Abstract
The thymus is the crucial tissue where thymocytes develop from hematopoietic precursors that originate from the bone marrow and differentiate to generate a repertoire of mature T cells able to respond to foreign antigens while remaining tolerant to self-antigens. Until recently, most of the knowledge on thymus biology and its cellular and molecular complexity have been obtained through studies in animal models, because of the difficulty to gain access to thymic tissue in humans and the lack of in vitro models able to faithfully recapitulate the thymic microenvironment.
This review focuses on recent advances in the understanding of human thymus biology in health and disease obtained through the use of innovative experimental techniques (eg. single cell RNA sequencing, scRNAseq), diagnostic tools (eg. next generation sequencing), and in vitro models of T-cell differentiation (artificial thymic organoids) and thymus development (eg. thymic epithelial cell differentiation from embryonic stem cells or induced pluripotent stem cells).
Keywords: Thymus, T-cell development, Inborn errors of immunity, T-cell lymphopenia, Thymic development, Thymic stroma
1. Introduction
The thymus is the lymphoid organ specialized in T cell development; it contains cells of different embryonic origin that form a meshwork with a well-defined tissue architecture [1, 2]. During embryogenesis, the thymus primordium originates together with the parathyroid glands from the third endodermal pharyngeal pouches (PP), surrounded by neural crest cells (NCC) [3]. The main genetic regulatory network driving embryonic thymus development comprises a series of transcription factors including HOXA3, PAX1, PAX9, EYA1, SIX1 and TBX1 [4, 5]. These genes are regulated by signaling molecules released by the NCC, including retinoic acid (RA), proteins of the Wingless-int (WNT) family, bone morphogenic proteins (BMP), fibroblast growth factors (FGF) and sonic hedgehog (SHH) proteins (Reviewed in [6]). All these molecules are involved in driving the development of the thymic primordium, starting from the third PP, while HOXA3 and EYA1, and possibly CHD7, play also a role in the development of neural crest derived mesenchymal cells [7]. A recent report in mice has indicated the transcription factor Foxi3 as a new player in the pathway involved in thymus organogenesis [8]. FOXI3 is expressed in the third PP endoderm and mice FOXI3-null lack thymus and parathyroid glands. FOXI3 appears to act downstream of TBX1 and regulates PAX9, but it might interact also with HOXA3 [8]. All these first steps of thymus organogenesis are independent from FOXN1 expression, while the second phase of thymus development is FOXN1-dependent [9]. In this phase, FOXN1 induces the expression of several target genes that play a crucial role in thymocyte recruitment (eg. DLL4, CXCL12, CCL25) and thymic epithelial cell (TEC) maintenance and differentiation in cortical TEC (cTEC) and medullary TEC (mTEC) [10–13].
The main cellular component of the thymus are the developing thymocytes, which derive from bone marrow hematopoietic stem cells that continuously colonize the thymus, entering through blood vessels at the cortico-medullary junction [4, 5]. Other cell types of hematopoietic origin also reside in thymus and include dendritic cells (DC), natural killer cells (NK) and B cells, but these represent a minimal fraction of cells when compared to developing T cells. Non-hematopoietic stromal cells represent another important cell component of the thymus and are mainly represented by TECs, which are the cell type studied more in detail over the years. TECs can be divided in two main subsets, cortical (cTECs) and medullary TECs (mTECs), which share the same endodermal origin but play different roles and are located in distinct areas within the thymus [14]. In particular, cTECs are involved in the early phases of thymocyte development, which take place in the thymus cortex, and actively participate in the positive selection leading to the generation of CD4+ CD8+ double positive (DP) cells. The thymocytes then move to the medulla, where they complete their maturation to CD4+ or CD8+ single positive (SP) cells and undergo negative selection to eliminate self-reactive T cells. The mTECs play a crucial role in supporting the later stages of thymocyte development and in particular a specialized subset of mTECs mediate the negative selection process, through the expression of tissue restricted antigens (TRAs) induced by the transcriptional activator AutoImmune Regulator (AIRE). TRAs are presented to SP thymocytes; among these, SP cells with high affinity to self-antigens are eliminated or differentiate into regulatory T cells (Tregs), while thymocytes with low affinity to self-antigen survive and leave the thymus with a phenotype of naïve or recent thymic emigrant (RTE) cells to reach the peripheral tissues.
2. Recent advances in human thymus biology
The study of the human thymus has been hampered for many years not only because of the limited availability of this tissue, but also because of the lack of experimental techniques able to dissect the complexity of the thymus. Indeed, we know that the thymus is composed of many cell types; studies using flow cytometry, histology or bulk RNA sequencing (RNAseq) could only capture in part this complexity, leaving many questions unanswered. The more extensive knowledge on human hematopoietic cell markers previously allowed a more detailed dissection of these cells in human thymus, when compared to the stromal cell compartment. However, in the last few years, use of innovative techniques such as single cell RNA sequencing (scRNAseq) and spatial transcriptomics have provided a tremendous amount of new insights into the complexity of this tissue, both in hematopoietic and stromal cell compartments. Indeed, several studies [15–18] have indicated that the diversity and complexity of cTEC and mTEC subsets is much greater, although the function and spatial localization of the novel described TEC subsets has yet to be fully understood. Recently, there has also been a better appreciation of the importance of other non-epithelial stromal cell types, including endothelial cells, fibroblasts and other mesenchymal cells, in the development and function of the thymus [2, 4, 17, 19, 20]. All these cell types contribute to correct function of the thymus and their interactions, also called “lympho-stromal cross-talk” are crucial for the correct development and function of the thymus and for the generation of a diverse and self-tolerant repertoire of T lymphocytes.
Park and colleagues [16] generated a human thymus cell atlas using dissociated cells from 15 prenatal samples (ranging from 7 to 17 post conception weeks) and 9 post-natal tissues (ranging from 3 months to 35 years). Most of the cells in these samples were sorted using the hematopoietic cell markers CD45 or CD3, while only 3 samples were enriched using the epithelial cell marker EpCAM, prior to the single cell capture. In their dataset, the authors identified more than 40 different cell types or cell states, which expressed specific marker genes. In particular, they showed that cell states in the thymus dynamically change in terms of abundance and gene expression profiles through the development from fetal to post-natal life. The authors were able to establish computationally the trajectory of human T cell development from early progenitors to the different subsets of mature T cell types; using this trajectory, they built a list of putative transcription factors that guide T cell determination. Moreover, they identified a novel subset of unconventional T cells, among the CD8αα T cell subset, characterized by the expression of GNG4 and located in the peri-medullary region of the thymus. They also defined novel subpopulations of thymic fibroblasts and epithelial cells, and characterized their tissue location. Two different subsets of thymic fibroblasts (Fb) were identified in this study, expressing distinct gene sets and likely playing different roles: Fb1 cells express genes such as COLEC11 and ALDH1A2 and are postulated to play a role in sustaining epithelial cell growth, while Fb2 cells express extracellular matrix genes and semaphorins and are supposed to participate in vascular development. Finally, the authors identified several subsets of TECs. A previous study using scRNAseq in murine TECs by Bornstein and colleagues [15] had shown for the first time that the complexity of TECs had been greatly underestimated until then. This study also described the presence of 4 distinct sub-populations of mTECs. Using the annotations from Bornstein and colleagues’ murine dataset, Park and colleagues were able to discover the presence of conserved subsets of TECs across species, including PSMB11-positive cortical TEC (cTEC), KRT14-positive mTECI, AIRE-positive mTECII and KRT1-expressing mTECIII. They also distinguished a subset of mTEC expressing the markers POU2F3 and DCLK1, listed as the specific markers of the Tuft-like mTECIV subset newly identified in murine thymus [15, 21], although in the human thymus these markers are not uniquely expressed by Tuft-like mTECs. Finally, this study identified for the first time in humans clusters of TECs that were identified as neuroendocrine and myoid cells, respectively, based on their gene expression profile. This report provided for the first time evidence of the complexity of the stromal compartment in the human thymus; however, stromal cells represented only the minority of the single cell dataset, since the EpCAM+ cell enrichment was performed in few samples [16]. A subsequent study by Bautista and colleagues [17] aimed at dissecting in greater detail the cellular heterogeneity of the human stromal cell compartment. In this manuscript, the authors performed scRNAseq on enriched CD45-negative cells isolated after enzymatic digestion from 5 human thymic samples ranging from fetal to adult. They identified several stromal clusters, including epithelial, mesenchymal, pericytic, endothelial and mesothelial cell clusters. Gene expression analysis revealed that mesenchymal cell clusters expressed many ligands and regulators of WNT, BMP, Transforming Growth Factor beta (TGF-β), Insulin-like Growth Factor (IGF), and Fibroblast Growth Factor (FGF) signaling pathways, which have been all shown in mouse models as crucial in sustaining development and function of TECs, which express specific receptors that bind such factors [22–29]. Additionally, expression of some of the ligands, such as KGF, BMP4 and FRZB, in mesenchymal cells was increased in post-natal and adult samples, indicating a different role for these cells in supporting TEC development over-time. Pericytes and mesothelial cells were also found to express many ligands of the above-mentioned signaling pathways, while they were exclusively expressing genes such as INHBA (encoding for the subunits of Activin A) and the Activin antagonist follistatin (FST), respectively, both of which have been recently shown to play important roles in TEC differentiation and maintenance [23]. In summary, these data clearly show how these various non-epithelial stromal subsets play a complementary role in supporting human TEC development and function. On the other hand, endothelial cells were found to express extracellular matrix and adhesion molecules, such us fibronectin (FN1) and LGALS3, which could play a role in attracting and regulating migration of hematopoietic progenitors. In this report the authors were also able to explore in detail the complexity of the human TEC compartment and defined novel markers for some of the TEC subsets. Upon re-clustering of epithelial cells, they described two different clusters containing cells expressing cortical TEC (cTEC) markers, cTEChi and cTEClo, characterized by high and low levels of cTEC functional markers, respectively, and several other clusters expressing mTEC markers. In addition to the mTEC subsets already described by Park et al [16], such as CCL21+ mTECs, AIRE+ mTECs, KRT1+ mTECs, neuroendocrine, and myoid mTECs, the authors identified novel clusters of TECs, such as myelin-expressing TECs, ciliated TECs and ionocytes. CFTR+ ionocytes are particularly intriguing, since these cells had not been previously identified in the human thymus, whereas they are a well-known component of the lung epithelium, where they arise from basal cells and give rise to neuroendocrine and tuft-like cells. In the human thymus, all these cell subsets were shown to be located in close proximity in association with Hassall’s corpuscles; this could suggest the presence in the thymus of a common progenitors for these subsets. The authors also introduced a novel subset of TECs, the immature TECs, which express TEC identity genes but lack genes characteristic of cTECs and mTECs, and may represent TECs that have lost their differentiated phenotype. In fact, immature TECs were found especially enriched in the adult thymic sample, to the detriment of functional TECs. Characterization of this subset could be important to determine the genes and pathways playing a role in thymic involution. Another interesting hypothesis suggested by Bautista and colleagues is that in addition to AIRE+ mTECs, other TECs likely participate in tolerance induction by expressing tissue restricted antigens (TRA), which could then be presented by antigen presenting cells, such as DC. They propose that myoid cells could play a role in inducing tolerance to muscle antigens, since they found that genes encoding for the acetylcholine receptor (CHRNA1) and titin (TTN), which are associated to the neuromuscular autoimmune disease myasthenia gravis are more abundantly expressed by myoid, ciliated and neuroendocrine cells as compared to AIRE-mTEC. Figure 1 shows the new subsets of TECs described in this section and lists some of their representative markers.
