Abstract
Membranes contain lipids that are composed of fatty acids (FA) and a polar head. Membrane homeostasis is crucial for optimal bacterial growth and interaction with the environment. Bacteria synthesize their FAs via the FASII pathway. Gram-positive bacteria can incorporate exogenous FAs which need to be phosphorylated to become substrate of the lipid biosynthetic pathway. In many species including staphylococci, streptococci and enterococci, this phosphorylation is carried out by the Fak complex, which is composed of two subunits, FakA and FakB. FakA is the kinase. FakB proteins are members of the DegV family, proteins known to bind FAs. Two or three FakB types have been identified depending on the bacterial species and characterized by their affinity for saturated and/or unsaturated FAs. Some species such as Streptococcus pyogenes, which causes a wide variety of diseases ranging from mild non-invasive to severe invasive infections, possess an uncharacterized additional DegV protein. We identify here this DegV member as a fourth FakB protein, named FakB4. The fakB4 gene is co-regulated with FASII genes suggesting an interaction with endogenous fatty acids. fakB4 deletion has no impact on membrane phospholipid composition nor on the percentage of other major lipids. However, the fakB4 mutant strain produced more lipids and more extracellular membrane vesicles than the wild-type strain. This suggests that FakB4 is involved in endogenous FA binding and controls FA storage or catabolism resulting in a limitation of extracellular FA release via membrane vesicles.
Introduction
Cell membranes commonly comprise a lipid bilayer composed of a polar head and an apolar body constituted of fatty acids (FA). FA structure and length are decisive for membrane topology and properties such as fluidity, permeability and integrity. These features are crucial for the adaptation of bacteria to various environments [1]. Most bacteria synthesize FAs via the fatty acid synthesis pathway FASII. This pathway is not essential in the Gram-positive bacteria that use exogenous FAs (eFAs) to synthesize their lipids [2–5]. Incorporation of eFAs into the lipids of Gram-positive bacteria involve a FA kinase (Fak) complex that phosphorylates eFAs, yielding acyl-PO4. The acyl-PO4 is used by PlsY and PlsC for lipid synthesis or transformed in an acyl-ACP, which is used by PlsC or elongated via FASII [6]. The Fak complex is composed of two proteins; FakA provides the kinase activity and interacts with FakB proteins; FakB proteins are members of the DegV family [7, 8]. DegV proteins bind FA with high affinity indicating that DegV proteins may take part in lipid transport and in FA metabolic processes [9]. Whereas single fakA genes are found in Firmicutes, degV genes are multiple; two fakB have been identified in Staphylococcus aureus, three in Streptococcus pneumoniae, and four in Streptococcus suis, Enterococcus faecalis and Enterococcus faecium [7, 10–12]. In S. aureus, FakB1 and FakB2 bind exclusively saturated and preferentially unsaturated FAs, respectively. In S. pneumoniae FakB1, FakB2 and FakB3 bind saturated, mono-unsaturated and poly-unsaturated FAs, respectively. In E. faecalis FakB1 binds preferentially saturated FAs, whereas the three others bind unselectively FAs [11]. Although FakB1 and FakB2 are well conserved, the third FakB may be more related to S. pneumoniae FakB3 or to S. suis FakB4. In the case of enterococci, FakB1, FakB2 and FakB4 are present but the last FakB, termed FakB5, does not resemble the FakB3s [12]. The role of S. suis FakB4 could not be studied by a biochemical approach because it appeared as inclusion bodies in vitro [12]. S. pyogenes, a Gram-positive human pathogen responsible for more than 500,000 deaths annually worldwide [13], also possesses four degV genes, encoding the three characterized FakB proteins as well as a fourth uncharacterized DegV protein [12, 14].
To decipher the role of this fourth DegV protein, encoded by the gene M28_Spy1638, we compared amino acid sequence with the known FakB proteins and determined whether this gene is coregulated with the FASII genes [14]. We also constructed a fakB4 mutant strain to investigate potential impacts on bacterial FA and lipid compositions. Finally, we characterized potential structural effects of the absence of FakB4.
Results
The S. pyogenes uncharacteized degV gene encodes a FakB protein and is controlled by FabT
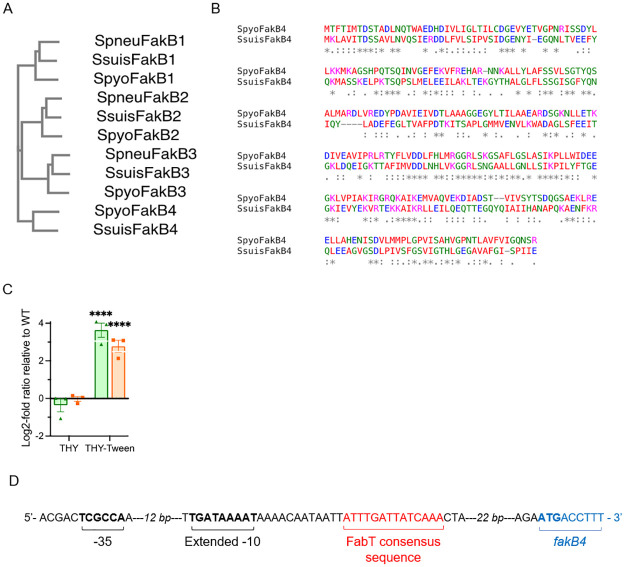
In S. pyogenes M28PF1, the M28_Spy1638 gene product belongs to the DegV protein family. To determine whether it is a FakB protein and to which FakB protein it is most closely related, we constructed a phylogenetic tree using the FakB sequences from S. pneumoniae TIGR4 and S. suis 05ZYH33 (Fig 1A). The protein is most related to S. suis FakB4. The S. pyogenes DegV1638 protein has the same number of amino acid residues as the other FakB proteins, whereas S. suis FakB4 sequence lacks one fourth of the amino acid residues, from the 5th to the 69th. Excluding the truncated sequence, these proteins share 58.62% identity. We consequently named degV1638 gene fakB4 and the encoded protein FakB4. To give a clearer picture of the protein relatedness and the similarity between the all FakB proteins, we determined the identity percentages between all homologous FakB proteins (Table 1) and aligned the two FakB4 proteins (Fig 1B).
