ABSTRACT
Human sapoviruses (HuSaVs), like human noroviruses (HuNoV), belong to the Caliciviridae family and cause acute gastroenteritis in humans. Since their discovery in 1976, numerous attempts to grow HuSaVs in vitro were unsuccessful until 2020, when these viruses were reported to replicate in a duodenal cancer cell-derived line. Physiological cellular models allowing viral replication are essential to investigate HuSaV biology and replication mechanisms such as genetic susceptibility, restriction factors, and immune responses to infection. In this study, we demonstrate replication of two HuSaV strains in human intestinal enteroids (HIEs) known to support the replication of HuNoV and other human enteric viruses. HuSaVs replicated in differentiated HIEs originating from jejunum, duodenum and ileum, but not from the colon, and bile acids were required. Between 2h and 3 to 6 days postinfection, viral RNA levels increased up from 0.5 to 1.8 log10-fold. Importantly, HuSaVs were able to replicate in HIEs independent of their secretor status and histo-blood group antigen expression. The HIE model supports HuSaV replication and allows a better understanding of host-pathogen mechanisms such as cellular tropism and mechanisms of viral replication.
IMPORTANCE Human sapoviruses (HuSaVs) are a frequent but overlooked cause of acute gastroenteritis, especially in children. Little is known about this pathogen, whose successful in vitro cultivation was reported only recently, in a cancer cell-derived line. Here, we assessed the replication of HuSaV in human intestinal enteroids (HIEs), which are nontransformed cultures originally derived from human intestinal stem cells that can be grown in vitro and are known to allow the replication of other enteric viruses. Successful infection of HIEs with two strains belonging to different genotypes of the virus allowed discovery that the tropism of these HuSaVs is restricted to the small intestine, does not occur in the colon, and replication requires bile acid but is independent of the expression of histo-blood group antigens. Thus, HIEs represent a physiologically relevant model to further investigate HuSaV biology and a suitable platform for the future development of vaccines and antivirals.
KEYWORDS: sapovirus, norovirus, human intestinal enteroids, histo-blood group antigens, enteric virus, enteric viruses
INTRODUCTION
Sapovirus (SaV) is a genus of small RNA viruses within the Caliciviridae family, which also contains the Norovirus (NoV) genus. HuSaV was first discovered in 1976, by electron microscopic observation of fecal samples from patients with gastroenteritis in the United Kingdom (1), but the most studied strain originated in the early 1980s, from samples from children in a Japanese orphanage located in Sapporo (2). HuSaVs contain a single-stranded, positive-sense, poly-A tailed RNA genome of 7.1 kb to 7.7 kb, encoding two open reading frames (ORF) that are packaged in an icosahedral capsid ranging from 30 to 38 nm in diameter (3). ORF1 encodes a polyprotein that is proteolytically cleaved into at least six nonstructural proteins and possibly the major structural protein VP1 (4), which self-assembles to form a particle of 180 copies of VP1 (5). ORF2 encodes a minor structural protein, VP2 (6). A subgenomic RNA comprising the coding sequence of VP1 and VP2 has been identified in a porcine strain; thus, VP1 may arise from the cleavage of the polyprotein, or from the translation of this subgenomic RNA (3). Caliciviridae share a capsid structure, with VP1 divided in two domains: a S domain, mostly conserved between strains, and a P domain, with a subdomain P2 displaying the most variable region in amino acid sequences, primarily accounting for antigenic and immunogenic diversity (5). VLP production does not require VP2 owing to the self-assembling properties of VP1 dimers, and HuSaV VP2 structure and function are still largely uncharacterized. HuSaVs are classified into four genogroups: GI, GII, GIV and GV (7). Other SaV genogroups have been identified in various animal species such as minks, bats, sea lions, dogs, rodents, or chimpanzees (8).
HuSaV infection causes classical acute gastroenteritis (AGE) symptoms that are considered milder or similar to those caused by HuNoV and human rotavirus (RV) (9). Molecular detection is required to properly identify the causative pathogen and is infrequently performed for HuSaVs. Yet, HuSaVs are one of the common causes of viral gastroenteritis globally, causing ~3.4% of cases based on a meta-analysis of 106 studies around the world (10). In recent studies, HuSaVs have been detected more frequently than human RV in countries with childhood vaccination programs targeting RV (11–13). Infection is likely to occur across the human population at a young age, usually before 2 years (14, 15). Viral progeny is excreted in feces, up to 1011 copies per grams of stool (16), and HuSaV transmission occurs through the fecal-oral route via contact with HuSaV-positive feces, vomitus, contaminated surfaces, contaminated food and/or drinks. Yet, as opposed to the marked winter seasonality of HuNoV in the Northern Hemisphere, seasonality is less apparent for HuSaV. Studies on different continents show differing seasonal patterns, but correlation with high rainfall, low temperature, and floods is suspected (9). In sewage, HuSaV may be detected year-round (17).
Similar to HuNoV, in vitro replication of HuSaV was a challenge for more than 40 years from when the virus was first isolated. A porcine sapovirus (PoSaV), the Cowden strain, can be cultivated in porcine LLC-PK1 cells in a bile-dependent manner (18). More recently, Takagi et al. succeeded in propagating GI.1 and GII.3 HuSaV strains in HuTu80 (duodenum) and NEC8 (ileocecum) cancer cell lines, in the presence of bile acids, further demonstrating the important role of this cofactor in HuSaV infection (19). However, due to the previous lack of a reliable culture system, there is still little information about the HuSaV viral life cycle and tropism.
Studies of tissues obtained from infected piglets suggest that replication of the PoSaV Cowden strain occurs in the small intestine, especially in the duodenum and jejunum (20). PoSaV requires sialic acids linked to O-glycoproteins as receptors, but the HuSaV tropism and receptor(s) remain unknown (21). Regarding HuNoV, epidemiological studies have demonstrated that some HuNoV strains are, at least partially, restricted to people with the secretor phenotype (22), who express histo-blood group antigens (HBGAs) in their intestinal mucosa. Most HuNoV strains bind to HBGAs (23, 24). Conversely, HuSaV has not previously been shown to interact with HBGAs (25).
Human intestinal enteroids (HIEs) have become an essential tool to study HuNoV replication in human cells, overcoming decades of unsuccessful attempts at creating a cellular model allowing culture of these viruses (26). HIEs are derived from LGR5+ stem cells isolated from intestinal crypts, maintained in a 3-dimensional environment, and can be grown to form multicellular structures recapitulating small intestine tissues or as two-dimensional monolayers. Using this physiological model has allowed a better understanding of HuNoV–host interactions and confirmed the dependency of several HuNoV strains for the expression of HBGA to initiate successful infection (27–30). This model also allows replication of enteric viruses, including rotavirus, enterovirus, coronavirus, adenovirus, astrovirus (31).
Here, we undertook the study to evaluate whether HIEs support the replication of HuSaVs, to examine whether replication requires bile acids, and to investigate HuSaV tropism and the requirement of HBGAs for infection using HIEs from different segments of the gut and from different human donors with different HBGA phenotypes. Our results show that the HIE model is fit to investigate HuSaV–host interactions.
RESULTS
HIE cultures support HuSaV replication.
To evaluate if HIEs could support HuSaV replication, we inoculated differentiated J2 HIE monolayers with HuSaV-positive stool filtrates at different concentrations in the presence of the bile acid GCDCA, similar to the protocol used previously for HuNoVs (32). Of the five stool filtrates evaluated, two (S611 and S513) showed 0.5 to 1.8 log10, i.e., 3 to 63-fold, increases in genome copies from 1 or 2 hpi to 72 hpi, respectively, depending on the amount of virus in the inoculum (Fig. 1A). We confirmed that the S611 strain replicated successfully on J2 HIE monolayers more than 40 times over a 2-year period, with a geometric mean fold change in viral genome of 32 between 1 or 2 hpi and 72 hpi (P < 0.0001, Wilcoxon signed-rank test) (Fig. 1B). Due to a lower titer, S513 was used in 9 experiments on J2 HIE monolayers, and showed replication in 6, with a mean fold change of 7.7 between 1 or 2h and 72 hpi (P = 0.01, Wilcoxon signed-rank test), lower than S611 in the same HIE line (Fig. 1C). In most experiments, monolayers were also inoculated with HuNoV GII.4 TCH11-64 strain as a control (Fig. 1D), for which the mean fold change was much higher (4,388 mean fold change; P < 0.0001, 1h or 2h versus 72 hpi, Wilcoxon signed-rank test) than for the two tested HuSaV strains. Together, these results demonstrate that two strains of HuSaV replicated repeatedly in J2 HIEs, albeit with lower fold changes after 3 days of infection compared to a HuNoV GII.4 strain.
FIG 1.
