ABSTRACT
Porcine enteric alphacoronavirus (PEAV) is a new bat HKU2-like porcine coronavirus, and its endemic outbreak has caused severe economic losses to the pig industry. Its broad cellular tropism suggests a potential risk of cross-species transmission. A limited understanding of PEAV entry mechanisms may hinder a rapid response to potential outbreaks. This study analyzed PEAV entry events using chemical inhibitors, RNA interference, and dominant-negative mutants. PEAV entry into Vero cells depended on three endocytic pathways: caveolae, clathrin, and macropinocytosis. Endocytosis requires dynamin, cholesterol, and a low pH. Rab5, Rab7, and Rab9 GTPases (but not Rab11) regulate PEAV endocytosis. PEAV particles colocalize with EEA1, Rab5, Rab7, Rab9, and Lamp-1, suggesting that PEAV translocates into early endosomes after internalization, and Rab5, Rab7, and Rab9 regulate trafficking to lysosomes before viral genome release. PEAV enters porcine intestinal cells (IPI-2I) through the same endocytic pathway, suggesting that PEAV may enter various cells through multiple endocytic pathways. This study provides new insights into the PEAV life cycle.
IMPORTANCE Emerging and reemerging coronaviruses cause severe human and animal epidemics worldwide. PEAV is the first bat-like coronavirus to cause infection in domestic animals. However, the PEAV entry mechanism into host cells remains unknown. This study demonstrates that PEAV enters into Vero or IPI-2I cells through caveola/clathrin-mediated endocytosis and macropinocytosis, which does not require a specific receptor. Subsequently, Rab5, Rab7, and Rab9 regulate PEAV trafficking from early endosomes to lysosomes, which is pH dependent. The results advance our understanding of the disease and help to develop potential new drug targets against PEAV.
KEYWORDS: Rab proteins, endocytic pathway, endosome trafficking, porcine enteric alphacoronavirus, single-virus tracking
INTRODUCTION
Porcine enteric alphacoronavirus (PEAV), also known as swine acute diarrhea syndrome/swine enteric alphacoronavirus (SADS-CoV/SeACoV), was first identified in China in February 2017 (1). The clinical symptoms are similar to those caused by other known porcine enteric coronaviruses (2, 3). The piglet mortality rate is 90%, and the disease has killed 24,693 piglets (4). Although the epidemic was effectively controlled, there were still reports of outbreaks in 2019 (5), and in 2021 SADS-CoV was detected in swine diarrhea samples in Guangxi (6). Current antiviral treatment strategies cannot completely protect against various porcine coronaviruses, and there is no cross-protection between viruses. A systematic and comprehensive dissection of the process of PEAV entering the host is urgently needed to understand the pathogenic mechanism of PEAV better.
PEAV is an enveloped, single-stranded, positive-sense RNA virus with a genome length of ~27 kb (1). It contains a 5′ untranslated region (5′ UTR), open reading frame 1a/1b (ORF1a/1b), spike (S), nonstructural protein 3 (NS3a)/ORF3, an envelope protein (E), matrix protein (M), nucleocapsid (N), nonstructural protein 7a (NS7a), and 3′ UTR (4, 7). The S protein amino acid homology of PEAV (the smallest S protein among coronaviruses) and HKU2 is 87% (1). Similar to the S protein of the porcine epidemic diarrhea virus (PEDV), which binding to its receptor in an acidic environment causes conformational changes resulting in viral endocytosis, which drives the fusion of the viral envelope with the cell membrane and releases the viral genome into the cytoplasm. Therefore, the same type of mechanism is likely to occur for the PEAV S protein (8). Efforts to understand the mechanism of PEAV infection include determining viral genome structure, S glycoprotein cryo-electron microscopy (cryo-EM) structural elucidation (9), and PEAV infection receptor exploration (10, 11). However, little is known about the endocytic pathway, endosomal trafficking, and viral fusion.
Coronaviruses utilize multiple endocytic pathways to enter host cells, including clathrin-mediated (12), caveola-mediated (13), and macropinocytosis (14). Porcine epidemic diarrhea coronavirus (PEDV) is another swine enteric coronavirus, and its entry into cells is via clathrin-mediated endocytosis (CME) independent of caveola-mediated endocytosis (CavME) (15). However, a recent study demonstrated via live-cell imaging that PEDV could enter through clathrin- and caveola-dependent pathways (16). Similarly, porcine delta coronavirus (PDCoV) can enter IPI-2I cells via clathrin-mediated- and macropinocytosis pathways (14). These studies suggest that coronaviruses may hijack multiple endocytic pathways to enter host cells. Recent studies showed that bile acids promote the caveola-associated entry of SADS-CoV into porcine intestinal enteroids (17). However, the detailed mechanism of PEAV entry into host cells remains unclear.
The dynamics of the vesicle network in endocytic pathways are regulated by Rab proteins, small GTPases of the Ras superfamily, and their effectors (18). Rab proteins’ extensive and conserved functions regulate intracellular material transport and even innate immunity (19, 20). Rab5 regulates the transport of newly endocytosed vesicles from the plasma membrane to early endosomes (21). Rab7 regulates early endosomal maturation and trafficking from late endosomes to lysosomes (22). Rab9 promotes late endosome entry into the trans-Golgi network (23), while Rab11 involves vesicle cycling (24). Various viruses often use these Rab proteins to regulate the viral endosome transport process, including human immunodeficiency virus (HIV) (23), herpes simplex virus (HSV) (25), classical swine fever virus (CSFV) (26), and coronaviruses, including PDCoV (14) and PEDV (16). However, which type of Rab protein allows entry of PEAV into Vero cells is unknown.
Here, we used chemical inhibitors, RNA interference, and expression of dominant-negative (DN) plasmids to examine which cellular molecules are involved in PEAV entry. Furthermore, the dynamics of fluorescently labeled Rab5, Rab7, Rab9, Rab11, and PEAV were imaged in living cells at the single-particle level. The entry of PEAV into Vero/IPI-2I cells depends on three endocytic pathways: caveolae, clathrin, and macropinocytosis. Rab5, Rab7, and Rab9 are involved in the viral transport process and are transported from the early endosome to the lysosome in a low-pH-dependent environment. This work extends our understanding of the early stages of PEAV infection.
RESULTS
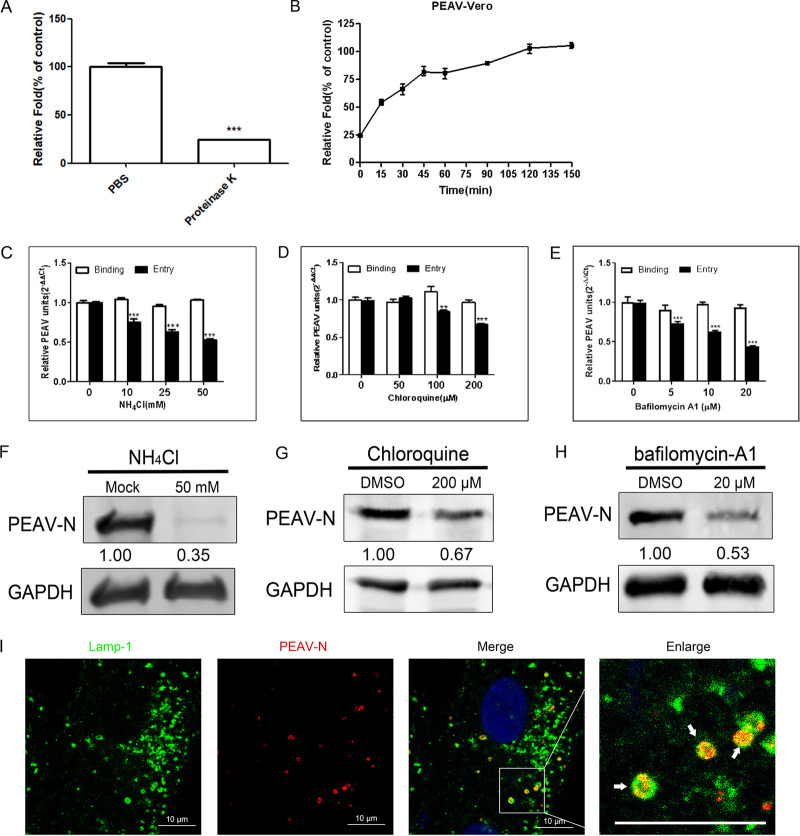
PEAV entry is pH-dependent.
Proteinase K treatment of infected cells resulted in the detachment of most attached PEAV particles (75.8%) but not internalized PEAV particles (Fig. 1A). PEAV rapidly increased at 0 to 45 min postinfection and gradually stabilized, reaching a plateau at 120 min postinfection (Fig. 1B). Therefore, the condition of infection for 1 h at 37°C was selected to study the effect of inhibitors on virus entry. As expected, NH4Cl (Fig. 1C), chloroquine (Fig. 1D), and bafilomycin-A1 (Fig. 1E) inhibited PEAV entry but did not inhibit PEAV binding compared to that of the control group by real-time PCR. Consistently, the expression level of PEAV viral protein was significantly reduced after NH4Cl (Fig. 1F), chloroquine (Fig. 1G), and bafilomycin-A1 (Fig. 1H) treatment. The endosome-lysosome system is involved in regulating endocytic vesicle pH (27). Confocal microscopy showed that PEAV significantly colocalized with the late endosomal marker protein Lamp-1 (Fig. 1I). These results suggest that PEAV entry into Vero cells is pH dependent.
FIG 1.
PEAV entry depends on pH. (A) Vero cells were inoculated with PEAV (MOI = 1) (1 h, 4°C), treated with proteinase K (5 min, 20°C), washed, and lysed. Extracted viral RNA levels were detected using qRT-PCR. PBS was used as a control. (B) Vero cells were inoculated with PEAV (MOI = 1) (1 h, 4°C), washed, and incubated with preheated maintenance medium (37°C, 0 to 150 min). Cells were treated with proteinase K for different durations, and RNA levels were detected using qRT-PCR. (C to E) Vero cells were pretreated (37°C, 15 min) with different concentrations of endosomal acidification inhibitors (NH4Cl, chloroquine, and bafilomycin-A1), followed by incubation with PEAV (MOI = 1) to allow binding (1 h, 4°C) and entry (37°C, 1 h) in the presence of drugs. All results are presented as the mean ± SD from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (F to H) Vero cells were inoculated with PEAV (MOI = 5) in the presence of the NH4Cl (50 mM), chloroquine (200 μM), and bafilomycin-A1 (20 μM) for a prolonged infection period (2.5 h, 37°C). Western blotting was performed to detect the early expression of PEAV N protein. Relative protein content was calculated using ImageJ software. (I) Vero cells were inoculated with PEAV (MOI = 10) (1 h, 4°C), washed, incubated (30 min, 37°C), and fixed (4% PFA, 15 min, room temperature). Virus particles and lysosomes were stained with anti-PEAV N and anti-Lamp-1 monoclonal antibodies, respectively, and their respective localizations were observed using a confocal microscope. The white arrow shows that PEAV colocalizes with lysosomal vesicles. Scale bar = 10 μm.
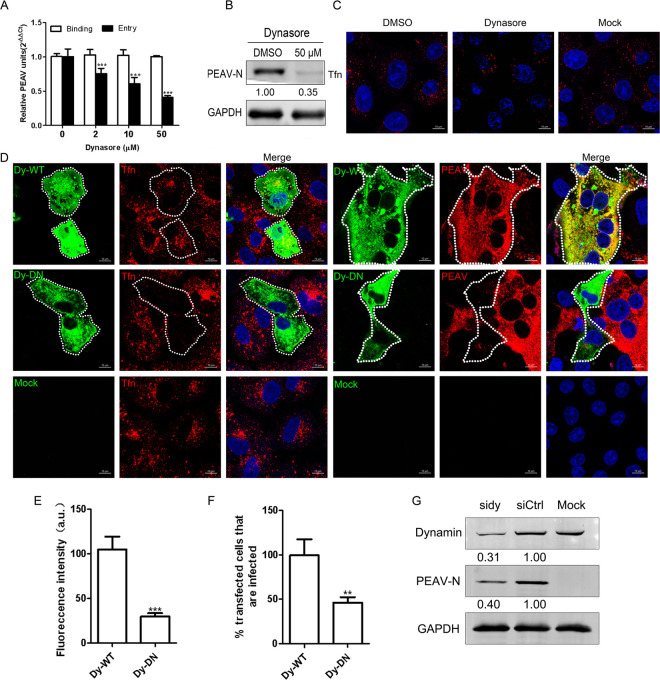
PEAV entry is dynamin dependent.
