ABSTRACT
The monoclonal antibodies (MAbs) NCI05 and NCI09, isolated from a vaccinated macaque that was protected from multiple simian immunodeficiency virus (SIV) challenges, both target an overlapping, conformationally dynamic epitope in SIV envelope variable region 2 (V2). Here, we show that NCI05 recognizes a CH59-like coil/helical epitope, whereas NCI09 recognizes a β-hairpin linear epitope. In vitro, NCI05 and, to a lesser extent, NCI09 mediate the killing of SIV-infected cells in a CD4-dependent manner. Compared to NCI05, NCI09 mediates higher titers of antibody-dependent cellular cytotoxicity (ADCC) to gp120-coated cells, as well as higher levels of trogocytosis, a monocyte function that contributes to immune evasion. We also found that passive administration of NCI05 or NCI09 to macaques did not affect the risk of SIVmac251 acquisition compared to controls, demonstrating that these anti-V2 antibodies alone are not protective. However, NCI05 but not NCI09 mucosal levels strongly correlated with delayed SIVmac251 acquisition, and functional and structural data suggest that NCI05 targets a transient state of the viral spike apex that is partially opened, compared to its prefusion-closed conformation.
IMPORTANCE Studies suggest that the protection against SIV/simian-human immunodeficiency virus (SHIV) acquisition afforded by the SIV/HIV V1 deletion-containing envelope immunogens, delivered by the DNA/ALVAC vaccine platform, requires multiple innate and adaptive host responses. Anti-inflammatory macrophages and tolerogenic dendritic cells (DC-10), together with CD14+ efferocytes, are consistently found to correlate with a vaccine-induced decrease in the risk of SIV/SHIV acquisition. Similarly, V2-specific antibody responses mediating ADCC, Th1 and Th2 cells expressing no or low levels of CCR5, and envelope-specific NKp44+ cells producing interleukin 17 (IL-17) also are reproducible correlates of decreased risk of virus acquisition. We focused on the function and the antiviral potential of two monoclonal antibodies (NCI05 and NCI09) isolated from vaccinated animals that differ in antiviral function in vitro and recognize V2 in a linear (NCI09) or coil/helical (NCI05) conformation. We demonstrate that NCI05, but not NCI09, delays SIVmac251 acquisition, highlighting the complexity of antibody responses to V2.
KEYWORDS: CH59, NCI05, NCI09, V2, monoclonal antibodies, simian immunodeficiency virus, trogocytosis
INTRODUCTION
The prime-boost vaccine regimen based on ALVAC-HIV and gp120 formulated in alum afforded a significant but modest 31.2% decrease in the risk of clinical human immunodeficiency virus (HIV) acquisition 3.5 years following immunization, with an estimated 60.5% decrease in risk in the first 6 months (1, 2). The primary correlate of reduced risk in the RV144 phase III trial that produced these results was the level of IgG binding to the variable region 1 (V1)/V2 variable loops of gp120 scaffolded on gp70 (3). Strikingly, neutralizing antibody responses did not correlate with a reduced risk of HIV acquisition, suggesting a nonneutralizing effector function of V1/V2 IgG (3). In vaccinees with low IgA levels, antibody-dependent cellular cytotoxicity (ADCC) was a secondary correlate of reduced risk of HIV acquisition (3). COMPASS analysis further revealed that polyfunctional CD4+ T cells expressing tumor necrosis factor alpha (TNF-α), gamma interferon (IFN-γ), interleukin 4 (IL-4), IL-2, and/or CD40L also correlated with a reduced risk of infection (4). Peptide array binding analyses of RV144 plasma identified V2 CRF01_AE as the only linear epitope significantly associated with a reduced risk of infection independent of IgA and neutralizing antibody responses (5). Furthermore, 31 of 32 vaccinees in RV144 exhibited antibody responses to the cyclic V2 peptide spanning the HXB2 HIV clone gp120 amino acid positions 169 to 184 (6). Positions K169 and I181 were identified as sites of virus selection in HIV variant transmission to RV144 vaccinees who became infected (7).
V1 and V2 together form a disulfide-bonded, folded domain positioned at the apex of the trimeric envelope glycoprotein spike, which has been visualized in complex with neutralizing antibodies as a flexible 5-stranded β-sandwich or β-barrel structure (8). V1/V2 provides integrity to the closed trimer structure while occluding the coreceptor binding site in V3 and upon CD4 binding undergoes a large 40-Å displacement (9). Positions 169 and 181 in V2 are respectively part of the cryptic RDK and LDI/V binding motifs for the gut-homing host α4β7 integrin receptor (10, 11). Like the mucosal addressin ligand of α4β7, MAdCAM, V2 has nanomolar affinity for α4β7 in the presence of divalent cations and can be inhibited by monoclonal antibodies and mimetics targeting α4β7 (12). Simian immunodeficiency virus (SIV) and HIV preferentially home to the gastrointestinal (GI) tract during acute infection, presumably due to the preponderance of activated CD4+ T cells, and these viruses disrupt the integrity of the gastrointestinal tract lining, leading to microbial translocation and chronic immune activation (13). The conserved high affinity of V2 for α4β7, the damage to gut-associated lymphoid tissue (GALT) early in infection, and the evidence of α4β7 incorporation into virions suggests the hypothesis that the gut-homing activity of HIV and SIV is an important step for the establishment of virus infection (10–14).
Further insight into the role of anti-V2 binding, nonneutralizing antibodies has been provided by the recent demonstration that V2 binding to α4β7 on CD4+ T cells costimulates CD4+ cells and, by upregulating CCR5 expression, facilitates HIV/SIV infection (11, 15). Interestingly, one of these studies demonstrated that not all antibodies to V2 are able to inhibit V2 costimulation (15). Nonneutralizing monoclonal antibodies (MAbs) obtained from volunteers in the RV144 phase III trial or from macaques vaccinated with ALVAC-SIV gp120/alum that recognize the HIV or SIV V2 in an α-helical conformation inhibit α4β7 engagement or V2 costimulation of CD4+ T cells and decrease HIV/SIV replication in vitro (14). In contrast, antibodies recognizing V2 in the β-strand conformation do not.
The vaccine efficacy of the RV144 trial was recapitulated and confirmed in two independent macaque studies using ALVAC-based vaccine modalities with SIV immunogens, with anti-V2 IgG levels associated with a decreased risk of acquisition following intrarectal exposure to SIVmac251 (16, 17). Additional macaque studies with optimized vaccine regimens, wherein the prime was replaced with DNA and/or the V1 envelope region was deleted to avoid the elicitation of anti-V1 interfering antibodies, demonstrated that antibodies recognizing the V2 in an α-helical (but not the β-barrel) conformation bind to virions and mediate ADCC, associated with a decreased risk of virus acquisition (18). In the same studies, anti-V2 polyclonal responses to SIV peptide probes encompassing positions 169 and 181 of V2 were also associated with a transient decrease in virus burden in plasma and mucosal tissues in vaccinated animals that became infected. These results point to V2 being a region of virus vulnerability, likely because of its exposure on the apex of virions and on the surfaces of SIV-infected cells. However, it is noteworthy that in all of these vaccine studies, the decreased risk of SIVmac251 acquisition was associated not only with V2-specific antibodies and V2-specific ADCC but also with the frequency of CD14+ monocytes and Th1/Th2 cells in blood and NKp44+ IL-17+ cells in the mucosa (16–20; M. Bissa).
NCI05 and NCI09 are MAbs that recognize specific conformations of the cryptic α4β7 binding site of SIVmac251 V2 but are unable to neutralize the challenge virus (18). We tested the effects of NCI05 and NCI09 on SIVmac251 acquisition and found that, compared to untreated controls, both MAbs were unable to decrease the risk of viral acquisition when delivered in vivo by passive transfer in the absence of other innate and cellular vaccine-elicited immune responses. However, our investigation did reveal that these MAbs, which target differing V2 conformations, also differ in their ability to mediate immune evasion in vitro and the timing of virus acquisition in vivo.
RESULTS
Distinct V2 structures recognized by NCI05 and NCI09.
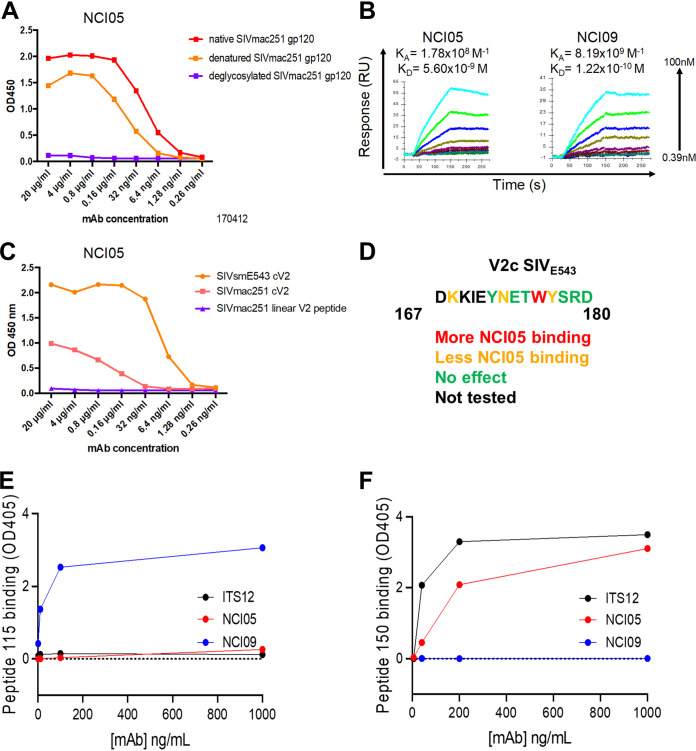
We obtained two V2-specific MAbs, NCI05 and NCI09, from a vaccinated macaque that resisted 22 low-dose intrarectal challenges of SIVmac251 for over a year. These MAbs recognize native gp120 on SIVmac251 -infected cells and monomeric gp120/gp140 from SIVmac251 and SIVsmE660, and they bind to virions (18). NCI05 binds to native and denatured SIVmac251 gp120, and its binding is abolished by deglycosylation (Fig. 1A). The binding affinities (KD) measured by surface plasmon resonance (SPR) with deglycosylated SIVM766 gp120 were 5.6 nM and 0.122 nM for NCI05 and NCI09, respectively (Fig. 1B). NCI05 recognizes the cyclic V2 of SIVSME543 better than NCI09, but it does not bind the linear SIVmac251 V2 peptide (Fig. 1C), suggesting that NCI05 recognizes a conformational epitope. NCI05 and NCI09 both recognize elements in the V2b region, which harbors a cryptic/allosteric α4β7 binding motif (10, 18). Crystallography revealed that NCI09 recognizes a linear SIVmac251 V2158–173 peptide in a β-hairpin conformation (18).
FIG 1.
Anti-V2 MAb epitopes, binding, neutralization, and kinetics. (A) Binding of NCI05 to deglycosylated SIVmac251 gp120, denatured SIVmac251 gp120, and native SIVmac251 gp120 measured by ELISA. (B) Surface plasmon resonance measuring NCI05 and NCI09 binding to deglycosylated SIVmac251-M766 gp120 ligand. (C) Binding of NCI05 to SIVsmE543 gp120, SIVmac251 cyclic V2 (cV2), and SIVmac251 linear V2 peptide measured by ELISA. (D) SIVE543 V2 amino acid sequence with colored lettering showing amino acids which augment (red) or decrease (orange) NCI05 binding. (E and F) ELISA showing binding of ITS12, NCI05, and NCI09 to (E) linear peptide 115 and (F) coil/helical peptide 150.
