition, requiring substantial sequence change to produce a shift in the TD. Oligoprobes are able to discriminate single point mutations using the temperature at which 50% of oligoprobe-target duplexes separate (41). This temperature is called the melting temperature (TM) and it is dependent upon the concentration of the dsDNA, its length, nucleotide sequence and the solvent composition, and is often confused with TD (Fig. 1A) (42).
Linear oligoprobes
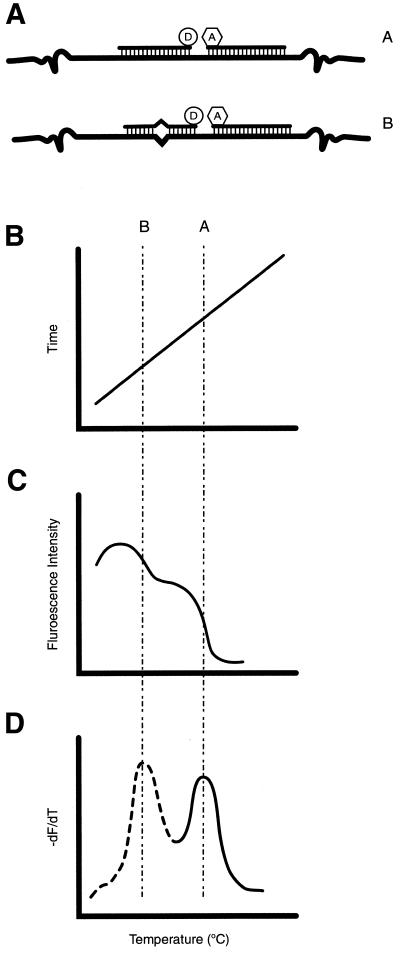
The use of a pair of adjacent, fluorogenic hybridisation oligoprobes was first described in the late 1980s (43,44) and, now known as ‘HybProbes’, they have become the method of choice for the LightCycler™ (Roche Molecular Biochemicals, Germany), a capillary-based, microvolume fluorimeter and thermocycler with rapid temperature control (20,45). The upstream oligoprobe is labelled with a 3′ donor fluorophore (FITC) and the downstream probe is commonly labelled with either a LightCycler Red 640 or Red 705 acceptor fluorophore at the 5′ terminus so that when both oligoprobes are hybridised, the two fluorophores are located within 10 nt of each other, sometimes attracting the name ‘kissing’ probes (Figs 2C, D and 3C). The plastic and glass composite capillaries are optically clear and act as cuvettes for fluorescence analysis, as well as facilitating rapid heat transfer. Capillaries are rotated past a blue light-emitting diode and fluorescence is monitored by three photodetection diodes with different wavelength filters. The temperature is varied by rapidly heating and cooling air using a heating element and fan which produce ramp rates of 20°C/s, prolonging polymerase survival (46). Additionally, because the oligoprobes are not significantly hydrolysed during amplification (47) and the LightCycler is able to monitor the changes in fluorescence emission during denaturation of the adjacent oligoprobes from their amplicon, this system can perform single tube genotyping. This capability, which makes use of fluorescent melting curve analysis, provides significant information about the sequence to which the oligoprobes are binding. Mutation(s) under one or both oligoprobes can be determined by the decrease in melting temperature that they incur due to destabilisation of the oligoprobe/target duplex (Fig. 4). This has imparted significant improvements in speed upon the diagnosis of genetic disease as well as a growing number of multiplex PCR approaches for the detection of related viral pathogens. Despite the fact that the hybridisation does not reach equilibrium using these ramp rates, the apparent TM values are both reproducible and characteristic of a given probe/target duplex (48). However, the capillaries are fragile and require some experience to handle (49).
Figure 3.
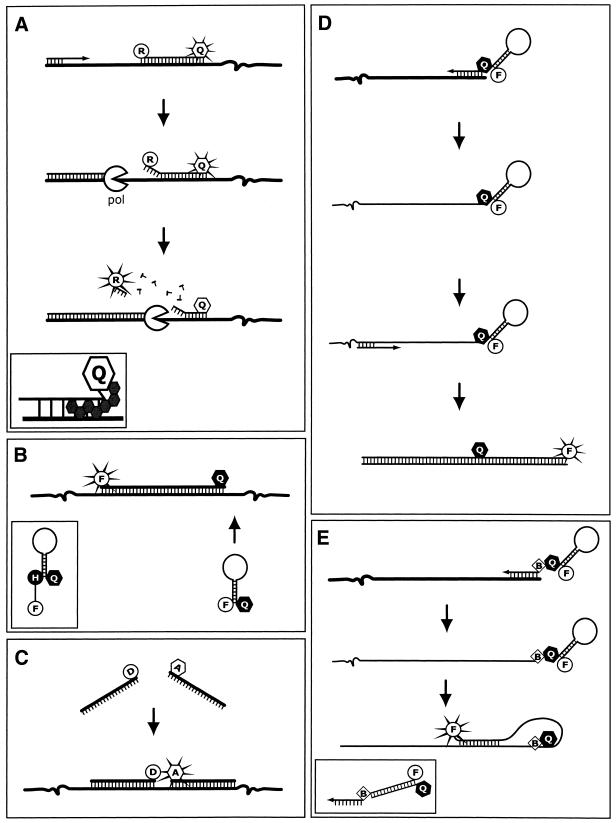
Oligoprobe chemistries. (A) 5′ Nuclease oligoprobes. As the DNA polymerase (pol) progresses along the relevant strand, it displaces and then hydrolyses the oligoprobe via its 5′→3′ endonuclease activity. Once the reporter (R) is removed from the extinguishing influence of the quencher (Q, open), it is able to release excitation energy at a wavelength that is monitored by the instrument and different from the emissions of the quencher. Inset shows the NFQ and MGB molecule that make up the improved MGB nuclease-oligoprobes. (B) Hairpin oligoprobes. Hybridisation of the oligoprobe to the target separates the fluorophore (F) and non-fluorescent quencher (Q, closed) sufficiently to allow emission from the excited fluorophore, which is monitored. Inset shows a wavelength-shifting hairpin oligoprobe incorporating a harvester molecule. (C) Adjacent oligoprobes. Adjacent hybridisation results in a FRET signal due to interaction between the donor (D) and acceptor (A) fluorophores. This bimolecular system acquires its data from the acceptor’s emissions in an opposite manner to the function of nuclease oligoprobe chemistry. (D) Sunrise primers. The opposite strand is duplicated so that the primer’s hairpin structure can be disrupted. This separates the labels, eliminating the quenching in a similar manner to the hairpin oligoprobe. (E) Scorpion primers. The primer does not require extension of the complementary strand; in fact it blocks extension to ensure that the hairpin in the probe is only disrupted by specific hybridisation with a complementary sequence designed to occur downstream of its own, nascent strand. Inset shows a duplex scorpion that exchanges the stem–loop structure for a primer element terminally labelled with the fluorophore and a separate complementary oligonucleotide labelled with a quencher at the 5′ terminus.
Figure 4.
Fluorescent melting curve analysis. At the completion of a real-time PCR using a pair of adjacent oligoprobes, the reaction can be cooled to a temperature below the expected TM of the oligoprobes then heated above 85°C at a fraction of a degree per second (B). During heating, the emissions of the acceptor can be constantly acquired (C). Software calculates the negative derivative of the fluorescence over time, producing clear peaks that indicate the TM of the oligoprobe-target melting transition (D). When one or more mutations are present under one or both oligoprobes (A), this TM is shifted and this shift can be used diagnostically to discriminate single nucleotide polymorphisms in the template. Ideally, one of the oligoprobes, the anchor, is designed to bind to a stable sequence region, whereas the other, sensor, will span the mismatch. Mismatches near the centre of the probe and flanked by G:C pairs are more destabilising than mismatches near the ends of the oligoprobe flanked by A:T pairs.
When comparing signals from the different chemistries, the destruction of nuclease oligoprobes continues despite a plateau in product accumulation whereas SYBR green 1 fluorescence in the no template control generally increases non-specifically during later cycles. Adjacent oligoprobe fluorescence begins to decrease as the rate of collision between the growing numbers of complementary amplicon strands increases favouring the formation of dsDNA over the hybridisation of oligoprobe to its target DNA strand. Additionally, there is the possibility that some oligoprobe is consumed by sequence-related endonuclease activity (50,51). All three oligoprobe chemistries (SYBR Green I, nuclease and adjacent oligoprobes) seem capable of detecting amplified product with approximately the same sensitivity (20).
Combinations of the above approaches are now appearing as more users of the instrumentation become familiar with the concepts behind real-time PCR and contribute to the literature. If a sequence-specific, fluorophore-labelled linear oligoprobe is added to a SYBR green 1 mix, currently called the Bi-probe system, FRET will occur and an additional layer of specificity can be obtained (44,52,53). An assay using a BODIPY®FL-labelled oligoprobe was adapted to run in the LightCycler using a β-globin target sequence (54). The probe was designed so that the fluorophore was located on a terminal cytosine and was quenched by proximity with a complementary guanine. The assay demonstrated that quenching varies linearly with the concentration of template across a defined concentration range. The commonly used fluorophore FITC is inherently quenched by deoxyguanosine nucleotides. The level of quenching can be increased if more guanines are present or a single guanine is located in the first overhang position, 1 nt beyond the fluorophore-labelled terminus of the probe. This approach to amplicon detection is easier to design than fluorogenic oligoprobes, simpler to synthesise and use in real-time PCR and does not require a DNA polymerase with nuclease activity (55).
The light-up probe is a peptide nucleic acid to which the asymmetric cyanine fluorophore thiazole orange is attached (56). When hybridised with a nucleic acid target, either as a duplex or triplex, depending on the oligoprobe’s sequence, the fluorophore becomes strongly fluorescent. These probes do not interfere with the PCR, do not require conformational change, are sensitive to single nucleotide mismatches allowing fluorescence melting analysis, and because a single reporter is used, a direct measurement of fluorescence can be made instead of the measurement of a change in fluorescence between two fluorophores (56,57). However, non-specific fluorescence has been reported during later cycles using these probes (58).
5′ Nuclease oligoprobes
In the late 1980s homogeneous assays were few and far between, but rapid advances in thermocycler instrumentation and the chemistry of nucleic acid manipulation have since made these assays commonplace. The success of these assays revolves around a signal changing in some rapid and measurable way upon hybridisation of a probe to its target (59). By using an excess, the time required for hybridisation of an oligoprobe to its target, especially when the amount of that target has been increased by PCR or some other amplifying process, is significantly reduced (41,59).
In 1991, Holland et al. (6) described a technique that was to form the foundation for homogeneous PCR using fluorogenic oligoprobes. Amplicon was detected by monitoring the effect of Taq DNA polymerase’s 5′→3′ endonuclease activity on specific oligoprobe/target DNA duplexes. The radiolabelled products were examined using thin layer chromatography and the presence or absence of hydrolysis was used as an indicator of duplex formation. These oligoprobes contained a 3′ phosphate moiety, which blocked their extension by the polymerase, but otherwise had no affect on the amplicon’s yield.
The desirable criteria for an oligoprobe label are (i) easy attachment of the label to DNA, (ii) detectability at low concentrations, (iii) detectability using simple instrumentation, (iv) production of an altered signal upon specific hybridisation, (v) biological safety, (vi) stability at elevated temperatures and (vii) an absence of interference with the activity of the polymerase (6,18).
