Abstract
Background
The DNA single-strand annealing proteins (SSAPs), such as RecT, Redβ, ERF and Rad52, function in RecA-dependent and RecA-independent DNA recombination pathways. Recently, they have been shown to form similar helical quaternary superstructures. However, despite the functional similarities between these diverse SSAPs, their actual evolutionary affinities are poorly understood.
Results
Using sensitive computational sequence analysis, we show that the RecT and Redβ proteins, along with several other bacterial proteins, form a distinct superfamily. The ERF and Rad52 families show no direct evolutionary relationship to these proteins and define novel superfamilies of their own. We identify several previously unknown members of each of these superfamilies and also report, for the first time, bacterial and viral homologs of Rad52. Additionally, we predict the presence of aberrant HhH modules in RAD52 that are likely to be involved in DNA-binding. Using the contextual information obtained from the analysis of gene neighborhoods, we provide evidence of the interaction of the bacterial members of each of these SSAP superfamilies with a similar set of DNA repair/recombination protein. These include different nucleases or Holliday junction resolvases, the ABC ATPase SbcC and the single-strand-binding protein. We also present evidence of independent assembly of some of the predicted operons encoding SSAPs and in situ displacement of functionally similar genes.
Conclusions
There are three evolutionarily distinct superfamilies of SSAPs, namely the RecT/Redβ, ERF, and RAD52, that have different sequence conservation patterns and predicted folds. All these SSAPs appear to be primarily of bacteriophage origin and have been acquired by numerous phylogenetically distant cellular genomes. They generally occur in predicted operons encoding one or more of a set of conserved DNA recombination proteins that appear to be the principal functional partners of the SSAPs.
Introduction
Homologous DNA recombination is a fundamental process in the biochemistry of DNA repair and replication, which contributes to the generation of the genetic diversity critical for natural selection. An important step in the recombination process is the pairing of homologous double-stranded DNAs followed by the exchange of DNA strands between the paired molecules. Experimental studies have shown that members of the archetypal RecA family of recombinases are central to this reaction in all extant forms of life [1,2].
Studies in Escherichia coli have shown that, although RecA is the principle protein involved in pairing and strand exchange, unrelated proteins, that have a much more restrictive phyletic distribution, can also promote similar reactions in a RecA dependent or RecA-independent manner [3]. These alternative or additional mediators of homologous recombination include the well-characterized prophage RecT, phage λ Redβ and phage P22 ERF proteins [4,5]. Similarly, in yeast and vertebrates, the RAD52 protein is involved in the pairing and strand exchange reaction and can promote recombination in a RAD51 (the eukaryotic RecA homolog)-dependent or independent manner [6]. The RecT protein works in conjunction with the RecE-nuclease [7] and was initially described in genetic studies on the complemention of mutations in the RecBCD pathway of DNA repair [8-10]. Biochemically, RecT has been shown to bind single-stranded (ss) DNA 3' overhang regions generated by the RecE nuclease, and promote strand exchange between homologous DNA partners by assisting the pairing of complementary single-stranded regions [4,10]. The reaction catalyzed by the RecT/RecE system is similar to that described for the phage λ exonuclease (exo/Redβ) and the single-strand annealing protein Redβ. The similarity between these two systems is further extended by the observation that the RecT/E system can complement mutations in the λ exo/Redβ system [10,12]. In eukaryotes, RAD52 protein has been shown to exhibit properties similar to those of RecT and Redβ proteins: it binds ssDNA and promotes strand exchange via the pairing of complementary single strands [6,13]. In vitro studies on quaternary structures have shown that the single strand annealing proteins (SSAPs), RecT, Redβ, ERF and RAD52, form similar helical super-structures [14-17]. This has led to the proposal that RecT, Redβ, ERF and the eukaryotic RAD52 function in an analogous fashion, and even are "structural homologs" [14].
However, no sequence or secondary structural similarities have been noticed between different SSAPs and current understanding of their evolutionary history and phyletic range remains poor. Here, we describe the results of an in-depth sequence analysis of these proteins and delineate their evolutionary relationships and phyletic horizon in available genomes. We show that, in spite of the functional similarities, and the similar quaternary structures, there are three distinct superfamilies of SSAPs, namely the RecT/Redβ, RAD52 and ERF, that appear to be evolutionarily unrelated to each other. These superfamilies show a wide distribution in viral and cellular genomes, but appear to have originally evolved in large DNA bacteriophages. Through an analysis of the contextual information provided by the predicted operons, in which the SSAPs occur, we predict several previously undetected functional connections of these proteins, which might shed new light on the corresponding DNA repair/recombination pathways.
