ABSTRACT
The lack of robust immunocompetent animal models for hepatitis C virus (HCV) impedes vaccine development and studies of immune responses. Norway rat hepacivirus (NrHV) infection in rats shares HCV-defining characteristics, including hepatotropism, chronicity, immune responses, and aspects of liver pathology. To exploit genetic variants and research tools, we previously adapted NrHV to prolonged infection in laboratory mice. Through intrahepatic RNA inoculation of molecular clones of the identified variants, we here characterized four mutations in the envelope proteins responsible for mouse adaptation, including one disrupting a glycosylation site. These mutations led to high-titer viremia, similar to that observed in rats. In 4-week-old mice, infection was cleared after around 5 weeks compared to 2 to 3 weeks for nonadapted virus. In contrast, the mutations led to persistent but attenuated infection in rats, and they partially reverted, accompanied by an increase in viremia. Attenuated infection in rat but not mouse hepatoma cells demonstrated that the characterized mutations were indeed mouse adaptive rather than generally adaptive across species and that species determinants and not immune interactions were responsible for attenuation in rats. Unlike persistent NrHV infection in rats, acute resolving infection in mice was not associated with the development of neutralizing antibodies. Finally, infection of scavenger receptor B-I (SR-BI) knockout mice suggested that adaptation to mouse SR-BI was not a primary function of the identified mutations. Rather, the virus may have adapted to lower dependency on SR-BI, thereby potentially surpassing species-specific differences. In conclusion, we identified specific determinants of NrHV mouse adaptation, suggesting species-specific interactions during entry.
IMPORTANCE A prophylactic vaccine is required to achieve the World Health Organization’s objective for hepatitis C virus elimination as a serious public health threat. However, the lack of robust immunocompetent animal models supporting hepatitis C virus infection impedes vaccine development as well as studies of immune responses and viral evasion. Hepatitis C virus-related hepaciviruses were discovered in a number of animal species and provide useful surrogate infection models. Norway rat hepacivirus is of particular interest, as it enables studies in rats, an immunocompetent and widely used small laboratory animal model. Its adaptation to robust infection also in laboratory mice provides access to a broader set of mouse genetic lines and comprehensive research tools. The presented mouse-adapted infectious clones will be of utility for reverse genetic studies, and the Norway rat hepacivirus mouse model will facilitate studies of hepacivirus infection for in-depth characterization of virus-host interactions, immune responses, and liver pathology.
KEYWORDS: animal models, HCV, hepacivirus, hepatitis C virus, model virus, cell culture, neutralizing antibodies, species adaptation
INTRODUCTION
Hepatitis C virus (HCV) is an important human pathogen leading to chronic infection of ~60 million individuals at increased risk of hepatitis, liver cirrhosis, and hepatocellular carcinoma (1). Significant improvements to HCV therapy have been implemented over the last decade (2), but no vaccine is available. The lack of robust immunocompetent small animal models supporting experimental HCV infection has impeded progress in understanding viral pathology, immune responses, and vaccine development. Naturally, HCV infects only humans, whereas chimpanzees can be experimentally infected but are no longer available to research (3). GB virus B (GBV-B), a related virus that can infect small New World monkeys, never became a widely used model due to its typical acute resolving course of infection and sparse availability of reagents (4). More recently, a plethora of HCV-related hepaciviruses have been discovered in bats, cows, horses, monkeys, and rodents (5, 6). Of particular interest are equine hepacivirus (EqHV), which is the closest genetic relative to HCV and a causative agent of hepatitis and potentially other liver disease in horses (7), and rodent hepacivirus (RHV), which could serve as a small animal model (8). RHV from Norway rats (NrHV or RHV-rn) typically leads to high-titer persistent infection in laboratory rats with characteristics reminiscent of HCV infection in patients, including liver tropism, hepatitis, and liver steatosis as well as intrahepatic innate immune activation and lymphocyte infiltration (9).
The NrHV prototype isolate RHV-rn1 has a positive single-stranded genomic RNA of 9,656 nucleotides containing a single open reading frame (ORF) encoding a polyprotein of 2,958 amino acids (aa). Its predicted genomic organization resembles that of HCV, with cleavage of the polyprotein into structural (core, E1, and E2) and nonstructural (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) mature proteins. As for HCV, the RHV 5′ untranslated region (UTR) possesses two binding sites for the liver-specific microRNA, miR-122, which is required for replication (9–11). Intrahepatic inoculation of RHV-rn1 genomic RNA leads to productive infection in rats (9). Further, the rat model has been used to evaluate T-cell-based vaccine candidates that partially prevent chronic infection (12, 13) and to study the lack of effector functions and exhaustion of CD8+ T cells during persistency (14). Robust replication was not observed in cell culture after transfection of in vitro-transcribed genomic RNA (11), preventing larger-scale analysis of neutralizing antibodies (NAbs). This limitation was, however, overcome using subgenomic selectable replicons, which led to the identification of replication-enhancing mutations and rat hepatoma cells with increased permissiveness, enabling studies of the intracellular part of the viral life cycle in vitro (11). This recently also allowed establishment of infectious culture systems, which were used to demonstrate NrHV dependency on the HCV entry factor homolog, scavenger receptor B-I (SR-BI), and to establish neutralization assays in rat hepatoma cells (15).
NrHV infection of mice would allow exploitation of a broader set of genetic variants and research tools, but high-titer infection with rat-derived virus is short-lived in immunocompetent mice, with clearance after 2 to 3 weeks in 4-week-old mice and already after 1 week in older mice. We therefore adapted NrHV through passage in the severely immunodeficient NRG (NOD-Rag1−/− IL2Rγ−/−) mice (10). These NOD-congenic mice harbor Rag1 null and IL2Rγ null mutations, leading to a lack of B, T, and NK lymphocytes. The mouse-adapted virus had prolonged infection of up to 5 weeks in wild-type (WT) mice. The immunological features of NrHV infection in mice resemble those of human viral hepatitis, with intrahepatic expansion of virus-specific CD4+ and CD8+ T cells, associated liver injury, and T-cell-dependent acute clearance. Persistent infection could be achieved following transient depletion of CD4+ T cells prior to infection or by continued CD8+ T-cell depletion. In the former case, persistent infection was associated with exhaustion of hepatic CD8+ T cells. Secondary infections were cleared more rapidly in a CD8+ T-cell-dependent manner (10).
To understand the molecular determinants of NrHV mouse adaptation, we here developed infectious clone-based reverse genetic systems and used these to deconvolute species adaptation. We identified a robust mouse-adapted clone, replication competent in mouse hepatoma cell culture but attenuated in rat cells in vivo and in vitro. Using a recently established neutralization assay (15), we observed that long-term infection in rats but not acute resolving infection in mice was associated with emergence of NAbs. The development and characterization of mouse-adapted infectious clones will provide a valuable platform to advance NrHV mouse models, thereby providing tools for better understanding of hepaciviral infection and associated immune responses, liver pathology, and guidance of HCV vaccine development.
RESULTS
Intrahepatic inoculation of NrHV genomic RNA launches infection in immunocompromised mice.
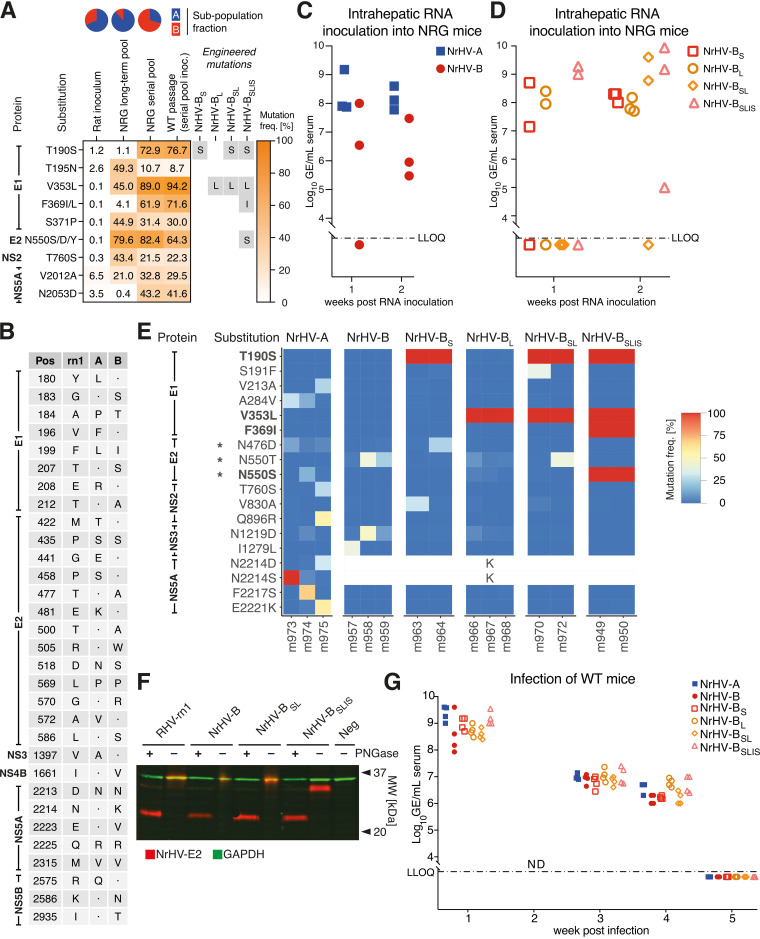
To adapt rat-derived NrHV (RHV-rn1) to mice, we previously set up long-term or serial passage infections in NRG mice to obtain pools of mouse-adapted viruses (10). The polyclonal rat serum inoculum contained two subpopulations, “A” and “B,” each of which during NRG mouse infection accumulated several putative adaptive ORF mutations, predominantly in the E1 and E2 envelope proteins (Fig. 1A). Sequencing of the complete UTRs from the NRG serial passage did not identify further putative adaptive mutations. To allow functional studies of NrHV mouse adaptation, we first generated complete consensus clones for NrHV-A and -B based on the previously reported rat infectious RHV-rn1 clone backbone (9) but with 18 and 21 nonsynonymous differences, respectively (Fig. 1B). Intrahepatic inoculation of in vitro-transcribed (IVT) RNA from these molecular clones led to robust infection in NRG mice with viral serum RNA titers of ~108 genome equivalents (GE)/mL for NrHV-A and 106 to 107 GE/mL for NrHV-B at week 2, at which point the experiment was terminated (Fig. 1C).
