Abstract
We have recently reported that liposomes in combination with histidine (HK)-containing polymers enhanced the expression of luciferase in transfected cells. In transformed or malignant cell lines, branched HK polymers (combined with liposome carriers) were significantly more effective than the linear HK polymer in stimulating gene expression. In the current study, we found that the linear HK polymer enhanced gene expression in primary cell lines more effectively than the branched polymers. The differences in the optimal carrier (linear versus branched) were not due to initial cellular uptake, size of the complexes or level of gene expression. There was, however, a strong association between the optimal type of HK polymer and the pH of endocytic vesicles (P = 0.0058). By altering the percentage of histidines carrying a positive charge, the endosomal pH of a cell may determine the amount of DNA released from the linear or branched HK polymer. In the two cell lines in which the linear HK was the optimal polymer, the endocytic vesicles were strongly acidic with a pH of <5.0. Conversely, in the four cell lines in which the branched polymers were optimal transfection agents, the pH of endocytic vesicles was >6.0. Furthermore, binding data support the relationship between DNA release from the optimal HK polymer and endosomal pH. The interplay between optimal HK polymers and the endosomal pH may lead to improved gene-delivery polymers tailored to a particular cell.
INTRODUCTION
Non-viral carriers offer the potential of delivering genes to targets in a highly efficient, safe and specific manner. There are several non-viral candidates for gene delivery including liposomes, peptides, proteins and synthetic polymers (1–6). A problem associated with non-viral carriers, however, is their low in vitro and in vivo transfection efficiency (7,8). As most non-viral carriers of DNA depend on endocytosis for their import into cells, one strategy for increasing gene expression has been to discover polymers/peptides that disrupt endosomes. pH-buffering polymers represent an important class of these endosomolytic carriers, and such polymers (6,9–13) have been shown to increase transfection in vitro and in vivo. Once endocytized, pH-buffering polymers such as PEI are thought to enhance transfection by acting as ‘proton sponges’ resulting in passive chloride influx. This series of events may lead to endosomal swelling and disruption (14). Disruption of endosomes by these pH-buffering carriers appears to be dependent on the vacuolar proton pump, V-type H(+) ATPase, as bafilomycin A1, a specific inhibitor of the proton pump, reduces transfection (14).
Similar to other pH-buffering polymers, we found that pH-buffering co-polymers composed of histidine and lysine (HK polymers) in combination with cationic liposomes, were highly efficient carriers of plasmids. The potential advantages of the HK polymer compared with many other pH-buffering agents include: (i) a peptide-base bond that is likely to be metabolizable; (ii) a well-defined sequence and structure; and (iii) the ability to vary the ratio of histidine and lysine. In addition to the ability of histidine to buffer and disrupt endosomes, there are several other mechanisms by which the HK co-polymer may increase the transfection of liposome carriers. For example, the lysine moiety of the co-polymer may neutralize, in part, the negative charge of the plasmid DNA and the cationic liposomes may neutralize the remainder of DNA charge and provide a scaffold for the polymer:DNA complex. Furthermore, a reduction in size of the complexes containing HK polymers may contribute to increased gene expression (15,16).
In most cell lines, we found that branched polymers were clearly superior at augmenting transfection compared with the linear polymers (16). This enhancement of gene expression by branched polymers compared with linear polymers may partly be due to the reduced size of the complexes because of DNA condensation. Surprisingly, we have found recently that in a few cell lines the linear HK polymers were more effective transfection agents than the branched HK polymers. As a result, the differences in the optimal transfection HK polymers (linear or branched) among different cell lines cannot be attributed solely to the reduction in size of transfection complexes or their ability to disrupt endosomes.
An alternative mechanism that may explain this variability of optimal HK polymers between cells may be differences in the endosomal pH in the cells. By affecting the percentage of histidines in the polymer that contains a positive charge, and thus the release of negatively charged DNA, the pH of endocytic vesicles may be the critical factor that determines the optimal HK polymer (linear versus branched) in a particular cell type. In this study, we established a strong correlation between the optimal HK polymer (linear or branched) and the pH of endocytic vesicles in several cell lines.
MATERIALS AND METHODS
Cells
MDA-MB-435 (a gift from Dr Erik Thompson, Lombardi Cancer Center, Washington, DC), MDA-MB-231 [American Tissue Culture Collection (ATCC), Manassas, VA], CRL-5800 (ATCC), Chinese hamster ovary (CHO) cells, bovine aortic endothelial cells (BAE; Coriell Cell Repositories, Camden, NJ) and normal human dermal fibroblasts (NHDF; Clonetics, San Diego, CA) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum and 2 mM glutamine. MDA-MB-435 and MDA-MB-231 cells are undifferentiated estrogen-independent breast cancer lines obtained from unrelated human donors, whereas the CRL-5800 cell line was isolated from a human non-small cell lung cancer. Transfection and endocytic pH experiments on the two primary cell lines, BAE and NHDF, were carried out between passages 3 and 6 of these cells.
Polymers
The polymers were synthesized in the biopolymer core facility at the University of Maryland as described previously (15,16). The sequences of polymers derived from the HK or HHK series were as follows: (i) HK (19mer) │K-H-K-H-K-H-K-H-K-G-K-H-K-H-K-H-K-H-K│; (ii) HK2b (39mer) │K-H-K-H-K-H-K-H-K-G-K-H-K-H-K-H-K-H-K│2 K; (iii) HK3b (59mer) │K-H-K-H-K-H-K-H-K-G-K-H-K-H-K-H-K-H-K│3 K-K; (iv) HK4b (79mer) │K-H-K-H-K-H-K-H-K-G-K-H-K-H-K-H-K-H-K│4 K-K-K; (v) HHK (20mer) │K-H-K-H-H-K-H-H-K-H-H-K-H-H-K-H-H-K-H-K│; (vi) HHK2b (41mer) │K-H-K-H-H-K-H-H-K-H-H-K-H-H-K-H-H-K-H-K│2 K; (vii) HHK3b (62mer) │K-H-K-H-H-K-H-H-K-H-H-K-H-H-K-H-H-K-H-K│3 K-K; and (viii) HHK4b (83mer) │K-H-K-H-H-K-H-H-K-H-H-K-H-H-K-H-H-K-H-K│4 K-K-K. Sequences i–iv represent increasing complexity of the HK series whereas sequences v–viii represent increasing complexity of the HHK series; furthermore, the designations 2b, 3b and 4b indicate the number of branches emanating from a lysine core.
