Abstract
Swine pathogenic infection caused by Escherichia coli, known as swine colibacillosis, represents an epidemiological challenge not only for animal husbandry but also for health authorities. To note, virulent E. coli strains might be transmitted, and also cause disease, in humans. In the last decades, diverse successful multidrug-resistant strains have been detected, mainly due to the growing selective pressure of antibiotic use, in which animal practices have played a relevant role. In fact, according to the different features and particular virulence factor combination, there are four different pathotypes of E. coli that can cause illness in swine: enterotoxigenic E. coli (ETEC), Shiga toxin-producing E. coli (STEC) that comprises edema disease E. coli (EDEC) and enterohemorrhagic E. coli (EHEC), enteropathogenic E. coli (EPEC), and extraintestinal pathogenic E. coli (ExPEC). Nevertheless, the most relevant pathotype in a colibacillosis scenario is ETEC, responsible for neonatal and postweaning diarrhea (PWD), in which some ETEC strains present enhanced fitness and pathogenicity. To explore the distribution of pathogenic ETEC in swine farms and their diversity, resistance, and virulence profiles, this review summarizes the most relevant works on these subjects over the past 10 years and discusses the importance of these bacteria as zoonotic agents.
Keywords: swine colibacillosis, AMR bacteria, E. coli pathotypes, prevalence, epidemiology
1. Introduction
Porcine infection caused by Escherichia coli (E. coli), so-called swine colibacillosis, is responsible for a wide range of problems, such as neonatal diarrhea, post-weaning diarrhea (PWD), edema disease (ED), septicemia, polyserositis, coliform mastitis, and urinary tract infection [1]. Among the huge diversity, certain strains of E. coli, named enterotoxigenic E. coli (ETEC), are able to cause intestinal disease, which results in neonatal diarrhea, PWD, and ED. These porcine infections are the most threatening for the swine industry worldwide due to significant economic losses associated with morbidity, mortality, decreased weight gain, the rising cost of treatments, vaccinations, and feed supplements [1,2,3].
PWD and ED may occur separately or together either in an individual outbreak or in the same pig [1]. Within 2–3 weeks after weaning, the piglets are more susceptible to microbial infections, owing to the existence of an immature immune system associated with sow milk removal and resulting from the interruption of the nutritive intake of immunoglobulin present in the milk [2,4]. Therefore, this period is crucial and usually associated with the most severe form of enteric E. coli infection, manifested by sudden death or severe diarrhea [1].
As an effort to promote health and growth performance, diverse approaches have been used to prevent and treat swine colibacillosis, with antibiotics being the most commonly used strategy [2,4]. Consequently, due to the growing selective pressure of antibiotic use to treat these E. coli infections, the emergence of the antimicrobial resistance (AMR) phenomenon has limited treatment options for pig producers and an increased public health concern because of the potential transfer of AMR genetic determinants directly by contact and indirectly into the food chain, water, and manure, among others [1,5]. It is important to note that E. coli has a great capacity to acquire resistance genes, mainly through horizontal gene transfer [6], in which the mobile genetic elements, such as plasmids, transposons, and gene cassettes in class 1 and class 2 integrons, seem to play a main role in the dissemination [5]. Furthermore, E. coli behaves as a donor and as a recipient of resistance genes and thus can exchange those genes with other bacteria and act as a reservoir of AMR genes [5]. Accordingly, extended-spectrum β-lactamases, carbapenemases, 16S rRNA methylases, plasmid-mediated quinolone resistance (PMQR) genes, and mcr genes constitute the most problematic genetic determinant classes of AMR in E. coli [5].
Hence, this review assembled diverse studies of importance regarding infections caused by ETEC and their epidemiologic and antimicrobial consequences at the global level during the past 10 years to bring useful and organized information about the distribution, diversity, resistance, and virulence profiles according to the pathogenic serotypes expressed by ETEC in swine farms.
2. Etiology
According to the taxonomy, the German pediatrician Theodor Escherich (1857–1911) gave the origin to the name of the genus Escherichia. This genus belongs to the family of Enterobacteriaceae, which contemplates the Gram-negative facultatively anaerobic rods, where the species Escherichia coli fits since they are Gram-negative, peritrichously flagellated rods of variable length and a diameter of about 1 μm [1].
Over the decades, the subdivision of species into types has been carried out by the development of several classification systems. Among these, serotyping (described in Table 1) is a recognized typing system to classify E. coli strains [1]. Nevertheless, since certain porcine pathogenic E. coli belong to a limited number of serotypes, this method is less used today for diagnostic purposes [1]. Thus, the serotyping technique has been substituted by the direct detection of genes coding for bacterial determinants involved in their pathogenesis, called virulence factors. Therefore, the term pathotype is applied to the classification of E. coli typologies according to the combinations of virulence factors [3].
Common E. coli pathotypes include Shiga toxin-producing E. coli (STEC) that contains two groups, edema disease E. coli (EDEC) and enterohemorrhagic E. coli (EHEC), enteropathogenic E. coli (EPEC), and extraintestinal pathogenic E. coli (ExPEC), with the most relevant E. coli pathotype in porcine, the enterotoxigenic E. coli (ETEC) [1]. It is important to highlight that EHEC and EPEC pathotypes are associated with the “attaching and effacing” (A/E) lesion development [1]. Nevertheless, EPEC is found in pigs with PWD, whereas EHEC is highly pathogenic in humans, and some zoonoses of this pathotype are sporadically recovered [1]. Therefore, this review is focused on ETEC due to its high pathogenicity in porcine, being the one responsible for the most cases, in number and severity, of swine enteric colibacillosis. Besides that, ETEC was classified as the most important multi-drug resistant (MDR) bacteria in pig production by the European Food Safety Authority (EFSA) [7]. Hence, in the following subchapter, the mechanisms of virulence and the pathogenesis for ETEC are detailed.
2.1. ETEC Virulence Factors and Their Impact on Trigger Colibacillosis Infection
E. coli is both a harmless commensal bacterium in the intestines of several mammals, as well as a dangerous pathogen [5]. Still, a small proportion of strains are pathogenic and can cause severe to life-threatening intestinal and extra-intestinal infections in humans and animals [5,7,8,9,10]. The pathogenicity of the strains is characterized by the presence of certain virulence factor combinations in particular adhesins and toxin secretions. As it has been described, the role of adhesins and surface proteins called fimbriae is to enable the adherence of ETEC to specific receptors on the brush borders of the small intestine’s enterocytes [8]. Regarding fimbriae, there are five common antigenically different types found in pigs: F4 (K88), F5 (K99), F41, F6 (987P), and F18 [11]. The first four fimbria types are responsible for mediating adhesion in neonates, while F18 is not associated with neonatal colibacillosis; however, it is common in postweaning colibacillosis as is F4. It is also important to highlight that hemolysis is a common trait for pathogenic F4 and F18 isolates [4]. The adhesion of ETEC through fimbriae conducts the release of toxins inside the epithelial cell that promote the secretion of water and electrolytes into the intestinal lumen [1,2,8]. ETEC constitutes the most relevant and pathogenic strains in porcine, where the following groups of toxins are produced, namely the two major classes of enterotoxins, heat-stable toxin (ST) and heat-labile toxin (LT), as well as the enteroaggregative heat-stable toxin 1 (EAST1), as described in Table 1. STs are divided into Sta (also nominated STI, ST1, or StaP) and STb (also nominated STII or ST2) [1,2]. Similarly, LT toxins are divided into two groups, LTI and LTII. On the other hand, EAST1 is widespread among porcine ETEC, but its role in this illness remains controversial [1]. It is important to note that EAST1 alone does not seem capable of developing the disease; however, together with LT, it takes effect [1,2]. Additionally, it is important to note that Shiga toxin (Stx or VT), namely the Shiga toxin type 2e (Stx2e), E. coli is also found in some ETEC strains, being commonly found in ETEC strains expressing LT and/or ST toxins [11]. This toxin is a causative factor of edema in swine, and it has been also associated with diarrhea commonly found in colibacillosis [1,11].
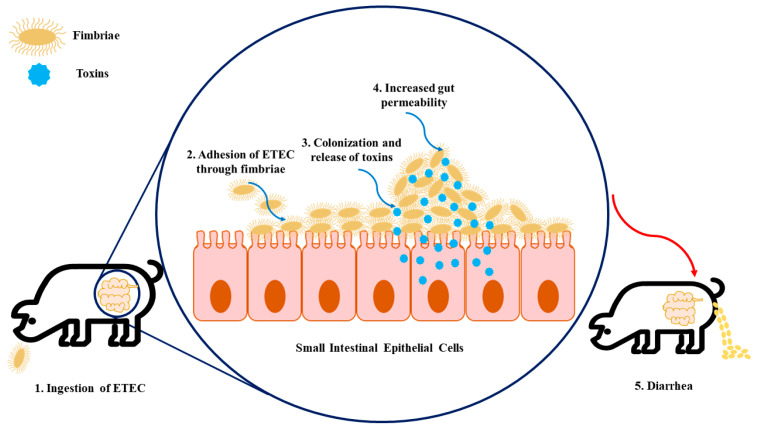
Regarding the route of contamination, ETEC is firstly ingested through the oral route and then passes through the stomach. When it reaches the intestine, in the presence of suitable environmental conditions, ETEC proliferates and causes disease [1,8]. At this point, they colonize the small intestine following the attachment of fimbria adhesins to specific receptors present on the epithelium as well as in the mucus coating the epithelium of the small intestine (see Figure 1). This promotes the production and release of enterotoxins, as mentioned above, inside the epithelial cell that stimulates the secretion of water and electrolytes into the intestinal lumen, which leads to diarrhea, weight loss, and possibly death [1,3,8]. To note, the combinations of different ETEC virulence factors (adhesins and toxins) are associated with the development of different diseases, namely neonatal diarrhea and PWD, as shown in Table 1.
Figure 1.
The infection model of enterotoxigenic Escherichia coli (ETEC) on intestinal epithelial cells. (1) Firstly, the swine ingest ETEC, enabling its transition to the gastrointestinal tract. (2) The fimbriae expressed by ETEC allow the adhesion of bacteria to specific receptors present in the intestinal epithelial cells. (3) Colonization arises in the small intestinal mucosa, which leads to the production of toxins. (4) These enterotoxins promote water and electrolyte loss into the intestinal lumen, resulting in increased gut permeability. (5) As a consequence of increased gut permeability and massive water loss, diarrhea, weight loss, and mortality can happen.