FIG 1.
Intrahepatic inoculation of NrHV genomic RNA in immunocompromised mice identifies mouse-adaptive mutations. (A) Variant frequency table for NrHV whole ORF sequencing of the rat inoculum used for long-term or serial passage in NRG mice, of the resulting serum pools, and of WT mice inoculated with the serial pool. The corresponding fractions of the NrHV subpopulations “A” (blue) and “B” (red) are shown at the top. The table includes mutations present at <10% in the rat inoculum and either >40% in the long-term pool (equivalent to >50% of subpopulation A, constituting ~85%) or >30% in the serial pool (equivalent to >50% of subpopulation B, constituting ~65%). Mutations selected for further study are highlighted in gray. (B) Amino acid differences between the RHV-rn1 (GenBank accession no. KX905133), NrHV-A (MF113386), and NrHV-B (ON758386) clones. Numbering is according to the NrHV polyprotein. Dots indicate identity with the RHV-rn1 reference. The NrHV-A and -B UTRs are identical to those of RHV-rn1 except for the single nucleotide substitution G271A. (C) Intrahepatic inoculation of IVT genomic RNA of NrHV-A and NrHV-B consensus clones into 4- to 8-week-old NRG mice. (D) Intrahepatic inoculation of IVT genomic RNA of NrHV-BS, NrHV-BL, NrHV-BSL, and NrHV-BSLIS into 4- to 8-week-old NRG mice. (E) NrHV whole ORF sequencing analysis of serum from NRG mice at week 2. Shown are substitutions with a frequency of >20% in at least one sample against the respective reference genome, NrHV-A or NrHV-B. Engineered mutations are highlighted in boldface. Asterisks indicate residues of predicted glycosylation sites. The blanked position 2214 represents sequence differences between NrHV-A and NrHV-B. The full data set is available at GEO (accession no. GSE225619). (F) Immunoblot of whole-cell lysate derived from HEK-293T cells expressing the E2 protein of RHV-rn1, NrHV-B, NrHV-BSL, or NrHV-BSLIS with or without deglycosylation by PNGase F amidase treatment. (G) Infection of 4-week-old C57BL/6J WT mice with week 2 serum from RNA-inoculated NRG mice. LLOQ, lower limit of quantification; MW, molecular weight; NRG, NOD-Rag1−/−IL2Rγ−/−.
To understand the molecular determinants for mouse adaptation, we engineered putative adaptive mutations into NrHV-B, which exhibited the simpler mutational landscape with four dominating mutations, all in E1-E2, after serial passage in NRG mice (Fig. 1A) (10). The addition of the individual E1 mutations T190S or V353L (NrHV-BS and NrHV-BL, respectively) led to a 10- to 100-fold increase in viremia compared to that of the parental backbone 2 weeks after intrahepatic RNA inoculation in NRG mice (Fig. 1D). A combination of these two mutations (NrHV-BSL) or additional combination with the mutations F369I and N550S (NrHV-BSLIS) further increased viremia ~10-fold to ~109 GE/mL, except for two outlier animals with an apparent insufficient RNA launch.
To survey the selection pressure on the different variants, we did high-throughput sequencing of the entire ORF on week 2 serum from the intrahepatically inoculated NRG mice. For NrHV-A and -B, but also for variants with one or two engineered mutations, further selection pressure was observed with amino acid substitutions with a frequency of >20% in most mice at this early time point (Fig. 1E). This was particularly pronounced at the predicted glycosylation sites N476 and N550. No changes were observed for NrHV-BSLIS, which already contained the N550S mutation. NrHV-BSLIS E2, recombinantly expressed in HEK-293T cells, indeed was observed at a lower molecular weight on an immunoblot than variants containing the original N550 (Fig. 1F), consistent with removal of glycosylation at this site. All E2 variants displayed identical electrophoretic mobilities after removal of N-linked glycans by peptide-N-glycosidase (PNGase) F amidase treatment.
All NrHV variants could be passaged to C57BL/6J WT mice, leading to serum titers of 108 to 109 GE/mL 1 week after infection of 4-week-old mice (Fig. 1G). The highest titers were observed for the NrHV-BSLIS variant and for NrHV-A, which, however, had already adapted at this point (Fig. 1E). Infection was cleared after around 5 weeks, resembling infection with polyclonal NRG-adapted serum (10). NrHV-BSLIS thus exhibited higher viral titers and lower adaptation pressure in mice than the parental NrHV-A and -B clones.
Intrahepatic inoculation into WT mice validates NrHV mouse adaptation.
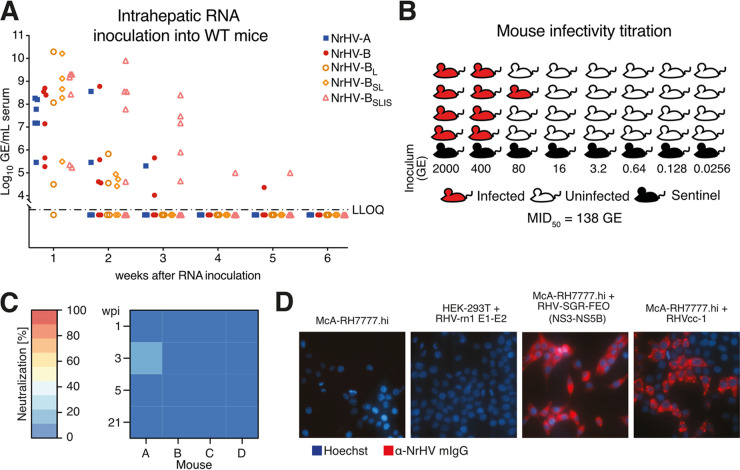
To define the function of the selected mutations in mice, we performed intrahepatic inoculation of 8-week-old immunocompetent WT mice. Whereas the NrHV-A and -B infections were cleared after 2 to 3 weeks, most NrHV-BSLIS animals remained positive 3 weeks postinoculation, with a single animal remaining positive for at least 5 weeks (Fig. 2A). This course of NrHV-BSLIS infection resembled what was seen upon inoculation of WT mice with polyclonal NRG-adapted serum (10). At week 3 for the longest-infected animal, no further mutations had accumulated, and passage of week 5 serum from this animal to naive WT mice did not recapitulate the prolonged infection. Clearance of NrHV-BL and NrHV-BSL infection was similar to that of NrHV-A and -B, suggesting that all four analyzed envelope mutations were necessary for efficient adaptation of NrHV to mice.
FIG 2.
Validation of NrHV mouse-adaptive mutations in WT mice and analysis of antibody responses. (A) Intrahepatic inoculation of IVT genomic RNA of NrHV variants into 8-week-old C57BL/6J WT mice. (B) Schematic of mouse infectivity titration of the mouse-adapted NrHV challenge stock. Mice of each inoculum dose were cohoused with a sentinel animal that did not become infected. The MID50 was calculated according to the Reed-Muench method for establishment of 50% endpoint titers. (C) Neutralization potential of C57BL/6J WT sera at the indicated weeks postinfection (wpi) with week 2 serum from NrHV-B RNA-inoculated NRG mice. Percentage neutralization of RHVcc-1 infection relative to naive preinfection serum is shown. Each data point represents triplicate experiments. (D) Serum from C57BL/6J WT mice collected 6 wpi with NrHV-BSLIS was used to visualize viral antigen in HEK-293T cells expressing RHV-rn1 E1-E2 and in McA-RH7777.hi cells containing a subgenomic replicon (SGR) expressing the NS3-NS5B proteins or infected with RHVcc-1.
We next passaged serum from NrHV-BSLIS RNA-inoculated WT mice twice to naive mice to generate a large virus stock with a titer of 2.2 × 107 GE/mL. No further coding changes were observed in the complete genome of this stock, except for an ~30% presence of A698V in p7. To characterize the infectivity of the NrHV-BSLIS stock, we injected 5-fold serial dilutions, from 2,000 to 0.0256 GE, each into four replicate WT mice and determined a 50% mouse infectious dose (MID50) of 138 GE (Fig. 2B). Well-characterized mouse-adapted NrHV challenge stocks like this will be important resources for studies of vaccine protection from viral challenge in this HCV surrogate model.
To understand whether clearance of infection in WT mice was associated with emergence of neutralizing antibodies, we applied a recently developed neutralization assay using a cell culture-adapted RHV-rn1 strain, RHVcc-1 (15). Curiously, no neutralizing activity was observed in sera from week 1, 3, 5, or 21 from the four WT mice infected with NrHV-B virus (Fig. 2C). Antibodies to nonstructural proteins, however, did develop, as serum from week 6 post-NrHV-BSLIS infection specifically stained McA-RH7777.hi rat hepatoma cells infected with culture-adapted RHVcc-1 virus or transfected with the RHV subgenomic replicon encoding NS3-NS5B but not HEK-293T cells recombinantly expressing E1-E2 (Fig. 2D).
NrHV mouse-adaptive mutations cause attenuation in rats.