Preparation of liposomes and plasmids
Preparation of the 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) liposomes and plasmids has been described previously (17). In brief, DH5α bacteria (Life Technologies, Gaithersburg, MD) containing the plasmids encoding luciferase (PCI-Luc) were grown in Superbroth to mid-log phase. The plasmids were then purified by using Qiagen columns. An analytical gel of each plasmid (cut and uncut) was performed to ensure that there was no contamination with other nucleic acids. Liposomes were composed of DOTAP lipids (Avanti, Birmingham, AL). After hydration of the lipids, the liposomes were sonicated until clear with a Branson 1210 bath sonicator in the presence of argon. The liposomes were then extruded through 100 nm polycarbonate membranes with LipsoFast-Basic (Avestin Inc., Ottawa, ON). The final concentration of the liposome stock was 1 mg/ml.
Transfection of cells
From 2 × 104 to 4.5 × 104 cells were initially plated in 24-well plates and incubated for 48 h. The cells, which were 50–70% confluent, were then transfected with PCI-Luc. In transfection experiments, the co-polymer (from 0.375 to 7.5 nmol) was incubated with PCI-Luc (0.75 µg) for 30 min in OptiM. Cationic liposomes (1.5 µg) were then added, gently mixed and allowed to stand for an additional 30 min. The resulting polymer:liposome:DNA complex was then diluted with OptiM. Four hours after transfection the complexes were removed and DMEM with 10% serum was added. Incubation was continued for another 48 h for MDA-MB-231, MDA-MB-435, CRL-5800, CHO and NHDF cells and 24 h for BAE cells. Luciferase levels were measured with the direct current Turner 20/20 luminometer (Turner Design, Sunnyvale, CA) as described previously (15,16). Relative light units were converted to picograms (pg) of protein of luciferase by using recombinant luciferase (Promega, Madison, WI) as a standard. Duplicate measurements were performed for each concentration and three separate experiments were conducted.
Uptake studies
MDA-MB-231 and BAE cells were plated into 24-well plates and incubated for 48 h. The cells, which were between 50 and 70% confluent, were then incubated with the polymer:DNA:liposome complexes for 30 min and 4 h. At the desired time point, the medium was removed, cells were then washed with PBS and 50 mM of methylamine was added to the cells and incubation was continued for 1 h. The cell fluorescent density was recorded at 520 nm using the CytoFluor 2350 fluorescent multi-well reader (Millipore, Bedford, MS), set at an excitation wavelength of 485 nm. The fluorescent density was then converted to nanomoles of polymer using fluoresceinylated polymers as standard. For preparation of complexes, the fluoresceinylated polymers (7.5 nmol of HK and 0.375 nmol of HHK4b) were incubated with PCI-Luc (0.75 µg) in OptiM for 30 min. Cationic liposomes (1.5 µg) were then added, gently mixed and allowed to stand for an additional 30 min. To ensure intracellular uptake, cells exposed to the labeled complex for 30 min and 4 h were also examined with a fluorescence microscope (Diaphot-TMD, Nikon, Tokyo, Japan).
Confocal microscopy pH measurement
Cells were plated in 8-well coverglass bottom chambers (Lab-Tek, Naperville, IL). After 24 h when the cells were 30–50% confluent, they were incubated in 1 mg/ml of the pH indicator LysoSensor Yellow/Blue dextran (Molecular Probes, Eugene, OR) for 12 h (18) and then washed with cold PBS. LysoSensor is an UV-excited fluorophore with emission maxima at 450 and 530 nm that are proportional to pH. This dextran-conjugated indicator is taken up by vesicles that are part of the endocytic pathway and are comprised of endosomes and lysosomes. Higher 530/450 nm ratios correlate with a lower pH. The labeled cells were observed with a laser scanning confocal microscope (Zeiss LSM410, Thornwood, NY), using excitation at 360 nm and an emission filter at 450 nm and a long pass emission filter at 515 nm. With the dextran-labeled LysoSensor Yellow/Blue dye, the ratio of 530/450 nm emission was calculated for endocytic vesicles by using the MetaMorph Image processing system (Version 4.5r5, Universal Imaging Corp., PA) and averaged for each image (18). The 530/450 nm ratio was calculated on a minimum of 50 intracellular vesicles in each of the four images analyzed per experiment and each experiment was repeated three times. Dye emission at 530 nm was assigned the color red (R), whereas the emission at 450 nm was assigned the color green (G).
To establish a pH titration curve (18–20), CHO cells were first incubated with the pH-sensitive dye overnight and then treated with 10 µM monensin and 20 µM nigericin. The treated cells were equilibrated for 10 min with a reaction buffer consisting of 5 mM NaCl, 115 mM KCl, 1.2 mM MgSO4 and 25 mM 4-morpholineethanesulfonic acid (MES) buffer titrated between pH 4.5 and 7.0. After these incubations, the 530/450 nm intensity ratio of the endocytic vesicles was determined as described above.
For the pH-sensitive dextran, DM-NERF (Molecular Probes) dye (21), a laser scanning confocal microscope (Zeiss LSM 510) equipped with an argon laser was used to measure the endocytic vesicle pH of cells. In contrast to LysoSensor Yellow/Blue, whose emissions are pH-dependent, it is the excitations of DM-NERF (maxima 488 and 514 nm) that are pH dependent. The dye-labeled cells were excited at 488 and 514 nm and their emissions at 560 nm using a barrier filter were recorded. Consequently, for the DM-NERF indicator, the more acidic the endosome the stronger the 488 nm excitation/540 nm emission pair will be compared with the 514 nm excitation/540 nm emission pair. pH titration curves were established, and ratiometric calculations were made as described above for the LysoSensor indicator.
