Abstract
Heterologous markers are important tools required for the molecular dissection of gene function in many organisms, including Saccharomyces cerevisiae. Moreover, the presence of gene families and isoenzymes often makes it necessary to delete more than one gene. We recently introduced a new and efficient gene disruption cassette for repeated use in budding yeast, which combines the heterologous dominant kanr resistance marker with a Cre/loxP-mediated marker removal procedure. Here we describe an additional set of four completely heterologous loxP-flanked marker cassettes carrying the genes URA3 and LEU2 from Kluyveromyces lactis, his5+ from Schizosaccharomyces pombe and the dominant resistance marker bler from the bacterial transposon Tn5, which confers resistance to the antibiotic phleomycin. All five loxP–marker gene–loxP gene disruption cassettes can be generated using the same pair of oligonucleotides and all can be used for gene disruption with high efficiency. For marker rescue we have created three additional Cre expression vectors carrying HIS3, TRP1 or bler as the yeast selection marker. The set of disruption cassettes and Cre expression plasmids described here represents a significant further development of the marker rescue system, which is ideally suited to functional analysis of the yeast genome.
INTRODUCTION
In 1996 the complete sequence of the genome of the budding yeast Saccharomyces cerevisiae was published (1) and functional analysis of the more than 6000 genes found in this organism entered a new phase. Subsequently new tools were developed to speed up this process. Gene disruption is one of the most powerful techniques available for the study of gene function, and a breakthrough was achieved when it became apparent that only very short sequences of yeast DNA on either side of a marker gene are needed for efficient integration into the yeast genome by homologous recombination. This permits the use of gene disruption cassettes created by PCR (2,3). Thus, for the first time, large-scale, systematic gene disruption projects became feasible (4,5). The next important step was the introduction of the completely heterologous kanr marker, which confers resistance to the antibiotic G418 (6). This dominant marker dispenses with the need for yeast strains that are auxotrophic for the markers normally used for gene disruption. Moreover, because the kanr gene (which is driven by the Ashbya gossypii TEF2 promoter and terminated by the A.gossypii TEF2 terminator) shows no homology to the yeast genome, recombination between the yeast genome and internal parts of the kanMX disruption cassette (which would result in misintegration) is minimal, thus maximising the frequency of correct integration. Consequently, the kanMX gene disruption cassette was chosen for large-scale gene disruption projects, as well as for use in the genome-wide gene knockout project (5,7,8).
In functional studies in S.cerevisiae it is often necessary to delete more than one gene. This is partly due to the fact that substantial portions of the yeast genome are duplicated (9,10). Moreover, as many cellular functions are maintained by more than one isoenzyme, it is often necessary to create multiple gene disruptions to uncover gene function (11–13). One prominent example is the hexose transporter family: in this case, concurrent knockout of at least 20 transporter genes was required to block uptake of hexose completely (14).
Marker rescue and reuse offers a convenient and efficient way to introduce multiple gene disruptions. Removal of a previously integrated disruption cassette is most easily achieved if the respective cassette is flanked by directly repeated sequences 400–500 bp in length. Mitotic recombination between the repeated sequences results in excision of the marker and leaves a single repeat in the genome (15). This principle has been used with a variety of marker genes (16–20). The main problems encountered with this approach are: (i) the low rate of mitotic recombination, which makes it necessary to screen for rare instances of marker loss (except in cases where counter-selection is possible, e.g. URA3; 21); (ii) the fact that each marker rescue event leaves an additional repeat sequence in the genome which serves as an unwanted recombination target for integration of the gene disruption cassette in the next round of gene deletion.
Recently we designed a new and efficient gene disruption cassette for repeated use by combining the advantages of the kanMX gene disruption cassette with those of the Cre/loxP recombination system of bacteriophage P1 (22). The kanMX cassette is flanked by two direct repeats of the 34 bp loxP sequence, yielding the loxP–kanMX–loxP cassette, which can be converted into a highly efficient gene disruption cassette by the addition of short DNA sequences homologous to the genomic locus of interest using PCR (Fig. 1). Once correctly integrated into the genome the kanMX marker can be efficiently rescued by transformation with a plasmid carrying the gene for Cre recombinase under control of the GAL1 promoter and induction of Cre expression by shifting the cells to galactose-containing medium. Cre-induced recombination results in loss of the kanMX cassette, leaving behind a single loxP site at the original integration site (Fig. 1) (22). Since its introduction the loxP–kanMX–loxP cassette has been widely used (see for example 23–26). Many gene families have now been analysed in this way: for example, the above mentioned hexose transporter family (14) and a series of gene families of unknown function investigated within the framework of the EUROFAN project (27; J.H.Hegemann and U.Gueldener, in preparation). Moreover, a modified loxP–kanMX–loxP cassette has been used to control gene expression in a novel way. For this purpose the loxP–kanMX–loxP cassette is embedded in the ACT1 intron and, after genomic integration of this ‘maxi-intron’ into the 5′-terminal region of the gene of interest, expression of this gene is completely inhibited. Gene expression is restored only after Cre-mediated site-specific recombination has deleted the kanMX unit and thus created a small spliceable intron carrying a single loxP site (28). In a different application the loxP–kanMX–loxP marker cassette was used to repeatedly add an epitope to different gene products at their C-termini (29). Finally the loxP–kanMX–loxP marker has been incorporated into an integration cassette that can be used to fuse GFP to any protein of interest (30; U.Gueldener and J.H.Hegemann, in preparation). Meanwhile the loxP–kanMX–loxP/Cre system has also been adapted successfully for use in the yeast Kluyveromyces lactis (31).