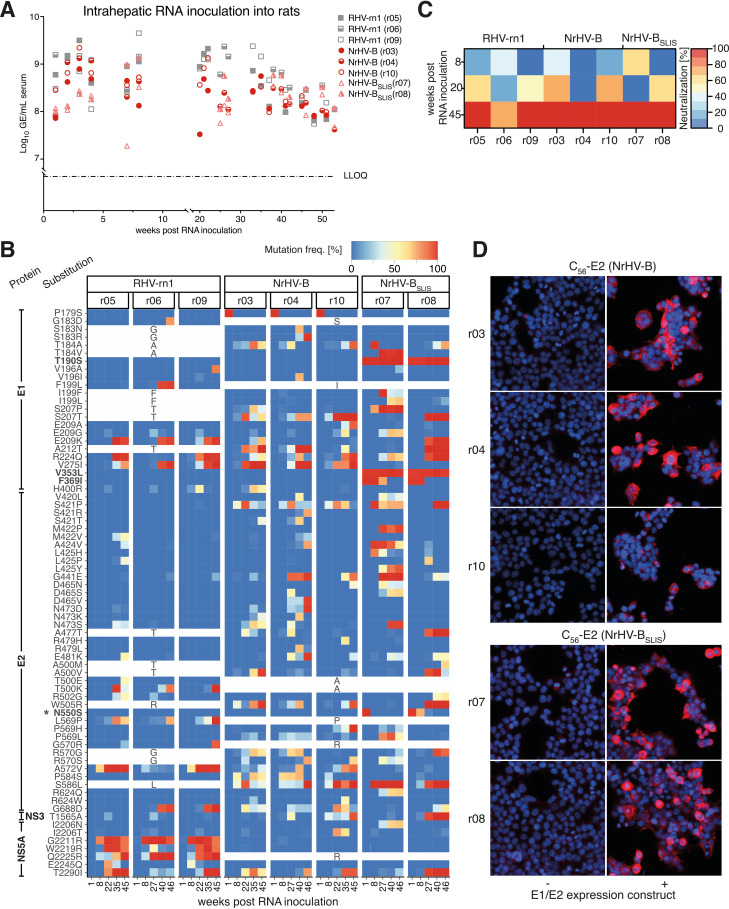
To determine whether the identified substitutions indeed were species specific, we next inoculated inbred Lewis rats intrahepatically with in vitro-transcribed RNA from NrHV-B and NrHV-BSLIS and compared the results to those of the previously established RHV-rn1 clone (9). During the acute phase, similar serum titers of ~109 GE/mL were observed for RHV-rn1 and NrHV-B, except for week 1, where the NrHV-B serum titer was ~5-fold lower (Fig. 3A). For this experiment, we used an NrHV-B clone having S179, given that this was observed for the minor subpopulation “B” of the original rat inoculum used for mouse adaptation (10). The rapid increase in viremia to levels comparable to those of RHV-rn1 correlated with the amino acid substitution S179P. Proline at position 179 is also present in the NrHV-B clone used in mice. NrHV-BSLIS infection was attenuated, with titers remaining around 108 GE/mL throughout the experiment, except for one rat with titers of ~109 GE/mL at week 7 to 8 (Fig. 3A). Although different groups of mutations were observed at week 8 between the NrHV-BSLIS-infected animals, any correlation with the different viral titers observed could not be identified. The N550S mutation consistently reverted early during NrHV-BSLIS infection, whereas the F369I mutation reverted after week 8. The T190S and V353L mutations remained present throughout the experiment (Fig. 3B). Persistent infection was established in all animals, with titers plateauing at ~108 GE/mL from week 20 onwards, except for the RHV-rn1-infected animals, where titers remained at ~109 GE/mL until around week 30.
FIG 3.
NrHV mouse adaptation causes attenuated but persistent infection in rats. (A) Intrahepatic inoculation of IVT genomic RNA of NrHV variants into 10-week-old inbred Lewis rats. (B) NrHV whole ORF sequencing analysis of serum from Lewis rats. Polyprotein positions with a combined frequency of one or several mutations of >50% in at least two rats are included. Individual substitutions with minimal contribution (<20%) at such positions were not included. Engineered mutations are highlighted in boldface. An asterisk indicates the predicted glycosylation site. Blanked positions represent sequence differences between RHV-rn1 (reference for r05, r06, and r09) and NrHV-B (reference for others). The full data set is available at GEO (accession no. GSE225619). (C) Percentage neutralization of RHVcc-1 infection relative to naive preinfection serum at indicated time points post-RNA inoculation of Lewis rats with RHV-rn1, NrHV-B, or NrHV-BSLIS. Each data point represents triplicate experiments. (D) Representative microscopy images of HEK-293T cells expressing an NrHV-B C56-E2 construct after staining with sera collected from rat r03, r04, or r10 (inoculated with NrHV-B) or expressing NrHV-BSLIS C56-E2 and stained with r07 or r08 serum (inoculated with NrHV-BSLIS) taken 20 weeks post-RNA inoculation. Images were taken 2 days posttransfection at ×200 magnification. LLOQ, lower limit of quantification. Data for r05, r06, and r09 were previously published (15) but are included here for direct comparison.
Extensive selection pressure particularly in E1-E2, including in the E2 N terminus (predicted from residue 418), was observed in all animals from around week 20 onwards (Fig. 3B). This correlated with the emergence of neutralizing antibodies after 20 to 45 weeks of infection in rats (Fig. 3C). At week 20 post-NrHV-B or NrHV-BSLIS infection, E1/E2-specific antibodies, however, were present in all rats irrespective of their neutralization potential, as serum stained the recombinantly expressed envelope proteins in HEK-293T cells (Fig. 3D). Finally, selection pressure was observed in the NS5A region of residues 2211 to 2245 for RHV-rn1, similar to previous observations (15), but not for NrHV-B or NrHV-BSLIS, suggesting continuous evolution of RHV-rn1 in this region (Fig. 3B). In aggregate, these data suggested that the envelope mutations present in NrHV-BSLIS are specifically mouse adaptive and do not provide a general advantage to the virus across species.
NrHV mouse-adaptive mutations attenuate infection in rat hepatoma cell culture.
To determine whether the attenuated phenotype of NrHV-BSLIS in rats was due to species adaptation or rat-specific immune responses, we transfected McA-RH7777.hi rat hepatoma cell cultures with NrHV-A, -B, or -BSLIS with and without the previously identified cell culture-adaptive mutations (cc), i.e., L586S (already present in NrHV-B), S1757A, and T2373A. The culture-adapted RHVcc-1 and the original RHV-rn1 clone were included as controls (15). RHVcc-1 spread to the majority of the cells within 8 days, followed by NrHV-B(cc) at around day 14 (Fig. 4A). NrHV-A(cc) and NrHV-BSLIS(cc) followed after around 40 days of infection. The unadapted RHV-rn1 and NrHV-B finally spread after around 70 days, and the experiment was terminated. No spread was observed for NrHV-A and NrHV-BSLIS. Similar findings were observed in a replicate experiment of the most fit variants, RHVcc-1 and NrHV-B(cc). After passage to naive cells, NrHV-B(cc) of the two experiments had acquired either Q342P (E1) or F609S (E2) (Table 1), the latter of which was previously identified as a culture-adaptive mutation engineered in RHVcc-2 (15). Despite the obvious attenuation of NrHV-BSLIS(cc), its mouse-adaptive mutations did not revert, and in addition to nonstructural mutations, it rather accumulated four additional mutations in the envelope proteins, presumably compensating for the SLIS mutations. NrHV-A(cc) accumulated 12 ORF mutations, suggesting less benefit from the RHVcc-1-derived culture-adaptive mutations. The unadapted RHV-rn1 and NrHV-B accumulated 6 and 4 coding mutations, respectively, including F609S (NrHV-B) and the replicon-derived mutation D2374G (RHV-rn1) (11). Engineering of F609S into NrHV-B(cc) led to efficient replication and virus production after genomic RNA transfection (Fig. 4B), and we named this novel robust cell culture clone NrHV-B(cc)-1. Infection of McA-RH7777.hi rat hepatoma cells with serum-derived NrHV-B (rat derived) or NrHV-BSLIS (rat or mouse derived) virus did lead to antigen-positive cells, but those were few and infection did not spread in the culture, emphasizing the prerequisite of cell culture adaptation for robust infection and functional assays in vitro (Fig. 4C). Hence, the SLIS mutations were disadvantageous in rat hepatoma cell culture.
FIG 4.
NrHV mouse-adaptive mutations attenuate infection in rat cell culture. (A) Percentage of NrHV antigen-positive McA-RH7777.hi rat hepatoma cells after transfection with NrHV-A, NrHV-B, NrHV-BSLIS, or the RHV-rn1 control without or with (cc) engineered RHVcc-1-derived cell culture-adaptive mutations (15). (B) RHV infectivity titers in supernatant after transfection of McA-RH7777.hi cells with genomic RNA of RHVcc-1, NrHV-B(cc), culture-adapted NrHV-B [NrHV-B(cc)-1], or replication-deficient RHV-pol(–). Data points represent mean values ± SD from technical triplicates. (C) Infection of McA-RH7777.hi cells at a multiplicity of infection of 10 (GE/cell) using a 1:9 dilution of C57BL/6 WT serum taken 2 weeks after infection with NrHV-BSLIS. Sentinel mouse serum at the same dilution was used as a negative control. Representative microscopy images were taken 2 days postinfection at ×200 magnification. (D) Coelectroporation of AML12 mouse hepatoma cells with 0.325 μM miR-122 mimic and genomic RNA of NrHV-B(cc), NrHV-BSLIS(cc), or RHVcc-1. RHV antigen-positive cells are shown in representative microscopy images taken at ×200 magnification on day 6 (left) and quantified over time (right). Additional miR-122 mimic was supplemented every 4 days.
TABLE 1.
Mutations identified after propagation in cell culturea
| Protein | Position | Reference | Mutation in: |
Adaptation | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RHVcc-1 | RHV-rn1 | NrHV-A(cc) | NrHV-B(cc) 1st | NrHV-B(cc) 2nd | NrHV-B | NrHV-BSLIS(cc) | ||||
| Core | 156 | I | M | I/M | M | (Mouse) | ||||
| E1 | 179 | P | Sb | Sb | Sb | |||||
| 190 | T | S | Mouse | |||||||
| 320 | A | V | ||||||||
| 342 | Q | Q/P | P | |||||||
| 344 | G | S | ||||||||
| 353 | V | L | Mouse | |||||||
| 369 | F | I | Mouse | |||||||
| 381 | F | L | ||||||||
| 393 | C | S | ||||||||
| E2 | 445 | L | P | |||||||
| 493 | G | E | ||||||||
| 550 | N | S | Mouse | |||||||
| 586 | L | S | S | Sc | Sc | Sc | Sc | Culture | ||
| 609 | F | S | S | Culture | ||||||
| 637 | N | K | N/K | (Culture) | ||||||
| NS2 | 863 | A | V | |||||||
| NS3 | 1398 | K | R | |||||||
| 1518 | D | D/H | ||||||||
| 1520 | P | L | ||||||||
| NS4A | 1610 | E | G | |||||||
| NS4B | 1623 | Q | R | |||||||
| 1757 | S | A | A | A | A | A | Culture | |||
| 1862 | M | V | ||||||||
| NS5A | 1900 | H | Y | |||||||
| 1993 | V | V/I | ||||||||
| 2107 | A | A/T | ||||||||
| 2147 | E | E/D | ||||||||
| 2182 | S | T | ||||||||
| 2295 | L | L/S | ||||||||
| 2297 | P | P/S | ||||||||
| 2304 | P | S | ||||||||
| 2320 | Q | R | ||||||||
| 2362 | S | T | ||||||||
| 2373 | T | A | A | A | A | A | Culture | |||
| 2374 | D | G | Culture | |||||||
| NS5B | 2392 | Q | R | (Culture) | ||||||
| 2527 | S | S/T | ||||||||
| 2579 | T | A | ||||||||
| 2916 | E | E/K | ||||||||
| 2945 | I | M | ||||||||
Engineered mutations are underlined and in boldface. Mutations demonstrated to provide adaptation to mice or cell culture (infectious virus or replicon) are indicated. Parentheses indicate that the mutation was observed but not tested in reverse genetic studies.
