Abstract
YABBY is among the specific transcription factor (TF) gene family in plants and plays an important role in the development of the leaves and floral organs. Its specific roles include lateral organ development, the establishment of dorsoventral polarity, and response to abiotic stress. Potato is an important crop worldwide and YABBY genes are not still identified and characterized in potato. So, little has been known about YABBY genes in potato until now. This study was carried out to perform genome-wide analysis, which will provide an in-depth analysis about the role of YABBY genes in potato. There have been seven StYAB genes identified, which are found to be located on seven different chromosomes. Through multiple sequence analyses, it has been predicted that the YABBY domain was present in all seven genes while the C2-C2 domain was found to be absent only in StYAB2. With the help of cis-element analysis, the involvement of StYAB genes in light, stress developmental, and hormonal responsiveness has been found. Furthermore, expression analysis from RNA-seq data of different potato organs indicated that all StYAB genes have a role in the vegetative growth of the potato plant. In addition to this, RNA-seq data also identified StYAB3, StYAB5, and StYAB7 genes showing expression during cadmium, and drought stress, while StYAB6 was highly expressed during a viral attack. Moreover, during the attack of Phytophthora infestans on a potato plant StYAB3, StYAB5, StYAB6, and StYAB7 showed high expression. This study provides significant knowledge about the StYAB gene structures and functions, which can later be used for gene cloning, and functional analysis; this information may be utilized by molecular biologists and plant breeders for the development of new potato lines.
Keywords: expression pattern, genome-wide analysis, potato, transcription factor, YABBY gene family
1. Introduction
YABBY transcription factors (TF) are plant specific and belong to a subfamily of zinc finger protein. These gene family members have two regions, N-terminal zinc finger region (C2-C2), and the C-terminal YABBY region; both domains contain highly conserved amino acids (AA) that are specifically involved in DNA binding [1]. The YABBY gene family encodes six genes in Arabidopsis thaliana, which are further categorized into five subfamilies as: FILAMENTOUS FLOWER (FIL)/YAB1, CRABS CLAW (CRC), INNER NO OUTER (INO), YAB2, and YAB5. The YAB2 gene played a role in flower development, CRC genes have a role in carpel polarity and the development of the nectary, and INO genes deal with the outer integument development of ovule, while YAB5/YAB3 are associated with vegetative growth of leaves and cotyledons [2,3]. The YABBY TF also plays an important role in the lateral growth [4] and expansion of leaves [5], the flower development [6], the carpel development [7], the development of ovules [8], the development of veins in leaves [9], shattering of the seeds in cereals [10], the plant response to abiotic stress [11], and fruit ripening [12,13]. The YABBY gene has been found to be affected by hormones such as auxin, abscisic acid (ABA), and gibberellins [11,14].
Based on the crucial roles of YABBY genes in plant development, the genome-wide characterization and expression of YABBY genes have been studied in many plants (both angiosperms and gymnosperms). Until now, YABBY genes have been identified and characterized in different plant species, and the number of genes varied from 6 to 23. The number of YABBY genes reported are 9 in Ananas comosus [6], 7 in Vitis vinifera [12], 11 in Cucurbita moschata, 12 in Cucurbita maxima, 11 in Cucurbita pepo [15], 16 in Phyllostachys edulis [2], 11 in Brassica rapa, 11 in Brassica oleracea, 12 in Brassica nigra, 19 in Brassica juncea, 21 in Brassica napus [16,17], 17 in Glycine max [11], 12 in Gossypium arboreum, 12 in Gossypium raimondii, 23 in Gossypium hirsutum [3], 21 in Triticum aestivum [18], 8 in Phaseolus vulgaris (Common bean) [19], 9 in Solanum lycopersicum [20], 8 in Oryza sativa [21,22], 13 in Zea mays [23], and 9 in Lactuca sativa [24].
The expression patterns of YABBY genes in the vegetative tissues of monocot species are quite diverse. For instance, maize ZYB9 and ZYB14, which are FIL/YAB3-like genes, are expressed adaxially and are believed to regulate lateral outgrowth [25]. In rice, a well-studied monocot species, YABBY gene functions have been extensively explored. The homolog of CRC, OsDL, has been found to influence both the leaves and flower development [26]. Additionally, during rice domestication, the YABBY genes OsSh1 and ObSh3 were found to be required for seed shattering. OsYAB1 has been identified as a regulator of meristem development and of the maintenance of the stamens and carpels [27]. OsYAB3 (TOB3), OsYAB4 (TOB2), and OsYAB5 (TOB1) are enriched in lateral organ primordia and play a crucial role in rice spikelet development [28,29]. These findings demonstrate that YABBY genes have diverse functions in plant growth and development, with each subgroup exhibiting both differentiated and conserved functions. In core eudicots, the vegetative YABBY genes have been found to participate in various aspects of leaf development, and some of these genes also regulate shoot apical meristem development and phyllotaxy. Although YABBY genes are only expressed in abaxial regions, in Zea mays adaxial-specific expression is observed during the leaf primordia initiation [30]. Moreover, the CRC gene in eudicots is limited to the nectaries and carpels while its ortholog in monocots is involved in the development of the leaf midrib [31]. Therefore, the evolutionary history of YABBY genes seems to coincide with the emergence of leaves in seed plants [27], highlighting the need to comprehend the inter-species and intra-species relationships of YABBY genes. Potato (Solanum tuberosum) is an essential component of the human diet and its consumption is increasing day by day [32]. Potato is the world’s most important crop followed by maize, rice, and wheat. This vital crop has a very high potential and nutritional value [33]. S. tuberosum is a rich source of minerals, proteins, fats, vitamins, and energy. Potatoes are not only limited to food sources but also used drastically in industrial products such as potato chips, potato starch and powder flakes and pellets, and certain other value-added products [34]. Potato crop often faces the issue of low yields, because it is a root crop and is obtained as a result of vegetative growth [35]. As YABBY genes played a crucial role in vegetative development and also played a role in abiotic stresses, many of yield losses can be minimized if YABBY genes have been identified and functionally characterized in potato crops. Despite the immense importance of potato as a food crop, no significant research regarding its YABBY TFs has been reported until now. The objective of the study was the identification of the genes belonging to the YABBY TF family in potato genome and inter-operating their functions with the help of RNA-seq data through various bioinformatics tools. The results of genome-wide identification and characterization will be utilized for functional analysis and cloning of the members of this gene family.
2. Materials and Methods
2.1. Database Search and Retrieval of Sequences
The amino acid (AA) sequence of the YABBY domain was retrieved from Pfam database (accessed on 2 December 2021) and the YABBY domain (PF04690) in these sequences was identified. This domain (PF04690) was used for the identification of StYAB genes in potato genome database at Phytozome v13 (https://phytozome-next.jgi.doe.gov, accessed on 2 December 2021 using BLAST-P (Protein-basic local alignment search tool) program. The retrieved sequences of AA were verified on NCBI CDD (Conserved Domain Database) (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) accessed on 2 December 2021 with the default parameters. The proteins lacking a real YABBY domain (PF04690) (https://pfam.xfam.org/family/PF04690) accessed on 2 December 2021 in their sequences were excluded.
2.2. Determination of Physio-Chemical Properties of YABBY Proteins
The ProtParam tool (accessed on 2 December 2021) (http://web.expasy.org/protparam/) was used to predict the protein length (AA residues), molecular weight, and pI value of YABBY proteins. The information on gene IDs, chromosomal positions, sequences of gene, and protein was taken from Phytozome. These YABBY encoding genes were renamed based on the order of their physical position. The subcellular localization of StYAB genes were predicted with the help of WoLF PSORT (accessed on 2 December 2021) (https://wolfpsort.hgc.jp/).
