Abstract
There are about 50 copies of each chromosome in the somatic macronucleus of the ciliated protozoan Tetrahymena. Approximately 0.8% of the adenine residues in the macronuclear DNA of Tetrahymena are methylated to N6-methyladenine. The degree of methylation varies between sites from a very low percentage to >90%. In this study a correlation was found between nucleosome positioning and DNA methylation. Eight GATC sites with different levels of methylation were examined. There was a direct correlation between the degree of methylation and proximity to linker DNA at these sites. Although methylation occurs preferentially in linker DNA, the patterns and extent of methylation in a histone H1 knockout strain were virtually indistinguishable from those in wild-type cells.
INTRODUCTION
The nuclear genomes of various eukaryotes differ widely with respect to methylation of the DNA. The genomes of some eukaryotes, including Caenorhabditis and Saccharomyces, have little or no detectable methylation. The Drosophila genome, which was long thought to lack methylation, has recently been shown to have low levels of 5-methylcytosine (1). Mammalian and plant genomes have significant levels of 5-methylcytosine. Cytosine methylation is essential for mammalian development (2), where its major biological roles are the regulation of gene expression (3) and transcriptional silencing of transposons (4). Some algae, including Chlamydomonas (5) and Volvox, have both 5-methylcytosine and N6-methyladenine in their nuclear DNA. In Volvox methylcytosine has a role in transgene silencing and the function of adenine methylation is unknown (6). The nuclear genomes of the ciliated protozoa Tetrahymena and Paramecium are unusual for eukaryotes in that there is no detectable methylcytosine, but a fraction of the adenine residues are methylated to N6-methyladenine (7,8).
In the somatic macronucleus of Tetrahymena ∼0.8% of the adenine residues are methylated. Because the genome is ∼75% A-T, this amounts to approximately one methylated adenine per 165 bp of DNA. In vivo, methylation occurs at the sequence 5′-NAT-3′ (9). A small fraction of the methylated adenines occur in the sequence GATC, and can be assayed by digestion with restriction enzyme isoschizomers that are differentially sensitive to adenine methylation.
Methylation in Tetrahymena is site specific (10). Some sites are uniformly methylated at most or all of the 50 copies of the genome in the macronucleus. Others are only partially methylated, i.e. both strands are modified on a fraction of the molecules that is characteristic of the site. The pattern of methylation is consistent between clonal cell lines (11) and does not vary detectably with the physiological state of the cell or the transcriptional activity of nearby genes (12).
Sequence of the target site is not sufficient for DNA methylation in Tetrahymena. In transformation experiments a site that is uniformly methylated on the chromosome was not methylated when a fragment containing the site was inserted into the rDNA minichromosome (13). This was true regardless of the methylation state of the DNA at the time it was introduced into the cell. Thus methylation in Tetrahymena is subject to a position effect.
Previous analysis of bulk chromatin suggested that there was a relationship between chromatin structure and DNA methylation in Tetrahymena. The proportion of methylated adenine residues was significantly decreased in DNA extracted from nuclei following digestion with micrococcal nuclease (MNase), suggesting that the methylation occurred preferentially in linker DNA (14). The goal of this study was to investigate that question in more detail, by determining the location of specific methylated sites with respect to nucleosome positioning. Analysis of eight specific sites from three different regions of the genome showed that DNA methylation occurs preferentially in linker DNA. Nevertheless, internucleosomal methylation is independent of the presence or absence of the H1 linker histone.
MATERIALS AND METHODS
Accession numbers
The sequences of the genomic clones have been deposited in the GenBank database. The accession numbers are: cyd1, L34029; cyd2, AF416723; Tlr1rB-B, L25254.
Strains
CU428.1, Mpr/Mpr [6-methylpurine-sensitive (6-mp-s), VII] of inbreeding line B was obtained from Peter Bruns. ΔH1, a knockout strain of the macronuclear HHO gene (15), was a generous gift of X. Shen and M. A. Gorovsky. Cells were grown in PPYS (16) supplemented with penicillin/streptomycin. For ΔH1, 200 µg/ml paromomycin was added to the medium for both strain maintenance and cell culture.
Indirect end-labeling
DNA isolation, standard Southern blots, chromatin preparation and MNase digestions were done by standard methods as described previously (17). For indirect end-labeling experiments, small fragments of DNA were separated on 1.8% agarose gels in TEA buffer (40 mM Tris–acetate, pH 7.8, 1 mM EDTA). The gels were pre-run for 450 V h, with recirculation of the buffer, to remove residue from the agarose which otherwise produced a hybridization artifact in the top half of the gel. After the pre-run, the buffer in the chamber was replaced with fresh buffer. The samples were loaded and the gels were run at 80 V until the bromophenol blue dye front was 2 cm from the bottom of the gel. The gels were processed to maximize the retention and hybridization efficiency of small DNA fragments (18). Regions of the gel that were to be blotted were not stained. The lane containing the DNA size markers was cut from the gel and stained with ethidium bromide to monitor separation of the fragments. The DNA was denatured by soaking the gel for 30 min in 0.4 N NaOH and transferred to Genescreen plus hybridization membrane (NEN Life Sciences) in the same buffer for 4 h without a buffer reservoir. The filters were rinsed twice for 1–2 min in 2× standard saline citrate (SSC), pH 7.0, to neutralize the pH and the DNA was crosslinked to the filter with a Stratagene crosslinker.