NrHV-B (but not NrHV-BSLIS) was engineered with 179S, given that S was found in the original rat inoculum (GenBank accession no. ON758386).
The NrHV-B sequence already has 586S.
With a specific role for the SLIS mutations in mouse adaptation, these would be expected not to attenuate replication in mouse hepatocytes. Permissiveness of mouse hepatoma cell cultures to RHV-rn1 replication is low (11) but can be enhanced by exogenous addition of miR-122 (16). We therefore coelectroporated miR-122 mimic and RNA transcripts of RHVcc-1, NrHV-B(cc), and NrHV-BSLIS(cc) into AML12 mouse hepatoma cells. RHVcc-1 infection spread faster than infection with NrHV-B(cc), which was similar to that of NrHV-BSLIS(cc), demonstrating that the SLIS mutations do not attenuate replication in mouse hepatoma cells in vitro (Fig. 4D). AML12 cells did not produce viral particles infectious for AML12 or McA-RH7777.hi cells despite exogenous miR-122 delivery. They furthermore were resistant to infection with McA-RH7777.hi cell-derived virus, suggesting a primary mechanism of cell-to-cell spread in the electroporation experiment (17). In aggregate, these in vitro data were consistent with a primary role for the SLIS mutations in NrHV adaptation to mice.
NrHV mouse-adaptive mutations do not compensate for species-specific differences in SR-BI interaction.
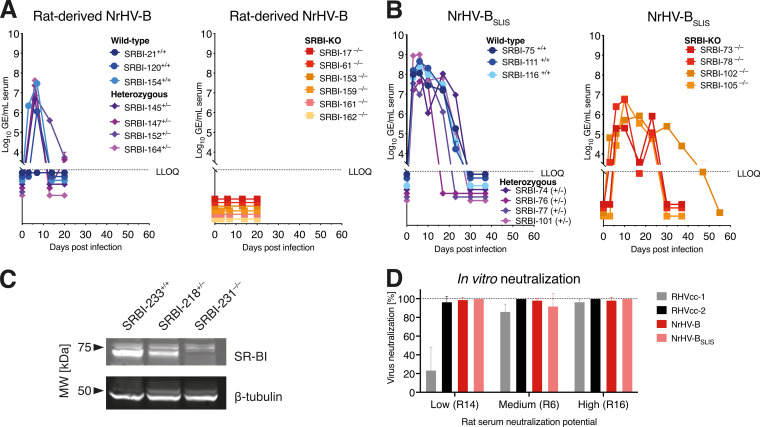
The localization of the SLIS mutations in the E1-E2 envelope proteins suggested a potential role in adapting viral entry to mouse cells. Since we recently identified the rat homolog of the HCV coreceptor SR-BI as an important NrHV host factor (15), we hypothesized that the SLIS mutations would enable interactions with mouse SR-BI. Since direct assessment of this in vitro is challenged by the inability to culture NrHV-BSLIS without additional envelope mutations, we set out to investigate this in vivo. Inoculation with rat serum-derived NrHV-B led to infection of WT but not SR-BI knockout (SR-BI−/−) mice (Fig. 5A), suggesting that NrHV-B apparently was already able to use mouse SR-BI without adaptation. Inoculation with mouse serum-derived NrHV-BSLIS, in contrast, led to infection of SR-BI−/− mice (15) (Fig. 5B), although infection was delayed and attenuated compared to that of WT mice. The absence of SR-BI in the liver of knockout mice was confirmed by immunoblotting (Fig. 5C). These data suggested that the SLIS mutations, rather than adapting to mouse SR-BI, may reduce SR-BI dependency, which would constitute an alternative way to overcome any species-specific barrier in SR-BI interaction. This was supported by the fact that SR-BI knock-down did not affect the number of positive McA-RH7777.hi cells after infection with NrHV-BSLIS derived from rat serum (r08, week 1). We note that certain glycan-disruptive mutations in HCV E2 strongly decreased SR-BI dependency, and this correlated with a higher sensitivity to neutralizing antibodies (18). We therefore set out to compare the sensitivities of NrHV-B and NrHV-BSLIS to rat sera with low, medium, or high levels of neutralizing antibodies. However, since NrHV-B already exhibited high sensitivity to neutralization, any further effect by the SLIS mutations could not be assessed (Fig. 5D). Increased neutralization sensitivity by culture adaptation was previously observed for RHVcc-2 (15) and for HCV (19–21), and this apparently also affected NrHV-B. In aggregate, our data would be consistent with NrHV mouse adaptation through decreased SR-BI dependency, although substantiating this through in vitro experiments was challenged by the inability to culture unadapted NrHV-B and NrHV-BSLIS and the inherent enhanced sensitivity to neutralizing antibodies introduced by culture adaptation of this NrHV strain.
FIG 5.
NrHV mouse-adaptive mutations do not compensate for species-specific differences in SR-BI interaction. (A) Infection of WT+/+, heterozygous+/−, or SR-BI−/− knockout mice with 106 GE of NrHV-B virus derived from rat serum (week 1 post-intrahepatic infection). The age of mice in this experiment was 9 to 15 months (mean, 11 months). (B) Infection of WT+/+, heterozygous+/−, or SR-BI−/− knockout mice with 104 GE of NrHV-BSLIS virus derived from mouse serum. Data in this panel were previously published (15) but are included here for direct comparison. The age of mice in this experiment was 2 months. (C) Immunoblot detection of SR-BI in total liver protein extracts prepared from WT+/+, heterozygous+/−, or SR-BI−/− knockout mice, demonstrating the absence of immunoreactive SR-BI in knockout mouse liver. (D) Percentage neutralization of RHVcc-1, RHVcc-2, NrHV-B (second experiment stock), or NrHV-BSLIS (see Table 1 for changes to viral ORF) using rat sera with low (R14), medium (R6), or high (R16) neutralization potential against RHVcc-1, as previously determined (15). Comparison was made in triplicate against naive preinfection sera from R16 (all sera at week 45 postinfection).
DISCUSSION
The establishment of experimental NrHV infection in rats and mice as models for HCV provides strong complementary platforms, each with specific advantages. Inbred rat strains, like the Lewis rats used here, become persistently infected, whereas some level of spontaneous clearance is observed in outbred strains like Holtzman rats (9). Considering this parameter, infection thereby mimics HCV in patients and NrHV infection of rats serves as an interesting challenge model for vaccine development. In particular, a first goal of an HCV vaccine would be to prevent chronicity (22), which could be studied in the rat model. Accordingly, NrHV in rats could be useful for an initial assessment and comparison of vaccine platforms and a combination of such. Given the narrow host tropism of HCV, and after discontinuation of chimpanzee research, no challenge model is available, although vaccine-induced immune responses can be studied in mice (23). A human challenge model has been proposed but is still a pending solution to ethical and other concerns (24). Shortcomings using NrHV to model HCV infection include nonidentical epitopes between NrHV and HCV and the few reported NrHV strains spanning limited antigenic diversity in comparison to those of HCV (25). The innate immune responses, liver inflammation, delayed seroconversion compared to that of most acute viral infections, and spontaneous T-cell dysfunction, are all representative features of HCV infection mimicked in the rat model (9, 14, 15). For certain aspects of HCV infection in vivo, including assessment of antiviral drugs, neutralizing antibodies, receptor usage, and to some extent pathology, existing animal models are useful. For example, human liver chimeric mice can be infected with clinical strains of HCV, and ongoing efforts also attempt complementation with human immune cells in this model (23). Transgenic mice exogenously expressing the HCV human entry factors have been engineered, but infection levels are low, leading to concerns of accurate representation of pathology and immune responses (6). Finally, mouse-adapted HCV allows some studies of infection in vivo (26).
For functional studies, mouse models are advantageous, in particular due to the abundance of genetic strains, knockout models, and immunological reagents, allowing detailed dissection of immune responses to infection (10, 27). Genetically manipulated mouse strains have already been shown to allow direct interrogation of NrHV-host interactions in vivo, as has been demonstrated for the host factors miR-122 (10) and SR-BI (15). Transient depletion of CD4+ T cells prior to infection further allows establishment of chronic infection, enabling studies of long-term pathology and immune exhaustion in murine models (10). Reverse genetic systems represent valuable tools for such studies, facilitating manipulation of viral RNA and protein sequences responsible for interactions with host factors, including specific epitopes for T cells and antibodies. Furthermore, this study provides a genetically robust mouse model challenge virus with a defined MID50 specific infectivity of 138 GE. This compares well with values obtained for other hepacivirus models, including 2 to 100 GE for HCV in chimpanzees (28–30) and ~10 GE for EqHV in horses (31). With the current study, molecular clones leading to robust infection in both mice and rats (9) are now available.
