Abstract
Great strides in understanding the molecular underpinnings of RNA catalysis have been achieved with advances in RNA structure determination by NMR spectroscopy and X-ray crystallography. Despite these successes the functional relevance of a given structure can only be assessed upon comparison with biochemical studies performed on functioning RNA molecules. The hairpin ribozyme presents an excellent case study for such a comparison. The active site is comprised of two stems each with an internal loop that forms a series of non-canonical base pairs. These loops dock into each other to create an active site for catalysis. Recently, three independent structures have been determined for this catalytic RNA, including two NMR structures of the isolated loop A and loop B stems and a high-resolution crystal structure of both loops in a docked conformation. These structures differ significantly both in their tertiary fold and the nature of the non-canonical base pairs formed within each loop. Several of the chemical groups required to achieve a functioning hairpin ribozyme have been determined by nucleotide analog interference mapping (NAIM). Here we compare the three hairpin structures with previously published NAIM data to assess the convergence between the structural and functional data. While there is significant disparity between the interference data and the individual NMR loop structures, there is almost complete congruity with the X-ray structure. The only significant differences cluster around an occluded pocket adjacent to the scissile phosphate. These local differences may suggest a role for these atoms in the transition state, either directly in chemistry or via a local structural rearrangement.
INTRODUCTION
Nucleotide analog interference mapping (NAIM) is an efficient chemical group mutagenesis strategy used to identify the subset of atoms important for the function of an RNA molecule (1,2). The method identifies chemical groups involved in secondary or tertiary hydrogen bonds, ligand binding, active site chemistry or other roles required for RNA activity. These data should be useful for analyzing the functional relevance of three-dimensional RNA structures because chemical groups that display an effect on RNA activity when modified or deleted should be the same groups structurally implicated for their involvement in activity (3,4). If there is only a weak correlation between the structural and biochemical data, then it suggests that the interactions observed in the structure are not relevant to the functioning RNA or that the structure does not represent the active state. In either case, NAIM analysis is highly complementary to structure determination, as it can ascribe functional relevance to contacts observed within a structure.
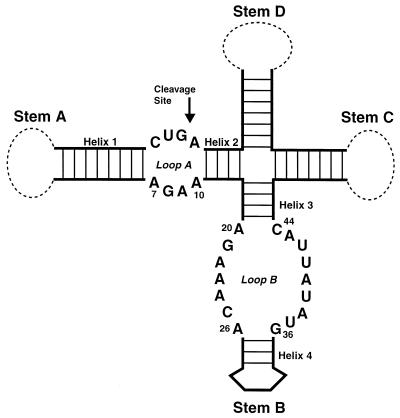
The intent of this report is to compare previously published interference data with recent structures obtained for the hairpin ribozyme, a small catalytic RNA motif that performs a reversible self-cleavage reaction. In nature, the ribozyme functions in the processing of replication intermediates in the ‘life’ cycle of a tobacco ringspot nepovirus satellite RNA (5). In this context, the ribozyme folds into an extended cruciform structure (Fig. 1) (6). The minimum elements of the hairpin RNA required for catalytic activity in vitro are just two arms of this cruciform, which include four short helical elements (helix 1–4) and two internal loop regions, A and B (7,8). The ribozyme performs a cleavage reaction at a specific site within the symmetric loop A, yielding two products with a 5′-hydroxyl group and a 2′,3′-cyclic phosphate, respectively (5). The products of this reaction are similar to three other naturally occurring catalytic RNAs, the hammerhead, VS and HDV ribozymes, although significant differences in secondary structure imply that each molecule has its own distinct catalytic strategy for arriving at these products (9).
Figure 1.
Schematic secondary structure of the hairpin ribozyme in its natural context. The hairpin ribozyme motif is in bold line. The remainder of the satellite RNA sequence is represented by dashed lines. The four stems of the ribozyme are labeled. The cleavage site is marked with an arrow.
Three structures have been reported for the hairpin ribozyme. These include two NMR structures of the isolated stems containing loop A and loop B (10,11) and a high-resolution crystal structure of the docked complex with a 2′-O-methyl inhibitor in place of the 2′-OH nucleophile (12). These structures differ significantly in both their tertiary structure fold and in the nature of the non-canonical base pairs formed in each loop. Such dramatic differences raise a significant question: which if any of these structures represent the hairpin ribozyme in its active conformation? Previously, we performed NAIM analysis on the hairpin with several nucleotide analogs of A, C, G and U (13). These included analogs that eliminated or modified functional groups on either the heterocyclic base or the ribose sugar. Here, we utilize these data to assess the functional relevance of the available hairpin ribozyme structures.