Short fragments to be used as probes for indirect end-labeling experiments were synthesized by PCR using primers between the primary restriction site and the proximal GATC site (Table 1). The probes were radiolabeled by a random primer labeling protocol, substituting the PCR primers for random primers.
Table 1. Primers for PCR synthesis of indirect end-labeling probes.
RESULTS
Adenine methylation occurs preferentially in linker DNA
Nucleosomes are specifically positioned with respect to DNA sequence in the macronuclear genome of Tetrahymena. This was originally suggested by bulk genome analysis (19) and was recently confirmed by indirect end-labeling (17). Alignment of the nucleosome positioning maps with the location of methylated and unmethylated GATC sites, as determined by genomic restriction mapping and DNA sequence analysis, suggested that the methylated GATC sites were preferentially located in linker DNA and unmethylated sites were generally in DNA associated with nucleosome cores. In order to directly test the hypothesis that there is a relationship between chromatin structure and DNA methylation, the positions of nucleosomes relative to methylated and unmethylated GATC sites were mapped by indirect end labeling (20).
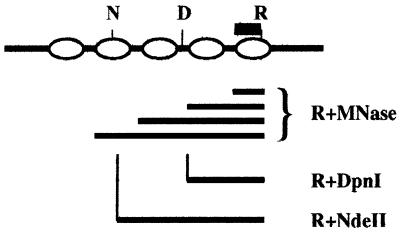
The strategy for mapping of methylated GATC sites relative to nucleosome position is shown in Figure 1. Total nuclei were isolated from growing Tetrahymena cells and subjected to limited digestion with MNase. DNA was subsequently isolated from the treated nuclei and the fragments were displayed on agarose gels, producing the characteristic ladder pattern of DNA fragments in multiples of ∼200 bp, due to preferential digestion of the DNA in the linker region of chromatin. Although no attempt was made to remove micronuclei, the pattern is typical of the nucleosome repeat length in macronuclear DNA because macronuclei contain about 10 times as much DNA as micronuclei and macronuclei are preferentially recovered at the centrifugation step of nuclear isolation.
Figure 1.
Strategy for localization of methylated adenine relative to nucleosome position via indirect end labeling. Lines, macronuclear DNA; ovals, nucleosomes; bar, probe for indirect end labeling; R, primary restriction site; D, DpnI; N, NdeII; bracketed lines, fragments generated by digestion with MNase and R. Fragments generated by digestion with the primary restriction enzyme and an enzyme that recognizes a site in linker DNA co-migrate on an agarose gel with one of the fragments in the MNase ladder digested with the primary restriction enzyme. Fragments generated by digestion with the primary restriction enzyme and an enzyme that recognizes a site in nucleosome core DNA migrate between ladder fragments.
An aliquot of the purified MNase-digested DNA was subsequently digested to completion with an appropriate primary restriction enzyme, designated R in Figure 1, and Southern blots of the DNA were hybridized with probes complementary to sequences very close to the restriction site. The fragments detected by autoradiography of the film were those with the primary restriction site at one end and the other end in linker DNA. The ladder pattern of the DNA digested with the restriction enzyme may be shifted relative to that of the MNase fragments, depending on the position of the restriction site with respect to linker DNA.
In order to determine the position of methylated and unmethylated GATC sites relative to linker DNA, macronuclear DNA was digested with the same primary restriction enzyme plus enzymes that are sensitive to adenine methylation. DpnI, for example, digests DNA at the palindromic site GATC only if the adenine residues on both strands are methylated. Thus digestion with DpnI as the second enzyme generates fragments with the primary restriction site at one end and a site containing methyladenine at the other end. If the methylated GATC sites are located in linker DNA, these fragments are expected to co-migrate with those generated by the restriction enzyme and MNase. Similarly, NdeII digests DNA only at unmethylated GATC sites. Thus if unmethylated sites are preferentially located in nucleosome core DNA, fragments generated by digestion with the primary restriction enzyme and NdeII will migrate between the fragments in the ladder generated with MNase and the primary restriction enzyme.
Although the average internucleosomal distance in the macronuclei of Tetrahymena is ∼200 bp (21), the length of linker DNA between adjacent nucleosomes varies considerably (17). Indirect end labeling obviates the necessity to estimate the precise position of nucleosomes and, by assaying co-migration of DNA fragments, tests directly for localization of methylated restriction sites in linker DNA.
Judging from the size of restriction fragments in restricted genomic DNA, the vast majority of GATC sites in the Tetrahymena macronucleus are unmethylated. For this study indirect end-labeling analysis was done on sites that were previously identified as methylated and associated unmethylated or partially methylated sites. cyd1 and cyd2 were identified in a screen for methylated GATC sites (11). Tlr1.rB-B was originally isolated not on the basis of methylation state, but because it contains the right boundary of an element that is deleted from the macronuclear genome (22).