We previously adapted NrHV to infect laboratory mice (10), but the putative mutations responsible for species adaptation were not characterized. In addition, the presence of two subpopulations, A and B, in the original rat inoculum complicated conclusions. We here delineated a defined set of mutations allowing adaptation of NrHV-B to WT mice by introducing T190S, V353L, and F369I in E1 and N550S in E2. Substitutions at several of the residues involved, including residues 190 and 550, were also observed in SCID mice infected with culture-derived RHVcc-1 (15), further validating the importance of these residues in mouse adaptation. The S179 mutation observed in subpopulation B of the original rat inoculum (10) and included in the NrHV-B clone launched in rats rapidly changed to P, coinciding with restoration of the ~5-fold titer attenuation compared to that of RHV-rn1 observed at week 1. This suggested that this position was not species dependent but potentially responding to immune pressure in the specific rat. The mouse-adapted variant, NrHV-BSLIS, was attenuated in rats, followed by reversion of F369I and N550S, suggesting specific mouse adaptation rather than general adaptation across species by these mutations. Given that all mouse-adaptive mutations are in the envelope proteins, the observed reversion in rats is compatible with selection pressure from NAbs. In WT mice, only very low levels of neutralizing activity but pronounced T-cell responses were observed (10), mirroring cases of resolving HCV infection in chimpanzees in the absence of NAbs (32). Low or absent levels of NAbs in mice would be consistent with the lack of efficient antibody-mediated selection pressure on the SLIS residues. The rapid reversion of N550S to restore the glycosylation site in rats would align with a role in shielding important immune recognition epitopes, potentially by making the virus less sensitive to NAbs. For HCV, specific E1-E2 glycans stabilize a closed, difficult-to-neutralize, envelope conformation, whereas others have the opposite effect (18). For HCV, 5 to 6 and 11 N-linked glycans are typically predicted in E1 and E2, respectively, whereas for NrHV, the numbers are 3 and 4 (33). Due to poor sequence conservation in E2, it is difficult to directly identify glycan homologs, but the glycan localized closest to NrHV N550 is HCV N623 (H77 reference numbering; accession no. AF009606), and mutagenesis of this strongly increases sensitivity to neutralization (18), supporting the above-mentioned hypothesis. T-cell responses in addition to NAbs may play a role in selection pressure in immunocompetent rats, including toward previously defined epitopes in the envelope and nonstructural proteins (14). The T190S mutation for example is adjacent to the E1 191–199 major histocompatibility complex class I (MHC-I) epitope, in which substitutions occurred in several rats. The same was observed in the E2 439–456 MHC-I epitope.
The observed infection of SR-BI−/− mice by NrHV-BSLIS and not NrHV-B is consistent with decreased dependency on SR-BI. Remarkably, a strong inverse correlation between neutralization sensitivity and SR-BI dependency during HCV entry was previously reported (18). This is governed by the flexible E2 N-terminal hypervariable region 1 (HVR1) and by the presence of N-linked glycans determining the equilibrium between open and closed envelope conformations, making the virus more or less susceptible to neutralization. The existence of an HVR1 has not been defined for NrHV, although during long-term infection in the current study, a number of changes were observed in the E2 N-terminal region. Recent analysis, on the other hand, suggested that hepaciviruses other than HCV in general lack an E2 N-terminal HVR1 (34). Nonetheless, the role of N-linked glycans may be conserved, and N550S accordingly may not only increase sensitivity to neutralizing antibodies but concomitantly decrease SR-BI dependency, a strategy that would be advantageous for NrHV mouse adaptation if the barrier to adapting to mouse SR-BI is high and levels of neutralizing antibodies raised in mice are low, which is suggested by the present and previous studies (10). Of note, however, it cannot be excluded that the lack of NrHV-B infection in SR-BI−/− mice was a result of the generally lower fitness of NrHV-B in mice compared to NrHV-BSLIS, an aspect also reflected in the requirement for a higher inoculation dose for NrHV-B (Fig. 5A and B). Furthermore, the mice used for the NrHV-B experiment were significantly older than the mice used for the NrHV-BSLIS experiment (mean of 47 versus 8 weeks), which could impact interpretations, although previous observations suggest that age has an impact primarily on infection for very young animals (≤4 weeks) (10). NrHV-BSLIS could not be cultured without the acquisition of a number of additional envelope mutations, preventing direct interpretation of neutralization sensitivity and SR-BI dependency experiments in vitro. For example, single culture-adaptive mutations were previously demonstrated to greatly change neutralization susceptibility (15, 19–21). Nonetheless, the disruption of predicted NxS/T glycosylation sites (X refers to any amino acid except proline) was associated with mouse adaptation in several other cases, including E2 N476 for NrHV-A and NrHV-BS, E2 N550 (or S552L) for RHVcc-1 infection of SCID mice, and E1 N263 (through S265P) after passage of SCID mouse serum to cell culture (15). N550S may therefore exemplify a more general mechanism of adaptation. Alternative explanations of mouse adaptation remain, including disruption of glycosylation sites to enable interaction with other entry receptors. The role of the homologs of the HCV coreceptors, CD81, claudin-1, and occludin, remains to be determined for other hepaciviruses (6). Attempts to adapt HCV to infect mouse hepatocytes also led to selection of envelope mutations (26). These mediated interaction with mouse CD81, which forms a species barrier of HCV infection in mice (35), but also reduced SR-BI dependency and increased sensitivity to NAbs (26). Although this virus could enter the mouse liver in vivo, robust infection was not observed, even in innate immunocompromised models (36). Thus, parallels exist in attempts to adapt HCV and NrHV to mice, which may inform further species adaptation.
The attenuated growth of NrHV-BSLIS also in rat cell culture further confirmed that its rat in vivo attenuation was due to species adaptation and not rat-specific immune responses. Surprisingly, the SLIS mutations did not revert in culture, and additional mutations instead accumulated. Unfortunately, this prevented direct studies of the mouse-adapted NrHV-BSLIS variant in culture. Nonetheless, these cell culture experiments through further adaptation led to the establishment of the culture-adapted NrHV-B(cc)-1, thereby providing a cell culture infectious clone in a background different from that of the RHV-rn1-based RHVcc-1 (15). Further exploration of NrHV feral isolates will allow a full understanding of diversity and thereby studies, e.g., of the broadness of immune responses. The ability to study NrHV infection also in mouse hepatocyte culture would be advantageous for species- and tropism-specific analyses. AML12 cells complemented with exogenous miR-122 led to efficient RHVcc-1 replication, but these cells were not competent for infectious virus production and were not susceptible to NrHV infection. Similar to HCV replication in HepG2 cells, a combined exogenous delivery of miR-122 and entry factors accordingly may be necessary (37).
In conclusion, this study identified and characterized adaptive mutations responsible for mouse adaptation of NrHV. Our data would be consistent with selection of mouse-adapted variants with a lower dependency on SR-BI and possibly also increased sensitivity to neutralizing antibodies. The resulting reverse genetic systems will be valuable tools for studies of this virus in vitro and in vivo and complete this toolbox for both rats and mice. Given similarities to HCV in the course of infection, immune responses, and liver pathology, the rodent infection models using NrHV could prove valuable for understanding hepaciviral immunity and pathology and as a vaccine challenge model in order to identify the most promising concepts to advance in human HCV vaccination studies.
MATERIALS AND METHODS
Ethics statement.
Animal infection experiments were done either at The Rockefeller University in accordance with the NIH Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee (IACUC), protocol 21056-H, or at the Department of Experimental Medicine, University of Copenhagen, under protocols 2017-15-0201-01288 and 2022-15-0201-01292 approved by the national Animal Experiments Inspectorate. All work was consistent with an affirmative response to the ARRIVE 10 questionnaire.
Animal work.
C57BL/6J and NOD.Cg-Rag1 IL2rgtmlWjl/Sz (NRG) mice were purchased from the Jackson Laboratory. For intrahepatic inoculation of in vitro-transcribed (IVT) RNA of NrHV clones, C57BL/6J or NRG mice were anesthetized using isoflurane and shaved in the abdominal region, and 10 μg of IVT NrHV genomic RNA diluted in 100 μL of phosphate-buffered saline (PBS) was percutaneously injected at two different hepatic locations, each location receiving a volume of 50 μL. For infection experiments, C57BL/6J mice were infected with 104 NrHV GE by retro-orbital or tail vein injection. Differences in route of infection were determined by different institutional requirements. To assess viral titers and neutralizing antibodies, mice were bled via the retro-orbital route at multiple time points post-RNA inoculation and serum was stored at −80°C until analysis.
RNA launch in rats was conducted in female LEWOrl/Rj rats (Janvier) by percutaneous intrahepatic injection under isoflurane anesthesia of 10 μg IVT NrHV genomic RNA dissolved in 100 μL sterile PBS distributed in two hepatic locations, each receiving 5 μg. Blood was sampled from rats by tail vein cannulation under isoflurane anesthesia according to guidelines for blood sampling.
The role of mouse adaptation for SR-BI interaction was assessed in WT and SR-BI-deficient mice (Scarb1tm1Kri/J) (38) obtained from the Jackson Laboratory. Male and female mice were maintained in the B6;129S outbred background. Age-matched scarb1+/+, scarb1+/−, and scarb1−/− male and female mice (age range, 7 to 63 weeks) were infected with 104 GE of the mouse serum-derived NrHV-BSLIS challenge stock via tail vein injection or with 106 GE of rat serum-derived NrHV-B (r10; 1 week postinfection [wpi]) intraperitoneally under isoflurane anesthesia. Blood was sampled by temporal vein puncture in mice under isoflurane anesthesia according to guidelines for blood sampling. For genotyping of mice, DNA was extracted from fresh mouse tail tip biopsy specimens by using the SYBR green Extract-N-Amp tissue PCR kit (Sigma). The presence of the disrupted or the WT SR-BI allele was detected by PCR amplification using primer TS-O-01391 (TGAAGGTGGTCTTCAAGAGCAGTCCT) in combination with either TS-O-01392 (TATCCTCGGCAGACCTGAGTCGTGT; WT specific) or TS-O-01393 (GATTGGGAAGACAATAGCAGGCATGC; mutant specific). The PCR was carried out using Q5 hot-start high-fidelity 2× master mix (New England Biolabs), 0.5 μM each primer, and 2 μL of DNA extracted from mouse tail. The cycling parameters were 98°C for 30 s, followed by 40 cycles of 98°C for 10 s, 57°C for 15 s, and 72°C for 80 s, with a final extension at 72°C for 3 min.
Cells and antibodies.
McA-RH7777.hi cells, highly permissive for NrHV replication (11, 15), and HEK-293T cells were maintained in Dulbecco's modified Eagle medium (DMEM; Thermo Fisher), supplemented with 10% fetal bovine serum (FBS), and, for McA-RH7777.hi cells, with further addition of 100 U/mL penicillin and 100 μg/mL streptomycin (Pen Strep; Sigma). AML12 cells were maintained in DMEM-F12 medium, supplemented with 10% FBS, 1× insulin-transferrin-selenium supplement (Gibco), 40 ng/mL dexamethasone, and the above-specified antibiotics. All cell lines were kept in a 5% CO2 atmosphere at 37°C and passaged every 2 to 3 days using trypsin-EDTA (Sigma).
