Abstract
DAP-like kinase (Dlk, also termed ZIP kinase) is a leucine zipper-containing serine/threonine-specific protein kinase with as yet unknown biological function(s). Interaction partners so far identified are either transcription factors or proteins that can support or counteract apoptosis. Thus, Dlk might be involved in regulating transcription or, more generally, survival or apoptosis. Here we report on a new interaction partner, the rat homolog of Schizosaccharomyces pombe CDC5 protein, a presumptive transcription and splicing factor involved in the G2/M transition. In vitro, rat CDC5 forms complexes with, but is not phosphorylated by, Dlk. Rather, it was phosphorylated by an associated kinase which was identified as CK2. The interaction domain of Dlk was mapped to the leucine zipper, while that of CDC5 was mapped to the C-terminal region between residues 500 and 802. In vivo, both proteins co-localize perfectly in distinct speckle-like structures in the nucleus, some of which overlap with promyelocytic leukemia protein. Interestingly, splicing factor SC35, which also resides in speckles, was partially displaced upon overexpression of either CDC5 or Dlk, perhaps due to phosphorylation by Dlk. Together with previous data, these results suggest that Dlk might play a role in coordinating specific transcription and splicing events.
INTRODUCTION
Protein phosphorylation is perhaps the widest used regulatory tool of eukaryotic organisms and almost every process from DNA replication to metabolic reactions is regulated by reversible phosphorylation. To deal with the specific demands of such diverse processes a huge number of kinases exists. However, as most of the novel enzymes were discovered by virtue of their sequence homology or as interaction partners in two-hybrid screens, their biological role is not readily evident.
We and others have recently identified a new kinase, Dlk, for DAP-like kinase (1), or ZIP kinase, for zipper-interacting protein kinase (2). This kinase belongs to the novel subfamily of DAP kinases (death-associated protein kinases) (3,4) which exhibit a high degree of sequence identity in their catalytic domains but differ greatly in their non-catalytic parts. Due to this relationship we will use the term ‘Dlk’ throughout this paper. All the DAP-related kinases have been implicated in apoptosis, but a direct role in this process was only demonstrated for DAP kinase itself, which is involved in interferon γ- and TNFα/Fas-induced cell death (5,6).
Dlk, which normally resides in the nucleus in association with nuclear speckles, does not induce apoptosis to a significant extent (1). However, C-terminal truncation or co-expression with pro-apoptotic protein Par-4 results in relocation of Dlk to the actin cytoskeleton and efficient induction of apoptosis (7,8). On the other hand, the function of nuclear Dlk remains obscure. A unique property of Dlk not shared by the other DAP-related kinases is its leucine zipper at the C-terminus, which enables it to interact with the transcription factors ATF-4 (2,8) and AATF (apoptosis-antagonizing transcription factor) (9). In vitro, Dlk phosphorylates the myosin light chain (MLC) and core histones H3, H4 and H2A (1). These latter findings point to a role in transcription, perhaps chromatin remodeling and/or splicing. To search for interaction partners that might serve as regulators or targets of Dlk and that might illuminate its function we employed the two-hybrid system (8). One of the putative interaction partners was the rat homolog of the Schizosaccharomyces pombe CDC5 protein, which is described in more detail in this paper.
To avoid confusion, we want to point out here that the CDC5 genes of S.pombe and Saccharomyces cerevisiae encode proteins with completely different functions. The S.cerevisiae CDC5 gene encodes a polo-like kinase (plo1 in S.pombe) (10,11) which plays a crucial role in regulating diverse mitotic processes, such as centrosome maturation, spindle formation and the metaphase/anaphase transition (reviewed in 12,13). On the other hand, the CDC5 gene of S.pombe encodes a putative transcription and splicing factor that is involved in regulating the G2/M transition (14,15). The S.cerevisiae homolog of the latter is Cef1 (16). The S.pombe CDC5 homolog is structurally and functionally highly conserved throughout evolution and the homologs from Arabidopsis thaliana (17), Drosophila melanogaster and even human are capable of complementing the gene defect in yeast (18).
The CDC5 protein is a nuclear phosphoprotein which, in mammalian species, consists of 802 amino acids. It contains, as characteristic elements, two so-called myb repeats and one myb-like repeat (18,19). In the transcription factor Myb these elements form helix–turn–helix structures and represent the DNA-binding domain. This relationship to Myb proteins suggested that CDC5 might also be a transcription factor. In support of this assumption, CDC5 can bind to DNA and, when fused to the Gal4 DNA-binding domain (BD), it can act as a transactivator of Gal4-responsive reporter genes (20). More convincingly, a specific target sequence of CDC5 (GATTTAACATAA) was elucidated and this sequence element renders a reporter gene CDC5-responsive (21). However, CDC5 was also shown to be involved and, in fact, to be essential for pre-mRNA splicing in both yeast and mammalian splicing systems (16,22–24). Thus, CDC5 might be involved in both transcription and splicing and might actually provide a link between the two processes.
In this paper we describe the identification of rat CDC5 as an interaction partner of Dlk. Both proteins co-localize perfectly in speckle-like structures which partially overlap with promyelocytic leukemia (PML) protein. Interestingly, overexpression of CDC5 or Dlk resulted in partial displacement of splicing factor SC35 from nuclear speckles, perhaps resulting from competition for the same binding partners or from phosphorylation of SC35 by Dlk.
MATERIALS AND METHODS
Cell culture and transfection
Rat embryo fibroblasts, line REF52.2, were used for expression studies. Cells were grown as monolayers in Dulbecco’s minimal essential medium (Gibco BRL, Karlsruhe, Germany) supplemented with 10% fetal bovine serum (Biochrome Seromed, Berlin, Germany) and antibiotics. Transfections were performed with Lipofectamine (Gibco BRL) according to the manufacturer’s protocol. Insect SF9 (Spodoptera frugiperda) cells were used for the baculovirus expression system and were grown in serum-free SF900 medium containing 50 µg/ml gentamycin (Gibco BRL) at 27°C.
Two-hybrid assays
The two-hybrid screen was performed with the conventional two-hybrid system (25) with yeast strain Y190 and the Gal4 BD or transactivation domain (AD) proteins as fusion partners. Thus, a Gal4 BD–Dlk fusion protein was employed as bait and a cDNA library from SV40-transformed rat cells SV52 fused to the Gal4 AD was used as prey (8,9). To determine the interaction domains, deletion mutants of Dlk or CDC5 were expressed as Gal4 BD or Gal4 AD fusion proteins, respectively, upon transformation of yeast Y190 or AH109 cells. A functional interaction between the hybrid proteins was indicated by activation of the Gal4-responsive reporter gene His3 or Ade2, respectively, and β-galactosidase. A detailed description has been given previously (8).