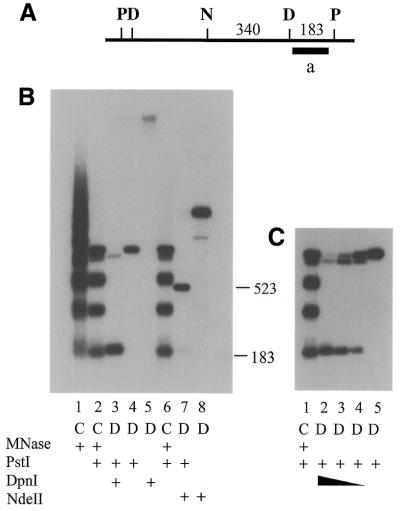
The methylated site cyd2 (23) and two additional GATC sites are present on a 0.89 kb PstI fragment (Fig. 2A). Since the right end PstI site is located in or near linker DNA (17), digestion with PstI produced only a slight shift in the ladder of MNase fragments (Fig. 2B). Digestion of the MNase fragments with PstI was apparently complete, because no ladder fragments >0.89 kb were detected. In macronuclear DNA digested with DpnI the major fragment detected with the indirect end-labeling probe was a large fragment that extends from the cyd2 site to a site more than 6 kb to the right in Figure 2A. Two fragments were seen in DNA digested with both DpnI and PstI. The relative intensity of the two fragments indicated that the vast majority of the molecules were methylated at the cyd2 site. The 183 bp PstI–DpnI fragment co-migrated with the first fragment in the MNase/PstI ladder, showing that the methylated GATC site, cyd2, is in linker DNA.
Figure 2.
Indirect end-labeling of the cyd2 region. (A) Restriction map of the cyd2 region. P, PstI; D, DpnI; N, NdeII. The methylated site closest to probe a is the site formerly designated cyd2. Numbers above the line indicate the distance in base pairs between restriction sites where the DNA has been cloned and sequenced. The black bar indicates the hybridization probe for the experiments in (B) and (C). (B) Indirect end labeling of chromatin and DNA in the cyd2 region. DNA isolated from macronuclei (lanes D) was digested with PstI, DpnI and/or NdeII. Chromatin (lanes C) was digested with micrococcal nuclease and the DNA was purified before digestion with restriction enzymes. (C) Partial digestion of DNA from the cyd2 region. Labeling is the same as in (B). The DpnI used to digest the DNA in lanes 3 and 4 was diluted 1:10 and 1:100, respectively, relative to that in lane 2. The numbers to the right of (B) indicate the sizes of the restriction fragments in lanes 3 and 7.
A small proportion of the macronuclear DNA molecules were resistant to digestion with DpnI at the cyd2 site because they were unmethylated or hemi-methylated (11). A faint band was present migrating slightly faster than the PstI fragment, suggesting that there was a second methylated GATC site between cyd2 and the left PstI site (Fig. 2A). Since there are no PstI fragments detectable in this lane, most or all of the molecules that are unmethylated at cyd2 are methylated at this second GATC site. To test this hypothesis, DNA digested with PstI was partially digested with DpnI (Fig. 2C). As expected, with incomplete digestion at cyd2 the relative intensity of the larger PstI–DpnI fragment increased. Since that fragment co-migrated with one of the MNase/PstI ladder fragments, the second DpnI site is also located in linker DNA.
The GATC site between the two methylated sites mapped in Figure 2B is unmethylated. To determine the position of the unmethylated GATC site relative to nucleosomes, the positions of fragments in the MNase/PstI ladder were compared to those of macronuclear DNA digested with PstI and NdeII (Fig. 2B). Digestion of the PstI fragment with NdeII generated a 522 bp PstI–NdeII fragment. That fragment migrated between the second and third bands of the PstI/MNase ladder, indicating that the unmethylated GATC site was in DNA associated with the nucleosome core. The major band in DNA digested with NdeII alone places an unmethylated GATC site ∼800 bp to the right of the PstI site. The faint band in DNA digested with NdeII is the size expected for the fragment between the right-most NdeII site and digestion of a small proportion of the molecules at cyd2. This was in accord with the presence of the faint band in DNA digested with PstI + DpnI and was ascribed to the presence of a few molecules that are unmethylated at cyd2.
In summary, the two fragments generated by digestion of DNA with PstI and DpnI in Figure 2 are closely aligned with the PstI/MNase fragments in the adjacent lanes, indicating that these methylated sites are in or near linker DNA. Conversely, the PstI–NdeII fragment does not co-migrate with any of the PstI/MNase fragments, suggesting that the unmethylated site is in nucleosome core DNA.
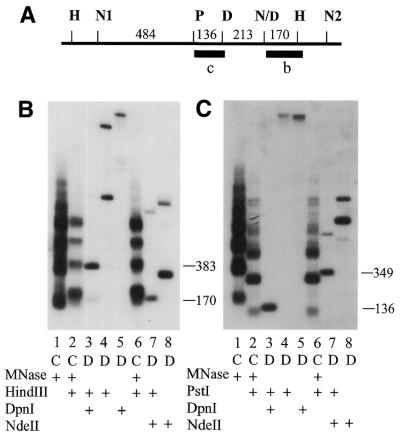
The data in Figure 3 show a close association of a methylated GATC site from a different region of the genome with linker DNA and indicate that proximity to the linker correlates with the degree of methylation at any particular site. For analysis of the cyd1 site, DNA was digested with MNase and HindIII and hybridized with probe b (Fig. 3A and B). The major band in Figure 3B, lane 3, co-migrated with the second ladder fragment in the DNA digested with MNase and HindIII, indicating that this methylated GATC site is located in linker DNA.