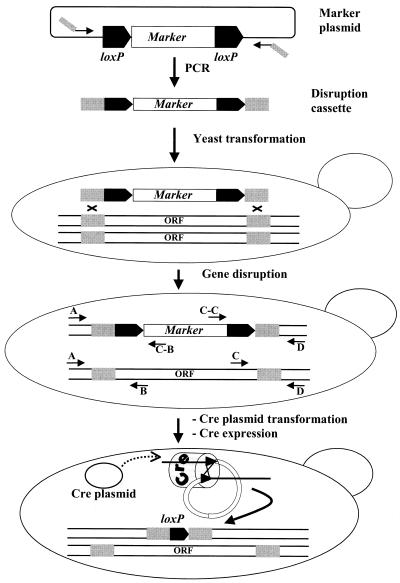
Figure 1.
The loxP/Cre gene disruption and marker rescue procedure. The gene disruption cassette consists of a selection marker gene (marker), usually conferring drug resistance or prototrophy, flanked by two 34 bp loxP sequences as direct repeats located adjacent to 45 bp of sequence flanking the chromosomal target sequence (ORF) to be deleted. The disruption cassette is produced by PCR using oligonucleotides comprising 19 or 22 3′ nucleotides complementary to sequences in the template (marker plasmid) flanking the disruption cassette and 45 5′ nucleotides that anneal to sites upstream or downstream of the genomic target sequence to be deleted. After transformation of the linear disruption cassette into yeast cells, selected transformants are checked by PCR for correct integration of the cassette and concurrent deletion of the chromosomal target sequence. The verification PCRs are done using combinations of primers complementary to sequences within the cassette (C-B, cassette B primer; C-C, cassette C primer) and to sequences within or flanking the target sequence (A, B, C and D). In a diploid yeast cell (shown here) in which one allele of the target sequence has been replaced by a disruption cassette, all PCRs indicated will produce products of the expected size. Finally, expression of the Cre recombinase results in removal of the marker gene, leaving behind a single loxP site at the chromosomal locus.
One limitation of the Cre/loxP system so far has been its restriction to the kanr marker gene. This has meant that the system could not be used in the background of the kanMX deletion strains created in the genome-wide knockout project (8). Furthermore, it would be interesting in certain cases to follow a deleted gene or genes in genetic crosses (by following the disruption cassette markers) before the markers are excised by inducing Cre expression.
In this paper we describe the construction of additional loxP–marker gene–loxP cassettes for repeated gene disruption. Most importantly we introduce a new dominant drug resistance cassette carrying the bler gene from the bacterial transposon Tn5, which confers resistance to the antibiotic phleomycin (Phleo). Finally, construction of additional Cre expression plasmids carrying HIS3, TRP1 or the dominant bler marker gene allows a much wider application of this powerful marker rescue system and significantly increases the number of new markers of the Cre/loxP system (22,27,31), thus further enhancing the utility of the Cre/loxP system in functional genome research.
MATERIALS AND METHODS
Strains and media
Escherichia coli strain XL1-Blue was obtained from Stratagene (Heidelberg, Germany). The yeast strain CEN.PK2-1C was used for in vivo recombination to generate plasmid pSH65 and for gene disruption experiments. All culture media were prepared as described previously (32). For selection of G418 resistance after yeast transformations, cells were plated onto YPD plates containing 200 µg/ml G418 sulphate (Gibco BRL, Germany; batch 11811-031, activity may be batch-dependent) (dissolve in water and add to autoclaved YPD cooled to 60°C, to a final active concentration of 200 µg/ml) after an initial period of growth (2 h) in non-selective medium. Selection for phleomycin resistance was performed on YPD plates containing 7.5 µg/ml phleomycin (PHLEL0100; Cayla, France) (dissolve in water and add to autoclaved YPD cooled to 60°C). Systematic testing revealed that in this case preincubation in non-selective medium did not enhance transformation efficiency. For selection for the kanr gene in E.coli, bacteria were plated on YT plates containing 50 µg/ml kanamycin sulphate (M6750; Sigma, Germany) (6). Pfu polymerase (M7741) was obtained from Promega (Germany); Taq polymerase was prepared as described (33).