Figure 1: Subsets of thymic epithelial cells described in human thymus and their representative markers.
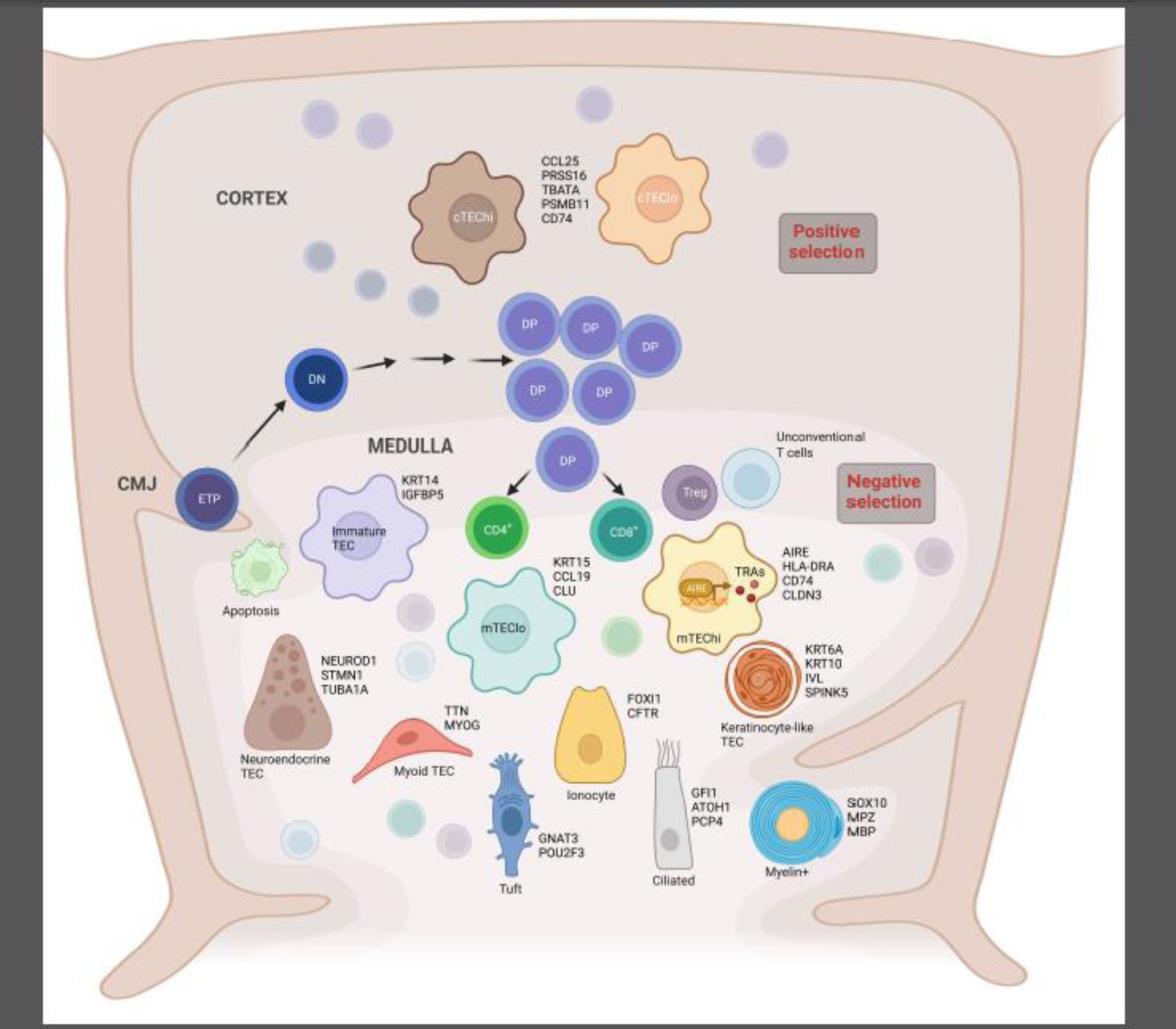
Simplified schematic representation of T cell development in human thymus, indicating the early stages of development from the entrance of bone marrow-derived early thymic progenitors (ETP) at the cortico-medullary junction (CMJ), through the different steps of their positive selection in the cortex, from CD4− CD8− double negative (DN) to CD4+ CD8+ double positive (DP). The thymocytes then move to the medulla where they complete their maturation, undergo negative selection to eliminate self-reactive T cell specificities and give rise to the different subsets of mature T cells: CD4+, CD8+, T regulatory cells (Treg) and unconventional T cells. Cortical thymic epithelial cell (cTEC) and medullary TEC (mTEC) subsets recently described in human thymus are represented here. For each epithelial cell subset, some of the specific markers are listed.
Of note, a report published by Campinoti and colleagues [30] has taken on the task of identifying and characterizing the still elusive epithelial stem/progenitor cells in human post-natal thymus. By culturing TEC obtained after enzymatic dissociation of human thymus, they were able to identify a subset of cells possessing the ability to expand extensively upon weekly passages. These cells could derive both from cortical and medullary TEC, were found to express high levels of CD49f and CD90, and presented a hybrid epithelial-mesenchymal phenotype. Most importantly, when these cells were combined with thymic interstitial cells in a rat thymic decellularized extracellular matrix structure, they were able to reconstitute a 3D structure reproducing the native thymus. The resulting scaffolds were also capable of supporting human T cell development from hematopoietic progenitors, both in vitro and in vivo. These hybrid epithelial-mesenchymal cells, acting as progenitor human post-natal stromal cells, could prove very useful for prospective applications aiming at thymus regeneration for the treatment of thymic defects.
Other interesting and novel observations on the human thymus where recently published in a human atlas integrating scRNAseq and spatial transcriptomic data from 9 immune organs collected pre-natally between 4 and 17 weeks post-conception [31]. This report confirmed that T cell progenitor cells are only found in fetal thymus, corroborating the notion that the thymus is absolutely necessary for the development of T cells and that, in conditions in which it is absent, there is complete T cell lymphopenia [32]. Another intriguing part of this study explored the origin of unconventional T cell subsets, which has not yet been fully clarified. Unconventional T cells were found to express the innate marker ZBTB16 (PLZF) and could be separated in 3 subsets: Type 3 innate T cells or Th17-like T cells (RORC+ and CCR6+), Type1 innate T cells or NKT cells (EOMES+ and TBX21+) and CD8αα T cells. Interestingly, conventional mature T cells spatially co-localized with mTECs in the inner medulla, while CD8αα T cells and Type 1 innate T cells co-localized with DC at the cortico-medullary junction. Regulatory T cells (Treg) and Type 3 innate T cells were found in both locations. CD8αα T cells and Type 1 innate T cells may thus undergo negative selection processes mediated by DC and not by mTECs, as is the case for conventional T cells. Additionally, the authors proposed that also the process of positive selection could be different for unconventional T cells. Indeed, by evaluating the TCR repertoire in single T cells, they observed that usage of V-J genes within the T cell receptor α (TRA) locus in unconventional T cells had a pattern that was intermediate between what detected in double positive (DP) and conventional T cells, suggesting that fewer recombination rounds occur in unconventional T cells before positive selection. These observations led the authors to hypothesize that unconventional T cells may originate after positive selection on neighboring DP T cells, as opposed to positive selection on cTECs as is the case for conventional T cells.
Importantly, all this extraordinary amount of novel knowledge on the complexity of the human thymus has been derived from studies performed on normal thymic samples. More limited data are available on thymic cell composition and spatial distribution in patients with defects of thymus development and function. This is because such defects are quite rare and, in most severe cases, no thymic tissue can be visualized and recovered. Moreover, ethical reasons limit availability of thymic tissues from patients in which the thymus is present, although reduced in size or not fully functional. Finally, post-mortem samples are often collected some time after death, and may not be adequate to perform studies such as scRNAseq. For these reasons, the only thymic tissues available from patients with thymic defects are limited to those in which the immunodeficiency is associated to a cardiac abnormality requiring cardio-thoracic surgery, eg. DiGeorge Syndrome (DGS), CHARGE Syndrome and Trisomy 21. In these cases, the thymus is removed in its entirety or partially at the time of surgery to gain access to the heart. While several studies have reported on abnormalities of thymus architecture in patients with these forms of immunodeficiencies [33–38], no reports of scRNAseq and/or spatial transcriptomics on thymic tissue obtained from patients carrying thymic defects have been published yet. Data obtained from these pathological samples will be extremely informative and crucial in providing additional insights into human thymus development and function.
3. Novel diagnostic tools to identify thymic defects
Limited availability of human thymic tissue from healthy individuals and from patients carrying genetic defects affecting thymus development and/or function represents a significant challenge to identify and characterize novel thymic defects. For these reasons, discovering novel genetic causes of thymic abnormalities has been extremely problematic, and many thymic defects remain undiagnosed even today. However, in the most recent years the increasing worldwide availability of newborn screening (NBS) for severe combined immunodeficiencies (SCID) using tests evaluating T Cell Receptor Excision Circles (TRECs) [39], has allowed the identification, very early in life, of patients with severe T-cell lymphopenia. Of note, in published reports, DGS and other thymic defects have been identified in about 1:20000–60000 infants with a positive TREC-based newborns screening assay; of these, complete athymia has been reported in about 5% of the cases [40–49]. However, the TREC assay does not identify all cases of 22q11.2del syndrome [50–52]. Next generation sequencing techniques, targeted to genes known to cause Inborn Errors of Immunity (IEI) or spanning across the whole exome (WES) or even the whole genome (WGS), have become more accessible to clinicians and affordable, allowing the discovery of a growing number of novel causes of thymus abnormalities (see Table 1). Several of these disorders are due to heterozygous gene and chromosomal defects, including 22q11.2del syndrome, TBX1 and FOXI3 deficiencies. In some cases, multiple inheritance patterns have been identified. For example, FOXN1 deficiency may occur as a fully penetrant autosomal recessive trait, or as a heterozygous condition with variable clinical penetrance. Importantly, thymus-intrinsic defects are often associated with multi-organ clinical manifestations. Finally, non-genetic causes can lead to abnormalities of thymic development, most notably poorly controlled maternal diabetes [53, 54].