Isolation of the complete CDC5 cDNA and construction of expression plasmids
For generation of the complete coding sequence of rat CDC5 the Long Expand PCR System (Roche Molecular Biochemicals, Mannheim, Germany) was employed using a cDNA library from 208F rat fibroblasts as template. The forward primer (5′-CCCGGGTACCAAGATGCCCCGGATTATGA-3′) and reverse primer (5′-CCCGGGATCCTCTGTGCTTCAGAACTTTG-3′) were derived from the 5′- and 3′-termini of the published sequence of rat CDC5 (italic, GenBank accession no. AF000578.2), extended by KpnI and BamHI restriction sites, respectively. The PCR product was completely sequenced and cloned into the respective expression vectors: pEGFP, pDsRED, pGAD424 (Clontech, Heidelberg, Germany), pCMV-Tag2 (Stratagene Europe) or pVL1392HIS (1). CDC5 deletion mutants were generated as described previously for Dlk (7). Where necessary, restriction site linkers for cloning were provided by PCR. Molecular biological techniques followed standard protocols (26,27).
In vitro translation
For in vitro translation the coding sequence of CDC5 was excised by partial restriction with EcoRI/BamHI and the fragment of 2436 bp was cloned into the EcoRI and BamHI sites of pBluescript SK+. In vitro translation was performed in the TNT-T7/T3 Coupled Reticulocyte Transcription/Translation System (Promega, Heidelberg, Germany), as recommended by the manufacturer.
Generation of recombinant baculoviruses and purification of His-tagged proteins by affinity chromatography
The generation and propagation of recombinant baculovirus has been described (1). For expression and purification of His-tagged Dlk, CDC5 or AATF-protein, SF9 cells were infected with the respective recombinant viruses for 3 days and sequentially extracted as described (1), with the following modifications: cells were lysed with isotonic lysis buffer [10 mM NaPO4, 140 mM NaCl, 3 mM MgCl2, 5 mM β-mercaptoethanol, protease inhibitors (Roche) and 0.5% Nonidet P-40, pH 9.0] and the chromatin digested with 0.1 mg/ml DNase I at 30°C for 15 min. The nuclei were pelleted by low speed centrifugation and the supernatant containing soluble proteins was discarded. The nuclear pellet was resuspended in buffer J (10 mM Tris pH 7.5, 140 mM NaCl, 1% Nonidet P-40, 1% Na deoxycholate, 0.1% SDS, 5 mM β-mercaptoethanol, 0.01% w/v aprotinin), diluted 3-fold with PBS, pH 9, and incubated with Ni–NTA–agarose (Qiagen, Hilden, Germany) without further centrifugation. The beads were extensively washed with PBS and IMAC-50 (20 mM Tris–HCl pH 8, 0.5 M NaCl, 10% glycerol, 1 mM PMSF, 5 mM β-mercaptoethanol and 50 mM imidazole) and either used as such for affinity chromatography or the bound proteins were eluted with 500 mM imidazole, 500 mM NaCl, 1 mM DDT, 10% glycerol and aprotinin and stored at –70°C. For binding studies, protein bound to Ni–NTA–agarose was incubated with in vitro translated and [35S]methionine-labeled CDC5 at 4°C for 2 h. The beads were further washed and bound proteins were eluted, analyzed by SDS–PAGE and visualized by fluorography.
Kinase assays
Kinase assays were performed with purified His-tagged Dlk and purified substrates essentially as described (1). Reactions were stopped with 2× SDS–PAGE sample buffer containing 10 mM EDTA and subjected to SDS–PAGE and autoradiography. Purified His-tagged ASF/SF2 or GST–SC35 were kindly provided by Dr S. Stamm (Institute of Biochemistry, Erlangen, Germany) or by Dr R. Lührmann (Max Planck Institute of Biophysical Chemistry, Göttingen, Germany), respectively.
Fluorescence microscopy analyses
Antibodies. Anti-PML monoclonal antibody (mAb) 5E10 (28) and mAb104, directed against splicing factors, were kindly provided by Dr S. Stamm; polyclonal anti-U5-116 kD antiserum, directed against a member of the snRNP family (29), was a gift from Dr R. Lührmann; anti-Sm antibodies were a kind gift from Dr B. Wirth (Institute of Human Genetics, Bonn, Germany); anti-SC35 Mab was purchased from BD Biosciences (Heidelberg, Germany), anti-SF2/ASF and anti-PCNA were from Santa Cruz and anti-FLAG M-2 antibody from Stratagene. Secondary Cy3-conjugated antibodies were from Dianova (Hamburg, Germany).
Immunofluorescence. Cells were fixed, permeabilized and stained with primary and secondary antibodies as described (7,8). Briefly, cells were washed in PBS, fixed in 3% paraformaldehyde in PBS for 15 min, permeabilized in 0.2% Triton X-100 in PBS and then incubated in blocking solution (1% BSA in PBS) for 30 min. Incubations with the primary and secondary antibodies were performed for 60 or 30 min, respectively, at appropriate dilutions in PBS, at room temperature. Between each step cells were washed two or three times in PBS. After final washing the cells were briefly rinsed in water, after which the coverslips were mounted in Permafluor mounting medium (Immunotech). The immunostained cells were examined with an Axiophot fluorescence microscope (Zeiss) with a 63× oil immersion objective.
For visualization of GFP or dsRED fusion proteins cells were fixed as described above, without permeabilization. For staining of nuclei with 4,6-diamidino-2-phenylindole (DAPI) cells were fixed and permeabilized as above and then stained with DAPI at a final concentration of 1 µg/ml for 15 min at room temperature. Final washing steps and mounting was performed as described above.
RESULTS
Using the conventional yeast two-hybrid system with Gal4–Dlk as bait, we isolated several cDNAs that coded for putative interaction partners of Dlk (8,9). One of these, a clone of 795 bp, was identical to the 3′-portion of the rat homolog of the S.pombe CDC5 gene, with an open reading frame of 264 codons. This clone was apparently incomplete since the human and mouse ‘CDC5-like’ cDNAs were reported to be 3.4 kb, coding for proteins of 802 amino acids (18,19). In contrast, the rat CDC5-like cDNA was initially reported to be 1907 bp, coding for a polypeptide of 523 residues (30), while an updated version comprised 2847 bp with an open reading frame of 802 amino acids (GenBank accession no. AF000578.2). Since the human CDC5-like protein seems to play similar roles in the G2→M transition and can complement mutant CDC5 in S.pombe, it was regarded as a true homolog and designated hCDC5 (20). Accordingly, we will refer to the rat homolog as rCDC5 or, for simplicity, CDC5. (Please note the difference between S.pombe and S.cerevisiae CDC5; see Introduction.)
To obtain the complete coding region of rCDC5 we performed PCR using rCDC5-specific oligonucleotides as primers and a cDNA library from 208F rat fibroblasts as template. The forward primer was derived from the 5′-portion of the published sequence just upstream of the start codon and the reverse primer from the 3′-portion just downstream of the stop codon (see Materials and Methods), omitting the untranslated regions. This reaction revealed a product of 2440 bp, as predicted. Sequence analysis revealed 100% identity with the rCDC5 cDNA in the database with a continuous open reading frame of 802 codons. The sequence showed 98% identity with human CDC5 (18,19) with only 15 amino acid exchanges, six of which are conservative, while the others are not expected to cause significant alterations in structure or function.