Co-localization experiment
BAE cells were plated in 8-well coverglass bottom chambers. After 48 h, when the cells were 50 and 70% confluent, the fluoresceinylated HHK4b:DNA:liposome complexes (prepared as described above) and/or 0.5 mg/ml AlexaFluoR 594 dextran (Molecular Probes) were added to cells. Two hours later, cells were incubated with 15% FCS in OptiM for 10 min and washed with PBS. These probes were observed with the LSM410 using 488 nm excitation and 515–545 nm emission filters for fluorescein, and 568 nm excitation and a 590 nm long pass emission filter for AlexaFluoR 594.
Stability of the polymer:DNA complexes at different pH
First, 200 µl of MagnaBind™ streptavidin beads (Pierce, Rockford, IL) were washed with 3× 200 µl vol of PBS, magnetically separated using an external magnetic field (MPC-E-1, Dynal, Norway) and aliquots were placed in two fresh tubes; then 3 µg of biotinylated DNA in 20 µl of Tris–EDTA pH 7.4 was added to the washed beads and incubation for 2 h was continued at room temperature. Biotinylated DNA was prepared using 0.75 µg of Biotin-Chem-link (Roche, Indianapolis, IN) to label 100 µg of PCI-Luc. At the end of the incubation, the beads with the attached biotinylated DNA were washed with 3× 200 µl vol of PBS, then 15 nmol of fluoresceinylated HK or 3 nmol of fluoresceinylated HHK4b was added to the DNA-bound beads, and incubated for another 2 h. After washing the beads three times with aliquots of PBS, both HK:DNA- and HHK4b:DNA-bound beads were placed in two fresh tubes. MES buffer (pH 6.5 or 4.5) was added to each of them. Thirty minutes later, the supernatant in each tube was collected and the free fluorescent HK polymer (free HK) was measured with the CytoFluor 2350 multi-well reader. Then, the bound beads were washed twice with PBS and the washes were also collected. To strip off the remainder of the polymer from the DNA-labeled beads, 100 µl of 2 M sodium chloride was added to the bound beads for two 10-min time periods and the supernatant was collected (bound HK). Finally, additional PBS was added to each fraction so that the final volume was 300 µl. The fluorescence intensity of these fractions was measured at 520 nm by using CytoFluo 2350 at an excitation wavelength of 485 nm. To compensate for non-specific binding to streptavidin beads, similar measurements were made on beads with no bound DNA. The relative association of the polymer with DNA at pH 4.5 and 6.5 was calculated as follows: bound HK / total HK (free + bound) where HK represents the difference between HKpH 4.5 or 6.5, DNA:resin – HKpH 4.5 or 6.5, resin only. As a result, the greater the association number, the less polymer is released from DNA. These binding experiments of the different HK polymer with DNA were repeated three times.
RESULTS
The optimal gene-delivery carrier, linear or branched polymers in combination with liposomes, varies in MDA-MB-231 and BAE cells
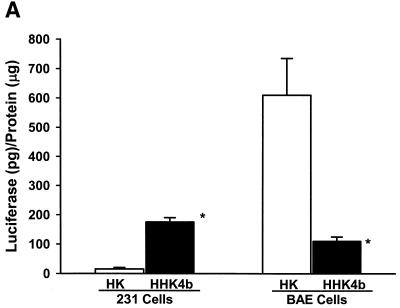
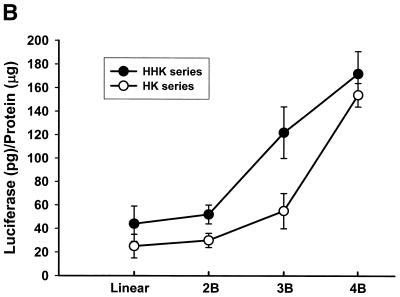
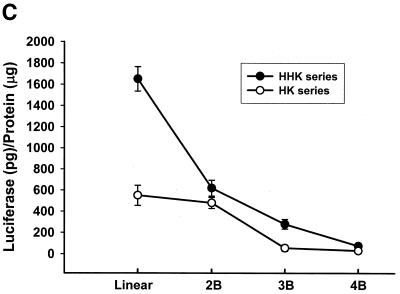
We have shown previously that the highly branched HHK4b polymer is more effective than the linear HK polymer in increasing transfection efficiency in several cell lines, including MDA-MB-231 cells (16). Surprisingly, the linear HK polymer in combination with liposomes was found to enhance gene expression significantly in the BAE cell line compared with the HHK4b:liposome combination (Fig. 1A). To further investigate the effect of polymer complexity on transfection in MDA-MB-231 and BAE cells, we compared eight polymers with various degrees of branching derived from the HK or HHK series (HK series: HK, HK2b, HK3b, HK4b; HHK series: HHK, HHK2b, HHK3b and HHK4b). In MDA-MB-231 cells, there was a strong positive correlation between polymers with increasing branching and transfection (Fig. 1B) (for HHK series, r = 0.97, P = 0.015). In contrast, a different pattern of transfection with branching polymers was observed in BAE cells (for HHK series, r = –0.94, P = 0.029). We found that polymers with greater complexity correlated inversely with transfection (Fig. 1C). In BAE cells, the linear HHK polymer enhanced transfection 24-fold more than the HK4B polymer. Furthermore, the maximal percent difference in transfection enhancement in BAE cells was observed between the linear HHK and HK polymers (64.5%), whereas in MDA-MB-231 cells, the maximal difference in enhancement was between the HHK3b and HK3B polymer (39%).
Figure 1.