Table 1:
Congenital Thymus Disorders
| Disease | Genetic defect | Inheritance | OMIM | Immune cells | Additional features |
|---|---|---|---|---|---|
| DiGeorge/velocardio-facial syndrome Chromosome 22q11.2 deletion syndrome (22q11.2DS) | Large deletion (1.5–3Mb) typically in chromosome 22 (TBX1) | AD | 602054 | T cells are decreased or normal 5% have low TRECs at NBS B cells are normal |
CHD Hypoparathyroidism Velopalatal insufficiency Facial dysmorphisms Intellectual disability [59, 85–92] |
| DiGeorge/velocardio-facial syndrome | Unknown | Sporadic | T cells are decreased or normal B cells are normal |
||
| TBX1 deficiency | TBX1 | AD | 602054 | T cells are decreased or normal In some cases low TRECs at NBS B cells are normal |
|
| TBX2 deficiency | TBX2 | AD | T cells are decreased or normal | CHD Craniofacial dysmorphisms Developmental defects Skeletal malformations Endocrine abnormalities [93, 94] |
|
| CHARGE syndrome |
CHD7
SEMA3E |
AD AD |
608892 608166 |
T cells are decreased or normal In some cases low TRECs at NBS B cells are normal |
CHD Coloboma of the eye Choanal atresia Intellectual disability Genital and ear anomalies [95–98] |
| Unknown | |||||
| Winged helix nude FOXN1 deficiency | FOXN1 | AR | 601705 | T cells are decreased B cells are normal |
Athymia Congenital alopecia Nail dystrophy [99–103] |
| FOXN1 haploinsufficiency | FOXN1 | AD | 600838 | Severe T cell lymphopenia at birth T cell numbers normalized in adults B cell numbers can be normal or low |
Recurrent respiratory tract infections Skin involvement (eczema, dermatitis) Nail dystrophy [55, 57] |
| Chromosome 10p13-p14 deletion syndrome (10p13-p14DS) | Del10p13-p14 | AD | 601362 | T cells are decreased B cells are normal |
CHD in some cases Hypoparathyroidism Renal disease Deafness Growth retardation Facial dysmorphisms [104–107] |
| Chromosome 11q deletion syndrome (Jacobsen syndrome) | 11q23del | AD | 147791 | T cells are decreased B cells are decreased Immunoglobulins and antibody responses are decreased NK cells are decreased |
Recurrent respiratory tract infections Multiple warts Facial dysmorphism Growth retardation [108] |
| Chromosome 2p11.2 microdeletion |
2p11.2del
(FOXI3) |
AD | T cells are decreased | Transient hypocalcemia Asymmetric crying face [58] | |
| FOXI3 haploinsufficiency | FOXI3 | AD | T cells are decreased | ||
| Immunoskeletal dysplasia with neurodevelopmental abnormalities (EXTL3 deficiency) | EXTL3 | AR | 617425 | T cells are decreased B cells are normal Eosinophilia |
Short stature Cervical spinal stenosis Neurodevelopmental defects [74, 109, 110] |
| Immunodeficiency with multiple intestinal atresias | TTC7A | AR | 609332 | Variable T cell numbers May have low TRECs at NBS B cell numbers are normal or low Hypogammaglobulinemia |
Recurrent infections Multiple intestinal atresias [111–114] |
| Otofaciocervical syndrome type 2 (OTFCS2) | PAX1 | AR | 615560 | Severe T cell lymphopenia Low TRECs B cells are normal |
Athymia Ear abnormalities Winged scapulae, abnormal clavicles [75, 76] |
| Autoimmune Polyendrocrinopathy with Candidiasis and Ectodermal Dystrophy (APECED, APS-1) | AIRE | AR or AD | 240300 | Normal T and B cell numbers | Multiple autoimmune manifestations Dental enamel hypoplasia Alopecia areata Enteropathy Pernicious anemia Chronic mucocutaneous candidiasis [115–117] |
AD: autosomal dominant; AR: autosomal recessive; CHD: congenital heart defect; NBS: newborn screening; TREC: T cell receptor excision circles
Recently, this approach allowed the identification of a novel cause of severe T-cell lymphopenia at birth caused by a defect in thymic stromal cells: FOXN1 haploinsufficiency [55, 56]. In particular, we and others have described a series of pediatric patients with marked T cell lymphopenia and low TREC levels at birth, who were found to carry heterozygous loss-of-function FOXN1 variants. FOXN1 is a master gene regulator of TEC function; bi-allelic loss-of-function variants in the FOXN1 gene lead to the nude/SCID phenotype, characterized by thymic aplasia, alopecia and nail dystrophy [57]. This is a severe condition that requires thymus implantation. By contrast, clinical and immunological abnormalities tend to improve spontaneously over-time in individuals with heterozygous loss-of-function FOXN1 variants, so that definitive therapeutic interventions are not required in the majority of these subjects. Three of the subjects included in our report underwent hematopoietic stem cell transplantation (HSCT) before being diagnosed with FOXN1 haploinsufficiency, but none of them had clinical benefit and one died from complications related to the transplant [55], consistent with the notion that FOXN1 haploinsufficiency causes a thymus stromal intrinsic defect that cannot be corrected by HSCT.
Using the same approach, a few years ago patients from 5 kindreds with low TRECs that presented overlapping microdeletions on chromosome 2p11.2 spanning the FOXI3 gene were described [58]. FOXI3 haploinsufficiency was postulated to be the cause of T-cell lymphopenia in these patients. FOXI3 is involved in the same pathway critical for thymus development in which also TBX1 and FOXN1 are key players, and patients with haploinsufficiency in all these genes share similar phenotypes [55, 56, 59, 60]. Additionally, heterozygous Foxi3-mutant mice show a smaller thymus [58]. We recently confirmed that FOXI3 haploinsufficiency is another cause of T-cell lymphopenia at birth [61]. We reported two unrelated subjects with low TREC levels at birth and T-cell lymphopenia, demonstrated heterozygous loss-of-function variants in the FOXI3 gene in both of them. We confirmed that the T-cell lymphopenia was not caused by an intrinsic defect of hematopoietic cells, by using an in vitro T-cell differentiation assay based on an artificial thymic organoid (ATO) platform [62, 63]. Indeed, CD34+ cells from the peripheral blood of one of the subjects carrying the FOXI3 variant were able to efficiently differentiate into mature T cells in vitro, with kinetics and absolute numbers comparable to those of CD34+ cells isolated from a normal control. T-cell lymphopenia in subjects carrying FOXI3 variants may improve over time in which case no definitive therapy is required); however, because of the low number of patients identified so far, the natural history of the disease and its severity remain to be fully defined.
This is an important example of how critical is for the timely and correct management of patients to understand whether subjects presenting with severe and persistent T cell lymphopenia and low TREC carry genetic defects affecting the hematopoietic cells or the thymic stromal cells. Discriminating the hematopoietic-intrinsic versus -extrinsic nature of the defect would indeed allow to choose the most effective treatment for the patient. In a T-cell lymphopenic infant with a heterozygous FOXN1 variant of uncertain significance, the ATO assay could help by indicating in a reasonably timely manner whether the T-cell lymphopenia is caused by a hematopoietic or a thymic defect. During the past years, many groups have worked on the development of efficient in vitro assays for T cell differentiation, taking advantage of the Notch ligand signaling, mediated by stromal cell engineered to express DLL4 or DLL1 or through binding of these ligands to cell culture plates [64, 65]. Recently Seet and colleagues [63] published a serum-free 3D artificial thymus organoid (ATO) system, generated by aggregating CD34+ cells with a murine stromal cell line expressing the Notch ligands DLL1 or DLL4, that could efficiently and reproducibly generate mature TCRαβ+ CD3+ T cells in less than two months. We and others [62, 66] tested the ATO system using CD34+ cells isolated from peripheral blood or bone marrow of patients carrying known mutations in genes causing T-cell lymphopenia of different severity and could establish that this system was able to reliably discriminate the hematopoietic-intrinsic or - extrinsic nature of the defect and to recapitulate the block in the T cell development in case of hematopoietic autonomous defects. Furthermore, the ATO system demonstrated that mouse models might not faithfully recapitulate the equivalent human conditions, as in the case of RAG deficiency. Indeed, in Rag-deficient mice, T cell development is blocked at double-negative 3 (DN3 stage), while CD34+ cells from RAG-deficient patients were able to differentiate up to double positive (DP) cells when cultured in the ATO system [62, 66]. Notably, the only case in which the ATO system could not reproduce the hematopoietic cell-autonomous block in T cell development was represented by adenosine deaminase (ADA) deficiency. CD34+ cells from these patients are able to differentiate into mature TCRαβ+ CD3+ cells in the ATO platform, likely because the stromal cell line included in this system produces ADA, allowing for cross-correction of the hematopoietic cell defect. We subsequently used the ATO system to confirm the hematopoietic or thymic stromal nature of diseases with novel genetic causes, such as POLD1 [67], SASH3 [68] and FOXI3 deficiency [61]. We believe that this system provides a powerful assay that can quickly guide decisions on clinical interventions in cases of infants with life-threatening severe T-cell lymphopenia of unknown genetic etiology.
4. Novel tools to model thymic epithelial cell development
Although the ATO system has proved to be very useful in determining whether the cause of T-cell lymphopenia might be hematopoietic or thymic stromal-based, this system can only be used to study the precise block in development in specific hematopoietic intrinsic defects. CD34+ cells of patients with thymic defects are able to efficiently differentiate into mature T cells when cultured in the ATO system, in which T-cell differentiation is supported by the murine stromal cell line expressing the Notch ligand and by the cytokines and supplements provided in the culture medium. Thus, in order to model defects affecting thymic stromal cells, alternative assays aimed at reproducing TEC development need to be generated. In the past years, knowledge on the signaling pathways driving thymus development has increased considerably and has led to the generation of many different protocols for the induction of TEC progenitor cells (TEPs) and TECs starting from embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs).
One of the first successful protocols was published by Lai and Jin [69], who were able to generate TEPs in vitro starting from murine ESCs using a combination of FGF7, FGF10, BMP4 and EGF. They cultured mESC cells in 3D and 2D conditions, and obtained the highest frequencies (about 25%) and absolute numbers of EpCAM+ cells in 2D cultures in the presence of all the factors. These cells expressed in large majority (between 68 and 82%) both cTEC and mTEC markers, such as Keratin 8 (K8) and Keratin 5 (K5), and importantly also showed expression of TEC genes Pax1, Pax9 and Foxn1, although at levels lower than those found in EpCAM+ cells isolated from mouse embryonic thymi. Purified mESC-derived EpCAM+ cells were able to differentiate into mature cTECs and mTECs when reaggregated with CD4−CD8− CD45+ thymocytes and transplanted in vivo under the kidney capsule of syngeneic mice. Grafts harvested 6 weeks after transplants showed evidence of thymopoietic activity, since they contained CD4 and CD8 DP and single positive (SP) cells. Interestingly, the authors showed that mESC-derived EpCAM+ cells, when injected in the thymus of lethally irradiated mice prior to bone marrow transplant, were capable of increasing thymocyte reconstitution and peripheral naïve T cell numbers.