Table 1.
Virulence factors of enterotoxigenic Escherichia coli (ETEC) associated with swine enteric colibacillosis.
| Adhesins | Toxins | Serotypes | Disease |
|---|---|---|---|
| F5, F6, F41 | STa | O8, O9, O20, O64, O101 | Neonatal diarrhea |
| F4 | STa, STb, LT, EAST1, α-hemolysin b | O8, O138, O141, O145, O147, O149, O157 | Neonatal diarrhea Diarrhea in young pigs preweaning |
| F4, AIDA a, unknown | STa, STb, LT, EAST1, α-hemolysin b | O8, O138, O139, O141, O147, O149, O157 | PWD |
| F18, AIDA a | STa, STb, LT, Stx (or VT) c, EAST1, α-hemolysin b | O8, O138, O139, O141, O147, O149, O157 | PWD |
a AIDA is a non-fimbrial adhesin involved in diffuse adherence; nevertheless, the mechanism of AIDA in swine colibacillosis is not yet elucidated. However, this non-fimbrial adhesin has been associated with ETEC strains from weaned pigs with PWD [2]. b α-hemolysin is a pore-forming cytolysin associated with ETEC strains that cause diarrhea in animals [12]. c Shiga toxins (Stx or VT) are cytotoxins produced by Shiga-toxin-producing E. coli (STEC), and in swine, the most important STECs are those that cause edema disease (ED) [1].
2.1.1. Neonatal ETEC
Neonatal diarrhea caused by ETEC is only associated with STa and might have one or more of the fimbriae associated, including F4 (K88), F5 (K99), F6 (987P), and F41 [1]. Of note, F4 is usually related to the colonization of the length of the jejunum and ileum, whereas F5, F6, and F41 mostly colonize the posterior jejunum and ileum [2]. Recently, Dubreuil and colleagues reviewed the prevalence of F4-, F5-, and F6-fimbriated ETEC from neonatal diarrhea and concluded that they exhibited both temporal and geographic variations [13]. This illness usually occurs in the first four days after the piglet is born and is characterized by whitish-yellow diarrhea, with a watery or creamy consistency, in large quantities [2]. Once ETEC has adhered to the epithelium, it binds to specific receptors in the apical region of the epithelial cells in the jejunal region and begins to produce the enterotoxin Sta, culminating in the hypersecretion of electrolytes and fluids in the small intestine. Eventually, if the large intestine is unable to reabsorb these excess fluids, the piglet enters a state of dehydration (and in more severe cases, metabolic acidosis) and eventually dies [1].
2.1.2. Postweaning ETEC
The enterotoxins STa, STb, LT, and EAST-1 are typically produced individually or together in the ETECs that cause diarrhea in postweaning or older suckling pigs [1]. The fimbriae involved in this disease are mainly F4 and F18, and both possess several variant subtypes based on antigenic differences. In this respect, F4 and F18 evidenced the following subtypes, ab, ac, and ad and ab and ac, respectively [2]. Post-weaning diarrhea usually appears 2 to 3 weeks after weaning and is characterized by the appearance of watery diarrhea that can vary from yellowish-grey or a slightly pinkish color, which lasts for up to a week [1]. The ETEC responsible for this condition generally colonizes the duodenal and jejunal portions of the small intestine, thus inducing the hypersecretion of fluids, very similar to that described in neonatal diarrhea [1].
3. Global Epidemiology of Swine Enteric Colibacillosis: Prevalence, Diversity, and Outbreaks
Enteric colibacillosis in swine is related to high morbidity and mortality [1,14]. It has been reported that mortality can reach up to 70% in neonatal piglets with severe watery diarrhea, 1.5–2% in post-weaned and/or grow-finish pigs with moderate diarrhea, and up to 25% in untreated pigs with severe to moderate diarrhea [1,7]. In fact, this remarkable swine infection is widespread, taking place in both industrialized and developing countries and in temperate, subtropical, and tropical climates [1,14].
It is important to note that, as mentioned briefly above, this infection requires the presence, by ingestion, of ETEC and specific predisposing environmental conditions and host factors. Thus, these strains proliferate in the intestine and cause illness due to specific virulence factors, as reported in Section 2.1. The degree of ETEC colonization determines the occurrence of the disease [2]. Interestingly, it was already shown that ETEC strains were present in 16.6% of non-diarrhoeic pigs during the piglets’ suckling period, 66% in the nursery phase, and 17.3% in the finisher population. Furthermore, ETEC strains can be shed in the feces of healthy pigs [2,7].
ETECs can be found in fecal-contaminated feed, water, and soil and the environment of the pig barn. Long survival times in the environment are achieved by low temperatures and enough moisture, among other factors [1]. In slurry samples, a porcine ETEC O139:K82 strain remained viable for more than 11 weeks [1]. The spread of pathogenic E. coli is supposed to mainly occur via other pigs and contaminated barn environments. In addition, other transmission modes, namely via aerosols, have been reported [1]. Importantly, it was shown that airborne transmission between pigs in wire cages 1.5 m apart was repeatedly observed in transmission experiments with an F4-ETEC strain [1]. In addition, other possible modes of transmission are contaminated feed and water, contaminated trucks that transport pigs, and possibly other animal species. As a result of this transmission cycle, the same strain is usually found in many sick pigs and often in consecutive batches of pigs. To control the transmission of this infection, it is necessary to use strict hygienic measures [1] since routine cleaning and disinfection are usually insufficient to break the cycle of infection by ETEC [15].
Complete information on the prevalence is scarce, which makes the comparison between countries difficult. However, previous analysis already suggested some differences, as shown in Table 2.
Table 2.
Prevalence of ETEC in several countries from studies published since 2010.
| Country | Prevalence of ETEC (%) (n = Number of Isolates) |
Sampling Information/Origin | Period | Reference |
|---|---|---|---|---|
| Argentina | 15.2 (n = 990) | 11 farms with no history or clinical signs of colibacillosis | 2015 | [16] |
| Australia | 58.8 (n = 325) | 22 pig herds | 2013–2014 | [17] |
| Belgium and the Netherlands | 36.4 (n = 160) | 88 farms | 2012–2014 | [18] |
| France | 64.8 (n = 455) | 91 farms | 2012–2014 | [18] |
| Germany | 47.1 (n = 99) | 17 farms | 2012–2014 | [18] |
| Italy | 81.0 (n = 159) | 84 farms | 2012–2014 | [18] |
| Poland | 30 (n = 386) a | 70 pig herds | 2011–2013 | [19] |
| South Africa | 72.0 (n = 228) | 8 piggeries of different sizes (16–650 sow units) and production systems: large-scale commercial (>250 sow units), medium-scale commercial (51–250 sow units), and emerging small-scale pig farms (<50 sow units) | 2015–2016 | [20] |
| South Africa | 18.6 (n = 263) | 263 neonatal and post-weaned pigs | 2013 | [21] |
| Spain | 86.5 (n = 186) | 50 different Spanish farms | 2005–2017 | [22] |
| Spain | 67.0 (n = 499) | 179 outbreaks | 2008–2018 | [23] |
| Switzerland | 50.4 (n = 131) | 115 pigs suffering from diarrhea | 2014–2015 | [24] |
a Prevalence of enterotoxigenic E coli with fimbriae F4 (ETEC-F4).
Regarding the diversity and heterogeneity of ETEC, it is important to highlight that ETEC populations in pig fecal microbiota and in the farm environment are very dynamic and show high levels of diversity [2,25]. Numbers in the large intestine average around 107 colony-forming units (CFU)/g of contents; nevertheless, E. coli contributes less than 1% to the total bacterial count [1].
A recent study across Europe concerning ETEC pathotypes demonstrated a higher prevalence of F4 compared to F18 isolates in Belgium and the Netherlands, France, and Italy, as can be seen in Table 2 [18]. In contrast, in Spain, a different tendency was observed, with F18 being the prevalent adhesin [23]. In fact, the reported association of F18 isolates with PWD in other countries varied widely, as can be seen in Table 3, from 15.4% in Australia [26], 35% in Slovakia [27], 39.3% in Denmark [28], and 53% in the United States [29] to 61.9% in Poland [30]. Furthermore, a high prevalence of F18 was reported in Japan (62.9%), where, in addition, most isolates carried the stx2e gene (60.1%), which describes a very different pathogenic profile concerning other geographic areas and represents a high risk to swine production [31]. It is important to highlight that, in Spain, around 10% of Stx2e-positive isolates were found [23] similar to what was reported in other European countries [18].
It is also remarkable that, in Spain, according to García-Meniño and colleagues, the most common virulence profiles within each pathotype were LT, STb, and F4 and LT, STa, STb, and F18 (37.3% and 18.6% of the 161 ETEC isolates, respectively) [22]. According to the area of study, there is a divergence in the combination of virulence factors harboring the ETEC; in 2010, LT, STb, and F18 were the most predominant genotypes in the United States, for instance [29]. As expected, it was also previously described that the distribution of enterotoxins/fimbriae can also vary over time in a region [2,21]. An example is the United States, where, in 2001–2002, LT, STb, and F4 were the most prevalent genotype [32], which differ from the previous study [29].
It is also important to mention that LT and ST are well-known enterotoxins responsible for the diarrhea symptom, while it has been proposed that Stx2e is responsible for the severe neurological damage observed in swine edema disease [31]. Although enough data do not exist to assess the differences in disease severity between Stx-producing ETEC and other virulence factor-containing isolates, it has been reported that swine infected with E. coli-producing enterotoxins and Stx2e commonly exhibit diarrhea as an initial symptom, which is followed by lethal neurological symptoms [33].
Table 3.
