Abstract
The precise mapping and quantification of DNA methylation as an epigenetic parameter during development and in diseased tissues is of great importance for functional genomics. Here we describe a rapid, quantitative method to assess methylation levels at specific CpG sites using PCR products of bisulfite-treated genomic DNA. Using single nucleotide primer extension (SNuPE) assays in combination with ion pair reverse phase high performance liquid chromatography (IP RP HPLC) separation techniques, methylated and unmethylated CpGs can be discriminated and quantified based on the different masses and hydrophobicities of the extended primer products. The assay is linear, highly reproducible and several sites can be measured simultaneously in one reaction. It can be semi-automated and eliminates the need for cloning and sequencing of individual bisulfite PCR products.
INTRODUCTION
Epigenetic control of gene expression plays a crucial role in functional genomics. The precise measurement of epigenetic changes is of particular interest in the fields of developmental biology, cancer, multifactorial diseases and aging. Many methods have been developed for the detection of methylation, among which the bisulfite-based sequencing technique is the most precise and sensitive, providing sequence-independent information (1,2). A number of successful modifications of the original protocols have been published, mostly improving technical handling and sensitivity of the method. However, little progress has been made to circumvent the laborious steps of cloning and sequencing (3,4). Although a bisulfite-based single nucleotide primer extension (SNuPE) assay has been described for direct quantification, this protocol still requires radioactive quantification following gel separation and hence cannot be applied for high throughput performance (5).
Here we present a simple SNuPE assay in combination with an ion pair reverse phase high performance liquid chromatography (IP RP HPLC) separation method to rapidly quantify the methylation status at several CpG sites simultaneously. The basis for the method is a combination of bisulfite PCR modification, primer extension and IP RP HPLC separation of the extended primer products. After bisulfite treatment of the DNA (5) and PCR amplification of the region to be analyzed, the PCR products are purified to remove non-extended oligos and dNTPs. Following the purification step, specific primers just 5′ to a CpG site(s) are hybridized to the denatured single-stranded PCR product. The annealed primers are then extended using Sequenase in the presence of both ddCTP and ddTTP. The incorporation of ddTTP indicates that the original chromosomal DNA is unmethylated and conversely the incorporation of ddCTP indicates that the CpG is methylated (Fig. 1). In order to quantitate the incorporation of either ddTTP or ddCTP the extension products are directly (without any further purification) loaded on a dHPLC column (WAVE DNA Fragment Analysis System; Transgenomics). The amounts of ddTTP and ddCTP extended products are quantified by integrating the areas of the peaks and calculation of their percentage ratios using the WaveMaker program v.4.1 (Transgenomics) (see also Materials and Methods).
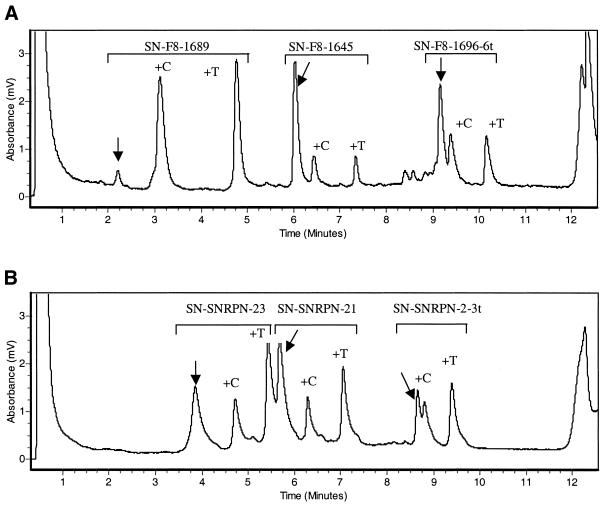
Figure 1.
Chromatograms of the multiplexed SNuPE reaction. (A) In the Factor VIII exon 14 region, representing a theoretical 50% methylated and 50% unmethylated. (B) In the SNRPN region. For each of the oligos used the unextended primer is indicated by a vertical arrow followed by the +C and +T extended primers representing the methylated (extended by ddCTP) and the unmethylated (extended by ddTTP) portions, respectively. All unlabeled peaks represent impurities from the initial unreacted primers.