Plasmid construction
Positioning of new heterologous marker genes between two loxP sites. Plasmid pUG6 carrying the loxP–kanMX–loxP module has been described previously (22). For construction of plasmid pUG27 (carrying loxP–his5+–loxP) the BglII–SacI DNA fragment from plasmid pFA6a-HIS3MX6 (18), carrying the Schizosaccharomyces pombe his5+ ORF (which complements the S.cerevisiae his3 mutation) plus the A.gossypii TEF2 promoter and terminator, was cloned into BglII + SacI-cleaved plasmid pUG6, replacing the kanMX unit between the two loxP sites. For construction of plasmid pUG66 (carrying loxP–bler–loxP) the vector pUT332, containing the bler gene from transposon Tn5 (34), was used as the template in a PCR with the oligonucleotide primers 1104 (5′-GCCGTAAGCCATGGCCGACCAAGCGACGCCCAAC-3′) and 1106 (5′-GGCGCCGGAGTACTGATCATGAGATGCCTGCAAGC-3′) (restriction sites underlined) to amplify the bler open reading frame. The resulting PCR product was cut with NcoI and ScaI and ligated with NcoI + ScaI-cleaved pUG6, thus replacing the kanr coding sequence. For construction of plasmid pUG72 (carrying loxP–URA3–loxP; originally named pJJH726) the K.lactis URA3 gene including the promoter and terminator was amplified from genomic DNA of the K.lactis type strain CBS2359 with oligonucleotides KlURA3-SacI (5′-GGCGAGCTCGTTTTATTTAGGTTCTATCGAGG-3′) and KlURA3-BamHI (5′-CCGCGGATCCCAATACAACAGATCACGTG-3′). The resulting PCR product of ∼1.4 kb was cleaved with SacI and BamHI and ligated to BglII + SacI-cleaved pUG6, thus replacing the complete kanMX unit. For construction of plasmid pUG73 (carrying loxP–LEU2–loxP; originally named pJJH727) the K.lactis LEU2 gene plus promoter and terminator sequences was amplified by PCR using K.lactis genomic DNA and the oligonucleotides KlLEU2-SacI (5′-GGCGAGCTCGCTGTGAAGATCCCAGCAAAGG-3′) and KlLEU2-BamHI (5′-GGCGGGATCCGCAGGCTAACCGGAACCTG-3′). The PCR product of ∼2.2 kb was digested with SacI and BamHI and ligated into BglII + SacI-cut pUG6.
Construction of new Cre expression plasmids. Plasmid pSH47 (GAL1-cre, URA3) has been described previously (22). For construction of plasmid pSH62 (GAL1-cre, HIS3) a 1.4 kb EcoRI–XhoI DNA fragment of pSH47 carrying the cre open reading frame was ligated with EcoRI + XhoI-cleaved p413GAL1 (35). For construction of plasmid pSH63 (GAL1-cre, TRP1) the 1.4 kb EcoRI–XhoI DNA fragment of pSH47 carrying the cre gene was ligated to EcoRI + XhoI-cut p414GAL1 (35). For construction of plasmid pSH65 (GAL1-cre, ble) the vector pUT332, containing the bler gene from transposon Tn5 plus the S.cerevisiae TEF1 promoter (34), was used as template in a PCR with the oligonucleotide primers 1095 (5′-AGAAGGTTTTGGGACGCTCGAAGGCTTTAATTTGCGGCCGCGTTGTAAAACGACGGCCAG-3′) and 1096 (5′-AGTGAGCGCGCGTAATACGACTCACTATAGGGCGAATTGGGCGTACACGCGTCTGTACAG-3′). At their 3′-ends the oligonucleotides (underlined) are homologous to plasmid pUT322, while the 5′-ends of the oligonucleotides are homologous to the sequences to the left and right of the KpnI restriction site in pSH47. The ∼1.0 kb PCR product carrying the bler gene plus the S.cerevisiae TEF1 promoter was transformed into yeast strain CEN.PK2 together with the KpnI-cleaved plasmid pSH47, selecting for uracil prototrophy after homologous recombination of both DNA molecules in yeast. Genomic DNA from individual yeast transformants was isolated (32), transformed into E.coli and plasmid DNA was prepared. The correct plasmid, called pSH64, was identified by restriction enzyme and sequence analysis. Subsequently pSH64 was cut with NcoI (site located within the URA3 coding sequence) and the ends were filled-in with Pfu polymerase and religated to inactivate the URA3 gene, yielding pSH65.
The complete DNA sequences of all pUG and pSH plasmids have been deposited in GenBank and the corresponding accession numbers can be found in Figures 2 and 5.
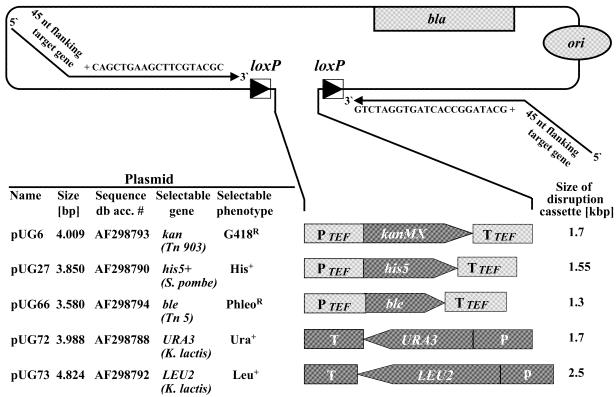
Figure 2.
The series of loxP–marker gene–loxP gene disruption cassettes. The marker plasmids pUG6, pUG27, pUG66, pUG72 and pUG73 serve as templates for PCR to generate the five different gene disruption cassettes. All marker genes (selectable genes) are derived from organisms other than S.cerevisiae and are expressed from the A.gossypii TEF2 promoter and terminated by the TEF2 terminator (6), except for the two K.lactis genes, URA3 and LEU2, which retain their own regulatory sequences. kanr and bler are dominant resistance marker genes conferring resistance to G418 and phleomycin (selectable phenotype), respectively. Each marker gene including promoter and terminator is flanked by loxP sites. Since the vector backbone is the same in all cases, each of the five disruption cassettes can be amplified by PCR using the same two primers. The complete vector sequences have been deposited at GenBank under the accession numbers listed in the figure (Sequence db acc. #).
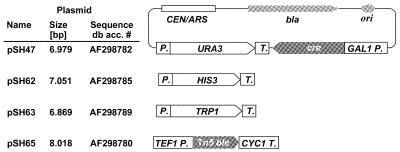
Figure 5.