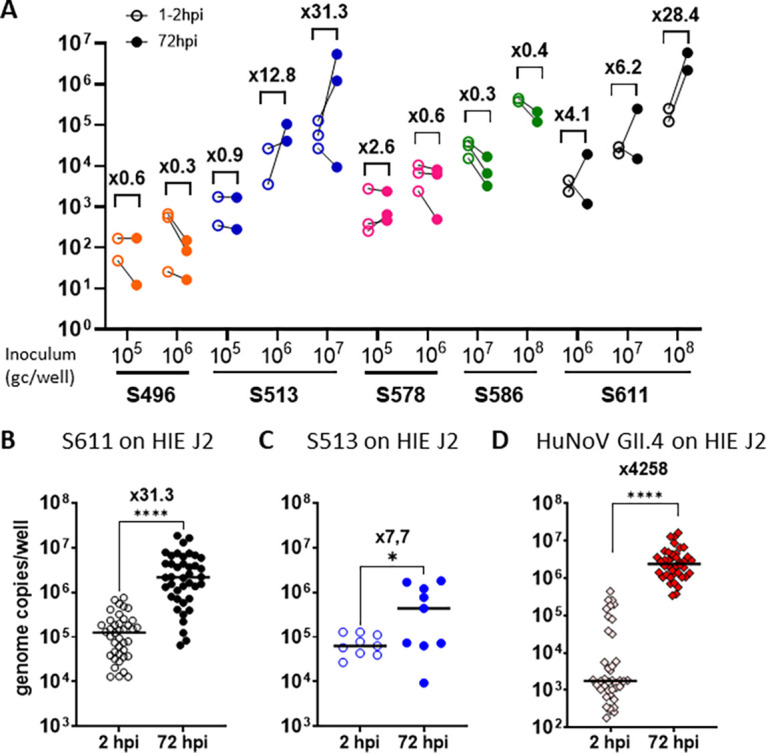
Two human sapovirus strains replicate in jejunal human intestinal enteroids (HIE). (A) Five HuSaV-positive stools filtrates were used to infect differentiated monolayers of HIE J2 cells in the presence of 500 μM bile acid GCDCA, with inocula of 1.105 to 1.108 HuSaV genome copies per well (horizontal axis). HuSaV genome quantities in each well (vertical axis) were measured by qRT-PCR at 1 or 2h (empty circles) and 72h (full disks) postinfection (pi). Differences between HuSaV titers at 2 and 72 hpi were not statistically significant, n = 2 to 3 experiments, P > 0.05, Wilcoxon test. (B) An inoculum of 1 × 108 gc/well of HuSaV strain S611 was used in repeated experiments to infect differentiated J2 monolayers with 500 μM GCDCA and showed a mean genome fold change of 31.3 (n = 40 experiments, P < 0.0001, Wilcoxon test). (C) An inoculum of 1x107 gc/well of HuSaV strain S513 was also used to infect differentiated J2 monolayers with 500 μM GCDCA and showed a geometrical mean genome amplification of 7.7 (n = 9 experiments, P = 0,012, Wilcoxon test). (D) In parallel with HuSaV infections, differentiated J2 monolayers were also infected with 1.105 genome copies per well of HuNoV GII.4 TCH11-64 as a control with 500 μM GCDCA and showed efficient replication with a geometric mean fold increase of 4258 (n = 40 experiments, P < 0.0001, Wilcoxon test). (A, B, C, D) each dot is the mean of three technical replicates for one independent experiment at 2h (empty circles or pink diamonds) and 72h postinfection (plain disks and red diamonds), horizontal lines are the geometrical mean of virus genome copies per well considering all experiments, and the number above indicates the mean fold change in viral genome between the two time points.
We next investigated the kinetics and magnitude of S611 replication in HIEs. To assess if the low fold changes observed at 72 hpi were due to a more protracted course of infection for HuSaV in HIEs, we measured the fold increase after 6 days (144h) of infection (Fig. 2A). Viral genome concentrations were higher at 72 hpi and 144 hpi than at 2 hpi (P < 0.05 for both, Wilcoxon signed rank test), with fold changes in viral genome of 9.8 and 17.1 at 72h and 144 hpi, respectively, confirming successful viral replication. There was a small but significant difference between the two late time points (P value = 0.047). We next tested the replication of the S611 strain in the HuTu80 duodenal cell line that was recently shown to support replication of HuSaV GI.1, with various amounts of virus in the inoculum. Like in HIEs, S611 replicated in HuTu80 cultures, with fold changes in viral genome copies of ~2 log10 between 1 day and 3 or 6 days postinfection (Fig. 2B), lower than the fold increase previously reported for another GI.1 strain (19). Of note, we were able once to use the S611 fresh after collection, before any freeze-thaw cycle, on J2 HIEs in a single experiment that showed a high fold increase (×1,115) of the viral genome in 72 hpi (Fig. S1). The kinetics of S611 replication in the D109 HIE line showed an increase in viral genome copies between 1 and 72h postinfection, followed by a plateau until 6 days (144h) postinfection (Fig. 2C). To control for the exposure of HIEs to fecal filtrates, in the absence of viral replication, we inoculated cultures with heat-inactivated virus, where the viral genome copies showed a slow but steady decline as expected (Fig. 2C). In parallel, the viability of the cell culture was monitored using a metabolic assay and was not affected by the HuSaV replication, nor by exposure to the inactivated virus (Fig. 2D). These results show that in our experimental conditions, HuSaV S611 replicates at low levels and does not alter the viability of HIEs.
FIG 2.
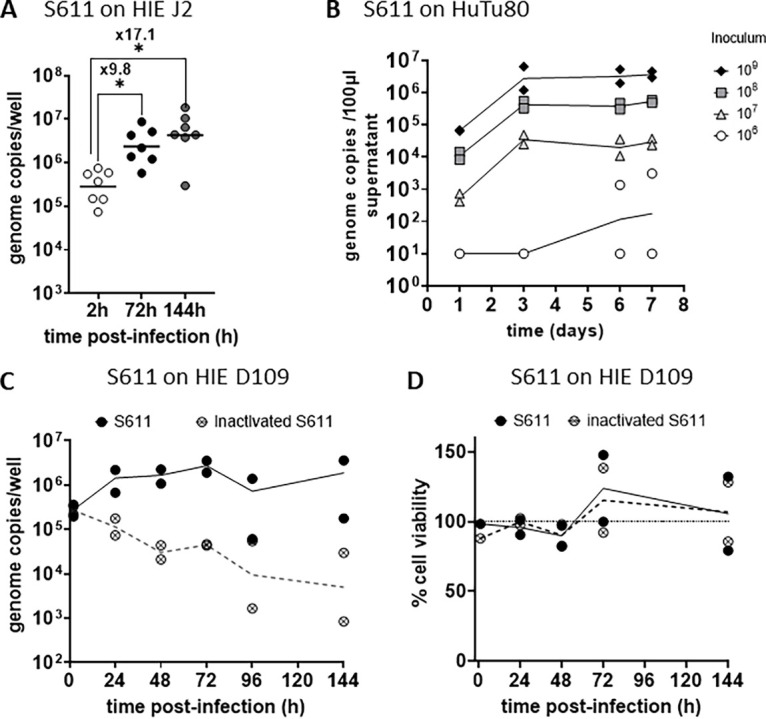
Kinetics and magnitude of HuSaV S611 replication in HIE. (A) Differentiated J2 monolayers were infected with 1.108 cg/well of HuSaV S611 with 500 μM GCDCA and the viral genome quantified after 2h, 72h and 144h. Each dots represents the mean of three technical replicates in an experiment (n = 7). In comparison with 2 hpi, viral replication was detected at 72 hpi (P < 0.05, Wilcoxon test) and 144 hpi (P < 0.05, Wilcoxon test) with geometrical mean fold changes in viral genome of 9.8 and 17.1, respectively. (B) HuTu80 were infected with S611 at four different concentrations from 1.105 to 1.109 cg/well with 500 μM GCDCA. Mean viral titers of three technical replicates were measured in the culture supernatants at 1, 3, 6, and 7 days postinfection and are plotted for each experiment (n = 2), with lines connecting the geometrical means. (C) Differentiated D109 monolayers were inoculated with 1.108 cg/well native (plain black circles) or heat-inactivated (crossed circles) S611 with 500 μM GCDCA and the viral genome quantified after 2h, 24h, 48h, 72h, 96h, and 144h. The mean of technical replicates is plotted as a circle for each experiment (n = 2) and geometrical means are connected with a plain (native virus) or a dotted (inactivated virus) line. (D) The viability of the differentiated D109 monolayers either infected by native (plain black circles) or heat-inactivated (crossed circles) S611 with 500 μM GCDCA for n = 2 experiments in comparison with an uninfected control (100%, dotted line).
Replication of S611 HuSaV strain in jejunal HIE is dependent on bile acid and cell differentiation.