The prevalence of the most known fimbriae and toxins in ETEC strain isolates from swine.
| Country (n = Number of Isolates) |
Percentage (Number) of Positive Isolates (%) | Reference | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fimbriae | Toxins | |||||||||
| F4 | F5 | F6 | F18 | F41 | LT | STa | STb | Stx2e | ||
| Australia (n = 104) | 38.5–96.3 | - | - | 0–15.4 | - | 62.1–92.3 | 64.8–92.3 | 83.7–100 | - | [26] |
| Belgium and The Netherlands (n = 100) | 51.0 | 1.0 | 1.0 | 42.0 | - | 14.0 | 22.0 | 30.0 | 5.0 | [18] |
| Denmark (n = 219) | 44.7 | - | 0.9 | 39.3 | - | 61.6 | 26.5 | 77.6 | - | [28] |
| France (n = 91) | 47.3 | - | - | 35.2 | - | 45.1 | 40.7 | 76.9 | 19.8 | [18] |
| Germany (n = 64) | 14.1 | - | - | 14.1 | - | 9.4 | 26.6 | 57.8 | 3.1 | [18] |
| Italy (n = 84) |
59.3 | 1.2 | 1.2 | 38.1 | 1.2 | 56.0 | 63.1 | 71.4 | 9.5 | [18] |
| Poland (n = 40) | 22.5 | - | - | 61.9 | - | 22.5 | 72.5 | 77.5 | 17.5 | [30] |
| Spain (n = 181) | 38.2 | 4.8 | 1.1 | 43.5 | 2.7 | 66.1 | 50.5 | 74.7 | 13.5 | [22] |
| Spain a (n = 277) | 27.7–40.5 | 16.7 | 11.9 | 51.5 | 16.7 | - | - | - | 10 | [25] |
| Slovakia (n = 101) |
19 | 0.9 | 5 | 35 | 0.9 | 20 | 26 | 46 | 5 | [27] |
| Uganda b (n = 83) | 8.4 | - | - | - | - | - | 1.2 | 26.5 | 2.4 | [34] |
| United States (n = 175) | 41.7 | - | - | 53.1 | - | 52.6 | 38.2 | 96 | - | [29] |
| Zimbabwe (n = 1984) | 28.4 | 22.3 | 1.5 | 25.4 | 22.3 | 50 | 73 | 16 | 27 | [35] |
a All isolates were positive for genes encoding enterotoxins (STa and/or STa and/or STb). b A prevalence of 16% of AIDA was detected. Note: “-”, means not found.
4. Antimicrobial Prevalence in Enteric Colibacillosis Treatment
The high morbidity and mortality rates are not the only problems associated with enteric colibacillosis in swine production but also the cost associated with its treatment with antibiotics [2]. Moreover, the spread of AMR determinants combined with the decrease in the available antimicrobial treatments is currently a global problem. Normally, antibiotics should be administered only to pigs that show clinical signs of colibacillosis; however, when the mortality increases in the farm production, the prophylactic treatment is applied to all animals [22]. However, the use of antimicrobials for the growth promotion of food animals has been banned in several countries [36]. It is noteworthy that, in Europe, according to Regulation (EU) 2019/61 on Veterinary Medicines and Regulation (EU) 2019/4 on Medicated Feed, antibiotics shall not be applied routinely, nor used for prophylaxis, unless in exceptional cases. It should only be applied for metaphylaxis when the risk of spreading infection is very high and there are no other options. Similarly, in the USA, since 2017, growth promotion uses of medically important (to human health) antibiotics are not allowed. Only therapeutic use (treatment, control, prevention) for a specific animal health condition is allowed under the direction of a veterinarian [37,38]. Additionally, in Brazil, since 1998, the use of several antimicrobial classes as growth promoters is prohibited, and recently, in 2016, colistin, a last-resort treatment for multidrug-resistant Gram-negative infections, was also banned [39].
In fact, it has been reported that the level of AMR in the gut microbiota increases with the number of antimicrobials used [40]. Due to the emergence of antimicrobial-resistant bacteria and the spread of AMR genes, antimicrobial resistance has become a global problem in the swine industry [5]. To overcome this problem, several countries have been monitoring AMR [5].
One factor that contributes to promoting the AMR phenomenon is the fact that E. coli presents a high capacity to acquire and pass antimicrobial resistance genes via horizontal gene transfer [5]. In fact, E. coli isolates revealed an alarming scenario with high resistance to a different antimicrobial class, as is the case of penicillins, aminoglycosides, tetracyclines, sulphonamides, fluoroquinolones, and phenicols [2]. In 2019, E. coli was considered one of the major pathogens responsible for the deaths associated with AMR [41]. Therefore, there is an increasing trend in the detection of AMR among ETEC from pigs with enteric colibacillosis [2]. Table 4 presents several studies from different countries on the resistance profile to different antimicrobial classes of E. coli isolated from swine with the disease. In Spain, ETEC isolates from a swine farm presented high levels of resistance to antibiotics commonly used for the treatment, such as in the case of ampicillin (Table 4) [22,23]. Similar values of resistance to ampicillin were observed in several regions in China [42,43,44,45]. Tetracycline is another antibiotic widely used in veterinary medicine that has been reported in several studies with a huge AMR in different parts of the world. A recent study developed in Denmark revealed a percentage of around 57% resistance to tetracycline of 90 ETEC isolates from pigs [46]. Furthermore, in the same study, antibiotics of other classes presented a resistance higher than 50%, namely spectinomycin, streptomycin, sulfamethoxazole, and trimethoprim (Table 4).
The variation of resistance in ETEC isolates in different countries emphasizes the importance of performing antimicrobial susceptibility testing in farm productions to select the correct antimicrobial agent for the treatment of ETEC. Moreover, it is not allowed in several countries to use antimicrobials for growth production since healthy pigs can serve as reservoirs for resistant E. coli and resistant bacteria can be transferred from the animals to humans by direct contact or by the food chain or indirectly through the environment [5,47].
Table 4.
The prevalence of antimicrobial resistance among E. coli isolates from swine over the world.
| Antimicrobial Class/Other Designations | Antimicrobial Agents | % Resistant Rates (n = Swine Isolates) | Country/City | Year/Time Range of the Study | Reference |
|---|---|---|---|---|---|
| Penicillins | Ampicillin | 85.9 (n = 608) | China/Shanghai | 2009–2021 | [48] |
| 75.4 (n = 481) | Spain/Lugo | 2006–2016 | [23] | ||
| 71.9 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| 84.8 (n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 27.9 (n = 129) | China/Tibet | 2012 | [43] | ||
| 60.7 (n = 89) | Denmark/Frederiksberg C | 2014 | [46] | ||
| 81.4 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 60.86 (n = 23) | Bangladesh/Tangail | 2018 | [50] | ||
| 86.4 (n = 118) | Korea | 2016–2017 | [40] | ||
| 48.3 (n = 90) | Denmark | 2018–2019 | [51] | ||
| 89.1 (n = 55) | United States | 2013–2014 | [44] | ||
| 34.5 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Ampicillin-sulbactam | 64.6 (n = 481) | Spain/Lugo | 2006–2016 | [23] | |
| Ticarcillin | 73.8 (n = 481) | Spain/Lugo | 2006–2016 | [23] | |
| 81.4 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| β-lactam combination agents | Amoxicillin/clavulanic acid | 42.3 (n = 608) | China/Shanghai | 2009–2021 | [48] |
| 84.63 (n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 11.76 (n = 135) | Santa Catarina/ Brazil |
2016–2017 | [52] | ||
| 33.5 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 82.6 (n = 23) | Bangladesh/Tangail | 2018 | [50] | ||
| 5.1 (n = 118) | Korea | 2016–2017 | [40] | ||
| 1.1 (n = 90) | Denmark | 2018–2019 | [51] | ||
| 9.5 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Ampicillin/ sulbactam |
70.8 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| 5 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Penicillins + β-lactamase inhibitors | Piperacillin/ tazobactam |
0.6 (n = 161) | Spain/Lugo | 2005–2017 | [22] |
| Cephalosporins | Ceftiofur | 22.5 (n = 608) | China/Shanghai | 2009–2021 | [48] |
| 52.63 (n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 25 (n = 135) | Santa Catarina/ Brazil |
2016–2017 | [52] | ||
| 10.9 (n = 129) | China/Tibet | 2012 | [43] | ||
| 25.5 (n = 55) | United States | 2013–2014 | [44] | ||
| Cefepime | 9.2 (n = 481) | Spain/Lugo | 2006–2016 | [23] | |
| 7.5 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 2.5 (n = 118) | Korea | 2016–2017 | [40] | ||
| 4.2 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Cefazolin | 60.82 (n = 455) | China/Beijing | 2014–2016 | [42] | |
| 10.6 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 10.2 (n = 118) | Korea | 2016–2017 | [40] | ||
| Cefuroxime | 8.7 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| Cefotaxime | 10.6 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| 9.1 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Ceftazidime | 5 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| 3 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Cephalothin | 64.4 (n = 118) | Korea | 2016–2017 | [40] | |
| 41.7 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Cefoxitin | 3.4 (n = 118) | Korea | 2016–2017 | [40] | |
| 1.8 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Ceftriaxone | 6 (n = 168) | China/Shenzhen | 2009–2014 | [45] | |
| Carbapenems | Ceftazidime | 1.9 (n = 608) | China/Shanghai | 2009–2021 | [48] |
| 1.5 (n = 481) | Spain/Lugo | 2006–2016 | [23] | ||
| 5.9 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| Meropenem | 0.3 (n = 608) | China/Shanghai | 2009–2021 | [48] | |
| Aminoglycosides | Kanamycin | 63.74 (n = 455) | China/Beijing | 2014–2016 | [42] |
| 3.6 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Spectinomycin | 65.7 (n = 608) | China/Shanghai | 2009–2021 | [48] | |
| 2.3 (n = 129) | China/Tibet | 2012 | [43] | ||
| 18 (n = 89) | Denmark/Frederiksberg C | 2014 | [46] | ||
| 43.6 (n = 55) | United States | 2013–2014 | [44] | ||
| 55.6 (n = 90) | Denmark | 2018–2019 | [51] | ||
| Gentamicin | 37.2 (n = 608) | China/Shanghai | 2009–2021 | [48] | |