2.3. Gene Structure Analysis
For the prediction of the gene structure, genomic sequences, and 1000 bp upstream promoter, the sequences of StYAB genes were retrieved from Phytozome v13 (accessed on 2 December 2021). Furthermore, the gff3 file of the potato genome was retrieved from the PLAZA genome database (https://bioinformatics.psb.ugent.be/plaza/) accessed on 2 December 2021. These sequences were further used to visualize the gene structure using the gene structure display server (GSDS v2.0) (accessed on 3 December 2021) (http://gsds.cbi.pku.edu.cn/) to find the intron-exon arrangement, PlantCare database (accessed on 4 December 2021) (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) for cis-regulatory elements detection, and Multiple EM for motif elicitation (MEME) program (http://meme.nbcr.net/meme/) accessed on 4 December 2021 for motif recognition.
2.4. Comparative Phylogenetic Analysis
Phylogenetic analysis was performed between the genomes of S. tuberosum, A. thaliana, S. lycopersicum, C. sativus, and C. maxima. The AA sequences of YABBY proteins were aligned using MUSCLE [36]. These protein sequences were used to generate the phylogenetic tree in MEGA 11 software using the neighbor-joining (NJ) method, with a bootstrapping value set at 1000 replications. It was further modified using iTOL (https://itol.embl.de/) accessed on 5 December 2021.
2.5. Gene Duplication and Synteny Analysis
The divergence time of StYAB genes was estimated with the help of Ka/Ks values. The protein sequence alignment was made using MUSCLE and the number of Ka/Ks substitution rates were determined using Tbtools v 1.108. Default parameters were used as described in the package manual. Ka/Ks ratios of paralogous genes were calculated in order to estimate the molecular evolution rate of each gene pair. The time of divergence (DT) was estimated by putting the Ks value in T = Ks/2λ equation, where λ is equal to 2.6 × 10−9 [37]. MCScanX v 1.0 (Multiple Collinearity Scan toolkit) was adopted to analyze the gene duplication events, with the default parameters. To find the synteny relationship between the paralogous genes of potato and orthologous genes of Arabidopsis, tomato, and chili, syntenic and dual syntenic maps were constructed using Tbtools [38].
2.6. Gene Ontology Enrichment Analysis
Gene ontology (GO) term enrichment analysis was performed to further validate the functions of StYAB genes using GO annotations [31]. Uniprot (Online database) (accessed on 8 December 2021) (https://www.uniprot.org/) was used to find out the molecular functions of genes as well as the various biological processes. Furthermore, an online web tool ShinyGo v0.741 (accessed on 9 December 2021) (http://bioinformatics.sdstate.edu/go/) was used for the analysis to better understand the role of StYAB genes in potatoes.
2.7. Expression Analysis
Expression data of all seven StYAB genes were taken from different plant organs (unfertilized leaves, floral buds, axial regulator, developing tubers, petioles, and stems) and under different biological stresses (viral attack, Phytophthora infestans attack on leaves, cadmium stress, drought stress, and meristem dormancy). Expression analysis of StYAB genes was performed using previously generated RNA-seq data. Data related to the growth stages and the behavior of the different organs under various biotic and abiotic stresses was retrieved from three databases, i.e., NCBI gene expression omnibus (GEO) (accessed on 15 December 2021), expression atlas (accessed on 17 December 2021), and expressed sequence tags (EST) (accessed on 20 December 2021). For the expression profiling, reads per kilobases per million mapped read (RPKM) values from the RNA-seq data were log2 transformed [39]. The data for expression in different organs and stresses were downloaded from previously published papers [40,41,42,43,44].
2.8. MiRNA Analysis
The micro-RNA (miRNA) sequences related to StYAB genes in potato were identified from psRNATarget (accessed on 25 December 2021) (https://www.zhaolab.org/psRNATarget/) using the CDS sequences of all the StYAB genes with the default parameters. The putative functions of the identified miRNA were retrieved from previously performed in vitro and in vivo experiments.
3. Results
3.1. Identification of the YABBY Genes in Potato
There were seven StYAB genes identified in potato. The StYAB genes encoding the protein’s length vary from 142 to 219 AA and the molecular weight varies from 16.15737 to 24.48169 kD, with StYAB2 being the smallest and StYAB3 being the longest protein (Table 1). In total, 33 out 56 AAs in the conserved YABBY domain were found to be 100% conserved in all the YABBY domain sequences of potato (Figure S1), while the remaining 23 AA were found to be variable in all the StYAB proteins. Furthermore, sub-cellular localization of StYAB genes showed that StYAB7, StYAB3, StYAB4, and StYAB2, were localized in a nucleus, while StYAB6 and StYAB1 were probably localized in the extracellular structures. The StYAB5 genes were localized in the chloroplast (Table 2 and Figure S2).
Table 1.
Information about the discovered seven YABBY genes in potato genome. Accession number, chromosome number and location, gene direction, amino acid sequence length, molecular weight, isoelectric point (Pi value), no. of introns and exons.
| YABBY Gene Name | Accession Number | Chromosome Number | Chromosome Location (bp) |
Direction | No. of Amino Acids | Molecular Weight (kD) | Pi Value | No. of Introns | No. of Exons | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phytozome V13 (6.1) | PLAZA Genome Database | Start | End | ||||||||
| StYAB 1 | Soltu.DM.07G003610 | PGSC0003DMT400079392 | 7 | 4307036 | 4311510 | F | 179 | 20.16703 | 8.77 | 6 | 7 |
| StYAB 2 | Soltu.DM.05G008980 | PGSC0003DMT400072893 | 5 | 9003101 | 9004644 | F | 142 | 16.15737 | 8.25 | 4 | 5 |
| StYAB 3 | Soltu.DM.08G025380 | PGSC0003DMT400012083 | 8 | 55208205 | 55211393 | R | 219 | 24.48169 | 7.22 | 6 | 7 |
| StYAB 4 | Soltu.DM.01G031660 | PGSC0003DMT400066793 | 1 | 71511821 | 71516043 | R | 218 | 24.16953 | 7.23 | 6 | 7 |
| StYAB 5 | Soltu.DM.11G024670 | PGSC0003DMT400007731 | 11 | 44742927 | 44749556 | R | 178 | 20.14184 | 9.24 | 5 | 6 |
| StYAB 6 | Soltu.DM.12G026680 | PGSC0003DMT400007474 | 12 | 56595139 | 56601086 | R | 194 | 22.09788 | 8.17 | 6 | 7 |
| StYAB 7 | Soltu.DM.06G029310 | PGSC0003DMT400015197 | 6 | 54473907 | 54479995 | F | 191 | 21.272 | 8.79 | 5 | 6 |
Table 2.
Gene ontology enrichment analysis of StYAB genes their GO functions, sub-cellular localization signal genes expression, orthologs in arabidopsis, and their functions are presented in the table.