Cre expression plasmids. All plasmids carry the CEN6/ARSH4 module to ensure stability in yeast and a Cre expression cassette consisting of the cre open reading frame flanked by the galactose-inducible GAL1 promoter and the CYC1 terminator from S.cerevisiae. The vectors differ in the type of selection marker used. The URA3, HIS3 and TRP1 marker genes are from S.cerevisiae and retain their own regulatory sequences, while the bler gene from transposon Tn5 is expressed from the S.cerevisiae TEF1 promoter.
Plasmid requests
The vectors pUG6 (kanr), pUG27 (his5+), pUG66 (bler), pUG72 (URA3) and pUG73 (LEU2) carrying the various loxP–marker gene–loxP gene disruption cassettes as well as the Cre expression plasmids pSH47 (yeast selection marker URA3), pSH62 (HIS3), pSH63 (TRP1) and pSH65 (bler) can be obtained individually (pUG6, accession no. P30114; pUG27, accession no. P30115; pUG66, accession no. P30116; pUG72, accession no. P30117; pUG73, accession no. P30118; pSH47, accession no. P30119; pSH62, accession no. P30120; pSH63, accession no. P30121; pSH65, accession no. P30122) or as a package (DEL-MARKER-SET) from Euroscarf (Frankfurt, Germany; http://www.uni-frankfurt.de/fb15/mikro/euroscarf/). The package includes the heterologous gene disruption cassettes from the McCusker group (19,20). Companies should contact Johannes H. Hegemann.
PCR-mediated generation of loxP–marker gene–loxP gene disruption cassettes and their integration into the yeast genome
All loxP–marker gene–loxP disruption cassettes presented in this work can be generated by PCR using oligonucleotides carrying the same 19 and 22 3′ nucleotides, while the 40 5′ nucleotides must be homologous to sequences to the left or right of the gene to be deleted (see Fig. 2 and Table 1). As a test case for the new disruption cassettes the ADE2 and YOR387c genes were deleted using the oligonucleotides listed in Table 1. For each disruption cassette one preparative PCR was performed as described previously (22). The PCR conditions were: 95°C for 5 min (hot start); 25 cycles of 94°C for 40 s, 58°C for 1 min and 68°C for 2 min; a final step at 68°C for 15 min. The PCR product was purified and the cassettes used for yeast transformation as described before (22).
Table 1. List of oligonucleotides (5′→3′)a.
aFor nomenclature of verification primers A–D see Figure 1.
bLower case letters indicate nucleotides homologous to sequences left and right of the cloned disruption cassettes (see Fig. 2).
cPrimers kan-B and kan-C can be used for the disruption cassette markers kanr, his5+ and bler.
RESULTS AND DISCUSSION
Construction of new loxP-flanked gene disruption cassettes
The recently introduced loxP–kanMX–loxP gene disruption cassette combines the advantages of the heterologous, dominant kanr marker with the capacity for efficient gene disruption by Cre-mediated recombination between the two loxP sites, resulting in efficient marker rescue (22) (Fig. 1). In order to expand the Cre/loxP technology and maximise the utility of this system, we have constructed additional loxP–marker gene–loxP cassettes. The four newly created loxP-flanked marker cassettes, like the original loxP–kanMX–loxP cassette, completely lack homology to the genome of the budding yeast, which maximises the probability of recovering the desired homologous integration event (Fig. 2). Construction of the new gene disruption cassettes started from the widely used loxP–kanMX–loxP cassette cloned in plasmid pUG6 (22). For construction of the loxP–his5+–loxP cassette the kanMX unit located between the two loxP sites in plasmid pUG6 was replaced by the S.pombe his5+ unit obtained from plasmid pFA6a-HIS3MX6 (18), yielding plasmid pUG27. The S.pombe his5+ gene complements the S.cerevisiae his3 mutation. The overall degree of identity between the S.pombe his5+ and S.cerevisiae HIS3 DNA sequences is 59%, with several blocks exhibiting higher homology (70–76% identity). This, however, is too low to allow efficient homologous integration of the S.pombe gene at the HIS3 locus (18).
For construction of the heterologous loxP–URA3–loxP and loxP–LEU2–loxP cassettes, both genes plus their promoter and terminator sequences were isolated by PCR from K.lactis genomic DNA and cloned between the two loxP sequences in plasmid pUG6, replacing the kanMX unit and resulting in the plasmids pUG72 and pUG73. The two K.lactis genes complement the corresponding ura3 and leu2 mutations in S.cerevisiae (36,37). The K.lactis genes are 73 (URA3) and 77% (LEU2) identical to their S.cerevisiae homologues. This is too low to permit efficient recombination between the corresponding K.lactis and S.cerevisiae genes, in particular as the longest stretch of DNA sequence identity between corresponding genes is 17 bp in the case of URA3 and 18 bp for LEU2. The loxP–URA3–loxP cassette should be particularly useful, because (i) most S.cerevisiae laboratory strains are ura3– and (ii) 5-fluoroorotic acid-containing media can be readily used for counter-selection (21).
Finally, the bler gene from transposon Tn5 was used to create the new heterologous dominant drug resistance marker cassette loxP–bler–loxP. The bler gene renders prokaryotes and lower eukaryotes resistant to phleomycin, which belongs to the metallo-glycopeptide group of antibiotics of the bleomycin family (38). Cells expressing the bler gene become resistant to phleomycin as a result of tight binding of the antibiotic by the Ble protein. The Ble protein appears not to be toxic to S.cerevisiae. Phleomycin is produced in fermentation cultures by Streptomyces verticillus (39). It has previously been shown that expression of the bler gene from an episomal or an integrative plasmid allows direct selection of phleomycin-resistant S.cerevisiae transformants (34,38,40). The bler gene was isolated by PCR from plasmid pUT332 (34) and placed between the A.gossypii TEF promoter and terminator in plasmid pUG6, yielding plasmid pUG66 (Fig. 2).
