Abstract
Cefiderocol is a new siderophore cephalosporin that is effective against multidrug-resistant Gram-negative bacteria, including carbapenem-resistant strains. The aim of this study was to evaluate the activity of this new antimicrobial agent against a collection of pathogens using broth microdilution assays and to analyze the possible mechanism of cefiderocol resistance in two resistant Klebsiella pneumoniae isolates. One hundred and ten isolates were tested, comprising 67 Enterobacterales, two Acinetobacter baumannii, one Achromobacter xylosoxidans, 33 Pseudomonas aeruginosa and seven Stenotrophomonas maltophilia. Cefiderocol showed good in vitro activity, with an MIC < 2 μg/mL, and was able to inhibit 94% of the tested isolates. We observed a resistance rate of 6%. The resistant isolates consisted of six Klebsiella pneumoniae and one Escherichia coli, leading to a resistance rate of 10.4% among the Enterobacterales. Whole-genome sequencing analysis was performed on two cefiderocol-resistant Klebsiella pneumoniae isolates to investigate the possible mutations responsible for the observed resistance. Both strains belonged to ST383 and harbored different resistant and virulence genes. The analysis of genes involved in iron uptake and transport showed the presence of different mutations located in fhuA, fepA, iutA, cirA, sitC, apbC, fepG, fepC, fetB, yicI, yicJ, and yicL. Furthermore, for the first time, to the best of our knowledge, we described two Klebsiella pneumoniae isolates that synthesize a truncated fecA protein due to the transition from G to A, leading to a premature stop codon in the amino acid position 569, and a TonB protein carrying a 4-amino acid insertion (PKPK) after Lysine 103. In conclusion, our data show that cefiderocol is an effective drug against multidrug-resistant Gram-negative bacteria. However, the higher resistance rate observed in Enterobacterales underlines the need for active surveillance to limit the spread of these pathogens and to avoid the risks associated with the emergence of resistance to new drugs.
Keywords: resistance, cefiderocol, multi-drug resistance (MDR), whole-genome sequencing (WGS), iron transporters
1. Introduction
Antibiotic resistance is one of the main threats to human health, and for this reason, the World Health Organization (WHO) has recognized it as a global priority that needs to be addressed [1]. During the last two decades, there has been an increased prevalence of multidrug-resistant (MDR) Gram-negative bacteria, such as third-generation cephalosporin- and carbapenem-resistant Enterobacterales (CRE), carbapenem-resistant (CR) Pseudomonas aeruginosa, and CR Acinetobacter baumannii, which has led to higher morbidity and mortality rates [2,3,4,5,6]. The diffusion of carbapenem-resistant Enterobacterales, and in particular, of enterobacteria such as Klebsiella pneumoniae producing carbapenemase (KPC), New Delhi metallo-β-lactamase (NDM), and oxacillinase(OXA)-48-like enzymes, is a major concern for public health due to the reduced available treatment options and high case-fatality rates [7,8,9].
Italy has the highest number of infections and deaths due to multidrug-resistant bacteria in Europe [10]. There has been an alarming increase in resistance to carbapenems in several bacterial species in Italy, including Klebsiella pneumoniae (29.5% of isolates), Pseudomonas aeruginosa (15.9% of isolates), and Acinetobacter baumannii (80.8% of isolates). Among these species, Klebsiella pneumoniae producing the KPC enzyme is the most widespread bacterial strain resistant to antimicrobials in Italy. Since 2019, the appearance of isolates carrying NDM genes have been documented [11].
To overcome the issue of emerging antibiotic resistance, new drugs are needed. Cefiderocol, the first injectable siderophore cephalosporin approved by the FDA, was developed for this purpose [12]. Cefiderocol is a cephalosporin with high affinity for PBP 3 and its structure is similar to cefepime. Like other β-Lactams, it binds to bacterial penicillin binding proteins (PBP), inhibiting peptidoglycan synthesis. However, it is different from other β-Lactams in that it enters the bacterial periplasmic space through its siderophore-like activity. This drug presents an intrinsic structural stability not only to Ambler class A (e.g., KPC), C (e.g., AmpC), and D (e.g., OXA-48), but also to Amber class B β-lactamases (e.g., NDM, VIM, and IMP). In fact, cefiderocol is the first active antimicrobial against metallo-β-lactamases. Its molecular structure derives from ceftazidime and cefepime, with a carboxypropanoxyimino group in the side chain at position 7 of the cephem nucleus, improving stability to β-lactamases similar to ceftazidime, and a pyrrolidinium group at position 3 of the side chain, avoiding recognition by β-lactamases similar to cefepime. Additionally, cefiderocol has a catechol group in position 3 of the side chain, which is responsible for the siderophore-like property. This catechol group is recognized by transporters such as CirA and Fiu in E. coli or PiuA in P. aeruginosa, allowing cefiderocol to accumulate in the periplasmic space (“Trojan horse”) and avoid resistance mechanisms such as loss of porins. Altogether, these structural features enable this new antibiotic to be active against MDR Gram-negative bacteria such as Pseudomonas aeruginosa, Acinetobacter baumannii, Stenotrophomonas maltophilia, and Enterobacterales that producing β-lactamases, including AmpC and extended-spectrum β-lactamases, or carbapenemases. In particular, cefiderocol activity against KPC-producing Klebsiella pneumoniae is comparable or higher than that of ceftazidime/avibactam [13]. Overall, cefiderocol appears to cause minor adverse effects such as diarrhea (4%), infusion site pain (3%), headache (3%), nausea (2%) [12].
In light of the emergence of bacteria resistant to different antibiotic classes, the aim of the current study was to evaluate in vitro susceptibility of different multi-drug resistant Gram-negative bacteria isolates responsible for different infections to cefiderocol. Although cefiderocol seems promising for treating severe infections caused by MDR bacteria such as complicated urinary infections and lower respiratory infections, some cases of resistance to this molecule due to mutations in iron transporter genes are documented in the literature [14,15]. Therefore, in order to analyze the possible mechanism of cefiderocol resistance, we performed whole-genome sequencing (WGS) and relative genomic analysis of two cefiderocol-resistant Klebsiella pneumoniae isolates.
2. Results
2.1. Antimicrobial Activity of Cefiderocol against MDRGram-Negative Bacteria
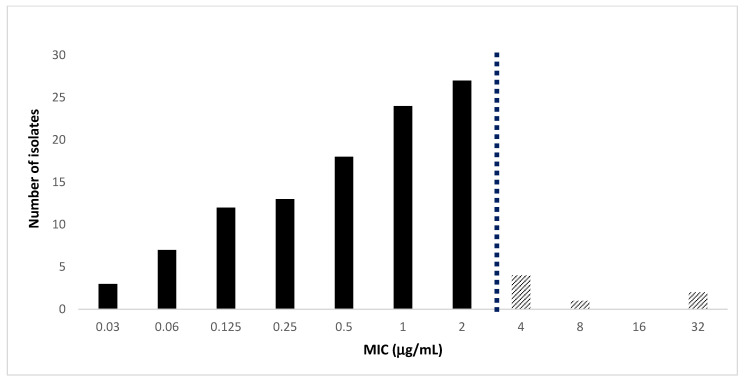
For all the clinical isolates included in this study, MIC distributions of cefiderocol are shown in Figure 1. According to the EUCAST criteria, 94% of the isolates were susceptible, while six percent of them were resistant.
Figure 1.
Distribution of cefiderocol MICs for all of the isolates according to EUCAST breakpoints. Black column, susceptible isolates; striped columns, resistant isolates. The dashed bar separates susceptible and resistant isolates.
Resistant isolates consisted of six Klebsiella pneumoniae and one Escherichia coli strains.