| Gene ID | GO Function | Sub-Cellular Localization | Gene Expression |
Ortholog in Arabidopsis | References | ||
|---|---|---|---|---|---|---|---|
| Molecular Function | Biological Process | Gene Name | Function | ||||
| StYAB1 | Abaxial cell fate specification | Leaf development | Extra chromosomal | Unfertilized leaves and floral buds | YABBY 5 | Axial regulator | [45,46] |
| StYAB2 | Cell fate commitment, floral bud determinacy, and style development | Leaf and flower development | Nucleus | Axial regulator | INO | Outer integument growth | [47] |
| StYAB3 | Fruit development, polarity specification of abaxial-adaxial surface, regulation of shoot apical meristem, and leaf development | Leaf, flower, and fruit development | Nucleus | Phytophthora treated adult leaves, cadmium stress, dormant and non-dormant potato tubers, photoperiod effect, phloem associated cells | YABBY1/FIL | Axial regulator | [45] |
| StYAB4 | Fruit development, polarity specification of abaxial-adaxial surface, regulation of shoot apical meristem, and leaf development | Leaf, flower, and fruit development | Nucleus | Axial regulator | YABBY3 | Axial regulator | [47] |
| StYAB5 | Abaxial cell fate commitment | Leaf development | Chloroplast | Floral buds, cadmium stress, dormant and non-dormant potato tubers, photoperiod effect, phloem associated cells | YABBY2 | Putative axial regulator | [48,49] |
| StYAB6 | Abaxial cell fate commitment | Leaf development | Extra chromosomal | Developing tubers, cadmium stress, dormant and non-dormant potato tubers, photoperiod effect, phloem associated cells | YABBY5 | Axial regulator | [45,50] |
| StYAB7 | Abaxial cell fate commitment | Leaf development | Nucleus | Floral buds, cadmium stress, dormant and non-dormant potato tubers, photoperiod effect, phloem associated cells | YABBY2 | Putative axial regulator | [48,50] |
3.2. Gene Structures and Recognition of Conserved Cis-Elements and Motifs
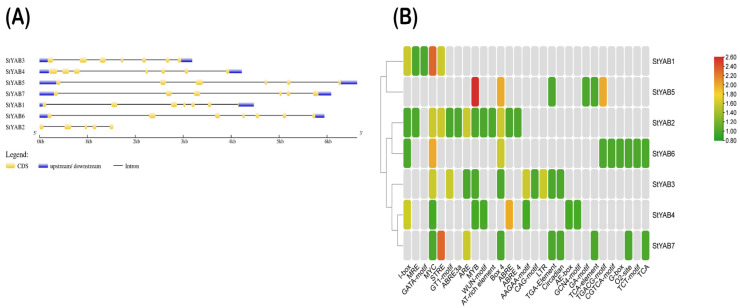
The gene structure prediction showed that four genes contain seven exons and six introns (57.14%), two genes had six exons and five introns (28.57%), and one gene had five exons and four introns (14.28%) (Table 1 and Figure 1A).
Figure 1.
Predicted gene structures and cis-regulatory elements in StYAB. (A) Predicted gene structure of seven StYAB genes. The yellow regions show the exon regions, black lines show the introns, while the blue portions show the upstream/downstream sequences. (B) Cis-regulatory elements in putative StYAB promoters, which are associated with different plant developmental processes.
In total, 62 cis-elements were found in the StYAB genes (Figure 1A). These include light-responsive (24), stress-responsive (6), development- and metabolism-responsive (18), and hormone-responsive (14) elements. Overall, 38% of the elements were found to be responsive to light, indicating that StYAB genes play an important role in the light stress response. Six cis-elements were found to be related to a stress response, which predicts its role in stress-related activity. Moreover, a total of 18 cis-elements were found to be related to the development and metabolism of plants (Table S1). It makes up 29% of the cis-elements and it hints towards the role of StYAB genes in the growth and development of potato plants. Furthermore, three hormones related to cis-elements were identified, including Auxin, MeJA, and ABA, signifying the potential targets to study the role of hormones under stress conditions. It has been found that most hormone-related regions are dispersed in all seven genes. This signifies the role of these genes in the hormonal response (Figure 1B and Table S1).
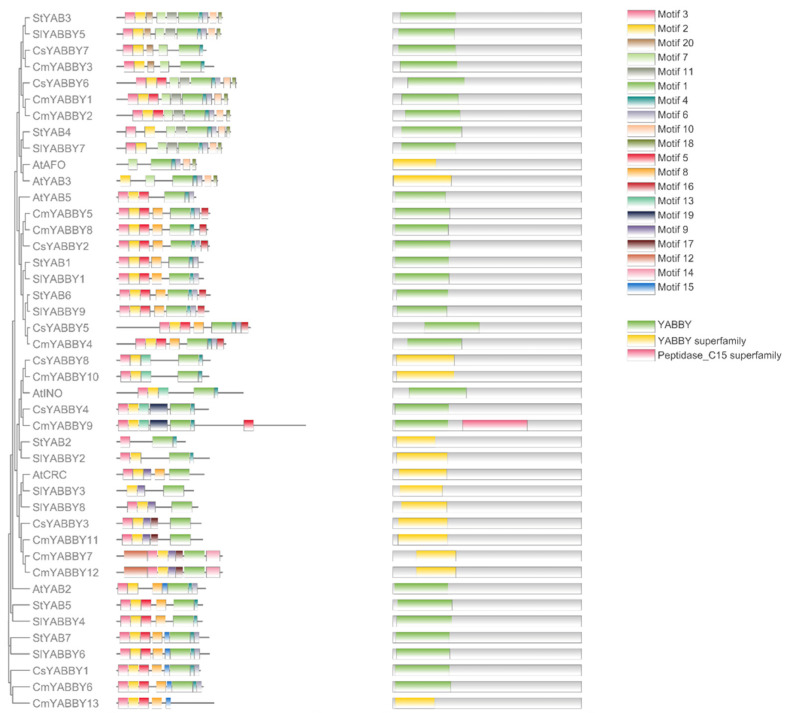
The distribution of 15 motifs on 7 YABBY proteins of potato by using MEME version 4.9.0 and interlinking it with phylogenetic tree to develop a good understanding of their association. The bars represent motifs with different color codes for different types of motifs (Figure 2).
Figure 2.
The distribution of 15 motifs on seven StYAB proteins. The motifs were found in potato by using MEME version 4.9.0 and inter-linking it with a phylogenetic tree to develop a good understanding of their association. The bars represent motifs with different color codes for the different types of motifs.
3.3. Phylogenetic Analysis of the YABBY Gene Family
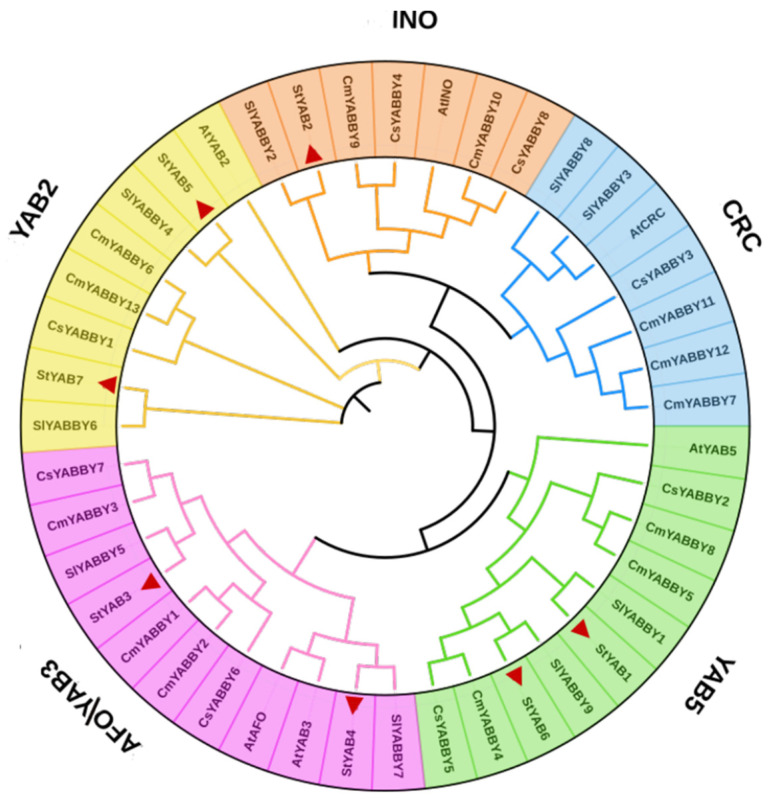
The results of the phylogenetic analysis depicted that StYAB genes can be categorized into five subgroups as in Arabidopsis, i.e., INO, CRC, YAB5, AFO/YAB3, and YAB2. Among these subgroups, AFO/YAB3 is the biggest group containing 11 genes, YAB5 is second with 10 genes, YAB2 is third with 8 genes, INO is fourth with 7 genes, and CRC is the smallest with 7 genes. No potato gene was reported in the CRC group (Figure S3 and Table S2). It was also found that all the StYAB genes showed the close similarity with S. lycopersicum in all the identified groups (Figure 3). It is inferred from the phylogenetic analysis that StYAB genes might be directly evolved from SlYAB genes.