A year later, another report from Inami and colleagues [70] published an improved protocol for the generation of TEPs starting from human ESCs and iPSCs. The most significant change they apported to the protocol of Lai and Jin was the addition of an initial step of differentiation of 4 days with Activin A and Lithium Chloride (LiCl) to induce definitive endoderm phenotype, prior to adding the factors used in the previous protocol (FGF7, FGF8, FGF10 and BMP4) for the induction of TEP. Finally, after 12 days of culture, they added an extra step of 4 days, involving the use of RANKL, and aimed at promoting further maturation of TEP. The addition of RANKL greatly increased the expression of the TEC genes Pax1, K5 and Foxn1 and also showed induction of low levels of Aire, indicating that the exposure of TEPs to RANKL induced their further maturation towards the mTEC lineage. Unfortunately, this study did not provide evidence for the ability of the TECs obtained through this protocol to regenerate a thymus microenvironment or to possess thymopoietic activity, in vitro or in vivo.
Another interesting approach to readily identify and select hESC-derived TEP was published a few years later by Soh and colleagues [71]; this approach entailed the generation of FOXN1-GFP reporter hESC lines, in order to readily identify FOXN1 expressing cells by using GFP. The authors were able to efficiently generate TECs from these hESC reporter lines by culturing them as embryoid bodies and adding Activin A for the first 4 days, and then FGF7 starting at day 14, and replenishing it once a week up to 35 days. Starting from day 12, differentiation cultures showed a progressive upregulation of pharyngeal pouch markers (HOXA3 and PAX9) and epithelial cell markers (IVL and FOXN1), while endodermal markers (SOX17 and FOXA2) were downregulated. However, functional assessment of FOXN1+ hESC-derived TEP, performed co-culturing them with ProT cells did not provide evidence of thymopoietic activity, suggesting that the TEPs obtained through this protocol may be functionally inadequate to promote T cell differentiation.
Great advancement in the protocols for the generation of TEPs were made in two reports published by Parent and colleagues [72] and Sun and colleagues [73]. They were able to develop multi-step protocols during which the expression of markers characteristic of the several stages of differentiation from hESC to TEPs, mimicking the in vivo thymus organogenesis, could be monitored over time.
Both protocols showed that the introduction of Retinoic Acid (RA), which was previously shown to be a key molecule in the early formation of pharyngeal pouches [6], was critical for the anteriorization of the definitive endoderm and the induction of the markers of the third Pharyngeal Pouch Endoderm (PPE), such as HOXA3, TBX1 and EYA1. Further directed differentiation from PPE to TEP was achieved by Sun and colleagues by using BMP4 and WNT3, while Parent and colleagues in addition to these factors also used the Hedgehog inhibitor Cyclopamine (CPM) and FGF8. Both protocols showed efficient generation of TEPs, which could further mature into functional TECs able to support T cell differentiation upon transplantation into athymic mice.
Importantly, the protocol developed by Parent and colleagues was shown to be efficacious in modeling the effects of mutations in two genes recently discovered to cause thymic stromal cell defects, EXTL3 and PAX1 [74, 75]. EXTL3 is a glycosyltransferase involved in the synthesis of heparan sulfate, and homozygous missense variants in this gene were demonstrated to cause T-cell lymphopenia, severe skeletal dysplasia, and developmental delay [74]. Volpi and colleagues generated iPSC from fibroblasts obtained from a patient carrying a homozygous variant in EXTL3 and evaluated the ability of these cells to generate TEP, in comparison to an iPSC line generated from a normal control. They could demonstrate that TEP differentiated from the EXTL3-mutated patient’s iPSC had a decreased expression of TEC-specific genes, such as FOXN1, K5 and EYA1, while retained higher expression of SOX17, a gene that in the control line reached a peak at the DE stage and was subsequently downregulated [74]. A similar analysis was performed in TEP differentiated from iPSC lines generated from patients carrying PAX1 mutations [75]. PAX1 is a transcription factor that plays a critical role during embryogenesis, as it is expressed in the pharyngeal pouches from which the thymus, tonsils, parathyroid glands, thyroid, and middle ear derive [9]. Mutations in this gene cause a rare syndrome called otofaciocervical syndrome type 2 (OTFCS2), characterized by facial dysmorphism, ear anomalies and hearing loss, skeletal malformations, mild intellectual disability and severe T-cell lymphopenia [75, 76]. Gene expression profile of TEPs obtained from PAX1-mutant patients showed that several genes were decreased in comparison to TEPs obtained from normal donors, including genes crucial for TEC development such as FOXN1, TP63 and BMP4, but also genes involved in skeletal, cartilage, pharyngeal, neural crest and ear development, that would account for the broad range of malformations presented by patients carrying PAX1 mutations [75].
Additional approaches to improve efficiency of TEP generation took advantage of proof-of-principle studies showing that Foxn1 over-expression could alone induce reprogramming of murine fibroblasts into functional TEC, able to support T cell differentiation both in vitro and in vivo [77] and that culture of ESCs with recombinant HOXA3 and FOXN1 would significantly enhance TEP induction [78]. Based on these evidence, Otsuka and colleagues [79] described a protocol of TEP differentiation using an iPSCs line engineered to constitutively express Foxn1 and demonstrated that Foxn1 expression enhanced the differentiation of cells expressing TEC markers, along with the up-regulation of the endogenous Foxn1 gene. At about the same time, a report from Chhatta and colleagues [80] showed that transduction of a lentiviral vector encoding for a codon-optimized Foxn1 gene in PPE cells obtained from human iPSCs, by using a protocol adapted from Parent and colleagues [72], could significantly improve the functionality of the TEP generated. Both reports showed that forced expression of Foxn1 could significantly enhance TEP ability to support T cell differentiation and induce tolerance [79, 80].
Further optimization of the protocols for TEP differentiation were recently published by Ramos and colleagues [81] and Gras-Peña and colleagues [82]. Both reports introduced the use of Sonic Hedgehog (SHH) activation during the step of pharyngeal endoderm induction, and this resulted in increased expression of PAX9, PAX1, and TBX1 genes. Interestingly, Gras-Peña and colleagues introduced in their protocol the use of Noggin, a BMP4 antagonist, between the pharyngeal endoderm and the TEP induction steps and prior to adding again BMP4. The introduction of Noggin increased significantly the expression of both FOXN1 and PAX9 in TEP at the end of the differentiation [82]. Remarkably, the cells obtained at the end of this protocol, expressed many thymic marker genes, including FOXN1, PAX1 and AIRE, at levels comparable to those found in fetal thymus, and when transplanted in vivo in immunodeficient mice, mixed with human thymic mesenchymal cells, could further mature and transiently support human T-cell development, but were not able to organize in a thymus-like structure [82]. In a crucial experiment performed by Ramos and colleagues, at the end of their TEP differentiation protocol, they reaggregated these cells and transplanted them in vivo in athymic nude mice. Fourteen to nineteen weeks after transplant, they harvested the thymic grafts and performed bulk and scRNAseq analyses. They could demonstrate that TEC differentiate in vivo from TEP, and that their transcription profile is similar to that of primary post-natal TEC. However, even with this improved protocol they could not retrieve clusters of more mature cTECs and mTECs, indicating that there is still room for improvement in TEP differentiation protocols. Nonetheless, scRNAseq analysis of the TECs isolated from the thymic grafts suggested a previously unrecognized role for NOTCH pathway in human TEC development, in addition to providing a list of target genes that could be important in human thymus development and could be exploited to further improve protocols for the generation of TEC in vitro.
A critical improvement in TEC generation could arise from the use of 3D culture systems. Indeed, it is known that the 3D structure is fundamental for the maintenance and functionality of TEC [83], and Zeleniak and colleagues [84] recently demonstrated how performing TEP generation from iPSC maintained in 3D alginate capsules led to increased expression of TEC markers. More importantly, when these cells were introduced in decellularized murine thymic scaffold, together with human CD34+ hematopoietic progenitor cells, they were able to mature further into both cTECs and mTECs and could support generation of mature CD4+ and CD8+ T cells, both in vitro and in vivo.
In summary, the results obtained on TEP differentiation over the years have shown that much progress has been made in generating cells that can now express TEC gene markers at levels comparable to primary human TEC, and that these cells can be used to recapitulate gene defects affecting TEC development. However, further improvement in the differentiation protocols is still needed in order to achieve maturation of these cells in vitro, able to recapitulate the complexity of mature cTEC and mTEC subsets that constitute the human thymus. Attaining this goal would not only allow to model in vitro defects of thymus development affecting later stages of TEC maturation (eg. AIRE deficiency) but could also generate TEC that could be used in in vitro T cell differentiation assays, such as the ATO system, instead of murine stromal cell lines, thus allowing the generation of thymic organoids reproducing more thoroughly the human thymus.
5. Conclusions and future prospective
The extensive new knowledge achieved in the recent years in molecular and cellular features of the human thymus and the consequent better understanding of the inter-cellular cross-talk among cell subsets, in terms of interactions and spatial localization, will provide critical information for the development of in vitro models able to faithfully recapitulate the human thymus microenvironment. It can be anticipated that this will allow not only a more efficient generation of T cells in vitro, but also provide better models to assess the impact of novel gene defects on thymus development and function. Ultimately, all the knowledge that would be achieved through these improved models will provide crucial novel insights into thymic development and function that could be exploited to develop future strategies of thymus engineering for the treatment of thymic defects.
Acknowledgements
This research was supported by the Division of Intramural Research Program of the National Institute of Allergy and Infectious diseases, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest disclosure
The authors declare no financial conflict of interest.