Dynamin is involved in the entry of PEDV (28). To test whether PEAV requires dynamin, we used dynasore (a noncompetitive inhibitor of the GTPase activity of dynamin). We found that dynasore significantly inhibits PEAV entry, but not the binding, in a dose-dependent manner (Fig. 2A). The expression of early viral proteins of PEAV was also significantly decreased under 50 μM dynasore treatment (Fig. 2B). Transferrin (Tfn) uptake depends on dynamin (29, 30), which is widely used during the study of viral endocytosis (26, 31). Our results show that dynasore significantly inhibits the uptake of Tfn by Vero cells (Fig. 2C). To confirm the role of dynamin further, we constructed a dominant-negative (DN) mutant of dynamin. As expected, Tfn uptake (~75.3%) (Fig. 2E) and PEAV infection (~53%) (Fig. 2F) were significantly reduced in DN dynamin-overexpressing Vero cells compared with wild-type (WT) cells (Fig. 2D). Similarly, knockdown of endogenous dynamin significantly decreased the expression of PEAV N protein (Fig. 2G). These results suggest that dynamin is required for PEAV entry.
FIG 2.
PEAV entry requires a dynamin. (A) Vero cells were pretreated with dynasore (15 min, 37°C), followed by PEAV (MOI = 1) inoculation to allow binding (1 h, 4°C) and entry (1 h, 37°C) in the presence of drugs. (B) In the presence of dynasore (50 μM), cells were inoculated with PEAV (MOI = 5) (2.5 h, 37°C), followed by Western blotting to detect the early expression of PEAV N protein. Relative amounts of protein were calculated using ImageJ software. (C) Vero cells were pretreated with dynasore (50 μM) or dimethyl sulfoxide (DMSO) (negative control) (15 min, 37°C), followed by incubation with 20 μg/mL Alexa Fluor 568-labeled Tfn (15 min, 37°C), and Tfn uptake was examined using confocal microscopy. (D) Vero cells were transfected with dynamin (WT or DN) plasmid (24 h, 37°C) and then incubated with 20 μg/mL Alexa Fluor 568-labeled Tfn (30 min, 37°C). Cells were fixed (4% PFA, 15 min, room temperature) or inoculated with PEAV (MOI = 5) (6 h, 37°C) and fixed and then immunodetected with anti-PEAV N monoclonal antibody. (E) Total fluorescence intensity per cell was calculated in the cells transfected with dynamin DN plasmid. (F) At least 300 transfected cells were scored positive (+) or negative (−) for PEAV infection; values are expressed as a percentage of infected cells observed in the control sample. (G) Cells were transfected with siDynamin or siCtrl (48 h, 37°C) and inoculated with PEAV (MOI = 5). Viral N protein expression was quantified using Western blotting at 2.5 h postinfection (hpi). The relative protein content was calculated using ImageJ software. All results are presented as the mean ± SD from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Scale bar = 10 μm.
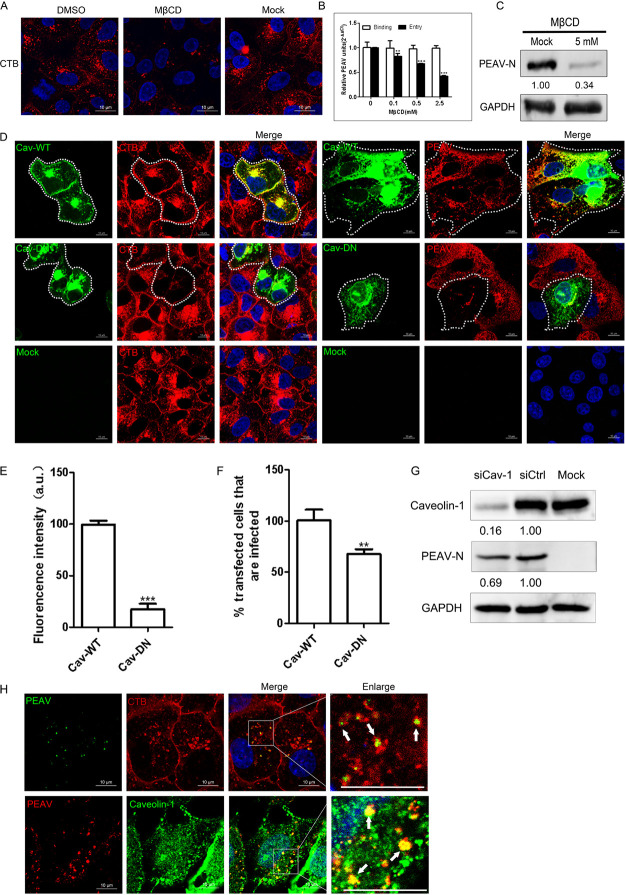
PEAV entry depends on caveolae and cholesterol.
Endocytosis mediated by caveolae is dependent on dynamin (32). However, caveolin-1 and plasma membrane cholesterol are essential for forming and maintaining caveolae (33). Therefore, we further evaluated the role of caveolae and cholesterol in PEAV entry. Early studies confirmed that cholera toxin subunit B (CTB) depends on caveola-mediated endocytosis (34). Confocal microscopy showed that methyl-β-cyclodextrin (MβCD), an inhibitor that depletes cholesterol, can significantly reduce CTB uptake by Vero cells (Fig. 3A) and inhibit PEAV entry and viral protein expression but does not affect virus binding compared with the control group (Fig. 3B and C). We also found that cells transfected with DN caveolin-1 showed significantly reduced uptake of CTB by 82.2% (Fig. 3E) and infection with PEAV by 32.7% (Fig. 3F) compared with cells transfected with WT caveolin-1 (Fig. 3D). Similarly, knockdown of endogenous caveolin-1 also significantly reduced the expression level of PEAV N protein (Fig. 3G). In addition, using confocal microscopy, we observed that PEAV was colocalized with CTB and endogenous caveolin-1 (Fig. 4H). These results confirm that PEAV entry into Vero cells depends on caveolae and cholesterol.
FIG 3.
PEAV entry depends on caveolae and cholesterol. (A) Vero cells were pretreated with MβCD (5 mM, 15 min, 37°C), followed by incubation with 20 μg/mL Alexa Fluor 555-labeled CTB (15 min, 37°C) in the presence of drugs. DMSO was used as a negative control, and Tfn uptake was observed using confocal microscopy. (B) Vero cells were pretreated with MβCD (15 min, 37°C) and then inoculated with PEAV (MOI = 1) to allow binding (1 h, 4°C) and entry (1 h, 37°C) in the presence of drugs. (C) Cells were inoculated with PEAV (MOI = 5) (2.5 h, 37°C) in the presence of MβCD (5 mM). The early expression of PEAV N protein was detected using Western blotting, and the relative amounts of protein were calculated using ImageJ software. (D) Vero cells were transfected with caveolin-1 WT or DN plasmid (24 h, 37°C), followed by incubation with 20 μg/mL Alexa Fluor 555-labeled CTB (30 min, 37°C) or inoculation with PEAV (MOI = 5) (6 h, 37°C). Cells were fixed (4% PFA, 15 min, room temperature) and then immunodetected using anti-PEAV N monoclonal antibody. (E) Total fluorescence intensity per cell was calculated in the cells transfected with caveolin-1 DN plasmid. (F) Percentage of infected cells observed in WT/DN samples. Over 300 transfected cells were analyzed as described above. (G) Cells were transfected with siCaveolin-1 or siCtrl for (48 h, 37°C) and inoculated with PEAV (MOI = 5) in the presence of EIPA (50 μM) inhibitors. Viral N protein expression was quantified at 2.5 hpi using Western blotting. The relative protein content was calculated by ImageJ software. (H) Vero cells were seeded in confocal glass bottom dishes until 90% confluence, inoculated with PEAV (MOI = 10) (1 h, 4°C), and washed. Cells were then incubated in a maintenance medium with or without 20 μg/mL (Alexa Fluor 555-labeled CTB) (15 min, 37°C), fixed (4% PFA, 15 min, room temperature), immunodetected using anti-PEAV N, and observed using confocal microscopy. Virions and caveolin-1 were stained with anti-PEAV N and anti-caveolin-1 monoclonal antibodies. Confocal microscopy revealed PEAV colocalization with CTB or caveolin-1 (white arrows). All results are presented as the mean ± SD from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Scale bar = 10 μm.
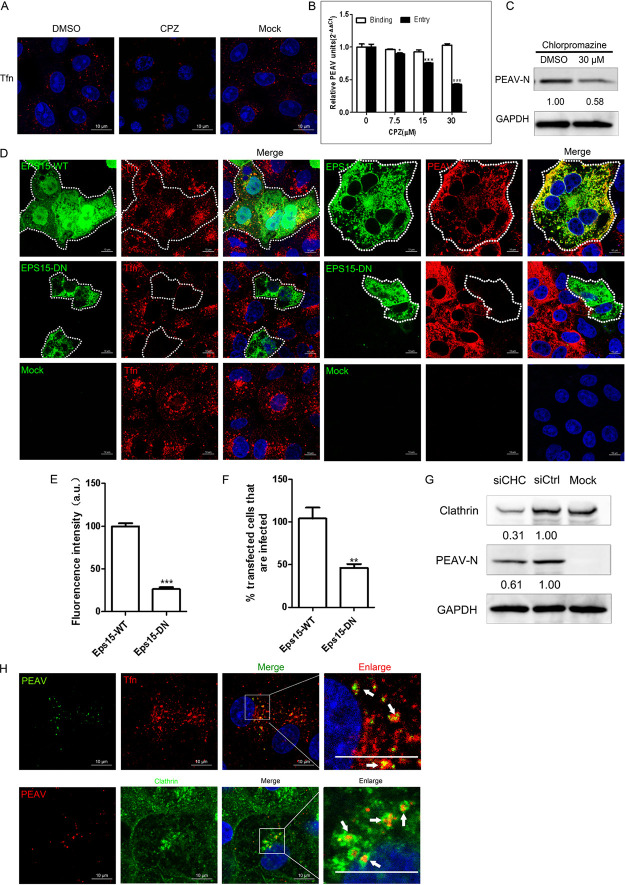
FIG 4.
PEAV entry is clathrin dependent. (A) Vero cells were pretreated with CPZ (30 μM, 15 min, 37°C), followed by incubation with 20 μg/mL Alexa Fluor 568-labeled Tfn (15 min, 37°C). DMSO was used as a negative control, and the uptake of Tfn was measured using confocal microscopy. (B) Vero cells were pretreated with CPZ (15 min, 37°C), followed by inoculation with PEAV (MOI = 1) to allow binding (1 h, 4°C) and entry (1 h, 37°C) in the presence of drugs. (C) In the presence of CPZ (30 μM) (D), cells were inoculated with PEAV (MOI = 5) (2.5 h, 37°C) and immunoblotted to detect the early expression of PEAV N protein. Relative amounts of protein were calculated using ImageJ software. (D) Vero cells were transfected with Eps15 (WT or DN) plasmids (24 h, 37°C), and then incubated with 20 μg/mL Alexa Fluor 568-labeled Tfn (30 min, 37°C) and fixed (4% PFA, 15 min, room temperature) or inoculated with PEAV (MOI = 5) (6 h, 37°C) and fixed (4% PFA, 15 min, room temperature), followed by immunodetection using anti-PEAV N monoclonal antibody. (E) Total fluorescence intensity per cell was calculated in transfected Eps15 DN plasmid cells. (F) Percentage of infected cells observed in WT/DN samples. Over 300 transfected cells were analyzed as described above. (G) Clathrin HC knockdown inhibited PEAV infection. Cells were transfected with siClathrin HC or siCtrl (48 h, 37°C) and inoculated with PEAV (MOI = 5) in the presence of EIPA inhibitors (2.5 h, 37°C), and viral N protein expression was quantified using Western blotting. Relative amounts of protein were calculated using ImageJ software. (H) Vero cells were seeded with PEAV (MOI = 10) (1 h, 4°C), washed, incubated in a maintenance medium with or without 20 μg/mL Alexa Fluor 568-labeled Tfn (15 min, 37°C), and then fixed (4% PFA, 15 min, room temperature). Virions and clathrin were stained with anti-PEAV N and anticlathrin monoclonal antibodies. Confocal microscopy revealed PEAV colocalization with Tfn or clathrin (white arrows). All results are presented as the mean ± SD from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Scale bar = 10 μm.
