Abstract
The RAD52 epistasis group genes are involved in homologous recombination, and they are conserved from yeast to humans. We have cloned a novel human gene, RAD54B, which is homologous to yeast and human RAD54. Human Rad54B (hRad54B) shares high homology with human Rad54 (hRad54) in the central region containing the helicase motifs characteristic of the SNF2/SWI2 family of proteins, but the N-terminal domain is less conserved. In yeast, another RAD54 homolog, TID1/RDH54, plays a role in recombination. Tid1/Rdh54 interacts with yeast Rad51 and a meiosis-specific Rad51 homolog, Dmc1. The N-terminal domain of hRad54B shares homology with that of Tid1/Rdh54, suggesting that Rad54B may be the human counterpart of Tid1/Rdh54. We purified the hRad54 and hRad54B proteins from baculovirus-infected insect cells and examined their biochemical properties. hRad54B, like hRad54, is a DNA-binding protein and hydrolyzes ATP in the presence of double-stranded DNA, though its rate of ATP hydrolysis is lower than that of hRad54. Human Rad51 interacts with hRad54 and enhances its ATPase activity. In contrast, neither human Rad51 nor Dmc1 directly interacts with hRad54B. Although hRad54B is the putative counterpart of Tid1/Rdh54, our findings suggest that hRad54B behaves differently from Tid1/Rdh54.
INTRODUCTION
Homologous recombination is the main pathway for DNA double-strand break (DSB) repair in yeast, and it has been revealed that the recombinational repair pathway also plays an important role in higher eukaryotes (1,2). In Saccharomyces cerevisiae, the RAD52 epistasis group genes are involved in the recombinational repair of DNA damage, including DSBs, as well as playing a role in meiotic recombination (3). It has been demonstrated that the RAD52 epistasis group genes are well conserved structurally and functionally throughout evolution (4). Recently, we cloned a novel human gene, RAD54B (hRAD54B), which shares homology with yeast and human RAD54 (ScRAD54 and hRAD54, respectively) (5).
Rad54 is a member of the SNF2/SWI2 family of DNA-dependent ATPases. Members of this family are involved in various functions, such as transcriptional regulation (SNF2, MOT1 and BRM), chromosome stability (lodestar), nucleotide excision repair (ERCC6 and Rad16) and recombination (Rad54) (6). ScRad54 and hRad54 have double-stranded (ds) DNA-dependent ATPase activities, which are important for their in vitro and in vivo functions (7–12). Using the energy of ATP hydrolysis, ScRad54 remodels DNA and stimulates homologous DNA pairing by ScRad51 in vitro (7–11). Homozygous RAD54 mutants in mouse and chicken are highly radiation- and methyl methanesulfonate-sensitive and have reduced levels of homologous recombination (13,14).
Human RAD54B encodes a protein of 910 amino acids. The central part, which contains the seven helicase motifs found in members of the SNF2/SWI2 family, is well conserved between hRad54 and hRad54B, while the N-terminal region is less conserved except for the first 10 amino acids (5). The expression pattern of RAD54B coincides with those of members of the RAD52 epistasis group. Recently, we found that hRad54B associates with human Rad51 (hRad51) in mammalian cells through the N-terminal domain of hRad54B (15). In addition, hRad54B formed nuclear foci that co-localize with hRad51, hRad54 and Brca1.
There is another RAD54 homolog in S.cerevisiae, called TID1/RDH54 (ScTID1/RDH54) (16–18). ScTid1/Rdh54 interacts with yeast Rad51 (ScRad51) and a meiosis-specific Rad51 homolog, Dmc1 (ScDmc1) (16,19). In vitro, ScTid1/Rdh54 shows similar biochemical properties to ScRad54 through its ATPase activity (19), though its in vivo functions are different. In contrast to RAD54, which is required for sister chromatid-based repair, genetic assays have shown that TID1/RDH54 is required for mitotic interchromosomal recombination as well as for meiotic recombination (17,18,20). Homologs of TID1/RDH54 in eukaryotes other than S.cerevisiae have not been identified so far.
Here we demonstrate the biochemical properties of hRad54B. We purified the hRad54 and hRad54B proteins from baculovirus-infected insect cells and compared the biochemical properties of these proteins. hRad54B is a DNA-binding protein and a dsDNA-dependent ATPase like hRad54, though its rate of ATP hydrolysis is lower than that of hRad54. A sequence analysis showed that hRad54B shares homology with ScTid54/Rdh54 in the N-terminal domain, though we could not detect the direct interaction of hRad54B with hRad51 and hDmc1, suggesting that hRad54B indirectly associates with hRad51 in vivo. Our data suggest that hRad54B is the human counterpart of ScTid1/Rdh54, though its function is different from that of ScTid1/Rdh54.
MATERIALS AND METHODS
Plasmid constructions
Human RAD54, RAD54B and DMC1 cDNAs were cloned from a human testis cDNA library (Clontech Laboratories). The constructs expressing hRad54 and hRad54B containing single amino acid substitutions at positions 189 and 332, respectively, were generated by site-directed mutagenesis and sequenced to ensure that no mutation other than those intended had been introduced. These proteins are referred to as hRad54K189R and hRad54BK332R, respectively. The cDNAs encoding the hRad54, hRad54K189R, hRad54B and hRad54BK332R proteins were subcloned into pFastBac HTc (Life Technologies, Inc.) to yield plasmids pFastBac–hRad54, pFastBac–hRad54K189R, pFastBac–hRad54B and pFastBac–hRad54BK332R, respectively. The hRad51 expression vector was constructed as described (21). The hDmc1 expression vector was constructed by subcloning the hDMC1 cDNA into the T7 polymerase expression vector pET-15b (Novagen).