Figure 3.
Indirect end labeling of the cyd1 region. (A) Restriction map of the cyd1 region. H, HindIII; D, DpnI; N, NdeII; N/D; partially methylated site at which molecules that are methylated on both strands are digested by DpnI and unmethylated molecules are digested by NdeII. Bars indicate probes for the experiments shown in (B) and (C). (B) Indirect end labeling experiment of the cyd1 region in DNA digested with HindIII and hybridized with probe b. Arrangement of the lanes and labeling is as in Figure 2B. The larger band in lane 4 is due to partial digestion of the DNA in this lane with HindIII. (C) Indirect end-labeling of the cyd1 region in DNA digested with PstI and hybridized with probe c. The numbers to the right of (B) and (C) indicate the size of the restriction fragments in lanes 3 and 7.
A faint band was also present in lane 3 that migrated close to, but slightly below, the first MNase/HindIII ladder fragment, suggesting that there was a low level of methylation at the GATC site labeled N/D in Figure 3A. This was confirmed by digestion of the DNA with MNase and NdeII (lane 7). The major fragment of 171 bp is produced by digestion of molecules that are unmethylated at this site. However, there is an additional, larger fragment in this lane. This was consistent with the hypothesis that N/D is partially methylated. Molecules that were resistant to digestion at N/D due to methylation were found in fragments of 1015 bp resulting from digestion at N1 and HindIII.
The fragment resulting from digestion at HindIII and N/D runs slightly faster than the bottom fragment of the MNase/HindIII ladder (Fig. 3B, lane 7), suggesting that the GATC site is just to the right of a linker region. In order to obtain a better estimate of the proximity of site N/D to linker DNA, the experiment was repeated on DNA digested with PstI and probed from the left end with probe c (Fig. 3A and C). In this experiment the smaller of the two fragments in lane 7 (from PstI to N/D) runs slightly behind the MNase/HindIII ladder fragment, consistent with the model that the GATC site lies near the right end of the linker DNA (Fig. 3A). The fragment at ∼680 bp suggests that there is another unmethylated site, N2, ∼180 bp to the right of the HindIII site, and confirms the partial methylation of N/D. Comparison of Figure 3B, lane 3, with Figure 3C, lane 7, shows that the site that is more completely methylated is in closer alignment with the fragment in the chromatin. Thus the DpnI site is closer to the center of the linker DNA than the N/D site, at which the majority of the molecules are unmethylated.
The correlation of methylation of GATC sites with proximity to linker DNA in the chromatin was extended by examining a third region of the genome that has one methylated and two partially methylated GATC sites. The experiment in Figure 4 showed three fragments hybridizing to the probe in DNA digested with HindIII as the primary enzyme and DpnI. The data show that the two sites marked N/D are partially methylated. All molecules that are not methylated at one of these sites are digested at the DpnI site, D. This was expected, because the D site was previously shown to be a fully methylated (11). All three of these fragments align fairly closely to fragments in the chromatin ladder. Similar to the experiment in Figure 3, comparison of lanes 3 and 7 shows that the proximity of the various fragments in the digested DNA with those in the chromatin ladder fragment correlated with the degree of methylation. That observation is less meaningful when comparing several fragments in the same lane because the resolution of the fragments is better in the lower end of the gel. Nonetheless, the results are consistent with the observation that the more closely centered a GATC site is on the linker DNA, the higher the percentage of methylated molecules.
Figure 4.
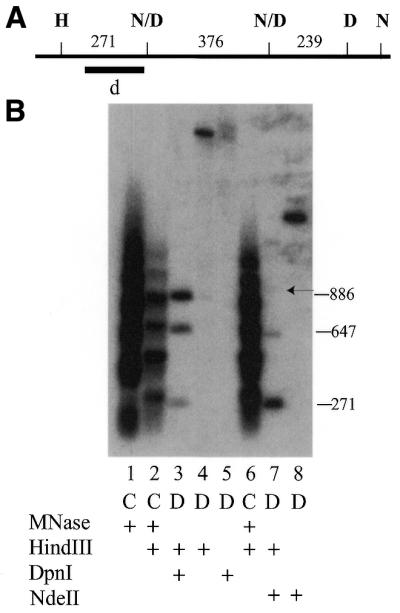
Indirect end labeling of the Tlr1.rB-B region. (A) Partial restriction map of Tlr1.rB-B. D, DpnI; N/D, partially methylated sites. The black bar indicates the probe for the experiment in (B). (B) Indirect end-labeling experiment of GATC sites in the Tlr1.rB-B region. Arrangement of the lanes and labeling is as in Figure 2B. The numbers to the right of (B) indicate the size of the restriction fragments in lanes 3 and 7.