PEAV entry is clathrin dependent.
Dynamin is dependent on caveola-mediated endocytosis and clathrin-mediated endocytosis (35). PEDV mediates endocytosis using both caveolae and clathrin (16, 28). Tfn uptake depends on clathrin-mediated endocytosis (36). To test whether PEAV entry is clathrin dependent, we pretreated cells with 30 μM chlorpromazine (CPZ) (an inhibitor of lattice polymerization of clathrin) and found that cellular uptake of Tfn was significantly reduced (Fig. 4A). CPZ did not inhibit PEAV binding, but significantly inhibited PEAV entry and viral protein expression, compared with the control group (Fig. 4B and C). Eps15 is a crucial component of clathrin-mediated endocytosis (37, 38). The role of clathrin in PEAV entry was further evaluated by cell transfection with the Eps15 WT and DN plasmid, and the homologous domain of WT was deleted to form enhanced green fluorescent protein (EGFP)-tagged Eps15 Δ29/295 (Eps15 DN). Tfn uptake (~73.3%) (Fig. 4E) and PEAV infection (~57.8%) (Fig. 4F) were significantly reduced in DN mutant-transfected cells compared with WT-transfected cells (Fig. 4D). Meanwhile, PEAV N protein expression decreased with the knockdown of the clathrin heavy chain compared with the small interfering RNA (siRNA) control (siCtrl) (Fig. 4G). Confocal microscopy showed that PEAV colocalizes with Tfn and endogenous clathrin in Vero cells (Fig. 5H). These results suggest that the entry of PEAV into Vero cells depends on clathrin-mediated endocytosis.
FIG 5.
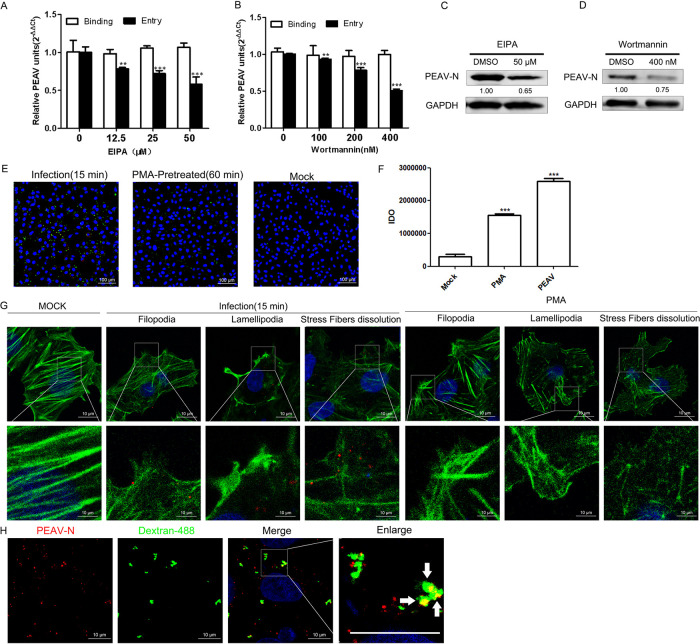
PEAV entry is dependent on macropinocytosis. (A and B) Vero cells were pretreated with EIPA (A) or wortmannin (B) (15 min, 37°C), followed by inoculation with PEAV (MOI = 1) to allow binding (1 h, 4°C) and entry (1 h, 37°C) in the presence of drugs. (C and D) In the presence of EIPA (50 μM) (C) and wortmannin (400 nM) (D), cells were inoculated with PEAV (MOI = 5) (2.5 h, 37°C) and immunoblotted to detect the early expression of PEAV N protein. Relative amounts of protein were calculated using ImageJ software. (E) Vero cells were inoculated with PEAV (MOI = 5) (1 h, 4°C), pretreated with PMA (positive control) (200 nM, 60 min, 37°C) or maintenance medium (negative control) (1 h, 4°C), and then incubated in maintenance medium containing 0.5 mg/mL Alexa Fluor 488-labeled dextran (15 min, 37°C). Dextran uptake was visualized using immunofluorescence microscopy. (F) Dextran uptake by Vero cells is represented by dextran fluorescence IDO values measured with ImageJ software. The mean ± SD values represent three individual pictures (**, P < 0.01; ***, P < 0.001). (G) Vero cells were inoculated with PEAV (MOI = 5) (1 h, 4°C), washed, incubated (15 min, 37°C), and fixed (4% PFA, 15 min, RT). Vero cells were pretreated with PMA (positive control) (200 nM, 60 min, 37°C) or maintenance medium (negative control) (1 h, 4°C), fixed (4% PFA, 15 min, room temperature), and then incubated with anti-PEAV N (red), and cytoskeletal changes were observed via probing actin with Alexa Fluor 488 phalloidin (green). (H) Vero cells were inoculated with PEAV (MOI = 5) (1 h, 4°C), washed, and incubated in a maintenance medium containing 0.5 mg/mL Alexa Fluor 488-labeled dextran (15 min, 37°C). Cells were fixed (4% PFA, 15 min, room temperature) and immunodetected with an anti-PEAV N protein antibody (red). Confocal microscopy revealed PEAV and dextran colocalization (white arrows).
PEAV entry is macropinocytosis dependent.
PDCoV has extensive host tropism (39, 40) and enters the cell via macropinocytosis (14). Cells lacking APN (PDCoV functional entry receptor) are still susceptible to PDCoV (41, 42). PEAV has a wide range of cellular tropisms (11). Therefore, PEAV, besides dependence on caveola- and clathrin-mediated endocytosis, may also have a receptor-independent entry mode. To test whether PEAV requires macropinocytosis, we used the inhibitors of macropinocytosis activation, 5-(N-ethyl-N-isopropyl) amiloride (EIPA) and wortmannin. The results show that EIPA (Fig. 5A) and wortmannin (Fig. 5B) significantly inhibit PEAV entry, but not the binding, in a dose-dependent manner. The expression of early viral proteins of PEAV was also significantly decreased under 50 μM EIPA (Fig. 5C) and 400 nM wortmannin (Fig. 5D) treatment.
We hypothesized that PEAV activates macropinocytosis during its internalization, thereby allowing cells to increase the uptake of fluorescent-labeled dextran, a macropinocytosis-dependent glycan (43). Dextran uptake by Vero cells was at a low level (Fig. 5E). However, phorbol 12-myristate 13-acetate (PMA), a drug that induces extensive plasma membrane remodeling, which increases dextran uptake, during PEAV infection of the treatment group significantly increased the dextran uptake of Vero cells (Fig. 5F). Inoculation of Vero cells with PEAV or treatment with PMA (positive control) resulted in cells forming filopodia and lamellipodia, and stress fiber dissolution occurred (Fig. 5G). These features are characteristic of micropinocytosis. In addition, PEAV particles colocalized with dextran during PEAV and dextran co-uptake experiments (Fig. 5H). These results indicate that PEAV can enter Vero cells via macropinocytosis.
The role of Rab proteins in PEAV cell entry.
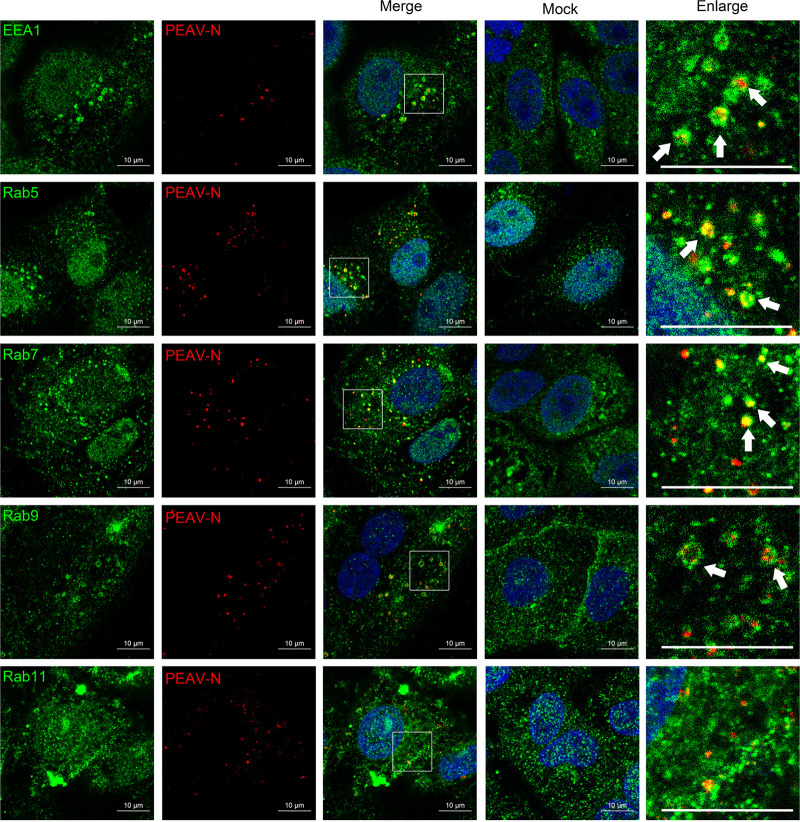
Rab proteins regulate endosome transport in cells in specific subcellular compartments. Here, we examined the colocalization of PEAV with EEA1, Rab5, Rab7, Rab9, and Rab11 at different infection times by confocal microscopy. Most PEAV particles colocalize with EEA1 and Rab5 (15 min) and Rab7 and Rab9 (30 min), but not Rab11 (Fig. 6). The DN plasmids of Rab5(S34N), Rab7(T22N), Rab9(S22N), and Rab11(S25N) were constructed based on Vero cells to inhibit the function of Rab protein. These plasmids were widely reported in envelope virus studies (26, 44). Cells overexpressing Rab5(S34N), Rab7(T22N), and Rab9(S22N) significantly decreased the expression of PEAV N protein, while cells overexpressing Rab11(S25N) did not significantly change the expression of PEAV N protein compared with cells overexpressing wild-type Rab5, Rab7, Rab9, and Rab11 (Fig. 7A and B). In addition, PEAV infection was severely reduced when the expression of Rab5, Rab7, and Rab9, but not Rab11, was temporarily silenced by specific siRNA in Vero cells compared with the control (Fig. 7C). The results suggest that PEAV infection depends on Rab5, Rab7, and Rab9.
FIG 6.
PEAV colocalizes with EEA1 and Rab proteins. Vero cells were inoculated with PEAV (MOI = 10) (1 h, 4°C), washed, incubated at 37°C for 15 min (Rab5) or 30 min (Rab7, Rab9, and Rab11), and fixed (4% PFA, 15 min, room temperature). Cells were immunodetected using anti-PEAV N (red), anti-EEA1, anti-Rab5, anti-Rab7, anti-Rab9, and anti-Rab11 (green) and observed using confocal microscopy. The white arrows indicate the colocalization of PEAV with EEA1, Rab5, Rab7, and Rab9. Scale bar = 10 μm.
FIG 7.