Fig 1. The M28_Spy1638 gene encodes a FakB4 protein and belongs to the FabT regulon.
A) Phylogenetic tree of the three S. pneumoniae (Spneu), four S. suis (Ssuis), three S. pyogenes (Spyo), FakB proteins and the M28_Spy1638 product, named FakB4; the branch lengths are, from top to bottom, 0.17461, 0.15163, 0.20011, 0.13539, 0.13176, 0.17447, 0.15335, 0.1527, 0.18335, 0.22949, 0.21848. B) Sequence comparison of S. pyogenes and S. suis FakB4 proteins. C) Relative expression of fabT, green, and fakB4 orange, in the mFabT versus the WT strain. Strains were grown in THY or THY-Tween 80 medium and RNAs were quantified by qRT-PCR. Expression was normalized to that of gyrA; relative gene expression is expressed as the log2-fold ratio in the mFabT versus the WT strain. 2-way ANOVA, Bonferroni post-test, ****p<0.0001. D) Cartoon representing the regulatory region of the fakB4 gene with -35 and extended -10 boxes, previously published [18], FabT consensus sequence and fakB4 translation initiation site indicated in black, red and blue, respectively.
Table 1. Protein identity to S. pyogenes homologous FakB protein.
| Species | FakB1 | FakB2 | FakB3 | FakB4 | ||||
|---|---|---|---|---|---|---|---|---|
| Identity | Length | Identity | Length | Identity | Length | Identity | Length | |
| S. pyogenes | 100% | 282 | 100% | 283 | 100% | 280 | 100% | 286 |
| S. pneumoniae | 57.39% | 282 | 63.89% | 279 | 61.72% | 281 | ||
| S. suis | 61.94% | 283 | 65.72% | 277 | 61.46% | 296 | 58.62% | 222 |
M5005_Spy1650, the fakB4 homolog in a M1 strain of S. pyogenes, is one of the six genes that is controlled by FabT, the FASII transcriptional repressor of the FASII genes, in all conditions tested [15]. FabT is present in streptococci, enterococci and lactococci. FabT belongs to the MarR family of regulators [for review, [16]]. The expression of the other fakB genes is not controlled by FabT in the M1 strain [15] and a FASII coregulation may shed light on the function of the protein. We therefore tested whether fakB4 present in the M28PF1 strain is coregulated with FASII genes. We compared by qRT-PCR, the relative expressions of fabT, that is self-repressed, and fakB4 in a fabT mutant strain (mFabT, S3 Table) versus in the WT strain. The mFabT strain expresses a FabT variant in which the histidine in position 105 was replaced by a tyrosine. The FabTH105Y mutation leads to derepression of the FASII genes (to be published elsewhere), as previously described for a S. pyogenes fabT mutant strain [15]. FabT co-repressors are long chain acyl-Acyl Carrier proteins (acyl-ACP) [17]. Strains were therefore grown in THY and in THY-Tween 80; Tween 80 provides, the long chain unsaturated fatty acid 18:1Δ9 (Fig 1C). Neither fabT nor fakB4 gene expression was affected by the FabTH105Y mutation when the strains were grown in THY, the role of FabT being limited without the presence of the co-repressor. In contrast, fabT expression was derepressed in the mFabT strain in THY-Tween 80 (12.44-fold), indicating that the FabTH105Y protein displays a hampered repression capacity. The expression of the fakB4 gene was similarly affected by the fabT mutation in THY-Tween 80 (6.88-fold). We hypothesized that fakB4 may be directly controlled by FabT. Consensus FabT binding sequences are found along the FASII locus, which includes fabT [[16], and references herein]. We sought whether one was also present downstream from the fakB4 transcription start site previously described [18]. Using ANTTTGATTATCAAATT as a probe, we found the sequence, ATTTTGATTATCAAA, that shows 100% identity on 15 out of 17 nucleotides and that is localized between the transcription and the translation start sites (Fig 1B).
fakB4 is therefore coregulated with the FASII genes in both S. pyogenes strains in which it has been studied and likely directly controlled by FabT. This suggests that, in contrast to FakB1, FakB2 and FakB3, FakB4 may not be involved in the incorporation of exogenous FA into the membrane lipids.
FakB4 does not play a major role in exogenous FA incorporation
To further characterize the role of FakB4 we constructed an isogenic mutant strain in which fakB4 is interrupted by the insertion of a plasmid containing a fakB4 internal fragment (see Material and methods). We hypothesized that the growth characteristics of the WT and mFakB4 strain would be identical in rich laboratory medium, but may differ in FA-containing medium, if FakB4 was involved in specific FA uptake (S1 Fig). Both strains had exactly the same growth characteristics at 37 °C in THY, THY-Tween 80 or THY-FBS.