NrHV-specific IgG (anti-NrHV) was purified from sera derived from four C57BL/6 mice at week 6 after infection with NrHV-BSLIS, after the mice had cleared the infection. NrHV anti-E2 monoclonal IgG was isolated from mice immunized with purified NrHV E2 ectodomain (aa 418 to 597) recombinantly expressed in HEK-293T cells, as previously described (15).
Extraction, quantification, and deep sequencing of viral RNA.
For the extraction of NrHV genomic RNA from infected rodents, 25 μL of rat or mouse serum was diluted in 225 μL of PBS, added to a 2-mL Phasemaker tube (Thermo Fisher Scientific), and mixed with 750 μL of TRIzol LS reagent (Thermo Fisher Scientific). After addition of 200 μL of chloroform, vigorous shaking for 15 s, 3 min of incubation at room temperature, and centrifugation at 12,000 × g for 15 min at 4°C, the aqueous phase was mixed with 450 μL of anhydrous ethanol and transferred to an RNA Clean & Concentrator-5 column (Zymo Research) for downstream RNA purification and concentration. The complete NrHV ORF was amplified and subjected to library preparation and deep-sequencing analysis as previously described (10). The obtained reads were analyzed using the previously published VirVarSeq analysis pipeline (39) with a set fixed quality value of 30 to call variation at the codon level. This package is available at http://sourceforge.net/projects/virtools/?source=directory. All samples had a minimum coverage of 4,000 reads per position over the entire ORF. Variant calling frequency tables from VirVarSeq from samples of relevance were merged and filtered in RStudio. For NRG mice in which variation was less, substitutions with a frequency of >20% in at least one sample were kept. For rats in which variation was greater, positions with a combined frequency of one or several mutations of >50% in at least two rats were kept. Individual substitutions with minimal contribution (<20%) at such positions were not included. The presented heat maps were made from the resulting tables using ggplot2 in RStudio. The raw data and processed frequency tables were deposited at GEO (accession no. GSE225619).
For NrHV RNA quantification, one-step reverse transcription quantitative PCR (RT-qPCR) with TaqMan fast virus 1-step master mix (Thermo Fisher Scientific) was performed using a cobas z 480 analyzer (Roche) with the following protocol: 50°C for 30 min, 95°C for 5 min, and then 40 cycles of 95°C for 15 s, 56°C for 30 s, and 60°C for 45 s. The NrHV NS3-specific primers used for this protocol were TS-O-00561 (sense; TACATGGCTAAGCAATACGG), TS-O-00562 (antisense; AAGCGCAGCACCAATTCC), and TS-O-00563 (probe; [6FAM]CTCACGTACATGACGTACGGCATG[BHQ1a-6FAM] [where BHQ is black hole quencher and 6FAM is 6-carboxyfluorescein]). An NrHV standard curve was generated from IVT NrHV genomic RNA. After in vitro transcription and rigorous DNase treatment, the NrHV genomes were quantified and diluted to concentrations ranging from 5 × 107 to 5 GE/μL.
5′- and 3′-end determination.
Total RNA was purified from serum by TRIzol LS (Thermo Fisher Scientific) extraction. A modified version of the 5′ RACE system for rapid amplification of cDNA ends, version 2.0 (Invitrogen), was applied to determine the NrHV 5′ end. For first-strand cDNA synthesis, purified total RNA was preincubated with 2 μM gene-specific primer TS-O-00259 (GATGGTTTACAGCGGAAACG) and 10 mM deoxynucleoside triphosphate (dNTP) mix at 65°C for 5 min and then 1 min on ice. Next, the RT reaction was carried out by adding Superscript III reverse transcriptase (Invitrogen) and RNasin Plus RNase inhibitor (Promega), followed by incubation in a 30-min gradient from 50°C to 55°C, 5 min at each 1°C increment, followed by inactivation at 70°C for 15 min. Input RNA was degraded by the addition of RNase H and RNase T1 mix for 20 min at 37°C. The resulting cDNA was purified by S.N.A.P. column purification and tailed with 2 mM dCTP, by incubation at 94°C for 2 min, before the addition of terminal deoxynucleotidyl transferase (TdT) and incubation for another 10 min at 37°C and 10 min at 65°C. The 5′ end was amplified by nested PCR amplification using Q5 hot-start high-fidelity 2× master mix. First, C-tailed cDNA was amplified in PCR 1 using 10 μM 5′ RACE abridged anchor primer (AAP) as a forward primer and 10 μM TS-O-001674 (AATGTTTCGCTGCTGACC) as a reverse primer and later reamplified in PCR 2 using 10 μM abridged universal amplification primer (AUAP) as a forward primer and TS-O-01719 (TTCGCTGCTGACCTGCTCTGGC) as a reverse primer.
For 3′-end determination, purified RNA was tailed with 10 mM ATP by incubation with yeast poly(A) polymerase (USB Affymetrix) and RNasin plus RNase inhibitor (Promega) at 37°C for 10 min. NrHV cDNA was then synthesized by preincubation of the reaction mixture with TS-O-00178 (AAP-T20NV, GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTTTTTVN) and 10 mM dNTP mix at 65°C for 5 min, and then holding the mixture at 48°C, while adding the RT mixture of Superscript III reverse transcriptase (Invitrogen), 0.1 M dithiothreitol (DTT), and RNasin plus RNase inhibitor (Promega), before allowing the incubation to continue on a 48°C to 55°C gradient, followed by inactivation at 70°C for 15 min. Input RNA was degraded by incubation with RNase H and RNase T1 mix for 20 min at 37°C. cDNA from 3′-end A-tailed RNA was amplified with Q5 hot-start high-fidelity 2× master mix in PCR 1 using 10 μM AUAP and 10 μM RHV-9020-F (AGCATACACGCCCAGGGAAA) and further reamplified in PCR 2, using AUAP and TS-O-00258 (CCAGGCGTTGCATGATAACAC). The same cycling parameters were used in PCR 1 and PCR 2 for both 5′- and 3′-end determination: 98°C for 1 min, followed by 40 cycles of 98°C for 30 s, 52°C for 40 s, and 72°C for 1 min, and a final extension step at 72°C for 5 min. Finally, the amplified 5′- and 3′-end PCR fragments were purified from a 1% agarose gel by using a Zymoclean gel DNA recovery kit (Zymo Research) and sequenced by Sanger sequencing (Macrogen Europe).
Construction of molecular clones.
Consensus sequences of the rat-derived NrHV subpopulations A and B have been previously defined (10) and deposited in the GenBank database (NrHV-A, accession no. MF113386; NrHV-B, accession no. ON758386). Infectious molecular clones pNrHV-A and pNrHV-B were derived by fusing 3 or 4 PCR-amplified NrHV fragments from subclones of respective consensus sequence within the amplified PCR fragment (InFusion cloning; TaKaRa Bio). NrHV subclones were generated by TOPO XL-2 cloning (Thermo Fisher Scientific) of the whole NrHV ORF amplified from genomic RNA extracted from mouse or rat serum samples, as described above. The NrHV-A and NrHV-B ORF consensus sequences were engineered into the original pRHV-rn1 (9), to make the final pNrHV-A and pNrHV-B plasmid clones. Specific mutations were engineered using site-specific megaprimer PCR mutagenesis. Mutagenic forward primers and fully sequence-matched reverse primers were used to amplify 300- to 2,000-bp PCR products (Q5 hot-start high-fidelity DNA polymerase; New England Biolabs). The megaprimer PCR products were column purified (Zymo Research) and used as primer for the second round of PCR, using the following cycling parameters: 98°C for 30 s, followed by 20 cycles of 98°C for 10 s, 48°C for 1 min, and 72°C for 20 min, with a final extension at 72°C for 20 min. The parental methylated template DNA was digested with DpnI for 1 h at 37°C (Thermo Fisher), and unmethylated PCR-amplified plasmids were transformed into One Shot TOP10 chemically competent bacteria (Invitrogen). Bacterial colonies resistant to ampicillin were picked and grown at 30°C for 72 h in 150 mL of LB medium, supplemented with 100 μg/mL ampicillin. Plasmid DNA was extracted using the GenElute HP plasmid midiprep kit (Sigma). As for bacterial lysis, 1.5-fold of the recommended volumes of buffers (lysis, neutralization, and binding) were used prior to binding the plasmid DNA on the column. Plasmid DNA was extracted and purified using the EndoFree plasmid maxi kit (Qiagen) when intended for use as the template to generate IVT RNA for intrahepatic inoculation in rodents. All plasmid constructs were confirmed by restriction analysis and Sanger sequencing (Macrogen Europe).
RNA in vitro transcription of NrHV clones.
Genomic NrHV RNA was produced from molecular consensus clones containing 5′-end-flanking T7 promoter sequence and an MluI restriction site at the 3′ end of the NrHV genome mediating authentic 5′ and 3′ ends (9). RNA was transcribed from 2.5 μg of MluI-linearized NrHV plasmid using the RiboMAX T7 RNA polymerase kit (Promega). Template DNA was degraded for 30 min at 4°C with RQ1 RNase-free DNase. For in vivo inoculation, subsequent RNA purification was carried out on RNeasy mini columns, including an additional on-column DNase I digestion step (Qiagen). The integrity of the viral genomic RNA was assessed by formaldehyde agarose gel electrophoresis, and quantities were measured using the Qubit RNA broad-range assay kit (Thermo Fisher Scientific).
Infectivity titration and neutralization assay.