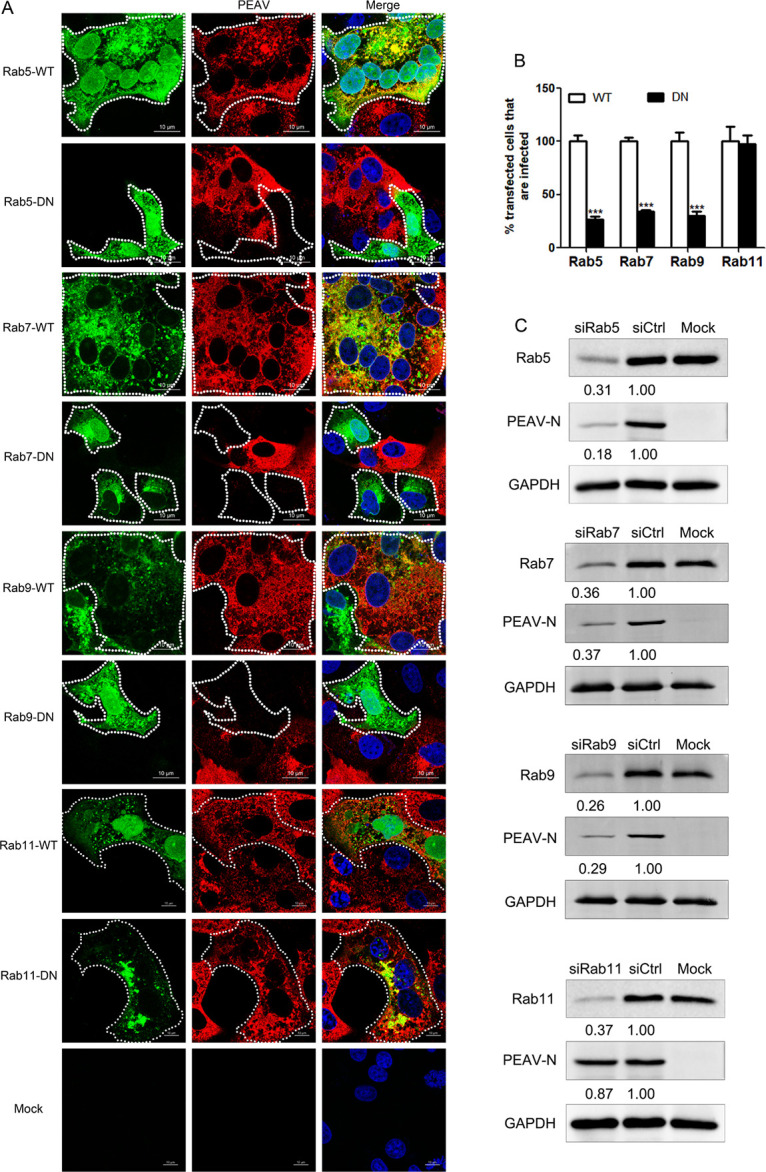
PEAV entry depends on Rab5, Rab7, and Rab9. (A) Vero cells were transfected with Rab5, Rab7, and Rab9 (WT or DN) plasmids (24 h, 37°C), followed by PEAV inoculation (MOI = 5) (6 h, 37°C), and fixed, and immunodetected using the anti-PEAV N monoclonal antibody. Scale bar = 10 μm. (B) Infection of transfected cells with Rab5, Rab7, and Rab9 DN cells was quantified as a percentage of the infection rate observed in WT samples. Over 300 transfected cells were included in the quantification. All results are presented as the mean ± SD from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (C) Cells were transfected with siRab5, siRab7, and siRab9 or siCtrl (48 h, 37°C) and inoculated with PEAV (MOI = 5) (2.5 h, 37°C). Viral N protein expression was quantified by Western blotting. Relative amounts of protein were calculated using ImageJ software.
Purified PEAV particles were covalently labeled with Alexa Fluor 647 NHS ester to avoid nonspecific binding of lipophilic labeling dyes such as DiD to lipid-rich membranes after entering cells. Viruses stained with anti-PEAV N protein, and obvious colocalization was observed by confocal microscopy (Fig. 8A). Labeled PEAV particles showed a typical coronavirus structure by transmission electron microscopy (Fig. 8B). A single viral dynamic transport assay performed using Vero cells transfected with EGFP-labeled Rab5, Rab7, and Rab9 plasmids and incubated with labeled virus showed that labeled PEAV was cotransported in the vesicular structure formed by Rab5, Rab7, and Rab9 proteins (Fig. 8C). Taken together, these results reveal that PEAV trafficking in endosomes is dependent on Rab5, Rab7, and Rab9.
FIG 8.
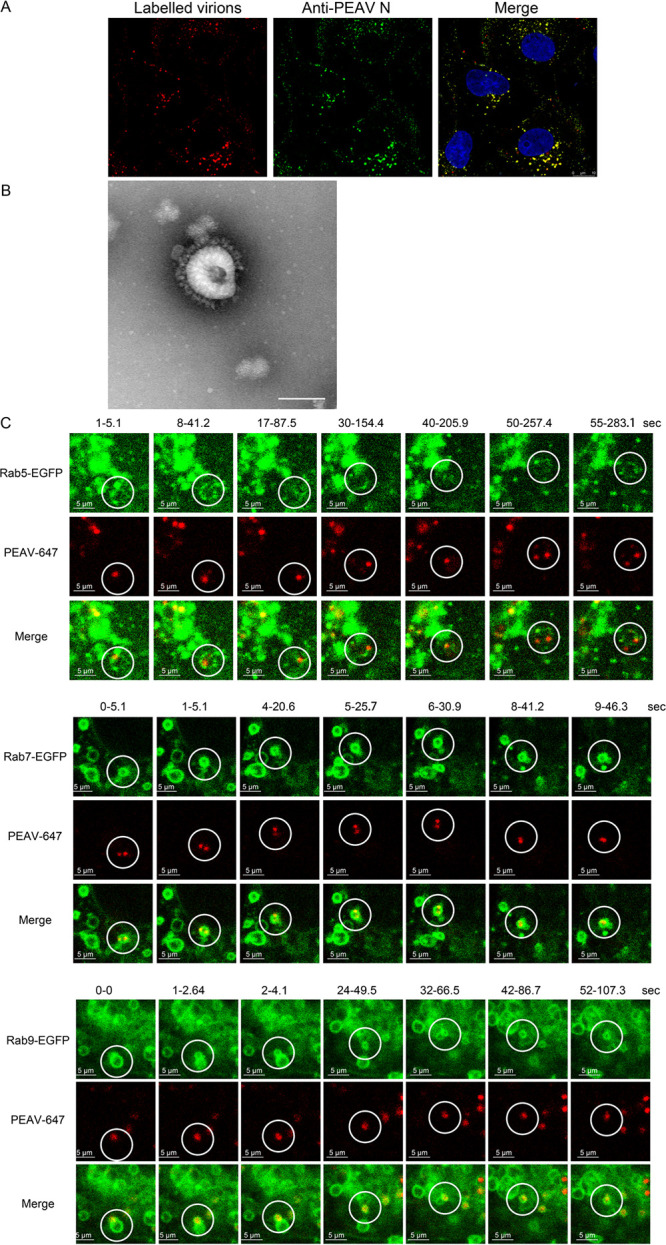
PEAV is dynamically transported in Rab5, Rab7, and Rab9 vesicles. (A) Vero cells were inoculated with Alexa Fluor 647 NHS ester-labeled PEAV (MOI = 20) (1 h, 4°C), washed, incubated (30 min, 37°C), and fixed (4% PFA, 15 min, RT). The cells were immunodetected using an anti-PEAV N protein antibody (green) and observed using confocal microscopy. (B) Labeled PEAV was prepared with negative staining and observed under a transmission electron microscope. Scale bar = 100 nm. (C) Vero cells were transfected with WT Rab plasmid (2.5 μg, 24 h, 37°C), and then inoculated with Alexa Fluor 647 NHS ester-labeled PEAV (MOI = 10) (11 h, 4°C), washed, and incubated in maintenance medium at 37°C for 15 min (Rab5) or 30 min (Rab7 and Rab9). Single-virus tracking images were acquired using a live-cell workstation with the light scanning module of a Leica TCS SP8 STED 3×confocal microscope. Scale bar = 5 μm.
PEAV entry into IPI-2I cells depends on caveolae, clathrin, and macropinocytosis.
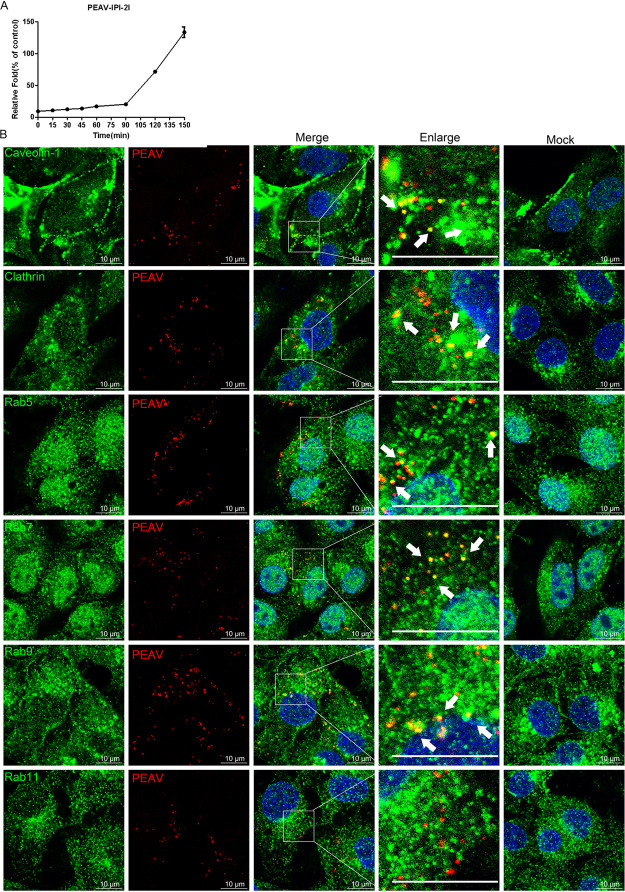
Viruses may enter different types of cells by different pathways. Recently, an ileal epithelial cell line (IPI-2I) has been used to study enteroviruses, including PEDV, PDCoV, and PEAV (14, 45, 46). Surprisingly, PEAV entry into IPI-2I cells showed low efficiency at 0 to 90 min postinfection, with only ~20% internalization at 90 min. The gene expression level of the PEAV N rapidly increased without an obvious peak period with increasing infection time (Fig. 9A). We speculate that the low efficiency of PEAV adsorption or entry into IPI-2I cells is due to the difference in the content of the cell surface adhesion factor and entry receptor. Based on this, although the entry is still taking place, the successfully internalized virus particles that have started replicating will likely interfere in determining a peak period. Further studies are required to prove this and pinpoint the peak entry time.
FIG 9.
PEAV colocalizes with clathrin, caveolin-1, and Rabs (Rab5, Rab7, and Rab9). (A) Vero cells were inoculated with PEAV (MOI = 1) (1 h, 4°C), washed, incubated with preheated maintenance medium (0 to 150 min, 37°C), and treated with proteinase K (5 min, 20°C), and the lysates were subsequently collected for quantification of RNA by RT-qPCR. (B) Vero cells were inoculated with PEAV (MOI = 20) (1 h, 4°C), washed, and incubated at 37°C for 60 min (caveolin-1, clathrin, and Rab5) or 90 min (Rab7, Rab9, and Rab11), and fixed (4% PFA, 15 min, room temperature). Immunodetection of anti-PEAV N (red), anti-caveolin-1, anticlathrin, anti-Rab5, anti-Rab7, anti-Rab9, and anti-Rab11 (green) antibodies was performed using confocal microscopy. The white arrows indicate the colocalization of PEAV with caveolin-1, clathrin, Rab5, Rab7, and Rab9. Scale bar = 10 μm.
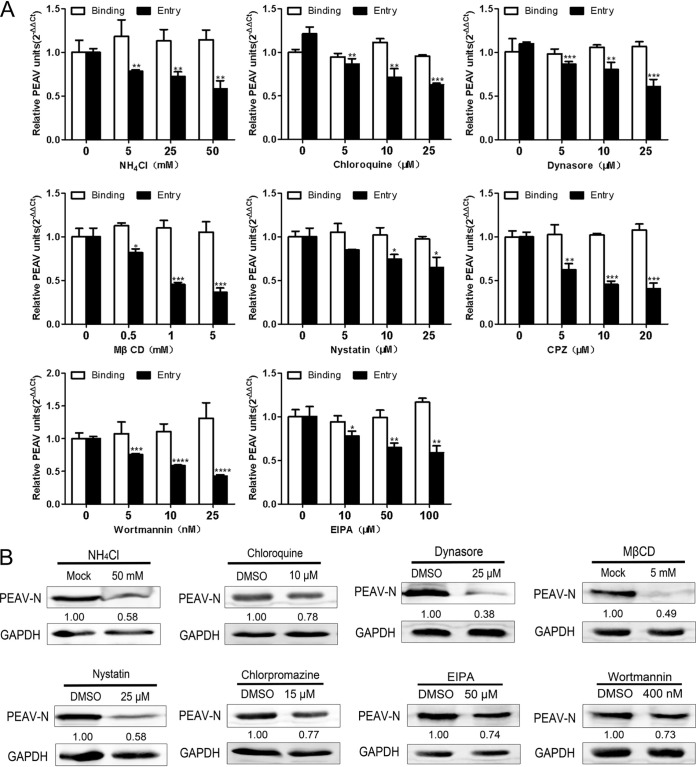
Analysis of the endocytosis pathway of PEAV entry into IPI-2I cells showed that PEAV significantly colocalized with caveolin-1, clathrin, and Rab5 at 60 min and Rab7 and Rab9 at 90 min but not with Rab11 (Fig. 9B). This result is consistent with that observed in Vero cells. Endocytosis inhibitors, including endosome acidification (chloroquine and NH4Cl), and inhibitors of dynamin (dynasore), cholesterol (MβCD and nystatin), clathrin (CPZ), and macropinocytosis (EIPA, wortmannin) significantly reduced the entry of PEAV into IPI-2I cells in a dose-dependent manner (Fig. 10A). This is consistent with the results obtained with Vero cells. At the same time, these inhibitors significantly reduced the level of viral protein expression at subtoxic doses (Fig. 10B). These results suggest that the endocytic pathway is the same in Vero and IPI-2I cells, although with different levels of entry efficiency. The virus uses three endocytic pathways: caveolae, clathrin, and macropinocytosis. PEAV entry also requires dynamin, cholesterol, and a low-pH environment.