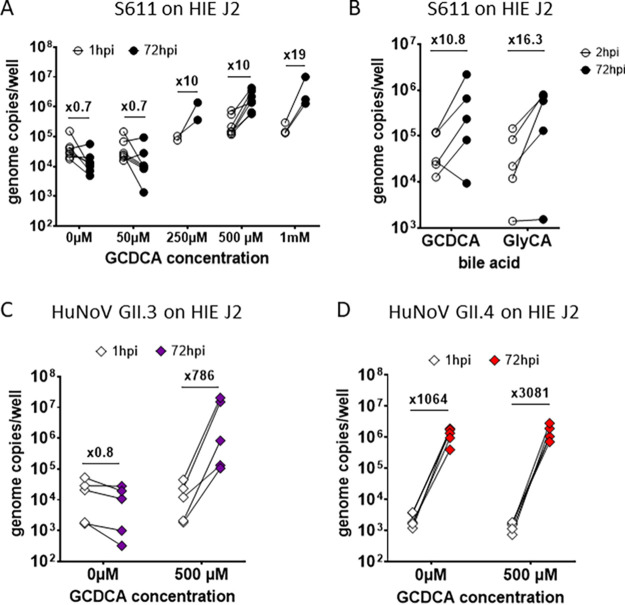
We next assessed whether bile acids were necessary for HuSaV S611 replication in HIEs as previously shown for replication of the PoSaV Cowden strain (33) and in previous work conducted with HuSaV in the HuTu80 human duodenal cell line (19). J2 HIE monolayers were inoculated with HuSaV S611 in the presence of increasing concentrations (from 0 μM to 1 mM) of the bile acid GCDCA. We observed that GCDCA was necessary for a productive infection of J2 HIE monolayers by HuSaV S611, with fold changes in viral genome copies >3 appearing when GCDCA concentrations were between 250 μM and 1 mM (Fig. 3A). A concentration of 500 μM was set for the next experiments.
FIG 3.
Bile acids are a necessary cofactor of HuSaV infection in HIE. (A) Differentiated J2 monolayers were infected with 1.108 cg/well of S611 in the presence of the bile acid GCDCA at concentrations ranging from 50 μM to 1 mM, or in the absence of bile acid. The viral titer was measured and is depicted as the mean of three technical replicates for n = 2 to 6 experiments at 1 hpi (white circles) and 72 hpi (black circles) and the geometrical mean fold change between the two points indicated above. (B) The bile acid GlyCA (right side) was also used in comparison to GCDCA (left side) with concentrations of 500 μM on differentiated J2 monolayers infected with 1.108 cg/well of S611, n = 5 experiments. (C) and (D) Experiments in (A) were also conducted with 1.106 cg/well of HuNoV GII.3 (purple) or 1.105 cg/well of HuNoV GII.4 (red), with GCDCA concentrations of 500 μM or without GCDCA.
We also tested the effect of another bile acid, sodium glycocholate (GlyCA), as it was used in the study of HuSaV replication in HuTu80 cells (19). HuSaV S611 replicated similarly with 500 μM GCDCA (mean fold change of 10.8 between 2h and 72h pi) or GlyCA (mean fold change of 16.3) (Fig. 3B).
HuNoV bile-dependent strain (GII.3) and partially independent strain (GII.4) were also inoculated in parallel as controls and recapitulated results obtained previously (26) (Fig. 3C and D).
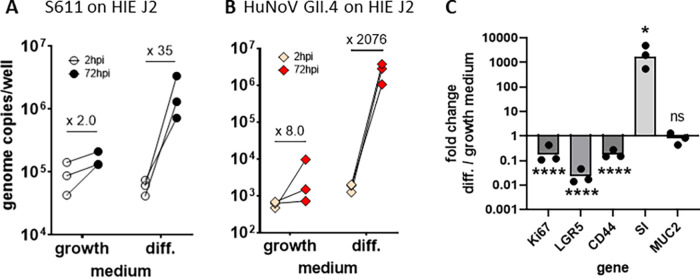
We then took advantage of the HIEs to investigate whether HuSaV infects intestinal stem cells and/or differentiated cells, such as enterocytes that are a target of HuNoV. J2 HIEs were plated as monolayers and either infected the following day and kept in stem-cell growth medium (Intesticult OGM) or induced to differentiate using a medium with reduced concentrations of niche growth factors for 4 days before being infected. HuSaV replicated to higher fold changes in differentiated cells compared to in undifferentiated cells (Fig. 4A). Similar results, albeit with higher fold increases, were observed with HuNoV GII.4 (Fig. 4B). The expression of five genes known to be modulated during differentiation was assessed by qRT-PCR. Relative expression of stem cell and proliferation markers (Ki67, LGR5, CD44) was lower under the condition with differentiation medium (mean fold changes of 0.22, 0.036, 0.19, respectively; P values <0.0001, paired t test). Conversely, Sucrase-isomaltase (SI) was markedly induced (fold change of 2592, P value <0.05, paired t test), indicating that cells differentiated into enterocytes. The mucus 2 (Muc2) marker of goblet cells was not affected (fold change 0.92, P value = 0.74, paired t test). Altogether these results show that HIE differentiation is necessary for a consistent replication of HuSaV.
FIG 4.
HuSaV replicate in differentiated jejunal HIE. Comparison of the mean viral titer per well of S611 HuSaV (A) and GII.4 HuNoV (B) at 2 hpi (white bars) and 72 hpi (black bars) in J2 monolayers cultured and infected in growth or differentiation (diff.) medium with 500 μM GCDCA, for n = 3 experiments, 3 technical replicates per experiment. Geometric mean fold changes between the two time points are indicated above for each condition. (C) Fold change in relative expression levels of 5 genes (normalized on GAPDH) between HIE J2 monolayers cultured in differentiation versus growth medium in the experiments presented in (A). *, P < 0.05, ****, P < 0.0001, paired t test.
HuSaV replicates in cells of the three portions of small intestine from different donors.
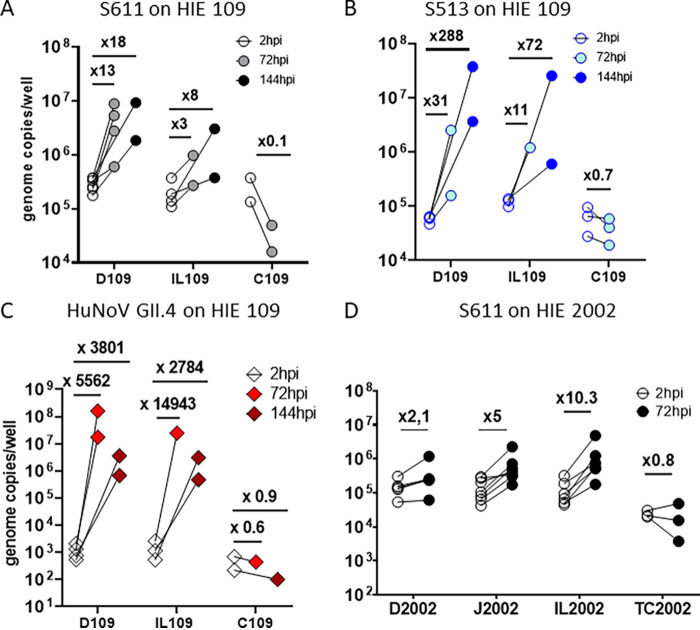
To assess the tropism of HuSaV in the human gut, we used HIEs originating from different segments of the intestine of the same donor. In donor 109, we observed the replication of both S611 (Fig. 5A) and S513 (Fig. 5B) in duodenal (D109) and ileal (IL-109) cells, after 72 or 144 hpi, but not in cells from the colon (C109). The HuNoV GII.4 TCH11-64 strain showed the same pattern (Fig. 5C), as previously reported (29). S611 replicated poorly in IL-109, whereas S513 showed higher fold increases in ileal cells. In HIEs from donor 2002 (Fig. 5D), for which cultures from all segments from a single individual were available, S611 replicated in cells from the jejunum (J2002) and the ileum (IL-2002), but not from the duodenum (D2002) or the transverse colon (TC2002). From these results, we conclude that HuSaV can replicate in HIEs from different donors, and in cells from the 3 portions of the small intestine, but that strain-related or donor-related factors may modulate this tropism.
FIG 5.
HuSaV replicates in human cells from the three segments of the small intestine. Differentiated HIE monolayers from different segments of the small intestine (J for jejunum, D for duodenum, IL for ileum, C for colon and TC for terminal colon) originating from two donors, 109 (A, B, C) and 2002 (D), were infected with 1.108 cg/well of HuSaV S611 (A, D – white, gray, black circles), 1.107 cg/well of HuSaV S513 (B – blue circles) or 1.105 cg/well of HuNoV GII.4 as a control (C – pink and red diamonds) with 500 μM GCDCA. The mean viral titer per well for three technical replicates is depicted as circles for 2h, 72h or 144 hpi, for n = 2 to 6 experiments. Above each condition is displayed the corresponding viral genome fold change between the two time points.
HuSaV replication in HIE is independent of secretor status.