Gene disruption properties of the new loxP–marker gene–loxP cassettes
An important property of the new cloned gene disruption cassettes is that, like the original loxP–kanMX–loxP cassette (22), each of the marker genes plus regulatory elements is flanked by two loxP sequences embedded in the same vector backbone (see Fig. 2 and Materials and Methods). This allows the use of the same primer sequences to amplify all of the gene disruption cassettes.
To test the fidelity of the new cassettes for targeted gene disruption all cassettes were used to delete two genes: ADE2 and YOR387c. The 2 × 5 gene disruption cassettes were amplified in parallel using primers 1134 and 1135 (for ADE2) and 1017 and 1018 (for YOR387c) (Table 1). The 5′-ends of the primers contain 45 nt stretches that are homologous to sequences upstream of the ATG start codon and downstream of the stop codon, respectively, of the target genes, followed by a 19 nt segment homologous to sequences to the left of the loxP site and a 22 nt motif that anneals to the right of the loxP site of the disruption cassettes (Figs 1 and 2). Comparable amounts of the various disruption cassettes were generated by PCR and purified (Fig. 3). Approximately 2 µg of the gene disruption cassettes were transformed into strain CEN.PK2-1C and transformants were selected on the corresponding SD dropout plates, on YPD + 7.5 µg/ml phleomycin (for the loxP–bler–loxP cassette) or on YPD + 200 µg/ml G418 (for the loxP–kanMX–loxP cassette) plates, as described before (22). Transformation of the five different disruption cassettes in all cases yielded comparable numbers of transformants.
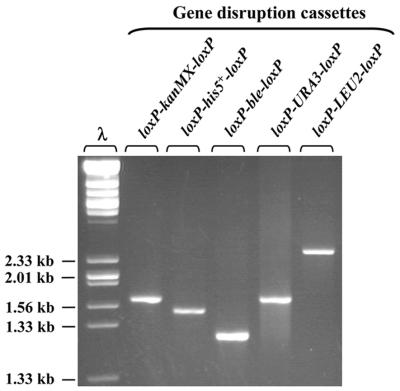
Figure 3.
For deletion of the ADE2 gene, five different gene disruption cassettes were produced in parallel PCRs using the primers 1134 and 1135 (see Table 1). The sizes of the cassettes vary between 1300 and 2500 bp (2% of each PCR product was loaded).
For selection of phleomycin-resistant colonies we first determined the usable range of antibiotic concentrations and found that 7.5 µg/ml phleomycin (in YPD) was optimal for selection of loxP–bler–loxP transformants. At this concentration no spontaneous phleomycin-resistant colonies were found. In contrast to what has been described for transformation with a bler gene-carrying multicopy plasmid (40), we could not improve the transformation efficiency of the loxP–bler–loxP cassette further by incubating the transformation mix in liquid YPD medium for several hours (up to 10 h was tested) prior to plating on YPD plates containing phleomycin. However, we did not try to enhance the transformation efficiency of the loxP–bler–loxP cassette further by adding glycerol to the transformation plates (38).
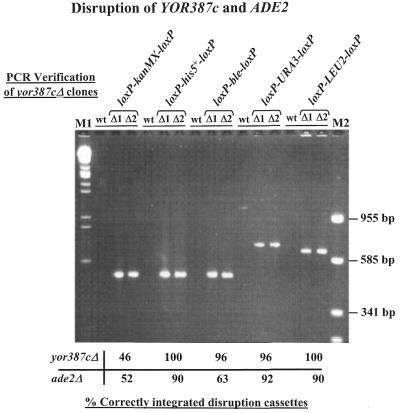
To confirm correct integration of the various disruption cassettes into the ADE2 and YOR387c loci, 24 transformants from each plate were subjected to colony PCR using combinations of the corresponding target gene-specific (primers A–D) and disruption cassette-specific primers (primers C-B and C-C) (Fig. 1 and Table 1). A typical example of the results of the PCR analysis of putative yor387c deletants using target gene-specific primer A and one of the three cassette-specific primers C-B is shown in Figure 4. The PCR products obtained from the various deletion strains are between 350 and 600 bp in length and thus exhibit the calculated cassette-specific sizes predicted for correct integration. The frequency of correctly integrated cassettes at the YOR387c locus varied between 46% for the loxP–kanMX–loxP cassette and 100% for the loxP–his5+–loxP and loxP–LEU2–loxP cassettes. Analysis of the ade2 transformants revealed a similar level of correctly integrated cassettes: between 52% (loxP–kanMX–loxP) and 92% (loxP–URA3–loxP) of the transformants had undergone the correct homologous recombination event (Fig. 4). As expected, all ade2 deletion strains characterised by PCR also produced red colonies on indicator plates due to accumulation of a biosynthetic intermediate product, thus confirming the PCR results (data not shown). In summary, PCR analysis of putative yor387c and ade2 deletants revealed that all five cassettes disrupted their target genes with similar efficiency: at least 50% of the transformants had undergone the predicted homologous recombination event. Similar efficiencies for this type of gene disruption have been observed by others, mainly due to the fact that the gene disruption cassettes used in these cases consist of completely heterologous DNA sequences (6,19,22).