Purification of the human recombination proteins
pFastBac–hRad54, pFastBac–hRad54K189R, pFastBac–hRad54B and pFastBac–hRad54BK332R were transformed into DH10Bac Escherichia coli cells to allow site-specific transposition into bacmid bMON14272 (BAC-TO-BAC Baculovirus Expression System, Life Technologies, Inc.). High molecular weight recombinant bacmids were isolated and transfected into Sf9 cells to produce virus stocks that were amplified as described by the manufacturer. For protein production, 6.0 × 108 Sf9 cells were infected with the recombinant baculoviruses. After 2 days the cells were collected by low speed centrifugation and washed twice with ice-cold phosphate-buffered saline. All the purification steps were carried out at 4°C. The cells were lysed in 25 ml of 54-lysis buffer [20 mM Tris–HCl pH 7.5, 100 mM KCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 10% glycerol, 1 mM phenylmethylsulfonyl fluoride and 1% Nonidet P-40] for 30 min after a brief sonication. The lysates were clarified by centrifugation, and the supernatant was filtrated through 0.45 µm filter. This crude extract (fraction I) was loaded onto a 10-ml phosphocellulose column (Whatman P11) equilibrated with 54 P-cell wash buffer (20 mM Tris–HCl pH 7.5, 100 mM KCl, 1 mM EDTA, 1 mM DTT, 10% glycerol). To elute hRad54B, a 100 ml linear gradient of 0.1–1.0 M KCl was applied, and the eluate was collected in 2.5 ml fractions. The fractions containing hRad54B, which were eluted at ~0.5 M KCl, were identified by SDS–polyacrylamide gel electrophoresis (PAGE) followed by Coomassie staining, pooled and dialyzed against 2 × 0.5 l of 54 HisTrap wash buffer (20 mM Tris–HCl pH 7.5, 500 mM KCl, 5 mM β-mercaptoethanol, 10% glycerol). This fraction (fraction II) was then loaded onto a 1-ml HiTrap Chelating column (Amersham Pharmacia Biotech) charged with nickel ion equilibrated in 54 HisTrap wash buffer using the ÄKTA FPLC system (Amersham Pharmacia Biotech). The column was sequentially washed with 6 ml of 54 HisTrap wash buffer containing 40 and 200 mM imidazole, respectively. The 200 mM imidazole elution step contained hRad54B, and the fractions containing hRad54B were pooled and dialyzed against 2 × 0.3 l of 54 P-cell wash buffer (fraction III). Fraction III was loaded onto a MonoS PC 1.6/5 column (Amersham Pharmacia Biotech) equilibrated with 54 P-cell wash buffer using the SMART system (Amersham Pharmacia Biotech). hRad54B was eluted with a 0.6 ml linear gradient from 0.1 to 1.0 M KCl. The protein (fraction IV) was eluted at ~0.3 M KCl, and aliquots were frozen in liquid N2 and stored at –80°C. hRad54BK332R, hRad54 and hRad54K189R were purified in exactly the same manner, as was hRad54B.
hRad51 was overexpressed in E.coli strain JM109 (DE3) and purified to near homogeneity as described (22). hDmc1 was also overexpressed in E.coli and purified in a similar manner as was hRad51; the details of this process will be described elsewhere.
DNA substrates
φX174 viral (+) strand was purchased from New England Biolabs, and the replicative form was from Life Technologies, Inc. The 50mer oligonucleotides used for single-stranded (ss) DNA/dsDNA-binding and ATPase assays were: 50–1, 5′-ATTTCATGCTAGACAGAAGAATTCTCAGTAACTTCT-TTGTGCTGTGTGTA-3′; 50–2, the exact complement of 50–1; SAT-1, 5′-ATTTCATGCTAGACAGAAGAATTCTCAGTAACTTCTTTGTGCTGTGTGTA-3′. The 5′-end of 50–1 was labeled with T4 polynucleotide kinase in the presence of [γ-32P]ATP. 5′-32P-labeled 50–1 was annealed to 50–2 to create a blunt-ended, double-stranded 50mer.
Antibodies
A rabbit polyclonal antiserum against chicken Rad54 was a gift from Dr A. Shinohara (Osaka University, Osaka, Japan). A rabbit polyclonal antiserum against Rad54B was obtained as described previously (15). These antibodies were used for western blotting as described previously (15).
ATPase assay
Standard reaction mixtures contained 20 mM KPO4 pH 7.0, 40 mM KCl, 4 mM MgCl2, 1 mM DTT, 100 µg/ml bovine serum albumin, 1 mM ATP, 0.25 µCi of [γ-32P]ATP (>5000 Ci/mmol), 60 µM (in nucleotides) of pUC19 and 150 nM of hRad54, hRad54K189R, hRad54B or hRad54BK332R protein in a 20 µl volume. Other conditions concerning the presence or absence of MgCl2, ATP concentration, the presence or absence of DNA, ssDNA [poly(dA)] or dsDNA (pUC19) and the concentrations of the purified proteins are indicated in the figure legends. The reaction mixtures were incubated at 30°C, and the reactions were terminated by the addition of EDTA to 167 mM. Released phosphate was separated from ATP by thin layer chromatography on polyethyleneimine cellulose (Merck) using 1 M formic acid and 0.3 M LiCl as a running buffer. Hydrolysis was quantified with the use of the BAS2000 image analyzer (Fuji Film Corp.). Background hydrolysis observed in the absence of protein (∼4%) was subtracted.
To examine the effect of hRad51 and hDmc1 on the ATPase activities of hRad54 and hRad54B, hRad51 or hDmc1 (240 nM) was incubated with ssDNA (SAT-1, 2 µM) at 37°C for 10 min in standard reaction mixture prior to the addition of hRad54, hRad54K189R, hRad54B or hRad54BK332R (120 nM). Reactions were initiated by the addition of dsDNA (pGsat4, 30 µM).
ssDNA-binding assay
Single-stranded φX174 (20 µM) or a 32P-labeled single-stranded 50mer oligonucleotide (50–1, 100 nM) was mixed with hRad54 or hRad54B in 10 µl of standard reaction buffer, containing 20 mM Tris–HCl pH 7.5, 1 mM MgCl2, 1 mM DTT, 2 mM ATP, 20 mM creatine phosphate, 12 U/ml creatine kinase and 100 µg/ml bovine serum albumin. The reaction mixtures were incubated at 37°C for 10 min and were analyzed by 0.8% agarose gel (single-stranded φX174) or by non-denaturing 10% polyacrylamide gel (single-stranded 50mer) electrophoresis in TAE buffer (40 mM Tris, 40 mM acetic acid, 1 mM EDTA pH 8.0).
dsDNA-binding assay
Superhelical φX174 (20 µM) or a 32P-labeled double-stranded 50mer oligonucleotide (50–1 + 50–2, 200 nM) was mixed with hRad54 or hRad54B in 10 µl of standard reaction buffer, which was identical to that used in the ssDNA-binding assay. The reaction mixtures were incubated at 37°C for 10 min and were analyzed by 0.8% agarose gel (double-stranded φX174) or by non-denaturing 10% polyacrylamide gel (double-stranded 50mer) electrophoresis in 0.5× TBE buffer (for agarose gel) or TBE buffer (for polyacrylamide gel) (90 mM Tris, 64.6 mM boric acid, 2 mM EDTA, pH 8.0).