An innovative approach used nick-translation PCR in combination with dual-fluorophore labelled oligoprobes (14). In the first truly homogenous assay of its kind, one fluorophore was added to the 5′ terminus and one to the middle of a sequence specific oligonucleotide probe. When in such close proximity, the 5′ reporter fluorophore (6-carboxy-fluoroscein) transferred laser-induced excitation energy by FRET to the 3′ quencher fluorophore (6-carboxy-tetramethyl-rhodamine; TAMRA), which reduced the lifetime of the reporter’s excited state by taking its excess energy and emitting it as a fluorescent signal of its own (Fig. 2A and B). TAMRA emitted the new energy at a wavelength that was monitored but not utilised in the presentation of data. However, when the oligoprobe hybridised to its template, the fluorophores were released due to hydrolysis of the oligoprobe component of the probe/target duplex. Once the labels were separated, the reporter’s emissions were no longer quenched and the instrument monitored the resulting fluorescence. These oligoprobes have been called 5′ nuclease, hydrolysis or TaqMan® oligoprobes (Fig. 3A). Nuclease oligoprobes have design requirements that are applicable to the other linear oligoprobe chemistries, including (i) a length of 20–40 nt, (ii) a GC content of 40–60%, (iii) no runs of a single nucleotide, particularly G, (iv) no repeated sequence motifs, (v) an absence of hybridisation or overlap with the forward or reverse primers and (vi) a TM at least 5°C higher than that of the primers, to ensure the oligoprobe has bound to the template before extension of the primers can occur (60).
This technology, however, required the development of a platform to excite and detect fluorescence as well as perform thermal cycling. A charge-coupled device had been described in 1992 for the quantification of conventional reverse transcription (RT)–PCR products (61). In 1993 this approach was combined with a thermal cycler resulting in the first real-time PCR fluorescence excitation and detection platform (29). To date, the ABI Prism® 7700 sequence detection system (Perkin Elmer Corporation/Applied Biosystems, USA) has been the main instrument used for 5′ nuclease oligoprobes. Non-PCR related fluorescence fluctuations have been normalised using a non-participating or ‘passive’ internal reference fluorophore (6-carboxy-N,N,N′,N′-tetramethylrhodamine; ROX). The corrected values, obtained from a ratio of the emission intensity of the reporter signal and ROX, are called RQ+. To further control amplification fluctuations, the fluorescence from a ‘no-template’ control reaction (RQ–) is subtracted from RQ+ resulting in the ΔRQ value that indicates the magnitude of the signal generated for the given PCR (62).
The fractional cycle number at which the real-time fluorescence signal mirrors progression of the reaction above the background noise was used as an indicator of successful target amplification (63). This threshold cycle (CT) is defined as the PCR cycle in which the gain in fluorescence generated by the accumulating amplicon exceeds 10 standard deviations of the mean baseline fluorescence, using data taken from cycles 3 to 15 (Fig. 1B) (64). The CT is proportional to the number of target copies present in the sample (17).
A recent improvement to the nuclease oligoprobe has resulted in the minor groove binding (MGB) oligoprobes (Fig. 3A, inset). This chemistry replaces the standard TAMRA quencher with an NFQ and incorporates a molecule that stabilises the oligoprobe-target duplex by folding into the minor groove of the dsDNA (65). This allows the use of very short (14 nt) oligoprobes, which are ideal for detecting single nucleotide polymorphisms (SNPs). A related use of dual labelled oligonucleotide sequences has been to provide the signal-generating portion of the DzyNA–PCR system (66). Here, the reporter and quencher are separated after cleavage of the probe by a DNAzyme, which is created during PCR as the complement of an antisense DNAzyme sequence included in the 5′ tail of one of the primers. Upon cleavage, the dual labelled substrate releases the fluorophores and generates a signal in an analogous manner to the 5′ nuclease probe.
Hairpin oligoprobes
Molecular beacons were the first hairpin oligoprobes and are a variation of the dual-labelled nuclease oligoprobe (Fig. 3B). The hairpin oligoprobe’s fluorogenic labels are called fluorophore and quencher, and they are positioned at the termini of the oligoprobe. The labels are held in close proximity by distal stem regions of homologous base pairing deliberately designed to create a hairpin structure which results in quenching either by FRET or a direct energy transfer by a collisional mechanism due to the intimate proximity of the labels (Fig. 2E and F) (67). In the presence of a complementary sequence, designed to occur within the bounds of the primer binding sites, the oligoprobe will hybridise, shifting into an open configuration. The fluorophore is now spatially removed from the quencher’s influence and fluorescent emissions are monitored during each cycle (68). The occurrence of a mismatch between a hairpin oligoprobe and its target has a greater destabilising effect on the duplex than the introduction of an equivalent mismatch between the target and a linear oligoprobe. This is because the hairpin structure provides a highly stable alternate conformation. Therefore, hairpin oligoprobes have been shown to be more specific than the more common linear oligoprobes making them ideal candidates for detecting SNPs (67). The quencher, 4-(4′-dimethylamino-phenylazo)-benzene (DABCYL), differs from that described for the nuclease oligoprobes because it is an NFQ.
The wavelength-shifting hairpin probe is a recent improvement to this chemistry which makes use of a second, harvesting fluorophore. The harvester passes excitation energy acquired from a blue light source and releases it as fluorescent energy in the far-red wavelengths. The energy can then be used by a receptive ‘emitter’ fluorophore that produces light at characteristic wavelengths (Fig. 3B, inset). This offers the potential for improved multiplex real-time PCR and SNP analysis (Fig. 4), using currently available instruments (69). Because the function of these oligoprobes depends upon correct hybridisation of the stem, accurate design is crucial to their function (47).
Self-fluorescing amplicon
The self-priming amplicon is similar in concept to the hairpin oligoprobe, except that the label becomes irreversibly incorporated into the PCR product (Fig. 3D and E). Two approaches have been described: sunrise primers (now commercially called Amplifluor™ hairpin primers) and scorpion primers (31,70). The sunrise primer consists of a 5′ fluorophore and a DABCYL NFQ. The labels are separated by complementary stretches of sequence that create a stem when the sunrise primer is closed. At the 3′ terminus is a target-specific primer sequence. The sunrise primer’s sequence is intended to be duplicated by the nascent complementary strand and, in this way, the stem is destabilised, the two fluorophores are held ~20 nt (70 Å) apart and the fluorophore is free to emit its excitation energy for monitoring (70). This system could suffer from non-specific fluorescence due to duplication of the sunrise primer sequence during the formation of primer-dimer.
The scorpion primer is almost identical in design except for an adjacent hexethylene glycol molecule that blocks duplication of the signalling portion of the scorpion. In addition to the difference in structure, the function of scorpion primers differs slightly in that the 5′ region of the oligonucleotide is designed to hybridise to a complementary region within the amplicon. This hybridisation forces the labels apart disrupting the hairpin and permitting emission in the same way as hairpin probes (31).
VIRAL QUANTITATION
The majority of diagnostic PCR assays reported to date have been used in a qualitative, or ‘yes/no’ format. The development of real-time PCR has brought true quantitation of target nucleic acids out of the pure research laboratory and into the diagnostic laboratory.
Determining the amount of template by PCR can be performed in two ways: as relative quantitation and as absolute quantitation. Relative quantitation describes changes in the amount of a target sequence compared with its level in a related matrix. Absolute quantitation states the exact number of nucleic acid targets present in the sample in relation to a specific unit (71). Generally, relative quantitation provides sufficient information and is simpler to develop. However, when monitoring the progress of an infection, absolute quantitation is useful in order to express the results in units that are common to both scientists and clinicians and across different platforms. Absolute quantitation may also be necessary when there is a lack of sequential specimens to demonstrate changes in virus levels, no suitably standardised reference reagent or when the viral load is used to differentiate active versus persistent infection.
A very accurate approach to absolute quantitation by PCR is the use of competitive co-amplification of an internal control nucleic acid of known concentration and a wild-type target nucleic acid of unknown concentration, with the former designed or chosen to amplify with an equal efficiency to the latter (72–76). However, while conventional competitive PCR is relatively inexpensive, real-time PCR is far more convenient, reliable and better suited to quick decision making in a clinical situation (77,78). This is because conventional, quantitative, competitive PCR (qcPCR) requires significant development and optimisation to ensure reproducible performance and a predetermined dynamic range for both the amplification and detection components (79).
Although a comparison of absolute standard curves, relative standard curves and CT values produces similar final values (80), the general belief remains that an internal control in combination with replicates of each sample are essential for reliable quantitation by PCR (38,39). Unfortunately, real-time PCR software with the ability to calculate the concentration of an unknown by comparing signals generated by an amplified target and internal control is only beginning to emerge. This issue will hopefully be addressed in upcoming commercial releases (81). Therefore, the next best approach to quantitation by PCR is the use of an external standard curve. This approach relies upon titration of an identically amplified template, in a related sample matrix, within the same experimental run. While the external standard curve is the more commonly described approach, it suffers from uncontrolled and unmonitored inter-tube variations. Because of this omission, such experiments should be described as semi-quantitative. Despite this sub-optimal approach, fluorescence data is generally collected from PCR cycles that span the linear amplification portion of the reaction where the fluorescent signal and the accumulating DNA are proportional. Because the emissions from fluorescent chemistries are temperature dependent, data is generally acquired only once per cycle at the same temperature in order to monitor amplicon yield (45). The CT of the sample at a specific fluorescence value can then be compared with similar data collected from a series of standards by the calculation of a standard curve. The determination of the CT depends upon the sensitivity and ability of the instrument to discriminate specific fluorescence from background noise, the concentration and nature of the fluorescence-generating component and the amount of template initially present.
Real-time PCR offers significant improvements to the quantitation of viral load because of its enormous dynamic range that can accommodate at least eight log10 copies of nucleic acid template (33,52,77,82–89). This is made possible because the data are chosen from the linear phase (LP; Fig. 1B) of amplification where conditions are optimal, rather than the end-point where the final amount of amplicon present may have been affected by inhibitors, poorly optimised reaction conditions or saturation by inhibitory PCR by-products and double-stranded amplicon. The result of taking data from the end-point is that there may not be a relationship between the initial template and final amplicon concentrations.
Real-time PCR is also an attractive alternative to conventional PCR for the study of viral load because of its low inter-assay and intra-assay variability (77,87,90) and its equivalent or greater analytical sensitivity in comparison with traditional viral culture, or conventional single-round, and nested PCR (77,85,91–96). Real-time PCR has been reported to be at least as sensitive as Southern blot (92). However, these reports could be an over-estimate due to the choice of smaller targets, which amplify more efficiently, or due to the use of different or improved primers for the real-time assays because the use of software to design optimised primers and oligoprobes is more common.
When this increased sensitivity and broad dynamic range are combined, it is possible to quantitate template from samples containing a large range of concentrations, as is often the case in patient samples. This avoids the need for dilution of the amplicon prior to conventional detection or repeat of the assay using a diluted sample because the first test result falls outside the limits of the assay. These are problems encountered when using some conventional qcPCR assay kits, which cannot encompass high viral loads whilst maintaining suitable sensitivity (52,97–99). The flexibility of real-time PCR is also demonstrated by its ability to detect one target in the presence of a vast excess of another target in duplexed assays (84).