Scanning mutagenesis of a SIVE543 V2167–180 peptide (DKKIEYNETWYSRD) showed that binding to NCI05 was highly dependent on W176 and weakly dependent on K168, N173, and Y177 (Fig. 1D). We therefore designed an extended peptide encompassing V2 residues 164 to 180, but with an R166S mutation to knock out binding of NCI09; the resulting peptide (peptide 150) bound NCI05 at <10 ng/mL, suggesting that the epitope of NCI05 lies inside this region of V2, independent of the glycosylation state of N173, which is part of an NxT glycosylation motif. Thus, we were able to design peptides 150 (also referred to as V2c; amino acid sequence, L164KSDKKIEYNETWYSRD180) and 115 (G163LKRDKTKEYN173) and confirm that they bind specifically to NCI05 and NCI09, respectively (Fig. 1E and F).
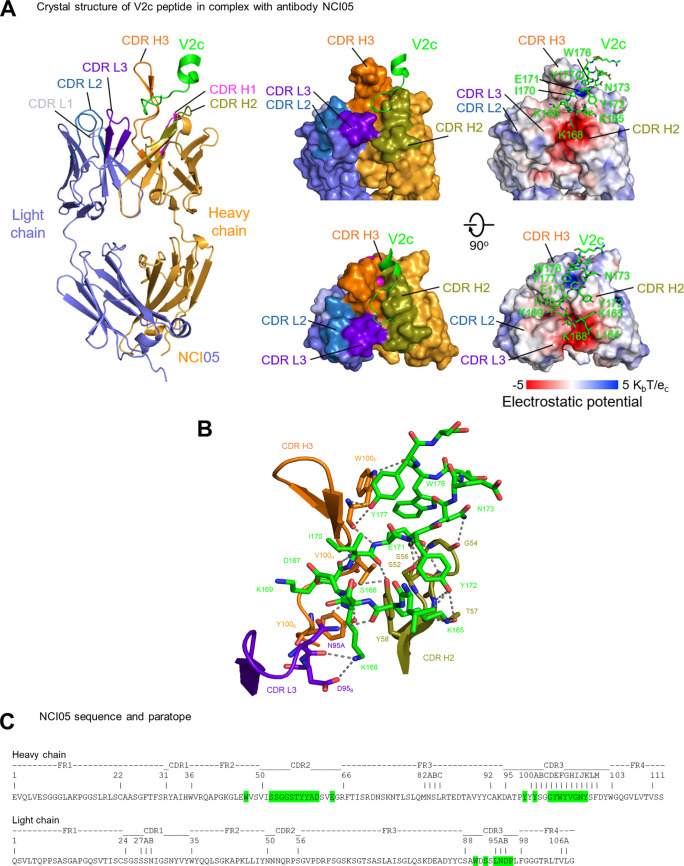
To elucidate the mode of binding of NCI05 to the V2 peptide 150, we determined a crystal structure at 2.35 Å of Fab NCI05 in complex with peptide 150 (Fig. 2A; also, see Table S1 in the supplemental material). Well-defined electron density (Fig. S1A) allowed placement of residues 164 to 179 of peptide 150 and all of the complementarity determinant regions (CDRs) of NCI05. CDRs H2, H3, and L3 of NCI05 form a three-way groove with the junction at the interface of these three CDRs. Peptide 150 binds to NCI05 in a U-shaped coil conformation with its N terminus lying in the groove between CDRs H2 and L3. The peptide turns into the groove between CDRs H2 and H3 at residues K168 and K169 and rotates out of the groove at N173 to pack in a helical conformation alongside the upright CDR H3 (Fig. 2A, middle). The interface buries ~850 Å2 of epitope surface and ~810 Å2 of paratope surface (Table S2).
FIG 2.
NCI05 binds to the coil/helical conformation of V2. (A) Crystal structure of NCI05 in complex with V2 peptide V2c at 2.35 Å resolution. (Left) the refined structure of antibody and V2 peptide complex with NCI05 heavy chain (orange), light chain (gray), and V2 peptide (green). CDRs are colored as follows: H1, magenta; H2, olive; H3, orange; L1, light blue; L2, navy blue; and L3, purple. (Center) Two 90° rotations of the surface representation show that the V2c peptide binds to the groove formed by CDRs H2, H3, and L3. (Right) The electrostatic potential surface is shown in the same views, with the V2c peptide in sticks representation. (B) Detailed interactions between NCI05 and the V2c peptide. The peptide and NCI05 paratope residues are shown in sticks representation, with other regions of NCI05 shown as cartoon. Hydrogen bonds and salt bridges between the interacting atoms are indicated with gray dashed lines. (C) Sequence and paratope of NCI05. Heavy- and light-chain paratope residues are highlighted in green. NCI05 residues were numbered according to Kabat nomenclature.
Electrostatic potential analysis indicated that the groove between CDR H2 and CDR L3 is highly negatively charged due to the clustering of CDR L3 D95B (Kabat numbering of antibody residues), CDR H2 D61, and CDR E64 in this region, providing a suitable binding surface for the positively charged peptide 150 N-terminal residues K165, K168, and K169 (Fig. 2A, right), where a salt bridge is formed between K168 and CDR L3 D95B (Fig. 2B and C; Table S2). A positively charged surface is also observed in the groove between CDRs H2 and H3 where peptide 150 E181 interacts (Fig. 2A, right).
Most residues in the 17-residue peptide 150, except the N-terminal L164, E174-T175, and C-terminal S178-R179-D180, contact NCI05 in the structure. Twenty-one hydrogen bonds are formed between the peptide antigen and NCI05 (Fig. 2B; Table S2). K168 at the turn of the bound peptide is one of the key binding residues, which provides 190 Å2 binding surface and forms hydrogen bonds to both heavy-chain CDR H3 Y100K and light-chain CDR L3 N95A in addition to the salt bridge to D95B, consistent with the observed reduction in binding when mutated to alanine (Fig. 2B and C; Table S2). E183 of peptide 150 inserts into the positively charged pocket between the CDR H2 and H3 and makes three main and side chain hydrogen bonds to CDR H3 S52 and S56. W176 and Y177 at the C terminus of peptide 150 interact with the upright CHR H3 strand by providing 170 Å2 binding surface and form 2 hydrogen bonds with W100F, with W-W interactions generally interpreted as strong binding hot spots. Observed refined structure of electron density in the crystal structure clearly defined a N-linked glycan site at N100J of NCI05 CDR H3 in which the fucose moiety interacts with K169 of peptide 150 (Fig. S1A). Overall, peptide 150 interacts with NCI05 through electrostatic complementarity and extensive hydrogen bonds in an extended random coil conformation for the central binding region, but with a strong aromatic group hot spot between W176-Y177 and the CDR H3.
The V2 peptide recognized by NCI05 resembles that recognized by NCI09 at residues 168 to 172 in the center of V2c but differs in both the N and C termini (Fig. S1B). The C terminus of the NCI05-bound peptide 150 makes a turn at N173 to contact NCI05 CDR H3, mainly through aromatic residues W176 and Y177 in a helical conformation (Fig. 2B). The superposition of V2 bound by NCI05 and NCI09 over their commonly recognized peptide region (residues 168 to 172) indicated that the antibodies had different approaching angles to the peptide but recognized the same face on the V2 loop (Fig. S1C). Comparison of diverse anti-HIV and -SIV antibody-bound conformations of this V2 region reveals the conformational dynamics of this V2 segment and suggests that the NCI09-targeted epitope is a β-hairpin outlier among the diverse antibody targets, which mostly exhibit coil/helical conformations. However, the conformation bound by NCI09 is more similar to the unbound prefusion conformation (Fig. S1B and D). The superposition of the central V2c amino acids (residues 169 to 171) that resemble the prefusion conformation in both the NCI09- and NCI05-bound conformations (18, 21) (Fig. S1B) suggests that NCI05 and NCI09 binding are incompatible with the prefusion, apex-stabilized closed state (Fig. S1E).
The backbone conformation adopted by V2164–180 bound to NCI05 is noticeably dissimilar from the segment’s β-strand conformation observed in the prefusion conformation of the SIV viral spike, in which the V1/V2 domain spike apex is stabilized by the neutralizing antibody PGT145 (PDB 8DVD) (Fig. S1B) (21). It is noteworthy that the strands turn at different locations and the side chain of K168 also points to opposite sides of the V2 loop (18, 21) (Fig. S1B), suggesting that the NCI05 and NCI09 binding were incompatible with the prefusion closed state (Fig. S1E). Previous studies have revealed that CD4 binding to prefusion Env can induce a partially open state that has the potential to flip the V1/V2 into an open conformation for binding to antibodies like NCI05 (Fig. S1F). At present, however, this concept remains speculative.
Notably, the backbone of the α-helical turn in the NCI05-recognized conformation is identical in location and structure to that recognized by the human anti-V2 antibody CH59. The remainder of V2 bound to both antibodies were random coils, resulting in an overall very similar conformation between CH59-bound HIV-V2 and NCI05-bound SIV V2 (Fig. S1G). NCI09-bound V2 is also nearly completely dissimilar to those bound by NCI05, CH59, and CH58, which all have some or substantial local similarity to each other in terms of both backbone V2 conformation and V2 contacts with the antibodies (Fig. S1H).
Functional activity of the V2-specific MAbs NCI05 and NCI09 in vitro.
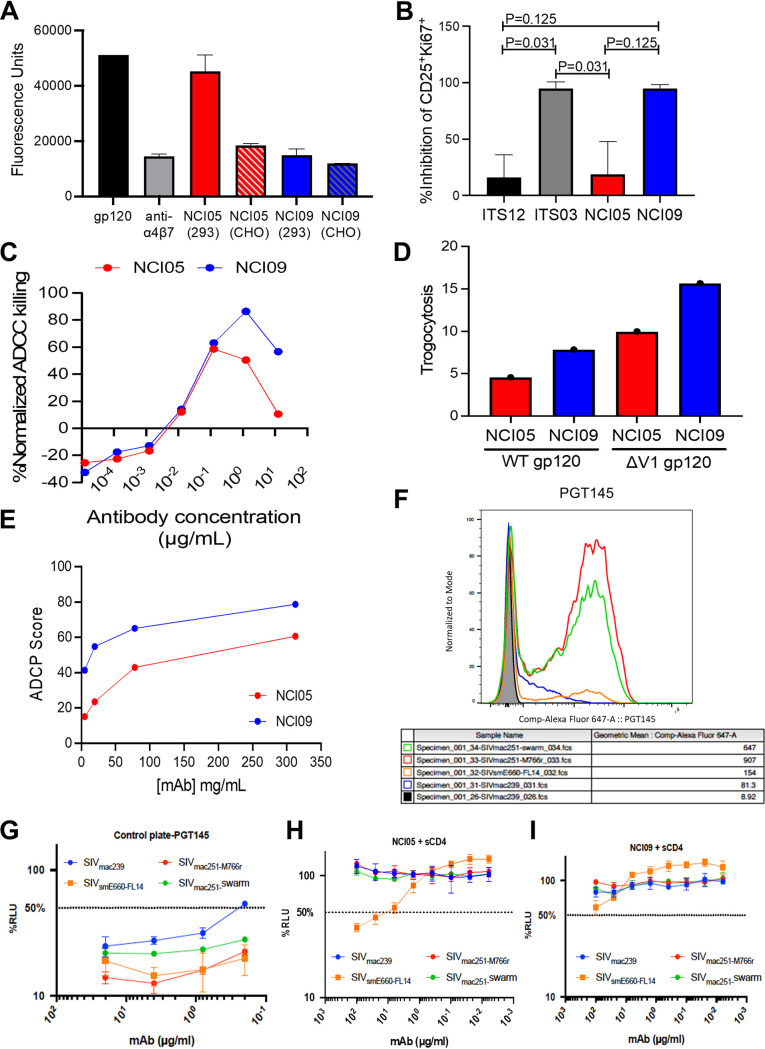
NCI05 and NCI09 do not neutralize tier 2 SIVmac251 (18). When NCI05 and NCI09 are expressed in CHO cells, they equally inhibit SIVgp120 binding to the α4β7 integrin cells (Fig. 3A). However, expression of NCI05 in 293 cells results in a loss of inhibitory activity, likely related to the differential glycosylation of this antibody in the two cell types (Fig. 3A). We therefore tested the effects of NCI05 and NCI09 on V2-mediated costimulation of T cells, since both HIV and SIV V2 bind to α4β7 and deliver costimulatory signals to CD4+ T cells, which, in the case of HIV, can be blocked by MAb CH58 recognizing V2 in an α-helix conformation (15). Interestingly, NCI09 decreased the percentage of activated and proliferating CD25+ Ki67+ CD4+ T cells following costimulation with CD28, whereas NCI05 did not (Fig. 3B; Fig. S2). Comparison of the ADCC mediated by the two antibodies demonstrated no difference at low antibody doses, but at high doses, NCI09 killing was higher than that of NCI05 (Fig. 3C). NCI09 also mediated a higher level of trogocytosis measured with both wild-type and V1 deletion-containing gp120 (Fig. 3D) and higher antibody-dependent cellular phagocytosis (ADCP) (Fig. 3E).