Out of the 56 Klebsiella pneumoniae strains, 37 isolates were KPC producers, three isolates were OXA-48 producers, one isolate produced NDM, 14 isolates co-produced two carbapenemases (OXA-48 and NDM), and one exhibited a membrane impermeability phenotype. Out of the eight Escherichia coli strains, four isolates were NDM producers, three isolates were AmpC producers, and one isolate was ESBL positive. Out of the three Enterobacter spp. strains, two isolates were VIM producers and two isolates coproduced OXA-48 and KPC carbapenemases. Concerning non-fermenting Gram-negative bacteria, out of the 33 Pseudomonas aeruginosa strains, four isolates were IMP producers, two isolates showed membrane impermeability, and one isolate produced VIM. For 26 isolates, the mechanism of resistance was not identified by the implemented routine assays. For Acinetobacter baumannii, Achromobacter xylosoxidans, and Stenotrophomonas maltophilia, the antimicrobial resistance mechanisms were not evaluated.
All of the isolates displayed a multidrug antimicrobial resistance phenotype because they were resistant to different antibiotic classes (cefepime, ceftriaxone, ceftazidime, aztreonam, amikacin, ciprofloxacin, trimethoprim/sulfamethoxazole, imipenem, meropenem, ceftazidime/avibactam, and ceftolozane/tazobactam). MIC50 and MIC90 values of all tested antibiotics against the bacterial isolates are shown in the Table 1.
Table 1.
In vitro activity of antibiotics against Gram-negative bacteria collected during the study period.
| Organisms (N° of Isolates) | Drug | MIC Range (μg/mL) | MIC50 | MIC90 | Susceptible a (%) | Resistant a (%) |
|---|---|---|---|---|---|---|
| K. pneumoniae (n = 56) | CFDC | 0.03–4 | 1 | 2 | 89 | 11 |
| AN | <1–32 | 32 | 32 | 27 | 73 | |
| AMC | >16 | >16 | >16 | 0 | 100 | |
| FEP | 0.12 to >16 | >16 | 32 | 0 | 100 | |
| CT | >8 to >16 | >16 | >16 | 0 | 100 | |
| CAZ | >32 | >32 | >32 | 0 | 100 | |
| CZA | 0.5 to >16 | >16 | >16 | 29 | 71 | |
| CIP | <0.06 to >2 | >2 | >2 | 4 | 96 | |
| GEM | <1 to >8 | 2 | >8 | 52 | 48 | |
| IPM | <0.25 to >8 | >8 | >8 | 2 | 98 | |
| MEM | 4 to >128 | >8 | >128 | 2 | 98 | |
| TZP | >64 | >64 | >64 | 0 | 100 | |
| CST | <0.5 to >8 | 0.5 | 1 | 98 | 2 | |
| SXT | <20 to >160 | >160 | >160 | 20 | 80 | |
| E. coli (n = 8) | CFDC | 0.03–32 | NC | NC | 88 | 12 |
| AN | 2–16 | NC | NC | 88 | 12 | |
| AMC | >16 | NC | NC | 0 | 100 | |
| FEP | 0.12 to >16 | NC | NC | 12 | 88 | |
| CT | <0.25 to >16 | NC | NC | 50 | 50 | |
| CAZ | 8 to >32 | NC | NC | 0 | 100 | |
| CZA | 0.12 to >8 | NC | NC | 50 | 50 | |
| CIP | >2 | NC | NC | 0 | 100 | |
| GEM | <1 to >8 | NC | NC | 63 | 37 | |
| IPM | <0.25 to >8 | NC | NC | 50 | 50 | |
| MEM | <0.25 to >8 | NC | NC | 50 | 50 | |
| TZP | <4 to >64 | NC | NC | 37 | 63 | |
| CST | 0.5 | NC | NC | 100 | 0 | |
| SXT | <20 to >160 | NC | NC | 12 | 88 | |
| Enterobacter spp. (n = 3) | CFDC | 0.12–2 | NC | NC | 100 | 0 |
| AN | <1 | NC | NC | 100 | 0 | |
| AMC | >16 | NC | NC | 0 | 100 | |
| FEP | >16 | NC | NC | 0 | 100 | |
| CT | >16 | NC | NC | 0 | 100 | |
| CAZ | >32 | NC | NC | 0 | 100 | |
| CZA | >8 | NC | NC | 0 | 100 | |
| CIP | 1 to >2 | NC | NC | 0 | 100 | |
| IPM | >8 | NC | NC | 0 | 100 | |
| MEM | >8 | NC | NC | 0 | 100 | |
| TZP | >64 | NC | NC | 0 | 100 | |
| CST | 0.5 | NC | NC | 100 | 0 | |
| SXT | >160 | NC | NC | 0 | 100 | |
| P. aeruginosa (n = 33) | CFDC | 0.03–2 | 1 | 2 | 100 | 0 |
| AN | <1 to >32 | 4 | >32 | 84 | 16 | |
| FEP | 4 to >16 | >16 | >16 | 0 | 100 | |
| CT | 0.5 to >64 | >8 | >16 | 40 | 60 | |
| CAZ | 2 to >32 | >32 | >32 | 0 | 100 | |
| CZA | 2 to >64 | >8 | >8 | 28 | 72 | |
| CIP | 0.5 to >2 | >2 | >2 | 0 | 100 | |
| IPM | 2 to >8 | >8 | >8 | 0 | 100 | |
| MEM | 1 to >8 | >8 | >8 | 0 | 100 | |
| TZP | 32 to >64 | >64 | >64 | 0 | 100 | |
| CST | <0.5 to >8 | 0.5 | 2 | 94 | 6 | |
| A. baumannii (n = 2) | CFDC | <=2 | NC | NC | 100 | 0 |
| AN | >32 | NC | NC | 0 | 100 | |
| FEP | >16 | NC | NC | 0 | 100 | |
| TIG | 1 | NC | NC | NA | NA | |
| CAZ | >32 | NC | NC | 0 | 100 | |
| ATM | >256 | NC | NC | NA | NA | |
| CIP | >2 | NC | NC | 0 | 100 | |
| IPM | >8 | NC | NC | 0 | 100 | |
| MEM | >8 to >256 | NC | NC | 0 | 100 | |
| TZP | >64 | NC | NC | 0 | 100 | |
| CST | 0.5 | NC | NC | 100 | 0 | |
| A. xylosoxidans (n = 1) | CFDC | 0.06 | NC | NC | 100 | 0 |
| AN | >16 | NC | NC | NA | NA | |
| AMC | >16 | NC | NC | NA | NA | |
| FEP | >16 | NC | NC | NA | NA | |
| CT | >64 | NC | NC | NA | NA | |
| CAZ | 16 | NC | NC | NA | NA | |
| CZA | 16 | NC | NC | NA | NA | |
| CIP | >1 | NC | NC | NA | NA | |
| GEM | >8 | NC | NC | NA | NA | |
| MEM | 8 | NC | NC | 0 | 100 | |
| TZP | >128 | NC | NC | 0 | 100 | |
| CST | 2 | NC | NC | NA | NA | |
| SXT | <1 | NC | NC | NA | NA | |
| S. maltophilia (n = 7) | CFDC | 0.5 to <2 | NC | NC | 100 | 0 |
| AN | >32 | NC | NC | NA | NA | |
| CT | >32 | NC | NC | NA | NA | |
| CAZ | >32 | NC | NC | 0 | 100 | |
| CZA | >16 | NC | NC | NA | NA | |
| MEM | >32 to >64 | NC | NC | NA | NA | |
| CST | >8 | NC | NC | NA | NA | |
| SXT | 0.01 to >4 | NC | NC | 71 | 29 |
Drug abbreviations: CFDC, cefiderocol; AN, amikacin; AMC, amoxicillin-clavulanic acid; FEP, cefepime; CT, ceftolozane-tazobactam; CAZ, ceftazidime; CZA, ceftazidime-avibactam; CIP, ciprofloxacin; GEM, gentamicin; IPM, imipenem; MEM, meropenem; TZP, piperacillin-tazobactam; CST, colistin; SXT, trimethoprim-sulfamethoxazole; ATM, aztreonam. NA, not applicable; NC, not calculated because the number of isolates was <10. a EUCAST breakpoints.