Viral load is also a useful indicator of the extent of active infection, virus–host interactions and the response to antiviral therapy, all of which can play a role in the treatment regimen selected (100,101). Conventional quantitative PCR has already proven the benefits of applying nucleic acid amplification to the monitoring of viral load as a useful marker of disease progression and as a component of studies into the efficacy of antiviral compounds (74,100,102–104). The severity of some diseases has been shown to correlate with the viral load making real-time PCR quantitation useful to study not simply the presence of a virus but the role of viral reactivation or persistence in the progression of disease (78,82,91,105–112).
An example of the benefits which real-time PCR has brought to the quantitative detection of human cytomegalovirus (CMV) is seen in patients who are immunosuppressed following solid organ or bone marrow transplantation. Although qualitative detection of CMV DNA by PCR has been used as an indicator for the success of antiviral therapy, quantitative assays are preferred in order to monitor patient’s therapeutic responses. Moreover, since it has been postulated that the monitoring of viral replication over time is a more reliable indicator of a developing viral disease than the determination of absolute viral amounts at a single point of time, several quantitative assays have been established and evaluated to increase diagnostic accuracy. Quantitative competitive assays based on end-point analysis have displayed detection limits of 5 × 101 genome equivalents (ge) per assay and a dynamic range of 5 × 101–5 × 104 ge/assay (113). Hybridisation-based assays covered approximately the same dynamic range of four orders of magnitude with detection limits of 20 ge/assay (114,115). Although these assays possess dynamic ranges that may be sufficient for most clinical applications, they display a high inter- and intra-assay variability, up to 40% (115).
In contrast, one of the first published real-time PCR assays for the detection of CMV DNA could be performed in <90 min, spanned a dynamic range of six to seven orders of magnitude with a detection limit of at least 10 ge/assay and an inter-assay and intra-assay variability of <10% and <5%, respectively, using plasma samples from bone marrow transplant patients (116).
MULTIPLEX REAL-TIME PCR
Multiplexing (using multiple primers to allow amplification of multiple templates within a single reaction) is a useful application of conventional PCR (117). However, its transfer to real-time PCR has confused its traditional terminology. The term multiplex real-time PCR is more commonly used to describe the use of multiple fluorogenic oligoprobes for the discrimination of multiple amplicons. The transfer of this technique has proven problematic because of the limited number of fluorophores available (14) and the common use of a monochromatic energising light source. Although excitation by a single wavelength produces bright emissions from a suitably selected fluorophore, this restricts the number of fluorophores that can be included (69). Recent improvements to the design of the hairpin primers, and hairpin and nuclease oligoprobes as well as novel combinations of fluorophores such as in the bi-probe and light-up probe systems, have promised the ability to discriminate an increasing number of targets.
The discovery and application of the non-fluorescent quenchers has liberated some wavelengths that were previously occupied by the emissions from the early quenchers themselves. This breakthrough has permitted the inclusion of a greater number of spectrally discernable oligoprobes per reaction, and highlighted the need for a single non-fluorescent quencher, which can quench a broad range of emission wavelengths (e.g. 400–600 nm). Early real-time PCR systems contained optimised filters to minimise overlap of the emission spectra from the fluorophores. Despite this, the number of fluorophores that could be combined and clearly distinguished was limited when compared with the discriminatory abilities of conventional multiplex PCR. More recent real-time PCR platforms have incorporated either multiple light-emitting diodes to span the entire visible spectrum, or a tungsten light source, which emits light over a broad range of wavelengths. When these platforms incorporate high quality optical filters it is possible to apply any current real-time PCR detection chemistries on the one machine. Nonetheless, these improvements generally allow only four-colour oligoprobe multiplexing, of which one colour is ideally set aside for an internal control to monitor inhibition and perhaps even act as a co-amplified competitor. Some real-time PCR designs have made use of single or multiple nucleotide changes between similar templates to allow their differentiation by TM (Fig. 4) thus avoiding the need for multiple fluorophores (49,91,118–122). This approach has been used far more commonly in the detection of human genetic diseases where as many as 27 possible nucleotide substitutions have been detected using only one or two fluorophores (48,123–128).
To date, there have only been a handful of truly multiplexed real-time PCR assays described in the literature and few of these have been applied to the diagnosis of infectious disease. Some of these approaches cannot, technically, be considered real-time, homogenous assays because they require interruption of the procedure to transfer template, their fluorescence is detected by end-point analysis or the assays are not performed within the same tube. One of the best viral, multiplex, real-time PCR protocols can discriminate between four retroviral target sequences (129), however, conventional multiplex PCR using end-point detection can easily discriminate more than five different amplified sequences, indicating a greater flexibility when compared with real-time PCR (130–133).
Despite these limitations, a number of assays have been applied to the detection of several viral genomes at once, including the use of non-specific label, SYBR green 1 to detect herpes simplex viruses (HSV), varicella zoster virus (VZV) or CMV, in separate tubes (134) or, by adaptation of a conventional multiplex PCR, to identify HSV-1 and HSV-2, VZV and enteroviruses within a single capillary by applying fluorescent melting curve analysis (121). Another single-tube multiplexed nuclease oligoprobe RT–PCR was capable of simultaneously detecting Influenza A and B in patient’s respiratory samples with improved sensitivity compared with conventional or shell-viral culture (95).
Future developments of novel chemistries such as combinatorial fluorescence energy transfer tags (135), and improvements to the design of real-time instrumentation and software will greatly enhance the future of multiplex real-time PCR.
APPLICATIONS TO VIROLOGY
Real-time PCR has been extremely useful for studying viral agents of infectious disease and helping to clarify disputed infectious disease processes. Most of the assays presented in the literature allow an increased frequency of virus detection compared with conventional techniques, which makes the implementation of real-time PCR attractive to many areas of virology.
Of course, real-time PCR has proven increasingly valuable for general virological studies, although increasingly, these applications are difficult to review due to the nature of their use as a tool rather than the focus of published study. Such studies have investigated the role of viruses in a range of diseases by simply confirming the presence or absence of the virus (136,137) or, in the future, by monitoring the levels of specific gene activity (138) as a result of growth under manipulated conditions. Altered viral entry or replication, caused by the modification of target tissues, can also be followed using real-time PCR as can links between virus replication and the expression of cellular genes (139–141).
Real-time PCR has enhanced the speed and scope of measuring viral strain and titre differences in patients displaying different syndromes due to varieties of the same virus (106). Also, epidemiological studies have been improved in speed and scope through the use of real-time PCR because it can reliably measure the amount of two nucleic acid targets within a single reaction (91,142). New chemistries have allowed better discrimination of multiple viral genotypes within a single reaction vessel (143) and provided an alternative to methods of virus detection based on morbidity and mortality assays.
The use of real-time PCR has provided insight into the role(s) some compounds have on PCR inhibition as well as shedding light on the efficiency of different nucleic acid extraction methods, from a diverse range of sample types (144–146). This ability to utilise template from a range of sample types fulfils a requirement for an ideal detection system, which is the ability to apply a single technology across many fields. This flexibility is highlighted by the detection of viral nucleic acids derived, in different ways, from plants (147–149), animals (86,89,94,150–152), urban sludge (85,153), tissue culture (23,77,96,154–160), various solid tissues (108,118,161–166), cerebrospinal fluid (49,167,168), peripheral blood mononuclear cells (82,93,105,112,169–171), plasma (81,88,90,172–176), serum (30,33,36,87,97,99,109,177–181), swabs (182,183), saliva (106) and urine (78,122,184,185). Also, chronic conditions such as sarcoma (186–189), carcinoma (92,111), cervical intraepithelial neoplasia (190–192), and lymphoproliferative disorders (193,194) can be relatively easily studied to investigate direct or indirect links with viral infection. These studies have targeted viruses which include the flaviviruses (33,36,96,109,195), hepadnaviridae (52,97), herpesviruses (21,77,78,82–84,91,92,98,105,106,108, 111,112,144,167,171), orthomyxoviruses (95), parvoviruses (88), papovaviruses (122,143,145), paramyxoviruses (94), picornaviruses (85,86), retroviruses (90,93,156,196,197) and TT virus (137). Viral load monitoring by real-time PCR has also proven beneficial for studying patients following organ transplant (21,83,107,198–201).
This technology has now become an essential tool in the thorough assessment of viral gene therapy vectors prior to their use in clinical trials. Nuclease oligoprobes have been most commonly reported for these studies, which assess the biodistribution, function and purity of these drug preparations (164,197,202–205).
Additionally, the study of emerging viruses has been complimented by the use of real-time PCR as a tool to demonstrate links between unique viral sequences and patient clinical signs and symptoms (94,96,150,160,206,207).
The speed and flexibility of real-time PCR has also proven useful to commercial interests for the screening of microbial contamination of large-scale reagent preparations produced from eukaryotic expression systems (208,209).
CONCLUSIONS AND SUMMARY
Advances in the development of fluorophores, nucleotide labelling chemistries, and the novel applications of oligoprobe hybridisation have provided real-time PCR technology with a broad enough base to ensure its acceptance. Recently, instrumentation has appeared that is capable of incredibly short cycling times combined with the ability to detect and differentiate multiple amplicons. New instruments are also flexible enough to allow the use of any of the chemistries described in this review making real-time nucleic acid amplification an increasingly attractive and viable proposition for the routine diagnostic laboratory. In many cases these laboratories perform tissue culture to isolate virus and serological methods to confirm the identity of the isolate, which may take a considerable, and clinically relevant, amount of time.
The familiarity that leads to comfortable routine use of a technology is now apparent in the inclusion of the fluorogenic oligoprobe chemistries in many laboratories. According to the literature, the most widely used format is the 5′ endonuclease oligoprobe although that is most likely due to its commercial maturity. The more recently developed oligoprobe chemistries have been used in a number of innovative applications and it is apparent from the rate and content of real-time PCR-related publications that they are becoming more widely accepted.
Recent developments in multiplex real-time PCR have suggested a future in which easy identification, genotyping and quantitation of viral targets in single, rapid reactions will be commonplace. Of course, this technology is by no means restricted to virology, as significant achievements have appeared in the area of mutation detection, applying all the benefits described above to enhance the detection of genetic disease and, where applicable, allow quantitation of the extent of such genetic changes.
Micro- and macro-array technology will surely play some chimeric role in the real-time PCR of future research, but for now there is significant potential for routine, research and commercial interests to redesign existing systems for greater sophistication, flexibility and the ability to generate high quality quantitative data. The development of assays capable of real-time PCR that can discriminate as many targets as conventional multiplex PCR assays, whilst producing quantitative data at a greatly increased speed, will consolidate fluorogenic nucleic acid amplification as a routine tool for the laboratory of tomorrow.
Acknowledgments
ACKNOWLEDGEMENTS
The Royal Children’s Hospital Foundation Grant number 922-034, which was sponsored by the Woolworth’s ‘Care for Kids’ campaign, supported this work. This is publication number 139 from the Sir Albert Sakzewski Virus Research Centre.