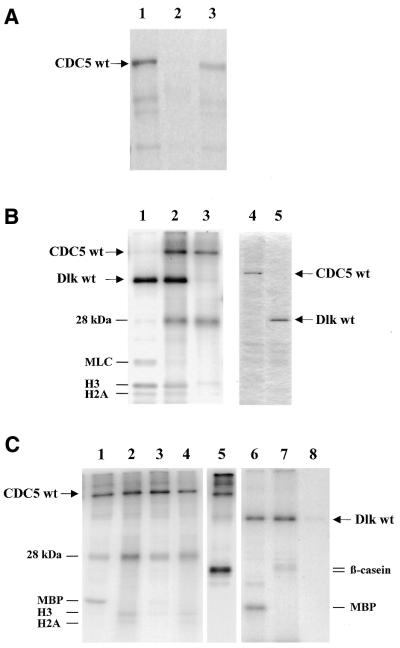
To confirm the interaction between CDC5 and Dlk, as revealed by the two-hybrid system, CDC5 was translated in vitro in a coupled transcription/translation system and allowed to bind to affinity purified His-tagged Dlk. In vitro translation revealed a product of 105 kDa, as predicted (Fig. 1A, lane 1). This protein bound specifically to His–Dlk (lane 3) but not to control beads saturated with proteins from control extracts (lane 2).
Figure 1.
CDC5 binds to Dlk but is not a substrate of Dlk in vitro. (A) CDC5 was synthesized and labeled with [35S]methionine in vitro and employed in a binding reaction using baculovirus-expressed His–Dlk or proteins from uninfected cells adsorbed to Ni–NTA–agarose as matrix. Bound proteins were eluted and analyzed by SDS–PAGE on a 10% polyacrylamide gel and fluorography, as described in the Materials and Methods. Lane 1, input CDC5; lanes 2 and 3, CDC5 bound to control beads or His–Dlk, respectively. The input represented 1/10 of the translation product used for binding. (B) In vitro phosphorylation of CDC5. His–CDC5 and His–Dlk were isolated from recombinant baculovirus-infected SF9 cells by affinity purification on Ni–NTA–agarose. Purified proteins were separated on a 10% SDS–polyacrylamide gel and stained with Coomassie blue (lanes 4 and 5) or subjected to in vitro phosphorylation individually (lanes 1 and 3) or in combination (lane 2). A mixture of MLC and histones was included as exogenous substrates. Phosphorylated products were analyzed by SDS–PAGE (13.5% polyacrylamide) and autoradiography, as outlined under Materials and Methods, with exposure for 4 h. (C) CDC5 is phosphorylated by associated CK2. Purified CDC5 or Dlk were subjected to kinase reactions in the presence of ATP (lanes 1, 2 and 5–7) or GTP (lanes 3, 4 and 8). MBP (lanes 1, 3, 6 and 8), a MLC/histone mix (lanes 2 and 4) or dephosphorylated β-casein (lanes 5 and 7) were included as substrates. SDS–PAGE analyses were on a 13.5% (lanes 1–5) or 10% polyacrylamide gel (lanes 6–8). Autoradiography was for 12 h. The 28 kDa band most likely represents the β-subunit of CK2 (33).
CDC5 is not a substrate of Dlk in vitro
CDC5 is a phosphoprotein and was shown to be a substrate of CDKs (31,32). Additionally, CDC5 contains consensus phosphorylation sites for CK2, PKA, PKC and MAP kinase (19). To see whether CDC5 is also phosphorylated by its interaction partner Dlk, it was expressed as a His-tagged protein in the baculovirus system, purified by affinity chromatography on Ni–NTA–agarose (Fig. 1B, lane 4) and subjected to in vitro phosphorylation using purified His–Dlk (Fig. 1B, lane 5) as kinase. As shown in Figure 1B (lane 3), CDC5 was efficiently phosphorylated in the absence of Dlk as well as in its presence (lane 2). These results suggested that CDC5 was phosphorylated by a co-purified kinase but not, or only weakly, by Dlk. In addition to CDC5 and Dlk there was one major phosphorylated protein band at 28 kDa. This was not seen when purified Dlk was employed in an autophosphorylation reaction (lane 1). On the other hand, exogenously added substrates (MLC and histones) were only phosphorylated by Dlk (Fig. 1B, lanes 1 and 2) and not by the CDC5-associated kinase (lane 3).
To obtain clues about the nature of the co-purified kinase we employed some commonly used substrates, myelin basic protein (MBP) or a mixture of MLC and histones, in the kinase reactions. As shown in Figure 1C (lanes 1 and 2), MBP was efficiently phosphorylated by the CDC5-associated kinase while MLC was not and the histones only weakly, thus clearly distinguishing it from Dlk, which phosphorylates MBP (Fig. 1C, lane 6), but also MLC and core histones H3, H4 and H2B (Fig. 1B, lane 1). Since protein kinase CK2 is often found as a contaminating kinase, we employed GTP as the phosphate donor, which is diagnostic for CK2. Additionally, we employed β-casein as substrate. The phosphorylation patterns of purified CDC5 with its associated proteins was identical with GTP (Fig. 1C, lanes 3 and 4), although phosphorylation of MBP was somewhat weaker. Additionally, β-casein was efficiently phosphorylated (lane 5). In contrast, Dlk was not able to use GTP (lane 8) and β-casein was only weakly phosphorylated (Fig. 1C, lane 7).
We next sought to distinguish CDC5 phosphorylation by CK2 from that by Dlk by employing a CK2-specific inhibitor, heparin. Heparin (10 nM) efficiently blocked phosphorylation of CDC5 both with ATP or GTP as phosphate donor and β-casein as substrate. However, Dlk, which was not inhibited under these conditions, was still incapable of phosphorylating CDC5 (data not shown). Likewise, when we tried to dissociate most of the CK2 activity from CDC5, the latter was still not phosphorylated by Dlk (not shown). These data show that recombinant CDC5 protein purified from SF9 cells is associated with and efficiently phosphorylated by CK2. The phosphorylated 28 kDa band presumably represents the regulatory β subunit of insect CK2, which is phosphorylated by the catalytic α subunit (33). On the other hand, CDC5 does not seem to be a substrate of Dlk, at least not in vitro.
