SUMMARY
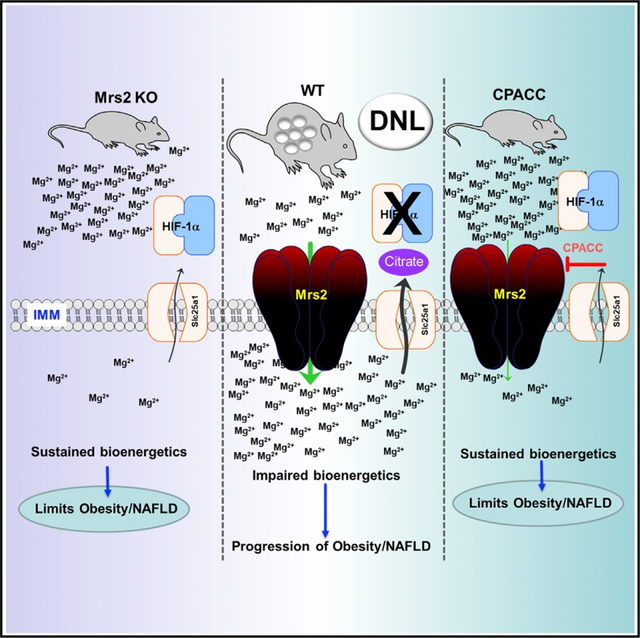
The most abundant cellular divalent cations, Mg2+ (mM) and Ca2+ (nM-μM), antagonistically regulate divergent metabolic pathways with several orders of magnitude affinity preference, but the physiological significance of this competition remains elusive. In mice consuming a Western diet, genetic ablation of the mitochondrial Mg2+ channel Mrs2 prevents weight gain, enhances mitochondrial activity, decreases fat accumulation in the liver, and causes prominent browning of white adipose. Mrs2 deficiency restrains citrate efflux from the mitochondria, making it unavailable to support de novo lipogenesis. As citrate is an endogenous Mg2+ chelator, this may represent an adaptive response to a perceived deficit of the cation. Transcriptional profiling of liver and white adipose reveals higher expression of genes involved in glycolysis, β-oxidation, thermogenesis, and HIF-1α-targets, in Mrs2−/− mice that are further enhanced under Western-diet-associated metabolic stress. Thus, lowering mMg2+ promotes metabolism and dampens diet-induced obesity and metabolic syndrome.
In brief
Madaris et al. show that limiting mitochondrial Mg2+ enhances bioenergetics in hepatocytes and thereby prevents Western-diet-induced NAFLD, microvascular rarefaction, and spontaneous tumor prevalence, and Mrs2 channel deletion reprograms whole-body energy metabolism that is driven by HIF-1α transcriptional regulation in liver and adipose tissues in a citrate metabolite-dependent manner.
Graphical Abstract
INTRODUCTION
Mitochondrial dysfunction and altered metabolite flux are among the cellular hallmarks of metabolic syndrome. Mitochondria are critical hubs for cellular metabolism, and anomalies have been linked to disorders including obesity, diabetes, and cardiovascular disease.2–5 Ion channels are vital regulators of numerous cellular signals that control mitochondrial bioenergetics, proliferation, and cell fate.3,6–10 In multicellular organisms, mitochondrial Ca2+ (mCa2+) uptake is a key driver of cellular bioenergetics. On the other hand, overstimulation leading to excess Ca2+ induces mitochondrial permeability pore activation, driving energetic collapse and cell death.9,11,12 Although mitochondrial Ca2+ modulates cellular function, the influence of Mg2+ on mitochondrial energetics is largely unexplored. Thus, to understand bioenergetics, we must improve our understanding of the links between mitochondrial divalent ion fluxes and cellular function.
The major mCa2+ uptake machinery, the mitochondrial calcium uniporter (MCU), is essential to shape cytosolic Ca2+ dynamics and promote mitochondrial respiration.13,14 Indeed, genetic ablation of MCU in murine liver, heart, and skeletal muscle causes metabolic aberrations such as triglyceride accumulation, lowered ketone body production, and increased total body fat.4,15 Conservation of MCU”s role in governing cellular metabolism is demonstrated by global knockout of MCU in zebrafish.4 Interestingly, several pieces of evidence suggest that, in addition to directly opposing actions of Ca2+, intracellular Mg2+ (iMg2+) can impair the activity of MCU and thereby lower mCa2+.3,16,17 Unlike MCU, the mitochondrial Mg2+ (mMg2+) uptake machinery, MRS2, is conserved in both anaerobic and aerobic species.3,10,18 iMg2+ is essential for a wide range of cellular processes, including membrane integrity, charge neutralization, stabilization of nucleic acids, and biological activity of nucleotides and serves as a cofactor for numerous cytosolic and mitochondrial metabolic enzymes.3,19 However, the molecular signals that rely on mMg2+ dynamics have not been fully elucidated. Remarkably, the mammalian glycolytic end-product, L-lactate, triggers a dynamic transfer of free Mg2+ ions between the endoplasmic reticulum (ER) and mitochondria to shape bioenergetics and cellular metabolism.10 Deficiency of Mrs2 in mice is protective from organ dysfunction in an in vivo model of septic shock and allows maintenance of proper mitochondrial bioenergetics.10 Despite these advances, the molecular events that link iMg2+ dynamics and cellular metabolism are largely unknown.
The liver plays a dominant role in shaping glucose and lipid metabolism during feeding and fasting. This cyclic process is controlled by hormones, ligand-gated GPCR signals, and ion channels.20–22 In hepatocytes, glucagon, vasopressin, and epinephrine stimulate GPCR-linked intracellular Ca2+ (iCa2+) mobilization, which enhances the activity of mitochondrial enzymes involved in oxidative metabolism.23 Over decades, several Mg2+ transporters have been proposed to regulate cellular Mg2+ homeostasis in mammals; however, the role of this cation in balancing carbohydrate and fat metabolism has remained ambiguous.3 Since the basal “free” Mg2+ dynamically controls iCa2+, any aberration to this signaling cascade may contribute to metabolic diseases such as diabetes, obesity, non-alcoholic fatty liver disease (NAFLD), hepatic steatohepatitis (NASH), and even progression to liver cirrhosis and hepatocellular carcinoma (HCC).3 Thus, further interrogation of the influence of iMg2+ on long-term Western diet (WD)-induced hepatic steatosis and obesity is urgently needed. Here we set out to define the mechanistic link between iMg2+ dynamics and whole-body metabolism in the setting of an obesogenic diet.
RESULTS
Lowering mMg2+ mitigates long-term Western-diet-induced obesity and metabolic syndrome
The highly conserved bacterial CorA homologue, Mrs2, is an inner mitochondrial membrane-localized Mg2+ selective uptake machinery whose biological function is not well understood.10 Here, we investigated whether long-term WD-induced metabolic syndrome could be prevented by perturbation of mMg2+ dynamics. Fourteen-week-old male mice were subjected to WD feeding for up to 52 weeks. The WD is composed of 40% (kcal) fat in comparison to 17% in the standard chow diet (CD). This protocol leads to obesity and deterioration of organ function.24,25 Remarkably, consumption of the WD for 30 weeks caused obesity in wild-type (WT) but not Mrs2−/− mice (Figure 1A). We continued the dietary protocol up to 52 weeks; even under this long-term dietary stress, loss of Mrs2 prevented WD-induced weight gain (Figure 1B). WD-fed WT mice but not Mrs2−/− developed hepatomegaly typical of obesity and NAFLD (Figures 1B and 1C). MRI body composition analysis revealed reciprocal changes in fat and lean body mass with WD feeding in WT or Mrs2−/− mice (Figures 1D and 1E). Intriguingly, the CD groups exhibited a normal fat/lean mass ratio highlighting a metabolic switch in the Mrs2−/− mice upon consumption of an obesogenic diet.
Figure 1. Blocking mitochondrial Mg2+ uptake prevents diet-induced obesity.
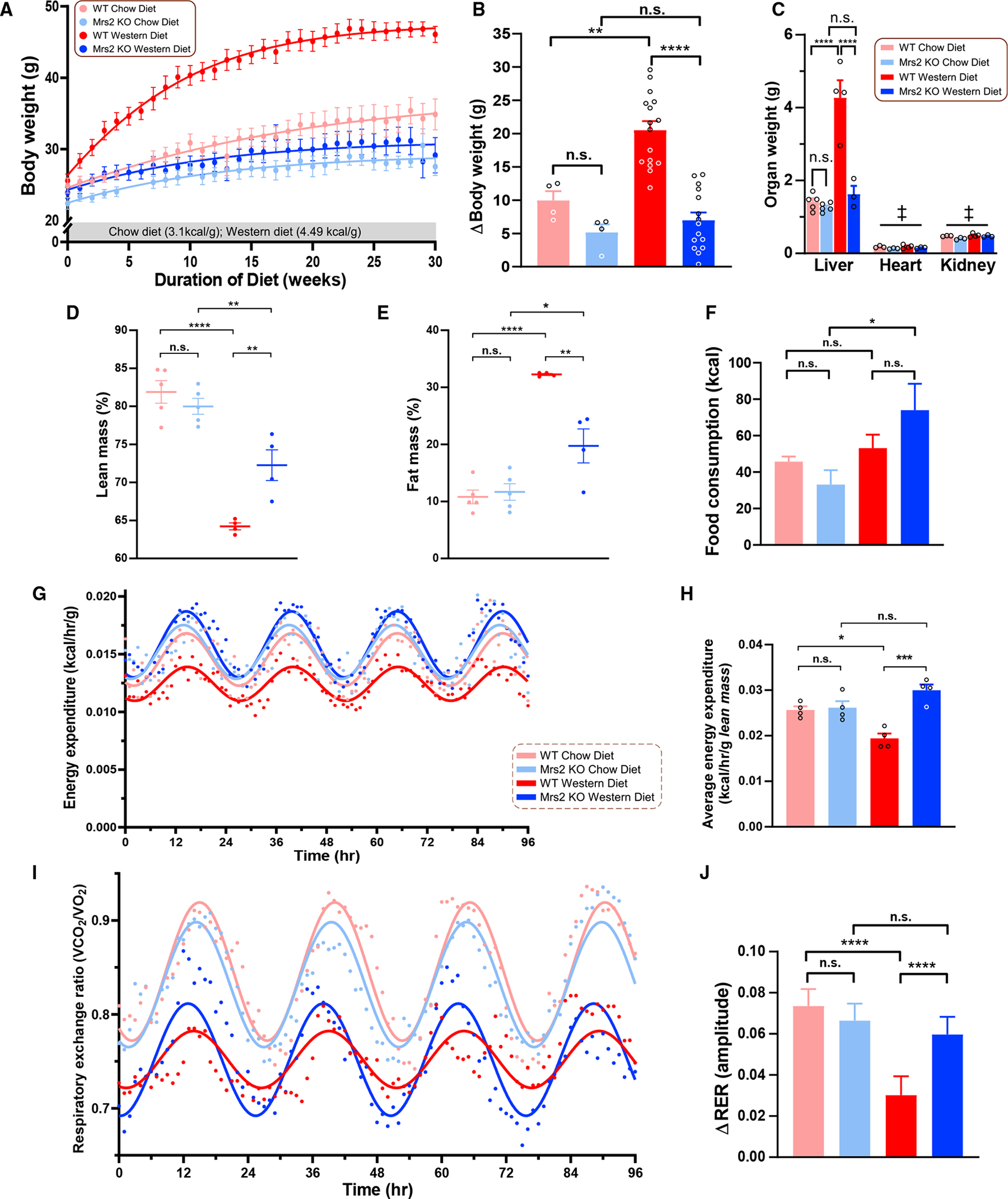
(A) Body weight change over a 30-week diet period. Data fit with a Gompertz growth curve (n = 4–16 mice/group).
(B) Weight gained during the dietary regimen.
(C) Liver, heart, and kidney weights after 1 year diet period. ‡ = Not significant, multiple comparisons are done the same as the liver. n = 3–4 mice (organs) per group.
(D and E) Percent lean mass (D) and fat mass (E) of indicated groups. n = 3–4 mice.
(F) Measurement of food intake of 1 year CD- or WD-fed mice during indirect calorimetry with Promethion system.
(G) Energy expenditure of CD- or WD-fed (1 year) WT or Mrs2−/− mice. Data fit with a fixed-frequency sinusoidal equation y = B + A sin(0.25x + ϕ). n = 4–5 mice.
(H) Lean mass-normalized energy expenditure data derived from (G).
(I) Respiratory exchange ratio (RER) of CD- or WD-fed (1 year) WT or Mrs2−/− mice. Data fit with a fixed-frequency sinusoidal equation y = B + A sin(0.25x+ϕ). n = 4–5 mice.
(J) The change in RER from light to dark cycle. All data shown as mean ± SEM; ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, n.s. = not significant.
Next, mice were placed in indirect calorimetry chambers to examine energy expenditure and behavior.26,27 The difference in weight gain in the WD-fed mice (Figures 1A and 1B) did not appear to be attributable to changes in food intake, water consumption, or locomotion, as WD-fed Mrs2−/− mice displayed no significant changes in these parameters compared with their WT counterparts (Figures 1F, S1A, and S1B). Mice were subjected to an overnight (16 h) fast followed by ad libitum feeding for 4 h then measurement of blood parameters. In agreement with the elevated dietary cholesterol (2%), WD-fed mice have elevated plasma cholesterol despite the differences in body weight (Figures S1C and S1D). As typically seen in obese mice, the fasting blood glucose levels were higher in WT WD, but the Mrs2−/− mice exhibited normal glucose that was comparable to that of mice on CD (Figure S1E). Fasting insulin levels were relatively similar between the groups but trended higher in the obese mice (Figure S1E). Although refed glucose levels were similar among the four groups, they were slightly elevated in the CD groups (Figure S1F), whereas post-prandial insulin levels were increased in WD mice (Figure S1F). Energy expenditure was lower in WT WD mice compared with Mrs2−/− WD mice (Figures 1G, 1H, and S1G). However, rectal body temperature was similar in all groups (Figure S1H). Further, the ANCOVA analysis supports these findings (Figures S1I and S1J).26,27 Mice consuming WD had an overall decrease in respiratory exchange ratio (RER), as is expected with increased reliance on fat-based respiration.28 WD-fed WT mice had dampened RER oscillations between the light and dark cycles; however, Mrs2−/− mice preserved oscillations in RER during dietary stress (Figures 1I and 1J). Together, these data demonstrate that lowering [Mg2+]m opposes changes associated with diet-induced metabolic syndrome.
Deletion of Mrs2 prevents WD-induced liver steatosis, microvascular rarefaction, and spontaneous tumor prevalence
Consumption of high-fat diet (HFD) contributes to liver steatosis and hepatocellular carcinoma.29 The plasma ALT (alanine transaminase) level, a marker of liver damage, was elevated in WT WD but not in Mrs2−/− WD mice or the CD controls (Figure 2A).30 Fatty deposition, neoplastic lesions, and enlargement are all characteristic of NAFLD and these changes were absent in Mrs2−/− WD mice (Figures 2B and S2A). Further, microscopic examination of the livers from WD-fed mice revealed steatosis and fibrosis in WT but not in Mrs2−/− mice or CD controls (Figure 2C [top] and Figure S2B; Figure 2C [bottom] and Figure S2C). Remarkably, WT WD mice frequently developed liver tumors, whereas Mrs2−/− WD mice displayed significantly lower incidence (Figures 2D and S2D). Microvascular rarefaction (microvascular reduction) is a phenomenon that occurs commonly during aging-associated cardiovascular diseases.31 Staining for the CD31+ endothelial marker32 revealed decreased capillary density in WT WD mice (Figures 2E and 2F). In contrast, WD failed to cause microvascular rarefaction in Mrs2−/− mice (Figures 2E, 2F and S2E). Closer examination using electron microscopy revealed numerous large lipid droplets in WT WD-fed mice that were absent in Mrs2−/− WD hepatocytes (Figure S2F). Since WD-induced obesity is associated with chronic kidney disease, we evaluated the cortex for fibrosis by examining collagen deposition. Masson’s trichrome staining of coronal sections showed collagen accumulation (blue color) in WD-fed WT mice, while collagen deposition was nearly absent in WD-fed Mrs2−/− mice (Figure S2G). The histologic evaluation of inguinal white adipose tissue (iWAT) revealed an expansion of adipocytes in WT WD, but it is abrogated in the Mrs2−/− WD mice (Figure S2H). Although WT WD liver tissue exhibited hepatocellular carcinoma, the iWAT did not show apparent neoplastic foci (Figure S2H). These data indicate that the deletion of Mrs2 channel preserves microvascular density while preventing diet-induced NAFLD and HCC progression.
Figure 2. Mrs2 deletion prevents the progression of NAFLD and development of HCC.