References
- [1].Rodewald HR, Thymus organogenesis, Annu Rev Immunol 26 (2008) 355–88. [DOI] [PubMed] [Google Scholar]
- [2].Manley NR, Richie ER, Blackburn CC, Condie BG, Sage J, Structure and function of the thymic microenvironment, Front Biosci (Landmark Ed) 16(7) (2011) 2461–77. [DOI] [PubMed] [Google Scholar]
- [3].Blackburn CC, Manley NR, Developing a new paradigm for thymus organogenesis, Nat Rev Immunol 4(4) (2004) 278–89. [DOI] [PubMed] [Google Scholar]
- [4].Gordon J, Manley NR, Mechanisms of thymus organogenesis and morphogenesis, Development 138(18) (2011) 3865–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Manley NR, Condie BG, Transcriptional regulation of thymus organogenesis and thymic epithelial cell differentiation, Prog Mol Biol Transl Sci 92 (2010) 103–20. [DOI] [PubMed] [Google Scholar]
- [6].Provin N, Giraud M, Differentiation of Pluripotent Stem Cells Into Thymic Epithelial Cells and Generation of Thymic Organoids: Applications for Therapeutic Strategies Against APECED, Front Immunol 13 (2022) 930963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Giardino G, Borzacchiello C, De Luca M, Romano R, Prencipe R, Cirillo E, Pignata C, T-Cell Immunodeficiencies With Congenital Alterations of Thymic Development: Genes Implicated and Differential Immunological and Clinical Features, Front Immunol 11 (2020) 1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hasten E, Morrow BE, Tbx1 and Foxi3 genetically interact in the pharyngeal pouch endoderm in a mouse model for 22q11.2 deletion syndrome, PLoS Genet 15(8) (2019) e1008301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Farley AM, Morris LX, Vroegindeweij E, Depreter ML, Vaidya H, Stenhouse FH, Tomlinson SR, Anderson RA, Cupedo T, Cornelissen JJ, Blackburn CC, Dynamics of thymus organogenesis and colonization in early human development, Development 140(9) (2013) 2015–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zuklys S, Handel A, Zhanybekova S, Govani F, Keller M, Maio S, Mayer CE, Teh HY, Hafen K, Gallone G, Barthlott T, Ponting CP, Hollander GA, Foxn1 regulates key target genes essential for T cell development in postnatal thymic epithelial cells, Nat Immunol 17(10) (2016) 1206–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nowell CS, Bredenkamp N, Tetelin S, Jin X, Tischner C, Vaidya H, Sheridan JM, Stenhouse FH, Heussen R, Smith AJ, Blackburn CC, Foxn1 regulates lineage progression in cortical and medullary thymic epithelial cells but is dispensable for medullary sublineage divergence, PLoS Genet 7(11) (2011) e1002348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chen L, Xiao S, Manley NR, Foxn1 is required to maintain the postnatal thymic microenvironment in a dosage-sensitive manner, Blood 113(3) (2009) 567–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nehls M, Kyewski B, Messerle M, Waldschutz R, Schuddekopf K, Smith AJ, Boehm T, Two genetically separable steps in the differentiation of thymic epithelium, Science 272(5263) (1996) 886–9. [DOI] [PubMed] [Google Scholar]
- [14].Abramson J, Anderson G, Thymic Epithelial Cells, Annu Rev Immunol 35 (2017) 85–118. [DOI] [PubMed] [Google Scholar]
- [15].Bornstein C, Nevo S, Giladi A, Kadouri N, Pouzolles M, Gerbe F, David E, Machado A, Chuprin A, Toth B, Goldberg O, Itzkovitz S, Taylor N, Jay P, Zimmermann VS, Abramson J, Amit I, Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells, Nature 559(7715) (2018) 622–626. [DOI] [PubMed] [Google Scholar]
- [16].Park JE, Botting RA, Dominguez Conde C, Popescu DM, Lavaert M, Kunz DJ, Goh I, Stephenson E, Ragazzini R, Tuck E, Wilbrey-Clark A, Roberts K, Kedlian VR, Ferdinand JR, He X, Webb S, Maunder D, Vandamme N, Mahbubani KT, Polanski K, Mamanova L, Bolt L, Crossland D, de Rita F, Fuller A, Filby A, Reynolds G, Dixon D, Saeb-Parsy K, Lisgo S, Henderson D, Vento-Tormo R, Bayraktar OA, Barker RA, Meyer KB, Saeys Y, Bonfanti P, Behjati S, Clatworthy MR, Taghon T, Haniffa M, Teichmann SA, A cell atlas of human thymic development defines T cell repertoire formation, Science 367(6480) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bautista JL, Cramer NT, Miller CN, Chavez J, Berrios DI, Byrnes LE, Germino J, Ntranos V, Sneddon JB, Burt TD, Gardner JM, Ye CJ, Anderson MS, Parent AV, Single-cell transcriptional profiling of human thymic stroma uncovers novel cellular heterogeneity in the thymic medulla, Nat Commun 12(1) (2021) 1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Michelson DA, Hase K, Kaisho T, Benoist C, Mathis D, Thymic epithelial cells co-opt lineage-defining transcription factors to eliminate autoreactive T cells, Cell 185(14) (2022) 2542–2558 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Foster K, Sheridan J, Veiga-Fernandes H, Roderick K, Pachnis V, Adams R, Blackburn C, Kioussis D, Coles M, Contribution of neural crest-derived cells in the embryonic and adult thymus, J Immunol 180(5) (2008) 3183–9. [DOI] [PubMed] [Google Scholar]
- [20].Nitta T, Takayanagi H, Non-Epithelial Thymic Stromal Cells: Unsung Heroes in Thymus Organogenesis and T Cell Development, Front Immunol 11 (2020) 620894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Miller CN, Proekt I, von Moltke J, Wells KL, Rajpurkar AR, Wang H, Rattay K, Khan IS, Metzger TC, Pollack JL, Fries AC, Lwin WW, Wigton EJ, Parent AV, Kyewski B, Erle DJ, Hogquist KA, Steinmetz LM, Locksley RM, Anderson MS, Thymic tuft cells promote an IL-4-enriched medulla and shape thymocyte development, Nature 559(7715) (2018) 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Barsanti M, Lim JM, Hun ML, Lister N, Wong K, Hammett MV, Lepletier A, Boyd RL, Giudice A, Chidgey AP, A novel Foxn1(eGFP/+) mouse model identifies Bmp4-induced maintenance of Foxn1 expression and thymic epithelial progenitor populations, Eur J Immunol 47(2) (2017) 291–304. [DOI] [PubMed] [Google Scholar]
- [23].Lepletier A, Hun ML, Hammett MV, Wong K, Naeem H, Hedger M, Loveland K, Chidgey AP, Interplay between Follistatin, Activin A, and BMP4 Signaling Regulates Postnatal Thymic Epithelial Progenitor Cell Differentiation during Aging, Cell Rep 27(13) (2019) 3887–3901 e4. [DOI] [PubMed] [Google Scholar]
- [24].Balciunaite G, Keller MP, Balciunaite E, Piali L, Zuklys S, Mathieu YD, Gill J, Boyd R, Sussman DJ, Hollander GA, Wnt glycoproteins regulate the expression of FoxN1, the gene defective in nude mice, Nat Immunol 3(11) (2002) 1102–8. [DOI] [PubMed] [Google Scholar]
- [25].Gordon J, Patel SR, Mishina Y, Manley NR, Evidence for an early role for BMP4 signaling in thymus and parathyroid morphogenesis, Dev Biol 339(1) (2010) 141–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Patel SR, Gordon J, Mahbub F, Blackburn CC, Manley NR, Bmp4 and Noggin expression during early thymus and parathyroid organogenesis, Gene Expr Patterns 6(8) (2006) 794–9. [DOI] [PubMed] [Google Scholar]
- [27].Bleul CC, Boehm T, BMP signaling is required for normal thymus development, J Immunol 175(8) (2005) 5213–21. [DOI] [PubMed] [Google Scholar]
- [28].Swann JB, Krauth B, Happe C, Boehm T, Cooperative interaction of BMP signalling and Foxn1 gene dosage determines the size of the functionally active thymic epithelial compartment, Sci Rep 7(1) (2017) 8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tsai PT, Lee RA, Wu H, BMP4 acts upstream of FGF in modulating thymic stroma and regulating thymopoiesis, Blood 102(12) (2003) 3947–53. [DOI] [PubMed] [Google Scholar]
- [30].Campinoti S, Gjinovci A, Ragazzini R, Zanieri L, Ariza-McNaughton L, Catucci M, Boeing S, Park JE, Hutchinson JC, Munoz-Ruiz M, Manti PG, Vozza G, Villa CE, Phylactopoulos DE, Maurer C, Testa G, Stauss HJ, Teichmann SA, Sebire NJ, Hayday AC, Bonnet D, Bonfanti P, Reconstitution of a functional human thymus by postnatal stromal progenitor cells and natural whole-organ scaffolds, Nat Commun 11(1) (2020) 6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Suo C, Dann E, Goh I, Jardine L, Kleshchevnikov V, Park JE, Botting RA, Stephenson E, Engelbert J, Tuong ZK, Polanski K, Yayon N, Xu C, Suchanek O, Elmentaite R, Dominguez Conde C, He P, Pritchard S, Miah M, Moldovan C, Steemers AS, Mazin P, Prete M, Horsfall D, Marioni JC, Clatworthy MR, Haniffa M, Teichmann SA, Mapping the developing human immune system across organs, Science 376(6597) (2022) eabo0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Collins C, Sharpe E, Silber A, Kulke S, Hsieh EWY, Congenital Athymia: Genetic Etiologies, Clinical Manifestations, Diagnosis, and Treatment, J Clin Immunol 41(5) (2021) 881–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Marcovecchio GE, Bortolomai I, Ferrua F, Fontana E, Imberti L, Conforti E, Amodio D, Bergante S, Macchiarulo G, D’Oria V, Conti F, Di Cesare S, Fousteri G, Carotti A, Giamberti A, Poliani PL, Notarangelo LD, Cancrini C, Villa A, Bosticardo M, Thymic Epithelium Abnormalities in DiGeorge and Down Syndrome Patients Contribute to Dysregulation in T Cell Development, Front Immunol 10 (2019) 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Marcovecchio GE, Ferrua F, Fontana E, Beretta S, Genua M, Bortolomai I, Conti A, Montin D, Cascarano MT, Bergante S, D’Oria V, Giamberti A, Amodio D, Cancrini C, Carotti A, Di Micco R, Merelli I, Bosticardo M, Villa A, Premature Senescence and Increased Oxidative Stress in the Thymus of Down Syndrome Patients, Front Immunol 12 (2021) 669893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Levin S, Schlesinger M, Handzel Z, Hahn T, Altman Y, Czernobilsky B, Boss J, Thymic deficiency in Down’s syndrome, Pediatrics 63(1) (1979) 80–7. [PubMed] [Google Scholar]