Results
RecT and Redβ are evolutionarily related and define a widespread family of DNA recombination proteins
Several lines of evidence, including genetic analyses, and similarities in biochemistry and quaternary structures, suggest that the E. coli RecT and phage λ Redβ proteins are functionally equivalent as mediators of single-strand exchange in DNA recombination [10,12]. However, no sequence similarity has been detected between these proteins leaving their actual evolutionary relationships unresolved. In order to gain a better understanding of their functions and origins, we undertook a detailed sequence analysis of these two proteins using iterative sequence profile searches with the PSI-BLAST program with a inclusion threshold of .01 iterated until convergence. Such searches, with Redβ proteins from different lambdoid bacteriophages as queries, retrieved not only other obvious Redβ homologs, but also the RecT protein family. For example, searches initiated with the Redβ homolog, PF161 protein (Genbank gi: 9836834 amino acids 1 to 188) from Borrelia hermsii[18], detected the E. coli RecT protein in the 5th iteration with significant expectation (e) values (3 × 10-3). Subsequent iterations retrieved several more RecT-related proteins from diverse sources. Further, transitive searches with the proteins detected in the above searches resulted in the identification of more divergent homologs, such as a protein termed the 'enterohemolysin associated protein' (EHAP1) from E. coli[19] and its orthologs in Salmonella (Fig. 1). An examination of the pairwise alignments generated by these searches showed that all these proteins shared a characteristic set of residues, including two highly conserved aromatic residues at the N- and C-termini, respectively, and two consecutive acidic residues near the C-terminus. These observations strongly suggested that RecT and Redβ, along with several other proteins, could be unified into a single protein superfamily with a core conserved domain of approximately 200 amino acid residues.
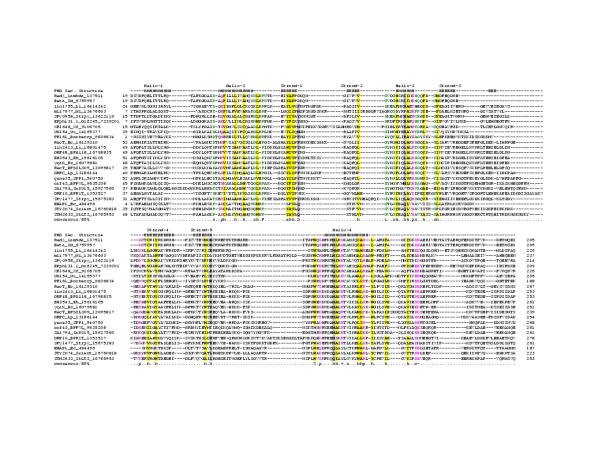
Figure 1.
Multiple sequence alignment of the RecT/Redβ superfamily of proteins. Proteins are denoted with their gene names, species abbreviation and gi numbers. The coloring reflects the amino acid conservation at 85% consensus. The consensus abbreviations and coloring scheme are as follows: h: hydrophobic residues (L,I,Y,F,M,W,A,C,V), l: aliphatic (L,I,A,V) and a: aromatic (F,Y,W,H) residues shaded yellow; o: alcohol (S,T), colored blue, c: charged (K,E,R,D,H) residues, +: basic (K/R/H) residues, -: acidic (D,E) residues, and p: polar (S,T,E,C,D,R,K,H,N,Q) residues colored purple; s: small (S,A,C,G,D,N,P,V,T) and u:tiny (G,A,S) residues, colored green; b: big (L,I,F,M,W,Y,E,R,K,Q) residues shaded gray. Secondary structure assignments are as follows: H: Helix, E: Extended (Strand). Species abbreviations are as follows: Bh: Bacillus halodurans, Borhercp: Borrelia hermsii circular plasmid, BPA118: Bacteriophage A118, BPbIL309: Bacteriophage bIL309, BPphi31_1: Bacteriophage phi31.1, BPPVL: Bacteriophage PVL, BPR1T: Bacteriophage R1T, Bs: Bacillus subtilis, ec: Escherichia coli, lambda: Bacteriophage λ, Li: Listeria innocua, Lp: Legionella pneumophila, Ml: Mesorhizobium loti, Salent: Salmonella enterica subsp. enterica serovar Typhi, SaN315: Staphylococcus aureus N315 subsp. aureus N315,Sd: Shigella dysenteriae, SPP1: Bacteriophage SPP1, StLT2: Salmonella typhimurium LT2, Strpy: Streptococcus pyogenes, Uu: Ureaplasma urealyticum, Xf; Xylella fastidiosa
A multiple alignment of all members of the RecT/Redβ superfamily was generated using the T_coffee program followed by adjustments based on the PSI-BLAST search results. This alignment was used to predict their secondary structure using the JPRED and PHD methods; these predictions pointed to an α + β domain with a core of five β-strands and five α-helices (Fig. 1). Some of the strongest conservation is concentrated in the long helices, and the pattern includes some charged or polar residues, suggesting that they are probably exposed and participate in the protein-protein and protein-DNA interactions that are typical of this superfamily (helices 2,3, 4 in Fig. 1). The conserved, regularly spaced hydrophobic residues in the RecT/Redβ superfamily are predicted to be buried, allowing these domains to assume a globular structure. Experimental studies have shown that the strand transfer reaction mediated by RecT and its binding to dsDNA are sensitive to Mg+2 concentrations and it was proposed that the levels of free Mg+2 could regulate RecT activity [4]. Similarly, Redβ has been shown to promote single strand annealing in a Mg+2-dependent manner [20]. In this context, the conservation of the two C-terminal acidic residues in the majority of members of this superfamily suggests that these might be involved in the coordination of Mg+2 and implies that the metal ion-dependent conformational switching is likely to be a generic feature of this family.
