ABSTRACT
Antigen epitope identification is a critical step in the vaccine development process and is a momentous cornerstone for the development of safe and efficient epitope vaccines. In particular, vaccine design is difficult when the function of the protein encoded by the pathogen is unknown. The genome of Tilapia lake virus (TiLV), an emerging virus from fish, encodes protein functions that have not been elucidated, resulting in a lag and uncertainty in vaccine development. Here, we propose a feasible strategy for emerging viral disease epitope vaccine development using TiLV. We determined the targets of specific antibodies in serum from a TiLV survivor by panning a Ph.D.-12 phage library, and we identified a mimotope, TYTTRMHITLPI, referred to as Pep3, which provided protection against TiLV after prime-boost vaccination; its immune protection rate was 57.6%. Based on amino acid sequence alignment and structure analysis of the target protein from TiLV, we further identified a protective antigenic site (399TYTTRNEDFLPT410) which is located on TiLV segment 1 (S1). The epitope vaccine with keyhole limpet hemocyanin (KLH-S1399–410) corresponding to the mimotope induced the tilapia to produce a durable and effective antibody response after immunization, and the antibody depletion test confirmed that the specific antibody against S1399–410 was necessary to neutralize TiLV. Surprisingly, the challenge studies in tilapia demonstrated that the epitope vaccine elicited a robust protective response against TiLV challenge, and the survival rate reached 81.8%. In conclusion, this study revealed a concept for screening antigen epitopes of emerging viral diseases, providing promising approaches for development and evaluation of protective epitope vaccines against viral diseases.
IMPORTANCE Antigen epitope determination is an important cornerstone for developing efficient vaccines. In this study, we attempted to explore a novel approach for epitope discovery of TiLV, which is a new virus in fish. We investigated the immunogenicity and protective efficacy of all antigenic sites (mimotopes) identified in serum of primary TiLV survivors by using a Ph.D.-12 phage library. We also recognized and identified the natural epitope of TiLV by bioinformatics, evaluated the immunogenicity and protective effect of this antigenic site by immunization, and revealed 2 amino acid residues that play important roles in this epitope. Both Pep3 and S1399–410 (a natural epitope identified by Pep3) elicited antibody titers in tilapia, but S1399–410 was more prominent. Antibody depletion studies showed that anti-S1399–410-specific antibodies were essential for neutralizing TiLV. Our study demonstrated a model for combining experimental and computational screens to identify antigen epitopes, which is attractive for epitope-based vaccine development.
KEYWORDS: antigen epitope, Tilapia lake virus, epitope vaccine, phage display technology
INTRODUCTION
Antigen selection is a key step in vaccine design. Initially, vaccines were prepared with the whole virus as an antigen, including live attenuated vaccines and inactivated vaccines. A vaccine against smallpox, a live attenuated vaccine that saved countless lives and led to the eradication of the disease in 1977, has been on the market since 1796 (1). Widespread vaccination with injectable inactivated polio vaccine and live attenuated oral polio vaccine has brought poliovirus infinitely close to eradication (2). One of the first vaccines to be used in the 2019 coronavirus disease (COVID-19) pandemic was an inactivated vaccine, which has contributed to the control of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (3, 4). However, live attenuated and inactivated vaccines do not target a specific antigen, and their vaccine targets are ambiguous. With the analysis of the genome sequence of SARS-CoV-2 and the study of vaccines against SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV), the spike (S) protein on the virus surface may be an ideal target for vaccine development (3–6). Until now, the development of a SARS-CoV-2 vaccine based on S protein as antigen target was the most widely studied example, and the vaccine types included mRNA vaccine, DNA vaccine, recombinant subunit vaccines, and virus vector-based vaccines (6). With continued research developments, it was found that the receptor-binding domain (RBD) on the S protein binds to angiotensin-converting enzyme 2 on host cells, which leads to SARS-CoV-2 invading cells; therefore, the vaccine target was focused on RBD (4, 7–9). The development of monoclonal antibodies targeting the S protein stem helix has encouraged researchers to design antigens based on the heptad repeat 1 (HR1) and heptad repeat 2 (HR2) regions (10, 11). Notably, protective epitope peptide vaccines against the S protein have been developed, and antibodies targeting these peptides can effectively neutralize SARS-CoV-2 and its variants (12–14). The refinement of antigen targets does not reduce the efficacy of vaccines but makes the preparation and safety of vaccines more reliable. Antigenic epitopes, also known as antigenic determinants, are the smallest antigen regions required to trigger the required immune response and are a sensible way to achieve immune focus and develop safe vaccines (15). Therefore, epitope-based approaches to vaccine design are probably the most ideal for development of vaccines in the future.
Revolutionary developments in information technology and molecular biology, as well as the growth of genomic data repositories, have led to an increase in the popularity of epitope-based vaccine design among researchers. Bioinformatics-based vaccine design has been referred to as reverse vaccinology (16). In fact, the first vaccine based on reverse vaccinology, for meningococcus B, was recently licensed (17, 18). Currently, the development of epitope-based peptide vaccines with the help of immunoinformatics platforms has been used in a variety of diseases, for example, SARS-CoV-2 (19), Kaposi sarcoma (20), yellow fever virus (21), MERS-CoV (22), and Theileria parasites (23). However, most epitope vaccines based on immunoinformatics have not been validated in vivo, and different epitope prediction servers and software used will also lead to different results. Technological innovations in structural biology provide fine-grained molecular structure information, and structure-guided vaccine antigen design is on the rise, marking the advent of an era of “reverse vaccinology 2.0” (24). Based on the structural characterization of HIV-1 broad-spectrum neutralizing antibody VRC34.01 in complex with envelope protein, it was found that the antibody directed most of its binding energy to 8 amino acid residues in the conserved N-terminal region of HIV-1 fusion peptides (25). This binding feature encouraged researchers to develop fusion peptide-based immunogens targeting the 8 amino acid residues at the N terminus of the fusion peptide as a vaccine target, and these immunogens elicited monoclonal antibodies in mice were capable of neutralizing 31% of a cross-clade panel of 208 HIV-1 strains (26). In addition, the B cell epitopes of SARS-CoV-2 spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins were simulated and predicted by a structure-based method, and the immunogenicity of the epitopes was verified by immunizing mice; it was found that 11 predicted epitopes could induce neutralizing antibodies, 6 of which were immunodominant epitopes (27). Structure-based immunogens require first the isolation and resolution of the monoclonal antibody structure, and second the understanding of the target antigen of the virus. However, these requirements are undoubtedly disastrous for vaccine antigen design for an emerging viral disease. In addition, the cost of organism structure resolution is enormous, and only a small fraction of protein structures is currently resolved. Therefore, epitope identification based on refined protein structure is currently inapplicable in most diseases, particularly emerging diseases.
Phage display technology entails display of recombinant peptides or proteins on the surface of phage particles and then of select target peptides or proteins through the interaction between phages and selected fixed ligands; it is the first genetic engineering technology used to study protein-ligand interactions, which were first reported by Smith in 1985 (28). At present, phage display technology has been widely used in epitope screening, which provides a favorable tool for the development of epitope vaccines. For example, the genome fragment phage display library of Ebola virus (EBOV) was spanned to identify the immunodominant sites on the glycoprotein of EBOV, and it was determined that these epitopes could induce EBOV neutralizing antibodies and protect against lethal doses of EBOV challenge (29). In addition, the commercial random phage display peptide library was used to screen and identify the B cell epitope of a bovine diarrhea virus core protein and a novel mimic epitope peptide for duck hepatitis A virus (30, 31). Guo et al. used COVID-19 serum to screen the phage display peptide library and found B cell epitopes located on the NP protein and the S protein, whose amino acid residues were TLPK and VGG, respectively (32). Among them, the VGG residue sequence overlapped highly with the recognition epitope of a previously discovered SARS-CoV-2 neutralizing antibody, P2B-2F6 (33), which indicated that the epitope vaccine developed based on the VGG residue may have enormous application potential (32). Random peptide libraries (RPLs) and natural peptide libraries are two different types of phage display library; the peptide sequences displayed by RPLs are encoded by synthetic random degenerate oligonucleotides, and the advantage of these display libraries is that it can be applicable to all pathogens and it is positive for rapid screening and identification of epitopes of some emerging diseases (34, 35). However, in most cases, the amino acid composition obtained by RPL screening is not exactly the same as the natural epitope in the target pathogen. However, these peptides unrelated to pathogens mimic continuous or discontinuous epitopes in function, so they are also called mimotopes (36, 37). Fortunately, with the development of bioinformatics, we can infer the epitopes corresponding to specific pathogen target proteins according to the amino acid sequence of the simulated epitopes using a sequence alignment software program and then further identify these epitopes through biological characterization.
Tilapia lake virus (TiLV) is an emerging viral disease of aquatic animals which mainly affects Nile tilapia (Oreochromis niloticus) and hybrid tilapias. Studies have shown that TiLV is an enveloped, negative, single-stranded RNA virus with 10 segments in its genome (38–43), each with an open reading frame, which may encode a total of 14 proteins (44). Genomic homology analysis showed that segment 1 had weak homology with the PB1 subunit of influenza C virus (38), segments 1, 2, 4, and 5 had strong homology with Dhori virus, evolution of segments 6, 7, and 8 was similar to that of influenza virus, and segment 3 showed weak homology with infectious Salmon anemia virus (44). Unfortunately, however, the function of the protein encoded by TiLV is currently poorly understood, which seriously hampers the development of a vaccine against this virus. Although studies have described a TiLV vaccine (45–48), the antigen is uncertain and there is a gap in the antigenic epitopes of TiLV. In this study, our strategy was to use a Ph.D.-12 phage library to screen phage-positive clones that bound with serum antibodies and to comprehensively evaluate the possibility of synthetic peptides displayed by phage-positive clones as immunogens through an enzyme-linked immunosorbent assay (ELISA), in vivo immunogenicity evaluation, and the analysis of the ability to provide protection for lethal TiLV. Subsequently, discovering and designing the natural epitope of TiLV relied on an in silico study and determination of the reliability of the real epitope by ELISA, an amino acid mutation study, and antibody reaction test induced in tilapia. To reveal the importance of this epitope as a vaccine antigen of TiLV, we conducted tilapia immunization and TiLV challenge studies on the antigen sites identified in our study to establish a proof of concept, which can guide rational vaccine design against TiLV. Furthermore, this study provides an epitope peptide-based antigen design strategy for emerging diseases, including viral or bacterial diseases, to deal with infectious diseases.
RESULTS
Identification of TiLV-specific mimotopes by Ph.D.-12 phage library.
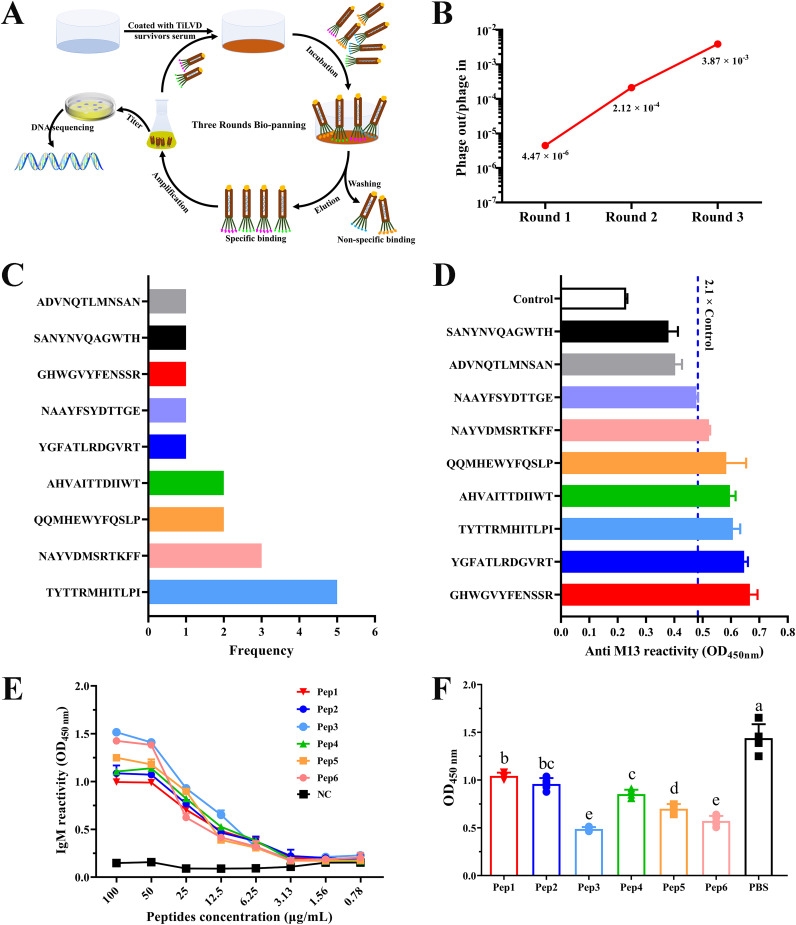
As shown in Fig. 1A, a Ph.D.-12 phage display library was used for the screening of serum samples derived from fish recovered after infection with TiLV tilapia to identify TiLV-specific mimotopes. Notably, to identify convalescent-phase serum TiLV-specific IgM antibody titers, we performed an ELISA on convalescent-phase sera and TiLV-negative sera prior to biopanning. The results showed that the antibody titer of IgM in convalescent-phase serum was about 1,300, which was significantly higher than that in negative serum (see Fig. S1 in the supplemental material). Subsequently, 3 rounds of consecutive biopanning were carried out with convalescent-phase serum as bait, and the number of enriched phages increased from 6.70 × 105 in the first round of panning to 5.80 × 107 after the third round of panning, and the enrichment efficiency increased from the initial 4.47 × 10−6 to 3.87 × 10−3 (Table 1, Fig. 1B), indicating that the phage that specifically binds to the antibody was effectively enriched and its enrichment factor was 865-fold. After 3 rounds of affinity panning, 22 independent phage clones were randomly picked for purification to complete DNA sequencing. The sequencing results found that all phages contained 9 different peptide sequences, among which the peptide sequence TYTTRMHITLPI had 5 occurrences, NAYVDMSRTKFF had 3 occurrences, and both QQMHEWYFQSLP and AHVAITTDIIWT had 2 occurrences, and others were found in a single occurrence (Fig. 1C, Table 2).