REFERENCES
- 1.Freymuth F., Eugene,G., Vabret,A., Petitjean,J., Gennetay,E., Brouard,J., Duhamel,J.F. and Guillois,B. (1995) Detection of respiratory syncytial virus by reverse transcription–PCR and hybridization with a DNA enzyme immunoassay. J. Clin. Microbiol., 33, 3352–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mullis K.B. and Faloona,F. (1987) Specific synthesis of DNA in vitro via a polymerase-catalysed chain reaction. Methods Enzymol., 155, 335–350. [DOI] [PubMed] [Google Scholar]
- 3.Niubo J., Perez,J.L., Carvajal,A., Ardanuy,C. and Martin,R. (1994) Effect of delayed processing of blood samples on performance of cytomegalovirus antigenemia assay. J. Clin. Microbiol., 32, 1119–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guatelli J.C., Gingeras,T.R. and Richman,D.D. (1989) Nucleic acid amplification in vitro: detection of sequences with low copy numbers and application to diagnosis of human immunodeficiency virus type 1 infection. Clin. Microbiol. Rev., 2, 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kidd I.M., Clark,D.A. and Emery,V.C. (2000) A non-radioisotopic quantitative competitive polymerase chain reaction method: application in measurement of human herpesvirus 7 load. J. Virol. Methods, 87, 177–181. [DOI] [PubMed] [Google Scholar]
- 6.Holland P.M., Abramson,R.D., Watson,R. and Gelfand,D.H. (1991) Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus.Proc. Natl Acad. Sci. USA, 88, 7276–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Vliet G.M.E., Hermans,C.J. and Klatser,P.R. (1993) Simple colorimetric microtiter plate hybridization assay for detection of amplified Mycobacterium leprae DNA. J. Clin. Microbiol., 31, 665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller G.H., Huang,D.-P., Shih,J.W.-K. and Manak,M.M. (1990) Detection of hepatitis B virus DNA in serum by polymerase chain reaction amplification and microtiter sandwich hybridization. J. Clin. Microbiol., 28, 1411–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kemp D.J., Churchill,M.J., Smith,D.B., Biggs,B.A., Foote,S.J., Peterson,M.G., Samaras,N., Deacon,N.J. and Doherty,R. (1990) Simplified colorimetric analysis of polymerase chain reactions: detection of HIV sequences in AIDS patients. Gene, 94, 223–228. [DOI] [PubMed] [Google Scholar]
- 10.Kox L.F.F., Noordhoek,G.T., Kunakorn,M., Mulder,S., Sterrenburg,M. and Kolk,A.H.J. (1996) Microwell hybridization assay for detection of PCR products from Mycobacterium tuberculosis complex and the recombinant Mycobacterium smegmatis strain 1008 used as an internal control. J. Clin. Microbiol., 34, 2117–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dekonenko A., Ibrahim,M.S. and Schmaljohn,C.S. (1997) A colorimetric PCR-enzyme immunoassay to identify hantaviruses. Clin. Diag. Virol., 8, 113–121. [DOI] [PubMed] [Google Scholar]
- 12.Watzinger F., Hörth,E. and Lion,T. (2001) Quantitation of mRNA expression by competititive PCR using non-homologous competitors containing a shifted restriction site. Nucleic Acids Res., 29, e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lomeli H., Tyagi,S., Pritchard,C.G., Lizardi,P.M. and Kramer,F.R. (1989) Quantitative assays based on the use of replicatable hybridization probes. Clin. Chem., 35, 1826–1831. [PubMed] [Google Scholar]
- 14.Lee L.G., Connell,C.R. and Bloch,W. (1993) Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res., 21, 3761–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak K.J., Flood,S.J.A., Marmaro,J., Giusti,W. and Deetz,K. (1995) Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl., 4, 357–362. [DOI] [PubMed] [Google Scholar]
- 16.Heid C.A., Stevens,J., Livak,K.J. and Williams,P.M. (1996) Real time quantitative PCR. Genome Res., 6, 986–994. [DOI] [PubMed] [Google Scholar]
- 17.Gibson U.E.M., Heid,C.A. and Williams,P.M. (1996) A novel method for real time quantitative RT–PCR. Genome Res., 6, 995–1001. [DOI] [PubMed] [Google Scholar]
- 18.Matthews J.A. and Kricka,L.J. (1988) Analytical strategies for the use of DNA probes Anal. Biochem., 169, 1–25. [DOI] [PubMed] [Google Scholar]
- 19.Wittwer C.T., Fillmore,G.C. and Garling,D.J. (1990) Minimizing the time required for DNA amplification by efficient heat transfer to small samples. Anal. Biochem., 186, 328–331. [DOI] [PubMed] [Google Scholar]
- 20.Wittwer C.T., Ririe,K.M., Andrew,R.V., David,D.A., Gundry,R.A. and Balis,U.J. (1997) The LightCycler™: a microvolume multisample fluorimeter with rapid temperature control. Biotechniques, 22, 176–181. [DOI] [PubMed] [Google Scholar]
- 21.Nitsche A., Steuer,N., Schmidt,C.A., Landt,O., Ellerbrok,H., Pauli,G. and Siegert,W. (2000) Detection of human cytomegalovirus DNA by real-time quantitative PCR. J. Clin. Microbiol., 38, 2734–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gyllensten U.B. and Erlich,H.A. (1988) Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc. Natl Acad. Sci. USA, 85, 7652–7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poddar S.K. (2000) Symmetric vs asymmetric PCR and molecular beacon probe in the detection of a target gene of adenovirus. Mol. Cell. Probes, 14, 25–32. [DOI] [PubMed] [Google Scholar]
- 24.Stryer L. and Haugland,R.P. (1967) Energy transfer: a spectroscopic ruler. Proc. Natl Acad. Sci. USA, 58, 719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clegg R.M. (1992) Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol., 211, 353–388. [DOI] [PubMed] [Google Scholar]
- 26.Förster T. (1948) Zwischenmolekulare energiewanderung und fluoreszenz. Ann. Phys., 6, 55–75. [Google Scholar]
- 27.Selvin P. (1995) Fluorescence resonance energy transfer. Methods Enzymol., 246, 300–334. [DOI] [PubMed] [Google Scholar]
- 28.Didenko V.V. (2001) DNA probes using fluorescence resonance energy transfer (FRET): designs and applications. Biotechniques, 31, 1106–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higuchi R., Fockler,C., Dollinger,G. and Watson,R. (1993) Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (NY), 11, 1026–1030. [DOI] [PubMed] [Google Scholar]
- 30.Martell M., Gómez,J., Esteban,J.I., Sauleda,S., Quer,J., Cabot,B., Esteban,R. and Guardia,J. (1999) High-throughput real-time reverse transcription–PCR quantitation of hepatitis C virus RNA. J. Clin. Microbiol., 37, 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitcombe D., Theaker,J., Guy,S.P., Brown,T. and Little,S. (1999) Detection of PCR products using self-probing amplicons and fluorescence. Nat. Biotechnol., 17, 804–807. [DOI] [PubMed] [Google Scholar]
- 32.Higuchi R., Dollinger,G., Walsh,P.S. and Griffith,R. (1992) Simulutaneous amplification and detection of specific DNA sequences. Biotechnology (NY), 10, 413–417. [DOI] [PubMed] [Google Scholar]
- 33.Ishiguro T., Saitoh,J., Yawata,H., Yamagishi,H., Iwasaki,S. and Mitoma,Y. (1995) Homogeneous quantitative assay of hepatitis C virus RNA by polymerase chain reaction in the presence of a fluorescent intercalater. Anal. Biochem., 229, 207–213. [DOI] [PubMed] [Google Scholar]
- 34.Tseng S.Y., Macool,D., Elliott,V., Tice,G., Jackson,R., Barbour,M. and Amorese,D. (1997) An homogeneous fluorescence polymerase chain reaction assay to identify Salmonella. Anal. Biochem., 245, 207–212. [DOI] [PubMed] [Google Scholar]
- 35.Morrison T.M., Weis,J.J. and Wittwer,C.T. (1998) Quantification of low-copy transcripts by continuous SYBR green I monitoring during amplification. Biotechniques, 24, 954–962. [PubMed] [Google Scholar]
- 36.Komurian-Pradel F., Paranhos-Baccalà,G., Sodoyer,M., Chevallier,P., Mandrand,B., Lotteau,V. and André,P. (2001) Quantitation of HCV RNA using real-time PCR and fluorimetry. J. Virol. Methods, 95, 111–119. [DOI] [PubMed] [Google Scholar]
- 37.Chou Q., Russell,M., Birch,D.E., Raymond,J. and Bloch,W. (1992) Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications. Nucleic Acids Res., 20, 1717–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halford W.P. (1999) The essential prerequisites for quantitative RT–PCR. Nat. Biotechnol., 17, 835. [DOI] [PubMed] [Google Scholar]
- 39.Halford W.P., Falco,V.C., Gebhardt,B.M. and Carr,D.J.J. (1999) The inherent quantitative capacity of the reverse transcription–polymerase chain reaction. Anal. Biochem., 266, 181–191. [DOI] [PubMed] [Google Scholar]
- 40.Pfaffl M. (2001) In Meuer,S., Wittwer,C. and Nakagawara,K.-I. (eds), Rapid Cycle Real-Time PCR: Methods and Applications. Springer, Berlin, pp. 281–291.
- 41.Wetmur J.G. (1991) DNA probes: applications of the principles of nucleic acid hybridization. Crit. Rev. Biochem. Mol. Biol., 26, 227–259. [DOI] [PubMed] [Google Scholar]
- 42.Ririe K.M., Rasmussen,R.P. and Wittwer,C.T. (1997) Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem., 245, 154–160. [DOI] [PubMed] [Google Scholar]
- 43.Heller M.J. and Morrison,L.E. (1985) In Kingsbury,D.T. and Falkow,S. (eds), Rapid Detection and Identification of Infectious Agents. Academic Press, New York, pp. 245–256.