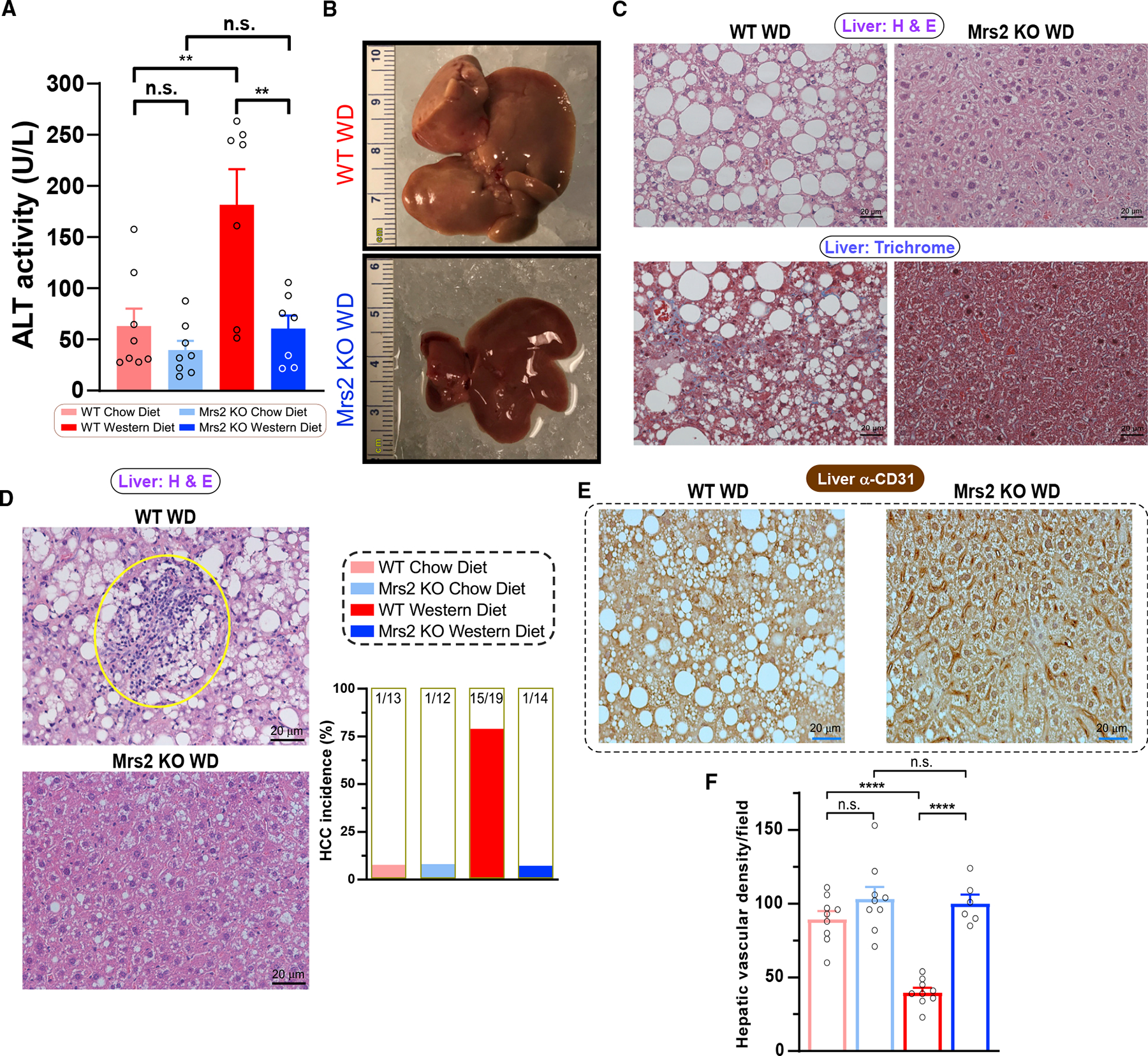
(A) ALT levels in the plasma after 1-year diet period. n = 7–8 mice.
(B) Representative liver from WD-fed (1 year) mice, WT (top) and Mrs2−/− (bottom). Scale indicates centimeter.
(C) Representative liver tissue sections stained with H&E (top) and Masson’s trichrome (bottom) from WT and Mrs2−/− fed WD. n = 3 mice.
(D) Representative images of H&E-stained liver sections from 1-year WD-fed mice (WT, top and Mrs2−/−, bottom). Note a focus of immune cell infiltrates (circled) an indicator of tumor nodules. Bar graph represents the hepatocellular carcinoma (HCC) incidence in these four groups. n = 12–19 mice/group.
(E) Immunohistochemical (IHC) analysis (α-CD31/PECAM1) of sinusoidal vasculature in liver from WD-fed WT (left) and Mrs2−/− mice (right). n = 3 mice.
(F) Quantification of hepatic microvascular density from Figure 2E n = 3 mice. Nine images per group were used for quantification. All data shown as mean ± SEM; ****p < 0.0001, **p < 0.01, n.s. = not significant.
HIF-1α pathway as a switchable link in WD-induced obesity
To identify the causal link between iMg2+ dynamics and metabolic disease progression, we performed global RNA profiling of liver and adipose tissues (Figures S3A–S3C). The panoramic transcriptomic changes in liver from WT WD and Mrs2−/− WD showed that ≈5,500 protein-coding genes were differentially regulated, of which more than 200 are involved in metabolic diseases and cellular processes such as glucose and fatty acid metabolism and mitochondrial bioenergetics, consistent with the antagonistic role of Mg2+ against Ca2+-dependent signaling (Figure 3A).10,33 However, comparison of livers from WT and Mrs2−/− CD revealed that only eight genes were significantly changed, of which six were differentially regulated in both dietary conditions (Figures S3D and S3E). As presented in Figures 2C and 2D, WT WD liver exhibited a significant elevation of tumorigenesis, inflammation, and fibrosis markers when compared with Mrs2−/− WD (Figures 3B and S3F).34–36 Although few individual genes were altered in liver from CD-fed mice, the Kyoto Encyclopedia of Genes and Genome (KEGG) pathway analysis revealed significant patterns in numerous gene sets (pathways) (Figure S3D and S3F left). Unbiased transcription factor analysis using the ChIP-X Enrichment Analysis (ChEA) program demonstrated HIF1-α as a likely candidate responsible for a subset of the transcriptional changes seen in WD-fed animals (Figure S3G).37 Remarkably, HIF1-α-dependent signaling was observed to be differentially regulated in WD-fed mice (Figures S4G and S4H). Furthermore, KEGG-based analysis of WD liver tissue revealed a substantial repression of thermogenesis, oxidative phosphorylation, fatty acid catabolism, glycolysis, TCA cycle, and the hypoxia-inducible pathways in WT but not Mrs2−/− mice (Figure S3H). These data suggest a procatabolic state in Mrs2−/− liver tissue during chronic WD consumption.
Figure 3. Blunting mMg2+ dynamics enhances hepatic oxidative metabolism profiles and adipocyte browning under Western diet regimen.
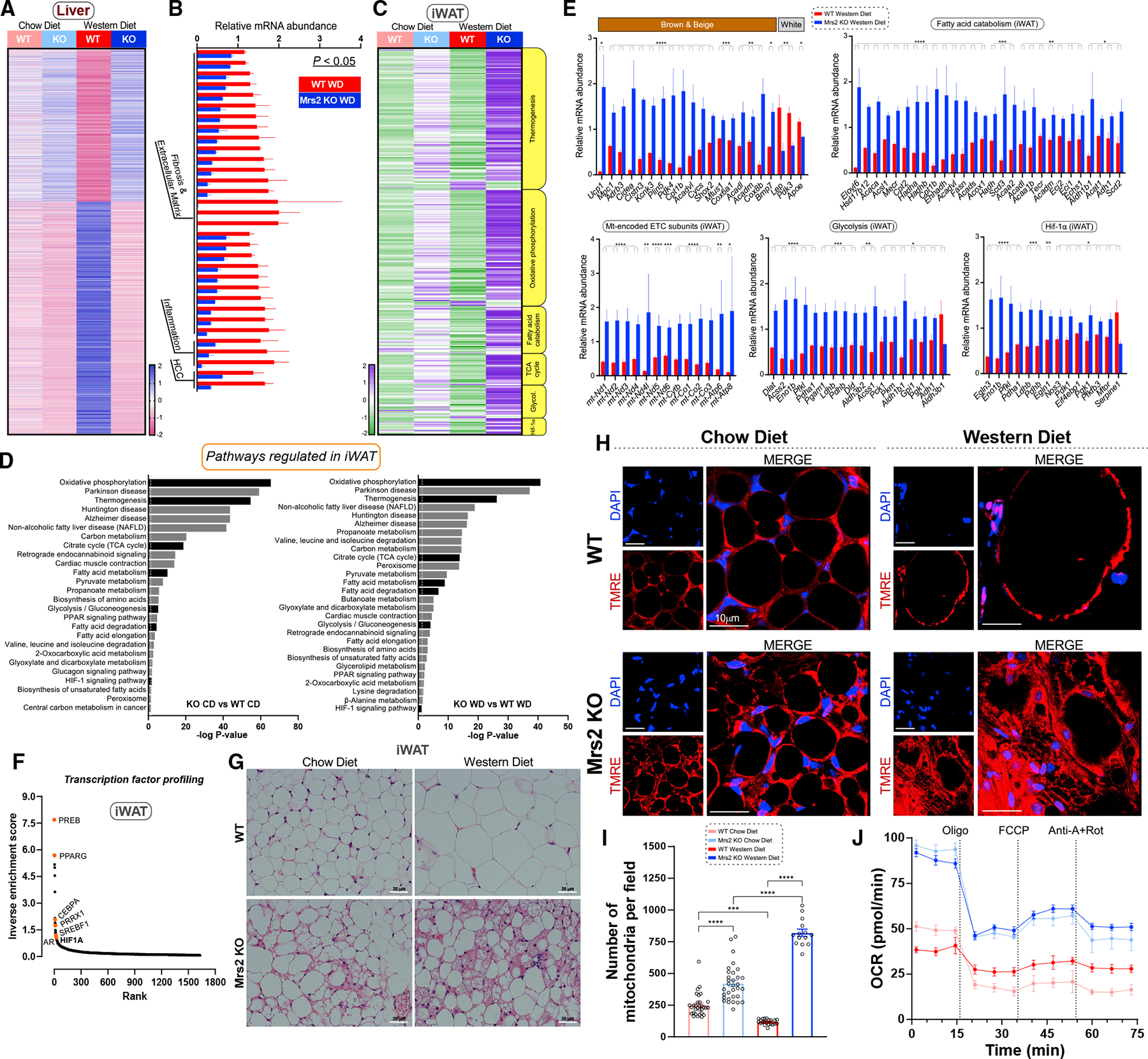
(A) Heatmap depicts differentially expressed genes (adj. p < 0.05) in liver tissue from CD- or WD-fed Mrs2−/− vs. WT mice. Values shown as row Z scores. n = 3 mice.
(B) Expression of key marker genes associated with fibrosis/extracellular matrix, inflammation, and HCC (liver; WD-fed mice). Significance was determined by an adjusted p-value of p < 0.05. The FPKM was normalized and expressed as relative mRNA abundance.
(C) Targeted KEGG analysis of iWAT from WT and Mrs2−/− mice fed CD or WD (see STAR Methods). Values shown as row Z scores. n = 3 mice/group.
(D) KEGG gene set enrichment analysis of iWAT with all significantly (adjusted p < 0.05) differentially regulated pathways shown in chow diet comparison (left) and Western diet comparison (right). Pathways of interest are indicated (black). The dotted line indicates significant threshold.
(E) Targeted differential enrichment analysis of adipose tissue from WD-fed WT and Mrs2−/−mice. Graphs indicate key adipose genes, mitochondrial encoded electron transport chain (ETC) subunits, and differentially regulated genes from KEGG pathways indicated as fatty acid catabolism, glycolysis, and HIF1-α signaling. All data are normalized like (B), n = 4 mice. p-values extracted from differential gene expression analysis. Data are mean ± SEM; ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05.
(F) Transcription factor analysis of WD iWAT using rank-based ChEA software. Select transcription factors highlighted. Y axis depicts mathematical inverse of the enrichment score 3 100.
(G) Representative H&E staining of adipose tissue from WT and Mrs2−/− either fed with CD or WD. n = 4 mice/group.
(H and I) Adipose tissue explants were stained with indicator (TMRE) and DAPI. Live images were acquired and mitochondrial density was quantified (I). n = 3 mice. Fourteen to 32 images per group were used for analysis.
(J) iWAT homogenate from WT and Mrs2−/− mice were subjected to OCR measurement. n = 3 mice.
We next explored changes in the transcriptional profiles of inguinal white adipose tissue (iWAT). Although gene expression profiles were changed considerably in this tissue, we focused on the major metabolic pathways and HIF1-α signaling (Figures 3C–3E). As in liver, the ChEA unbiased transcription factor analysis of iWAT revealed HIF1-α as a strong candidate for controlling the transcriptional program (Figure 3F).37 Since a massive elevation of thermogenic candidates was observed under CD as well as WD in the iWAT of Mrs2−/− mice, we performed histologic analysis that revealed a browning of iWAT in Mrs2−/− CD that was augmented in Mrs2−/− WD mice (Figure 3G). We next measured vascular density in iWAT and was found to be significantly increased in Mrs2−/− WD (Figure S3I). As expected, brown/beige markers, mt-encoded OXPHOS and ATP synthase subunits were significantly elevated in Mrs2−/− WD (Figure 3E). Similarly, the RNA-seq analyses showed upregulation of beige markers and fatty acid catabolism genes in Mrs2−/− WD mice (Figure 3E). Quantification of Ucp1 and Lep gene expression in iWAT tissue by RT-qPCR mirrored the RNA-seq findings (Figure S3J). Upon examination of the glycolytic pathway, HIF-1α controlled glycolytic transcripts were significantly upregulated in Mrs2−/− WD mice (Figure 3E). We further examined mitochondrial density and bioenergetics in iWAT explants. Consistently, WD-fed WT animals had hypertrophic, unilocular adipocytes with a minimal mitochondrial network; meanwhile, Mrs2−/− WD had smaller, multilocular, adipocytes with a heavy mitochondrial network (Figures 3H and 3I). The iWAT homogenate of Mrs2−/− had higher oxygen consumption rate (OCR) when compared with WT (Figures 3J and S3K). Overall, our RNA-seq data highlight a beiging effect and a potential contribution of HIF-1α-driven regulation of transcriptional programming in iWAT (Figures 3C–3F and S3I–S3K).38–41 Comparing our transcriptional data with Mg2+ binding proteins revealed substantial overlap, suggesting direct regulation and adaptive mechanisms in Mrs2−/− mice (Figures S3L and S3M).42
WD-induced suppression of mitochondrial fatty acid flux and Ca2+ uptake machinery is preventedin Mrs2−/− mice
To understand the molecular changes that facilitate glucose and lipid oxidation in Mrs2−/− mice in response to WD, we isolated hepatocytes from WT and Mrs2−/− mice fed control or WD for 52–54 weeks. Overnight, cultured hepatocytes were co-stained with the neutral lipid indicator BODIPY and ΔΨm TMRE to quantify lipid droplets and mitochondrial ratio. The WT WD hepatocytes exhibited more and larger lipid droplets when compared with other groups (Figures 4A–4C and S4A). A closer examination of the lipid droplets and mitochondria revealed a preservation of lipid droplet-mitochondria homeostasis in Mrs2−/− WD, but not in WT WD. Specifically, lipid droplets occupied less than 10% of the extra-nuclear region in CD groups and Mrs2−/− WD but increased to ~20% in WT WD (Figure 4B). Upon visualization of hepatocytes, we observed a unique mitochondrial BODIPY signal phenotype in Mrs2−/− fed WD (Figures 4C and S4A), reminiscent of fat-droplet associated mitochondria that have been reported recently.43 Colocalization analysis showed mitochondrial localization of BODIPY in the Mrs2−/− WD group (Figure 4D). Having observed mitochondrial BODIPY accumulation, the abundance of the mitochondrial fatty acid transporter carnitine palmitoyltransferase-1A was assessed in liver tissues. Remarkably, the CPT1A protein abundance was suppressed in WT WD, but its level was substantially maintained in Mrs2−/− suggesting higher b-oxidation to fuel bioenergetics (Figure 4E, upper panel).
Figure 4. Loss of Mrs2 enhances mitochondrial bioenergetics in hepatocytes from mice fed a Western diet.
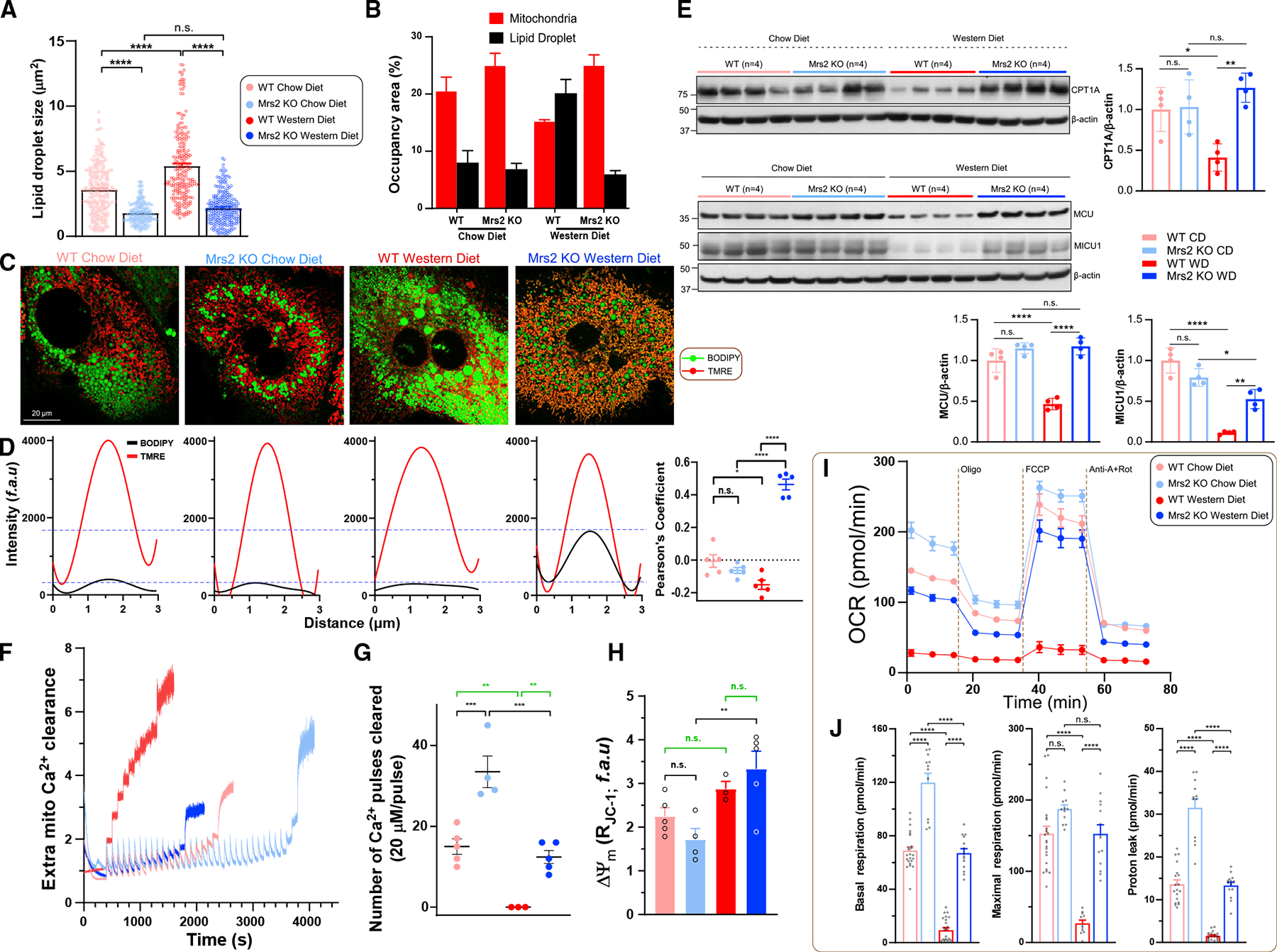
(A and B) Quantification of hepatocyte lipid droplet size and comparison of occupancy by mitochondria (red) and lipid droplet (black) areas in 1 year diet fed mice. n = 3–4 mice.
