Abstract
Infrared and UV spectroscopies have been used to study the assembly of a hairpin nucleotide sequence (nucleotides 3–30) of the 5′ non-coding region of the hepatitis C virus RNA (5′-GGCGGGGAUUAUCCCCGCUGUGAGGCGG-3′) with a RNA 20mer ligand (5′-CCGCCUCACAAAGGUGGGGU-3′) in the presence of magnesium ion and spermidine. The resulting complex involves two helical structural domains: the first one is an intermolecular duplex stem at the bottom of the target hairpin and the second one is a parallel triplex generated by the intramolecular hairpin duplex and the ligand. Infrared spectroscopy shows that N-type sugars are exclusively present in the complex. This is the first case of formation of a RNA parallel triplex with purine motif and shows that this type of targeting RNA strands to viral RNA duplexes can be used as an alternative to antisense oligonucleotides or ribozymes.
INTRODUCTION
The nucleotide sequence of the genome of hepatitis C virus, the main causative agent of chronic non-A, non-B hepatitis, has been elucidated (1,2). An option for disrupting viral gene expression at the level of transcription uses synthetic oligonucleotides capable of hybridizing with double-stranded nucleic acids (3). Such triplex-forming oligonucleotides (TFOs) can lead to the so-called purine (YR⋅R) and pyrimidine (YR⋅Y) triplexes, depending on whether they consist of purines or pyrimidines. If the oligonucleotide ligands are of pyrimidine type, their binding to nucleic acid double helices is pH dependent because cytosines must be protonated to form two hydrogen bonds with guanine. However, triplex formation involving purine ligands does not depend on pH (3). Examples of both structural motifs have been identified with DNA (3,4–6). Examples of pyrimidine (YR⋅Y) triplexes are well known (7–9) in RNA, but no detailed structure involving purine motifs has been reported so far. In addition, poly(rA) and poly(rG) have been reported to bind to poly(rG·rC) duplex (10,11), but no structural data were given regarding triple helix formation.
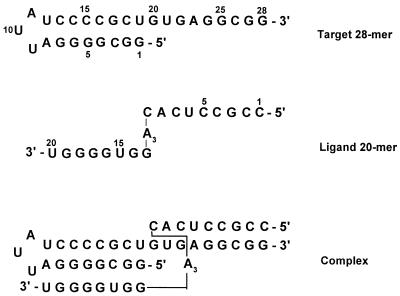
Although the RNA of hepatitic C virus can be written as single strands, self-pairing between adjacent or remote sequences gives rise to double-stranded regions. The well conserved 5′ non-coding region contains a hairpin (nucleotides 3–30) (1) the duplex stem of which comprises 8 bp (Fig. 1). Despite the hairpin stem including a purine-rich tract suitable as a target for TFOs, it seems relatively short to lead to insufficiently stable triplexes because the length of triplexes is usually not less than 9 or 10 nt (12). One way to increase the stability of ligand–duplex complexes is to form, simultaneously, a double strand with a single-stranded region at the bottom of a nucleic acid hairpin, and a triple helix against the duplex region (13), whereby the required binding free energy could be gained. This is why we have prepared a complex as described in Figure 1. The ligand oligonucleotide has two domains; the first one (nucleotides 1–9) is complementary to the single-stranded sequence at the bottom of the hairpin, and the second one (nucleotides 13–20) is designed to form a triplex with the duplex stem of the target hairpin.
Figure 1.
Sequences of the target and ligand oligonucleotides.
Fourier transform infrared spectroscopy has proven to be well suited for the characterization of nucleic acid conformations (14,15), providing information on interactions involving specific groups. We confirm here, by infrared spectroscopy, the in vitro formation by this TFO of the triple helix, which also provides structural data concerning the nucleoside conformations (14,15). The formation of the above complex involving the triplex and duplex domains has also been followed by UV absorption spectrophotometry (thermal denaturation) and gel retardation electrophoresis.
