ABSTRACT
Merkel cell polyomavirus (MCPyV) has been associated with approximately 80% of Merkel cell carcinoma (MCC), an aggressive and increasingly incident skin cancer. The link between host innate immunity, viral load control, and carcinogenesis has been established but poorly characterized. We previously established the importance of the STING and NF-κB pathways in the host innate immune response to viral infection. In this study, we further discovered that MCPyV infection of human dermal fibroblasts (HDFs) induces the expression of type I and III interferons (IFNs), which in turn stimulate robust expression of IFN-stimulated genes (ISGs). Blocking type I IFN downstream signaling using an IFN-β antibody, JAK inhibitors, and CRISPR knockout of the receptor dramatically repressed MCPyV infection-induced ISG expression but did not significantly restore viral replication activities. These findings suggest that IFN-mediated induction of ISGs in response to MCPyV infection is not crucial to viral control. Instead, we found that type I IFN exerts a more direct effect on MCPyV infection postentry by repressing early viral transcription. We further demonstrated that growth factors normally upregulated in wounded or UV-irradiated human skin can significantly stimulate MCPyV gene expression and replication. Together, these data suggest that in healthy individuals, host antiviral responses, such as IFN production induced by viral activity, may restrict viral propagation to reduce MCPyV burden. Meanwhile, growth factors induced by skin abrasion or UV irradiation may stimulate infected dermal fibroblasts to promote MCPyV propagation. A delicate balance of these mutually antagonizing factors provides a mechanism to support persistent MCPyV infection.
IMPORTANCE Merkel cell carcinoma is an aggressive skin cancer that is particularly lethal to immunocompromised individuals. Though rare, MCC incidence has increased significantly in recent years. There are no lasting and effective treatments for metastatic disease, highlighting the need for additional treatment and prevention strategies. By investigating how the host innate immune system interfaces with Merkel cell polyomavirus, the etiological agent of most of these cancers, our studies identified key factors necessary for viral control, as well as conditions that support viral propagation. These studies provide new insights for understanding how the virus balances the effects of the host immune defenses and of growth factor stimulation to achieve persistent infection. Since virus-positive MCC requires the expression of viral oncogenes to survive, our observation that type I IFN can repress viral oncogene transcription indicates that these cytokines could be explored as a viable therapeutic option for treating patients with virus-positive MCC.
KEYWORDS: DNA tumor virus, Merkel cell carcinoma, Merkel cell polyomavirus, persistent infection, virus-host interactions
INTRODUCTION
Merkel cell polyomavirus (MCPyV) is the most recently discovered human oncogenic virus (1). It has been associated with Merkel cell carcinoma (MCC), a highly aggressive form of skin cancer (1–10). MCPyV is also a widespread virus, detected on the skin of most healthy adults (11–13). Serological studies have established that MCPyV infects most humans during early childhood and that viral prevalence increases as populations age (14–16). The vast majority of MCPyV infections are asymptomatic (17), but some result in the development of MCC (1–4).
Although MCC cases are rare, the incidence of MCC has tripled over the last 20 years (18–21) and increased by more than 95% in the United States since 2000 (19). MCC is one of the most aggressive skin cancers, with a nearly 50% mortality rate (exceeding the rate of melanoma) and a 5-year survival rate of less than 45% (1–8, 22, 23). MCC metastasizes rapidly, and currently there are no effective treatments for cases of metastatic cancers (24). Chemotherapies have thus far failed to produce durable responses in patients with metastatic disease (25, 26). Recently, anti-PD1 and anti-PD-L1 treatments have shown exciting results in some MCC patients, but the durability of responses is variable, and a significant proportion of patients do not respond (27–29). Given the high prevalence of MCPyV infection and the increasing incidence of MCC diagnoses (21), a better understanding of the mechanisms that drive MCPyV oncogenesis is needed in order to develop more effective strategies to prevent and treat this highly lethal skin cancer.
MCPyV has a circular, double-stranded DNA genome of ~5 kb that carries an efficient repertoire of genes (3, 22, 30). The viral genome is divided into early and late regions by a noncoding regulatory region (NCRR) containing the viral origin of replication and bidirectional promoters responsible for early and late gene transcription (3, 31, 32). The early region encodes large tumor antigen (LT), small tumor antigen (sT), 57kT, and a microRNA and contains an alternate LT open reading frame (ALTO) (3, 32–34). The late region encodes the major and minor capsid proteins, VP1 and VP2 (35, 36). Of the early genes, the roles of LT and sT in promoting viral replication and host cell proliferation have been best described (22, 32, 37–40).
MCPyV integration into the host genome is a key event that drives virus-positive MCC development (22, 41, 42). Integrated MCPyV genome is detectable in more than 80% of MCC tumors, in which the virus maintains the expression of native sT and a truncated LT (LTT) (1, 43–45). These are the major oncogenes that support MCPyV-induced tumorigenesis (1, 22, 30, 37, 44–51). Despite our knowledge of the MCPyV genome and the functions of its viral oncogenes, many aspects of its virology and infectious cycle are poorly understood. Hence, it is unclear what mechanisms and prerequisite conditions trigger MCPyV genome integration and MCC development.
MCPyV maintains asymptomatic infection in most of the general population (11–13), but in elderly and immunosuppressed individuals, infection has a higher chance to trigger MCC development (52, 53). Epidemiological studies have established a strong link between immunosuppression, elevated MCPyV genome loads, and increased risk for MCC (53, 54). Factors that can dampen antiviral immunity, such as advanced age, chronic UV exposure, and immunosuppression, have been shown to significantly increase the risk of developing MCPyV-positive MCC (18, 53). MCPyV DNA prevalence and viral load on sun-exposed skin increase dramatically in individuals over the age of 40 and remain high for older age groups (55). Within the same age range group, individuals who are immunocompromised due to HIV/AIDS, chronic lymphocytic leukemia, or treatment for autoimmunity or organ transplantation are at the greatest risk for MCC development (9, 56–59). Additionally, MCPyV DNA is more likely to be detected on the skin of HIV-positive men than healthy controls, and those with poorly controlled HIV infection frequently maintain higher MCPyV DNA loads than individuals with better-controlled infections (55). Furthermore, viral oncogenesis is more rapid and aggressive in HIV-positive and immunosuppressed patients (60), suggesting that the elevated MCPyV DNA loads associated with immunosuppression may contribute to the increased likelihood of MCC development. Collectively, these epidemiological studies suggest that MCPyV can strike a delicate balance with human hosts to avoid eradication and maintain persistent infection, but failure of the immune system to control MCPyV infection may enable unimpeded propagation of MCPyV. Rampant MCPyV infection could then lead to more frequent replication errors, which can in turn stimulate genome integration and ultimately promote MCPyV-induced tumorigenesis. Little is known, however, about the host immune response elicited by MCPyV. The way MCPyV interfaces with the host immune response to either establish long-term asymptomatic infection or integrate and trigger oncogenesis in different settings is also poorly understood.
Our abilities to probe the host immune responses that keep MCPyV infection in check have been predominantly limited by a dearth of tools and systems with which to study MCPyV infection. Using ex vivo skin sections and in vitro cultures of cells isolated from human foreskins, we previously discovered that human dermal fibroblasts (HDFs) have the capability to support MCPyV infection activities (61–63). The in vitro and ex vivo MCPyV infection models established in our studies afford a great opportunity to investigate the host innate immune response to MCPyV infection and to determine what impact those responses have on the course of MCPyV proliferation. Using this system, our additional studies demonstrated that MCPyV infection activates the cGAS-STING as well as the NF-κB signaling pathways to induce robust expression of key interferon (IFN)-stimulated genes (ISGs) and inflammatory cytokines in HDFs (64). We also discovered that the PYHIN protein IFI16 upregulates inflammatory cytokines in response to MCPyV infection (64).
In the present study, we sought to further characterize the molecular events leading to the MCPyV-induced ISG response. We also examined the impact of the MCPyV-induced ISG response on viral infection fate. We discovered that type I IFN can restrict viral infection at the early transcription stage, but growth factors that are often enriched in skin wounds can antagonize this effect to stimulate viral infection. Together, our data provide new insights for understanding how the intricate MCPyV-host relationship provides a molecular mechanism to support viral persistence.
RESULTS
Visualization of MCPyV infection-induced ISG expression at the single-cell level.
In our previous study, we discovered that MCPyV infection induces a robust innate immune response in HDFs driven by the cGAS-STING and NF-κB signaling pathways (64). Additionally, we found that MCPyV infection causes the upregulated expression of key ISGs and inflammatory cytokines (64). Based on this finding, we sought to further investigate the possible role of ISGs in regulating the innate immune response to MCPyV infection in this study.
ISGs are normally induced by the activation of IFN downstream signaling. IFNs secreted from the virus-infected cell are known to function via autocrine and paracrine modalities through binding of a heterodimeric cell surface receptor complex, which activates the JAK-STAT signaling pathway to stimulate the transcription of ISGs in the nucleus (65–67). Our previous studies detected global, population-level changes in innate immune response signaling in response to MCPyV infection (64). To more accurately probe the questions of how the innate immune response in infected HDFs is regulated and whether presumed cytokine signaling spreads the innate immune response to neighboring cells, the activation of innate immune effectors was examined at the single-cell level in infected HDFs by immunofluorescence (IF) staining.
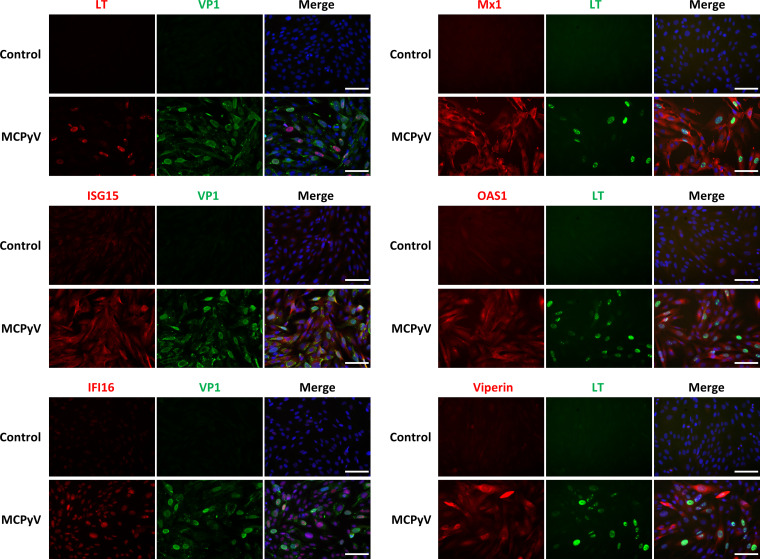
Expression of several key ISGs downstream of type I and type III IFN (Mx1, ISG15, OAS1, IFI16, and viperin) was induced at the protein level by MCPyV infection (Fig. 1). We also observed that the ISGs examined in our studies were broadly expressed among cells with both detectable and undetectable expression of viral proteins (Fig. 1). This result implies that a potential antiviral response is spread to all cells through cytokine signaling, which may then control viral infection.
FIG 1.
Visualization of MCPyV infection-induced ISGs at the single-cell level. HDFs treated with live MCPyV or heat-inactivated MCPyV (control) were immunostained using antibodies for VP1, LT, and the indicated ISGs. The cells were also counterstained with DAPI (4′,6-diamidino-2-phenylindole). Bars, 100 μm.
MCPyV infection induces expression of IFN-β and other innate immune cytokines.