(C) Representative confocal images of hepatocytes were acquired after staining with lipid/fatty acid marker BODIPY-488 and indicator TMRE. n = 3 isolations.
(D) Spatial overlap and intensity profiles of mitochondrial colocalization of BODIPY and TMRE signals. n = 3 isolations.
(E) Western analysis of mitochondrial carnitine palmitoyltransferase 1A and MCU complex, MCU and MICU1 protein abundance in liver tissues harvested fromWT and Mrs2−/− mice. n = 4 mice.
(F–H) Assessment of MCU-mediated mCa2+ uptake (F), retention capacity (G), (H). n = 4 mice.
(I) OCR measurement of hepatocytes from WT and Mrs2−/− normalized to total protein content. n = 3 mice.
(J) Basal and maximal respiration and proton leak from (I). Data represent individual wells from three different hepatocyte isolations in each group. All data shown as mean ±SEM; ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, n.s. = not significant.
Since MCU-mediated mitochondrial Ca2+ uptake is essential for metabolic activities including β-oxidation, TCA cycle, and electron transport chain complex, MCU and MICU1 protein abundance was determined in liver.44 WT WD showed a striking low abundance of MCU and MICU1 while Mrs2−/− WD MCU complex abundance was comparable to WT or Mrs2−/− CD mice (Figure 4E, lower panel). To further establish that MCU-mediated Ca2+ uptake and mitochondrial Ca2+ handling is necessary for proper bioenergetics, MCU complex activity was measured in hepatocytes. Using a permeabilized hepatocyte model, we found that the mCa2+ uptake and Ca2+ retention capacity were increased in Mrs2−/− CD when compared with WT CD (Figures 4F and 4G). In addition, we found that WT WD exhibited complete abrogation of mCa2+ uptake but Mrs2−/− WD hepatocytes significantly retained these activities (Figures 4F and 4G). Despite the impaired handling of Ca2+ and fatty acids, was not compromised in WT WD (Figure 4H). Consistent with RNA-seq datasets, the WD regimen exhibited suppression of Tom20 and OXPHOS complex abundance, suggesting loss of mitochondrial density that was rescued in Mrs2−/− WD (Figure S4B). Based on these results, we hypothesized that the mitochondrial OCR had succumbed in WT WD hepatocytes. Overnight cultured hepatocytes from these cohorts were tested for basal respiration, maximal O2 consumption, and mitochondrial proton uncoupling using a Seahorse flux analyzer. Basal, uncoupler-induced maximal, and proton leak were significantly suppressed in WT WD hepatocytes as compared with the other three groups (Figures 4I and 4J). In addition, the extracellular acidification rate, an index of glycolysis, was partially maintained in Mrs2−/− WD as compared with WT WD hepatocytes (Figure S4C). These results indicate that WD-induced lipid accumulation in the liver could be due in part to decreased mitochondrial fatty acid utilization, consequent remodeling of hepatic nuclear encoded transcripts, and protein complex activities that are iMg2+-dependent.
Efflux of the DNL precursor and endogenous Mg2+ chelator citrate is blunted in Mrs2−/− mice
Citrate is the major precursor of de novo lipogenesis (DNL) in metazoans; we measured the plasma citrate levels from these four cohorts.45 Blood samples were drawn from these mice before harvesting organs for RNA-seq and histological studies. The plasma citrate concentration was not significantly affected by WD in WT mice; however, the plasma citrate was considerably lower in Mrs2−/− mice, regardless of diet (Figure S5A). To assess the contribution of liver in citrate efflux, extracellular citrate levels were measured in WT and Mrs2−/− hepatocytes. Extracellular citrate accumulation was blunted in Mrs2−/− hepatocyte culture media (Figure S5B). To complement Mrs2−/−, we used a dominant-negative-like mutant Mrs2 that lacks mMg2+ uptake.10 WT hepatocytes expressing mutant Mrs2 exhibited a lower citrate accumulation, indicating a tendency of low citrate efflux due to lowered [Mg2+]m (Figure S5C).10 The differences in citrate levels in plasma and media from in vitro cultured hepatocyte prompted us to examine mitochondrial citrate efflux, since the TCA cycle is the major source. To examine the mitochondrial citrate concentration ([mcitrate]), the newly created mitochondria-targeted genetically encoded citrate sensor mitoCitron was transiently constituted in HepG2 cells.46 Upon stimulation with high glucose (17 mM), the mitoCitron signal was rapidly increased, followed by a dissipation that corresponds to citrate production and subsequent utilization or efflux from the mitochondria (Figure S5D). We next expressed both cytosolic and mitochondria citrate sensors in WT and Mrs2−/− hepatocytes. Mitochondrial citrate production was higher in Mrs2−/− hepatocyte mitochondria; however, the appearance of cytosolic citrate in Mrs2−/− hepatocytes was greatly reduced following glucose stimulation (Figures S5E and S5F). In addition, we reconstituted the loss-of-function Mrs2 mutant in WT hepatocytes to measure the cytosolic and mitochondrial citrate concentration. Consistent with Mrs2−/−, the Mrs2 mutant expressing hepatocytes exhibited lower cytosolic citrate flux when compared with control (Figures S5G and S5H).10 Furthermore, we evaluated citrate dynamics in HepG2 cells and obtained data consistent with the primary hepatocytes (Figures S5I and S5J). Expression of Slc25a1 (the mitochondrial citrate carrier) was observed to be similar in cells from CD-fed animals and only slightly reduced in WD-fed conditions (Figure S5K). RNA-seq analysis of liver tissue from these animals correlated perfectly with the cellular data (Figure S5L). Thus, the decreased citrate efflux from Mrs2−/− mitochondria is not attributed to reduced Slc25a1 expression; these data support the model that mMg2+ could directly regulate SLC25A1-mediated citrate efflux.
HIF-1α is stabilized in Mrs2−/− mice
To examine the intrinsic role of Mrs2, we previously challenged Mrs2−/− mice with LPS to induce an inflammatory response. We noticed that there was a barely detectable HIF-1α stabilization in control but a significant HIF-1α protein accumulation following LPS stimulation in Mrs2−/− hepatocytes.10 Having observed the remarkable response against an inflammatory stimulus, we compared the HIF-1α stabilization between genotypes following stimulation with LPS or prolyl hydroxylase (PHD) inhibitor cobalt chloride (CoCl2). Hepatocytes derived from Mrs2−/− mice showed higher accumulation of HIF-1α (Figures 5A–5C). We envisioned that loss of Mrs2 might control metabolite fluxes that regulate HIF-1α stability under normoxic conditions. HIF-1α is the most commonly studied transcription factor that responds to oxygen tension and its stabilization controls numerous metabolic cascades, including glycolysis and oxidative phosphorylation.47–49 We conducted a targeted metabolite screen to identify whether these metabolites potentially altered in Mrs2−/− control HIF-1α stabilization. WT hepatocytes were treated with the PHD inhibitor FG-4952 for 6 h in the presence or absence of metabolites.50 Of the conditions tested, the presence of citric acid destabilized HIF-1α (Figure 5D) and its effect was observed at a concentration ≥6 mM (Figure 5E). In hepatocytes, CoCl2, FG-4592, and DMOG all promoted HIF-1α stabilization, with higher concentrations causing increased stabilization (Figures S6A–S6C). We next explored whether citric-acid-mediated HIF-1α destabilization is a hydroxylation-dependent mechanism by reconstitution of hydroxylation-dead mutant HIF-1α in COS-7 cells.51 As expected, 10 mM citric acid completely blunted FG-4592-dependent stabilization (Figure 5F, top panel). In contrast, constitutively stabilized HIF-1α mutant was unaffected by the presence of citric acid, suggesting that the hydroxylated HIF-1α is the target (Figure 5F, bottom panel). We next tested the potency of citric acid and sodium citrate derivatives. WT or Mrs2−/− hepatocytes were treated for 6 h. HIF-1α stabilization was less affected by di- or tri-sodium citrate than by citric acid (Figure 5G). This suggests that pH may play a role in the destabilization of HIF-1α. Nevertheless, lactic acid and citric acid alter intracellular pH but HIF-1α destabilization is citrate-dependent (Figures 5D and 5F). The citric-acid-mediated HIF-1α destabilization was observed across a range of inducers (Figure 5H) and the intracellular chelation of Mg2+ ions suppressed FG-4952-induced HIF-1α stabilization (Figure 5I). Furthermore, we showed that blockade of mitochondrial pyruvate entry with UK5099 (UK) did not prevent HIF-1α stabilization (Figure 5I). We evaluated the effect of higher levels of Mg2+ on citrate-mediated HIF-1α destabilization and found it to be weakly iMg2+ dependent; high levels of Mg2+ (10 mM) were sufficient to promote low levels of HIF-1α stabilization (Figure 5J, green highlight). To determine whether the citrate precursor oxaloacetate (OAA) or citrate product α-ketoglutarate (α-KG) exert similar functions in human kidney cell line (HK-2), cells were treated with OAA, pyruvate, citric acid, a-KG, succinate, and malate after six hours of CoCl2 exposure. Citric acid but no other metabolites exerted HIF-1α destabilization (Figures 5K and S6D). We next tested whether citric-acid-induced HIF-1α destabilization still occurs in hepatocytes pretreated with FG-4562. We found that the citric acid elicited HIF-1α destabilization, suggesting that the citrate effect is specific and robust (Figure S6E). To evaluate whether the citric-acid-mediated HIF-1α destabilization was at the post-translational level, WT hepatocyte homogenate was treated with FG-4562 for 6 h prior to citric acid exposure. Citric-acid-induced HIF-1α destabilization still occurs in hepatocyte homogenate, suggesting that its role is at the post-translational level (Figure S6F). Since citric acid destabilizes HIF-1α, we next asked FG-4592-mediated HIF-1α stabilization is transient in intact cells that was partly due to intracellular citrate accumulation. As predicted, we found that HIF-1α abundance decreased over time in hepatocytes treated with FG-4592 (100 mM) (Figure 5L, left panel) and correlates with citrate accumulation (Figure 5L, right panel). We next tested HIF-1α abundance in liver tissues harvested from the dietary cohorts. HIF-1α protein abundance was prominent in both Mrs2−/− CD and Mrs2−/− WD, whereas HIF-1α protein was nearly completely absent in WT WD (Figure 5M). Collectively, these results suggest that HIF-1α signaling is a key driver in glycolysis, OXPHOS, β-oxidation, and thermogenesis in Mrs2−/− mice (Figures 3, 4, and S3).
Figure 5. Loss of Mrs2 stabilizes and citric acid destabilizes HIF-1α.
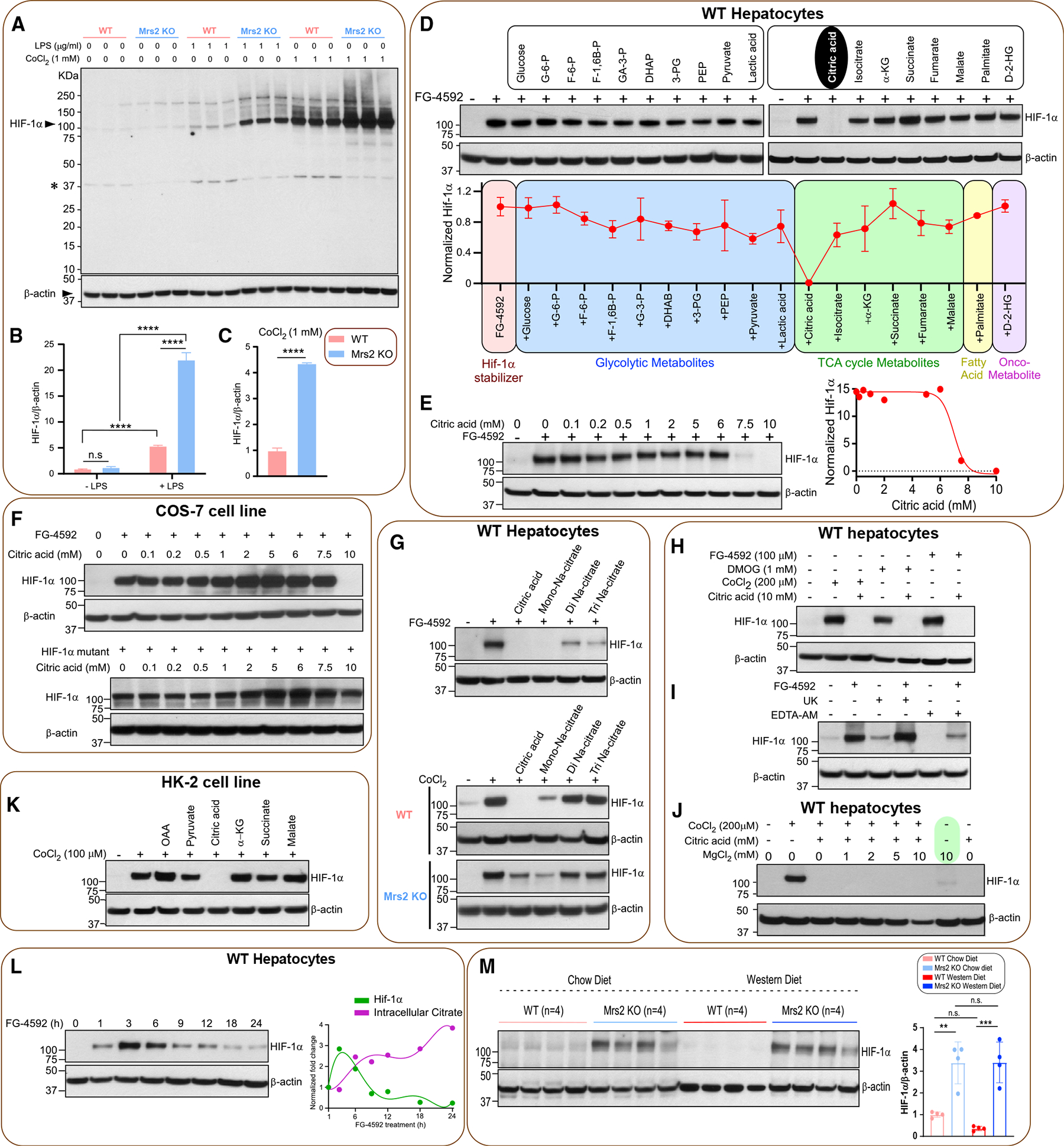
(A) Hepatocytes were treated with LPS or CoCl2 for 6 h. Total lysates were subjected to Western blot analysis to determine HIF-1α protein abundance. n = 3 isolations.
(B and C) ImageJ analysis of HIF-1α protein abundance.
(D) WT hepatocytes were treated with PHD inhibitor FG-4592 (100 mM) for 6 h with or without metabolites. Total cell lysates were probed for HIF-1α protein abundance (see STAR Methods) and quantified (bottom panel) (n = 2).
(E) Dose curve for citrate-induced HIF-1α destabilization. Right panel shows the normalized protein abundance (n = 2).
(F) Top panel shows citrated induced HIF-1α degradation in the presence of various doses of citrate in COS-7 cells (n = 2). Bottom panel depicts the effect of citrate on HIF-1α mutant protein stabilization (n = 2).
(G) Top panel shows the effect of citrate derivatives on FG4592-mediated HIF-1α stabilization (n = 2). Bottom panel shows the effect of citrate on CoCl2-mediated HIF-1α stabilization in WT or Mrs2−/− hepatocytes (n = 2).
(H) Effect of citrate on FG-4952, DMOG, or CoCl2-dependent HIF-1α stabilization in WT hepatocytes (n = 2).
(I) Effect of mitochondrial pyruvate transport blocker UK5099 or iMg2+ chelator EDTA-AM on HIF-1α stabilization (n = 2).
(J) Effect of MgCl2 supplementation on HIF-1α stabilization under normoxia (n = 2).
(K) Effect of TCA cycle citrate precursor OAA or a-KG on HIF-1α destabilization (n = 2).
(L) Intracellular accumulation of citrate elicits HIF-1α destabilization. Right panel depicts reciprocal action of citrate on FG-4592-dependent HIF-1α stabilization (n = 3).
(M) Western blot analysis of HIF-1α protein stabilization in liver tissues harvested from the 1-year diet period of WT and Mrs2−/− mice (n = 4/group). Densitometric analysis of HIF-1α protein from (M). All data shown as mean ± SEM; ****p < 0.0001, ***p < 0.001, **p < 0.01, n.s. = not significant.
Bacterial CorAblocker selectively inhibits Mrs2 channel activity
Bacterial CorA forms a pentameric complex that selectively drives Mg2+ uptake in an electrogenic manner. CorA is highly selectively inhibited by cobalt and ruthenium derivatives (i.e., cobalt(III)hexaammine, ruthenium(III)hexaammine, chloropentaammine cobalt(III)chloride [CPACC; half maximal inhibitory concentration (IC50) ~100 μM], chloropentaammine ruthenium(III)chloride) with an IC50 ~3–100 μM (Figure 6A).52 Mrs2 is the eukaryotic homologue of CorA and contains two transmembrane domains with a well-conserved F/Y-G-M-N motif; therefore, we tested whether CorA blockers alter Mrs2-mediated mMg2+ uptake.18 Digitonin-permeabilized hepatocytes were used to measure the inhibitory effect and determine its IC50. Permeabilized cells were treated with compound followed by a 1-mM bolus of MgCl2. A 1-mM Mg2+ bolus was added after baseline recording, and the extramitochondrial Mg2+ clearance was used as a readout for Mrs2-mediated mMg2+ uptake using Mag-Green as a Mg2+ indicator (Figures S7A–S7D).10 Of all four compounds tested, ruthenium derivatives did not exhibit any inhibitory effect on Mrs2 activity (Figure S7A). Remarkably, CPACC exerted complete inhibition of mMg2+ uptake at 10 mM (Figures 6B, S7B and S7C). Inhibition by cobalt(III)hexaammine in intact hepatocytes was not observed (Figure S7D). We found CPACC (IC50 = 0.175 μM) to be over 500 times more effective at inhibiting mMg2+ in permeabilized hepatocytes in comparison with bacterial CorA (Figure 6B).52 We next investigated Mrs2 inhibition by CPACC in intact hepatocytes loaded with MagGreen-AM. Mitochondrial Mg2+ uptake was measured by confocal microscopy following L-lactate stimulation and was significantly inhibited in a dose-dependent manner (Figures 6C and 6D). We next examined whether CPACC had any effect on MCU-mediated mCa2+ uptake in permeabilized cells. Importantly, CPACC treatment did not inhibit mCa2+ uptake, revealing the specificity of CPACC on Mrs2 (Figure 6E). The availability of the compound to intact cells was measured using graphite furnace atomic absorption spectroscopy (GFAAS) and bicinchoninic acid in parallel. It is important to note that CPACC accumulates in the cell in a dose-dependent manner (Figure S7E). Encouraged by the potent inhibitory effect of CPACC and cellular permeability, we next measured the cytotoxicity of CPACC in cells. CPACC was effectively non-toxic in HeLa or HEK293T cells up to 2 mM and 500 μM, respectively (Figure 6F).