MATERIALS AND METHODS
Oligonucleotides
The oligonucleotides were obtained from Xeragon (Switzerland). They were purified by HPLC and desalted, then lyophilized and stored frozen in 10 mM Tris–borate, 10 mM MgCl2 and 2 mM spermidine (pH 8.0). Their concentrations were determined on the basis of their absorbance (16). Triple helix complex formation was performed by equimolar mixing of the target hairpin and ligand, at a total concentration of 100 µM in nucleotides, then heating to 80°C for 2 min in 10 mM Tris–borate, 10 mM MgCl2 and 2 mM spermidine (pH 8.0), and slowly cooling (5°C/h) to 4°C. The resulting stock solution was then allowed to stand at 4°C for ∼18 h. The 9 bp duplex was also prepared in this way by equimolar mixing of the two oligonucleotides having the nucleotides 20–28 and 1–9 sequences of the target hairpin and ligand, respectively.
Oligonucleotide melting studies
The melting temperatures of the oligonucleotide samples were measured by UV spectroscopy, using a Shimadzu UV-2100 spectrophotometer. The temperature of the cell holder (sample and reference) was varied by circulating water through a thermostat device. Measurement of the temperature was performed directly in the reference cell.
Aliquots of the oligonucleotide stock solutions were taken in order to prepare buffered solution samples (10 mM Tris–borate, 10 mM MgCl2 and 2 mM spermidine), with 50 µM total concentration in nucleotides. As described above for the triple helix complex formation, the solution samples of the isolated target hairpin and ligand were incubated at 80°C for 2 min, then slowly annealed (5°C/h) from 80 to 4°C and stored overnight at 4°C. The solution samples were then heated to 85°C again at 1°C/min intervals. The melting curves of the 9 bp duplex, target hairpin and ligand were also recorded using solution samples at 50 mM total concentration in nucleotides and heating to 85°C at 1°C/min intervals. The absorption was monitored at 260 nm.
Infrared spectroscopy
FTIR measurements were performed using a Perkin Elmer 1725X Fourier spectrophotometer equiped with a DGTS detector. About 1–1.5 µl droplets of concentrated buffered D2O solutions containing either target hairpin, ligand or triple helix complex (4 mM in single strands) were deposited in water-jacketed cells sealed by ZnSe windows. The resolution was set to 2 cm–1 and 64 scans were accumulated. Spectral data were treated with the Galaxy Spectra Calc. program which includes baseline correction, smoothing and solvent substraction. The phosphate symmetric stretching vibration appearing near 1085 cm–1 was used as internal standard for spectral normalization.
Gel retardation electrophoresis
Part of the above buffered solutions of hairpin alone or ligand bound to the hairpin, used for melting studies, were employed for gel retardation assays with the aim of establishing hairpin–ligand complex formation. Samples were loaded onto a 12% polyacrylamide gel (43:1 acrylamide:bisacrylamide) at 10°C, which had been prepared with 50 mM Tris–borate, 10 mM MgCl2 and 2 mM spermidine (pH 8.2). Samples were electrophoresed in this buffer at 6 V/cm at 10°C before being visualized.
Molecular modeling
Conformational energy minimizations of the triple helix segments were performed using the AMBER 4.1 suite of computer programs. No water molecules were explicitly taken into account in these calculations. Their effect was simulated by the use of a sigmoidal distance-dependent dielectric function. Triple helices were built from a library of ribonucleotides by specifying helicoidal parameters in agreement with the Cambridge convention. Starting geometries have been built by introducing into theoretical models the infrared experimental data concerning sugar conformations. For the triple helix, as the infrared spectra showed the presence of only N-type sugars, triplex models have been constructed including N-type sugars in the Watson–Crick duplex and in the third strand.