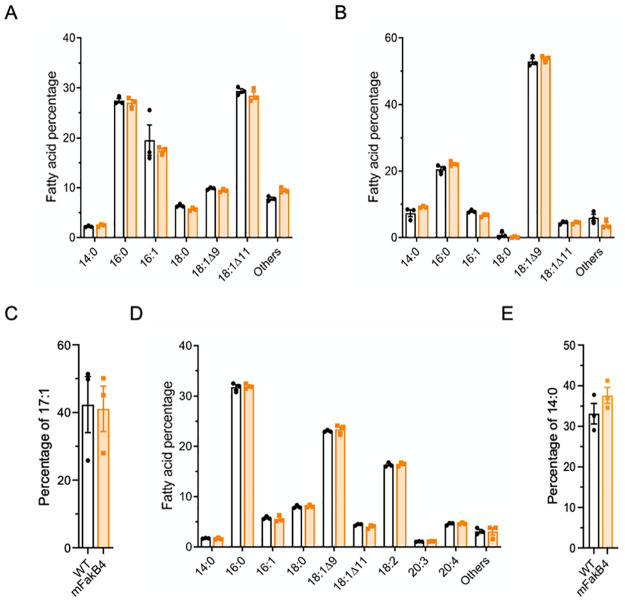
The Fak complexes, containing the kinase FakA interacting with FakB isoforms, are responsible for eFA phosphorylation and transfer to the lipid synthesis pathway [7]. The deletion of given FakB proteins leads to default in the incorporation of specific FAs into the lipids. To test whether FakB4 is involved in eFA incorporation, we determined the composition of membrane lipids from mFakB4 and WT strains grown in various media (Fig 2A, 2B and 2D, S1 Table). The FA distribution was similar in WT and mFakB4 strains grown in THY (Fig 2A, S1 Table). The addition of Tween 80 strongly modified the FA distribution in the WT and the mFakB4 strains that remained similar; there was a strong diminution of 16:0 and 16:1 and a rise of 18:1Δ9, the major Tween 80 component, signing the eFA incorporation and FASII arrest (Fig 2B, S1 Table). This suggested that eFAs are similarly incorporated by both strains and that FakB4 is not required for 18:1Δ9 incorporation. A similar incorporation by both strains was checked by assessing the incorporation of 17:1, that is not produced by the bacteria (Fig 2C). The percentage in both strain membranes was the same confirming the absence of eFA incorporation defect in the mFakB4 strain. However, 17:1 and 18:1Δ9, provided by Tween 80, are monounsaturated FAs. We reasoned that if FakB4 was specific for other FAs, a difference may have been unnoticed. We consequently also determined the FA composition after growth in the presence of human plasma that also contains saturated and poly-unsaturated FAs (Fig 2D, S1 Table). Again, the membrane composition was different from those found in THY and THY-Tween 80 grown bacteria, but again similar in both strains. The concentration of poly-unsaturated 18:2 and 20:4 strongly rose, indicating that, as already shown for Staphylococcus aureus, S. pneumoniae, S. agalactiae and E. faecalis, S. pyogenes incorporated poly-unsaturated FAs [2, 3, 10, 19]. The saturated FA brought by plasma is 16:0, a FA that is synthesized by S. pyogenes, limiting the conclusion on fakB4 impact on C16:0 incorporation. We thus also assessed exogenous 14:0 incorporation, a FA that is less produced by S. pyogenes. The percentage of exogenous C14:0 incorporated into membrane phospholipids is similar in both fakB4 and WT strains (Fig 2E). These results indicate that the mFakB4 strain had no membrane FA composition defect whatever the FA environment tested and that FakB4 is not involved in eFA incorporation into the membrane phospholipids.
Fig 2. The WT and mFakB4 strains show identical relative FA compositions.
A, B, D) Membrane fatty acid percentage of WT and mFakB4 strains grown in A) THY, B) THY-Tween 80, D) THY-Plasma; C, E) Membrane C) 17:1 or E) 14:0 percentage in WT and mFakB4 strains cultivated in THY-17:1 or C14:0; A-D, N = 3 (S1 Table). 2-way ANOVA, Bonferroni post-test.
FakB4 controls the lipid quantity
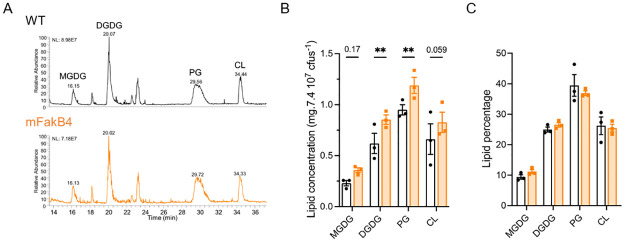
The FA distribution being unmodified in the mFakB4 strain, we analyzed the global lipid contents in the strains (Fig 3A–3C, S2 Table). Contrary to FA analysis that yields an FA distribution, the global lipid analysis is normalized per cfu (Fig 3A and 3B). The digalactosyldiacylglycerol and phosphatidylglycerol concentrations per cfus were higher in the mFakB4 than in the WT strain and there was a trend to similar increases for the monogalactosyldiacylglycerol and the cardiolipins. Importantly, the ratios between the different lipid classes were, like those of the FAs, identical between both strains (Fig 3C, S2 Table). This suggests that the absence of FakB4 has a global impact on the four lipid families. We controlled that the lipid excess in the mFakB4 strain was not due to bacterial cell lysis that would produce cellular debris, modifying the OD/cfu ratio by determining that ratio during growth (S2 Fig). There was no difference in this ratio between the strains during growth. Altogether, these results indicated that the absence of FakB4 did not affect FA or lipid distribution but led to an increased lipid quantity per bacterial unit.
Fig 3. The WT and mFakB4 strains show different lipid quantities, but identical relative lipid composition.
Lipid content in the WT and mFakB4 strains grown in THY, by class of lipids: A-B) lipid composition; A) HPLC-MS profile representing the main lipid classes, B) lipid concentration, C) ratio of lipid classes; N = 3 (S2 Table). Bars and symbols, white, WT strain; orange, mFakB4 strain; MGDG, monogalactosyldiacylglycerol; DGDG, digalactosyldiacylglycerol; PG, phosphatidylglycerol; CL, cardiolipin. 2-way ANOVA, Bonferroni post-test, **p<0.01; p values are indicated when between 0.05 <p<0.2.