Figure 6. CPACC selectively inhibits Mrs2 activity.
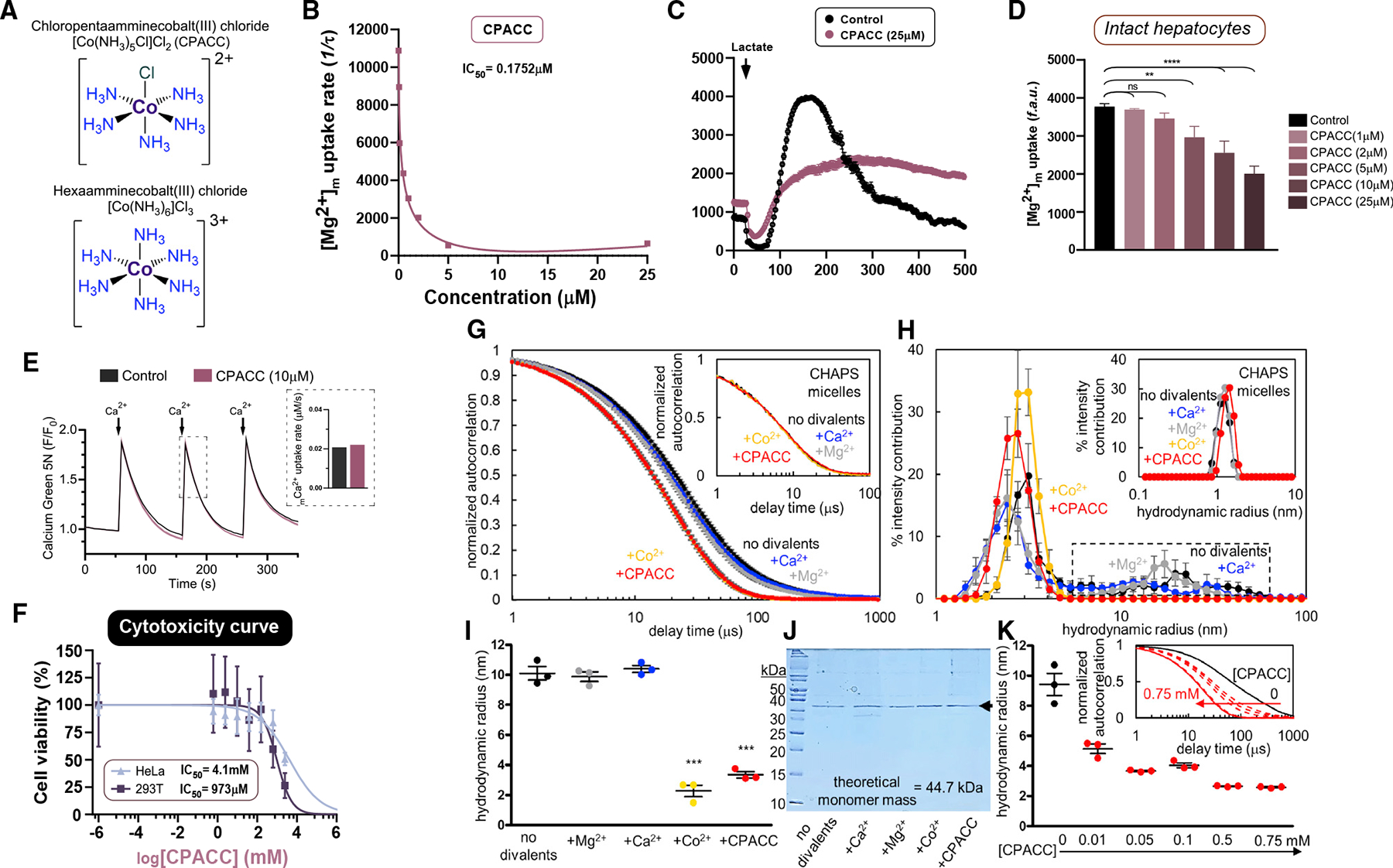
(A) Chemical structures of chloropentaammine cobalt(III) chloride [Co(NH3)5Cl]Cl2 (CPACC) and hexaamminecobalt(III) chloride [Co(NH3)6]Cl3.
(B) Dose-response curve of permeabilized hepatocytes pulsed with 1 mM MgCl2 with increasing concentrations of CPACC. n = 3.
(C and D) Effect of CPACC on Mrs2-mediated mMg2+ uptake in intact hepatocytes (C) and its quantification (D). n = 3–4. ****p < 0.0001, **p < 0.01, n.s. = not significant.
(E) Spectrofluorimetric measurement of MCU-mediated Ca2+ uptake in control (n = 1) or 10 μM CPACC (n = 3) treated HeLa cells. Quantification of mCa2+ uptake rate (inset).
(F) Cytotoxicity dose-response curves for CPACC in HeLa and HEK293T cells. n = 3 (replicates; n = 6).
(G) Autocorrelation functions of human MRS258–443 (0.5 mg/mL) with and without 5 mM MgCl2, CaCl2 and CoCl2 and 0.5 mM CPACC. Inset shows micelles without protein.
(H) Hydrodynamic radii distribution derived from (G). Dashed box highlights the disappearance of higher order oligomers only in the presence of CoCl2 and CPACC. Inset shows micelles without protein.
(I) Weight-averaged hydrodynamic radii based on cumulants deconvolution of autocorrelation functions shown in (G).
(J) Coomassie blue-stained SDS-PAGE gel (15% [w/v]) confirming the divalent cations and CPACC do not affect MRS258–443 integrity.
(K) Cumulants-determined weight-averaged hydrodynamic radii of MRS258–443 (0.5 mg/mL) as a function of increasing CPACC, highlighting the concentration-dependent decrease in particle size. Inset shows a systematic shift to earlier decay times with increasing CPACC levels. Mean ± SEM; n = 3. ***p < 0.001 for + Co2+ and +CPACC compared with no divalent cation, +Mg2+ and +Ca2+ groups.
Next, we assessed whether CPACC directly affects the assembly of purified human MRS2 using dynamic light scattering (DLS). DLS has been used to determine whether the complex stability of proteins/channels is altered by molecular chaperones or drugs.53 Because of the high sensitivity of DLS to changes in complex sizes, we evaluated full-length Mrs2 assembly in the presence of Ca2+ (5 mM), Mg2+ (5 mM), Co2+ (5 mM) or the MRS2 blocker CPACC (0.5 mM) (Figures 6G–6K). Remarkably, we found that Co2+ and the Co2+ derivative CPACC, but not Mg2+ or Ca2+, potently decreased the full-length MRS2 complex sizes as determined from the earlier decaying autocorrelation functions, without affecting protein integrity (Figures 6G–6K). A concentration-response assessment confirmed robust decreases in hydrodynamic radii at all CPACC concentrations tested and as low as 10 mM (Figure 6K). Together, these data indicate that CPACC directly binds to Mrs2 and inhibits the activity.
CPACC treatment reduces lipid droplet size and promotes adipose browning, thus lowering whole body weight
Given the potent inhibition of mMg2+ uptake and low toxicity of CPACC, we hypothesized that CPACC could mimic the Mrs2−/− cellular phenotype. WT hepatocytes isolated from over 12-month-old mice were treated with 50 μM CPACC for 48 h. Control and CPACC-treated hepatocytes were stained with BODIPY and TMRE for intracellular tracing using confocal imaging. CPACC-treated hepatocytes displayed a marked reduction of lipid droplet size and appeared to dissipate or smear (Figures 7A, 7B, S7F, and S7G). Importantly, no negative effect on mitochondrial membrane potential was observed (Figures 7C and S7F). We next asked if CPACC treatment affects the mitochondrial OCR. Freshly isolated WT hepatocytes were treated with 5–25 μM concentrations of CPACC for 16 h before measurement of OCR. CPACC increased basal mitochondrial respiration without a significant effect on maximal OCR (Figures 7D and S7H). Thus, decreased hepatic lipid droplet size by blockade of Mrs2 may result in part from enhanced glucose and fatty acid oxidation, although our data do not preclude the possibility of CPACC-dependent partial stabilization of HIF-1α driving glycolysis and thermogenesis (Figures 3 and S3). Since we observed an enhancement of basal OCR in CPACC-treated hepatocytes, we also measured the extracellular acidification rate in control and CPACC-treated hepatocytes and found it to be higher in the CPACC condition, indicating higher glycolytic activity (Figure S7I). Based on the prominent browning/beiging in Mrs2−/− iWAT, we examined the expression of beige markers, Ucp1 and Cidea, and white adipose marker, Lep, in the iWAT of the CPACC-treated HFD-fed WT mice. Twelve-week HFD-fed WT mice were treated with CPACC (20 mg/kg) or vehicle via intraperitoneal injection every 3 days over 6 weeks. Our results demonstrate a transition from WAT to beige adipose (Figure 7E). Similar to Mrs2−/− mice, the RT-qPCR analysis revealed a glycolytic transcriptional signature of HIF-1α activation in iWAT of CPAAC-treated HFD mice (Figure 7E). After demonstrating that pharmacologic blockade of Mrs2 by CPACC reduced lipid droplet size and enhanced mitochondrial function, we asked whether in vivo administration of CPACC prevents weight gain. CPACC administration restricted body weight gain (Figures 7F and 7G). Plasma ALT levels from CPACC-treated mice were lower than those in the control group, suggesting improved liver function (Figure 7H). Since Mrs2−/− mice have lower levels of plasma citrate, we asked if CPACC treatment lowers plasma citrate in HFD mice. Indeed, CPACC-treated mice had reduced plasma citrate levels (Figure 7I). Together, these data suggest that limiting mMg2+ promotes oxidative metabolism in multiple tissues.
Figure 7. Limiting Mrs2-mediated Mg2+ uptake by CPACC reduces lipid droplet size and promotes mitochondrial function, thus preventing weight gain in vivo.
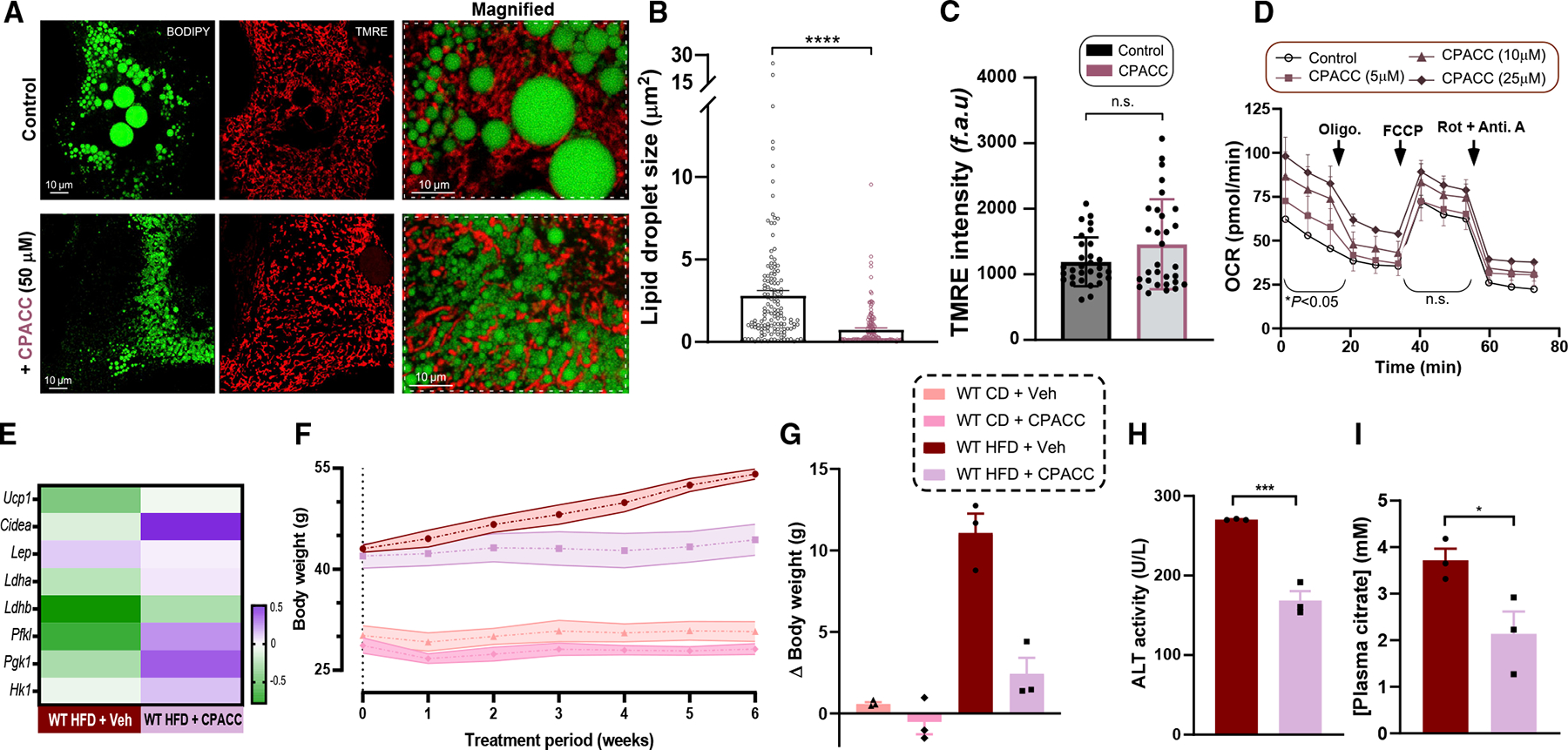
(A) Visualization of hepatocytes isolated from 12-month-old WT mice using BODIPY-488 (left) and TMRE (middle), with or without CPACC (50 μM) treatment for 48 h.
(B and C) Quantification of lipid droplet size and from (A). n = 3–4.
(D) OCR rate in WT hepatocytes with or without CPACC treatment. n = 3.
(E) Heatmap of normalized fold change of mRNA in iWAT from HFD-fed WT with or without intraperitoneal (i.p.) CPACC treatment. n = 3 mice.
(F) WT mice were fed HFD or CD for 14 weeks followed by CPACC (20 mg/kg) or vehicle for additional 6 weeks i.p. every 3 days. The body weight was measured weekly. n = 3 mice.
(G) Body weight change during the 6-week treatment period.
(H and I) Plasma ALT (H) and citrate (I) levels of HFD-fed WT with or without CPACC treatment. n = 3 mice. Data shown as mean ± SEM; ****p < 0.0001, ***p < 0.001, *p < 0.05, n.s. = not significant.
DISCUSSION
Four major cations, Na+, K+, Mg2+, and Ca2+, control numerous cellular functions including membrane polarization, bioenergetics, metabolism, transcription, and proliferation.3,7,8 Upon cellular stimulation, highly compartmentalized Ca2+ ions rapidly mobilize to initiate “on” reactions. In contrast, the antagonistic cellular Mg2+ dynamics carry out “off” reactions, in some cases through direct competitive binding.14,54–56 Although this concept was proposed several decades ago, the control and physiological consequences of iMg2+ dynamics remain poorly understood. In addition, cellular Mg2+ is an intrinsic component of numerous metabolic enzymes, phosphometabolites, and metabolic precursors such as citrate. For instance, GPCR, nicotinic, and NMDA receptors, L-typeCa2+ channels, ryanodine receptors, and ion pumps activity were modulated by Mg2+ and aberrations contribute to several prevalent diseases such as hypertension, heart disease, diabetes, ischemia/reperfusion injury, preeclampsia, cancer, chronic obstructive pulmonary disease, asthma, neurological diseases, Parkinson, and Alzheimer’s diseases.3,57,58
Recently, it was shown that the glycolytic end-product L-lactate acts as an activator of ER Mg2+ release, and the elevated free iMg2+ was subsequently taken up by mitochondria through a highly conserved Mg2+ selective transport machinery, Mrs2.10 However, the physiological function of MRS2-mediated Mg2+ uptake and its effect on bioenergetics and whole-body metabolic consequences thereof have not been determined. To decipher the contribution of mMg2+ in physiology and disease contexts, we used a mouse model of long-term WD-induced metabolic disease progression. Our study reveals striking protection from the metabolic consequences of a diet that is high in fat and sugar. To identify pathways that facilitate diet-induced metabolic disorders, we fed WD to control and global Mrs2−/− mice for more than 52 weeks. Our findings reveal both qualitatively and quantitatively different mechanisms in diet-regulated metabolic syndrome that are Mg2+ dependent. Upon WD consumption, Mrs2−/− mice exhibited no body weight gain, near-complete ablation of energy storage expansion, prevention of spontaneous HCC tumor incidence, and retention of microvascular density with normal liver function (Figures 1 and 2). Remarkably, Mrs2−/− mice maintained normal circadian oscillations of respiration, whereas this pattern is disrupted in WT WD mice. Having observed such a significant physiological outcome, we tested mechanisms that might contribute to protection from WD, including thermogenic activation, increased glucose and fatty oxidation, and oxidative phosphorylation. Our histological, biochemical, and global transcriptomic regulation analyses fully support the absence of metabolic syndrome phenotype in Mrs2−/− mice.