In summary, fragments generated by indirect end labeling of DNA digested at seven specific methylated or partially methylated sites migrated in parallel with fragments generated by digestion with MNase. The proximity of a GATC site to linker DNA correlated well with the degree of methylation. We conclude that adenine methylation occurs preferentially in linker DNA in Tetrahymena.
Methylation patterns are independent of histone H1
There are at least two models that could account for preferential methylation in linker DNA. First, the linker DNA might be more accessible to the methyltransferase (MTase). Second, the MTase might be recruited to the internucleosomal regions by association with proteins that are specifically localized in the linker DNA. In the macronucleus of Tetrahymena, linker DNA is associated with histone H1, encoded by the single copy gene HHO (24). A knockout strain of the HHO gene, ΔH1, has been constructed. In the knockout strain the chromatin of the macronucleus is visibly less condensed, at the level of the light microscope, than the chromatin of a wild-type cell (15). Nonetheless, nucleosome phasing in the ΔH1 strain is the same as in wild-type cells (17). In order to determine whether the presence of histone H1 was required to establish the normal methylation pattern, methylation in a strain containing the wild-type complement of histone H1 was compared with that in the histone H1 knockout strain.
The methylation state of GATC sites was assessed by digestion of macronuclear DNA with the restriction enzyme isoschizomers Sau3A, NdeII and DpnI. The analysis of three GATC sites in the cyd2 region is shown in Figure 5. The uniformly methylated cyd2 site (D2) was mapped with respect to XbaI restriction sites by genomic Southern blot analysis (data not shown). Digestion of genomic DNA with XbaI and DpnI showed that cyd2 and a second GATC site in this region are uniformly methylated, since the vast majority of the molecules were digested with DpnI at both of the sites. A low level of hybridization to fragments at 4.2 and 2.5 kb is visible in long exposures. This is consistent with the observation that a few molecules are resistant to DpnI digestion at this site (Fig. 2B). Previous experiments suggest that these fragments are due to the presence of a small percentage of hemi-methylated molecules, which are resistant to digestion with both DpnI and NdeII (E. E. Capowski, unpublished data). A third GATC site, N, is unmethylated, as shown in Figure 2, and thus was not digested by DpnI. The patterns of hybridizing fragments in DNA from CU428.1 and ΔH1 are very similar, except that fragments attributed to hemi-methylated DNA molecules in strain CU428.1 were not detectable in DNA from ΔH1, even though the intensity of the other fragments was very similar.
Figure 5.
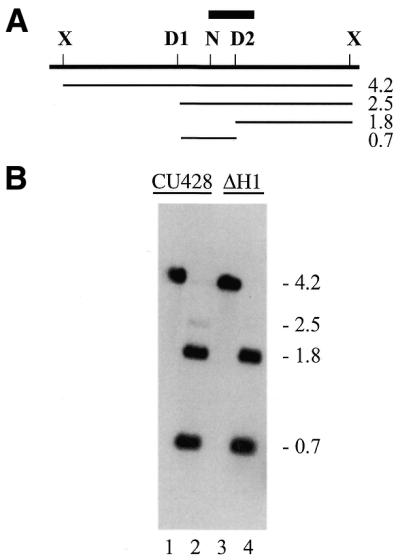
Methylation of the cyd2 region in the presence and absence of histone H1. (A) Restriction map of the cyd2 region. X, XbaI; N, NdeII; D, DpnI; bar, probe for the Southern blot in (B). Lines and numbers below the map indicate the position and size of the restriction fragments in (B). (B) Southern blot of macronuclear DNA. DNA in lanes 1 and 3 was digested with XbaI; in lanes 2 and 4 with XbaI + DpnI.
Analysis of the uniformly methylated GATC site cyd1 is shown in Figure 6. The DNA in lanes 1 and 3 was digested with HindIII. The DNA in lanes 2 and 4 was digested with HindIII and DpnI. The vast majority of the molecules in both CU428.1 and the ΔH1 strain were cut at the cyd1 site, labeled D. The small fraction of DNA molecules resistant to digestion by DpnI can be attributed to hemi-methylation. Two GATC sites in the cyd1 region were resistant to digestion with DpnI in the DNA from strain CU428.1, indicating that they lack methylation. [The low level of methylation at the NdeIIb site near cyd1 (Fig. 3B, lane 3) would be expected to generate faint fragments of 170 and 213 bp, which were not detectable under these conditions.] The two sites that are unmethylated in DNA from wild-type cells are also unmethylated in the DNA of the ΔH1 cell line.
Figure 6.
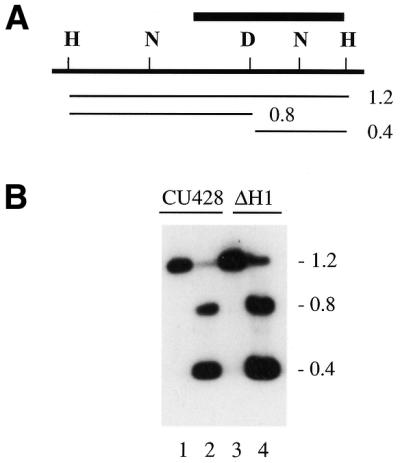
Methylation of the cyd1 region in the presence and absence of histone H1. (A) Restriction map of the cyd1 region. H, HindIII; N, NdeII; D, DpnI; bar, probe for the Southern blot in (B). Lines and numbers below the map indicate the position and size of the restriction fragments in (B). (B) Southern blot of macronuclear DNA. DNA in lanes 1 and 3 was digested with HindIII; in lanes 2 and 4 with HindIII + DpnI.