FIG 10.
PEAV entry into IPI-2I cells depends on caveolae, clathrin, and macropinocytosis and requires dynamin, cholesterol, and low pH. (A) IPI-2I cells or Vero cells were pretreated with the following category of inhibitors: endosome acidification (chloroquine, ammonium chloride), dynamin (dynasore), cholesterol (MβCD and nystatin), clathrin (CPZ), and macropinocytosis (EIPA, wortmannin) (15 min, 4°C). This treatment was followed by inoculation with PEAV (MOI = 1) to allow binding (1 h, 4°C) and entry (1 h, 37°C) in the presence of drugs. (B) IPI-2I cells were pretreated using a subtoxic inhibitor dose of chloroquine, ammonium chloride, dynasore, MβCD, nystatin, CPZ, EIPA, or wortmannin. Cells were inoculated with PEAV (MOI = 5) for a prolonged infection period (2.5 h, 37°C) in the presence of drugs and further incubated in a drug-free maintenance medium (3.5 h, 37°C), followed by immunoblotting to detect early PEAV N protein expression. Relative amounts of protein were calculated using ImageJ software. All results are presented as the mean ± SD from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
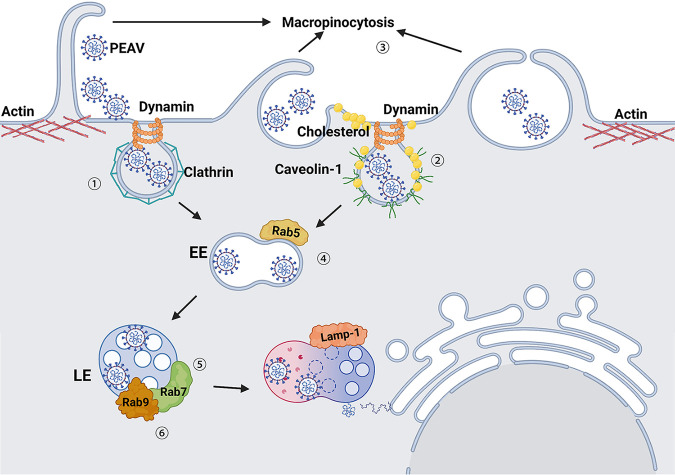
Envelope viruses have evolved a variety of endocytosis pathways to break through the host cell membrane barrier and successfully release viral genomes into the cytoplasm (47, 48). Understanding how viruses hijack cellular endocytosis to establish infection can expand our understanding of the viral infection mechanism and contribute to development of antiviral drugs. This study used a systematic approach to dissect the entry pathway of PEAV-infected host cells (Fig. 11). PEAV infection uses three different endocytosis pathways: caveolae, clathrin, and macropinocytosis. After internalization, PEAV is transported from the early endosome to the lysosome regulated by Rab5, Rab7, and Rab9 proteins, completes membrane fusion in a pH-dependent manner, and releases the genome into the cytoplasm.
FIG 11.
Schematic model of PEAV entry into host cells. (Step 1) PEAV entry by clathrin-mediated endocytosis is dependent on dynamin. (Step 2) PEAV entry by caveola-mediated endocytosis requires cholesterol. (Step 3) PEAV activates macropinocytosis and depends on macropinocytosis endocytosis. (Step 4) After PEAV internalization, transport depends on Rab5 to regulate entry into early endosomes. (Step 5) Rab7 regulates early endosomal PEAV transport to the late endosomes or lysosomes. (Step 6) Rab9 regulates late endosomal entry into the Golgi apparatus. Finally, the virus core penetration is complete, and the genome is released.
Hijacking the endocytic pathway is key for establishing viral infection. Viruses usually enter cells via certain endocytosis pathways. However, many coronaviruses can enter cells via multiple endocytosis pathways, such as severe acute respiratory syndrome coronavirus (SARS-CoV) (12, 49), mouse hepatitis coronavirus (MHV) (50), and infectious bronchitis coronavirus (IBV) (51). In this work, clathrin-mediated endocytosis inhibitors (chlorpromazine and dynasore), overexpression of Eps15 and dynamin II DN plasmids, or knockdown of clathrin heavy chain (HC) and dynamin II significantly reduced the entry of PEAV and subsequent viral protein expression. Furthermore, confocal microscopy revealed that PEAV particles were clustered in the vesicles coated with clathrin and colocalized with Tfn, a marker dependent on clathrin uptake. These results suggest that PEAV enters host cells via an endocytosis pathway mediated by clathrin, which requires dynamin involvement. Because of the cage structure formed by the clathrin scaffold, with an inner diameter of ~50 to 100 nm (52), this pathway is usually used for endocytosis by small- or medium-size viruses (53). Vesicles coated with clathrin can also be deformed to adapt to the entry of relatively large viruses, such as African swine fever virus (ASFV) (icosahedron of 200 nm) (54) or bullet-like vesicular stomatitis virus (VSV) (180 by 70 nm) (55, 56). Coronaviruses, such as PEAV, normally have an inner diameter of 50 to 100 nm and are classified as small- to medium-size viruses. Therefore, it is not surprising that PEAV enters via clathrin-mediated endocytosis. Other porcine coronaviruses also depend on clathrin-mediated endocytosis, including PEDV (15), PDCoV (14), and transmissible gastroenteritis virus (TGEV) (57). Recent research shows that by endocytosis, a single virus particle of PEDV (16) and TGEV (58) can enter the same host cell, depending on two endocytosis pathways: clathrin and caveolae. Therefore, we speculated that PEAV might have similar characteristics. Addition of the inhibitor MβCD to deplete cholesterol significantly reduced the cell entry of PEAV and subsequent viral protein expression, indicating that caveola endocytosis may be involved in the entry of PEAV. Caveolae are usually defined as 60- to 80-nm pits (59), so they are typically used by small viruses (~45 nm in diameter), such as simian virus 40 (SV40) (60) and classical swine fever virus (CSFV) (26). PEAV also entered cells by the caveola-mediated endocytosis pathway, despite its diameter reaching 100 nm. Knockout of endogenous caveolin-1 or expression of caveolin-1 DN significantly reduced the entry of PEAV and subsequent viral protein expression. We speculate that entry may occur due to the wide range of particle diameter sizes (50 to 100 nm) of the coronavirus, and the virus can use different particle diameters to induce different endocytosis pathways in cells. This phenomenon may also exist in other coronaviruses, which explains the controversial endocytosis pathway of PEDV, which is only dependent on clathrin-mediated or the common mediation of caveolae and clathrin (15, 28). However, more research is needed to prove this.
Macropinocytosis is an actin-dependent endocytosis process that involves the nonspecific uptake of fluids, solutes, or particles in response to the activation of cell surface receptors (61, 62). A variety of viruses use macropinocytosis for entry, such as vaccinia virus (43), human cytomegalovirus (63), and Ebola virus (64). Recent studies showed that the porcine delta coronavirus (PDCoV) activates macropinocytosis at the early stage of infection. Researchers speculate that this is related to the extensive cell tropism of PDCoV (14). PEAV replicates in up to 21 cell lines, including those of humans, monkeys, pigs, bats, and mice (11). Therefore, receptor-independent endocytosis may cause PEAV's extensive host tropism, and it may activate macropinocytosis. Macropinocytosis activation depends on Na+/H+ exchange and phosphatidylinositol 3-kinase (PI3K) activation (65). Inhibition of micropinocytosis activation by EIPA and wortmannin significantly reduced the entry of PEAV and the subsequent expression of viral proteins. Furthermore, dextran uptake by cells significantly increased and virus colocalization with dextran was observed after PEAV infection.
Meanwhile, macropinocytosis activation can disturb the plasma membrane (65). We found that actin skeleton rearrangement can be induced at the early stage of PEAV infection, and there are obvious morphological changes after macropinocytosis activation, such as filopodia, lamellipodia, and stress fiber dissolution, indicating that PEAV can enter cells by inducing macropinocytosis. Our study confirmed that PEAV could enter cells via three endocytosis pathways—caveolae, clathrin, and micropinocytosis—showing that PEAV has the potential for extensive cytotropism. Our study also confirmed that PEAV can still enter cells through the three endocytic pathways even under low-dose infection conditions (multiplicity of infection [MOI] = 0.1 by qRT-PCR or MOI = 5 by confocal microscopy) (see Fig. S1A to D in the supplemental material). A side-by-side comparison of these pathways (caveolae, clathrin, and micropinocytosis) shows that macropinocytosis may be the main pathway (~43.78%) (Fig. S1E to F). In addition, the entry of PEAV into the pig ileum epithelial cell line IPI-2I also depends on these three endocytosis pathways. The efficiency of PEAV entering IPI-2I cells is low; therefore, inclusion of some special components in the intestinal environment may be necessary to improve entry efficiency, such as bile acid (17). The metabolic demands of immortalized cells differ from those of primary or naturally healthy cells in an organism, which may affect the types of cell entry routes available to a virus. This study further evaluated the endocytic pathway of PEAV entry into primary jejunal epithelial cells (Fig. S2C). As expected, PEAV could enter cells by three endocytosis pathways, including caveolae, clathrin, and micropinocytosis, and macropinocytosis may be the main pathway (~45.1%) (Fig. S2D). Our results suggest that PEAV potentially hijacks multiple endocytosis pathways in different cells. However, this result depends on the antibodies’ specific recognition ability, which requires analysis using more sophisticated statistical methods.
Rab GTPases play an important role in regulating the transport of endosomes and act as organizers of almost all membrane transport processes in eukaryotic cells (19). Viruses entering host cells via endocytosis can hijack Rab protein and small GTPases to achieve their transport and localization to specific subcellular chambers (53). The role of Rab protein in coronavirus infection remains unclear. Knockdown of early endosome-related genes (EEA1 and Rab5), late endosome-related genes (Rab7A and Rab7B), and late endosome to lysosomal maturation-related genes (VPS39) significantly inhibits the entry of mouse hepatitis virus (MHV) into host cells (66). Similarly, PEDV endocytosis depends on the endosomal pathway, wherein Rab5 and Rab7 are involved in the endosomal transport of the virus and eventually reach lysosomes (16). This study found that PEAV particles and early endosome EEA1 colocalize, indicating that the virus depends on endosome transport after entry into host cells.
The expression of DN plasmid or knockdown of endogenous Rab5, Rab7, and Rab9 significantly reduced PEAV infection, correlating with confocal microscopy results that PEAV particles colocalize with endogenous Rab5, Rab7, and Rab9. However, static images and whole-cell analysis cannot show the dynamic transport process of PEAV particles. The single-virus fluorescent labeling experiment demonstrated that PEAV particles were cotransported in Rab5-, Rab7-, and Rab9-labeled vesicles. Increased infection time showed that more PEAV particles were gathered in vesicles containing Rab9 and Lamp-1 proteins by confocal microscopy. We speculate that PEAV is similar to PEDV (28) or PDCoV (14), which depends on an acidic pH endosome environment to realize membrane fusion and release the genome, as proven by the significant decrease of PEAV internalization and subsequent viral protein expression following treatment with inhibitors of endosomal acidification (NH4Cl, chloroquine, and bafilomycin-A1). In general, PEAV depends on the acidic pH environment of the endosome, regulates endosome transport through Rab5, Rab7, and Rab9, and finally reaches the lysosome after endocytosis.