FIG 1.
Biopanning and identification of affinity peptides from TiLV convalescent-phase serum by phage-displayed peptide library. (A) Schematic representation of the strategy of biopanning for serum-specific affinity peptides. TiLV convalescent-phase serum was used as the target, and three rounds of affinity panning were performed using phage display 12-mer peptide libraries. After biopanning, immunopositive phage clones were further identified by DNA sequencing and ELISA. (B) Monitoring the output/input ratios of phage in sequential rounds of biopanning. (C) The corresponding amino acid sequences of 22 randomly selected phage clones after DNA sequencing and the frequency of occurrence of each sequence. Seventeen of the 22 phage clones were sequenced normally. After obtaining the sequencing results, we merged the phage clones with the same sequence for subsequent ELISA determination. (D) Determination of the reactivity of selected phage clones with TiLV convalescent-phase serum by ELISA. When the OD450 (phage clone) was ≥ 2.1 × OD450 (control), the reaction was considered immunopositive, and the clone was used for subsequent peptide synthesis. According to the OD450 (from high to low values), the immunopositive clones were named Pep1, Pep2, Pep3, Pep4, Pep5, and Pep6. (E) Variation curves of the reactions between different concentrations of synthetic peptides and TiLV convalescent-phase serum by ELISA. The synthetic peptides were serially diluted, and the reactivity with serum antibody (1:100 diluted) was measured by absorbance at 450 nm. Each dilution was tested in 3 replicates, and the values in the curve are the mean absorbance ± standard deviation (SD). (F) Synthetic peptides competitively inhibited the reaction of TiLV convalescent-phase serum with TiLV. Each well was coated with 6.0 × 104 copies of TiLV, serum was diluted 1:100, and the concentration of synthetic peptide was 100 μg/mL. The values shown are means ± SD with 5 replicates. Statistical significance was tested by one-way ANOVA and Tukey’s multiple-comparison tests. Significant differences (P < 0.05) are indicated by different lowercase letters.
TABLE 1.
Enrichment of specifically binding phages during three rounds of biopanning
| Biopanning round no. | Phage input (PFU) | Phage output (PFU) | Phage output/input |
|---|---|---|---|
| 1 | 1.50 × 1011 | 6.70 × 105 | 4.47 × 10−6 |
| 2 | 2.50 × 1010 | 5.30 × 106 | 2.12 × 10−4 |
| 3 | 1.50 × 1010 | 5.80 × 107 | 3.87 × 10−3 |
TABLE 2.
Translated amino acid sequences of positive phage clone DNA sequences
| Nucleotide sequence | Amino acid sequence | Frequencya |
|---|---|---|
| ACGTATACTACGCGTATGCATATTACGCTTCCGATT | TYTTRMHITLPI | 5 |
| AATGCTTATGTTGATATGTCGCGTACTAAGTTTTTT | NAYVDMSRTKFF | 3 |
| CAGCAGATGCATGAGTGGTATTTTCAGAGTTTGCCG | QQMHEWYFQSLP | 2 |
| GCTCATGTGGCGATTACTACGGATATTATTTGGACT | AHVAITTDIIWT | 2 |
| TATGGTTTTGCTACGCTGCGGGATGGGGTTAGGACG | YGFATLRDGVRT | 1 |
| AATGCTGCGTATTTTTCTTATGATACGACGGGTGAG | NAAYFSYDTTGE | 1 |
| GGGCATTGGGGTGTTTATTTTGAGAATAGTTCGCGG | GHWGVYFENSSR | 1 |
| TCGGCGAATTATAATGTGCAGGCTGGGTGGACGCAT | SANYNVQAGWTH | 1 |
| GCGGATGTGAATCAGACTTTGATGAATTCTGCGAAT | ADVNQTLMNSAN | 1 |
The frequency refers to the number of times the sequence appears in 17 phage clones.
To determine the binding activity of phage clones carrying these 9 peptide epitopes to serum antibodies, we performed a phage ELISA. The results showed that among the 9 peptides displayed by phage, 6 phage clones showed positive reactions, that is, the optical density at 450 nm (OD450) of the sample was greater than 2.1× that of the control (Fig. 1D). Among all the identified positive phage clones, their binding activities were denoted, from high to low, as Pep1, Pep2, Pep3, Pep4, Pep5, and Pep6. To this end, we synthesized the six corresponding peptide molecules and reverified the reactive ability of the synthetic peptides to serum with ELISA. The results showed that the 6 synthetic peptides had reactivity with TiLV-positive serum and showed a significant dose effect (Fig. 1E). In addition, the specific binding of different synthetic peptides to anti-TiLV serum was further explored by competitive ELISA, and it was found that 6 synthetic peptides could competitively inhibit the specific binding of TiLV to anti-TiLV serum, among which Pep3 and Pep6 had the most significant inhibitory effect (Fig. 1F).
Immunization of tilapia with mimotopes induced humoral immunity in vivo.
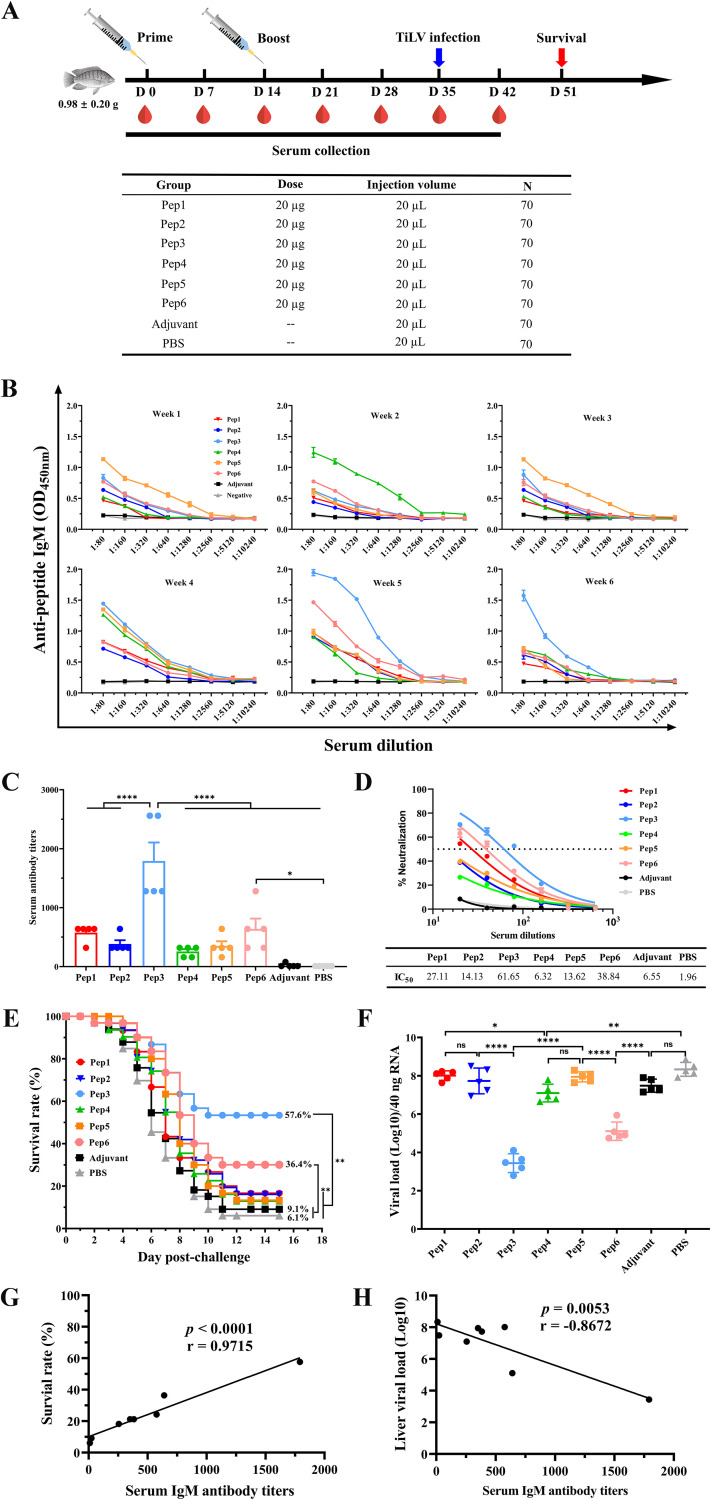
To understand changes in epitope-specific serum antibody responses induced by mimotope vaccination, serum antibody levels of epitope-specific IgM were measured at 7, 14, 21, 28, 35, and 42 days after primary vaccination (Fig. 2A). At 7 days after the prime vaccination, the serum antibody level in the Pep5-immunized group was the highest and then decreased; after 14 days of the booster immunization, the serum antibody level was still in the rising stage, but then it declined sharply (Fig. 2B). In addition, we observed that the serum antibody levels of the Pep4-immunized group reached the highest level 14 days after immunization, regardless of whether it was prime or booster (Fig. 2B). However, high levels of epitope-specific IgM in the Pep3-immunized group only appeared after 35 and 42 days of primary vaccination and peaked at 35 days after primary vaccination, subsequently showing a downward trend (Fig. 2B). Nonetheless, compared with other immunized groups, the Pep3-immunized group still had the highest serum antibody level at 42 days after primary vaccination (Fig. 2B). Similarly, the serum antibody level in the Pep6-immunized group reached the highest level at 35 days after the initial immunization, but unlike the Pep3-immunized group, it decreased sharply at 42 days of after immunization (Fig. 2B). Notably, neither the phosphate-buffered saline (PBS) nor the adjuvant control group produced specific serum IgM antibodies throughout.
FIG 2.
Synthetic peptides elicited a humoral immune response in tilapia after vaccination. (A) Schematic diagram of tilapia immunization, sample collection, and challenge scheme. Tilapias (70 per group) were vaccinated intraperitoneally with 20 μg per fish of synthetic peptide mixed with Freund’s adjuvant on D0 and D14, respectively. After the prime immunization, blood was collected by broken neck at D7, D14, D21, D28, D35, and D42. On D35, 33 fish were randomly selected for challenge intraperitoneally with 9.0 × 107 copies of TiLV per fish, and the survival rate was calculated for each group at D51. (B) Peptide-specific antibody responses in sera of each group at different time points after vaccination were analyzed by ELISA. Serum samples were serially diluted 2-fold (initial as 1:80), and the peptide-specific antibody response of the serum was measured by the absorbance at 450 nm. Each well was coated with 2 μg of peptide. The values shown are mean absorbance ± SD for each dilution with triplicates. (C) TiLV-specific antibody endpoint titers of different groups of serum at challenge time points (D35) were determined by ELISA, for which plates were coated with TiLV at 6.0 × 104 copies per well. The dilution method for serum was the same as that described for panel B. Endpoint titers of the serum samples are shown as the reciprocal of the highest dilution when the OD450 for samples was ≥2× the OD450 of the negative control. (D) Dose-response neutralization of live TiLV titration curves and IC50 values of serum sample with different vaccination groups at D35. Serum was diluted from 1:20 to 1:640 at a 2-fold ratio, and the test was repeated 3 times per serum sample. Results are expressed as means ± SD. (E) Fish health was monitored daily, and dead fish were collected until day 15 postchallenge. The P values for survival rate are based on a log-rank (Mantel-Cox) test (n = 33). (F) Quantification of TiLV virus in liver tissue on 8 day postchallenge by qRT-PCR. Data are means ± SD (n = 5 per group). (G and H) Correlation between TiLV-specific antibody titers in serum and survival rate within 15 days postchallenge (G) or viral load quantified by qRT-PCR in liver tissue on day 8 postchallenge (H). Positive Pearson correlations were observed between TiLV-specific antibody titers and survival rate (r = 0.9715; P < 0.0001) (G) or inverse Pearson correlation with viral load (r = −0.8672; P = 0.0053) (H). Statistical significance was tested by one-way ANOVA and Tukey’s multiple-comparison tests. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, no significant difference.