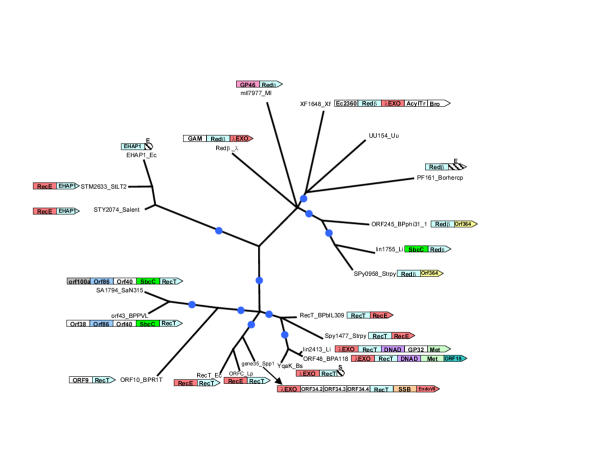
Phylogenetic analyses of the RecT/Redβ superfamily using the least squares and maximum likelihood methods distinguished three distinct groups, namely the RecT-like, the Redβ-like and the EHAP1-like families (Fig. 2). Previously, the RecT proteins have been known from very few bacteria and Redβ has only been detected in λ and closely related phages. However, we showed that the Redβ family is widespread in bacteria, such as Borrelia hermsii, Xylella, Ureaplasma, Listeria, Streptococcus pyogenes, Mesorhizobium loti. The RecT family is predominantly seen in the low-GC Gram-positive bacteria, such as Bacillus, Streptococcus, Lactococcus and Listeria, and their phages (Fig. 2). E. coli and Legionella pneumophila are the only two γ-proteobacteria that possess this protein, suggesting that they might have acquired RecT via a relatively recent horizontal transfer from Gram-positive bacteria. The sporadic distribution of the RecT and Redβ family proteins in bacterial genomes and their presence in phages suggest that these proteins ultimately are of phage origin and have been co-opted by the bacterial DNA recombination/repair systems. Consistent with this, practically all the bacterial members of these families appear to belong to prophages or their remnants, as they are mostly in the neighborhood of what appear to be clearly phage-derived genes. The EHAP1-like family is extremely divergent and represented thus far only in E. coli and the closely related Salmonella. Practically all members of the RecT/Redβ superfamily are single-domain proteins showing extended similarity to each other throughout their globular regions. The only exceptions are the E. coli EHAP1 and the PF161 protein encoded in the Borrelia hermsii circular plasmid, which are fused to C-terminal fragments of the ERF protein (see below).
Figure 2.
Maximum likelihood tree for the RecT/Redβ superfamily of proteins. The internal branches with RELL bootstrap support >70% are indicated by blue circles. Proteins are designated by their gene names and species abbreviations as in Fig. 1. The gene neighborhoods of the RecT/Redβ superfamily genes are shown in association with the corresponding branches whenever they they contained genes for proteins with plausible functional connections with SSAPs. The hatched boxes represent fragments of ERF (Indicated by E) and SSB (indicated by S) genes encoding C-terminal regions as described in the text. Gene abbreviations are as follows: GP46: GP46 of bacteriophage PSA, GP32:GP32 of bacteriophage PSA, ORF15:ORF15 of Streptococcus thermophilus bacteriophage 7201, ORF40: ORF40 of bacteriophage PVL ORF86:ORF86 of Staphylococcus aureus temperate phage φSLT, Orf100a:0rf100a of Staphylococcus aureus temperate phage φSLT, ORF364: ORF364 of bacteriophage φ31.1, Ec2360: b2360 of E. coli, AcylTr: N-Acyltransferase, Bro: Bro-N domain fused to XF0704, Met: DNA Methyltransferase
ERF defines a superfamily of SSAPs that are evolutionarily distinct from the RecT/Redβ super family
The ERF protein of phage P22 is involved in the circularization of the linear dsDNA phage genome upon entry into the host cell [21-23]. Experimental studies have shown that, mutations in ERF are complemented by Redβ and that in vitro ERF adopts quaternary structures analogous to those of Redβ and RecT [14,17,24,25]. However, in the comprehensive analysis of the RecT/Redβ superfamily no statistically significant similarity could be detected between these proteins and the ERF proteins. To explore the evolutionary affinities of the ERF domains, we carried out a sequence profile analysis as described above for the RecT case using transitive PSI-BLAST analysis. As a result of these searches, homologs of ERF encoded in several bacterial and phage genomes from diverse taxa were identified. The alignments generated in these searches consistently pointed to a region of approximately 150 amino acids that is conserved in all these proteins, with a characteristic motif of the form: GuXXoYhp + YXhXXhh (where G is glycine, Y-tyrosine, u is a tiny residue, h-hydrophobic, p is a polar residue, o is an alcohol residue, + is a basic residue, and X is any residue; Fig. 3). This suggested that ERF was the prototype of a family of conserved bacterial domains.
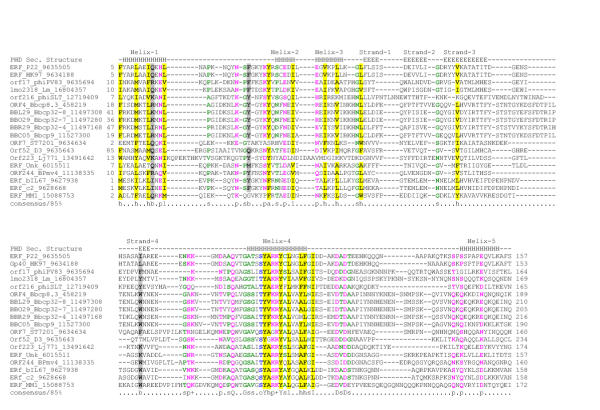
Figure 3.