The tested HuSaV strains replicated in HIE cell monolayers from three different donors: 2, 109 and 2002, all secretor positive (Table 1). We next investigated if secretor phenotype and blood type (HBGA phenotype) could impact HuSaV replication, as it does restrict several HuNoV strains. S611 replicated successfully in 3 of 7 experiments in jejunal HIE J10 (secretor and Lewis negative) and in 6 of 7 experiments in J11 (secretor positive) (Fig. 6A, left panel), whereas HuNoV GII.4 did not replicate in J10 (Fig. 6A, right panel). Interestingly, S611 initial binding and viral genome fold changes in 72 hpi appeared to be lower on the J10 HIE line than in the two secretor positive lines.
TABLE 1.
List of HIE lines used in this study
| HIE line | Tissue of origin | Phenotype |
Reference | |
|---|---|---|---|---|
| Secretor status | ABO | |||
| J2 | Jejunum | Sec + | B, Lewis b | Ettayebi et al., 2016 (26) |
| J11 | Jejunum | Sec + | Lewis b | |
| J2002 | Jejunum | Sec + | Lewis b | |
| D2002 | Duodenum | |||
| IL2002 | Ileum | Ettayebi et al., unpublished data | ||
| TC2002 | Transverse colon | |||
| J4FUT2KI | Jejunum | Sec + | Lewis b | Haga et al., 2020 (34) |
| D109 | Duodenum | Sec + | A, Lewis b | Rajan et al., 2018 (35) |
| IL109 | Ileum | |||
| C109 | Colon | |||
| J2FUT2KO | Jejunum | Sec - | Lewis a | Haga et al., 2020 (34) |
| J4 | Jejunum | Sec - | Lewis a | Haga et al., 2020 (34) |
| J10 | Jejunum | Sec - | A | Rimkute et al., 2020 (30) |
FIG 6.
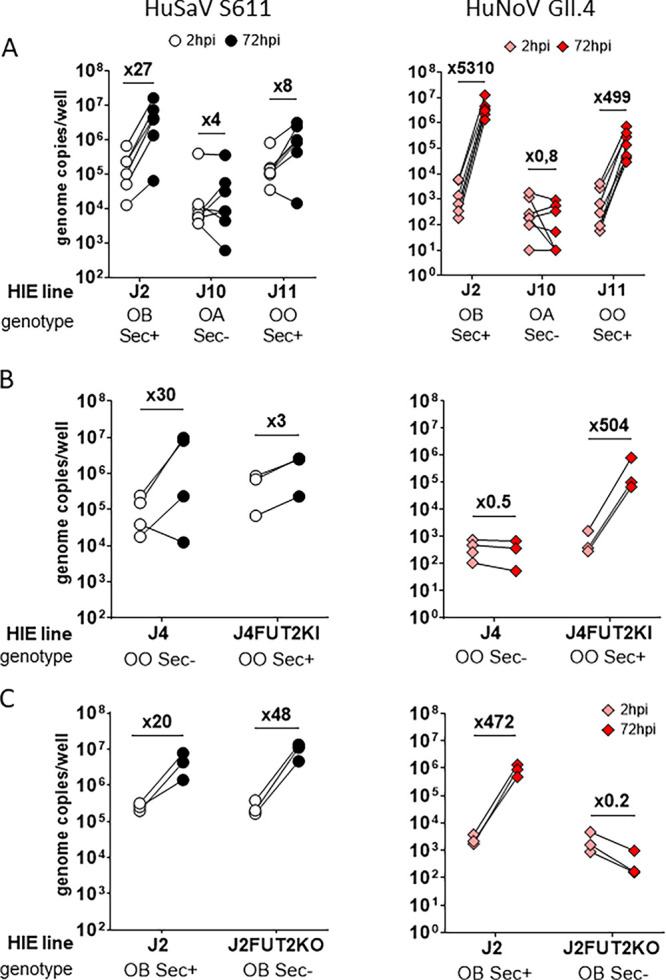
HuSaV replicate in HIE independently of HBGA genotype. Differentiated monolayers of jejunal HIE from different donors were infected with 1.108 cg/well of HuSaV S611 (white and black) or 1.105 cg/well of HuNoV GII.4 (pink and red) with 500 μM GCDCA in each case and the mean viral titer per well measured for three technical replicates and n = 3 to 7 experiments at 2 hpi (white, pink) and 72 hpi (black, red). (A) Three jejunal HIE cells lines, J2, J10, J11 with contrasted HBGA genotype (OB Sec+, Sec-, and OO Sec+, respectively). (B) J4 cells (OO Sec- genotype) and the corresponding knock-in, J4FUT2KI (OO Sec+). (C) J2 cells (OB Sec+) and the corresponding knockout, J2FUT2KO (OB Sec-).
Next, we tested the dependency to the secretor phenotype in isogenic cells, using two HIE lines and their genetically modified counterparts: J4, a secretor negative line, with the J4FUT2KI knock-in, and J2, a secretor positive, with the J2FUT2KO knockout. HuSaV S611 was able to replicate in all HIEs independently of the expression of FUT2 (Fig. 6B and C). As observed previously (26, 34), HuNoV did not show replication in secretor negative J10, J4 and J2FUT2KO HIE lines. These results show that S611 HuSaV strain can replicate in both secretor positive and negative jejunum HIEs from different donors, with 5 secretor positive and 3 secretor negative HIE lines being tested. As expected, GII.4 HuNoV showed no replication in the 3 secretor negative HIE lines (Fig. 6A and B and C, right panels). Of note, most of the tested HIEs are O blood group but S611 also replicated in A and B type group HIEs (D-IL-109 for A group, and J2 for B group, Table 1).
DISCUSSION
Prior attempts to grow HuSaV, mostly in primary kidney and intestine cells of human or simian origin, have been reported, but many have produced either inconclusive or nonreproducible results (35–37). On the other hand, the PoSaV Cowden strain replicates successfully in porcine kidney cells (18, 38), and its replication is dependent on bile acids with sialic acids acting as a binding factor. Recently, Takagi et al. reported efficient replication and passaging of GI.1 and GII.3 HuSaV strains in a HuTu80 adenocarcinoma cell line of duodenal origin in the presence of bile acids, and a reverse genetics system also produced GII.3 virions after transfection of HuTu80 cells (6, 19). Our work focused on HIEs, a physiological model of human intestine in which several enteric viruses can replicate. The genetically diverse range of donors and genetically modified HIE lines also allows investigation of viral tropism and cofactors required for replication.
In this work, we were able to successfully grow two HuSaV strains in HIEs: S611 (GI.1) and S513 (GI.2) (Table 2). These two genotypes cause the majority of cases in Europe (39, 40), and were the most frequently detected in HuSaV-positive fecal samples in a 2021 study in Spain (11). These two strains showed consistent viral replication in the HIE J2 line of jejunal origin, over a time frame of 2 years for S611, characterized by a moderate multiplication. Indeed, the viral genome fold changes within 3 or 6 days postinfection did not reach levels as high as those achieved by GII.4 HuNoV in HIEs (2 to 3 log10) or HuSaV in HuTu80 (19) (from 2 to 6 log10), which precluded serial propagation experiments in our study. However, in our hands, S611 replication in the HuTu80 cell line was also moderate (1 to 2 log10), which suggests that this may be a characteristic of the strains used in this study. Indeed, previous work has shown that some HuNoV strains also display a low or moderate viral replication in HIEs (26, 29, 41). A unique experiment conducted with fresh S611, before any freezing, showed a higher 3 log10-fold (x1115) increase in the J2 HIE line (Fig. S1), but additional fresh stool samples containing virus were not available to evaluate the repeatability of this observation. Hence, our results showing moderate fold changes in viral genomes for HuSaV S611 and S513 need to be repeated with more strains, and if possible, fresh stool samples, before drawing conclusions on the magnitude of HuSaV replication in HIE in general. In addition, kinetic experiments showing that the viral replication mostly occurs in the first day of infection suggest that HuSaV S611 did not propagate efficiently in HIE cultures, and was limited to one replication cycle, as also observed for several HuNoV strains (42). Finally, although most experiments showed replication of S611 (38 out of 40 experiments) and S513 (6 out of 9), in some cases with similar inoculum and experimental conditions, the viruses did not grow. This has been observed previously for some HuNoV strains with low or moderate amplification (41). Similarly, heterogeneity in viral fold change was also reported for a HuNoV strain across repeated experiments (29). Here, HuNoV GII.4 was included in most experiments to control for the ability of the cell cultures to sustain an infection (see Materials and Methods). Thus, the resulting heterogeneity in HuSaV replication may be linked to a possible heterogeneity of the stool samples where the viral particles may form aggregates or vesicle-cloaked clusters, as reported for HuNoV (43, 44).
TABLE 2.