In order to analyze the TiLV-specific antibody response in vivo before virus challenge, we measured TiLV-specific IgM antibody titers in serum at 35 days after of primary immunization using TiLV particles as antigen. TiLV-specific serum antibody titer in the Pep3-immunized group was the highest, reaching 1,792, which was significantly higher than that in the adjuvant control group (Fig. 2C). In addition, the TiLV-specific serum antibody titers in the Pep3-immunized group were significantly higher than those in the groups immunized with Pep6, Pep5, Pep4, Pep2, or Pep1. However, although the serum antibody titer of the Pep6-immunized group was 640, it was only significantly different from the PBS-immunized group. Furthermore, to evaluate the neutralization activity of mimotope-peptide immune sera, virus neutralization tests were used to determine the reactivity of sera against live TiLV. Among the 6 mimotope peptides, 3 could generate neutralizing antibodies, including Pep1, Pep3, and Pep6. The sera from Pep1-, Pep3-, and Pep6-immunized groups were incubated with TiLV to infect CIK cells, and we observed that CIK cells remained highly active 7 days after infection, as by determined a 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay, which strongly suggested that serum from these groups was effective in neutralizing TiLV to prevent the virus from damaging cells. It is worth mentioning that the 50% inhibitory concentration (IC50, expressed as the reciprocal of the dilution multiple of the serum with 50% neutralization activity) of these immune group sera against TiLV were 27.11, 38.84, and 61.65, respectively (Fig. 2D). These results showed that Pep1, Pep3, and Pep6 produced antibodies capable of neutralizing TiLV after immunization.
In addition to TiLV-specific serum antibody titers, we also determined immune-related enzyme activity in serum at 35 days after primary immunization. Compared with the adjuvant and blank control groups, the activities of acid phosphatase (ACP), alkaline phosphatase (AKP) and total antioxidant capacity (T-AOC) in all immunization groups were significantly increased (Fig. S2). For superoxide dismutase (SOD) activity, except for the Pep4 immunization group, the SOD activities of other immunization groups were all significantly increased compared to those in the adjuvant and blank control groups (Fig. S2D). In addition, the ACP, AKP, SOD, and T-AOC activities of the Pep3-immunized group showed the highest enzyme activity levels among all groups.
Immunization of tilapia with mimotopes provides protection against TiLV challenge.
At 35 days after of primary immunization, the tilapias were challenged with TiLV at a lethal dose by intraperitoneal injection to assess the immunoprotective effects following mimotope immunization (Fig. 2A). The cumulative mortality with each group within 15 days was calculated (Fig. 2E), and the TiLV loading in tilapia liver was determined by reverse transcription-quantitative PCR (RT-qPCR) (Fig. 2F). Vaccination with mimotope Pep3 provided 57.6% protection against TiLV challenge, and the group vaccinated with Pep6 showed 36.4% protection (Fig. 2E). Other mimotope peptides encompassing Pep5, Pep2, Pep4, or Pep1 only endowed inefficient protection from lethality after TiLV challenge. The protective effect afforded by mimotope peptides appeared to be achieved by mediating neutralizing antibody titers and other mechanisms, because protection rates were positively correlated with serum IgM antibody and neutralizing antibody levels (Fig. 2G and H). Furthermore, viral loads in the Pep3- and Pep6-immunized groups were significantly reduced relative to those in the control and adjuvant groups, and the lowest levels were observed in the Pep3-immunized group (Fig. 2F). Remarkably, Pep3 mimotope peptide provided 57.6% protection in tilapia against lethal TiLV challenge after two doses of immunization, and this group stood out in all groups due to the highest neutralizing antibody titers.
Identification of TiLV natural epitope with mimotope peptide Pep3.
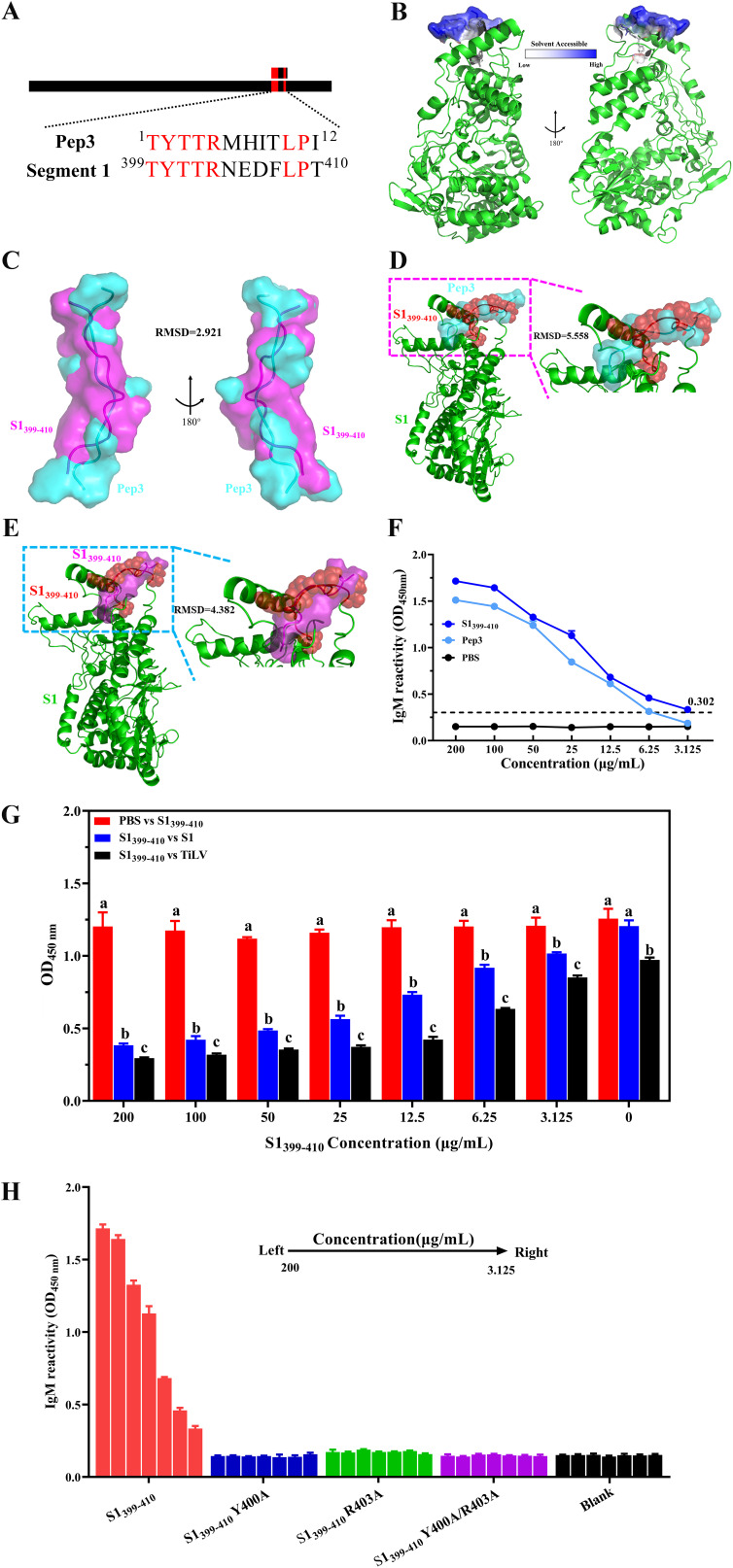
Mimotopes inevitably introduce exogenous sequences, which may have potential risks in clinical applications (49). It is crucial to identify the true epitope within the pathogen by mimotope, so we identified the natural epitope of TiLV corresponding to Pep3 by sequence alignment. We found that Pep3 shared a high degree of sequence similarity with residues 399 to 410 located on the protein encoded by TiLV segment 1 (S1) (Fig. 3A), and the sequence identity reached 58.3% (Fig. S3A). However, the recognition rates of the amino acid sequence of the Pep3 mimotope peptide with other protein sequences encoded by TiLV were lower than 50% (Fig. S3B to I). Therefore, we guessed that the S1399–410 amino acid residues might represent the natural epitope of TiLV.
FIG 3.
Recognition and characterization of the TiLV natural epitope. (A) Sequence alignment between Pep3 and segment 1 (S1). Red indicates consensus amino acid residues, and black represents unmatched amino acid residues. The identity between Pep3 and the S1399–410 region of S1 was 58.33%. S1 protein sequence was derived from NCBI (https://www.ncbi.nlm.nih.gov/) accession number YP_009246481. (B) Folding of the S1399–410 region in the S1 protein structure and its surface accessibility. S1 protein structure was predicted using RoseTTAFold in Robetta and modified using PyMOL. S1399–410 is shown as stick and surface; other regions are shown as cartoons. The relative solvent accessibility of each amino acid residue in the S1399–410 region was calculated by PyMOL, with darker colors indicating higher solvent accessibility. (C) Structural alignment between Pep3 and S1399–410 peptide by PyMOL. Both Pep3 and S1399–410 are presented as cartoon and surface, with Pep3 shown in cyan and S1399–410 showed in magenta. (D and E) Alignment studies between Pep3 (D) or S1399–410 (E) peptide structures and S1protein structure using PyMOL. The region aligned with Pep3/S1399–410 in S1 protein is indicated by a red sphere and cartoon, and the Pep3 (cyan) and S1399–410 (magenta) peptide are represented by cartoon and surface. (F) Reactivity of synthetic peptides with positive serum. Synthetic peptides were coated in ELISA plates at different concentrations, three replicates per concentration, and anti-TiLV serum was diluted 1:100. Values are expressed as means ± SD. (G) S1399–410 peptide competitively inhibited the reactivity of anti-TiLV serum antibody with S1 protein of TiLV virus. S1399–410 peptide was added to the coated plate after being coincubated with anti-TiLV serum (1:100 dilution) at 37°C for 2 h. (H) Reactivity identification of S1399–410 peptide and its mutant peptide with anti-TiLV serum antibody. Serum was diluted 1:100. Data are from an experiment performed in triplicate and are shown as means ± SD. Statistical significance was tested by one-way ANOVA and Tukey’s multiple-comparison tests. Significant differences (P < 0.05) are indicated by different lowercase letters.
The structure prediction of the S1 protein showed that a long loop is formed in the S1399–410 region, and most of its amino acids are on the solvent-accessible surface (Fig. 3B). These structural features are based on the most common peptide vaccine design. Further structural comparison revealed that the S1399–410 region had a more similar spatial structure to the Pep3 peptide, with a root mean square deviation (RMSD) of 2.921 (Fig. 3C). However, when comparing the spatial conformation of Pep3 and S1399–410 peptides with S1, the structural deviation between S1399–410 and S1 was smaller, with an RMSD of 4.382 (Fig. 3D and E). The results of these structural analyses fully demonstrated that the S1399–410 peptide is to a certain extent closer in conformation to the corresponding residues of S1.
To identify this epitope, we first tested using TiLV convalescent-phase serum samples to verify that the S1399–410 region could bind to the antibodies of serum samples. Here, the corresponding S1399–410 synthetic peptide was still reactive with TiLV convalescent-phase sera at a concentration of 3.125 μg/mL, while the lowest reactive concentration of Pep3 peptide was 6.25 μg/mL (Fig. 3F). In addition, competitive binding verified that the S1399–410 synthetic peptide inhibited the binding of serum antibodies to S1 protein and TiLV virions in a dose-dependent manner with the S1399–410 synthetic peptide (Fig. 3G). Moreover, in an attempt to identify the amino acids necessary for the S1399–410 epitope to bind to serum antibodies, we synthesized an alanine mutant peptide of S1399–410 (Table S1), and the mutations of these amino acid residues were determined by the three-dimensional (3D) structural characteristics of S1 protein. We observed that the S1399–410 residues formed a loop structure (Fig. 3D) in the 3D structure of the S1 protein and formed an ideal solvent-exposed surface (Fig. 3B), and the selected mutated amino acid residues were located at the vertices of this structure. These resulting features may be an important area for antibody binding. Based on the view of the S1 full-length protein structure, we assumed that the loop structure formed in the S1399–410 region is an important structural basis for antibody binding. To directly test whether these assumed amino acid residues are involved in serum antibody recognition, we synthesized mutant peptides in which amino acid residue Tyr at position 400 or amino acid residue Arg at position 403, or both, in the S1399–410 subunit were replaced by Ala (Table S1). We tested the effect of these amino acid residue mutations on serum antibody binding and found that mutations in either the Tyr residue or the Arg residue or both did significantly reduce the binding capacity of the epitope to anti-TiLV serum antibody (Fig. 3H). In short, these studies demonstrated that the S1399–410 subunit is a candidate epitope of TiLV and binds to anti-TiLV serum antibody through its formation of loop structure dependence.