Cefiderocol MICs ranged from 0.12 to 32 μg/L for Enterobacterales (from 0.03 to 2 μg/mL for Klebsiella pneumoniae, from 0.03 to 32 μg/mL for Escherichia coli, and from 0.12 to 2 μg/mL for Enterobacter spp.). Against Klebsiella pneumoniae isolates, the cefiderocol concentration able to inhibit 90 percent of the tested isolates was 2 μg/mL, and the susceptibility rate to cefiderocol was higher than those of the other antibiotics such as ceftozolane-tazobactam, ceftazidime, and ceftazidime-avibactam, but was lower than colistin. For non-fermenting Gram-negative bacteria, cefiderocol MICs ranged from 0.03 to 2 μg/mL (from 0.03 to 3 μg/mL for Pseudomonas aeruginosa, <2 μg/mL for Acinetobacter baumannii, 0.06 μg/mL for Achromobacter xilosoxidans, and from 0.5 to <2 μg/mL for Stenotrophomonas maltophilia). All non-fermenting Gram-negative bacteria were fully susceptible to cefiderocol.
2.2. Genomic Characteristics of Two Cefiderocol Resistant Klebsiella pneumoniae Isolates
To analyze the molecular mechanism underlining cefiderocol resistance, we performed a whole-genome sequencing analysis of two Klebsiella pneumoniae isolates (BSKP542 and BSKP713) responsible for invasive infections with a cefiderocol MIC of 4 μg/mL. The isolates were resistant to all of the tested antibiotics except for colistin and the aztreonam-avibactam combination.
The full length of BSKP542 and BSKP713 was 5,722,475 bp and 5,727,485 bp, respectively. Both isolates had 56.67% CG and 83 contigs of size ≥ 5000 bp. Both harbored the polysaccharide capsule KL30 and lipopolysaccharide O antigen O1V2 loci. BSKP542 and BSKP 713 were both assigned to the sequence type 383 (gapA-infB-mdh-pgi-phoE-rpoB-tonB allele number 2-6-1-3-8-1-18).
The genome of both isolates consisted of a chromosomal backbone, carrying all the virulence factors and some resistance genes, and five plasmids (Table 2a,b). In particular, 46 putative virulence factors and two efflux pump genes were detected in both genomes, while 35 and 33 resistance genes were identified in the BSKP542 and BSKP713 genomes, respectively. A virulence score of 0 was assigned to both our isolates because neither carried yersiniabactin (ybt), aerobactin (iuc), or colibactin (clb) [16]. The plasmids identified with an identity >91.6% were: pHAD28 (KU674895), pNDM-MAR (JN420336.1), pKpQIL-IT (JN233705), pKPN3 (CP000648), pOXA-48 (JN626286), and pNDM-5-IT (MG649062), which were previously described by Tian et al. in 2020 [17].
Table 2.
(a) Antimicrobial resistance (AMR) determinants of the two cefiderocol-resistant Klebsiella pneumoniae isolates. (b) Virulence determinants of the two cefiderocol-resistant Klebsiella pneumoniae isolates.
| (a) | |||||
|---|---|---|---|---|---|
| Isolates | ST | Serotype | AMR Determinants | ||
| Gene Function | Detected Genes | Hypothetical Location | |||
| BSKP542 | 383 | HL30, O1V2 | aminoglycoside resistance | aac (6′)-1b’ | pNDM-MAR |
| aadA2 | pNDM-5-IT | ||||
| aph(3′)-VI | chromosome | ||||
| armA | chromosome | ||||
| beta-lactam resistance | bla NDM-1 | pNDM-MAR | |||
| bla CTX-M15 | pNDM-MAR | ||||
| bla TEM-1b | pKpQIL-IT | ||||
| bla Tem-1c | pKpQIL-IT | ||||
| bla OXA-48 | pOXA-48 | ||||
| bla NDM-5 | pNDM-5-IT | ||||
| bla SHV-26 | chromosome | ||||
| bla SHV-78 | chromosome | ||||
| bla SHV-98 | chromosome | ||||
| bla SHV-179 | chromosome | ||||
| bla SHV-145 | chromosome | ||||
| bla SHV-194 | chromosome | ||||
| bla SHV-199 | chromosome | ||||
| bla OXA-9 | chromosome | ||||
| bla CTX-M14b | chromosome | ||||
| ompK36 | chromosome | ||||
| ompK37 | chromosome | ||||
| macrolide resistance | mph(A) | pNDM-5-IT | |||
| mph(E) | pNDM-5-IT | ||||
| msr(E) | chromosome | ||||
| quinolone resistance | qnrB19 | pHAD28 | |||
| qnrB1 | pNDM-MAR | ||||
| qnrS1 | chromosome | ||||
| parC | chromosome | ||||
| gyrA | chromosome | ||||
| tetracycline resistance | tet(A) | pNDM-5-IT | |||
| fosfomycin resistance | fosA | chromosome | |||
| phenicol resistance | catA1 | pNDM-MAR | |||
| trimethoprim resistance | dfrA5 | chromosome | |||
| sulphonamide resistance | sul1 | pNDM-5-IT | |||
| sul2 | chromosome | ||||
| BSKP713 | 6339 | HL30, O1V2 | aminoglycoside resistance | aac (6′)-1b’ | pNDM-MAR |
| aadA2 | pNDM-5-IT | ||||
| aph(3′)-VI | chromosome | ||||
| armA | chromosome | ||||
| beta-lactam resistance | bla NDM-1 | pNDM-MAR | |||
| bla CTX-M15 | pNDM-MAR | ||||
| bla TEM-1b | pKpQIL-IT | ||||
| bla Tem-1c | pKpQIL-IT | ||||
| bla OXA-48 | pOXA-48 | ||||
| bla NDM-5 | pNDM-5-IT | ||||
| bla SHV-26 | chromosome | ||||
| bla SHV-78 | chromosome | ||||
| bla SHV-98 | chromosome | ||||
| bla SHV-179 | chromosome | ||||
| bla SHV-145 | chromosome | ||||
| bla SHV-194 | chromosome | ||||
| bla SHV-199 | chromosome | ||||
| bla OXA-9 | chromosome | ||||
| bla CTX-M14b | chromosome | ||||
| ompK36 | chromosome | ||||
| ompK37 | chromosome | ||||
| macrolide resistance | mph(A) | pNDM-5-IT | |||
| mph(E) | pNDM-5-IT | ||||
| msr(E) | chromosome | ||||
| quinolone resistance | qnrB19 | pHAD28 | |||
| qnrB1 | pNDM-MAR | ||||
| qnrS1 | chromosome | ||||
| tetracycline resistance | tet(A) | pNDM-5-IT | |||
| fosfomycin resistance | fosA | chromosome | |||
| phenicol resistance | catA1 | pNDM-MAR | |||
| trimethoprim resistance | dfrA5 | chromosome | |||
| sulphonamide resistance | sul1 | pNDM-5-IT | |||
| sul2 | chromosome | ||||
| (b) | |||||
| Isolates | Virulence Determinants | ||||
| Gene Function | Detected Genes | ||||
| BSKP542 and BSKP713 | Adhesion | ||||
| type 1 fimbriae | mrkABCDFHJ | ||||
| type 3 fimbriae | fimABCDEFGHIK | ||||
| type 4 pili | pilW | ||||
| fimbrial adherence determinants | stbABCDE | ||||
| Iron uptake | |||||
| aerobactin | iutA | ||||
| ent siderophore | entABCDEFS; fepABCDG; fes | ||||
| salmochelin | iroEN | ||||
| Regulatory system | rcsAB | ||||
| Secretion system | T6SS (I-III) | ||||
| Efflux pump genes | |||||
| RND efflux pump | arcAB | ||||
Due to the short-read output, we were unable to retrieve the complete antibiotic resistance plasmid sequences from both genomes. In particular, blaNDM-1, blaCTX-M-15, qnrB1, aac(6′)-1b’, and cat genes, which are respectively responsible for carbapenem, β-lactam, quinolone, aminoglycoside, and chloramphenicol resistance, were probably located on the pNDM-MAR plasmid from Klebsiella pneumoniae, showing 99.5% of identity and 100% of identical sites with BSKP542 and BSKP713 genomes (Table 2a). The pHAD28 plasmid from Salmonella enterica, subspecies enterica serovar Hadar strain HAD28 shared a minor identity percentage (91.6%) with our isolate genomes and probably carried the qnrB19 gene coding for quinolones resistance. The pKpQIL-IT plasmid from the Klebsiella pneumoniae strain ST258 shared the highest identity and percentage of identical sites (both 100%) with the BSKP542 and BSKP713 genomes. Moreover, the pKpQIL-IT plasmid probably harbored the blaTEM-1 gene accounting for β-lactams resistance. Additionally, the blaOXA-48 gene was presumably localized on the pOXA-48 plasmid, characterized by 100% identity and 74.3% identical sites with the Klebsiella pneumoniae isolates of study. Although the pKPN3 plasmid was present in both isolates, it did not carry any antimicrobial resistance genes. The pNDM-5-IT plasmid, first described in Escherichia coli strain Ec001, shared 90.4% and 90.6% of identical sites withBSKP713 and BSKP542, respectively. The pNDM-5-IT plasmid also harbored many genes responsible for resistance to tetracycline, aminoglycosides, carbapenems, suphonamides, and macrolides, including tet(A), aadA2, blaNDM-5, sul1, mphE, and mphA (Table 2a).