The FakB4 strain expels extracellular vesicles
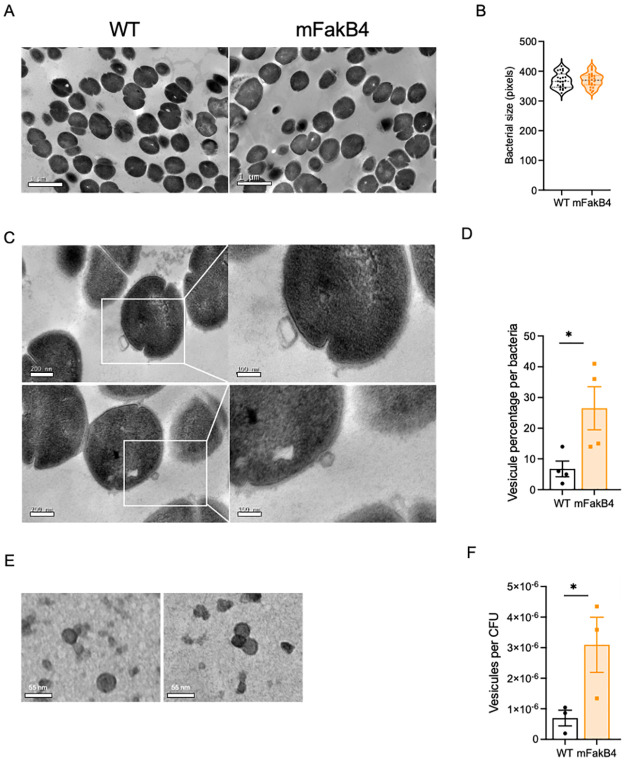
FA availability sets cell envelope capacity which in turn affects cell size [20]. We consequently hypothesized that the lipid increase observed in the mFakB4 strain may impact the bacterial morphology or the cell size. WT and mFakB4 strains grown in THY were consequently observed by transmission electron microscopy (TEM) and the diameter of bacteria measured (Fig 4A and 4B). Both strains displayed similar bacterial morphology and cell size. A close examination of the TEM images of the surface of S. pyogenes during the exponential phase indicated the presence of structures composed of double layers, extracellular membrane vesicles (EMV), particularly at the surface of the mFakB4 strain (Fig 4A). Their relative abundance in WT and mFakB4 strains was compared; there were 2.5-fold more EMVs at the surface of the mFakB4 strain than of the WT strain (Fig 4C and 4D, S3A Fig). These data suggest that extracellular vesicles attached to the bacterial surface contribute to the lipid surplus measured in the mFakB4 strain. We wondered whether EMVs could also be found in the culture supernatant and their abundance impacted by the fakB4 mutation. Pelleted supernatants were therefore analyzed by TEM. EMVs were present and there were three times more EMVs in the supernatant from mFakB4 strain than in that of the WT strain (Fig 4E and 4F, S3B and S3C Fig). These data indicated that vesicle expulsion during the exponential phase of growth is increased in a S. pyogenes strain devoid of FakB4.
Fig 4. The mFakB4 strain secretes extracellular vesicles.
A, C) TEM images of bacterial sections, left WT, right mFakB4; A) 4000X, C) 15000X, inset 30000X (scale = 200 nm; inset scale = 100 nm); B, D) Image analysis, B) bacterial diameter length, N = 4, 24 independent images, D) percentage of vesicles per bacteria, N = 4, 67 independent images. E) TEM images, of extracellular membrane vesicles contained in mFakB4 culture supernatant (scale = 40 nm); F) Image analysis N = 3, 30 independent images. B, D, F) One-way ANOVA, Bonferroni post-test, *p<0.05.
Discussion
FakB proteins, members of the DegV family of proteins, bind eFAs and form a complex with FakA kinase that phosphorylate these FAs yielding substrates for PlsX or PlsY and further incorporation into lipids [6, 7, 10]. The specific roles of FakB1, FakB2 and, when present, FakB3, have been characterized in S. aureus, S. pneumoniae and S. suis [7, 10, 12]. However, some species possess a fourth DegV protein whose role has not been determined [12]. In the present work, we show that, in S. pyogenes, this fourth DegV protein is a FakB protein, which does not impact eFA incorporation in membrane lipids. The fakB4 gene is coregulated with the FASII genes, and is therefore less expressed in the presence of eFAs than in their absence. This suggests that, whereas FakB1, FakB2 and FakB3 are involved in eFA incorporation, the role of FakB4 may be connected to endogenous FAs found in the cytosol [21].
The mFakB4 mutant strain produced more lipids than its wild-type counter-part. Microbial cell size is known to depend on FA availability [20]. Yet, the enhanced lipid abundance in the mFakB4 mutant strain did not result in an increased cell size but in an enhanced EMV production. This suggests that FakB4-FA binding disrupts this dependency. EMV production has been reported in S. pyogenes [22–25]. It is negatively regulated by the CovRS, a two-component system that controls the expression of 15% of S. pyogenes genes and that is turned down in response to sub-lethal concentrations of the antimicrobial peptide LL-37, a host produced peptide [22, 26]. In agreement with this regulation, extracellular membrane vesicles production is triggered by LL-37 [23]. This indicates that EMV production is stimulated in the host, where S. pyogenes is in the presence of eFAs. EMV production may be a FA detoxification mechanism. In S. pyogenes, EMVs are more abundant during late exponential or early stationary growth phase [22]. In contrast, in the fakB4 mutant strain, the EMVs were more abundant than in the WT already in mid-exponential phase, in laboratory growth medium. This suggests that FakB4 limits EMV production during exponential phase through endogenous FA-binding and that EMV production likely compensates excess endogenous FAs toxicity.
The mFakB4 higher lipid production indicates that more acyl molecules are available in a fakB4 mutant than in the wild-type strain. This strongly supports that, like the other members of the DegV family, FakB4 binds acyl molecules. It further indicates that, in contrast to FakB1—, FakB2- and FakB3-bound acyl molecules, the FakB4-bound acyl moieties are not subsequently transferred to the phospholipid biosynthetic pathway. Thus, FakB4 may have an important role in protecting the bacteria from FA excess. This can be carried out by two mechanisms. FakB4, by binding the supernumerary FA molecules, stores them. Alternatively, when FA molecules are in excess, FakB4 steers them towards a catabolic pathway that needs to be further characterized. Various FA detoxification mechanisms have been described [for review, [27]]. Among them, β-oxidation counteracts excess FAs in many bacteria. However, S. aureus that is unable to carry out this reaction possesses FarE, an efflux pump, which displays a detoxification function [28]. None of these mechanisms have been described in streptococci. S. pyogenes possesses a hydratase, the myosin cross-reactive antigen, that catalyzes the hydration of cis-9 and cis-12 double bonds [29]. Its gene is overexpressed in a fabT mutant strain in conditions that yield membrane composition modifications [15]. The hydration of unsaturated FAs prevents their accumulation which is detrimental to the bacteria. However, this mechanism produces saturated FAs, which may thus accumulate inside the bacterium and be toxic. FakB4, by taking care of these FAs, annihilate the toxicity without requiring their export.