- [36].Larocca LM, Lauriola L, Ranelletti FO, Piantelli M, Maggiano N, Ricci R, Capelli A, Morphological and immunohistochemical study of Down syndrome thymus, Am J Med Genet Suppl 7 (1990) 225–30. [DOI] [PubMed] [Google Scholar]
- [37].Lima FA, Moreira-Filho CA, Ramos PL, Brentani H, Lima Lde A, Arrais M, Bento-de-Souza LC, Bento-de-Souza L, Duarte MI, Coutinho A, Carneiro-Sampaio M, Decreased AIRE expression and global thymic hypofunction in Down syndrome, J Immunol 187(6) (2011) 3422–30. [DOI] [PubMed] [Google Scholar]
- [38].Skogberg G, Lundberg V, Lindgren S, Gudmundsdottir J, Sandstrom K, Kampe O, Anneren G, Gustafsson J, Sunnegardh J, van der Post S, Telemo E, Berglund M, Ekwall O, Altered expression of autoimmune regulator in infant down syndrome thymus, a possible contributor to an autoimmune phenotype, J Immunol 193(5) (2014) 2187–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chan K, Puck JM, Development of population-based newborn screening for severe combined immunodeficiency, J Allergy Clin Immunol 115(2) (2005) 391–8. [DOI] [PubMed] [Google Scholar]
- [40].Chien YH, Chiang SC, Chang KL, Yu HH, Lee WI, Tsai LP, Hsu LW, Hu MH, Hwu WL, Incidence of severe combined immunodeficiency through newborn screening in a Chinese population, J Formos Med Assoc 114(1) (2015) 12–6. [DOI] [PubMed] [Google Scholar]
- [41].Argudo-Ramirez A, Martin-Nalda A, Gonzalez de Aledo-Castillo JM, Lopez-Galera R, Marin-Soria JL, Pajares-Garcia S, Martinez-Gallo M, Garcia-Prat M, Colobran R, Riviere JG, Quintero Y, Collado T, Ribes A, Garcia-Villoria J, Soler-Palacin P, Newborn Screening for SCID. Experience in Spain (Catalonia), Int J Neonatal Screen 7(3) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Vogel BH, Bonagura V, Weinberg GA, Ballow M, Isabelle J, DiAntonio L, Parker A, Young A, Cunningham-Rundles C, Fong CT, Celestin J, Lehman H, Rubinstein A, Siegel S, Weiner L, Saavedra-Matiz C, Kay DM, Caggana M, Newborn screening for SCID in New York State: experience from the first two years, J Clin Immunol 34(3) (2014) 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kwan A, Abraham RS, Currier R, Brower A, Andruszewski K, Abbott JK, Baker M, Ballow M, Bartoshesky LE, Bonilla FA, Brokopp C, Brooks E, Caggana M, Celestin J, Church JA, Comeau AM, Connelly JA, Cowan MJ, Cunningham-Rundles C, Dasu T, Dave N, De La Morena MT, Duffner U, Fong CT, Forbes L, Freedenberg D, Gelfand EW, Hale JE, Hanson IC, Hay BN, Hu D, Infante A, Johnson D, Kapoor N, Kay DM, Kohn DB, Lee R, Lehman H, Lin Z, Lorey F, Abdel-Mageed A, Manning A, McGhee S, Moore TB, Naides SJ, Notarangelo LD, Orange JS, Pai SY, Porteus M, Rodriguez R, Romberg N, Routes J, Ruehle M, Rubenstein A, Saavedra-Matiz CA, Scott G, Scott PM, Secord E, Seroogy C, Shearer WT, Siegel S, Silvers SK, Stiehm ER, Sugerman RW, Sullivan JL, Tanksley S, Tierce M.L.t., Verbsky J, Vogel B, Walker R, Walkovich K, Walter JE, Wasserman RL, Watson MS, Weinberg GA, Weiner LB, Wood H, Yates AB, Puck JM, Bonagura VR, Newborn screening for severe combined immunodeficiency in 11 screening programs in the United States, JAMA 312(7) (2014) 729–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Barbaro M, Ohlsson A, Borte S, Jonsson S, Zetterstrom RH, King J, Winiarski J, von Dobeln U, Hammarstrom L, Newborn Screening for Severe Primary Immunodeficiency Diseases in Sweden-a 2-Year Pilot TREC and KREC Screening Study, J Clin Immunol 37(1) (2017) 51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rechavi E, Lev A, Simon AJ, Stauber T, Daas S, Saraf-Levy T, Broides A, Nahum A, Marcus N, Hanna S, Stepensky P, Toker O, Dalal I, Etzioni A, Almashanu S, Somech R, First Year of Israeli Newborn Screening for Severe Combined Immunodeficiency-Clinical Achievements and Insights, Front Immunol 8 (2017) 1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Liao HC, Liao CH, Kao SM, Chiang CC, Chen YJ, Detecting 22q11.2 Deletion Syndrome in Newborns with Low T Cell Receptor Excision Circles from Severe Combined Immunodeficiency Screening, J Pediatr 204 (2019) 219–224 e1. [DOI] [PubMed] [Google Scholar]
- [47].Amatuni GS, Currier RJ, Church JA, Bishop T, Grimbacher E, Nguyen AA, Agarwal-Hashmi R, Aznar CP, Butte MJ, Cowan MJ, Dorsey MJ, Dvorak CC, Kapoor N, Kohn DB, Markert ML, Moore TB, Naides SJ, Sciortino S, Feuchtbaum L, Koupaei RA, Puck JM, Newborn Screening for Severe Combined Immunodeficiency and T-cell Lymphopenia in California, 2010–2017, Pediatrics 143(2) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hale JE, Platt CD, Bonilla FA, Hay BN, Sullivan JL, Johnston AM, Pasternack MS, Hesterberg PE, Meissner HC, Cooper ER, Barmettler S, Farmer JR, Fisher D, Walter JE, Yang NJ, Sahai I, Eaton RB, DeMaria A, Notarangelo LD, Pai SY, Comeau AM, Ten Years of Newborn Screening for Severe Combined Immunodeficiency (SCID) in Massachusetts, J Allergy Clin Immunol Pract 9(5) (2021) 2060–2067 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wakamatsu M, Kojima D, Muramatsu H, Okuno Y, Kataoka S, Nakamura F, Sakai Y, Tsuge I, Ito T, Ueda K, Saito A, Morihana E, Ito Y, Ohashi N, Tanaka M, Tanaka T, Kojima S, Nakajima Y, Ito T, Takahashi Y, TREC/KREC Newborn Screening followed by Next-Generation Sequencing for Severe Combined Immunodeficiency in Japan, J Clin Immunol 42(8) (2022) 1696–1707. [DOI] [PubMed] [Google Scholar]
- [50].Lingman Framme J, Borte S, von Dobeln U, Hammarstrom L, Oskarsdottir S, Retrospective analysis of TREC based newborn screening results and clinical phenotypes in infants with the 22q11 deletion syndrome, J Clin Immunol 34(4) (2014) 514–9. [DOI] [PubMed] [Google Scholar]
- [51].Fronkova E, Klocperk A, Svaton M, Novakova M, Kotrova M, Kayserova J, Kalina T, Keslova P, Votava F, Vinohradska H, Freiberger T, Mejstrikova E, Trka J, Sediva A, The TREC/KREC assay for the diagnosis and monitoring of patients with DiGeorge syndrome, PLoS One 9(12) (2014) e114514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Barry JC, Crowley TB, Jyonouchi S, Heimall J, Zackai EH, Sullivan KE, McDonald-McGinn DM, Identification of 22q11.2 Deletion Syndrome via Newborn Screening for Severe Combined Immunodeficiency, J Clin Immunol 37(5) (2017) 476–485. [DOI] [PubMed] [Google Scholar]
- [53].Novak RW, Robinson HB, Coincident DiGeorge anomaly and renal agenesis and its relation to maternal diabetes, Am J Med Genet 50(4) (1994) 311–2. [DOI] [PubMed] [Google Scholar]
- [54].Digilio MC, Marino B, Formigari R, Giannotti A, Maternal diabetes causing DiGeorge anomaly and renal agenesis, Am J Med Genet 55(4) (1995) 513–4. [DOI] [PubMed] [Google Scholar]
- [55].Bosticardo M, Yamazaki Y, Cowan J, Giardino G, Corsino C, Scalia G, Prencipe R, Ruffner M, Hill DA, Sakovich I, Yemialyanava I, Tam JS, Padem N, Elder ME, Sleasman JW, Perez E, Niebur H, Seroogy CM, Sharapova S, Gebbia J, Kleiner GI, Peake J, Abbott JK, Gelfand EW, Crestani E, Biggs C, Butte MJ, Hartog N, Hayward A, Chen K, Heimall J, Seeborg F, Bartnikas LM, Cooper MA, Pignata C, Bhandoola A, Notarangelo LD, Heterozygous FOXN1 Variants Cause Low TRECs and Severe T Cell Lymphopenia, Revealing a Crucial Role of FOXN1 in Supporting Early Thymopoiesis, Am J Hum Genet 105(3) (2019) 549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Du Q, Huynh LK, Coskun F, Molina E, King MA, Raj P, Khan S, Dozmorov I, Seroogy CM, Wysocki CA, Padron GT, Yates TR, Markert ML, de la Morena MT, van Oers NS, FOXN1 compound heterozygous mutations cause selective thymic hypoplasia in humans, J Clin Invest 129(11) (2019) 4724–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Giardino G, Sharapova SO, Ciznar P, Dhalla F, Maragliano L, Radha Rama Devi A, Islamoglu C, Ikinciogullari A, Haskologlu S, Dogu F, Hanna-Wakim R, Dbaibo G, Chou J, Cirillo E, Borzacchiello C, Kreins AY, Worth A, Rota IA, Marques JG, Sayitoglu M, Firtina S, Mahdi M, Geha R, Neven B, Sousa AE, Benfenati F, Hollander GA, Davies EG, Pignata C, Expanding the Nude SCID/CID Phenotype Associated with FOXN1 Homozygous, Compound Heterozygous, or Heterozygous Mutations, J Clin Immunol 41(4) (2021) 756–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bernstock JD, Totten AH, Elkahloun AG, Johnson KR, Hurst AC, Goldman F, Groves AK, Mikhail FM, Atkinson TP, Recurrent microdeletions at chromosome 2p11.2 are associated with thymic hypoplasia and features resembling DiGeorge syndrome, J Allergy Clin Immunol 145(1) (2020) 358–367 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Paylor R, Glaser B, Mupo A, Ataliotis P, Spencer C, Sobotka A, Sparks C, Choi CH, Oghalai J, Curran S, Murphy KC, Monks S, Williams N, O’Donovan MC, Owen MJ, Scambler PJ, Lindsay E, Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome, Proc Natl Acad Sci U S A 103(20) (2006) 7729–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zweier C, Sticht H, Aydin-Yaylagul I, Campbell CE, Rauch A, Human TBX1 missense mutations cause gain of function resulting in the same phenotype as 22q11.2 deletions, Am J Hum Genet 80(3) (2007) 510–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Ghosh R, Bosticardo M, Singh S, Similuk M, Delmonte OM, Pala F, Peng C, Jodarski C, Keller MD, Chinn IK, Groves AK, Notarangelo LD, Walkiewicz MA, Chinen J, Bundy V, FOXI3 haploinsufficiency contributes to low T-cell receptor excision circles and T-cell lymphopenia, J Allergy Clin Immunol (2022). [DOI] [PMC free article] [PubMed]