- 44.Cardullo R.A., Agrawai,S., Flores,C., Zamecnik,P.C. and Wolf,D.E. (1988) Detection of nucleic acid hybridization by nonradiative fluoresence resonance energy transfer. Proc. Natl Acad. Sci. USA, 85, 8790–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wittwer C.T., Herrmann,M.G., Moss,A.A. and Rasmussen,R.P. (1997) Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques, 22, 130–138. [DOI] [PubMed] [Google Scholar]
- 46.Weis J.H., Tan,S.S., Martin,B.K. and Wittwer,C.T. (1992) Detection of rare mRNAs via quantitative RT–PCR. Trends Genet., 8, 263–264. [DOI] [PubMed] [Google Scholar]
- 47.Bustin S.A. (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol., 25, 169–193. [DOI] [PubMed] [Google Scholar]
- 48.Gundry C.N., Bernard,P.S., Herrmann,M.G., Reed,G.H. and Wittwer,C.T. (1999) Rapid F508del and F508C assay using fluorescent hybridization probes. Genet. Test, 3, 365–370. [DOI] [PubMed] [Google Scholar]
- 49.Schalasta G., Arents,A., Schmid,M., Braun,R.W. and Enders,G. (2000) Fast and type-specific analysis of herpes simplex virus types 1 and 2 by rapid PCR and fluorescence melting-curve-analysis. Infection, 28, 85–91. [DOI] [PubMed] [Google Scholar]
- 50.Wilhelm J., Pingoud,A. and Hahn,M. (2001) Comparison between Taq DNA polymerase and its Stoffel fragment for quantitative real-time PCR with hybridization probes. Biotechniques, 30, 1052–1062. [DOI] [PubMed] [Google Scholar]
- 51.Lyamichev V., Brow,M.A.D. and Dahlberg,J.E. (1993) Structure-specific endonucleolytic cleavage of nucleic acids by eubacterial DNA polymerases. Science, 260, 778–783. [DOI] [PubMed] [Google Scholar]
- 52.Brechtbuehl K., Whalley,S.A., Dusheiko,G.M. and Saunders,N.A. (2001) A rapid real-time quantitative polymerase chain reaction for hepatitis B virus. J. Virol. Methods, 93, 105–113. [DOI] [PubMed] [Google Scholar]
- 53.Walker R.A., Saunders,N., Lawson,A.J., Lindsay,E.A., Dassama,M., Ward,L.R., Woodward,M.J., Davies,R.H., Liebana,E. and Threlfall,E.J. (2001) Use of a LightCycler gyrA mutation assay for rapid identification of mutations conferring decreased susceptibility to ciprofloxacin in multiresistant Salmonella enterica serotype typhimurium DT104 isolates. J. Clin. Microbiol., 39, 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kurata S., Kanagawa,T., Yamada,K., Torimura,M., Yokomaku,T., Kamagata,Y. and Kurane,R. (2001) Fluorescent quenching-based quantitative detection of specific DNA/RNA using a BODIPY®FL-labeled probe or primer. Nucleic Acids Res., 29, e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crockett A.O. and Wittwer,C.T. (2001) Fluorescein-labelled oligonucleotides for real-time PCR: using the inherent quenching of deoxyguanosine nucleotides. Anal. Biochem., 290, 89–97. [DOI] [PubMed] [Google Scholar]
- 56.Svanvik N., Westman,G., Wang,D. and Kubista,M. (2001) Light-up probes: thiazole orange-conjugated peptide nucleic acid for the detection of target nucleic acid in homogensous solution. Anal. Biochem., 281, 26–35. [DOI] [PubMed] [Google Scholar]
- 57.Isacsson J., Cao,H., Ohlsson,L., Nordgren,S., Svanvik,N., Westman,G., Kubista,M., Sjöback,R. and Sehlstedt,U. (2000) Rapid and specific detection of PCR products using light-up probes. Mol. Cell. Probes, 14, 321–328. [DOI] [PubMed] [Google Scholar]
- 58.Svanvik N., Sehlstedt,U., Sjöback,R. and Kubista,M. (2000) Detection of PCR products in real time using light-up probes. Anal. Biochem., 287, 179–182. [DOI] [PubMed] [Google Scholar]
- 59.Morrison L.E., Halder,T.C. and Stols,L.M. (1989) Solution-phase detection of polynucleotides using interacting fluorescent labels and competitive hybridization. Anal. Biochem., 183, 231–244. [DOI] [PubMed] [Google Scholar]
- 60.Landt O. (2001) In Meuer,S., Wittwer,C. and Nakagawara,K. (eds), Rapid Cycle Real-time PCR: Methods and Applications. Springer Verlag, Germany, pp. 35–41.
- 61.Nakayama H., Yokoi,H. and Fujita,J. (1992) Quantification of mRNA by non-radioactive RT–PCR and CCD imaging system. Nucleic Acids Res., 20, 4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gelmini S., Orlando,C., Sestini,R., Vona,G., Pinzani,P., Ruocco,L. and Pazzagli,M. (1997) Quantitative polymerase chain reaction-based homogeneous assay with fluorogenic probes to measure c-cerB-2 oncogene amplification. Clin. Chem., 43, 752–758. [PubMed] [Google Scholar]
- 63.Wilhelm J., Hahn,M. and Pingoud,A. (2001) Influence of DNA target melting behavior on real-time PCR quantification. Clin. Chem., 46, 1738–1743. [PubMed] [Google Scholar]
- 64.Jung R., Soondrum,K. and Neumaier,M. (2000) Quantitative PCR. Clin. Chem. Lab. Med., 38, 833–836. [DOI] [PubMed] [Google Scholar]
- 65.Kutyavin I.V., Afonina,I.A., Mills,A., Gorn,V.V., Lukhtanov,E.A., Belousov,E.S., Singer,M.J., Walburger,D.K., Lokhov,S.G., Gall,A.A. et al. (2000) 3′-Minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res., 28, 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Todd A.V., Fuery,C.J., Impey,H.L., Applegate,T.L. and Haughton,M.A. (2000) DzyNA–PCR: use of DNAzymes to detect and quantify nucleic acid sequences in a real-time fluorescent format. Clin. Chem., 46, 625–630. [PubMed] [Google Scholar]
- 67.Tyagi S., Bratu,D.P. and Kramer,F.R. (1998) Multicolor molecular beacons for allele discrimination. Nat. Biotechnol., 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 68.Tyagi S. and Kramer,F.R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol., 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 69.Tyagi S., Marras,S.A.E. and Kramer,F.R. (2000) Wavelength-shifting molecular beacons. Nat. Biotechnol., 18, 1191–1196. [DOI] [PubMed] [Google Scholar]
- 70.Nazarenko I.A., Bhatnager,S.K. and Hohman,R.J. (1997) A closed tube format for amplification and detection of DNA based on energy transfer. Nucleic Acids Res., 25, 2516–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Freeman W.M., Walker,S.J. and Vrana,K.E. (1999) Quantitative RT–PCR: pitfalls and potential. Biotechniques, 26, 112–125. [DOI] [PubMed] [Google Scholar]
- 72.Orlando C., Pinzani,P. and Pazzagli,M. (1998) Developments in quantitative PCR. Clin. Chem. Lab. Med., 36, 255–269. [DOI] [PubMed] [Google Scholar]
- 73.Becker-Andre M. and Hahlbrock,K. (1989) Absolute mRNA quantification using the polymerase chain reaction (PCR). A novel approach by a PCR aided transcript titration assay (PATTY). Nucleic Acids Res., 17, 9437–9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Clementi M., Menzo,S., Manzin,A. and Bagnarelli,P. (1995) Quantitative molecular methods in virology. Arch. Virol., 140, 1523–1539. [DOI] [PubMed] [Google Scholar]
- 75.Gilliland G., Perrin,S. and Bunn,H.F. (1990) PCR Protocols: A Guide to Methods and Applications. Academic Press, San Diego, CA, pp. 60–69.
- 76.Siebert P.D. and Larrick,J.W. (1992) Competitive PCR. Nature, 359, 557–558. [DOI] [PubMed] [Google Scholar]
- 77.Locatelli G., Santoro,F., Veglia,F., Gobbi,A., Lusso,P. and Malnati,M.S. (2000) Real-time quantitative PCR for human herpesvirus 6 DNA. J. Clin. Microbiol., 38, 4042–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tanaka N., Kimura,H., Iida,K., Saito,Y., Tsuge,I., Yoshimi,A., Matsuyama,T. and Morishima,T. (2000) Quantitative analysis of cytomegalovirus load using a real-time PCR assay. J. Med. Virol., 60, 455–462. [DOI] [PubMed] [Google Scholar]
- 79.Ferré F. (1992) Quantitative or semi-quantitative PCR: reality versus myth. PCR Methods Appl., 2, 1–9. [DOI] [PubMed] [Google Scholar]
- 80.Johnson M.R., Wang,K., Smith,J.B., Heslin,M.J. and Diasio,R.B. (2000) Quantitation of dihydropyrimidine dehydrogenase expression by real-time reverse transcription polymerase chain reaction. Anal. Biochem., 278, 175–184. [DOI] [PubMed] [Google Scholar]
- 81.Kleiber J., Walter,T., Haberhausen,G., Tsang,S., Babiel,R. and Rosenstraus,M. (2000) Performance characteristics of a quantitative, homogeneous TaqMan RT–PCR test for HCV RNA. J. Mol. Diagn., 2, 158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kimura H., Morita,M., Yabuta,Y., Kuzushima,K., Kato,K., Kojima,S., Matsuyama,T. and Morishima,T. (1999) Quantitative analysis of Epstein–Barr virus load by using a real-time PCR assay. J. Clin. Microbiol., 37, 132–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Najioullah F., Thouvenot,D. and Lina,B. (2001) Development of a real-time PCR procedure including an internal control for the measurement of HCMV viral load. J. Virol. Methods, 92, 55–64. [DOI] [PubMed] [Google Scholar]
- 84.Ryncarz A.J., Goddard,J., Wald,A., Huang,M.-L., Roizman,B. and Corey,L. (1999) Development of a high-throughput quantitative assay for detecting herpes simplex virus DNA in clinical samples. J. Clin. Microbiol., 37, 1941–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Monopoeho S., Mignotte,B., Schwartzbrod,L., Marechal,V., Nicolas,J.-C., Billaudel,S. and Férré,V. (2000) Quantification of enterovirus RNA in sludge samples using single tube real-time RT–PCR. Biotechniques, 29, 88–93. [DOI] [PubMed] [Google Scholar]
- 86.Alexandersen S., Oleksiewicz,M.B. and Donaldson,A.I. (2001) The early pathogenesis of foot-and-mouth disease in pigs infected by contact: a quantitative time-course study using TaqMan RT–PCR. J. Gen. Virol., 82, 747–755. [DOI] [PubMed] [Google Scholar]
- 87.Abe A., Inoue,K., Tanaka,T., Kato,J., Kajiyama,N., Kawaguchi,R., Tanaka,S., Yoshiba,M. and Kohara,M. (1999) Quantitation of hepatitis B virus genomic DNA by real-time detection PCR. J. Clin. Microbiol., 37, 2899–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gruber F., Falkner,F.G., Dorner,F. and Hämmerle,T. (2001) Quantitation of viral DNA by real-time PCR applying duplex amplification, internal standardization and two-colour fluorescence detection. Appl. Environ. Microbiol., 67, 2837–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moody A., Sellers,S. and Bumstead,N. (2000) Measuring infectious bursal disease virus RNA in blood by multiplex real-time quantitative RT–PCR. J. Virol. Methods, 85, 55–64. [DOI] [PubMed] [Google Scholar]
- 90.Schutten M., van den Hoogen,B., van der Ende,M.E., Gruters,R.A., Osterhaus,A.D.M.E. and Niesters,H.G.M. (2000) Development of a real-time quantitative RT–PCR for the detection of HIV-2 RNA in plasma. J. Virol. Methods, 88, 81–87. [DOI] [PubMed] [Google Scholar]