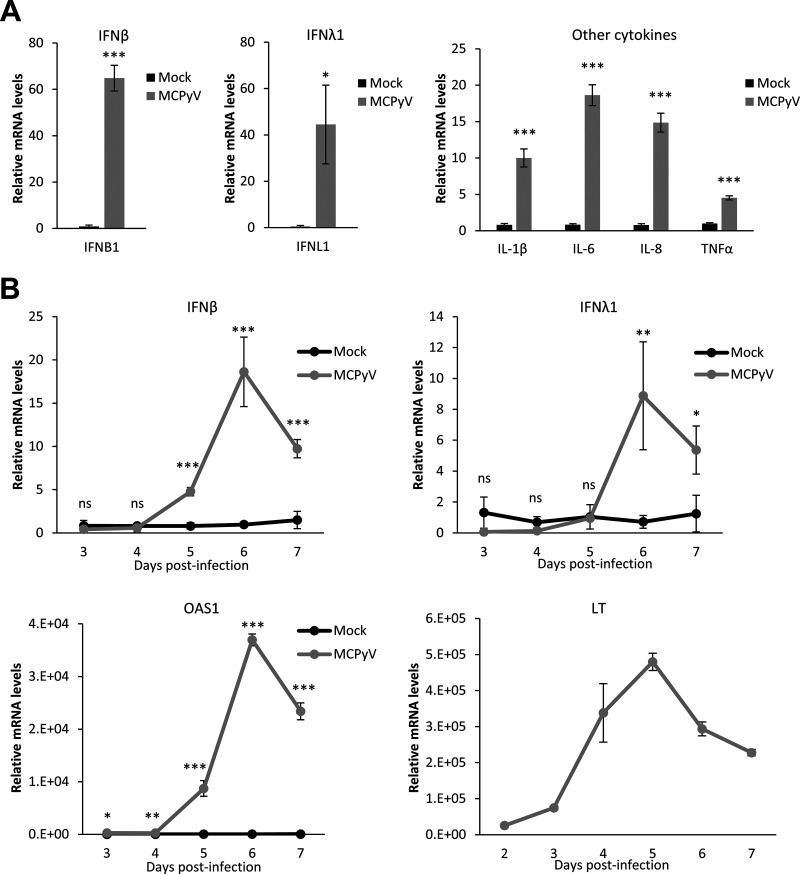
Because MCPyV infection induced a robust induction of ISGs in both our current and previous studies (Fig. 1) (64), we decided to further examine the level of IFNs released by MCPyV-infected cells. In our prior study (64), we found that MCPyV infection causes a modest induction of type I IFNs. In that study, HDFs treated with heat-inactivated MCPyV stocks were used as the negative control for HDFs infected with live virus in order to specifically examine the effects of viral activity and not of other components of the viral preparation on the innate immune response (64). We subsequently discovered that our heat-inactivation protocol does not completely kill MCPyV and the resulting MCPyV stocks could still induce type I IFN expression in HDFs (data not shown), contributing to the apparently low IFN induction by live virus in our prior measurements (64). In this study, we decided to reexamine the complete innate immune response to MCPyV treatment in HDFs using untreated cells—rather than cells treated with heat-inactivated MCPyV stocks—as the negative control in all experiments. In this modified infection protocol, primary HDFs were untreated or treated with live MCPyV virions in the absence of serum and presence of collagenase for 2 days to promote viral entry and then treated with fetal bovine serum (FBS) to initiate viral gene expression. By examining the relative expression of IFNs and inflammatory cytokines in MCPyV-infected HDFs, we discovered that the expression of both IFN-β and IFN-λ1 was stimulated significantly in MCPyV-treated cells compared to untreated cells (Fig. 2A). The cycle threshold (CT) values obtained from the reverse transcription-quantitative PCR (RT-qPCR) analysis for other IFNs (IFN-α, IFN-λ2-4, IFN-ω, and IFN-γ) were all above 35, suggesting that the expression levels of these IFNs were not stimulated by MCPyV infection. MCPyV infection also stimulated the transcription of inflammatory cytokines such as interleukin 1β (IL-1β), IL-6, IL-8, and tumor necrosis factor alpha (TNF-α), recapitulating our previous findings (Fig. 2A) (64). Furthermore, expression of IFN-β, IFN-λ, and the key downstream ISGs such as OAS1 increased dramatically during the course of viral infection, with the peak of expression coinciding with the peak of viral early gene expression (Fig. 2B). This result suggests that the IFN and ISG responses are primarily induced by MCPyV DNA replication and/or transcription but not by the inoculating virions or components of the virus preparation (Fig. 2B).
FIG 2.
MCPyV infection induces the expression of IFN-β and other innate immune cytokines. (A) mRNA levels of IFN-β, IFN-λ1, and other cytokines were measured by RT-qPCR in HDFs infected with MCPyV on day 6 postinfection. The values for the mRNA molecules in mock-infected HDFs were set as 1. (B) mRNA levels of IFN-β, IFN-λ1, OAS1, and LT were measured by RT-qPCR in HDFs infected with MCPyV on day 3 to day 7 postinfection. One of the values for mock-infected HDFs on day 3 was set as 1. No LT was detected in the mock-infected HDFs. Error bars indicate standard deviations from three independent experiments. ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant.
Treatment with IFN-β antibody and JAK inhibitor represses expression of MCPyV infection-induced ISGs but does not relieve MCPyV replication activities.
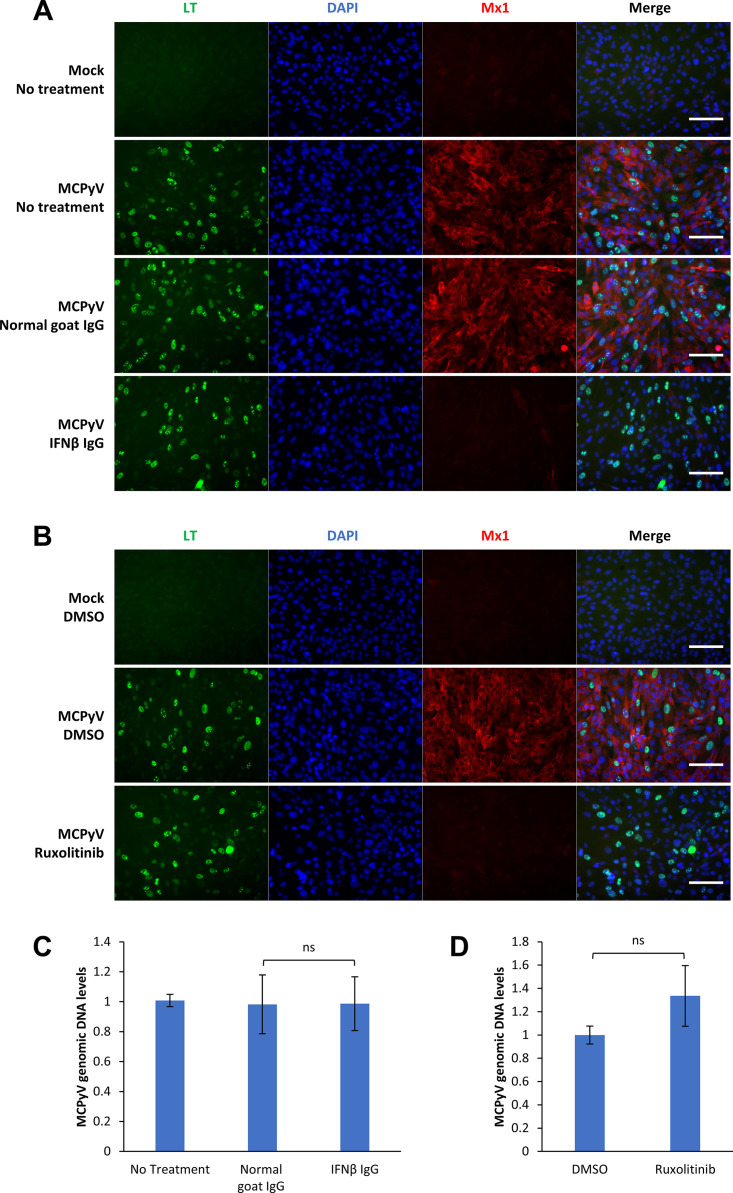
The broad activation of ISGs in MCPyV-infected HDFs indicates that the JAK-STAT signaling pathway downstream of IFN may contribute to the induction and spread of the antiviral response between cells during infection. To determine whether JAK-STAT signaling downstream of IFNs and the IFN receptors mediates an antiviral response to MCPyV, HDFs were treated with either an IFN-β antibody or the JAK inhibitor ruxolitinib on days 2 and 3.5 of infection and collected on day 5 of infection for IF or RT-qPCR analysis (Fig. 3). Consistent with previous observations (Fig. 1) (64), MCPyV infection stimulated robust ISG expression as demonstrated by significantly increased Mx1 expression at the protein level (Fig. 3A and B). Both IFN-β antibody and ruxolitinib treatments efficiently repressed Mx1 expression in MCPyV-infected cells, suggesting that they are effective at abrogating MCPyV-induced IFN signaling (Fig. 3A and B). Because the IFN-β antibody (inhibiting type I IFN only) and ruxolitinib (inhibiting signaling downstream of both type I and III IFN) were both highly effective at repressing Mx1 expression, our data suggest that type I IFN is the primary cytokine driving ISG expression in infected HDFs (Fig. 3A and B). However, successful ablation of ISG induction by IFN-β antibody and ruxolitinib does not appear to have a significant impact on restoring MCPyV infection activity (Fig. 3C and D). From these data, we conclude that IFN-β drives the expression of infection-induced ISGs, but the ISG response does not appear to play an important role in repressing MCPyV infection in our current system.
FIG 3.
Treatment with IFN-β antibody and JAK inhibitor represses MCPyV infection-induced ISGs but does not relieve MCPyV replication. (A) MCPyV-infected or mock-infected HDFs were either untreated or treated with normal goat IgG or IFN-β IgG on day 2 and day 3.5 postinfection. Cells were fixed on day 5 postinfection, immunostained for LT and Mx1, and counterstained with DAPI. (B) MCPyV-infected or mock-infected HDFs were treated with DMSO or 1 μM ruxolitinib on day 2 and day 3.5 postinfection. Cells were fixed on day 5 postinfection, immunostained for LT and Mx1, and counterstained with DAPI. Bars, 100 μm. (C) MCPyV-infected or mock-infected HDFs were either untreated or treated with normal goat IgG or IFN-β IgG on day 2, day 3.5, and day 5 postinfection. Cells were harvested on day 6 postinfection for DNA extraction. MCPyV genomic DNA levels were determined by qPCR quantification and normalized to the GAPDH genomic DNA levels. The value for one of the untreated, MCPyV-infected groups was set to 1. (D) MCPyV-infected or mock-infected HDFs were treated with DMSO or 1 μM ruxolitinib on day 2, day 3.5 and day 5 postinfection. Cells were harvested on day 6 postinfection for DNA extraction. MCPyV genomic DNA levels were determined by qPCR quantification and normalized to the GAPDH genomic DNA levels. The value for one of the DMSO-treated, MCPyV-infected groups was set to 1. Error bars represent the standard deviations from three independent experiments. ns, not significant.