Figure 3.
Phylogenetic relationship among the YABBY genes of S. tuberosum, A. thaliana, S. lycopersicum, C. sativus, and C. maxima was studied. S. tuberosum genes are marked with a red triangle. The evolutionary history was inferred using the NJ method with 1000 bootstrap replications. This analysis involved 43 YABBY genes. The evolutionary analyses were conducted in MEGA 11.
3.4. StYAB Gene Location on Chromosomes and Duplication Assessment
The chromosomal localization of the StYAB genes predicted that these genes were distributed on different chromosomes. StYAB1, StYAB2, StYAB3, StYAB4, StYAB5, StYAB6, and StYAB7 were present on chromosomes 7, 5, 8, 1, 11, 12, and 6 respectively (Table 1; Figure S4).
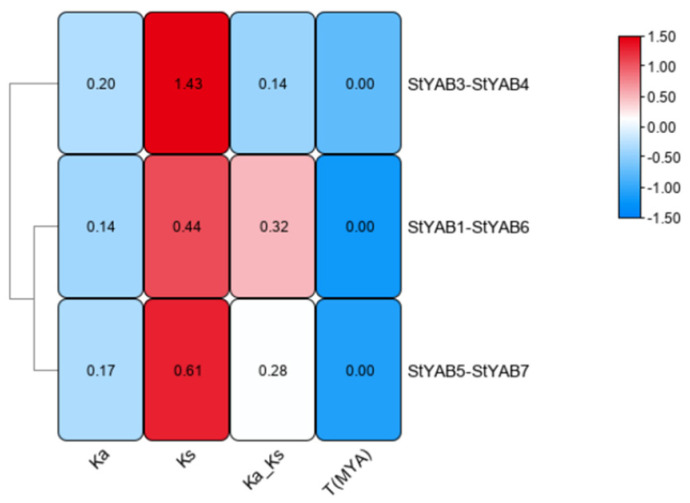
This estimated ratio was 0.14 between StYAB3 and StYAB4, 0.28 between StYAB5 and StYAB7, and 0.32 between StYAB1 and StYAB6. The estimated date of duplication was 275.3137046 million years ago (MYA) in StYAB3–StYAB4, 117.7436777 (MYA) in StYAB5–StYAB7, and 85.22345807 (MYA) in StYAB1–StYAB6 (Figure 4; Table S3).
Figure 4.
The time of gene duplication was estimated for the different paralogous pairs of the potato StYAB genes on the basis of Ks and Ka values. The analyses were conducted using Tbtools v 1.108. Ka/Ks represents the ratio of nonsynonymous (Ka) versus synonymous (Ks) mutations.
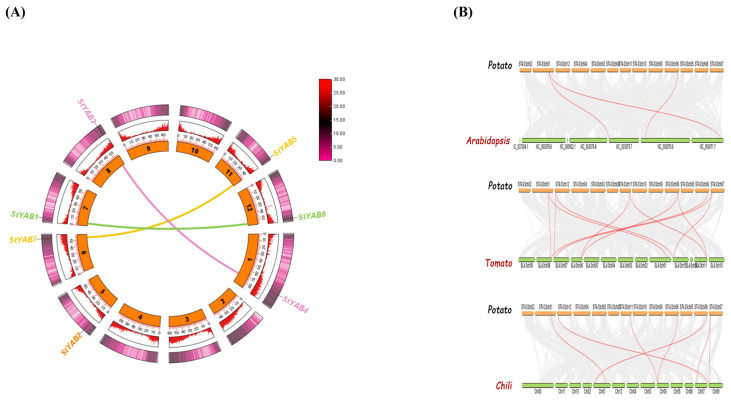
The syntenic analysis showed that StYAb1, StYAB3, and StYAB7 are the paralogs of StYAB6, StYAB4, and StYAB5, respectively, while the StYAB2 gene happened to be a singleton as no duplicate has been found (Figure 5A). The dual synteny analysis showed that Arabidopsis had 3 duplicated genes. In between potato and tomato, 11 duplicated genes were found, while 7 genes were found duplicated in between potato and chili (Figure 5B).
Figure 5.
(A) The genome-wide synteny analysis of StYAB genes showing the dominance of segmental duplication and rare occurrence of tandem duplication. The joining lines show the duplicated StYAB genes in the genome. (B) Dual synteny analysis of potato-Arabidopsis, potato-tomato, and potato-chili. The orange bars represent the chromosomes of potato while the green bars represent the chromosomes of Arabidopsis, tomato, and chili, respectively. The red lines show the duplicated genes in the respective genomes.
3.5. Enrichment Analysis and Ortholog Identification
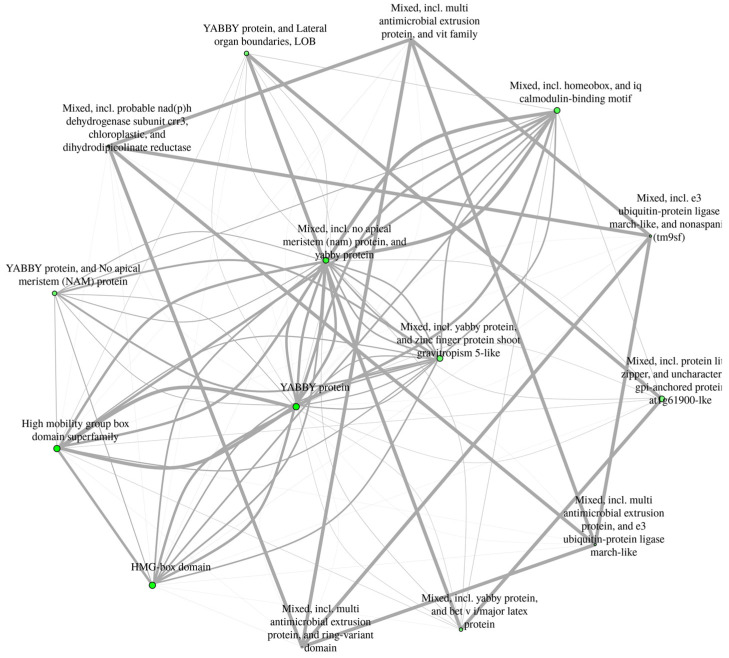
The GO analysis showed that the StYAB genes are involved in the diverse cellular functions and biological processes. All seven StYAB genes are involved in the abaxial cell fate specification and cellular development. Moreover, it was found that StYAB genes further participate in leaf, shoot, and meristem development (Table 2, Figure 6 and Figure S5).
Figure 6.
Static network enrichment graph, showing the network of StYAB gene functions. The darker nodes are more the significantly enriched gene sets. The bigger nodes represent the larger gene sets. The thicker edges represent more overlapped genes.