We have also found a notable reprograming of adipose transcriptional profiles that favors thermogenesis, glycolysis, fatty acid oxidation, and oxidative phosphorylation in Mrs2−/−, and that transcriptional reprograming was heightened under WD regimen (Figures 3C–3E). Our data indicate that a retrograde signal that emanates from mitochondria controls transcriptional profiles in an Mg2+-dependent manner. We found a marked suppression of mitochondrial citrate efflux and stabilization of HIF-1α in Mrs2−/− mice. Since differences in citrate efflux from Mrs2−/− mitochondria are not attributed to Slc25a1 abundance, at least at the transcript level, we speculate that mMg2+ could regulate SLC25A1 activity directly. In support of the model that reduced citrate efflux could have beneficial metabolic effects, the genetic ablation of Slc25a1 prevented obesity and NAFLD implicating cytosolic efflux of citrate in metabolic syndrome progression.59 In addition, these findings shed light on how intracellular divalent cation dynamics control diet-induced metabolic dysregulation and disease progression. HIF-1α may be a key mediator of altered iMg2+ dynamics in multiple tissues and may have additional benefits beyond those we have measured. For instance, it has been shown that aerobic or ischemic-mediated HIF-1α stabilization exerts protection against coronary artery disease.40 Aerobic stabilization of HIF-1α transcriptionally controls several monovalent and divalent cation channels, and mitochondria localized SOD2 that prevents oxidative stress and hypertension implying a potentially profound influence of mMg2+ and HIF-1α-dependent signaling across cellular metabolism.40
Our findings reveal that blockade of mMg2+ uptake by both genetic and pharmacologic interventions diminishes weight gain and hepatic lipid accumulation (Figures 4 and 7) while inducing a more thermogenic transcriptional profile in adipocytes (Figures 3 and 7). We find that a cobalt derivative (CPACC) inhibits mMg2+ uptake at low micromolar ranges in hepatocytes and phenocopies effects of Mrs2 deletion, including protection from weight gain in vivo and reducing hepatocyte lipid accumulation ex vivo. Since CorA channel interacts with the fully hydrated Mg2+ ion, it is hypothesized that amine groups of the divalent and trivalent cations mimic the size and shape of a hydrated Mg2+ cation that exerts the inhibition of Mg2+ ion permeation.14,18 Indeed, our studies demonstrate that CPACC alters the physical structure of Mrs2 to restrict the flux of Mg2+ (Figure 7). This proof-of-concept approach will aid in designing Mg2+ channel modulators that buffer WD-induced adipose expansion and prevent metabolic dysregulation. Taken together, our findings reveal a mechanistic link between iMg2+ dynamics and whole-body energy metabolism in mammals.
Limitations of the study
Although our study provides a comprehensive understanding of the role of the highly conserved, Mg2+ selective mitochondrial channel MRS2 in bioenergetics and whole-body metabolism, several limitations should be highlighted. We performed prolonged dietary stress to recapitulate human metabolic syndrome; however, acute dietary stress may have less chance for indirect effects and thus help clarify which consequences of Mrs2 ablation are primary. We recognize the limitation of using global knockout of Mrs2, which does not allow examination of individual tissue’s role in controlling metabolism. MRS2 channel is ubiquitously expressed and its impact on other organs, including brain, cardiac, kidney, lung, and skeletal muscle need a thorough investigation. Third, the reciprocity of cellular and organellar divalent cation (Ca2+ and Mg2+) dynamics and its physiological consequences remain to be fully established. Our study identified an inorganic-based blocker of MRS2 and its utility in vivo but future studies warrant developing a small molecule modulator of MRS2 that can be used in human studies. Nevertheless, our study unveils the link between Mg2+ channel and prolonged dietary-stress-induced obesity and metabolic diseases.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Madesh Muniswamy (muniswamy@uthscsa.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed materials transfer agreement.
Data and code availability
RNA-seq data of liver and iWAT have been deposited in the NCBI Gene Expression Omnibus (GEO)1 and are publicly available as of the data of publication. The accession number is listed in the key resources table. Additional information regarding RNA-seq data and analysis can be found in Data S1. All other data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Total OXPHOS Rodent WB Antibody Cocktail | Abcam | Cat# ab110413 |
| CPT-1a | Abcam | Cat# 234111 |
| CPT-2 | Abcam | Cat# 181114 |
| HIF-1α | Cell Signaling, MA, USA | Cat# 14179S |
| Hydroxy HIF-1α | Cell Signaling | Cat# 3434S |
| MCU | Cell Signaling | Cat #D2Z3B |
| MICU1 | Cell Signaling | Cat# D4P8Q |
| β-Actin | Santa Cruz Biotechnology | Cat# SC47778 |
| α-Mouse Secondary (HRP-conjugated) | Cell Signaling | Cat# 5127S |
| α-Rabbit Secondary (HRP-conjugated) | Cell Signaling | Cat# 911968 |
| Goat Anti-Mouse CD-31/PECAM-1 (unconjugated) | R&D Systems Inc., MN, USA | Cat# AF3628 |
| Horse Anti-Goat IgG (H + L) Biotin Conjugated RTU Secondary |
Vector Laboratories, CA, USA | BP-9500-50 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Ad-mRFP | Vector Biolabs, PA, USA | Cat# 1660 |
| Ad-Mrs2(mut).mRFP.FLAG [Mutant Mrs2] | Vector Biolabs | N/A |
| BL21-CodonPlus Competent Cells (Escherichia coli) | Agilent Technologies, CA, USA | Cat# 230280 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Western Diet (WD) | Research Diets | Cat# D09100310 |
| Standard Chow Diet (CD) | Envigo | Cat# 7012 |
| High Fat Diet (HFD) | Research Diets | Cat# D12492 |
| Human MRS2 (MRS258–443) | This Study and Daw et al.10 | NCBI acc. NP_065713. |
| HisPurTM Ni-NTA Resin | ThermoFisher Scientific | Cat# 88223 |
| Ndel Restriction Enzyme | New England Biolabs | Cat# R0111S |
| Xhol Restriction Enyme | New England Biolabs | Cat# R0146S |
| HaltTM Protease and Phosphatase Inhibitor Cocktail | ThermoFisher Scientific | Cat# 78441 |
| RestoreTM PLUS Western Blot Stripping Buffer | ThermoFisher Scientific | Cat# 46430 |
| NuPAGETM MOPS SDS Running Buffer (20X) | ThermoFisher Scientific | Cat# NP0001 |
| SuperSignalTM West Pico PLUS Chemiluminescent Substrate | ThermoFisher Scientific | Cat# 34578 |
| Bis-Tris Polyacrylamide Gel (4%–12%) | ThermoFisher Scientific | Cat# WG1402BOX |
| TaqManTM Fast Advanced Master Mix | ThermoFisher Scientific | Cat# 4444557 |
| SuperScipt® VILOTM | ThermoFisher Scientific | Cat# 11754-050 |
| Lipopolysaccharide (LPS) (E. coli 0111:B4) | Sigma Aldrich | Cat# F3665 |
| FG-4592 (Roxadustat) | Cayman Chemical, MI, USA | Cat# 15294 |
| DMOG | Sigma Aldrich | Cat# D3695-10MG |
| Cobalt(II) Chloride (CoCl2) | Fluka (Sigma Aldrich) | Cat# 60818 |
| Glucose | Sigma Aldrich | Cat# G8644 |
| Glucose-6-Phosphate | Sigma Aldrich | Cat# G7879 |
| Fructose-6-Phosphate | Sigma Aldrich | Cat# F3627 |
| Fructose-1,6-Bisphosphate | Sigma Aldrich | Cat# F6803 |
| Glyceraldehyde-3-Phosphate | Sigma Aldrich | Cat# G5251 |
| Dihydroxyacetone Phosphate | Sigma Aldrich | Cat# D7137 |
| 3-Phosphoglyceic Acid | Sigma Aldrich | Cat# P8877 |
| Phospho(enol)pyruvic Acid | Sigma Aldrich | Cat# P7127 |
| Pyruvate | Sigma Aldrich | Cat# P2256 |
| Lactic Acid | Sigma Aldrich | Cat# L1750 |
| Citric Acid | Sigma Aldrich | Cat# C1909 |
| Isocitric Acid | ThermoFisher | Cat# 205010250 |
| α-Ketoglutarate | Sigma Aldrich | Cat# K2000 |
| Succinate | Sigma Aldrich | Cat# S2378 |
| Fumarate | Chem Cruz Biochemicals (Santa Cruz) | Cat# SC-215873 |
| Malate | Sigma Aldrich | Cat# 1613881 |
| Palmitate | Sigma Aldrich | Cat# P9767 |
| D-2-hydroxyglutarate (D-2-HG) | Toronto Research Chemicals | Cat# H942595 |
| Oxaloacetic Acid (OAA) | Sigma Aldrich | Cat# O7753 |
| Mono Sodium Citrate | Sigma Aldrich | Cat# 71497 |
| Di Sodium Citrate | Sigma Aldrich | Cat# 71635 |
| Tri Sodium Citrate | Sigma Aldrich | Cat# 54641 |
| EDTA-AM | AAT Bioquest, CA, USA | Cat# 19010 |
| UK5099 (Mitochondrial Pyruvate Carrier Inhibitor) | CalbioChem (MilliPore Sigma) | Cat# 5048170001 |
| Magnesium Chloride (MgCl2) | Sigma Aldrich | Cat# M1028 |
| Chloropentaammine Cobalt(III) Chloride (CPACC) | This Study | N/A |
| Hexaammine Cobalt(II) Chloride | This Study | N/A |
| Chloropentaammine Ruthenium(III) Chloride | This Study | N/A |
| Hexaammine Ruthenium(III) Chloride | This Study | N/A |
| Magnesium GreenTM Pentapotassium Salt | ThermoFisher Scientific | Cat# M3733 |
| Magnesium GreenTM, AM | ThermoFisher Scientific | Cat# M3735 |
| MitotrackerTM Deep Red FM | ThermoFisher Scientific | Cat# M22426 |
| Tetramethylrhodamine, Ethyl Ester, Perchlorate (TMRE) | ThermoFisher Scientific | Cat# T669 |
| BODIPYTM 493/503 | ThermoFisher Scientific | Cat# D3922 |
| FURA-FF Pentapotassium Salt | Cayman Chemical | Cat# 20415 |
| Calcium Green 5N | ThermoFisher Scientific | Cat# C3737 |
| JC-1 | Sigma Aldrich | Cat# 7176 |
| Digitonin | Sigma Aldrich | Cat# D141 |
| ATP Dipotassium Salt Hydrate | Sigma Aldrich | Cat# A8937 |
| ATP Magnesium Salt | Sigma Aldrich | Cat# A9187 |
| Calcium chloride | Sigma Aldrich | Cat# 2115 |
| Thapsigargin | Thermo Fisher Scientific | Cat# T7458 |
| Seahorse XF Calibrant Solution | Seahorse Bioscience (Agilent) | Cat# 100840-000 |
| Glucose | Sigma-Aldrich | Cat# G8644 |
| L-Glutamine, 200mM | Gibco (ThermoFisher Scientific) | Cat# 250030 |
| Sodium Pyruvate, 100mM | Gibco (ThermoFisher Scientific) | Cat# 11360 |
| HEPES (1M) | Gibco (ThermoFisher Scientific) | Cat# 15630 |
| Rotenone | Sigma-Aldrich | Cat# 557368 |
| Antimycin A | Sigma Aldrich | Cat# A8674 |
| Oligomycin | Sigma Aldrich | Cat# 75351 |
| FCCP | Sigma Aldrich | Cat# C2920 |
| Normal Horse Serum | Vector Laboratories | Cat# S-2000-20 |
| Fetal Bovine Serum, Characterized, Heat Inactivated | Hyclone (GE Life Sciences) UT, USA | Cat# SH30396.03 |
| Antibiotic-Antimycotic, 100X | Gibco (ThermoFisher Scientific) | Cat# 15240 |
| Saline (0.9% Sodium Chloride Injection USP) | Braun, PA, USA | Cat# L8002 |
| EGTA, OmniPur (powder) | MilliporeSigma | Cat# 4100-50GM |
| Collagenase (Type 4) | Worthington Biochemical Corp., NJ, USA | Cat# LS004188 |
|
| ||
| Critical commercial assays | ||
|
| ||
| PierceTM BCA Protein Assay Kit | ThermoFisher Scientific | Cat# 23225 |
| Citrate Assay Kit | Abcam | Cat# ab83396 |
| Alanine Transaminase Kit | Abcam | Cat# ab105134 |
| Trichloroacetic Acid Precipitation Kit | Abcam | Cat# ab204708 |
| RNeasy Plus Universal Mini Kit | QIAGEN, MD, USA | Cat# 73404 |
| Agilent Seahorse XF Cell Mito Stress Test Kit | Agilent Technologies, CA, USA | Cat# 103015-100 |
| Agilent Seahorse XFe96 FluxPak | Agilent Technologies | Cat# 102416-100 |
| VECTASTAIN Elite ABC-HRP Kit | Vector Laboratories | Cat# PK-6100 |
| Avidin/Biotin Blocking Kit | Vector Laboratories | Cat# SP02001 |
| LiquiColor Enzymatic Cholesterol Assay Kit | Stanbio, TX, USA | Cat# 1010225 |
|
| ||
| Deposited data | ||
|
| ||
| RNA-seq of liver and iWAT | This study | GEO: GSE216538 |
| from WT and Mrs2−/− mice | ||
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Human: HEK 293 Cell Line | ATCC | Cat# CRL-1573 |
| Human: HeLa Cell Line | ATCC | Cat# CCL-2 |
| Human: HepG2 Cell Line | ATCC | Cat# HB-8065 |
| Human: HK-2 Cell Line | ATCC | Cat# CRL-2190 |
| Cercopithecus aethiops: | ATCC | Cat# CRL-1651 |
| COS-7 Cell Line | ||
| Mouse: Primary Hepatocytes | This Study | N/A |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| C57BL/6J Mice | The Jackson Laboratory, USA | C57BL/6J |
| Mrs2 −/− (Mrs2 knockout) Mice | Daw et al.10 | Mrs2 KO |
|
| ||
| Oligonucleotides | ||
|
| ||
| Ucp1 (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm01244861_m1 |
| Cidea (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm00432554_m1 |
| Lep (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm00434759_m1 |
| Ldha (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm01612132_g1 |
| Ldhb (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm05874166_g1 |
| Pfkl (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm00435605_mH |
| Pgk1 (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm00435617_m1 |
| Hk1 (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm00439335_m1 |
| Actb (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm01205647_g1 |
|
| ||
| Recombinant DNA | ||
|
| ||
| pCMV Citron (Cyto) | Addgene (Zhao et al.46) | Cat# 134303 |
| pCMV MitoCitron | Addgene (Zhao et al.46) | Cat# 134305 |
| pET-28a (+) | Novagen (Millipore Sigma) | Cat# 69864-3 |
| HA-HIF1α P402A/P564A/N803A-pcDNA3 | Addgene (Yan et al., 201760) | Cat# 87261 |
|
| ||
| Software and algorithms | ||
|
| ||
| Canvas 11.0 | ACD Systems | https://www.canvasgfx.com/en/support/canvas-11/ |
| GraphPad Prism version 8 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Leica Application Suite X (LAS X) | Leica Microsystems Inc., IL, USA | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| ImageJ | Schneider et al., 202161 | https://imagej.nih.gov/ij/ |
| Agilent Seahorse Wave Desktop Software (v2.6.1) | Agilent Technologies, CA, USA | https://www.agilent.com/en/products/cell-analysis/software-download-for-wave-desktop |
| Felix GX Software (v4) | PTI - Horiba, Canada | https://www.horiba.com/int/products/detail/action/show/Product/pti-felixgx-1652/ |
| Promethion Software | Sable Systems NV, USA | https://www.sablesys.com/products/promethion-line/promethion-software/ |
| R (v4.0.4) | R Core Team | https://www.R-project.org/ |
| Hisat2 (v2.0.5) | Kim et al.62; Kim et al.63; Pertea et al.64; Zhang et al.65 | http://daehwankimlab.github.io/hisat2/ |
| FeatureCounts (v1.5.0-p3) | Liao et al.66 | http://www.bioconductor.org/packages/release/bioc/html/Rsubread.html |
| DESeq2 (v1.20.0) | Love et al.67 | https://github.com/mikelove/DESeq2 |
| GSEA (Gene Set Enrichment Analysis) Tool (v4.2.2) | Subramanian et al.68; Mootha et al.69 | https://www.gsea-msigdb.org/gsea/index.jsp |
| ClusterProfile R Tool (v3.14) | Yu et al., 201270 | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| KEGG (Kyoto Encyclopedia of Genes and Genome) | Kanehisa and Goto34 | https://www.genome.jp/kegg |
| Dynamics Software (v7.8.1.3) | Wyatt Technology Corp., CA, USA | https://www.wyatt.com/products/software/dynamics.html |
| ChEA (ChIP-X Enrichment Analysis) software | Lachmann et al.37 | https://maayanlab.doud/Harmonizome/resource/ChIP-X+Enrichment+Analysis |
| BioVenn | Hulsen et al.71 | https://www.biovenn.nl/ |
| DeepVenn | Hulsen72 | https://www.deepvenn.com |
| SankeyMATIC Plot Generator | Steve Bogart (Open-Source Freeware) | https://sankeymatic.com |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
HEK293 (ATCC CRL-1573), HeLa (ATCC CCL-2), and COS-7 (ATCC CRL-1651) cells were grown in high glucose complete growth medium (high glucose Dulbecco’s modified Eagle’s medium, HG-DMEM). HepG2 (ATCC HB-8065, male) cells were grown in normal glucose complete growth medium (Dulbecco’s modified Eagle’s medium, DMEM) These cell lines were supplemented with 10% (v/v) FBS, 1% (v/v) antimycotic-antibacterial cocktail (Gibco). HK-2 (ATCC CRL2190) cells were maintained in specialized serum-free Keratinocyte-SFM media supplemented with required EGF and BGE (Gibco). All cells kept in a 37°C, 5% CO2 humidified incubator. Cells lines were detached using Trypsin-EDTA 0.05%, (HEK293 and HK-2) or 0.25% (COS-7, HeLa, HepG2). All transfected cells were grown in corresponding complete growth media supplemented with puromycin (2 μg/mL) or G418 (500 μg/mL). Cell line’s sex are both female (HEK293 and HeLa) and male (COS-7, HepG2, and HK-2).
Animal models
Wild-type (WT) and Mrs2−/− (KO) C57BL/6J mice (male) were housed and maintained in our animal breeding facility with prior approval and accordance with the Institutional Animal Care and Use Committee (IACUC). Animals were housed in temperature controlled (20–22°C) rooms with an automatic 12-h light and 12-h dark cycle. Mice were fed a Western Diet (WD; Research Diets D09100310), a standard Chow Diet (CD; Envigo 7012), or a High Fat Diet (HFD; Research Diets D12492) with water available ad libitum. For most experiments, WT and KO mice were maintained on the WD or the CD starting at 12 weeks of age with termination at 12 months. In a subset of experiments, WT mice were switched to an HFD (or remained on CD) starting at 6 weeks of age. After 6 weeks of consistent diet, these HFD- and CD-fed mice were administered control (saline) or CPACC (20 mg/kg BW in saline) i.p. every 3 days. Body weight, health, and food were measured regularly for all groups. Body temperature was recorded using a rectal thermometer (Digi-Sense). When needed, blood was collected in K3/EDTA coated tubes and plasma prepared by centrifugation at 1,000×g for 10 min at 4°C.