List of viral strains used to perform infection experiments
| Virus | Strain | Titer (gc/mL)a | Genotype | Accession number or ref |
|---|---|---|---|---|
| HuNoV | TCH11-64 | 5.8 × 109 | GII.4 | (26, 29) |
| TCH04-577 | 1.07 × 1010 | GII.3 | GU930737 (26, 29) | |
| HuSaV | S496 | 1.2 × 109 | GIV.1 | OP654149 |
| S513 | 5.81 × 108 | GI.2 | OP654151 | |
| S578 | 2.89 × 108 | GI.2 | OP654152 | |
| S586 | 7.2 × 1010 | GII.3 | OP654150 | |
| S611 | 3.66 × 1010 | GI.1 | OP654153 |
gc/mL: genome copies/mL of fecal filtrate.
Despite these moderate levels of replication, the HIE model was still adequate to investigate HuSaV biology. We first confirmed that bile acids are required for efficient replication of HuSaV S611 in HIEs. Here, GCDCA was mainly used, similarly to the HuNoV replication model (26), but could be replaced by GlyCA (Fig. 3B), as was done on HuTu80 cells (19). Some strains of HuNoV being able to replicate in HIEs independently of bile acid (Fig. 3D) (26), other HuSaV samples should be tested in this model as well to generalize this observation. Based on HuNoV work, bile acids are thought to promote viral replication by enhancing endosomal uptake, acidification, and ceramide levels on the apical membrane (32). Work on PoSaV also showed similar effects, as bile acids are required for virions to escape endosomes into the cytoplasm and for the production of ceramide (45).
Likewise, intestinal stem-cell differentiation was necessary to support efficient replication of HuSaV S611 in jejunal HIE (Fig. 4A), as observed previously for HuNoV (Fig. 4B) (29). Moreover, since this differentiation was mostly characterized by a strong induction in SI expression, a marker of enterocytes, rather than Muc2, a marker of goblet cells (Fig. 4C), our results suggest that enterocytes may be the main cell type sustaining HuSaV replication in HIE cultures, although the contribution of other cell types cannot be excluded.
Using the HIE model also allowed us to investigate the potential tropism of HuSaV in the human intestine, with cells originating from jejunum, duodenum, ileum and colon of two different donors. We observed the replication of both HuSaV strains in HIEs derived from the three segments of the small intestine, with variations between the cultures from the two tested donors. D2002 showed only one experiment with >3-fold change in viral genome at 72 hpi, but efficient replication occurred in duodenal cells from the other donor, D109 cells. Conversely, in the colonic HIE (C109 and TC2002), viral titers of HuSaV S611 or S513 decreased over 3 days (Fig. 5A and D), indicating that cells derived from this section of the intestine do not support measurable viral replication. Our data strongly suggest that the tested HuSaV strains replicate in the small intestine but not in the colon. Likewise, PoSaV is believed to mainly infect small intestinal tissues based on histological localization of virus-induced damage in gnotobiotic piglets (46). This is also consistent with the fact that the small intestine is physiologically exposed to bile acids, a necessary cofactor for replication of the PoSaV Cowden strain and the HuSaV strains tested to date.
HBGA polymorphisms in the human population are known to drive sensitivity or resistance to infection with HuNoV and other enteric pathogens (47). Epidemiological data suggest that this is not the case for HuSaV (48, 49), but these studies were conducted on small cohorts (with number of HuSaV infected patients between 33 and 42) and some did not observe the usual secretor-associated trend when looking at HuNoV infections (50). Here, we took advantage of HIEs originating from donors expressing various HBGAs to assess the dependency of HuSaV on secretor or ABO phenotypes. S611 replicated both in secretor-positive (J2, J11, J4FUT2KI) and secretor negative (J10, J4, J2FUT2KO) jejunal HIE cells and in HIE from A, B, and O blood group donors. Some small differences can be noted, such as a reduced viral binding at 2 hpi, less experiments showing viral replication (3 out of 7 experiments) and lower viral replication when infecting the secretor-negative J10 HIEs compared to secretor positive J2. However, his could be due to other unknown differences between J2 and J10, which were derived from different donors and could have different levels of expression of the viral receptor or other cellular cofactors. To exclude these, we then used isogenic cell lines differing only with the expression of FUT2. J4 HIEs, initially secretor negative with homozygous se428/se428 in the FUT2 gene, supported S611 replication, with viral binding at 2 hpi similar to that of J2 cells. When knocked-in for FUT2 (J4FUT2KI), these cultures remained infected by S611. Our results are also in agreement with previous studies made with GI and GV HuSaV VLPs that showed no interactions between VLPs and human HBGAs from saliva or synthetic carbohydrates (25).
Further epidemiological studies conducted on a larger scale could totally eliminate a possible link between secretor status and susceptibility to HuSaV infection, although data presented in this paper and previous studies strongly suggest that HuSaV infection and replication are independent from intestinal HBGAs.
Due to a lower viral concentration and volume of sample, S513 could not be used in all experiments, which would have allowed more comparisons between the two HuSaV GI genotypes. In 4 different cell lines tested with both viruses (J2, D109, IL-109 and C109), the two strains behaved similarly. Three other tested strains did not replicate on HIE J2 (Fig. 1A), which could be due to their lower titers, especially for S496 and S578. Thus, additional strains will need to be tested to investigate whether other GI genotypes or other HuSaV genogroups also replicate in HIEs.
Initially overshadowed by more pathogenic viral AGE agents, HuSaVs have increasingly been detected in the human population, possibly due to the more widespread use of molecular diagnosis tools and/or to an actual epidemiological change, following RV vaccine implementation. Recent research highlight some unique characteristics of the SaV genus in comparison with the more studied NoV (5, 51). Most of the HuSaV biology remains to be explored, and we show here that HIEs represent a physiologically relevant model for further investigation of HuSaVs replication cycle and virus-host interactions.
MATERIALS AND METHODS
Viral strains.
HuSaV-positive or HuNoV-positive 10% fecal suspensions were prepared in PBS, 0.22 μm-filtered, aliquoted and stored at −80°C, as described previously (26), before being used to inoculate cells. Viral titers were determined by quantitative, one-step reverse transcription and PCR (qRT-PCR) on extracted nucleic acids, as described previously for HuNoV (52). Table 2 describes the viral strains used for subsequent experiments. The HuSaV S611 strain was heat-inactivated for 15 min at 60°C before being used to inoculate HIE monolayers in some experiments.
Viral strain sequencing.
Stool fecal suspensions were incubated with 2000 U of OmniCleave Endonuclease (Epicentre, Madison, US) for 1 h at 37°C to eliminate free nucleic acids, followed by nucleic acid extraction as in (53). A final step of RNA purification and concentration was performed using a Zymo-spin column (RNA Clean & Concentrator, Zymo Research, Irvine, USA). Libraries were prepared by synthesizing cDNA using the Superscript IV kit (Life Technologies) with random hexamers according to the manufacturer’s instructions (Life Technologies). cDNA first strand was physically fragmented (Ultrasonicator M220, Covaris) for 2.5 min. and afterwards, the NEBNext Ultra II RNA Library Prep kit for Illumina (E7770L –NEB) was used for Illumina pools. Sequencing was performed on an Illumina MiSeq to generate 2 × 250 bp reads. Viral reads were assembled into full genomes and genotyped using the Genome Detective online tool (https://www.genomedetective.com/).
HIE cells.
HIEs used in this study were obtained from healthy tissues of human donors who gave informed consent, according to a protocol approved by the Baylor College of Medicine Institutional Review Board (BCM IRB). The HIE cultures were maintained as described previously (29) in agreement with the ethical evaluation committee of the French Institute of medical research and Health (CEEI,). The tissue origin of the stem cells is designated using letters: J for jejunum, D for duodenum, IL for ileum and C for colon, and the number indicates the donor. In addition, a genetically modified J2 HIE line (J2FUT2KO) that is an isogenic J2 line with the FUT2 gene responsible for the secretor phenotype knocked out and J4 cells genetically modified to express a functional FUT2 gene (J4FUT2KI), as described previously, were also used for experiments (34).
HIE culture.
All HIEs were grown in a 30 μL dome of Matrigel growth-factor-reduced basement membrane matrix (Corning) surrounded with 500 μL of Intesticult Intestinal Organoid Growth Medium (OGM; StemCell) supplemented with antibiotics (0.3% PenStrep) in 24-well plates (Nunclon, ThermoScientific). Cells were incubated at 37°C and 5% CO2. Medium was changed every 2 to 3 days, and cells were passaged every week using trypsin 0.05%- EDTA (Gibco, ThermoFisher) and repeated pipetting to dissociate cells within the enteroids, before being centrifuged, resuspended in Matrigel and plated.