Multiple sequence alignment of the ERF protein superfamily. The coloring reflects the amino acid conservation at 85% consensus. The coloring scheme and secondary structure assignment abbreviations are as in Fig. 1. Species abbreviations are as follows: Bbcp32-4: Borrelia burgdorferi cicular plasmid (cp) 32-4, Bbcp32-7: B. burgdorferi cp 32-7, Bbcp32-8: B. burgdorferi cp 32-8, Bbcp8.3: B. burgdorferi cp 8.3, Bbcp9: B burgdorferi cp 9, bIL67: bacteriophage IL67, c2: Lactococcus phage c2, D3: Pseudomonas phage D3, HK97: bacteriophage HK97, Lj771: Lactobacillus johnsonii prophage Lj771, Lm: Listeria monocytogenes, MM1: Streptococcus pneumoniae bacteriophage MM1, BPmv4: Bacteriophage mv4, P22: Bacteriophage P22, phiPV83: Bacteriophage φPV83, phiSLT: Staphylococcus aureus temperate phage φSLT, ST7201: Streptococcus thermophilus bacteriophage 7201, Unk: Unknown.
Secondary structure prediction based on the multiple alignment of the ERF domain suggests a globular α + β fold with five helices and three or four strands (Fig. 3). The above-mentioned motif that is typical of this family is associated with helix 4 of this domain; given the presence of conserved basic residues, it may be critical for DNA-binding and strand-transfer activity of the ERF-like proteins. Additionally, in the loop between helices 4 and 5 of the ERF domain there is a universally conserved acidic motif of the form DXD. Analogous to the RecT superfamily, this acidic dyad might coordinate a divalent cation and undergo a conformational change dependent on metal-binding. However, the average size of the core domains, the patterns of conserved residues, and the predicted secondary structures of the RecT/Redβ and ERF domains show no correspondence to each other, implying that there is no direct evolutionary link between these protein groups.
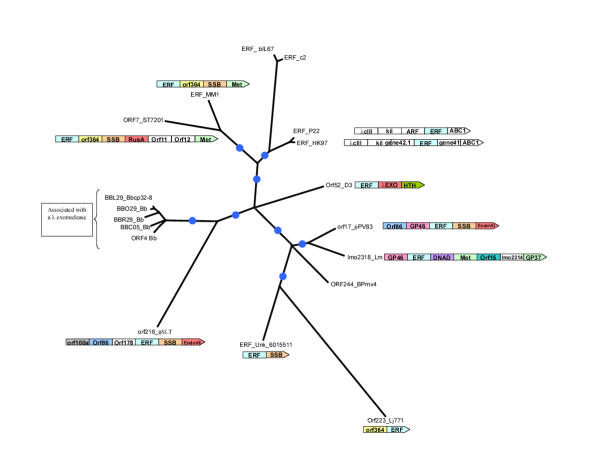
ERF homologs are encoded by the genomes of several temperate phages of Gram-positive bacteria and γ-proteobacteria; additionally, we detected members of this superfamily in Listeria and in all the circular plasmids and one linear plasmid of Borrelia burgdorferi (Fig. 4). Thus, like the RecT/Redβ superfamily, the ERF family is likely to have emerged in the temperate phages, and was disseminated to the Borrelia circular plasmids and some bacterial genomes via prophages.
Figure 4.
Maximum likelihood tree for the ERF superfamily of proteins. The designations, gene names and species abbreviations are as in Fig. 2A. The internal branches with RELL bootstrap support >70% are indicated by blue circles. The gene neighborhoods of the ERF proteins are shown whenever they contained gene coding for proteins with potential functional relevance. Gene abbreviations are as in Fig. 1B
Detection of bacterial homologs of RAD52 and identification of an aberrant HhH domain in these proteins
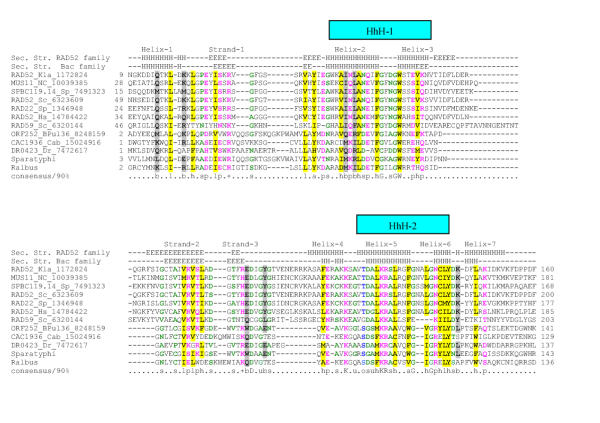
The baker's yeast protein RAD52 and its paralog RAD59 define a small family of proteins thus far represented in fungi, vertebrates and the early-branching ameboid eukaryote, Entamoeba histolytica. Rad52 functions in conjunction with the RecA ortholog, the RAD51 recombinase in double-strand break repair and meiotic recombination [6]. RAD52 binds ssDNA during recombination and also shows a quaternary organization similar to those of RecT/Redβ and ERF [16,26]. However, RAD52-like proteins showed no detectable sequence similarity with either the ERF or the RecT/Redβ-like proteins. Sequence searches initiated with the conserved globular region of the eukaryotic RAD52 proteins readily detected their homologs from other eukaryotes and, at convergence, also retrieved from the database certain bacterial proteins, such as DR0423 from Deinococcus and CAC1936 from Clostridium respectively, with border-like statistical significance (e ~ .05). These bacterial proteins form a small family that is additionally represented in Salmonella paratyphi A, the temperate bacteriophage u136 of Lactococcus lactis (ORF252-encoded protein) and a Shiga toxin-converting phage from E. coli. Iterative profile searches initiated with CAC1936 from Clostridium acetobutylicum and its S. paratyphi A ortholog correspondingly retrieved S. cerevisiae RAD52 and its eukaryotic homologs, with borderline e-values at convergence (~0.043). The alignment between these bacterial proteins and the eukaryotic Rad52 homologs was co-linear throughout the entire length of their shared globular region and the Gibbs sampling procedure detected two motifs of greater than 20 residues, with a probability of chance occurrence in these proteins less than 10-18 (Fig. 5). In addition to the similar conservation pattern, separate secondary structure predictions for both the eukaryotic RAD52 family and their potential bacterial homologs showed a complete concordance of the predicted structural elements between RAD52 and the bacterial proteins, strongly suggesting that they all belong to a single homologous superfamily (hereinafter the RAD52 superfamily).