MATERIALS AND METHODS
Bisulfite treatment
Bisulfite treatment of DNA was performed by a bead-based method as described by Engemann et al. (6). The primers used for amplification of bisulfite-treated lymphocyte DNA from Factor VIII exon 14 containing eight CpG sites at codons 1602, 1620, 1645, 1648, 1652, 1689 and 1696, were as described by El-Maarri et al. (7), while the primers used to amplify the 5′-region of the SNRPN gene were as described by El-Maarri et al. (2).
Primer extension reaction
Primers used for the primer extension reaction were as follows: SN-F8-1645, 5′-aat tta tta gtt ttg aaa-3′; SN-F8-1689, 5′-gat gaa aat tag agt ttt-3′; SN-F8-1696, 5′-ttt ttt agt ttt taa aag aaa ata-3′; SN-SNRPN-2-3t, 5′-ttt ggg att ttt gta ttg-3′; SN-SNRPN-21, 5′-taa ggt tag ttg tgt-3′; SN-SNRPN-23, 5′-ga tag ttt ggg gag-3′. The underlined t residues do not correspond to the DNA sequence and were added to increase the masses and hydrophobicities and thus the retention times of these oligos. The primers used in the SNuPE reactions are 5′ to CpG sites in codons 1645, 1689 and 1696 of the Factor VIII gene and CpG sites 2, 21 and 23 in the 5′-region of the SNRPN gene, as described by El-Maarri et al. (2).
The reaction was carried out as described by Hoogendoorn et al. (8). Briefly the reaction was carried out in 20 µl total volume containing ∼50 ng purified PCR product, 50 µM each ddCTP and ddTTP (Amersham), 12.5 pmol each primer and 3 U ThermoSequenase (Amersham). The thermal cycling was as follows: initial denaturation step of 3 min at 95°C, followed by 50 cycles of 30 s at 42°C and 2 min at 60°C.
dHPLC analysis
The HPLC analysis was performed on a WAVE DNA Fragment Analysis System from Transgenomics. An aliquot of 10 µl of the SNuPE reaction was loaded. The oven temperature was set to 50°C and elution was with a gradient of acetonitrile made by mixing buffers A and B, consisting of 0.1 M triethyammonium acetate (TEAA) buffer and 0.1 M TEAA buffer with 25% acetonitrile, respectively. Each run was made up of a gradient of 30–41% buffer B for the Factor VIII gene and 24–43% buffer B for the SNRPN gene over 10 min, followed by re-equilibration of the column by injection of 30% buffer B for 1 min. The eluted DNA was detected with a UV detector at 260 nm.
Calculation of methylation extent
The degree of methylation at a given CpG site was determined according to the term %Meth = (AC/AC + AT) × 100, where AC and AT are the surface areas under the peaks corresponding to the ddCTP-extended primer and the ddTTP-extended primer, respectively, as calculated by Wave Maker v.4.1 (Transgenomics).
RESULTS
At 50°C, using TEAA as the ion-pairing reagent, separation of short extended oligos by dHPLC is based on differences in both their length (mass) and hydrophobicity. As a consequence the incorporation, during the SNuPE reaction, of the more hydrophilic ddTTP increases the retention time compared to oligos extended by ddCTP (Fig. 1). Therefore, the order of appearance of the oligos is unextended oligo followed by the ddCTP- and ddTTP-extended oligos, respectively. Separation of the above three oligos is good enough to give three distinguishable peaks that can be accurately measured.