Effect of linear and branched derivatives of histidine–lysine co-polymers on transfection efficiency in MDA-MB-231 and BAE cells. The co-polymers were mixed with 0.75 µg of DNA (PCI-luc) before 1.5 µg of liposomes was added; 24–48 h after transfection, luciferase activity was determined. (A) Transfection efficiency of 7.5 nmol of the linear HK or 0.375 nmol of the highly branched HHK4b in MDA-MB-231 and BAE cells (*P = 0.05, unpaired t-test). (B) Transfection efficiency of four branched HK polymers (linear HK, HK2b, HK3b and HK4b) or four HHK polymers (linear HHK, HHK2b, HHK3b and HHK4b) at concentrations of 7.5, 0.375, 0.375 and 0.375 nmol in MDA-MB-231 cells. (C) Similarly, transfection efficiency of polymers as described in (B) for BAE cells. The data represent the mean ± SEM of luciferase at each concentration of polymer (n = 3).
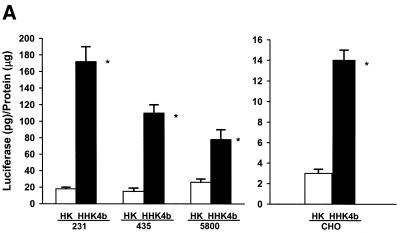
Different gene expression patterns with HK and HHK4b polymers observed in several cell lines
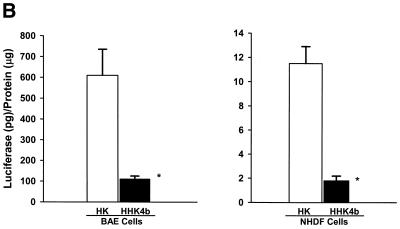
In addition to MDA-MB-231 and BAE cells that showed discrepant polymer transfection patterns, we screened several cell lines and determined that cells could be separated into two groups according to their response to HK and HHK4b. One group of cells (MDA-MB-435, CRL-5800 and CHO) behaved similarly to MDA-MB-231 cells. That is, the highly branched HHK4b polymer was more effective than the linear HK in enhancing the transfection efficiency of liposomes (Fig. 2A). On the other hand, linear HK was more efficient than HHK4b in increasing gene expression in two primary cell lines (BAE and NHDF cells) (Fig. 2B). Notably, we observed that the optimal HK carrier (linear or branched) for a particular cell did not change over a wide range of concentrations tested. Similar to other investigators, we investigated if bafilomycin, a H+ vacuolar proton pump inhibitor, could affect transfection and increase our understanding of polymer complexity and transfection. With these bafilomycin studies, however, we found marked inter-experimental variation in the transfection results in several cell lines.
Figure 2.
Comparison of gene expression in various cell lines with HK or HHK4b in combination with liposomes. After the HK (7.5 nmol) or HHK4b (0.375 nmol) polymers were mixed with DNA and liposomes, these complexes were incubated with the different cell lines for 4 h. Luciferase activity was measured 24–48 h later. Transfection efficiency was compared in the transformed cell lines, MDA-MB-231, MDA-MB-435, CHO and CRL-5800 (A), as well as the primary cell lines, BAE and NHDF (B). *P < 0.05, HHK4b versus HK, unpaired t-test. The data represent the mean ± SEM of luciferase at each concentration of polymer (n = 3).
Uptake is not a major factor in the transfection differences in MDA-MB-231 and BAE cells by HK and HHK4b polymers
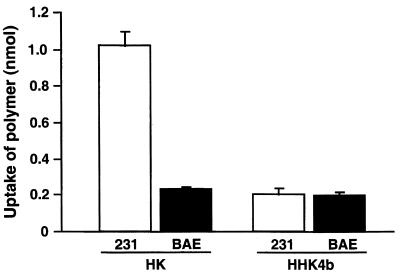
As differential uptake of HK or HHK4b complexes in MDA-MB-231 and BAE cells may explain the differences in optimal carriers, we compared the quantities of these complexes taken into these cells in 30 min (Fig. 3). The linear HK complexes had a higher rate of uptake in BAE and in MDA-MB-231 cells than did HHK4b complexes. In contrast to the uptake results, the HHK4b enhanced gene expression significantly in MDA-MB-231 cells compared with the linear HK polymer. The amount of uptake at 4 h for the HK and HHK4b complexes was similar to the 30 min measurement (data not shown). Confocal microscopy of the cells exposed to the labeled HK and HHK4b complexes for 30 min showed intracellular accumulation in discrete regions consistent with entrapment in endocytic vesicles (data not shown). As the uptake of these complexes in MDA-MB-231 cells did not correlate with the optimal transfection polymer, uptake does not appear to be a major factor for the different optimal carriers in either cell line.
Figure 3.
Uptake of HK and HHK4b-associated complexes in BAE and MDA-MB-231 cells. The fluoresceinylated polymers (7.5 nmol HK or 0.375 nmol HHK4b) were initially incubated with plasmid DNA (0.75 µg) for 30 min in OptiM, and cationic liposomes (1.5 µg) were then added to form the ternary complex. BAE and MDA-MB-231 cells were incubated with the complexes for 30 min as described in the Materials and Methods. Cell fluorescent density was recorded at 520 nm by using the fluorescent multi-well reader at an excitation wavelength of 485 nm. Data represent the mean ± SEM of replicate experiments.
The pH of endocytic vesicles correlates with the effects of HK and HHK4b on gene expression
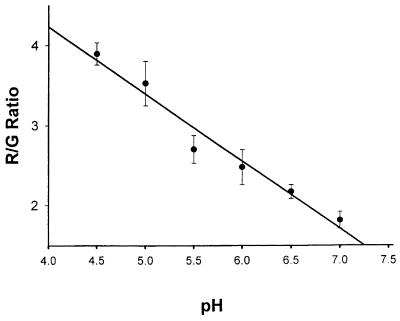
Once uptake of the transfection complex occurs by endocytosis, translocation of DNA from endosomes to cytosol plays an important role in transfection efficiency. Endosomal pH, by altering the affinity of DNA binding to the HK-containing polymer, will probably affect this translocation and, thus, transfection efficiency. We investigated whether cells in which the highly branched HHK4b is the optimal polymer had a higher endocytic pH compared with the cells optimally transfected with the linear polymer. As a result, we examined the pH of endocytic vesicles, using the pH-sensitive dye, LysoSensor Yellow/Blue. This dextran-linked dye accumulates in endocytic vesicles (endosomes and lysosomes) through endocytosis and exhibits pH-dependent dual-emission spectra. At acidic pH, the dye emits maximally at 530 nm, whereas at neutral pH the dye emits maximally at 450 nm. Thus, the ratio of 530 (assigned R) to 450 nm (assigned G) provides an accurate measurement of endocytic pH. In comparing these ratiometric measurements with those obtained from pH standards, the LysoSensor Yellow/Blue dye can distinguish pH values ranging from 4.5 to 7.0 with an accuracy of ∼0.5 U (Fig. 4).