This work investigated the PEAV-dependent endocytic pathway into Vero cells from multiple perspectives: pharmacological inhibitors, RNA knockdown, overexpression of DN functional plasmids, and colocalization of endocytic marker proteins. PEAV entered into Vero, IPI-2I, and primary jejunal epithelial cells through three endocytic pathways: caveola- and clathrin-mediated endocytosis and macropinocytosis. Moreover, macropinocytosis may be the main pathway to enter cells. In addition, single-virus tracking proved that the dynamic transport process of PEAV entry into the cell depends on an endosomal acidic pH environment for transport from the early endosome to the lysosome and complete productive infection. The process depends on Rab5, Rab7, and Rab9 proteins after endocytosis. In conclusion, we comprehensively studied the early events of PEAV entry, providing a better understanding of the cell entry mechanism of bat HKU2-like coronavirus and providing insights for development of new antiviral agents.
MATERIALS AND METHODS
Cells and viruses.
Vero cells (ATCC CCL-81) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, NY, USA), supplemented with 10% fetal bovine serum (Gibco, Invitrogen). IPI-2I cells were obtained from the China Center for Type Culture Collection. All cells were kept at 37°C in a humidified atmosphere of 5% CO2. PEAV strain GDS04, a gift from Cao Yongchang of Sun Yat-sen University, was propagated in medium containing 0.3% tryptone phosphate broth (TPB) and 10 μg/mL trypsin (Gibco) in Vero cells, and passage 16 (P16) was used in the study. The prepared virus supernatant was centrifuged at 10,000 × g for 15 min, the supernatant was centrifuged at 100,000 × g at 4°C for 1.5 h with a Beckmann SW32Ti rotor, and the virus was suspended in HNE buffer solution (5 mM HEPES, 150 mM NaCl, and 0.1 mM EDTA [pH 7.4]). The virus suspension was added to a sucrose gradient (10% to 60%) and centrifuged in the same rotor at 120,000 × g for 2 h at 4°C. The virus bands were collected from the 40 to 60% sucrose layer and centrifuged at 40,000 × g for 1.5 h to remove sucrose. Finally, the purified virus was resuspended in HNE buffer and stored at −80°C for future use.
Virus infection and labeling.
Vero cells were grown to 100% confluence, washed with phosphate-buffered saline (PBS) to remove the serum, and inoculated with medium containing different multiplicities of infection (MOIs) of viruses containing 0.3% TPB and 10 μg/mL trypsin (virus maintenance medium). The virus supernatant (with cell fragments removed) was used in all binding and entry experiments in the study. EIPA treatment (50 μM) was used to eliminate interference from macropinocytosis to eliminate the interference between various endocytosis pathways partially. In RNA knockdown assays, additional EIPA was added to viral inoculation and maintenance medium, which was then changed to a drug-free medium after 2 h for maintenance. The virus from the 40 to 60% sucrose layer was labeled with Alexa Fluor 647 NHS ester (100 μM) (1 h, 20°C) in the dark. The virus was vortexed every 15 min, unincorporated dye was removed using an Amicon Ultra-15 centrifugal filter unit (10 kDa) (Merck Millipore, Cork, Ireland) by centrifugation at 6,000 × g for 30 min at 4°C, and the wash step was repeated three times using 2 mL PBS each time. Finally, labeled PEAVs were filtered through a 0.22-μm-pore filter (Millipore). An appropriate amount of the labeled virus was taken for negative staining, and the morphology was observed with a transmission electron microscope (FEI/Talos L120C). Finally, the labeled purified virus was aliquoted and stored at −80°C.
Antibodies, inhibitors, and reagents.
Monoclonal antibody (1:500) was used for the indirect fluorescent antibody (IFA) test, and polyclonal antibody (1:1,000) was used for Western blotting against PEAV N protein. Anti-caveolin-1, anti-clathrin heavy chain (HC), anti-dynamin II, anti-Rab5, and anti-Rab11 (ab2910, ab21679, ab65556, ab18211, and ab3612, respectively) were purchased from Abcam (1:200). Anti-clathrin HC, anti-Rab7, and anti-Rab9 (66487-1-Ig, 55469-1-AP, and 11420-1-AP, respectively) were purchased from Proteintech (1:200). Anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:5,000) and secondary antibodies (1:300) conjugated to Alexa Fluor 488 and 594 were purchased from Proteintech. IRDye 800CW goat anti-mouse or anti-rabbit IgG was purchased from Licor (1:15,000). Methyl-β-cyclodextrin (MβCD; C4555), chlorpromazine hydrochloride (CPZ; C0982), 5-(N-ethyl-N-isopropyl)amiloride (EIPA; A3085), ammonium chloride (A9434), chloroquine phosphate (PHR1258), and proteinase K were purchased from Sigma-Aldrich. Dynasore (ab120192) was purchased from Abcam. Nystatin (S1934), wortmannin (S2758), phorbol 12-myristate 13-acetate (PMA; S7791), and bafilomycin-A1 (S1413) were purchased from Selleck Chemicals. DNA/RNase-free H2O, the CCK8 cell viability detection kit, and phalloidin actin 488 fluorescent dye were purchased from Beyotime Biotechnology (Shanghai, China). Alexa Fluor 568-labeled transferrin (Tfn; T23365), Alexa Fluor 555-labeled cholera toxin subunit B (CTB; C22843), and Alexa Fluor 488-labeled dextran (D22910) were purchased from Thermo Fisher Scientific.
DNA constructs and transfection conditions.
The following plasmid sequences were amplified through PCR using Vero cell cDNA and then inserted into pCAGGS-EGFP: caveolin-1, XM_007982676.2; Eps15, XM_007978767.2; dynamin II, XM_007995249.2; Rab5, XM_008009261.2; Rab7, XM_007985441.2; Rab9, XM_007991060.2; and Rab11, XM_008016207.2. DN plasmid was obtained by referring to previous research to construct mutations or deletions of the corresponding functional domain: caveolin-1 DN (deletion of positions 82 to 178), Eps15 D29/295 (deletion of the homologous [EH] domain) (16), dynamin II DN (K44A) (67), Rab5 DN (S34N), Rab7 DN (T22N) (68), Rab9 DN (S22N) (69), and Rab11 DN (S25N) (70). The primers used to construct the plasmids are listed in Table 1.
TABLE 1.
Primers used to construct plasmids
| Plasmid | Sequence (5′→3′) |
|
|---|---|---|
| Forward | Reverse | |
| Cav1-WT | GGCGGTGGCGGATCGCTCGAGATGTCGGGGGGCAAATACG | TTGGCAGAGGGAAAAAGATCTTTATATTTCTTTCTGCATGTTGATGC |
| Cav1-dn | GGCGGTGGCGGATCGCTCGAGATGGATGGCATCTGGAAGGC | TTGGCAGAGGGAAAAAGATCTTTATATTTCTTTCTGCATGTTGATGC |
| Eps15-WT | GGCGGTGGCGGATCGCTCGAGATGGCTGCGGCGGCCCAG | TTGGCAGAGGGAAAAAGATCTTCATGCTTCTGAGATCTCAGATTTG |
| Eps15-dn | TCTGCAGTTGAGCTCGCCACTTTGCAGAAGAACAT | GCAAAGTGGCGAGCTCAACTGCAGAAGTTC |
| Rab5-WT | GGCGGTGGCGGATCGCTCGAGATGGCTAATCGAGGAGCAACAA | TTGGCAGAGGGAAAAAGATCTTTAATTACTACAACACTGACTCCTGGTTG |
| Rab5-dn | TGGCAAAAACAGCCTAGTGCTTCGTTTTGTGAA | CTAGGCTGTTTTTGCCAACAGCAGACTCTCCC |
| Rab7-WT | GGCGGTGGCGGATCGCTCGAGATGACCTCTAGGAAGAAAGTGTTGC | TTGGCAGAGGGAAAAAGATCTTCAGCAACTGCAGGTTTCCG |
| Rab7-dn | GGAGTTGGTAAGAACTCACTCATGAACCAGTATGTGAACAA | GAGTTCTTACCAACTCCAGAATCTCCCAGGAT |
| Rab9-WT | GGCGGTGGCGGATCGCTCGAGATGGCAGGAAAATCATCGCTT | TTGGCAGAGGGAAAAAGATCTTCAACAGCACGAGGAGCTGG |
| Rab9-dn | GGAAGAGTAACCTGATGAACAGATACGTGACTAACAAGT | CATCAGGTTACTCTTCCCAACTCCACCATCTC |
| Rab11-WT | GGCGGTGGCGGATCGCTCGAGATGGGCACCCGCGACGAC | TTGGCAGAGGGAAAAAGATCTTTAGATGTTCTGACAGCACTGCACC |
| Rab11-dn | GAACAATCTCCTGTCTCGATTTACTCGAAATG | GAGACAGGAGATTGTTCTTTCCAACACCAGAATCTCCAA |
| Dyn2-WT | GGCGGTGGCGGATCGCTCGAGATGGGCAACCGCGGGATG | TTGGCAGAGGGAAAAAGATCTCTAGTCGAGCAGGGATGGCTC |
| Dyn2-dn | GCCAGCTCAGTGCTCGAGAACTTCGTGGGCCG | TCGAGCACTGAGCTGGCGCCGGCGCTCTGGCCGCC |
| qPEAV-N | CTGACTGTTGTTGAGGTTAC | TCTGCCAAAGCTTGTTTAAC |
| GAPDH | CCTTCCGTGTCCCTACTGCCAAC | GACGCCTGCTTCACCACCTTCT |
Vero cells were cultured up to 80% confluence and transfected using Lipofectamine 3000 transfection reagent and Opti-MEM (Thermo Fisher Scientific) for transient expression analysis using 2.5 μg transfection endocytosis-related plasmid for 24 h. Viral or Tfn/CTB uptake into cells was then performed.
Quantitative real-time PCR.
Viral RNA was extracted using the Fastagen kit according to the manufacturer's instructions. The RNA concentration was measured using a NanoDrop 2000 spectrophotometer. According to the manufacturer's instructions, cDNA was synthesized using 500 or 1,000 ng total RNA using iScript reverse transcriptase (Thermo Fisher Scientific). A 1:5 dilution of cDNA was used to perform quantitative real-time PCR (qRT-PCR) using a Light Cycler 480 real-time PCR system (Roche Diagnostics, Indianapolis, IN, USA). Data are presented as the threshold cycle (2−ΔΔCT) value from quadruplicate samples, and the gene coding for GAPDH was used as a reference gene.
Measurement of virus internalization.
Viral internalization measurements were made using a previous method with minor modifications (71). Cells were inoculated in a 35-mm cell culture dish, and PEAV (MOI = 1) was inoculated at 4°C for 1 h. Unattached viruses were washed using PBS, the viral maintenance medium was replaced, and the cells were incubated at 37°C. The cells were washed with PBS at 0, 15, 30, 45, 60, 90, 120, and 150 min and treated with proteinase K (0.5 mg/mL) for 5 min at 4°C to remove the adsorbed (but not internalized) virus. Proteinase K solution was discarded, and the cells were washed three times with PBS. Cells for RNA separation were collected by centrifugation at 1,000 × g for 2 min at 4°C. A sample without proteinase K treatment was added at 0 min to represent the total amount of virus successfully adsorbed. The level of internalized viral RNA was analyzed using qRT-PCR.
Cell viability assay.
Vero or IPI-2I cells were seeded into 96-well plates and grown to 100% confluence. Different concentrations of drug inhibitors were added, and the cells were incubated at 37°C for 165 min. The medium of IPI-2I cells was changed to drug-free medium and incubated for a further 6 h. According to the manufacturer's instructions, the CCK-8 kit was used to determine cell viability. The absorbance values at 450 nm were measured to determine cell viability. An average of 4 to 8 readings were taken. Cell survival rate (%) = (optical density [OD] of experimental group/OD of the control group) × 100%. Similarly, cells inoculated on a 96-well plate were transfected with corresponding siRNAs or the siRNA control (siCtrl) for 48 h, and a CCK-8 kit was used to determine cell viability. The effect of inhibitors or siRNA cell viability is shown in (Fig. S3).
Binding and entry assay.