HAIRPIN RIBOZYME STRUCTURES
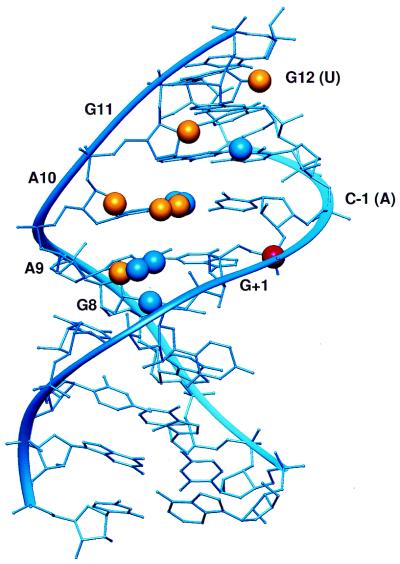
Two independent NMR structures have been determined for the hairpin ribozyme. The structure of helix 1, loop A, and helix 2 (termed stem A) was solved in isolation from the rest of the ribozyme by Cai and Tinoco (11) (Fig. 2). In this structure, the bases of loop A form a series of four non-canonical pairs resulting in an extended helix. One base, U+2, flips out of the helix and is oriented toward the solvent.
Figure 2.
NAIM compared with the stem A NMR structure. Sites of interference are denoted by spheres. Blue spheres represent sites of interference consistent with the structure. The yellow spheres represent sites that cannot be interpreted in comparison with the structure. The scissile phosphate is denoted by a red sphere.
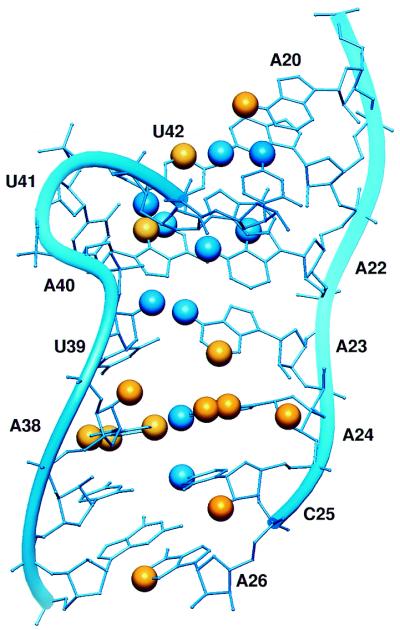
The other half of the ribozyme was also determined by NMR. Butcher and coworkers (10) reported the structure of helix 3, loop B, and helix 4 (termed stem B) (Fig. 3). Similar to the loop A structure, the nucleotides in loop B form a series of non-canonical base pairs with two bulged uridines, U39 and U41. The geometry of the non-canonical pairing disrupts the continuous stack, so that helix 3 stacks upon four pairs in the top of loop B, while helix 4 stacks upon the three pairs at the bottom of loop B. As a result, this stem adopts a bent structure with a narrow minor groove that appears to have some conformational flexibility. Though stem A and stem B each form an independently folded unit, the authors suggest that conformational changes may be necessary to achieve the active, docked structure.
Figure 3.
NAIM compared with the NMR structure of stem B. The diagram is labeled as in Figure 2.
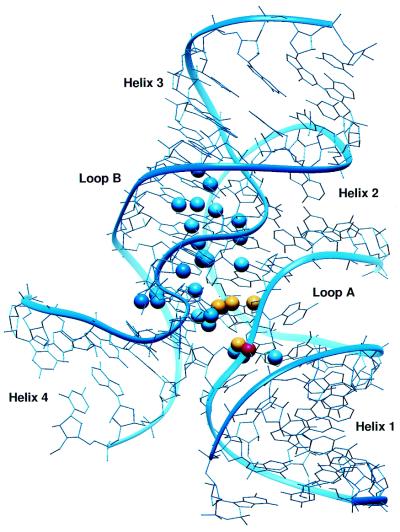
Rupert and Ferre-D’Amare recently reported the 2.4 Å structure of the intact ribozyme in which loop A is docked into loop B (Fig. 4) (12). In this context the secondary structures in loop A and loop B are significantly different than in the isolated stems. For example, all of the non-canonical base pairs observed in the NMR structure of stem A adopt a different conformation in the docked structure. Instead of being flipped out of the helix, U+2 is in the helical stack and pairs with G8, while G+1 adopts an unusual 2′-endo sugar pucker and syn base configuration and base pairs with C25 in loop B (12,14). Likewise, several differences are observed in the non-canonical base pairs observed in stem B. These changes are necessary to facilitate specific tertiary interactions between loops A and B.