RESULTS AND DISCUSSION
Thermal behavior of oligonucleotide samples
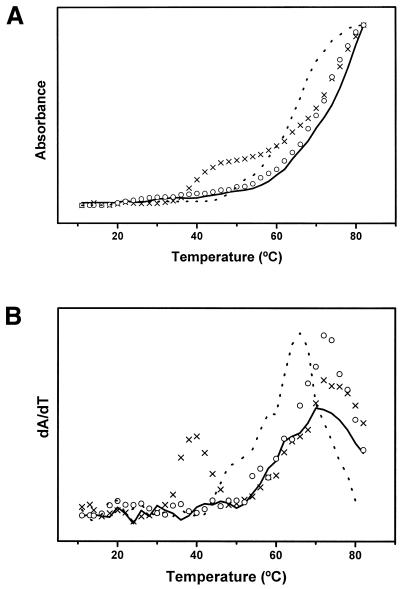
The melting curve obtained for the hairpin target 28mer is presented in Figure 2. Only one transition is detected with a midpoint located at 70°C. Some previous thermal denaturation studies dealing with oligonucleotide folding in solution reported two-step melting profiles (17–19). The first transition occurring at low temperatures was concentration dependent and was attributable to an intermolecular conversion from duplex to hairpin. The second one, independent of oligonucleotide concentration, was assigned to the conversion from hairpin to coil. In our investigation the possible duplex–hairpin transition is not detected. This may be explained by the fact that the low oligonucleotide concentration used here favors the existence of intramolecular hairpins rather than intermolecular duplexes.
Figure 2.
(A) UV melting curves of the target 28mer hairpin (solid line), ligand 20mer (dotted line), 9 bp duplex (circles) and complex (crosses). (B) Derivatives of the 260 nm absorbance versus temperature.
The melting curve for the ligand oligonucleotide (Fig. 2) also shows only one transition occurring at 63°C. This transition is probably generated by disruption of the Watson–Crick intramolecular duplex that can be formed through base pairing between the 3–9 and 14–20 nt sequences of the ligand 20mer. This Tm is lower than that of the target hairpin as expected from the number of the GC base pairs and duplex fragment lengths (Fig. 1). In contrast, the melting profile of the hairpin–ligand complex seems to be biphasic. However, on the assumption of the complex structure shown in Figure 1, three transitions should be expected: the one corresponding to the third-strand separation in the triplex fragment and two more transitions generated by the intra- and intermolecular duplex fragments where the hairpin target is involved. We have recorded the melting curve of an independent duplex, which is equivalent to the hairpin bottom one in the complex, thus consisting of the 1–9 and 20–28 nt sequences of the ligand and target hairpin, respectively (Fig. 1). The observed Tm at 72°C for this independent 9 bp duplex is practically coincident with the highest Tm of the complex, which suggests that the hairpin–ligand interactions involve formation of double helix through the above hairpin nucleotides 20–28 and ligand nucleotides 1–9 sequences, as reflected in the proposed complex structure (Fig. 1). This Tm is very near that of the hairpin intramolecular duplex disruption (70°C), whereby these two transition temperatures are not resolved for the target hairpin–ligand complex. The concentration independent first transition (39°C) of the complex curve can be assigned to third-strand dissociation of the triple helix structure in agreement with infrared spectroscopy, as described later. On the other hand, although in this case the assays of gel retardation electrophoresis are not specifically indicative of triplex formation, they do support the intermolecular association between the target hairpin and ligand, because the hairpin–ligand complex was found to be substantially retarded in its mobility compared with the isolated oligonucleotides (Fig. 3). It is, therefore, reasonable to exclude G-duplex or tetraplex formation from guanine-rich ligand chains, because otherwise the electrophoretic spot of free hairpin would have appeared, which is not the case.
Figure 3.
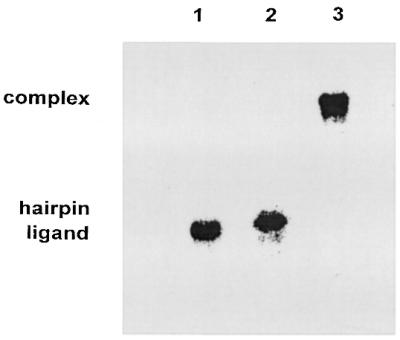
Gel mobility shift assay for complex formation by the target 28mer hairpin and the ligand 20mer. Lane 1, ligand; lane 2, hairpin; lane 3, complex.