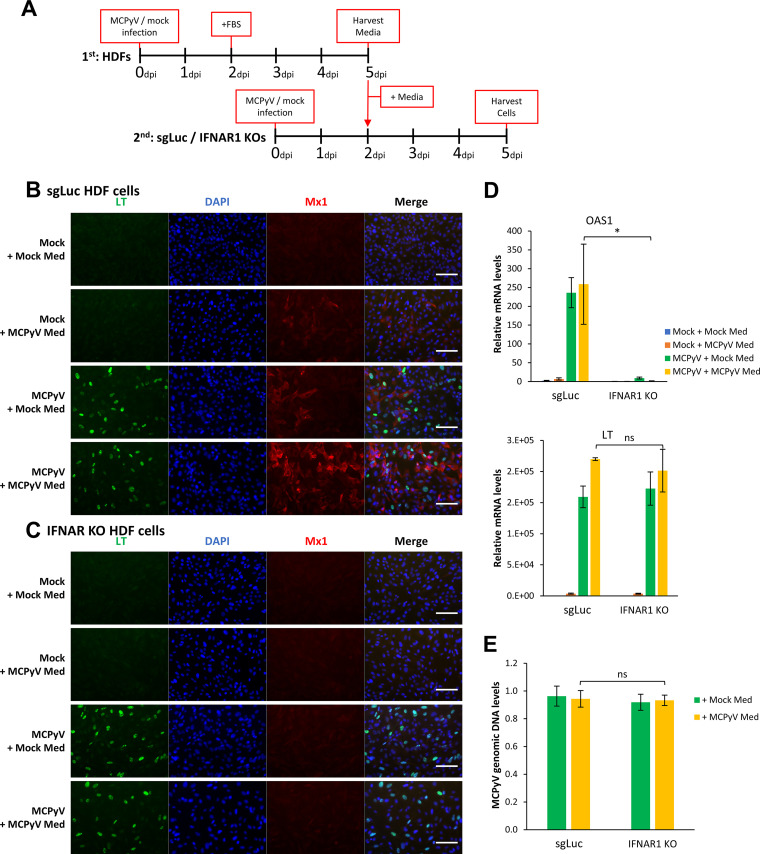
IFNAR1 knockout inhibits MCPyV-induced ISG expression but does not relieve MCPyV replication activities.
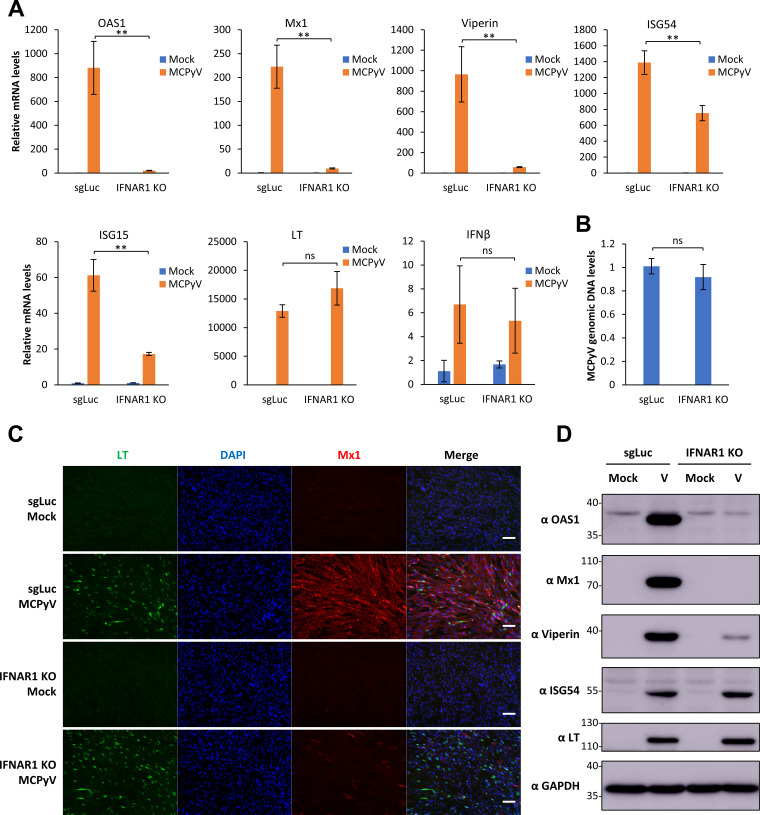
To further examine the function of IFN downstream signaling in controlling MCPyV infection, we applied lentiCRISPR/Cas9 technology to generate stable knockouts (KOs) of IFN-α/β receptor subunit 1 (IFNAR1) in HDFs using three different single guide RNAs (sgRNAs) (Fig. S1). Of the three stable cell lines generated, the IFNAR1 KO sg2 cell line was selected for further experiments based on its ability to effectively repress IFNAR1 expression and ISG response to exogenous type I IFN compared to control sgLuc HDFs (encoding sgRNA targeting luciferase) (see Fig. S1A and B in the supplemental material). These IFNAR1 KO cells, which are incapable of “receiving” IFN signaling, were used to assess the role of type I IFN signaling in preventing MCPyV infection. IFNAR1 KO HDFs and control sgLuc HDFs were treated with MCPyV and analyzed on day 5 or 6 postinfection (Fig. 4). In the control sgLuc HDFs, MCPyV infection induced robust expression of ISGs such as OAS1, Mx1, viperin, and ISG15, at both the mRNA level and the protein level (Fig. 4A, C, and D). In response to MCPyV infection, IFNAR1 KO effectively abrogated the induction of these ISGs while displaying little effect on the IFN-β expression induced by infection (Fig. 4A, C, and D). However, inhibition of paracrine IFN-β signaling by IFNAR1 KO did not significantly affect MCPyV transcription or replication as indicated by the levels of MCPyV LT mRNA and of viral genome copy number (Fig. 4A, LT panel, and Fig. 4B). These results further suggest that the IFN downstream IFNAR1 signaling pathway is unlikely to play an important role in influencing MCPyV infection.
FIG 4.
IFNAR1 knockout inhibits MCPyV-induced ISG expression but does not relieve MCPyV infection. (A) sgLuc or IFNAR1 KO (sg2) stable HDFs were mock infected or infected with MCPyV. mRNA levels of ISGs, LT, and IFN-β were measured by RT-qPCR on day 5 postinfection. One of the values from mock-infected sgLuc HDFs was set as 1. (B) MCPyV-infected sgLuc or IFNAR1 KO stable HDFs were harvested for DNA extraction on day 6 postinfection. MCPyV genomic DNA levels were determined by qPCR quantification and normalized to the GAPDH genomic DNA levels. The value for one of the MCPyV-infected sgLuc HDF groups was set as 1. Error bars represent the standard deviations from three independent experiments. (C) sgLuc or IFNAR1 KO stable HDFs were mock infected or infected with MCPyV. Cells were fixed on day 5 postinfection, immunostained for LT and Mx1, and counterstained with DAPI. Bars, 100 μm. (D) Whole-cell lysates were extracted from mock- or MCPyV-infected sgLuc or IFNAR1 KO HDFs on day 5 postinfection, resolved with SDS-PAGE, and immunoblotted with the indicated antibodies. **, P < 0.01; ns, not significant.
ISG54 knockout does not significantly affect MCPyV infection.
Unlike the other ISGs examined, ISG54 induction by MCPyV infection was not significantly inhibited in IFNAR1 KO HDFs (Fig. 4A and D), confirming an IFN-independent mechanism for inducing ISG54 expression (68). Because ISG54 has the ability to promote apoptosis (69), we next assessed its possible role as the main factor mediating the antiviral response to MCPyV infection. To accomplish this, lentiCRISPR/Cas9 technology was applied to generate stable ISG54 KO HDFs using three sgRNAs. Among these cells, the ISG54 KO sg1 cells were selected for further study based on their significantly repressed level of ISG54 expression (Fig. S2A). Still, after treatment with MCPyV, ISG54 KO HDFs did not show increased MCPyV infection and replication. Instead, these cells had a moderately reduced capacity for supporting MCPyV replication (Fig. S2B). These data suggest that ISG54 does not play a significant role in antagonizing MCPyV infection.
Paracrine IFN signaling and downstream ISG production are not essential for controlling MCPyV infection.
To this point, our data have shown that inhibiting IFN signaling in HDFs does not significantly affect MCPyV’s replicative capacity (Fig. 3 and 4). However, the inhibitors and CRISPR KO used in these experiments were applied in cells that had already been treated and possibly infected with MCPyV; these methods of inhibiting IFN signaling after establishing MCPyV infection may therefore be insufficient to block the antiviral response. We also wanted to consider the possibility that the MCPyV-induced innate immune response signals to other cells in a paracrine manner to block viral infection in naive cells. However, the paracrine response cannot be tested using our current MCPyV infection system in HDFs, which supports only a single round of infection, because the FBS present in the system inhibits further spread of infection to neighboring cells (61). Additionally, the infected HDFs may not produce or release enough newly packaged viral particles to establish infection in neighboring cells. If IFN signaling controls MCPyV infection by stimulating an antiviral state in uninfected cells and inhibiting the second round of infection, then our prior experiments and the current infection system, which examined only the first round of infection, did not accurately address this possibility.
To better recapitulate the paracrine effect of MCPyV infection, we modified our infection protocol to mimic two rounds of infection in HDFs (Fig. 5A). Normal HDFs were infected per our usual protocol for the first round of infection, and on day 5, we harvested the medium from the cells. This medium, containing cytokines and viral particles released by cells during the first round of infection, was then applied to IFNAR1 KO or sgLuc HDFs, which were treated or mock infected with MCPyV. These treated IFNAR1 KO/sgLuc HDFs from the second round of MCPyV infection were then harvested on day 5 for subsequent analysis (Fig. 5A).
FIG 5.
Simulation of serial infections to determine if paracrine IFN signaling and downstream ISG production are important for controlling MCPyV infection. (A) Schematic visualization of the experimental process mimicking the paracrine effect of MCPyV infection. The media of mock- and MCPyV-infected HDFs were harvested on day 5 of infection and used to treat mock or newly MCPyV-infected sgLuc or IFNAR1 KO cells on day 2 postinfection. (B) sgLuc stable HDFs treated as indicated in panel A were immunostained for LT and Mx1 and counterstained with DAPI. (C) IFNAR1 KO HDFs treated as indicated in panel A were immunostained for LT and Mx1 and counterstained with DAPI. Bars, 100 μm. (D) Using cells treated as indicated in panel A, mRNA levels of OAS1 and LT were measured by RT-qPCR. One of the values from mock-infected sgLuc cells treated with the medium collected from mock-infected HDFs was set as 1. (E) MCPyV-infected sgLuc or IFNAR1 KO cells treated as indicated in panel A were harvested for DNA extraction on day 5 postinfection. MCPyV genomic DNA levels were determined by qPCR quantification and normalized to the GAPDH genomic DNA levels. The value for one of the MCPyV-infected sgLuc HDF samples treated with medium collected from mock-infected HDF was set as 1. Error bars represent the standard deviation from three independent experiments. *, P < 0.05; ns, not significant.
In the control sgLuc HDFs, extracellular IFN released from the first round of infection was modestly effective at stimulating downstream ISG expression in neighboring cells (Fig. 5B and D, compare Mock + MCPyV Med with Mock + Mock Med), while release of viral particles did not occur at a sufficient level to induce infection in naive cells (Fig. 5B, Mock + MCPyV Med). Also in these cells, fresh MCPyV added in the second round of infection led to robust LT expression and further enhanced ISG induction (Fig. 5B and D, compare MCPyV + Mock Med and MCPyV + MCPyV Med results with Mock + Mock Med and Mock + MCPyV Med results). IFNAR1 KO was effective at inhibiting downstream ISG production induced by IFNs from both the first and second rounds of infection (Fig. 5C and the IFNAR KO data in Fig. 5D). However, these cells did not show any significant alteration in viral transcription or replication (see the LT signal in Fig. 5B and C, mRNA in Fig. 5D, and viral genome copy number in Fig. 5E). These results indicate that ISG induction downstream of paracrine IFN signaling is not sufficient to block MCPyV infection.
Type I IFN treatment inhibits MCPyV postentry activities.