Figure 5.
Multiple sequence alignment of the RAD52 protein superfamily. The coloring reflects the consensus at 90% conservation. The coloring scheme and secondary structure assignment abbreviations are as in Fig. 1. Species abbreviations are as follows: BPul36: Bacteriophage ul36. Cab: Clostridium acetobutylicum, Dr: Deinococcus radiodurans, Hs: Homo sapiens, Kla: Kluyveromyces lactis, NC: Neurospora crassa, Sc: Saccharomyces cerevisiae, Sp: Schizosaccharomyces pombe, Sparatyphi: Salmonella paratyphi A, Ralbus: Ruminococcus albus. The Shiga toxin-converting phage RAD52-like protein (gi: 17977996) is nearly identical to the Salmonella paratyphi A RAD52 like protein. The RAD52-like proteins from Bacteriophage ul36 (gi: 8248159) and Ruminococcus albus are respectively adjacent to genes encoding the single-strand binding protein and the λ-type exonuclease.
The secondary structure predictions showed that the Rad52 superfamily proteins adopt a structure with interspersed α-helices and β-strands (Fig. 5). Additionally, fold predictions using 3DPSSM (E-value=.0085, corresponding to a 90% confidence in the prediction) and the hybrid fold method (Z-score = 19.5) predicted the presence of a potential Helix-hairpin-Helix (HhH) fold in members of the RAD52 superfamily. The HhH domain is a small nucleic acid-binding module comprised of two helices joined by a central loop (hairpin), which functions as the DNA-binding moiety of numerous repair and recombination proteins[27,28]. Two HhH modules are predicted in the core conserved domain of the RAD52 family, the first one bounded by the predicted helices 2 and 3, and the second one bounded by helices 5 and 6 (Fig. 5). Although these predicted HhH modules are very divergent in sequence from the typical versions, the hairpin in both HhH modules of the RAD52 family proteins is bounded by small residues, typically glycine; this conforms to the signature motif characteristic of the classical HhH modules [28,29]. However, in the case of the RAD52 superfamily the predicted HhH modules appear to have been welded into a large globular superstructure that maintained its evolutionary distinctness over time. The conservation pattern and predicted structural elements of the RAD52 superfamily are distinct from those predicted for the ERF and RecT/Redβ superfamilies (Fig 1, 3, 5), supporting the lack of a direct evolutionary relationship between these proteins.
The RAD52 superfamily shows a sporadic phyletic distribution, and even in the crown-group eukaryotes, might have been secondarily lost in certain lineages, such as plants, nematodes and insects. The sporadic distribution of this family among phylogenetically distant bacteria, along with its presence in several prophages, suggests that, like the RecT/Redβ and ERF superfamilies, at least the bacterial RAD52-proteins might be of predominantly phage origin. The core of the eukaryotic recombination system appears to have been inherited from the system present in the common ancestor shared with the archaea [29]. However, RAD52 is thus far absent in all archaeal genomes and is restricted to a single orthologous group in the eukaryotes [29]. Thus, it appears plausible that eukaryotic RAD52 was ultimately derived through lateral transfer either from a bacterial genome or directly from a viral source, at a point at least predating the divergence of the crown group eukaryotes and Entamoeba.
Contextual information from gene neighborhoods provides details regarding functional interactions of the SSAPs with DNA recombination pathways
The clustering of functionally related genes in prokaryotic genomes into co-transcribed and co-regulated units, operons, often allows functional assignments through the principle of 'guilt by association' [30-32]. Generally, genes whose products physically interact to form a complex or are involved in successive steps in a biochemical pathway form operons that are conserved over large evolutionary distances [30]. On previous occasions, we have used gene neighborhoods or operons to predict novel DNA repair complexes and their components [33]. Accordingly, a similar approach was applied to the three families of SSAPs (RecT/Redβ, ERF, Rad52), to shed light on their functional links.