Vero cells or IPI-2I cells were seeded in a 6-well plate and pretreated with a subtoxic dose of drug inhibitor at 37°C for 15 min, followed by inoculation with PEAV (MOI = 1) (1 h, 4°C) in the presence of drugs to facilitate virus binding without internalization. The cells were lysed, and RNA was extracted as binding according to the manufacturer's instructions. For entry, the cells were pretreated with drugs at 37°C for 15 min, PEAV (MOI = 1) was inoculated in the presence of drugs at 4°C for 1 h, and then PBS was used to wash the cells three times to remove the nonattached virus. A change to virus maintenance medium containing drugs at 37°C for 1 h was performed to allow virus internalization. Then, the cells were washed with GN buffer (pH 3) for 5 min to remove the noninternalized virus particles on the cell surface and then washed with precooled PBS three times. The cells were lysed according to the manufacturer's instructions, and RNA was extracted as binding.
RNA interference.
All siRNAs were purchased from RiboBio (Guangzhou, China). Vero cells were seeded in 12-well plates at 50 to 60% confluence on the day of transfection. According to the manufacturer's instructions, the corresponding siRNA (50 pmol) was transfected with liposome RNAiMAX. Knockdown was verified by immunoblotting 48 h after transfection. The indicated siRNAs are listed in Table 2.
TABLE 2.
siRNA duplexes used in this study
| siRNA | Sequence |
|---|---|
| siRab5 | GTCTGCTGTTGGCAAATCA |
| siRab7 | GATGGTGGATGACAGACTA |
| siRab9 | CGACTATCCTTACTTTGAA |
| siRab11 | GGTTTGTCGTTCATTGAGA |
| siCaveolin-1 | GCATTTACTTCGCCATTCT |
| siDynamin | GACATCGAGTACCAGATCA |
| siClathrin HC | CCTTATCCTCACCGCAATT |
Immunofluorescence assay.
Vero cells or IPI-2I cells were first grown on a 14-mm glass bottom cell culture dish (Cellvis) until 70% confluence, followed by transfection with plasmids, incubation for 24 h, then inoculation with PEAV. The cells were fixed with 4% paraformaldehyde (PFA) at different times. The samples were incubated at 20°C for 15 min, washed with PBS three times, and permeabilized with 0.3% Triton X-100 for 5 min. The cells were stained with specific antibodies at 4°C overnight or 37°C for 2 h. All antibodies were diluted in 2% (wt/vol) bovine serum albumin (BSA) in PBS. Cells were washed three times with PBS and incubated with the corresponding secondary antibody (1 h, 37°C). All secondary antibodies were diluted in PBS. Cell nuclei were stained with 1 μg/mL DAPI (4′,6-diamidino-2-phenylindole), and actin was stained with 1:100 Alexa Fluor 488 phalloidin. Fluorescent images were acquired using the light scanning module of a Leica TCS SP8 STED 3× confocal microscope.
Western blotting.
Vero cells or IPI-2I cells were cultured in a 6- or 12-well plate. Cells were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer for 30 min at 4°C. Cell lysates were collected by centrifugation at 10,000 × g for 10 min at 4°C, and proteins in the lysates were separated using SDS-PAGE. Protein transfer was performed onto polyvinylidene fluoride (PVDF) membranes (0.45-μm pore) (Merck Millipore) and probed with the indicated antibodies. GAPDH was used as a loading control.
Fluid-phase uptake assay.
Vero cells were grown in a 24-well plate (Nest) or a 14-mm glass bottom cell culture dish (Cellvis) to 90% confluence. Cells were inoculated with PEAV (MOI = 5), 200 nM PMA (positive control), or incubation maintenance medium (negative control) for 1 h at 4°C. PMA or nonattachment virus was washed with PBS and incubated in a maintenance medium containing 0.5 mg/mL dextran labeled with Alexa Fluor 488 at 37°C for 30 min. Noninternalized dextran was removed by acid washing (0.1 M sodium acetate, 50 mM NaCl [pH 5.5]), cells were fixed in 4% PFA, and immunofluorescence assays were performed. Finally, dextran and PEAV co-uptake or colocalization was measured using the light scanning module of a Leica TCS SP8 STED 3× confocal microscope (×20 or ×63 magnification).
Ruffling and blebbing assay.
Vero cells were grown on a 14-mm glass bottom cell culture dish until 90% confluence. Cells were inoculated with PEAV (MOI = 5), incubation maintenance medium (1 h, 4°C), or 200 nM PMA (1 h, 37°C) as a positive control. Subsequently, cells were washed with PBS and incubated in a maintenance medium (30 min, 37°C). Cells were fixed in 4% PFA and assayed by immunofluorescence using Alexa Fluor 488 phalloidin to detect actin according to the manufacturer's instructions. Fluorescent images were acquired using the light scanning module of a Leica TCS SP8 STED 3× confocal microscope.
Single-virus tracking.
Vero cells were grown on a 14-mm glass bottom cell culture dish (Cellvis) until 70% confluence, followed by transfection with 2.5 μg wild-type Rab plasmid (24 h, 37°C) using Lipofectamine and Opti-MEM, followed by inoculation with PEAV labeled with Alexa Fluor 647 NHS ester (MOI = 10) (1 h, 4°C). The medium was replaced with preheated maintenance medium (37°C), and the light scanning module of the Leica living cell workstation was also preheated before the experiment. The stable cell imaging environment used 5% carbon dioxide at 37°C. The excitation and emission wavelengths were 488 nm and 647 nm, respectively. Rab5, Rab7, and Rab9 were detected using real-time virus tracking images at 15 and 30 min, respectively.
Statistical analysis.
All data are presented as the mean ± standard deviation (SD). A Student's t test was used to compare the data from pairs of treated and untreated groups. Statistical significance in the figures is indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001). All statistical analyses and calculations were performed using Prism 5 (GraphPad Software, Inc., La Jolla, CA).
ACKNOWLEDGMENTS
This research was funded by the National Natural Science Foundation of China (32102646), Natural Science Foundation of Guangdong Province (2020A1515110315), Start-up Research Project of Maoming Laboratory (2021TDQD002), and China Agriculture Research System of MOF and MARA (cars-35).
Footnotes
Supplemental material is available online only.
Contributor Information
Lang Gong, Email: gonglang@scau.edu.cn.
Tom Gallagher, Loyola University Chicago—Health Sciences Campus.
REFERENCES
- 1.Gong L, Li J, Zhou Q, Xu Z, Chen L, Zhang Y, Xue C, Wen Z, Cao Y. 2017. A new bat-HKU2-like coronavirus in swine, China, 2017. Emerg Infect Dis 23:1607–1609. 10.3201/eid2309.170915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong N, Fang L, Zeng S, Sun Q, Chen H, Xiao S. 2015. Porcine deltacoronavirus in mainland China. Emerg Infect Dis 21:2254–2255. 10.3201/eid2112.150283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun D, Wang X, Wei S, Chen J, Feng L. 2016. Epidemiology and vaccine of porcine epidemic diarrhea virus in China: a mini-review. J Vet Med Sci 78:355–363. 10.1292/jvms.15-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P, Fan H, Lan T, Yang XL, Shi WF, Zhang W, Zhu Y, Zhang YW, Xie QM, Mani S, Zheng XS, Li B, Li JM, Guo H, Pei GQ, An XP, Chen JW, Zhou L, Mai KJ, Wu ZX, Li D, Anderson DE, Zhang LB, Li SY, Mi ZQ, He TT, Cong F, Guo PJ, Huang R, Luo Y, Liu XL, Chen J, Huang Y, Sun Q, Zhang XL, Wang YY, Xing SZ, Chen YS, Sun Y, Li J, Daszak P, Wang LF, Shi ZL, Tong YG, Ma JY. 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556:255–258. 10.1038/s41586-018-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou L, Li QN, Su JN, Chen GH, Wu ZX, Luo Y, Wu RT, Sun Y, Lan T, Ma JY. 2019. The re-emerging of SADS-CoV infection in pig herds in Southern China. Transbound Emerg Dis 66:2180–2183. 10.1111/tbed.13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun Y, Xing J, Xu ZY, Gao H, Xu SJ, Liu J, Zhu DH, Guo YF, Yang BS, Chen XN, Zheng ZZ, Wang H, Lang G, Holmes EC, Zhang GH. 2022. Re-emergence of severe acute diarrhea syndrome coronavirus (SADS-CoV) in Guangxi, China, 2021. J Infect 85:e130–e133. 10.1016/j.jinf.2022.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Q, Wang HY. 2021. Porcine enteric coronaviruses: an updated overview of the pathogenesis, prevalence, and diagnosis. Vet Res Commun 45:75–86. 10.1007/s11259-021-09808-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, van Kuppeveld F, He Q, Rottier P, Bosch BJ. 2016. Cellular entry of the porcine epidemic diarrhea virus. Virus Res 226:117–127. 10.1016/j.virusres.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J, Qiao S, Guo R, Wang X. 2020. Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution. Nat Commun 11:3070. 10.1038/s41467-020-16876-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards CE, Yount BL, Graham RL, Leist SR, Hou YJ, Dinnon KR, Sims AC, Swanstrom J, Gully K, Scobey TD, Cooley MR, Currie CG, Randell SH, Baric RS. 2020. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc Natl Acad Sci USA 117:26915–26925. 10.1073/pnas.2001046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang YL, Qin P, Wang B, Liu Y, Xu GH, Peng L, Zhou J, Zhu SJ, Huang YW. 2019. Broad cross-species infection of cultured cells by bat HKU2-related swine acute diarrhea syndrome coronavirus and identification of its replication in murine dendritic cells in vivo highlight its potential for diverse interspecies transmission. J Virol 93:e01448-19. 10.1128/JVI.01448-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue Y, Tanaka N, Tanaka Y, Inoue S, Morita K, Zhuang M, Hattori T, Sugamura K. 2007. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J Virol 81:8722–8729. 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nomura R, Kiyota A, Suzaki E, Kataoka K, Ohe Y, Miyamoto K, Senda T, Fujimoto T. 2004. Human coronavirus 229E binds to CD13 in rafts and enters the cell through caveolae. J Virol 78:8701–8708. 10.1128/JVI.78.16.8701-8708.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang P, Zhang J, Zhang H, Xia S, Ren J, Tian L, Bai D, Fang L, Xiao S. 2021. Porcine deltacoronavirus enters porcine IPI-2I intestinal epithelial cells via macropinocytosis and clathrin-mediated endocytosis dependent on pH and dynamin. J Virol 95:e134521. 10.1128/JVI.01345-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JE, Cruz DJ, Shin HJ. 2014. Clathrin- and serine proteases-dependent uptake of porcine epidemic diarrhea virus into Vero cells. Virus Res 191:21–29. 10.1016/j.virusres.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Wang J, Hou W, Shan Y, Wang S, Liu F. 2021. Dynamic dissection of the endocytosis of porcine epidemic diarrhea coronavirus cooperatively mediated by clathrin and caveolae as visualized by single-virus tracking. mBio 12:e00256-21. 10.1128/mBio.00256-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang QY, Yang YL, Tang YX, Qin P, Wang G, Xie JY, Chen SX, Ding C, Huang YW, Zhu SJ. 2022. Bile acids promote the caveolae-associated entry of swine acute diarrhea syndrome coronavirus in porcine intestinal enteroids. PLoS Pathog 18:e1010620. 10.1371/journal.ppat.1010620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jordens I, Marsman M, Kuijl C, Neefjes J. 2005. Rab proteins, connecting transport and vesicle fusion. Traffic 6:1070–1077. 10.1111/j.1600-0854.2005.00336.x. [DOI] [PubMed] [Google Scholar]
- 19.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. 2005. Rab conversion as a mechanism of progression from early to late endosomes. Cell 122:735–749. 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 20.Su C, Zheng C. 2021. When Rab GTPases meet innate immune signaling pathways. Cytokine Growth Factor Rev 59:95–100. 10.1016/j.cytogfr.2021.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Mendoza P, Diaz J, Torres VA. 2014. On the role of Rab5 in cell migration. Curr Mol Med 14:235–245. 10.2174/1566524014666140128111347. [DOI] [PubMed] [Google Scholar]