Isolation and culture of primary murine hepatocytes
Primary adult murine hepatocytes were isolated using portal vein perfusion. After cannulation of the portal vein, the liver is perfused with wash media (35 mM HEPES and 0.75 mM EGTA) followed by digestion media (DMEM, GIBCO, Cat#12320, GIBCO) containing freshly added Collagenase D (380 μg/mL, Worthington) to dissociate extracellular matrix. After perfusion, liver lobes were gently dissected and dissociated in isolation media (DMEM supplemented with 1% (v/v) FBS). The crude hepatocytes were filtered to 100 μm and subjected to three steps of centrifugation-wash cycles (50×g, 4°C, 5 min). After each spin, the pellet was washed in 25mL of isolation media. The cell’s final resuspension is in hepatocyte growth media (Williams E media (Sigma, #W4128) containing 10% (v/v) FBS, 1% (v/v) antibiotic-antimycotic solution, and 200mM L-glutamine). Finally, cells are counted using trypan-blue exclusion and seeded in hepatocyte growth media according to planned experimental procedures. Hepatocytes are grown in pre-coated collagen culture dishes (Corning BioCoat); or for experiments using confocal microscopy, seeded on in-house collagen coated 25-mm glass coverslips. After 4–8 h, cell attachment is visually examined, and media is replaced with fresh hepatocyte growth media.
Whole-body respirometry measurement and analysis, body composition, and blood chemistry
Mouse metabolic studies were performed at the Penn Diabetes Research Center Rodent Metabolic Phenotyping Core (University of Pennsylvania). A standard 12h light/dark cycle was maintained throughout the study in a dedicated temperature controlled (20–22°C) housing room. Mice were acclimated for 5 days before recording. Indirect calorimetry data were recorded using a computer-controlled Promethion Core system (Sable Systems, Las Vegas, NV). Each cage is thermally controlled and equipped with an XYZ beam break array (BXYZ-R, Sable Systems), a voluntary running wheel (WHEEL-M), and mass measurement modules (2 mg resolution) for food intake and water intake. All animals had ad libitum access to specified diet and water for 5 days of the study. On the final day, mice were overnight fasted from 1900 to 0900 using a computer-controlled script for automated access to food hopper (Promethion AC-2 Access Control Module) in order to restrict feeding at designated time intervals during the calorimetry run. Respiratory gases are measured with an integrated fuel cell oxygen analyzer, NDIR CO2 analyzer, capacitive water vapor partial pressure analyzer and barometric pressure analyzer (CGF, Sable Systems). Gas sensors were calibrated using 100% N2 as the zero reference. Oxygen consumption (VO2) and carbon dioxide (VCO2) production are measured for each mouse at 5 min intervals for 20 s resulting in a 3 min cycle time. Respiratory Exchange Ratio (RER) is calculated as the ratio of VCO2/VO2. Energy expenditure is calculated using the abbreviated Weir equation: Kcal/h = 60*(0.003941*VO2+0.001106*VCO2).73 Consecutive adjacent infrared beam breaks were counted and converted to distance, with a minimum movement threshold set at 1 cm/s. Voluntary wheel revolutions were also measured continuously as revolutions converted to distance. Data acquisition and instrument control were coordinated by IM-3 software and the obtained raw data were processed using MacroInterpreter v.2.38 (Sable Systems) using an analysis script detailing all aspects of data transformation (One-click macro v.2.45; Sable Systems). Analysis of indirect calorimetric data was done using CalR, Excel, and GraphPad Prism v8.26,27 RER and energy expenditure time point data were fit with a fixed-frequency sinusoidal equation y = B + A sin(0.25x+ϕ). Normalization to lean mass where indicated. EchoMRI nuclear magnetic resonance spectrometer (Echo Medical Systems, Houston, TX) was used to measure whole body lean and fat mass. Blood chemistry parameters were determined as followed: plasma cholesterol with an enzymatic assay (Stanbio), glucose using a ReliON glucometer, and insulin using ELISA.
METHOD DETAILS
Liver, iWAT, and kidney tissue preparation and processing
Liver, inguinal white adipose, and kidneys were dissected from mice and gently washed in cold, Ca2+ and Mg2+ free DPBS. Excess fatty tissue was excised from the kidneys and the lymph nodes were removed from the subcutaneous iWAT. Kidneys were sliced longitudinally and the iWAT and liver (left lateral lobe) were sliced transversely. Each tissue was fixed in 10% neutral buffered formalin for histological examination. Additional liver slices were fixed for transmission electron microscopy (2% paraformaldehyde and 2% glutaraldehyde in 0.1M Na cacodylate buffer, pH 7.4). Adipose explants for confocal microscopy were made using a 2mm punch. Remaining liver tissue was cut into smaller pieces, rinsed well with ice-cold DPBS (Ca2+/Mg2+ free) to rinse blood from the tissue, and flash frozen in liquid nitrogen; the remaining iWAT was also flash frozen. Before RNA isolation of the liver and iWAT, the tissue was crushed cryogenically to homogenize the tissue.
Histology and quantification
Tissues were fixed in 10% neutral buffered formalin and washed in 70% ethanol overnight before paraffin embedment and generation of unstained slides. Paraffin embedment, mounting, slide preparation, and standard staining was done by the Histology Laboratory in UT Health San Antonio Department of Pathology & Laboratory Medicine with detailed protocols on file. Briefly, tissues are dried, paraffin embedded, sectioned (longitudinally, kidney; and, transverse, liver and iWAT), and placed on slides. For H&E and Masson’s trichome staining, sections were processed according to the Histology Laboratory’s established protocols. Unstained liver tissue sections were used to conduct IHC (immunohistochemistry) in-house. Sections were deparaffinized then rehydrated, followed by antigen retrieval (0.01M citrate buffer). Endogenous peroxidase blocking was done using 3% H2O2 for 10 min. Avidin and biotin blocking was conducted using Vector Laboratories Avidin/Biotin Blocking Kit (SP02001). Nonspecific binding was blocked by 1 h incubation in normal horse serum (NHS, Vector Laboratories, S-2000-20) containing 0.1% Tween 20 in 0.01M PBS and 10% BSA. Unconjugated goat anti-mouse CD-1/PECAM-1 (14 μg/mL; R&D Systems, AF3628) primary was applied overnight at 4°C in 10% NHS. Sections were incubated with biotinylated ready-to-use horse anti-goat IgG secondary (Vector Laboratories, BP-9500-50) for 1 h at room temperature. Secondary development was done using the VECTASTAIN Elite ABC-HRP kit (Vector Laboratories PK-6100) and DAB reagent according to manufacturer’s recommendations. Developed sections were counterstained with Mayer’s Hematoxylin. Thorough washing with PBS was conducted after each step. Slides were imaged using an Olympus light microscope and imaged at 20x magnification. Vascular density was quantified from H&E-stained slides using ImageJ.74 A digital filter was placed over selected images for vascular clarity and the number of micro vessels observed in each image was recorded. 2–3 slides (different mice) per group were used.
Plasma alanine transaminase activity and citrate measurements
Liver function and damage was determined by measuring alanine transaminase (ALT). The ALT activity in the plasma was determined using the Alanine Transaminase Activity Assay Kit (Abcam ab105134) following manufacturer’s protocol. The plasma ALT levels over two different time points were obtained and corresponding ALT activity was determined by reading in a plate reader at 570 nm. Plasma and extracellular citrate levels were determined using the Citrate Assay Kit (Abcam ab83396) by reading in a plate reader at 570 nm following the manufacturer’s protocol. When required, deproteination was accomplished using a trichloroacetic acid precipitation kit (Abcam ab204708) for tissue and cells, or 10,000 MWCO spin column for plasma.
Gene expression studies (RT-qPCR, RNA-seq, and analysis)
Total RNA was isolated from liver tissue, iWAT, or hepatocytes using a kit following manufacturer’s instructions (Qiagen). RT-qPCR was conducted utilizing TagMan probes: Ucp1 (Mm01244861_m1), Cidea (Mm00432554_m1), Lep (Mm00434759_m1), Ldha (Mm01612132_g1), Ldhb (Mm05874166_g1), Pfkl (Mm00435605_mH), Pgk1 (Mm00435617_m1), Hk1 (Mm00439335_m1), and Actb (Mm01205647_g1). The ΔΔCt method was used to analyze the data with Actb as the housekeeping gene. RNA-seq data generation was contracted with NovoGene. Libraries were constructed using poly-T magnetic beads following by fragmentation, cDNA synthesis, adaptor ligation, and finally PCR. Quality control was conducted by removing low quality reads or those containing the adaptor or poly-N strings. Reads were aligned to the reference genome using Hisat2 v2.0.5.62–65 FeatureCounts v1.5.0-p3 was used to count mapped reads and FPKM values were subsequently calculated to correct for gene length and sequencing depth.66 Consistency between biological replicates was confirmed by correlation analysis. DESeq2 v1.20.0 was used for differential expression analysis67; p-values were adjusted using the Benjamini-Hochberg method and statistical significance was considered by an adjusted p-value < 0.05. Gene set enrichment analysis (GSEA) was done using the publicly available GSEA analysis tool from Broad Institute (http://www.broadinstitute.org/gsea/index.jsp).68,69 The R package ClusterProfile was used to test for significant enrichment of KEGG pathways.34 In the heatmaps in Figures 3C and S3H, the following KEGG pathways are utilized as the query for the respective pathway: “thermogenesis” (thermogenesis; mmu04714), “oxidative phosphorylation” (oxidative phosphorylation; mmu00190), “fatty acid catabolism” (fatty acid metabolism; mmu01212 and fatty acid degradation; mmu00071), “TCA cycle” (citrate cycle (TCA cycle); mmu00020), “glycolysis” (glycolysis/gluconeogenesis; mmu00010), “HIF-1a” (Hif1 signaling pathway; mmu04066).34 Following NovoGene’s preliminary analysis, further in-house analysis was conducted. Normalized FPKM values for heatmaps were generated using the STANDARDIZE formula in Microsoft Excel to generate a row Z score and each gene was normalized individually for comparison within each gene across the four groups. Relative mRNA abundance was calculated by normalizing FPKM values in Excel, specifically using the average FPKM for the gene across the four groups. Each gene was normalized individually. Unbiased transcription factor analysis was conducted using ChEA software.37 Area-proportional Venn diagrams were created with BioVenn and DeepVenn.71,72 Finally, the Sankey plot was generated using the SankeyMATIC tool (https://sankeymatic.com/). Detailed information for the RNA-Seq data and analysis can be found in an Excel workbook (Data S1) in the supplementary files.
Immunoblotting
Crushed tissues were homogenized in lysis buffer (ThermoFisher) on ice for 1 min (20–30 strokes) using an automatic Dounce homogenizer set to 2000 RPM. Whole cell lysate was prepared in lysis buffer (Abcam) using 3 sets of 3-s sonication (power level 3) intervals on ice. Protein concentration was estimated using Pierce BCA assay kit and samples were prepared and heated for 90C for 5 min. Equal amounts of protein were loaded and separated on 4%–12% Bis-Tris polyacrylamide gel (Thermo Fisher Scientific), transferred to a PVDF membrane, blocked for 1–2 h using 5% fat free skim milk, washed and finally, probed with corresponding antibodies as specified below. Antibodies were from Cell Signaling Technology (HIF-1α and hydroxy HIF-1α dilution 1:3000, MCU dilution 1:5000, MICU1 dilution 1:3000), Abcam (CPT-1a, CPT-2, OXPHOS cocktail dilution 1:3000), ZYMED Laboratories (β-actin; dilution 1:1,000), and Amersham (secondary antibodies conjugated with peroxidase). Development was done using X-ray film using a series of timed exposures and ImageJ was used for densitometric analysis on scanned film images.74
Mitochondrial oxygen consumption rate
Primary murine hepatocytes were plated on in-house collagen coated 96-well Seahorse XF Cell Culture Microplates (Agilent) at a density of 4 × 105 cells/well. After initial processing, equivalent weights of iWAT were gently homogenized in 750μL ICM buffer (mM, 120 KCl, 10 NaCl, 1 KH2PO4, 20 HEPES-Tris, pH 7) before seeding in uncoated Seahorse microplates. In some experiments, hepatocytes were treated with 5, 10 and 25 mM of chloropentammine cobalt(III) chloride (CPACC) for 1 h at 37°C. Media was changed to Seahorse XF Cell Mito Stress Test Kit (Agilent) assay media supplemented with glucose, glutamine, pyruvate, and HEPES with concentrations equivalent to that of the hepatocyte growth media 1 h before the experiment start time. After media change, per manufacturer instructions, cells and tissue homogenate were placed in a CO2-free incubator for 1 h. Oxygen consumption rate (OCR) was measured at 37°C in an XF96 extracellular flux analyzer (Seahorse Bioscience, Agilent) calibrated using Seahorse XF Calibrant solution (Seahorse Bioscience, Agilent) in a CO2-free incubator overnight. Respiratory chain inhibitors (2 μM oligomycin, 5 μM FCCP, and a mixture of 1 μM antimycin A and 1 μM rotenone) were added at the indicated time points. Data were collected using Agilent Seahorse Wave 2.6.1 Desktop software and analyzed using GraphPad Prism version 8.75,76
Confocal microscopic examination of cellular lipid localization and mitochondrial membrane potential maintenance
Primary murine hepatocytes were stained with the lipid indicator BODIPY (ex/em 493/503 nm, 1 μg/mL) and mitochondrial membrane potential indicator tetramethylrhodamine, ethyl ester (TMRE, ex/em 556/610 nm, 100 nM) in serum-free conditions for 30 min (5% CO2) and washed before imaging.4 For CPACC-treated experiments, hepatocytes were acquired from 12-month-old mice and cells were treated with CPACC (50mM) for 12 h. Fresh iWAT explants were stained with TMRE (20nM) and DAPI (0.5 μg/mL) for 30 min (5% CO2) and washed before imaging (63x). All confocal microscopic images were acquired using a Leica SP8 confocal microscope (Manheim, Germany) coupled with a temperature-controlled environmental chamber. Leica Application Suite X was used to quantify lipid-mitochondrial colocalization using line scan analysis followed by smooth curve fitting. ImageJ was used to calculate Pearson’s and Mander’s colocalization coefficients. The lipid droplet size and mitochondrial membrane potential in hepatocytes were quantified using both Leica Application Suite X and ImageJ. The mitochondrial density in iWAT explants was quantified utilizing modules in KNIME software.77 Briefly, the LASX image files were imported to the Files and Folders module, read with Image Reader (Table), and then channel separated using Splitter. The TMRE channel was then processed with a background subtraction ImageJ macro (rolling = 50) followed by a Gaussian blur macro (σ = 2) and Watershed macro (iterations = 1; count = 1). Finally, the Analyze Particle macro (size = 0 to infinity; circularity 0–1) to obtain the mitochondrial area and number. All statistical analyses were done using GraphPad Prism v8.
Metabolite screening for Hif1-a stabilization
Primary murine hepatocytes from WT or Mrs2−/− mice were treated with either LPS (0 or 1 μg/mL) or CoCl2 (1mM) for 6 h. In most experiments, cells were treated with FG-4592 (0–200μM), CoCl2 (0–200μM), or DMOG (dimethyloxallyl glycine, 0–1mM) for 6 h as indicated. An additional experiment utilized FG-4592-treated (6 h) hepatocytes that were lysed (lysis buffer, ThermoFisher) and then treated with the indicated (0–10mM) dose of citrate (Figure S7F). In the targeted metabolite screen, hepatocytes were cotreated (6 h) with FG-4592 (100μM) and the designated metabolite: glucose, glucose-6-phosphate (G-6-P), fructose-6-phosphate (F-6-P), fructose-1,6-bisphosphate (F-1,6-BP), r-glyceraldehyde-3-phosphate (GA-3-P), dihydroxyacetone phosphate (DHAP), 3-phosphoglycerate (3-PG), phosphoenol pyruvate (PEP), pyruvate (pyruvic acid), lactic acid, citric acid, isocitrate, α-ketoglutarate (α-KG),succinate,fumarate,ormalate(10mM);or,palmitateor2-hydroxyglutarate(D-2-HG)(100μM).Additionalscreeningwasconducted utilizing HK-2 cells treated with CoCl2 with or without a metabolite(10mM): oxaloacetate (OAA), pyruvate, citric acid, α-KG, succinate, or malate. Citric acid derivatives (mono, di, and tri sodium salts) were used at a concentration of 10mM in WT and Mrs2−/− hepatocytes. The time-dependent nature of citrate-mediated Hif1-α stabilization was examined by utilizing hepatocytes treated with FG-4592 (0–24hrs) with or without citric acid (10mM). The role of Mg2+ in Hif1-α stabilization was further explored by treated cells with EDTA-AM (43μM) or MgCl supplementation (0–10mM). UK-5099 (100mM) was used to explore the impact of pyruvate on Hif1-α stabilization. COS-7 cells were transfected with mutant Hif1-α construct (addgene#87261) and treated with citric acid (0–10mM) (Figure 6F).49 Cells were rinsed with DPBS (Ca2+/Mg2+ free) then scraped in lysis buffer followed by generation of whole lysate.
Evaluation of dose-dependent Mrs2 inhibition by confocal live cell imaging system
Primary murine hepatocytes were treated with varying doses (1,2,5,10.25 mM) of CPACC, or control, for 12 h. To visualize Mg2+ and the mitochondria, cells were stained with 2.5mM Magnesium Green-AM (ex/em 488/510 nm) and 1 mM MitoTracker Deep Red FM (ex/em 644/665 nm) for 30 min in their normal growth conditions. Using the Leica SP8 confocal microscope, time lapse images were collected every 3 s. After 30 s of baseline recording, the cells were stimulated with lactate (5 mM). Data were quantified using Leica Application Suite X.
Visualization of citrate flux in live cells using confocal microcopy
Primary murine hepatocytes were grown and seeded as previously described. HepG2 cells were grown on 0.1% gelatin coated glass coverslips. After overnight growth, cells were transiently transfected with 2 μg of genetically encoded biosensors, CMV-Citron or CMV mito-Citron1 (deposited by Robert Campbell46 at Addgene, #134303 and #134305; GFP ex/em 488/510 nm) using Lipofectamine 3000 transfection reagent. After 24hr, transfected hepatocytes were infected with adenoviruses (Vector BioLabs), Ad-RFP and Ad-Mrs2(mut)mRFP-FLAG (MOI 10). After 48 h of infection, the cells were washed and imaged using the Leica SP8 Confocal microscope under 603 oil immersion. After 30 s of baseline recording, cells were stimulated with 20 mM of glucose (hepatocytes) or 16.7 mM (HepG2) and the corresponding fluorescence emissions in the cytosol and mitochondria were recorded. The citrate transient-generated fluorescence emission was quantified using Leica Application Suite X and analyzed using GraphPad Prism v8.