Figure 4.
Standard calibration curve for the LysoSensor Yellow/Blue pH indicator dye. CHO cells were incubated for 12 h with LysoSensor Yellow/Blue dextran, and equilibrated with MES calibration buffers (pH from 4.5 to 7.0) containing nigericin and monensin. Indicator emissions at 450 and 530 nm from endocytic vesicles were measured with confocal microscopy using a 360 nm excitation wavelength. The 530/450 nm fluorescence ratio (R/G) of these endocytic vesicles was measured as described in the Materials and Methods.
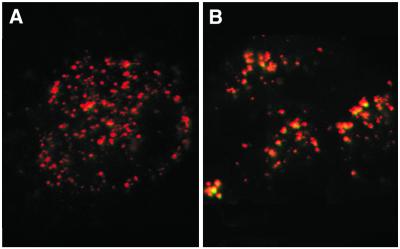
After preparing the standard pH calibration curve, the pH of endocytic vesicles was calculated in several cell lines. Since endosomes increase their acidity progressively until they merge with lysosomes, we not surprisingly observed some heterogeneity in the pH of endocytic vesicles within a cell (Fig. 5). Nevertheless, the primary BAE cell line with a higher 530/450 nm (R/G) ratio had a significantly lower pH (pH 4.48) than MDA-MB-231 cells (pH 6.19) (Fig. 5 and Table 1). In addition to BAE cells, the primary cell line, NHDF, had a significantly lower pH (pH 4.99) than the transformed MDA-MB-231, CHO (pH 6.57) and CRL-5800 (pH 6.86) cells. As there may be several endocytic entry pathways in cells (e.g. clathrin-dependent and caveolae), we also determined whether dextran-conjugated dyes co-localized with HK-containing complexes; we found that HK-containing complexes and the dextran dyes co-localized in endocytic vesicles (Fig. 6). Thus, the data suggest that the pH of endocytic vesicles as measured with dextran-linked dyes (LysoSensor Yellow/Blue and DM-NERF) is similar to the pH milieu surrounding the HK-containing complexes.
Figure 5.
Endocytic pH as indicated by LysoSensor Yellow/Blue Dextran dye in BAE and MDA-MB-231 cells. After BAE and 231 cells were incubated for 12 h with the indicator dye, these cells were examined with confocal microscopy. The emissions 530 and 450 nm were assigned the colors red (R) and green (G), respectively. The more acidic the vesicles of the endocytic pathway, the greater the R/G ratio. See Materials and Methods for further details. These results show that the endocytic vesicles in the primary BAE cell line (A) are substantially lower than the pH of those in the transformed MDA-MB-231 cell line (B).
Table 1. The pH of endocytic vesicles from different cell lines.
| Cells | Endocytic pH | Optimal polymer linear HK versus HHK4b |
|---|---|---|
| BAE | 4.48 ± 0.156 | Linear HK |
| NHDF | 4.99 ± 0.182 | Linear HK |
| MDA-MB-231 | 6.19 ± 0.238 | HHK4b |
| MDA-MB-435 | 6.07 ± 0.283a | HHK4b |
| CHO | 6.57 ± 0.168 | HHK4b |
| CRL-5800 | 6.86 ± 0.140 | HHK4b |
After cells became 30–50% confluent, LysoSensor Yellow/Blue or NERF dextran was added and incubated with the cells for 12 h. Excess dye was removed with cold PBS. The labeled cells were observed with a laser confocal microscope. Dual emission was measured at 450 and 530 nm for LysoSensor Yellow/Blue-labeled cells. The 530/450 nm fluorescence ratio was calculated and pH was obtained from the pH calibration curve.
aBecause of interfering autofluorescence in MDA-MB-435, DM-NERF dye was used to measure the pH of endocytic vesicles in these cells. In MDA-MB-435 cells labeled with DM-NERF, dual excitations were performed at 488/514 nm with their respective emissions recorded at 560 nm. The values are expressed as mean ± SEM of these experiments (n = 3). See Materials and Methods for further details.
Figure 6.
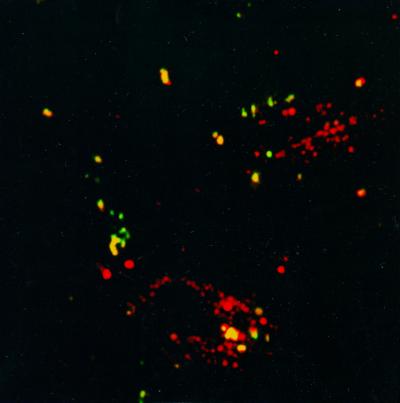
Co-localization of dextran and polymer:liposome:DNA complexes within endocytic vesicles. After BAE cells were 50 and 70% confluent, fluoresceinylated HHK4b:DNA:liposome complexes and AlexaFluoR 594 dextran were added to cells. Two hours later, cells were incubated with 15% FCS in OptiM for 10 min and washed with PBS. These probes were observed with the LSM410 using 488 nm excitation and 515–545 nm emission filters for fluorescein (complexes appear green), and 568 nm excitation and a 590 nm long pass emission filter for AlexaFluoR 594 (dextran appears red). The polymer complexes co-localized with the dextran within the endocytic vesicles.
We were unable to evaluate the pH of the MDA-MB-435 cell line with the LysoSensor Yellow/Blue dye because of the low endocytic fluorescence intensity and the green autofluorescence of these cells. Therefore, we used the pH-sensitive dye DM-NERF to measure the endocytic pH in MDA-MB-435 cells. DM-NERF is a dextran-linked fluorescent dye with dual-excitation peaks at 514/488 nm; the ratio of emissions at 560 nm resulting from these excitation wavelengths correlates with pH. This approach established that MDA-MB-435 cells (pH 6.07) have a higher endosomal pH compared with BAE cells (Table 1).