Figure 4.
PCR analysis to confirm correct integration of the gene disruption cassettes at the YOR387c and ADE2 loci (see Fig. 1). PCR was carried out on transformants obtained in the YOR387c disruption experiment, using target gene-specific primer A (primer 1025) and the disruption cassette-specific primer C-B (for kanr, his5+ and bler the kan-B primer 363; for URA3 the Ura-B primer 1115; for LEU2 the Leu2-B primer 1136). The products obtained from two yor387cΔ mutants (Δ1 and Δ2) disrupted with each of the various disruption cassettes and from wild-type (wt) cells were then subjected to agarose gel electrophoresis. For both genes 24 transformants obtained with each of the gene disruption cassettes were analysed in similar experiments. The percentages of correctly integrated cassettes at YOR387c and ADE2 are indicated below the gel. DNA fragment size markers: M1, EcoRI + HindIII-cleaved λ DNA; M2, Bsp143I-cleaved pUC19 (the sizes of selected marker DNA fragments are shown on the right).
Our results show that the two heterologous K.lactis genes URA3 and LEU2 exhibit excellent selection marker qualities for this type of gene deletion. In fact, the URA3 gene from this yeast has previously been used for gene disruption experiments by others (27,41). Likewise, the his5+ gene from S.pombe proved to be an efficient marker for gene disruption, as has been reported by others for a non-loxP version of the his5+ gene (18,42).
The results of the gene knockout experiment show that the bler gene is an efficient heterologous dominant resistance marker: 63 and 96% of the transformants proved to have correctly integrated the loxP–bler–loxP gene disruption cassette at the ADE2 and YOR387c loci, respectively. The transformation efficiency of this cassette is comparable to that of the second dominant resistance marker, loxP–kanMX–loxP, and to that of the other heterologous auxotrophic markers used here (data not shown). Moreover, ade2Δ strains carrying the bler marker showed no obvious growth disadvantage when compared with ade2Δ strains containing one of the other marker genes, suggesting that these markers are probably phenotypically neutral. However, this needs to be tested more thoroughly in the future. To exclude the possibility that high level expression of the phleomycin resistance protein has adverse effects on cellular functions in S.cerevisiae, the marker can be easily removed by activating the Cre-mediated recombination process to delete the bler gene.
Finally, it is important to note that no cross-resistance with other currently used dominant resistance markers has been reported. Therefore, phleomycin can be used to select cells that are already resistant to other selective agents, for example G418. This makes the dominant loxP–bler–loxP cassette a very useful tool when working with the kanr-based KO strain collection produced by the Transatlantic Yeast Consortium during the EU project EUROFAN 2. Our laboratory routinely uses both dominant markers to produce double deletion strains.
New Cre expression plasmids for marker rescue
Expression of Cre recombinase in yeast strains carrying one (or more) of the loxP–marker gene–loxP cassettes results in efficient recombination between the loxP sites, removing the marker gene and leaving behind a single loxP site at the chromosomal locus (22). To increase the flexibility of this system, we have exchanged the URA3 selection marker in the original cre expression vector pSH47 for HIS3 (resulting in plasmid pSH62) and TRP1 (pSH63), as well as for the dominant resistant marker bler (pSH65) (Fig. 5). The cre expression cassette consisting of the inducible GAL1 promoter, the cre gene and the CYC1 terminator is not affected by these modifications. The new vectors were tested for efficiency of marker rescue and were found to induce the same rates of marker loss as described for the original pSH47 vector (22; data not shown). In the meantime these new cre expression vectors have also been successfully used by others (see for example 28). In our routine work we no longer induce Cre expression by shifting loxP–marker gene–loxP marked cells transformed with a cre plasmid to galactose medium. Instead, we grow these cells overnight in liquid YPD medium (with glucose as carbon source) and then plate ∼200 cells on YPD plates. Replica plating onto YPD and YPD plus the drug (or onto the corresponding selective medium in the case of the auxotrophic markers) results in the identification of cells which have lost the marker gene from the disruption cassette (∼5% of the total). This is probably due to residual activity of the GAL1 promoter in the presence of glucose.
Several loxP-flanked marker genes can be removed from the genome simultaneously (data not shown). However, care must be taken not to induce Cre expression too strongly (e.g. by using multicopy plasmids or inducing Cre expression for prolonged times), as this will result in unintended recombination events between various loxP sites (27).
Swapping of cassette markers
An additional advantage of the loxP-flanked kanMX, bler and his5+ marker genes is their use for marker cassette exchange. The presence of completely identical DNA sequences of ∼300 bp to the left and right of the marker gene in each cassette (see Fig. 2) allows for easy replacement of each of the marker genes by any of the others. For this purpose only two oligonucleotides have to be designed which allow amplification of the marker gene of interest plus the promoter, terminator and loxP sequences. Transformation of this cassette into a yeast strain carrying one of the other two cassettes integrated into the genome replaces the marker gene within the integrated cassette. Successful cassette swapping can be verified by PCR analysis using target gene-specific (A and D) and new cassette-specific primers. To exclude unintended targeting events, swapping experiments should be restricted to single deletion strains that do not carry plasmid pSH65 (carrying S.cerevisiae TEF1 sequences).