| 47.7 (n = 481) | Spain/Lugo | 2006–2016 | [23] | ||
| 7.7 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| 57.31 (n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 32.35 (n = 135) | Santa Catarina/ Brazil |
2016–2017 | [52] | ||
| 6.9 (n = 129) | China/Tibet | 2012 | [43] | ||
| 14.6 (n = 89) | Denmark/Frederiksberg C | 2014 | [46] | ||
| 58.4 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 36.4 (n = 118) | Korea | 2016–2017 | [40] | ||
| 32.7 (n = 55) | United States | 2013–2014 | [44] | ||
| 6.7 (n = 90) | Denmark | 2018–2019 | [51] | ||
| 5.4 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Tobramycin | 47.7 (n = 481) | Spain/Lugo | 2006–2016 | [23] | |
| 6.2 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| 54.7 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| Streptomycin | 40.35 (n = 455) | China/Beijing | 2014–2016 | [42] | |
| 16.2 (n = 129) | China/Tibet | 2012 | [43] | ||
| 29.2 (n = 89) | Denmark/Frederiksberg C | 2014 | [46] | ||
| 86.4 (n = 118) | Korea | 2016–2017 | [40] | ||
| 68.9 (n = 90) | Denmark | 2018–2019 | [51] | ||
| 18.5 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Amikacin | 15.2 (n = 455) | China/Beijing | 2014–2016 | [42] | |
| 1.2 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Apramycin | 14.6 (n = 89) | Denmark/Frederiksberg C | 2014 | [46] | |
| 8.9 (n = 90) | Denmark | 2018–2019 | [51] | ||
| Neomycin | 50 (n = 118) | Korea | 2016–2017 | [40] | |
| 49.1 (n = 55) | United States | 2013–2014 | [44] | ||
| 25.6 (n = 90) | Denmark | 2018–2019 | [51] | ||
| Tetracyclines | Doxycycline | 85.9 (n = 608) | China/Shanghai | 2009–2021 | [48] |
| 62.7 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| Tetracycline | 91.6 (n = 608) | China/Shanghai | 2009–2021 | [48] | |
| 67.7 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| 83.63 (n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 40.4 (n = 129) | China/Tibet | 2012 | [43] | ||
| 47.2 (n = 89) | Denmark/Frederiksberg C | 2014 | [46] | ||
| 65.21 (n = 23) | Bangladesh/Tangail | 2018 | [50] | ||
| 86.4 (n = 118) | Korea | 2016–2017 | [40] | ||
| 56.7 (n = 90) | Denmark | 2018–2019 | [51] | ||
| 21.4 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Minocycline | 41.5 (n = 481) | Spain/Lugo | 2006–2016 | [23] | |
| 52.2 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| Chlortetracycline | 80 (n = 55) | United States | 2013–2014 | [44] | |
| Oxytetracycline | 94.5 (n = 55) | United States | 2013–2014 | [44] | |
| Sulfonamides | Sulfisoxazole | 85.4 (n = 608) | China/Shanghai | 2009–2021 | [48] |
| Sulphaamethoxazole | 75.2 (n = 608) | China/Shanghai | 2009–2021 | [48] | |
| 69.7 (n = 89) | Denmark/Frederiksberg C | 2014 | [46] | ||
| 67.8 (n = 90) | Denmark | 2018–2019 | [51] | ||
| Sulfadimethoxine | 61.8 (n = 55) | United States | 2013–2014 | [44] | |
| Fluoroquinolones | Enrofloxacin | 41.3 (n = 608) | China/Shanghai | 2009–2021 | [48] |
| 72.51135(n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 54.41 (n = 135) | Santa Catarina/Brazil | 2016–2017 | [52] | ||
| 58.2 (n = 55) | United States | 2013–2014 | [44] | ||
| Ofloxacin | 39 (n = 608) | China/Shanghai | 2009–2021 | [48] | |
| Ciprofloxacin | 61.5 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| 26.3 (n = 118) | Korea | 2016–2017 | [40] | ||
| 9.8 (n = 41, farm 1); 8.8% (n = 34, farm 2); 21.7% (n = 23, farm 3); 39.6% (n = 48, farm 4); 3.4% (n = 58, farm 5); 50% (n = 24, farm 6); 70% (n = 10, farm 7) |
Germany Mecklenburg–Western Pomerania | 2018 | [53] | ||
| 12.3 (n = 481) | Spain/Lugo | 2006–2016 | [23] | ||
| 16.4 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| 60.82 (n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 7.8 (n = 129) | China/Tibet | 2012 | [43] | ||
| 47.82 (n = 23) | Bangladesh/Tangail | 2018 | [50] | ||
| 3.6 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Levofloxacin | 55.3 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| 3.6 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Polymyxins | Colistin | 21.9 (n = 608) | China/Shanghai | 2009–2021 | [48] |
| 76.4 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 5.9 (n = 118) | Korea | 2016–2017 | [40] | ||
| Phosphonic | Fosfomycin | 4.6 (n = 481) | Spain/Lugo | 2006–2016 | [23] |
| 2.0 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| 1.9 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| Phenicols | Florfenicol | 77.78 (n = 455) | China/Beijing | 2014–2016 | [42] |
| 27.9 (n = 129) | China/Tibet | 2012 | [43] | ||
| 40 (n = 55) | United States | 2013–2014 | [44] | ||
| 92.6 (n = 608) | China/Shanghai | 2009–2021 | [48] | ||
| Chloramphenicol | 58.5 (n = 481) | Spain/Lugo | 2006–2016 | [23] | |
| 18.5 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| 76.61 (n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 57.8 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 88.1 (n = 118) | Korea | 2016–2017 | [40] | ||
| 16.7 (n = 90) | Denmark | 2018–2019 | [51] | ||
| 1.2 (n = 168) | China/Shenzhen | 2009–2014 | [46] | ||
| Trimethoprim | 69.7 (n = 89) | Denmark/Frederiksberg C | 2014 | [47] | |
| 53.3 (n = 90) | Denmark | 2018–2019 | [51] | ||
| 13.1 (n = 168) | China/Shenzhen | 2009–2014 | [46] | ||
| Folate pathway inhibitors | Trimethoprim-sulfamethoxazole | 72.3 (n = 481) | Spain/Lugo | 2006–2016 | [42] |
| 49.5 (n = 694) | Austria/Vienna | 2016–2018 | [49] | ||
| 85.55 (n = 455) | China/Beijing | 2014–2016 | [43] | ||
| 75 (n = 135) | Santa Catarina/Brazil | 2016–2017 | [52] | ||
| 19.4 (n = 129) | China/Tibet | 2012 | [44] | ||
| 59.6 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 56.8 (n = 118) | Korea | 2016–2017 | [40] | ||
| 30.9 (n = 55) | United States | 2013–2014 | [44] | ||
| 13.1 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Quinolone | Nalidixic acid | 60 (n = 481) | Spain/Lugo | 2006–2016 | [23] |
| 90.05 (n = 455) | China/Beijing | 2014–2016 | [42] | ||
| 19.4 (n = 129) | China/Tibet | 2012 | [43] | ||
| 87.6 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| 73.91 (n = 23) | Bangladesh/Tangail | 2018 | [50] | ||
| 61.9 (n = 118) | Korea | 2016–2017 | [40] | ||
| 8.9 (n = 90) | Denmark | 2018–2019 | [51] | ||
| 77.4 (n = 168) | China/Shenzhen | 2009–2014 | [45] | ||
| Levofloxacin | 10.8 (n = 481) | Spain/Lugo | 2006–2016 | [23] | |
| Norfloxacin | 24.6 (n = 118) | Korea | 2016–2017 | [40] | |
| Monobactam | Aztreonam | 2.2 (n = 694) | Austria/Vienna | 2016–2018 | [49] |
| 8.1 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| Glycylcyclines | Tigecycline | 1.9 (n = 161) | Spain/Lugo | 2005–2017 | [22] |
| Quindoxin | Olaquindox | 39.77 (n = 455) | China/Beijing | 2014–2016 | [42] |
| Polymyxin | 20.47 (n = 455) | China/Beijing | 2014–2016 | [42] | |
| Nitrofurans | Nitrofurantoin | 2.34 (n = 455) | China/Beijing | 2014–2016 | [42] |
| 9.3 (n = 161) | Spain/Lugo | 2005–2017 | [22] | ||
| ESBL-producing isolates | 10.6 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| MDR (≥3 categories) | 91.3 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| MDR (≥6 categories) | 59 (n = 161) | Spain/Lugo | 2005–2017 | [22] | |
| Macrolide | Azithromycin | 78.26 (n = 23) | Bangladesh/Tangail | 2018 | [50] |
| Erythromycin | 47.82 (n = 23) | Bangladesh/Tangail | 2018 | [50] | |
| Tilmicosin | 100 (n = 55) | United States | 2013–2014 | [44] | |
| Lincomycin | Clindamycin | 100 (n = 55) | United States | 2013–2014 | [44] |
| 3-MDR | Isolates resistant to penicillin, and cephalosporins, and at least one other class of antibiotics | 36.6 (n = 41, farm 1); 32.4 (n = 34, farm 2); 87 (n = 23, farm 3); 95.8 (n = 48, farm 4); 22.4 (n = 58, farm 5); 95.8 (n = 24, farm 6); 90 (n = 10, farm 7) |
Germany Mecklenburg–Western Pomerania | 2018 | [53] |
| 5-MDR | Isolates resistant to penicillin and cephalosporins and at least three other classes of antibiotics | 4.9 (n = 41, farm 1); 5.9 (n = 34, farm 2); 17.4 (n = 23, farm 3); 14.6 (n = 48, farm 4); 1.7 (n = 58, farm 5); 8.3 (n = 24, farm 6) 0 (n = 10, farm 7) |
Germany Mecklenburg–Western Pomerania | 2018 | [53] |
5. Prevalence of AMR-Associated Resistance Genes in ETEC
In swine production, the occurrence of AMR among ETEC has been a longstanding problem [54,55]. It is also noteworthy that the tendency of porcine ETEC to express a multidrug-resistant (MDR) phenotype has increased during the last decade [26,54]. As a result of the continuous use of antimicrobials, it is plausible that at least some MDR ETEC will probably develop pan-resistance, which means a phenotype with resistance to all commonly applied drugs plus resistance to vital antimicrobials such as fluoroquinolones and third- and fourth-generation cephalosporins [56]. It has been demonstrated that the combination of specific virulence-associated plasmids with resistance-associated plasmids is directly related to the fitness of drug-resistant ETEC in the swine production environment [26]. F-type plasmids generally encode for virulence genes, whereas the IncFII-like IncFV, IncA/C, and Incl1 plasmids encode for resistance genes [56,57]. Plasmids encoding for virulent enterotoxins (for example, LT, STb) and the ones associated with antibiotic resistance are commonly transferred together [56,58]. Additionally, antibiotic resistance genes (ARGs) and virulence genes (VGs) were statistically associated with MDR-ETEC isolates from Canada [54]. In addition, a common plasmid harboring both enterotoxin VGs and tetracycline ARG (tetB) was demonstrated in F18-positive O141 [59] and O149:H10 [60] strains [26]. Lastly, for O149:H10 strains, an enhanced virulence was evidenced [60]. Based on these findings, Martínez and Baquero hypothesized that the application of antibiotics may potentially promote the transmission of virulence genes between bacteria [61].