For infectivity titration assays, 96-well microplates were coated for 2 h at 37°C with 50 μL of laminin (Sigma) at a concentration of 10 μg/mL, corresponding to approximately 1.5 μg/cm2. Unbound laminin was discarded, and the wells were washed three times with PBS before seeding of 13,500 permissive McA-RH7777.hi rat hepatoma cells per well. The following day, the cells were infected with serially diluted supernatant to be titrated (10-fold dilutions; lowest dilution, 1:2) and incubated for 48 h under standard growth conditions. The cells were fixed with methanol for 5 min at room temperature, and viral antigen was visualized by immunofluorescence staining using mouse anti-NrHV IgG and Alexa Fluor 594 goat anti-mouse IgG (A-11005; Thermo Fisher Scientific). Imaging was performed at ×50 magnification using a Carl Zeiss Axio Vert.A1 microscope. Culture supernatant infectivity titers were calculated following manual counting of focus-forming units (FFU). One FFU was defined as a cluster of more than one infected cell at a distance of at least two noninfected cells from any other FFU. Data points represent means ± standard deviations (SD) from triplicate wells.
For the neutralization assay, McA-RH7777.hi rat hepatoma cells were seeded as specified above. The next day, RHVcc-1 adjusted to 25,000 FFU/mL was incubated for 1 h at 37°C with heat-inactivated serum (56°C for 30 min). Subsequently, the culture medium was discarded from the 96-well plates, and the virus-antibody suspension (100 μL) was transferred to the McA-RH7777.hi cells and incubated for 4 h at 37°C. Following removal of the virus-antibody mixture, the cells were washed twice with PBS supplemented with 10% FBS and incubated for 48 h under standard growth conditions. Infected cells were visualized as described above for the infectivity titration assay. Percent neutralization was calculated by relating FFU counts to the mean of three replicate negative-control samples. Preimmune serum of the respective animal was used as a negative control.
Recombinant NrHV E1-E2 expression and immunoblotting.
For recombinant NrHV E1-E2 expression, a cytomegalovirus (CMV) promoter-driven expression construct encoding the last 56 residues of NrHV core and complete E1 and E2 envelope protein sequences was cloned, as previously done for HCV (40). The day before transfection with 5 μg plasmid DNA using Lipofectamine 2000, 3 × 105 HEK-293T cells per well were seeded into 6-well plates coated with poly-l-lysine (Sigma). Two days after transfection, the HEK-293T cells were gently dislodged with trypsin-EDTA and lysed for 10 min on ice in 100 μL radioimmunoprecipitation (RIPA) buffer, supplemented with Halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). Chromosomal DNA was digested with 3 μL of RQ1 RNase-free DNase (Promega) at 37°C for 5 min prior to removal of cell debris by centrifugation at 14,000 × g for 15 min at 4°C. The total protein concentration was quantified using the Pierce BCA protein assay kit (Thermo Fisher Scientific). To remove envelope N-linked glycans, 30 μg of protein was mixed with glycoprotein denaturing buffer (NEB) and denatured at 99°C for 10 min. Thereafter, GlycoBuffer 2 (NEB), NP-40, and PNGase F (NEB) were added, and the amidase reaction was performed at 37°C for 1 h. For detection of SB-BI in liver tissue, 100 mg of liver was collected into 1 mL of ice-cold RIPA buffer supplemented with 2 mM phenylmethylsulfonyl fluoride (PMSF), 1× protease inhibitor cocktail, and 1 mM orthovanadate and homogenized vigorously. The lysate was clarified and assessed for protein content as described above. For gel fractionation, a total of 30 μg of protein from HEK-293T cells or 20 μg of protein from tissue was resuspended in NuPAGE LDS sample buffer and NuPAGE sample reducing agent and subsequently loaded onto 1-mm NuPAGE 10% bis-Tris gels. The fluorescent Precision Plus Protein WesternC blotting standard (Bio-Rad) was diluted 1:10 in 1× sample loading buffer, as specified above, and 5 μL was loaded as a protein molecular weight reference. The proteins were fractionated by SDS-PAGE for 1 h at 150 V in a XCell SureLock mini electrophoresis cell filled with 1× NuPAGE MOPS (morpholinepropanesulfonic acid) SDS running buffer. Subsequent wet electroblotting protein transfer onto an Immuno-Blot low-fluorescence polyvinyl difluoride (PVDF) membrane (Bio-Rad) was carried out for 1.5 h at 35 V in an XCell II blot module filled with 1× NuPAGE transfer buffer. All reagents, materials, and devices used for SDS-PAGE were purchased from Invitrogen unless otherwise stated. The membrane was blocked overnight at 4°C with blocking buffer for fluorescent Western blotting (Rockland). For immunodetection, anti-NrHV-E2 mouse monoclonal IgG (clone 3G2, 1:1,000 [15]) and anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) rabbit monoclonal IgG (1:10,000, ab181602; Abcam) were applied in blocking buffer for 4 h at room temperature with agitation. For detection of SR-BI, membranes were reacted with rabbit anti-SR-BI (Novus Biologicals NB400-104SS) diluted 1:1,000 and mouse anti-β-tubulin as a loading control (Thermo Fisher, MA5-16308) diluted 1:2,000. Species-specific Alexa Fluor Plus 647 (A32728; Thermo Fisher Scientific)- or Alexa Fluor Plus 488 (A32731; Thermo Fisher Scientific)-conjugated antibodies were diluted in PBS supplemented with 0.1% Tween 20 (1:5,000) and used for visualization of antigen. PBS supplemented with 0.1% Tween 20 was used to wash off unbound primary and secondary antibodies (three 5-min wash steps). The membrane was fixed in anhydrous methanol, air dried, and imaged utilizing an ImageQuant LAS 4000 biomolecular imaging system (GE Healthcare).
Transfection, electroporation, and complementation with miR-122 mimic.
McA-RH7777.hi rat hepatoma cells were transfected with Lipofectamine 2000 (Thermo Fisher). A total of 300,000 cells per well were seeded into 6-well plates precoated with a minimal volume of 0.1% gelatin (Sigma). Transfection was done the following day for 4 h at 37°C in DMEM supplemented with 3% FBS. Opti-MEM was used solely for the formation of transfection complexes, using 5 μg RNA. Following transfection, the cells were washed twice with prewarmed PBS and the medium was replaced with DMEM supplemented with 10% FBS.
For electroporation and miR-122 complementation, AML12 mouse hepatoma cells were washed twice with ice-cold PBS, strained (40 μm), and resuspended at a density of 1.5 × 107 cells/mL in ice-cold CytoMix buffer (120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4/KH2PO4 [pH 7.6], 25 mM HEPES [pH 7.6], 2 mM EGTA [pH 7.6], 5 mM MgCl2 [pH adjusted with KOH]) (41). Subsequently, 400 μL of the cell suspension was mixed with 5 μg RNA and miR-122 mimic (0.325 μM in 400 μL; nucleotide sequence, UGGAGUGUGACAAUGGUGUUUG) and transferred to a 4-mm electroporation cuvette (Bio-Rad). The following parameters were used for controlled exponential decay pulse electroporation: voltage, 270 V; capacitance, 975 μF; and resistance, ∞ ohms, typically within a time constant of 20 to 30 ms (Gene Pulser Xcell electroporation system; Bio-Rad). Following electroporation, the cells were immediately resuspended in prewarmed medium and transferred to a 10-cm culture-grade dish. Following electroporation and recovery, the AML12 cells were maintained in 6-well plates and additional miR-122 mimic (0.325 μM, final concentration) was supplemented every 4 days, using Lipofectamine RNAiMAX transfection reagent (Thermo Fisher) according to the manufacturer’s instructions.
ACKNOWLEDGMENTS
We thank Arash Grakoui for providing NrHV anti-E2 monoclonal antibodies, Amit Kapoor and members of the authors’ laboratories for fruitful discussion, Birgitte Børsting for help with the E1-E2 expression constructs, and the Department of Infectious Diseases, Hvidovre Hospital, and Department of Immunology and Microbiology, University of Copenhagen, for support. We further acknowledge Lotte Mikkelsen and Anna-Louise Sørensen (Department of Infectious Diseases, Hvidovre Hospital) and Louise B. Christensen (Department of Clinical Microbiology, Hvidovre Hospital) for laboratory assistance and the Department of Clinical Microbiology, Hvidovre Hospital, for access to MiSeq equipment and the Department of Experimental Medicine, University of Copenhagen, for rodent husbandry and care.
This study was supported by the European Research Council (starting grant no. 802899 to T.K.H.S), the Independent Research Fund Denmark (advanced grant no. 4004-00598 to J.B. and grant no. 1030-00426 to T.K.H.S), the Danish Cancer Society (grant no. R204-A12639 to J.B. and T.K.H.S.), the Novo Nordisk Foundation (Distinguished Investigator grant no. NNF19OC0054518 and NNF19OC0055462 to J.B.), and U.S. Public Health Service/National Institutes of Health award R01AI131688 to C.M.R. R.W. was supported by an Early Postdoc.Mobility Fellowship (no. P2BEP3_178527) and a Postdoc.Mobility Fellowship (no. P400PB-183952) from the Swiss National Science Foundation.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Troels K. H. Scheel, Email: tscheel@sund.ku.dk.
J.-H. James Ou, University of Southern California.
REFERENCES
- 1.WHO. 2017. Global hepatitis report 2017. https://www.who.int/news-room/fact-sheets/detail/hepatitis-c. Accessed 20 April 2022.