3.6. Expression Analysis of StYAB Gene
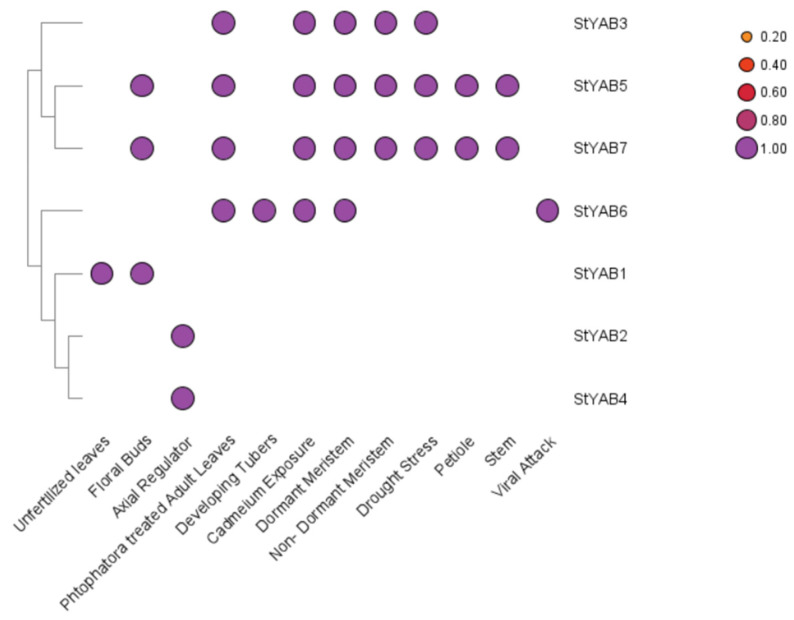
The results of the expression analysis showed that StYAB genes are highly expressed in different organs such as unfertilized leaves, floral buds, developing tubers, dormant meristems, petioles, and stems [40,41,42]. The differential expression of specific StYAB are shown in Figure 7. These results showed that the StYAB genes were involved in the vegetative development of potato plants. The different StYAB genes also showed their expression under different stress conditions (viral attack, Phytophthora infestans attack on leaves, cadmium stress, drought stress, and meristem dormancy) [40,41,42,43,44]. The result of RNA-seq data showed that StYAB3, StYAB5, and StYAB7 were differentially expressed in response to cadmium stress and drought stress. The StYAB6 was highly expressed when there was any viral attack. Moreover, the StYAB3, StYAB5, StYAB6, and StYAB7 were highly expressed under Phytophthora infestans infection. These results suggested that the StYAB genes played a significant role under the different abiotic and biotic stress conditions (Figure 7 and Figures S6–S11).
Figure 7.
Heat map shows the expression profile of the StYAB genes in different organs and under different biological stresses of potato.
3.7. MiRNA Targets in POTATO
A total of 10 miRNA sequences targeting 5 StYAB genes (StYAB3, StYAB4, StYAB5, StYAB6, and StYAB7) were detected. The expectation value observed was from 4 to 5 and the minimum length of the miRNA was 21 bp to a maximum of 24 bp. The miRNA sequences targeting StYAB1 and StYAB2 were not found (Table 3 and Table S4).
Table 3.
StYAB genes targeting putative miRNA functions along with their targeted genes.
| Gene | Targeting miRNA | Function | Reference |
|---|---|---|---|
| StYAB3 | Stu-miR172d-5p | Enhance nodulation | [51] |
| StYAB4 | stu-miR5303g, stu-miR5303i, stu-miR5303h, stu-miR1886g-5p, stu-miR5303j, | Long Non-RNA development during tuber sprouting | [52] |
| StYAB5 | stu-miR8037 | No target site reported | [53] |
| StYAB6 | stu-miR7992-5p | Targets nuclear inclusion protein | [54] |
| StYAB7 | stu-miR390-3p, stu-miR1886g-5p | Promotes lateral growth | [55] |
4. Discussion
Potato (Solanum tuberosum L.) is a highly important crop species worldwide, with global production exceeding 388 million metric tons per year, it is considered a promising crop for reducing human hunger and poverty due to its high yield potential and nutritional properties. Despite being predominantly grown by resource-poor farmers in developing countries, potato crops are susceptible to yield losses caused by various abiotic and biotic stresses. Transcription factors (TFs) are important regulatory elements that enable plants to respond to external environmental stresses, with families such as NAC, bZIP, MYB, YABBY, NF-YB, and AP2 being particularly relevant in minimizing the yield losses [45,46]. Among these families, the YABBY TF family has received a lot of attention in plant research, especially in relation to its involvement in early embryonic development, lateral organ development, the establishment of the near-distal axis polarity of leaves, and plant development and stress response [47].
While YABBY genes have been extensively studied in dicotyledonous plants such as Arabidopsis, relatively little is known about monocot YABBY genes, including those of potato. Thus, a genome-wide study was conducted to identify and characterize the StYAB genes in potato. The study identified seven StYAB genes in total and found that their gene structure varied in terms of the number of exons and introns, potentially leading to functional variability. Furthermore, we observed different subcellular localizations for the StYAB genes, with four localized in the nucleus, two in extracellular structures, and one in the chloroplast. An analysis of the StYAB gene promoters revealed the presence of various cis-regulatory elements related to plant growth and development, light, and stress response, further supporting the idea that these genes are involved in different vegetative growth and have a role in mitigating different biotic and abiotic stress conditions.
To better understand the possible functions of StYAB genes in the Solanaceae family, expression patterns of different genes were analyzed under different stress conditions. It was found that StYAB1, StYAB3, StYAB2, and StYAB5 exhibited similar levels of expression to their homologs in Arabidopsis and rice, indicating functional conservation. Additionally, while only one homolog of the INO group was observed in S. tuberosum, we found that StYAB2, the homolog of the INO group, was expressed in processes related to axial regulation. These results suggest that StYAB genes have diverse functions in plant growth and development, and may play a role in mitigating the different stress conditions. However, further extensive studies are necessary to better understand the specific functions of StYAB genes.
The YABBY TF family is a subgroup of the zinc finger protein superfamily, and its members contain two structural domains: the zinc-finger domain and YABBY domain [48]. The amino acids in these domains are linked with DNA specificity and are highly conserved [49]. While Arabidopsis, tomato, and other dicotyledonous species have five sub-groups, including CRC, FIL/YAB3, INO, YAB2, and YAB5, monocotyledon rice has only four sub-groups, lacking YAB5. During evolution, YABBY TFs in monocots, dicots, and different subpopulations have undergone functional differentiation. In this study, StYAB genes were classified into five groups as per in A. thaliana, i.e., INO, YAB2, YAB5, AFO, and CRC. However, no member of the StYAB gene family was detected in the CRC group.
Gene duplication is a common evolutionary event that occurs when an organism’s genome acquires an additional copy of a pre-existing gene. This can occur through various mechanisms, such as unequal crossing over, retro-transposition, and whole-genome duplication. Gene duplication plays a critical role in evolution, as it creates opportunities for functional divergence and innovation. The duplicated genes can acquire new functions through various mechanisms, including sub-functionalization and neo-functionalization, leading to an increase in the complexity and diversity of organisms [50].
Synteny analysis is a powerful tool used to study gene duplication events and their evolutionary consequences. It involves the comparison of the order and arrangement of genes on different chromosomes, or within a genome. By identifying the syntenic regions, researchers can detect the gene duplications and infer the evolutionary history of the gene families [56]. The synteny analysis can also reveal the mechanisms of gene duplication, such as segmental or tandem duplication, and provide insights into the functional diversification of the gene families.
This can be predicted from their chromosome location. This means genes that are present on the same chromosome can possibly be the result of tandem duplication, whereas those genes that are located on different chromosomes might possibly be the result of segmented duplication. In the case of StYAB genes, segmented duplication was predominantly observed.