Notably, the genes encoding the three evolutionarily distinct SSAPs co-occurred with similar sets of DNA repair/recombination-related proteins (Figs. 2,4). In at least one case, each of them was found adjacent to the gene for the single-strand-binding protein (SSB), an OB-fold protein that binds ssDNA (Figs. 2,4). This association ties in with the function of the SSAPs in single-strand annealing, suggesting that they closely interact with SSB. It has been suggested in the case of RecT that it may compete with SSB for binding single strand overhangs and thereby make them available for the annealing process [3]. Similar interactions between other SSAPs and SSB, that probably coats the ssDNA generated by nucleases, appear likely. Genes for SSAPs from all the 3 distinct superfamilies may also occur adjacent to or in the vicinity of genes encoding nucleases or Holliday junction resolvases (HJRs). Genes for RecT/Redβ superfamily proteins are associated with genes encoding a λ-type exonuclease (LE) of the type II restriction enzyme fold, RecE, which also might be a divergent member of this fold, and a nuclease of the Endonuclease VII (EndoVII) fold [7,34] (Fig. 2). The ERF superfamily genes are associated with a RusA superfamily nuclease/HJR and EndoVII fold nucleases (Fig. 4) [34]. Furthermore, the Borrelia plasmids that encode ERF, also almost always additionally encode a λ-type exonuclease, even if it is not the adjacent gene. In a single instance, in the Gram-positive bacterium Ruminococcus albus, the gene encoding a RAD52 superfamily protein occurs adjacent to a gene for a λ-type exonuclease. These nucleases probably contribute to the repair process, in which SSAPs are involved, by providing the initial break in the dsDNA and/or in digesting the nicked target to generate ssDNA.
The RecT and Redβ family proteins often co-occur with the SbcC gene that encodes an ABC ATPase with a large coiled-coil segment. These proteins are known to cooperate with SbcD, nuclease of the calcineurin-like phosphoesterase superfamily and to degrade dsDNA in the 3' → 5' direction generating ssDNA [35,36]. It seems likely that RecT/Redβ proteins, at least in certain cases, function in conjunction with the SbcCD-pathway, by utilizing the single-stranded regions generated by the SbcCD nuclease. Additionally, several genes, whose functions are less clear, tend to co-occur with the genes coding for the SSAPs. These include DNA methyltransferases and the primosomal protein DnaD from low-GC Gram-positive bacteria [37] that co-occur with both ERF and RecT superfamily members (Figs. 2,4). The poorly characterized phage-or prophage-specific genes that are frequently observed in these neighborhoods include ORF15 (Streptococcus thermophilus bacteriophage 7201), ORF86, ORF100a (Staphylococcus aureus temperate phage φSLT) and ORF364 (bacteriophage φ31.1) (Figs. 2,4). Secondary structure predictions indicate a high α-helical content for these proteins. It is likely that these α-helical proteins are phage innovations that could function as adaptors in the recombination pathway either as accessory protein-protein interacting domains or as DNA-binding domains.
Evidence for convergent operon evolution and in situ non-orthologous displacement of genes in operons encoding SSAPs
A superposition of the gene neighborhood information upon the phylogenetic trees for the SSAP superfamilies provides insights into the evolutionary processes that led to the emergence of the operons that include the SSAP genes. As discussed above, the RecT/Redβ superfamily clearly splits into three distinct families (Fig. 2). The phylogenetic tree shows that SbcC co-occurs with the SSAP once within Redβ-family and once within the RecT-family. An examination of the tree and the respective gene neighborhoods suggests that independent juxtaposition of SbcC with Redβ-like and RecT-like genes on two separate occasions is the most parsimonious explanation. The alternative explanation, namely that the gene coding for the common ancestor of the Redβ and RecT already co-occurred with the sbcC gene is far less likely because it would require over 10 independent losses of this, apparently, functionally advantageous organization in different bacterial and bacteriophage lineages. Likewise, the observation that, in one or more cases, genes encoding each of the SSAPs co-occur in the same predicted operon with SSB or a λ-type exonuclease, suggests that similar operon structures may also emerge independently in evolution. Thus, the same or analogous operon organizations may emerge convergently on multiple occasions, probably due to the selective pressure arising from the strong interactions between the SSAPs and their functional partners such as SbcC, SSB and LE.
The distribution of the gene neighborhoods, in which a member of the RecT superfamily occurs next to the RecE on the phylogenetic tree of the RecT/Redβ superfamily, indicates that the RecE-RecT combination was probably the ancestral state for at least the RecT and EHAP1 families (Fig. 2). This implies that, on at least two occasions, the gene for λ endonuclease displaced the functionally analogous recE gene and became the adjacent gene to RecT (Fig. 2). That this displacement might have occurred by in situ insertion of a non-orthologous gene is suggested by the detection, on three separate occasions, of unusual remnants of pre-existing genes. The RecT/Redβ superfamily members, namely EHAP1 from the enterobacteria and PF161 from Borrelia hermsii, contain a small, C-terminal fragment of the core conserved domain of the ERF superfamily, which is located C-terminal of their bona fide RecT/Redβ domains. These fragments of the ERF protein are closely related to other ERF domains from related organisms and are unlikely to fold into the native conformation characteristic of the full-length ERF domain. For example the ERF fragment fused to the EHAP1 RecT/Redβ domain is closely related to the P22 phage ERF domain. This suggests that in each of these cases a RecT superfamily gene was inserted in frame into a pre-existing ERF gene leaving behind only a non-functional fragment of it (Fig. 2). In a very similar case, the bacterial RAD52-like protein from a Shiga toxin encoding temperate phage is fused to an extreme C-terminal fragment that is nearly identical to the C-terminal most portion of the P22 ERF protein. In this case, it appears that the pre-existing ERF gene was displaced through the insertion of a bacterial RAD52-like gene. Interestingly, and in the same vein, the RecT proteins from Bacillus species contain a short C-terminal acidic module that is missing in other RecT proteins, but is highly similar to the C-terminal region of SSBs, particularly those from Gram-positive bacteria (data not shown). This suggests that, at some stage in their evolution, the Bacillus recT gene protein has recombined with the gene coding for SSB, which might even have resulted in a functional replacement of an SSB with an SSAP.