For infection experiments, 3D grown enteroids were dissociated for 4 min at 37°C in a water bath with Trypsin-EDTA 0.05% (Invitrogen) and mixed by repeated pipetting in order to obtain a single cell suspension. A minimum of 1x105 cells per well were plated in 96-wells plates (Nunclon, ThermoScientific) coated with 33 μg/mL human collagen type IV (Sigma-Aldrich) in molecular-grade water. After 24 to 48 h of culture at 37°C and 5% CO2 with OGM supplemented with Y-27632 (10 μM, Sigma-Aldrich), medium was replaced with differentiation medium made of 50% Intestinal Organoid Growth Medium basal component, and 50% complete medium without growth factor (CMGF-), which is advanced DMEM/F12 medium (ThermoFisher) supplemented with 2 mM GlutaMax (Life technologies) and 10 mM HEPES, for 4 days to induce cellular differentiation.
Infection experiments.
Infections were performed on confluent monolayers 4 days after initiating differentiation. When not specified, the viral inoculum concentration used per well was as described in Table 3. Viral inocula were prepared with CMGF- supplemented with various concentrations of glycochenodeoxycholic acid (GCDCA, Sigma-Aldrich), ranging from 5 μM to 1 mM, and 500 μM when not specified. Cells were incubated with viral inoculum for 1 to 2 h. Inocula were then removed, and cells were washed 3 times with CMGF-. 100 μL of differentiation medium with GCDCA were then added to each well, and for each set of infections, one plate was immediately frozen at −20°C. The other plate was incubated at 37°C under 5% CO2 for 3 to 7 days, then frozen at −20°C until undergoing RNA extraction.
TABLE 3.
Viral concentrations of inocula used for infection experiments
| Virus | Strain | Concn / well | Genotype |
|---|---|---|---|
| HuNoV | TCH11-64 | 1 × 105 | GII.4 |
| TCH04-577 | 1 × 106 | GII.3 | |
| HuSaV | S611 | 1 × 108 | GI.1 |
| S513 | 1 × 107 | GI.2 |
HuTu80 culture and infection.
The HuTu80 cell line (ATCC, number HTB-40) was cultured in DMEM supplemented with 2 mM Glutamax, 0,3% PenStrep and 5% FBS at 37°C under 5% CO2. Monolayers were made by dissociating cells from a confluent flask and seeding 24-well plates with 5 × 105 cells in 0.5 mL of culture medium. Before infection, the medium was replaced with 0.5 mL of virus growth media, which is a 3% FBS culture media supplemented with the bile acid GCDCA at concentrations varying from 250 μM to 1 mM. The viral inoculum was then added to each well, and cells were washed twice with 2% FBS DMEM 1 day after inoculation. One mL of virus growth medium was then added to each well, and 100 μL of supernatant was harvested at each time point and frozen until RNA extraction. The protocol was adapted from (19).
RNA extraction from cell cultures.
RNA was extracted from cell cultures in 96-well plates with the NucliSens kit (bioMérieux) using the EasyMag automated system. The plates were left at room temperatures to thaw, and then 100 μL of lysis buffer was added to each well and incubated for 10 min. The 200 μL-content of each well was then homogenized and transferred to an EasyMag well containing 2 mL of lysis buffer, and 50 μL of magnetic silica was added. The manufacturer’s program was then used for nucleic acid extraction with 100 μL elution buffer.
Viral quantification by qRT-PCR.
The genomic viral titers were measured by qRT-PCR, using the RNA UltraSense One-Step Quantitative RT-PCR kit. For HuSaV, the forward primers were SaV1F, 5′-TTG GCC CTC GCC ACC TAC-3′ and SaV124F, 5′-GAY CAS GCT CTC GCY ACC TAC-3′, the reverse primer was SaV1245R, 5′CCC TCC ATY TCA AAC ACT A-3′ and the probe was SaV124TP, 5′-FAM-CCR CCT ATR AAC CA-MGBNFQ (54). For HuNoV, the primers were used as previously described (55). Initial denaturation was carried out at 55°C for 15 min followed by 5 min at 95°C, and then 45 cycles consisting of 15 s at 95°C followed by 30 s at 60°C and 30 s at 65°C were performed.
Quantification of viral copy numbers was calculated with a 2 × 6 points standard curve made by 10-fold serial dilutions of synthetic DNA (Integrated DNA Technologies), with the target sequence between nucleotide 4626 and 5683 of HuSaV strain Mc114 (GenBank AY237422.3), or the sequence between nucleotide 4191 and 5863 of HuNoV GII.4 Houston virus (56) (GenBank EU310927).
Gene expression assay by fluidic qPCR.
To assess the differentiation of intestinal cells at the time of infection, the expression of 6 genes was measured in RNA extracted 2 h postinfection (hpi) using a microfluidic qPCR system (Standard Biotools). Primer and probes for gene expression assays (GEA, Applied Biosystems) were chosen to target a reference gene (GAPDH), markers of proliferating (Ki67) and stem cells (LGR5, CD44) and two differentiation markers, Sucrase-Isomaltase (SI) for enterocytes and Mucine 2 (Muc2) for goblet cells. Briefly, 7 μL of RNA was reverse-transcribed using the SuperScript II kit (Thermo Scientific), as per the manufacturer’s instructions, in 20 μL reaction mixtures with 2.5 μM random hexamers and nonamers (Thermo Scientific). Complementary DNA (1.25 μL per sample) was preamplified in 5 μL reaction mixtures using 1 μL of Preamp Master Mix (Standard Biotools) with 1.25 μL of 0.2× primers-probe assays pooled, through 2 min denaturation at 95°C followed by 14 cycles of 15s denaturation at 95°C and 4 min. hybridation-elongation at 60°C. Immediately after the preamplification, PCR products were diluted (1:5) by adding 20 μL of TE and stored at 4°C prior to use in qPCR. For the qPCR assay, a Flex Six integrated fluidic circuit (Standard Biotools) was loaded on one side with 12 × 3 μL reaction mixtures based on the UltraSense One-Step Quantitative RT-PCR kit (Thermo Scientific) in a BioMark device (Standard Biotools), together with 1× GE Sample Loading Reagent (Standard Biotools) and 1.35 μL of preamplified DNA, and on the other side with 3 μL of assay mix containing 1.5 μL of one gene expression assay (10× final) and 1.5 μL assay loading reagent (Standard Biotools), in duplicate for each of the six assays. The qPCR program included a thermal mix with 30 min at 25°C and 1h at 70°C, 5 min. denaturation at 95°C and 45 cycles of 15s denaturation at 95°C, 30s hybridation at 60°C and 30s elongation at 65°C with fluorescence acquisition. It was run in a BioMark device (Standard Biotools). Results were analyzed using the BioMark analysis software (Standard Biotools).
Evaluation of cell viability during HuSaV infection.
Infected HIE monolayers were incubated with 25 μL of Blue Cell Titer (Promega) diluted in 75 μL of differentiation medium, for 3 h. Fluorescence was then measured with a TECAN Infinite 200 Pro (Tecan Group Ltd.) at an excitation wavelength of 560 nm and emission wavelength of 590 nm. Next, the wells were washed twice with CMGF- and the differentiation medium was replaced. Wells without cells were used as baseline absorbance values. The percentage of viable cells was calculated by comparing the absorbance of the measured wells for each condition (inoculated with live or heat-treated S611) to the corresponding control wells, which contained cells that were not infected and were considered to display 100% viability value at each time point.
Controls and statistical analysis.
Viral replication was evaluated by calculating the geometric mean fold change for each set of experiments, as the mean of triplicate 2^ΔCt measured between a late time point (usually 72 hpi) and the first time point (1 to 2h pi) for each replicate. Hence, a geometrical mean fold change above 1 corresponds to an increase in viral genome quantities over time, and below 1, to a decrease. The virus was considered to replicate when the geometrical mean fold change was equal or greater than 3 (~0.5log10). For most experiments with HuSaV, HIE monolayers were inoculated with HuNoV GII.4 (TCH11-64) in parallel as a positive control. Three experiments were excluded from analysis when HuNoV unexpectedly did not replicate (fold increase 0.5 log10 or less), indicating that the culture conditions were suboptimal. Statistical analyses were run using GraphPad Prism version 9.4.0.
ACKNOWLEDGMENTS
We are grateful to Jacques Le Pendu for helpful discussions and insight.
This research was funded by the French Ministry for Agriculture, Direction Générale de l’Alimentation (to F.S.L.G.) and in part by PO1-AI057788 from the National Institutes of Health (to M.K.E. and R.L.A.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
F.S.L.G., M.K.E., and R.L.A. obtained funding; G.E.-N., C.L.M., J.S., X.-L.Z., K.E., and M.D. performed experiments; G.E.-N., J.S., and M.D. analyzed the results; G.E.-N., C.L.M., and M.D. wrote the first version of the manuscript; G.E.-N., K.E., R.L.A., F.S.L.G., M.K.E., and M.D. contributed to the final manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Marion Desdouits, Email: marion.desdouits@ifremer.fr.
Susana López, Instituto de Biotecnologia/UNAM.