The serum antibody induced after immunizing tilapia with candidate vaccine S1399–410 is necessary for neutralizing TiLV.
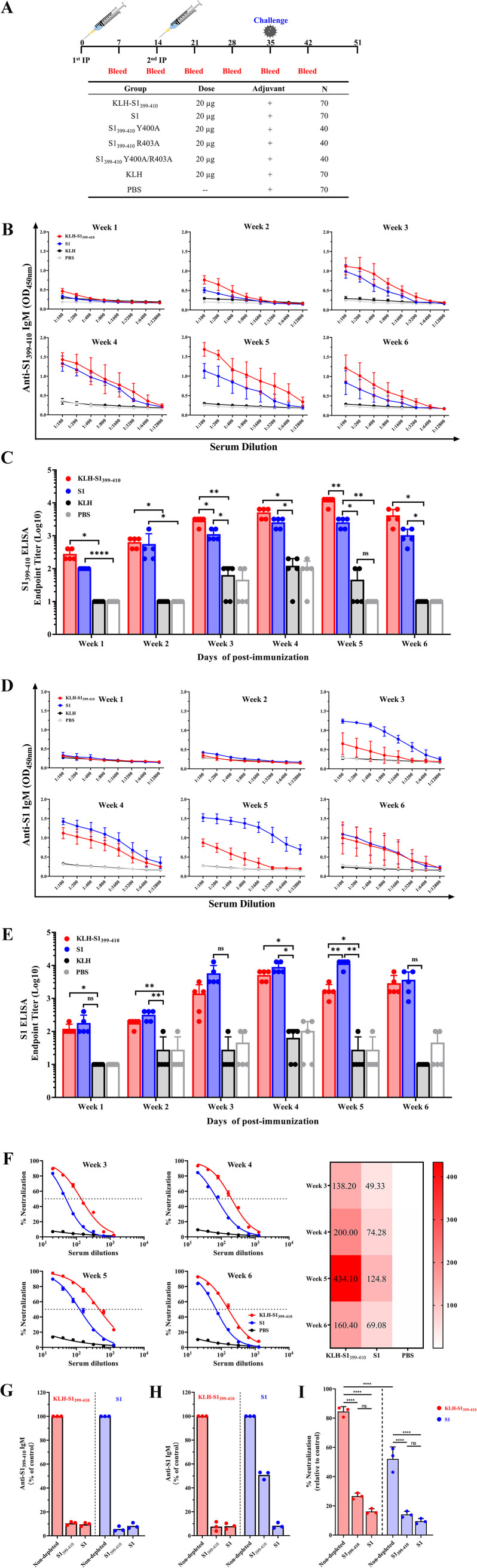
In order to directly test the ability of the S1399–410 epitope to induce an active immune response in tilapia, the S1399–410 epitope peptide was synthesized by solid phase and conjugates with a keyhole limpet hemocyanin (KLH) carrier protein. Tilapia (70 fish each group) were intraperitoneally injected with 20 μg of the KLH-conjugated S1399–410 epitope peptide (Fig. 4A) at day 0 (D0) and D14. In addition, the control group was only vaccinated with 20 μL of PBS–20 μg KLH (negative-control group) or injected with 20 μg full-length S1 protein of TiLV as the positive-control group (Fig. 4A).
FIG 4.
Antibodies targeting the epitope of S1399–410 effectively neutralized TiLV. (A) Schedule for immunization, sample collection, and challenge of tilapia. Tilapias (70 fish per group for nonmutated peptides, 40 fish per group for mutated peptides) were prime vaccinated at D0, with a 14-day interval before a booster vaccination at a dose of 20 μg per fish of synthetic peptide vaccine, which was mixed with Freund's adjuvant and administered by intraperitoneal injection. At D35, 33 fish were randomly selected for the TiLV challenge test (the mutant peptide vaccine did not perform), and survival rates were calculated at D51. (B) Specific antibody analysis against the S1399–410 epitope peptide in serum was collected at different time points in the KLH-S1399–410, S1, and PBS immunization groups. Serum samples (5 fish per group) were serially diluted 2-fold (initial as 1:100) and the peptide-specific antibody response of the serum was measured by the absorbance at 450 nm. Each well was coated with 2 μg of peptide. The values are presented as means absorbance ± SD for each dilution with triplicates. (C) Endpoint titers of antibodies targeting S1399–410 in serum samples at different time points, shown as the reciprocal of the highest dilution when the OD450 (samples)was ≥2× OD450 of preimmunization serum. (D) Specific antibody analysis against the S1 protein in serum collected at different time points in the KLH-S1399–410, S1, and PBS immunization groups. Serum samples (5 fish per group) were serially diluted 2-fold (initial as 1:100), and the S1-specific antibody response of the serum was measured by the absorbance at 450 nm. Each well was coated with 0.5 μg of S1 protein. The values are mean absorbance ± SD for each dilution with triplicates. (E) Endpoint titers of antibodies targeting S1 protein in serum samples at different time points, shown as the reciprocal of the highest dilution when the OD450 (samples) was ≥2× OD450 of preimmunization serum. (F) Dose-response neutralization of live TiLV titration curves and IC50 heat map of serum samples with different vaccination groups at D21, D28, D35, and D42. Serum was diluted from 1:20 to 1:1,280 at 2-fold ratios, and the test was repeated 3 times per serum sample (5 fish mix serum). V are expressed as means ± SD. (G and H) Depletion or nondepletion of antibodies to specific peptide or protein in the serum at D35 after primary immunization was validated by ELISA. Anti-S1399–410 IgM reactive after depletion of serum antibodies specific to S1399–410 (left) or S1 protein (right) (G). Anti-S1 IgM reactive after depletion of serum antibodies specific to S1399–410 (left) or S1 protein (right) (H). (I) Neutralization efficacy of pooled sera not depleted or depleted of specific peptide or protein antibodies against live TiLV. Data are expressed as percent neutralization relative to a blank group of unincubated serum and virus. Data are from an experiment performed in triplicate. Data are expressed as means. Statistical significance was tested by one-way ANOVA and Tukey’s multiple-comparison tests. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, no significant difference.
After the initial immunization, the serum of each immune group was collected at D7, D14, D21, D28, D35, and D42, and the immune response of serum to epitope peptide (S1399–410) and full-length protein S1 was determined by ELISA. At D35 after immunization with the S1399–410 epitope vaccine, a high antibody response against the epitopes was produced in the sera of immunized fish, with antibody titers exceeding 10,000 (Fig. 4B and C). Similarly, the serum antibody induced by the S1 immunization group also peaked at D35, and the anti-S1 antibody titers reached 12,800 (Fig. 4D and E). It is worth noting that the serum antibody titer against S1399–410 IgM in the S1399–410–immunized group was still significantly higher than that in the S1-immunized group at D42 (Fig. 4C). As expected, no specific antibodies against the S1399–410 peptide or S1 protein were produced in the serum of any of the mutant peptide vaccination groups (Fig. S4). Although the serum antibody titer against S1399–410 was significantly higher than that of the KLH-PBS group at 5 weeks after immunization with an S1399–410 R403A variant (i.e., with an Arg-to-Ala change at position 403), its titer was only about 100 (Fig. S4C).
To estimate the inhibitory ability of the sera from the immunized groups on D21, D28, D35, and D42 after the second immunization, a viral inhibition test was carried out. The neutralizing titer in serum had a similar trend to the specific antibody titer, with peak serum neutralization titers for S1399–410 epitope vaccine and S1 subunit vaccine at D35, and their IC50s were 434.10 and 124.80 (reciprocal dilutions), respectively (Fig. 4F). Unsurprisingly, the sera of the PBS control group failed to produce antibodies that specifically bound to S1399–410 or S1 and did not neutralize TiLV at any time point.
To further validate that serum antibodies induced by the S1399–410 epitope were necessary to neutralize TiLV, we conducted an antibody depletion assay against S1399–410. For the groups immunized with S1399–410 and S1, the specific-S1399–410 and S1 antibody levels in serum were depleted by ELISA, and the depletion efficiency was 89.5% and 91.7%, respectively (Fig. 4G). Interestingly, we found that the specific antibody against S1 was depleted in the S1-immunized group, while the depletion efficiency of serum antibody against S1399–410 was only 50.8% (Fig. 4H). Compared with the nondepleted sera controls, the ability of S1399–410 and S1 immune group serum to neutralize TiLV virus both significantly decreased after the depletion of serum antibody targeting S1399–410 (Fig. 4I). In sum, antibodies targeting the S1399–410 epitope are critical for neutralizing TiLV, whereas the anti-S1399–410 IgM antibody content was lower in the S1-immunized group than in the KLH-S1399–410-immunized group.
Protection against a lethal dose of TiLV in tilapia after immunization with S1399–410.
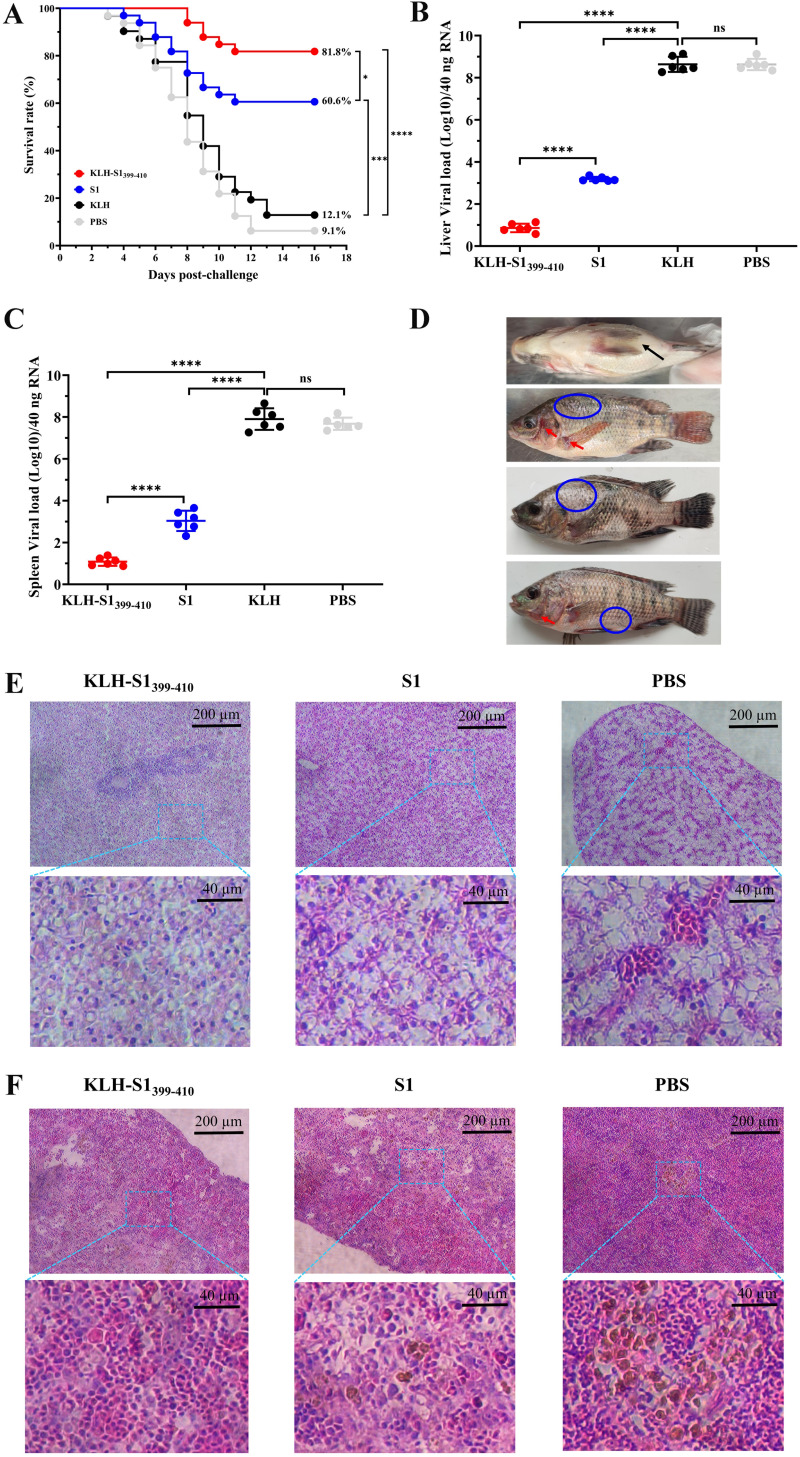
At D35 after primary immunization, 33 tilapia were randomly selected from each group, transferred to another clean aquarium, and challenged with a lethal dose of TiLV through intraperitoneal injection to evaluate vaccine immune protection efficacy. The health of the fish was monitored, and number of dead fish was recorded on a daily basis until 16 days after challenge. We found that the PBS and KLH control groups started to die on day 4 after challenge, the S1-immunized group began to die on day 5 after challenge, and in the S1399–410-immunized group we did not observe death until day 8 after challenge. In addition, the S1399–410-immunized group provided the best protection effect, with a survival rate of 81.8%, while the survival rate of the S1 immunized group was only 60.6% (Fig. 5A), which may have been related to the specific antibody titer in tilapia during the challenge. The viral loads of liver and spleen of dead tilapia were measured at 8 days postchallenge. The results showed that the viral loads of S1- and S1399–410-immunized groups were significantly lower than those of the control group, and the viral load of the S1399–410 group was significantly lower than that of the S1 group (Fig. 5B and C). We observed that the dead tilapia had typical symptoms of infection with TiLV, including gill cover, basal part of pectoral fin congestion, abdominal swelling, and scale loss (Fig. 5D). In addition, we tested the dead tilapia for TiLV specifically, and the results also confirmed that the tilapia death was caused by infection with TiLV (data not shown). The histopathological observations showed that, compared with the control group, the tilapia immunized with the vaccine had reduced damage to the liver and spleen from TiLV (Fig. 5E and F). Briefly, hepatocytes showing fibrosis with polarized nuclei and aggregation were observed in the S1-immune group (middle), whereas the PBS group (right) showed severe hepatocyte fibrosis, nuclear polarization, and syncytial liver symptoms and hepatic congestion (Fig. 5E). Spleen tissue showed few (S1 immunization group) or large aggregates (PBS group) of the melanomacrophage center and vacuolated nucleus (Fig. 5F). In conclusion, the S1399–410 immunization group provided excellent immune protection (81.8%), which was significantly higher than in the S1 immunization group, while the control group did not provide effective protection against a lethal dose of TiLV.