Spectrofluorimetric measurement of mitochondrial Mg2+ and Ca2+ dynamics
Fluorescence measurements were conducted in a multiwavelength excitation dual wavelength emission spectrofluorometer (Delta RAM, PTI, HORIBA) similar to prior studies.4,10 Cells were washed with Ca2+ & Mg2+ free DPBS, pH 7.4. Following centrifugation (50× g, 4°C, 5 min), approximately 4–5×106 cells were resuspended and permeabilized using 40 μg/mL digitonin in 1.5 mL of intracellular medium (ICM) (mM, 120 KCl, 10 NaCl, 1 KH2PO4, 20 HEPES-Tris, pH 7). Suspension was additionally supplemented with succinate (5mM), ATP, and a fluorescent dye. Magnesium measurements were performed using K+/ATP (1.5 mM) and Mag Green (0.5mM); meanwhile, calcium measurements were performed using Mg2+/ATP (1.5mM) and Fura-2FF (1 μM). Mag-Green has an excitation of 505nm and emissions of 535nm and 595nm based on Mg2+ binding. Fura2-FF has excitations 340nm and 380nm based on Ca2+ binding and emits at 510nm. Changes in extramitochondrial Mg2+ ([Mg2+]out) or Ca2+ ([Ca2+]out) was used as an indicator of mitochondrial uptake of these ions. After a 400 s background acquisition period, cells were pulsed with either a single bolus of 1 mM Mg2+ or multiple 20 μM Ca2+ pulses followed by the mitochondrial uncoupler, FCCP (2 μM). To measure the inhibition of mMg2+ uptake, permeabilized hepatocytes in ICM were supplemented with different concentrations (0.05, 0.1, 0.5, 1,2,5,10.25 μM) of CPACC, or control, and pulsed with a bolus of 1 mM Mg2+ at 450s followed by FCCP (10 μM) at 1000s. This set of experiments was also conducted using hexaammine Co(III) chloride at the same dosages; additionally, ruthenium ion based inhibition of mMg2+ was investigated using hexaammine Ru(III) chloride (5mm) and chloropentammine Ru(III) chloride (5μM). The mMg2+ uptake rate was calculated from the linear portion of the traces immediately after Mg2+ addition. All experiments were done at 37°C with constant stirring.
Cellular uptake assay
Hepatocytes were treated with varying doses of CPACC for 30 min, cells were washed in Ca2+ and Mg2+ free DPBS, and cell pellets flash-frozen. Pellets were suspended in ice-cold lysis buffer (0.1% Triton X-100 and 0.2% HNO3 in ultrapure water). The suspension was vortexed for 30s and incubated on ice for 45 min. The cell lysate was centrifuged, and the supernatant was transferred to a clean tube prior to analysis. The Co content of the lysate was determined using graphite furnace atomic absorption spectroscopy (GFAAS) and was normalized to the protein content of the sample, which was determined using the bicinchoninic acid (BCA) assay kit following manufacturer instructions (ThermoFisher). Results are reported as the average mass ratio of Co to protein (pg/μg) in each sample ±SEM.
MRS2 cloning, expression, and purification
The full length human MRS2 (NCBI accession NP_065713.1), identified as MRS258–443 (i.e., residues 58–443) and excluding the mitochondrial targeting sequence, was cloned into pET-28a using NdeI and XhoI restriction sites. The sequence and frame of the MRS258–443 coding insert within the pET-28a vector was confirmed by Sanger DNA sequencing. Transformed BL21(DE3) codon + Escherichia coli were grown in Luria-Bertani (LB) broth supplemented with 60 μg/mL kanamycin to an optical density (600nm) of ≈0.6–0.8 at 37°C. Expression was induced upon addition 300 μM isopropyl β-D-1-thiogalactopyranoside (IPTG), and growth was continued overnight. The protein was purified from the harvested cells under native conditions according to the HisPur nickel-nitriloacetic acid (Ni-NTA) manufacturer guidelines (ThermoFisher). After elution from the Ni-NTA beads using buffer containing 20 mM Tris, 150 mM NaCl, 1 mM DTT, 10 mM CHAPS, 325 mM imidazole, pH 8.0, the 6×His-MRS258–443 was dialyzed in the same buffer without imidazole and thrombin digested (≈1 U/mg protein) to remove the 6×His tag. A final size exclusion chromatography (SEC) purification step was performed through a Superdex 200 10/300 GL column connected to an AKTA Pure FPLC (GE Healthcare). The buffer for the SEC and all experiments was 20 mM Tris, 150 mM NaCl, 1 mM DTT, 10 mM CHAPS, pH 8.0.
Dynamic light scattering (DLS)
A Dynapro Nanostar (Wyatt) equilibrated to 20°C was used for all DLS experiments. Protein samples at ~0.5 mg/mL in 20 mM Tris, 150 mM NaCl, 1 mM DTT, 10 mM CHAPS, pH 8.0 without or with 5 mM CaCl2, 5 mM MgCl2, 5 mM CoCl2 or 0.5 mM CPACC were centrifuged at 12,000 ×g for 5 min before a 5 μL aliquot of the supernatant was removed and loaded into a JC501 cuvette (Wyatt). The sample was equilibrated for 5 min at 20°C before 10 autocorrelation function acquisitions of 5s each were recorded and averaged. Two aliquots (technical replicates) from each sample were averaged, and each experimental condition was performed on three independent/individual protein expression preparations (biological replicates). Autocorrelation functions were deconvoluted using the regularization algorithm to extract polydisperse distributions of hydrodynamic radii and cumulants algorithm to extract weight-averaged monodisperse hydrodynamic radii, using the accompanying Dynamics (v7.8.1.3) software (Wyatt). The dependence of hydrodynamic size on CPACC concentration was determined using ~0.5 mg/mL protein supplemented with 0.01, 0.05, 0.1, 0.5, or 0.75 mM CPACC, using a similar experimental setup as described above and cumulants analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical tests and data representation
Unless otherwise indicated, statistical analysis is conducted using one-way ANOVA with Tukey’s multiple comparisons. Where indicated, two-tailed Student’s t-test was used. GraphPad Prism version 8 was used for statistical testing and regression analysis. Data are represented as mean ± SEM unless otherwise indicated.
Supplementary Material
Highlights.
Mitochondrial Mg2+ channel Mrs2 rheostats MCU Ca2+ signals to maintain bioenergetic circuit
DNL precursor and cellular Mg2+ chelator citrate curbs HIF-1α signal and oxidative metabolism
Lowering mMg2+ mitigates prolonged dietary-stress-induced obesity and metabolic syndrome
Mrs2 channel blocker CPACC reduces lipid accumulation and promotes browning and weight loss
ACKNOWLEDGMENTS
We thank Robert Campbell and William G. Kaelin Jr for sharing the cytoCitron1 (addgene#134303) and mito-Citron1 (addgene#134305) plasmids and HIF1-α mutant (addgene#87261) constructs. We also thank the Department of Pathology and Laboratory Medicine histology core facility at UTHSA for histology preparation/staining. This research was funded by the National Institutes of Health (R35GM145294, R01GM109882, R01DK135179, and R01HL142673) to M.M. This work was partly supported by DOD/DHP-CDMRP PR181598P-1 and San Antonio Partnership for Precision Therapeutics (SAPPT) to M.M., CIHR-438225 to P.B.S., and National Science Foundation (CHE-1750295) to J.J.W. T.R.M. is supported by the NIH (R01GM109882-S1 and T32 AG 021890). We thank the Rodent Metabolic Phenotyping Core, supported in part by the Penn Diabetes Research Center grant (P30-DK19525) and S10-OD025098.
Footnotes
DECLARATION OF INTERESTS
M.M. is an inventor on a patent filed by UTHSA on CPACC as a Mrs2 blocker for Mg2+ in physiology and disease. J.A.B. is a consultant to Pfizer.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112155.
REFERENCES
- 1.Edgar R, Domrachev M, and Lash AE (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu S, Yang L, Li P, Hofmann O, Dicker L, Hide W, Lin X, Watkins SM, Ivanov AR, and Hotamisligil GS (2011). Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature 473, 528–531. 10.1038/nature09968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Baaij JHF, Hoenderop JGJ, and Bindels RJM (2015). Magnesium in man: implications for health and disease. Physiol. Rev. 95, 1–46. 10.1152/physrev.00012.2014. [DOI] [PubMed] [Google Scholar]
- 4.Tomar D, Jaña F, Dong Z, Quinn WJ 3rd, Jadiya P, Breves SL, Daw CC, Srikantan S, Shanmughapriya S, Nemani N, et al. (2019). Blockade of MCU-mediated Ca(2+) uptake perturbs lipid metabolism via PP4-dependent AMPK dephosphorylation. Cell Rep. 26, 3709–3725.e7. 10.1016/j.celrep.2019.02.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Picard M, Wallace DC, and Burelle Y (2016). The rise of mitochondria in medicine. Mitochondrion 30, 105–116. 10.1016/j.mito.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glancy B, and Balaban RS (2012). Role of mitochondrial Ca2+ in the regulation of cellular energetics. Biochemistry 51, 2959–2973. 10.1021/bi2018909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clapham DE (2007). Calcium signaling. Cell 131, 1047–1058. 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 8.Berridge MJ, Bootman MD, and Roderick HL (2003). Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 4, 517–529. 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 9.Rizzuto R, De Stefani D, Raffaello A, and Mammucari C (2012). Mitochondria as sensors and regulators of calcium signalling. Nat. Rev. Mol. Cell Biol. 13, 566–578. 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- 10.Daw CC, Ramachandran K, Enslow BT, Maity S, Bursic B, Novello MJ, Rubannelsonkumar CS, Mashal AH, Ravichandran J, Bakewell TM, et al. (2020). Lactate elicits ER-mitochondrial Mg(2+) dynamics to integrate cellular metabolism. Cell 183, 474–489.e17. 10.1016/j.cell.2020.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamer KJ, and Mootha VK (2015). The molecular era of the mitochondrial calcium uniporter. Nat. Rev. Mol. Cell Biol. 16, 545–553. 10.1038/nrm4039. [DOI] [PubMed] [Google Scholar]
- 12.O”Rourke B, Ashok D, and Liu T (2021). Mitochondrial Ca(2+) in heart failure: not enough or too much? J. Mol. Cell. Cardiol. 151, 126–134. 10.1016/j.yjmcc.2020.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirichok Y, Krapivinsky G, and Clapham DE (2004). The mitochondrial calcium uniporter is a highly selective ion channel. Nature 427, 360–364. 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- 14.McCormack JG, Halestrap AP, and Denton RM (1990). Role of calcium ions in regulation of mammalian intramitochondrial metabolism. Physiol. Rev. 70, 391–425. [DOI] [PubMed] [Google Scholar]
- 15.Garbincius JF, and Elrod JW (2022). Mitochondrial calcium exchange in physiology and disease. Physiol. Rev. 102, 893–992. 10.1152/physrev.00041.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rutter GA, Osbaldeston NJ, McCormack JG, and Denton RM (1990). Measurement of matrix free Mg2+ concentration in rat heart mitochondria by using entrapped fluorescent probes. Biochem. J. 271, 627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SK, Shanmughapriya S, Mok MCY, Dong Z, Tomar D, Carvalho E, Rajan S, Junop MS, Madesh M, and Stathopulos PB (2016). Structural insights into mitochondrial calcium uniporter regulation by divalent cations. Cell Chem. Biol. 23, 1157–1169. 10.1016/j.chembiol.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moomaw AS, and Maguire ME (2008). The unique nature of mg2+ channels. Physiology 23, 275–285. 10.1152/physiol.00019.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li FY, Chaigne-Delalande B, Kanellopoulou C, Davis JC, Matthews HF, Douek DC, Cohen JI, Uzel G, Su HC, and Lenardo MJ (2011). Second messenger role for Mg2+ revealed by human T-cell immunodeficiency. Nature 475, 471–476. 10.1038/nature10246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sveidahl Johansen O, Ma T, Hansen JB, Markussen LK, Schreiber R, Reverte-Salisa L, Dong H, Christensen DP, Sun W, Gnad T, et al. (2021). Lipolysis drives expression of the constitutively active receptor GPR3 to induce adipose thermogenesis. Cell 184, 3502–3518.e33. 10.1016/j.cell.2021.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachman ES, Dhillon H, Zhang CY, Cinti S, Bianco AC, Kobilka BK, and Lowell BB (2002). betaAR signaling required for diet-induced thermogenesis and obesity resistance. Science 297, 843–845. 10.1126/science.1073160. [DOI] [PubMed] [Google Scholar]
- 22.Rosen ED, and Spiegelman BM (2014). What we talk about when we talk about fat. Cell 156, 20–44. 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Assimacopoulos-Jeannet F, McCormack JG, and Jeanrenaud B (1986). Vasopressin and/or glucagon rapidly increases mitochondrial calcium and oxidative enzyme activities in the perfused rat liver. J. Biol.Chem. 261, 8799–8804. [PubMed] [Google Scholar]
- 24.Winzell MS, and Ahrén B (2004). The high-fat diet–fed mouse: a model for studying mechanisms and treatment of impaired glucose tolerance and type 2 diabetes. Diabetes 53, S215–S219. 10.2337/diabetes.53.suppl_3.S215. [DOI] [PubMed] [Google Scholar]
- 25.Basaranoglu M, Basaranoglu G, Sabuncu T, and Sentürk H (2013). Fructose as a key player in the development of fatty liver disease. World J. Gastroenterol. 19, 1166–1172. 10.3748/wjg.v19.i8.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller TD, Klingenspor M, and Tschop MH (2021). Revisiting energy expenditure: how to correct mouse metabolic rate for body mass. Nat. Metab. 3, 1134–1136. 10.1038/s42255-021-00451-2. [DOI] [PubMed] [Google Scholar]
- 27.Mina AI, LeClair RA, LeClair KB, Cohen DE, Lantier L, and Banks AS (2018). CalR: a web-based analysis tool for indirect calorimetry experiments. Cell Metab. 28, 656–666.e1. 10.1016/j.cmet.2018.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghijselings IE, Bockstael B, De Waele E, and Jonckheer J (2022). Effect of extracorporeal carbon dioxide removal on respiratory quotient measured by indirect calorimetry: Unravelling the mystery. Exp Physiol 107, 424–428. 10.1113/EP090282. [DOI] [PubMed] [Google Scholar]
- 29.Loomba R, Friedman SL, and Shulman GI (2021). Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 184, 2537–2564. 10.1016/j.cell.2021.04.015. [DOI] [PubMed] [Google Scholar]
- 30.Reid AE (2001). Nonalcoholic steatohepatitis. Gastroenterology 121, 710–723. 10.1053/gast.2001.27126. [DOI] [PubMed] [Google Scholar]
- 31.Ungvari Z, Toth P, Tarantini S, Prodan CI, Sorond F, Merkely B, and Csiszar A (2021). Hypertension-induced cognitive impairment: from pathophysiology to public health. Nat. Rev. Nephrol. 17, 639–654. 10.1038/s41581-021-00430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grunewald M, Kumar S, Sharife H, Volinsky E, Gileles-Hillel A, Licht T, Permyakova A, Hinden L, Azar S, Friedmann Y, et al. (2021). Counteracting age-related VEGF signaling insufficiency promotes healthy aging and extends life span. Science 373, eabc8479. 10.1126/science.abc8479. [DOI] [PubMed] [Google Scholar]
- 33.Ramachandran K, Maity S, Muthukumar AR, Kandala S, Tomar D, Abd El-Aziz TM, Allen C, Sun Y, Venkatesan M, Madaris TR, et al. (2022). SARS-CoV-2 infection enhances mitochondrial PTP complex activity to perturb cardiac energetics. iScience 25, 103722. 10.1016/j.isci.2021.103722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanehisa M, and Goto S (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M (2019). Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, and Tanabe M (2021). KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 49, D545–D551. 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, and Ma’ayan A (2010). ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 26, 2438–2444. 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iliopoulos O, Levy AP, Jiang C, Kaelin WG Jr., and Goldberg MA (1996). Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc. Natl. Acad. Sci. USA 93, 10595–10599. 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang C, Jiang L, Zhang H, Shimoda LA, DeBerardinis RJ, and Semenza GL (2014). Analysis of hypoxia-induced metabolic reprogramming. Methods Enzymol. 542, 425–455. 10.1016/B978-0-12-416618-9.00022-4. [DOI] [PubMed] [Google Scholar]
- 40.Semenza GL (2014). Hypoxia-inducible factor 1 and cardiovascular disease. Annu. Rev. Physiol. 76, 39–56. 10.1146/annurev-physiol-021113-170322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bishop T, and Ratcliffe PJ (2020). Genetic basis of oxygen sensing in the carotid body: HIF2alpha and an isoform switch in cytochrome c oxidase subunit 4. Sci. Signal. 13, eaba1302. 10.1126/scisignal.aba1302. [DOI] [PubMed] [Google Scholar]