Conclusion
We have constructed four new gene disruption cassettes with the marker genes bler, his5+, URA3 and LEU2, consisting of completely heterologous DNA flanked by loxP sequences. In addition, the Cre expression plasmid has been equipped with three other selection markers (HIS3, TRP1 and bler). The set of five different gene disruption cassettes and four different Cre expression vectors presented here will significantly extend the tool kit for functional analysis of the S.cerevisiae genome. In fact, some of the new vectors have already been used successfully in a variety of experiments (see for example 28,43; J.Heinisch, unpublished results).
The availability of the two dominant marker genes kanr and bler and a Cre expression vector carrying bler as the yeast selection marker facilitates the creation of multiple gene knockouts not only in commonly used laboratory strains, but also in prototrophic industrial or wild-type strains of S.cerevisiae.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Ute Gengenbacher and Sabine Klein for technical assistance. Dr Paul Hardy is thanked for critical reading of the manuscript. We are grateful to Dr Yde Steensma for plasmid pUT332. This work was supported by a contract awarded to J.H.H. in the context of the EUROFAN project of the EC (BIO4-CT95-0080) and a grant from the Deutsche Forschungsgemeinschaft (He1880/4-1) to J.H.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +49 211 81 13733; Fax: +49 211 81 13567; Email: hegemann@uni-duesseldorf.de Present addresses: U. Gueldener, MIPS, GSF, Ingolstaedter Landstrasse 1, 85764 Neuherberg, GermanyJ. Heinisch, Universität Hohenheim, Institut für Lebensmitteltechnologie (150), Fachgebiet Gärungstechnologie, Garbenstrasse 25, 70599 Stuttgart, Germany AF298780, AF298782, AF298785, AF298788–AF298790, AF298792–AF298794
REFERENCES
- 1.Goffeau A., Barrell,B.G., Bussey,H., Davis.R.W., Dujon,B., Feldmann,H., Galibert,F., Hoheisel,J.D., Jacq,C., Johnston,M. et al. (1996) Life with 6000 genes. Science, 274, 546, 563–567. [DOI] [PubMed] [Google Scholar]
- 2.Baudin A., Ozier-Kalogeropoulos,O., Denouel,A., Lacroute,F. and Cullin,C. (1993) A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res., 21, 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McElver J. and Weber,S. (1992) Flag N-terminal epitope overexpression of bacterial alkaline phosphatase and Flag C-terminal epitope tagging by PCR one-step targeted integration. Yeast, 8 (special issue), S627. [Google Scholar]
- 4.Niedenthal R., Riles,L., Güldener,U., Klein,S., Johnston,M. and Hegemann,J.H. (1999) Systematic analysis of S. cerevisiae chromosome VIII genes. Yeast, 15, 1775–1796. [DOI] [PubMed] [Google Scholar]
- 5.Entian K.-D., Schuster,T., Hegemann,J.H., Becher,D., Feldmann,H., Guldner,U., Gotz,R., Hansen,M., Hollenberg,C.P., Jansen,G. et al. (1999) Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol. Gen. Genet., 262, 683–702. [DOI] [PubMed] [Google Scholar]
- 6.Wach A., Brachat,A., Pohlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- 7.Dujon B. (1998) European Functional Analysis Network (EUROFAN) and the functional analysis of the Saccharomyces cerevisiae genome. Electrophoresis, 19, 617–624. [DOI] [PubMed] [Google Scholar]
- 8.Winzeler E.A., Shoemaker,D.D., Astromoff,A., Liang,H., Anderson,K., Andre,B., Bangham,R., Benito,R., Boeke,J.D., Bussey,H. et al. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]
- 9.Wolfe K.H. and Shields,D.C. (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature, 387, 708–713. [DOI] [PubMed] [Google Scholar]
- 10.Mewes H.W., Albermann,K., Bahr,M., Frishman,D., Gleissner,A., Hani,J., Heumann,K., Kleine,K., Maierl,A., Oliver,S.G. et al. (1997) Overview of the yeast genome. Nature, 387, 7–65. [Erratum, Nature (1997), 387, 737] [DOI] [PubMed] [Google Scholar]
- 11.Dolinski K., Muir,S., Cardenas,M. and Heitman,J. (1997) All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 13093–13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.During-Olsen L., Regenberg,B., Gjermansen,C., Kielland-Brandt,M.C. and Hansen,J. (1999) Cysteine uptake by Saccharomyces cerevisiae is accomplished by multiple permeases. Curr. Genet., 35, 609–617. [DOI] [PubMed] [Google Scholar]
- 13.Reifenberger E., Freidel,K. and Ciriacy,M. (1995) Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol. Microbiol., 16, 157–167. [DOI] [PubMed] [Google Scholar]
- 14.Wieczorke R., Krampe,S., Weierstall,T., Freidel,K., Hollenberg,C.P. and Boles,E. (1999) Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett., 464, 123–128. [DOI] [PubMed] [Google Scholar]
- 15.Alani E., Cao,L. and Kleckner,N. (1987) A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics, 116, 541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Replogle K., Hovland,L. and Rivier,D.H. (1999) Designer deletion and prototrophic strains derived from Saccharomyces cerevisiae strain W303-1a. Yeast, 15, 1141–1149. [DOI] [PubMed] [Google Scholar]
- 17.Schneider B.L., Steiner,B., Seufert,W. and Futcher,A.B. (1996) pMPY-ZAP: a reusable polymerase chain reaction-directed gene disruption cassette for Saccharomyces cerevisiae. Yeast, 12, 129–134. [DOI] [PubMed] [Google Scholar]