- 91.Kearns A.M., Turner,A.J.L., Taylor,C.E., George,P.W., Freeman,R. and Gennery,A.R. (2001) LightCycler-based quantitative PCR for rapid detection of human herpesvirus 6 DNA in clinical material. J. Clin. Microbiol., 39, 3020–3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Capone R.B., Pai,S.I., Koch,W.M. and Gillison,M.L. (2001) Detection and quantitation of human papillomavirus (HPV) DNA in the sera of patients with HPV-associated head and neck squamous cell carcinoma. Clin. Cancer Res., 6, 4171–4175. [PubMed] [Google Scholar]
- 93.Leutenegger C.M., Klein,D., Hofmann-Lehmann,R., Mislin,C., Hummel,U., Boni,J., Boretti,F., Guenzburg,W.H. and Lutz,H. (1999) Rapid feline immunodeficiency virus provirus quantitation by polymerase chain reaction using the TaqMan fluorogenic real-time detection system. J. Virol. Methods, 78, 105–116. [DOI] [PubMed] [Google Scholar]
- 94.Smith I.L., Halpin,K., Warrilow,D. and Smith,G.A. (2001) Development of a fluorogenic RT–PCR assay (TaqMan) for the detection of Hendra virus. J. Virol. Methods, 98, 33–40. [DOI] [PubMed] [Google Scholar]
- 95.van Elden L.J.R., Nijhuis,M., Schipper,P., Schuurman,R. and van Loon,A.M. (2001) Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J. Clin. Microbiol., 39, 196–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lanciotti R.S., Kerst,A.J., Nasci,R.S., Godsey,M.S., Mitchell,C.J., Savage,H.M., Komar,N., Panella,N.A., Allen,B.C., Volpe,K.E. et al. (2000) Rapid detection of west nile virus from human clinical specimens, field-collected mosquitoes and Avian samples by a TaqMan reverse transcriptase–PCR assay. J. Clin. Microbiol., 38, 4066–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Weinberger K.M., Wiedenmann,E., Böhm,S. and Jilg,W. (2000) Sensitive and accurate detection of hepatitis B virus DNA using a kinetic fluorescence detection system (TaqMan PCR). J. Virol. Methods, 85, 75–82. [DOI] [PubMed] [Google Scholar]
- 98.Schaade L., Kockelkorn,P., Ritter,K. and Kleines,M. (2000) Detection of cytomegalovirus DNA in human specimens by LightCycler PCR. J. Clin. Microbiol., 38, 4006–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kawai S., Yokosuka,O., Kanda,T., Imazeki,F., Maru,Y. and Saisho,H. (1999) Quantification of hepatitis C virus by TaqMan PCR: comparison with HCV amplicor monitor assay. J. Med. Virol., 58, 121–126. [DOI] [PubMed] [Google Scholar]
- 100.Clementi M. (2000) Quantitative molecular analysis of virus expression and replication. J. Clin. Microbiol., 38, 2030–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Limaye A.P., Jerome,K.R., Kuhr,C.S., Ferrenberg,J., Huang,M.-L., Davis,C.L., Corey,L. and Marsh,C.L. (2000) Quantitation of BK virus load in serum for the diagnosis of BK virus-associated nephropathy in renal transplant recipients. J. Infect. Dis., 183, 1669–1672. [DOI] [PubMed] [Google Scholar]
- 102.Holodniy M., Katzenstein,D., Sengupta,S., Wang,A.M., Casipit,C., Schwartz,D.H., Konrad,M., Groves,E. and Merigan,T.C. (1991) Detection and quantification of human immunodeficiency virus RNA in patient serum by use of the polymerase chain reaction. J. Infect. Dis., 163, 862–866. [DOI] [PubMed] [Google Scholar]
- 103.Kaneko S., Murakami,S., Unoura,M. and Kobayashi,K. (1992) Quantitation of hepatitis C virus RNA by competitive polymerase chain reaction. J. Med. Virol., 37, 278–282. [DOI] [PubMed] [Google Scholar]
- 104.Menzo S., Bagnarelli,P., Giacca,M., Manzin,A., Varaldo,P.E. and Clementi,M. (1992) Absolute quantitation of viremia in human immunodeficiency virus infection by competitive reverse transcription and polymerase chain reaction. J. Clin. Microbiol., 30, 1752–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ohyashiki J.K., Suzuki,A., Aritaki,K., Nagate,A., Shoji,N., Ohyashiki,K., Ojima,T., Abe,K. and Yamamoto,K. (2000) Use of real-time PCR to monitor human herpesvirus 6 reactivation after allogeneic bone marrow transplantation. Int. J. Mol. Med., 6, 427–432. [DOI] [PubMed] [Google Scholar]
- 106.Furuta Y., Ohtani,F., Sawa,H., Fukuda,S. and Inuyama,Y. (2001) Quantitation of varicella-zoster virus DNA in patients with ramsay hunt syndrome and zoster sine herpete. J. Clin. Microbiol., 39, 2856–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Limaye A.P., Huang,M.-L., Leisenring,W., Stensland,L., Corey,L. and Boeckh,M. (2001) Cytomegalovirus (CMV) DNA load in plasma for the diagnosis of CMV disease before engraftment in hematopoietic stem-cell transplant recipients. J. Infect. Dis., 183, 377–382. [DOI] [PubMed] [Google Scholar]
- 108.Lallemand F., Desire,N., Rozenbaum,W., Nicolas,J.-C. and Marechal,V. (2000) Quantitative analysis of human herpesvirus 8 viral load using a real-time PCR assay. J. Clin. Microbiol., 38, 1404–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Laue T., Emmerich,P. and Schmitz,H. (1999) Detection of dengue virus RNA inpatients after primary or secondary dengue infection by using the TaqMan automated amplification system. J. Clin. Microbiol., 37, 2543–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chang L.-J., Urlacher,V., Iwakuma,T., Cui,Y. and Zucali,J. (1999) Efficacy and safety anlyses of a recombinant human immunodeficieny virus type 1 derived vector system. Gene Ther., 6, 715–728. [DOI] [PubMed] [Google Scholar]
- 111.Lo Y.M.D., Chan,L.Y.S., Lo,K.-W., Leung,S.-F., Zhang,J., Chan,A.T.C., Lee,J.C.K., Hjelm,N.M., Johnson,P.J. and Huang,D.P. (1999) Quantitative analysis of cell-free Epstein–Barr virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res., 59, 1188–1191. [PubMed] [Google Scholar]
- 112.Hawrami K. and Breur,J. (1999) Development of a flurogenic polymerase chain reaction assay (TaqMan) for the detection and quantitation of varicella zoster virus. J. Virol. Methods, 79, 33–40. [DOI] [PubMed] [Google Scholar]
- 113.Roberts T.C., Brennan,D.C., Buller,R.S., Gaudreault-Keener,M., Schnitzler,M.A., Sternhell,K.E., Garlock,K.A., Singer,G.G. and Storch,G.A. (1998) Quantitative polymerase chain reaction to predict occurrence of symptomatic cytomegalovirus infection and assess response to ganciclovir therapy in renal transplant recipients. J. Infect. Dis., 178, 626–636. [DOI] [PubMed] [Google Scholar]
- 114.Rollag H., Sagedal,S., Holter,E., Degre,M., Ariansen,S. and Nordal,K.P. (1998) Diagnosis of cytomegalovirus infection in kidney transplant recipients by a quantitative RNA–DNA hybrid capture assay for cytomegalovirus DNA in leukocytes. Eur. J. Clin. Microbiol. Infect. Dis., 17, 124–127. [DOI] [PubMed] [Google Scholar]
- 115.Boivin G., Handfield,J., Murray,G., Toma,E., Lalonde,R., Lazar,J.G. and Bergeron,M.G. (1997) Quantitation of cytomegalovirus (CMV) DNA in leukocytes of human immunodeficiency virus-infected subjects with and without CMV disease by using PCR and the SHARP Signal Detection System. J. Clin. Microbiol., 35, 525–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nitsche A., Steuer,N., Schmidt,C.A., Landt,O. and Siegert,W. (1999) Different real-time PCR formats compared for the quantitative detection of human cytomegalovirus DNA. Clin. Chem., 45, 1932–1937. [PubMed] [Google Scholar]
- 117.Chamberlain J.S., Gibbs,R.A., Ranier,J.E., Nguyen,P.N. and Caskey,C.T. (1988) Deletion screening of the Duchenne muscular dystrophy locis via multiplex DNA amplification. Nucleic Acids Res., 16, 11141–11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Espy M.J., Uhl,J.R., Mitchell,P.S., Thorvilson,J.N., Svien,K.A., Wold,A.D. and Smith,T.F. (2000) Diagnosis of herpes simplex virus infections in the clinical laboratory by LightCycler PCR. J. Clin. Microbiol., 38, 795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Espy M.J., Ross,T.K., Teo,R., Svien,K.A., Wold,A.D., Uhl,J.R. and Smith,T.F. (2000) Evaluation of lightcycler PCR for implementation of laboratory diagnosis of herpes simplex virus infections. J. Clin. Microbiol., 38, 3116–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Loparev V.N., McCaustland,K., Holloway,B.P., Krause,P.R., Takayama,M. and Schmid,D.S. (2000) Rapid genotyping of varicella-zoster virus vaccine and wild-type strains with fluorophore-labeled hybridization probes. J. Clin. Microbiol., 38, 4315–4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Read S.J., Mitchell,J.L. and Fink,C.G. (2001) LightCycler multiplex PCR for the laboratory diagnosis of common viral infections of the central nervous system. J. Clin. Microbiol., 39, 3056–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Whiley D.M., Mackay,I.M. and Sloots,T.P. (2001) Detection and differentiation of human polyomaviruses JC and BK by LightCycler PCR. J. Clin. Microbiol., 39, 4357–4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schütz E., von Ashen,N. and Oellerich,M. (2000) Genotyping of eight thiopurine methyltransferase mutations: Three-color multiplexing, ‘two-color/shared’ anchor and fluorescence-quenching hybridization probe assays based on thermodynamic nearest-neighbour probe design. Clin. Chem., 46, 1728–1737. [PubMed] [Google Scholar]
- 124.Lay M.J. and Wittwer,C.T. (1997) Real-time fluorescence genotyping of factor V Leiden during rapid-cycle PCR. Clin. Chem., 43, 2262–2267. [PubMed] [Google Scholar]
- 125.Herrmann M.G., Dobrowolski,S.F. and Wittwer,C.T. (2000) Rapid β-globin genotyping by multiplexing probe melting temperature and color. Clin. Chem., 46, 425–429. [PubMed] [Google Scholar]
- 126.Lee L.G., Livak,K.J., Mullah,B., Graham,R.J., Vinayak,R.S. and Woudenberg,T.M. (1999) Seven-color, homogeneous detection of six PCR products. Biotechniques, 27, 342–349. [DOI] [PubMed] [Google Scholar]
- 127.Bernard P.S. and Wittwer,C.T. (2000) Homogeneous amplification and variant detection by fluorescent hybridization probes. Clin. Chem., 46, 147–148. [PubMed] [Google Scholar]
- 128.Elenitoba K.S.J., Bohling,S.D., Wittwer,C.T. and King,T.C. (2001) Multiplex PCR by multicolor fluorimetry and fluorescence melting curve analysis. Nature Med., 7, 249–253. [DOI] [PubMed] [Google Scholar]
