Abstract
CRISPR cell/gene therapy has been designed largely based on a single reference human genome. A new study reveals how human genetic diversity could lead to off-target effects and presents a novel tool to identify these risks.
Just as the benefits from a single treatment of CRISPR genome editors can be life-changing and durable,1 the risks could be significant and irreversible. Adverse events in early-stage trials of CRISPR are starting to be reported, including a recent tragic death2 of a muscular dystrophy patient dosed with a CRISPR activation editor. While it is too early to determine whether the cause of death was related to genome editing, the genome editing community, along with clinicians and patients, eagerly awaits the development of new tools to measure the benefits and risks of genome editing strategies. One particularly vexing question in the field is whether trial participants should be genotyped before enrollment in these studies and how to interpret the resulting sequence data to ensure safety and efficacy. Now, a decade after the first reports of CRISPR-based genome editing, a new study by Cancellieri and colleagues3 in this issue of Nature Genetics reports on the development of a novel tool to quantify the potential of human genetic diversity to alter genome editing outcomes.
CRISPR-based genome editors work by binding to a target sequence with high specificity. The high specificity of the editor can be exploited by design to develop allele-specific genome editors that correct or disrupt targeted mutations,4,5 but when an individual genetic variant is not targeted by the editor nor anticipated in the design of an editor, it may be a serious concern. In molecular and cellular assays, the genome editor binds not only to the intended target sequence but also binds off-target sequences in the genome that have one or more base mismatches from the 17-20 base target sequence. These off-target sites are typically nominated from a single human reference genome (e.g., Genome assembly GRCh38.p14). Then, a variety of preclinical assays are used to assay the ability of the editor to bind and modify the nominated off-target sites: in test tubes with DNA isolated from human cells, in immortalized human cell lines, more sophisticated human biological systems like organoids, human cadaver tissues, small animal models, and large animal models6 (Fig. 1). Human genetic diversity is poorly represented in these preclinical approaches. This concern is not just theoretical: in cancer research, a new analysis of the cell models of the Cancer Dependency Map7 uncovered a bias against African genetic ancestry-associated germline variants.
Figure 1. Assaying human genetic diversity can advance the design of human therapeutics involving genome editing.
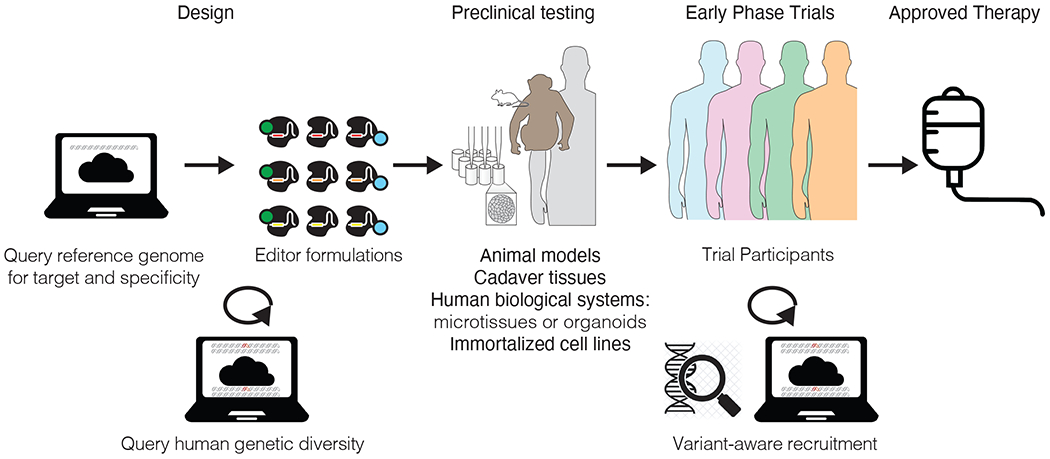
Development first starts with a design phase where a target is identified, and off-target sites are computationally identified against a reference human genome. With new tools to assay human genetic diversity (bottom), new editors can be designed in a variant-aware manner to mitigate the potential for off-target effects. After empirically assaying the potential for on-target editing as well as off-target effects in a variety of models in the preclinical testing, a candidate formulation can enter early-phase clinical trials. Participants could be included or excluded based on the presence of the variant in the participant’s genome that has high off-target potential. These variant-aware decisions should be made using DNA tests to ensure the presence of a variant in an individual, rather than using proxies for the variant like race or ethnicity.
A single-nucleotide polymorphism (SNP) could increase the propensity of the editor to bind to the polymorphism and modify that allele in an off-target manner.8,9 Other variants, like copy number variation and tandem repeats, in theory, could also produce additional high-affinity targets for the editor. Multiple breaks in the genome by nuclease-based editors can lead to translocations and large-scale chromosomal aberrations, even chromothripsis10. The functional consequences of these genomic modifications have been predicted to lead to dysfunction of the edited cells and cancer, particularly if tumor suppressive pathways are affected.