Altogether this study indicates that the activities of FakB4 on the one hand, and of the three other FakB proteins, on the other, are entailed in different growth conditions; thus, these proteins target FAs from different origins and exhibit different roles. Further investigations on the FakB4 interactome will shed light on S. pyogenes lipid metabolism.
Materials and methods
Bacterial strains and culture conditions
The strains used in this study are described in S3 Table. S. pyogenes strains were grown under static condition at 37 °C in Todd Hewitt broth supplemented with 0.2% Yeast Extract (THY) or on THY agar plates. To study, the role of eFA addition, the medium was supplemented with 0.1% Tween 80 (THY-Tween 80) (Sigma-Aldrich, P1754), essentially composed of 18:1Δ9, with 100 μM 17:1 (Larodan, Sweden), with 10% Fetal Bovine Serum (THY-FBS), essentially composed of, approx., 43% saturated, 23% mono-unsaturated and 8% polyunsaturated FAs (Gibco), or with 10% human plasma (Etablissement Français du Sang) (THY-Plasma). For all experiments, strains were prepared as follows. Overnight cultures were diluted to an OD600nm = 0.05 and grown in THY to the exponential phase (OD600nm comprised between 0.4 and 0.5). The mFakB4 strain was grown in the presence of 10 μg.ml-1 erythromycin. E. coli strains were grown in LB or on LB agar plates supplemented with 150 μg.ml-1 erythromycin when appropriate.
In silico analysis
S. pyogenes FakB protein sequences were extracted from the S. pyogenes M28PF1 strain, named WT in this study, using the Geneious software, searching for “DegV”. The FakB sequences of the S. pneumoniae TIGR4 strain (WP_000762061, WP_000161399, WP_000219939) and S. suis 05ZYH33 (SSU05_0827, SSU05_1650, SSU05_0561, SSU05_1899) were collected from NCBI. Sequence alignments and the phylogenetic tree, a Neighbour-joining tree without distance corrections were produced using Clustal W (1.83) multiple sequence alignment in EBI tools (https://www.ebi.ac.uk).
The presence of putative FabT binding sequences on the M28PF1 genome was sought with the ANTTTGATTATCAAATT sequence as a probe, accepting up to 2 mismatches using Geneious prime Biomatters development, www.geneious.com [16].
RNA isolation
S. pyogenes strains were cultured at 37 °C in THY or THY-Tween 80, and cells were harvested at exponential growth phase (OD600 comprised between 0.4 and 0.5). After adding 2 volumes of RNA protect* (Qiagen), total RNA was extracted as previously described [30]. The bacteria from three independent cultures for each condition were lyzed by a 15 mg.ml-1 lysozyme, 300 U.ml-1 mutanolysine treatment for 30 min at 20 °C followed by two cycles of Fast-prep (power 6, 30 s) at 4 °C, then using the Macherey-Nagel RNA extraction kit, as indicated by the supplier. RNA integrity was analyzed using an Agilent Bioanalyzer (Agilent Biotechnologies) and a PCR. Absence of DNA was further confirmed by PCR.
First-strand cDNA synthesis, quantitative PCR qPCR
Five hundred nanograms of total RNA was used for first-strand cDNA synthesis using SuperScriptTM II reverse transcriptase and random primers according to the manufacturer’s instructions (Invitrogen, Life technologies).
Quantitative PCR was carried out with SYBR Green PCR kits (Applied Biosystems, Life technologies) using four pairs of primers (S4 Table). gyrA and rpoB were used as the housekeeping reference genes. Relative quantification of specific gene expression was calculated with the 2−ΔΔCt method using gyrA as the reference gene and expressed in log2-fold change. Each assay was performed in triplicate on each sample as previously described [30].
Strain construction
The primers used for the generation of the plasmids and verifying the different strains are described in S4 Table. The mFabT strain harbors a point mutation in fabT; the mFakB4 strain corresponds to an insertion interrupting the fakB4 gene. They were obtained by homologous recombination of the plasmid pG1-mFabT or single cross-over with the plasmid pG1-DegVint2, respectively, following the same protocol as described previously [31, 32]. Regarding the fabT mutation, the plasmid pG1-mFabT was obtained by amplifying nucleotides -222 to +805, relative to the translation start site from the chromosome of a spontaneous mutant from our collection, CCH1963, where the C in position 313 was replaced by a T, leading to the replacement of the histidine in position 105 by a tyrosine. The strain was entirely sequenced as described previously, and no other mutation was found compared with the parent strain M28PF1 except for the point mutation in fabT [14]. For the mFakB4 strain, an internal fragment, from nucleotides 296 to 601 was amplified and cloned in pG1 using the In Fusion cloning kit® (Clonetech) giving rise to the plasmid pG1-DegVint2. The plasmid was then inserted by a single cross-over event in M28PF1 chromosome, resulting in the interruption of fakB4. The absence of spurious mutations in the strains was verified by whole genome sequencing. This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAQMHW000000000. The version described in this paper is version JAQMHW010000000.
Fatty acid analysis
Strains were grown in THY, THY-Tween 80, THY-Plasma or THY-17:1 until OD600nm = 0.4–0.5. Fatty acids were extracted and analyzed as previously described [2, 3, 5]. Briefly, analyses were performed in a split-splitless injection mode on an AutoSystem XL Gas Chromatograph (Perkin-Elmer) equipped with a ZB-Wax capillary column (30 m x 0.25 mm x 0.25 mm; Phenomenex, France). Data were recorded and analyzed by TotalChrom Workstation (Perkin-Elmer). FA peaks were detected between 12 and 40 min of elution, and identified by comparing to retention times of purified esterified FA standards (Mixture ME100, Larodan, Sweden). Results are shown as percent of specific FA as calculated from their proportions compared to total peak areas (TotalChrom Workstation; Perkin Elmer).