2.3. Analysis of Mutations and Alterations in Genes Involved in Iron Uptake and Transport Detected in Two Cefiderocol-Resistance Klebsiella pneumoniae Isolates
In order to identify possible mutations which could contribute to cefiderocol resistance, the genes involved in iron uptake and transport were compared to reference genomes. All of the analyzed genes are reported in the Table 3.
Table 3.
Analysis of possible mutations in genes involved in iron uptake and transport systems.
| Genes | Function | Mutations/Alterations | |
|---|---|---|---|
| BSKP542 | BSKP713 | ||
| fhuA | Iron uptake [18] | V176F, I178V, I212L, G269D, V609G | V176F, I178V, I212L, G269D, V609G |
| fepA | Iron uptake [18] | P531A | P531A |
| fbpA | Iron uptake [18] | WT | WT |
| efeO | Iron uptake [18] | WT | WT |
| exbB | Iron uptake [18] | WT | WT |
| exbD | TonB-dependent energy transduction system reported to affect the function of Iron transporters [18] |
WT | WT |
| fiuA | Iron uptake [18] | Absent | Absent |
| fur | Iron uptake [18] | WT | WT |
| iutA | Iron uptake [18] | E160K, S285P | E160K, S285P |
| baeS | Encodes a sensor kinase protein of the two-component BaeSR signal transduction system [18,19] | L366H | L366H |
| envZ | Two-component transcriptional regulator reported to affect the expression of iron transporters [18,19] |
R289C | R289C |
| cirA | Encodes receptor which preferentially transports catecholate siderophores [18,19] | A134V, N558D | A134V, N558D |
| feoA | Ferrous iron uptake [18] | WT | WT |
| sitC * | Iron/manganese ABC transporter permease subunit [18] | Y11H, R166C | Y11H, R166C |
| apbC | Iron-sulfur cluster carrier protein [18] | P334S, I339T | P334S, I339T |
| fepG | Iron-enterobactin ABC transporter permease [18] | V63M, G206S | V63M, G206S |
| fepC | Iron-enterobactin ABC transporter ATP-binding protein [18] | T9A, I182N | T9A, I182N |
| fetB | Iron export ABC transporter permease subunit FetB [18] | WT | WT |
| fetA | Iron ABC transporter ATP-binding protein FetA [18] | S27N | S27N |
| fecA | TonB energy transducing system-dependent ferric citrate uptake receptor [20] | A9V, V16L, premature STOP codon in position aa569 | A9V, V16L, premature STOP codon in position aa569 |
| tonB | Component of inner membrane protein complex providing energy to TonB dependent transporters [19] |
G60A, A80V, insertion of 4 aa in position 103 (PKPK), P168A, E219Q | G60A, A80V, insertion of 4 aa in position 103 (PKPK), P168A, E219Q |
| fiu | Encodes receptor that preferentially transports catecholate siderophores [18,19] | D387N | D387N |
| ompR | Two-component transcriptional regulator reported to affect the expression of iron transporters [19] |
A72V | A72V |
| yicI | Transporter family [18,19] | S30N, L323Q, K358N, G624C, H653R | S30N, L323Q, K358N, G624C, H653R |
| yicJ | Transporter family [18,19] | P102L, F181Y | P102L, F181Y |
| yicL | Transporter family [18,19] | WT | WT |
| chrA * | Heavy metals transporter [19,20] | A245V, G344A | A245V, G344A |
Reference sequence, Klebsiella pneumoniae ATCC 13883; * reference sequence, Klebsiella pneumoniae CP071027.
Mutations in these genes are associated with cefiderocol resistance in the literature [18,19,20]. Several missense mutations were observed in a number of genes involved in iron uptake, such as fhuA, fepA, and iutA. All of the mutations identified were the same in both isolates. Furthermore, the same missense mutations in both isolates were detected for some genes involved in iron transport, such as cirA, sitC, apbC, fepG, fepC, fetB, yicI, yicJ, and yicL. Two mutations, (L366H) and (R289C), were found in the baeS and envZ genes, which are responsible for the production of sensor kinases for two component systems able to regulate the expression of cirA and fiu iron uptake genes [19,21]. Mutations in these two genes have been previously associated with cefiderocol resistance [19].
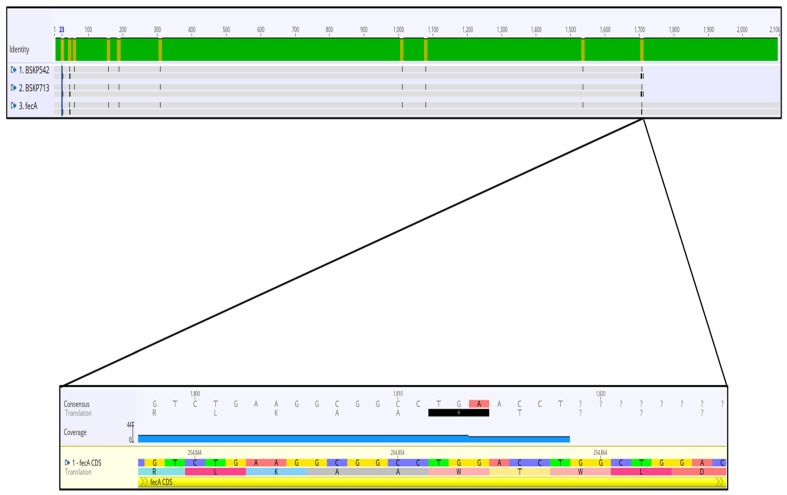
Interestingly, in addition to other missense mutations, we found a premature stop codon (G569A) in the fecA gene leading to a truncated protein (Figure 2) and an insertion of four amino acids (PKPK) after lysine in the tonB protein (Table 3). FecA transports ferric iron across the outer membrane, and tonB confers the ability to acquire iron through all TonB-dependent iron uptake systems.
Figure 2.
Presence of nonsense mutations in the fecA gene in BSKP713 and BSKP542 isolates. A nucleotide mutation from G to A in position 254,858 generated a TGA stop codon in the fecA gene in both of our isolates.