FIG 5.
Epitope-based synthetic peptide vaccine acted efficiently as prophylactic against TiLV infection. (A) Survival rate of different immunization groups after 15 days of intraperitoneal challenge with a lethal dose of TiLV. The survival curve was calculated by GraphPad Prism software. (B and C) Viral load of TiLV in liver (B) and spleen (C) tissues were quantified by qRT-PCR at 8 days after challenge. Data are means ± SD (n = 6 per group). (D) Representative figures showing typical symptoms of TiLV infection in tilapia, including abdominal swelling (black arrows), hyperemia (red arrows), and scales dropping off (blue circle). (E and F) Histopathological changes of different immune groups after 8 days of challenge were observed by H&E staining. Hepatocytes (E) showing fibrosis with polarized nuclei and aggregation were observed in the S1-immune group (middle), whereas the PBS group (right) showed severe hepatocyte fibrosis, nuclear polarization, and syncytial liver symptoms, and with hepatic congestion. Spleen tissue (F) showed few (S1 immunization group) or large aggregates (PBS group) of the melanomacrophage center (MMC) and vacuolated nucleus. Statistical significance was tested by one-way ANOVA and Tukey’s multiple-comparison tests. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05.
DISCUSSION
In the present study, we reported that a synthetic peptide vaccine (KLH-S1399–-410), which can confer 81.8% protection against lethal TiLV challenge, is an effective strategy to prevent TiLV infection. In a prime-boost immunization regimen, the synthetic peptide vaccine containing a single TiLV epitope conjugated with KLH carrier protein showed excellent immunogenicity in tilapia and generated durable neutralizing antibody responses. Our research mainly revealed two key findings. First, based on the 3D structural characteristics of S1 protein and alanine mutation studies, S1399–410 is an epitope of TiLV which is closely related to the reactivity of anti-TiLV serum antibodies. Second, the specific antibody induced by S1399–410 is necessary for neutralizing TiLV and induced a long-lasting antibody reaction in tilapia against TiLV infection.
The library capacity of random phage display peptide library is huge, and it can be used to screen almost any target of interest (50, 51). In terms of antigen screening, a random phage display peptide library is a powerful tool. After screening the library, simulated epitope peptides with high affinity to target substances can be obtained, and these peptides have been reported to be used in vaccines to activate the immune response in the body to fight diseases (52–54). In this study, we selected six peptides with high affinity from a Ph.D.-12 phage library by biopanning with anti-TiLV serum as the target. Further analysis showed that peptide Pep3 could induce specific immune response in tilapia to protect the fish from challenge with a lethal dose of TiLV and inhibited virus infection in vivo (Fig. 2E and F). However, many mimic peptides identified by random phage display peptide library have not been validated in vivo for their contribution to resistance to viral infection (32, 55). In our study, we evaluated the immune responses elicited in vivo by different peptide targets identified to be immune-responsive with specific antibodies in the sera of TiLV survivors by using synthetic peptide vaccines (Fig. 2). Vaccination after Pep3 synthetic peptide vaccine induced high levels of peptide-specific antibodies in tilapia, and these antibodies were associated with high levels of virus neutralization (Fig. 2C, D, and H). It is worth mentioning that the dynamic changes in antibody levels induced by Pep3 had a similar trend as the prime-boost vaccination strategy of vaccines developed based on TiLV segment 8 (47). However, unfortunately, the protection provided by the Pep3 synthetic peptide vaccine was inferior, with a survival rate of 57.6% (Fig. 2E). This may have been due to the short half-life of linear peptides in vivo (56, 57); unfortunately, we did not investigate this further in the present study. However, we found that the spatial conformation of Pep3 peptide overlapped with S1 worse than S1399–410, which may reveal some reasons why Pep3 had a weaker immune effect than S1399–410 (Fig. 3D and E).
In order to identify the natural epitopes of TiLV, we performed further studies on Pep3. It is extremely impractical when working with random peptide libraries to expect to obtain affinity peptides with amino acid residues that are exactly the same as those of viral proteins, and so sequence alignment is required to further identify the native epitopes of viral proteins corresponding to affinity peptides (32, 55, 58). In the present study, we found a high degree of similarity between the Pep3 peptide and residues 399 to 410 of TiLV S1 protein, and so we speculated that the S1399–410 region is a natural epitope of TiLV (Fig. 3A). S1 structural analysis showed that a long loop structure was formed in the S1399–410 region and most of the amino acid residues were exposed to solvent-accessible surfaces (Fig. 3B). Studies have shown that linear epitopes are commonly found in protein loops, which are prime candidates for peptide vaccine design (59). For example, the peptide consensus residues VGG, which are bio-enriched by phage display technology for COVID-19 serum, are in a ring region located in the RBM region of the spike protein (32). Similar loop structures have also been used as design targets for peptide vaccines in other diseases (54, 60–63). These structural characteristics indicated that it might be used as a TiLV antigen epitope, which was crucial to induce specific neutralizing antibodies (54, 61).
To address the immunoreactivity of the S1399–410 region to TiLV-positive serum, we compared synthesized S1399–410 peptide with full-length recombinant S1 protein by ELISA, and we found that the synthetic peptide S1399–410 interacted with TiLV serum antibody and, through competitive inhibition of the reactivity of the S1 protein, with TiLV serum (Fig. 3G). Furthermore, mutation analyses demonstrated that whether S1399–410 reacts effectively with serum antibody may depend on the amino acid residues at positions 400 and 403 in the S1 protein, which are located at the long loop structure (Fig. 3D). It was found that tilapia has the highest serum antibody titer 19 days after being infected with TiLV (64). In order to verify that S1399–410 can induce a specific antibody reaction in tilapia, we vaccination tilapia by intraperitoneal injection through prime-boost immunization. We found that at D35 after the initial immunization, the serum antibody titer of the S1399–410-immunized group reached the highest level, with an average titer of more than 10,000 (Fig. 4B). Moreover, there was still a high serum antibody reaction at D42, which indicated that KLH-S1399–410 can simulate natural infection with TiLV to produce a lasting antibody reaction after immunization. However, the production of serum antibody must be related to the antibody neutralizing the virus (60, 65, 66). The antibody depletion assay proved that the antibody produced by S1399–410 after immunizing tilapia was necessary to neutralize TiLV, as the virus neutralization potency was significantly reduced when anti-S1399–410 antibody was depleted. In most cases, the serum antibody reaction is directly related to the immune protection rate of the vaccine (67). As confirmed by this study, the higher the serum antibody titer before the TiLV challenge, the better the immune protection effect (Fig. 5). Consistent with our expectation, the mutant synthetic peptide failed to elicit serum antibody response after immunization, proving the important role of amino acid residues Y400 and R403 on S1399–410 in induction of TiLV neutralizing antibody.
Of course, there were also certain limitations in this study, such as the challenge test in this study only evaluated the immune protective efficacy of the vaccine against a single strain, which may adversely affect the promotion of the vaccine. Second, the S1 protein where S1399–410 is located was used in the design of the positive-control group instead of the antigen previously developed as a vaccine, which failed to reflect the superiority of KLH-S1399–410 epitope vaccine to a certain extent. Third, we did not evaluate the cellular immune response generated in vivo after vaccine immunization. Those above-mentioned limitations of our study will be our focus in future research. In addition, whether single-epitope tandem-expressed antigens can elicit a stronger humoral and/or cellular immune response also deserves further investigation.
In general, this study provided evidence to prove that the synthetic peptide vaccine based on S1399–410 regions coupled with KLH triggered specific neutralizing antibody responses and had a high protective effect on reducing the viral load in tissues and lethality after TiLV infection. A graphical abstract of the current study is shown in Fig. 6. Moreover, the strategy of vaccine antigen development proposed in this study provided a possible reference for emerging diseases in the future, as it could be used to select an effective antigen for vaccination against disease in a background of unknown function of pathogen proteins.
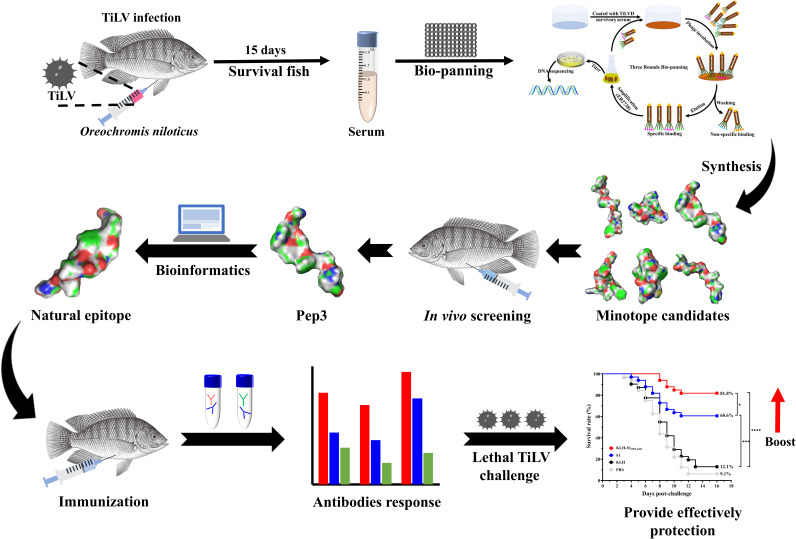
FIG 6.
Graphical abstract of the current study, in which we designed an epitope vaccine for tilapia against Tilapia lake virus via prime-boost vaccination.
MATERIALS AND METHODS
Phage display peptide library, cell culture, and virus.
The Ph.D.-12 phage display kit used in the current study was purchased from New England Biolabs (catalog number E8110) and contained a tube of the Ph.D.-12 phage library. Ctenopharyngodon idellus kidney (CIK) cells were donated by Lingbing Zeng from Yangtze River Fisheries Research, Institute of Chinese Academy of Fishery Science (Wuhan, Hubei, China). CIK cells were cultured at 28°C in 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum containing 100 U/mL penicillin and 100 μg/mL streptomycin. TiLV was isolated and stored by our laboratory, and we named it TiLV-CQ-2021.
Biopanning of Ph.D.-12 phage library.