Thus it appears likely that functionally equivalent genes may displace their analogs in operons via insertion into the same position.
Conclusions
We show that functionally similar SSAPs belong to at least three evolutionarily distinct superfamilies. We unify the Redβ and RecT proteins and their homologs, which have not been reported as being related at the sequence level, into a single superfamily, supporting the notion that these proteins share a similar mechanism of action. The second superfamily typified by the ERF proteins is predominantly found in bacteriophages and is also present on all circular plasmids from Borrelia, suggesting a role in the recombination of these plasmids. The third superfamily, typified by the yeast RAD52 protein and previously detected only in eukaryotes, was shown to include bacterial and phage homologs and to contain a modified HhH domain. By comparing the gene neighborhoods of the SSAPs, we show that the predicted operons that include the SSAP genes evolve according to the "LEGO" principle. In these operons, the SSAP genes are linked to the genes for various DNAses and DNA repair related proteins, such as SSB and SbcC, which implies functional connections between the encoded proteins. Evidence is presented of convergent emergence of similar SSAP-encoding operons in different lineages and of in situ non-orthologous displacement of functionally similar genes in these operons.
Materials and Methods
Sequence searches of the non-redundant (NR) and the unfinished genomes databases, were done using the gapped BLAST and PSI-BLAST programs [38]. Iterative PSI-BLAST searches used for in-depth sequence analysis were done with the profile inclusion cutoff expectation value (E value) set at 0.1. Multiple sequence alignments were generated using the T_Coffee program [39] and the output was adjusted using PSI-BLAST search results and secondary structure predictions, which were conducted using the PHD [40,41] and Jpred [42] programs. Fold predictions were done using the 3-D position specific score matrix (3DPSSM) [43] and the Hybrid fold method [44]. Phylogenetic analysis was carried out using the neighbor-joining algorithm, with subsequent local rearrangements using the maximum likelihood algorithm [45]. The robustness of tree topology was assessed with 10000 Resampling of Estimated Log Likelihoods (RELL) bootstrap replicates. The MOLPHY and Phylip software packages were used for the analyses [46,47].
Authors' contributions
Author 1 (LMI) contributed to the discovery process, preparation of the manuscript and multiple sequence alignments. Author 2 (EVK) contributed to the analysis of the predicted operons encoding RecT and SSB proteins from Gram positive bacteria, Author 3 (LA) contributed to the discovery process, preparation of the manuscript and conceived the study.
Contributor Information
Lakshminarayan M Iyer, Email: lakshmin@ncbi.nlm.nih.gov.
Eugene V Koonin, Email: koonin@ncbi.nlm.nih.gov.
L Aravind, Email: aravind@ncbi.nlm.nih.gov.
References
- Roca AI, Cox MM. RecA protein: structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci. 1998;3:D570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot P, Kolodner RD. DNA strand invasion promoted by Escherichia coli RecT protein. J Biol Chem. 1998;273:12274–12280. doi: 10.1074/jbc.273.20.12274. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HW, Julin DA. Structure and function of the Escherichia coli RecE protein, a member of the RecB nuclease domain family. J Biol Chem. 2001;276:46004–46010. doi: 10.1074/jbc.M108627200. [DOI] [PubMed] [Google Scholar]
- Barbour SD, Nagaishi H, Templin A, dark AJ. Biochemical and genetic studies of recombination proficiency in Escherichia coli. II. Rec+ revertants caused by indirect suppression of rec-mutations. Proc NatI Acad Sci U SA. 1970;67:128–135. doi: 10.1073/pnas.67.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner SR, Nagaishi H, Templin A, dark AJ. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc NatI Acad Sci USA. 1971;68:824–827. doi: 10.1073/pnas.68.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R, Hall SD, Luisi-DeLuca C. Homologous pairing proteins encoded by the Escherichia coli recE and recT genes. Mol Microbiol. 1994;11:23–30. doi: 10.1111/j.1365-2958.1994.tb00286.x. [DOI] [PubMed] [Google Scholar]
- Hall SD, Kane MF, Kolodner RD. Identification and characterization of the Escherichia coli RecT protein, a protein encoded by the recE region that promotes renaturation of homologous single-stranded DNA. J Bacteriol. 1993;175:277–287. doi: 10.1128/jb.175.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SD, Kolodner RD. Homologous pairing and strand exchange promoted by the Escherichia coli RecT protein. Proc Natl Acad Sci USA. 1994;91:3205–3209. doi: 10.1073/pnas.91.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen UH, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passy SI, Yu X, Li Z, Radding CM, Egelman EH. Rings and filaments of beta protein from bacteriophage lambda suggest a superfamily of recombination proteins. Proc Natl Acad Sci U S A. 1999;96:4279–4284. doi: 10.1073/pnas.96.8.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thresher RJ, Makhov AM, Hall SD, Kolodner R, Griffith JD. Electron microscopic visualization of RecT protein and its complexes with DNA. J Mol Biol. 1995;254:364–371. doi: 10.1006/jmbi.1995.0623. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- Poteete AR, Sauer RT, Hendrix RW. Domain structure and quaternary organization of the bacteriophage P22 Erf protein. J Mol Biol. 1983;171:401–418. doi: 10.1016/0022-2836(83)90037-2. [DOI] [PubMed] [Google Scholar]