Taken together, the data show that in cell lines in which the optimal HK polymer was linear, the pH was lower in these endocytic vesicles. Conversely, in cell lines in which the branched polymer was optimal, the pH was found to be significantly higher. The association between linear or branched polymers and the pH of vesicles from the endocytic pathway was significant (P = 0.0058, r2 = 0.8792). We expect that delineation of the various endocytic pathways and precise location within the endosomes in which DNA dissociates may further improve the correlation. Current methodologies do not allow simultaneous identification of the specific location in the endocytic pathway and measurement of the pH of acidic vesicles. Despite the technical limitations and the complexity of the endocytosis pathway that could mask relationships between endocytic pH and transfection, there was a statistically significant association between the branching of the polymer and the endocytic pH.
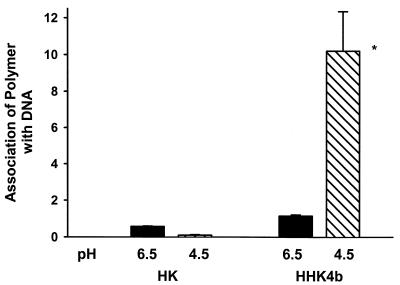
Stability of the polymer:DNA complex is pH dependent
As the optimal HK polymer appears to depend on the endosomal pH, we measured the binding of the polymer to DNA at pH 6.5 and 4.5. The more positive the association number, the greater the affinity between the polymer and DNA at that particular pH. Moreover, the association number correlated inversely with the amount of polymer released from the DNA. As shown in Figure 7, we found that compared with the linear polymer, the branched polymer was released from DNA at a reduced rate. More important, the binding of the branched polymer, HHK4b, was much stronger at a highly acidic pH than at a more basic pH. This is consistent with our hypothesis that at more acidic pH the branched polymer binds to the DNA more tightly and, as a result, there is decreased gene expression because endosomes merge with lytic lysosomes. Similar measurements with the linear HK polymer further support the importance of the relationship between polymer:DNA binding and endosomal pH. Compared with the branched HHK4b polymer, the linear HK polymer binds with significantly less affinity to DNA at both pH 4.5 and 6.5. Furthermore, the linear HK polymer binds to DNA with less affinity at pH 4.5 than at 6.5 (Fig. 7) or 7.4. Thus, at a strongly acidic endosomal pH, the linear HK polymer would readily release the DNA, thereby increasing gene expression compared with the highly branched HHK4b polymer.
Figure 7.
Stability of polymer:DNA complexes at pH 6.5 and 4.5. The amount of HK or HHK4b associated with DNA-bound beads was measured after 30 min of incubation at pH 6.5 and 4.5. The greater association numbers correlate with reduced release of the polymer from the DNA. The polymers were labeled with flurorescein and the amount of bound polymer was measured with a flurorescent microplate reader (485 nm excitation/530 nm emission). The graph indicates that the branched polymers bind DNA with substantially higher affinity at low pH than linear polymers. *P < 0.01, HHK4b (pH 4.5) versus HHK4b (pH 6.5) or HK (pH 6.5, 4.5).
DISCUSSION
When several HK polymers were tested as transfection agents on different cell lines, we recognized an additional layer of mechanistic complexity. Although the linear HK polymer stimulates plasmid transfection in all cell lines tested, different HK polymers stimulate transfection efficiency to varied degrees. The branched HK polymer is clearly superior to the linear HK polymer in many malignant or transformed cells, whereas the linear HK polymer is significantly better than the branched HK polymer at enhancing the transfection of plasmid DNA in two primary cell lines. Therefore, this study was designed to investigate the mechanisms that might explain the varied cell specificity of transfection enhancement with the linear and branched HK carriers.
One mechanism for different optimal carriers as gene-delivery agents may involve the interplay of the polymer:DNA binding affinities with the degree of acidity within the endosome. Our data showed a positive correlation between degree of branching and transfection efficiency in malignant cells; in contrast, there was a negative correlation between branching and transfection efficiency in primary cells. Although these findings suggest that binding between the polymers and DNA is involved in transfection enhancement, an additional factor affecting binding is required to explain the differential cellular transfection of HK polymers. Our experiments with fluorescent indicators showed that in primary cell lines, for which the optimal polymer is linear HK, the vesicles of the endocytic pathway were strongly acidic; in contrast, in malignant or transformed cells with a higher pH, the branched HK polymers were the optimal carriers. Our finding that endocytic vesicles of malignant cells are more basic than primary cell lines is in agreement with findings of other investigators (19,22). Nevertheless, more cell lines are required to establish that malignant or primary cell lines have a specific transfection profile with HK polymers. More importantly, these two groups of cells identified by different optimal HK transfection polymers have enabled us to recognize the relationship among endocytic pH, complexity of the polymer and transfection.
The mechanism underlying the varied endocytic pH between the cell lines in this and other studies is unknown. Acidification of endosomes and lysosomes is controlled by a vacuolar H(+) ATPase and chloride channels (23,24); in addition to specific inhibitors of the vacuolar type H(+) ATPase (e.g. bafilomycin), a number of factors (e.g. intracellular pH, ion composition, membrane potential) have been identified that may affect the pH of these endocytic vesicles (25–29). Currently, neither extrinsic factors (e.g. chloride concentration, intracellular pH) nor phenotypic variation in the proton pump and its associated ion channels can be dismissed as the cause for different endocytic pH in cells. Notably, the ability to regulate the endosomal pH may result in different optimal carriers within the same cell line.