FIG 3.
Gp120/gp140 binding, neutralization, inhibition of gp120-α4β7 binding, inhibition of gp120 T-cell costimulation, ADCC, ADCP, and trogocytosis by MAbs NCI05 and NCI09. (A) Binding of SIVmac251-M766 gp120 to α4β7-expressing RPMI8866 cells in the presence of vedolizumab (positive control) and NCI MAbs produced in either 293 or CHO cells. (B) Inhibition of gp120-induced T-cell activation by ITS12 (negative control), ITS03 (positive control), NCI05, and NCI09. (C) Titration curves of normalized ADCC by NCI05 and NCI09 with ΔV1 gp120-coated CEM.NKR-CCR5 target cells. (D) Trogocytosis by CD14+ monocytes incubated with ΔV1 and wild-type gp120-coated CEM.NKR-CCR5 target cells labeled with PKH26 in the presence of NCI05 and NCI09. (E) ADCP by THP-1 cells measured in the presence of ΔV1 and wild-type gp120-coated CEM.NKR-CCR5 target cells and titrations of NCI05 and NCI09 antibodies. (F) Binding of PGT145 to V2 of various SIV-infected cells in the absence of soluble CD4. (G) ADCC mediated by KHYG-1 NK cells recognizing PG145 bound to V2 of various SIV-infected CEM.NKR-CCR5-sLTR-Luc cells in the absence of soluble CD4. (H and I) ADCC mediated by (H) NCI05 and (I) NCI09 against various SIV-infected CEM.NKR-CCR5-sLTR-Luc cells in the presence of sCD4.
Next, we tested NCI05 and NCI09 for Env binding and ADCC against cells infected with the infectious molecular clones SIVmac239, SIVsmE660-FL14, and SIVmac251-M766r and with uncloned SIVmac251 swarm in the presence and absence of soluble CD4 (sCD4). A broadly neutralizing antibody to HIV-1 (PGT145) that was previously shown to cross-react with a conserved proteoglycan epitope at the V2 apex of SIV Env and to mediate ADCC against SIV-infected cells was included as a positive control (22), and an irrelevant dengue-virus specific antibody dengue virus type 3 virus (DEN3) was included as a negative control. As expected, PGT145 stained Env and mediated efficient ADCC against cells infected with all four SIV strains (Fig. 3F and G). We observed a low level of Env staining for NCI05 on the surfaces of SIVsmE660-FL14-infected cells that was increased by the addition of sCD4 (Fig. S3A and B). Accordingly, this antibody mediates measurable ADCC only in the presence of sCD4 (Fig. 3H; Fig. S3C). NCI09 weakly bound to the SIVmac251 swarm used in our challenge experiments (Fig. S3D and E) and had a detectable, low level of ADCC against SIVsmE660-FL14-infected cells in the presence of sCD4 (Fig. 3I; Fig. S3F). An antibody to dengue virus type 3 used as a control demonstrated neither envelope cell surface staining nor ADCC (Fig. S3G and S4A). Thus, the V2 epitopes for NCI05 and NCI09 are exposed on the surfaces of cells infected with SIVsmE660-FL14 upon CD4 engagement but not on cells infected with SIVmac239, SIVmac251-M766r, or SIVmac251.
Systemic and mucosal distribution of NCI05 and NCI09 antibodies in vivo.
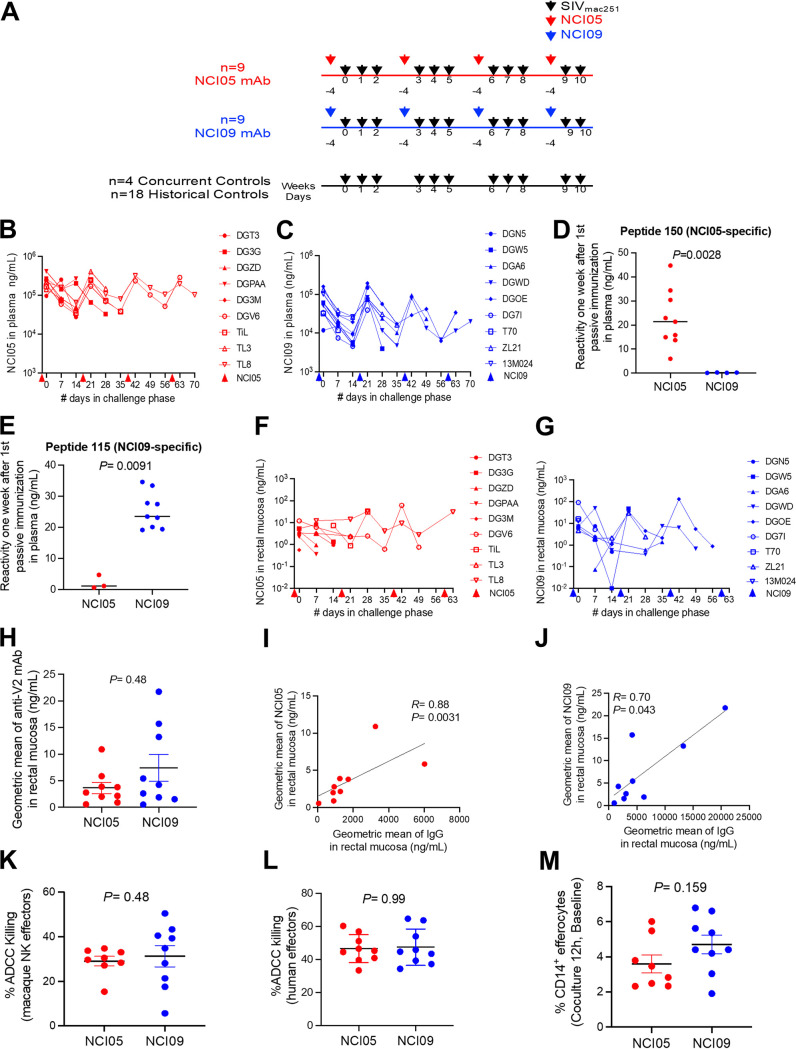
The functional differences between NCI05 and NCI09 provided the opportunity to compare their antiviral activities in the SIVmac251 macaque model. We treated two groups of 9 macaques each with NCI05 or NCI09 at a dose of 20 mg/kg of body weight, beginning at 4 days prior to the first SIVmac251 challenge to allow for tissue distribution of the MAbs (Fig. 4A). Administration was continued during challenge exposure to SIVmac251 every 3 weeks for a total of four doses of antibody in each group (Fig. 4A). Plasma levels of NCI05 and NCI09 measured by ELISA were similarly sustained over time in both animal groups (Fig. 4B and C). As expected, the sera from animals administered NCI05 specifically recognized the coil/helical peptide 150, and plasma from animals treated with NCI09 recognized peptide 115 (Fig. 4D and E). The levels of NCI05 and NCI09 in rectal secretions were equivalent over time in the two groups (Fig. 4F and G), as the geometric mean of NCI05 and NCI09 (in nanograms per milliliter; measured by enzyme-linked immunosorbent assay [ELISA] reactivity to gp120) did not differ significantly between the groups (Fig. 4H). In both groups, the concentration (in nanograms per milliliter) of NCI05 and NCI09 correlated directly with the total IgG level (Fig. 4I and J).
FIG 4.
Mucosal and plasma NCI05 and NCI09 levels in passively immunized animals. (A) Study design of passive immunization (NCI05 in red, NCI09 in blue) and 1:250 intrarectal SIVmac251 challenges (black arrows). (B and C) Concentrations of (B) NCI05 and (C) NCI09 measured in the plasma by ΔV1 gp120 ELISA during the SIVmac251 challenge phase. Colored arrows below the x axis indicate the times of passive immunization 4 days prior to each challenge. (D and E) ELISA detection of V2 MAb binding in the plasma of animals 1 week after the first passive immunization with NCI05 and NCI09 to (D) peptide 150 (biotin-GGG-LKSDKKIEYNETWYSRD) and (E) peptide 115 (biotin-GGG-GLKRDKTKEYN), respectively. (F and G) Mucosal concentrations of (F) NCI05 and (G) NCI09 measured by ΔV1 gp120 ELISA during the SIVmac251 challenge phase. (H) Geometric means of V2 MAbs measured in the rectal mucosa of the NCI05 and NCI09 groups. (I and J) Spearman rank correlation between the geometric mean of total mucosal IgG and mucosal (I) NCI05 and (J) NCI09. (K) Percent ADCC killing in plasma from passively immunized macaques mediated by macaque NKG2A+ NK effectors from the NCI05 and NCI09 groups. (L) Percent ADCC killing in plasma from passively immunized macaques mediated by human PBMC effectors. (M) Efferocytosis by CD14+ efferocytes in PBMCs collected prior to passive immunization from the NCI05 and NCI09 groups.
Equivalent NK and monocyte functions prior to administration of NCI05 and NCI09.
We performed ADCC assays with NCI05 or NCI09 on target cells coated with SIVmac251-M766 gp120ΔV1Δ protein, an immunogen with a predominant V2 α-helix conformation (18), together with NKG2A+ effector cells purified from macaques in the two groups prior to antibody administration. This study was designed to exclude possible natural differences in NK effector function in the animals. We found no differences in ADCC activity with the peripheral blood mononuclear cells (PBMCs) of the animals to be inoculated with NCI05 or NCI09 using the matching antibody (Fig. 4K). Next, we tested the plasma collected at 1 week following antibody infusion to measure ADCC, using the standard assay with human PBMCs as effectors and target cells coated with the SIVmac251-M766 gp120ΔV1 protein. We observed equivalent ADCC killing activity between the two groups (Fig. 4L). To further probe natural immunity in each group, we measured CD14+ cell-mediated efferocytosis prior to antibody treatment, since vaccine-induced epigenetic changes in CD14+ cells and efferocytosis were found to correlate with a decreased risk of virus acquisition in vaccinated macaques (Bissa et al., submitted). We observed no difference in the ability of CD14+ cells from macaques of either group to engulf apoptotic neutrophils (Fig. 4M). Collectively, these data document that prior to MAb treatment, the animal groups shared equivalent NK effector activity and CD14+-cell-mediated efferocytosis.
Mucosal antibody levels, serum ADCC, and efferocytosis correlate with delayed virus acquisition in NCI05-treated animals.