The strength of these off-target interactions between the variant allele and the editor is scored in this new study. The team developed a web-based computational tool, called CRISPRme, to nominate variants in an individual’s genome (or to those in variant databases) that may confer higher specificity to the SpCas9 editor. Then, the team applied the tool to predict the potential editing of these off-target variants for several clinically-relevant genome editing targets. For a candidate hematopoietic stem/progenitor cell therapy with an edited BCL11A enhancer that could be used to treat sickle cell disease and β-thalassemia, intriguingly, the most likely off-target site, overlooked by prior analyses, is introduced by a SNP common in African-ancestry individuals at approximately 5%. The team provides experimental evidence of its off-target potential in a cell product manufactured from primary cells of a variant carrier and lowers this potential by using an alternate editor, a higher fidelity Cas9. The high-fidelity editor has reduced editing efficiency, so the dosing and kinetics of editing may need to be carefully considered for patients with these variants conferring off-target potential. Constitutive expression of the editor is standard in viral vectors that deliver CRISPR machinery into the nucleus of a cell, leading to durable editor activity that accumulates a high potential for off-target effects. Pulsed nonviral delivery of the editor could provide a shorter burst of editing activity, with a lower potential for off-target effects. The authors provide a helpful discussion of these and other design strategies to mitigate off-target potential due to human genetic variation.
These results in themselves are immediately important for several ongoing trials for sickle cell disease and β-thalassemia. Furthermore, the work has implications for nearly any genome editing trial underway, as first-in-human clinical studies with genome-editing drugs are underway for treating retinal disorders,11 cardiovascular disease12, cancer13, and many rare diseases. The US Food and Drug Administration recently drafted guidance on product design, product testing, preclinical safety assessment, and clinical trial design for cell/gene therapy products incorporating genome editing of human somatic cells.14 Considerations of human genetic diversity may be incorporated into the final guidance. Participation in future clinical trials for CRISPR cell/gene therapies could incorporate inclusion and exclusion criteria based on the nominated off-target variants. During market authorization by a country’s or region’s health agency for a successful candidate in these trials, the patient population who benefits from these drugs could be limited to those who were included in the trial and exclude those who were not. Reimbursement by insurance is also likely to track decisions during market authorization. Therefore, variant-aware considerations in the design of editors can lead to significant implications for access in the clinic to these therapies. Furthermore, adding this personalized level of profiling of genome editors will likely only add to the development costs of trials. The price per-dose of gene therapies, which currently range from 0.4-2.8 million USD, is already of concern to many. However, the costs of doing these variant-aware assays are likely to go down with sequencing advances, and the costs of ignoring these risks may ultimately be higher, especially if patients and the public hear of unfortunate adverse events that could have been avoided by the use of these new approaches.
Finally, genetic diversity in this study is strictly defined as variants in the DNA sequence and is not interchangeable for race or ethnicity, which are socially-constructed categories. Using new tools such as the one recently reported3, it will be essential to produce genome-editing drugs that account for human genetic diversity, while ensuring broad access to these new potentially-curative cell/gene therapies.
Acknowledgments.
US National Science Foundation EEC-1648035, AWD-101645-G3/RJ375-G3, US National Institute for Health R35GM119644, U01EY032333, UH3NS111688
Competing interests.
K.S. receives sponsored research support from Spotlight Therapeutics and Synthego and is an advisor to Notch Therapeutics and Andson Biotech.
References
- 1.Graham F Daily briefing: Why protests haven’t lead to a surge in COVID-19. Nature Publishing Group UK 10.1038/d41586-020-01928-y (2020) doi: 10.1038/d41586-020-01928-y. [DOI] [PubMed] [Google Scholar]
- 2.Science News Staff. News at a glance: A new antibiotic, COVID-19 in Antarctica, and a Venus mission deferred. Science (2022) doi: 10.1126/science.adf7363. [DOI] [Google Scholar]
- 3.Cancellieri Samuele, Zeng Jing, Linda Yingqi Lin Manuel Tognon, My Anh Nguyen Jiecong Lin, Bombieri Nicola, Maitland Stacy A., Ciuculescu Marioara-Felicia, Katta Varun, Tsai Shengdar Q., Armant Myriam, Wolfe Scot A., Giugno Rosalba, Bauer Daniel E., Pinello Luca. Human genetic diversity alters off-target outcomes of therapeutic gene editing. Nat. Genet. TBD, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinha D et al. Human iPSC Modeling Reveals Mutation-Specific Responses to Gene Therapy in a Genotypically Diverse Dominant Maculopathy. Am. J. Hum. Genet 107, 278–292 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keough KC et al. AlleleAnalyzer: a tool for personalized and allele-specific sgRNA design. Genome Biol . 20, 167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saha K et al. The NIH Somatic Cell Genome Editing program. Nature 592, 195–204 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misek SA et al. Germline variation contributes to false negatives in CRISPR-based experiments with varying burden across ancestries . bioRxiv 2022.11.18.517155 (2022) doi: 10.1101/2022.11.18.517155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lessard S et al. Human genetic variation alters CRISPR-Cas9 on- and off-targeting specificity at therapeutically implicated loci. Proc. Natl. Acad. Sci. U. S. A 201714640 (2017) doi: 10.1073/pnas.1714640114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott DA & Zhang F Implications of human genetic variation in CRISPR-based therapeutic genome editing. Nat. Med 23, 1095–1101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leibowitz ML et al. Chromothripsis as an on-target consequence of CRISPR-Cas9 genome editing. Nat. Genet 1–11 (2021) doi: 10.1038/s41588-021-00838-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeder ML et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med 25, 229–233 (February 2019). [DOI] [PubMed] [Google Scholar]
- 12.Musunuru K et al. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature 593, 429–434 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Foy SP et al. Non-viral precision T cell receptor replacement for personalized cell therapy. Nature 1–0 doi: 10.1038/s41586-022-05531-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Center for Biologics Evaluation & Research. Human Gene Therapy Products Incorporating Human Genome Editing. U.S. Food and Drug Administration; https://www.fda.gov/regulatory-information/search-fda-guidance-documents/human-gene-therapy-products-incorporating-human-genome-editing. [Google Scholar]