Figure 4.
NAIM compared with the crystal structure of the docked ribozyme. The diagram is labeled as in Figure 2.
SUPERPOSITION OF NAIM DATA ON THE HAIRPIN STRUCTURES
In an effort to systematically compare the data obtained on active hairpin ribozymes with the available hairpin structures, we superimposed the interference data collected with all of the analogs onto the coordinates of each structure (Figs 2–4). The interferences used in this comparison are listed in Table 1. We used the following criterion to assess the correlation between each structure and the biochemical data. (i) A given interference is considered to be agreement if the analog deletes a functional group involved in an interaction within the structure. For example, the N2 amine of G21 interacts with the N7 of A22 in the loop B NMR structure. Both of these groups lead to interference when modified, hence this contact is consistent with the interference data. (ii) An interference resulting from incorporation of a bulky nucleotide analog matches the structure if it creates a steric clash, which implies that the structure cannot sufficiently accommodate the analog modification. For example, the N2 amine of G8 makes two contacts via its N2 amine in the crystal structure to the 2′-OH of A-1 and the O2 of U+2. Consistent with these contacts, methylation of this amine disrupts activity. (iii) An analog may affect the local conformation of the nucleotide, for example disruption of the sugar pucker. An example of this occurs at U41 with interference from 2′-fluoro substitution, an analog that favors a C-3′-endo conformation. U41 base adopts C-2′-endo in both the NMR and the crystal structure and hence 2′-fluoro interference is consistent with both structures. For interferences that meet these criteria, the affected functional groups are designated with blue colored spheres in Figures 2–4. If none of these criteria are met within the structure, the functional groups that show interference are designated with yellow colored spheres.
Table 1. NAIM analysis of the hairpin ribozyme.
| Position | Analog effects | Comment |
|---|---|---|
| G8 | Ino, m2G | |
| A9 | m6A, DAP, Pur, 2AP, c3A, n8A | n8A is an enhancement, attributed to the N1 position (17) |
| A10 | Pur, 2AP, dA, FA, c3A, n8A | n8A interference is pH dependent, attributed to N1 ionization (17) |
| G11 | Ino, m2G, dG, FG | |
| U12 | dU, FU | |
| G21 | Ino, m2G | |
| A20 | m6A | |
| A22 | 7dA, m6A | |
| A23 | m6A, DAP, 2AP | |
| A24 | DAP, Pur, FA, c3A | |
| C25 | dC, 5mFC, Zeb | |
| A26 | DAP, 2AP | |
| A38 | 7dA, DAP, Pur, 2AP, dA, FA | |
| U39 | FU, m5U | |
| A40 | DAP, 2AP, c3A | |
| U41 | FU | |
| U42 | 5mU, dU | dU is an enhancement, attributed to increased conformational flexibility (13) |
| A43 | 7dA | |
| C44 | Zeb |
Ino, inosine; m2G, N2-methylguanosine; dG, 2′-deoxyguanosine; FG, 2′-deoxy-2′-fluoroguanosine; m6A, N6-methyladenosine; DAP, 2,6-diaminopurine riboside; Pur, purine riboside; 2AP, 2-aminopurine riboside; c3A, 3-deazaadenosine; n8A, 8-azaadenosine; dA, 2′-deoxyadenosine; FA, 2′-deoxy-2′-fluoroadenosine, 7dA, 7-deazaadenosine; dC, 2′-deoxycytidine; 5mFC, 5-methyl-2′-fluorocytidine; Zeb, Zebularine; dU, 2′-deoxyuridine; FU, 2′-deoxy-2′-fluorouridine; 5mU, 5-methyluridine (ribothymidine).
STEM A AND B STRUCTURES ARE LARGELY INCONSISTENT WITH THE NAIM DATA
Even a cursory inspection of the figures shows that the isolated stem A and stem B NMR structures are not well matched with the NAIM data (Figs 2 and 3). In the structure of loop A, most of the interferences (>50%) cannot be explained by the structure. Similarly, the loop B NMR structure does not match well with the interference pattern.