The panning procedure was performed as described by the vendor with modifications. Briefly, the wells of a microtiter plate (NEST, China) were coated overnight at 4°C with 150 μL of anti-TiLV serums (1:100 diluted in 0.1 M NaHCO3 [pH 8.6]) in a humidified container. Blocking buffer (0.5% bovine serum albumin [BSA] in 0.1 M NaHCO3 [pH 8.6]) was then added for 2 h at 4°C to block the wells. After washing six times with Tris-buffered saline with 0.1% Tween 20 (TBST; TBS is 50 mM Tris-HCl, 150 mM NaCl [pH 7.5]), the wells were incubated for 1 h at room temperature with 10 μL (1.50 × 1011) library phages diluted in 100 μL of TBST. The wells were next washed 10 times with TBST to remove any unbound phage. The bound phage were eluted with 100 μL of elution solution (0.2 M glycine [pH 2.2]) for 30 min at room temperature and neutralized with 15 μL of 1 M Tris-HCl (pH 9.1). The titers of eluted phage were estimated, and Escherichia coli ER2738 was infected with the bound phage for amplification.
In the second and third rounds of biopanning, 1:200 and 1:300 dilutions of anti-TiLV serum were incubated with 2.50 × 1010 and 1.50 × 1010 amplified phages selected in the first biopanning round for enrichment of high-affinity binding. The concentration of Tween 20 in the TBST was increased from 0.1% (vol/vol) in the first round to 0.5% (vol/vol) in the other two rounds of biopanning. In addition, for the bound phage titers were determined but phage were not amplified in the third round. According to the library manufacturer’s instructions, single colonies were selected, isolated, and amplified.
Selection, amplification, and phage DNA sequencing of positive clones.
After three rounds of biopanning, 22 phage clones (blue plaques) were randomly chosen from selection plates. These were individually expanded in E. coli ER2738 and stored in aseptic glycerol at 20°C.
The amplified positive phage clones were precipitated by 200 μL PEG–NaCl (20% [wt/vol] polyethylene glycol 8000, 2.5 M NaCl). After centrifugation at 12,000 rpm for 10 min, the supernatant was carefully discarded. The precipitated phage were resuspended in 100 μL iodide buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 4 M NaI) with 250 μL of ethanol and then incubated for 10 min at room temperature. After centrifugation at 12,000 rpm for 10 min at 4°C, phage single-strand DNA (ssDNA) was precipitated in the bottom of EP tubes. Ethanol 70% was used to wash the precipitated ssDNA. After drying under a vacuum, the ssDNA was resuspended in 30 μL TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Finally, the exogenous ssDNA was sent to Sangon Biotech Co. Ltd. (Shanghai, China) for sequencing. The sequencing primer was as follows: 5′-CCCTCATAGTTAGCGTAACG-3′ (−96 gIII).
Analysis of the binding capacity of phage clones by ELISA.
The positive clones were amplified in 1-mL volumes. The supernatants were reserved for subsequent use. The ELISA plates were coated with monoclonal anti-TiLV serum (1:100 diluted in 0.1 M NaHCO3 [pH 8.6]) at 4°C overnight and blocked with BSA (5 mg/mL in 0.1 M NaHCO3 [pH 8.6]) for 2 h at 4°C. Phage clones were added to the antibody-coated wells and incubated for 1 h at room temperature. The primary phage library was used as the negative control. After washing 6 times with washing buffer, 1:5,000-diluted horseradish peroxidase (HRP)-conjugated anti-M13 major coat protein antibody (sc-53004; Santa Cruz Biotechnology, Japan) in blocking buffer was added to the wells and incubated for 1 h at room temperature with agitation. The wells were washed again, and peroxidase activity was detected by adding 3,3′,5,5′-tetramethylbenzidine (TMB) substrate. The OD450 was subsequently recorded using a microplate reader. Each phage clone experiment was performed in triplicate.
Immunoassays of synthetic peptides by indirect ELISA and competitive ELISA.
Peptides Pep1, Pep2, Pep3, Pep4, Pep5, and Pep6 were synthesized (purity of >80%) by SynthBio (Hefei, Anhui, China). The amino acid sequence of each peptide is shown in Table 2. For indirect ELISA, 96-well plates were coated with 100 μL of peptide solution (100, 50, 25, 12.5, 6.25, 3.13, 1.56, and 0.78 μg/mL) and placed at 4°C overnight. Plates were washed 3 times with 0.05% Tween 20 in PBST, followed by blocking with 2% BSA for 2 h at 37°C. After washing, diluted TiLV-positive serum samples were added to the plate at 100 μL/well (1:100 dilution) and incubated at 37°C for 1.5 h. After washing, mouse anti-tilapia IgM antibody (1:4,500) purchased from Frdbio Bioscience & Technology (Wuhan, Hubei, China) was added to the plate, at 100 μL per well, and incubated at 37°C for 1 h. A 100-μL volume of HRP-conjugated goat anti-mouse IgG (D110087; Sangon Biotech, Shanghai, China) at 1:5,500 dilution was added to the plate and incubated at 37°C for 1 h. TMB substrate (Salarbio, Beijing, China) was added and followed by stop solution, and then the absorbance at 450 nm was determined.
For the competitive ELISA, 96-well plates were coated with 6 × 104 copies of TiLV. After blocking, the same volume of a 100-μg/mL peptide solution was incubated with TiLV-positive serum (1:100 dilution) for 1 h at 37°C. The peptide-serum combinations were added to TiLV-coated 96-well plates and incubated at 37°C for 1 h. Other procedures were exactly the same as those for the indirect ELISA.
Immunization.
All experimental animal care and study procedures were handled according to the guidelines of the Animal Ethical and Welfare Committee, Northwest A&F University, China.
For the mimotopes vaccine immunization program (Fig. 2A), 8 groups of Nile tilapia (Oreochromis niloticus) weighing approximately 0.98 ± 0.20 g, 70 fish per group, were immunized intraperitoneally with 20 μg of the peptide emulsified in complete Freund's adjuvant (1:1 [vol/vol]). Among them, the adjuvant group was a 1:1 mixture of adjuvant and PBS, while the blank group was injected with PBS only. Two weeks later, all fish were boosted by intraperitoneal injection with peptide emulsified in incomplete Freund's adjuvant. At 1 to 6 weeks after primary immunization, fish serum (5 fish/group) was collected for further studies.
For the natural epitope vaccine immunization program (Fig. 4A), the candidate natural epitope peptide-KLH conjugate was dissolved at 2 mg/mL in sterile PBS solution, then mixed with complete Freund's adjuvant in a 1:1 volume ratio, and emulsified using a syringe and repeated pipetting. Nile tilapias (Oreochromis niloticus) weighing approximately 0.93 ± 0.24 g were prebled before immunization, and then 20 μg peptide equivalent was injected intraperitoneally to complete the primary immunization, followed by a booster immunization of the same dose at 14-day intervals. Tilapia were collected by neck dislocation at 7, 14, 21, 28, 35, and 42 days after the primary immunization, and serum was prepared and then stored at −80°C until detection.
Determination of serum antibody titers in vaccinated fish by enzyme-linked immunosorbent assay.
Serum antibody titers after animal immunization were determined by ELISA. For immune serum of mimotope vaccine, 96 well plates were coated with containing 2 μg peptide/ 6.0 × 104 copies TiLV virions solution and maintained. The collected serum was diluted by fold and added at 100 μL/well, followed by mouse anti-tilapia IgM (Frdbio, Wuhan, Hubei, China) as the primary antibody and goat anti-mouse IgG (D110087; Sangon Biotech, Shanghai, China) as the secondary antibody. The specific operation method was the same as that for the indirect ELISA method described above. The cutoff threshold was set to be at least 2 times higher than the absorbance value of the negative serum sample, and serum antibody titers were determined at the final dilution with a value above the cutoff threshold. The serum antibody titers were regarded as 10 when the cutoff threshold was not reached at the lowest dilution.
For immune serum of natural epitope vaccine, all peptides or recombinant proteins were adjusted to a final concentration of 20 μg/mL with coating solution, coated on ELISA plates at 100 μL/well, and incubated overnight at 4°C. The next day, 300 μL/well of 2% BSA solution as blocked solution was placed at 37°C for 2 h and then washed three times with 0.05% PBST solution. Tilapia sera from different groups were diluted 1:200 with PBS, then added in 100-μL volumes to the wells and incubated at 37°C for 1 h. Plates were washed 5 times with PBST (0.05% Tween 20) and incubated with 100 μL of mouse anti-tilapia IgM antibody diluted in 5% skim milk for 1 h at 37°C. After washing the plate in the same way, 100 μL/well of HRP-conjugated goat anti-mouse IgG diluted in 5% skim milk was added and incubated at 37°C for 1 h. A 100-μL volume of TMB was added to each well and plates were placed in the dark at room temperature for color development; 2 M H2SO4 was used to terminate the reaction, and the absorbance at 450 nm was measured on an ELISA reader. The calculation method of serum antibody end-point titers was the same as above.
Virus neutralization test.
Serum samples collected from the same group at different time points were pooled into one virus neutralization test sample. Serum samples were filtered through a sterile 0.22-μm filter and then heat inactivated at 42°C for 30 min; heat-inactivated serum (triplicate) was prediluted 20-fold with DMEM, and a 2-fold dilution series was then prepared using DMEM with 0.15% BSA to obtain dilutions ranging from 1:20 to 1:1,280. Diluted serum or DMEM (control) was added to 96-well plates at 50 μL per well, 3 wells per dilution. Then, 50 μL of TiLV was added to each well and incubated at 25°C for 4 h. After incubation, mixture was added to the precultured monolayer CIK cells with 100 μL/well and incubated at 25°C for 7 days. Cell viability was measured by MTT assay to evaluate serum neutralization to TiLV, and the assay was performed according to the method provided by the manufacturer. Wells with only viral fluid in CIK cells were considered to have a neutralizing ability of 0% for TiLV, while wells with only cell culture medium were considered to have completely preserved cell viability with 100%. Thus, when cell viability was 50%, the reciprocal of its serum dilution factor served as the IC50.
Bioinformatics mining of TiLV natural epitopes.
Inspired by the above studies, in order to identify the native epitopes of TiLV, we performed bioinformatics analysis. Alignment of the most immunogenic peptide sequences with TiLV-encoded protein sequences used the Jalview program (68) for the determination of linear epitopes. According to the alignment results, the Robetta server (69) was used to predict the structure of the target protein. For the 3D structure of the peptide, we used the I-TASSER server (70–72) to predict the structure. Peptide-peptide or peptide-protein conformational alignment was performed using PyMOL software. All protein or peptide 3D structures were visualized by PyMOL (Schrödinger).
Depletion test of peptide affinity antibodies in serum.
Following previously described principles, we performed a serum-peptide affinity depletion assay (60, 73, 74). Synthetic peptides were added to ELISA plates at 50 μg/well and incubated at 37°C for 1 h. After blocking with 2% BSA solution, the plate was washed 3 times with PBS containing 0.1% Tween 20, followed by a PBS wash to remove residual Tween 20. The collected serum from the immune group was diluted (1:100) with DMEM and added to the wells of the plate at 50 μL/well and then incubated at room temperature for 20 min to fully adsorb the antibodies. Unbound fractions were recovered after 10 cycles of repeated adsorption. The free fraction recovered in each round was verified by ELISA for serum antibody depletion levels. The antibody-depleted samples were mixed with TiLV and incubated at 25°C for 2 h. A serum neutralization assay was performed to verify the neutralizing activity of antibody-depleted serum.
Vaccine protection against virus challenge.
At 3 weeks after the booster immunization, 33 fish were randomly selected from each group for intraperitoneal injection challenge with TiLV virus (TiLV-CQ-2021) solution at 9.0 × 107 copies TiLV/injection. A heating rod was added to each aquarium to maintain the water temperature at 29 ± 1°C, and the clinical features of disease and mortality were monitored daily. TiLV target tissues such as liver and spleen were collected daily from dead fish until no fish died in each group. Cumulative survival was recorded to assess the protective effect of the vaccine, and survival rate was calculated using the following formula: survival rate = [1 − (mortality of immunized group)/(total number in immunized group)] × 100%.
Viral loads in the liver and spleen of dead fish on day 8 postchallenge in each group were determined by qRT-PCR using virus-specific primers (TiLV-112F, 5′-CTGAGCTAAAGAGGCAATATGGATT-3′; TiLV-112R, 5′-CGTGCGTACTCGTTCAGTATAAGTTCT-3′) to determine fish death from TiLV infection and to determine the antiviral capacity of different immune groups. Briefly, liver and spleen were each homogenized in 1 mL TRIzol, and total RNA was extracted according to the manufacturer's instructions. The extracted RNA was used to measure the RNA concentration using a Nanodrop spectrophotometer, and the concentration was adjusted to 50 ng/μL. Subsequently, 200 ng of RNA was reverse transcribed into 20 μL cDNA using a reverse transcription kit to serve as a template for RT-qPCR. RT-qPCR detection was performed using the CFX96 real-time PCR detection system (Bio-Rad, USA). The total reaction volume was 20 μL, which contained 10 μL of 2× ChamQ SYBR qPCR master mix, 0.3 μM forward and reverse primers, 4 μL of cDNA template, and 0.4 μL of 50× ROX reference dye I, with double-distilled H2O added to obtain the final volume. The qPCR conditions were denaturation at 95°C for 3 min followed by 40 cycles of 95°C for 10 s and 60°C for 30 s (75).