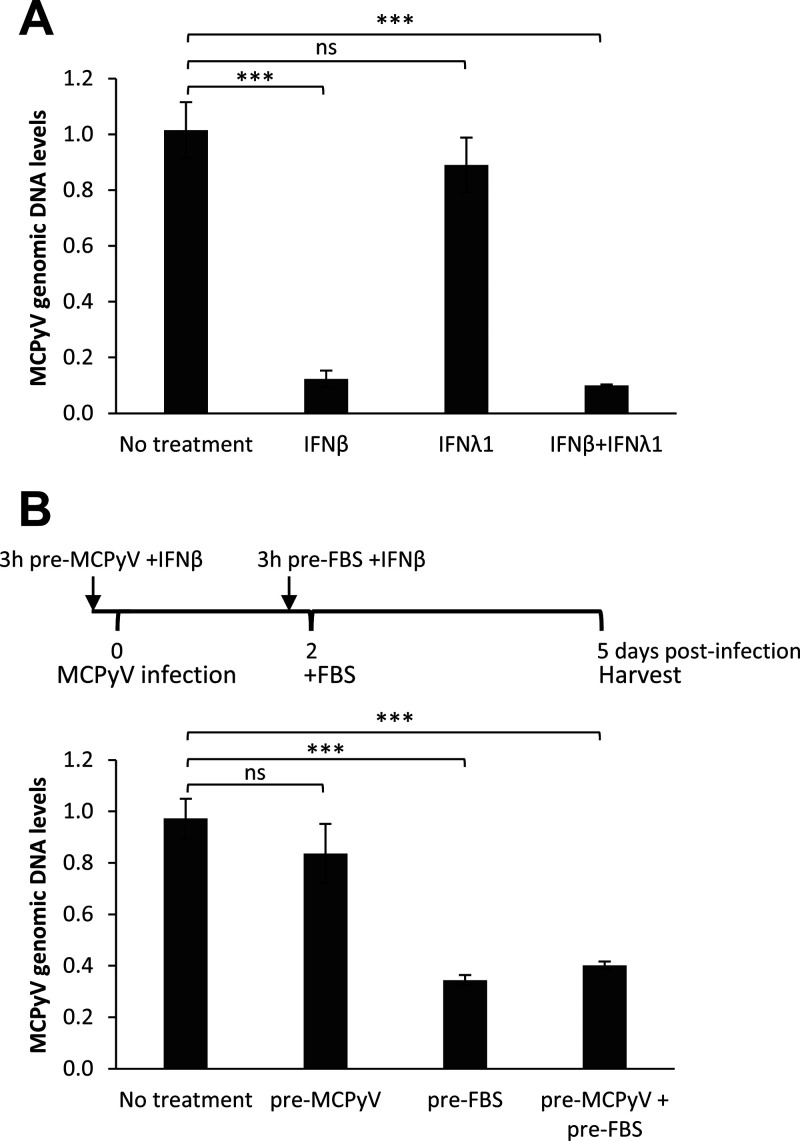
Inhibition of ISG signaling downstream of IFN had thus far not affected MCPyV activity. One possible explanation for our results is that, instead of maintaining an antiviral state in cells, type I/III IFN may exert a more direct effect on viral activity at a specific point during the viral life cycle. To directly test whether type I or III IFN affects MCPyV activities, we treated HDFs with MCPyV and either IFN-β, IFN-λ, or both on day 2 of infection prior to the addition of FBS to the cells. Treatment with IFN-β, but not IFN-λ, significantly repressed viral replication (Fig. 6A), indicating that type I IFN is primarily responsible for the impact on MCPyV infection.
FIG 6.
Treatment with IFN-β, but not IFN-λ1, inhibits MCPyV postentry activities. (A) MCPyV-infected HDFs were either untreated or treated with 1,000 U/mL IFN-β, 1,000 U/mL IFN-λ1, or both for 30 min before FBS was added to the medium on day 2 postinfection. Cells were harvested for DNA extraction on day 5 postinfection. MCPyV genomic DNA levels were determined by qPCR quantification and normalized to the GAPDH genomic DNA levels. The value for one of the MCPyV-infected HDF groups with no IFN treatment was set as 1. (B) HDFs infected with MCPyV were either untreated or treated with 100 U/mL IFN-β at the time points indicated. Cells were harvested for DNA extraction on day 5 postinfection. MCPyV genomic DNA levels were determined by qPCR quantification and normalized to the GAPDH genomic DNA levels. The value for one of the MCPyV-infected HDF samples with no IFN treatment was set as 1. Error bars represent the standard deviations from three independent experiments. ***, P < 0.001; ns, not significant.
To further investigate which stage of the MCPyV life cycle is sensitive to IFN-β’s inhibition effect, treatment was repeated at two different time points: prior to viral infection and prior to the initiation of viral gene expression and replication, as marked by the addition of FBS. Interestingly, treatment of HDFs with IFN-β prior to infection had no significant effect on viral replication, but IFN-β added to the cells 2 days posttreatment was able to significantly inhibit viral replication activities (Fig. 6B). This observation indicates that type I IFN specifically inhibits MCPyV after viral entry has already occurred.
Type I IFN treatment inhibits MCPyV early transcription.
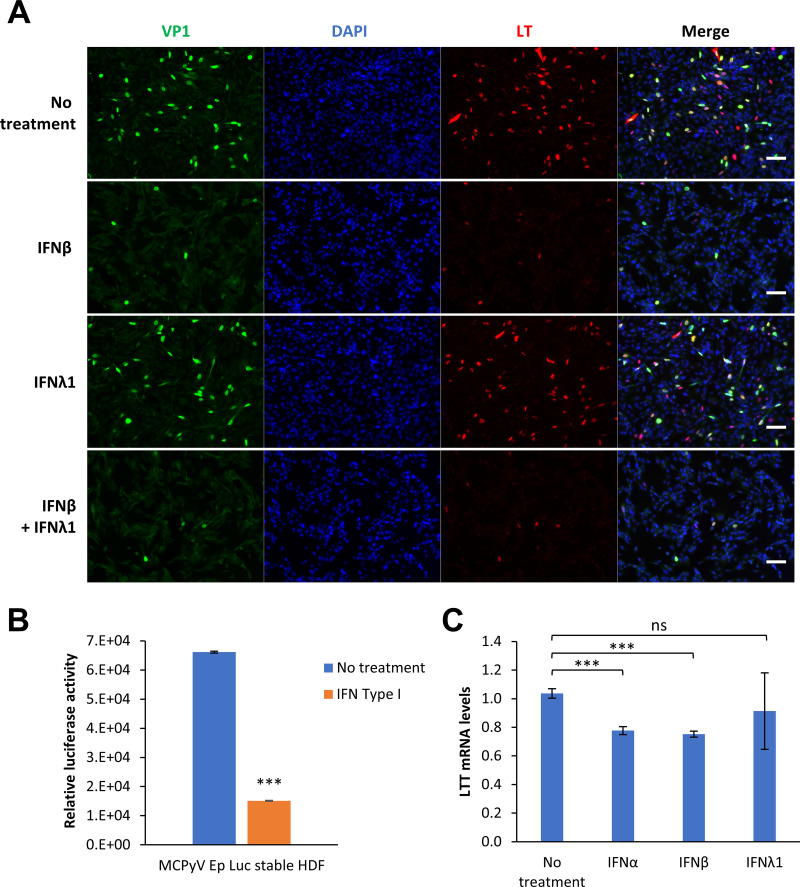
To elucidate which aspects of the MCPyV viral life cycle are suppressed by IFN, we first performed IF analysis of MCPyV-infected and IFN-treated HDFs to examine viral protein expression. Treatment with IFN-β and not IFN-λ on day 2 postinfection significantly repressed the expression of both MCPyV LT and VP1 at the protein level (Fig. 7A). Since protein translation occurs downstream of transcription, and since MCPyV replication is dependent on LT expression, we also investigated whether type I IFN antagonizes MCPyV infection at the level of viral transcription. Stable HDFs expressing either a luciferase reporter or the MCPyV LTT and sT genes, all under the regulation of the MCPyV early promoter, were used to examine the direct effects of IFN on MCPyV promoter-driven gene expression (Fig. 7B and C). Treatment of both of these cell types with type I IFNs inhibited MCPyV early promoter-driven expression. In contrast, ISRE-driven luciferase reporter gene expression in stable HDFs is stimulated by IFN (Fig. S3A). Our studies therefore demonstrate that type I IFNs antagonize MCPyV at the level of early viral transcription, a crucial process during the viral life cycle (Fig. 7B and C).
FIG 7.
Type I IFN treatment inhibits MCPyV early transcription. (A) Treatment with IFN-β, but not IFN-λ1, inhibits MCPyV activities. MCPyV-infected HDFs were either untreated or treated with 1,000 U/mL IFN-β, 1,000 U/mL IFN-λ1, or both for 30 min before FBS was added to the medium on day 2 postinfection. Cells were fixed on day 5 postinfection, immunostained for VP1 and LT, and counterstained with DAPI. Bars, 100 μm. (B) HDFs stably expressing an MCPyV early promoter-driven luciferase reporter were either untreated or treated with 500 U/mL type I IFN for 20 h. Cell lysates were harvested for luciferase assays and normalized to the protein levels measured by Bradford assays. (C) HDFs stably expressing MCPyV early promoter-driven LTT/ST were untreated or treated with 1,000 U/mL IFN-α, IFN-β, or IFN-λ1 for 19 h. LTT mRNA levels were measured by RT-qPCR and normalized to GAPDH mRNA levels. The value for one of the no-treatment groups was set as 1. Error bars represent the standard deviations from three independent experiments. ***, P < 0.001; ns, not significant.
Growth factor-mediated stimulation of MCPyV infection provides a mechanism to support viral persistence.