2.4. Phylogenetic Analysis
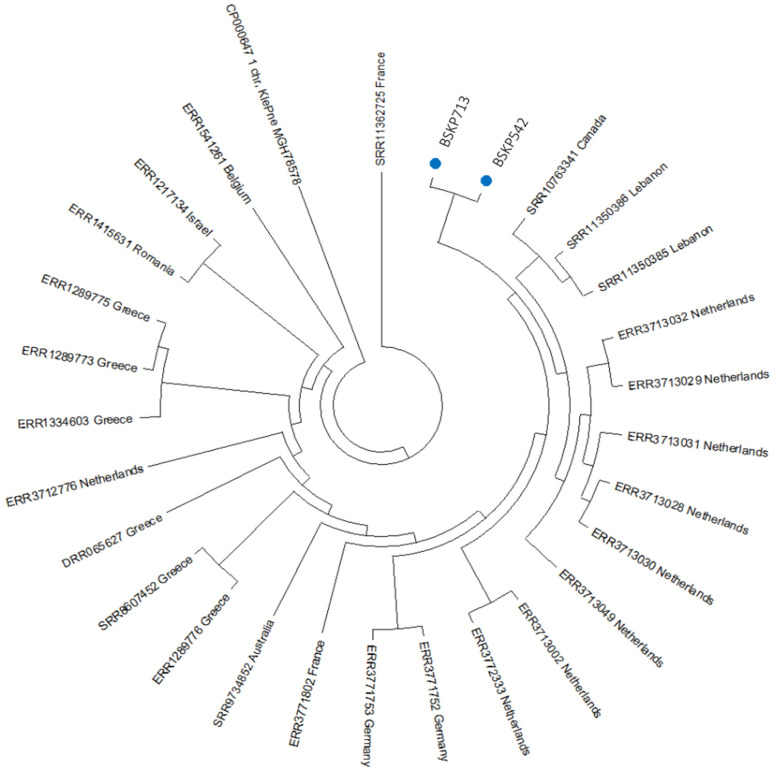
To deepen our understanding of the evolutionary relationships of BSKP542 and BSKP713 isolates among all the other Klebsiella pneumoniae isolates belonging to ST383 available in the Pathogen Watch database, we implemented a parsimony phylogenetic tree (Figure 3). Interestingly, our isolates clustered together, and they shared a common ancestor with a cluster formed by one isolate from Canada, two from Lebanon, and six from the Netherlands. Of note, 80.7% of isolates belonging to ST 383 originated in Europe, 14.2% were from Asia, 3.8% from North America, and 3.8% from Australia.
Figure 3.
Phylogenetic tree showing evolutionary relationships among BSKP542 and BSKP713 isolated in this study and all the other isolates that also belonged to ST383. Our isolates are shown with blue circles. Klebsiella pneumoniae ATCC 700721 was employed as an outgroup.
3. Discussion
This study shows that cefiderocol has an antimicrobial activity superior to that of comparators, including the new beta-lactam/beta-lactamase inhibitor combination drugs, such as ceftazidime-avibactam or ceftolozane-tazobactam, against isolates of ESBL and/or carbapenemase-producing Enterobacterales, carbapenem-non susceptible Pseudomonas aeruginosa, MDR Acinetobacter baumannii, and MDR Stenotrophomonas maltophilia. The cefiderocol susceptibility rates based on the EUCAST breakpoints for Klebsiella pneumoniae, Escherichia coli, Enterobacter spp, Pseudomonas aeruginosa, Acinetobacter baumannii, Stenotrophomonas maltophilia, and Achromobacter xylosoxidans were 88%, 89%, 100%, 100%, 100%, 100%, and 100%, respectively. A concentration of 2 μg/mL inhibited 94% of the isolates.
The activity of cefiderocol was lower among isolates belonging to the Enterobacterales group, and all of the resistant isolates were included. The overall observed resistance rate was 6.3%, but among the Enterobacterales, it reached 10.4%, a higher percentage compared that reported by other studies [22,23,24,25]. However, several studies have shown that the resistance rate of metallo-β-lactamases (MBL)-producing Enterobacterales is higher compared to non-MBL producers [26,27,28]. The resistance rate reported for NDM-producing pathogens ranged from 48.6% to 59%, with intervals of MIC90 values from 32 μg/mL to >64 μg/mL [26,27,29]. In addition to NDM-producers, VIM-producer strains also showed high resistance rates [26,27].
The carbapenemases produced by our resistant isolates were KPC (4 isolates) with MICs ranging from 4 μg/mL to >32 μg/mL, NDM (one isolate) with a MIC of 32 mg/mL, and NDM+OXA-48 (two isolates) with a MIC of 4 μg/mL. Resistance rates of 8.9% and 16.4%, based on the EUCAST breakpoint, were found in two studies from Europe in KPC-producing pathogens [26,27].
For Enterobacterales that produced β-lactamase OXA-48, a resistance rate of 7.1% and 11.8% was reported, while AmpC-positive pathogens were found to be susceptible to cefiderocol; this last finding was further confirmed by our study [26,27].
Our data show that cefiderocol had excellent activity against Pseudomonas aeruginosa with a MIC90 value of 2 μg/mL, consistent with the results of other studies [30,31,32,33,34]. We did not find a correlation between the presence of a particular carbapenemase and an increase in MICs against cefiderocol, as found in other studies [26,27].
Cefiderocol has a good activity also against Stenotrophomonas maltophilia, with MIC values ranging from 0.5 to <2 μg/mL. This range was higher than the one found in other studies, which ranged from 0.12 to 0.5 μg/mL [22,23,34]. However, due to the multiple resistance mechanisms present in this microorganism such as aminoglycoside-modifying enzymes, the L1 metallo-β-lactamase, the L2 serine active site β-lactamase [35], and the limited choice of effective drugs [36], cefiderocol seems to be a promising therapeutic option.
Even though only a few isolates were included in this study, Achromobacter xylosoxidans and Acinetobacter baumannii were susceptible to cefiderocol with MIC values of 0.06 μg/mL and <2 μg/mL, respectively.
Although the short-term usage of cefiderocol has been reported to cause strains with reduced susceptibility or resistance, primary resistance is infrequent. It is recommended to test susceptibility before the beginning of therapy, and to detect MIC changes, it is strongly recommended to test for strains isolated from patients treated with cefiderocol
Different resistant mechanisms have been proposed. For example, amino acid changes in the R2 loop of AmpC β-lactamases have been associated with a decreased susceptibility to cefiderocol [37,38]. Mutations in genes encoding TonB-dependent receptors, which are important for cefiderocol import, such as PirA, PiuA, and PiuD, have been detected in Acinetobacter baumannii and Pseudomonas aeruginosa [39,40]. The production of NDM and PER lactamases has been reported to contribute to an increase in cefiderocol MICs, but not to cefiderocol resistance [41]. Furthermore, mutations in the cirA gene, encoding the catecholate receptor, have been detected in resistant NDM-producing Enterobacter cloacae [42].
Because our strains showed cefiderocol resistance regardless of the beta lactamases harbored, we explored different iron transporter genes to identify mutations that could be associated with cefiderocol resistance. Our results revealed the presence of various mutations located in different genes involved in iron uptake and transport, such as fhuA, fepA, iutA, cirA, sitC, apbC, fepG, fepC, fetB, yicI, yicJ, and yicL. These mutations were the same in both sequenced strains. Furthermore, we identified, for the first time to the best of our knowledge, a nonsense mutation in the fecA gene that gives rise to a premature stop codon and a truncated protein. This protein is a ferric iron citrate receptor, which represents another pathway by which bacteria may acquire iron. Its function is to transport ferric iron across the outer membrane, and it has been reported that the fecA system contributes to the virulence of enterobactin-mutant Escherichia coli [43]. Moreover, we found a 4-aminoacid insertion in the tonB protein. Generally, the TonB-dependent receptors are located in the outer membrane of Gram-negative bacteria and are important for the entry of cefiderocol into the periplasmic space.
Future studies are needed to confirm and elucidate the role of cefiderocol resistance in all these newly described mutations, and in particular, those mutations found in the fecA and TonB genes. Both isolates belonged to ST-383, a sequence type predominantly found in Europe.
This study presents many limitations. First, it included isolates from a single center and from only one geographic region. Moreover, our collection was small, and some Gram-negative bacteria were represented by only a few isolates. Larger studies with a greater number of isolates from more centers and different geographical areas are necessary to generalize these results.
4. Materials and Methods
4.1. Isolates
A total of 110 clinical isolates were included in this study. Only one isolate per patient infection episode was included. The strains were isolated from different clinical specimens collected between 2021 and 2022 in the Laboratory of Microbiology, Spedali Civili, Brescia, Italy. The samples consisted of needle aspirate, n = 3; bronchoalveolar lavage, n = 13; broncho aspirate, n = 7; biopsy specimens, n = 5; blood cultures, n = 26; sputum samples, n = 5; peritoneal fluid, n = 1, intra-abdominal fluid, n = 4; bile fluid, n = 1; pus, n = 1; skin swabs, n = 6; tracheal aspirate, n = 19; wounds, n = 8; and urine, n = 11.