A standard plasmid containing the TiLV genomic segment 3 cDNA was constructed as previously described (75). A 491-bp cDNA fragment from segment 3 of the TiLV genome was amplified using specific primers (40) (nested ext-2, TTGCTCTGAGCAAGAGTACC; nested ext-1, TATGCAGTACTTTCCCTGCC) and cloned into the pMD19-T vector (TaKaRa). The recombinant plasmid was named 19T-S3 (491 bp) and was transformed into E. coli strain DH5α. Plasmid DNA was extracted from E. coli and DNA sequenced using a miniplasmid extraction kit. The concentration of the recombinant plasmid was determined by Nanodrop 2000 spectrophotometer, and its plasmid concentration was 53.46 ng/μL, equivalent to 1.65 × 1010 copies/μL.
Histopathology analysis.
Liver and spleen tissues from tilapia viscera were washed with sterile PBS to remove surface blood stains and then fixed with 4% paraformaldehyde. Samples were dehydrated, embedded in paraffin, sectioned, and stained with hematoxylin-eosin (H&E) by Y&KBio (Xi'an, Shannxi, China). The pathological changes of the tissue sections were then observed under an electron optical microscope.
Statistical analysis.
GraphPad Prism version 9.0 (GraphPad Prism Software, USA) was used to perform all statistical analyses. Correlations were calculated with a Person two-tailed test. P values were analyzed by one-way analysis of variance (ANOVA) with Dunett’s multiple-comparison test, and P values of <0.05 were considered significant with a 95% confidence interval.
Data availability.
The segment 3 of the TiLV genome sequence has been submitted to the National Center for Biotechnology Information and assigned GenBank accession number OM417602.
ACKNOWLEDGMENTS
This study was funded by the National Natural Science Foundation of China (grant number 32173011 and 41966004).
Conceptualization, Bin Zhu; Formal Analysis, Yu-Ming Gong, Xue-Feng Wei, and Yang Li; Investigation, Bin Zhu; Methodology, Bin Zhu, Peng-Fei Li, Yu-Ming Gong, Yu-Ying Zheng, and Xue-Feng Wei; Project Administration, Bin Zhu; Resources, Bin Zhu; Software, Yu-Ming Gong, Yu-Ying Zheng, and Qing Yu; Visualization, Yu-Ming Gong and Xue-Feng Wei; Writing – Original Draft, Yu-Ming Gong; Writing – Review & Editing, Yu-Ming Gong, Xue-Feng Wei, and Qing Yu.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Peng-Fei Li, Email: pfli2014@126.com.
Bin Zhu, Email: zhubin1227@126.com.
Stacey Schultz-Cherry, St. Jude Children's Research Hospital.
REFERENCES
- 1.Brouwers TJ, Van der Zeijst BAM. 2021. Vaccine production, safety, and efficacy, p 281–288. In Bamford DH, Zuckerman M (ed), Encyclopedia of virology, 4th ed. Academic Press, Oxford, United Kingdom. [Google Scholar]
- 2.Bandyopadhyay AS, Zipursky S. 2023. A novel tool to eradicate an ancient scourge: the novel oral polio vaccine type 2 story. Lancet Infect Dis 23:e67–e71. 10.1016/S1473-3099(22)00582-5. [DOI] [PubMed] [Google Scholar]
- 3.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei Y-Y. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 383:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amanat F, Krammer F. 2020. SARS-CoV-2 vaccines: status report. Immunity 52:583–589. 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367:1260–1263. 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, Wang W, Chen Z, Lu S, Yang F, Bi Z, Bao L, Mo F, Li X, Huang Y, Hong W, Yang Y, Zhao Y, Ye F, Lin S, Deng W, Chen H, Lei H, Zhang Z, Luo M, Gao H, Zheng Y, Gong Y, Jiang X, Xu Y, Lv Q, Li D, Wang M, Li F, Wang S, Wang G, Yu P, Qu Y, Yang L, Deng H, Tong A, Li J, Wang Z, Yang J, Shen G, Zhao Z, Li Y, Luo J, Liu H, Yu W, Yang M, Xu J, Wang J, Li H, Wang H, et al. 2020. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 586:572–577. 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 9.He Q, Mao Q, Peng X, He Z, Lu S, Zhang J, Gao F, Bian L, An C, Yu W. 2022. Immunogenicity and protective efficacy of a recombinant protein subunit vaccine and an inactivated vaccine against SARS-CoV-2 variants in non-human primates. Signal Transduct Target Ther 7:69. 10.1038/s41392-022-00926-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pang W, Lu Y, Zhao YB, Shen F, Fan CF, Wang Q, He WQ, He XY, Li ZK, Chen TT, Yang CX, Li YZ, Xiao SX, Zhao ZJ, Huang XS, Luo RH, Yang LM, Zhang M, Dong XQ, Li MH, Feng XL, Zhou QC, Qu W, Jiang S, Ouyang S, Zheng YT. 2022. A variant-proof SARS-CoV-2 vaccine targeting HR1 domain in S2 subunit of spike protein. Cell Res 32:1068–1085. 10.1038/s41422-022-00746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinto D, Sauer MM, Czudnochowski N, Low JS, Tortorici MA, Housley MP, Noack J, Walls AC, Bowen JE, Guarino B, Rosen LE, di Iulio J, Jerak J, Kaiser H, Islam S, Jaconi S, Sprugasci N, Culap K, Abdelnabi R, Foo C, Coelmont L, Bartha I, Bianchi S, Silacci-Fregni C, Bassi J, Marzi R, Vetti E, Cassotta A, Ceschi A, Ferrari P, Cippà PE, Giannini O, Ceruti S, Garzoni C, Riva A, Benigni F, Cameroni E, Piccoli L, Pizzuto MS, Smithey M, Hong D, Telenti A, Lempp FA, Neyts J, Havenar-Daughton C, Lanzavecchia A, Sallusto F, Snell G, Virgin HW, Beltramello M, et al. 2021. Broad betacoronavirus neutralization by a stem helix–specific human antibody. Science 373:1109–1116. 10.1126/science.abj3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li T, Kan Q, Ge J, Wan Z, Yuan M, Huang Y, Xie Q, Yang Y, Shao H, Li X, Ye L, Qin A, Bu Z, Liu P, Ye J. 2021. A novel linear and broadly neutralizing peptide in the SARS-CoV-2 S2 protein for universal vaccine development. Cell Mol Immunol 18:2563–2565. 10.1038/s41423-021-00778-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farfán-Castro S, García-Soto MJ, Comas-García M, Arévalo-Villalobos JI, Palestino G, González-Ortega O, Rosales-Mendoza S. 2021. Synthesis and immunogenicity assessment of a gold nanoparticle conjugate for the delivery of a peptide from SARS-CoV-2. Nanomedicine (Lond) 34:102372. 10.1016/j.nano.2021.102372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Lai D, Zhang H, Jiang H, Tian X, Ma M, Qi H, Meng Q, Guo S, Wu Y, Wang W, Yang X, Shi D, Dai J, Ying T, Zhou J, Tao S. 2020. Linear epitopes of SARS-CoV-2 spike protein elicit neutralizing antibodies in COVID-19 patients. Cell Mol Immunol 17:1095–1097. 10.1038/s41423-020-00523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skwarczynski M, Toth I. 2016. Peptide-based synthetic vaccines. Chem Sci 7:842–854. 10.1039/C5SC03892H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rappuoli R. 2000. Reverse vaccinology. Curr Opin Microbiol 3:445–450. 10.1016/s1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- 17.Serruto D, Bottomley MJ, Ram S, Giuliani MM, Rappuoli R. 2012. The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: immunological, functional and structural characterization of the antigens. Vaccine 30:B87–B97. 10.1016/j.vaccine.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Ryan M, Stoddard J, Toneatto D, Wassil J, Dull PM. 2014. A multi-component meningococcal serogroup B vaccine (4CMenB): the clinical development program. Drugs 74:15–30. 10.1007/s40265-013-0155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacharya M, Sharma AR, Patra P, Ghosh P, Sharma G, Patra BC, Lee S-S, Chakraborty C. 2020. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): immunoinformatics approach. J Med Virol 92:618–631. 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chauhan V, Rungta T, Goyal K, Singh MP. 2019. Designing a multi-epitope based vaccine to combat Kaposi Sarcoma utilizing immunoinformatics approach. Sci Rep 9:2517. 10.1038/s41598-019-39299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tosta SFdO, Passos MS, Kato R, Salgado Á, Xavier J, Jaiswal AK, Soares SC, Azevedo V, Giovanetti M, Tiwari S, Alcantara LCJ. 2021. Multi-epitope based vaccine against yellow fever virus applying immunoinformatics approaches. J Biomol Struct Dyn 39:219–235. 10.1080/07391102.2019.1707120. [DOI] [PubMed] [Google Scholar]
- 22.Tahir Ul Qamar M, Saleem S, Ashfaq UA, Bari A, Anwar F, Alqahtani S. 2019. Epitope-based peptide vaccine design and target site depiction against Middle East respiratory syndrome coronavirus: an immune-informatics study. J Transl Med 17:362. 10.1186/s12967-019-2116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kar PP, Srivastava A. 2018. Immuno-informatics analysis to identify novel vaccine candidates and design of a multi-epitope based vaccine candidate against Theileria parasites. Front Immunol 9:2213. 10.3389/fimmu.2018.02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rappuoli R, Bottomley MJ, D'Oro U, Finco O, De Gregorio E. 2016. Reverse vaccinology 2.0: human immunology instructs vaccine antigen design. J Exp Med 213:469–481. 10.1084/jem.20151960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong R, Xu K, Zhou T, Acharya P, Lemmin T, Liu K, Ozorowski G, Soto C, Taft JD, Bailer RT, Cale EM, Chen L, Choi CW, Chuang G-Y, Doria-Rose NA, Druz A, Georgiev IS, Gorman J, Huang J, Joyce MG, Louder MK, Ma X, McKee K, O'Dell S, Pancera M, Yang Y, Blanchard SC, Mothes W, Burton DR, Koff WC, Connors M, Ward AB, Kwong PD, Mascola JR. 2016. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352:828–833. 10.1126/science.aae0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu K, Acharya P, Kong R, Cheng C, Chuang GY, Liu K, Louder MK, O’Dell S, Rawi R, Sastry M. 2018. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat Med 24:857–867. 10.1038/s41591-018-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu S, Xie X-X, Zhao L, Wang B, Zhu J, Yang T-R, Yang G-W, Ji M, Lv C-P, Xue J, Dai E-H, Fu X-M, Liu D-Q, Zhang L, Hou S-J, Yu X-L, Wang Y-L, Gao H-X, Shi X-H, Ke C-W, Ke B-X, Jiang C-G, Liu R-T. 2021. The immunodominant and neutralization linear epitopes for SARS-CoV-2. Cell Rep 34:108666. 10.1016/j.celrep.2020.108666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith GP. 1985. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315–1317. 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 29.Khurana S, Ravichandran S, Hahn M, Coyle EM, Stonier SW, Zak SE, Kindrachuk J, Davey RT, Dye JM, Chertow DS. 2020. Longitudinal human antibody repertoire against complete viral proteome from Ebola virus survivor reveals protective sites for vaccine design. Cell Host Microbe 27:262–276.e4. 10.1016/j.chom.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang R, Yang Y, Lan J, Lin S, Xie Z, Zhang X, Jiang S. 2020. A novel peptide isolated from a phage display peptide library modeling antigenic epitope of DHAV-1 and DHAV-3. Vaccines 8:121. 10.3390/vaccines8010121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Ding X, Zhu L, Zhang G. 2021. The identification of a B-cell epitope in bovine viral diarrhea virus (BVDV) core protein based on a mimotope obtained from a phage-displayed peptide library. Int J Biol Macromol 183:2376–2386. 10.1016/j.ijbiomac.2021.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Guo JY, Liu IJ, Lin HT, Wang MJ, Chang YL, Lin SC, Liao MY, Hsu WC, Lin YL, Liao JC. 2021. Identification of COVID-19 B-cell epitopes with phage-displayed peptide library. J Biomed Sci 28:1–13. 10.1186/s12929-021-00740-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, Yu J, Shan S, Zhou B, Song S, Tang X, Yu J, Lan J, Yuan J, Wang H, Zhao J, Zhang S, Wang Y, Shi X, Liu L, Zhao J, Wang X, Zhang Z, Zhang L. 2020. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584:115–119. 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 34.Devlin JJ, Panganiban LC, Devlin PE. 1990. Random peptide libraries: a source of specific protein binding molecules. Science 249:404–406. 10.1126/science.2143033. [DOI] [PubMed] [Google Scholar]