It is possible that the failure of some functional groups to appear relevant within the NMR structures could result from a lack of tertiary contacts to the opposing stem in the functional docked structure. For example, several interferences at positions A9-U12 do not reflect contacts within the secondary structure of the stem, but they cluster in the minor groove surface of the loop A secondary structure. This may reflect a pre-ordered loop B binding surface.
However, there is reason to doubt that the stem A NMR structure represents a pre-folded structural domain that utilizes this minor groove surface to interact with stem B in the functioning ribozyme. Several functional groups expected to be necessary for ribozyme activity based on the geometry of stem A fail to show interference with analogs that delete or modify them. While these are negative data, they may indicate that certain aspects of the NMR structure do not adequately reflect the docked, active structure. For example, the N2 exocyclic amine of G+1 was shown to be critical for ribozyme activity (15,16). Site-specific inosine substitution at this position reduces ribozyme cleavage activity to below detectable levels. In the NMR structure, this amine interacts with the N7 of A9. Analogs that delete or modify the N7 moiety do not interfere with ribozyme activity at A9. If the sole role of G+1 amine is to form a single H-bond interaction with the A9 N7, then analogs that disrupt this position should affect ribozyme activity at a level comparable with the G+1 inosine effect. While it is possible that the G+1 amine may form an additional contact in the active structure, these data argue that the stem A structure might not reflect that conformation.
Unlike the interferences in stem A, those in stem B do not cluster into a discreet region of the structure. Instead, they are scattered across the minor and major grooves throughout the length of the molecule. As such, these interferences are not likely to reflect the formation of tertiary contacts missing in the isolated loop B stem structure. It is more likely that the secondary structure of loop B undergoes a rearrangement prior to forming the docked structure.
THE CRYSTAL STRUCTURE MATCHES THE NAIM DATA EXCEPT NEAR THE SCISSILE PHOSPHATE
In contrast to the isolated stem structures, the crystal structure of the intact ribozyme is almost fully consistent with the NAIM data (Fig. 4). Approximately 85% of the interferences can be explained by features in this structure. Interference is observed at most of the functional groups involved in the non-canonical secondary structure in stems A and B, as well as those in tertiary interface. Bulky analogs are accommodated on the surface of this structure, but not in the tightly packed interface between the internal loops.
All of the unexplained interferences cluster around the scissile phosphate in a pocket formed by the conserved residues G+1, A9, A10, C25 and A38 (12). All of the functional groups that point into this cavity are required for catalysis, including the N2 amine of G+1, the N6 amine of A38, and the N1 imino group of A10 (2,13,15). Furthermore, occupation of this cavity by methylation of the N6 amine of A9 significantly disrupts activity (13). Previous work indicated that ionization of A10 is important for ribozyme catalysis, that the N6 amine of A38 is indispensable for catalysis, and that these groups are likely to be involved in the stabilization of the catalytic transition state (17). These few exceptions may reflect local disturbances in the structure necessary to accommodate the 2′-O-methyl substitution at the cleavage site or transition state specific effects on ribozyme chemistry. They may provide clues into the mechanism of catalysis by the hairpin ribozyme.
INTERPRETATION AND ANALYSIS
Several lines of evidence suggest that the crystal structure of the docked complex more accurately reflects the biologically relevant structure than either NMR structure. (i) The strong correlation between the NAIM data and the crystal structure evident from Figure 4 in this analysis. (ii) The nucleophile, reactive phosphate, and 5′-oxy leaving group are in an in-line configuration (12). This specific geometry is necessary for the SN2 phosphodiester reaction, but is not the geometry of the scissile phosphate within the loop A structure (11). (iii) The crystal structure matches the artificial phylogenetic data on the ribozyme. All of the conserved nucleotides in the hairpin sequence are involved in specific secondary and tertiary structure contacts (18). (iv) Several specific tertiary interactions predicted by crosslinking and mutagenesis experiments are observed in the crystal structure (8,14,19). (v) The structure is consistent with the majority of solution modification experiments and site-specific analog substitution data (8,16,20–24). (vi) Hampel and Burke have shown that a highly reactive photo-crosslink indicative of the ‘loop E-like’ motif present in the loop B NMR structure is inhibitory to docking and ribozyme catalysis and that the crosslink is lost upon formation of the active structure (21).