- Stevenson B, Porcella SF, Oie KL, Fitzpatrick CA, Raffel SJ, Lubke L, Schrumpf ME, Schwan TG. The relapsing fever spirochete Borrelia hermsii contains multiple, antigen-encoding circular plasmids that are homologous to the cp32 plasmids of Lyme disease spirochetes. Infect Immun. 2000;68:3900–3908. doi: 10.1128/IAI.68.7.3900-3908.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroeher UH, Bode L, Beutin L, Manning PA. Characterization and sequence of a 33-kDa enterohemolysin (Ehly 1) – associated protein in Escherichia coli. Gene. 1993;132:89–94. doi: 10.1016/0378-1119(93)90518-8. [DOI] [PubMed] [Google Scholar]
- Kmiec E, Holloman WK. Beta protein of bacteriophage lambda promotes renaturation of DNA. J Biol Chem. 1981;256:12636–12639. [PubMed] [Google Scholar]
- Botstein D, Matz MJ. A recombination function essential to the growth of bacteriophage P22. J Mol Biol. 1970;54:417–440. doi: 10.1016/0022-2836(70)90119-1. [DOI] [PubMed] [Google Scholar]
- Weaver S, Levine M. Recombinational circularization of Salmonella phage P22 DNA. Virology. 1977;76:29–38. doi: 10.1016/0042-6822(77)90278-1. [DOI] [PubMed] [Google Scholar]
- Poteete AR. Location and sequence of the erf gene of phage P22. Virology. 1982;119:422–429. doi: 10.1016/0042-6822(82)90101-5. [DOI] [PubMed] [Google Scholar]
- Poteete AR, Fenton AC. Lambda red-dependent growth and recombination of phage P22. Virology. 1984;134:161–167. doi: 10.1016/0042-6822(84)90281-2. [DOI] [PubMed] [Google Scholar]
- Poteete AR, Fenton AC. Efficient double-strand break-stimulated recombination promoted by the general recombination systems of phages lambda and P22. Genetics. 1993;134:1013–1021. doi: 10.1093/genetics/134.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak AZ, Larquet E, Stasiak A, Muller S, Engel A, Van Dyck E, West SC, Egelman EH. The human Rad52 protein exists as a heptameric ring. Curr Biol. 2000;10:337–340. doi: 10.1016/S0960-9822(00)00385-7. [DOI] [PubMed] [Google Scholar]
- Shao X, Grishin NV. Common fold in helix-hairpin-helix proteins. Nucleic Acids Res. 2000;28:2643–2650. doi: 10.1093/nar/28.14.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty AJ, Serpell LC, Ponting CP. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Walker DR, Koonin EV. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization, and prediction of gene function using genomic context. Genome Res. 2001;11:356–372. doi: 10.1101/gr.GR-1619R. [DOI] [PubMed] [Google Scholar]
- Huynen M, Snel B, Lathe W, 3rd, Bork P. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 2000;10:1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci. 1998;23:324–328. doi: 10.1016/S0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11:1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Makarova KS, Koonin EV. Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, Kirkham LA, Leach DR. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Natl Acad Sci U S A. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, de Leau ES, Leach DR. DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res. 1999;27:1039–1046. doi: 10.1093/nar/27.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruand C, Ehrlich SD, Janniere L. Primosome assembly site in Bacillus subtilis. Embo J. 1995;14:2642–2650. doi: 10.1002/j.1460-2075.1995.tb07262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Rost B, Schneider R, Sander C. Protein fold recognition by prediction-based threading. J Mol Biol. 1997;270:471–480. doi: 10.1006/jmbi.1997.1101. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C, Schneider R. PHD – an automatic mail server for protein secondary structure prediction. Comput AppI Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–3. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- Kelley LA, MacCallum RM, Sternberg MJ. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J Mol Biol. 2000;299:499–520. doi: 10.1006/jmbi.2000.3741. [DOI] [PubMed] [Google Scholar]
- Fischer D. Pacific Symposium on Biocomputing, Hawaii. Singapore: World scientific;; 2000. Hybrid fold recognition: combining sequence derived properties with evolutionary information. pp. 119–130. [PubMed] [Google Scholar]
- Wolf YI, Rogozin IB, Grishin NV, Tatusov RL, Koonin EV. Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol Biol. 2001;1:8. doi: 10.1186/1471-2148-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi J, Hasegawa M. MOLPHY: Programs for Molecular Phylogenetics Tokyo: Institute of Statistical Mathematics; 1992.
- Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]