- 42.Consortium UniProt (2021). UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489. 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Benador IY, Veliova M, Mahdaviani K, Petcherski A, Wikstrom JD, Assali EA, Acín-Pérez R, Shum M, Oliveira MF, Cinti S, et al. (2018). Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion. Cell Metab. 27, 869–885.e6. 10.1016/j.cmet.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alevriadou BR, Patel A, Noble M, Ghosh S, Gohil VM, Stathopulos PB, and Madesh M (2021). Molecular nature and physiological role of the mitochondrial calcium uniporter channel. Am. J. Physiol. Cell Physiol. 320, C465–C482. 10.1152/ajpcell.00502.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinkosky SL, Groot PHE, Lalwani ND, and Steinberg GR (2017). Targeting ATP-citrate lyase in hyperlipidemia and metabolic disorders. Trends Mol. Med. 23, 1047–1063. 10.1016/j.molmed.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 46.Zhao Y, Shen Y, Wen Y, and Campbell RE (2020). High-performance intensiometric direct- and inverse-response genetically encoded biosensors for citrate. ACS Cent. Sci. 6, 1441–1450. 10.1021/acscentsci.0c00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Semenza GL (2013). HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Invest. 123, 3664–3671. 10.1172/JCI67230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lum JJ, Bui T, Gruber M, Gordan JD, DeBerardinis RJ, Covello KL, Simon MC, and Thompson CB (2007). The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes Dev. 21, 1037–1049. 10.1101/gad.1529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaelin WG Jr., and Ratcliffe PJ (2008). Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402. 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 50.Buckley DL, Gustafson JL, Van Molle I, Roth AG, Tae HS, Gareiss PC, Jorgensen WL, Ciulli A, and Crews CM (2012). Small-molecule inhibitors of the interaction between the E3 ligase VHL and HIF-1alpha. Angew. Chem. Int. Ed. Engl. 51, 11463–11467. 10.1002/anie.201206231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan Q, Bartz S, Mao M, Li L, and Kaelin WG Jr. (2007). The hypoxia-inducible factor 2alpha N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Mol. Cell Biol. 27, 2092–2102. 10.1128/MCB.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kucharski LM, Lubbe WJ, and Maguire ME (2000). Cation hexaammines are selective and potent inhibitors of the CorA magnesium transport system. J. Biol. Chem. 275, 16767–16773. 10.1074/jbc.M001507200. [DOI] [PubMed] [Google Scholar]
- 53.Kitamura A, Kubota H, Pack CG, Matsumoto G, Hirayama S, Takahashi Y, Kimura H, Kinjo M, Morimoto RI, and Nagata K (2006). Cytosolic chaperonin prevents polyglutamine toxicity with altering the aggregation state. Nat. Cell Biol. 8, 1163–1170. 10.1038/ncb1478. [DOI] [PubMed] [Google Scholar]
- 54.Midgley PJ, Rutter GA, Thomas AP, and Denton RM (1987). Effects of Ca2+ and Mg2+ on the activity of pyruvate dehydrogenase phosphate phosphatase within toluene-permeabilized mitochondria. Biochem. J. 241, 371–377. 10.1042/bj2410371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hunter DR, Haworth RA, and Southard JH (1976). Relationship between configuration, function, and permeability in calcium-treated mitochondria. J. Biol. Chem. 251, 5069–5077. [PubMed] [Google Scholar]
- 56.Clapham DE (1995). Calcium signaling. Cell 80, 259–268. 10.1016/0092-8674(95)90408-5. [DOI] [PubMed] [Google Scholar]
- 57.Herroeder S, Schönherr ME, De Hert SG, and Hollmann MW (2011). Magnesium–essentials for anesthesiologists. Anesthesiology 114, 971–993. 10.1097/ALN.0b013e318210483d. [DOI] [PubMed] [Google Scholar]
- 58.Valdivia HH, Kaplan JH, Ellis-Davies GC, and Lederer WJ (1995). Rapid adaptation of cardiac ryanodine receptors: modulation by Mg2+ and phosphorylation. Science 267, 1997–2000. 10.1126/science.7701323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan M, Mosaoa R, Graham GT, Kasprzyk-Pawelec A, Gadre S, Parasido E, Catalina-Rodriguez O, Foley P, Giaccone G, Cheema A, et al. (2020). Inhibition of the mitochondrial citrate carrier, Slc25a1, reverts steatosis, glucose intolerance, and inflammation in preclinical models of NAFLD/NASH. Cell Death Differ. 27, 2143–2157. 10.1038/s41418-020-0491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan Q, Bartz S, Mao M, Li L, and Kaelin WG Jr. (2007). The hypoxia-inducible factor 2alpha N-terminal and C-terminal transactivation domains cooperate to promote renal tumorigenesis in vivo. Mol Cell Biol 27, 2092–2102. 10.1128/MCB.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim D, Paggi JM, Park C, Bennett C, and Salzberg SL (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pertea M, Kim D, Pertea GM, Leek JT, and Salzberg SL (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Park C, Bennett C, Thornton M, and Kim D (2021). Rapid and accurate alignment of nucleotide conversion sequencing reads with HISAT-3N. Genome Res. 31, 1290–1295. 10.1101/gr.275193.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 67.Love MI, Huber W, and Anders S (2014). Moderated estimation of foldchange and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrå le M, Laurila E, et al. (2003). PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273. 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 70.Yu G, Wang LG, Han Y, and He QY (2012). clusterProfiler: an Rpackage for comparing biological themes among gene clusters. OMICS 16, 284–287. 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hulsen T, de Vlieg J, and Alkema W (2008). BioVenn – a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genom. 9, 488. 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hulsen T (2022). DeepVenn - a web application for the creation of area-proportional Venn diagrams using the deep learning framework Tensorflow.js. Preprint at arXiv. 10.48550/arXiv.2210.04597. [DOI] [Google Scholar]
- 73.Weir JB (1949). New methods for calculating metabolic rate with special reference to protein metabolism. J. Physiol. 109, 1–9. 10.1113/jphysiol.1949.sp004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tomar D, Dong Z, Shanmughapriya S, Koch DA, Thomas T, Hoffman NE, Timbalia SA, Goldman SJ, Breves SL, Corbally DP, et al. (2016). MCUR1 is a scaffold factor for the MCU complex function and promotes mitochondrial bioenergetics. Cell Rep. 15, 1673–1685. 10.1016/j.celrep.2016.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Irrinki KM, Mallilankaraman K, Thapa RJ, Chandramoorthy HC, Smith FJ, Jog NR, Gandhirajan RK, Kelsen SG, Houser SR, May MJ, et al. (2011). Requirement of FADD, NEMO, and BAX/BAK for aberrant mitochondrial function in tumor necrosis factor alpha-induced necrosis. Mol. Cell Biol. 31, 3745–3758. 10.1128/MCB.05303-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Berthold MR, Cebron N, Dill F, Gabriel TR, Kötter T, Meinl T, Ohl P, Thiel K, and Wiswedel B (2009). KNIME - the Konstanz information miner: version 2.0 and beyond. SIGKDD Explor. Newsl. 11, 26–31. 10.1145/1656274.1656280. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data of liver and iWAT have been deposited in the NCBI Gene Expression Omnibus (GEO)1 and are publicly available as of the data of publication. The accession number is listed in the key resources table. Additional information regarding RNA-seq data and analysis can be found in Data S1. All other data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Total OXPHOS Rodent WB Antibody Cocktail | Abcam | Cat# ab110413 |
| CPT-1a | Abcam | Cat# 234111 |
| CPT-2 | Abcam | Cat# 181114 |
| HIF-1α | Cell Signaling, MA, USA | Cat# 14179S |
| Hydroxy HIF-1α | Cell Signaling | Cat# 3434S |
| MCU | Cell Signaling | Cat #D2Z3B |
| MICU1 | Cell Signaling | Cat# D4P8Q |
| β-Actin | Santa Cruz Biotechnology | Cat# SC47778 |
| α-Mouse Secondary (HRP-conjugated) | Cell Signaling | Cat# 5127S |
| α-Rabbit Secondary (HRP-conjugated) | Cell Signaling | Cat# 911968 |
| Goat Anti-Mouse CD-31/PECAM-1 (unconjugated) | R&D Systems Inc., MN, USA | Cat# AF3628 |
| Horse Anti-Goat IgG (H + L) Biotin Conjugated RTU Secondary |
Vector Laboratories, CA, USA | BP-9500-50 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Ad-mRFP | Vector Biolabs, PA, USA | Cat# 1660 |
| Ad-Mrs2(mut).mRFP.FLAG [Mutant Mrs2] | Vector Biolabs | N/A |
| BL21-CodonPlus Competent Cells (Escherichia coli) | Agilent Technologies, CA, USA | Cat# 230280 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Western Diet (WD) | Research Diets | Cat# D09100310 |
| Standard Chow Diet (CD) | Envigo | Cat# 7012 |
| High Fat Diet (HFD) | Research Diets | Cat# D12492 |
| Human MRS2 (MRS258–443) | This Study and Daw et al.10 | NCBI acc. NP_065713. |
| HisPurTM Ni-NTA Resin | ThermoFisher Scientific | Cat# 88223 |
| Ndel Restriction Enzyme | New England Biolabs | Cat# R0111S |
| Xhol Restriction Enyme | New England Biolabs | Cat# R0146S |
| HaltTM Protease and Phosphatase Inhibitor Cocktail | ThermoFisher Scientific | Cat# 78441 |
| RestoreTM PLUS Western Blot Stripping Buffer | ThermoFisher Scientific | Cat# 46430 |
| NuPAGETM MOPS SDS Running Buffer (20X) | ThermoFisher Scientific | Cat# NP0001 |
| SuperSignalTM West Pico PLUS Chemiluminescent Substrate | ThermoFisher Scientific | Cat# 34578 |
| Bis-Tris Polyacrylamide Gel (4%–12%) | ThermoFisher Scientific | Cat# WG1402BOX |
| TaqManTM Fast Advanced Master Mix | ThermoFisher Scientific | Cat# 4444557 |
| SuperScipt® VILOTM | ThermoFisher Scientific | Cat# 11754-050 |
| Lipopolysaccharide (LPS) (E. coli 0111:B4) | Sigma Aldrich | Cat# F3665 |
| FG-4592 (Roxadustat) | Cayman Chemical, MI, USA | Cat# 15294 |
| DMOG | Sigma Aldrich | Cat# D3695-10MG |
| Cobalt(II) Chloride (CoCl2) | Fluka (Sigma Aldrich) | Cat# 60818 |
| Glucose | Sigma Aldrich | Cat# G8644 |
| Glucose-6-Phosphate | Sigma Aldrich | Cat# G7879 |
| Fructose-6-Phosphate | Sigma Aldrich | Cat# F3627 |
| Fructose-1,6-Bisphosphate | Sigma Aldrich | Cat# F6803 |
| Glyceraldehyde-3-Phosphate | Sigma Aldrich | Cat# G5251 |
| Dihydroxyacetone Phosphate | Sigma Aldrich | Cat# D7137 |
| 3-Phosphoglyceic Acid | Sigma Aldrich | Cat# P8877 |
| Phospho(enol)pyruvic Acid | Sigma Aldrich | Cat# P7127 |
| Pyruvate | Sigma Aldrich | Cat# P2256 |
| Lactic Acid | Sigma Aldrich | Cat# L1750 |
| Citric Acid | Sigma Aldrich | Cat# C1909 |
| Isocitric Acid | ThermoFisher | Cat# 205010250 |
| α-Ketoglutarate | Sigma Aldrich | Cat# K2000 |
| Succinate | Sigma Aldrich | Cat# S2378 |
| Fumarate | Chem Cruz Biochemicals (Santa Cruz) | Cat# SC-215873 |
| Malate | Sigma Aldrich | Cat# 1613881 |
| Palmitate | Sigma Aldrich | Cat# P9767 |
| D-2-hydroxyglutarate (D-2-HG) | Toronto Research Chemicals | Cat# H942595 |
| Oxaloacetic Acid (OAA) | Sigma Aldrich | Cat# O7753 |
| Mono Sodium Citrate | Sigma Aldrich | Cat# 71497 |
| Di Sodium Citrate | Sigma Aldrich | Cat# 71635 |
| Tri Sodium Citrate | Sigma Aldrich | Cat# 54641 |
| EDTA-AM | AAT Bioquest, CA, USA | Cat# 19010 |
| UK5099 (Mitochondrial Pyruvate Carrier Inhibitor) | CalbioChem (MilliPore Sigma) | Cat# 5048170001 |
| Magnesium Chloride (MgCl2) | Sigma Aldrich | Cat# M1028 |
| Chloropentaammine Cobalt(III) Chloride (CPACC) | This Study | N/A |
| Hexaammine Cobalt(II) Chloride | This Study | N/A |
| Chloropentaammine Ruthenium(III) Chloride | This Study | N/A |
| Hexaammine Ruthenium(III) Chloride | This Study | N/A |
| Magnesium GreenTM Pentapotassium Salt | ThermoFisher Scientific | Cat# M3733 |
| Magnesium GreenTM, AM | ThermoFisher Scientific | Cat# M3735 |
| MitotrackerTM Deep Red FM | ThermoFisher Scientific | Cat# M22426 |
| Tetramethylrhodamine, Ethyl Ester, Perchlorate (TMRE) | ThermoFisher Scientific | Cat# T669 |
| BODIPYTM 493/503 | ThermoFisher Scientific | Cat# D3922 |
| FURA-FF Pentapotassium Salt | Cayman Chemical | Cat# 20415 |
| Calcium Green 5N | ThermoFisher Scientific | Cat# C3737 |
| JC-1 | Sigma Aldrich | Cat# 7176 |
| Digitonin | Sigma Aldrich | Cat# D141 |
| ATP Dipotassium Salt Hydrate | Sigma Aldrich | Cat# A8937 |
| ATP Magnesium Salt | Sigma Aldrich | Cat# A9187 |
| Calcium chloride | Sigma Aldrich | Cat# 2115 |
| Thapsigargin | Thermo Fisher Scientific | Cat# T7458 |
| Seahorse XF Calibrant Solution | Seahorse Bioscience (Agilent) | Cat# 100840-000 |
| Glucose | Sigma-Aldrich | Cat# G8644 |
| L-Glutamine, 200mM | Gibco (ThermoFisher Scientific) | Cat# 250030 |
| Sodium Pyruvate, 100mM | Gibco (ThermoFisher Scientific) | Cat# 11360 |
| HEPES (1M) | Gibco (ThermoFisher Scientific) | Cat# 15630 |
| Rotenone | Sigma-Aldrich | Cat# 557368 |
| Antimycin A | Sigma Aldrich | Cat# A8674 |
| Oligomycin | Sigma Aldrich | Cat# 75351 |
| FCCP | Sigma Aldrich | Cat# C2920 |
| Normal Horse Serum | Vector Laboratories | Cat# S-2000-20 |
| Fetal Bovine Serum, Characterized, Heat Inactivated | Hyclone (GE Life Sciences) UT, USA | Cat# SH30396.03 |
| Antibiotic-Antimycotic, 100X | Gibco (ThermoFisher Scientific) | Cat# 15240 |
| Saline (0.9% Sodium Chloride Injection USP) | Braun, PA, USA | Cat# L8002 |
| EGTA, OmniPur (powder) | MilliporeSigma | Cat# 4100-50GM |
| Collagenase (Type 4) | Worthington Biochemical Corp., NJ, USA | Cat# LS004188 |
|
| ||
| Critical commercial assays | ||
|
| ||
| PierceTM BCA Protein Assay Kit | ThermoFisher Scientific | Cat# 23225 |
| Citrate Assay Kit | Abcam | Cat# ab83396 |
| Alanine Transaminase Kit | Abcam | Cat# ab105134 |
| Trichloroacetic Acid Precipitation Kit | Abcam | Cat# ab204708 |
| RNeasy Plus Universal Mini Kit | QIAGEN, MD, USA | Cat# 73404 |
| Agilent Seahorse XF Cell Mito Stress Test Kit | Agilent Technologies, CA, USA | Cat# 103015-100 |
| Agilent Seahorse XFe96 FluxPak | Agilent Technologies | Cat# 102416-100 |
| VECTASTAIN Elite ABC-HRP Kit | Vector Laboratories | Cat# PK-6100 |
| Avidin/Biotin Blocking Kit | Vector Laboratories | Cat# SP02001 |
| LiquiColor Enzymatic Cholesterol Assay Kit | Stanbio, TX, USA | Cat# 1010225 |
|
| ||
| Deposited data | ||
|
| ||
| RNA-seq of liver and iWAT | This study | GEO: GSE216538 |
| from WT and Mrs2−/− mice | ||
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Human: HEK 293 Cell Line | ATCC | Cat# CRL-1573 |
| Human: HeLa Cell Line | ATCC | Cat# CCL-2 |
| Human: HepG2 Cell Line | ATCC | Cat# HB-8065 |
| Human: HK-2 Cell Line | ATCC | Cat# CRL-2190 |
| Cercopithecus aethiops: | ATCC | Cat# CRL-1651 |
| COS-7 Cell Line | ||
| Mouse: Primary Hepatocytes | This Study | N/A |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| C57BL/6J Mice | The Jackson Laboratory, USA | C57BL/6J |
| Mrs2 −/− (Mrs2 knockout) Mice | Daw et al.10 | Mrs2 KO |
|
| ||
| Oligonucleotides | ||
|
| ||
| Ucp1 (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm01244861_m1 |
| Cidea (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm00432554_m1 |
| Lep (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm00434759_m1 |
| Ldha (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm01612132_g1 |
| Ldhb (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm05874166_g1 |
| Pfkl (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm00435605_mH |
| Pgk1 (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm00435617_m1 |
| Hk1 (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm00439335_m1 |
| Actb (RT-qPCR Tagman Probe) | ThermoFisher Scientific | Mm01205647_g1 |
|
| ||
| Recombinant DNA | ||
|
| ||
| pCMV Citron (Cyto) | Addgene (Zhao et al.46) | Cat# 134303 |
| pCMV MitoCitron | Addgene (Zhao et al.46) | Cat# 134305 |
| pET-28a (+) | Novagen (Millipore Sigma) | Cat# 69864-3 |
| HA-HIF1α P402A/P564A/N803A-pcDNA3 | Addgene (Yan et al., 201760) | Cat# 87261 |
|
| ||
| Software and algorithms | ||
|
| ||
| Canvas 11.0 | ACD Systems | https://www.canvasgfx.com/en/support/canvas-11/ |
| GraphPad Prism version 8 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Leica Application Suite X (LAS X) | Leica Microsystems Inc., IL, USA | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| ImageJ | Schneider et al., 202161 | https://imagej.nih.gov/ij/ |
| Agilent Seahorse Wave Desktop Software (v2.6.1) | Agilent Technologies, CA, USA | https://www.agilent.com/en/products/cell-analysis/software-download-for-wave-desktop |
| Felix GX Software (v4) | PTI - Horiba, Canada | https://www.horiba.com/int/products/detail/action/show/Product/pti-felixgx-1652/ |
| Promethion Software | Sable Systems NV, USA | https://www.sablesys.com/products/promethion-line/promethion-software/ |
| R (v4.0.4) | R Core Team | https://www.R-project.org/ |
| Hisat2 (v2.0.5) | Kim et al.62; Kim et al.63; Pertea et al.64; Zhang et al.65 | http://daehwankimlab.github.io/hisat2/ |
| FeatureCounts (v1.5.0-p3) | Liao et al.66 | http://www.bioconductor.org/packages/release/bioc/html/Rsubread.html |
| DESeq2 (v1.20.0) | Love et al.67 | https://github.com/mikelove/DESeq2 |
| GSEA (Gene Set Enrichment Analysis) Tool (v4.2.2) | Subramanian et al.68; Mootha et al.69 | https://www.gsea-msigdb.org/gsea/index.jsp |
| ClusterProfile R Tool (v3.14) | Yu et al., 201270 | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| KEGG (Kyoto Encyclopedia of Genes and Genome) | Kanehisa and Goto34 | https://www.genome.jp/kegg |
| Dynamics Software (v7.8.1.3) | Wyatt Technology Corp., CA, USA | https://www.wyatt.com/products/software/dynamics.html |
| ChEA (ChIP-X Enrichment Analysis) software | Lachmann et al.37 | https://maayanlab.doud/Harmonizome/resource/ChIP-X+Enrichment+Analysis |
| BioVenn | Hulsen et al.71 | https://www.biovenn.nl/ |
| DeepVenn | Hulsen72 | https://www.deepvenn.com |
| SankeyMATIC Plot Generator | Steve Bogart (Open-Source Freeware) | https://sankeymatic.com |