Mapping of the interaction domains
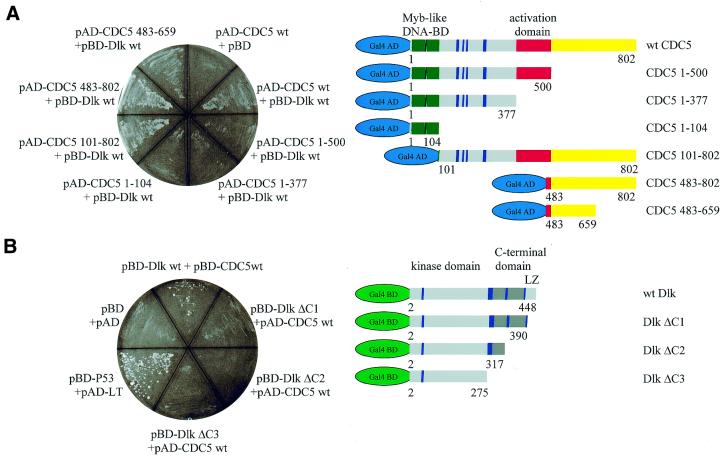
Having confirmed the interaction between Dlk and CDC5 in vitro, we wanted to determine the interaction domains between the two proteins. For this purpose we employed a series of deletion mutants of both CDC5 and Dlk in the yeast two-hybrid interaction assay. CDC5 deletion mutants were constructed by PCR, providing suitable linkers for cloning, and fused to the Gal4 AD, as shown in Figure 2A (right). The Dlk mutants fused to the Gal4 BD have been described previously (8) (see Fig. 2B, right). These constructs were co-expressed in various combinations in yeast cells and the transformed cells were scored for adenine autotrophy and β-galactosidase activity. Appropriate controls were included. Mapping the interaction domain in Dlk (shown in Fig. 2B, left) revealed that full-length (wt) CDC5 interacted with full-length Dlk (wt) but not with DlkΔC1, lacking the leucine zipper, or any of the shorter deletion mutants (ΔC2 and ΔC3), indicating that the leucine zipper of Dlk was required. On the other hand, Dlk (wt) interacted with full-length CDC5 and with deletion mutant CDC5(101–802), lacking the N-terminal 100 residues, but not with any of the mutants lacking 302 or more residues from the C-terminus [mutants CDC5(1–500), CDC5(1–377) and CDC5(1–104); Fig. 2A, left]. Considering that the original two-hybrid clone comprised the coding region from codon 427 to 690, these results placed the interaction domain of CDC5 with Dlk between residues 501 and 690. To confirm this conclusion we generated additional deletion constructs comprising residues 483–659 and 483–802. Both constructs exhibited interaction with Dlk, although interaction of the larger one was somewhat stronger (Fig. 2A, left), suggesting that residues between 500 and 659 and between 660 and 802 contribute to the interaction.
Figure 2.
Mapping of the interaction domains of Dlk and CDC5 by two-hybrid assay. Full-length and deletion constructs of CDC5 (A) and Dlk (B) fused to the Gal4 AD or BD domains, respectively, (see schemes on the right) were transfected in various combinations into yeast AH109, as indicated. Shown are yeast colonies grown on adenine-deficient agar plates as a result of a functional interaction of the Dlk and CDC5 proteins. (A) Full-length Gal4 BD–Dlk was co-transfected with Gal4 AD–CDC5 variants. Co-transfection of Gal4 BD with Gal4 AD–CDC5 wt served as a negative control. (B) Interaction of full-length Gal4 AD–CDC5 with Gal4 BD–Dlk variants. pBD + pAD served as a negative and pBDp53 + pADLT as a positive control. In the schematics of CDC5, the myb repeats are shown in green, the activation domain (20) in red, the putative NLSs in blue and the C-terminal part containing the interaction domain in yellow. The schematics of the Dlk variants indicate the locations of the catalytic domain and the leucine zipper (in light gray), the non-catalytic domain in dark gray and the putative NLSs in blue; the C-terminal most NLS is the functional one (7).
Subcellular localization of CDC5 and interaction with Dlk in vivo
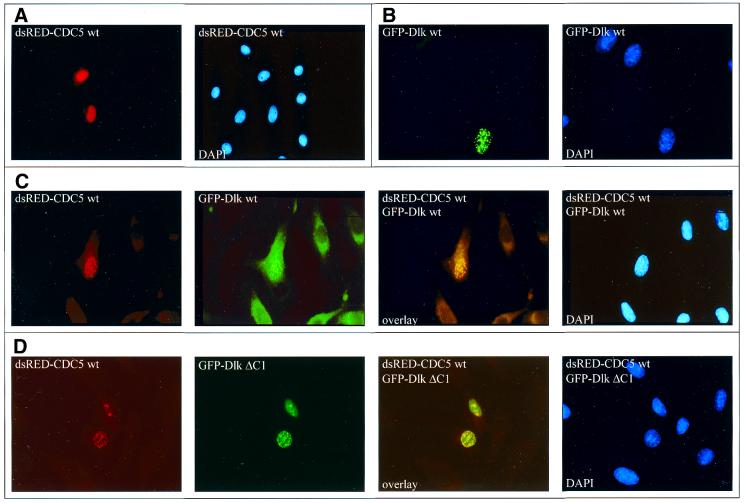
We next investigated a possible interaction of the two proteins in vivo by co-expression and fluorescence co-localization. Dlk was expressed as a GFP-tagged fusion protein and CDC5 as a dsRED- or FLAG-tagged fusion protein. For fluorescence studies, both constructs were transfected at the minimal amounts required for detection to minimize artefacts of overexpression. CDC5 alone usually exhibited a punctuate nuclear staining pattern (Fig. 3A, C or D, left), in agreement with previous reports (22). This staining pattern resembled that of Dlk (Fig. 3B) (1). Indeed, upon co-transfection both proteins co-localized perfectly in nuclear dots (Fig. 3C). This strict co-localization of CDC5 was not seen with mutant DlkΔC1, lacking the leucine zipper. In this case, DlkΔC1 generally displayed a more diffuse nuclear distribution, as seen previously (7), while CDC5 still occurred in speckles (Fig. 3D). This result corroborated the two-hybrid data that the interaction of Dlk with CDC5 is mediated by its leucine zipper. On the other hand, the association of CDC5 with speckles apparently did not depend on Dlk.
Figure 3.
Co-expression and subcellular localization of Dlk and CDC5. Rat embryo REF52.2 fibroblasts were transfected with expression plasmids coding for dsRED–CDC5 (A), GFP–Dlk (B) or co-transfected with dsRED–CDC5 and GFP–Dlk wt (C) or GFP–DlkΔC1 (D) (see scheme in Fig. 2B). After 20 h cells were fixed and stained with DAPI and inspected with a Zeiss Axioplan fluorescence microscope. Each set of images shows the individual fluorescent proteins, DAPI stains of the same cells and, in the case of transfected cells (C and D), merged pictures of dsRED–CDC5 and GFP–Dlk or dsRED–CDC5 and GFP–DlkΔC1, respectively.
Mapping of the nuclear localization signal
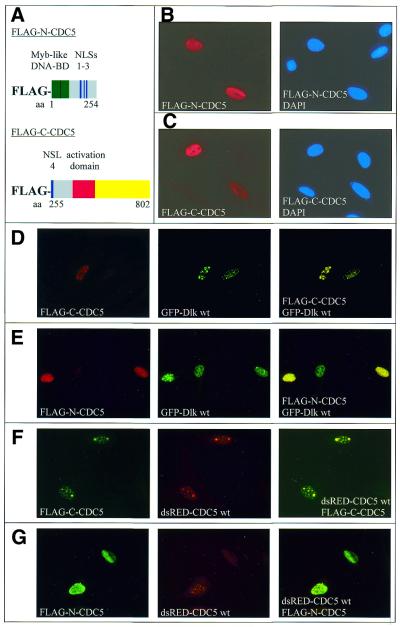
CDC5 contains four putative nuclear localization signals (NLSs) between amino acids 165 and 271. Deletion mutants encompasing residues 1–254 (FLAG–N-CDC5) and 255–802 (FLAG–C-CDC5) were constructed, thereby separating NLSs 1–3 from NLS 4 (see scheme in Fig. 4A). The deletion constructs were expressed as FLAG-tagged proteins and their subcellular localization was examined by indirect immunofluorescence analyses. As shown in Figure 4B and C, respectively, both the N-terminal (FLAG–N-CDC5) and C-terminal constructs (FLAG–C-CDC5) were efficiently translocated to the nucleus, indicating that one or more of the putative NLSs 1–3 on one side and NLS 4 on the other are sufficient to direct nuclear transport. However, both deletion proteins displayed a diffuse rather than a speckled distribution, suggesting that full-length CDC5 is required for association with speckles.