Infrared spectroscopy and triplex formation
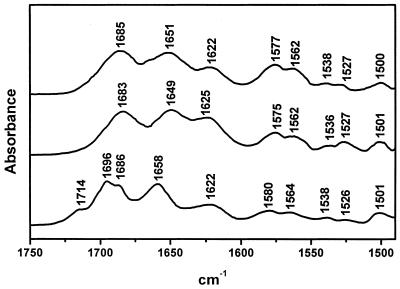
The infrared spectra of nucleic acids in the 1800–1500 cm–1 range consist of absorption bands originating from the in-plane double-bond vibrations of the bases. These bands are sensitive to base interactions involving hydrogen bonding and/or stacking. This spectral region was studied in heavy water solution due to the presence of a strong interfering water absorption band around 1640 cm–1. Figure 4 includes the infrared spectrum of the RNA hairpin where two intense bands close to 1685 and 1651 cm–1 are observed. The band at 1685 cm-1 has been assigned to the C6=O6 stretching of base-paired guanines plus C2=O2 stretching of uracils (20,21), and the 1651 cm–1 band has been assigned to νC2=O2 vibrations of cytosines plus νC4=O4 of uracils. Upon melting, the band near 1685 cm-1 decreases in intensity and shifts to ∼1660 cm–1. This band is due to overlapping contribution of the νC6=O6 vibrations of free or non-base-paired guanines, the νC2=O2 vibrations of free cytosines and the νC4=O4 vibrations of free uracils. Consequently, the carbonyl band located at 1685 cm-1 can be attributed to the duplex stem of the target hairpin (Fig. 1). The same can be said for the infrared spectrum of the ligand 20mer, which shows a strong band near 1683 cm-1 (Fig. 4). The duplex in this case can be formed through antiparallel base pairing of the 3–9 and 14–20 nt sequences in the ligand 20mer.
Figure 4.
Infrared spectra recorded in D2O solutions at 15°C; target 28mer hairpin (top), ligand (middle) and complex (bottom).
The band around 1622 cm–1 is generated by the νC=C and νC=N vibrations of adenine along with a minor contribution of some ring vibrations of cytosine (14,20). The two bands located in the 1580–1560 cm–1 range arise mainly from the guanine νC=N motions (22) and show more intensity in the target hairpin. This can be explained on the basis of the fact that the hairpin guanine-rich sequence (nucleotides 20–28) is single stranded, which involves increasing intensity (22,23) when compared with the base-paired guanine-rich (nucleotides 14–20) sequence of the ligand 20mer.
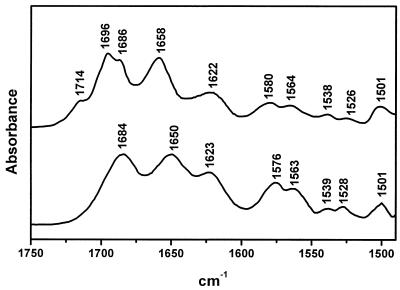
Some characteristic spectral features are observed in the infrared spectrum of the target hairpin–ligand complex (Fig. 4). Compared with the spectra of the pure components, two new bands appear at 1714 and 1696 cm–1. These two bands are present in the infrared spectra of nucleic acid triple helices, including G*G⋅C triplets and third-strand parallel orientation, relative to the target purine chain in the duplex (24,25). On the other hand, the guanine bands in the 1580–1560 cm–1 range show weaker intensities when compared with the isolated components, which supports that the guanine bases in the 20–28 nt sequence of the target hairpin are Watson–Crick paired. The infrared spectra recorded at different temperatures allow us to assign the 39°C UV transition to the third-strand dissociation of the triple helix structure. In fact, the 1714 and 1696 cm–1 bands, which are attributable to triplex formation, disappear upon heating above 40°C (Fig. 5). A complex solution heated, for instance, at 48°C shows that the above bands are absent, and the guanine νC6=O6 vibration is located at a frequency (1684 cm–1) that is characteristic of duplexes, as described above. The dissociation of the guanine-rich third strand involves the known intensity increase observed for the guanine bands near 1580 and 1565 cm–1. Hence, the transition at 39°C occurs through the third-strand dissociation and maintenance of the duplex segments in the complex. The ultraviolet spectroscopy melting curves and these infrared results, taken together, confirm complex formation including triplex and duplex domains as suggested in Figure 1.