The clinical isolates included 67 Enterobacterales, 2 Acinetobacter baumannii, 1 Achromobacter xylosoxidans, 33 Pseudomonas aeruginosa and 7 Stenotrophomonas maltophilia. Of the 67 Enterobacterales isolated, 56 were Klebsiella pneumoniae, 8 were Escherichia coli, and 3 were Enterobacter species. The isolates were identified by MALDI-TOF (Matrix Assisted Laser Desorption/Ionization; BioMérieux, Florence, Italy).
4.2. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility tests were performed using an automatic microdilution broth (VITEK®2 system, BioMérieux). The Etest method (BioMérieux) was implemented for imipenem, meropenem, ceftazidime/avibactam, and ceftolozane/tazobactam susceptibility analysis, whereas the broth microdilution method was performed for colistin susceptibility testing, as recently recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [44] and the Clinical and Laboratory Standards Institute (CLSI) [45].
Cefiderocol susceptibility was evaluated using an iron-depleted cation-adjusted Mueller-Hinton broth and plate (ID-CAMHB), with the both microdilution method performed using SensititreTM Cefiderocol MIC panel CMP1SHIH (Thermo Fisher Scientific, Monza, Italy) according to the manufacturer’s instructions. Briefly, 0.4 mL of a 1:20 diluted 0.5 McFarland bacterial solution was added to the ID-CAMHB tube provided in the kit. Subsequently, 100 µL of the obtained solution was dispensed into each well of the panel (range of cefiderocol concentration from 0.03 to 32 μg/mL). The panels were incubated at 35 °C for 20 h in ambient air before reading the MIC endpoints. ID-CAMHB did not significantly affect the growth of any quality controls or test organisms. The MIC of cefiderocol was read depending on the presence of strong growth in the ID-CAMHB growth control (i.e., a button of approximately 2 mm or greater). The cefiderocol MIC was read as the first panel of the well in which the isolate growth was significantly reduced (i.e., a button of <1 mm or light/faint turbidity) relative to the growth observed in the ID-CAMHB growth control well. The results were interpreted according to the latest EUCAST breakpoint (www.eucast.org/clinical_breakpoints, accessed on 2 January 2023) guidelines. The EUCAST PK/PD (non-species-related) susceptibility breakpoint of cefiderocol was applied to Acinetobacter spp. and S. maltophilia (MIC ≤ 2 μg/mL) because their species-specific clinical breakpoints were not available in EUCAST [46].
4.3. Phenotypic Detection of Carbapenemases
The O.K.N.V.I. Resist-5 immunochromatographic assay (Coris BioConcept, Gembloux, Belgium) was performed to efficiently detect class A (KPC), class B (VIM, IMP and NDM), and class D (OXA-48) carbapenemases according to the manufacturer’s instructions [47,48].
4.4. Whole Genome Sequencing (WGS)
Two Klebsiella pneumoniae isolates (BSKP542 and BSKP713) underwent whole-genome sequencing (WGS) to identify their genomic characteristics. Bacterial DNA was extracted from the bacterial colonies using a QIAmp DNA Mini Kit ® (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The extracted bacterial DNA was quantified using the Qubit DNA HS Assay Kit (Thermo Fisher Scientific).
Whole genome libraries of the 2 bacterial isolates were prepared using the Nextera DNA Flex kit (Illumina, San Diego, CA, USA), and deep sequencing was performed on an Illumina MiSeq platform. Reads were checked for bacterial species confirmation using Kraken2 software [49]. Reads were trimmed with Trimmomatic version 0.39 [50] and assembled using SPAdes version 3.13.0 [51].
The online tool Pathogenwatch (https://pathogen.watch, accessed on 21 December 2021) was implemented to determine the in silico multi locus sequence typing and to analyze assemblies that passed quality control, with more than 95% of Klebsiella core genes detected. Sequence type definition was confirmed using the Bacterial Isolates Genome Sequence Database of the Pasteur Institute (https://bigsdb.pasteur.fr/klebsiella/, accessed on 19 April 2022). Antimicrobial resistance genes, virulence genes (including the derived virulence score), capsule, and lipopolysaccharide-type genes were identified using Kleborate via Pathogenwatch. To confirm the Pathogenwatch data, ResFinder 3.2 was used to detect antimicrobial resistance genes (https://cge.cbs.dtu.dk/services/ResFinder/, accessed on 24 May 2022). The presence of plasmids was detected using PlasmidFinder 2.0 (https://cge.cbs.dtu.dk/services/PlasmidFinder-2.0/, accessed on 9 November 2022). Virulent factors were determined by blasting the assembled the isolate genome against the Virulent Factor Database (VFDB) using the VFanalyser tool (http://www.mgc.ac.cn/VFs/, accessed on 7 January 2022). The presence of mobile genomic elements in plasmids was assessed using the Mobile Element Finder (https://cge.cbs.dtu.dk/services/MobileElementFinder/, accessed on 9 November 2022), then confirmed manually by mapping the raw reads using Bowtie2 implemented in Geneious software (v. 11.1.5) (Biomatters Ltd., Auckland, New Zealand). The kSNP3 program was implemented to build a parsimony phylogenetic tree [52] of BSKP542 and BSKP713 isolates, along with all the other isolates of ST-383 available in Pathogenwatch.
5. Conclusions
Our data show that cefiderocol is an effective drug against most of the analyzed multidrug-resistant Gram-negative bacteria, with a susceptibility rate of 94%. However, the presence of a higher resistance rate in carbapenem-resistant Enterobacterales highlights the importance of active surveillance, including genomic surveillance through rapid molecular testing. Such testing can quickly identify and differentiate between carbapenemases genes, allowing for appropriate therapy and avoiding the spread of these difficult-to-treat microorganisms. Maintaining the activity of novel drugs will consequently reduce the risks associated with emerging drug resistance.
Author Contributions
Conceptualization, M.A.D.F.; methodology, M.P., F.G., S.C. and G.P.; bioinformatic analysis, A.B.; formal analysis, all authors; data curation, A.B., M.P. and F.G.; writing—original draft preparation, M.P., A.B. and M.A.D.F.; writing—review and editing, all authors; supervision, M.A.D.F. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Ethical approval was not required for this study because the clinical samples were obtained within routine microbiological diagnostics. In this study, no patient specimens were analyzed, only bacterial isolates from routine diagnostic cultures. Samples were coded, anonymized, and preserved as part of routine diagnostics (standard care). The procedures performed in this study were in accordance with the ethical standards of the Institutional and National Research Committee, and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed Consent Statement
Patient consent was waived because clinical strains used in this study were retrospectively collected and no patient identification information was presented.
Data Availability Statement
Data are available at EBI under study accession n. PRJEB60070.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.WHO [(accessed on 17 November 2021)];2022 Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance.