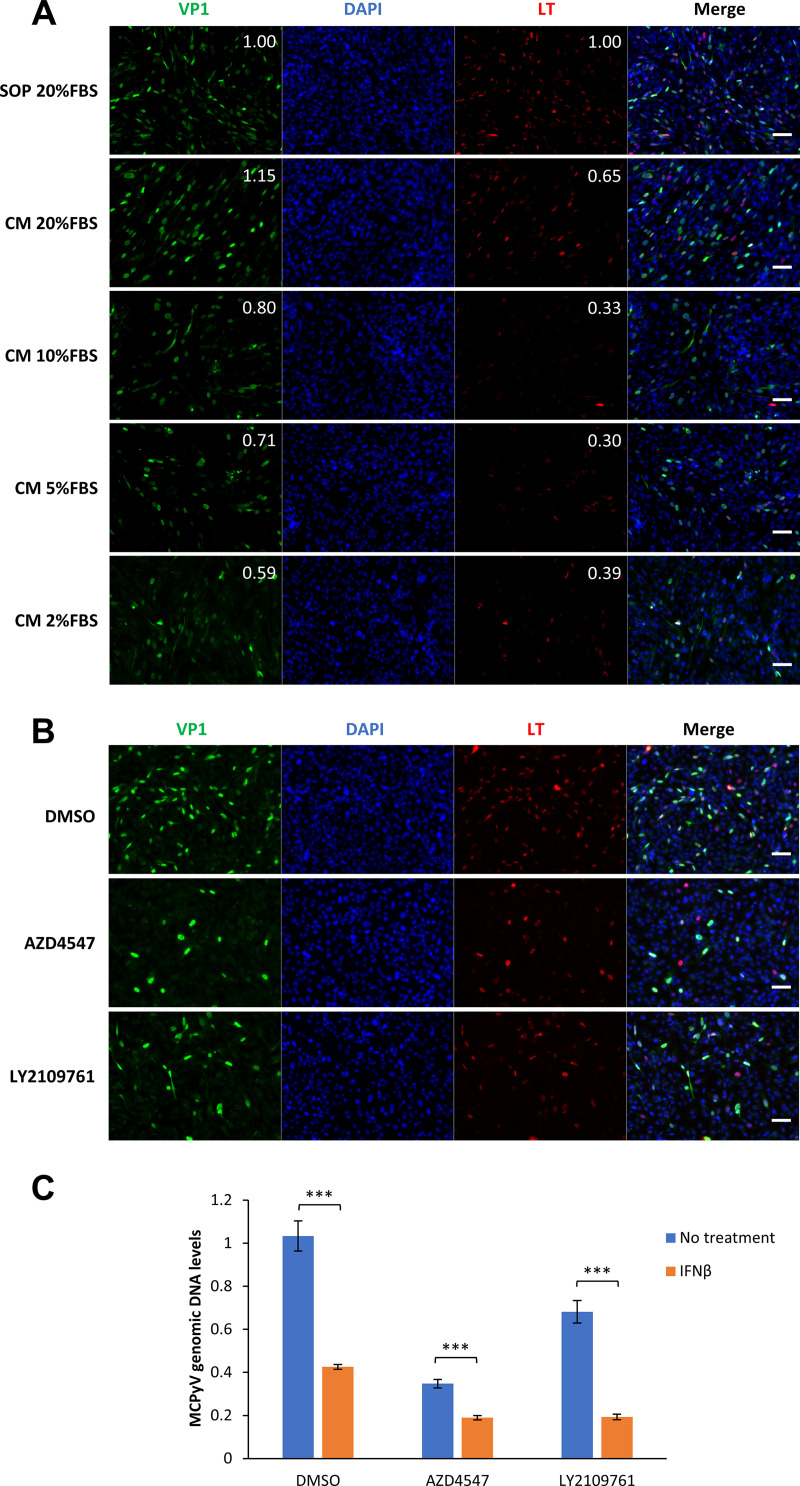
Throughout our studies, we observed that while IFN treatment could significantly inhibit MCPyV replication and transcription, it did not completely abrogate these viral activities (Fig. 6 and 7). These results suggest that other factors in our infection system may potentially antagonize type I IFN during MCPyV infection. A key component of our MCPyV infection system is the addition of FBS to cells after they have been treated with MCPyV in serum-free medium (61). We found that FBS can inhibit viral entry but promote viral gene expression (61). FBS contains many growth factors, such as insulin-like growth factor 1 (IGF-1), transforming growth factor beta (TGF-β), and fibroblast growth factor 2 (FGF-2), which are among the cocktail of growth factors enriched near proliferative fibroblasts at a wound site in damaged human skin (70–72). We therefore tested if FBS or its contained growth factor elements were involved in stimulating MCPyV activity during infection.
As we described previously (61), FBS added at a 20% final concentration to MCPyV-treated cells was sufficient to stimulate robust MCPyV gene expression, but reducing FBS concentration to 2 to 10% caused a significant reduction in LT and VP1 expression (Fig. 8A). This observation supports the idea that some of its components could directly upregulate MCPyV infectious activity in HDFs. To probe which growth factors contained in FBS are involved, we utilized inhibitors against the receptors of each growth factor to determine whether downregulation of a specific growth factor activity has any repressive effects on MCPyV and its proliferation. We found that blocking the function of the TGF-β and FGF receptors with the respective inhibitors LY2109761 and AZD4547 significantly repressed MCPyV protein expression and replication (Fig. 8B and C). These studies therefore identify TGF-β and FGFs as the key growth factors that stimulate MCPyV gene expression and replication. Importantly, we found that combining IFN-β with either AZD4547 or LY2109761 further inhibited MCPyV replication, providing further support for our hypothesis that the growth factors induced in wounded or UV-irradiated skin promote MCPyV propagation by antagonizing the IFN response (Fig. 8C).
FIG 8.
Growth factors in FBS stimulate MCPyV infection. (A) HDFs were treated with MCPyV. On day 2 postinfection, one group of the cells was maintained in the infection medium and supplemented with 20% FBS according to our standard infection protocol (SOP). For the other two groups of cells, the medium was changed to fresh medium (CM) supplemented with 20%, 10%, 5%, or 2% FBS. Cells were fixed on day 5 postinfection, immunostained for VP1 and LT, and counterstained with DAPI. The brightness of the IF signal was quantified using the IntDen (integrated density) function of ImageJ software. The values for the SOP photos were set as 1. The relative brightness of VP1 and LT signals was calculated by normalizing each value to the DAPI signal (top right corners). Bars, 100 μm. (B) MCPyV-infected or mock-infected HDFs were treated with DMSO, 1 μM AZD4547, or 2 μM LY2109761 on day 2 postinfection. Cells were fixed on day 5 postinfection, immunostained for VP1 and LT, and counterstained with DAPI. Bars, 100 μm. (C) MCPyV-infected HDFs were either untreated or treated with 100 U/mL IFN-β for 3 h on day 2 postinfection. The cells were then treated with 20% FBS and either DMSO, 1 μM AZD4547, or 2 μM LY2109761. Cells were harvested for DNA extraction on day 5 postinfection. MCPyV genomic DNA levels were determined by qPCR quantification and normalized to the GAPDH genomic DNA levels. The value for one of the MCPyV-infected, DMSO-treated samples without IFN treatment was set as 1. Error bars represent the standard deviations from three independent experiments. ***, P < 0.001.
DISCUSSION
Like many human cancers with a viral etiology, MCC develops much more frequently in immunosuppressed individuals (52, 53), underscoring the critical role of host immunity in viral oncogenic control. MCPyV is one of the most lethal tumorigenic viruses in immunocompromised patients, but how the virus interfaces with the host immune system to achieve persistent infection and how inadequate control of MCPyV infection encourages MCC tumorigenesis remain poorly understood.
In previous studies, we applied the MCPyV infection model established in our lab to investigate the largely unknown innate immune response to MCPyV infection. We showed that MCPyV infection generates pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) that activate STING- and NF-κB-mediated induction of cytokines and ISGs (64). In the current study, we further discovered that MCPyV infection of primary normal HDFs stimulates expression of type I and III IFNs, which in turn stimulate robust induction of its downstream ISGs (Fig. 1 and 2). Due to the role of IFNs and ISGs in stimulating the antiviral state of host cells (73), we assessed the signaling pathways responsible for IFN-induced ISG induction and its functional impact on MCPyV infection. We found that treatment with IFN-β antibody, JAK inhibitors, and CRISPR KO of IFNAR1 could all dramatically repress MCPyV infection-induced ISG expression, but none of these approaches significantly elevated MCPyV replication activities (Fig. 3 to 5). These findings suggest that IFN-induced ISG production in response to MCPyV infection is not essential for limiting viral infection in HDFs. Instead, we found that type I IFN exerts a more direct effect on MCPyV infection at the postentry stage by repressing early viral transcription (Fig. 6 and 7). This finding is significant because MCPyV early gene transcription is critical for supporting both the MCPyV life cycle and MCC tumorigenesis. In addition to the effect of IFNs on MCPyV infection, we found that growth factors such as TGF-β and FGFs, which are present in FBS but also physiologically upregulated in the dermis during healing of skin wounds and UV-induced skin damage (70–72, 74), could significantly stimulate MCPyV proliferation (Fig. 8).
Together, our data support a hypothetical model in which MCPyV maintains persistent infection in healthy individuals. In this model, healthy immune responses such as IFN production induced by viral activity may restrict viral propagation to reduce MCPyV burden within the host. At the same time, growth factors induced by UV irradiation or abrasion of the skin may stimulate infected dermal fibroblasts to promote MCPyV propagation. A delicate balance of these mutually antagonizing factors would allow the virus to persist at a low level in infected cells. In immunocompromised patients and those affected by MCC risk factors, such as excessive exposure to UV radiation and wounding (42), disturbance of the virus-host balance could alter the course of MCPyV propagation to stimulate pathological rampant MCPyV replication and oncogene overexpression, which can in turn promote viral DNA integration and MCPyV-induced tumorigenesis.
Our studies also revealed a cross talk between the innate immune response and growth factor-induced signaling pathways. We previously found that MCPyV infection could activate the STING/TBK1/IRF3 and NF-κB pathways (64). In addition to its antiviral properties, it has been well documented that the NF-κB pathway can also induce the expression of key growth factors, such as IGF-1, TGF-β, FGFs, and epidermal growth factors (EGFs), and stimulate the activities of the growth factor receptors (75–80). These observations suggest that even in healthy unwounded skin, growth factor signaling pathways can be stimulated inside infected cells by activated NF-κB to promote viral propagation. Conversely, these growth factors and related receptor kinases can also negatively regulate NF-κB-associated pathways, providing a mechanism by which these growth factors may antagonize IFN signaling to promote MCPyV infection (81, 82).
The findings herein also pose questions regarding the role of ISGs in MCPyV infection. Previous studies have shown that overexpression of T antigens encoded by SV40, JC polyomavirus (JCPyV), or BK polyomavirus (BKPyV) in mouse embryonic fibroblasts leads to upregulation of ISGs and an antiviral state (83, 84). Similarly, JCPyV infection of human renal proximal tubular epithelial cells triggers IFN secretion and subsequent ISG induction, which could at least partially control viral infection (85). BKPyV infection of microvascular endothelial cells also stimulates an IFN-mediated ISG induction that could dampen viral infection (86). However, it is unclear why blocking the induction of ISGs in response to MCPyV infection does not appear to have a significant impact on viral activity. One explanation is that MCPyV has evolved strategies to successfully counteract the ISG-mediated host immune response. Another possibility is that some ISGs are induced by IFNs in a manner that is independent of the IFNR-JAK-STAT canonical pathway and therefore are not repressed by IFN-β antibody, JAK inhibitors, or IFNAR1 KO. ISG54 is one such ISG, which appeared to be nonessential for blocking MCPyV infection (Fig. S2). Still, it is possible that other ISGs not explored here are involved in controlling MCPyV infection and need to be investigated. Finally, it could also be that the amount of type I IFNs and ISGs induced by MCPyV infection may be too small to block viral replication and transcription (Fig. S3B).
Our observation that type I IFN can repress MCPyV early transcription is consistent with a previous study showing that type I and II IFNs exert direct inhibitory effects on MCC cell lines in vitro and in vivo through modulation of MCPyV LT expression (87). Our findings add to the existing literature supporting the ability of IFNs to control the transcriptional regulation of specific genes such as T cell coinhibitory receptors, programmed death ligand 1 (PD-L1), nitric oxide synthase 2 (NOS2), and triggering receptors expressed on myeloid cells (TREM) (88–91). However, the manner in which IFNs inhibit the MCPyV early promoter remains to be fully investigated. Nevertheless, because MCPyV-positive MCC cells are dependent on the expression of viral oncogenes to survive, our findings as well as those presented by Willmes et al. (87) indicate that type I IFN may be a promising therapeutic option for treating patients with MCPyV-positive MCC.
In summary, by characterizing the MCPyV immune signaling pathways and examining their impact on viral infection, our studies provide new clues for understanding how an oncogenic virus such as MCPyV strikes a fine balance with the host defense and growth control mechanisms to achieve persistent infection. Our findings also shed new light on how the antiviral and anticancer properties of type I IFNs may be explored as a new strategy for preventing and treating MCPyV-positive MCC.
MATERIALS AND METHODS
Cell culture.
HDFs were isolated from human foreskin tissues as described in our previous studies (61, 63) and were maintained in DMEM (Invitrogen) supplemented with 10% FBS (HyClone), 1× nonessential amino acids (Gibco), and 1× GlutaMAX (Gibco).
MCPyV infection.