Protein–protein binding assay
An Affi-Gel 15 slurry (250 µl; Bio-Rad) was washed twice with 0.5 ml of Affi-Gel buffer, containing 20 mM HEPES–KOH, 0.1 M KCl, 0.5 mM EDTA, 2 mM β-mercaptoethanol, 10% glycerol and 0.05% Triton X-100. The washed beads were mixed with 0.5 ml of 4.0 mg/ml hRad51 or 4.7 mg/ml hDmc1 and were incubated at 4°C for 5 h. Afterward, the supernatant was removed, and the conjugated beads were washed six times with 0.5 ml of Affi-Gel buffer. The final concentration of hRad51 and hDmc1 conjugated to Affi-Gel 15, determined by the amounts of unbound hRad51 and hDmc1, was 3.3 mg/ml each. Ethanolamine was added to the final concentration of 100 mM and incubated at 4°C for 1 h to block residual active ester sites, followed by washing three times with 0.5 ml of Affi-Gel buffer. The Affi-Gel 15-protein matrices were adjusted to 33% slurries with Affi-Gel buffer and were stored at 4°C.
For the binding assay, 33 µg of hRad51 or hDmc1 (30 µl of the slurry) was mixed with 20 µg of hRad54 or hRad54B. The proteins were mixed at room temperature for 90 min. The unbound proteins were removed, and the Affi-Gel protein beads were washed six times with 0.5 ml of Affi-Gel buffer. SDS–PAGE sample buffer (10 µl of a 2-fold stock) was mixed directly with the washed beads. After heating the mixture at 98°C for 2 min, proteins were visualized by 9% SDS–PAGE. Bands were visualized by Coomassie staining.
RESULTS
Purification of the human Rad54 and Rad54B proteins
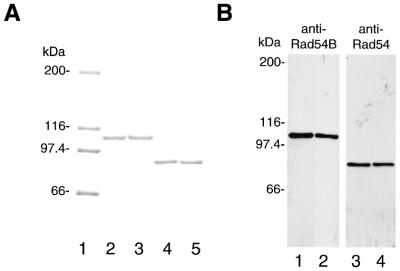
To explore the biochemical properties of hRad54B, we purified the hRad54B protein from baculovirus-infected Sf9 insect cells. First, we constructed a cDNA encoding hRad54B containing an N-terminal polyhistidine tag. In addition, we generated a cDNA expression construct encoding hRad54B containing a single amino acid substitution at position 332 (referred to as hRad54BK332R hereafter). This invariant lysine residue is in the putative Walker A nucleotide binding motif and was changed to an arginine residue using site-directed mutagenesis. Conversion of the equivalent lysine residue of ScRad54, ScTid1/Rdh54 or hRad54 into an arginine residue severely impairs nucleotide triphosphate hydrolysis (8,11,12,19). We also constructed a cDNA encoding wild-type hRad54 or hRad54 containing a lysine-to-arginine substitution in the Walker A motif (referred to as hRad54K189R). All the proteins were tagged at the N-terminus with six histidines, expressed in Sf9 cells and purified to near homogeneity, as described in the Materials and Methods. A sample of the final purification step of each protein is shown in Figure 1A. The nature of the purified proteins was confirmed by means of western blotting using anti-Rad54B or anti-Rad54 antiserum (Fig. 1B). Sizing experiments in Superdex 200 with protein standards have suggested that hRad54B in solution behaves as a monomer (data not shown).
Figure 1.
Purification of the hRad54B and hRad54 proteins. (A) A 7.5% SDS–PAGE containing 1 µg of purified hRad54B (lane 2), hRad54BK332R (lane 3), hRad54 (lane 4) and hRad54K189R (lane 5) was stained with Coomassie Blue. The size of the protein molecular mass markers in lane 1 is indicated in kDa. (B) Western blot analysis of the purified proteins. hRad54B (lane 1) and hRad54BK332R (lane 2) were visualized using anti-Rad54B antiserum, and hRad54 (lane 3) and hRad54K189R (lane 4) were visualized using anti-Rad54 antiserum. Each lane was loaded with 200 ng of purified protein. Molecular mass standards (in kDa) are shown.
The human Rad54B protein is a dsDNA-dependent ATPase
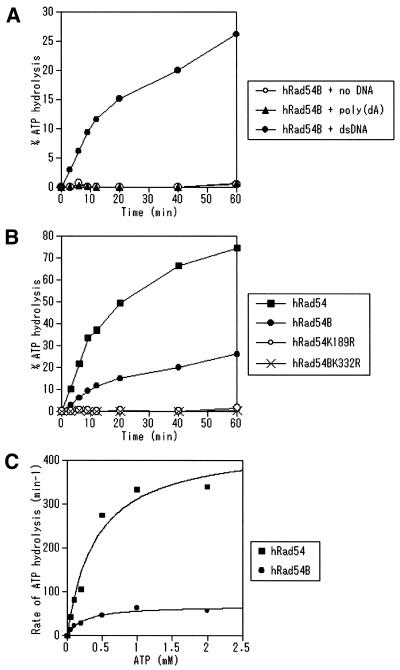
We tested whether hRad54B possesses ATPase activity because Walker type nucleotide binding motifs are present in this protein. It has been reported that ScRad54, ScTid1/Rdh54 and hRad54 hydrolyze ATP in a dsDNA-dependent manner (7–12,19). Like hRad54, hRad54B showed ATPase activity in the presence of dsDNA (Fig. 2A). The ATPase activity of hRad54B was dependent on the presence of magnesium (data not shown). Poly(dA), a ssDNA that does not form a secondary structure, failed to stimulate the ATPase activity of hRad54B (Fig. 2A). Next, we compared the ATPase activities of hRad54, hRad54B, hRad54K189R and hRad54BK332R. As shown in Figure 2B, the ATPase activity of hRad54B is lower than that of hRad54. hRad54BK332R, like hRad54K189R, was defective in ATP hydrolysis (Fig. 2B), identifying the Walker A box as part of the active site of the DNA-dependent ATPase. These results also suggest that no significant contaminating ATPase activity was present in our preparation. The kinetics of ATP hydrolysis by hRad54B in the presence of 60 µM of dsDNA was examined and compared with that by hRad54 (Fig. 2C). The Km for the ATPase activity of hRad54B was determined based on the data shown in Figure 2C, and is 220 µM, while the Km for that of hRad54 is 410 µM. The Vmax for the ATPase activities of hRad54B and hRad54 are 69 and 440 min–1, respectively. We examined the rates of ATP hydrolysis of these proteins under various concentrations of dsDNA, and found that the rates of ATP hydrolysis of both proteins were saturated at 60 µM of dsDNA (data not shown). These data indicate that the rate of ATP hydrolysis by hRad54B is several-fold lower than that by hRad54.