- 35.Mullen LM, Nair SP, Ward JM, Rycroft AN, Henderson B. 2006. Phage display in the study of infectious diseases. Trends Microbiol 14:141–147. 10.1016/j.tim.2006.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehmann D, Sodoyer R, Leterme S. 2004. Characterization of BoHV-1 gE envelope glycoprotein mimotopes obtained by phage display. Vet Microbiol 104:1–17. 10.1016/j.vetmic.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Geysen HM, Rodda SJ, Mason TJ. 1986. A priori delineation of a peptide which mimics a discontinuous antigenic determinant. Mol Immunol 23:709–715. 10.1016/0161-5890(86)90081-7. [DOI] [PubMed] [Google Scholar]
- 38.Bacharach E, Mishra N, Briese T, Zody MC, Kembou Tsofack JE, Zamostiano R, Berkowitz A, Ng J, Nitido A, Corvelo A, Toussaint NC, Abel Nielsen SC, Hornig M, Del Pozo J, Bloom T, Ferguson H, Eldar A, Lipkin WI. 2016. Characterization of a novel Orthomyxo-like virus causing mass die-offs of tilapia. mBio 7:e00431-16. 10.1128/mBio.00431-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Del-Pozo J, Mishra N, Kabuusu R, Cheetham S, Eldar A, Bacharach E, Lipkin WI, Ferguson HW. 2017. Syncytial hepatitis of tilapia (Oreochromis niloticus L.) is associated with Orthomyxovirus-like virions in hepatocytes. Vet Pathol 54:164–170. 10.1177/0300985816658100. [DOI] [PubMed] [Google Scholar]
- 40.Eyngor M, Zamostiano R, Kembou Tsofack JE, Berkowitz A, Bercovier H, Tinman S, Lev M, Hurvitz A, Galeotti M, Bacharach E, Eldar A. 2014. Identification of a novel RNA virus lethal to tilapia. J Clin Microbiol 52:4137–4146. 10.1128/JCM.00827-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Surachetpong W, Janetanakit T, Nonthabenjawan N, Tattiyapong P, Sirikanchana K, Amonsin A. 2017. Outbreaks of Tilapia lake virus infection, Thailand, 2015–2016. Emerg Infect Dis 23:1031–1033. 10.3201/eid2306.161278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong HT, Senapin S, Gangnonngiw W, Nguyen VV, Rodkhum C, Debnath PP, Delamare-Deboutteville J, Mohan CV. 2020. Experimental infection reveals transmission of Tilapia lake virus (TiLV) from tilapia broodstock to their reproductive organs and fertilized eggs. Aquaculture 515:734541. 10.1016/j.aquaculture.2019.734541. [DOI] [Google Scholar]
- 43.Aich N, Paul A, Choudhury TG, Saha H. 2021. Tilapia lake virus (TiLV) disease: current status of understanding. Aquac Fish 7:7–17. 10.1016/j.aaf.2021.04.007. [DOI] [Google Scholar]
- 44.Acharya V, Chakraborty HJ, Rout AK, Balabantaray S, Behera BK, Das BK. 2019. Structural characterization of open reading frame-encoded functional genes from Tilapia lake virus (TiLV). Mol Biotechnol 61:945–957. 10.1007/s12033-019-00217-y. [DOI] [PubMed] [Google Scholar]
- 45.Yu N, Zeng W, Xiong Z, Liu Z. 2022. A high efficacy DNA vaccine against Tilapia lake virus in Nile tilapia (Oreochromis niloticus). Aquac Rep 24:101166. 10.1016/j.aqrep.2022.101166. [DOI] [Google Scholar]
- 46.Mai TT, Kayansamruaj P, Taengphu S, Senapin S, Costa JZ, del-Pozo J, Thompson KD, Rodkhum C, Dong HT. 2021. Efficacy of heat-killed and formalin-killed vaccines against Tilapia tilapinevirus in juvenile Nile tilapia (Oreochromis niloticus). J Fish Dis 44:2097–2109. 10.1111/jfd.13523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng W, Wang Y, Chen X, Wang Q, Bergmann SM, Yang Y, Wang Y, Li B, Lv Y, Li H, Lan W. 2021. Potency and efficacy of VP20-based vaccine against Tilapia lake virus using different prime-boost vaccination regimens in tilapia. Aquaculture 539:736654. 10.1016/j.aquaculture.2021.736654. [DOI] [Google Scholar]
- 48.Gong YM, Wei XF, Zhou GQ, Liu MZ, Li PF, Zhu B. 2022. Mannose functionalized biomimetic nanovaccine enhances immune responses against Tilapia lake virus. Aquaculture 560:738535. 10.1016/j.aquaculture.2022.738535. [DOI] [Google Scholar]
- 49.He C, Yang J, Hong W, Chen Z, Peng D, Lei H, Alu A, He X, Bi Z, Jiang X. 2022. A self-assembled trimeric protein vaccine induces protective immunity against Omicron variant. Nat Commun 13:5459. 10.1038/s41467-022-33209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith GP. 2019. Phage display: simple evolution in a petri dish (Nobel lecture). Angew Chem Int Ed Engl 58:14428–14437. 10.1002/anie.201908308. [DOI] [PubMed] [Google Scholar]
- 51.Barderas R, Benito-Peña E. 2019. The 2018 Nobel Prize in Chemistry: phage display of peptides and antibodies. Anal Bioanal Chem 411:2475–2479. 10.1007/s00216-019-01714-4. [DOI] [PubMed] [Google Scholar]
- 52.Yang L, Cen J, Xue Q, Li J, Bi Y, Sun L, Liu W. 2013. Identification of rabies virus mimotopes screened from a phage display peptide library with purified dog anti-rabies virus serum IgG. Virus Res 174:47–51. 10.1016/j.virusres.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 53.Xin ZT, Liu C, Gao YP, Mao CQ, Zhao A, Zhang J, Shao NS, Ling SG, Xue YN. 2003. Identification of mimotopes by screening of a bacterially displayed random peptide library and its use in eliciting an immune response to native HBV-preS. Vaccine 21:4373–4379. 10.1016/s0264-410x(03)00436-5. [DOI] [PubMed] [Google Scholar]
- 54.Hernandez DN, Tam K, Shopsin B, Radke EE, Kolahi P, Copin R, Stubbe FX, Cardozo T, Torres VJ, Silverman GJ. 2020. Unbiased identification of immunogenic Staphylococcus aureus leukotoxin B-cell epitopes. Infect Immun 88:e00785-19. 10.1128/IAI.00785-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tarafdar S, Virata ML, Yan H, Zhong L, Deng L, Xu Y, He Y, Struble E, Zhang P. 2022. Multiple epitopes of hepatitis B virus surface antigen targeted by human plasma-derived immunoglobulins coincide with clinically observed escape mutations. J Med Virol 94:649–658. 10.1002/jmv.27278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathur D, Prakash S, Anand P, Kaur H, Agrawal P, Mehta A, Kumar R, Singh S, Raghava GP. 2016. PEPlife: a repository of the half-life of peptides. Sci Rep 6:36617. 10.1038/srep36617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moynihan KD, Holden RL, Mehta NK, Wang C, Karver MR, Dinter J, Liang S, Abraham W, Melo MB, Zhang AQ, Li N, Gall SL, Pentelute BL, Irvine DJ. 2018. Enhancement of peptide vaccine immunogenicity by increasing lymphatic drainage and boosting serum stability. Cancer Immunol Res 6:1025–1038. 10.1158/2326-6066.CIR-17-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang M, Zhai L, Yu W, Wei Y, Wang L, Liu S, Li W, Li X, Yu S, Chen X, Zhang H, Chen J, Feng Z, Yu L, Cui Y. 2018. Identification of a protective B-cell epitope of the Staphylococcus aureus GapC protein by screening a phage-displayed random peptide library. PLoS One 13:e0190452. 10.1371/journal.pone.0190452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malonis RJ, Lai JR, Vergnolle O. 2019. Peptide-based vaccines: current progress and future challenges. Chem Rev 120:3210–3229. 10.1021/acs.chemrev.9b00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poh CM, Carissimo G, Wang B, Amrun SN, Lee CYP, Chee RSL, Fong SW, Yeo NKW, Lee WH, Torres-Ruesta A. 2020. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat Commun 11:2806. 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hernandez DN, Tam K, Shopsin B, Radke EE, Law K, Cardozo T, Torres VJ, Silverman GJ. 2020. Convergent evolution of neutralizing antibodies to Staphylococcus aureus γ-hemolysin C that recognize an immunodominant primary sequence-dependent B-cell epitope. mBio 11:e00460-20. 10.1128/mBio.00460-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen X, Li C, Lin W, Li T, Li X, Bai X, Wulin S, Zhang Q, Li S, Liu M, Liu J-H, Zhang Y. 2020. A novel neutralizing antibody targeting a unique cross-reactive epitope on the hi loop of domain II of the envelope protein protects mice against duck Tembusu virus. J Immunol 204:1836–1848. 10.4049/jimmunol.1901352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mo X, Li X, Yin B, Deng J, Tian K, Yuan A. 2019. Structural roles of PCV2 capsid protein N-terminus in PCV2 particle assembly and identification of PCV2 type-specific neutralizing epitope. PLoS Pathog 15:e1007562. 10.1371/journal.ppat.1007562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tattiyapong P, Dechavichitlead W, Waltzek TB, Surachetpong W. 2020. Tilapia develop protective immunity including a humoral response following exposure to Tilapia lake virus. Fish Shellfish Immunol 106:666–674. 10.1016/j.fsi.2020.08.031. [DOI] [PubMed] [Google Scholar]
- 65.Lee J, Klenow L, Coyle EM, Golding H, Khurana S. 2018. Protective antigenic sites in respiratory syncytial virus G attachment protein outside the central conserved and cysteine noose domains. PLoS Pathog 14:e1007262. 10.1371/journal.ppat.1007262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang B-Z, Hu Y-F, Chen L-L, Yau T, Tong Y-G, Hu J-C, Cai J-P, Chan K-H, Dou Y, Deng J, Wang X-L, Hung IF-N, To KK-W, Yuen KY, Huang J-D. 2020. Mining of epitopes on spike protein of SARS-CoV-2 from COVID-19 patients. Cell Res 30:702–704. 10.1038/s41422-020-0366-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zeng W, Wang Y, Hu H, Wang Q, Bergmann SM, Wang Y, Li B, Lv Y, Li H, Yin J, Li Y. 2021. Cell culture-derived Tilapia lake virus-inactivated vaccine containing montanide adjuvant provides high protection against viral challenge for tilapia. Vaccines 9:86. 10.3390/vaccines9020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview Version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, Wang J, Cong Q, Kinch LN, Schaeffer RD, Millán C, Park H, Adams C, Glassman CR, DeGiovanni A, Pereira JH, Rodrigues AV, van-Dijk AA, Ebrecht AC, Opperman DJ, Sagmeister T, Buhlheller C, PavkovKeller T, Rathinaswamy MK, Dalwadi U, Yip CK, Burke JE, Garcia KC, Grishin NV, Adams PD, Read RJ, Baker D. 2021. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373:871–876. 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng W, Zhang C, Li Y, Pearce R, Bell EW, Zhang Y. 2021. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep Methods 1:100014. 10.1016/j.crmeth.2021.100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER suite: protein structure and function prediction. Nat Methods 12:7–8. 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang J, Zhang Y. 2015. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:W174–W181. 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kam Y-W, Lum F-M, Teo T-H, Lee WWL, Simarmata D, Harjanto S, Chua C-L, Chan Y-F, Wee J-K, Chow A, Lin RTP, Leo Y-S, Le Grand R, Sam I-C, Tong J-C, Roques P, Wiesmüller K-H, Rénia L, Rötzschke O, Ng LFP. 2012. Early neutralizing IgG response to Chikungunya virus in infected patients targets a dominant linear epitope on the E2 glycoprotein. EMBO Mol Med 4:330–343. 10.1002/emmm.201200213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee CYP, Carissimo G, Chen Z, Lum FM, Abu Bakar F, Rajarethinam R, Teo TH, Torres-Ruesta A, Renia L, Ng LF. 2020. Type I interferon shapes the quantity and quality of the anti-Zika virus antibody response. Clin Transl Immunol 9:e1126. 10.1002/cti2.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tattiyapong P, Sirikanchana K, Surachetpong W. 2018. Development and validation of a reverse transcription quantitative polymerase chain reaction for Tilapia lake virus detection in clinical samples and experimentally challenged fish. J Fish Dis 41:255–261. 10.1111/jfd.12708. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4 and Table S1. Download jvi.00050-23-s0001.docx, DOCX file, 0.7 MB (695KB, docx)
Data Availability Statement
The segment 3 of the TiLV genome sequence has been submitted to the National Center for Biotechnology Information and assigned GenBank accession number OM417602.