- 129.Vet J.A.M., Majithia,A.R., Marras,S.A.E., Tyagi,S., Dube,S., Poiesz,B.J. and Kramer,F.R. (1999) Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc. Natl Acad. Sci. USA, 96, 6394–6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Quereda C., Corral,I., Laguna,F., Valencia,M.E., Tenorio,A., Echevarria,J.E., Navas,E., Martín-Davilla,P., Moreno,A., Moreno,V. et al. (2000) Diagnostic utility of a multiplex herpesvirus PCR assay performed with cerebrospinal fluid from human immunodeficiency virus-infected patients with neurological disorders. J. Clin. Microbiol., 38, 3061–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kehl S.C., Henrickson,K.J., Hua,W. and Fan,J. (2001) Evaluation of the hexaplex assay for detection of respiratory viruses in children. J. Clin. Microbiol., 39, 1696–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Echevarria J.E., Erdman,D.D., Swierkosz,E.M., Holloway,B.P. and Anderson,L.J. (1998) Simultaneous detection and identification of human parainfluenza viruses 1, 2 and 3 from clinical samples by multiplex PCR. J. Clin. Microbiol., 36, 1388–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Henegariu O., Heerema,N.A., Dloughy,S.R., Vance,G.H. and Vogt,P.H. (1997) Multiplex PCR: critical parameters and step-by-step protocol. Biotechniques, 23, 504–511. [DOI] [PubMed] [Google Scholar]
- 134.Nicoll S., Brass,A. and Cubie,H.A. (2001) Detection of herpes viruses in clinical samples using real-time PCR. J. Virol. Methods, 96, 25–31. [DOI] [PubMed] [Google Scholar]
- 135.Tong A.K., Li,Z., Jones,G.S., Russo,J.J. and Ju,J. (2001) Combinatorial fluorescence energy transfer tags for multiplex biological assays. Nat. Biotechnol., 19, 756–759. [DOI] [PubMed] [Google Scholar]
- 136.Kato T., Mizokami,M., Mukaide,M., Orito,E., Ohno,T., Nakano,T., Tanaka,Y., Kato,H., Sugauchi,F., Ueda,R. et al. (2000) Development of a TT virus DNA quantification system using real-time detection PCR. J. Clin. Microbiol., 38, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Iriyama M., Kimura,H., Nishikawa,K., Yoshioka,K., Wakita,T., Nishimura,N., Shibata,M., Ozaki,T. and Morishima,T. (1999) The prevalence of TT virus (TTV) infection and its relationship to hepatitis in children. Med. Microbiol. Immunol. (Berl.), 188, 83–89. [DOI] [PubMed] [Google Scholar]
- 138.Liu J., Feldman,P. and Chung,T.D.Y. (2002) Real-time monitoring in vitro transcription using molecular beacons. Anal. Biochem., 300, 40–45. [DOI] [PubMed] [Google Scholar]
- 139.Kennedy M.M., O’Leary,J.J., Oates,J.L., Lucas,S.B., Howells,D.D., Picton,S. and McGee,J.O. (1998) Human herpes virus 8 (HHV-8) in Kaposi’s sarcoma: lack of association with Bcl-2 and p53 protein expression. J. Clin. Pathol., 51, 155–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kennedy M.M., Biddolph,S., Lucas,S.B., Howells,D.D., Picton,S., McGee,J.O. and O’Leary,J.J. (1999) CD40 upregulation is independent of HHV-8 in the pathogenesis of Kaposi’s sarcoma. J. Clin. Pathol., 52, 32–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kennedy M.M., Biddolph,S., Lucas,S.B., Howells,D.D., Picton,S., McGee,J.O., Silva,I., Uhlmann,V., Luttich,K. and O’Leary,J. (1999) Cyclin D1 expression and HHV-8 in Kaposi sarcoma. J. Clin. Pathol., 52, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zerr D.M., Huang,M.-L., Corey,L., Erickson,M., Parker,H.L. and Frenkel,L.M. (2000) Sensitive method for detection of human herpesvirus 6 and 7 in saliva collected in field studies. J. Clin. Microbiol., 38, 1981–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jordens J.Z., Lanham,S., Pickett,M.A., Amarasekara,S., Aberywickrema,I. and Watt,P.J. (2000) Amplification with molecular beacon primers and reverse line blotting for the detection and typing of human papillomaviruses. J. Virol. Methods, 89, 29–37. [DOI] [PubMed] [Google Scholar]
- 144.Niesters H.G., van Esser,J., Fries,E., Wolthers,K.C., Cornelissen,J. and Osterhaus,A.D. (2000) Development of a real-time quantitative assay for detection of Epstein–Barr virus. J. Clin. Microbiol., 38, 712–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Biel S.S., Held,T.K., Landt,O., Niedrig,M., Gelderblom,H.R., Siegert,W. and Nitsche,A. (2000) Rapid quantification and differentiation of human polyomavirus DNA in undiluted urine from patients after bone marrow transplantation. J. Clin. Microbiol., 38, 3689–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Petrik J., Pearson,G.J.M. and Allain,J.-P. (1997) High throughput PCR detection of HCV based on semiautomated multisample RNA capture. J. Virol. Methods, 64, 147–159. [DOI] [PubMed] [Google Scholar]
- 147.Skaf J.S., Schultz,M.H., Hirata,H. and de Zoeten,G.A. (2000) Mutational evidence that VPg is involved in the replication and not the movement of Pea enation mosaic virus-1. J. Gen. Virol., 81, 1103–1109. [DOI] [PubMed] [Google Scholar]
- 148.Roberts C.A., Dietzgen,R.G., Heelan,L.A. and Maclean,D.J. (2000) Real-time RT–PCR fluorescent detection of tomato spotted wilt virus. J. Virol. Methods, 88, 1–8. [DOI] [PubMed] [Google Scholar]
- 149.Eun A.J.-C., Seoh,M.-L. and Wong,S.W. (2000) Simultaneous quantitation of two orchid viruses by the TaqMan® real-time RT–PCR. J. Virol. Methods, 87, 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Halpin K., Young,P.L., Field,H.E. and Mackenzie,J.S. (2000) Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J. Gen. Virol., 81, 1927–1932. [DOI] [PubMed] [Google Scholar]
- 151.Egli C., Thür,B., Liu,L. and Hofmann,M.A. (2001) Quantitative TaqMan® RT–PCR for the detection and differentiation of European and North American strains of porcine reproductive and respiratory syndrome virus. J. Virol. Methods, 98, 63–75. [DOI] [PubMed] [Google Scholar]
- 152.Gut M., Leutenegger,C.M., Huder,J.B., Pedersen,N.C. and Lutz,H. (1999) One-tube fluorogenic reverse transcription–polymerase chain reaction for the quantitation of feline coronaviruses. J. Virol. Methods, 77, 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Monopoeho S., Maul,A., Mignotte-Cadiergues,B., Schwartzbrod,L., Billaudel,S. and Ferré,F. (2001) Best viral elution method available for quantification of enteroviruses in sludge by both cell culture and reverse transcription. Appl. Environ. Microbiol., 67, 2484–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Garin D., Peyrefitte,C., Crance,J.-M., Le Faou,A., Jouan,A. and Bouloy,M. (2001) Highly sensitive Taqman® PCR detection of Puumala hantavirus. Microbes Infect., 3, 739–745. [DOI] [PubMed] [Google Scholar]
- 155.O’Doherty U., Swiggard,W.J. and Malim,M.H. (2000) Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol., 74, 10074–10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Klein D., Janda,P., Steinborn,R., Salmons,B. and Günzburg,W.H. (1999) Proviral load determination of different feline immunodeficiency virus isolates using real-time polymerase chain reaction: influence of mismatches on quantification. Electrophoresis, 20, 291–299. [DOI] [PubMed] [Google Scholar]
- 157.Hadfield T.L., Turell,M., Dempsey,M.P., David,J. and Park,E.J. (2001) Detection of West Nile virus in mosquitoes by RT–PCR. Mol. Cell. Probes, 15, 147–150. [DOI] [PubMed] [Google Scholar]
- 158.Saldanha C.E., Lubinski,J., Martin,C., Nagashunmugam,T., Wang,L., van der Keyl,H., Tal-Singer,R. and Friedman,H.M. (2000) Herpes simplex virus type 1 glycoprotein E domains involved in virus spread and disease. J. Virol., 74, 6712–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Poddar S.K. (1999) Detection of adenovirus using PCR and molecular beacon. J. Virol. Methods, 82, 19–26. [DOI] [PubMed] [Google Scholar]
- 160.Gibb T.R., Norwood,D.A.,Jr, Woollen,N. and Henchal,E.A. (2001) Development and evaluation of a fluorogenic 5′-nuclease assay to identify Marburg virus. Mol. Cell. Probes, 15, 259–266. [DOI] [PubMed] [Google Scholar]
- 161.Pevenstein S.R., Williams,R.K., McChesney,D., Mont,E.K., Smialek,J.E. and Straus,S.E. (1999) Quantitation of latent varicella-zoster virus and herpes simplex virus genomes in human trigeminal ganglia. J. Virol., 73, 10514–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Espy M.J., Rys,P.N., Wold,A.D., Uhl,J.R., Sloan,L.M., Jenkins,G.D., Ilstrup,D.M., Cockerill,F.R.,III, Patel,R., Rosenblatt,J.E. et al. (2001) Detection of herpes simplex virus DNA in genital and dermal specimens by LightCycler PCR after extraction using the IsoQuick, MagNA Pure and BioRobot 9604 methods. J. Clin. Microbiol., 39, 2233–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Espy M.J., Teo,R., Ross,T.K., Svien,K.A., Wold,A.D., Uhl,J.R. and Smith,T.F. (2000) Diagnosis of Varicella-Zoster Virus infections in the clinical laboratory by LightCycler PCR. J. Clin. Microbiol., 38, 3187–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Josefsson A.M., Magnusson,P.K.E., Ylitalo,N., Sørensen,P., Qwarforth-Tubbin,P., Andersen,P.K., Melbye,M., Adami,H.-O. and Gyllensten,U. (2000) Viral load of human papilloma virus 16 as a determinant for development of cervical carcinoma in situ: a nested case-control study. Lancet, 355, 2189–2193. [DOI] [PubMed] [Google Scholar]
- 165.Johnston C. (2000) Quantitative tests for human papillomavirus. Lancet, 355, 2179–2180. [DOI] [PubMed] [Google Scholar]
- 166.Watanabe M., Lee,B.-J., Kamitani,W., Kobayashi,T., Taniyama,H., Tomonaga,K. and Ikuta,K. (2001) Neurological diseases and viral dynamics in the brains of neonatally borna disease virus-infected gerbils. Virology, 282, 65–76. [DOI] [PubMed] [Google Scholar]
- 167.Peter J.B. and Sevall,J.S. (2001) Review of 3200 serially received CSF samples submitted for type-specific HSV detection by PCR in the reference laboratory setting. Mol. Cell. Probes, 15, 177–182. [DOI] [PubMed] [Google Scholar]
- 168.Kessler H.H., Mühlbauer,G., Rinner,B., Stelzl,E., Berger,A., Dörr,H.-W., Santner,B., Marth,E. and Rabenau,H. (2000) Detection of herpes simplex virus DNA by real-time PCR. J. Clin. Microbiol., 38, 2638–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kimura H., Kido,S., Ozaki,T., Tanaka,N., Ito,Y., Williams,R.K. and Morishima,T. (2000) Comparison of quantitations of viral load in varicella and zoster. J. Clin. Microbiol., 38, 2447–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ohga S., Nomura,A., Takada,H., Thara,K., Kawakami,K., Yanai,F., Takahata,Y., Tanaka,T., Kasuga,N. and Hara,T. (2001) Epstein–Barr virus (EBV) load and cytokine gene expression in activated T cells of chronic active EBV infection. J. Infect. Dis., 183, 1–7. [DOI] [PubMed] [Google Scholar]