- 22.Vonderheit A, Helenius A. 2005. Rab7 associates with early endosomes to mediate sorting and transport of Semliki Forest virus to late endosomes. PLoS Biol 3:e233. 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray JL, Mavrakis M, McDonald NJ, Yilla M, Sheng J, Bellini WJ, Zhao L, Le Doux JM, Shaw MW, Luo CC, Lippincott-Schwartz J, Sanchez A, Rubin DH, Hodge TW. 2005. Rab9 GTPase is required for replication of human immunodeficiency virus type 1, filoviruses, and measles virus. J Virol 79:11742–11751. 10.1128/JVI.79.18.11742-11751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilcke M, Johannes L, Galli T, Mayau V, Goud B, Salamero J. 2000. Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-Golgi network. J Cell Biol 151:1207–1220. 10.1083/jcb.151.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollinshead M, Johns HL, Sayers CL, Gonzalez-Lopez C, Smith GL, Elliott G. 2012. Endocytic tubules regulated by Rab GTPases 5 and 11 are used for envelopment of herpes simplex virus. EMBO J 31:4204–4220. 10.1038/emboj.2012.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang YN, Liu YY, Xiao FC, Liu CC, Liang XD, Chen J, Zhou J, Baloch AS, Kan L, Zhou B, Qiu HJ. 2018. Rab5, Rab7, and Rab11 are required for caveola-dependent endocytosis of classical swine fever virus in porcine alveolar macrophages. J Virol 92:e00797-18. 10.1128/JVI.00797-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Duve C. 2005. The lysosome turns fifty. Nat Cell Biol 7:847–849. 10.1038/ncb0905-847. [DOI] [PubMed] [Google Scholar]
- 28.Wei X, She G, Wu T, Xue C, Cao Y. 2020. PEDV enters cells through clathrin-, caveolae-, and lipid raft-mediated endocytosis and traffics via the endo-/lysosome pathway. Vet Res 51:10. 10.1186/s13567-020-0739-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vallee RB, Herskovits JS, Aghajanian JG, Burgess CC, Shpetner HS. 1993. Dynamin, a GTPase involved in the initial stages of endocytosis. Ciba Found Symp 176:185–197. 10.1002/9780470514450.ch12. [DOI] [PubMed] [Google Scholar]
- 30.Okamoto PM, Herskovits JS, Vallee RB. 1997. Role of the basic, proline-rich region of dynamin in Src homology 3 domain binding and endocytosis. J Biol Chem 272:11629–11635. 10.1074/jbc.272.17.11629. [DOI] [PubMed] [Google Scholar]
- 31.Zhao R, Shi Q, Han Z, Fan Z, Ai H, Chen L, Li L, Liu T, Sun J, Liu S. 2021. Newcastle disease virus entry into chicken macrophages via a pH-dependent, dynamin and caveola-mediated endocytic pathway that requires Rab5. J Virol 95:e2288-20. 10.1128/JVI.02288-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henley JR, Krueger EW, Oswald BJ, McNiven MA. 1998. Dynamin-mediated internalization of caveolae. J Cell Biol 141:85–99. 10.1083/jcb.141.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf AA, Fujinaga Y, Lencer WI. 2002. Uncoupling of the cholera toxin-G(M1) ganglioside receptor complex from endocytosis, retrograde Golgi trafficking, and downstream signal transduction by depletion of membrane cholesterol. J Biol Chem 277:16249–16256. 10.1074/jbc.M109834200. [DOI] [PubMed] [Google Scholar]
- 34.Montesano R, Roth J, Robert A, Orci L. 1982. Non-coated membrane invaginations are involved in binding and internalization of cholera and tetanus toxins. Nature 296:651–653. 10.1038/296651a0. [DOI] [PubMed] [Google Scholar]
- 35.Shpetner HS, Herskovits JS, Vallee RB. 1996. A binding site for SH3 domains targets dynamin to coated pits. J Biol Chem 271:13–16. 10.1074/jbc.271.1.13. [DOI] [PubMed] [Google Scholar]
- 36.Hanover JA, Willingham MC, Pastan I. 1984. Kinetics of transit of transferrin and epidermal growth factor through clathrin-coated membranes. Cell 39:283–293. 10.1016/0092-8674(84)90006-0. [DOI] [PubMed] [Google Scholar]
- 37.Benmerah A, Bayrou M, Cerf-Bensussan N, Dautry-Varsat A. 1999. Inhibition of clathrin-coated pit assembly by an Eps15 mutant. J Cell Sci 112:1303–1311. 10.1242/jcs.112.9.1303. [DOI] [PubMed] [Google Scholar]
- 38.Benmerah A, Poupon V, Cerf-Bensussan N, Dautry-Varsat A. 2000. Mapping of Eps15 domains involved in its targeting to clathrin-coated pits. J Biol Chem 275:3288–3295. 10.1074/jbc.275.5.3288. [DOI] [PubMed] [Google Scholar]
- 39.Liang Q, Zhang H, Li B, Ding Q, Wang Y, Gao W, Guo D, Wei Z, Hu H. 2019. Susceptibility of chickens to porcine deltacoronavirus infection. Viruses 11:573. 10.3390/v11060573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jung K, Hu H, Saif LJ. 2017. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch Virol 162:2357–2362. 10.1007/s00705-017-3351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoian A, Rowland R, Petrovan V, Sheahan M, Samuel MS, Whitworth KM, Wells KD, Zhang J, Beaton B, Cigan M, Prather RS. 2020. The use of cells from ANPEP knockout pigs to evaluate the role of aminopeptidase N (APN) as a receptor for porcine deltacoronavirus (PDCoV). Virology 541:136–140. 10.1016/j.virol.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu K, Zhou Y, Mu Y, Liu Z, Hou S, Xiong Y, Fang L, Ge C, Wei Y, Zhang X, Xu C, Che J, Fan Z, Xiang G, Guo J, Shang H, Li H, Xiao S, Li J, Li K. 2020. CD163 and pAPN double-knockout pigs are resistant to PRRSV and TGEV and exhibit decreased susceptibility to PDCoV while maintaining normal production performance. eLife 9:e57132. 10.7554/eLife.57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mercer J, Helenius A. 2008. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science 320:531–535. 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- 44.Liu CC, Zhang YN, Li ZY, Hou JX, Zhou J, Kan L, Zhou B, Chen PY. 2017. Rab5 and Rab11 are required for clathrin-dependent endocytosis of Japanese encephalitis virus in BHK-21 cells. J Virol 91:e01113-17. 10.1128/JVI.01113-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, Fang L, Liu S, Ke W, Wang D, Peng G, Xiao S. 2019. Susceptibility of porcine IPI-2I intestinal epithelial cells to infection with swine enteric coronaviruses. Vet Microbiol 233:21–27. 10.1016/j.vetmic.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Han Y, Shi H, Chen J, Zhang X, Wang X, Zhou L, Liu J, Zhang J, Ji Z, Jing Z, Ma J, Shi D, Feng L. 2020. Swine acute diarrhea syndrome coronavirus-induced apoptosis is caspase- and cyclophilin D-dependent. Emerg Microbes Infect 9:439–456. 10.1080/22221751.2020.1722758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schelhaas M. 2010. Come in and take your coat off—how host cells provide endocytosis for virus entry. Cell Microbiol 12:1378–1388. 10.1111/j.1462-5822.2010.01510.x. [DOI] [PubMed] [Google Scholar]
- 48.Yamauchi Y, Helenius A. 2013. Virus entry at a glance. J Cell Sci 126:1289–1295. 10.1242/jcs.119685. [DOI] [PubMed] [Google Scholar]
- 49.Wang H, Yang P, Liu K, Guo F, Zhang Y, Zhang G, Jiang C. 2008. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res 18:290–301. 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pu Y, Zhang X. 2008. Mouse hepatitis virus type 2 enters cells through a clathrin-mediated endocytic pathway independent of Eps15. J Virol 82:8112–8123. 10.1128/JVI.00837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang H, Yuan X, Sun Y, Mao X, Meng C, Tan L, Song C, Qiu X, Ding C, Liao Y. 2019. Infectious bronchitis virus entry mainly depends on clathrin mediated endocytosis and requires classical endosomal/lysosomal system. Virology 528:118–136. 10.1016/j.virol.2018.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McMahon HT, Boucrot E. 2011. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 12:517–533. 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 53.Mercer J, Schelhaas M, Helenius A. 2010. Virus entry by endocytosis. Annu Rev Biochem 79:803–833. 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- 54.Wang N, Zhao D, Wang J, Zhang Y, Wang M, Gao Y, Li F, Wang J, Bu Z, Rao Z, Wang X. 2019. Architecture of African swine fever virus and implications for viral assembly. Science 366:640–644. 10.1126/science.aaz1439. [DOI] [PubMed] [Google Scholar]
- 55.Cureton DK, Massol RH, Saffarian S, Kirchhausen TL, Whelan SP. 2009. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog 5:e1000394. 10.1371/journal.ppat.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cureton DK, Massol RH, Whelan SP, Kirchhausen T. 2010. The length of vesicular stomatitis virus particles dictates a need for actin assembly during clathrin-dependent endocytosis. PLoS Pathog 6:e1001127. 10.1371/journal.ppat.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu W, Zhang S, Shen Y, Yang Q. 2018. Epidermal growth factor receptor is a co-factor for transmissible gastroenteritis virus entry. Virology 521:33–43. 10.1016/j.virol.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Li Y, Wang S, Liu F. 2020. Dynamics of transmissible gastroenteritis virus internalization unraveled by single-virus tracking in live cells. FASEB J 34:4653–4669. 10.1096/fj.201902455R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parton RG, Simons K. 2007. The multiple faces of caveolae. Nat Rev Mol Cell Biol 8:185–194. 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- 60.Pelkmans L, Kartenbeck J, Helenius A. 2001. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat Cell Biol 3:473–483. 10.1038/35074539. [DOI] [PubMed] [Google Scholar]
- 61.Meier O, Boucke K, Hammer SV, Keller S, Stidwill RP, Hemmi S, Greber UF. 2002. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J Cell Biol 158:1119–1131. 10.1083/jcb.200112067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.West MA, Bretscher MS, Watts C. 1989. Distinct endocytotic pathways in epidermal growth factor-stimulated human carcinoma A431 cells. J Cell Biol 109:2731–2739. 10.1083/jcb.109.6.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee JH, Pasquarella JR, Kalejta RF. 2019. Cell line models for human cytomegalovirus latency faithfully mimic viral entry by macropinocytosis and endocytosis. J Virol 93:e01021-19. 10.1128/JVI.01021-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, Halfmann P, Kawaoka Y. 2010. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog 6:e1001121. 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mercer J, Helenius A. 2009. Virus entry by macropinocytosis. Nat Cell Biol 11:510–520. 10.1038/ncb0509-510. [DOI] [PubMed] [Google Scholar]
- 66.Burkard C, Verheije MH, Wicht O, van Kasteren SI, van Kuppeveld FJ, Haagmans BL, Pelkmans L, Rottier PJ, Bosch BJ, de Haan CA. 2014. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog 10:e1004502. 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Damke H, Baba T, Warnock DE, Schmid SL. 1994. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol 127:915–934. 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krzyzaniak MA, Zumstein MT, Gerez JA, Picotti P, Helenius A. 2013. Host cell entry of respiratory syncytial virus involves macropinocytosis followed by proteolytic activation of the F protein. PLoS Pathog 9:e1003309. 10.1371/journal.ppat.1003309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mahanty S, Ravichandran K, Chitirala P, Prabha J, Jani RA, Setty SR. 2016. Rab9A is required for delivery of cargo from recycling endosomes to melanosomes. Pigment Cell Melanoma Res 29:43–59. 10.1111/pcmr.12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cayouette S, Bousquet SM, Francoeur N, Dupre E, Monet M, Gagnon H, Guedri YB, Lavoie C, Boulay G. 2010. Involvement of Rab9 and Rab11 in the intracellular trafficking of TRPC6. Biochim Biophys Acta 1803:805–812. 10.1016/j.bbamcr.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 71.Zhu YZ, Xu QQ, Wu DG, Ren H, Zhao P, Lao WG, Wang Y, Tao QY, Qian XJ, Wei YH, Cao MM, Qi ZT. 2012. Japanese encephalitis virus enters rat neuroblastoma cells via a pH-dependent, dynamin and caveola-mediated endocytosis pathway. J Virol 86:13407–13422. 10.1128/JVI.00903-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods and Fig. S1 to S3. Download jvi.00210-23-s0001.pdf, PDF file, 0.6 MB (600.8KB, pdf)