Interestingly, a similar pattern of segmented duplication was observed in the YABBY gene family of Oryza sativa. This finding is consistent with the results of StYAB gene synteny analysis in potato, as well as in other plant species such as C. arietinum and Olea europaea. These results highlight the importance of gene duplication events in shaping the evolution of plant genomes and the diversification of gene families. Further investigation of the functional divergence and neofunctionalization of YABBY genes in these plant species could shed light on their role in the development and evolution of plant structures. It was further confirmed with the help of synteny analysis (Figure 5A and Figure S5). The same results were observed in the case of C. arietinum [51] and Olea europaea [52]. Gene duplication is the main driver of genome expansion in species. Variations in the gene family members across different eukaryotic species might be because of the process of evolution [53]. Moreover, the dual synteny blocks of potato were constructed with three crops, i.e., Arabidopsis, tomato, and chili, separately to determine the connection between them. The dual syntenic block of potato and Arabidopsis genomes predicted that two genes in chromosome 1 and one gene of chromosome 6 have duplicated genes in Arabidopsis as it was shown by joining threads between the respective chromosomes (Figure 5b). The dual syntenic block between the potato and tomato genome predicted that both species share 11 duplicated genes from different chromosomes as shown by joining threads (Figure 5b). Lastly, the dual synteny of potato and chili showed that they share seven similar genes on different chromosomes, as shown in (Figure 5b).
MicroRNAs (miRNAs) are important regulatory entities of plants. They have a role in the regulation of almost all biological processes of plants, such as plant growth and development, during biotic and abiotic stress. They are highly conserved and are very specific in function. The Stu-miR390-3p member of family miR390 was detected, targeting StYAB7. This family has a role in promoting lateral growth [54], which means that StYAB7 has a role in primary growth and inhibits lateral growth. Four members of the miR5303 family, stu-miR5303g, stu-miR5303h, stu-miR5303i, and stu-miR5303j, were detected targeting our StYAB4 gene, and play important roles in long RNA development during tuber sprouting [55], which depicts that StYAB4 has a role that somewhat inhibits tuber sprouting in potato. One member from the miR172 family was detected, stu-miR172d-5p, targeting our gene StYAB3 [57]. This miRNA (stu-miR172d-5p) promotes nodulation in potato by inhibiting StYAB3. One member from the miR1886 family, i.e., stu-miR1886g-5p, was detected targeting two genes StYAB4 and StYAB7. This miRNA has a role in plant growth and development [58]. Furthermore, two miRNA families, i.e., miR7992 and miR8037, with one member each detected in potato, targeted StYAB6 and StYAB5 respectively. miR7992-5p targets nuclear inclusion protein [59], while no role of miR8037 was found (Table 3).
5. Conclusions
This study identified seven StYAB genes in Solanum tuberosum. Based on structural analysis, it was found that StYAB genes have five to seven exons. Based on motif analysis and promoter analysis, it was found that cis-regulatory elements related to light, and developmental and hormonal responsiveness were present in the promoter of StYAB genes, which conferred their role in the vegetative development of potatoes. Apart from that, some cis-regulatory elements related to biotic and abiotic stress were also found. Along with the presence of cis-regulatory elements, expression analysis results demonstrated that StYAB1, StYAB2, and StYAB4 had a role in the vegetative development of the potato plant. Additionally, StYAB genes showed expression during different abiotic and biotic stresses. StYAB3, StYAB5, StYAB6, and StYAB7 showed expression during the Phytophthora infestans attack, while StYAB6 also showed higher expression levels during the virus attack on potato plants. Furthermore, StYAB3, StYAB5, and StYAB7 also showed expression during abiotic stresses, i.e., cadmium and drought stress. However, this in-silico study represents a comprehensive genome-wide knowledge of StYAB genes in potato, but further investigation such as gene cloning and functional analysis are required to validate these genes and their functional roles against the different physiological and biological processes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14040824/s1, Figure S1: The sequence logos are based on alignments of StYAB gnese; Figure S2: Heat Map representing sub-cellular localization of all seven StYAB genes; Figure S3: YABBY genes distribution chart representing, total number of YABBY genes from different species in 5 sub-groups based on phylogenetic analysis; Figure S4: Distribution of StYAB genes on potato chromosomes, lines predicting the possible gene duplication on different chromosomes; Figure S5: Fold Enrichment chart representing the overlapping StYAB genes functions; Figure S6: Heat map shows the expression profile of the StYAB genes in leaves and Roots of potato cultivar under different levels of Cadmium stress i.e., Control = 0 mg per kg, T1 = 1 mg per kg and T2 = 5 mg per kg); Figure S7: Heat map shows the expression profile of the StYAB genes in Dormant and Non- Dormant potato cultivar; Figure S8: Heat map shows the expression profile of the StYAB genes in leaves of four different potato cultivars under drought stress; Figure S9: Heat map shows the expression profile of the StYAB genes in petiole of potato cultivars under different photoperiod; Figure S10: Heat map shows the expression profile of the StYAB genes in phloem associated cells of petiole and lower stem of potato cultivar; Figure S11: Heat map shows the expression profile of the StYAB genes in tubers of two potato cultivars (Phytophthora Infestans resistant and susceptible) after different time of inoculating infection in both cultivars (0 h, 24 h, and 48 h); Table S1: Putative Cis-regulatory elements found in promoter regions of StYAB genes; Table S2: StYAB genes distribution among groups based on phylogenetic analysis; Table S3: Ka/Ks ratio duplicated gene pairs in potato; Table S4: miRNA targets prediction of StYAB genes. The miRNA data was downloaded from psRNATarget (Online Web tool).
Author Contributions
All authors contributed to the study’s conception and design. Material preparation, data collection, and analysis were performed by H.S.-U.-D.M., M.S., H.A., A.A., J.T. and M.A.(Muhammad Awais). The first draft of the manuscript was written by H.S.-U.-D.M., M.A.J. and H.A.; Q.A., A.A., J.T., M.A. (Muhammad Ashfaq), G.J. and R.S. reviewed the manuscript. All authors commented on previous versions of the manuscript. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work is supported by the University of Birmingham and University of the Punjab.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Hussain Q., Asim M., Zhang R., Khan R., Farooq S., Wu J. Transcription Factors Interact with ABA through Gene Expression and Signaling Pathways to Mitigate Drought and Salinity Stress. Biomolecules. 2021;11:1159. doi: 10.3390/biom11081159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma R., Huang B., Huang Z., Zhang Z. Genome-wide identification and analysis of the YABBY gene family in Moso Bamboo (Phyllostachys edulis (Carriere) J. Houz) PeerJ. 2021;9:e11780. doi: 10.7717/peerj.11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z., Gong Q., Wang L., Jin Y., Xi J., Li Z., Qin W., Yang Z., Lu L., Chen Q., et al. Genome-Wide Study of YABBY Genes in Upland Cotton and Their Expression Patterns under Different Stresses. Front. Genet. 2018;9:33. doi: 10.3389/fgene.2018.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hussain Q., Zheng M., Ashraf M.F., Khan R., Yasir M., Farooq S., Zhang R., Wu J. Genome-wide identification, characterization and expression analysis of the ABA receptor PYL gene family in response to ABA, photoperiod, and chilling in vegetative buds of Liriodendron chinense. Sci. Hortic. 2022;303:111200. doi: 10.1016/j.scienta.2022.111200. [DOI] [Google Scholar]