Some studies have established an association between ETEC infection and the presence of associated-antimicrobial resistance genes, as shown in Table 5. According to our analysis, in resistance genes linked to the antimicrobial class of aminoglycosides, there is a heterogeneous distribution at the level of prevalence and type of gene along the countries explored. In Denmark, the most prevalent genes were aph and aadA with very similar values of 64.4% and 63.3%, respectively [51]. Similarly, Australia showed a similar prevalence for the aadA gene with 58.6% [56]. However, in this country, the gene ant(3)-I was the most predominant, with 93.3% [26]. For Korea and Switzerland [26], aac(3)-III and strB were the most common genes. In the study carried out in Korea, Choi et al. [62] evaluated the presence of aminoglycoside-resistant genes in pathogenic and non-pathogenic E. coli isolates from pigs during 2004–2007 and concluded that the prevalence of gentamicin/apramycin resistance-associated genes was much higher in diseased pigs than in healthy pigs.
Regarding the class of -lactams, the gene blaTEM is very widespread and is associated with a significant prevalence around the world but with different subtypes such as blaTEM-1, blaTEM-1-A, blaTEM-1-B, and blaTEM-30. This is not surprising given the fact that extended-spectrum -lactam (ESBL) and AmpC -lactamase production constitute an important resistance mechanism in members of the Enterobacteriaceae family, in which E. coli is included [63]. In the literature, IncI1 plasmids are evidenced for the possession of genes encoding for antimicrobial resistance, namely ESBL genes [63,64,65]. Based on this knowledge, Johnson and colleagues [66] evaluated the presence of Incl1-associated genes (ardA, pill, and repl) and four genes related to -lactam resistance (blaCTX, blaCMY-2, blaNDM-1, and blaTEM) in ETEC isolates from commercial farms of the United States, corresponding to 88 cases of postweaning diarrhea and 111 cases of neonatal diarrhea. Of the cases examined, 60–66% and 37–40% harbored IncI1 plasmid-associated genes for PWD and ND, respectively. Regarding the -lactam resistance genes, PWD isolates held blaCMY-2 and blaTEM at a prevalence of 41% and 50.9%, respectively, whereas the ND isolates held these genes at rates of 25.3% and 22.8%, respectively [66].
Concerning the phenicol class, Switzerland [24] is highlighted with a higher prevalence of catA1 and catIII, 67% and 50% for the respective genes, whereas for Denmark [51] and Australia [26], these genes are expressed at low rates.
Finally, relating to the sulfonamides class, sul1 and sul2 genes are the most common for Denmark [51], Switzerland [24], and Australia [26,56], with values above 30% of prevalence.
At a global overview, it is important to note that Australian porcine ETEC was different from isolates of other parts of the world due to the geographical isolation and decades of prohibition of the importation of livestock and fresh meat. For instance, it is very relevant to note that this is a unique country that never permitted the application of fluoroquinolones and gentamicin in food-producing animals [67]. In addition, the usage of third-generation cephalosporins for mass medication is very restricted, and fourth-generation cephalosporins are not registered for application [26,68]. Consequently, these isolates are absent of resistance to the following critically relevant antimicrobials, third-generation cephalosporins, and fluoroquinolones [62].
Table 5.
The prevalence of resistance genes among the ETEC isolates from swine over the world.
| Antibiotic Group | Gene | % Prevalence (n = Swine Isolates) | Country/City | Year/Time Range of the Study | Reference |
|---|---|---|---|---|---|
| Aminoglycosides | aph (phosphotransferases) | 64.4% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] |
| aphA1 | 27.1% (N = 70) | Australia | 1999–2005 | [56] | |
| aadA (nucleotidyltransferases) | 63.3% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] | |
| 58.6% (n = 70) | Australia | 1999–2005 | [56] | ||
| aac (acetyltransferases) | 10% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] | |
| aac(3)-II | 18.3% (N = 71) a | Korea | 2004–2007 | [62] | |
| aac(3)-III | 31% (N = 71) a | ||||
| aac(3)-IV | 47.1% (N = 70) | Australia | 1999–2005 | [56] | |
| ant(2″)-I | 7% (N = 71) a | Korea | 2004–2007 | [62] | |
| armA | 2.8% (N = 71) a | ||||
| ant(3)-I | 93.3% (N = 104) | Australia | 1999–2005 | [26] | |
| aac(3)-IV | 47.1% (N = 104) | ||||
| aphA-I | 27.9% (N = 104) | ||||
| strA | 8% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| 50% (N = 70) | Australia | 1999–2005 | [56] | ||
| strB | 16% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| 55.7% (N = 70) | Australia | 1999–2005 | [56] | ||
| Beta-lactams | blaTEM-1-A | 4.4% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] |
| blaTEM-1-B | 46.7% (N = 90) | ||||
| blaTEM-30 | 1.1% (N = 90) | ||||
| blaTEM-1 | 87% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| blaTEM | 43.3% (N = 104) | Australia | 1999–2005 | [26] | |
| 38% (N = 199) | United States | 2007–2008 | [66] | ||
| 40% (N = 70) | Australia | 1999–2005 | [56] | ||
| blaCMY-2 | 34% (N = 199) | United States | 2007–2008 | [66] | |
| Lincosamides | Inu(F) | 5.6 % (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] |
| Inu(G) | 5.6 % (N = 90) | ||||
| Macrolides | mdf(A) | 100% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] |
| mph(A) | 8.9% (N = 90) | ||||
| mph(B) | 7.8% (N = 90) | ||||
| erm(B) | 10% (N = 90) | ||||
| ereA | 7.1% (N = 70) | Australia | 1999–2005 | [56] | |
| Phenicols | catA1 | 3.3% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] |
| 67% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| cmlA1 | 8.9% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] | |
| floR | 5.6% (N = 90) | ||||
| catAIII | 50% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| catI | 9.6% (N = 104) | Australia | 1999–2005 | [26] | |
| catII | 1% (N = 104) | ||||
| cmlA | 31.7% (N = 104) | ||||
| 12.9% (N = 70) | Australia | 1999–2005 | [56] | ||
| Polymyxins | mcr-1 | 26.4% (N = 186) | Spain | 2006–2017 | [69] |
| mcr-4 | 72.8% (N = 186) | ||||
| mcr-5 | 3.6% (N = 186) | ||||
| Sulfonamides | sul1 | 33.3 % (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] |
| 57% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| 65.4% (N = 104) | Australia | 1999–2005 | [26] | ||
| 57.1% (N = 70) | Australia | 1999–2005 | [62] | ||
| sul2 | 46.7% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] | |
| 64% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| 20.2% (N = 104) | Australia | 1999–2005 | [26] | ||
| 21.4% (N = 70) | Australia | 1999–2005 | [62] | ||
| sul3 | 10% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] | |
| 31% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| Tetracycline | tet(A) | 44.4% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] |
| 65% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| 44.2% (N = 104) | Australia | 1999–2005 | [26] | ||
| 35.7% (N = 70) | Australia | 1999–2005 | [56] | ||
| tet(B) | 14.4% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] | |
| 23% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| 28.8% (N = 104) | Australia | 1999–2005 | [26] | ||
| 7.1% (N = 70) | Australia | 1999–2005 | [56] | ||
| tet(C) | 35% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| 16.3% (N = 104) | Australia | 1999–2005 | [26] | ||
| 5.7% (N = 70) | Australia | 1999–2005 | [56] | ||
| tet(D) | 3% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| tet(E) | 2% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| tet(X) | 1.1% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] | |
| Trimethoprim | dfrA1 | 37.8% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] |
| 59% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| dfrA5 | 2.2% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] | |
| 7% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| dfrA7 | 7% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| dfrA12 | 8.9% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] | |
| 10% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| dfrA13 | 7% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| dfrA14 | 5.6% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] | |
| 5% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| dfrA17 | 2.2% (N = 90) | Denmark | F4-positive isolates: 2018, 2019, and 1989–1992 F18 isolates: 2019 and with a strain recovered in the 1970s |
[51] | |
| 5% (N = 119) b | Switzerland | 2014–2015 | [24] | ||
| dfrA19 | 7% (N = 119) b | Switzerland | 2014–2015 | [24] | |
| dhfrI | 1.9% (N = 104) | Australia | 1999–2005 | [26] | |
| dhfrV | 31.7% (N = 104) | Australia | 1999–2005 | [26] | |
| 25.7% (N = 70) | Australia | 1999–2005 | [56] | ||
| dhfrXIII | 30.8% (N = 104) | Australia | 1999–2005 | [26] |
a Pathogenic E.coli isolates; b enterovirulent E. coli from pigs (where NETEC = 66).
Horizontal Gene Transfer
In addition to the identification of ETEC-associated AMR genes and their prevalence, it is demanding to understand how the ETEC-associated VGs emerge and disseminate across species since both classes of genes (VGs, AMR) are intrinsically related, as was explained above. It has been described that variability in VG and colonization factor combinations highlight the genomic diversity within the ETEC pathogroup [70]. These data suggest that ETEC consists of a genetically heterogeneous group of strains that gained the ETEC-associated virulence genes by horizontal gene transfer. In fact, it has been shown that strains within a single pathogroup can originate from distinct genetical backgrounds [70,71,72,73]. Multi-locus sequence typing (MLST) has shown that ETEC strains originate from different evolutionary lineages, proposing that the acquisition of the elt or est genes may be enough to make an ETEC strain [74]. This hypothesis is also supported by the study carried out by Chen and colleagues, which shows that the prototypical ETEC strain H10407 chromosome is almost identical to the chromosome of E. coli K-12 strain MG1655, suggesting that the major event in the emergence of ETEC from E. coli is, thus, the acquisition of virulence plasmids carrying elt or est [75]. Contrary to these current notions, recent evidence, based on the sequence analysis of a representative collection of isolates of ETEC isolated between 1980 and 2011, showed that persistent plasmid-chromosomal background combinations exist in certain phylogenetic lineages [76]. Due to these divergences, additional research is needed to understand the gene transfer between strains to improve the prevention as well as treatment. As such, a more detailed understanding of what actually constitutes a naturally occurring ETEC strain is vital.