Figure 5.
Infrared spectra of the complex recorded in D2O solutions at 15°C (top) and 48°C (bottom).
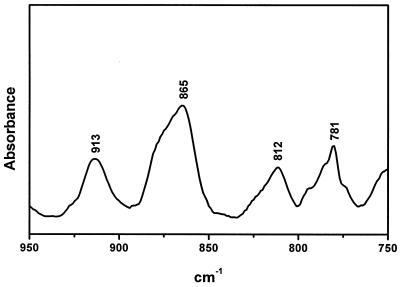
The spectrum of the complex in the 900–750 cm–1 region is shown in Figure 6, where bands characteristic of the sugar conformations are observed. The absorption band at 812 cm–1 is due to sugar ring vibrations (7,8) and is a marker for the C(3′)-endo-anti sugar pucker, and the same can be said for the 865 cm–1 band. RNA oligonucleotides classically contain sugars in this nucleoside conformation (N-type geometry associated with A family-type RNA conformation) and are usually observed around 866 and 814 cm-1, while the S-type sugars [C(2′)-endo-anti, B family-form geometry] are detected by an absorption located near 834 cm-1 (7,8). Consequently, the spectrum of the target hairpin–ligand complex presents, as expected, exclusively N-type sugar absorptions observed at the above bands.
Figure 6.
Infrared spectrum of the complex in D2O solution at 15°C.
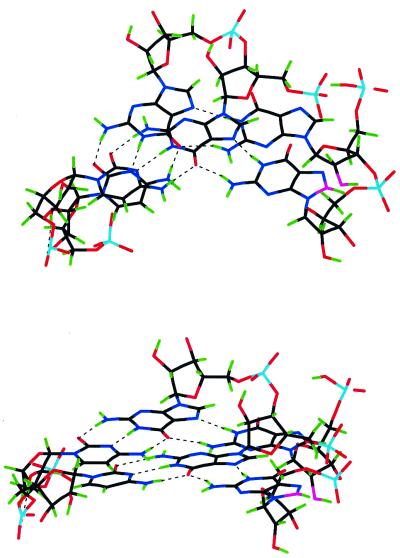
In conclusion, apart from the fact that the potential formation of the above complex in vivo may be of biological relevance, there is an important point concerning triple helix formation. We report here the first case of formation of a purine motif RNA triplex with the third-strand oligonucleotide attached in a parallel fashion to the corresponding purine target, this binding probably being stabilized by the adjacent duplex in the above complex. Hence, these spectroscopic results are indicative that RNA third strands can not be excluded in purine motif RNA triplex formation. Although both parallel and antiparallel triplexes with purine motif have been reported for DNA (24,26,27), previous attempts to obtain some of the above structures for RNA were not successful, since RNA strands were excluded from RNA purine triple helices (28) because they were not formed through incubation at room temperature. However, the addition of spermidine and incubation at cooler temperatures in the experiments reported here seems to aid the formation of the purine motif. Moreover, in the above previous study (28) aimed at obtaining purine motif RNA triplexes only the third-strand antiparallel orientation was considered, and this structure seems to be hampered by steric factors as deduced from calculations of molecular energy minimization. In fact, the triplex stem with third-strand antiparallel orientation relative to the target purine chain shows the hydroxyl groups on the third strand pointing toward the neighboring nucleobase on the 5′-side of the same strand and are sterically unfavorable in the triple helix structure (Fig. 7). Thus, our molecular modeling study suggests that a RNA third-strand antiparallel orientation seems to be less favored than the parallel one. Consequently, and with the aim of inhibiting the gene expression of RNA virus, appropriate oligoribonucleotides can be used to target certain sequences in a RNA viral duplex leading to parallel purine motif RNA triplexes, and this targeting could be either alternative or simultaneous with that involving antisense oligonucleotides or ribozymes.