- 171.White I. and Campbell,T.C. (2000) Quantitation of cell-free cell-associated Kaposi’s sarcoma associated herpesvirus DNA by real-time PCR. J. Clin. Microbiol., 38, 1992–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mercier B., Burlot,L. and Ferec,C. (1999) Simultaneous screening for HBV DNA and HCV RNA genomes in blood donations using a novel TaqMan PCR assay. J. Virol. Methods, 77, 1–9. [DOI] [PubMed] [Google Scholar]
- 173.Nitsche A., Müller,C.W., Radonic,A., Landt,O., Ellerbrok,H., Pauli,G. and Siegert,W. (2001) Human herpesvirus 6A DNA is detected frequently in plasma but rarely in peripheral blood leukocytes of patients after bone marrow transplantation. J. Infect. Dis., 183, 130–133. [DOI] [PubMed] [Google Scholar]
- 174.Lei K.I.K., Chan,L.Y.S., Chan,W.Y., Johnson,P.J. and Lo,Y.M.D. (2000) Quantitative analysis of circulating cell-free Epstein–Barr virus (EBV) DNA levels in patients with EBV-associated lymphoid malignancies. Brit. J. Haematol., 111, 239–246. [DOI] [PubMed] [Google Scholar]
- 175.Quackenbush S.L., Casey,R.N., Murcek,R.J., Paul,T.A., Work,T.M., Limpus,C.J., Chaves,A., duToit,L., Perez,J.V., Aguirre,A.A. et al. (2001) Quantitative analysis of herpesvirus sequences from normal tissue and fibropapillomas of marine turtles with real-time PCR. Virology, 187, 105–111. [DOI] [PubMed] [Google Scholar]
- 176.Aberham C., Pendl,C., Gross,P., Zerlauth,G. and Gessner,M. (2001) A quantitative, internally controlled real-time PCR assay for the detection of parvovirus B19 DNA. J. Virol. Methods, 92, 183–191. [DOI] [PubMed] [Google Scholar]
- 177.Sauleda S., Reesink,H.J., Esteban,J.I., Hess,G., Esteban,R. and Guardia,J. (1999) Profiles of GBV-C/hepatitis G virus markers in patients coinfected with hepatitis C virus. J. Med. Virol., 59, 45–51. [PubMed] [Google Scholar]
- 178.Fujisawa T., Horiike,N., Michitaka,K. and Onji,M. (2000) Influence of RNA titre and amino acid changes in the NS5A region of GB virus C/hepatitis virus G virus on the effectiveness of interferon therapy. J. Gastroenterol. Hepatol., 15, 632–639. [DOI] [PubMed] [Google Scholar]
- 179.Houng H.-S.H., Chen,R.C.-M., Vaughn,D.W. and Kanesa-thasan,N. (2001) Development of a fluorogenic RT–PCR system for quantitative identification of dengue virus serotypes 1–4 using conserved and serotype-specific 3′ noncoding sequences. J. Virol. Methods, 95, 19–32. [DOI] [PubMed] [Google Scholar]
- 180.Saito T., Shinzawa,H., Uchida,T., Kawamata,O., Honma,S., Watanabe,H., Shao,L., Saito,K., Togashi,H. and Takahashi,T. (1999) Quantitative DNA analysis of low-level hepatitis B viremia in two patients with serologically negative chronic hepatitis B. J. Med. Virol., 58, 325–331. [DOI] [PubMed] [Google Scholar]
- 181.Pas S.D., Fries,E., De Man,R.A., Osterhaus,A.D.M.E. and Niesters,H.G. (2000) Development of a quantitative real-time detection assay for hepatitis B virus DNA and comparison with two commercial assays. J. Clin. Microbiol., 38, 2897–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Schweiger B., Zadow,I., Heckler,R., Timm,H. and Pauli,G. (2000) Application of a fluorogenic PCR assay for typing and subtyping influenza viruses in respiratory samples. J. Clin. Microbiol., 38, 1552–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Koenig M., Reynolds,K.S., Aldous,W. and Hickman,M. (2001) Comparison of Light-Cycler PCR, enzyme immunoassay and tissue culture for detection of Herpes Simplex Virus. Diagn. Microbiol. Infect. Dis., 40, 107–110. [DOI] [PubMed] [Google Scholar]
- 184.Schalasta G., Eggers,M., Schmid,M. and Enders,G. (2000) Analysis of human cytomegalovirus DNA in urines of newborns and infants by means of a new ultrarapid real-time PCR-system. J. Clin. Virol., 19, 175–185. [DOI] [PubMed] [Google Scholar]
- 185.Kearns A.M., Draper,B., Wipat,W., Turner,A.J.L., Wheeler,J., Freeman,R., Harwood,J., Gould,F.K. and Dark,J.H. (2001) LightCycler-based quantitative PCR for detection of cytomegalovirus in blood, urine and respiratory samples. J. Clin. Microbiol., 39, 2364–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kennedy M.M., Lucas,S.B., Jones,R.R., Howells,D.D., Picton,S.J., Hanks,E.E., McGee,J.O. and O’Leary,J. (1997) HHV8 and Kaposi’s sarcoma: a time cohort study. J. Clin. Pathol., 50, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kennedy M.M., Cooper,K., Howells,D.D., Picton,S., Biddolph,S., Lucas,S.B., McGee,J.O. and O’Leary,J.J. (1998) Identification of HHV8 in early Kaposi’s sarcoma: implications for Kaposi’s sarcoma pathogenesis. J. Clin. Pathol., 51, 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.O’Leary J.J., Kennedy,M., Luttich,K., Uhlmann,V., Silva,I., Russell,J., Sheils,O., Ring,M., Sweeney,M., Kenny,C. et al. (2000) Localisation of HHV-8 in AIDS related lymphadenopathy. J. Clin. Pathol., 53, 43–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.O’Leary J., Kennedy,M., Howells,D., Silva,I., Uhlmann,V., Luttich,K., Biddolph,S., Lucas,S., Russell,J., Bermingham,N. et al. (2000) Cellular localisation of HHV-8 in Castleman’s disease: is there a link with lymphnode vascularity? J. Clin. Pathol., 53, 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Josefsson A., Livak,K. and Gyllensten,U. (1999) Detection and quantitation of human papillomavirus by using the fluorescent 5′ exonuclease assay. J. Clin. Microbiol., 37, 490–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Swan D.C., Tucker,R.A., Holloway,B.P. and Icenogle,J.P. (1997) A sensitive, type-specific, fluorogenic probe assay for detection of human papillomavirus DNA. J. Clin. Microbiol., 35, 886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lanham S., Herbert,A. and Watt,P. (2001) HPV detection and measurement of HPV-16, telomerase and survivin transcripts in colposcopy clinic patients. J. Clin. Pathol., 54, 304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.MacKenzie J., Gallagher,A., Clayton,R.A., Perry,J., Eden,O.B., Ford,A.M., Greaves,M.F. and Jarrett,R.F. (2001) Screening for herpesvirus genomes in common acute lymphoblastic leukemia. Leukemia, 15, 415–421. [DOI] [PubMed] [Google Scholar]
- 194.Jabs W.J., Hennig,H., Kittel,M., Pethig,K., Smets,F., Bucsky,P., Kirchner,H. and Wagner,H.J. (2001) Normalized quantification by real-time PCR of Epstein–Barr virus load in patients at risk for posttransplant lymphoproliferative disorders. J. Clin. Microbiol., 39, 564–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Beames B., Chavez,D., Guerra,B., Notvall,L., Brasky,K.M. and Lanford,R.E. (2000) Development of a primary tamarin hepatocyte culture system for GB virus-B: a surrogate model for hepatitis C virus. J. Virol., 74, 11764–11772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lewin S.R., Vesanen,M., Kostrikis,L., Hurley,A., Duran,M., Zhang,L., Ho,D.D. and Markowitz,M. (1999) Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J. Virol., 73, 6099–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Choo C.K., Ling,M.T., Suen,C.K.M., Chan,K.W. and Kwong,Y.L. (2000) Retrovirus-mediated delivery of HPV16 E7 antisense RNA inhibited tumorigenicity of CaSki cells. Gynecol. Oncol., 78, 293–301. [DOI] [PubMed] [Google Scholar]
- 198.Hoshino Y., Kimura,H., Kuzushima,K., Tsurumi,T., Nemoto,K., Kikuta,A., Nishiyama,Y., Kojima,S., Matsuyama,T. and Morishima,T. (2000) Early intervention in post-transplant lymphoproliferative disorders based on Epstein–Barr viral load. Bone Marrow Transplant, 26, 199–201. [DOI] [PubMed] [Google Scholar]
- 199.Machida U., Kami,M., Fukui,T., Kazuyama,Y., Kinoshita,M., Tanaka,Y., Kanda,Y., Ogawa,S., Honda,H., Chiba,S. et al. (2000) Real-time automated PCR for early diagnosis and monitoring of cytomegalovirus infection after bone marrow transplantation. J. Clin. Microbiol., 38, 1536–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Gault E., Michel,Y., Nicolas,J.-C., Belabani,C., Nicolas,J.-C. and Garbarg-Chenon,A. (2001) Quantification of human cytomegalovirus DNA by real-time PCR. J. Clin. Microbiol., 39, 772–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tanaka N., Kimura,H., Hoshino,Y., Kato,K., Yoshikawa,T., Asano,Y., Horibe,K., Kojima,S. and Morishima,T. (2000) Monitoring four herpesviruses in unrelated cord blood transplantation. Bone Marrow Transplant, 26, 1193–1197. [DOI] [PubMed] [Google Scholar]
- 202.Gerard C.J., Arboleda,M.J., Solar,G., Mule,J.J. and Kerr,W.G. (1996) A rapid and quantitative assay to estimate gene transfer into retrovirally transduced hematopoietic stem/progenitor cells using a 96-well format PCR and fluorescent detection system universal for MMLV-based proviruses. Hum. Gene Ther., 7, 343–354. [DOI] [PubMed] [Google Scholar]
- 203.Hackett N.R., El Sawy,T., Lee,L.Y., Silva,I., O’Leary,J., Rosengart,T.K. and Crystal,R.G. (2000) Use of quantitative TaqMan real-time PCR to track the time-dependent distribution of gene transfer vectors in vivo. Mol. Ther., 2, 649–656. [DOI] [PubMed] [Google Scholar]
- 204.Sanburn N. and Cornetta,K. (1999) Rapid titer determination using quantitative real-time PCR. Gene Ther., 6, 1340–1345. [DOI] [PubMed] [Google Scholar]
- 205.Scherr M., Battmer,K., Blömer,U., Ganser,A. and Grez,M. (2001) Quantitative determination of lentiviral vector particle numbers by real-time PCR. Biotechniques, 31, 520–526. [DOI] [PubMed] [Google Scholar]
- 206.Lanciotti R.S. and Kerst,A.J. (2001) Nucleic acid sequence-based amplification assays for rapid detection of West Nile and St. Louis Encephalitis. J. Clin. Microbiol., 39, 4506–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gibb T.R., Norwood,D.A.,Jr, Woollen,N. and Henchal,E.A. (2001) Development and evaluation of a fluorgenic 5′ nuclease assay to detect and differentiate between Ebola virus subtypes Zaire and Sudan. J. Clin. Microbiol., 39, 4125–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Brorson K., Swann,P.G., Lizzio,E., Maudru,T., Peden,K. and Stein,K.E. (2001) Use of a quantitative product-enhanced reverse transcriptase assay to monitor retrovirus levels in mAb cell-culture and downstream processing. Biotechnol. Prog., 17, 188–196. [DOI] [PubMed] [Google Scholar]
- 209.de Wit C., Fautz,C. and Xu,Y. (2000) Real-time quantitative PCR for retrovirus-like particle formation in CHO cell culture. Biologicals, 28, 137–148. [DOI] [PubMed] [Google Scholar]