Figure 2.
The hRad54B protein is a dsDNA-dependent ATPase. (A) hRad54B (150 nM) was incubated with 1 mM ATP at 30°C in the absence of DNA or in the presence of either poly(dA) (60 µM) or dsDNA (60 µM) for the times indicated. The ATPase activities were assayed as described in Materials and Methods. (B) hRad54, hRad54B, hRad54K189R or hRad54BK332R (150 nM) was incubated with 1 mM ATP and dsDNA (60 µM) at 30°C for the times indicated. (C) The rate of ATP hydrolysis by hRad54B or hRad54 was determined at 30°C in the presence of dsDNA (60 µM) with a variety of ATP concentrations.
Rad54B binds both ssDNA and dsDNA
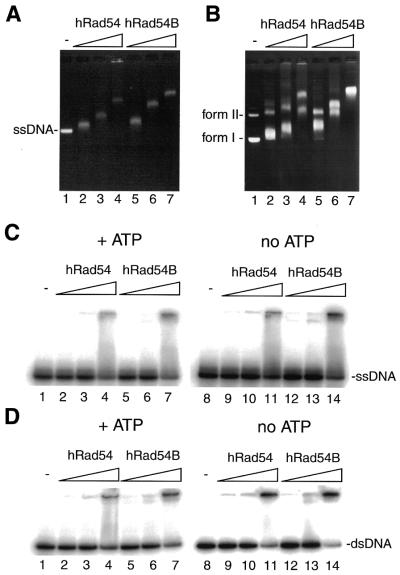
Next, we examined the DNA-binding properties of hRad54 and hRad54B. As shown in Figure 3A and B, hRad54 and hRad54B bound both circular ssDNA and superhelical dsDNA in a concentration-dependent manner to produce complexes that exhibited reduced mobility through agarose gels. ssDNA-binding activities of hRad54 and hRad54B were also examined using a 32P-labeled 50mer oligonucleotide as a substrate. The majority of the protein–ssDNA complexes formed by hRad54 and hRad54B failed to enter polyacrylamide gels (Fig. 3C). Large protein–DNA complexes formed by hRad54 and hRad54B were also observed when the oligonucleotide was annealed with its complementary strand to form duplex DNA (Fig. 3D). The bindings of hRad54 and hRad54B to ssDNA and dsDNA are independent of the presence of ATP, because protein–DNA complex formations were not affected by the absence of ATP (Fig. 3C and D, lanes 8–14). As expected, hRad54K189R and hRad54BK332R bound ssDNA/dsDNA with wild-type affinity (data not shown).
Figure 3.
DNA-binding activities of hRad54 and hRad54B. (A) Circular ssDNA binding by hRad54 and hRad54B. Circular ssDNA (20 µM) was incubated with hRad54 or hRad54B at 37°C for 10 min, and the reactions were analyzed by 0.8% agarose gel electrophoresis in 1× TAE buffer. The concentrations of hRad54 and hRad54B used in the DNA binding experiments were 0.2, 0.4 and 0.6 µM. (B) Superhelical dsDNA binding by hRad54 and hRad54B. Mixture of superhelical dsDNA (form I) and nicked circular dsDNA (form II) (20 µM) was incubated with hRad54 or hRad54B at 37°C for 10 min, and the reactions were analyzed by 0.8% agarose gel electrophoresis in 0.5× TBE buffer. The concentrations of hRad54 and hRad54B used in the DNA binding experiments were 0.2, 0.4 and 0.6 µM. (C) The ssDNA binding of hRad54 and hRad54B. A 32P-labeled single-stranded 50mer oligonucleotide (100 nM), which does not contain intramolecular base pairing, was incubated with hRad54 or hRad54B at 37°C for 10 min in the presence (lanes 1–7) or absence (lanes 8–14) of 2 mM ATP. The reactions were analyzed by non-denaturing 10% PAGE in TAE buffer. The concentrations of hRad54 and hRad54B were 50, 100 and 200 nM. (D) The dsDNA binding of hRad54 and hRad54B. A 32P-labeled double-stranded 50mer oligonucleotide (200 nM) was incubated with hRad54 or hRad54B at 37°C for 10 min in the presence (lanes 1–7) or absence (lanes 8–14) of 2 mM ATP. The reactions were analyzed by non-denaturing 10% PAGE in TAE buffer. The concentrations of hRad54 and hRad54B were 50, 100 and 200 nM.
Sequence homology of Rad54B and Tid1/Rdh54 in the N-terminal domain
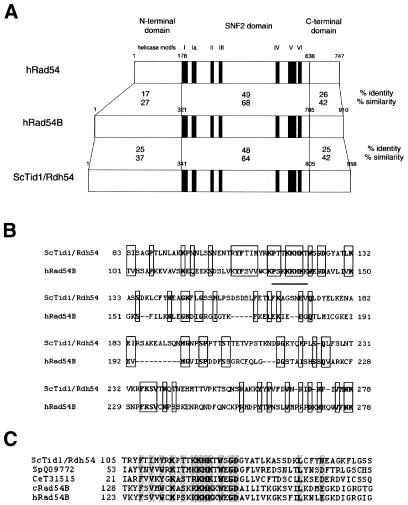
hRad54B shares high homology with hRad54, especially in the central region containing the seven helicase motifs characteristic of the SNF2/SWI2 family (this region is referred to as the SNF2 domain hereafter) (Fig. 4A). On the other hand, the N-terminal domain is less conserved except for the first 10 amino acids. A database search using BLAST revealed that the N-terminal domain of hRad54B shares homology with that of ScTid1/Rdh54, which is another Rad54 homolog in yeast (Fig. 4B) (16–18). The overall similarity between the N-terminal domains of hRad54B and ScTid1/Rdh54 is not significantly high (Fig. 4A), but the region containing a cluster of basic amino acid residues, which is predicted to be a nuclear localization signal, is well conserved (Fig. 4B). Putative nuclear localization signals also exist in the N-terminal domains of hRad54 and ScRad54, but they are different from those of hRad54B and ScTid1/Rdh54 (23). Sequence comparison revealed that this specifically conserved region between hRad54B and ScTid1/Rdh54 also exists in chicken Rad54B and a putative Tid1/Rdh54 homolog in Schizosaccharomyces pombe, which was pointed out by Dresser et al. (16) (Fig. 4C). We have also found that there is a SNF2/SWI2 family protein containing the similar region in Caenorhabditis elegans. The putative nuclear localization signal is conserved in all these proteins. These findings suggest that the structure of Tid1/Rdh54 is conserved among eukaryotes.