- 5.Eckardt N.A. YABBY Genes and the Development and Origin of Seed Plant Leaves. Plant Cell. 2010;22:2103. doi: 10.1105/tpc.110.220710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z., Li G., Cai M., Priyadarshani S.V.G.N., Aslam M., Zhou Q., Huang X., Wang X., Liu Y., Qin Y. Genome-Wide Analysis of the YABBY Transcription Factor Family in Pineapple and Functional Identification of AcYABBY4 Involvement in Salt Stress. Int. J. Mol. Sci. 2019;20:5863. doi: 10.3390/ijms20235863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarojam R., Sappl P.G., Goldshmidt A., Efroni I., Floyd S.K., Eshed Y., Bowman J.L. Differentiating Arabidopsis Shoots from Leaves by Combined YABBY Activities. Plant Cell. 2010;22:2113–2130. doi: 10.1105/tpc.110.075853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stahle M.I., Kuehlich J., Staron L., von Arnim A.G., Golz J.F. YABBYs and the Transcriptional Corepressors LEUNIG and LEUNIG_HOMOLOG Maintain Leaf Polarity and Meristem Activity in Arabidopsis. Plant Cell. 2009;21:3105–3118. doi: 10.1105/tpc.109.070458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X.-L., Yang Z.-P., Zhang J., Zhang L.-G. Ectopic Expression of BraYAB1-702, a Member of YABBY Gene Family in Chinese Cabbage, Causes Leaf Curling, Inhibition of Development of Shoot Apical Meristem and Flowering Stage Delaying in Arabidopsis thaliana. Int. J. Mol. Sci. 2013;14:14872–14891. doi: 10.3390/ijms140714872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin Z., Li X., Shannon L.M., Yeh C.-T., Wang M.L., Bai G., Peng Z., Li J., Trick H.N., Clemente T.E., et al. Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 2012;44:720–724. doi: 10.1038/ng.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao S.P., Lu D., Yu T.F., Ji Y.J., Zheng W.J., Zhang S.X., Chai S.C., Chen Z.Y., Cui X.Y. Genome-wide analysis of the YABBY family in soybean and functional identification of GmYABBY10 involvement in high salt and drought stresses. Plant Physiol. Biochem. 2017;119:132–146. doi: 10.1016/j.plaphy.2017.08.026. [DOI] [PubMed] [Google Scholar]
- 12.Zhang S., Wang L., Sun X., Li Y., Yao J., van Nocker S., Wang X. Genome-Wide Analysis of the YABBY Gene Family in Grapevine and Functional Characterization of VvYABBY4. Front. Plant Sci. 2019;10:1207. doi: 10.3389/fpls.2019.01207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartley G.E., Ishida B.K. Ethylene-sensitive and insensitive regulation of transcription factor expression during in vitro tomato sepal ripening. J. Exp. Bot. 2007;58:2043–2051. doi: 10.1093/jxb/erm075. [DOI] [PubMed] [Google Scholar]
- 14.Peng X., Wu Q., Teng L., Tang F., Pi Z., Shen S. Transcriptional regulation of the paper mulberry under cold stress as revealed by a comprehensive analysis of transcription factors. BMC Plant Biol. 2015;15:108. doi: 10.1186/s12870-015-0489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan J., Shen C., Xin J., Li Z., Li X., Zhou J. Genome-wide Characterization and Expression Analysis of YABBY Gene Family in Three Cultivars of Cucurbita Linn. And Their Response of Salt Stress in Cucurbita Moschata. 2020. [DOI]
- 16.Hou H., Wu P., Gao L., Zhang C., Hou X. Characterization and expression profile analysis of YABBY family genes in Pak-choi (Brassica rapa ssp. chinensis) under abiotic stresses and hormone treatments. Plant Growth Regul. 2019;87:421–432. doi: 10.1007/s10725-019-00475-5. [DOI] [Google Scholar]
- 17.Xia J., Wang D., Peng Y., Wang W., Wang Q., Xu Y., Li T., Zhang K., Li J., Xu X. Genome-Wide Analysis of the YABBY Transcription Factor Family in Rapeseed (Brassica napus L.) Genes. 2021;12:981. doi: 10.3390/genes12070981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buttar Z.A., Yang Y., Sharif R., Nan Wu S., Xie Y., Wang C. Genome Wide Identification, Characterization, and Expression Analysis of YABBY-Gene Family in WHEAT (Triticum aestivum L.) Agronomy. 2020;10:1189. doi: 10.3390/agronomy10081189. [DOI] [Google Scholar]
- 19.Inal B., Buyuk I., Ilhan E., Aras S. Genome-wide analysis of Phaseolus vulgaris C2C2-YABBY transcription factors under salt stress conditions. 3 Biotech. 2017;7:302. doi: 10.1007/s13205-017-0933-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Z., Van Houten J., Gonzalez G., Xiao H., van der Knaap E. Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol. Genet. Genom. 2013;288:111–129. doi: 10.1007/s00438-013-0733-0. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka W., Toriba T., Hirano H.Y. Three TOB1-related YABBY genes are required to maintain proper function of the spikelet and branch meristems in rice. New Phytol. 2017;215:825–839. doi: 10.1111/nph.14617. [DOI] [PubMed] [Google Scholar]
- 22.Toriba T., Harada K., Takamura A., Nakamura H., Ichikawa H., Suzaki T., Hirano H.Y. Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1. Mol. Genet Genom. 2007;277:457–468. doi: 10.1007/s00438-006-0202-0. [DOI] [PubMed] [Google Scholar]
- 23.Yu C., Zhi-hong L., Lei W. Characteristics and Expression Analysis of Transcription Factor YABBY Family in Maize. J. Agric. Sci. Technol. 2015;17:32–41. [Google Scholar]
- 24.Luo K., Zhang D., Zhai Z., Liu X., Zhou J., Zhang B., Li D. LsaYAB7, A Homologous Gene of FILAMENTOUS FLOWER, Participating In The Regulation of AdaxialAbaxial Polarity of Leaves In Lettuce. 2021. [DOI]
- 25.Juarez M.T., Twigg R.W., Timmermans M.C. Specification of adaxial cell fate during maize leaf development. Development. 2004;131:4533–4544. doi: 10.1242/dev.01328. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y.-Y., Hsiao Y.-Y., Chang S.-B., Zhang D., Lan S.-R., Liu Z.-J., Tsai W.-C. Genome-Wide Identification of YABBY Genes in Orchidaceae and Their Expression Patterns in Phalaenopsis Orchid. Genes. 2020;11:955. doi: 10.3390/genes11090955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin S., Li S., Gao Y., Bartholomew E.S., Wang R., Yang H., Liu C., Chen X., Wang Y., Liu X., et al. Genome-Wide Identification of YABBY Gene Family in Cucurbitaceae and Expression Analysis in Cucumber (Cucumis sativus L.) Genes. 2022;13:467. doi: 10.3390/genes13030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka W., Toriba T., Ohmori Y., Yoshida A., Kawai A., Mayama-Tsuchida T., Ichikawa H., Mitsuda N., Ohme-Takagi M., Hirano H.-Y.J.T.P.C. The YABBY gene TONGARI-BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice spikelet. Plant Cell. 2012;24:80–95. doi: 10.1105/tpc.111.094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang G.-Q., Liu K.-W., Li Z., Lohaus R., Hsiao Y.-Y., Niu S.-C., Wang J.-Y., Lin Y.-C., Xu Q., Chen L.-J.J.N. The Apostasia genome and the evolution of orchids. Nature. 2017;549:379–383. doi: 10.1038/nature23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohmori Y., Tanaka W., Kojima M., Sakakibara H., Hirano H.Y. WUSCHEL-RELATED HOMEOBOX4 is involved in meristem maintenance and is negatively regulated by the CLE gene FCP1 in rice. Plant Cell. 2013;25:229–241. doi: 10.1105/tpc.112.103432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borrego-Benjumea A., Carter A., Tucker J.R., Yao Z., Xu W., Badea A. Genome-Wide Analysis of Gene Expression Provides New Insights into Waterlogging Responses in Barley (Hordeum vulgare L.) Plants. 2020;9:240. doi: 10.3390/plants9020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korpan Y.I., Nazarenko E.A., Skryshevskaya I.V., Martelet C., Jaffrezic-Renault N., El’skaya A.V. Potato glycoalkaloids: True safety or false sense of security? Trends Biotechnol. 2004;22:147–151. doi: 10.1016/j.tibtech.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 33.Mondal M., Hossain M., Rasul M., Uddin S. Genetic diversity in potato (Solanum tuberosum L.) Bangladesh J. Bot. 2008;36:121–125. doi: 10.3329/bjb.v36i2.1499. [DOI] [Google Scholar]