REFERENCES
- 1.Madeley CR, Cosgrove BP. 1976. Caliciviruses in man. Lancet 307:199–200. doi: 10.1016/S0140-6736(76)91309-X. [DOI] [PubMed] [Google Scholar]
- 2.Chiba S, Sakuma Y, Kogasaka R, Akihara M, Horino K, Nakao T, Fukui S. 1979. An outbreak of gastroenteritis associated with calicivirus in an infant home. J Med Virol 4:249–254. doi: 10.1002/jmv.1890040402. [DOI] [PubMed] [Google Scholar]
- 3.Oka T, Wang Q, Katayama K, Saif LJ. 2015. Comprehensive review of human sapoviruses. Clin Microbiol Rev 28:32–53. doi: 10.1128/CMR.00011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oka T, Katayama K, Ogawa S, Hansman GS, Kageyama T, Ushijima H, Miyamura T, Takeda N. 2005. Proteolytic processing of sapovirus ORF1 polyprotein. J Virol 79:7283–7290. doi: 10.1128/JVI.79.12.7283-7290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyazaki N, Song C, Oka T, Miki M, Murakami K, Iwasaki K, Katayama K, Murata K. 2022. Atomic structure of the human sapovirus capsid reveals a unique capsid protein conformation in caliciviruses. J Virol 96:e00298-22. doi: 10.1128/jvi.00298-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li T-C, Kataoka M, Doan YH, Saito H, Takagi H, Muramatsu M, Oka T. 2022. Characterization of a human sapovirus genotype GII.3 strain generated by a reverse genetics system: VP2 is a minor structural protein of the virion. Viruses 14:1649. doi: 10.3390/v14081649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oka T, Mori K, Iritani N, Harada S, Ueki Y, Iizuka S, Mise K, Murakami K, Wakita T, Katayama K. 2012. Human sapovirus classification based on complete capsid nucleotide sequences. Arch Virol 157:349–352. doi: 10.1007/s00705-011-1161-2. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Zhang W, Cui L, Shen Q, Hua X. 2018. Metagenomic identification, genetic characterization and genotyping of porcine sapoviruses. Infect Genet Evol 62:244–252. doi: 10.1016/j.meegid.2018.04.034. [DOI] [PubMed] [Google Scholar]
- 9.Becker-Dreps S, González F, Bucardo F. 2020. Sapovirus: an emerging cause of childhood diarrhea. Curr Opin Infect Dis 33:388–397. doi: 10.1097/QCO.0000000000000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valcarce MD, Kambhampati AK, Calderwood LE, Hall AJ, Mirza SA, Vinjé J. 2021. Global distribution of sporadic sapovirus infections: a systematic review and meta-analysis. PLoS One 16:e0255436. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8376006/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Oliveira-Tozetto S, Santiso-Bellón C, Ferrer-Chirivella JM, Navarro-Lleó N, Vila-Vicent S, Rodríguez-Díaz J, Buesa J. 2021. Epidemiological and Genetic Characterization of Sapovirus in Patients with Acute Gastroenteritis in Valencia (Spain). Viruses 13:184. doi: 10.3390/v13020184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitkänen O, Vesikari T, Hemming-Harlo M. 2019. The role of the sapovirus infection increased in gastroenteritis after national immunisation was introduced. Acta Paediatr 108:1338–1344. doi: 10.1111/apa.14690. [DOI] [PubMed] [Google Scholar]
- 13.Halasa N, Piya B, Stewart LS, Rahman H, Payne DC, Woron A, Thomas L, Constantine-Renna L, Garman K, McHenry R, Chappell J, Spieker AJ, Fonnesbeck C, Batarseh E, Hamdan L, Wikswo ME, Parashar U, Bowen MD, Vinjé J, Hall AJ, Dunn JR. 2021. The changing landscape of pediatric viral enteropathogens in the post–rotavirus vaccine era. Clin Infect Dis off Publ Infect Dis Soc Am 72:576–585. doi: 10.1093/cid/ciaa100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farkas T, Deng X, Ruiz-Palacios G, Morrow A, Jiang X. 2006. Development of an enzyme immunoassay for detection of sapovirus-specific antibodies and its application in a study of seroprevalence in children. J Clin Microbiol 44:3674–3679. doi: 10.1128/JCM.01087-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Jahuira H, Gilman RH, Alva A, Cabrera L, Okamoto M, Xu H, Windle HJ, Kelleher D, Varela M, Verastegui M, Calderon M, Sanchez G, Sarabia V, Ballard SB, Bern C, Mayta H, Crabtree JE, Cama V, Saito M, Oshitani H. 2016. Etiological role and repeated infections of sapovirus among children aged less than 2 years in a cohort study in a peri-urban community of Peru. J Clin Microbiol 54:1598–1604. doi: 10.1128/JCM.03133-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harada S, Okada M, Yahiro S, Nishimura K, Matsuo S, Miyasaka J, Nakashima R, Shimada Y, Ueno T, Ikezawa S, Shinozaki K, Katayama K, Wakita T, Takeda N, Oka T. 2009. Surveillance of pathogens in outpatients with gastroenteritis and characterization of sapovirus strains between 2002 and 2007 in Kumamoto prefecture, Japan. J Med Virol 81:1117–1127. doi: 10.1002/jmv.21454. [DOI] [PubMed] [Google Scholar]
- 17.Kitajima M, Iker BC, Pepper IL, Gerba CP. 2014. Relative abundance and treatment reduction of viruses during wastewater treatment processes—identification of potential viral indicators. Sci Total Environ 488-489:290–296. doi: 10.1016/j.scitotenv.2014.04.087. [DOI] [PubMed] [Google Scholar]
- 18.Soliman M, Kim D-S, Kim C, Seo J-Y, Kim J-Y, Park J-G, Alfajaro MM, Baek Y-B, Cho E-H, Park S-I, Kang M-I, Chang K-O, Goodfellow I, Cho K-O. 2018. Porcine sapovirus Cowden strain enters LLC-PK cells via clathrin- and cholesterol-dependent endocytosis with the requirement of dynamin II. Vet Res 49:92. doi: 10.1186/s13567-018-0584-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takagi H, Oka T, Shimoike T, Saito H, Kobayashi T, Takahashi T, et al. 2020. Human sapovirus propagation in human cell lines supplemented with bile acids. Proc Natl Acad Sci USA 117:32078–32085. https://www.pnas.org/content/early/2020/11/25/2007310117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flynn WT, Saif LJ, Moorhead PD. 1988. Pathogenesis of porcine enteric calicivirus-like virus in four-day-old gnotobiotic pigs. Am J Vet Res 49:819–825. [PubMed] [Google Scholar]
- 21.Kim D-S, Hosmillo M, Alfajaro MM, Kim J-Y, Park J-G, Son K-Y, Ryu E-H, Sorgeloos F, Kwon H-J, Park S-J, Lee WS, Cho D, Kwon J, Choi J-S, Kang M-I, Goodfellow I, Cho K-O. 2014. Both α2,3- and α2,6-linked sialic acids on O-linked glycoproteins act as functional receptors for porcine sapovirus. PLoS Pathog 10:e1004172. doi: 10.1371/journal.ppat.1004172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nordgren J, Svensson L. 2019. Genetic Susceptibility to Human Norovirus Infection: an Update. Viruses 11:226. doi: 10.3390/v11030226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marionneau S, Ruvoën N, Le Moullac–Vaidye B, Clement M, Cailleau–Thomas A, Ruiz–Palacois G, Huang P, Jiang X, Le Pendu J. 2002. Norwalk virus binds to histo-blood group antigens present on gastroduodenal epithelial cells of secretor individuals. Gastroenterology 122:1967–1977. doi: 10.1053/gast.2002.33661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang P, Farkas T, Zhong W, Tan M, Thornton S, Morrow AL, Jiang X. 2005. Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J Virol 79:6714–6722. doi: 10.1128/JVI.79.11.6714-6722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shirato-Horikoshi H, Ogawa S, Wakita T, Takeda N, Hansman GS. 2007. Binding activity of norovirus and sapovirus to histo-blood group antigens. Arch Virol 152:457–461. doi: 10.1007/s00705-006-0883-z. [DOI] [PubMed] [Google Scholar]