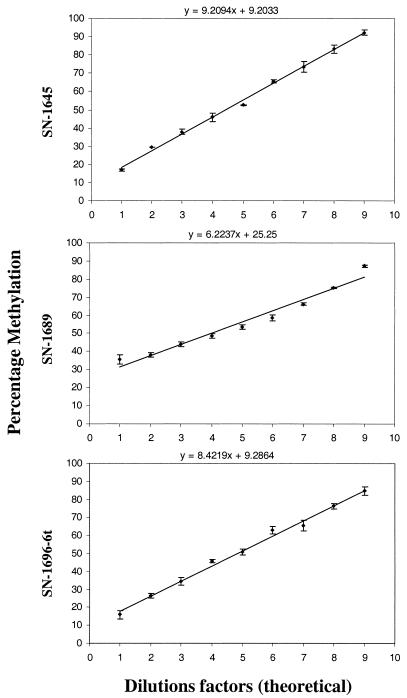
To test the accuracy and linearity of the reaction, PCR templates of the Factor VIII exon 14 region were analyzed as serial mixtures of methylated and unmethylated products, with a 10% increment in methylation (Fig. 2). Concordant with the expected 10% increment, the determined values exhibited a striking linearity for all three CpG sites studied. The slight non-linearity determined for one of the primer products may be due to increased non-specific annealing of its T-rich 3′-end to the rather A-rich sequence stretches of the bisulfite-treated PCR template. To avoid mispriming we recommend that the PCR fragment generated should be kept as short as possible and that the annealing and extension temperatures are optimized.
Figure 2.
The linearity curves for the three primers used in the SNuPE reaction (SN-F8-1645, SN-F8-1689 and SN-F8-1696-6t). The standard deviation of the data is shown for each measurement by a vertical bar. The theoretical equation of the linear response is shown above each graph.
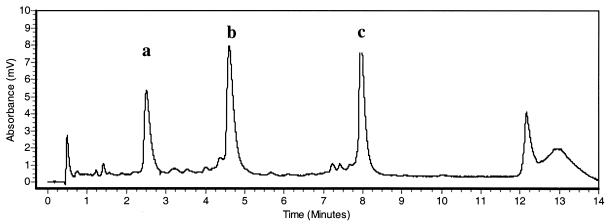
As a second test for the accuracy of our approach we analyzed different PCR products derived from the 5′-region of SNRPN exon 1. Using the SNuPE-IP RP HPLC assay we found that the methylation levels at three selected CpG sites were 33, 40 and 42%, respectively (average of six different samples, data not shown; see Fig. 1B for an example). These values were concordant with previous results obtained using cloning and sequencing approaches, which showed that the majority of CpGs in this region are methylated on ∼40% of the chromosomes (2). Initially all three primers used for analysis of the CpG sites in Factor VIII exon 14 (1645, 1689 and 1696) were designed with an identical length of 18 bases. Based on their different base compositions (thymines) the corresponding retention times were as follows: SN-F8-1645, 50% thymines, 5.36 min; SN-F8-1689, 36% thymines, 1.55 min; SN-F8-1696, 33% thymines, 2.81 min. One problem encountered in a parallel multiplex analysis was that the products of primers SN-F8-1689 and SN-F8-1696 partially co-migrated and clear separation and quantification of the peaks was impossible (data not shown). To overcome this problem, primers of variable length and hydrophobicity were designed. The addition of six thymidines to the 5′-end of oligo SN-F8-1696 shifted its retention time from 2.81 to 8.16 min under the same assay conditions, resulting in clean separation of the extended products (Fig. 3). We therefore recommend this modification as a general strategy to facilitate multiplex column separation of the SNuPE products.
Figure 3.
The retention times of the SN-F8-1696 oligos: (a) unmodified; (b) with six A residues at the 5′-end; (c) with six T residues at the 5′-end.
DISCUSSION
In recent years the bisulfite-based sequencing method has been by far the most widely used technique to analyze chromosomal methylation patterns. It provides sequence-independent information about the degree of methylation at a given CpG site and is sensitive enough to study methylation patterns in only a few cells. However, one major drawback of this method is that it requires laborious and time consuming cloning and sequencing steps. The described SNuPE-IP RP HPLC approach circumvents these laborious steps and provides a quick quantitative assay for a limited number of CpGs in the region of interest. The method will be extremely helpful to scrutinize methylation changes in larger numbers of samples at CpGs which are known to play a critical role. Since both the SNuPE-IP RP HPLC assay and the conventional cloning/sequencing method require complete bisulfite conversion and unbiased PCR amplification, a general problem for both methods may occur when using small amounts of starting material. Whereas the completeness of bisulfite conversion can be optimized by introducing several modifcations (6), a bias in the analysis may still arise when only a limited number of chromosomes are successfully amplified and hence the determined methylation levels do not represent the true methylation levels of all chromosomes. This can only be avoided by comparing results from independent bisulfite and PCR assays. Again, in this respect the SNuPE-IP RP HPLC approach would give a quick estimate of the reproductiveness of the different experiments.