Figure 4.
hRad54B shares homology with ScTid1/Rdh54 in the N-terminal domain. (A) A schematic alignment of the hRad54, hRad54B and ScTid1/Rdh54 proteins. These proteins were separated into three domains, and amino acid sequence identities and similarities among these proteins were calculated in each domain. The amino acid position of the boundary of each domain is indicated. The following amino acids were considered to be similar: D, E, N and Q; R, K and H; I, V, L and M; A, G, P, S and T; F, Y and W. No penalty was used to correct for gaps in the alignments. (B) Amino acid alignment of the N-terminal domain of hRad54B with that of ScTid1/Rdh54. Identical or similar amino acids are boxed and identical amino acids are shown in bold letters. Amino acid positions are shown. A putative nuclear localization signal is underlined. (C) Multiple sequence alignments of the conserved region in the N-terminal domain of S.cerevisiae Tid1/Rdh54 homologs. Sequences from S.cerevisiae (Sc) Tid1/Rdh54 (CAA85017), S.pombe (Sp) Tid1/Rdh54 homolog (Q09772), C.elegans (Ce) Tid1/Rdh54 homolog (T31515), chicken (c) Rad54B (AAG09308), and human (h) Rad54B (AF112481) are aligned (number in parentheses indicates the GenBank accession number for each protein). Identical or similar amino acids are shaded and identical amino acids are shown in bold. Amino acid positions are shown on the left. Homology searches against databases were performed using BLAST.
Rad54B does not interact directly with human Rad51 and Dmc1
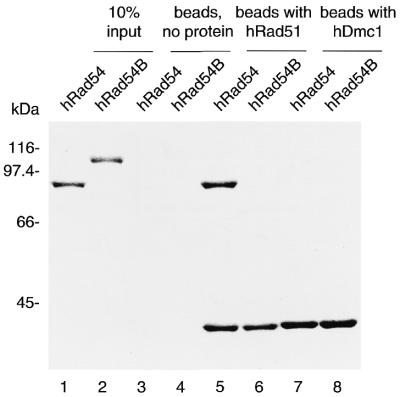
ScTid1/Rdh54 interacts with ScRad51 and a meiosis-specific Rad51 homolog, ScDmc1, in yeast (16,19). Since a sequence similarity exists between ScTid1/Rdh54 and hRad54B, it was of considerable interest to examine whether hRad54B interacts with hRad51 and hDmc1. We prepared hRad51- and hDmc1-conjugated affinity columns (Affi-Gel 15) to address this point. The Affi-Gel 15 beads with active ester sites blocked by ethanolamine were used as negative controls. The hRad54 protein that co-precipitated with the hRad51-conjugated Affi-Gel 15 beads was detected by 10% SDS–PAGE with Coomassie staining (Fig. 5, lane 5), showing the interaction of hRad54 with hRad51, as reported previously (24). On the other hand, we could not detect the interaction of hRad54B with hRad51 (Fig. 5, lane 6). Both hRad54 and hRad54B are deficient in the interaction with hDmc1 (Fig. 5, lanes 7 and 8). In the yeast two-hybrid assay, neither hRad51 nor hDmc1 interacts with hRad54B, further supporting the data shown in Figure 5 (data not shown and 15). These results indicate that hRad54B does not interact directly with hRad51 and hDmc1, which presents a striking contrast to ScTid1/Rdh54 (16,19).
Figure 5.
Protein–protein interaction assay of hRad54 and hRad54B with hRad51 and hDmc1. hRad54 or hRad54B (20 µg) was mixed with either hRad51 or hDmc1 (33 µg) that was covalently conjugated to an Affi-Gel 15 matrix. After 90 min of incubation at room temperature, the Affi-Gel 15 matrix was washed with binding buffer and was directly mixed with 2-fold SDS–PAGE sample buffer (10 µl). This mixture containing the Affi-Gel 15 matrix was boiled at 98°C for 2 min and was fractionated on 9% SDS–PAGE. Lanes 1 and 2 are one-tenth (2 µg) of the input proteins, lanes 3 and 4 are the negative control using the Affi-Gel 15 matrices with active ester sites blocked by ethanolamine, lanes 5 and 6 are the Affi-Gel 15 matrices conjugated with hRad51, and lanes 7 and 8 are the Affi-Gel 15 matrices conjugated with hDmc1. The bands were visualized by Coomassie staining. Molecular mass standards (in kDa) are shown.