6. Conclusions
It is indispensable to control the application of antimicrobials to treat ETEC-associated infections in swine farms in order to reduce the incidence of AMR and, consequently, the probability of zoonoses occurrence, which implies a public health concern derivate from the potential transfer of AMR genetic determinants directly by contact and indirectly into the food chain, water, manure, and others [5]. It is crucial to note that high levels of AMR in ETEC strains have been arising, namely in apramycin, neomycin, sulfonamide-trimethoprim, and colistin.
In Europe, the EFSA AHAW Panel (2021c) revealed clinical swine E. coli isolates with a high proportion of resistance to numerous antibiotics with a prevalence from 63% to 70% (namely to aminopenicillins, sulfonamides, and tetracycline) [77]. However, lower rates of resistance to clinically critical antibiotics (fluoroquinolones and third-generation cephalosporins) were detected. Although, the risk of development of a pan-resistant MDR ETEC remains and may constitute a huge problem in public health. Therefore, alternative therapeutics (prebiotics, probiotics, synbiotics, organic acids, phytogenic substances, bacteriophages, specific egg yolk antibodies, lactoferrin, antisense oligonucleotides, spray-dried animal plasma, and aptamers), as well as hygienic and sanitary measurements, should be applied in pig farming [2,4].
In sum, this knowledge about the distribution of pathogenic ETEC in swine farms and their diversity, resistance, and virulence profiles constitute a preliminary measure to adopt the best treatments. However, this review has some limitations, the assembled data can be outdated since the mutagenic capacity of E. coli constitutes an important aspect to take into account. In addition, some countries do not possess studies in the scope of this review. Therefore, from a future research perspective, it is crucial to evaluate alternative therapies in vitro and in vivo capable of decreasing the usage of antimicrobials, characterize serotypes of ETEC in Portugal since this information is scarce, and combine bioinformatic tools to complement the genetic composition characterization.
Author Contributions
The first draft of the manuscript was written by M.M.B., J.C., D.A. and A.M.C., and all authors (R.O., S.S., D.O.-M. and C.A.) commented on previous versions of the manuscript. J.C. and C.A. defined the content of the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors have no relevant financial or non-financial interests to disclose.
Funding Statement
This work was financially supported by the Project PTDC/CVT-CVT/4620/2021, funded by FEDER funds through COMPETE2020–Programa Operacional Competitividade e Internacionalização (POCI) and by national funds (PIDDAC) through FCT/MCTES. It was also supported by: LA/P/0045/2020 (ALiCE), UIDB/00511/2020 and UIDP/00511/2020 (LEPABE), funded by national funds through FCT/MCTES (PIDDAC). In addition, this work was also funded by FCT (Portuguese Foundation for Science and Technology), under strategic projects UIDP/CVT/00772/2020 (CECAV) and LA/P/0059/2020 (AL4AnimalS); and under the scope of the strategic funding of UIDB/04469/2020 unit (CEB). J.C. also thanks FCT for the CEEC Individual (2022.06886.CEECIND).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Fairbrother J.M., Nadeau É. Colibacillosis. In: Zimmerman J.J., Karriker L.A., Ramirez A., Schwartz K.J., Stevenson G.W., Zhang J., editors. Diseases of Swine. John Wiley & Son; Hoboken, NJ, USA: 2019. pp. 807–834. [Google Scholar]
- 2.Luppi A. Swine Enteric Colibacillosis: Diagnosis, Therapy and Antimicrobial Resistance. Porcine Health Manag. 2017;3:16. doi: 10.1186/s40813-017-0063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fairbrother J.M., Nadeau É., Gyles C.L. Escherichia coli in Postweaning Diarrhea in Pigs: An Update on Bacterial Types, Pathogenesis, and Prevention Strategies. Anim. Health Res. Rev. 2005;6:17–39. doi: 10.1079/AHR2005105. [DOI] [PubMed] [Google Scholar]
- 4.Castro J., Barros M.M., Araújo D., Campos A.M., Oliveira R., Silva S., Almeida C. Swine Enteric Colibacillosis: Current Treatment Avenues and Future Directions. Front. Vet. Sci. 2022;9:981207. doi: 10.3389/fvets.2022.981207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poirel L., Madec J.-Y., Lupo A., Schink A.-K., Kieffer N., Nordmann P., Schwarz S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018;6:4. doi: 10.1128/microbiolspec.ARBA-0026-2017. [DOI] [PubMed] [Google Scholar]
- 6.Javadi M., Bouzari S., Oloomi M. Escherichia coli—Recent Advances on Physiology, Pathogenesis and Biotechnological Applications. InTech; London, UK: 2017. Horizontal Gene Transfer and the Diversity of Escherichia coli; pp. 318–331. [Google Scholar]
- 7.Nielsen S.S., Bicout D.J., Calistri P., Canali E., Drewe J.A., Garin-Bastuji B., Gonzales Rojas J.L., Gortázar C., Herskin M., Michel V., et al. Assessment of Listing and Categorisation of Animal Diseases within the Framework of the Animal Health Law (Regulation (EU) No 2016/429): Antimicrobial-Resistant Escherichia coli in Dogs and Cats, Horses, Swine, Poultry, Cattle, Sheep and Goats. EFSA J. 2022;20:e07311. doi: 10.2903/j.efsa.2022.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gyles C.L., Fairbrother J.M. Escherichia coli. In: Gyles C.L., Prescott J.F., Songer J.G., Thoen C.O., editors. Pathogenesis of Bacterial Infections in Animals. Wiley-Blackwell; Hoboken, NJ, USA: 2010. pp. 267–279. [Google Scholar]
- 9.Loayza F., Graham J.P., Trueba G. Factors Obscuring the Role of E. coli from Domestic Animals in the Global Antimicrobial Resistance Crisis: An Evidence-Based Review. Int. J. Environ. Res. Public Health. 2020;17:3061. doi: 10.3390/ijerph17093061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valat C., Drapeau A., Beurlet S., Bachy V., Boulouis H.-J., Pin R., Cazeau G., Madec J.-Y., Haenni M. Pathogenic Escherichia coli in Dogs Reveals the Predominance of ST372 and the Human-Associated ST73 Extra-Intestinal Lineages. Front. Microbiol. 2020;11:580. doi: 10.3389/fmicb.2020.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim K., Song M., Liu Y., Ji P. Enterotoxigenic Escherichia coli Infection of Weaned Pigs: Intestinal Challenges and Nutritional Intervention to Enhance Disease Resistance. Front. Immunol. 2022;13:885253. doi: 10.3389/fimmu.2022.885253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burgos Y., Beutin L. Common Origin of Plasmid Encoded Alpha-Hemolysin Genes in Escherichia coli. BMC Microbiol. 2010;10:193. doi: 10.1186/1471-2180-10-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubreuil J.D., Isaacson R.E., Schifferli D.M. Animal Enterotoxigenic Escherichia coli. EcoSal Plus. 2016;7:1–47. doi: 10.1128/ecosalplus.ESP-0006-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubreuil J.D. Enterotoxigenic Escherichia coli and Probiotics in Swine: What the Bleep Do We Know? Biosci. Microbiota Food Health. 2017;36:75–90. doi: 10.12938/bmfh.16-030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hampson D.J., Fu Z.F., Robertson I.D. Investigation of the Source of Haemolytic Escherichia coli Infecting Weaned Pigs. Epidemiol. Infect. 1987;99:149–153. doi: 10.1017/S0950268800066966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moredo F.A., Piñeyro P.E., Márquez G.C., Sanz M., Colello R., Etcheverría A., Padola N.L., Quiroga M.A., Perfumo C.J., Galli L., et al. Enterotoxigenic Escherichia coli Subclinical Infection in Pigs: Bacteriological and Genotypic Characterization and Antimicrobial Resistance Profiles. Foodborne Pathog. Dis. 2015;12:704–711. doi: 10.1089/fpd.2015.1959. [DOI] [PubMed] [Google Scholar]
- 17.van Breda L.K., Dhungyel O.P., Ward M.P. Antibiotic Resistant Escherichia coli in Southeastern Australian Pig Herds and Implications for Surveillance. Zoonoses Public Health. 2018;65:e1–e7. doi: 10.1111/zph.12402. [DOI] [PubMed] [Google Scholar]
- 18.Luppi A., Gibellini M., Gin T., Vangroenweghe F., Vandenbroucke V., Bauerfeind R., Bonilauri P., Labarque G., Hidalgo Á. Prevalence of Virulence Factors in Enterotoxigenic Escherichia coli Isolated from Pigs with Post-Weaning Diarrhoea in Europe. Porc. Health Manag. 2016;2:20. doi: 10.1186/s40813-016-0039-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dors A., Czyzewska-Dors E., Wasyl D., Pomorska-Mól M. Prevalence and Factors Associated with the Occurrence of Bacterial Enteropathogens in Suckling Piglets in Farrow-to-Finish Herds. Vet. Rec. 2016;179:598. doi: 10.1136/vr.103811. [DOI] [PubMed] [Google Scholar]
- 20.Ogundare S.T., Fasanmi O.G., Fasina F.O. Risk Factors for Prevalence of Enterotoxigenic Escherichia coli (ETEC) in Diarrheic and Non-Diarrheic Neonatal and Weaner Pigs, South Africa. Biomed. Environ. Sci. 2018;31:149–154. doi: 10.3967/bes2018.018. [DOI] [PubMed] [Google Scholar]