A lower pH within endocytic vesicles of BAE and NHDF cells results in a higher percentage of histidines with a positive charge; consequently, the HK polymer is likely to bind with higher affinity to the DNA. Thus, with a pKa of 6.0 for histidine (30,31), 10% of the histidines in the HK polymer have a positive charge at pH 7.0, 50% of histidines have a charge at pH 6.0 and 90% of the histidines have a charge at pH 5. As the association between the branched HK polymer (HHK4b) and plasmid DNA may be sufficient to retard the dissociation within a more acidic endothelial endosomal vesicle, gene expression may be reduced because endosomes merge with the lytic lysosomes. This hypothesis was further supported by our binding data. In experiments using streptavidin beads:biotinylated plasmid DNA, the branched HHK4B polymer bound to DNA 20-fold more tightly at pH 4.5 compared with the linear HK polymer. As a result, the release of plasmid DNA from the linear HK polymer occurs readily; in contrast, the branched polymer releases DNA slowly at pH 4.5.
We have no doubt that there are additional levels of complexity essential for understanding transfection with the HK polymers. For example, at a more basic endocytic pH (e.g. pH 6–7) in which DNA dissociates from linear or branched HK polymers, other transfection-enhancing mechanisms are essential to enable the branched polymer to be a more effective carrier compared with the linear polymer. Two properties of branched polymers that might account for augmented transfection are increased DNA compaction and endosomal disruption. Increased condensation of DNA by cationic polymers is associated with increased transfection (32–34) and we found previously that the branched polymers decreased the size of liposome complexes by >2-fold compared with linear polymers (15,16). In addition, delineating the ability of the different HK polymers, once the DNA is released, to enhance fusion and disrupt endosomes may provide insight on these HK transfection polymers (35). We are currently investigating whether branched polymers once released from DNA disrupt endosomes significantly more than linear HK.
Although the size of these complexes (15,16) or enhanced endosomal disruption at acidic pH by the HK polymers may have important roles in gene expression, these mechanisms by themselves cannot explain the variation of optimal carriers in different cells. However, the binding and pH mechanisms postulated here are consistent and explain in part the various patterns of gene expression in different cells with these synthesized polymers. Notably, the transfection findings in this study were obtained with HK polymers (linear or branched) in combination with liposomes. Linear HK polymer in combination with liposomes was significantly more effective as a transfection carrier in certain cell lines compared with the branched HHK4b:liposome combination. Nevertheless, the linear HK polymers alone were very poor transfection carriers in all cells tested, whereas the branched HK carriers were effective carriers in cells that have endocytic vesicles with a higher pH. Thus, without liposomes the linear HK polymer would have been discounted as a transfection carrier in both endothelial cells and fibroblasts, and the relationship between branching and the pH of endocytic vesicles would have been difficult to discern.
These mechanistic studies may form the basis for developing improved carriers that are tailored for a particular cell type. In one primary cell line with more acidic vesicles, the linear HHK polymer with a greater buffering capacity than the linear HK polymer was significantly more effective at enhancing transfection. Furthermore, by adding a histidine-rich tail to the linear HK polymer, preliminary results in BAE cells show that this polymer in combination with liposomes enhanced transfection by >8-fold compared with the linear HK polymer. On the basis of these studies, we are currently developing carriers that will target specific cell types and produce higher gene expression.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Pamela Talalay for her careful reading and useful comments concerning the manuscript. We thank Dr Nicholas Ambulos of the Maryland Biopolymer Laboratory for synthesizing the peptides in this study. This work was supported by the National Institutes of Health (CA70394).
REFERENCES
- 1.Felgner P.L., Gadek,T.R., Holm,M., Roman,R., Chan,H.S., Wenz,M., Northrop,J.P., Ringold,G.M. and Danielsen,H. (1987) Lipofection: a highly efficient, lipid-mediated DNA transfection procedure. Proc. Natl Acad. Sci. USA, 84, 7413–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behr J.P., Demeneix,B., Loeffler,J.P. and Perez-Mutul,J. (1989) Efficient gene transfer into mammalian primary endocrine cells with lipopolyamine-coated DNA. Proc. Natl Acad. Sci. USA, 86, 6982–6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malone R.W., Felgner,P.L. and Verma,I.M. (1989) Cationic liposome-mediated RNA transfection. Proc. Natl Acad. Sci. USA, 86, 6077–6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morris M.C., Chaloin,L., Heitz,F. and Divita,G. (2000) Translocating peptides and proteins and their use for gene delivery. Curr. Opin. Biotechnol., 11, 461–466. [DOI] [PubMed] [Google Scholar]
- 5.Zaitsev S.V., Haberland,A., Otto,A., Vorob’ev,V.I., Haller,H. and Bottger,M. (1997) H1 and HMG17 extracted from calf thymus nuclei are efficient DNA carriers in gene transfer. Gene Ther., 4, 586–592. [DOI] [PubMed] [Google Scholar]
- 6.Boussif O., Lezouac’h,F., Zanta,M.A., Mergny,M.D., Scherman,D., Demeneix,B. and Behr,J.P. (1995) A versatile vector for gene and oligonucleotide transfer into cell in culture and in vivo: polyethylenimine. Proc. Natl Acad. Sci. USA, 92, 7297–7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marshall J. (1995) The trouble with vectors. Science, 269, 1051–1055. [Google Scholar]