Human Rad51 stimulates the ATPase activity of Rad54
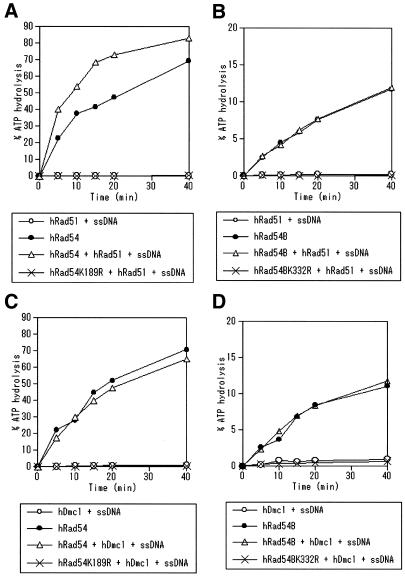
ATP hydrolysis by ScRad54 is required for efficient homologous DNA pairing by ScRad51 (8,11), while ScRad51 stimulates the ATPase activity of ScRad54 (9,10). ATP hydrolysis by ScTid1/Rdh54 is also indispensable for the promotion of D-loop formation by ScRad51 (19). These findings prompted us to examine the effects of hRad51 on the ATPase activities of hRad54 and hRad54B. We found that the hRad51 nucleoprotein filament (hRad51 and ssDNA) enhanced the ATPase activity of hRad54 (Fig. 6A). The ATPase activity of hRad51 was negligible compared with that of hRad54, and the ATPase activity of hRad54K189R remained low in the presence of the hRad51 nucleoprotein filament. These findings suggest that the observed increase in the ATP hydrolysis is attributable to the activity of hRad54. When hRad51 was added instead of the hRad51 nucleoprotein filament, the ATPase activity of hRad54 was similarly enhanced (data not shown). The RecA nucleoprotein filament showed no effect on the ATPase activity of hRad54, indicating the hRad51-specific stimulation of the ATPase activity of hRad54 (data not shown). On the other hand, the ATPase activity of hRad54B was not altered by the addition of the hRad51 nucleoprotein filament (Fig. 6B). It has been reported that hDmc1 forms ring structures that bind ssDNA and dsDNA (25,26), and that hDmc1 catalyzes the D-loop formation (27). Then, we also examined the effect of hDmc1 on the ATPase activities of hRad54 and hRad54B, but the hDmc1–ssDNA complex did not enhance the ATPase activities of hRad54 and hRad54B (Fig. 6C and D). These results and the data shown in Figure 5 suggest that the enhancement of ATPase activity requires the specific protein–protein interaction.
Figure 6.
Effects of hRad51 and hDmc1 on the ATPase activities of hRad54 and hRad54B. (A) hRad54 (120 nM) was incubated with 1 mM ATP and dsDNA (30 µM) without other proteins (closed circles), or with the hRad51–ssDNA complex (open triangles) at 30°C. The activity of the hRad51–ssDNA complex alone (open circles) and the activity of hRad54K189R in the presence of the hRad51–ssDNA complex (crosses) are also shown. (B) hRad54B (120 nM) was incubated with 1 mM ATP and dsDNA (30 µM) without other proteins (closed circles), or with the hRad51–ssDNA complex (open triangles) at 30°C. The activity of the hRad51–ssDNA complex alone (open circles) and the activity of hRad54BK332R in the presence of the hRad51–ssDNA complex (crosses) are also shown. (C) hRad54 (120 nM) was incubated with 1 mM ATP and dsDNA (30 µM) without other proteins (closed circles), or with the hDmc1–ssDNA complex (open triangles) at 30°C. The activity of the hDmc1–ssDNA complex alone (open circles) and the activity of hRad54K189R in the presence of the hDmc1–ssDNA complex (crosses) are also shown. (D) hRad54B (120 nM) was incubated with 1 mM ATP and dsDNA (30 µM) without other proteins (closed circles), or with the hDmc1–ssDNA complex (open triangles) at 30°C. The activity of the hDmc1–ssDNA complex alone (open circles) and the activity of hRad54BK332R in the presence of the hRad51–ssDNA complex (crosses) are also shown.
DISCUSSION
We have shown that the recently identified human Rad54 homolog, Rad54B, is a DNA-binding protein and a dsDNA-dependent ATPase such as hRad54. hRad54B shares homology with ScTid1/Rdh54, which is another Rad54 homolog in yeast, suggesting that hRad54B is the human counterpart of ScTid1/Rdh54. But unlike ScTid1/Rdh54, which interacts with yeast Rad51 and Dmc1, we could not detect the direct interaction of hRad54B with hRad51 and hDmc1, indicating the functional difference between ScTid1/Rdh54 and hRad54B.
Human RAD54B was cloned by the sequence similarity with hRAD54 (5). ScRad54, ScTid1/Rdh54, hRad54 and hRad54B belong to a subfamily of the SNF2/SWI2 family because they share high homology not only in the seven helicase motifs but also around these motifs in the SNF2 domain (5,23). There is a highly homologous region in the N-terminal domain of hRad54B and ScTid1/Rdh54, where little homology exists between hRad54 and hRad54B. This region contains a cluster of basic amino acid residues, which is a putative nuclear localization signal. We found that there are SNF2/SWI2 family proteins containing the similar region in other eukaryotes, indicating that the structure of ScTid1/Rdh54 is conserved among eukaryotes. These findings suggest that hRad54B is the human counterpart of ScTid1/Rdh54.
Human Rad54B was found to be a DNA-binding protein and to possess a dsDNA-dependent ATPase activity such as ScTid1/Rdh54, but there were two functional differences between hRad54B and ScTid1/Rdh54. First, hRad54B showed lower ATPase activity than did hRad54. In contrast to our results, previous studies have shown that the rate of ATP hydrolysis by ScRad54 and ScTid1/Rdh54 are comparable (1.27 × 103 min–1 at 1 mM ATP and 2.2 × 103 min–1 at 1.5 mM ATP, respectively) (7,19). Since we compared the ATPase activities of hRad54 and hRad54B under the same conditions using the proteins purified by the same procedure, it is likely that hRad54B is a less active ATPase than hRad54, showing a difference between species. Second, neither hRad51 nor hDmc1 interacted directly with hRad54B. We have previously reported that hRad54B associates with hRad51 in mammalian cells (15). One possibility is that hRad54B interacts with hRad51 indirectly and that there may be other proteins mediating the complex formation. Co-localization of hRad54B with hRad51, hRad54 and Brca1 in the nucleus supports the hypothesis that hRad54B is included in the recombination protein complex (15). Another possibility is that some post-translational modification, such as phosphorylation, stimulates the interaction between hRad54B and hRad51 (or hDmc1), as previously reported concerning the interaction between Rad51 and Rad52 (28).
In S.cerevisiae, both Rad54 and Tid1/Rdh54 play roles in homologous recombination. It has been reported that ScTid1/Rdh54 functions preferentially in recombination between homologous chromosomes, while ScRad54 is required primarily for sister chromatid-based repair (17,18,20). Human Rad54 has similar biochemical properties to its yeast counterpart and acts as a recombination protein. Biochemically, hRad54 possesses a robust ATPase activity and interacts with hRad51 (23,24), which enhances the ATPase activity of hRad54. Biologically, homozygous RAD54 mutants in mouse and chicken show high sensitivity to radiation and methyl methanesulfonate and have reduced levels of homologous recombination (13,14). In contrast, it is not clear whether hRad54B is involved in homologous recombination. But we have found that inactivation of hRAD54B in a colon cancer cell line resulted in severe reduction of targeted integration frequency (29), suggesting that hRAD54B also plays a role in homologous recombination. It is of interest whether hRad54B plays a role in meiotic recombination and mitotic interchromosomal recombination, which is considered to be very rare in animal cells.