Moreover, this method could also be used to determine the parental origin of methylation, however, two restrictions apply. The first is, as for all traditional methylation analysis methods, a known polymorphism is required. The second is that it should be in the vicinity of a CpG site. When this is the case, two oligos should be designed that correspond to the two alleles of the polymorphism, with about 5–7 bases flanking a CpG site. If the site is differentially methylated then only one of the two oligos will be extended by ddCTP (for methylated) and the other by ddTTP (unmethylated).
When performing the SNuPE-IP RP HPLC assay special attention should be given to primer design. Optimal primers should not be too short (between 15 and 18 bases) and should have a similar Tm of ∼40°C. Generally primers with degenerate positions (e.g. C or T) should be avoided. This causes difficulties in designing primers in CpG-dense regions, as the closeness of two successive CpGs may impede primer design. However, our data on the SNRPN region show that even in a CpG island, optimal primers can be selected and data comparable to cloning/sequencing analysis can be obtained. Furthermore, the content of T residues in the primer design should be carefully considered. Multiplexed primers should have different T residue contents, resulting in different hydrophobicities and column separation. If this cannot be done because of sequence constraints in the template we recommend artificially varying the hydrophobicity by adding stretches of T residues at the 3′-ends of the primers.
In conclusion, we have demonstrated that SNuPE-IP RP HPLC can be applied for the rapid analysis of methylation differences at specific CpG sites urgently needed in studies of carcinogenesis, imprinting diseases and aging. The assay is highly beneficial in comparing large numbers of samples in a relatively very short time. In combination with differential fluorescence labeling of the oligos, the assay will allow further multiplexing and facilitate the analysis of a substantial number of CpG sites in large series of samples in a short time at relatively low cost.
Acknowledgments
ACKNOWLEDGEMENT
Part of this work was supported by a BONFOR grant (O-145.0004).
REFERENCES
- 1.Frommer M., McDonald,L.E., Millar,D.S., Collis,C.M., Watt,F., Grigg,G.W., Molloy,P.L. and Paul,C.L. (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA, 89, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Maarri O., Buiting,K., Peery,E.G., Kroisel,P.M., Balaban,B., Wagner,K., Urman,B., Heyd,J., Lich,C., Brannan,C.I., Walter,J. and Horsthemke.,B. (2001) Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nature Genet., 3, 341–344. [DOI] [PubMed] [Google Scholar]
- 3.Xiong Z. and Laird,P.W. (1997) COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res., 12, 2532–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olek. A., Oswald.,J. and Walter.,J. (1996) A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res., 24, 5064–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalgo M.L. and Jones,P.A. (1997) Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE). Nucleic Acids Res., 12, 2529–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engemann S., El-Maarri,O., Hajkova,P., Oswald,J. and Walter,J. (2001) Bisulphite-based methylation analysis of imprinted genes. In Ward,A. (ed.), Methods in Molecular Biology. Humana Press, Totowa, NJ, Vol. 181, pp. 217–228. [DOI] [PubMed]
- 7.El-Maarri O., Olek,A., Balaban,B., Montag,M., van der Ven,H., Urman,B., Olek,K., Caglayan,S.H., Walter,J. and Oldenburg,J. (1998) Methylation levels at selected CpG sites in the factor VIII and FGFR3 genes, in mature female and male germ cells: implications for male-driven evolution. Am. J. Hum. Genet., 63, 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoogendoorn. B., Owen.,M.J., Oefner.,P.J., Williams,N., Austin,J. and O’Donovan,M.C. (1999) Genotyping single nucleotide polymorphisms by primer extension and high performance liquid chromatography. Hum. Genet., 104, 89–93. [DOI] [PubMed] [Google Scholar]