Figure 4.
Nuclear localization of deletion mutants of CDC5. (A) A schematic representation of deletion mutants of CDC5 used for mapping of NLSs. In this case, FLAG-tagged fusion proteins were employed to allow distinction from GFP- and dsRED-tagged proteins. Deletion mutants FLAG–N-CDC5 (residues 1–254) (B) and FLAG–C-CDC5 (residues 255–802) (C), or various combinations with GFP–Dlk (D and E) or dsRED–CDC5 wt (F and G) were expressed in REF52.2 cells. At 20 h post-transfection cells were fixed and processed for indirect immunofluorescence using anti-FLAG M-2 as primary and Cy3 conjugate as secondary antibody; additionally, all cells were stained with DAPI to visualize nuclei (not shown for double-transfected cells). Images on the right of (D)–(G) represent merged images.
To see how Dlk might behave in this setting, the CDC5 constructs were co-expressed with wild-type Dlk or mutant DlkΔC1. Interestingly, upon co-expression of wild-type Dlk, the C-terminal fragment (Fig. 4D), but not the N-terminal fragment (Fig. 4E), of CDC5 again appeared in speckles. This was not seen with mutant DlkΔC1, as expected (data not shown). Taken together, these results indicate that Dlk can target CDC5 to nuclear speckles.
It had been suggested that CDC5 forms homodimers, as deduced from the symmetry of the DNA-binding motif (21). This issue was investigated by employing the deletion mutants described above. If CDC5 is indeed able to form homodimers then co-expression of the truncation mutants with full-length CDC5 should result in targeting of the truncated proteins to speckles. This was indeed the case for the C-terminal (Fig. 4F) but not for the N-terminal fragment (Fig. 4G), thus showing that CDC5 forms dimers and that the interaction occurs within the C-terminal portion.
Overexpression of CDC5 and Dlk results in displacement of SC35 from nuclear speckles
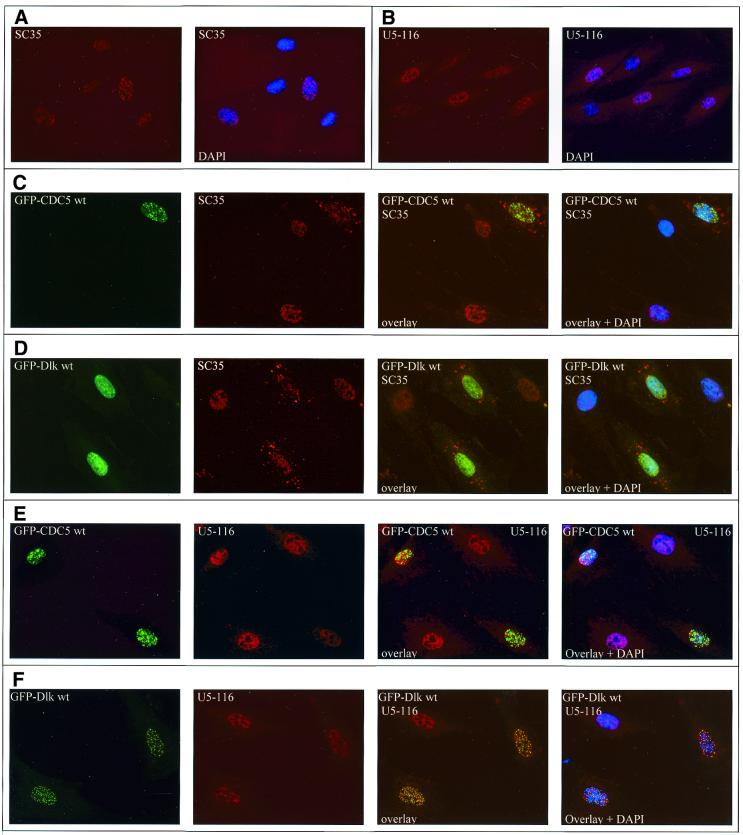
Human CDC5 has been reported to co-localize with splicing factor SC35 (22), a member of the SR protein family that is commonly used as a marker for spliceosomes and speckles (34,35). In contrast, no such co-localization with SC35 or U5-116 protein had been observed for Dlk (7). However, the interaction of Dlk with a bona fide splicing factor prompted us to reinvestigate this issue. CDC5 and Dlk were visualized as GFP fusion proteins upon transient transfection and endogenous SC35 or U5-116 were detected by indirect immunofluorescence with the respective antibodies. When SC35, U5-116, CDC5 or Dlk were examined individually (in untransfected versus transfected cells), they all displayed rather similar staining patterns [see Fig. 5A and B for SC35 and U5-116, respectively, and Fig. 5C and D (left) for CDC5 and Dlk, respectively]. However, the merged images of transfected cells revealed that the majority of CDC5- and Dlk-specific speckles were distinct from the SC35-specific (Fig. 5C and D) and U5-116-specific speckles (Fig. 5E and F). Strikingly, expression of both GFP–CDC5 and GFP–Dlk resulted in a gross redistribution of endogenous SC35, which appeared to be partially displaced from nuclear speckles and relocated to cytoplasmic structures (see SC35-specific staining in Fig. 5C and D). This effect was not seen with U5-116. Rather, both CDC5 and Dlk on the one hand and U5-116 on the other co-existed as distinct speckles in the nucleus (Fig. 5E and F). Thus, the displacement was specific for SC35. It could be due to competition of CDC5 or Dlk with SC35 for the same binding partners or, more likely, due to phosphorylation of SC35 (or other factors) by Dlk. To investigate this latter possibility, purified GST–SC35 and the related SR protein ASF/SF2 were subjected to a kinase reaction with Dlk in vitro. As shown in Figure 6 (lane 2), ASF/SF2 was efficiently phosphorylated, whereas GST–SC35 was only a poor substrate (not shown).
Figure 5.
Expression of CDC5 and Dlk results in displacement of splicing factor SC35. (A and B) The speckled nuclear distribution of SC35 and U5-116 is shown, as revealed by indirect immunofluorescence with the respective antibodies and Cy3-coupled secondary antibodies; the right image is merged with DAPI staining. (C and D) GFP–CDC5- and Dlk-transfected cells, respectively, were additionally stained for SC35. Please note that in transfected cells, SC35 is partially relocated to the cytoplasm. (E and F) GFP–CDC5- and Dlk-transfected cells were counterstained for U5-116.
Figure 6.