- 34.Koch M., Naumann M., Pawelzik E., Gransee A., Thiel H. The Importance of Nutrient Management for Potato Production Part I: Plant Nutrition and Yield. Potato Res. 2019;63:97–119. doi: 10.1007/s11540-019-09431-2. [DOI] [Google Scholar]
- 35.Goffart J.-P., Haverkort A., Storey M., Haase N., Martin M., Lebrun P., Ryckmans D., Florins D., Demeulemeester K.J.P.R. Potato Production in Northwestern Europe (Germany, France, the Netherlands, United Kingdom, Belgium): Characteristics, Issues, Challenges and Opportunities. Potato Res. 2022;65:503–547. doi: 10.1007/s11540-021-09535-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nasir M.W., Toth Z.J.A. Effect of drought stress on potato production: A review. Agronomy. 2022;12:635. doi: 10.3390/agronomy12030635. [DOI] [Google Scholar]
- 37.Edgar R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li G., Hou M., Liu Y., Pei Y., Ye M., Zhou Y., Huang C., Zhao Y., Ma H. Genome-wide identification, characterization and expression analysis of the non-specific lipid transfer proteins in potato. BMC Genom. 2019;20:375. doi: 10.1186/s12864-019-5698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 40.Gao L., Tu Z.J., Millett B.P., Bradeen J.M. Insights into organ-specific pathogen defense responses in plants: RNA-seq analysis of potato tuber-Phytophthora infestans interactions. BMC Genom. 2013;14:340. doi: 10.1186/1471-2164-14-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petek M., Rotter A., Kogovsek P., Baebler S., Mithofer A., Gruden K. Potato virus Y infection hinders potato defence response and renders plants more vulnerable to Colorado potato beetle attack. Mol. Ecol. 2014;23:5378–5391. doi: 10.1111/mec.12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mengist M.F., Byrne S.L., Griffin D., Milbourne D. A note on the early transcriptional response in leaves and root of potato plants to cadmium exposure. Ir. J. Agric. Food Res. 2021;60:27–32. doi: 10.15212/ijafr-2020-0124. [DOI] [Google Scholar]
- 43.Sprenger H., Kurowsky C., Horn R., Erban A., Seddig S., Rudack K., Fischer A., Walther D., Zuther E., Kohl K., et al. The drought response of potato reference cultivars with contrasting tolerance. Plant Cell Environ. 2016;39:2370–2389. doi: 10.1111/pce.12780. [DOI] [PubMed] [Google Scholar]
- 44.Campbell M., Suttle J., Douches D.S., Buell C.R. Treatment of potato tubers with the synthetic cytokinin 1-(alpha-ethylbenzyl)-3-nitroguanidine results in rapid termination of endodormancy and induction of transcripts associated with cell proliferation and growth. Funct. Integr. Genom. 2014;14:789–799. doi: 10.1007/s10142-014-0404-1. [DOI] [PubMed] [Google Scholar]
- 45.Lin X., Kaul S., Rounsley S., Shea T.P., Benito M.I., Town C.D., Fujii C.Y., Mason T., Bowman C.L., Barnstead M., et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- 46.Li X.-Q., Sveshnikov D., Zebarth B.J., Tai H., De Koeyer D., Millard P., Haroon M., Singh M. Detection of Nitrogen Sufficiency in Potato Plants Using Gene Expression Markers. Am. J. Potato Res. 2009;87:50–59. doi: 10.1007/s12230-009-9116-9. [DOI] [Google Scholar]
- 47.Zapata L., Ding J., Willing E.M., Hartwig B., Bezdan D., Jiao W.B., Patel V., Velikkakam James G., Koornneef M., Ossowski S., et al. Chromosome-level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms. Proc. Natl. Acad. Sci. USA. 2016;113:E4052–E4060. doi: 10.1073/pnas.1607532113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Theologis A., Ecker J.R., Palm C.J., Federspiel N.A., Kaul S., White O., Alonso J., Altafi H., Araujo R., Bowman C.L., et al. Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature. 2000;408:816–820. doi: 10.1038/35048500. [DOI] [PubMed] [Google Scholar]
- 49.Li X.Q., Griffiths R., De Koeyer D., Rothwell C., Gustafson V., Regan S., Flinn B. Functional genomic resources for potato. Can. J. Plant Sci. 2008;88:573–581. doi: 10.4141/CJPS07048. [DOI] [Google Scholar]
- 50.Flinn B., Rothwell C., Griffiths R., Lague M., DeKoeyer D., Sardana R., Audy P., Goyer C., Li X.Q., Wang-Pruski G., et al. Potato expressed sequence tag generation and analysis using standard and unique cDNA libraries. Plant Mol. Biol. 2005;59:407–433. doi: 10.1007/s11103-005-0185-y. [DOI] [PubMed] [Google Scholar]
- 51.Shahzad R., Harlina P.W., Ayaad M., Ewas M., Nishawy E., Fahad S., Subthain H., Amar M.H. Dynamic roles of microRNAs in nutrient acquisition and plant adaptation under nutrient stress: A review. Plant Omics. 2018;11:58–79. doi: 10.21475/poj.11.01.18.pne1014. [DOI] [Google Scholar]
- 52.Tiwari J.K., Buckseth T., Zinta R., Saraswati A., Singh R.K., Rawat S., Chakrabarti S.K. Genome-wide identification and characterization of microRNAs by small RNA sequencing for low nitrogen stress in potato. PLoS ONE. 2020;15:e0233076. doi: 10.1371/journal.pone.0233076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharif y., Zaynab M., Khan K.A., Li S. Identification and Expression Analysis of DUF4228 Domain Containing (DDP) Genes in Potato Under Abiotic and Phytohormone Stress. Mol. Biol. Rep. 2021 doi: 10.21203/rs.3.rs-902964/v1. [DOI] [Google Scholar]
- 54.Iqbal M.S., Hafeez M.N., Wattoo J.I., Ali A., Sharif M.N., Rashid B., Tabassum B., Nasir I.A. Prediction of Host-Derived miRNAs with the Potential to Target PVY in Potato Plants. Front. Genet. 2016;7:159. doi: 10.3389/fgene.2016.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hou X., Du Y., Liu X., Zhang H., Liu Y., Yan N., Zhang Z. Genome-Wide Analysis of Long Non-Coding RNAs in Potato and Their Potential Role in Tuber Sprouting Process. Int. J. Mol. Sci. 2017;19:101. doi: 10.3390/ijms19010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin T., Lashbrook C.C., Cho S.K., Butler N.M., Sharma P., Muppirala U., Severin A.J., Hannapel D.J. Transcriptional analysis of phloem-associated cells of potato. BMC Genom. 2015;16:665. doi: 10.1186/s12864-015-1844-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu Y., Feng Z., Liu X., Bian L., Xie H., Zhang C., Mysore K.S., Liang J. MiR393 and miR390 synergistically regulate lateral root growth in rice under different conditions. BMC Plant Biol. 2018;18:261. doi: 10.1186/s12870-018-1488-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hussain Q., Zheng M., Chang W., Ashraf M.F., Khan R., Asim M., Riaz M.W., Alwahibi M.S., Elshikh M.S., Zhang R., et al. Genome-Wide Identification and Expression Analysis of SnRK2 Gene Family in Dormant Vegetative Buds of Liriodendron chinense in Response to Abscisic Acid, Chilling, and Photoperiod. Genes. 2022;13:1305. doi: 10.3390/genes13081305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riaz M.W., Lu J., Shah L., Yang L., Chen C., Mei X.D., Xue L., Manzoor M.A., Abdullah M., Rehman S., et al. Expansion and Molecular Characterization of AP2/ERF Gene Family in Wheat (Triticum aestivum L.) Front. Genet. 2021;12:632155. doi: 10.3389/fgene.2021.632155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.