Although the biochemical data suggest that the NMR structures of stem A and stem B do not reflect the active conformation, it does not mean that these structures are incorrect. It is likely that each structure reflects the accurate, undocked conformation. In fact, this interpretation is suggested by crosslinks observed in stem B that are indicative of features observed in the NMR structure (21). Furthermore, the lack of observable imino NOEs for the loop region of stem B, as well as intermediate J-couplings for the majority of the bases, suggest that the structure of this molecule is unusually dynamic (10). If true, the hairpin ribozyme must undergo a complex series of rearrangements in order to form the docked complex. This is corroborated by FRET studies that suggest docking limits the rate of ligation chemistry (25).
While the NAIM data strongly support the functional relevance of the crystal structure, a handful of the most interesting interferences cluster in a cavity surrounding the scissile phosphate and are not explained by the structure. These groups do not appear to be making ground state contacts within the tertiary structure, but instead may participate in promoting the chemical transition. One possibility is that the cavity is occupied by a reaction molecule, such as a water, and that these groups are in position to coordinate and activate it for catalysis. While no water molecule was observed in the cavity within the hairpin crystal structure, the authors note that there is sufficient room for one to be accommodated (12). Alternately, ionization of A10 could stabilize charge in the pre-ligation ground state or in the transition state by interacting with a non-bridging phosphate oxygen of the scissile phosphate, while a subtle change in geometry could allow the amine of A38 to hydrogen bond to the 5′-hydroxyl nucleophile in the ligation reaction. Although this model would require alterations in the geometry of the bases that surround the reactive phosphate, it is possible that removal of the 2′-O-methyl inhibitor or subtle differences between the cleaved and ligated forms of the ribozyme could lead to these changes. In either case, movement of A10 and A38 by <3 Å would position these bases for a direct role in catalysis.
In summary, the NAIM data argue that the docked crystal structure very closely reflects the functioning state of the hairpin ribozyme. However, the crystal structure does not immediately elucidate the mechanism of hairpin ribozyme catalysis. Strikingly, the only positions of interference that do not correlate to the crystal structure lie in the vicinity of the active site. It is possible that these groups are involved in hairpin ribozyme catalysis via an as yet to be determined chemical pathway.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Michael Recht and Pete Funke for assistance with preparing figures. This work was supported by NSF grant CHE-0100057 to S.A.S.
REFERENCES
- 1.Strobel S.A. (1998) Ribozyme chemogenetics. Biopolymers, 48, 65–81. [DOI] [PubMed] [Google Scholar]
- 2.Grasby J.A. and Gait,M.J. (1994) Synthetic oligoribonucleotides carrying site-specific modifications for RNA structure–function analysis. Biochimie, 76, 1223–1234. [DOI] [PubMed] [Google Scholar]
- 3.Strobel S.A., Ortoleva-Donnelly,L., Ryder,S.P., Cate,J.H. and Moncoeur,E. (1998) Complementary sets of noncanonical base pairs mediate RNA helix packing in the group I intron active site. Nature Struct. Biol., 5, 60–66. [DOI] [PubMed] [Google Scholar]
- 4.Szewczak A.A., Ortoleva-Donnelly,L., Ryder,S.P., Moncoeur,E. and Strobel,S.A. (1998) A minor groove RNA triple helix within the catalytic core of a group I intron. Nature Struct. Biol., 5, 1037–1042. [DOI] [PubMed] [Google Scholar]
- 5.Buzayan J.M., Hampel,A. and Bruening,G. (1986) Nucleotide sequence and newly formed phosphodiester bond of spontaneously ligated satellite tobacco ringspot virus RNA. Nucleic Acids Res., 14, 9729–9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haseloff J. and Gerlach,W.L. (1989) Sequences required for self-catalysed cleavage of the satellite RNA of tobacco ringspot virus. Gene, 82, 43–52. [DOI] [PubMed] [Google Scholar]