HDFs or lentiCRISPRv2 sgRNA stable HDFs were infected with MCPyV using the method described previously (61, 63), which is outlined briefly here for convenience. HDFs were seeded with 1.2 × 104 cells each in a 96-well plate in DMEM–F-12 medium (11330-032; Thermo Fisher Scientific) containing 20 ng/mL EGF (236-EG-200; R&D Systems), 20 ng/mL basic FGF (bFGF) (PHG0368; Thermo Fisher Scientific), 3 μM CHIR99021 (13122; Cayman Chemical), and 1 mg/mL collagenase type IV (17104019; Thermo Fisher Scientific). The same amounts of MCPyV virions (108 to 109 viral genome equivalents) were added to each 96-well plate for viral infection, and 109 viral genome equivalents were used for each 96-well plate for Fig. 1 and Fig. 2 as well as the first HDF infection for Fig. 5. A dose of 108 viral genome equivalents per 96-well was used in the rest of the MCPyV infection experiments. After the cells were incubated at 37°C in 5% CO2 for 2 days, FBS was added to each well to reach a 20% final concentration. Depending on the experiment, the cells were incubated for 3 to 5 days before harvest.
To analyze MCPyV genomic DNA levels, harvested cell pellets were lysed in QuickExtract DNA extraction solution (Biosearch Technologies) according to the manufacturer’s instructions. The extracted DNA samples were then subjected to real-time qPCR using a QuantStudio 3 real-time PCR system (Applied Biosystems) with SYBR green master mix (Applied Biosystems). The MCPyV genomic DNA levels were detected with NCRR-targeted primers and normalized to the level of genomic glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Both NCRR and genomic GAPDH primer sequences can be found in reference 64.
Chemicals, antibodies, and reagents.
Ruxolitinib (Cayman Chemical), AZD4547 (LKT Laboratories), and LY2109761 (Sigma) powders were dissolved in dimethyl sulfoxide (DMSO) to a stock concentration of 10 mM. Normal goat IgG control (AB-108-C; R&D Systems) and human IFN-β antibody (AF814-SP; R&D Systems) lyophilized powder were reconstituted with sterile phosphate-buffered saline (PBS) to a stock concentration of 0.5 μg/μL for final use at 2 μg/mL for blocking the IFN pathway response. Recombinant human IFN-β (300-02BC; PeproTech) and recombinant human IFN-λ1 (300-02L; PeproTech) lyophilized powders were reconstituted with DMEM–F-12 medium supplemented with 1% FBS to 106 U/mL. Universal type I interferon (IFN-α; 11200-2; PBL Assay Science) was also diluted to 106 U/mL with DMEM–F-12 medium. Stock solutions of all chemicals, antibodies, or other reagents were stored in small aliquots at −80°C.
Recombinant lentivirus plasmid constructs.
sgRNAs were cloned into the lentiCRISPR v2 plasmid (Addgene) following protocols from the Feng Zhang lab (92). Oligonucleotides for the generation of sgRNA cassettes targeting the IFNAR1 or ISG54 genes were selected from a CRISPick search, the Brunello library, or the GeCKO v2 library (93–95) and are as follows: IFNAR1 sg1, 5′-GACCCTAGTGCTCGTCGCCG-3′; IFNAR1 sg2, 5′-TAGATGACAACTTTATCCTG-3′; IFNAR1 sg3, 5′-TCATTTACACCATTTCGCAA-3′; ISG54 sg1, 5′-GCACCTCAAAGGGCAAAACG-3′; ISG54 sg2, 5′-TTTCACCTGGAACTTGATGG-3′; and ISG54 sg3, 5′-AGCCACAATGTGCAACCTAC-3′.
For cloning of the pLenti-MCPyVEP-Luciferase-IRES-Puro construct, the firefly luciferase gene was PCR amplified from pGL3 (Promega) and subcloned into the pLenti-MCVEP-RFP-IRES-Puro vector (96) using the AgeI and ClaI sites.
For cloning of the pLenti-MCVEP-ST/LTT-IRES-Puro construct, the MCPyV early promoter (MCVEP) and the LTT/ST gene fragment were PCR amplified from MCPyV genomes religated from the pR17b plasmid (kindly provided by Christopher B. Buck [NCI]). The resulting fragment was subcloned using the XbaI/SpeI and MluI sites into the pLenti-IRES-Puro vector, which was modified from the pTRIPZ vector (Open Biosystems) by removing the fragments between the MluI and NotI sites.
For cloning of the pTRIPZ-ISRE-Luc construct, a synthesized promoter fragment including 10 copies of ISRE (interferon-sensitive response element with the sequence GATCAAAGTGAAAGG) and a minimal promoter sequence were cloned into the XbaI and AgeI sites to replace the inducible promoter region in the pTRIPZ vector to generate the pTRIPZ-ISRE-RFP construct. The firefly luciferase gene was PCR amplified from pGL4 (Promega) and subcloned into the AgeI/XmaI and MluI sites to replace the red fluorescent protein (RFP) gene in pTRIPZ-ISRE-RFP.
Generation of stable HDFs.
Stable cells were prepared as described previously (64, 96). To package lentiviruses, HEK293T cells at 90% confluence were transfected with the lentivirus construct together with psPAX2 and pMD.2G using Lipofectamine 2000 (Invitrogen). At 6 to 8 h posttransfection, the culture medium was changed to fresh medium. At 2 days posttransfection, lentiviruses in the supernatant were harvested, filtered through a 0.45-μm syringe filter, and concentrated with Lenti-X concentrator (TaKaRa) according to the manufacturer’s instructions. Early-passage HDFs were transduced with concentrated lentivirus supplemented with 8 μg/mL Polybrene (Sigma). Starting on day 2 after transduction, cells were selected using 1 μg/mL puromycin for 2 to 3 weeks until the cells proliferated.
RT-qPCR.
Total RNA was isolated using the NucleoSpin RNA XS kit (Macherey-Nagel) following the manufacturer’s instructions. Reverse transcription was performed using a 20-μL reaction mixture containing 350 ng of total RNA, oligo(dT) primers (Invitrogen), deoxynucleoside triphosphates (dNTPs; Invitrogen), and Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen). Real-time qPCR was performed using a QuantStudio 3 real-time PCR system (Applied Biosystems) with SYBR green master mix (Applied Biosystems). The mRNA level of each gene was normalized to the GAPDH mRNA level. The RT-qPCR primer sequences used in this study are as follows (also available in reference 64): IFN-β F, 5′-ACTGCCTCAAGGACAGGATG-3′; IFN-β R, 5′-AGCCAGGAGGTTCTCAACAA-3′; IFN-λ1 F, 5′-TTCCAAGCCCACCACAAC-3′; IFN-λ1 R, 5′-TCCCTCACCTGGAGAAGC-3′; LT F, 5′-TGACTTCTCTATGTTTGATGAGGTTGAC-3′; LT R, 5′-GACCCATACCCAGAGGAAGAG-3′; MMP-3 F, 5′-TGGGCCAGGGATTAATGGAG-3′; MMP-3 R, 5′-GGAACCGAGTCAGGTCTGTG-3′ (F indicates forward primers, and R indicates reverse primers).
Immunofluorescent staining.
Cells cultured on coverslips were fixed with 3% paraformaldehyde in PBS for 20 min. Immunofluorescent staining was performed as previously described (64). The following antibodies were used as primary antibodies: anti-MCPyV LT (1:250; sc-136172; Santa Cruz Biotechnology), anti-MCPyV VP1 (1:2,000; Christopher Buck laboratory), anti-Mx1 (1:1,000; 37849S; Cell Signaling Technology), and anti-IFI16 (1:1,000; ab55328; Abcam). Alexa Fluor 594 goat anti-mouse IgG (1:500; A11032; Thermo Fisher Scientific), Alexa Fluor 488 goat anti-rabbit IgG (1:500; A11034; Thermo Fisher Scientific), Alexa Fluor 594 goat anti-rabbit IgG (1:500; A11012; Thermo Fisher Scientific), and Alexa Fluor 488 goat anti-mouse IgG (1:500; A11029; Thermo Fisher Scientific) were used as secondary antibodies. All immunofluorescent images were captured using an inverted fluorescence microscope (IX81; Olympus) as described previously (64). The scale bars were added using ImageJ software.
Western blotting.
Preparation of whole-cell lysates and Western blot analyses were performed as described previously (64). The primary antibodies used in this study include anti-IFNAR1 (1:2,000; ab247992; Abcam), anti-ISG54 (1:1,000; HPA003408; Sigma), anti-OAS1 (1:500; sc-374656; Santa Cruz Biotechnology), anti-MCPyV LT (1:500; sc-136172; Santa Cruz Biotechnology), anti-Mx1 (1:2,000; 37849S; Cell Signaling Technology), antiviperin (1:2,000; 13996S; Cell Signaling Technology), and anti-GAPDH (1:2,000; 2118S; Cell Signaling Technology). Horseradish peroxidase (HRP)-linked anti-rabbit IgG (1:3,000; 7074S; Cell Signaling Technology) and HRP-linked anti-mouse IgG (1:3,000; 7076S; Cell Signaling Technology) were used as secondary antibodies. Western blots were developed using SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific), and images were captured using an Amersham Imager 600 (GE Healthcare).
Statistical analysis.
Statistical analysis was performed using the unpaired t test formula of Microsoft Excel software to compare the data from the control and experimental groups. A two-tailed P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
We thank all members of the Jianxin You laboratory for their feedback and support throughout this project.
This work was supported by the National Institutes of Health (NIH) grants R01CA187718, R21AR074073, R21AI149761, and 1R21CA267803 and the NCI Specialized Program of Research Excellence (SPORE) in Skin Cancer (P50-CA174523).
Footnotes
Supplemental material is available online only.
Contributor Information
Jianxin You, Email: jianyou@pennmedicine.upenn.edu.
Lawrence Banks, International Centre for Genetic Engineering and Biotechnology.