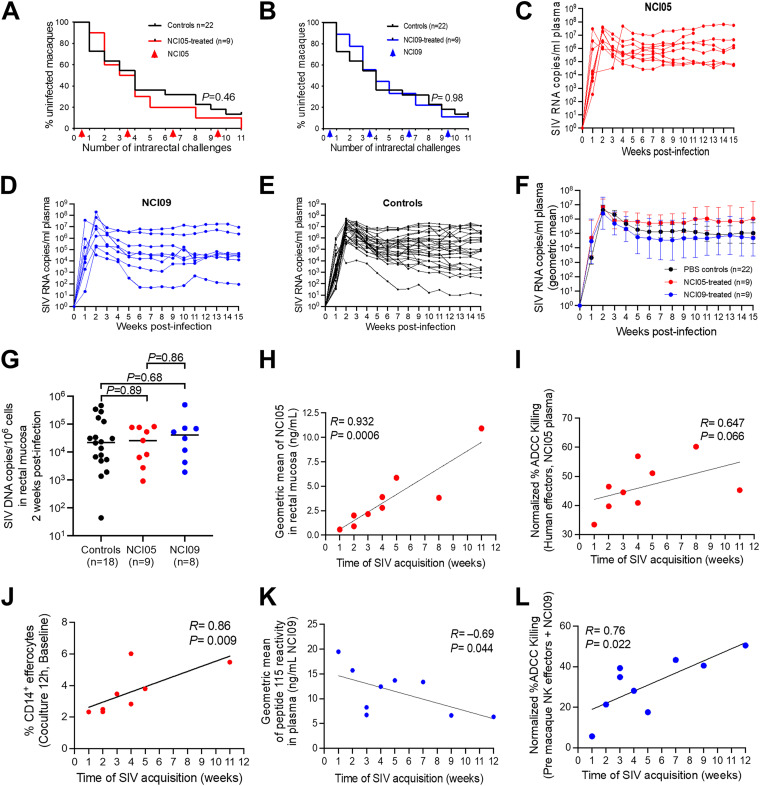
We exposed untreated or animals passively administered MAbs to SIVmac251 and assessed the per-exposure risk of virus acquisition compared to that of untreated controls. All 18 antibody-treated animals and 4 concurrent naive controls were exposed intrarectally to SIVmac251 on a weekly basis for a total of 11 weeks. As per study design, virological data from an additional 18 historical controls exposed to the identical virus stock in the same way in the same animal facility were used in the data analyses. We observed no difference in the per-challenge risk of SIVmac251 acquisition in control animals compared to that in animals treated with NCI05 or NCI09 (Fig. 5A and B). Similarly, no differences were observed in plasma virus levels in the acute or chronic phase of infection among the three groups (Fig. 5C to F), and mucosal viral DNA at 2 weeks postinfection did not differ among the three groups (Fig. 5G).
FIG 5.
Virus acquisition curves, viral load, and NCI05-specific and NCI09-specific ADCC correlations with time of virus acquisition. (A and B) Kaplan-Meier curves showing SIV acquisition rates in animals passively immunized with (A) NCI05 and (B) NCI09 and controls (black lines). (C to E) SIV viremia in individual animals following infection in (C) NCI05, (D) NCI09, and (E) control groups. (F) Geometric mean of SIV viremia in control, NCI05, and NCI09 groups. (G) SIV DNA measured in the rectal mucosa at 2 weeks postinfection in control, NCI05, and NCI09 groups. (H) Spearman rank correlation between the geometric mean of NCI05 measured in the rectal mucosa by ΔV1 gp120 ELISA and the time of SIV acquisition. (I) Normalized percentage of ADCC mediated by human PBMC effectors incubated with NCI05 plasma obtained 1 week after infection (J) Spearman rank correlation between the percent efferocytosis by CD14+ cells in macaque pre-PBMCs after 12 h and the time of SIV acquisition. (K) Spearman rank correlation between the geometric mean of NCI09 measured in plasma prior to infection by peptide 115 ELISA and the time of SIV acquisition. (L) Normalized percentage of ADCC mediated by macaque NKG2A+ NK effectors obtained prior to passive immunization and incubated with NCI09 plasma obtained 1 week after infection.
Next, we assessed whether NCI05 or NCI09 administration delayed the time to virus acquisition within each MAb-treated group. Strikingly, mucosal levels of NCI05 strongly correlated with a decreased risk of SIVmac251 acquisition (R = 0.932; P = 0.0006), and there was a similar trend for ADCC mediated by their plasma (R = 0.647; P = 0.066) (Fig. 5H and I, respectively). In addition, in the NCI05 group, the percentage of CD14+ cells mediating efferocytosis also correlated with delayed virus acquisition (Fig. 5J). Notably, ADCC did not correlate with time of acquisition in animals treated with NCI09 (Fig. S4C). The geometric mean of NCI09 in plasma prior to infection measured with peptide 115 correlated with faster virus acquisition (Fig. 5K), while the geometric mean of NCI05 in plasma measured with peptide 150 did not correlate with virus acquisition (Fig. S4B). Strikingly, however, ADCC measured from purified NKG2A+ cells (depleted of monocytes) prior to in vivo administration of NCI09 and with the addition of NCI09 in vitro correlated significantly with delayed virus acquisition (R = 0.76; P = 0.022) (Fig. 5L). This finding suggests that the ability of NCI09 to mediate a higher level of trogocytosis, a response mediated by monocytes, may have interfered with ADCC in vivo.
DISCUSSION
In humans, antibodies to the HIV envelope variable region 2 (V2) were associated with decreased risk of HIV acquisition in the RV144 HIV vaccine trial using the prime-boost ALVAC-HIV/gp120/alum regimen (3). Similarly, in macaques, SIV-based vaccines elicited anti-V2 antibodies that together with CD14+ monocytes, mucosal NKp44+ cells, and Th1 and Th2 cells correlated with a decreased risk of SIVmac251 acquisition (16–20; Bissa et al., submitted). A few studies using different technical approaches have directly assessed the innate ability of nonneutralizing antibodies to confer protection against SIV and simian-human immunodeficiency virus (SHIV) infection. Two early passive immunization studies with polyclonal anti-SIV antibodies in neonatal macaques reported 100% and 0% protection against oral SIVmac251 challenge, respectively (23, 24). Another study showed that passive immunization with nonneutralizing polyclonal IgG from an elite HIV controller with high ADCC functionality failed to protect macaques against intrarectal SHIV challenge (25). However, none of the studies reported a mapping of the fragment of antigen (Fab) specificities for polyclonal antibody preparations used in the passive immunizations. Two studies have demonstrated protection against single and mixed SHIV challenges in macaques following passive immunization with neutralizing anti-V2 monoclonal antibodies cloned from HIV-infected individuals (26, 27). In one study, macaques were passively immunized with a narrow-breadth neutralizing V2i MAb, 830A, cloned from a patient chronically infected with subtype B virus, which maintained ADCC functionality (28). The IgG1 of MAb 830A correlated with reduced levels of viral DNA in PBMCs isolated from inguinal, iliosacral, and axillary lymph nodes and of viral RNA in plasma following intrarectal SHIV challenge in macaques (28). However, no significant decrease in the risk of SHIV acquisition was reported (28). In another macaque study, systems serology was used to partition vaccinated animals into quartiles based on a protective, nonneutralizing, antibody quality signature, demonstrating that animals in the top quartiles were protected against SIVmac251 acquisition (29). Furthermore, in a separate cohort of vaccinated animals, passive transfer of purified polyclonal IgG purified from the top two quartiles of antibody quality was sufficient to significantly protect a low percentage of naive animals from intrarectal SIVmac251 challenge (29).
The key insights of our work reported here are, first, that SIV V2 can elicit antibodies that can either accelerate or delay viral transmission depending on the V2 structural conformation that they recognize and, second, that even antibodies to the same segment of V2 do not necessarily have the same functionality. Individual MAbs differ in their ability to inhibit T-cell activation, kill SIV-infected cells, and mediate trogocytosis, a monocyte-mediated mechanism in which antigens are nibbled from the surfaces of infected cells, thus aiding pathogen evasion of host responses (30).
The V2-specific MAbs NCI05 and NCI09 tested in the current study target overlapping peptides within the SIV V2 TGLKRDKTKEYNETWYSTD (the underlining of RDK indicates the cryptic α4β7 binding site) amino acid stretch, which encompasses both the canonical and cryptic α4β7 binding sites (10–12, 18). Both NCI05 and NCI09 are unable to neutralize the tier 2 SIVmac251 virus stock used in our challenge experiments (18). Structural analysis indicated that NCI09 targets a β-hairpin epitope within the N-terminal half of this segment, while NCI05 targets a coil/helical conformation occupying two-thirds of the C terminus of this V2 segment, with the coil/helical and β-hairpin epitopes overlapping. NCI05 mediates ADCC but has low neutralizing activity versus tier 1 SIVs and no neutralizing activity versus tier 2 virus. The ability of NCI05 to inhibit V2 binding to the α4β7 binding site is glycosylation dependent, and its ability to kill infected cells is triggered by CD4. These observations suggest that the N173 glycosylated form of V2c can also bind NCI05 (N173 points toward the solvent and the NCI05 CDR1/2 in the structure) and that the spike-open conformation triggered by CD4 binding favors the NCI05-bound conformation of V2 (Fig. S1F). We report here that CD4-induced conformations in V2 exposed on the trimeric envelope are susceptible to ADCC by NCI05 and much less so to that by NCI09. It is unclear, however, why NCI05 mediated ADCC in SIVsmE660-FL14-infected cells in the presence of sCD4 but not in cells infected by the SIVmac251 challenge virus. The robustness of the ADCC assay is highlighted by the high ADCC activity mediated by the positive control, PGT145, against cells infected with all of the SIV isolates tested. However, SIVsmE660-FL14 is the most antibody-sensitive infectious molecular clone of SIV tested in this assay, whereas the SIVmac251 challenge virus is the most genetically diverse swarm. Therefore, it should not be surprising that SIVsmE660-FL14 is more sensitive to NCI05 in the presence of sCD4. In HIV-1, coreceptor binding site (CoRBS) antibodies are also needed to stabilize CD4i conformation changes and facilitate the critical binding and ADCC of MAb A32 (31).
Treatment of macaques with NCI05 before SIVmac251 challenge exposure did not decrease the per-challenge risk of virus acquisition compared to that in untreated animals. A possible limitation of the passive immunization study with NCI05 is its low level of distribution from the subscapular injection site to the rectal mucosa, where antibodies targeting the V2 coil/helical conformation could have a prophylactic effect at sufficiently high concentrations. The ~120-kDa molecular weight of IgG1 probably affects the entry of MAbs to V2 into blood capillaries, which are easily permeable to molecules of <16 kDa (32), suggesting that in our study, IgG1 may have disseminated through the interstitial fluid to the central lymphatic ducts and then into the circulatory system and mucosal surfaces. Plasma concentrations of NCI05 and NCI09 were 3 orders of magnitude higher than in the rectal secretions, which demonstrates significant loss of IgG as these molecules traversed to the mucosal tissue. Thus, it is possible that the risk of SIV acquisition could be more effectively decreased with the modification of the dose, frequency, and route of NCI05 administration, as suggested by the findings that plasma-mediated ADCC activity and NCI05 levels in the mucosa correlated with delayed virus acquisition. This effect was unique to NCI05-treated macaques; the immune correlates of delayed acquisition were not recapitulated in animals treated with NCI09. The current data point to V2 in a coil/helical conformation distinct from that seen in complex with neutralizing antibodies as a key element in virus vulnerability. V1 deletion and the addition of a glycine-proline-glycine motif were previously shown to result in no vaccine efficacy, whereas deletion of V1 alone favors a putative α-helical structure of V2 and improves vaccine efficacy (18). The current work characterizes in detail the V2 structure recognized by NCI05 as a non-beta-strand structure exhibiting random coil and α-helical segments. Notably, the structure is substantially similar to that of the human antibody CH59, isolated from an RV144 vaccinee, which was similar to NCI05’s isolation from a protected macaque (33). This parallel suggests that macaque MAbs can be found and evaluated with SIVmac251 challenge in vivo to model the immunology of the corresponding MAb in protecting humans against HIV. Whether a macaque MAb can be found that parallels CH58, with its greater viral cross-reactivity and purely helical epitope, remains to be ascertained.