- 7.Hampel A. and Tritz,R. (1989) RNA catalytic properties of the minimum (–)sTRSV sequence. Biochemistry, 28, 4929–4933. [DOI] [PubMed] [Google Scholar]
- 8.Chowrira B.M. and Burke,J.M. (1991) Binding and cleavage of nucleic acids by the ‘hairpin’ ribozyme. Biochemistry, 30, 8518–8522. [DOI] [PubMed] [Google Scholar]
- 9.Doherty E.A. and Doudna,J.A. (2000) Ribozyme structures and mechanisms. Annu. Rev. Biochem., 69, 597–615. [DOI] [PubMed] [Google Scholar]
- 10.Butcher S.E., Allain,F.H. and Feigon,J. (1999) Solution structure of the loop B domain from the hairpin ribozyme. Nature Struct. Biol., 6, 212–216. [DOI] [PubMed] [Google Scholar]
- 11.Cai Z. and Tinoco,I.,Jr (1996) Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry, 35, 6026–6036. [DOI] [PubMed] [Google Scholar]
- 12.Rupert P.B. and Ferre-D’Amare,A.R. (2001) Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature, 410, 780–786. [DOI] [PubMed] [Google Scholar]
- 13.Ryder S.P. and Strobel,S.A. (1999) Nucleotide analog interference mapping of the hairpin ribozyme: implications for secondary and tertiary structure formation. J. Mol. Biol., 291, 295–311. [DOI] [PubMed] [Google Scholar]
- 14.Pinard R., Lambert,D., Walter,N.G., Heckman,J.E., Major,F. and Burke,J.M. (1999) Structural basis for the guanosine requirement of the hairpin ribozyme. Biochemistry, 38, 16035–16039. [DOI] [PubMed] [Google Scholar]
- 15.Chowrira B.M., Berzal-Herranz,A. and Burke,J.M. (1991) Novel guanosine requirement for catalysis by the hairpin ribozyme. Nature, 354, 320–322. [DOI] [PubMed] [Google Scholar]
- 16.Grasby J.A., Mersmann,K., Singh,M. and Gait,M.J. (1995) Purine functional groups in essential residues of the hairpin ribozyme required for catalytic cleavage of RNA. Biochemistry, 34, 4068–4076. [DOI] [PubMed] [Google Scholar]
- 17.Ryder S.P., Oyelere,A.K., Padilla,J.L., Klostermeier,D., Millar,D.P. and Strobel,S.A. (2001) Investigation of adenosine base ionization in the hairpin ribozyme by nucleotide analog interference mapping. RNA, 7, 1454–1463. [PMC free article] [PubMed] [Google Scholar]
- 18.Berzal-Herranz A., Joseph,S., Chowrira,B.M., Butcher,S.E. and Burke,J.M. (1993) Essential nucleotide sequences and secondary structure elements of the hairpin ribozyme. EMBO J., 12, 2567–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Earnshaw D.J., Masquida,B., Muller,S., Sigurdsson,S.T., Eckstein,F., Westhof,E. and Gait,M.J. (1997) Inter-domain cross-linking and molecular modelling of the hairpin ribozyme. J. Mol. Biol., 274, 197–212. [DOI] [PubMed] [Google Scholar]
- 20.Butcher S.E. and Burke,J.M. (1994) Structure-mapping of the hairpin ribozyme. Magnesium-dependent folding and evidence for tertiary interactions within the ribozyme–substrate complex. J. Mol. Biol., 244, 52–63. [DOI] [PubMed] [Google Scholar]
- 21.Hampel K.J. and Burke,J.M. (2001) A conformational change in the ‘loop E-like’ motif of the hairpin ribozyme is coincidental with domain docking and is essential for catalysis. Biochemistry, 40, 3723–3729. [DOI] [PubMed] [Google Scholar]
- 22.Chowrira B.M., Berzal-Herranz,A., Keller,C.F. and Burke,J.M. (1993) Four ribose 2′-hydroxyl groups essential for catalytic function of the hairpin ribozyme. J. Biol. Chem., 268, 19458–19462. [PubMed] [Google Scholar]
- 23.Schmidt S., Beigelman,L., Karpeisky,A., Usman,N., Sorensen,U.S. and Gait,M.J. (1996) Base and sugar requirements for RNA cleavage of essential nucleoside residues in internal loop B of the hairpin ribozyme: implications for secondary structure. Nucleic Acids Res., 24, 573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young K.J., Vyle,J.S., Pickering,T.J., Cohen,M.A., Holmes,S.C., Merkel,O. and Grasby,J.A. (1999) The role of essential pyrimidines in the hairpin ribozyme-catalysed reaction. J. Mol. Biol., 288, 853–866. [DOI] [PubMed] [Google Scholar]
- 25.Walter N.G. and Burke,J.M. (1997) Real-time monitoring of hairpin ribozyme kinetics through base-specific quenching of fluorescein-labeled substrates. RNA, 3, 392–404. [PMC free article] [PubMed] [Google Scholar]