REFERENCES
- 1.Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schadendorf D, Lebbe C, Zur Hausen A, Avril MF, Hariharan S, Bharmal M, Becker JC. 2017. Merkel cell carcinoma: epidemiology, prognosis, therapy and unmet medical needs. Eur J Cancer 71:53–69. doi: 10.1016/j.ejca.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 3.Gjoerup O, Chang Y. 2010. Update on human polyomaviruses and cancer. Adv Cancer Res 106:1–51. doi: 10.1016/S0065-230X(10)06001-X. [DOI] [PubMed] [Google Scholar]
- 4.Harms PW. 2017. Update on Merkel cell carcinoma. Clin Lab Med 37:485–501. doi: 10.1016/j.cll.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Agelli M, Clegg LX, Becker JC, Rollison DE. 2010. The etiology and epidemiology of merkel cell carcinoma. Curr Probl Cancer 34:14–37. doi: 10.1016/j.currproblcancer.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Agelli M, Clegg LX. 2003. Epidemiology of primary Merkel cell carcinoma in the United States. J Am Acad Dermatol 49:832–841. doi: 10.1016/s0190-9622(03)02108-x. [DOI] [PubMed] [Google Scholar]
- 7.Lemos B, Nghiem P. 2007. Merkel cell carcinoma: more deaths but still no pathway to blame. J Invest Dermatol 127:2100–2103. doi: 10.1038/sj.jid.5700925. [DOI] [PubMed] [Google Scholar]
- 8.Bhatia S, Afanasiev O, Nghiem P. 2011. Immunobiology of Merkel cell carcinoma: implications for immunotherapy of a polyomavirus-associated cancer. Curr Oncol Rep 13:488–497. doi: 10.1007/s11912-011-0197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engels EA, Frisch M, Goedert JJ, Biggar RJ, Miller RW. 2002. Merkel cell carcinoma and HIV infection. Lancet 359:497–498. doi: 10.1016/S0140-6736(02)07668-7. [DOI] [PubMed] [Google Scholar]
- 10.Kaae J, Hansen AV, Biggar RJ, Boyd HA, Moore PS, Wohlfahrt J, Melbye M. 2010. Merkel cell carcinoma: incidence, mortality, and risk of other cancers. J Natl Cancer Inst 102:793–801. doi: 10.1093/jnci/djq120. [DOI] [PubMed] [Google Scholar]
- 11.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. 2010. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7:509–515. doi: 10.1016/j.chom.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tolstov YL, Pastrana DV, Feng H, Becker JC, Jenkins FJ, Moschos S, Chang Y, Buck CB, Moore PS. 2009. Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. Int J Cancer 125:1250–1256. doi: 10.1002/ijc.24509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foulongne V, Sauvage V, Hebert C, Dereure O, Cheval J, Gouilh MA, Pariente K, Segondy M, Burguière A, Manuguerra J-C, Caro V, Eloit M. 2012. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One 7:e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen T, Hedman L, Mattila PS, Jartti T, Ruuskanen O, Soderlund-Venermo M, Hedman K. 2011. Serological evidence of Merkel cell polyomavirus primary infections in childhood. J Clin Virol 50:125–129. doi: 10.1016/j.jcv.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 15.Martel-Jantin C, Pedergnana V, Nicol JT, Leblond V, Tregouet DA, Tortevoye P, Plancoulaine S, Coursaget P, Touze A, Abel L, Gessain A. 2013. Merkel cell polyomavirus infection occurs during early childhood and is transmitted between siblings. J Clin Virol 58:288–291. doi: 10.1016/j.jcv.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Viscidi RP, Rollison DE, Sondak VK, Silver B, Messina JL, Giuliano AR, Fulp W, Ajidahun A, Rivanera D. 2011. Age-specific seroprevalence of Merkel cell polyomavirus, BK virus, and JC virus. Clin Vaccine Immunol 18:1737–1743. doi: 10.1128/CVI.05175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tolstov YL, Knauer A, Chen JG, Kensler TW, Kingsley LA, Moore PS, Chang Y. 2011. Asymptomatic primary Merkel cell polyomavirus infection among adults. Emerg Infect Dis 17:1371–1380. doi: 10.3201/eid1708.110079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stang A, Becker JC, Nghiem P, Ferlay J. 2018. The association between geographic location and incidence of Merkel cell carcinoma in comparison to melanoma: an international assessment. Eur J Cancer 94:47–60. doi: 10.1016/j.ejca.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulson KG, Park SY, Vandeven NA, Lachance K, Thomas H, Chapuis AG, Harms KL, Thompson JA, Bhatia S, Stang A, Nghiem P. 2018. Merkel cell carcinoma: current US incidence and projected increases based on changing demographics. J Am Acad Dermatol 78:457–463.E2. doi: 10.1016/j.jaad.2017.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzgerald TL, Dennis S, Kachare SD, Vohra NA, Wong JH, Zervos EE. 2015. Dramatic increase in the incidence and mortality from Merkel cell carcinoma in the United States. Am Surg 81:802–806. doi: 10.1177/000313481508100819. [DOI] [PubMed] [Google Scholar]
- 21.Hodgson NC. 2005. Merkel cell carcinoma: changing incidence trends. J Surg Oncol 89:1–4. doi: 10.1002/jso.20167. [DOI] [PubMed] [Google Scholar]
- 22.Liu W, MacDonald M, You J. 2016. Merkel cell polyomavirus infection and Merkel cell carcinoma. Curr Opin Virol 20:20–27. doi: 10.1016/j.coviro.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harms KL, Healy MA, Nghiem P, Sober AJ, Johnson TM, Bichakjian CK, Wong SL. 2016. Analysis of prognostic factors from 9387 Merkel cell carcinoma cases forms the basis for the new 8th edition AJCC Staging System. Ann Surg Oncol 23:3564–3571. doi: 10.1245/s10434-016-5266-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colunga A, Pulliam T, Nghiem P. 2017. Merkel cell carcinoma in the age of immunotherapy: facts and hopes. Clin Cancer Res 24:2035–2043. doi: 10.1158/1078-0432.ccr-17-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer JG, Blom A, Doumani R, Lewis C, Tarabadkar ES, Anderson A, Ma C, Bestick A, Parvathaneni U, Bhatia S, Nghiem P. 2016. Response rates and durability of chemotherapy among 62 patients with metastatic Merkel cell carcinoma. Cancer Med 5:2294–2301. doi: 10.1002/cam4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowey CL, Mahnke L, Espirito J, Helwig C, Oksen D, Bharmal M. 2017. Real-world treatment outcomes in patients with metastatic Merkel cell carcinoma treated with chemotherapy in the USA. Future Oncol 13:1699–1710. doi: 10.2217/fon-2017-0187. [DOI] [PubMed] [Google Scholar]
- 27.Nghiem P, Bhatia S, Lipson EJ, Sharfman WH, Kudchadkar RR, Brohl AS, Friedlander PA, Daud A, Kluger HM, Reddy SA, Boulmay BC, Riker AI, Burgess MA, Hanks BA, Olencki T, Margolin K, Lundgren LM, Soni A, Ramchurren N, Church C, Park SY, Shinohara MM, Salim B, Taube JM, Bird SR, Ibrahim N, Fling SP, Homet MB, Sharon E, Cheever MA, Topalian SL. 2019. Durable tumor regression and overall survival in patients with advanced Merkel cell carcinoma receiving pembrolizumab as first-line therapy. J Clin Oncol 37:693–702. doi: 10.1200/JCO.18.01896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Angelo SP, Russell J, Lebbé C, Chmielowski B, Gambichler T, Grob J-J, Kiecker F, Rabinowits G, Terheyden P, Zwiener I, Bajars M, Hennessy M, Kaufman HL. 2018. Efficacy and safety of first-line avelumab treatment in patients with stage IV metastatic Merkel cell carcinoma: a preplanned interim analysis of a clinical trial. JAMA Oncol 4:e180077. doi:10.1001/jamaoncol.2018.0077. doi: 10.1001/jamaoncol.2018.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becker JC, Stang A, Hausen AZ, Fischer N, DeCaprio JA, Tothill RW, Lyngaa R, Hansen UK, Ritter C, Nghiem P, Bichakjian CK, Ugurel S, Schrama D. 2017. Epidemiology, biology and therapy of Merkel cell carcinoma: conclusions from the EU project IMMOMEC. Cancer Immunol Immunother 67:341–351. doi: 10.1007/s00262-017-2099-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W, You J. 2020. Molecular mechanisms of Merkel cell polyomavirus transformation and replication. Annu Rev Virol 7:289–307. doi: 10.1146/annurev-virology-011720-121757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison CJ, Meinke G, Kwun HJ, Rogalin H, Phelan PJ, Bullock PA, Chang Y, Moore PS, Bohm A. 2011. Asymmetric assembly of Merkel cell polyomavirus large T-antigen origin binding domains at the viral origin. J Mol Biol 409:529–542. doi: 10.1016/j.jmb.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwun HJ, Guastafierro A, Shuda M, Meinke G, Bohm A, Moore PS, Chang Y. 2009. The minimum replication origin of Merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J Virol 83:12118–12128. doi: 10.1128/JVI.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo GJ, Chen CJ, Sullivan CS. 2009. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology 383:183–187. doi: 10.1016/j.virol.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Carter JJ, Daugherty MD, Qi X, Bheda-Malge A, Wipf GC, Robinson K, Roman A, Malik HS, Galloway DA. 2013. Identification of an overprinting gene in Merkel cell polyomavirus provides evolutionary insight into the birth of viral genes. Proc Natl Acad Sci USA 110:12744–12749. doi: 10.1073/pnas.1303526110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schowalter RM, Pastrana DV, Buck CB. 2011. Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry. PLoS Pathog 7:e1002161. doi: 10.1371/journal.ppat.1002161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schowalter RM, Reinhold WC, Buck CB. 2012. Entry tropism of BK and Merkel cell polyomaviruses in cell culture. PLoS One 7:e42181. doi: 10.1371/journal.pone.0042181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwun HJ, Shuda M, Feng H, Camacho CJ, Moore PS, Chang Y. 2013. Merkel cell polyomavirus small T antigen controls viral replication and oncoprotein expression by targeting the cellular ubiquitin ligase SCFFbw7. Cell Host Microbe 14:125–135. doi: 10.1016/j.chom.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsang SH, Wang R, Nakamaru-Ogiso E, Knight SA, Buck CB, You J. 2016. The oncogenic small tumor antigen of Merkel cell polyomavirus is an iron-sulfur cluster protein that enhances viral DNA replication. J Virol 90:1544–1556. doi: 10.1128/JVI.02121-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Li J, Schowalter RM, Jiao J, Buck CB, You J. 2012. Bromodomain protein Brd4 plays a key role in Merkel cell polyomavirus DNA replication. PLoS Pathog 8:e1003021. doi: 10.1371/journal.ppat.1003021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng H, Kwun HJ, Liu X, Gjoerup O, Stolz DB, Chang Y, Moore PS. 2011. Cellular and viral factors regulating Merkel cell polyomavirus replication. PLoS One 6:e22468. doi: 10.1371/journal.pone.0022468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Houben R, Schrama D, Becker JC. 2009. Molecular pathogenesis of Merkel cell carcinoma. Exp Dermatol 18:193–198. doi: 10.1111/j.1600-0625.2009.00853.x. [DOI] [PubMed] [Google Scholar]
- 42.Chang Y, Moore PS. 2012. Merkel cell carcinoma: a virus-induced human cancer. Annu Rev Pathol Mech Dis 7:123–144. doi: 10.1146/annurev-pathol-011110-130227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng J, Rozenblatt-Rosen O, Paulson KG, Nghiem P, DeCaprio JA. 2013. Merkel cell polyomavirus large T antigen has growth-promoting and inhibitory activities. J Virol 87:6118–6126. doi: 10.1128/JVI.00385-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. 2011. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest 121:3623–3634. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. 2008. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci USA 105:16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grundhoff A, Fischer N. 2015. Merkel cell polyomavirus, a highly prevalent virus with tumorigenic potential. Curr Opin Virol 14:129–137. doi: 10.1016/j.coviro.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 47.Wendzicki JA, Moore PS, Chang Y. 2015. Large T and small T antigens of Merkel cell polyomavirus. Curr Opin Virol 11:38–43. doi: 10.1016/j.coviro.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spurgeon ME, Lambert PF. 2013. Merkel cell polyomavirus: a newly discovered human virus with oncogenic potential. Virology 435:118–130. doi: 10.1016/j.virol.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, Moore PS, Becker JC. 2010. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol 84:7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verhaegen ME, Mangelberger D, Harms PW, Vozheiko TD, Weick JW, Wilbert DM, Saunders TL, Ermilov AN, Bichakjian CK, Johnson TM, Imperiale MJ, Dlugosz AA. 2015. Merkel cell polyomavirus small T antigen is oncogenic in transgenic mice. J Invest Dermatol 135:1415–1424. doi: 10.1038/jid.2014.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Houben R, Adam C, Baeurle A, Hesbacher S, Grimm J, Angermeyer S, Henzel K, Hauser S, Elling R, Brocker EB, Gaubatz S, Becker JC, Schrama D. 2012. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. Int J Cancer 130:847–856. doi: 10.1002/ijc.26076. [DOI] [PubMed] [Google Scholar]