Figure 7.
Model extracted from minimized structures of adjacent G*G⋅C triplets in an antiparallel triplex domain of the complex, to illustrate the position of the ligand hydroxyl groups. For sake of clarity, only two nucleotides of the three strands have been drawn. Two perspectives of the model are given in order to show the N-type sugar and anti-nucleoside conformation of the third strand located on the right part in each perspective. The oxygen atom of the ribose C(2′)OH group, which is very near the neighbor nucleobase on the 5′-side of the same strand, and the C8 atom of this nucleobase are in violet.
Acknowledgments
ACKNOWLEDGEMENT
We are grateful to Dirección General de Investigación, Comunidad Autónoma de Madrid, for financial support (Project 08.2/0030.1/99).
REFERENCES
- 1.Okamoto H., Okada,S., Sugiyama,Y., Kurai,K., Iizuka,H., Machida,A., Miyakawa,Y. and Mayumi,M. (1991) Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent regions. J. Gen. Virol., 72, 2697–2704. [DOI] [PubMed] [Google Scholar]
- 2.Okamoto H., Kurai,K., Okada,S.I., Yamamoto,K., Lizuka,H., Tanaka,T., Fukuda,S., Tsuda,F. and Mishiro,S. (1992) Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology, 188, 331–341. [DOI] [PubMed] [Google Scholar]
- 3.Praseuth D., Guieysse,A.L. and Hélène,C. (1999) Triple helix formation and the antigene strategy for sequence-specific control of gene expression. Biochim. Biophys. Acta, 1489, 181–206. [DOI] [PubMed] [Google Scholar]
- 4.Moser H.E. and Dervan,P.B. (1987) Sequence-specific cleavage of double helical DNA by triple helix formation. Science, 238, 645–650. [DOI] [PubMed] [Google Scholar]
- 5.Beal P. and Dervan,P.B. (1991) Second structural motif for recognition of DNA by oligonucleotide directed triple-helix formation. Science, 251, 1360–1363. [DOI] [PubMed] [Google Scholar]
- 6.Broitman S.L., Im,D.D. and Fresco,J.R. (1987) Formation of the triple-stranded polynucleotide helix, poly(A.A.U). Proc. Natl Acad. Sci. USA, 84, 5120–5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liquier J., Akhebat,A., Taillandier,E., Ceolin,F., Huynh-Dinh,T. and Igolen,J. (1991) Characterization by FTIR spectroscopy of the oligoribonucleotide duplexes r(A-U)6 and r(A-U)8. Spectrochim. Acta, 47A, 177–186. [Google Scholar]
- 8.Liquier J., Coffinier,P., Firon,M. and Taillandier,E. (1991) Triple helical polynucleotidic structures: sugar conformations determined by FTIR spectroscopy. J. Biomol. Struct. Dyn., 9, 437–445. [DOI] [PubMed] [Google Scholar]
- 9.Hoyne P.R., Gacy,A.M., McMurray,C.T. and Maher,L.J.,III (2000) Stabilities of intrastrand pyrimidine motif DNA and RNA triple helices. Nucleic Acids Res., 28, 770–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chastain M. and Tinoco,I.,Jr (1992) Poly(rA) binds poly(rG).poly(rC) to form a triple helix. Nucleic Acids Res., 20, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letai A.G., Palladino,M.A., Fromm,E., Rizzo,V. and Fresco,J.R. (1988) Specificity in information of triple-stranded nucleic acid helical complexes: studies with agarose-linked polyribonucleotide affinity columns. Biochemistry, 27, 9108–9112. [DOI] [PubMed] [Google Scholar]
- 12.Soyfer V.N. and Potaman,V.N. (1996) Triple-Helical Nucleic Acids. Springer, New York, Berlin.