- [62].Bosticardo M, Pala F, Calzoni E, Delmonte OM, Dobbs K, Gardner CL, Sacchetti N, Kawai T, Garabedian EK, Draper D, Bergerson JRE, DeRavin SS, Freeman AF, Gungor T, Hartog N, Holland SM, Kohn DB, Malech HL, Markert ML, Weinacht KG, Villa A, Seet CS, Montel-Hagen A, Crooks GM, Notarangelo LD, Artificial thymic organoids represent a reliable tool to study T-cell differentiation in patients with severe T-cell lymphopenia, Blood Adv 4(12) (2020) 2611–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Seet CS, He C, Bethune MT, Li S, Chick B, Gschweng EH, Zhu Y, Kim K, Kohn DB, Baltimore D, Crooks GM, Montel-Hagen A, Generation of mature T cells from human hematopoietic stem and progenitor cells in artificial thymic organoids, Nat Methods 14(5) (2017) 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Schmitt TM, Zuniga-Pflucker JC, Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro, Immunity 17(6) (2002) 749–56. [DOI] [PubMed] [Google Scholar]
- [65].Simons L, Ma K, de Chappedelaine C, Moiranghtem RD, Elkaim E, Olivre J, Susini S, Appourchaux K, Reimann C, Sadek H, Pelle O, Cagnard N, Magrin E, Lagresle-Peyrou C, Taghon T, Rausell A, Cavazzana M, Andre-Schmutz I, Generation of adult human T-cell progenitors for immunotherapeutic applications, J Allergy Clin Immunol 141(4) (2018) 1491–1494 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Bifsha P, Leiding JW, Pai SY, Colamartino ABL, Hartog N, Church JA, Oshrine BR, Puck JM, Markert ML, Haddad E, Diagnostic assay to assist clinical decisions for unclassified severe combined immune deficiency, Blood Adv 4(12) (2020) 2606–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Nichols-Vinueza DX, Delmonte OM, Bundy V, Bosticardo M, Zimmermann MT, Dsouza NR, Pala F, Dobbs K, Stoddard J, Niemela JE, Kuehn HS, Keller MD, Rueda CM, Abraham RS, Urrutia R, Rosenzweig SD, Notarangelo LD, POLD1 Deficiency Reveals a Role for POLD1 in DNA Repair and T and B Cell Development, J Clin Immunol 41(1) (2021) 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Delmonte OM, Bergerson JRE, Kawai T, Kuehn HS, McDermott DH, Cortese I, Zimmermann MT, Dobbs AK, Bosticardo M, Fink D, Majumdar S, Palterer B, Pala F, Dsouza NR, Pouzolles M, Taylor N, Calvo KR, Daley SR, Velez D, Agharahimi A, Myint-Hpu K, Dropulic LK, Lyons JJ, Holland SM, Freeman AF, Ghosh R, Similuk MB, Niemela JE, Stoddard J, Kuhns DB, Urrutia R, Rosenzweig SD, Walkiewicz MA, Murphy PM, Notarangelo LD, SASH3 variants cause a novel form of X-linked combined immunodeficiency with immune dysregulation, Blood 138(12) (2021) 1019–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lai L, Jin J, Generation of thymic epithelial cell progenitors by mouse embryonic stem cells, Stem Cells 27(12) (2009) 3012–20. [DOI] [PubMed] [Google Scholar]
- [70].Inami Y, Yoshikai T, Ito S, Nishio N, Suzuki H, Sakurai H, Isobe K, Differentiation of induced pluripotent stem cells to thymic epithelial cells by phenotype, Immunol Cell Biol 89(2) (2011) 314–21. [DOI] [PubMed] [Google Scholar]
- [71].Soh CL, Giudice A, Jenny RA, Elliott DA, Hatzistavrou T, Micallef SJ, Kianizad K, Seach N, Zuniga-Pflucker JC, Chidgey AP, Trounson A, Nilsson SK, Haylock DN, Boyd RL, Elefanty AG, Stanley EG, FOXN1 (GFP/w) reporter hESCs enable identification of integrin-beta4, HLA-DR, and EpCAM as markers of human PSC-derived FOXN1(+) thymic epithelial progenitors, Stem Cell Reports 2(6) (2014) 925–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Parent AV, Russ HA, Khan IS, LaFlam TN, Metzger TC, Anderson MS, Hebrok M, Generation of functional thymic epithelium from human embryonic stem cells that supports host T cell development, Cell Stem Cell 13(2) (2013) 219–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sun X, Xu J, Lu H, Liu W, Miao Z, Sui X, Liu H, Su L, Du W, He Q, Chen F, Shi Y, Deng H, Directed differentiation of human embryonic stem cells into thymic epithelial progenitor-like cells reconstitutes the thymic microenvironment in vivo, Cell Stem Cell 13(2) (2013) 230–6. [DOI] [PubMed] [Google Scholar]
- [74].Volpi S, Yamazaki Y, Brauer PM, van Rooijen E, Hayashida A, Slavotinek A, Sun Kuehn H, Di Rocco M, Rivolta C, Bortolomai I, Du L, Felgentreff K, Ott de Bruin L, Hayashida K, Freedman G, Marcovecchio GE, Capuder K, Rath P, Luche N, Hagedorn EJ, Buoncompagni A, Royer-Bertrand B, Giliani S, Poliani PL, Imberti L, Dobbs K, Poulain FE, Martini A, Manis J, Linhardt RJ, Bosticardo M, Rosenzweig SD, Lee H, Puck JM, Zuniga-Pflucker JC, Zon L, Park PW, Superti-Furga A, Notarangelo LD, EXTL3 mutations cause skeletal dysplasia, immune deficiency, and developmental delay, J Exp Med 214(3) (2017) 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Yamazaki Y, Urrutia R, Franco LM, Giliani S, Zhang K, Alazami AM, Dobbs AK, Masneri S, Joshi A, Otaizo-Carrasquero F, Myers TG, Ganesan S, Bondioni MP, Ho ML, Marks C, Alajlan H, Mohammed RW, Zou F, Valencia CA, Filipovich AH, Facchetti F, Boisson B, Azzari C, Al-Saud BK, Al-Mousa H, Casanova JL, Abraham RS, Notarangelo LD, PAX1 is essential for development and function of the human thymus, Sci Immunol 5(44) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Paganini I, Sestini R, Capone GL, Putignano AL, Contini E, Giotti I, Gensini F, Marozza A, Barilaro A, Porfirio B, Papi L, A novel PAX1 null homozygous mutation in autosomal recessive otofaciocervical syndrome associated with severe combined immunodeficiency, Clin Genet 92(6) (2017) 664–668. [DOI] [PubMed] [Google Scholar]
- [77].Bredenkamp N, Ulyanchenko S, O’Neill KE, Manley NR, Vaidya HJ, Blackburn CC, An organized and functional thymus generated from FOXN1-reprogrammed fibroblasts, Nat Cell Biol 16(9) (2014) 902–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Su M, Hu R, Jin J, Yan Y, Song Y, Sullivan R, Lai L, Efficient in vitro generation of functional thymic epithelial progenitors from human embryonic stem cells, Sci Rep 5 (2015) 9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Otsuka R, Wada H, Tsuji H, Sasaki A, Murata T, Itoh M, Baghdadi M, Seino KI, Efficient generation of thymic epithelium from induced pluripotent stem cells that prolongs allograft survival, Sci Rep 10(1) (2020) 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Chhatta AR, Cordes M, Hanegraaf MAJ, Vloemans S, Cupedo T, Cornelissen JJ, Carlotti F, Salvatori D, Pike-Overzet K, Fibbe WE, Hoeben RC, Mikkers HMM, Staal FJT, De novo generation of a functional human thymus from induced pluripotent stem cells, J Allergy Clin Immunol 144(5) (2019) 1416–1419 e7. [DOI] [PubMed] [Google Scholar]
- [81].Ramos SA, Morton JJ, Yadav P, Reed B, Alizadeh SI, Shilleh AH, Perrenoud L, Jaggers J, Kappler J, Jimeno A, Russ HA, Generation of functional human thymic cells from induced pluripotent stem cells, J Allergy Clin Immunol 149(2) (2022) 767–781 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Gras-Pena R, Danzl NM, Khosravi-Maharlooei M, Campbell SR, Ruiz AE, Parks CA, Suen Savage WM, Holzl MA, Chatterjee D, Sykes M, Human stem cell-derived thymic epithelial cells enhance human T-cell development in a xenogeneic thymus, J Allergy Clin Immunol 149(5) (2022) 1755–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Silva CS, Pinto RD, Amorim S, Pires RA, Correia-Neves M, Reis RL, Alves NL, Martins A, Neves NM, Fibronectin-Functionalized Fibrous Meshes as a Substrate to Support Cultures of Thymic Epithelial Cells, Biomacromolecules 21(12) (2020) 4771–4780. [DOI] [PubMed] [Google Scholar]
- [84].Zeleniak A, Wiegand C, Liu W, McCormick C, R. K, Alavi A, Guan H, Bertera S, Lakomy R, Tajima A, Cohen H, Wong S, Balikani L, Mizerak B, Bar-Joseph Z, Trucco M, Banerjee I, Fan Y, De novo construction of T cell compartment in humanized mice engrafted with iPSC-derived thymus organoids, Nat Methods 19(10) (2022) 1306–1319. [DOI] [PubMed] [Google Scholar]
- [85].McDonald-McGinn DM, Sullivan KE, Marino B, Philip N, Swillen A, Vorstman JA, Zackai EH, Emanuel BS, Vermeesch JR, Morrow BE, Scambler PJ, Bassett AS, 22q11.2 deletion syndrome, Nat Rev Dis Primers 1 (2015) 15071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Giardino G, Radwan N, Koletsi P, Morrogh DM, Adams S, Ip W, Worth A, Jones A, Meyer-Parsonson I, Gaspar HB, Gilmour K, Davies EG, Ladomenou F, Clinical and immunological features in a cohort of patients with partial DiGeorge syndrome followed at a single center, Blood 133(24) (2019) 2586–2596. [DOI] [PubMed] [Google Scholar]
- [87].Ryan AK, Goodship JA, Wilson DI, Philip N, Levy A, Seidel H, Schuffenhauer S, Oechsler H, Belohradsky B, Prieur M, Aurias A, Raymond FL, Clayton-Smith J, Hatchwell E, McKeown C, Beemer FA, Dallapiccola B, Novelli G, Hurst JA, Ignatius J, Green AJ, Winter RM, Brueton L, Brondum-Nielsen K, Scambler PJ, et al. , Spectrum of clinical features associated with interstitial chromosome 22q11 deletions: a European collaborative study, J Med Genet 34(10) (1997) 798–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Cancrini C, Puliafito P, Digilio MC, Soresina A, Martino S, Rondelli R, Consolini R, Ruga EM, Cardinale F, Finocchi A, Romiti ML, Martire B, Bacchetta R, Albano V, Carotti A, Specchia F, Montin D, Cirillo E, Cocchi G, Trizzino A, Bossi G, Milanesi O, Azzari C, Corsello G, Pignata C, Aiuti A, Pietrogrande MC, Marino B, Ugazio AG, Plebani A, Rossi P, Italian I Network for Primary, Clinical features and follow-up in patients with 22q11.2 deletion syndrome, J Pediatr 164(6) (2014) 1475–80 e2. [DOI] [PubMed] [Google Scholar]