Recently, the yeast SWI/SNF complex and other SWI2/SNF2 family protein-containing complexes were shown to generate superhelical torsion in DNA and chromatin (30), which may disrupt chromatin structure and increase the accessibility of DNA within chromatin. It was reported that human and yeast Rad54 alter DNA superhelicity using the energy of ATP hydrolysis (8–10,19,31). The dsDNA-dependent ATPase activities of hRad54 and hRad54B may play a role in the recombination process by making chromatin structure accessible for other recombination proteins or nucleoprotein filaments.
The RAD52 epistasis group genes are well conserved throughout evolution, but recent studies revealed that the role of each gene product is not necessarily the same in vertebrates as compared with yeast (2). For example, Rad52 is essential in yeast, while Rad52-deficient vertebrate cells show subtle phenotype (32,33), although DT40 cells deficient in both Rad52 and Xrcc3 are not viable (34). It has also been reported that the Rad51-paralog proteins, Xrcc3 and Rad51C, form a stable complex and catalyze the D-loop formation (35). Both ScRad54 and ScTid1/Rdh54 enhance the D-loop formation by ScRad51 (7,9,10,19). We have examined the effects of hRad54 and hRad54B on the D-loop formation by hRad51 and hDmc1 using the pGsat4 plasmid and the 50mer oligonucleotide, SAT-1, as substrates (22,35), but neither hRad54 nor hRad54B enhanced the D-loop formation by hRad51 and hDmc1 (K.Tanaka, W.Kagawa, T.Kinebuchi, H.Kurumizaka and K.Miyagawa, unpublished data). These findings suggest that hRad54 and hRad54B behave in a different way from ScRad54 and ScTid1/Rdh54 in the recombination process.
We have reported mutations in hRAD54 and hRAD54B in primary tumors (5,36). It is important to study whether these mutations affect the biochemical and biological functions of hRad54 and hRad54B. Recently, Lee et al. reported that ScTid1/Rdh54 is required for adaptation from G2/M arrest induced by a double-strand break in yeast (37), suggesting that hRad54B might play a role separate from homologous recombination in mitotic cells. Further investigations are required for a comprehensive understanding of the functions of hRad54 and hRad54B, and their roles in carcinogenesis.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Akira Shinohara (Osaka University, Osaka, Japan) for his gift of an antiserum against the chicken Rad54 protein, and Dr Shigeyuki Yokoyama (RIKEN Genomic Sciences Center, Yokohama, Japan) and Dr Takehiko Shibata (Cellular and Molecular Biology Laboratory, RIKEN, Saitama, Japan) for their helpful discussions. We are grateful to Mr Hiroshi Yokoyama (RIKEN Genomic Sciences Center, Yokohama, Japan), Dr Yuji Masuda and Miss Aiko Kinomura (Hiroshima University, Hiroshima, Japan) for their special technical assistance. This work was supported by Grants-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Johnson R.D. and Jasin,M. (2001) Double-strand-break-induced homologous recombination in mammalian cells. Biochem. Soc. Trans, 29, 196–201. [DOI] [PubMed] [Google Scholar]
- 2.Sonoda E., Takata,M., Yamashita,Y.M., Morrison,C. and Takeda,S. (2001) Homologous DNA recombination in vertebrate cells. Proc. Natl Acad. Sci. USA, 98, 8388–8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Game J.C. (1993) DNA double-strand breaks and the RAD50-RAD57 genes in Saccharomyces. Semin. Cancer Biol., 4, 73–83. [PubMed] [Google Scholar]
- 4.Petrini J.H., Bressan,D.A. and Yao,M.S. (1997) The RAD52 epistasis group in mammalian double strand break repair. Semin. Immunol., 9, 181–188. [DOI] [PubMed] [Google Scholar]
- 5.Hiramoto T., Nakanishi,T., Sumiyoshi,T., Fukuda,T., Matsuura,S., Tauchi,H., Komatsu,K., Shibasaki,Y., Inui,H., Watatani,M., Yasutomi,M., Sumii,K., Kajiyama,G., Kamada,N., Miyagawa,K. and Kamiya,K. (1999) Mutations of a novel human RAD54 homologue, RAD54B, in primary cancer. Oncogene, 18, 3422–3426. [DOI] [PubMed] [Google Scholar]
- 6.Eisen J.A., Sweder,K.S. and Hanawalt,P.C. (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res., 23, 2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petukhova G., Stratton,S. and Sung,P. (1998) Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature, 393, 91–94. [DOI] [PubMed] [Google Scholar]
- 8.Petukhova G., Van Komen,S., Vergano,S., Klein,H. and Sung,P. (1999) Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem., 274, 29453–29462. [DOI] [PubMed] [Google Scholar]
- 9.Van Komen S., Petukhova,G., Sigurdsson,S., Stratton,S. and Sung,P. (2000) Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell, 6, 563–572. [DOI] [PubMed] [Google Scholar]
- 10.Mazin A.V., Bornarth,C.J., Solinger,J.A., Heyer,W.-D. and Kowalczykowski,S.C. (2000) Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell, 6, 583–592. [DOI] [PubMed] [Google Scholar]
- 11.Solinger J.A., Lutz,G., Sugiyama,T., Kowalczykowski,S.C. and Heyer,W.-D. (2001) Rad54 protein stimulates heteroduplex DNA formation in the synaptic phase of DNA strand exchange via specific interactions with the presynaptic Rad51 nucleoprotein filament. J. Mol. Biol., 307, 1207–1221. [DOI] [PubMed] [Google Scholar]
- 12.Swagemakers S.M., Essers,J., de Wit,J., Hoeijmakers,J.H. and Kanaar,R. (1998) The human Rad54 recombinational DNA repair protein is a double-stranded DNA-dependent ATPase. J. Biol. Chem., 273, 28292–28297. [DOI] [PubMed] [Google Scholar]
- 13.Bezzubova O., Silbergleit,A., Yamaguchi-Iwai,Y., Takeda,S. and Buerstedde,J.M. (1997) Reduced X-ray resistance and homologous recombination frequencies in a RAD54-/- mutant of the chicken DT40 cell line. Cell, 89, 185–193. [DOI] [PubMed] [Google Scholar]