- 18.Wach A., Brachat,A., Alberti-Segui,C., Rebischung,C. and Philippsen,P. (1997) Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast, 13, 1065–1075. [DOI] [PubMed] [Google Scholar]
- 19.Goldstein A.L. and McCusker,J.H. (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast, 15, 1541–1553. [DOI] [PubMed] [Google Scholar]
- 20.Goldstein A.L., Pan,X. and McCusker,J.H. (1999) Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast, 15, 507–511. [DOI] [PubMed] [Google Scholar]
- 21.Boeke J.D., LaCroute,F. and Fink,G.R. (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet., 197, 345–346. [DOI] [PubMed] [Google Scholar]
- 22.Güldener U., Heck,S., Fiedler,T., Beinhauer,J.D. and Hegemann,J.H. (1996) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res., 24, 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmieri L., Rottensteiner,H., Girzalsky,W., Scarcia,P., Palmieri,F. and Erdmann,R. (2001) Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. EMBO J., 20, 5049–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makuc J., Paiva,S., Schauen,M., Kramer,R., Andre,B., Casal,M., Leao,C. and Boles,E. (2001) The putative monocarboxylate permeases of the yeast Saccharomyces cerevisiae do not transport monocarboxylic acids across the plasma membrane. Yeast, 18, 1131–1143. [DOI] [PubMed] [Google Scholar]
- 25.Schlumpberger M., Prusiner,S.B. and Herskowitz,I. (2001) Induction of distinct [URE3] yeast prion strains. Mol. Cell. Biol., 21, 7035–7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azad A.K., Stanford,D.R., Sarkar,S. and Hopper,A.K. (2001) Role of nuclear pools of aminoacyl-tRNA synthetases in tRNA nuclear export. Mol. Biol. Cell, 12, 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delneri D., Tomlin,G.C., Wixon,J.L., Hutter,A., Sefton,M., Louis,E.J. and Oliver,S.G. (2000) Exploring redundancy in the yeast genome: an improved strategy for use of the cre-loxP system. Gene, 252, 127–135. [DOI] [PubMed] [Google Scholar]
- 28.Cheng T.H., Chang,C.R., Joy,P., Yablok,S. and Gartenberg,M.R. (2000) Controlling gene expression in yeast by inducible site-specific recombination. Nucleic Acids Res., 28, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Antoni A. and Gallwitz,D. (2000) A novel multi-purpose cassette for repeated integrative epitope tagging of genes in Saccharomyces cerevisiae. Gene, 246, 179–185. [DOI] [PubMed] [Google Scholar]
- 30.Prein B., Natter,K. and Kohlwein,S.D. (2000) A novel strategy for constructing N-terminal chromosomal fusions to green fluorescent protein in the yeast Saccharomyces cerevisiae. FEBS Lett., 485, 29–34. [DOI] [PubMed] [Google Scholar]
- 31.Steensma H.Y. and Ter Linde,J.J. (2001) Plasmids with the Cre-recombinase and the dominant nat marker, suitable for use in prototrophic strains of Saccharomyces cerevisiae and Kluyveromyces lactis. Yeast, 18, 469–472. [DOI] [PubMed] [Google Scholar]
- 32.Kaiser C., Michaelis,S. and Mitchell,A. (1994) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Pluthero F.G. (1993) Rapid purification of high-activity Taq DNA polymerase. Nucleic Acids Res., 21, 4850–4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gatignol A., Dassain,M. and Tiraby,G. (1990) Cloning of Saccharomyces cerevisiae promoters using a probe vector based on phleomycin resistance. Gene, 91, 35–41. [DOI] [PubMed] [Google Scholar]
- 35.Mumberg D., Muller,R. and Funk,M. (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res., 22, 5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shuster J.R., Moyer,D. and Irvine,B. (1987) Sequence of the Kluyveromyces lactis URA3 gene. Nucleic Acids Res., 15, 8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y.P., Chen,X.J., Li,Y.Y. and Fukuhara,H. (1992) LEU2 gene homolog in Kluyveromyces lactis. Yeast, 8, 801–804. [DOI] [PubMed] [Google Scholar]
- 38.Gatignol A., Baron,M. and Tiraby,G. (1987) Phleomycin resistance encoded by the ble gene from transposon Tn 5 as a dominant selectable marker in Saccharomyces cerevisiae. Mol. Gen. Genet., 207, 342–348. [DOI] [PubMed] [Google Scholar]
- 39.Umezawa H. (1974) Chemistry and mechanism of action of bleomycin. Fed. Proc., 33, 2296–2302. [PubMed] [Google Scholar]
- 40.Wenzel T.J., Migliazza,A., Steensma,H.Y. and van den Berg,J.A. (1992) Efficient selection of phleomycin-resistant Saccharomyces cerevisiae transformants. Yeast, 8, 667–668. [DOI] [PubMed] [Google Scholar]
- 41.Storici F., Coglievina,M. and Bruschi,C.V. (1999) A 2-micron DNA-based marker recycling system for multiple gene disruption in the yeast Saccharomyces cerevisiae. Yeast, 15, 271–283. [DOI] [PubMed] [Google Scholar]
- 42.Longtine M.S., McKenzie,A.,III, Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- 43.Young M.E., Karpova,T.S., Brugger,B., Moschenross,D.M., Wang,G.K., Schneiter,R., Wieland,F.T. and Cooper,J.A. (2002) The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation. Mol. Cell. Biol., 22, 927–934. [DOI] [PMC free article] [PubMed] [Google Scholar]