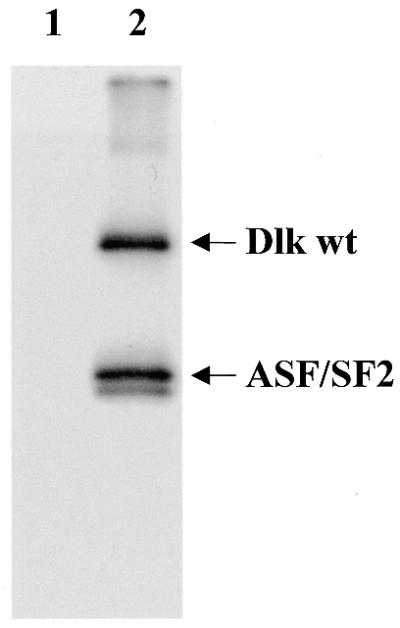
Dlk phosphorylates splicing factor ASF/SF2. His-tagged Dlk was incubated with His-tagged ASF/SF2 in a standard kinase reaction and the products were separated on a 10% SDS–polyacrylamide gel and autoradiographed for 6 h. Lane 1, as a control, ASF/SF2 incubated without Dlk; lane 2, autophosphorylated Dlk and phosphorylated ASF/SF2.
CDC5 co-localizes with PML bodies
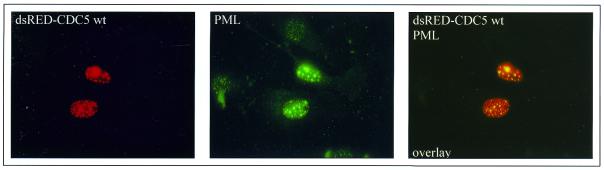
Dlk had been shown to partially co-localize with PML bodies (7). These structures represent assemblies of different proteins involved in cell cycle regulation, transcriptional regulation and apoptosis (reviewed in 36,37). As shown in Figure 7, CDC5 was associated with PML bodies to a similar extent as previously shown for Dlk. The significance of this association awaits further investigation.
Figure 7.
CDC5 exhibits partial co-localization with PML bodies. Expression of dsRED–CDC5 wt and indirect immunofluorescence of endogenous PML reveals partial co-localization of CDC5 with PML bodies.
DISCUSSION
In this paper we have identified a new interaction partner for Dlk, the rat homolog of S.pombe CDC5, as revealed by the two-hybrid system. The interaction of both proteins was verified by in vitro binding and intracellular co-localization. Unfortunately, we were not able to co-immunoprecipitate Dlk and CDC5, presumably because both proteins are tightly associated with nuclear structures and the rigorous extraction conditions required might have led to dissociation of the two proteins. However, the observation that both proteins co-localized perfectly in speckle-like structures and that Dlk was capable of recruiting CDC5 deletion mutants to speckles provides convincing evidence for an in vivo interaction.
The interaction domain in Dlk was mapped to the leucine zipper, as already shown for other interaction partners, ATF4 and AATF (2,8,9). Obviously, this interaction motif enables Dlk to participate in multiple interactions. On the other hand, the minimal interaction domain in CDC5 was mapped to a region between residues 500 and 659, but residues 660–802 seemed to contribute to this interaction, as it appeared to be stronger with the larger construct. In these regions no obvious interaction motif can be identified. Thus, leucine zippers can also interact with other structures. A reverse case is given by Par-4, yet another interaction partner of Dlk, which itself contains a leucine zipper (38) but, unexpectedly, interacts with the Arg-rich region of Dlk (8) and the zinc finger regions of WT-1 (39) and PKCζ (40). Other examples include complex formation between Fos and Rb (41) and ATF2 and protein kinase CK2 (42). It will be interesting to determine the interaction motifs more precisely to obtain some clues about the general principle underlying these interactions.
So far, Dlk had been implicated in apoptotic processes (2,4,7–9), which is strictly dependent on its cytoplasmic localization (7,8). On the other hand, the role of nuclear Dlk is not yet clear. Its interactions with several transcription factors (ATF4, AATF and Par4) and with PML bodies point to a role in transcription. However, firm evidence that Dlk is involved in transcriptional regulation is still lacking, mostly because information about the interacting transcription factors is also sparse. The interaction of Dlk with CDC5 again points to a role in transcription or splicing or both.
Whether CDC5 is a transcription or a splicing factor is not yet clear. The evidence for a function as a transcription factor is based on the identification of a DNA-binding motif and on transactivation assays with artificial reporter constructs (20,21). When we employed this assay we did not see a significant effect of Dlk on CDC5-mediated transactivation (H.Engemann, unpublished results). Clearly, proof of a role in transcription would require the identification of target genes. On the other hand, the evidence that CDC5 is involved in pre-mRNA splicing is based on yeast genetics (16,22,24), co-purification with splicing complexes (22,43) and splicing defects in CDC5-depleted in vitro systems (24). However, in these depletion experiments CDC5 was obviously complexed with other factors, since recombinant CDC5 alone was not capable of rescuing correct splicing. In fact, CDC5 seems to be part of a large complex of as many as 30 different proteins, most of which are known components of the splicing apparatus, as determined by mass spectrometry. Of these, six proteins appeared to form a tight core, called the CDC5 complex (24). Among the less tightly bound proteins were DNA-dependent protein kinase and phosphatases PP2c and PP1, the latter perhaps bound via its regulatory subunit NIPP-1, which was recently shown to directly interact with CDC5 (32). Dlk was not detected in the core complex, suggesting that its interaction with CDC5 may be weak. Additionally, the interaction might depend on the phosphorylation and/or physiological state of one or more of the interaction partners, as shown for NIPP-1 (32).
Surprisingly, Dlk did not phosphorylate CDC5, at least not in vitro. Rather, CDC5 was phosphorylated by a co-purified kinase, which was identified as CK2. This might be of functional significance, since CDC5 has 14 potential CK2 phosphorylation sites. It will be interesting to investigate which of the sites phosphorylated in vivo coincide with CK2 sites. Our negative in vitro results do not exclude phosphorylation of CDC5 by Dlk in vivo, which might require an appropriate microenvironment, such as the splicing complexes, or it might require phosphorylation by another kinase, such as Cdks (31,32). Alternatively, CDC5 might have a targeting function for Dlk to facilitate phosphorylation of associated proteins (see below). These issues need to be further investigated.
What might be the functional significance of the interaction between Dlk and CDC5? Both CDC5 and Dlk co-localized perfectly in structures reminiscent of speckles. However, we did not see co-localization of CDC5 or Dlk with SC35 or U5-116, which are markers for spliceosomes and speckles. Rather, SC35 was partially displaced from the nuclear speckles. Speckles are highly dynamic structures in which splicing factors are constantly assembled and disassembled to be recruited to sites of transcription and splicing (34,44). This process is regulated, in part, by phosphorylation/dephosphorylation of the factors involved (35,45). Several SR kinases have been identified by virtue of their phosphorylating SR proteins, e.g. SC35 and SF2/ASF in vitro, and their relocating SR proteins from speckles in vivo. These kinases include SRPK1 and 2 (46–48), Clk1–3 (49,50), Cdc2 (51) and topoisomerase I (52) (reviewed in 45). Thus, the effects seen with Dlk are quite similar to those observed with other SR kinases. Indeed, Dlk did phosphorylate SC35 and SF2/ASF in vitro, although phosphorylation of SC35 was rather weak, presumably because it was employed as a GST fusion protein. Whether the Dlk- and/or CDC5-induced relocation of SC35 is of functional consequence has to be further investigated. Although SC35 is a general splicing factor it can also influence alternative splicing of certain substrates, as shown for the IgM and HIV tat pre-mRNAs (53) and caspase 2 (Ich-1) transcripts (54). This can be achieved by changing its relative abundance. (55). Thus, altering the subcellular localization of SC35 by Dlk and CDC5 might be a regulatory mechanism for alternative splicing, presumably for a specific subset of transcripts. Of note in this context is a specific function of CDC5 in the G2/M transition (14,15,20). This function is not compatible with CDC5 being a general splicing factor. Taking into account that CDC5 might also act as transcription factor (20), it might participate either in transcription or splicing or both of G2/M-specific genes. In fact, CDC5 might actually provide a link between transcription and splicing, as suggested previously (20,21). Clearly, there are interesting and exciting issues to be investigated.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Gerd Landsberg for excellent technical assistance and Dr R. Lührmann (Göttingen, Germany) and Dr S. Stamm (Erlangen, Germany) for purified splicing factors to be employed in kinase reactions and antibodies against splicing factors and PML. This work was supported by the Deutsche Forschungsgemeinschaft, grant Sche246/12-1, and Graduiertenkolleg ‘Functional protein domains’.