- 21.Mohlatlole R.P., Madoroba E., Muchadeyi F.C., Chimonyo M., Kanengoni A.T., Dzomba E.F. Virulence Profiles of Enterotoxigenic, Shiga Toxin and Enteroaggregative Escherichia coli in South African Pigs. Trop. Anim. Health Prod. 2013;45:1399–1405. doi: 10.1007/s11250-013-0377-4. [DOI] [PubMed] [Google Scholar]
- 22.García-Meniño I., García V., Alonso M.P., Blanco J.E., Blanco J., Mora A. Clones of Enterotoxigenic and Shiga Toxin-Producing Escherichia coli Implicated in Swine Enteric Colibacillosis in Spain and Rates of Antibiotic Resistance. Vet. Microbiol. 2021;252:108924. doi: 10.1016/j.vetmic.2020.108924. [DOI] [PubMed] [Google Scholar]
- 23.García-Meniño I., García V., Mora A., Díaz-Jiménez D., Flament-Simon S.C., Alonso M.P., Blanco J.E., Blanco M., Blanco J. Swine Enteric Colibacillosis in Spain: Pathogenic Potential of Mcr-1 ST10 and ST131 E. coli Isolates. Front. Microbiol. 2018;9:2659. doi: 10.3389/fmicb.2018.02659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brand P., Brawand S.G., Perreten V. Pathotyping and Antibiotic Resistance of Porcine Enterovirulent Escherichia coli Strains from Switzerland (2014–2015) Schweiz. Arch. Tierheilkd. 2017;159:373–380. doi: 10.17236/sat00120. [DOI] [PubMed] [Google Scholar]
- 25.Marchant M., Moreno M.A. Dynamics and Diversity of Escherichia coli in Animals and System Management of the Manure on a Commercial Farrow-to-Finish Pig Farm. Appl. Environ. Microbiol. 2013;79:853–859. doi: 10.1128/AEM.02866-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith M.G., Jordan D., Chapman T.A., Chin J.J.C., Barton M.D., Do T.N., Fahy V.A., Fairbrother J.M., Trott D.J. Antimicrobial Resistance and Virulence Gene Profiles in Multi-Drug Resistant Enterotoxigenic Escherichia coli Isolated from Pigs with Post-Weaning Diarrhoea. Vet. Microbiol. 2010;145:299–307. doi: 10.1016/j.vetmic.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Vu Khac H.V., Holoda E., Pilipcinec E., Blanco M., Blanco J.E., Mora A., Dahbi G., López C., González E.A., Blanco J. Serotypes, Virulence Genes, and PFGE Profiles of Escherichia coli Isolated from Pigs with Postweaning Diarrhoea in Slovakia. BMC Vet. Res. 2006;2:10. doi: 10.1186/1746-6148-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frydendahl K. Prevalence of Serogroups and Virulence Genes in Escherichia coli Associated with Postweaning Diarrhoea and Edema Disease in Pigs and a Comparison of Diagnostic Approaches. Vet. Microbiol. 2002;85:169–182. doi: 10.1016/S0378-1135(01)00504-1. [DOI] [PubMed] [Google Scholar]
- 29.Post K.W., Bosworth B.T., Knoth Post DVM K.W., Bosworth B.T., Knoth J.L. Frequency of Virulence Factors in Escherichia Coli Isolated from Pigs with Postweaning Diarrhea and Edema Disease in North Caroline. Swine Health and Produc. 2000;8:1–2. [Google Scholar]
- 30.Osek J., Gallien P., Truszczynä M., Protz D. The Use of Polymerase Chain Reaction for Determination of Virulence Factors of Escherichia coli Strains Isolated from Pigs in Poland. Comp. Immunol. Microbiol. Infect. Dis. 1999;22:163–174. doi: 10.1016/S0147-9571(98)00083-6. [DOI] [PubMed] [Google Scholar]
- 31.Kusumoto M., Hikoda Y., Fujii Y., Murata M., Miyoshi H., Ogura Y., Gotoh Y., Iwata T., Hayashi T., Akiba M. Emergence of a Multidrug-Resistant Shiga Toxin-Producing Enterotoxigenic Escherichia coli Lineage in Diseased Swine in Japan. J. Clin. Microbiol. 2016;54:1074–1081. doi: 10.1128/JCM.03141-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis D.H. Enterotoxigenic Escherichia coli Infection in Pigs and Its Diagnosis. J. Swine Health Prod. 2002;10:171–175. [Google Scholar]
- 33.Baldo V., Salogni C., Giovannini S., D’Incau M., Boniotti M.B., Birbes L., Pitozzi A., Formenti N., Grassi A., Pasquali P., et al. Pathogenicity of Shiga Toxin Type 2e Escherichia Coli in Pig Colibacillosis. Front. Vet. Sci. 2020;7:545818. doi: 10.3389/fvets.2020.545818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikwap K., Larsson J., Jacobson M., Owiny D.O., Nasinyama G.W., Nabukenya I., Mattsson S., Aspan A., Erume J. Prevalence of Adhesin and Toxin Genes in E. coli Strains Isolated from Diarrheic and Non-Diarrheic Pigs from Smallholder Herds in Northern and Eastern Uganda. BMC Microbiol. 2016;16:178. doi: 10.1186/s12866-016-0796-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madoroba E., van Driessche E., de Greve H., Mast J., Ncube I., Read J., Beeckmans S. Prevalence of Enterotoxigenic Escherichia coli Virulence Genes from Scouring Piglets in Zimbabwe. Trop. Anim. Health Prod. 2009;41:1539–1547. doi: 10.1007/s11250-009-9345-4. [DOI] [PubMed] [Google Scholar]
- 36.Pissetti C., Kich J.D., Allen H.K., Navarrete C., de Freitas Costa E., Morés N., Cardoso M. Antimicrobial Resistance in Commensal Escherichia coli and Enterococcus spp. Isolated from Pigs Subjected to Different Antimicrobial Administration Protocols. Res. Vet. Sci. 2021;137:174–185. doi: 10.1016/j.rvsc.2021.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Public Health Producer Guide. Antibiotics on the Farm: What You Need to Know about New Regulations Overview FDA’s New Regulations. National Pork Board; Des Moines, IA, USA: 2015. 800-456-7675. [Google Scholar]
- 38.Holman D.B., Chénier M.R. Antimicrobial Use in Swine Production and Its Effect on the Swine Gut Microbiota and Antimicrobial Resistance. Can. J. Microbiol. 2015;61:785–798. doi: 10.1139/cjm-2015-0239. [DOI] [PubMed] [Google Scholar]
- 39.Diário Oficial da União Instrução Normativa N47, de 22 de Novembro de 2016. [(accessed on 11 January 2023)]; Available online: http://www.in.gov.br/autenticidade.html.
- 40.Do K.H., Byun J.W., Lee W.K. Virulence Genes and Antimicrobial Resistance of Pathogenic Escherichia coli Isolated from Diarrheic Weaned Piglets in Korea. J. Anim. Sci. Technol. 2020;62:543–552. doi: 10.5187/jast.2020.62.4.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murray C.J., Ikuta K.S., Sharara F., Swetschinski L., Robles Aguilar G., Gray A., Han C., Bisignano C., Rao P., Wool E., et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet. 2022;399:629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang G.Y., Guo L., Su J.H., Zhu Y.H., Jiao L.G., Wang J.F. Frequency of Diarrheagenic Virulence Genes and Characteristics in Escherichia coli Isolates from Pigs with Diarrhea in China. Microorganisms. 2019;7:308. doi: 10.3390/microorganisms7090308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li P., Wu D., Liu K., Suolang S., He T., Liu X., Wu C., Wang Y., Lin D. Investigation of Antimicrobial Resistance in Escherichia coli and Enterococci Isolated from Tibetan Pigs. PLoS ONE. 2014;9:e95623. doi: 10.1371/journal.pone.0095623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang F., Wu Z., Zheng Y., Frana T.S., Sahin O., Zhang Q., Li G. Genotypes and Antimicrobial Susceptibility Profiles of Hemolytic Escherichia coli from Diarrheic Piglets. Foodborne Pathog. Dis. 2019;16:94–103. doi: 10.1089/fpd.2018.2480. [DOI] [PubMed] [Google Scholar]
- 45.Li Y., Luo Q., Shi X., Lin Y., Qiu Y., Lv D., Jiang Y., Chen Q., Jiang M., Ma H., et al. Phenotypic and Genotypic Characterization of Clinical Enterotoxigenic Escherichia coli Isolates from Shenzhen, China. Foodborne Pathog. Dis. 2017;14:333–340. doi: 10.1089/fpd.2016.2233. [DOI] [PubMed] [Google Scholar]
- 46.Rosager W.N., Peter N.J., Erik Lind J.S., Svend H., Matthew D., Steen P.K. Comparison of Antimicrobial Resistance in E. coli Isolated from Rectal and Floor Samples in Pens with Diarrhoeic Nursery Pigs in Denmark. Prev. Vet. Med. 2017;147:42–49. doi: 10.1016/j.prevetmed.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 47.Stannarius C., Bürgi E., Regula G., Zychowska M.A., Zweifel C., Stephan R. Antimicrobial Resistance in Escherichia coli Strains Isolated from Swiss Weaned Pigs and Sows. Schweiz. Arch. Tierheilkd. 2009;151:119–125. doi: 10.1024/0036-7281.151.3.119. [DOI] [PubMed] [Google Scholar]
- 48.Lv C., Shang J., Zhang W., Sun B., Li M., Guo C., Zhou N., Guo X., Huang S., Zhu Y. Dynamic Antimicrobial Resistant Patterns of Escherichia coli from Healthy Poultry and Swine over 10 Years in Chongming Island, Shanghai. Infect. Dis. Poverty. 2022;11:98. doi: 10.1186/s40249-022-01025-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Renzhammer R., Loncaric I., Roch F.F., Pinior B., Käsbohrer A., Spergser J., Ladinig A., Unterweger C. Prevalence of Virulence Genes and Antimicrobial Resistances in E. coli Associated with Neonatal Diarrhea, Postweaning Diarrhea, and Edema Disease in Pigs from Austria. Antibiotics. 2020;9:208. doi: 10.3390/antibiotics9040208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rahman M.M., Ahmed P., Kar A., Sakib N., Shibly A.Z., Zohora F.T., Hasan M.N. Prevalence, Antimicrobial Resistance, and Pathogenic Potential of Enterotoxigenic and Enteropathogenic Escherichia coli Associated with Acute Diarrheal Patients in Tangail, Bangladesh. Foodborne Pathog. Dis. 2020;17:434–439. doi: 10.1089/fpd.2019.2741. [DOI] [PubMed] [Google Scholar]