- 2.Jean S.S., Lee W.S., Lam C., Hsu C.W., Chen R.J., Hsueh P.R. Carbapenemase-producing Gram-negative bacteria: Current epidemics, antimicrobial susceptibility and treatment options. Future Microbiol. 2015;10:407–425. doi: 10.2217/fmb.14.135. [DOI] [PubMed] [Google Scholar]
- 3.Jean S.S., Hsueh P.R., SMART Asia-Pacific Group Distribution of ESBLs, AmpC b-lactamses and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal and urinary tract infections in the Asia-Pacific region during 2008-14; results from the Study for Monitoring Antimicrobial Resistance Trends (SMART) J. Antimicrob. Chemother. 2017;72:166–171. doi: 10.1093/jac/dkw398. [DOI] [PubMed] [Google Scholar]
- 4.Lee C.H., Su T.Y., Ye J.J., Hsu P.C., Kuo A.J., Chia J.H., Lee M.H. Risk factors and clinical significance of bacteremia caused by Pseudomonas aeruginosa resistant only to carbapenems. J. Microbiol. Immunol. Infect. 2017;50:677–683. doi: 10.1016/j.jmii.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Tseng S.P., Wang S.F., Ma L., Wang T.Y., Yang T.Y., Siu L.K., Chuang Y.C., Lee P.S., Wang J.T., Wu T.L., et al. The plasmid-mediated fosfomycin resistance determinants and synergy of fosfomycin and meropenem in carbapenem-resistant Klebsiella pneumoniae isolates in Taiwan. J. Microbiol. Immunol. Infect. 2017;50:653–661. doi: 10.1016/j.jmii.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Ku Y.H., Chen C.C., Lee M.F., Chuang Y.C., Tang H.J., Yu W.L. Comparison of synergism between colistin, fosfomycin and tigecycline against extended-spectrum b-lactamase-producing Klebsiella pneumoniae isolates or with carbapenem resistance. J. Microbiol. Immunol. Infect. 2017;50:931–939. doi: 10.1016/j.jmii.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Lee C.R., Lee J.H., Park K.S., Kim Y.B., Jeong B.C., Lee S.H. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: Epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 2016;7:895. doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De la Calle C., Rodríguez O., Morata L., Marco F., Cardozo C., García-Vidal C., Río A.D., Feher C., Pellicé M., Puerta-Alcade P., et al. Clinical characteristics and prognosis of infections caused by OXA-48 carbapenemase producing Enterobacteriaceae in patients treated with ceftazidime-avibactam. Int. J. Antimicrob. Agents. 2019;53:520–524. doi: 10.1016/j.ijantimicag.2018.11.015. [DOI] [PubMed] [Google Scholar]
- 9.Syue L.S., Chen Y.H., Ko W.C., Hsueh P.R. New drugs for the treatment of complicated intra-abdominal infections in the era of increasing antimicrobial resistance. Int. J. Antimicrob. Agents. 2016;47:250–258. doi: 10.1016/j.ijantimicag.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 10.Cassini A., Diaz Högberg L., Plachouras D., Quattrocchi A., Hoxha A., Skov Simonsen G., Colomb-Cotinat M., Kretzschmar M.E., Devleeschauwer B., Cecchini M., et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019;19:56–66. doi: 10.1016/S1473-3099(18)30605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ISS 2021. [(accessed on 17 November 2022)]. Available online: https://www.epicentro.iss.it/antibiotico-resistenza/epidemiologia-italia.
- 12.El Labadidi R.M., Rizk J.G. Cefiderocol: A siderophore cephalosporin. Ann. Pharmacother. 2020;54:1215–1231. doi: 10.1177/1060028020929988. [DOI] [PubMed] [Google Scholar]
- 13.Zhanel G.G., Golden A.R., Zelenitsky S., Wiebe K., Lawrence C.K., Adam H.J., Idowu T., Domalaon R., Scweizer F., Zhanel M.A., et al. Cefiderocol: A siderophore cephalosporin with activity against carbapenem resistant and multi-drug resistant Gram-negative bacilli. Drugs. 2019;79:271–289. doi: 10.1007/s40265-019-1055-2. [DOI] [PubMed] [Google Scholar]
- 14.Ito A., Toba S., Sato T., Ota M., Takemura M., Nishikawa T., Toba S., Kohira N., Miyagawa S., Ishibashi N., et al. In vitro antibacterial properties of cefiderocol, a novel siderophore cephalosporin, against Gram-negative bacteria. Antimicrob. Agents Chemother. 2018;62:e01454-17. doi: 10.1128/AAC.01454-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McElheny C.L., Fowler E.L., Iovleva A., Shields R.K., Doy Y. In vitro evolution of cefiderocol resistance in an NDM-producing Klebsiella pneumoniae due to functional loss of CirA. Microbiol. Spectr. 2021;9:e0177921. doi: 10.1128/Spectrum.01779-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam M.M.C., Wick R.R., Watts S.C., Cerdeira L.T., Wyres K.L., Holt K.E. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 2021;132:4188. doi: 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian D., Wang B., Zhang H., Pan F., Wang C., Shi Y., Sun Y. Dissemination of the blaNDM-5 gene via IncX3-type plasmid among Enterobacteriaceae in children. mSphere. 2020;5:e00699-19. doi: 10.1128/mSphere.00699-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coppi M., Antonelli A., Niccolai C., Bartolini A., Bartolini L., Grazzini M., Mantengoli E., Farese A., Pieralli F., Mechi M.T., et al. Nosocomial outbreak by NDM-1-producing Klebsiella pneumoniae highly resistant to cefiderocol, Florence, Italy, August 2021 to June 2022. Euro. Surveill. 2022;27:2200795. doi: 10.2807/1560-7917.ES.2022.27.43.2200795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simner P.J., Beisken S., Bergman Y., Ante M., Posch A.E., Tamma P.D. Defining baseline mechanisms of cefiderocol resistance in the Enterobacterales. Microb. Drug Resist. 2022;28:161–170. doi: 10.1089/mdr.2021.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan P., Lu Y., Chen Z., Wu X., Hua X., Jiang Y., Zhou J., Yu Y. Emergence of high-level cefiderocol resistance in carbapenem-resistant Klebsiella pneumonia from bloodstream infections in patients with hematologic malignancies in China. Microbiol. Spectr. 2022;10:e0008422. doi: 10.1128/spectrum.00084-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karakonstantis S., Rousaki M., Kritsotakis E.I. Cefiderocol: Systematic review of mechanism of resistance, heteroresistance and in vivo emergence of resistance. Antibiotics. 2022;11:723. doi: 10.3390/antibiotics11060723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bassetti M., Echols R., Matsunaga Y., Ariyasu M., Doi Y., Ferrer R., Lodise T.P., Naas T., Niki Y., Paterson D.L., et al. Efficacy and safety of cefiderocol or best available therapy for the treatment od serious infections caused by carbapenem-resistant Gram-negative bacteria (CREDIBLE-CR): A randomized, open-label, multicentre, pathogen focused, descriptive phase 3 trial. Lancet Infect. Dis. 2020;21:226–240. doi: 10.1016/S1473-3099(20)30796-9. [DOI] [PubMed] [Google Scholar]
- 23.Karlowsky J.A., Hackel M.A., Tsuji M., Yamano Y., Echols R., Sahm D.F. In Vitro activity of cefiderocol, a siderophore cephalosporin, against Gram-negative bacilli isolated by clinical laboratories in North America and Europe in 2015–2016: SIDERO-WT-2015. Int. J. Antimicrob. Agents. 2019;53:456–466. doi: 10.1016/j.ijantimicag.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Yamano Y. In vitro activity against a broad range of clinically important Gram-negative bacteria. Clin. Infect. Dis. 2019;69:S544–S551. doi: 10.1093/cid/ciz827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albano M., Karau M.J., Schuetz A.N., Patel R. Comparison of agar dilution to broth microdilution for testing in vitro activity of cefiderocol against Gram-negative bacilli. J. Clin. Microbiol. 2020;59:e00966-20. doi: 10.1128/JCM.00966-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longshaw C., Manissero D., Tsuji M., Echols R., Yamano Y. In Vitro activity of the siderophore cephalosporin, cefiderocol, against molecularly charcterized, carbapenem-non-susceptible Gram-negative bacteria from Europe. JAC Antimicrob. Resist. 2020;2:dlaa060. doi: 10.1093/jacamr/dlaa060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mushtaq S., Sadouki Z., Vickers A., Livermore D.M., Woodford N. In Vitro activity of cefiderocol, a siderophore cephalosporin, against multidrug-resistant Gram-negative bacteria. Antimicrob. Agents Chemother. 