Lipid analysis
Strains were grown in THY until OD600nm = 0.4–0.5. The cultures were diluted to 7.4 107 cfus.mL-1. Lipid extractions were performed as previously described [5, 33, 34]. Lipids were identified following the method previously described [35]. The lipids separation was realized by normal phase HPLC (U3000 ThermoFisher Scientific) using a Inertsil Si 5μm column (150 x 2.1 mm I.D.) from GL Sciences Inc (Tokyo, Japan). Lipids were quantified using a Corona-CAD Ultra and identified by mass-spectrometry negative ionization and MS2/MS3 fragmentation (LTQ-Orbitrap Velos Pro). The concentration of each lipid class was determined as previously described using as standards DGDG, 840524P-5MG; MGDG, 840523P-5MG; CL (heart CA), 840012P-25MG; PG (egg), 841138P-25MG [36]. Lipids spectra were analyzed on Xcalibur™ software (ThermoFisher Scientific, version 4.2.47).
Bacterial morphology and size analysis
THY exponential phase cultures (0.4 < OD600nm < 0.5) were centrifuged and the pellets resuspended in 1 ml phosphate buffer saline (PBS) twice and incubated 30 min at 4 °C with a fixative solution (4% paraformaldehyde and 2.5% glutaraldehyde). Bacteria were then washed 3 times in PBS and subsequently treated as previously described, with slight modifications [20]. Secondary fixation was performed with 1% osmium tetroxide in PBS for 1 h at room temperature, then bacteria were centrifuged, the pellets washed 3 times in H2O and further dehydrated with increasing concentrations of ethanol (70%, 90%, and 100% twice). After dehydration, pellets were infiltrated with sequential 3:1, 1:1, and 1:3 solutions of resin (Embed 812 kit; Electron Microscopy Sciences, Hatfield, PA) and 100% ethanol for 1 h separately at room temperature. Finally, the samples were incubated with propylene oxide for 2 min and placed in the embedding molds with resin. The embedding resin blocks were polymerized in an oven at 60 °C for 24 h. Ultrathin sections (90 nm) were cut from the block surface using an ultramicrotome (Reichert Ultracut S ultramicrotome, Leica Microsystems) then stained with uranyl acetate and lead citrate (LFG, France) and observed by TEM (JEOL 1011, JEOL, Japan)) at 80 kV of acceleration voltage. Acquisitions were performed by an Orius 1000 CCD Camera (GATAN, USA) using Digital Micrograph software (GATAN, USA).
To determine the bacterial size, the larger bacteria from each independent image were selected as representative of those sliced through the sphere diameter. The pixels along the diameter were then quantified.
Extracellular vesicle isolation and quantification
Extracellular vesicles were isolated from S. pyogenes strains cultured at 37 °C in THY as described previously [22]. PBS-washed pellets were further processed as follows. Small sample droplets (20 μl) were deposited on 200 mesh formvar coated copper grids for 2 min. The excess of sample solution was removed by blotting the grid with filter paper (Whatman). Grids were incubated with a drop of staining solution (uranyle acetate) for 2 min and blotted with a filter paper. The grid was deposited on a filter paper. Samples were immediately examined in a JEOL 1011 transmission electron microscope (JEOL, Japan) with an ORIUS 1000 CCD camera (GATAN, France), operated with Digital Micrograph software (GATAN, France) for acquisition.
Supporting information
Wild-type, orange, and mFakB4, black, strains were precultured to mid-exponenetial phase, OD600 = 0.5 in THY and then diluted to on OD600 of 0.05 in A) THY, B) THY-Tween 80 and C) THY-fetal bovine serum. Growth was monitored in a Multiscan (Thermo Scientific). Note that the OD in the Mutliscan is roughly half of that in a classical spectrometer.
(DOCX)
WT and mFakB4 strains were grown in THY and samples were taken at different OD600 during growth; serial dilutions were plated. cfus were counted after incubating the plates 24 h at 37°C.
(DOCX)
A) TEM images of mFakB4 bacterial sections white arrows, vesicles (scale = 200 nm); B-C) TEM images of extracellular membrane vesicles (EMVs). B) Comparison of WT and mFakB4 culture supernatants (scale = 100 nm). C) Observation of EMVs contained in mFakB4 culture supernatant (scale = 55 nm); 15000X.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Alexandra Gruss for her constant interest and support. We thank Audrey Solgadi and Bastien Prost (UMS-IPSIT—Plateforme SAMM, Université Paris-Saclay) for performing HPLC-MS experiments and for their help with the lipid analysis; Sonia Abreu (LipSys2, Université Paris-Saclay) for total lipids separation development. We thank Maëva Gauduin, undergraduate in the laboratory, for technical help.
Data Availability
All relevant data are within the manuscript and its Supporting information files except for the stain sequences. The M28PF1 and mFakB4 sequence accession numbers are GenBank LR031521 and RR9468758, respectively.