- 51.García V., Gambino M., Pedersen K., Haugegaard S., Olsen J.E., Herrero-Fresno A. F4- and F18-Positive Enterotoxigenic Escherichia coli Isolates from Diarrhea of Postweaning Pigs: Genomic Characterization. Appl. Environ. Microbiol. 2020;86:e01913-20. doi: 10.1128/AEM.01913-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brisola M.C., Crecencio R.B., Bitner D.S., Frigo A., Rampazzo L., Stefani L.M., Faria G.A. Escherichia coli Used as a Biomarker of Antimicrobial Resistance in Pig Farms of Southern Brazil. Sci. Total Environ. 2019;647:362–368. doi: 10.1016/j.scitotenv.2018.07.438. [DOI] [PubMed] [Google Scholar]
- 53.Meissner K., Sauter-Louis C., Heiden S.E., Schaufler K., Tomaso H., Conraths F.J., Homeier-Bachmann T. Extended-Spectrum ß-Lactamase-Producing Escherichia coli in Conventional and Organic Pig Fattening Farms. Microorganisms. 2022;10:603. doi: 10.3390/microorganisms10030603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boerlin P., Travis R., Gyles C.L., Reid-Smith R., Janecko N., Lim H., Nicholson V., McEwen S.A., Friendship R., Archambault M. Antimicrobial Resistance and Virulence Genes of Escherichia coli Isolates from Swine in Ontario. Appl. Environ. Microbiol. 2005;71:6753–6761. doi: 10.1128/AEM.71.11.6753-6761.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X.M., Jiang H.X., Liao X.P., Liu J.H., Zhang W.J., Zhang H., Jiang Z.G., Lü D.H., Xiang R., Liu Y.H. Antimicrobial Resistance, Virulence Genes, and Phylogenetic Background in Escherichia coli Isolates from Diseased Pigs. FEMS Microbiol. Lett. 2010;306:15–21. doi: 10.1111/j.1574-6968.2010.01917.x. [DOI] [PubMed] [Google Scholar]
- 56.Shepard S.M., Danzeisen J.L., Isaacson R.E., Seemann T., Achtman M., Johnson T.J. Genome Sequences and Phylogenetic Analysis of K88- and F18-Positive Porcine Enterotoxigenic Escherichia coli. J. Bacteriol. 2012;194:395–405. doi: 10.1128/JB.06225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson T.J., Nolan L.K. Pathogenomics of the Virulence Plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 2009;73:750–774. doi: 10.1128/MMBR.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gomes T.A.T., Elias W.P., Scaletsky I.C.A., Guth B.E.C., Rodrigues J.F., Piazza R.M.F., Ferreira L.C.S., Martinez M.B. Diarrheagenic Escherichia coli. Braz. J. Microbiol. 2016;47:3–30. doi: 10.1016/j.bjm.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olasz F., Fekete P.Z., Blum-Oehler G., Boldogkoi Z., Nagy B. Characterization of an F18+ Enterotoxigenic Escherichia coli Strain from Post Weaning Diarrhoea of Swine, and of Its Conjugative Virulence Plasmid PTC. FEMS Microbiol. Lett. 2005;244:281–289. doi: 10.1016/j.femsle.2005.01.057. [DOI] [PubMed] [Google Scholar]
- 60.Goswami P.S., Gyles C.L., Friendship R.M., Poppe C., Kozak G.K., Boerlin P. Effect of Plasmid PTENT2 on Severity of Porcine Post-Weaning Diarrhoea Induced by an O149 Enterotoxigenic Escherichia coli. Vet. Microbiol. 2008;131:400–405. doi: 10.1016/j.vetmic.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 61.Martínez J.L., Baquero F. Interactions among Strategies Associated with Bacterial Infection: Pathogenicity, Epidemicity, and Antibiotic Resistance. Clin. Microbiol. Rev. 2002;15:647–679. doi: 10.1128/CMR.15.4.647-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi M.-J., Lim S.-K., Nam H.-M., Kim A.-R., Jung S.-C., Kim M.-N. Apramycin and Gentamicin Resistances in Indicator and Clinical Escherichia coli Isolates from Farm Animals in Korea. Foodborne Pathog. Dis. 2011;8:119–123. doi: 10.1089/fpd.2010.0641. [DOI] [PubMed] [Google Scholar]
- 63.Bortolaia V., Guardabassi L., Trevisani M., Bisgaard M., Venturi L., Bojesen A.M. High Diversity of Extended-Spectrum β-Lactamases in Escherichia coli Isolates from Italian Broiler Flocks. Antimicrob. Agents Chemother. 2010;54:1623–1626. doi: 10.1128/AAC.01361-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Diestra K., Juan C., Curiao T., Moyá B., Miró E., Oteo J., Coque T.M., Pérez-Vázquez M., Campos J., Cantón R., et al. Characterization of Plasmids Encoding BlaESBL and Surrounding Genes in Spanish Clinical Isolates of Escherichia coli and Klebsiella pneumoniae. J. Antimicrob. Chemother. 2009;63:60–66. doi: 10.1093/jac/dkn453. [DOI] [PubMed] [Google Scholar]
- 65.Cloeckaert A., Praud K., Doublet B., Bertini A., Carattoli A., Butaye P., Imberechts H., Bertrand S., Collard J.M., Arlet G., et al. Dissemination of an Extended-Spectrum-β-Lactamase BlaTEM-52 Gene-Carrying IncI1 Plasmid in Various Salmonella enterica Serovars Isolated from Poultry and Humans in Belgium and France between 2001 and 2005. Antimicrob. Agents Chemother. 2007;51:1872–1875. doi: 10.1128/AAC.01514-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson T.J., Shepard S.M., Rivet B., Danzeisen J.L., Carattoli A. Comparative Genomics and Phylogeny of the IncI1 Plasmids: A Common Plasmid Type among Porcine Enterotoxigenic Escherichia coli. Plasmid. 2011;66:144–151. doi: 10.1016/j.plasmid.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 67.Cheng A.C., Turnidge J., Collignon P., Looke D., Barton M., Gottlieb T. Control of Fluoroquinolone Resistance through Successful Regulation, Australia. Emerg. Infect. Dis. 2012;18:1453–1460. doi: 10.3201/eid1809.111515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jordan D., Chin J.J.C., Fahy V.A., Barton M.D., Smith M.G., Trott D.J. Antimicrobial Use in the Australian Pig Industry: Results of a National Survey. Aust. Vet. J. 2009;87:222–229. doi: 10.1111/j.1751-0813.2009.00430.x. [DOI] [PubMed] [Google Scholar]
- 69.García V., García-Meniño I., Mora A., Flament-Simon S.C., Díaz-Jiménez D., Blanco J.E., Alonso M.P., Blanco J. Co-Occurrence of Mcr-1, Mcr-4 and Mcr-5 Genes in Multidrug-Resistant ST10 Enterotoxigenic and Shiga Toxin-Producing Escherichia coli in Spain (2006–2017) Int. J. Antimicrob. Agents. 2018;52:104–108. doi: 10.1016/j.ijantimicag.2018.03.022. [DOI] [PubMed] [Google Scholar]
- 70.Sahl J.W., Steinsland H., Redman J.C., Angiuoli S.v., Nataro J.P., Sommerfelt H., Rasko D.A. A Comparative Genomic Analysis of Diverse Clonal Types of Enterotoxigenic Escherichia coli Reveals Pathovar-Specific Conservation. Infect. Immun. 2011;79:950–960. doi: 10.1128/IAI.00932-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rasko D.A., Rosovitz M.J., Myers G.S.A., Mongodin E.F., Fricke W.F., Gajer P., Crabtree J., Sebaihia M., Thomson N.R., Chaudhuri R., et al. The Pangenome Structure of Escherichia coli: Comparative Genomic Analysis of E. coli Commensal and Pathogenic Isolates. J. Bacteriol. 2008;190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cooper K.K., Mandrell R.E., Louie J.W., Korlach J., Clark T.A., Parker C.T., Huynh S., Chain P.S., Ahmed S., Carter M.Q. Comparative Genomics of Enterohemorrhagic Escherichia coli O145:H28 Demonstrates a Common Evolutionary Lineage with Escherichia coli O157:H7. BMC Genom. 2014;15:17. doi: 10.1186/1471-2164-15-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ogura Y., Ooka T., Iguchi A., Toh H., Asadulghani M., Oshima K., Kodama T., Abe H., Nakayama K., Kurokawa K., et al. Comparative Genomics Reveal the Mechanism of the Parallel Evolution of O157 and Non-O157 Enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. USA. 2009;106:17939–17944. doi: 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nyholm O., Halkilahti J., Wiklund G., Okeke U., Paulin L., Auvinen P., Haukka K., Siitonen A. Comparative Genomics and Characterization of Hybrid Shigatoxigenic and Enterotoxigenic Escherichia coli (STEC/ETEC) Strains. PLoS ONE. 2015;10:e0135936. doi: 10.1371/journal.pone.0135936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Q., Savarino S.J., Venkatesan M.M. Subtractive Hybridization and Optical Mapping of the Enterotoxigenic Escherichia coli H10407 Chromosome: Isolation of Unique Sequences and Demonstration of Significant Similarity to the Chromosome of E. coli K-12. Microbiology. 2006;152:1041–1054. doi: 10.1099/mic.0.28648-0. [DOI] [PubMed] [Google Scholar]
- 76.von Mentzer A., Connor T.R., Wieler L.H., Semmler T., Iguchi A., Thomson N.R., Rasko D.A., Joffre E., Corander J., Pickard D., et al. Identification of Enterotoxigenic Escherichia coli (ETEC) Clades with Long-Term Global Distribution. Nat. Genet. 2014;46:1321–1326. doi: 10.1038/ng.3145. [DOI] [PubMed] [Google Scholar]
- 77.Nielsen S.S., Bicout D.J., Calistri P., Canali E., Drewe J.A., Garin-Bastuji B., Gonzales Rojas J.L., Gortazar Schmidt C., Herskin M., Michel V., et al. Assessment of Animal Diseases Caused by Bacteria Resistant to Antimicrobials: Swine. EFSA J. 2021;19:e06955. doi: 10.2903/j.efsa.2021.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.