2020;64:e01582-20. doi: 10.1128/AAC.01582-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohira N., West J., Ito-Horiyama T., Nakamura R., Sato T., Rittenhouse S., Tsuji M., Yamano Y. In Vitro antimicrobial activity of a siderophore cephalosporin, s-649266, against enterobacteriaceae clinical isolates, including carbapenem-resistant strains. Antimicrob. Agents Chemother. 2016;60:729–734. doi: 10.1128/AAC.01695-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kazmierczak K.M., Tsuji M., Wise M.G., Hackel M., Yamano Y., Echols R., Sahm D.F. In Vitro activity of cefiderocol, a siderophore cephalosporin, against a recent collection of clinically relevant carbapenem-non-susceptibel Gram-negative bacilli, including serine carbapenemases-and metallo-beta-lactamase-producing isolates (Sidero-WT-2014 Study) Int. J. Antimicrob. Agents. 2019;53:177–184. doi: 10.1016/j.ijantimicag.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Iregui A., Khan Z., Landman D., Quale J. Activity of cefiderocol against enterobacterales, Pseudomonas aeruginosa, and Acinetobacter baumannii endemic to medical centers in New York City. Microb. Drug Resist. 2020;26:722–726. doi: 10.1089/mdr.2019.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Golden A.R., Adam H.J., Baxter M., Walkty A., Lagacé-Wiens P., Karlowsky J.A., Zhanel G.G. In Vitro activity of cefiderocol, a novel siderophore cephalosporin, against gram-negative bacilli isolated from patients in Canadian Intensive Care Units. Diagn. Microbiol. Infect Dis. 2020;97:115012. doi: 10.1016/j.diagmicrobio.2020.115012. [DOI] [PubMed] [Google Scholar]
- 32.Delgado-Valverde M., Conejo M.D.C., Serrano L., Fernández-Cuenca F., Pascual Á. Activity of cefiderocol against high-risk clones of multidrug-resistant Enterobacterales, Acinetobacter baumannii, Pseudomonas aeruginosa and Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2020;75:1840–1849. doi: 10.1093/jac/dkaa117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsueh S.C., Lee Y.J., Huang Y.T., Liao C.H., Tsuji M., Hsueh P.R. In Vitro activities of cefiderocol, ceftolozane/tazobactam, ceftazidime/avibactam and other comparative drugs against imipenem-resistant Pseudomonas aeruginosa and Acinetobacter baumannii, and Stenotrophomonas maltophilia, all associated with bloodstream infections in Taiwan. J. Antimicrob. Chemother. 2019;74:380–386. doi: 10.1093/jac/dky425. [DOI] [PubMed] [Google Scholar]
- 34.Hackel M.A., Tsuji M., Yamano Y., Echols R., Karlowsky J.A., Sahm D.F. In Vitro activity of the siderophore cephalosporin, cefiderocol, against carbapenem non susceptible and multidrug-resistant isolates of gram-negative bacilli collected worldwide in 2014 to 2016. Antimicrob. Agents Chemother. 2018;62:17. doi: 10.1128/AAC.01968-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang Y.T., Lin C.Y., Chen Y.H., Hsueh P.R. Update on infections caused by Stenotrophomans maltophilia with particular attention to resistance mechanisms and therapeutic options. Front Microbiol. 2015;6:893. doi: 10.3389/fmicb.2015.00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho S.Y., Kang C.I., Ha Y.E., Chung D.R., Lee N.Y., Peck K.R., Song J.H. Can levofloxacin be a useful alternatie to trimethoprim-sulfamethoxazole for treating Stenotrophomonas maltophilia bacteremia? Antimicrob. Agents Chemother. 2014;58:581–583. doi: 10.1128/AAC.01682-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawai A., McElheny C.L., Iovleva A., Kline E.G., Slui-Cremer N., Shields R.K., Doi Y. Structural basis of reduced susceptibility to ceftazidime-avibactam and cefiderocol in Enterobacter cloacae due to AmpC R2 loop deletion. Antimicrob. Agents Chemother. 2020;64:e00198-20. doi: 10.1128/AAC.00198-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shields R.K., Iovleva A., Kline E.G., Kawai A., McElheny C.L., Doi Y. Clinical evolution of AmpC-mediated ceftazidime-avibactam and cefiderocol resistance in Enterobacter cloacae complex following exposure to cefepime. Clin. Infect Dis. 2020;71:2713–2716. doi: 10.1093/cid/ciaa355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Streling A.P., Obaidi M.M., Lainhart W.D., Zangeneh T., Khan A., Dinh A.Q., Hanson B., Arias C.A., Millerm W.R. Evolution of cefiderocol non-susceptibility in Pseudomonas aeruginosa in a patient without previous exposure to the antibiotic. Clin. Infect. Dis. 2021;73:e4472–e4474. doi: 10.1093/cid/ciaa1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luscher A., Moynié L., Saint Auguste P., Bumann D., Mazza L., Pletzer D., Naismith J.H., Köhler T. TonB-dependent receptor repertoire of Pseudomonas aeruginosa for uptake of siderophore-drug conjugate. Antimicrob. Agents Chemother. 2018;62:e00097-18. doi: 10.1128/AAC.00097-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohira N., Hackel M.A., Ishioka Y., Kuroiwa M., Sahm D.F., Sato T., Maki H., Yamano Y. Reduced susceptibility mechanism to cefiderocol, a siderophore cephalosporin, among clinical isolates from a global surveillance programme (SIDERO-WT-2014) J. Glob. Antimicrob. Resist. 2020;22:738–741. doi: 10.1016/j.jgar.2020.07.009. [DOI] [PubMed] [Google Scholar]
- 42.Klein S., Boutin S., Kocer K., Fiedler M.O., Störzinger D., Weigand M.A., Tan B., Richter D., Rupp C., Mieth M., et al. Rapid development of cefiderocol resistance in carbapenem- resistant Enterobacter cloacae during therapy is associated with heterogeneous mutations in the catecholate siderophore receptor cirA. Clin. Infect. Dis. 2022;74:905–908. doi: 10.1093/cid/ciab511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang W.C., Wong M.Y., Wang S.H., Hashimoto M., Lin M.H., Lee M.F., Wu J.J., Wang M.C., Lin W.H., Jeng S.L., et al. The Ferric Citrate Uptake System Encoded in a Novel blaCTX–M–3- and blaTEM–1-Harboring Conjugative Plasmid Contributes to the Virulence of Escherichia coli. Front Microbiol. 2021;12:667782. doi: 10.3389/fmicb.2021.667782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.European Committee on Antimicrobial Susceptibility Testing (EUCAST) Recommendations for MIC Detemination of Colistin (Polymyxin E) as Recommended by the Joint CLSI-EUCAST Polymyxin Breakpoints Working Group. EUCAST 2016; org 1-1. [(accessed on 22 March 2016)]. Available online: www.eucast.org.
- 45.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2018. Twenty-Seven Informational Supplement M100-S28. [Google Scholar]
- 46.The European Committee on Antimicrobial Susceptibility Testing Breakpoints for Cefiderocol from EUCAST. Addendum (May 20202) to EUCAST Breakpoint Tables V.10.0. Breakpoints to Be Included in EUCAST Breakpoint Tables v 11.0. Jan, 2021. [(accessed on 26 September 2022)]. Available online: https://www.eucast.org/eucast_news/news_singleview?tx_ttnews%5Btt_news%5D=374&cHash=839808489fb6fb1033af6d8a1c5e7d07.
- 47.Brolund A., Lagerqvist N., Byfors S., Stuelens M.J., Monnet D.L., Albiger B., Kohlenberg A. Worsening epidemiological situation of carbapenemase-producing Enterobacteriaceae in Europe, assessment by national experts from 37 countries, July 2018. EuroSurveill. 2019;9:1900123. doi: 10.2807/1560-7917.ES.2019.24.9.1900123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong J., Kang D., Kim D. Performance evaluation of the newly developed in vitro rapid diagnostic test for detecting OXA-48-like, KPC-, NDM-, VIM- and IMP- type carbapenemases: The RESIST-5 O.K.N.V.I. multiplex lateral flow assay. Antibiotics. 2021;10:460. doi: 10.3390/antibiotics10040460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prjibelski A., Antipov D., Meleshko D., Lapidus A., Korobeynikov A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020;70:e102. doi: 10.1002/cpbi.102. [DOI] [PubMed] [Google Scholar]
- 52.Gardner S.N., Slezak T., Hall B.G. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment of reference genome. Bioinformatics. 2015;31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available at EBI under study accession n. PRJEB60070.