The current work does not address how the functional differences between NCI05 and NCI09 in vitro, such as trogocytosis and inhibition of T-cell costimulation (15), contribute mechanistically to their seemingly opposite effects on virus acquisition in vivo, underscoring the need for further studies to dissect how binding antibodies that protect against SIV acquisition may be a viable alternative to neutralizing antibodies. The finding that NCI09 seemingly accelerates virus acquisition was somewhat unexpected, given that this antibody inhibits T-cell activation in vitro. However, our data demonstrating that the absence of monocytes in the ADCC assay using purified NKG2A was key to establishing a correlation of the NKG2A ADCC level and time of virus acquisition and suggested the hypothesis that the ability of NCI09 to mediate ADCC may be counteracted by its efficient induction of trogocytosis. A limitation of this study, however, is that we could not assess whether passive administration of NCI05 or NCI09 mitigated the level of T-cell activation during the challenge exposure phase.
In conclusion, our work here demonstrates that antibodies such as NCI05, recognizing the V2 coil/helical conformation, are not alone sufficient to protect against SIVmac251 acquisition. This result is not surprising, given that the vaccine approach we used (16–20; Bissa et al., submitted) reproducibly engages multiple arms of the immune system to decrease the risk of virus acquisition, antibodies to V2 being only one of them. Systemic and mucosal immunity synergize with anti-V2 antibodies mediating ADCC, such as Th1 and Th2 cells expressing low levels of CCR5 and α4β7, envelope-specific mucosal memory NKp44+ cells producing IL-17, CD14+ cells mediating efferocytosis, and tolerogenic dendritic cells (DC-10), all immune responses working in concert to eliminate virus seeding and/or dissemination (16–20; Bissa et al., submitted). Like effective antiretroviral therapy, which targets different steps in the virus life cycle, the simultaneous engagement of different arms of immunity appears to be necessary to consistently prevent SIV/HIV infection in the absence of neutralizing antibodies.
MATERIALS AND METHODS
Animals and study design.
Colony-bred rhesus macaques (Macaca mulatta) were obtained from Covance Research Products (Alice, TX, USA) or the NIAID colony (Morgan Island, SC, USA). Animals were housed and cared for according to the standards of the Association and Accreditation of Laboratory Animal Care International.
NCI05 and NCI09 monoclonal antibodies were cloned in an IgG1 vector from a vaccinated animal, P770, which resisted SIV acquisition after 22 challenges and expressed in CHO cells as described previously (16, 18). Animals were injected via the subscapular route with phosphate-buffered saline (PBS) or 20 mg/kg of monoclonal antibody. Animals were randomized to three groups: 4 concurrent controls treated with PBS, 9 animals treated with 20 mg/kg NCI05, and 9 animals treated with 20 mg/kg NCI09. Four days after the first passive immunization, the animals were challenged intrarectally with 120 50% tissue culture infective doses (TCID50) of SIVmac251 on a weekly basis and continued to receive passive immunizations every 3 weeks to maintain antibody levels. Animals were monitored for SIV infection by digital droplet PCR (Advanced Bioscience Laboratories, Rockville, MD, USA). The acquisition rate for the control group was combined with that for 18 historical controls which were challenged with the same stock of SIVmac251 within 1 year of the study.
ΔV1 gp120 ELISA of plasma and rectal secretions. (i) Plasma.
High-affinity Immulon 2 HB plates were coated overnight with 100 ng of SIVmac251-M766 gp120ΔV1 at 4°C. Wells were washed once with PBS, blocked with 2% milk (Bio-Rad) in PBS for 1 h at room temperature, and washed three times with PBS plus 0.05% Tween 20. NCI05 and NCI09 standard curves and plasma from passively immunized animals were prepared in PBS using 1:2 dilutions. Fifty microliters of samples and standards was applied to each well and incubated overnight at 4°C on a rotating platform. Wells were washed five times with PBS plus 0.05% Tween 20, and 50 μL of 1:10,000 anti-monkey IgG conjugated to horseradish peroxidase (HRP; Abcam) was dispensed into the wells and incubated for 1 h with shaking at room temperature. Wells were washed five times with PBS plus 0.05% Tween 20. Wells were incubated with 50 μL of ELISA TMB (3,3′,5,5′-tetramethylbenzidine) substrate (Thermo Fisher Scientific) for 10 min and neutralized with TMB stop solution (KPL), and absorbance at 450 nm was measured with a Victor X4 plate reader (Perkin Elmer).
(ii) Rectal secretions.
Weck-cel sponges containing rectal secretions were obtained weekly beginning at 2 days after the first passive immunization. Swabs were physically inspected for blood, and all samples containing blood were discarded. Swabs were thawed on ice, transferred to a prechilled Ultrafree column containing a 5.0-μm polyvinylidene difluoride (PVDF) membrane, incubated with 300 μL of ice-cold extraction buffer containing 0.25% bovine serum albumin (BSA) (Sigma), protease inhibitor (Sigma), and 1× PBS (Invitrogen) for 10 min on ice, and centrifuged at 16,000 × g for 5 min at 4°C. The incubation and centrifugation steps were repeated for a total of two washes with a final centrifugation of 16,000 × g for 20 min at 4°C. The solution was then applied to an Amicon Ultra 30-kDa 0.5-mL centrifugal filter (Millipore) and concentrated to 400 μL by centrifuging at 14,000 × g. Samples were frozen at −80°C prior to ELISA. High-affinity Immulon 2 HB plates were coated overnight with 100 ng of SIVmac251-M766 gp120ΔV1 at 4°C. Wells were washed once with PBS, blocked with 2% BSA (KPL) in PBS for 2 h, and washed three times with PBS plus 0.05% Tween 20. Standard curves for NCI05 and NCI09 were prepared in using 1:2 dilutions in PBS ranging from 50 ng/mL to 0.95 ng/mL. Negative rectal swab controls from control animals were diluted 1:10 in 1% BSA to establish background signal and rectal samples from passively immunized animals were diluted 1:2 and 1:10 in 1% BSA. Fifty microliters of samples and standards was applied to each well and incubated overnight at 4°C on a rotating platform. Wells were washed five times with PBS plus 0.05% Tween 20, and 50 μL of 1:10,000 anti-monkey IgG conjugated to HRP (Abcam) was dispensed into the wells and incubated for 1 h with shaking at room temperature. Wells were washed five times with PBS plus 0.05% Tween 20. Wells were incubated with 50 μL of 1-Step Ultra TMB ELISA (Thermo Fisher Scientific) for 10 min and neutralized with TMB stop solution (KPL), and absorbance at 450 nm was measured with a Victor X4 plate reader (Perkin Elmer).
V2 peptide ELISA. (i) NCI05 and NCI09 reactivity to V2 peptides 115 and 150.
Briefly, streptavidin-coated plates were incubated for 3 h at room temperature (RT) in wash buffer (Tris-buffered saline [TBS], 0.1% BSA, 0.05% Tween 20) with the biotinylated peptides (0.1 μg/mL), followed by overnight incubation at 4°C with NCI05, NCI09, and ITS12 in serial dilutions. Plates were then incubated for 2 h at RT with goat anti-monkey IgG conjugated with alkaline phosphatase at 0.5 μg/mL. Plates were incubated with alkaline phosphatase substrate in developing buffer (PBS, 1 M diethanolamine (DEA), 0.24 M MgCl2·6H2O [pH 9.8]), and the optical density at 405 nm (OD405) was read.
(ii) Macaque plasma reactivity with V2 peptides 115 and 150.
Streptavidin plates (Thermo Fisher Scientific) were coated with 100 ng/well of biotinylated peptides 115 and 150 with shaking for 2 h at room temperature. Plates were washed three times with 1× TBS–0.1% BSA–0.05% Tween 20 wash buffer. Plasma samples obtained weekly beginning prior to passive immunization and weekly beginning 4 days after the first passive immunization were serially diluted in wash buffer and added to the wells along with NCI05 and NCI09 standard curves. Peptide-coated plates were incubated with plasma for 30 min with shaking at room temperature and washed three times with wash buffer. Plates were incubated with goat anti-monkey IgG conjugated to HRP (Abcam) diluted 1:10,000 in wash buffer for 30 min at room temperature and washed three times with wash buffer. Plates were incubated with 1-Step Ultra TMB ELISA substrate solution, neutralized with TMB stop solution (KPL), and read at 450 nm.
Surface Env staining.
CEM.NKR-CCR5-sLTR-Luc cells (2 × 106 cells) were infected with replication-competent SIVsmE660-FL14 (600 ng p27), SIVmac251-M766r (1,500 ng p27), and uncloned SIVmac251 (80 ng p27) or with vesicular stomatitis virus G protein (VSV G)-transcomplemented, vif deletion-containing SIVmac239 (40 ng) in the presence of 40 μg/mL Polybrene and centrifuged for 1.5 h at 1,200 × g. Antibody binding to Env was assessed by staining 4 to 6 days postinfection. Cells were treated with a Live/Dead NEAR IR viability dye (Invitrogen), washed in PBS containing 1% fetal bovine serum (FBS; staining buffer), and stained at room-temperature for 30 min with 5 μg/mL of Env-specific antibody or with a dengue virus-specific antibody (DEN3) as a negative control. For soluble CD4 (sCD4) experiments, cells were stained on ice for 30 min with anti-SIV Env (5 μg/mL) antibody and sCD4-183 (10 μg/mL; NIH AIDS Reagent Program). Cells were then washed and stained on ice with AF647-conjugated goat anti-human F(ab′)2 (Jackson Immunoresearch), followed by phycoerythrin (PE)-Cy7-conjugated anti-CD4 (clone OKT4; Biolegend). For intracellular SIV p27 Gag staining, cells were fixed in PBS with 2% paraformaldehyde, washed in staining buffer, and stained with AF488-conjugated anti-SIV Gag antibody (clone 552F12) in Perm/Fix Medium B (Invitrogen). Cells were washed, fixed in PBS with 2% paraformaldehyde, and analyzed on a BD FACSymphony flow cytometer. Antibody binding was assessed by Env staining on the surface of SIV-infected (CD4lowGag+) cells.
ADCC assay against SIV-infected cells.
ADCC was measured as described previously described (34, 35). CEM.NKR-CCR5-sLTR-Luc cells were infected with replication-competent SIVsmE660-FL14 (600 ng p27), SIVmac251-M766r (1,500 ng p27) and uncloned SIVmac251 (80 ng p27) or with VSV G-transcomplemented, vif deletion-containing SIVmac239 (40 ng) in the presence of 40 μg/mL Polybrene and centrifuged for 1.5 h at 1,200 × g. After 4 to 6 days of infection, the CEM.NKR-CCR5-sLTR-Luc cells were incubated for 8 h with an NK cell line (KHYG-1 cells) transduced with rhesus macaque CD16 at a 10:1 effector-to-target (E/T) ratio in the presence of antibodies. CD16+ KHYG-1 cells (105 cells/well) were incubated with CEM.NKR-CCR5-sLTR-Luc cells (104 cells/well) in triplicate wells of 96-well plates (0.2 mL/well). The assay was performed with or without sCD4-183 (10 μg/mL; NIH AIDS Reagent Program). ADCC was determined from the dose-dependent loss of luciferase activity. ADCC responses were calculated as the remaining luciferase activity (percent relative light units [RLU]) by dividing the difference in RLU between SIV-infected cells in the presence of antibody and uninfected cells without antibody (experimental − background) by the difference in RLU between SIV-infected cells and uninfected cells in the absence of antibody (maximal − background) and multiplying by 100.
ADCC assay with human PBMCs as effector and gp120-coated target cells.