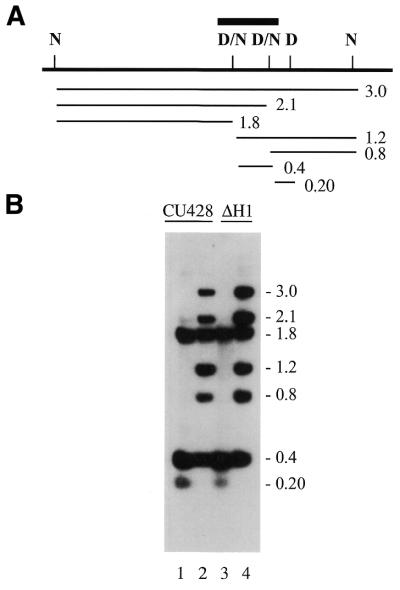
The third region of the genome examined, Tlr1r.B-B, contains one uniformly and two partially methylated GATC sites. This produces a complex pattern of fragments in DNA digested with methyl-sensitive enzymes (Fig. 7). Control DNA from strain CU428.1 (lane 1) was digested with Sau3A, which digests GATC sites regardless of the methylation state. The DNA in lane 2 was digested with NdeII to provide an indication of the methylation state of the various sites. The 3.0 kb fragments are molecules that are methylated at all three of the central GATC sites. Molecules at 2.1 kb are methylated at the left partially methylated site, but not at the right one, etc. The complex pattern of restriction fragments is not an artifact of partial digestion because digestion at the outer GATC sites is complete. Furthermore, the pattern is reproducible and is the same as the pattern obtained previously by digestion with MboI, which has the same methylation sensitivity as NdeII (11). The pattern of hybridizing fragments in DNA from the ΔH1 strain, shown in lanes 3 and 4, was not detectably different from that of strain CU428.1. Thus the absence of histone H1 made no difference as to whether a site was methylated or in the proportion of molecules methylated at a partially methylated site.
Figure 7.
Methylation of the Tlr1.rB-B region in the presence and absence of histone H1. (A) Restriction map of the Tlr1r region. N, NdeII; D, DpnI; N/D partially methylated GATC. The D, N/D, and N sites are all susceptible to digestion with Sau3a. Bar, probe for the Southern blot in (B). Lines and numbers below the map indicate the position and size of the restriction fragments in (B). (B) Southern blot of macronuclear DNA. Lanes 1 and 3, DNA digested with Sau3a; lanes 2 and 4, DNA digested with NdeII.
In summary, the methylation state of four methylated, two partially methylated and three unmethylated GATC sites from three different regions of the genome was the same in chromatin with and without histone H1. We conclude that although methylation occurs preferentially in linker DNA, histone H1 is not required for the maintenance of normal methylation patterns.
DISCUSSION
The position effect for methylation of nuclear DNA in Tetrahymena suggested that methylation may be related to chromatin structure (13). Early experiments with MNase digestion of bulk chromatin suggested that methyladenine was preferentially located in the linker DNA of Tetrahymena (14). We have confirmed that finding for eight specific methylated GATC sites in three different regions of the genome, showing that methylated and partially methylated sites are located in or near linker DNA and an unmethylated site is in DNA that is associated with the nucleosome core.
For technical reasons, all of the methylated AT sites examined in this study were in the restriction site GATC. Since methylated GATC sites contain a very small percentage of the total methylated adenines, it is worthwhile to consider whether these sites are representative of methylated sites as a whole. It is likely they are for two reasons. First, there is no evidence that GATC sites themselves are specifically positioned with respect to nucleosomes. In fact, the GATC sites examined in this study are in a variety of positions with respect to the nucleosome. One is in the nucleosome core and the others are in the linker DNA, with varying proximity to the nucleosome. Second, the experiments described here are consistent with an earlier study which showed that methyladenine residues in bulk chromatin were preferentially digested with MNase (14).
It is tempting to speculate that a causal relationship exists between methylation and the location of the target site in linker DNA. It is not known whether the MTase with de novo activity that establishes the pattern of methylation in the developing macronucleus and the MTase that maintains that pattern in vegetatively growing cells are one and the same. This may be the case, since there is strong genetic evidence that the maintenance MTase of Tetrahymena also has a de novo activity (11). This is in contrast to the semi-conservative mechanism that is thought to maintain stable methylation patterns in mammalian cells (25). If methylation in Tetrahymena is not maintained by a preference of the MTase for a hemi-methylated substrate, the faithful maintenance of methylation patterns must depend instead on some characteristic of the DNA sequence or chromatin structure that is consistent between clonal cell lines.