REFERENCES
- 1.Kögel D., Plöttner,O., Landsberg,G., Christian,S. and Scheidtmann,K.H. (1998) Cloning and characterization of Dlk, a novel serine/threonine kinase that is tightly associated with chromatin and phosphorylates core histones. Oncogene, 17, 2645–2654. [DOI] [PubMed] [Google Scholar]
- 2.Kawai T., Matsumoto,M., Takeda,K., Sanjo,H. and Akira,S. (1998) ZIP kinase, a novel serine/threonine kinase which mediates apoptosis. Mol. Cell. Biol., 18, 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inbal B., Shani,G., Cohen,O., Kissil,J.L. and Kimchi,A. (2000) Death-associated protein kinase-related protein 1, a novel serine/threonine kinase involved in apoptosis. Mol. Cell. Biol., 20, 1044–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kögel D., Prehn,J.H. and Scheidtmann,K.H. (2001) The DAP kinase family of pro-apoptotic proteins: novel players in the apoptotic game. Bioessays, 23, 352–358. [DOI] [PubMed] [Google Scholar]
- 5.Cohen O., Feinstein,E. and Kimchi,A. (1997) DAP-kinase is a Ca2+/calmodulin-dependent, cytoskeletal-associated protein kinase, with cell death-inducing functions that depend on its catalytic activity. EMBO J., 16, 998–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen O., Inbal,B., Kissil,J.L., Raveh,T., Berissi,H., Spivak-Kroizaman,T., Feinstein,E. and Kimchi,A. (1999) DAP-kinase participates in TNF-alpha- and Fas-induced apoptosis and its function requires the death domain. J. Cell Biol., 146, 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kögel D., Bierbaum,H., Preuss,U. and Scheidtmann,K.H. (1999) C-terminal truncation of Dlk/ZIP kinase leads to abrogation of nuclear transport and high apoptotic activity. Oncogene, 18, 7212–7218. [DOI] [PubMed] [Google Scholar]
- 8.Page G., Kögel,D., Rangnekar,V. and Scheidtmann,K.H. (1999) Interaction partners of Dlk/ZIP kinase: co-expression of Dlk/ZIP kinase and Par-4 results in cytoplasmic retention and apoptosis. Oncogene, 18, 7265–7273. [DOI] [PubMed] [Google Scholar]
- 9.Page G., Lödige,I., Kögel,D. and Scheidtmann,K.H. (1999) AATF, a novel transcription factor that interacts with Dlk/ZIP kinase and interferes with apoptosis. FEBS Lett., 462, 187–191. [DOI] [PubMed] [Google Scholar]
- 10.Kitada K., Johnson,A.L., Johnston,L.H. and Sugino,A. (1993) A multicopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is identified as CDC5. Mol. Cell. Biol., 13, 4445–4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohkura H., Hagan,I.M. and Glover,D.M. (1995) The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring and septum, can drive septum formation in G1 and G2 cells. Genes Dev., 9, 1059–1073. [DOI] [PubMed] [Google Scholar]
- 12.Glover D.M., Hagan,I.M. and Tavares,A.A. (1998) Polo-like kinases: a team that plays throughout mitosis. Genes Dev., 12, 3777–3787. [DOI] [PubMed] [Google Scholar]
- 13.Nigg E.A. (1998) Polo-like kinases: positive regulators of cell division from start to finish. Curr. Opin. Cell Biol., 10, 776–783. [DOI] [PubMed] [Google Scholar]
- 14.Nasmyth K. and Nurse,P. (1981) Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 182, 119–124. [DOI] [PubMed] [Google Scholar]
- 15.Ohi R., McCollum,D., Hirani,B., Den Haese,G.J., Zhang,X., Burke,J.D., Turner,K. and Gould,K.L. (1994) The Schizosaccharomyces pombe cdc5+ gene encodes an essential protein with homology to c-Myb. EMBO J., 13, 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai W.Y., Chow,Y.T., Chen,H.R., Huang,K.T., Hong,R.I., Jan,S.P., Kuo,N.Y., Tsao,T.Y., Chen,C.H. and Cheng,S.C. (1999) Cef1p is a component of the Prp19p-associated complex and essential for pre-mRNA splicing. J. Biol. Chem., 274, 9455–9462. [DOI] [PubMed] [Google Scholar]
- 17.Hirayama T. and Shinozaki,K. (1996) A cdc5+ homolog of a higher plant, Arabidopsis thaliana. Proc. Natl Acad. Sci. USA, 93, 13371–13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohi R., Feoktistova,A., McCann,S., Valentine,V., Look,A.T., Lipsick,J.S. and Gould,K.L. (1998) Myb-related Schizosaccharomyces pombe cdc5p is structurally and functionally conserved in eukaryotes. Mol. Cell. Biol., 18, 4097–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernstein H.S. and Coughlin,S.R. (1997) Pombe Cdc5-related protein. A putative human transcription factor implicated in mitogen-activated signaling. J. Biol. Chem., 272, 5833–5837. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein H.S. and Coughlin,S.R. (1998) A mammalian homolog of fission yeast Cdc5 regulates G2 progression and mitotic entry. J. Biol. Chem., 273, 4666–4671. [DOI] [PubMed] [Google Scholar]
- 21.Lei X.H., Shen,X., Xu,X.Q. and Bernstein,H.S. (2000) Human Cdc5, a regulator of mitotic entry, can act as a site-specific DNA binding protein. J. Cell Sci., 113, 4523–4531. [DOI] [PubMed] [Google Scholar]
- 22.Burns C.G., Ohi,R., Krainer,A.R. and Gould,K.L. (1999) Evidence that Myb-related CDC5 proteins are required for pre-mRNA splicing. Proc. Natl Acad. Sci. USA, 96, 13789–13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald W.H., Ohi,R., Smelkova,N., Frendewey,D. and Gould,K.L. (1999) Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol., 19, 5352–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ajuh P., Kuster,B., Panov,K., Zomerdijk,J.C., Mann,M. and Lamond,A.I. (2000) Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J., 19, 6569–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chien C.T., Bartel,P.L., Sternglanz,R. and Fields,S. (1991) The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl Acad. Sci. USA, 88, 9578–9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidmann,J.G., Smith,J.A. and Struhl,K. (1987) Current Protocols in Molecular Biology. John Wiley & Sons, New York, NY.