- [89].Repetto GM, Guzman ML, Delgado I, Loyola H, Palomares M, Lay-Son G, Vial C, Benavides F, Espinoza K, Alvarez P, Case fatality rate and associated factors in patients with 22q11 microdeletion syndrome: a retrospective cohort study, BMJ Open 4(11) (2014) e005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Du Q, de la Morena MT, van Oers NSC, The Genetics and Epigenetics of 22q11.2 Deletion Syndrome, Front Genet 10 (2019) 1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, Tokooya K, Jore BS, Lopez M, Pandita RK, Lia M, Carrion D, Xu H, Schorle H, Kobler JB, Scambler P, Wynshaw-Boris A, Skoultchi AI, Morrow BE, Kucherlapati R, TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome, Cell 104(4) (2001) 619–29. [DOI] [PubMed] [Google Scholar]
- [92].Yagi H, Furutani Y, Hamada H, Sasaki T, Asakawa S, Minoshima S, Ichida F, Joo K, Kimura M, Imamura S, Kamatani N, Momma K, Takao A, Nakazawa M, Shimizu N, Matsuoka R, Role of TBX1 in human del22q11.2 syndrome, Lancet 362(9393) (2003) 1366–73. [DOI] [PubMed] [Google Scholar]
- [93].Mesbah K, Rana MS, Francou A, van Duijvenboden K, Papaioannou VE, Moorman AF, Kelly RG, Christoffels VM, Identification of a Tbx1/Tbx2/Tbx3 genetic pathway governing pharyngeal and arterial pole morphogenesis, Hum Mol Genet 21(6) (2012) 1217–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Liu N, Schoch K, Luo X, Pena LDM, Bhavana VH, Kukolich MK, Stringer S, Powis Z, Radtke K, Mroske C, Deak KL, McDonald MT, McConkie-Rosell A, Markert ML, Kranz PG, Stong N, Need AC, Bick D, Amaral MD, Worthey EA, Levy S, Undiagnosed Diseases N, Wangler MF, Bellen HJ, Shashi V, Yamamoto S, Functional variants in TBX2 are associated with a syndromic cardiovascular and skeletal developmental disorder, Hum Mol Genet 27(14) (2018) 2454–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Pagon RA, Graham JM Jr., Zonana J, Yong SL, Coloboma, congenital heart disease, and choanal atresia with multiple anomalies: CHARGE association, J Pediatr 99(2) (1981) 223–7. [DOI] [PubMed] [Google Scholar]
- [96].Janssen N, Bergman JE, Swertz MA, Tranebjaerg L, Lodahl M, Schoots J, Hofstra RM, van Ravenswaaij-Arts CM, Hoefsloot LH, Mutation update on the CHD7 gene involved in CHARGE syndrome, Hum Mutat 33(8) (2012) 1149–60. [DOI] [PubMed] [Google Scholar]
- [97].Basson MA, van Ravenswaaij-Arts C, Functional Insights into Chromatin Remodelling from Studies on CHARGE Syndrome, Trends Genet 31(10) (2015) 600–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Bergman JE, Janssen N, van der Sloot AM, de Walle HE, Schoots J, Rendtorff ND, Tranebjaerg L, Hoefsloot LH, van Ravenswaaij-Arts CM, Hofstra RM, A novel classification system to predict the pathogenic effects of CHD7 missense variants in CHARGE syndrome, Hum Mutat 33(8) (2012) 1251–60. [DOI] [PubMed] [Google Scholar]
- [99].Frank J, Pignata C, Panteleyev AA, Prowse DM, Baden H, Weiner L, Gaetaniello L, Ahmad W, Pozzi N, Cserhalmi-Friedman PB, Aita VM, Uyttendaele H, Gordon D, Ott J, Brissette JL, Christiano AM, Exposing the human nude phenotype, Nature 398(6727) (1999) 473–4. [DOI] [PubMed] [Google Scholar]
- [100].Pignata C, Fiore M, Guzzetta V, Castaldo A, Sebastio G, Porta F, Guarino A, Congenital Alopecia and nail dystrophy associated with severe functional T-cell immunodeficiency in two sibs, Am J Med Genet 65(2) (1996) 167–70. [DOI] [PubMed] [Google Scholar]
- [101].Adriani M, Martinez-Mir A, Fusco F, Busiello R, Frank J, Telese S, Matrecano E, Ursini MV, Christiano AM, Pignata C, Ancestral founder mutation of the nude (FOXN1) gene in congenital severe combined immunodeficiency associated with alopecia in southern Italy population, Ann Hum Genet 68(Pt 3) (2004) 265–8. [DOI] [PubMed] [Google Scholar]
- [102].Markert ML, Marques JG, Neven B, Devlin BH, McCarthy EA, Chinn IK, Albuquerque AS, Silva SL, Pignata C, de Saint Basile G, Victorino RM, Picard C, Debre M, Mahlaoui N, Fischer A, Sousa AE, First use of thymus transplantation therapy for FOXN1 deficiency (nude/SCID): a report of 2 cases, Blood 117(2) (2011) 688–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Chou J, Massaad MJ, Wakim RH, Bainter W, Dbaibo G, Geha RS, A novel mutation in FOXN1 resulting in SCID: a case report and literature review, Clin Immunol 155(1) (2014) 30–32. [DOI] [PubMed] [Google Scholar]
- [104].Van Esch H, Groenen P, Fryns JP, Van de Ven W, Devriendt K, The phenotypic spectrum of the 10p deletion syndrome versus the classical DiGeorge syndrome, Genet Couns 10(1) (1999) 59–65. [PubMed] [Google Scholar]
- [105].Schuffenhauer S, Seidel H, Oechsler H, Belohradsky B, Bernsau U, Murken J, Meitinger T, DiGeorge syndrome and partial monosomy 10p: case report and review, Ann Genet 38(3) (1995) 162–7. [PubMed] [Google Scholar]
- [106].Shapira M, Borochowitz Z, Bar-El H, Dar H, Etzioni A, Lorber A, Deletion of the short arm of chromosome 10 (10p13): report of a patient and review, Am J Med Genet 52(1) (1994) 34–8. [DOI] [PubMed] [Google Scholar]
- [107].Fernandez RM, Sanchez J, Garcia-Diaz L, Pelaez-Nora Y, Gonzalez-Meneses A, Antinolo G, Borrego S, Interstitial 10p deletion derived from a maternal ins(16;10)(q22;p13p15.2): Report of the first familial case of 10p monosomy affecting to two familial members of different generations, Am J Med Genet A 170A(5) (2016) 1268–73. [DOI] [PubMed] [Google Scholar]
- [108].Mattina T, Perrotta CS, Grossfeld P, Jacobsen syndrome, Orphanet J Rare Dis 4 (2009) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Oud MM, Tuijnenburg P, Hempel M, van Vlies N, Ren Z, Ferdinandusse S, Jansen MH, Santer R, Johannsen J, Bacchelli C, Alders M, Li R, Davies R, Dupuis L, Cale CM, Wanders RJA, Pals ST, Ocaka L, James C, Muller I, Lehmberg K, Strom T, Engels H, Williams HJ, Beales P, Roepman R, Dias P, Brunner HG, Cobben JM, Hall C, Hartley T, Le Quesne Stabej P, Mendoza-Londono R, Davies EG, de Sousa SB, Lessel D, Arts HH, Kuijpers TW, Mutations in EXTL3 Cause Neuro-immuno-skeletal Dysplasia Syndrome, Am J Hum Genet 100(2) (2017) 281–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Guo L, Elcioglu NH, Mizumoto S, Wang Z, Noyan B, Albayrak HM, Yamada S, Matsumoto N, Miyake N, Nishimura G, Ikegawa S, Identification of biallelic EXTL3 mutations in a novel type of spondylo-epi-metaphyseal dysplasia, J Hum Genet 62(8) (2017) 797–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Samuels ME, Majewski J, Alirezaie N, Fernandez I, Casals F, Patey N, Decaluwe H, Gosselin I, Haddad E, Hodgkinson A, Idaghdour Y, Marchand V, Michaud JL, Rodrigue MA, Desjardins S, Dubois S, Le Deist F, Awadalla P, Raymond V, Maranda B, Exome sequencing identifies mutations in the gene TTC7A in French-Canadian cases with hereditary multiple intestinal atresia, J Med Genet 50(5) (2013) 324–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Bigorgne AE, Farin HF, Lemoine R, Mahlaoui N, Lambert N, Gil M, Schulz A, Philippet P, Schlesser P, Abrahamsen TG, Oymar K, Davies EG, Ellingsen CL, Leteurtre E, Moreau-Massart B, Berrebi D, Bole-Feysot C, Nischke P, Brousse N, Fischer A, Clevers H, de Saint Basile G, TTC7A mutations disrupt intestinal epithelial apicobasal polarity, J Clin Invest 124(1) (2014) 328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Lemoine R, Bigorgne A, Farin H, Saint Basile G, [TTC7A, a critical effector for the intestinal and immune system homeostasis], Med Sci (Paris) 30(6–7) (2014) 616–8. [DOI] [PubMed] [Google Scholar]
- [114].Chen R, Giliani S, Lanzi G, Mias GI, Lonardi S, Dobbs K, Manis J, Im H, Gallagher JE, Phanstiel DH, Euskirchen G, Lacroute P, Bettinger K, Moratto D, Weinacht K, Montin D, Gallo E, Mangili G, Porta F, Notarangelo LD, Pedretti S, Al-Herz W, Alfahdli W, Comeau AM, Traister RS, Pai SY, Carella G, Facchetti F, Nadeau KC, Snyder M, Notarangelo LD, Whole-exome sequencing identifies tetratricopeptide repeat domain 7A (TTC7A) mutations for combined immunodeficiency with intestinal atresias, J Allergy Clin Immunol 132(3) (2013) 656–664 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kisand K, Peterson P, Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy: known and novel aspects of the syndrome, Ann N Y Acad Sci 1246 (2011) 77–91. [DOI] [PubMed] [Google Scholar]
- [116].Puel A, Doffinger R, Natividad A, Chrabieh M, Barcenas-Morales G, Picard C, Cobat A, Ouachee-Chardin M, Toulon A, Bustamante J, Al-Muhsen S, Al-Owain M, Arkwright PD, Costigan C, McConnell V, Cant AJ, Abinun M, Polak M, Bougneres PF, Kumararatne D, Marodi L, Nahum A, Roifman C, Blanche S, Fischer A, Bodemer C, Abel L, Lilic D, Casanova JL, Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I, J Exp Med 207(2) (2010) 291–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Ferre EM, Rose SR, Rosenzweig SD, Burbelo PD, Romito KR, Niemela JE, Rosen LB, Break TJ, Gu W, Hunsberger S, Browne SK, Hsu AP, Rampertaap S, Swamydas M, Collar AL, Kong HH, Lee CR, Chascsa D, Simcox T, Pham A, Bondici A, Natarajan M, Monsale J, Kleiner DE, Quezado M, Alevizos I, Moutsopoulos NM, Yockey L, Frein C, Soldatos A, Calvo KR, Adjemian J, Similuk MN, Lang DM, Stone KD, Uzel G, Kopp JB, Bishop RJ, Holland SM, Olivier KN, Fleisher TA, Heller T, Winer KK, Lionakis MS, Redefined clinical features and diagnostic criteria in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy, JCI Insight 1(13) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]