- 52.Bertrand M, Mirabel X, Desmedt E, Vercambre-Darras S, Martin de Lassalle E, Bouchindhomme B, Guerreschi P, Martinot V, Mortier L. 2013. Merkel cell carcinoma: a new radiation-induced cancer? Ann Dermatol Venereol 140:41–45. doi: 10.1016/j.annder.2012.10.598. [DOI] [PubMed] [Google Scholar]
- 53.Heath M, Jaimes N, Lemos B, Mostaghimi A, Wang LC, Penas PF, Nghiem P. 2008. Clinical characteristics of Merkel cell carcinoma at diagnosis in 195 patients: the AEIOU features. J Am Acad Dermatol 58:375–381. doi: 10.1016/j.jaad.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wieland U, Silling S, Scola N, Potthoff A, Gambichler T, Brockmeyer NH, Pfister H, Kreuter A. 2011. Merkel cell polyomavirus infection in HIV-positive men. Arch Dermatol 147:401–406. doi: 10.1001/archdermatol.2011.42. [DOI] [PubMed] [Google Scholar]
- 55.Hashida Y, Kamioka M, Tanaka M, Hosokawa S, Murakami M, Nakajima K, Kikuchi H, Fujieda M, Sano S, Daibata M. 2016. Ecology of Merkel cell polyomavirus in healthy skin among individuals in an Asian cohort. J Infect Dis 213:1708–1716. doi: 10.1093/infdis/jiw040. [DOI] [PubMed] [Google Scholar]
- 56.Koljonen V, Kukko H, Pukkala E, Sankila R, Bohling T, Tukiainen E, Sihto H, Joensuu H. 2009. Chronic lymphocytic leukaemia patients have a high risk of Merkel-cell polyomavirus DNA-positive Merkel-cell carcinoma. Br J Cancer 101:1444–1447. doi: 10.1038/sj.bjc.6605306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hemminki K, Liu X, Ji J, Sundquist J, Sundquist K. 2012. Kaposi sarcoma and Merkel cell carcinoma after autoimmune disease. Int J Cancer 131:E326–E328. doi: 10.1002/ijc.27376. [DOI] [PubMed] [Google Scholar]
- 58.Penn I, First MR. 1999. Merkel's cell carcinoma in organ recipients: report of 41 cases. Transplantation 68:1717–1721. doi: 10.1097/00007890-199912150-00015. [DOI] [PubMed] [Google Scholar]
- 59.Buell JF, Trofe J, Hanaway MJ, Beebe TM, Gross TG, Alloway RR, First MR, Woodle ES. 2002. Immunosuppression and Merkel cell cancer. Transplant Proc 34:1780–1781. doi: 10.1016/s0041-1345(02)03065-8. [DOI] [PubMed] [Google Scholar]
- 60.Wieland U, Kreuter A. 2011. Merkel cell polyomavirus infection and Merkel cell carcinoma in HIV-positive individuals. Curr Opin Oncol 23:488–493. doi: 10.1097/CCO.0b013e3283495a5b. [DOI] [PubMed] [Google Scholar]
- 61.Liu W, Yang R, Payne AS, Schowalter RM, Spurgeon ME, Lambert PF, Xu X, Buck CB, You J. 2016. Identifying the target cells and mechanisms of Merkel cell polyomavirus infection. Cell Host Microbe 19:775–787. doi: 10.1016/j.chom.2016.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu W, Krump NA, MacDonald M, You J. 2018. Merkel cell polyomavirus infection of animal dermal fibroblasts. J Virol 92:e01610-17. doi: 10.1128/JVI.01610-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu W, Krump NA, Buck CB, You J. 2019. Merkel cell polyomavirus infection and detection. J Vis Exp 2019:58950. doi: 10.3791/58950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krump NA, Wang R, Liu W, Yang JF, Ma T, You J. 2021. Merkel cell polyomavirus infection induces an antiviral innate immune response in human dermal fibroblasts. J Virol 95:e02211-20. doi: 10.1128/JVI.02211-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Honda K, Takaoka A, Taniguchi T. 2006. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 66.Yarilina A, Ivashkiv LB. 2010. Type I interferon: a new player in TNF signaling. Curr Dir Autoimmun 11:94–104. doi: 10.1159/000289199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bonizzi G, Karin M. 2004. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol 25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 68.Weaver BK, Ando O, Kumar KP, Reich NC. 2001. Apoptosis is promoted by the dsRNA-activated factor (DRAF1) during viral infection independent of the action of interferon or p53. FASEB J 15:501–515. doi: 10.1096/fj.00-0222com. [DOI] [PubMed] [Google Scholar]
- 69.Stawowczyk M, Van Scoy S, Kumar KP, Reich NC. 2011. The interferon stimulated gene 54 promotes apoptosis. J Biol Chem 286:7257–7266. doi: 10.1074/jbc.M110.207068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martin P. 1997. Wound healing–aiming for perfect skin regeneration. Science 276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 71.Werner S, Grose R. 2003. Regulation of wound healing by growth factors and cytokines. Physiol Rev 83:835–870. doi: 10.1152/physrev.2003.83.3.835. [DOI] [PubMed] [Google Scholar]
- 72.Garoufalia Z, Papadopetraki A, Karatza E, Vardakostas D, Philippou A, Kouraklis G, Mantas D. 2021. Insulin-like growth factor-I and wound healing, a potential answer to non-healing wounds: a systematic review of the literature and future perspectives. Biomed Rep 15:66. doi: 10.3892/br.2021.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schoggins JW. 2019. Interferon-stimulated genes: what do they all do? Annu Rev Virol 6:567–584. doi: 10.1146/annurev-virology-092818-015756. [DOI] [PubMed] [Google Scholar]
- 74.Krämer M, Sachsenmaier C, Herrlich P, Rahmsdorf HJ. 1993. UV irradiation-induced interleukin-1 and basic fibroblast growth factor synthesis and release mediate part of the UV response. J Biol Chem 268:6734–6741. doi: 10.1016/S0021-9258(18)53311-1. [DOI] [PubMed] [Google Scholar]
- 75.Jimi E, Huang F, Nakatomi C. 2019. NF-κB signaling regulates physiological and pathological chondrogenesis. Int J Mol Sci 20:6275. doi: 10.3390/ijms20246275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee JG, Kay EP. 2012. NF-κB is the transcription factor for FGF-2 that causes endothelial mesenchymal transformation in cornea. Invest Ophthalmol Vis Sci 53:1530–1538. doi: 10.1167/iovs.11-9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vandermoere F, El Yazidi-Belkoura I, Adriaenssens E, Lemoine J, Hondermarck H. 2005. The antiapoptotic effect of fibroblast growth factor-2 is mediated through nuclear factor-κB activation induced via interaction between Akt and IκB kinase-β in breast cancer cells. Oncogene 24:5482–5491. doi: 10.1038/sj.onc.1208713. [DOI] [PubMed] [Google Scholar]
- 78.Armstrong K, Robson CN, Leung HY. 2006. NF-kappaB activation upregulates fibroblast growth factor 8 expression in prostate cancer cells. Prostate 66:1223–1234. doi: 10.1002/pros.20376. [DOI] [PubMed] [Google Scholar]
- 79.Rameshwar P, Narayanan R, Qian J, Denny TN, Colon C, Gascon P. 2000. NF-κB as a central mediator in the induction of TGF-β in monocytes from patients with idiopathic myelofibrosis: an inflammatory response beyond the realm of homeostasis. J Immunol 165:2271–2277. doi: 10.4049/jimmunol.165.4.2271. [DOI] [PubMed] [Google Scholar]
- 80.Cheng Y, Che X, Zhang S, Guo T, He X, Liu Y, Qu X. 2020. Positive cross-talk between CXC chemokine receptor 4 (CXCR4) and epidermal growth factor receptor (EGFR) promotes gastric cancer metastasis via the nuclear factor kappa B (NF-kB)-dependent pathway. Med Sci Monit 26:e925019. doi: 10.12659/MSM.925019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maddaluno L, Urwyler C, Rauschendorfer T, Meyer M, Stefanova D, Spörri R, Wietecha M, Ferrarese L, Stoycheva D, Bender D, Li N, Strittmatter G, Nasirujjaman K, Beer HD, Staeheli P, Hildt E, Oxenius A, Werner S. 2020. Antagonism of interferon signaling by fibroblast growth factors promotes viral replication. EMBO Mol Med 12:e11793. doi: 10.15252/emmm.201911793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Drafahl KA, McAndrew CW, Meyer AN, Haas M, Donoghue DJ. 2010. The receptor tyrosine kinase FGFR4 negatively regulates NF-kappaB signaling. PLoS One 5:e14412. doi: 10.1371/journal.pone.0014412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rathi AV, Cantalupo PG, Sarkar SN, Pipas JM. 2010. Induction of interferon-stimulated genes by simian virus 40 T antigens. Virology 406:202–211. doi: 10.1016/j.virol.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Giacobbi NS, Gupta T, Coxon AT, Pipas JM. 2015. Polyomavirus T antigens activate an antiviral state. Virology 476:377–385. doi: 10.1016/j.virol.2014.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Assetta B, De Cecco M, O’Hara B, Atwood WJ. 2016. JC polyomavirus infection of primary human renal epithelial cells is controlled by a type I IFN-induced response. mBio 7:e00903-16. doi: 10.1128/mBio.00903-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.An P, Saenz Robles MT, Duray AM, Cantalupo PG, Pipas JM. 2019. Human polyomavirus BKV infection of endothelial cells results in interferon pathway induction and persistence. PLoS Pathog 15:e1007505. doi: 10.1371/journal.ppat.1007505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Willmes C, Adam C, Alb M, Völkert L, Houben R, Becker JC, Schrama D. 2012. Type I and II IFNs inhibit Merkel cell carcinoma via modulation of the Merkel cell polyomavirus T antigens. Cancer Res 72:2120–2128. doi: 10.1158/0008-5472.CAN-11-2651. [DOI] [PubMed] [Google Scholar]
- 88.Sumida TS, Dulberg S, Schupp JC, Lincoln MR, Stillwell HA, Axisa P-P, Comi M, Unterman A, Kaminski N, Madi A, Kuchroo VK, Hafler DA. 2022. Type I interferon transcriptional network regulates expression of coinhibitory receptors in human T cells. Nat Immunol 23:632–642. doi: 10.1038/s41590-022-01152-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bazhin AV, von Ahn K, Fritz J, Werner J, Karakhanova S. 2018. Interferon-α up-regulates the expression of PD-L1 molecules on immune cells through STAT3 and p38 signaling. Front Immunol 9:2129. doi: 10.3389/fimmu.2018.02129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Levesque MC, Misukonis MA, O’Loughlin CW, Chen Y, Beasley BE, Wilson DL, Adams DJ, Silber R, Weinberg JB. 2003. IL-4 and interferon gamma regulate expression of inducible nitric oxide synthase in chronic lymphocytic leukemia cells. Leukemia 17:442–450. doi: 10.1038/sj.leu.2402783. [DOI] [PubMed] [Google Scholar]
- 91.Kasamatsu J, Deng M, Azuma M, Funami K, Shime H, Oshiumi H, Matsumoto M, Kasahara M, Seya T. 2016. Double-stranded RNA analog and type I interferon regulate expression of Trem paired receptors in murine myeloid cells. BMC Immunol 17:9. doi: 10.1186/s12865-016-0147-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE. 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, Zhang F. 2017. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc 12:828–863. doi: 10.1038/nprot.2017.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, Sullender ME, Vaimberg EW, Goodale A, Root DE, Piccioni F, Doench JG. 2018. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun 9:5416. doi: 10.1038/s41467-018-07901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu W, Kim GB, Krump NA, Zhou Y, Riley JL, You J. 2020. Selective reactivation of STING signaling to target Merkel cell carcinoma. Proc Natl Acad Sci USA 117:13730–13739. doi: 10.1073/pnas.1919690117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S3. Download jvi.01907-22-s0001.pdf, PDF file, 0.6 MB (581.4KB, pdf)