- 14.Essers J., Hendriks,R.W., Swagemakers,S.M., Troelstra,C., de Wit,J., Bootsma,D., Hoeijmakers,J.H. and Kanaar,R. (1997) Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell, 89, 195–204. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka K., Hiramoto,T., Fukuda,T. and Miyagawa,K. (2000) A novel human Rad54 homologue, Rad54B, associates with Rad51. J. Biol. Chem., 275, 26316–26321. [DOI] [PubMed] [Google Scholar]
- 16.Dresser M.E., Ewing,D.J., Conrad,M.N., Dominguez,A.M., Barstead,R., Jiang,H. and Kodadek,T. (1997) DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics, 147, 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein H.L. (1997) RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics, 147, 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinohara M., Shita-Yamaguchi,E., Buerstedde,J.-M., Shinagawa,H., Ogawa,H. and Shinohara,A. (1997) Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics, 147, 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petukhova G., Sung,P. and Klein,H. (2000) Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev., 14, 2206–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arbel A., Zenvirth,D. and Simchen,G. (1999) Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1. EMBO J., 18, 2648–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurumizaka H., Aihara,H., Kagawa,W., Shibata,T. and Yokoyama,S. (1999) Human Rad51 amino acid residues required for Rad52 binding. J. Mol. Biol. 291, 537–548. [DOI] [PubMed] [Google Scholar]
- 22.Kagawa W., Kurumizaka,H., Ikawa,S., Yokoyama,S. and Shibata,T. (2001) Homologous pairing promoted by the human Rad52 protein. J. Biol. Chem., 276, 35201–35208. [DOI] [PubMed] [Google Scholar]
- 23.Kanaar R., Troelstra,C., Swagemakers,S.M., Essers,J., Smit,B., Franssen,J.H., Pastink,A., Bezzubova,O.Y., Buerstedde,J.M., Clever,B., Heyer,W.-D. and Hoeijmakers,J.H. (1996) Human and mouse homologs of the Saccharomyces cerevisiae RAD54 DNA repair gene: evidence for functional conservation. Curr. Biol., 6, 828–838. [DOI] [PubMed] [Google Scholar]
- 24.Golub E.I., Kovalenko,O.V., Gupta,R.C., Ward,D.C. and Radding,C.M. (1997) Interaction of human recombination proteins Rad51 and Rad54. Nucleic Acids Res., 25, 4106–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masson J.-Y., Davis,A.A., Hajibagheri,N., Van Dyck,E., Benson,F.E., Stasiak,A.Z., Stasiak,A. and West,S.C. (1999) The meiosis-specific recombinase hDmc1 forms ring structures and interacts with hRad51. EMBO J., 18, 6552–6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Passy S.I., Yu,X., Li,Z., Radding,C.M., Masson,J.-Y. West,S.C. and Egelman,E.H. (1999) Human Dmc1 protein binds DNA as an octameric ring. Proc. Natl Acad. Sci. USA, 96, 10684–10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z., Golub,E.I., Gupta,R. and Radding,C.M. (1997) Recombination activities of HsDmc1 protein, the meiotic human homolog of RecA protein. Proc. Natl Acad. Sci. USA, 94, 11221–11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen G., Yuan,S.-S.F., Liu,W., Xu,Y., Trujillo,K., Song,B., Cong,F., Goff,S.P., Wu,Y., Arlinghaus,R., Baltimore,D., Gasser,P.J., Park,M.S., Sung,P. and Lee,E.Y.-H.P. (1999) Radiation-induced assembly of Rad51 and Rad52 recombination complex requires ATM and c-Abl. J. Biol. Chem., 274, 12748–12752. [DOI] [PubMed] [Google Scholar]
- 29.Miyagawa K., Tsuruga,T., Kinomura,A., Usui,K., Katsura,M., Tashiro,S., Mishima,H. and Tanaka,K. (2002) A role for RAD54B in homologous recombination in human cells. EMBO J., 21, 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Havas K., Flaus,A., Phelan,M., Kingston,R., Wade,P.A., Lilley,D.M.J. and Owen-Hughes,T. (2000) Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell, 103, 1133–1142. [DOI] [PubMed] [Google Scholar]
- 31.Tan T.L.R., Essers,J., Citterio,E., Swagemakers,S.M.A., de Wit,J., Benson,F.E., Hoeijmakers,J.H.J. and Kanaar,R. (1999) Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr. Biol., 9, 325–328. [DOI] [PubMed] [Google Scholar]
- 32.Rijkers T., van den Ouweland,J., Morolli,B., Rolink,A.G., Baarends,W.M., Van Sloun,P.P.H., Lohman,P.H.M. and Pastink,A. (1998) Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol., 18, 6423–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaguchi-Iwai Y., Sonoda,E., Buerstedde,J.-M., Bezzubova,O., Morrison,C., Takata,M., Shinohara,A. and Takeda,S. (1998) Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol. Cell. Biol., 18, 6430–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujimori A., Tachiiri,S., Sonoda,E., Thompson,L.H., Dhar,P.K., Hiraoka,M., Takeda,S., Zhang,Y., Reth,M. and Takata,M. (2001) Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J., 20, 5513–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurumizaka H., Ikawa,S., Nakada,M., Eda,K., Kagawa,W., Takata,M., Takeda,S., Yokoyama,S. and Shibata,T. (2001) Homologous-pairing activity of the human DNA-repair proteins Xrcc3·Rad51C. Proc. Natl Acad. Sci. USA, 98, 5538–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuda M., Miyagawa,K., Takahashi,M., Fukuda,T., Kataoka,T., Asahara,T., Inui,H., Watatani,M., Yasutomi,M., Kamada,N., Dohi,K. and Kamiya,K. (1999) Mutations in the RAD54 recombination gene in primary cancers. Oncogene, 18, 3427–3430. [DOI] [PubMed] [Google Scholar]
- 37.Lee S.E., Pellicioli,A., Malkova,A., Foiani,M. and Haber,J.E. (2001) The Saccharomyces recombination protein Tid1p is required for adaptation from G2/M arrest induced by a double-strand break. Curr. Biol., 11, 1053–1057. [DOI] [PubMed] [Google Scholar]