ADCC activity using plasma of infected animals was assessed as previously described using EGFP-CEM.NKR-CCR5-SNAP cells, which constitutively express enhanced green fluorescent protein (EGFP), as targets (36). Briefly, one million target cells were incubated with 50 μg of ΔV1 gp120 protein for 2 h at 37°C. The coated target cells were washed and labeled with SNAP-Surface Alexa Fluor 647 (New England Biolabs) as recommended by the manufacturer for 30 min at RT. Plasma samples, heat inactivated at 56°C for 30 min, were serially diluted (seven 10-fold dilutions starting at 1:10), and 100 μL was added to wells of a 96-well V-bottom plate (Millipore Sigma). Target cells (5,000; 50 μL) and human PBMCs (250,000; 50 μL) as effectors were added to each well to give an E/T ratio of 50:1. The plate was incubated at 37°C for 2 h followed by two PBS washes. The cells were resuspended in 200 μL of a 2% PBS–paraformaldehyde solution and acquired on an LSRII flow cytometer equipped with a high-throughput system (BD Biosciences). Specific killing was measured by loss of GFP from the SNAP-Surface Alexa Fluor 647-positive target cells. Target and effector cells cultured in the presence of R10 medium were used as background. NCI05 and NCI09 monoclonal antibodies were used as a positive control. Normalized percent killing was calculated as (killing in the presence of plasma − background)/(killing in the presence of positive control − background) × 100. The ADCC endpoint titer is defined as the reciprocal dilution at which the percent ADCC killing was greater than the mean percent killing of the background wells containing medium, target cells, and effector cells, plus three standard deviations.
Antibody-dependent cellular cytotoxicity assay using macaque NKG2A+ cells and gp120-coated cells.
NKG2A+ cells were isolated from cryopreserved PBMCs (10 × 106 cells) collected at baseline by using PE-conjugated CD159a antibody (no. IM3291U; Beckman Coulter) and anti-PE MicroBeads (Miltenyi Biotec). Briefly, CD14+ cells were removed, and the CD14− PBMCs were washed with 3 mL buffer and centrifuged at 2,000 rpm for 7 min. Pellets were resuspended in 100 μL buffer and 15 μL PE-conjugated CD159a antibody and incubated at 4°C for 15 min. At the end of the incubation, cells were washed with 3 mL buffer and incubated with 20 μL anti-PE microbeads and 80 μL buffer at 4°C for 15 min. Next, cells were washed with 3 mL buffer and resuspended in 500 μL of buffer. Positive selection was performed using an AutoMACSpro instrument (Miltenyi Biotec) following the Possel program. At the end of the separation, cells were counted and used for the assay. ADCC activity using NK effector cells from the animals was assessed using EGFP-CEM.NKR-CCR5-SNAP cells, which constitutively express EGFP, as targets. Briefly, one million target cells were incubated with 50 μg of ΔV1 GP120 protein for 2 h at 37°C. The coated target cells were washed and labeled with SNAP-Surface Alexa Fluor 647 (New England Biolabs) as recommended by the manufacturer for 30 min at RT. One hundred microliters of NCI05 or NCI09 antibodies was added at a 5-μg/mL concentration for the NCI05 or NCI09 passively immunized group into the wells of a 96-well V-bottom plate (Millipore Sigma). Target cells (5,000; 50 μL) and macaque NK cells (10,000; 50 μL) as effectors were added to each well to give an E/T ratio of 2:1. The plate was incubated at 37°C for 2 h followed by two PBS washes. The cells were resuspended in 200 μL of a 2% PBS–paraformaldehyde solution and acquired on a Symphony instrument equipped with a high-throughput system (BD Biosciences). Specific killing was measured by loss of GFP from the SNAP-Surface Alexa Fluor 647-positive target cells. Target and effector cells cultured in the presence of R10 medium were used as background. NCI05 and NCI09 monoclonal antibodies were used as a positive control. Normalized percent killing was calculated as (killing in the presence of plasma − background)/(killing in the presence of positive control − background) × 100.
Efferocytosis assay.
The frequency of CD14+ efferocytes in PBMCs was assessed with an efferocytosis assay kit (number 601770; Cayman Chemical Company). CD14+ cells were used as effectors, and apoptotic neutrophils as target cells. Ex vivo CD14+ monocyte cells rather than differentiated macrophages were used in the assay due to low cell availability. One animal in the NCI05 group was not tested due to lack of available PBMCs.
(i) Effector cells. CD14+ cells were isolated from cryopreserved PBMCs (10 × 106 cells) collected at baseline by using nonhuman primate CD14 MicroBeads (number 130-091-097; Miltenyi Biotec) and following the manufacturer’s instructions. Briefly, 10 × 106 PBMCs were thawed and incubated with 20 μL microbeads and 80 μL buffer at 4°C for 15 min. At the end of the incubation, cells were washed with 3 mL buffer and resuspended in 500 μL of buffer. Positive selection was performed using the AutoMACSpro instrument (Miltenyi Biotec) following the Possel program. At the end of the separation, cells were counted and stained with CytoTell Blue provided in the kit and following the manufacturer’s instructions. Briefly, cells were resuspended in buffer (107 cells/mL), an equal volume of buffer containing 2× CytoTell Blue (stock diluted 1:200 in buffer) was added, incubated at 37°C for 30 min, and washed three times with R10, resuspended in R10, and used for the efferocytosis assay.
(ii) Target cells. Two unrelated macaques were used as source of neutrophils as target cells. Neutrophils were isolated as previously described (37). Briefly, following isolation of PBMCs by Ficoll-Paque (GE Healthcare), an equal volume of a solution of 20% dextran in water was added to cellular pellets, gently mixed, incubated for 1 min followed by the addition of approximately 3 volumes of PBS, mixed again, and incubated in the dark for 50 to 60 min. At the end of incubation, the clear layers at the top of the tubes containing neutrophils were collected, washed with PBS, and centrifuged at 2,000 rpm for 10 min. Cells were treated with ACK lysing buffer (Quality Biological) for 5 min at 37°C, washed with R10, and counted. Neutrophils were stained with carboxyfluorescein succinimidyl ester (CFSE) provided in the kit and following the manufacturer’s instructions. Briefly, neutrophils were resuspended in buffer (107 cells/mL), and an equal volume of buffer containing 2× CFSE (stock diluted 1:200 in buffer) was added to cells, incubated at 37°C for 30 min, and washed three times with R10. The apoptosis of neutrophils was induced by treatment with the staurosporine apoptosis inducer provided in the kit. Briefly, isolated cells were resuspended in R10 containing staurosporine (stock diluted 1:1,000) and incubated at 37°C for 3 h. At the end of incubation, cells were washed twice with R10 and used for the efferocytosis assay. Apoptotic neutrophils from 2 animals were mixed before the coculture and used for the subsequent step.
Effector and apoptotic target cells were cultured alone (as controls) or cocultured at a ratio of one effector CD14+ cell to three target apoptotic neutrophils. Cells were incubated in an incubator at 37°C for 12 h.
(iii) CD14+ cells. At the end of the coculture, cells were washed with PBS, fixed with 1% paraformaldehyde in PBS, and acquired with a flow cytometer. Flow cytometry acquisitions were performed on a FACSymphony A5 instrument and examined using FACSDiva software (BD Biosciences) by acquiring all stained cells. Data were further analyzed using FlowJo v10.1 (TreeStar). The frequency of CD14+ efferocytes was determined as the frequency of positive cells for CytoTell Blue and CFSE on the single-cell gate (gating strategy: forward scatter [FSC]/side scatter [SSC]/single cells/CytoTell Blue+/CFSE+).
α4β7 binding assay.
The binding of α4β7 expressed by the RPMI8866 cell line to SIVmac766 gp120 in the absence or presence of SIV V2 MAbs was assessed by an adhesion assay (37). This assay was modified by culturing RPMI8866 cells in medium containing 1 μM retinoic acid (RA) for at least 7 days prior to use in adhesion assays. Inclusion of RA increases adhesion to gp120. Briefly, triplicate wells of a 96-well flat-bottom polypropylene plate (Greiner Bio-One) were coated overnight at 4°C with 2.0 μg of peptide-N-glycosidase (PNGase)-treated SIV gp120 diluted in 50 mM bicarbonate buffer, pH 9.6. The solution from the plates was discarded, and the plates were then blocked with blocking buffer (25 mM Tris, 2.7 mM potassium chloride, 150 mM sodium chloride, 0.5% BSA, 4 mM manganese chloride [pH 7.2]) for 1 h at 37°C. The solution was discarded, and plates were manually washed 4 times with blocking buffer. After blocking and washing, plates were incubated with 20 to 100 μg/mL of the designated anti-V2 MAbs in sample buffer (25 mM Tris, 2.7 mM KCl, 150 mM NaCl, 4 mM manganese chloride, 1% FBS [pH 7.2]) for 1 h at 37°C. RPMI8866 cells in a volume of 50 μL/well were preincubated for 40 min at 37°C with sample buffer in the absence or presence of 10 μg/mL of 2B4 (α4 MAb). Plates were then incubated with 50 μL/well of 2 × 105 RPMI8866 cells at 37°C (5% CO2) for 1 h and washed 5 times with PBS, followed by the addition of 100 μL of RPMI 1640 containing 1% FBS, 1% penicillin-streptomycin-glutamine and 25 mM HEPES with 10 μL/well of alamarBlue dye. Fluorescence (excitation, 560 nm, and emission, 590 nm) was measured immediately after the addition of the alamarBlue dye for 8 h.
T-cell costimulation assay.
CD4+ T-cell activation assays were carried out as previously described (15). Briefly, 96-well flat-bottom cell culture-treated plates were precoated 2 h at 4°C with 50 ng of anti-CD3 (clone OKT3; eBioscience) and followed by 200 ng of neutravidin in a total volume of 100 μL HEPES-buffered saline (HBS) overnight at 4°C. On stimulation day, neutravidin plates were coated with biotinylated SIVmac251-M766 gp120 at 37°C for 1 h. Antibodies to gp120 were added to coated plates for 1 h at 37°C before 200,000 freshly isolated CD4+ T cells in 100 μL complete RPMI (10% FBS) were added. In certain wells, retinoic acid (Sigma-Aldrich) was also added at a concentration of 10 nM. For activation assays, cells were stained for the activation markers CD25 and Ki67 on day 4. Flow cytometry analysis was carried out with FlowJo software.
On day 4 poststimulation, CD4+ T cells were harvested and stained for flow-cytometric analysis. Cells were stained with Live/Dead Aqua (Invitrogen). The following antibodies were used and purchased from BD Pharmingen: anti-CD25 FITC (clone M-A251), anti-Ki67 Alexa Fluor 647 (clone B56), anti-integrin β7 PE (clone FIB504), anti-CD45RO APC-H7 (clone UCHL1), and anti-CCR5 BV421 (clone 2D7/CCR5). For intracellular Ki67 staining, a BD Cytofix/Cytoperm kit was used following the manufacturer’s recommendations. Data were collected on a FACSCanto II (BD Biosciences) and analyzed using FlowJo and GraphPad Prism software.
Surface plasmon resonance spectroscopy.
The kinetics of anti-V2-loop monoclonal antibody binding to SIVmac251-M766 gp120 was assessed by surface plasmon resonance spectroscopy. The gp120 was immobilized to a CM5 biosensor surface (Cytiva) using amine-coupling chemistry to a surface density of ~750 resonance units. Twofold serial dilutions of the indicated antibodies were passed sequentially over the sensor surface for 2 min at 25 μL/min in HEPES-buffered saline supplemented with 3 mM EDTA, 0.005% Tween 20, and 0.05% soluble carboxymethyl dextran, followed by a 2-min dissociation phase using a Biacore 3000 biosensor (Cytiva). The surfaces were regenerated after each cycle by two brief injections of 4.5 M MgCl2.
Crystallization and structure determination.