It was previously shown that nucleosomes are specifically positioned with respect to DNA sequence in Tetrahymena (17). Thus localized chromatin structure is one characteristic that might serve to set a consistent methylation pattern. If nucleosome positioning determines methylation patterns, there are at least two hypotheses that could explain the preferential methylation of sequences in linker DNA. The first of these is that the MTase is recruited to the linker DNA by association with localized proteins. One obvious candidate for this role would be linker histones. However, the experiments described here rule out a requirement for histone H1 in recruitment of the Tetrahymena MTase, because methylation patterns are virtually identical in strains with and without histone H1.
Another possibility is that methylation of adenine occurs preferentially in linker DNA because that region is more accessible to the MTase. The observations of nucleosome positioning and methylation in the ΔH1 cell line are consistent with this model. Both nucleosome positioning (17) and methylation patterns (Figs 5–7) are maintained in the absence of histone H1. There was one slight difference between methylation in cells with the wild-type complement of histone H1 and the ΔH1 cell line. A small percentage of hemi-methylated molecules at the cyd2 site in wild-type cells (Figs 2B and 5B) were not detected in the cells lacking H1 histone (Fig. 5B). Although nucleosome positioning is maintained in ΔH1 cells, the chromatin is visibly less condensed at the level of the light microscope (15). Reduction in chromatin condensation may make the linker DNA slightly more accessible to the MTase than it is in wild-type cells.
If methylation is related to the location of a site in linker DNA, then what accounts for partially methylated sites? Since the clonal descendents of a single cell display identical partial methylation patterns, it is likely that partial methylation reflects variability of methylation state among the approximately 50 copies of the genome in a single macronucleus, rather than variability in the methylation pattern between individual cells (11).
One possibility is that association of the DNA with nucleosomes creates steric hindrance that diminishes the efficiency of methylation. This model is consistent with the direct correlation observed here between the proximity to nucleosome core DNA and the degree of methylation of individual sites.
Another factor that might contribute to partial methylation is variability in nucleosome positioning. In chromatin assembled in vitro, nucleosomes occupy a cluster of positions with 10 bp spacing, suggesting that the position of a nucleosome is a probability distribution rather than a unique location (26). If nucleosome positions are variable in vivo, it may be that partially methylated sites reflect a distribution of nucleosome positions, only some of which provide accessibility to the MTase.
The relationship between methylation and chromatin structure may be different for cytosine versus adenine methylation. First, early experiments indicated that in mouse L cells methylcytosine is enriched in the histone-protected nucleosome core sequences (27,28), rather than in linker DNA. Second, cytosine methylation changes the binding affinity of DNA sequences for nucleosome cores assembled on the DNA in vitro (29,30). Although these experiments have not been done in vivo, they suggest that cytosine methylation might play a role in nucleosome positioning. In transformation experiments in Tetrahymena the position effect took precedence over the methylation state of the transforming DNA, i.e. methylation was lost from a chromosomal site inserted in the rDNA (13). Thus the evidence suggests that adenine methylation is governed by chromatin structure, rather than the reverse.
The different relationships between chromatin structure and cytosine versus adenine methylation may be due in part to differences in the chromatin structure of the respective organisms. The most unusual characteristic of Tetrahymena chromatin is that histone H1 lacks the central, hydrophobic domain found in all other histone H1 proteins (24). However, histone H1 is not required to establish or maintain wild-type nucleosome positioning or DNA methylation patterns. Thus it seems more likely that differences in the enzymatic activity of the MTases and their association with chromatin may be related to differences in the biological roles of the two different methylated bases.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank G. Thomas Hayman for contributions to sequencing and helpful comments on the manuscript. T.A.V. was supported in part by an Arthur J. Schmitt fellowship and by Marquette University fellowships. This work was supported in part by grants GM52626 from the National Institutes of Health and MCB-9974885 from the National Science Foundation.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +1 414 288 1474; Fax: +1 414 288 7357; Email: kathleen.karrer@marquette.edu Present address: Teresa A. VanNuland, Abbott Laboratories, 200 Abbott Park Road, Abbott Park, IL 60064-3537, USA AF416723, L25254 and L34029
REFERENCES
- 1.Lyko F. (2001) DNA methylation learns to fly. Trends Genet., 17, 169–172. [DOI] [PubMed] [Google Scholar]