- 8.Xu M., Kumar,D., Srinivas,S., DeTolla,L.J., Yu,S.F., Stass,S.A. and Mixson,A.J. (1996) Parenteral gene therapy with p53 inhibits human breast tumors in vivo through a bystander mechanism without evidence of toxicity. Hum. Gene Ther., 8, 177–185. [DOI] [PubMed] [Google Scholar]
- 9.Haensler J. and Szoka,F.C. (1993) Polyamidoamine cascade polymers mediate efficient transfection of cells in culture. Bioconjug. Chem., 4, 372–379. [DOI] [PubMed] [Google Scholar]
- 10.Ferrari S., Moro,E., Pettenazzo,A., Behr,J.P., Zacchello,F. and Scarpa,M. (1997) ExGen 500 is an efficient vector for gene delivery to lung epithelial cells in vitro and in vivo. Gene Ther., 4, 1100–1106. [DOI] [PubMed] [Google Scholar]
- 11.Kukowska-Latallo J.F., Bielinska,A.U., Johnson,J., Spindler,R., Tomalia,D.A. and Baker,J.R.,Jr (1996) Efficient transfer of genetic material into mammalian cells using Starburst polyamidoamine dendrimers. Proc. Natl Acad. Sci. USA, 93, 4897–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Midoux P., Kichler,A., Boutin,V., Maurizot,J.C. and Monsigny,M. (1998) Membrane permeabilization and efficient gene transfer by a peptide containing several histidines. Bioconjug. Chem., 9, 260–267. [DOI] [PubMed] [Google Scholar]
- 13.Putnam D., Gentry,C.A., Pack,D.W. and Langer,R. (2001) Polymer-based gene delivery with low cytotoxicity by a unique balance of side-chain termini. Proc. Natl Acad. Sci. USA, 98, 1200–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kichler A., Leborgne,C., Coeytaux,E. and Danos,O. (2001) Polyethylenimine-mediated gene delivery: a mechanistic study. J. Gene Med., 3, 135–144. [DOI] [PubMed] [Google Scholar]
- 15.Chen Q.R., Zhang,L., Stass,S.A. and Mixson,A.J. (2000) Co-polymer of histidine and lysine markedly enhances transfection of liposomes. Gene Ther., 7, 698–704. [DOI] [PubMed] [Google Scholar]
- 16.Chen Q.R., Zhang,L., Stass,S.A. and Mixson,A.J. (2001) Branched co-polymers of histidine and lysine are efficient carriers of plasmids. Nucleic Acids Res., 29, 1334–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Q.R., Kumar,D., Stass,S.A. and Mixson,A.J. (1999) Liposomes complexed to plasmids encoding angiostatin and endostatin inhibit breast cancer in nude mice. Cancer Res., 59, 3308–3312. [PubMed] [Google Scholar]
- 18.Diwu Z., Chen,C.S., Zhang,C., Klaubert,D.H. and Haugland,R.P. (1999) A novel acidotropic pH indicator and its potential application in labeling acidic organelles of live cells. Chem. Biol., 6, 411–418. [DOI] [PubMed] [Google Scholar]
- 19.Jiang L.W., Maher,V.M., McCormick,J.J. and Schindler,M. (1990) Alkalinization of the lysosomes is correlated with ras transformation of murine and human fibroblasts. J. Biol. Chem., 265, 4775–4777. [PubMed] [Google Scholar]
- 20.Diwu Z. and Lown,J.W. (1993) Photosensitization with anticancer agents. 16. The photo-oxidation of hypocrellin A. A mechanism study using 18O labelling. J. Photochem. Photobiol. B, 18, 145–154. [DOI] [PubMed] [Google Scholar]
- 21.Lee R.J., Wang,S. and Low,P.S. (1996) Measurement of endosome pH following folate receptor-mediated endocytosis. Biochim. Biophys. Acta, 1312, 237–242. [DOI] [PubMed] [Google Scholar]
- 22.Altan N., Chen,Y., Schindler,M. and Simon,S.M. (1998) Defective acidification in human breast tumor cells and implications for chemotherapy. J. Exp. Med., 187, 1583–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cain C.C., Sipe,D.M. and Murphy,R.F. (1989) Regulation of endocytic pH by the Na+,K+-ATPase in living cells. Proc. Natl Acad. Sci. USA, 86, 544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Dyke R.W. (1996) Acidification of lysosomes and endosomes. Subcell. Biochem., 27, 331–360. [DOI] [PubMed] [Google Scholar]
- 25.Sundler R. (1997) Lysosomal and cytosolic pH as regulators of exocytosis in mouse macrophages. Acta Physiol. Scand., 161, 553–556. [DOI] [PubMed] [Google Scholar]
- 26.Tapper H. and Sundler,R. (1995) Bafilomycin A1 inhibits lysosomal, phagosomal and plasma membrane H(+)-ATPase and induces lysosomal enzyme secretion in macrophages. J. Cell Physiol., 163, 137–144. [DOI] [PubMed] [Google Scholar]
- 27.Tapper H. and Sundler,R. (1990) Role of lysosomal and cytosolic pH in the regulation of macrophage lysosomal enzyme secretion. Biochem. J., 272, 407–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lei G.H., Piao,Y.J., Wu,J.C., Bao,Y.Y., Huang,H. and Zhang,W. (1998) Effects of receptor-mediated endocytosis on macrophage membrane potential, cytosolic pH and lysosomal pH. Sheng Li Xue Bao, 50, 111–114. [PubMed] [Google Scholar]
- 29.Rybak S.L., Lanni,F. and Murphy,R.F. (1997) Theoretical considerations on the role of membrane potential in the regulation of endosomal pH. Biophys. J., 73, 674–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patchornik A., Berger,A. and Katchalski,E. (1957) Poly-L-histidine. J. Am. Chem. Soc., 79, 5227–5236. [Google Scholar]
- 31.Norland K.S., Gasman,G.D., Katchalsky,E. and Blout,E.R. (1963) Some optical properties of poly-l-benzyl-L-histidine and poly-L-histidine. Biopolymers, 1, 277–294. [Google Scholar]
- 32.Sorgi F.L., Bhattacharya,S. and Huang,L. (1997) Protamine sulfate enhances lipid-mediated gene transfer. Gene Ther., 4, 961–968. [DOI] [PubMed] [Google Scholar]
- 33.Gao X. and Huang,L. (1996) Potentiation of cationic liposome-mediated gene delivery by polycations. Biochemistry, 35, 1027–1036. [DOI] [PubMed] [Google Scholar]
- 34.Li S., Rizzo,M.A., Bhattacharya,S. and Huang,L. (1998) Characterization of cationic lipid-protamine-DNA (LPD) complexes for intravenous gene delivery. Gene Ther., 5, 930–937. [DOI] [PubMed] [Google Scholar]
- 35.Wang C.Y. and Huang,L. (1984) Polyhistidine mediates an acid-dependent fusion of negatively charged liposomes. Biochemistry, 23, 4409–4416. [DOI] [PubMed] [Google Scholar]