The V2Cmut peptide was dissolved in 100% dimethyl sulfoxide (DMSO) at 10 mg/mL. NCI05 Fab in 1× PBS was concentrated to 20 mg/mL in Amicon Ultra centrifugal filters (30-kDa molecular weight cutoff; Millipore Sigma) and incubated with V2Cmut peptide at a 1:2 molar ratio for 1 h at RT. The NCI05-V2Cmut complex was purified with a Superdex 10/300 GL size exclusion column in a buffer containing 150 mM NaCl, 5 mM HEPES and concentrated to 12 mg/mL. Crystals of NCI05-V2Cmut complex were screened using 576 conditions in Crystal Screen (Hampton Research) and Wizard Precipitant Synergy (Qiagen) screening kits with a Mosquito crystallization robot (SPT Labtech) by mixing 100 nL reservoir solution and 100 nL protein solution per drop. Crystal hits were obtained in a reservoir solution containing 0.2 M NaCl, 28.5% polyethylene glycol 8000, and 0.1 M phosphate-citrate (pH 4.2). Conditions were hand-optimized in 15-well plates in 2-μL drops to obtain diffraction quality crystals.
X-ray diffraction data were collected at the Advanced Photon Source (beamline SER-CAT ID22). A data set at a resolution of 2.35 Å was obtained with 30% ethylene glycol as a cryoprotectant. The diffraction data were indexed, integrated, and scaled with the HKL2000 package (38). The structures were determined by molecular replacement with Phaser (39) in the CCP4 program suite (40) with AlphaFold2-generated NCI05 Fab as the initial search model. Further refinement was carried out with PHENIX (41), starting with torsion angle simulated annealing with slow cooling. Iterative manual model building was carried out with COOT (42) with maps generated from combinations of standard positional, individual B-factor, and translation-libration-screw-rotation (TLS) model refinement algorithms. X-ray data and refinement statistics are summarized in Table S1.
Statistical analysis.
Statistics were calculated using GraphPad Prism. The Wilcoxon signed-rank test was used to compare paired measurements within individual animals and cell lines. The Mann-Whitney test was used to compare unpaired measurements between separate groups of animals. Spearman rank correlations were used to infer linear relationships between measured variables.
Data availability.
The coordinates and structural factors for antibody NCI05 in complex with peptide V2Cmut have been deposited in the Protein Data Bank (PDB ID 8FBW).
ACKNOWLEDGMENTS
We thank Matthew W. Breed and Joshua Kramer (Leidos Biomedical Research, NCI) for their contributions to animal care and technical procedures, Irene Kalisz (Advanced BioScience Laboratories, Inc.) for producing the monoclonal antibodies used here, and Brandon F. Keele (NCI) for sequencing the transmitted founder virus variants.
This work was supported by the Intramural Research program at the National Cancer Institute and the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, NIH, and by a cooperative agreement (WW81XWH-18-2-0040) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the U.S. Department of Defense (DOD). X-ray diffraction data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. SER-CAT is supported by its member institutions, and equipment grants (S10_RR25528, S10_RR028976 and S10_OD027000) from the National Institutes of Health. Use of sector 22 (Southeast Region Collaborative Access team) at the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract number W-31-109-Eng-38.
The views expressed are those of the authors and should not be construed to represent the positions of the NIH, the U.S. Army, the Department of Defense, or the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc.
J.D.S. and G.F. designed the study and wrote the manuscript with T.C. and T.Z. J.D.S. and I.S.D.C. conducted the passive immunization studies. J.D.S. performed all ELISA assays detecting V2 MAbs in plasma and rectal secretions. J.D.S. and A.G. processed rectal secretions. G.G. performed ELISA experiments suggesting recognition of a conformational epitope by NCI05 and cloned NCI05 and NCI09 with R.M. and M. Roederer. J.A. and C.C. performed SPR, α4β7, and CD4+ T-cell costimulation assays. T.C. and M.B.-F. designed coil/helical peptide 150 specific for NCI05. T.Z., D.J.V.W., and P.D.K. performed structural study. M.A.R. performed ADCC assays with gp120-coated human target cells, and N.C. and D.T.E. performed ADCC assays with SIV-infected cells in the presence of CD4. K.N. and D.P.-P. performed ADCP and trogocytosis assays. M.B. performed the efferocytosis assay. J.K. and M. Rao conducted virion capture assays and immunological assays.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Genoveffa Franchini, Email: franchig@mail.nih.gov.
Guido Silvestri, Emory University.
REFERENCES
- 1.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. MOPH-TAVEG Investigators. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220. 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 2.Robb ML, Rerks-Ngarm S, Nitayaphan S, Pitisuttithum P, Kaewkungwal J, Kunasol P, Khamboonruang C, Thongcharoen P, Morgan P, Benenson M, Paris RM, Chiu J, Adams E, Francis D, Gurunathan S, Tartaglia J, Gilbert P, Stablein D, Michael NL, Kim JH. 2012. Risk behaviour and time as covariates for efficacy of the HIV vaccine regimen ALVAC-HIV (vCP1521) and AIDSVAX B/E: a post-hoc analysis of the Thai phase 3 efficacy trial RV 144. Lancet Infect Dis 12:531–537. 10.1016/S1473-3099(12)70088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366:1275–1286. 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin L, Finak G, Ushey K, Seshadri C, Hawn TR, Frahm N, Scriba TJ, Mahomed H, Hanekom W, Bart P-A, Pantaleo G, Tomaras GD, Rerks-Ngarm S, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Michael NL, Kim JH, Robb ML, O'Connell RJ, Karasavvas N, Gilbert P, C De Rosa S, McElrath MJ, Gottardo R. 2015. COMPASS identifies T-cell subsets correlated with clinical outcomes. Nat Biotechnol 33:610–616. 10.1038/nbt.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gottardo R, Bailer RT, Korber BT, Gnanakaran S, Phillips J, Shen X, Tomaras GD, Turk E, Imholte G, Eckler L, Wenschuh H, Zerweck J, Greene K, Gao H, Berman PW, Francis D, Sinangil F, Lee C, Nitayaphan S, Rerks-Ngarm S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Robb ML, Michael NL, Kim JH, Zolla-Pazner S, Haynes BF, Mascola JR, Self S, Gilbert P, Montefiori DC. 2013. Plasma IgG to linear epitopes in the V2 and V3 regions of HIV-1 gp120 correlate with a reduced risk of infection in the RV144 vaccine efficacy trial. PLoS One 8:e75665. 10.1371/journal.pone.0075665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karasavvas N, Billings E, Rao M, Williams C, Zolla-Pazner S, Bailer RT, Koup RA, Madnote S, Arworn D, Shen X, Tomaras GD, Currier JR, Jiang M, Magaret C, Andrews C, Gottardo R, Gilbert P, Cardozo TJ, Rerks-Ngarm S, Nitayaphan S, Pitisuttithum P, Kaewkungwal J, Paris R, Greene K, Gao H, Gurunathan S, Tartaglia J, Sinangil F, Korber BT, Montefiori DC, Mascola JR, Robb ML, Haynes BF, Ngauy V, Michael NL, Kim JH, de Souza MS. MOPH-TAVEG Collaboration. 2012. The Thai phase III HIV type 1 vaccine trial (RV144) regimen induces antibodies that target conserved regions within the V2 loop of gp120. AIDS Res Hum Retroviruses 28:1444–1457. 10.1089/aid.2012.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, deCamp AC, Carrico C, Menis S, Magaret CA, Ahmed H, Juraska M, Chen L, Konopa P, Nariya S, Stoddard JN, Wong K, Zhao H, Deng W, Maust BS, Bose M, Howell S, Bates A, Lazzaro M, O'Sullivan A, Lei E, Bradfield A, Ibitamuno G, Assawadarachai V, O'Connell RJ, deSouza MS, Nitayaphan S, Rerks-Ngarm S, Robb ML, McLellan JS, Georgiev I, Kwong PD, Carlson JM, Michael NL, Schief WR, Gilbert PB, Mullins JI, Kim JH. 2012. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature 490:417–420. 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan R, Gorny MK, Zolla-Pazner S, Kong XP. 2015. The V1V2 region of HIV-1 gp120 forms a five-stranded beta barrel. J Virol 89:8003–8010. 10.1128/JVI.00754-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Cohen AA, Galimidi RP, Gristick HB, Jensen GJ, Bjorkman PJ. 2016. Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop. Proc Natl Acad Sci USA 113:E7151–E7158. 10.1073/pnas.1615939113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tassaneetrithep B, Tivon D, Swetnam J, Karasavvas N, Michael NL, Kim JH, Marovich M, Cardozo T. 2014. Cryptic determinant of alpha4beta7 binding in the V2 loop of HIV-1 gp120. PLoS One 9:e108446. 10.1371/journal.pone.0108446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA, McLaughlin S, Pascuccio M, Gopaul R, McNally J, Cruz CC, Censoplano N, Chung E, Reitano KN, Kottilil S, Goode DJ, Fauci AS. 2008. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol 9:301–309. 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 12.Lertjuthaporn S, Cicala C, Van Ryk D, Liu M, Yolitz J, Wei D, Nawaz F, Doyle A, Horowitch B, Park C, Lu S, Lou Y, Wang S, Pan R, Jiang X, Villinger F, Byrareddy SN, Santangelo PJ, Morris L, Wibmer CK, Biris K, Mason RD, Gorman J, Hiatt J, Martinelli E, Roederer M, Fujikawa D, Gorini G, Franchini G, Arakelyan A, Ansari AA, Pattanapanyasat K, Kong XP, Fauci AS, Arthos J. 2018. Select gp120 V2 domain specific antibodies derived from HIV and SIV infection and vaccination inhibit gp120 binding to alpha4beta7. PLoS Pathog 14:e1007278. 10.1371/journal.ppat.1007278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, Blazar BR, Rodriguez B, Teixeira-Johnson L, Landay A, Martin JN, Hecht FM, Picker LJ, Lederman MM, Deeks SG, Douek DC. 2006. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 12:1365–1371. 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 14.Guzzo C, Ichikawa D, Park C, Phillips D, Liu Q, Zhang P, Kwon A, Miao H, Lu J, Rehm C, Arthos J, Cicala C, Cohen MS, Fauci AS, Kehrl JH, Lusso P. 2017. Virion incorporation of integrin alpha4beta7 facilitates HIV-1 infection and intestinal homing. Sci Immunol 2:eaam7341. 10.1126/sciimmunol.aam7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goes LR, Sajani A, Sivro A, Olowojesiku R, Ray JC, Perrone I, Yolitz J, Girard A, Leyre L, Wibmer CK, Morris L, Gorini G, Franchini G, Mason RD, Roederer M, Mehandru S, Soares MA, Cicala C, Fauci AS, Arthos J. 2020. The V2 loop of HIV gp120 delivers costimulatory signals to CD4(+) T cells through integrin alpha4beta7 and promotes cellular activation and infection. Proc Natl Acad Sci USA 117:32566–32573. 