- 27.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Habor Laboratory Press, Cold Spring Habor, NY.
- 28.Stuurman N., de Graaf,A., Floore,A., Josso,A., Humbel,B., de Jong,L. and van Driel,R. (1992) A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci., 101, 773–784. [DOI] [PubMed] [Google Scholar]
- 29.Fabrizio P., Laggerbauer,B., Lauber,J., Lane,W.S. and Luhrmann,R. (1997) An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J., 16, 4092–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Too C.K. (1997) Differential expression of elongation factor-2, alpha4 phosphoprotein and Cdc5-like protein in prolactin-dependent/independent rat lymphoid cells. Mol. Cell. Endocrinol., 131, 221–232. [DOI] [PubMed] [Google Scholar]
- 31.Stukenberg P.T., Lustig,K.D., McGarry,T.J., King,R.W., Kuang,J. and Kirschner,M.W. (1997) Systematic identification of mitotic phosphoproteins. Curr. Biol., 7, 338–348. [DOI] [PubMed] [Google Scholar]
- 32.Boudrez A., Beullens,M., Groenen,P., Van Eynde,A., Vulsteke,V., Jagiello,I., Murray,M., Krainer,A.R., Stalmans,W. and Bollen,M. (2000) NIPP1-mediated interaction of protein phosphatase-1 with CDC5L, a regulator of pre-mRNA splicing and mitotic entry. J. Biol. Chem., 275, 25411–25417. [DOI] [PubMed] [Google Scholar]
- 33.Saxena A., Padmanabha,R. and Glover,C.V. (1987) Isolation and sequencing of cDNA clones encoding alpha and beta subunits of Drosophila melanogaster casein kinase II. Mol. Cell. Biol., 7, 3409–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang S. and Spector,D.L. (1996) Dynamic organization of pre-mRNA splicing factors. J. Cell. Biochem., 62, 191–197. [DOI] [PubMed] [Google Scholar]
- 35.Misteli T. and Spector,D.L. (1997) Protein phosphorylation and the nuclear organization of pre-mRNA splicing. Trends Cell Biol., 7, 135–138. [DOI] [PubMed] [Google Scholar]
- 36.Seeler J.S. and Dejean,A. (1999) The PML nuclear bodies: actors or extras? Curr. Opin. Genet. Dev., 9, 362–367. [DOI] [PubMed] [Google Scholar]
- 37.Zhong S., Salomoni,P. and Pandolfi,P.P. (2000) The transcriptional role of PML and the nuclear body. Nature Cell Biol., 2, E85–E90. [DOI] [PubMed] [Google Scholar]
- 38.Rangnekar V.M. (1998) Apoptosis mediated by a novel leucine zipper protein Par-4. Apoptosis, 3, 61–66. [DOI] [PubMed] [Google Scholar]
- 39.Johnstone R.W., See,R.H., Sells,S.F., Wang,J., Muthukkumar,S., Englert,C., Haber,D.A., Licht,J.D., Sugrue,S.P., Roberts,T. et al. (1996) A novel repressor, par-4, modulates transcription and growth suppression functions of the Wilms’ tumor suppressor WT1. Mol. Cell. Biol., 16, 6945–6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diaz-Meco M.T., Municio,M.M., Frutos,S., Sanchez,P., Lozano,J., Sanz,L. and Moscat,J. (1996) The product of par-4, a gene induced during apoptosis, interacts selectively with the atypical isoforms of protein kinase C. Cell, 86, 777–786. [DOI] [PubMed] [Google Scholar]
- 41.Nead M.A., Baglia,L.A., Antinore,M.J., Ludlow,J.W. and McCance,D.J. (1998) Rb binds c-Jun and activates transcription. EMBO J., 17, 2342–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamaguchi Y., Wada,T., Suzuki,F., Takagi,T., Hasegawa,J. and Handa,H. (1998) Casein kinase II interacts with the bZIP domains of several transcription factors. Nucleic Acids Res., 26, 3854–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neubauer G., King,A., Rappsilber,J., Calvio,C., Watson,M., Ajuh,P., Sleeman,J., Lamond,A. and Mann,M. (1998) Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nature Genet., 20, 46–50. [DOI] [PubMed] [Google Scholar]
- 44.Phair R.D. and Misteli,T. (2000) High mobility of proteins in the mammalian cell nucleus. Nature, 404, 604–609. [DOI] [PubMed] [Google Scholar]
- 45.Misteli T. (1999) RNA splicing: what has phosphorylation got to do with it? Curr. Biol., 9, R198–R200. [DOI] [PubMed] [Google Scholar]
- 46.Gui J.F., Tronchere,H., Chandler,S.D. and Fu,X.D. (1994) Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc. Natl Acad. Sci. USA, 91, 10824–10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuroyanagi N., Onogi,H., Wakabayashi,T. and Hagiwara,M. (1998) Novel SR-protein-specific kinase, SRPK2, disassembles nuclear speckles. Biochem. Biophys. Res. Commun., 242, 357–364. [DOI] [PubMed] [Google Scholar]
- 48.Wang H.Y., Lin,W., Dyck,J.A., Yeakley,J.M., Songyang,Z., Cantley,L.C. and Fu,X.D. (1998) SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J. Cell Biol., 140, 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colwill K., Feng,L.L., Yeakley,J.M., Gish,G.D., Caceres,J.F., Pawson,T. and Fu,X.D. (1996) SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J. Biol. Chem., 271, 24569–24575. [DOI] [PubMed] [Google Scholar]
- 50.Nayler O., Stamm,S. and Ullrich,A. (1997) Characterization and comparison of four serine- and arginine-rich (SR) protein kinases. Biochem. J., 326, 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okamoto Y., Onogi,H., Honda,R., Yasuda,H., Wakabayashi,T., Nimura,Y. and Hagiwara,M. (1998) cdc2 kinase-mediated phosphorylation of splicing factor SF2/ASF. Biochem. Biophys. Res. Commun., 249, 872–878. [DOI] [PubMed] [Google Scholar]
- 52.Rossi F., Labourier,E., Forne,T., Divita,G., Derancourt,J., Riou,J.F., Antoine,E., Cathala,G., Brunel,C. and Tazi,J. (1996) Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature, 381, 80–82. [DOI] [PubMed] [Google Scholar]
- 53.Mayeda A., Screaton,G.R., Chandler,S.D., Fu,X.D. and Krainer,A.R. (1999) Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol. Cell. Biol., 19, 1853–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang Z.H., Zhang,W.J., Rao,Y. and Wu,J.Y. (1998) Regulation of Ich-1 pre-mRNA alternative splicing and apoptosis by mammalian splicing factors. Proc. Natl Acad. Sci. USA, 95, 9155–9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duncan P.I., Stojdl,D.F., Marius,R.M., Scheit,K.H. and Bell,J.C. (1998) The Clk2 and Clk3 dual-specificity protein kinases regulate the intranuclear distribution of SR proteins and influence pre-mRNA splicing. Exp. Cell Res., 241, 300–308 [DOI] [PubMed] [Google Scholar]