- 13.Brossalina E. and Toulmé,J.J. (1993) A DNA hairpin as a target for antisense oligonucleotides. J. Am. Chem. Soc., 115, 796–797. [Google Scholar]
- 14.Dagneaux C., Gousset,H., Shchyolkina,A.K., Ouali,M., Letellier,R., Liquier,J., Florentiev,V.L. and Taillandier,E. (1996) Parallel and antiparallel A*A-T intramolecular triple helixes. Nucleic Acids Res., 24, 4506–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liquier J., Taillandier,E., Klinck,R., Guittet,E., Gouyette,C. and Huynh-Dinh,T. (1995) Spectroscopic studies of chimeric DNA-RNA and RNA 29-base intramolecular triple helixes. Nucleic Acids Res., 23, 1722–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fasman G.D. (1975) CRC Handbook of Biochemistry and Molecular Biology: Nucleic Acids, 3rd Edn. CRC Press, Boca Raton, FL, Vol. 1, p. 589.
- 17.Wemmer D.E., Chou,S.H., Hare,D.R. and Reid,B.R. (1985) Duplex-hairpin transitions in DNA: NMR studies on CGCGTATACGCG. Nucleic Acids Res., 13, 3755–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haasnoot C.A.G., Hilberts,C.W., van der Marel,G.A., van Boom,J.H., Singh,U., Pattabhiraman,N. and Kollman,P.A. (1986) On loop folding in nucleic acid hairpin-type structures. J. Biomol. Struct. Dyn., 4, 843–857. [DOI] [PubMed] [Google Scholar]
- 19.Xodo L.E., Manzini,G., Duadrifoglio,F., van der Marel,G.A. and van Boom,J.H. (1988) Oligonucleotide folding in solution: loop size and stability of B-hairpins. Biochemistry, 27, 6321–6326. [DOI] [PubMed] [Google Scholar]
- 20.Tsuboi M. (1969) Application of infrared spectroscopy to structure studies of nucleic acids. App. Spectrosc. Rev., 3, 45–90. [Google Scholar]
- 21.Taillandier E. and Liquier,J. (1992) Infrared spectroscopy of DNA. Methods Enzymol., 211, 307–335. [DOI] [PubMed] [Google Scholar]
- 22.Akhebat A., Dagneaux,C., Liquier,J. and Taillandier,E. (1992) Triple helical polynucleotide structures: an FTIR study of the C+·G·C triplet. J. Biomol. Struct. Dyn., 10, 577–588. [DOI] [PubMed] [Google Scholar]
- 23.Sarkar M., Dornberger,U., Rozners,E., Fritzsche,H., Strömberg,R. and Gräslund,A. (1997) FTIR spectroscopic studies of oligonucleotides that model a triple-helical domain in self-splicing group I introns. Biochemistry, 36, 15463–15471. [DOI] [PubMed] [Google Scholar]
- 24.Ouali M., Letellier,R., Sun,J.-S., Akhebat,A., Adnet,F., Liquier,J. and Taillandier,E. (1993) Determination of G*G·C triple-helix structure by molecular modeling and vibrational spectroscopy. J. Am. Chem. Soc., 115, 4264–4270. [Google Scholar]
- 25.White A.P. and Powell,J.W. (1995) Observation of the hydration-dependent conformation of the (dG)20·(dG)20·(dC)20 oligonucleotide triplex using FTIR spectroscopy. Biochemistry, 34, 1137–1142. [DOI] [PubMed] [Google Scholar]
- 26.Shchyolkina A.K., Borisova,O.F., Minyat,E.E., Timofeev,E.N., Il’icheva,I.A., Khomyakova,E.B. and Florontiev,V.L. (1995) Parallel purine-pyrimidine-purine triplex: experimental evidence for existence. FEBS Lett., 367, 81–84. [DOI] [PubMed] [Google Scholar]
- 27.Gondeau C., Maurizot,J.C. and Durand,M. (1998) Circular dichroism and UV melting studies on formation of an intramolecular triplex containing parallel T*A:T and G*G:C triplets: netropsin complexation with the triples. Nucleic Acids Res., 26, 4996–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semerad C.L. and Maher,L.J.,III (1994) Exclusion of RNA strands from a purine motif triple helix. Nucleic Acids Res., 22, 5321–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]