- 2.Li E., Bestor,T.H. and Jaenisch,R. (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell, 69, 915–926. [DOI] [PubMed] [Google Scholar]
- 3.Tajima S. and Suetake,I. (1998) Regulation and function of DNA methylation in vertebrates. J. Biochem., 123, 993–999. [DOI] [PubMed] [Google Scholar]
- 4.Bestor T.H. (2000) The DNA methyltransferases of mammals. Hum. Mol. Genet., 9, 2395–2402. [DOI] [PubMed] [Google Scholar]
- 5.Hattman S., Kenny,C., Berger,L. and Pratt,K. (1978) Comparative study of DNA methylation in three unicellular eucaryotes. J. Bacteriol., 135, 1156–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babinger P., Kobl,I., Mages,W. and Schmitt,R. (2001) A link between DNA methylation and epigenetic silencing in transgenic Volvox carteri. Nucleic Acids Res., 29, 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorovsky M.A., Hattman,D. and Pleger,G.L. (1973) [N6]methyladenine in the nuclear DNA of a eucaryote, Tetrahymena pyriformis. J. Cell Biol., 56, 697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cummings D.J., Tait,A. and Godard,J.M. (1974) Methylated bases in DNA from Paramecium aurelia. Biochim. Biophys. Acta, 374, 1–11. [DOI] [PubMed] [Google Scholar]
- 9.Bromberg S., Pratt,K. and Hattman,S. (1982) Sequence specificity of the DNA adenine methylase in the protozoan Tetrahymena thermophila. J. Bacteriol., 150, 993–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison G.S., Findly,R.C. and Karrer,K.M. (1986) Site-specific methylation of adenine in the nuclear genome of a eucaryote, Tetrahymena thermophila. Mol. Cell. Biol., 6, 2364–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capowski E.E., Wells,J.M., Harrison,G.S. and Karrer,K.M. (1989) Molecular analysis of methylation patterns in Tetrahymena thermophila. Mol. Cell. Biol., 9, 2598–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karrer K.M. and Stein-Gavens,S. (1990) Constancy of adenine methylation in Tetrahymena macronuclear DNA. J. Protozool., 37, 409–414. [DOI] [PubMed] [Google Scholar]
- 13.Karrer K.M. and VanNuland,T.A. (1998) Position effect takes precedence over target sequence in determination of adenine methylation patterns in the nuclear genome of a eukaryote, Tetrahymena thermophila. Nucleic Acids Res., 26, 4566–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pratt K. and Hattman,S. (1981) Deoxyribonucleic acid methylation and chromatin organization in Tetrahymena thermophila. Mol. Cell. Biol., 1, 600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen X., Yu,L., Weir,J.W. and Gorovsky,M.A. (1995) Linker histones are not essential and affect chromatin condensation in vivo. Cell, 82, 47–56. [DOI] [PubMed] [Google Scholar]
- 16.Gorovsky M.A., Yao,M.-C., Keevert,J.B. and Pleger,G.L. (1975) Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol., 9, 311–327. [DOI] [PubMed] [Google Scholar]
- 17.Karrer K.M. and VanNuland,T.A. (1999) Nucleosome positioning is independent of histone H1 in vivo. J. Biol. Chem., 274, 33020–33024. [DOI] [PubMed] [Google Scholar]
- 18.Reed K.C. and Mann,D.A. (1985) Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res., 13, 7207–7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pratt K. and Hattman,S. (1983) Nucleosome phasing in Tetrahymena macronuclei. J. Protozool., 30, 592–598. [DOI] [PubMed] [Google Scholar]
- 20.Wu C. (1980) The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to Dnase I. Nature, 286, 854–860. [DOI] [PubMed] [Google Scholar]
- 21.Gorovsky M.A., Glover,C., Johmann,C.A., Keevert,J.B., Mathis,D.J. and Samuelson,M. (1977) Histones and chromatin structure in Tetrahymena macro- and micronuclei. Cold Spring Harbor Symp. Quant. Biol., 42, 493–503. [DOI] [PubMed] [Google Scholar]
- 22.Wells J.M., Ellingson,J.L.E., Catt,D.M., Berger,P.J. and Karrer,K.M. (1994) A small family of elements with long inverted repeats is located near sites of developmentally regulated DNA rearrangement in Tetrahymena thermophila. Mol. Cell. Biol., 14, 5939–5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capowski E.E. (1989) Molecular analysis of N6-methyladenine patterns in the nuclear DNA of the ciliate, Tetrahymena thermophila. PhD dissertation, Brandeis University. [DOI] [PMC free article] [PubMed]
- 24.Wu M., Allis,C.D., Richman,R., Cook,R.G. and Gorovsky,M.A. (1986) An intervening sequence in an unusual histone H1 gene of Tetrahymena thermophila. Proc. Natl Acad. Sci. USA, 83, 8674–8678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bird A. (1999) DNA methylation de novo. Science, 286, 2287–2288. [DOI] [PubMed] [Google Scholar]
- 26.Meersseman G., Pennings,S. and Bradbury,E.M. (1992) Mobile nucleosomes—a general behavior. EMBO J., 11, 2951–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solage A. and Cedar,H. (1978) Organization of 5-methylcytosine in chromosomal DNA. Biochemistry, 17, 2934–2938. [DOI] [PubMed] [Google Scholar]
- 28.Razin A. and Cedar,H. (1977) Distribution of 5-methylcytosine in chromatin. Proc. Natl Acad. Sci. USA, 74, 2725–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Godde J.S., Kass,S.U., Hirst,M.C. and Wolffe,A.P. (1996) Nucleosome assembly on methylated CGG triplet repeats in the fragile X mental retardation gene 1 promoter. J. Biol. Chem., 271, 24325–24328. [DOI] [PubMed] [Google Scholar]
- 30.Davey C., Pennings,S. and Allan,J. (1997) CpG methylation remodels chromatin structure in vitro. J. Mol. Biol., 267, 276–288. [DOI] [PubMed] [Google Scholar]