Funding Statement
AF, RPH17043DJA, Dim One Health, https://www.dim1health.com/ CL, BioSPC, n°51809666, Université Paris Cité, https://u-paris.fr/ CL, FDT202106012831), Fondation pour la recherche médicale, https://www.frm.org/ QZ, SHSMU-ZDCX20212700, Shangai Jiao Tong University School of Medecine, https://www.shsmu.edu.cn/english/ QZ, bourse jeune chercheur, campus France, https://www.pologne.campusfrance.org/fr/bourse-sshn-sejour-de-recherche The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zhang YM, Rock CO. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol. 2008;6(3):222–33. Epub 2008/02/12. doi: 10.1038/nrmicro1839 . [DOI] [PubMed] [Google Scholar]
- 2.Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature. 2009;458(7234):83–6. Epub 2009/03/06. doi: 10.1038/nature07772 . [DOI] [PubMed] [Google Scholar]
- 3.Hays C, Lambert C, Brinster S, Lamberet G, du Merle L, Gloux K, et al. Type II Fatty Acid Synthesis Pathway and Cyclopropane Ring Formation Are Dispensable during Enterococcus faecalis Systemic Infection. J Bacteriol. 2021;203(20):e0022121. Epub 2021/07/27. doi: 10.1128/JB.00221-21 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsons JB, Frank MW, Subramanian C, Saenkham P, Rock CO. Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc Natl Acad Sci U S A. 2011;108(37):15378–83. Epub 2011/08/31. doi: 10.1073/pnas.1109208108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenanian G, Morvan C, Weckel A, Pathania A, Anba-Mondoloni J, Halpern D, et al. Permissive Fatty Acid Incorporation Promotes Staphylococcal Adaptation to FASII Antibiotics in Host Environments. Cell Rep. 2019;29(12):3974–82 e4. Epub 2019/12/19. doi: 10.1016/j.celrep.2019.11.071 . [DOI] [PubMed] [Google Scholar]
- 6.Parsons JB, Frank MW, Jackson P, Subramanian C, Rock CO. Incorporation of extracellular fatty acids by a fatty acid kinase-dependent pathway in Staphylococcus aureus. Mol Microbiol. 2014;92(2):234–45. Epub 2014/03/29. doi: 10.1111/mmi.12556 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parsons JB, Broussard TC, Bose JL, Rosch JW, Jackson P, Subramanian C, et al. Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc Natl Acad Sci U S A. 2014;111(29):10532–7. Epub 2014/07/09. doi: 10.1073/pnas.1408797111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meier M, Sit RV, Quake SR. Proteome-wide protein interaction measurements of bacterial proteins of unknown function. Proc Natl Acad Sci U S A. 2013;110(2):477–82. Epub 2012/12/26. doi: 10.1073/pnas.1210634110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schulze-Gahmen U, Pelaschier J, Yokota H, Kim R, Kim SH. Crystal structure of a hypothetical protein, TM841 of Thermotoga maritima, reveals its function as a fatty acid-binding protein. Proteins. 2003;50(4):526–30. Epub 2003/02/11. doi: 10.1002/prot.10305 . [DOI] [PubMed] [Google Scholar]
- 10.Gullett JM, Cuypers MG, Frank MW, White SW, Rock CO. A fatty acid-binding protein of Streptococcus pneumoniae facilitates the acquisition of host polyunsaturated fatty acids. J Biol Chem. 2019;294(44):16416–28. Epub 2019/09/19. doi: 10.1074/jbc.RA119.010659 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou Q, Zhu L, Cronan JE. The Enterococcus faecalis FabT transcription factor regulates fatty acid synthesis in response to exogenous fatty acids. Front Microbiol. 2022;13:877582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y, Zang N, Lou N, Xu Y, Sun J, Huang M, et al. Structure and mechanism for streptococcal fatty acid kinase (Fak) system dedicated to host fatty acid scavenging. Sci Adv. 2022;8(35):eabq3944. Epub 2022/09/03. doi: 10.1126/sciadv.abq3944 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5(11):685–94. Epub 2005/10/29. S1473-3099(05)70267-X [pii] doi: 10.1016/S1473-3099(05)70267-X . [DOI] [PubMed] [Google Scholar]
- 14.Longo M, De Jode M, Plainvert C, Weckel A, Hua A, Chateau A, et al. Complete Genome Sequence of Streptococcus pyogenes emm28 Clinical Isolate M28PF1, Responsible for a Puerperal Fever. Genome Announc. 2015;3(4). Epub 2015/07/18. doi: 10.1128/genomeA.00750-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eraso JM, Olsen RJ, Beres SB, Kachroo P, Porter AR, Nasser W, et al. Genomic Landscape of Intrahost Variation in Group A Streptococcus: Repeated and Abundant Mutational Inactivation of the fabT Gene Encoding a Regulator of Fatty Acid Synthesis. Infect Immun. 2016;84(12):3268–81. Epub 2016/09/08. doi: 10.1128/IAI.00608-16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambert C, Poyart C, Gruss A, Fouet A. FabT, a Bacterial Transcriptional Repressor That Limits Futile Fatty Acid Biosynthesis. Microbiol Mol Biol Rev. 2022;86(3):e0002922. Epub 2022/06/22. doi: 10.1128/mmbr.00029-22 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jerga A, Rock CO. Acyl-Acyl carrier protein regulates transcription of fatty acid biosynthetic genes via the FabT repressor in Streptococcus pneumoniae. J Biol Chem. 2009;284(23):15364–8. Epub 2009/04/21. doi: 10.1074/jbc.C109.002410 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosinski-Chupin I, Sauvage E, Fouet A, Poyart C, Glaser P. Conserved and specific features of Streptococcus pyogenes and Streptococcus agalactiae transcriptional landscapes. BMC Genomics. 2019;20(1):236. Epub 2019/03/25. doi: 10.1186/s12864-019-5613-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan B, Fung K, Ye S, Lai PM, Wei YX, Sze KH, et al. Linoleic acid metabolism activation in macrophages promotes the clearing of intracellular Staphylococcus aureus. Chem Sci. 2022;13(42):12445–60. Epub 2022/11/17. doi: 10.1039/d2sc04307f . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vadia S, Tse JL, Lucena R, Yang Z, Kellogg DR, Wang JD, et al. Fatty Acid Availability Sets Cell Envelope Capacity and Dictates Microbial Cell Size. Curr Biol. 2017;27(12):1757–67 e5. Epub 2017/06/13. doi: 10.1016/j.cub.2017.05.076 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sastre DE, Pulschen AA, Basso LGM, Benites Pariente JS, Marques Netto CGC, Machinandiarena F, et al. The phosphatidic acid pathway enzyme PlsX plays both catalytic and channeling roles in bacterial phospholipid synthesis. J Biol Chem. 2020;295(7):2148–59. Epub 2020/01/11. doi: 10.1074/jbc.RA119.011147 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Resch U, Tsatsaronis JA, Le Rhun A, Stubiger G, Rohde M, Kasvandik S, et al. A Two-Component Regulatory System Impacts Extracellular Membrane-Derived Vesicle Production in Group A Streptococcus. mBio. 2016;7(6). Epub 2016/11/03. doi: 10.1128/mBio.00207-16 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uhlmann J, Rohde M, Siemens N, Kreikemeyer B, Bergman P, Johansson L, et al. LL-37 Triggers Formation of Streptococcus pyogenes Extracellular Vesicle-Like Structures with Immune Stimulatory Properties. J Innate Immun. 2016;8(3):243–57. Epub 2015/12/08. doi: 10.1159/000441896 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beerens D, Franch-Arroyo S, Sullivan TJ, Goosmann C, Brinkmann V, Charpentier E. Survival Strategies of Streptococcus pyogenes in Response to Phage Infection. Viruses. 2021;13(4). Epub 2021/05/01. doi: 10.3390/v13040612 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murase K, Aikawa C, Nozawa T, Nakatake A, Sakamoto K, Kikuchi T, et al. Biological Effect of Streptococcus pyogenes-Released Extracellular Vesicles on Human Monocytic Cells, Induction of Cytotoxicity, and Inflammatory Response. Front Cell Infect Microbiol. 2021;11:711144. Epub 2021/08/06. doi: 10.3389/fcimb.2021.711144 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gryllos I, Tran-Winkler HJ, Cheng MF, Chung H, Bolcome R 3rd, Lu W, et al. Induction of group A Streptococcus virulence by a human antimicrobial peptide. Proc Natl Acad Sci U S A. 2008;105(43):16755–60. Epub 2008/10/22. 0803815105 [pii]. doi: 10.1073/pnas.0803815105 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kengmo Tchoupa A, Eijkelkamp BA, Peschel A. Bacterial adaptation strategies to host-derived fatty acids. Trends Microbiol. 2022;30(3):241–53. Epub 2021/07/06. doi: 10.1016/j.tim.2021.06.002 . [DOI] [PubMed] [Google Scholar]
- 28.Alnaseri H, Arsic B, Schneider JE, Kaiser JC, Scinocca ZC, Heinrichs DE, et al. Inducible Expression of a Resistance-Nodulation-Division-Type Efflux Pump in Staphylococcus aureus Provides Resistance to Linoleic and Arachidonic Acids. J Bacteriol. 2015;197(11):1893–905. Epub 2015/03/25. doi: 10.1128/JB.02607-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volkov A, Liavonchanka A, Kamneva O, Fiedler T, Goebel C, Kreikemeyer B, et al. Myosin cross-reactive antigen of Streptococcus pyogenes M49 encodes a fatty acid double bond hydratase that plays a role in oleic acid detoxification and bacterial virulence. J Biol Chem. 2010;285(14):10353–61. Epub 2010/02/11. doi: 10.1074/jbc.M109.081851 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plainvert C, Rosinski-Chupin I, Weckel A, Lambert C, Touak G, Sauvage E, et al. A Novel CovS Variant Harbored by a Colonization Strain Reduces Streptococcus pyogenes Virulence. J Bacteriol. 2023:e0003923. Epub 2023/03/16. doi: 10.1128/jb.00039-23 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Six A, Bellais S, Bouaboud A, Fouet A, Gabriel C, Tazi A, et al. Srr2, a multifaceted adhesin expressed by ST-17 hypervirulent Group B Streptococcus involved in binding to both fibrinogen and plasminogen. Mol Microbiol. 2015;97(6):1209–22. Epub 2015/06/23. doi: 10.1111/mmi.13097 . [DOI] [PubMed] [Google Scholar]
- 32.Weckel A, Ahamada D, Bellais S, Mehats C, Plainvert C, Longo M, et al. The N-terminal domain of the R28 protein promotes emm28 group A Streptococcus adhesion to host cells via direct binding to three integrins. J Biol Chem. 2018;293(41):16006–18. Epub 2018/08/29. doi: 10.1074/jbc.RA118.004134 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–7. Epub 1959/08/01. doi: 10.1139/o59-099 . [DOI] [PubMed] [Google Scholar]
- 34.Thedieck K, Hain T, Mohamed W, Tindall BJ, Nimtz M, Chakraborty T, et al. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol Microbiol. 2006;62(5):1325–39. Epub 2006/10/18. doi: 10.1111/j.1365-2958.2006.05452.x . [DOI] [PubMed] [Google Scholar]
- 35.Abreu S, Solgadi A, Chaminade P. Optimization of normal phase chromatographic conditions for lipid analysis and comparison of associated detection techniques. J Chromatogr A. 2017;1514:54–71. Epub 2017/08/05. doi: 10.1016/j.chroma.2017.07.063 . [DOI] [PubMed] [Google Scholar]
- 36.Moulin M, Solgadi A, Veksler V, Garnier A, Ventura-Clapier R, Chaminade P. Sex-specific cardiac cardiolipin remodelling after doxorubicin treatment. Biol Sex Differ. 2015;6:20. Epub 2015/10/20. doi: 10.1186/s13293-015-0039-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Wild-type, orange, and mFakB4, black, strains were precultured to mid-exponenetial phase, OD600 = 0.5 in THY and then diluted to on OD600 of 0.05 in A) THY, B) THY-Tween 80 and C) THY-fetal bovine serum. Growth was monitored in a Multiscan (Thermo Scientific). Note that the OD in the Mutliscan is roughly half of that in a classical spectrometer.
(DOCX)
WT and mFakB4 strains were grown in THY and samples were taken at different OD600 during growth; serial dilutions were plated. cfus were counted after incubating the plates 24 h at 37°C.
(DOCX)
A) TEM images of mFakB4 bacterial sections white arrows, vesicles (scale = 200 nm); B-C) TEM images of extracellular membrane vesicles (EMVs). B) Comparison of WT and mFakB4 culture supernatants (scale = 100 nm). C) Observation of EMVs contained in mFakB4 culture supernatant (scale = 55 nm); 15000X.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting information files except for the stain sequences. The M28PF1 and mFakB4 sequence accession numbers are GenBank LR031521 and RR9468758, respectively.