- 2.Pawlotsky JM. 2016. Hepatitis C virus resistance to direct-acting antiviral drugs in interferon-free regimens. Gastroenterology 151:70–86. 10.1053/j.gastro.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Bukh J. 2012. Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology 142:1279–1287.e3. 10.1053/j.gastro.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 4.Stapleton JT, Foung S, Muerhoff AS, Bukh J, Simmonds P. 2011. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J Gen Virol 92:233–246. 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartlage AS, Cullen JM, Kapoor A. 2016. The strange, expanding world of animal hepaciviruses. Annu Rev Virol 3:53–75. 10.1146/annurev-virology-100114-055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ploss A, Kapoor A. 2020. Animal models of hepatitis C virus infection. Cold Spring Harb Perspect Med 10:a036970. 10.1101/cshperspect.a036970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomlinson JE, Wolfisberg R, Fahnoe U, Patel RS, Trivedi S, Kumar A, Sharma H, Nielsen L, McDonough SP, Bukh J, Tennant BC, Kapoor A, Rosenberg BR, Rice CM, Divers TJ, Van de Walle GR, Scheel TKH. 2021. Pathogenesis, microRNA-122 gene-regulation, and protective immune responses after acute equine hepacivirus infection. Hepatology 74:1148–1163. 10.1002/hep.31802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Firth C, Bhat M, Firth MA, Williams SH, Frye MJ, Simmonds P, Conte JM, Ng J, Garcia J, Bhuva NP, Lee B, Che X, Quan PL, Lipkin WI. 2014. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. mBio 5:e01933-14. 10.1128/mBio.01933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trivedi S, Murthy S, Sharma H, Hartlage AS, Kumar A, Gadi SV, Simmonds P, Chauhan LV, Scheel TKH, Billerbeck E, Burbelo PD, Rice CM, Lipkin WI, Vandegrift K, Cullen JM, Kapoor A. 2018. Viral persistence, liver disease, and host response in a hepatitis C-like virus rat model. Hepatology 68:435–448. 10.1002/hep.29494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Billerbeck E, Wolfisberg R, Fahnoe U, Xiao JW, Quirk C, Luna JM, Cullen JM, Hartlage AS, Chiriboga L, Ghoshal K, Lipkin WI, Bukh J, Scheel TKH, Kapoor A, Rice CM. 2017. Mouse models of acute and chronic hepacivirus infection. Science 357:204–208. 10.1126/science.aal1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolfisberg R, Holmbeck K, Nielsen L, Kapoor A, Rice CM, Bukh J, Scheel TKH. 2019. Replicons of a rodent hepatitis C model virus permit selection of highly permissive cells. J Virol 93:e00733-19. 10.1128/JVI.00733-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartlage AS, Murthy S, Kumar A, Trivedi S, Dravid P, Sharma H, Walker CM, Kapoor A. 2019. Vaccination to prevent T cell subversion can protect against persistent hepacivirus infection. Nat Commun 10:1113. 10.1038/s41467-019-09105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atcheson E, Li W, Bliss CM, Chinnakannan S, Heim K, Sharpe H, Hutchings C, Dietrich I, Nguyen D, Kapoor A, Jarvis MA, Klenerman P, Barnes E, Simmonds P. 2020. Use of an outbred rat hepacivirus challenge model for design and evaluation of efficacy of different immunization strategies for hepatitis C virus. Hepatology 71:794–807. 10.1002/hep.30894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartlage AS, Walker CM, Kapoor A. 2020. Priming of antiviral CD8 T cells without effector function by a persistently replicating hepatitis C-like virus. J Virol 94:e00035-20. 10.1128/JVI.00035-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfisberg R, Thorselius CE, Salinas E, Elrod E, Trivedi S, Nielsen L, Fahnoe U, Kapoor A, Grakoui A, Rice CM, Bukh J, Holmbeck K, Scheel TKH. 2022. Neutralization and receptor use of infectious culture-derived rat hepacivirus as a model for HCV. Hepatology 76:1506–1519. 10.1002/hep.32535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka T, Moriishi K. 2022. Analysis of the functional interaction between rodent hepacivirus and miR-122. Abstr 28th International Symposium on Hepatitis C Virus, Flaviviruses, and Related Viruses, Ghent, Belgium.
- 17.Timpe JM, Stamataki Z, Jennings A, Hu K, Farquhar MJ, Harris HJ, Schwarz A, Desombere I, Roels GL, Balfe P, McKeating JA. 2008. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology 47:17–24. 10.1002/hep.21959. [DOI] [PubMed] [Google Scholar]
- 18.Prentoe J, Velazquez-Moctezuma R, Augestad EH, Galli A, Wang R, Law M, Alter H, Bukh J. 2019. Hypervariable region 1 and N-linked glycans of hepatitis C regulate virion neutralization by modulating envelope conformations. Proc Natl Acad Sci USA 116:10039–10047. 10.1073/pnas.1822002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grove J, Nielsen S, Zhong J, Bassendine MF, Drummer HE, Balfe P, McKeating JA. 2008. Identification of a residue in hepatitis C virus E2 glycoprotein that determines scavenger receptor BI and CD81 receptor dependency and sensitivity to neutralizing antibodies. J Virol 82:12020–12029. 10.1128/JVI.01569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathiesen CK, Prentoe J, Meredith LW, Jensen TB, Krarup H, McKeating JA, Gottwein JM, Bukh J. 2015. Adaptive mutations enhance assembly and cell-to-cell transmission of a high-titer hepatitis C virus genotype 5a core-NS2 JFH1-based recombinant. J Virol 89:7758–7775. 10.1128/JVI.00039-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuiani A, Chen K, Schwarz MC, White JP, Luca VC, Fremont DH, Wang D, Evans MJ, Diamond MS. 2016. A library of infectious hepatitis C viruses with engineered mutations in the E2 gene reveals growth-adaptive mutations that modulate interactions with scavenger receptor class B type I. J Virol 90:10499–10512. 10.1128/JVI.01011-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey JR, Barnes E, Cox AL. 2019. Approaches, progress, and challenges to hepatitis C vaccine development. Gastroenterology 156:418–430. 10.1053/j.gastro.2018.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burm R, Collignon L, Mesalam AA, Meuleman P. 2018. Animal models to study hepatitis C virus infection. Front Immunol 9:1032. 10.3389/fimmu.2018.01032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang TJ, Feld JJ, Cox AL, Rice CM. 2021. Controlled human infection model—fast track to HCV vaccine? N Engl J Med 385:1235–1240. 10.1056/NEJMsb2109093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartlage AS, Kapoor A. 2021. Hepatitis C virus vaccine research: time to put up or shut up. Viruses 13:1596. 10.3390/v13081596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bitzegeio J, Bankwitz D, Hueging K, Haid S, Brohm C, Zeisel MB, Herrmann E, Iken M, Ott M, Baumert TF, Pietschmann T. 2010. Adaptation of hepatitis C virus to mouse CD81 permits infection of mouse cells in the absence of human entry factors. PLoS Pathog 6:e1000978. 10.1371/journal.ppat.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raus S, Lopez-Scarim J, Luthy J, Billerbeck E. 2022. Hepatic iNKT cells produce type 2 cytokines and restrain antiviral T cells during acute hepacivirus infection. Front Immunol 13:953151. 10.3389/fimmu.2022.953151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shata MT, Tricoche N, Perkus M, Tom D, Brotman B, McCormack P, Pfahler W, Lee DH, Tobler LH, Busch M, Prince AM. 2003. Exposure to low infective doses of HCV induces cellular immune responses without consistently detectable viremia or seroconversion in chimpanzees. Virology 314:601–616. 10.1016/s0042-6822(03)00461-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katayama K, Kumagai J, Komiya Y, Mizui M, Yugi H, Kishimoto S, Yamanaka R, Tamatsukuri S, Tomoguri T, Miyakawa Y, Tanaka J, Yoshizawa H. 2004. Titration of hepatitis C virus in chimpanzees for determining the copy number required for transmission. Intervirology 47:57–64. 10.1159/000076643. [DOI] [PubMed] [Google Scholar]
- 30.Bukh J, Meuleman P, Tellier R, Engle RE, Feinstone SM, Eder G, Satterfield WC, Govindarajan S, Krawczynski K, Miller RH, Leroux-Roels G, Purcell RH. 2010. Challenge pools of hepatitis C virus genotypes 1–6 prototype strains: replication fitness and pathogenicity in chimpanzees and human liver-chimeric mouse models. J Infect Dis 201:1381–1389. 10.1086/651579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomer A, Delarocque J, Puff C, Nocke MK, Reinecke B, Baumgartner W, Cavalleri JMV, Feige K, Steinmann E, Todt D. 2022. Dose-dependent hepacivirus infection reveals linkage between infectious dose and immune response. Microbiol Spectr 10:e01686-22. 10.1128/spectrum.01686-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bukh J, Thimme R, Meunier JC, Faulk K, Spangenberg HC, Chang KM, Satterfield W, Chisari FV, Purcell RH. 2008. Previously infected chimpanzees are not consistently protected against reinfection or persistent infection after reexposure to the identical hepatitis C virus strain. J Virol 82:8183–8195. 10.1128/JVI.00142-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheel TK, Simmonds P, Kapoor A. 2015. Surveying the global virome: identification and characterization of HCV-related animal hepaciviruses. Antiviral Res 115:83–93. 10.1016/j.antiviral.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gomer A, Brown RJP, Pfaender S, Deterding K, Reuter G, Orton R, Seitz S, Bock CT, Cavalleri JMV, Pietschmann T, Wedemeyer H, Steinmann E, Todt D. 2022. Intra-host analysis of hepaciviral glycoprotein evolution reveals signatures associated with viral persistence and clearance. Virus Evol 8:veac007. 10.1093/ve/veac007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, Mu K, Jones CT, Schoggins JW, Catanese MT, Burton DR, Law M, Rice CM, Ploss A. 2011. A genetically humanized mouse model for hepatitis C virus infection. Nature 474:208–211. 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Schaewen M, Dorner M, Hueging K, Foquet L, Gerges S, Hrebikova G, Heller B, Bitzegeio J, Doerrbecker J, Horwitz JA, Gerold G, Suerbaum S, Rice CM, Meuleman P, Pietschmann T, Ploss A. 2016. Expanding the host range of hepatitis C virus through viral adaptation. mBio 7:e01915-16. 10.1128/mBio.01915-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narbus CM, Israelow B, Sourisseau M, Michta ML, Hopcraft SE, Zeiner GM, Evans MJ. 2011. HepG2 cells expressing microRNA miR-122 support the entire hepatitis C virus life cycle. J Virol 85:12087–12092. 10.1128/JVI.05843-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigotti A, Trigatti BL, Penman M, Rayburn H, Herz J, Krieger M. 1997. A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proc Natl Acad Sci USA 94:12610–12615. 10.1073/pnas.94.23.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verbist BM, Thys K, Reumers J, Wetzels Y, Van der Borght K, Talloen W, Aerssens J, Clement L, Thas O. 2015. VirVarSeq: a low-frequency virus variant detection pipeline for Illumina sequencing using adaptive base-calling accuracy filtering. Bioinformatics 31:94–101. 10.1093/bioinformatics/btu587. [DOI] [PubMed] [Google Scholar]
- 40.Bartosch B, Dubuisson J, Cosset FL. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med 197:633–642. 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van den Hoff MJ, Moorman AF, Lamers WH. 1992. Electroporation in 'intracellular' buffer increases cell survival. Nucleic Acids Res 20:2902. 10.1093/nar/20.11.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]