- 26.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng X-L, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK. 2016. Replication of human noroviruses in stem cell-derived human enteroids. Science 353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosmillo M, Chaudhry Y, Nayak K, Sorgeloos F, Koo BK, Merenda A, et al. 2020. Norovirus replication in human intestinal epithelial cells is restricted by the interferon-induced JAK/STAT signaling pathway and RNA polymerase II-mediated transcriptional responses. mBio 11:e00215-20. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7078467/. doi: 10.1128/mBio.00215-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Estes MK, Ettayebi K, Tenge VR, Murakami K, Karandikar U, Lin S-C, Ayyar BV, Cortes-Penfield NW, Haga K, Neill FH, Opekun AR, Broughman JR, Zeng X-L, Blutt SE, Crawford SE, Ramani S, Graham DY, Atmar RL. 2019. Human norovirus cultivation in nontransformed stem cell-derived human intestinal enteroid cultures: success and challenges. Viruses 11:638. doi: 10.3390/v11070638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ettayebi K, Tenge VR, Cortes-Penfield NW, Crawford SE, Neill FH, Zeng X-L, Yu X, Ayyar BV, Burrin D, Ramani S, Atmar RL, Estes MK. 2021. New insights and enhanced human norovirus cultivation in human intestinal enteroids. mSphere 6:e01136-20. doi: 10.1128/mSphere.01136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rimkutė I, Thorsteinsson K, Henricsson M, Tenge VR, Yu X, Lin SC, et al. 2020. Histo-blood group antigens of glycosphingolipids predict susceptibility of human intestinal enteroids to norovirus infection. J Biol Chem 20:15974–15987. doi: 10.1074/jbc.RA120.014855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crawford SE, Ramani S, Blutt SE, Estes MK. 2021. Organoids to dissect gastrointestinal virus–host interactions: what have we learned? Viruses 13:999. doi: 10.3390/v13060999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murakami K, Tenge VR, Karandikar UC, Lin S-C, Ramani S, Ettayebi K, Crawford SE, Zeng X-L, Neill FH, Ayyar BV, Katayama K, Graham DY, Bieberich E, Atmar RL, Estes MK. 2020. Bile acids and ceramide overcome the entry restriction for GII.3 human norovirus replication in human intestinal enteroids. Proc Natl Acad Sci USA 117:1700–1710. doi: 10.1073/pnas.1910138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang KO, Sosnovtsev SV, Belliot G, Kim Y, Saif LJ, Green KY. 2004. Bile acids are essential for porcine enteric calicivirus replication in association with down-regulation of signal transducer and activator of transcription 1. Proc Natl Acad Sci USA 101:8733–8738. doi: 10.1073/pnas.0401126101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haga K, Ettayebi K, Tenge VR, Karandikar UC, Lewis MA, Lin S-C, Neill FH, Ayyar BV, Zeng X-L, Larson G, Ramani S, Atmar RL, Estes MK. 2020. Genetic manipulation of human intestinal enteroids demonstrates the necessity of a functional fucosyltransferase 2 gene for secretor-dependent human norovirus infection. mBio 11:e00251-20. doi: 10.1128/mBio.00251-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oka T, Stoltzfus GT, Zhu C, Jung K, Wang Q, Saif LJ. 2018. Attempts to grow human noroviruses, a sapovirus, and a bovine norovirus in vitro. PLoS One 13:e0178157. doi: 10.1371/journal.pone.0178157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cubitt WD, McSwiggan DA, Moore W. 1979. Winter vomiting disease caused by calicivirus. J Clin Pathol 32:786–793. doi: 10.1136/jcp.32.8.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cubitt WD, Barrett ADTY. 1984. Propagation of human candidate calicivirus in cell culture. J Gen Virol 65:1123–1126. doi: 10.1099/0022-1317-65-6-1123. [DOI] [PubMed] [Google Scholar]
- 38.Flynn WT, Saif LJ. 1988. Serial propagation of porcine enteric calicivirus-like virus in primary porcine kidney cell cultures. J Clin Microbiol 26:206–212. doi: 10.1128/jcm.26.2.206-212.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varela MF, Rivadulla E, Lema A, Romalde JL. 2019. Human sapovirus among outpatients with acute gastroenteritis in Spain: a one-year study. Viruses 11:144. doi: 10.3390/v11020144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mann P, Pietsch C, Liebert UG. 2019. Genetic Diversity of Sapoviruses among Inpatients in Germany, 2008−2018. Viruses 11:726. doi: 10.3390/v11080726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costantini V, Morantz EK, Browne H, Ettayebi K, Zeng X-L, Atmar RL, Estes MK, Vinjé J. 2018. Human norovirus replication in human intestinal enteroids as model to evaluate virus inactivation. Emerg Infect Dis 24:1453–1464. doi: 10.3201/eid2408.180126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin SC, Qu L, Ettayebi K, Crawford SE, Blutt SE, Robertson MJ, et al. 2020. Human norovirus exhibits strain-specific sensitivity to host interferon pathways in human intestinal enteroids. Proc Natl Acad Sci USA 22:23782–23793. doi: 10.1073/pnas.2010834117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teunis PF, Moe CL, Liu P, E Miller S, Lindesmith L, Baric RS, Le Pendu J, Calderon RL. 2008. Norwalk virus: how infectious is it? J Med Virol 80:1468–1476. doi: 10.1002/jmv.21237. [DOI] [PubMed] [Google Scholar]
- 44.Santiana M, Ghosh S, Ho BA, Rajasekaran V, Du W-L, Mutsafi Y, De Jésus-Diaz DA, Sosnovtsev SV, Levenson EA, Parra GI, Takvorian PM, Cali A, Bleck C, Vlasova AN, Saif LJ, Patton JT, Lopalco P, Corcelli A, Green KY, Altan-Bonnet N. 2018. Vesicle-cloaked virus clusters are optimal units for inter-organismal viral transmission. Cell Host Microbe 24:208–220.e8. doi: 10.1016/j.chom.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shivanna V, Kim Y, Chang KO. 2014. The crucial role of bile acids in the entry of porcine enteric calicivirus. Virology 456–457:268–278. doi: 10.1016/j.virol.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo M, Hayes J, Cho KO, Parwani AV, Lucas LM, Saif LJ. 2001. Comparative pathogenesis of tissue culture-adapted and wild-type Cowden porcine enteric calicivirus (PEC) in gnotobiotic pigs and induction of diarrhea by intravenous inoculation of wild-type PEC. J Virol 75:9239–9251. doi: 10.1128/JVI.75.19.9239-9251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saikia K, Saharia N, Singh CS, Borah PP, Namsa ND. 2022. Association of histo-blood group antigens and predisposition to gastrointestinal diseases. J Med Virol 94:5149–5162. https://onlinelibrary.wiley.com/doi/abs/10.1002/jmv.28028. [DOI] [PubMed] [Google Scholar]
- 48.Bucardo F, Carlsson B, Nordgren J, Larson G, Blandon P, Vilchez S, Svensson L. 2012. Susceptibility of children to sapovirus infections, Nicaragua, 2005–2006. Emerg Infect Dis 18:1875–1878. doi: 10.3201/eid1811.111581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matussek A, Dienus O, Djeneba O, Simpore J, Nitiema L, Nordgren J. 2015. Molecular characterization and genetic susceptibility of sapovirus in children with diarrhea in Burkina Faso. Infect Genet Evol 32:396–400. doi: 10.1016/j.meegid.2015.03.039. [DOI] [PubMed] [Google Scholar]
- 50.Olivares AIO, Leitão GAA, Pimenta YC, Cantelli CP, Fumian TM, Fialho AM, da Silva E Mouta S, Delgado IF, Nordgren J, Svensson L, Miagostovich MP, Leite JPG, de Moraes MTB. 2021. Epidemiology of enteric virus infections in children living in the Amazon region. Int J Infect Dis 108:494–502. doi: 10.1016/j.ijid.2021.05.060. [DOI] [PubMed] [Google Scholar]
- 51.Tohma K, Kulka M, Coughlan S, Green KY, Parra GI. 2020. Genomic analyses of human sapoviruses detected over a 40-year period reveal disparate patterns of evolution among genotypes and genome regions. Viruses 12:516. doi: 10.3390/v12050516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Desdouits M, Polo D, Le Mennec C, Strubbia S, Zeng X-L, Ettayebi K, Atmar RL, Estes MK, Le Guyader FS. 2022. Use of human intestinal enteroids to evaluate persistence of infectious human norovirus in seawater. Emerg Infect Dis 28:1475–1479. doi: 10.3201/eid2807.220219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strubbia S, Phan MVT, Schaeffer J, Koopmans M, Cotten M, Le Guyader FS. 2019. Characterization of norovirus and other human enteric viruses in sewage and stool samples through next-generation sequencing. Food Environ Virol 11:400–409. doi: 10.1007/s12560-019-09402-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oka T, Katayama K, Hansman GS, Kageyama T, Ogawa S, Wu F-T, White PA, Takeda N. 2006. Detection of human sapovirus by real-time reverse transcription-polymerase chain reaction. J Med Virol 78:1347–1353. doi: 10.1002/jmv.20699. [DOI] [PubMed] [Google Scholar]
- 55.Polo D, Schaeffer J, Fournet N, Le Saux J-C, Parnaudeau S, McLeod C, Le Guyader FS. 2016. Digital PCR for quantifying norovirus in oysters implicated in outbreaks, France. Emerg Infect Dis 22:2189–2191. doi: 10.3201/eid2212.160841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Guyader FS, Parnaudeau S, Schaeffer J, Bosch A, Loisy F, Pommepuy M, Atmar RL. 2009. Detection and quantification of noroviruses in shellfish. Appl Environ Microbiol 75:618–624. doi: 10.1128/AEM.01507-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Download jvi.00383-23-s0001.tif, TIF file, 0.07 MB (71.1KB, tif)