10.1073/pnas.2011501117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaccari M, Gordon SN, Fourati S, Schifanella L, Liyanage NPM, Cameron M, Keele BF, Shen X, Tomaras GD, Billings E, Rao M, Chung AW, Dowell KG, Bailey-Kellogg C, Brown EP, Ackerman ME, Vargas-Inchaustegui DA, Whitney S, Doster MN, Binello N, Pegu P, Montefiori DC, Foulds K, Quinn DS, Donaldson M, Liang F, Loré K, Roederer M, Koup RA, McDermott A, Ma Z-M, Miller CJ, Phan TB, Forthal DN, Blackburn M, Caccuri F, Bissa M, Ferrari G, Kalyanaraman V, Ferrari MG, Thompson D, Robert-Guroff M, Ratto-Kim S, Kim JH, Michael NL, Phogat S, Barnett SW, Tartaglia J, Venzon D, Stablein DM, et al. 2016. Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. Nat Med 22:762–770. 10.1038/nm.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaccari M, Fourati S, Gordon SN, Brown DR, Bissa M, Schifanella L, Silva de Castro I, Doster MN, Galli V, Omsland M, Fujikawa D, Gorini G, Liyanage NPM, Trinh HV, McKinnon KM, Foulds KE, Keele BF, Roederer M, Koup RA, Shen X, Tomaras GD, Wong MP, Munoz KJ, Gach JS, Forthal DN, Montefiori DC, Venzon DJ, Felber BK, Rosati M, Pavlakis GN, Rao M, Sekaly RP, Franchini G. 2018. HIV vaccine candidate activation of hypoxia and the inflammasome in CD14(+) monocytes is associated with a decreased risk of SIVmac251 acquisition. Nat Med 24:847–856. 10.1038/s41591-018-0025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silva de Castro I, Gorini G, Mason R, Gorman J, Bissa M, Rahman MA, Arakelyan A, Kalisz I, Whitney S, Becerra-Flores M, Ni E, Peachman K, Trinh HV, Read M, Liu MH, Van Ryk D, Paquin-Proulx D, Shubin Z, Tuyishime M, Peele J, Ahmadi MS, Verardi R, Hill J, Beddall M, Nguyen R, Stamos JD, Fujikawa D, Min S, Schifanella L, Vaccari M, Galli V, Doster MN, Liyanage NPM, Sarkis S, Caccuri F, LaBranche C, Montefiori DC, Tomaras GD, Shen X, Rosati M, Felber BK, Pavlakis GN, Venzon DJ, Magnanelli W, Breed M, Kramer J, Keele BF, Eller MA, Cicala C, Arthos J, et al. 2021. Anti-V2 antibodies virus vulnerability revealed by envelope V1 deletion in HIV vaccine candidates. iScience 24:102047. 10.1016/j.isci.2021.102047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorini G, Fourati S, Vaccari M, Rahman MA, Gordon SN, Brown DR, Law L, Chang J, Green R, Barrenas F, Liyanage NPM, Doster MN, Schifanella L, Bissa M, Silva de Castro I, Washington-Parks R, Galli V, Fuller DH, Santra S, Agy M, Pal R, Palermo RE, Tomaras GD, Shen X, LaBranche CC, Montefiori DC, Venzon DJ, Trinh HV, Rao M, Gale M, Jr, Sekaly RP, Franchini G. 2020. Engagement of monocytes, NK cells, and CD4+ Th1 cells by ALVAC-SIV vaccination results in a decreased risk of SIVmac251 vaginal acquisition. PLoS Pathog 16:e1008377. 10.1371/journal.ppat.1008377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bissa M, Kim S, Galli V, Fourati S, Sarkis S, Arakelyan A, de Castro IS, Rahman MA, Fujiwara S, Vaccari M, Tomalka JA, Stamos JD, Schifanella L, Gorini G, Moles R, Gutowska A, Ferrari G, Lobanov A, Montefiori DC, Nelson GW, Cam MC, Chakhtoura M, Haddad EK, Doster MN, McKinnon K, Brown S, Venzon DJ, Choo-Wosoba H, Breed MW, Killoran KE, Kramer J, Margolis L, Sekaly RP, Hager GL, Franchini G. 2023. HIV vaccine candidate efficacy in female macaques mediated by cAMP-dependent efferocytosis and V2-specific ADCC. Nat Commun 14:575. 10.1038/s41467-023-36109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorman J, Wang C, Mason RD, Nazzari AF, Welles HC, Zhou T, Bess JW, Jr, Bylund T, Lee M, Tsybovsky Y, Verardi R, Wang S, Yang Y, Zhang B, Rawi R, Keele BF, Lifson JD, Liu J, Roederer M, Kwong PD. 2022. Cryo-EM structures of prefusion SIV envelope trimer. Nat Struct Mol Biol 29:1080–1091. 10.1038/s41594-022-00852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Bredow B, Andrabi R, Grunst M, Grandea AG, III, Le K, Song G, Berndsen ZT, Porter K, Pallesen J, Ward AB, Burton DR, Evans DT. 2019. Differences in the binding affinity of an HIV-1 V2 apex-specific antibody for the SIVsmm/mac envelope glycoprotein uncouple antibody-dependent cellular cytotoxicity from neutralization. mBio 10:e01255-19. 10.1128/mBio.01255-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Rompay KK, Berardi CJ, Dillard-Telm S, Tarara RP, Canfield DR, Valverde CR, Montefiori DC, Cole KS, Montelaro RC, Miller CJ, Marthas ML. 1998. Passive immunization of newborn rhesus macaques prevents oral simian immunodeficiency virus infection. J Infect Dis 177:1247–1259. 10.1086/515270. [DOI] [PubMed] [Google Scholar]
- 24.Florese RH, Van Rompay KK, Aldrich K, Forthal DN, Landucci G, Mahalanabis M, Haigwood N, Venzon D, Kalyanaraman VS, Marthas ML, Robert-Guroff M. 2006. Evaluation of passively transferred, nonneutralizing antibody-dependent cellular cytotoxicity-mediating IgG in protection of neonatal rhesus macaques against oral SIVmac251 challenge. J Immunol 177:4028–4036. 10.4049/jimmunol.177.6.4028. [DOI] [PubMed] [Google Scholar]
- 25.Dugast AS, Chan Y, Hoffner M, Licht A, Nkolola J, Li H, Streeck H, Suscovich TJ, Ghebremichael M, Ackerman ME, Barouch DH, Alter G. 2014. Lack of protection following passive transfer of polyclonal highly functional low-dose non-neutralizing antibodies. PLoS One 9:e97229. 10.1371/journal.pone.0097229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Julg B, Tartaglia LJ, Keele BF, Wagh K, Pegu A, Sok D, Abbink P, Schmidt SD, Wang K, Chen X, Joyce MG, Georgiev IS, Choe M, Kwong PD, Doria-Rose NA, Le K, Louder MK, Bailer RT, Moore PL, Korber B, Seaman MS, Abdool Karim SS, Morris L, Koup RA, Mascola JR, Burton DR, Barouch DH. 2017. Broadly neutralizing antibodies targeting the HIV-1 envelope V2 apex confer protection against a clade C SHIV challenge. Sci Transl Med 9:eaal1321. 10.1126/scitranslmed.aal1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Julg B, Liu PT, Wagh K, Fischer WM, Abbink P, Mercado NB, Whitney JB, Nkolola JP, McMahan K, Tartaglia LJ, Borducchi EN, Khatiwada S, Kamath M, LeSuer JA, Seaman MS, Schmidt SD, Mascola JR, Burton DR, Korber BT, Barouch DH. 2017. Protection against a mixed SHIV challenge by a broadly neutralizing antibody cocktail. Sci Transl Med 9:eaao4235. 10.1126/scitranslmed.aao4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hessell AJ, Shapiro MB, Powell R, Malherbe DC, McBurney SP, Pandey S, Cheever T, Sutton WF, Kahl C, Park B, Zolla-Pazner S, Haigwood NL. 2018. Reduced cell-associated DNA and improved viral control in macaques following passive transfer of a single anti-V2 monoclonal antibody and repeated simian/human immunodeficiency virus challenges. J Virol 92:e02198-17. 10.1128/JVI.02198-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alter G, Yu WH, Chandrashekar A, Borducchi EN, Ghneim K, Sharma A, Nedellec R, McKenney KR, Linde C, Broge T, Suscovich TJ, Linnekin T, Abbink P, Mercado NB, Nkolola JP, McMahan K, Bondzie EA, Hamza V, Peter L, Kordana N, Mahrokhian S, Seaman MS, Li W, Lewis MG, Lauffenburger DA, Hangartner L, Sekaly RP, Barouch DH. 2020. Passive transfer of vaccine-elicited antibodies protects against SIV in rhesus macaques. Cell 183:185–196.E14. 10.1016/j.cell.2020.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao S, Zhang L, Xiang S, Hu Y, Wu Z, Shen J. 2022. Gnawing between cells and cells in the immune system: friend or foe? A review of trogocytosis. Front Immunol 13:791006. 10.3389/fimmu.2022.791006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richard J, Pacheco B, Gohain N, Veillette M, Ding S, Alsahafi N, Tolbert WD, Prevost J, Chapleau JP, Coutu M, Jia M, Brassard N, Park J, Courter JR, Melillo B, Martin L, Tremblay C, Hahn BH, Kaufmann DE, Wu X, Smith AB, III, Sodroski J, Pazgier M, Finzi A. 2016. Co-receptor binding site antibodies enable CD4-mimetics to expose conserved anti-cluster A ADCC epitopes on HIV-1 envelope glycoproteins. EBioMedicine 12:208–218. 10.1016/j.ebiom.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald TA, Zepeda ML, Tomlinson MJ, Bee WH, Ivens IA. 2010. Subcutaneous administration of biotherapeutics: current experience in animal models. Curr Opin Mol Ther 12:461–470. [PubMed] [Google Scholar]
- 33.Liao HX, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, Kozink DM, Hwang KK, Chen X, Tsao CY, Liu P, Lu X, Parks RJ, Montefiori DC, Ferrari G, Pollara J, Rao M, Peachman KK, Santra S, Letvin NL, Karasavvas N, Yang ZY, Dai K, Pancera M, Gorman J, Wiehe K, Nicely NI, Rerks-Ngarm S, Nitayaphan S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Sinangil F, Kim JH, Michael NL, Kepler TB, Kwong PD, Mascola JR, Nabel GJ, Pinter A, Zolla-Pazner S, Haynes BF. 2013. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity 38:176–186. 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alpert MD, Heyer LN, Williams DEJ, Harvey JD, Greenough T, Allhorn M, Evans DT. 2012. A novel assay for antibody-dependent cell-mediated cytotoxicity against HIV-1- or SIV-infected cells reveals incomplete overlap with antibodies measured by neutralization and binding assays. J Virol 86:12039–12052. 10.1128/JVI.01650-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alpert MD, Harvey JD, Lauer WA, Reeves RK, Michael Piatak J, Carville A, Mansfield KG, Lifson JD, Li W, Desrosiers RC, Johnson RP, Evans DT. 2012. ADCC develops over time during peristent infection with live-attenuated SIV and is associate with complete protection against SIVmac251 challenge. PLoS Pathog 8:e1002890. 10.1371/journal.ppat.1002890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahman MA, Ko EJ, Enyindah-Asonye G, Helmold Hait S, Hogge C, Hunegnaw R, Venzon DJ, Hoang T, Robert-Guroff M. 2019. Differential Effect of Mucosal NKp44(+) Innate Lymphoid Cells and Deltagamma Cells on Simian Immunodeficiency Virus Infection Outcome in Rhesus Macaques. J Immunol 203:2459–2471. 10.4049/jimmunol.1900572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peachman KK, Karasavvas N, Chenine AL, McLinden R, Rerks-Ngarm S, Jaranit K, Nitayaphan S, Pitisuttithum P, Tovanabutra S, Zolla-Pazner S, Michael NL, Kim JH, Alving CR, Rao M. 2015. Identification of new regions in HIV-1 gp120 variable 2 and 3 loops that bind to alpha4beta7 integrin receptor. PLoS One 10:e0143895. 10.1371/journal.pone.0143895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collaborative Computational Project N. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763. 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4 and legends of Tables S1 and S2. Download jvi.01864-22-s0001.pdf, PDF file, 0.9 MB (895KB, pdf)
Table S1. Download jvi.01864-22-s0002.xlsx, XLSX file, 0.01 MB (11.6KB, xlsx)
Table S2. Download jvi.01864-22-s0003.xlsx, XLSX file, 0.01 MB (13.5KB, xlsx)
Data Availability Statement
The coordinates and structural factors for antibody NCI05 in complex with peptide V2Cmut have been deposited in the Protein Data Bank (PDB ID 8FBW).